Abstract
Cyclization reactions that create complex polycyclic scaffolds are hallmarks of alkaloid biosynthetic pathways. We present the discovery of three homologous cytochromes P450 from three monoterpene indole alkaloid-producing plants (Rauwolfia serpentina, Gelsemium sempervirens and Catharanthus roseus) that provide entry into two distinct alkaloid classes, the sarpagans and the β-carbolines. Our results highlight how a common enzymatic mechanism, guided by related but structurally distinct substrates, leads to either cyclization or aromatization.
Monoterpene indole alkaloids (MIAs) are not only known for their potent biological activities, such as anticancer, anti-malarial and anti-arrhythmic properties1,2, but also for their complex polycyclic structures. Understanding and manipulating the underlying biosynthetic pathways of MIAs provides an effective way to access this medicinal treasure trove. One key cyclization reaction in MIA biosynthesis is catalyzed by sarpagan bridge enzyme (SBE) (Supplementary Fig. 1), a reaction that marks the entry of the central MIA intermediate strictosidine into sarpagan, ajmalan and alstophyllan alkaloid classes3, including the class Ia anti-arrhythmic agent ajmaline4 (1) and anticancer compound koumine5. Additionally, SBE is likely the gateway to certain oxindole alkaloids (Gelsemium spp.)6. Although enzymatic activity for SBE was detected in plant extracts over 20 years ago7, the corresponding gene has not been discovered. Here, we describe the discovery of two cytochrome P450 genes from R. serpentina and Gelsemium sempervirens that encode SBE activity, namely the conversion of strictosidine-derived geissoschizine (2) to the sarpagan alkaloid polyneuridine aldehyde (PNA; 3). Intriguingly, we demonstrate that SBE also aromatizes the tetrahydro-β-carboline alkaloids tetrahydroalstonine (4) and ajmalicine (5) to the corresponding β-carboline alkaloids alstonine (6) and serpentine (7) (Supplementary Fig. 1). We hypothesize that a shared iminium intermediate is responsible for the unusual connection of cyclization and aromatization reaction modes. We also demonstrate how the discovery of SBE now enables heterologous production of an ajmalan alkaloid.
We hypothesized that SBE is a cytochrome P450 that acts on geissoschizine (2), the product of a soluble reductase and strictosidine aglycone (8), the central intermediate of all MIA pathways (Supplementary Fig. 1) based on previous work on R. serpentina cell suspension cultures7 and recent work on strychnos-type MIA8. We conducted a co-expression analysis of publicly available R. serpentina transcriptome data9 using a self-organizing map10 (Supplementary Fig. 2). Additionally, we hypothesized that G. sempervirens should contain a homologous enzyme since sarpagan alkaloids, such as the anticancer compound koumine, have been reported from this plant6. Sequence data from the recently published G. sempervirens transcriptomes and genome11 revealed that two of the R. serpentina candidates selected from the self-organizing map (Rs_CYP_12057 and Rs_CYP_3375) have homologues with high sequence similarity (61 and 51% amino acid sequence identity) to G. sempervirens genes.
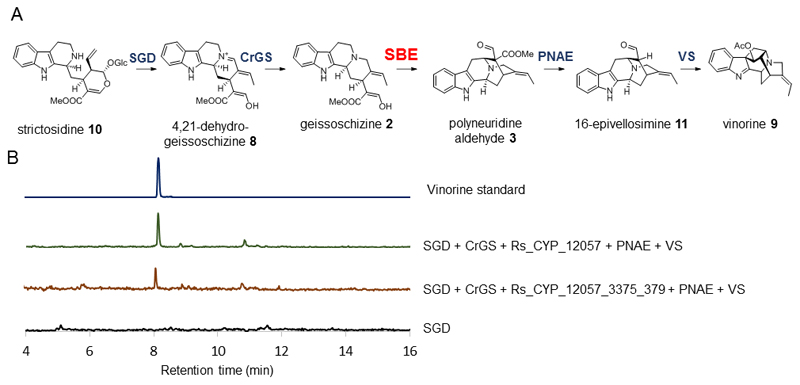
We used Agrobacterium-mediated transient expression in Nicotiana benthamiana to express SBE candidates. Since the expected PNA product may not be stable in planta, SBE candidates were expressed along with previously discovered pathway enzymes responsible for production of vinorine (9), which is derived from PNA and is a chemically stable intermediate in ajmaline biosynthesis (Fig. 1a). Thus, N. benthamiana was infiltrated with strictosidine (10) and A. tumefaciens strains encoding strictosidine glucosidase (SGD), geissoschizine synthase (CrGS, identified from Catharanthus roseus), polyneuridine aldehyde esterase (PNAE), vinorine synthase (VS) and SBE candidates for transient expression. To accelerate the screening process, up to three SBE candidates from R. serpentina and G. sempervirens were infiltrated simultaneously into N. benthamiana, and resulting plant tissue extracts were analyzed for the formation of vinorine. Liquid chromatography-mass spectrometry (LC-MS) analysis revealed the formation of vinorine (m/z 335) in plants that expressed one particular combination of SBE candidates in addition to SGD, CrGS, PNAE and VS (Fig. 1b). Deconvolution of this combination showed that it was Rs_CYP_12057, in the presence of SGD, CrGS, PNAE and VS, that was responsible for vinorine production (Fig. 1b).
Figure 1. Sarpagan bridge enzyme candidate screening using combinatorial expression in N. benthamiana.
a, Biosynthetic pathway from strictosidine to the sarpagan alkaloid vinorine. 4,21-dehydrogeissoschizine is an isomer of strictosidine aglycone. b, Reconstitution of the biosynthetic pathway to vinorine from strictosidine in N. benthamiana to identify SBE. Multiple-reaction monitoring (MRM) chromatograms showing in planta catalytic activity of Rs_CYP_12057 in combination with strictosidine substrate and other known enzymes to produce vinorine (SGD + CrGS + Rs_CYP_12057 + Rs_CYP_3375 + Rs_CYP_379 + PNAE + VS and SGD + CrGS + Rs_CYP_12057 + PNAE + VS) compared with the negative control (SGD only). Experiments were repeated three times independently with similar results.
We turned our attention to characterizing this SBE candidate more rigorously in vitro. However, geissoschizine is not available commercially, and CrGS primarily generates isositsirikine isomers8. Therefore, the related alkaloid geissoschizine methyl ether (12), together with structurally similar methyl ethers, were isolated from Uncaria rhynchophylla, which can be obtained as the Chinese drug Gou Teng12,13. These methyl ethers were chemically demethylated14 to provide milligram quantities of geissoschizine (Supplementary Fig. 3) and related compounds.
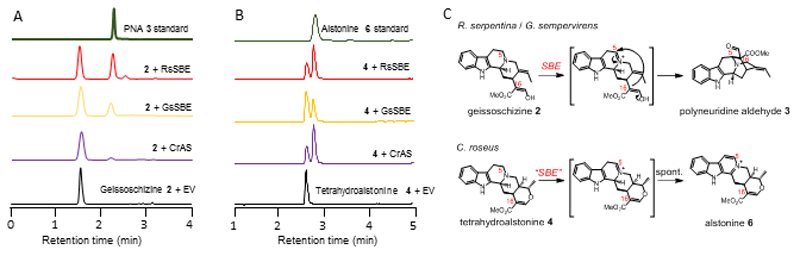
Next, CYP candidates were cloned into a yeast dual-expression vector that contained a cytochrome P450 reductase (CPR) to validate the in planta results. Geissoschizine (10 µM) was incubated with the yeast cultures for 24 hours. Only yeast cultures harboring the construct that encoded candidate Rs_CYP_12057 showed consumption of geissoschizine (m/z 353) and formation of a new product with m/z of 351 that had the same retention time and MS2 fragmentation pattern as the synthetic PNA standard15 (Fig. 2a). No other candidate gene from R. serpentina utilized geissoschizine as a substrate. Although R. serpentina is not amenable to gene silencing, a silencing protocol has been developed for the related species, Rauwolfia tetraphylla, where ajmaline is also found in aerial organs16. Silencing of the R. tetraphylla SBE homolog (93% identity) led to a statistically significant decrease in ajmaline production, substantiating the physiological role of this gene in alkaloid biosynthesis (Supplementary Fig. 4). The homologue of Rs_CYP_12057 from G. sempervirens (Gs_207-0.13; 62% amino acid sequence identity) also converted geissoschizine to PNA (Fig. 2a). The identification of SBE from G. sempervirens provides the first biochemical evidence that PNA is potentially involved in the biosynthesis of Gelsemium alkaloids.
Figure 2. The cyclization and aromatization catalytic function of recombinant RsSBE, GsSBE and CrAS depend on the substrate.
a,b, MRM chromatograms showing the in vivo catalytic activity of RsSBE (Rs_CYP_12057), GsSBE (Gs_207-0.13) and CrAS (CrCYP71AY1) using geissoschizine (2; a) and tetrahydroalstonine (4; b) (m/z 353.2) as substrates compared with the empty vector negative control (EV). The identity of the trace side product in the geissoschizine + RsSBE reaction (red chromatogram) could not be determined. c, Proposed reaction mechanism for cyclization (upper panel) and aromatization (lower panel) with geissoschizine (2) and tetrahydroalstonine (4) via a common iminium intermediate. All experiments were repeated three times independently with similar results.
Rs_CYP_12057 and Gs_207-0.13, which are homologues, phylogenetically belong to the same clade as CYP71AY1 from C. roseus (78% amino acid sequence identity). All three proteins are members of the CYP71AY family (Rs_CYP_12057, CYP71AY4 and Gs_207-0.13, CYP71AY5, respectively; Supplementary Fig. 5). Rs_CYP_12057 shows the expected localization to the endoplasmic reticulum (Supplementary Fig. 6). Although C. roseus does not produce sarpagan or ajmalan alkaloids, CrCYP71AY1 converted geissoschizine to PNA, albeit with much lower efficiency (Fig. 2a), as shown in previous work with cell-free extract7. An empty-vector control exhibited no detectable PNA formation under the same conditions (Fig. 2a). Microsomal fractions of yeast expressing these genes (Supplementary Fig. 7) were used to biochemically characterize these enzymes. Rs_12057 and Gs_207-0.13 exhibited Michaelis-Menten kinetics with an apparent KM value of 22.5 µM and 35.3 µM, respectively, for geissoschizine (Supplementary Fig. 8), while the activity of CrCYP71AY1 was so low that it could not be quantified accurately by steady-state kinetic assays. Due to the observed activity of Rs_CYP_12057 and Gs_207-0.13, these enzymes were named sarpagan bridge enzyme (SBE). Both RsSBE and GsSBE are highly expressed in root tissues, where the corresponding alkaloids are found in abundance, whereas CrCYP71AY1 is primarily expressed in stem tissue (Supplementary Fig. 9). The discovery of SBE now corroborates the previously proposed reaction sequence geissoschizine to PNA.
As the C5-C16 bond formation of SBE is a crucial entry point into several alkaloid classes (Supplementary Fig. 1) and could potentially be harnessed to generate molecular complexity for other scaffolds, we tested the substrate specificity of these enzymes using a range of MIAs (Supplementary Fig. 10). Surprisingly, desmethylhirsuteine and desmethylhirsutine, two alkaloids differing from geissoschizine only in the ethylidene side chain, were not turned over, suggesting a crucial role of the 19E ethylidene group in promoting a conducive conformation for downstream cyclizations17. Intriguingly, two heteroyohimbine alkaloids, tetrahydroalstonine and ajmalicine, were turned over, resulting in formation of new products with a mass suggesting a four electron oxidation (Fig. 2b,c). The reaction products of tetrahydroalstonine and ajmalicine were confirmed to be the corresponding fully aromatic β-carbolines, the anhydronium bases alstonine and serpentine, based on co-elution and MS2 MRM fragmentation pattern with reference compounds (Supplementary Fig. 11). The β-carbolines have a variety of biological activities; for example, alstonine is used as a remedy to treat psychosis and other nervous system disorders18. Geissoschizine methyl ether (m/z 367.2) was also oxidized to a product with a mass decreased by 4 atomic units (m/z 363.2), suggesting oxidation to the β-carboline, though limited substrate availability and low catalytic activity hindered rigorous characterization of this oxidized product (Supplementary Fig. 11). CrCYP71AY1 exhibited Michaelis-Menten enzyme kinetics with a KM of 19.6 µM for tetrahydroalstonine (Supplementary Fig. 8), and we designated this enzyme as alstonine synthase (AS). Collectively, these results highlight that all three SBE homologues from R. serpentina, G. sempervirens and C. roseus are, to a greater or lesser extent, capable of cyclization as well as aromatization reactions, depending on the identity of the starting substrate.
Our findings suggest a unifying mechanism for the action of SBE and the C. roseus homologue AS (Fig. 2c). We propose that SBE forms an iminium intermediate, by direct hydride abstraction or by step-wise oxidation, which can then follow two distinct reaction modes, cyclization or further oxidation. Cyclization occurs if a suitable nucleophile is correctly oriented to attack the iminium moiety. The side chain of geissoschizine (Fig. 2c), which exists primarily as the enol tautomer14,17, is an excellent nucleophile, allowing cyclization by a Mannich reaction, though an alternative radical mechanism cannot be ruled out. Alternatively, if no nucleophile is available for intramolecular cyclization (as in geissoschizine methyl ether, tetrahydroalstonine, or ajmalicine), the dihydropyridinium moiety that is generated by SBE could autoxidize spontaneously to the fully aromatic β-carboline, as has been previously observed in similar chemical systems19 (Fig. 2c). Earlier studies suggested a vacuolar peroxidase is responsible for the oxidation of ajmalicine20, and while we cannot exclude this possibility in planta, the shift in the substrate specificity of CrAS from geissoschizine to tetrahydroalstonine compared with RsSBE and GsSBE (Fig. 2a,b) supports a different biological role for CrAS. Moreover, the expression profile of CrAS (high in stem and roots) is in agreement with previous reports of β-carboline accumulation in C. roseus stem (and root) tissue20,21 (Supplementary Fig. 9). Such proposed function also illustrated how MIA-producing plants recruited and evolved the ADH/CYP71 biosynthetic module to synthesize strychnos, sarpagan and heteroyohimbine alkaloids. To explore the basis of the substrate specificity of CrAS, we made six point mutations based on sequence differences between CrAs and RsSBE that were also mapped onto a CYP homology model (Supplementary Fig. 12). Notably, PNA was produced by three CrAS mutants (S219A, E310W and V452I), suggesting that these residues are involved in mediating substrate recognition. This change in product profile suggests that these CYP71 enzymes may be amenable to protein engineering.
The indole alkaloids of the sarpagan–ajmalan type are found in approximately 100 plant species3. However, the biosynthetic gene leading to the intermediate PNA, at the crossroads of this pathway, has not been elucidated. Here we have shown how we rapidly identified this sarpagan bridge enzyme by combinatorial expression of ten CYP candidates in N. benthamiana, together with previously discovered pathway enzymes to push the production towards a stable product that could serve as a readout for enzymatic activity. This approach demonstrates that combinatorial expression is a productive strategy for gene discovery and pathway reconstitution of ajmaline and other strictosidine-derived alkaloids when unstable substrates or products are involved. These homologous cytochromes P450 from the three alkaloid producer plants R. serpentina, G. sempervirens and C. roseus cyclize geissoschizine and thereby provide the entry point to sarpagan, ajmalan, alstophyllan and Gelsemium oxindole alkaloids. Moreover, these enzymes are capable of aromatizing alternative substrates to form β-carboline alkaloids. This discovery highlights how enzymatic promiscuity in substrate specificity, along with the inherent reactivity of these alkaloid substrates, can create a suite of structurally diverse chemical products.
Methods
Plants and chemicals
R. serpentina seeds were a gift from Dr. Subhash Hiremath, Karnataka University, India. The seeds were germinated at 28/22°C light/dark in a growth chamber with a photoperiod of 16 h. The 4-6 weeks plantlets were transferred to individual pots and grown in the greenhouse at the same temperature and light conditions. G. sempervirens seeds were obtained from Plant World Seeds UK and plants were grown in a greenhouse. C. roseus Little Bright Eyes cultivar was grown as reported earlier22. Polyneuridine aldehyde was a kind gift from Drs. E. Poupon and L. Evanno (Univ. Paris-Sud, CNRS, Université Paris-Saclay, France). Vinorine (9) was purchased from Northernchem Inc. (Niagara Falls, Ontario, Canada). Tetrahydroalstonine (4) was purchased from Extrasynthese (Lyon, France). Serpentine (7) was obtained from ChemFaces Biochemical Co.,Ltd. Alstonine was produced by oxidation of tetrahydroalstonine (4) with Pd black following a procedure by Younai et al.23 (NMR spectrum see Supplementary Fig. 3). Geissoschizine methyl ether and the related methyl ether alkaloids hirsuteine, hirsutine and rhynchophylline were isolated from Uncaria rhynchophylla and demethylated as described below. These are known compounds and 1HNMR spectral data are provided. All other chemicals were of analytical grade from Sigma Aldrich.
Identification of cytochrome P450 (P450) candidates
Publicly available transcriptomic data of six different organs of Rauwolfia serpentina was filtered for contigs with FPKM (fragments per kilobase of exon per million fragments mapped) expression values higher than zero for more than half of the organs (i.e. transcripts with FPKM expression values of zero for more than half of the treatments or with zero expression variance across the samples were removed). Self-organizing maps were applied and visualized in R (RStudio 1.0.136, RStudio, Inc) with the Kohonen package24 as reported before10,22. The map was assigned to give about 50 contigs per node. Neighbouring nodes are related to each other by the similarity of their expression profile. The average expression profile of genes in the node is plotted within each node. To assess how well the generated self-organizing map fitted the data, two quality metrics were analyzed. The first was the within-node distance, which is defined as the mean distance from the weight vector of a node to the samples mapped to it. Therefore, the smaller the within-node distance, the more accurately the node's weight vector represents the samples mapped to the node. The other quality metric used was the internodal distance, defined as the sum of the distances from a node's weight vector to the weight vectors of its neighbouring nodes. The smaller the value, the more similar a node's weight vector is to the weight vectors of its surrounding nodes. In order for a node to be classed as high quality in this analysis, both of the described quality metrics for that node had to be in the lowest quartile compared with all nodes. Cytochrome P450 candidates that are in the same nodes or neighbouring nodes with similar expression patterns as previously reported genes were selected for cloning and testing for activity.
Phylogenetic analysis
Unrooted neighbour-joining phylogenetic tree for CYP candidates from this study and other reported CYPs from other organisms were performed using the Geneious Tree Builder program in the Geneious software package (Geneious 8.1.8, Biomatters). Abbreviations and accession numbers of amino acid sequences used to generate phylogenetic tree: R. serpentina CYP 12057 (RsSBE, CYP71AY4, MF537711), G. sempervirens Gs207 (GsSBE, CYP71AY5, MF537712); R. serpentina CYP5437 (KY926696), C. roseus cytochrome P450 (CYP71D1V1) CrCYP71D1V1, AEX07770.1; C. roseus CrCYP71BT1, AHK60840.1; C. roseus CYP71AY1, CrCYP71Y1, AHK60849.1; Solanum pennellii isoflavone 2'-hydroxylase-like, SpFH, XM_015218869.1; Theobroma cacao cytochrome CYP71D9, Tc71D9, XP_017975886; C. roseus geraniol 10-hydroxylase, CrG10H, Q8VWZ7.1; Nicotiana tabacum CYP82M1v4, ABC69375.1; C. roseus CYP72A224 7-deoxyloganic acid 7-hydroxylase, Cr7DLH, AGX93062.1; Papaver somniferum cytochrome P450 CYP82X1, PsCYP82X1; AFB74614.1; P. somniferum cytochrome P450 CYP82Y1, PsCYP82Y1, AFB74617.1; P. somniferum cytochrome P450 CYP82X2, PsCYP82X2, AFB74617.1, AFB74616.1; C. roseus tabersonine 16-hydroxylase, CrCYP71D12, FJ647194.1; Salvia miltiorrhiza cytochrome P450 CYP82V2, SmCYP82V2, AJD25203.1; C. roseus secologanin synthase, CrSLS, Q05047.
Candidate gene screening and reconstitution of ajmalan pathway in N. benthamiana
The full-length coding regions of ADH and CYP candidates were amplified using cDNA derived from total root RNA of R. serpentina using Takara Ex Taq DNA polymerase (Clontech), and primer sets listed in Supplementary Table 1. For transient expression and candidate gene screening in N. benthamiana, the full-length coding regions were cloned into BamHI and KpnI restriction sites of pTRBO25 using the In-fusion cloning system (Takara Clontech). pTRBO constructs were transformed into Agrobacterium tumefaciens GV3101 by electroporation. Transformants were selected on LB plates containing kanamycin, gentamicin and rifampicin. Cells were grown for 48 hrs at 28°C before harvested by centrifugation. The pellet was resuspended in infiltration buffer (10 mM NaCl, 1.75 mM CaCl2, 100 µM acetosyringone) and incubated at room temperature for 2 hrs. Agrobacterium suspensions (OD600 = 0.1 for each strain) were infiltrated into the abaxial side of 5 week old N. benthamiana leaves with a needleless 1 mL syringe. A. tumefaciens strains encoding strictosidine glucosidase (SGD), geissoschizine synthase (GS, identified from C. roseus), polyneuridine aldehyde esterase (PNAE), vinorine synthase (VS) and up to 3 SBE candidates were mixed and infiltrated into N. benthamiana for transient expression. Substrate (50 µM) and caffeine standard (100 µM) were infiltrated into the leaves 3 days post bacteria infiltration. Leaves were flash frozen in liquid N2 and stored at –70°C before processing.
N. benthamiana metabolite extraction and analysis
Frozen leaf tissues were ground into powder in liquid N2. 1000 µL of MeOH was added to each gram of tissue. The mixture was sonicated for 15 min and filtered before liquid chromatography-mass spectrometry (LC-MS) analysis. Elution of vinorine from combinatorial expression in N. benthamiana was monitored (multiple reaction monitoring, MRM) using four transitions m/z 335.2 > 243.2 (cone 36, collision 24), m/z 335.2 > 258.2 (cone 26, collision 32), m/z 335.1 > 275.2 (cone 36, collision 34), m/z 351.2 > 323.0 (cone 26, collision 16) on a Waters Acquity UPLC system (Milford, MA) coupled to a Xevo TQ-S mass analyzer.
Yeast expression vector construction
The full-length coding regions of CYP candidates were amplified using cDNA derived from total root RNA of R. serpentina using Takara Ex Taq DNA polymerase (Clontech), and primer sets listed in Supplementary Table 1. GsSBE was amplified from G. sempervirens root cDNA using the primers pESC-Leu2d-Gs207 fw/rv (Supplementary Table 1). CrAS was amplified from C. roseus stem cDNA using the primers pESC-Cr71AY1 (Supplementary Table 1). For heterologous expression of FLAG-tagged CYPs in yeast (Saccharomyces cerevisiae), the full-length coding regions of CYPs were cloned into SpeI restriction sites of the dual plasmid pESC-Leu2d::CPR26 yielding pESC-Leu2d::CYP/CPR using the In-fusion cloning system (Takara Clontech). The protease-deficient yeast strain YPL 154C:Pep4 KO strain27 was transformed with pESC-Leu2d::CYP/CPR. Yeast harboring pESC-Leu2d::CPR was used as the negative control.
Yeast culture, preparation of microsomes and immunoblot analysis
For routine yeast culture, the transgenic yeast strain was inoculated in 2 mL of synthetic complete (SC) medium lacking leucine (SC-Leu) containing 2% (w/v) glucose. The inoculum was cultured overnight at 30°C and 250 rpm. The culture was subsequently diluted 100-fold to an OD of 0.05 in SC-Leu supplemented with 2% (w/v) glucose and cultured for 16 h. Yeast was then harvested and sub-cultured for 24 h in SC-Leu containing 2% (w/v) galactose to induce recombinant protein production. Yeast cells were harvested by centrifugation and lysed in TES-B (0.6 M sorbitol in TE) using a Constant Systems cell disruptor at 35 kpsi and subsequently centrifuged at 11,000 g for 10 min at 4°C. The supernatant was then transferred to a new tube and centrifuged at 125,000 g for 90 min at 4°C. Finally, the pellet containing microsomes was resuspended with TEG buffer (20% (v/v) glycerol in TE). Recombinant enzymes were detected by immunoblot analysis using α-FLAG M2 (Genscript) detected with SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific) (Supplementary Fig. 9).
Enzyme assays
Standard enzyme assays were performed at 30°C for 30 min in 100 μL of 100 mM HEPES-NaOH, pH 7.5, containing 0.1 mg of total microsomal proteins, 10 μM substrate and 100 μM NADPH on a gyratory shaker with agitation (1000 rpm). Reactions were stopped by adding 800 μL methanol. A variety of alkaloids was used to test substrate specificity (see Supplementary Fig. 10, 11). The reaction mixture was extracted twice with methanol to precipitate and remove proteins. The supernatant was diluted with appropriate solvent for LC-MS/MS analysis. Steady-state enzyme kinetics was determined by varying the concentration of geissoschizine or tetrahydroalstonine between 0 and 300 μM at a fixed concentration of 200 μM NADPH (Supplementary Fig. 8). For quantification of alstonine and polyneuridine aldehyde, calibration curves were prepared and analyzed using eight calibration points in the range 0.006-0.08 ng/μL, in which the response was linear (R2=0.98 for both compounds). Data processing was done using MassLynx 4.1 and TargetLynx software (Waters). Four transitions were used to monitor the elution of polyneuridine aldehyde: m/z 351.2 > 158.0 (cone 26, collision 21), m/z 351.2 > 166.0 (cone 26, collision 24), m/z 351.2 > 319.0 (cone 26, collision 19), m/z 351.2 > 323.0 (cone 26, collision 16). To monitor the elution of alstonine, four transitions were used: m/z 349.2 > 206 (cone 36, collision 50), m/z 349.2 > 234.9 (cone 36, collision 34), m/z 349.2 > 263.1 (cone 36, collision 30), m/z 349.2 > 317.1 (cone 36, collision 26). Kinetic constants were calculated by fitting initial velocity versus substrate concentration to the Michaelis-Menten equation using GraphPad Prism 5.0 (GraphPad Software). Data represent mean of three technical replicates.
LC-MS/MS analysis
Enzyme assays and alkaloid profiles of N. benthamiana plants were analysed by ultraperformance liquid chromatography (UPLC) on a Waters Acquity UPLC system (Milford, MA) coupled to a Xevo TQ-S mass analyzer. For kinetic studies, chromatography was performed on a BEH Shield RP18 (50 x 2.1 mm; 1.7 μm) column at a flow rate of 0.6 mL.min-1. The column was equilibrated in solvent A (0.1% formic acid) and the following elution conditions were used: 0 min, 5% B (100% acetonitrile), from 0 to 3.5 min, 65% B; from 3.5 min to 3.75 min, 100% B; 3.75 min to 4.75, 100% B; 4.75 to 6 min, 5% B to re-equilibrate the column. Alternatively, the column was equilibrated in solvent A1 (0.1% NH4OH) from 0 to 3.5 min, 65% B; from 3.5 min to 3.75 min, 100% B; 3.75 min to 4.75, 100% B; 4.75 to 6 min, 5% B to re-equilibrate the column. The basic mobile phase improves the peak shape of polyneuridine aldehyde.
Virus Induced Gene Silencing
Silencing of RtSBE in R. tetraphylla was achieved according to the procedure described in Corbin et al., (2017)16. The RtSBE silencing fragment was amplified with primers 5’-CTGAGAGGATCCTACTTGAAGCAGTTGCGCCAGATTTATGC-3’ and 5’-CTGAGAGGATCCAAGCACGGTGTCAAGTTGCTTCAAC-3’ and the resulting 362-bp gene fragment was cloned into pTRV2 to generate the pTRV2-viSBE silencing plasmid. Gene silencing was achieved by through the biolistic-mediated inoculation of viral vectors. Around four weeks post-transformation, leaves from four wild-type plants, three plants transformed with empty pTRV2 vector and five plants transformed with pTRV2-SBE were collected. Collected leaves were frozen in liquid nitrogen, powdered using a mixer mill (Retsch MM400) and subjected to LCMS according to Parage et al. (2016)28 (ajmaline m/z 327, RT=4.95 min; reserpilline m/z 413, RT=10.67 min) and qRT-PCR analysis using primers 5’-CTCTATCATTTTGATTGGAAACTTCC-3’ and 5’-TAGTTATCCTTATTCCACGACTCGA-3’. The sample size was 10 plants per condition (10 for SBE, 10 for empty vector and 10 for wild type; the transformation efficiency in R. tetraphylla is low16,29; the remaining plants were not effectively transformed). The experiment was repeated for a total of twice. Statistical analyses were performed with student T-Test (two-sided). Early biosynthesis steps in Rauwolfia serpentina occur in roots; even though we infected R. tetraphylla leaves, TRV-based vectors can spread systemically and are capable of silencing genes in roots, which might contribute to the strong reduction in ajmaline content.
Intracellular localization of SBE
Determination of the subcellular localization of Rs_SBE and Rt_SBE was performed by subcloning the full length coding sequences into the pSCA-cassette vector as YFP fusions. The nuclear (nucleus-CFP), nucleocytosolic (CFP) and endoplasmic reticulum (ER-CFP) markers were described previously30,31. Transient transformation of C. roseus cells by particle bombardment and fluorescence imaging were performed following the procedures previously described30,31. C. roseus cells were bombarded with DNA-coated gold particles (1 μm) and 1,100 psi rupture disc at a stopping-screen-to-target distance of 6 cm, using the Bio-Rad PDS1000/He system and 400 ng of each plasmid per transformation. Cells were cultivated for 16 h to 38 h before being harvested and observed. The subcellular localization was determined using an Olympus BX-51 epifluorescence microscope equipped with an Olympus DP-71 digital camera and a combination of YFP and CFP filters. Transformation was repeated three times and for each more than 100 transformed cells were observed. The CellD software (Olympus) was used to control the camera for photos and color attribution. Photos were cropped with Photo-Paint (Corel) to conserve only cells in the figure.
Isolation of geissoschizine methyl ether, hirsuteine, hirsutine and rhynchophylline
The methyl ether alkaloids geissoschizine methyl ether, hirsuteine, hirsutine and rhynchophylline were isolated from Gou Teng (Ramulus Uncariae cum Uncis), i.e. dried plant material of Uncaria rhynchophylla used in Traditional Chinese Medicine. Plant material was obtained from tcm4u.co.uk. 193 g of the dried woody plant material was ground in a blender to a fine powder and extracted 3x with 1 L ethanol, yielding 23.3 g of crude extract after concentrating in vacuo. The crude extract was dissolved in 250 mL 0.01 m HCl and washed with 300 mL EtOAc. The EtOAc layer was then re-extracted with 250 mL 0.01 m HCl. Both aqueous layers were combined, basified with 1 m NaOH to pH 8-9 and extracted 4x with 300-450 mL CHCl3. The combined CHCl3 layers were concentrated in vacuo to yield 0.47 g crude alkaloid extract. The crude alkaloid extract was separated on silica (50 g, 30-70 µm) using a step-wise gradient elution with CHCl3/MeOH (95/5 (400 mL), 92/8, 90/10, 80/20, 70/30, 0/100; 200 mL each). Fractions containing the target compounds according to LCMS were separated by semipreparative HPLC on a Dionex Ultimate 3000 (Thermo Scientific) with UV detection at 254 nm using a BEH C18 OBD 130-5 column (250x10 mm, Waters XBridge) and a separation gradient (A: 0.1% NH4OH in H2O, B: acetonitrile, 40% to 70% B in 0 to 20 min) to give rhynchophylline (tR = 11.7 min), hirsuteine (tR = 12.6 min), hirsutine (tR = 13.7 min) and geissoschizine methyl ether (tR = 14.5 min). The identity of all compounds was confirmed by HRMS and NMR spectroscopy in comparison with literature data. See Supplementary Fig. 3 for NMR spectra, references and Supplementary Table 2 and 3 for 1H and 13C data, respectively.
Demethylation of geissoschizine methyl ether, hirsuteine, hirsutine and rhynchophylline
Demethylation of methyl ether alkaloids was performed according to literature32. Typically, 0.1-3 mg substrate was dissolved in 80 μL AcOH and 20 μL conc. HCl was added. The mixture was incubated at RT for 2-3 days and monitored by MS for appearance of the demethylated product (– m/z 14). Afterwards, the reaction was quenched by slow addition of 400 μL 4 m K2CO3 and extracted 3x with 300 μL EtOAc. The combined organic layers were concentrated in vacuo, redissolved in methanol and purified by semipreparative HPLC on a Dionex Ultimate 3000 (Thermo Scientific) with UV detection at 254 nm using a BEH C18 OBD 130-5 column (250x10 mm, Waters XBridge) and a separation gradient (A: 0.1% NH4OH in H2O, B: acetonitrile, 25% to 55% B in 0 to 20 min) (geissoschizine: tR = 7.5-8.6 min). The identity of geissoschizine was confirmed by HRMS and NMR spectroscopy in comparison with literature data14 (1H NMR spectrum see Supplementary Fig. 3, 1H NMR data see Supplementary Table 2, TopSpin (version 3.2) was used for analysis of NMR data).
RT-PCR of GsSBE
Expression of the SBE gene in G. sempervirens was tested by RT-PCR on cDNA obtained from RNA isolated from young leaves, old leaves, stem and roots. The aerial parts of 3-4 month old G. sempervirens plants were removed and separated into young leaves (bright green), old (mature) leaves (dark green), and stem. The remaining root ball was removed from soil, washed with tap water to remove soil particles and blotted dry. All tissues were flash frozen in liquid N2 and ground in a mortar while frozen, until fine powders were obtained. Approximately 50 mg of each tissue was used for RNA extraction using the RNeasy Plant Mini kit (Qiagen) according to the manufacturer’s instructions. RNA purity and quality was analyzed by spectrophotometry on a Nanodrop (Thermo Fisher) and agarose gel electrophoresis. cDNA synthesis was performed using the SuperScript III First-Strand Synthesis system (Thermo Fisher) with oligo(dT)20 primers following the manufacturer’s instructions. RT-PCR experiments were done using CloneAmp HiFi PCR premix (Clontech), 1 µL cDNA and 0.5 µL of each primer per 20 µL reaction. The primers GsSBE-RT fw/rv were used. As a positive control, primers designed for the homolog of the 7DLH gene of C. roseus, 7DLH-RT fw/rv (Supplementary Table 1), were used.
Gene expression in different plant organs
Gene expression analysis was carried out using publicly available data for C. roseus and R. serpentina9.
Site-directed mutagenesis of CrAS
To probe which residues are responsible for the specificity of the reaction of CrAS and RsSBE, a homology model of CrAS and RsSBE was built based on allene oxide synthase33 (PDB ID: 3DAM, 13% amino acid identity with CrAS) using the Phyre2 web portal for protein modeling, prediction and analysis (Structural Bioinformatics Group, Imperial College, London). Sequence alignment of CrAS was then used to identify the regions where sequence differences existed between the two proteins, and mapped them onto the homology model (Supplementary Figure 12). In most cases, the sequence changes were far from the putative heme and substrate binding sites. However, six mutations, including V125A, K211R, S219A, V308I, E310W, V452I, that are mostly on helices and loops that may be adjacent to the heme and substrate binding site of CrAS were created by overlap extension PCR as described before30. Mutant constructs were cloned into pESC-Leu2d and sequenced to verify the mutant gene sequence. Protein expression was confirmed by Western blot as described above. Enzyme assays were carried out with wildtype CrAS and other mutants as previously described.
Statistics and reproducibility
All information on statistical methods and reproducibility is shown in the corresponding figure legends.
Data Availability Statement
Gene sequences from this paper have been deposited to GenBank as: MF537711, R. serpentina CYP 12057 (RsSBE, CYP71AY4) and MF537712, G. sempervirens Gs_207-0.13 (GsSBE, CYP71AY5). C. roseus CYP71AY1 (CrAS, AHK60849.1) was deposited by ref. 34. All transcriptome data for R. serpentina and C. roseus are available at http://medicinalplantgenomics.msu.edu/.
Supplementary Material
Acknowledgements
T.T.T.D. is grateful to the EMBO Long Term Fellowship ALTF 739-2015. J.F. gratefully acknowledges DFG postdoctoral funding (FR 3720/1-1). This work was supported by grants from the European Research Council (311363), BBSRC (BB/J004561/1) (S.E.O.) and from the Région Centre, France (BioPROPHARM, CatharSIS grants) (V.C.). We thank Drs. Erwan Poupon and Laurent Evanno (Univ. Paris-Sud) for their generous gift of polyneuridine aldehyde standard. Rauwolfia serpentina seeds were a generous gift from Dr. Subhash Hiremath, Karnataka University, India. Dr. Dae-Kyun Ro generously provided pESC-Leu2d. Images of R. serpentina and C. roseus were provided by Dr. Tran Nguyen. We thank Dr. Lorenzo Caputi (John Innes Centre) for his assistance in building the homology model of CrAS and RsSBE and Drs. Lionel Hill and Gerhard Saalbach of the Molecular Analysis platform at John Innes Centre for their assistance in metabolic analysis.
Footnotes
Contributions
T.T.T.D., J.F. and S.E.O. designed the experiments and wrote the manuscript. T.T.T.D. characterized RsSBE, GsSBE and CrAS in vitro and in vivo, performed in planta combinatorial assay and analysis. J.F. performed all substrate purification, synthesis and product characterizations. C.L. contributed to N. benthamiana work. I.S.T.C. and V.C. performed VIGS and localization experiments.
Competing Interests
The authors declare no competing financial interests.
References
- 1.O’Connor SE, Maresh JJ. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep. 2006;23:532–547. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- 2.Cragg GM, Newman DJ. Natural products: A continuing source of novel drug leads. Biochim Biophys Acta - Gen Subj. 2013;1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Namjoshi OA, Cook JM. Sarpagine and related alkaloids. Alkaloids Chem Biol. 2016;76:63–169. doi: 10.1016/bs.alkal.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hashimoto Y, Hori R, Okumura K, Yasuhara M. Pharmacokinetics and antiarrhythmic activity of ajmaline in rats subjected to coronary artery occlusion. Br J Pharmacol. 1986;88:71–77. doi: 10.1111/j.1476-5381.1986.tb09472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, et al. Apoptotic effect of koumine on human breast cancer cells and the mechanism involved. Cell Biochem Biophys. 2015;72:411–6. doi: 10.1007/s12013-014-0479-2. [DOI] [PubMed] [Google Scholar]
- 6.Jin G-L, et al. Medicinal plants of the genus Gelsemium (Gelsemiaceae, Gentianales)--a review of their phytochemistry, pharmacology, toxicology and traditional use. J Ethnopharmacol. 2014;152:33–52. doi: 10.1016/j.jep.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt D, Stöckigt J. Enzymatic formation of the sarpagan-bridge: a key step in the biosynthesis of sarpagine- and ajmaline-type alkaloids. Planta Med. 1995;61:254–258. doi: 10.1055/s-2006-958067. [DOI] [PubMed] [Google Scholar]
- 8.Tatsis EC, et al. A three enzyme system to generate the Strychnos alkaloid scaffold from a central biosynthetic intermediate. Nat Commun. 2017;8:316. doi: 10.1038/s41467-017-00154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Góngora-Castillo E, et al. Development of transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLoS One. 2012;7:e52506. doi: 10.1371/journal.pone.0052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dang T-TT, Franke J, Tatsis E, O’Connor SE. Dual catalytic activity of a Cytochrome P450 controls bifurcation at a metabolic branch point of alkaloid biosynthesis in Rauwolfia serpentina. Angew Chem Int Ed Engl. 2017;56:9440–9444. doi: 10.1002/anie.201705010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stapleton JA, et al. Haplotype-Phased synthetic long reads from Short-Read Sequencing. PLoS One. 2016;11:e0147229. doi: 10.1371/journal.pone.0147229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuzurihara M, et al. Geissoschizine methyl ether, an indole alkaloid extracted from Uncariae ramulus et Uncus, is a potent vasorelaxant of isolated rat aorta. Eur J Pharmacol. 2002;444:183–9. doi: 10.1016/s0014-2999(02)01623-0. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z, et al. Geissoschizine methyl ether, a corynanthean-type indole alkaloid from Uncaria rhynchophylla as potential acetylcholinesterase inhibitor. Nat Prod Res. 2011;26:22–28. doi: 10.1080/14786419.2010.529811. [DOI] [PubMed] [Google Scholar]
- 14.Takayama H, Watanabe T, Seki H, Aimi N, Sakai S. Geissoschizine revisited -definite proof of its stereostructure. Tetrahedron Lett. 1992;33:6831–6834. [Google Scholar]
- 15.Ahamada K, Benayad S, Poupon E, Evanno L. Polyneuridine aldehyde: structure, stability overviews and a plausible origin of flavopereirine. Tetrahedron Lett. 2016;57:1718–1720. [Google Scholar]
- 16.Corbin C, et al. Virus-induced gene silencing in Rauwolfia species. Protoplasma. 2017;254:1813–1818. doi: 10.1007/s00709-017-1079-y. [DOI] [PubMed] [Google Scholar]
- 17.Eckermann R, Gaich T. The double-bond configuration of Corynanthean alkaloids and its impact on monoterpenoid indole alkaloid biosynthesis. Chemistry. 2016;22:5749–55. doi: 10.1002/chem.201505068. [DOI] [PubMed] [Google Scholar]
- 18.Elisabetsky E, Costa-Campos L. The alkaloid alstonine: a review of its pharmacological properties. Evidence-Based Complement Altern Med. 2006;3:39–48. doi: 10.1093/ecam/nek011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korytowski W, Felix CC, Kalyanaraman B. Mechanism of oxidation of 1-methyl-4-phenyl-2,3-dihydropyridinium (MPDP+) Biochem Biophys Res Commun. 1987;144:692–698. doi: 10.1016/s0006-291x(87)80020-7. [DOI] [PubMed] [Google Scholar]
- 20.Blom TJ, et al. Uptake and accumulation of ajmalicine into isolated vacuoles of cultured cells of Catharanthus roseus (L.) G. Don. and its conversion into serpentine. Planta. 1991;183:170–7. doi: 10.1007/BF00197785. [DOI] [PubMed] [Google Scholar]
- 21.Singh D, et al. Predominance of the serpentine route in monoterpenoid indole alkaloid pathway of Catharanthus roseus. Proc Indian Natl Sci Acad. 2008;74:97–109. [Google Scholar]
- 22.Payne RME, et al. An NPF transporter exports a central monoterpene indole alkaloid intermediate from the vacuole. Nat Plants. 2017;16208:1–9. doi: 10.1038/nplants.2016.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Younai A, Zeng B, Meltzer HY, Scheidt KA. Enantioselective syntheses of heteroyohimbine natural products: a unified approach through cooperative catalysis. Angew Chem Int Ed Engl. 2015;54:6900–4. doi: 10.1002/anie.201502011. [DOI] [PubMed] [Google Scholar]
- 24.Wehrens R, Buydens LMC. Self- and super-organizing maps in R: The kohonen package. J Stat Softw. 2007;21:1–19. [Google Scholar]
- 25.Lindbo JA. TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 2007;145:1232–40. doi: 10.1104/pp.107.106377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ro D, et al. Induction of multiple pleiotropic drug resistance genes in yeast engineered to produce an increased level of anti-malarial drug precursor, artemisinic acid. BMC Biotechnol. 2008;8:83. doi: 10.1186/1472-6750-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen DT, et al. Biochemical Conservation and evolution of germacrene A oxidase in Asteraceae. J Biol Chem. 2010;285:16588–16598. doi: 10.1074/jbc.M110.111757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parage C, et al. Class II Cytochrome P450 Reductase Governs the Biosynthesis of Alkaloids. Plant Physiol. 2016;172:1563–1577. doi: 10.1104/pp.16.00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valentine T, et al. Efficient virus-induced gene silencing in roots using a modified tobacco rattle virus vector. Plant Physiol. 2004;136:3999–4009. doi: 10.1104/pp.104.051466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stavrinides A, et al. Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity. Nat Commun. 2016;7:1–14. doi: 10.1038/ncomms12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guirimand G, et al. Optimization of the transient transformation of Catharanthus roseus cells by particle bombardment and its application to the subcellular localization of hydroxymethylbutenyl 4-diphosphate synthase and geraniol 10-hydroxylase. Plant Cell Rep. 2009;28:1215–34. doi: 10.1007/s00299-009-0722-2. [DOI] [PubMed] [Google Scholar]
- 32.Koike T, Takayama H, Sakai S. Synthetic studies on the picraline-type indole alkaloids-I: improved synthesis of C-Mavacurine-type compounds and a new skeletal rearrangement in a corynanthe-type derivative. Chem Pharm Bull (Tokyo) 1991;39:1677–1681. [Google Scholar]
- 33.Li L, Chang Z, Pan Z, Fu Z-Q, Wang X. Modes of heme binding and substrate access for cytochrome P450 CYP74A revealed by crystal structures of allene oxide synthase. Proc Natl Acad Sci. 2008;105:13883–13888. doi: 10.1073/pnas.0804099105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miettinen K, et al. The seco-iridoid pathway from Catharanthus roseus. Nat Commun. 2014;5:3606. doi: 10.1038/ncomms4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene sequences from this paper have been deposited to GenBank as: MF537711, R. serpentina CYP 12057 (RsSBE, CYP71AY4) and MF537712, G. sempervirens Gs_207-0.13 (GsSBE, CYP71AY5). C. roseus CYP71AY1 (CrAS, AHK60849.1) was deposited by ref. 34. All transcriptome data for R. serpentina and C. roseus are available at http://medicinalplantgenomics.msu.edu/.