Abstract
Background
The community-based Ontology of Biological and Clinical Statistics (OBCS) represents and standardizes biological and clinical data and statistical methods.
Methods
Both OBCS and the Vaccine Ontology (VO) were used to ontologically model various components and relations in a typical host response to vaccination study. Such a model was then applied to represent and compare three microarray studies of host responses to the yellow fever vaccine YF-17D. A literature meta-analysis was then conducted to survey yellow fever vaccine response papers and summarize statistical methods, using OBCS.
Results
A general ontological model was developed to identify major components in a typical host response to vaccination. Our ontology modeling of three similar studies identified common and different components which may contribute to varying conclusions. Although these three studies all used the same vaccine, human blood samples, similar sample collection time post vaccination, and microarray assays, statistically differentially expressed genes and associated gene functions differed, likely due to the differences in specific variables (e.g., microarray type and human variations). Our manual annotation of 95 papers in human responses to yellow fever vaccines identified 38 data analysis methods. These statistical methods were consistently represented and classified with OBCS. Eight statistical methods not available in existing ontologies were added to OBCS.
Conclusions
The study represents the first single use case of applying OBCS ontology to standardize, integrate, and use biomedical data and statistical methods. Our ontology-based meta-analysis showed that different experimental results might be due to different experimental assays and conditions, sample variations, and data analysis methods.
Keywords: OBCS, ontology, vaccine, host response to vaccination, statistical data analysis
INTRODUCTION
Statistics has always played a crucial role in protocol-driven biomedical research. In the era of “Big Data”, this role has become even more prominent, since statistics serves as the central tool in basic biological research and clinical trials. If the data, data processing and analysis, and statistical results of such information-driven research are to be understandable, all of these components should be annotated in a consistent and logically well-defined way. Unfortunately, most statistical studies in biological and clinical research have not been described in this way. Traditional statistical studies performed by independent investigators may have their own standards to process and analyze data. However, such standards are usually not available to other studies performed in different groups. This will affect big data analysis among different studies performed by multiple investigators, since these individually identified standards are only applicable to individual groups, with a lack of a universally standardized method that can be used across multiple related groups. In addition, different data types, statistical methods, and result types have inherent semantic relations, which are not presented in traditional standardized statistical studies. This creates an obstacle to the successful standardization of statistics in the sort of complex projects of analysis and automated inference now typical when very big (and heterogeneous) data are frequently generated and stored. The lack of such a formal representation prevents careful inspection, comparison and reproduction of statistical methods and statistical tests applied to data transformation and analysis, thereby reducing the ability to extract meaningful data from large data sets in the ways required by good scientific practice.
To address the obstacles described above, the Ontology of Biological and Clinical Statistics (OBCS; https://github.com/obcs/obcs) was recently developed as a community-based ontology of statistics in the biological and clinical domains [1]. An ontology is the human-and computer-interpretable representation of the types of entities in a specific scientific domain and of the relations between these types. Since the creation of the first version of the Gene Ontology in 1998 [2], hundreds of ontologies have been created and deposited in resources such as NCBO BioPortal [3] and Ontobee [4]. These ontologies achieved considerable success initially in biological data representation, and have led to a paradigm change in the way terminologies are treated now also in the field of health informatics [5]. The role of ontologies in biomedical data standardization, sharing, and analysis in the big data era has become increasingly important [5–8]. OBCS extends the Ontology of Biomedical Investigations (OBI) [9] and targets the domain of statistics in the biological and clinical fields. Co-developed by some 20 different disciplinary communities, OBI provides a set of logically defined terms covering a broad range of biomedical processes, experimental conditions, and data analysis methods [9]. As an ontology of the Open Biological and Biomedical Ontologies (OBO) Foundry ontology library [10], the OBCS development follows the OBO Foundry principles including openness and collaboration [10]. The original OBCS paper introduced the general OBCS development strategy, top level design, design pattern, and concise examples in biological and clinical areas [11]. With different demonstrations, OBCS allows not only consistent representation, classification, comparison and integration across heterogeneous data-sets, but also computer-assisted automatic reasoning.
In this paper, we will focus on the single domain of vaccinology and illustrate how OBCS can be used to represent and model the studies that have generated data of host immune responses to a specific type of vaccine. For such a study, it is ideal to have a domain-specific ontology. The Vaccine Ontology (VO) represents various entities of vaccines, vaccine components, vaccinations, host responses to vaccines, etc [12–14]. In conjunction with classes from VO, we discuss how OBCS is used to represent vaccine-specific statistical data process and analysis.
Our vaccine-specific study is focused on vaccines against yellow fever, a re-emerging infectious disease caused by the yellow fever virus (a RNA virus of the genus Flavivirus) [15,16]. Spread by the bite of an infected female mosquito, yellow fever is endemic to tropic regions of Africa and America and affects humans and nonhuman primates. Common symptoms include fever, chills, loss of appetite, nausea, muscle pains, and headaches. Liver damage, jaundice, and kidney problems often occur. The live attenuated yellow fever vaccine 17D (YF-17D) was developed by Theiler and Smith in 1937 [17], and substrains derived from the original 17D strain are still used for vaccinations today [16]. Theiler was awarded the Nobel Prize for his life-saving yellow fever vaccine research in 1951, so far the only Nobel Prize awarded for viral vaccine research [18]. Roukens and Visser reported in 2008 that vaccination with YF-17D induced low-grade viraemia in half of the vaccinees and elicited protective neutralizing antibody levels in 99% of the vaccinees [19]. As an effective vaccine against yellow fever viral infection, YF-17D has been used as a vaccine model for analysis of protective vaccine immune mechanism [20–23].
For a comprehensive OBCS application demonstration, OBCS, together with VO and other biomedical ontologies, are used in this study to represent metadata, experimental data, and statistical data analyses of example data sets from yellow fever vaccine research studies.
RESULTS
General ontological modeling of vaccine immune response study and data analysis using OBCS, OBI, and VO
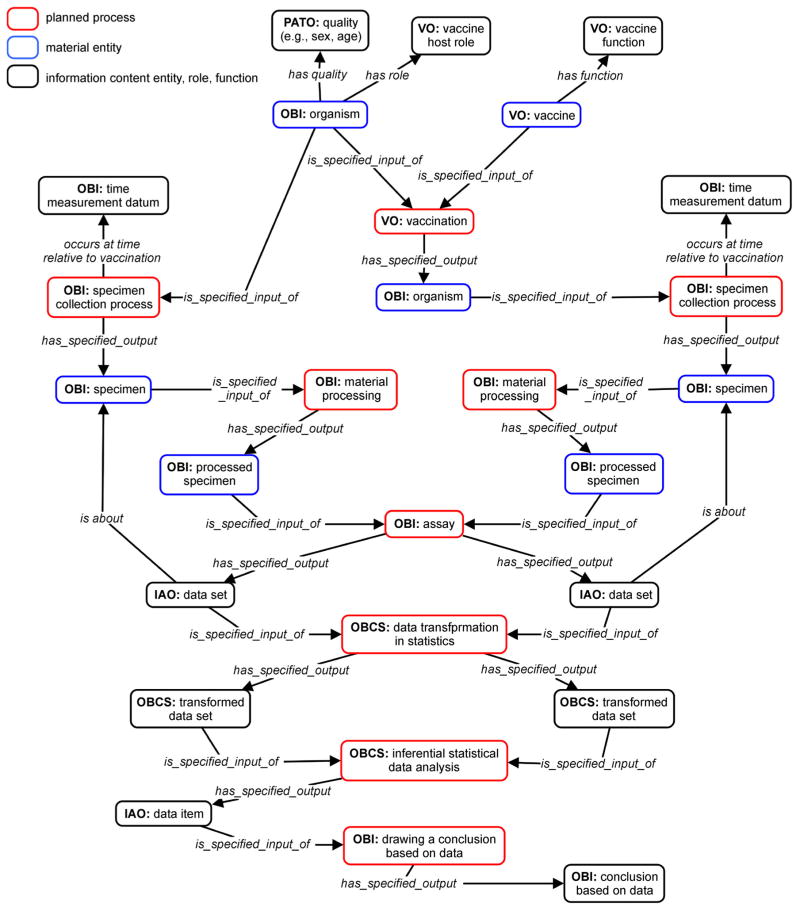
Figure 1 provides a general ontological representation of how OBCS combined with other ontologies is used to represent a typical study of host responses to vaccination and how data can be analyzed. Specifically, an organism (e.g., human) who has the vaccine host role (VO: vaccine host role) is first vaccinated with a specific vaccine, which has the vaccine function (including two subclass functions: preventive vaccine function and therapeutic vaccine function). At a time before or after the vaccination process, there is a specimen collection process (OBI: specimen collection process) which results in the output of a specimen. After material processing (OBI: material processing), the processed specimen’ (OBI: processed specimen) is used as an input of an experimental assay (OBI: assay). Such an assay will generate new data sets (IAO: data sets). The data sets can be used for data transformation in statistics (OBCS: data transformation in statistics). The resulting transformed data sets (OBCS: transformed data set), especially the comparison of transformed data sets which are about different specimens collected at different times relative to an vaccination, are then used for statistical data analysis (OBCS: statistical data analysis), which will generate a specific data item (IAO: data item) (e.g., p-value), which can be used to draw a conclusion (OBI: drawing a conclusion based on data) (Figure 1).
Figure 1. General modeling of vaccine immune response study and data analysis using VO and OBCS.
Ontology terms are indicated by using ontology name abbreviations as prefix (IAO: Information Artifact Ontology, OBCS: Ontology for Biological and Clinical Statistics, OBI: Ontology for Biomedical Investigations, PATO: Phenotypic quality, VO: Vaccine Ontology). Relations are italicized. The terms inside red, blue, and black boxes represent planned processes, material entities, and information content entity/ role/function, respectively. The two datasets (located on the left and right side the figure) are generated, each associated with one experimental condition (i.e., prior to or post vaccination).
As shown in the general data modeling, different components are involved. All these components and the relations among these components in Figure 1 are represented using terms in existing ontologies, mainly OBCS, OBI and VO. Many terms from OBI and IAO (The Information Artifact Ontology, an ontology used for representation of information entities, such as data, document, and protocol, https://github.com/information-artifact-ontology/IAO/) have already been imported to OBCS and/or VO. Note that in vaccine research, a typical study is to compare the host responses before and after vaccination. In this case, time relative to vaccination is an independent variable in a statistical analysis. The time immediately before a vaccination is usually defined as day 0. A time after vaccination can then be defined accordingly. It is also typical to analyze the host responses at different days after vaccination.
OBCS application in representing a typical study of host responses to vaccination
Gaucher et al. published a paper in 2008 that reports their applying functional genomics and polychromatic flow cytometry to define the signatures of human immune responses to yellow fever vaccine 17D (YF-17D), in a total of 43 volunteers at different time points (up to 1 year) after vaccination [22]. For the transcriptomics study, RNA samples obtained from the whole blood of 15 vaccinated individuals on day 0, 3, 7, 10, 14, 28, and 60 after vaccination were collected, amplified and hybridized onto Illumina microarray chips. Their data analysis identified 594 genes whose expression profiles changed significantly between day 0 and any time point after vaccination [22].
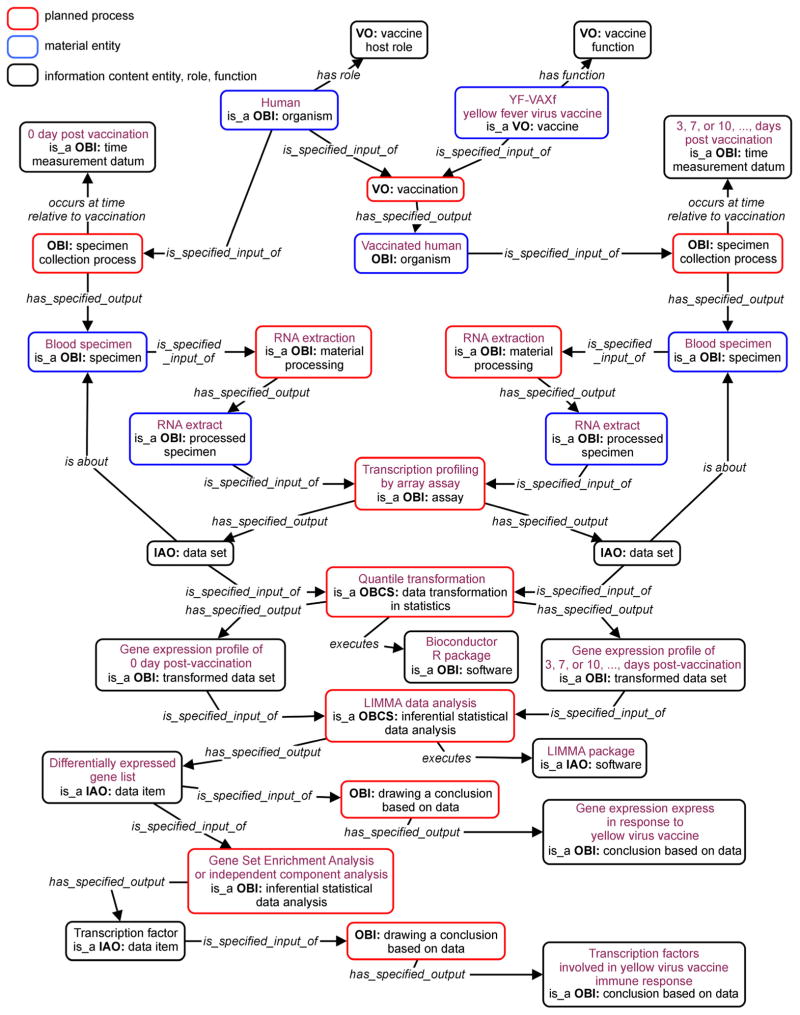
Figure 2 represents the functional genomics study performed by Gaucher et al. [22] that was generated based on the Figure 1 general model. In this use case, the vaccine is YF-VAX (made with a specific YF-17D strain), and the vaccine host is human. The time of specimen collection from blood specimen includes day 0 (before vaccination) and day 3,7,10, etc. after vaccination. The extracted RNAs were used for transcription profiling, which generated a large amount of gene expression data sets. After quantile transformation (a data transformation method), the results were used through LIMMA data analysis [24], which generated differentially expressed gene lists. Based on the gene lists and following Gene Set Enrichment Analysis (GSEA) [25], new results about transcription factors were generated and used to draw further conclusions based on data (Figure 2).
Figure 2. Ontology representation of a representative yellow fever vaccine study.
Following the Figure 1 general ontological model, the modeling is updated based on the study reported by Gaucher et al. [22]. More detail is described in the paper.
OBCS-based comparison of three studies of host responses to YF-17D vaccinations
In addition to the above study of human responses to YF-17D vaccination, there have been two similar studies [23,26]. Specifically, in the study of YF-VAX-vaccinated human responses by Scherer et al. [23], with 20 human subjects (11 females and 9 males) between the ages of 18 and 32 and in good health. Interestingly, their study identified 615 genes exhibiting significant changes in gene expression in peripheral blood mono-nuclear cells (PBMCs) at 4–7 days after vaccination with YF-VAX. The YF-VAX host response from this study was identified with a suppression of ribosomal and translation factors [23]. It appears that although many differentially expressed genes were shared, the results from this study differed significantly from the study by Gaucher et al. [22]. In the third study reported by Querec et al. [26], 15 healthy humans not previously vaccinated with YF-17D participated in the vaccination study. Blood samples were collected at days 0, 1, 3, 7 and 21 after vaccination. The transcriptional profiling of total PBMCs from these 15 subjects was conducted using Affymetrix Human Gen-ome U133 Plus 2.0 Arrays. Interestingly, only 97 genes were identified to be modulated by YF-17D vaccination [26].
These three publications all examined host immune responses to YF-17D vaccine using array approaches to detect gene expression changes in blood cells from different groups of human subjects. Surprisingly, the results from these three studies differed [22,23,26]. Particularly, the numbers of statistically differentially expressed genes in these studies were found different: 594 [22], 615 [23], and 97 [26] (Table 1). We hypothesized that such differences might be due to some differences in experimental designs, microarray data analysis methods, and experimental conditions. Table 1 compares these differences in the three studies in detail. While our comparative study showed lots of common information among three studies, our study also identified some differences. For example, the study by Gaucher et al. [22] used bead chips manufactured by Illumina; however, the study by Scherer et al. [23] used Agilent’s glass slide arrays that are Cy3/Cy5 two-color arrays, and Querec et al. [26] used Affymetrix Human Genome U133 Plus 2.0 microarrays. In addition, due to different kinds of array used, the raw data process software and following statistical analysis methods are different (shown in Table 1).
Table 1.
Detailed comparison between three yellow fever vaccine studies.
| # | Comparison item | Gaucher et al., 2008 | Scherer et al., 2007 | Querec et al., 2009 |
|---|---|---|---|---|
| 1 | Location of study | Montreal, Canada Lausanne, Switzerland Emory University, US |
University of Washington, US | Emory Vaccine Center, US |
| 2 | Human subjects | Montreal cohort, 20 volunteers Lausanne cohort, 13 volunteers Emory cohort, 10 volunteers |
20 healthy humans aged 18–40 | 15 healthy humans aged 18–45 |
| 3 | Vaccine* | Sanofi-Pasteur YF-VAX | Sanofi-Pasteur YF-VAX | YF-17D |
| 4 | Vaccination frequency | Single | Single | Single |
| 5 | Vaccination dose | 0.5 mL per dose | N/A | N/A |
| 6 | Vaccination route | Subcutaneous injection | N/A | N/A |
| 7 | Specimen | Blood samples | Blood samples | Blood samples |
| 8 | Sample preparation | 50 mL blood drawn, anti-coagulation using heparin, PBMCs prepared | 20 mL blood drawn, anti-coagulation using citrate, PBMCs prepared, resuspended in RNAlater, stored frozen at −20°C | Blood drawn, anti-coagulation using citrate, PBMCs prepared, lyzed with TRIzol (Invitrogen), stored at −80°C |
| 9 | Sample preparation data | Day 0, 3, 7, 10, 14, 28, 60 after vaccination | Day 0, 4–8 days after vaccination | Day 0, 1, 3, 7 and 21 after vaccination |
| 10 | RNA preparation | Qiagen PaxGene | RNeasy Mini Kits (Qiagen) | TRIzol (Invitrogen) |
| 11 | RNA amplification | Illumina RNA Amplification Kit | Low RNA Input Fluorescent Linear Amplification Kit | Two-round amplification using Affymetrix protocol |
| 12 | Process afterwards | cDNA synthesis → cRNA synthesis | Cy3/Cy5 two-color labeling, hybrization | Affymetrix GeneChip protocol |
| 13 | Microarry chip type | Illumina Human RefSeq-8 BeadChips v2 |
Agilent human 1 cDNA microarrays, and Human 1A Oligo Microarray Kit (V2) | Affymetrix Human Genome U133 Plus 2.0 Array |
| 14 | Microarry scanner | Illumina BeadStation 500GX scanner | Perkin Elmer ScanArray Express dual-laser microarray scanner | GeneChip Scanner 3000 (Affymetrix) |
| 15 | Raw data process software | Illumina BeadStudio v3 software | N/A | Affymetrix GCOS software |
| 16 | Gene expression filtering | Filter out probes with intensities below background | N/A | Filtering based on a threshold of normalized fold change |
| 17 | Gene expression data analysis method | BioConductor LIMMA | R LIMMA package A moderated empirical Bayes t-statistic and p-value |
R package |
| 18 | Gene annations | Unclear | UniGene build 133 | UniGene build 133 |
| 19 | # genes identified | 594 | 615 | 97 |
| 20 | Gene enrichment | GO enrichement for TF analysis | GO enrichement | Not done |
All three studies use YF-17D vaccine. YF-VAX is a brand name of the YF-17D vaccine manufactured at Sanofi-Pasteur.
Ontology-based meta-analysis of yellow fever vaccine investigation studies
The above two sections introduced three YF-17D vaccine studies. It is clear that they are similar, but also have some differences in their details. To identify similar studies, we performed a systematic literature data analysis annotation. By searching the keyword “human response to yellow fever vaccine” in PubMed, we found 245 related papers. After cleaning and filtering (excluding the invalid papers such as non-English papers, abstract papers, papers with unclear statistical methods, etc.), 95 valid papers were identified (Supplementary Table 1). We went through all the papers and represented the statistical analysis methods consistently using ontology terms. Our mapping of these methods to OBCS identified 8 terms unavailable in OBCS. These new terms were added to OBCS using the standard OBCS development procedure [1]. The latest OBCS release is available at http://purl.obolibrary.org/obo/obcs.owl. The ontology can be visualized on the ontology server Ontobee [4] at: http://www.ontobee.org/ontology/OBCS. The annotation resulted in a total of 38 statistical methods used in these papers (Table 1). These 38 statistical methods cover different types of data transformation and statistical data analysis.
To identify the relations among these 38 methods, we used the Ontofox tool [27] to generate an OBCS subset that contains these methods and their associated parent terms (Figure 3). In contrast to Table 1, the hierarchical display in Figure 3 clearly shows more specific relations among different methods. For example, Figure 3 classifies three specific ANOVA tests (one-way, two-way, and repeated measure ANOVA) under the parent ANOVA test, which is defined as a statistical hypothesis test. Figure 3 also groups five statistical tests under the parent test non-parametric test, which does not require the data to fit a normal distribution.
Figure 3. Hierarchy structure of OBCS statistical methods used in host response to yellow fever vaccine studies.
Specifically, all the statistical methods listed in Table 2 were retrieved from OBCS using the Ontofox tool. The Figure is a screenshot of the Protégé OWL editor visualization of the OBCS subset.
DISCUSSION
In this study, we focus on the specific vaccine host response domain, and demonstrate how the OBCS, combined with the domain-specific Vaccine Ontology (VO), can be used to represent experimental data and metadata, and statistical data analyses of host immune responses to various immunizations with yellow fever vaccine data sets. Our ontology-based methods start with an initial general modeling of host immune response to vaccine study (Figure 1), followed with one typical yellow fever vaccine study (Figure 2), then a comparison between three similar studies (Table 1), and eventually with a meta-analysis of all possible studies reported in PubMed (Table 2). From about 100 peer-reviewed papers, our manual survey and annotations identified 38 statistical methods used in these studies. Our study demonstrates that OBCS, combined with terms from OBI, VO, and other ontologies can be used to comprehensively model and represent experimental studies and related data analysis methods from a single biomedical domain, in a consistent and robust way.
Table 2.
Summarization of statistical methods used in yellow fever vaccine studies.
| # | Statistical method | Method classification | No. of papers |
|---|---|---|---|
| 1 | Network analysis | Data transformation | 2 |
| 2 | ROC curve analysis * | Data transformation | 1 |
| 3 | log2 transformation | Data transformation in statistics | 5 |
| 4 | Lowess transformation | Data transformation in statistics | 1 |
| 5 | Permutation | Data transformation in statistics | 1 |
| 6 | Quantile normalization (transformation) | Data transformation in statistics | 3 |
| 7 | Robust multi-array average normalization | Data transformation in statistics | 2 |
| 8 | Background correction data transformation | Error correction data transformation | 2 |
| 9 | BH false discovery rate correction method | Error correction data transformation | 4 |
| 10 | False discovery rate correction method | Error correction data transformation | 1 |
| 11 | Holm-Bonferroni family-wise error rate correction method | Error correction data transformation | 6 |
| 12 | ANOVA | Inferential statistical data analysis | 13 |
| 13 | Correlation statistical analysis | Inferential statistical data analysis | 1 |
| 14 | Gene Set Enrichment Analysis | Inferential statistical data analysis | 2 |
| 15 | ICA (independent component analysis) * | Inferential statistical data analysis | 1 |
| 16 | Intent-to-treat analysis * | Inferential statistical data analysis | 1 |
| 17 | Least square regression analysis * | Inferential statistical data analysis | 1 |
| 18 | LIMMA data analysis | Inferential statistical data analysis | 3 |
| 19 | Multiple linear regression analysis | Inferential statistical data analysis | 2 |
| 20 | One tailed test | Inferential statistical data analysis | 1 |
| 21 | One-way ANOVA | Inferential statistical data analysis | 10 |
| 22 | Pearson correlation coefficient calculation | Inferential statistical data analysis | 3 |
| 23 | PCA dimensionality reduction | Inferential statistical data analysis | 2 |
| 24 | Repeated measure ANOVA | Iinferential statistical data analysis | 2 |
| 25 | two-way ANOVA | Inferential statistical data analysis | 2 |
| 26 | Chi square test | Statistical hypothesis test | 16 |
| 27 | F test | Statistical hypothesis test | 2 |
| 28 | Fisher’s exact test | Statistical hypothesis test | 15 |
| 29 | Friedman test * | Statistical hypothesis test | 2 |
| 30 | Kolmogorov-Smimov test * | Statistical hypothesis test | 1 |
| 31 | Log-rank test * | statistical hypothesis test | 6 |
| 32 | Mann-Whitney U test | statistical hypothesis test | 20 |
| 33 | Non-parametric test | Statistical hypothesis test | 2 |
| 34 | Paired t-test | Statistical hypothesis test | 8 |
| 35 | Pearson’s Chi-square test * | Statistical hypothesis test | 2 |
| 36 | Student’s t-test | Statistical hypothesis test | 24 |
| 37 | Two tailed test | Statistical hypothesis test | 3 |
| 38 | Wilcoxon signed-rank test | Statistical hypothesis test | 9 |
The methods marked with * are 8 newly added methods.
Three published studies, one reported by Gaucher et al. 2008 [22], one by Scherer et al. 2007 [23], and the last by Querec et al. [26], all used microarrays to detect gene expression profiles in human subjects vaccinated with the same yellow fever vaccine (YF-17D) [22,23]. These studies all used microarray approaches to examine the gene expression profiles in blood cells from human subjects. Although the same vaccine (YF-Vax) and the same LIMMA data analysis were used in two studies [22,23], these two studies generated quite different results. While we can say this is likely due to human variation since the human subjects differ, it is also likely that there were many differences other than human variation. Our ontology-based modeling identified many differences in experimental operations (e.g., different microarray format) and the processing of raw data. Such a study confirmed our hypothesis that these three studies used different experimental designs, had different experimental conditions, and relied upon different data analysis methods and software. While such a conclusion of hypothesis testing might be considered trivial to vaccine research experts, the ontology-based systematic analysis provides a reasonable semantic platform and basis for more advanced computer-assisted analyses (e.g., associations between statistical methods and questions to be addressed, and identification of optimal statistical methods) in the future.
Our meta-analysis of various statistical methods in the yellow fever vaccine host response studies identified nearly 40 statistical methods. Some methods, such as Student’s t-test, Fisher’s exact test, Mann-Whitney U test, ANOVA, and Chi-square test, are commonly used (Table 2). Our OBCS-based classification clearly classifies these methods. The identification and classification of these statistical methods facilitate our better understanding of what and how statistical methods were used in reported yellow fever vaccine studies, and whether these statistical methods applied to similar experimental conditions/designs might have resulted in different results. In the future, we plan to extend our study to other biomedical scenarios and use cases and detect possible associations between statistical methods and experimental/clinical assays.
Although OBCS and VO are two different ontologies, they can be conveniently integrated and merged. One main reason is that both ontologies use the Basic Formal Ontology (BFO) as the upper level ontology [28]. BFO is the default upper level ontology for OBO library ontologies. Over 100 biomedical ontologies, including OBCS and OBI, also use BFO as their upper level ontology. Therefore, the usage of BFO makes all BFO-aligned ontologies well interoperable. Similar to the combined usage OBCS with VO, OBCS can be combined with other biomedical ontologies for the ontological representation of research in different biomedical domains.
Our vaccine case study demonstrates that OBCS, together with VO, can play an important role in standardizing, representing, and analyzing data in vaccine research to facilitate automated and reproducible comparisons of related studies. The OBCS ontology-based data representation can be applied to many areas. First, OBCS combined with a domain specific ontology can serve as a semantic framework that links all pieces of data and data analysis methods together in a logical manner. Second, such an ontology strategy can easily identify inconsistent results and whether the inconsistency is due to different analysis methods. Third, we can also apply the OBCS ontology to build up standardized statistical methods to analyze various data types. Fourth, an OBCS-based system can be used to generate a metadata ontology system to represent ontology-annotated experimental data. In the future, we plan to generate such a metadata ontology based on OBCS and other OBO Foundry ontologies and apply it to standardize all the terms and perform advanced data and metadata-analysis of large datasets combined from multiple studies. Such ontology-based approaches will enhance our discovery of new knowledge based on integration of data from multiple experiments in the same domain.
OBCS supports precision medicine. Precision medicine requires that specific details be modeled to identify the contributing factors to a specific health outcome. As shown in this study, OBCS-based ontology modeling allows the identification of specific variables and their values in various studies within the same research or clinical domain such as the host responses to yellow fever vaccinations shown in this study.
METHODS
Ontological modeling of host responses to vaccinations
The ontological modeling of host responses to vaccinations is primarily based on the OBCS and VO. The standard procedure in ontology modeling was used in this study [1,12–14]. After the general modeling was generated, we applied the modeling to studies of host responses to yellow fever vaccination identified from the PubMed literature database. The YF-17D vaccination was emphasized in this study. We also focused on the topic of different results obtained from similar experimental design, particularly the responses of human subjects to the same YF-17D vaccinations as reported from the three studies [22,23,26]. More studies were conducted as a meta-analysis described below.
Ontology-based meta-analysis of host responses to yellow fever vaccinations
The PubMed literature database was searched by using the keyword “human response to yellow fever vaccine”. Manual annotation was performed to identify statistical methods described in publications, which were then mapped to OBCS. Those terms that were not available in OBCS were added to OBCS using standard OBCS development procedure [9]. The Protégé OWL Editor [29] was used to edit OBCS. The HermiT reasoner (http://hermit-reasoner.com/) tool was used for reasoning over OBCS to detect possible inconsistencies or conflicts.
Generation of an OBCS subset containing related statistics terms used in the study
All related OBCS terms used in this study were output as a subset of OBCS using the Ontofox tool (http://ontofox.hegroup.org) [27]. The generated OBCS subset was displayed using the Protégé OWL Editor version 5.0 beta [30].
Supplementary Material
Acknowledgments
The development of VO and OBCS and the vaccine modeling research was supported by a grant from the USA National Institute of Allergy and Infectious Diseases (NIAID) (R01AI081062). We appreciate Mr. Omar Tibi’s proofreading and editorial changes of this manuscript.
Footnotes
The supplementary materials can be found online with this article at DOI 10.1007/s40484-017-0122-5.
COMPLIANCE WITH ETHICS GUIDELINES
The authors Jie Zheng, Hua Li, Qingzhi Liu, and Yongqun He declare they have no conflict of interests. All the data sets the authors used are from public repositories.
References
- 1.Zheng J, Harris MR, Masci AM, Lin Y, Hero A, Smith B, He Y. The Ontology of Biological and Clinical Statistics (OBCS) for standardized and reproducible statistical analysis. J Biomed Semantics. 2016;7:53. doi: 10.1186/s13326-016-0100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salvadores M, Alexander PR, Musen MA, Noy NF. BioPortal as a dataset of linked biomedical ontologies and terminologies in RDF. Semant Web. 2013;4:277–284. [PMC free article] [PubMed] [Google Scholar]
- 4.Ong E, Xiang Z, Zhao B, Liu Y, Lin Y, Zheng J, Mungall C, Courtot M, Ruttenberg A, He Y. Ontobee: A linked ontology data server to support ontology term dereferencing, linkage, query and integration. Nucleic Acids Res. 2017;45:D347–D352. doi: 10.1093/nar/gkw918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulz S, Balkanyi L, Cornet R, Bodenreider O. From concept representations to ontologies: a paradigm shift in health informatics? Healthc Inform Res. 2013;19:235–242. doi: 10.4258/hir.2013.19.4.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blake JA, Bult CJ. Beyond the data deluge: data integration and bio-ontologies. J Biomed Inform. 2006;39:314–320. doi: 10.1016/j.jbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Hoehndorf R, Schofield PN, Gkoutos GV. The role of ontologies in biological and biomedical research: a functional perspective. Brief Bioinform. 2015;16:1069–1080. doi: 10.1093/bib/bbv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform. 2009:67–79. [PMC free article] [PubMed] [Google Scholar]
- 9.Bandrowski A, Brinkman R, Brochhausen M, Brush MH, Bug B, Chibucos MC, Clancy K, Courtot M, Derom D, Dumontier M, et al. The ontology for biomedical investigations. PLoS One. 2016;11:e0154556. doi: 10.1371/journal.pone.0154556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng J, Harris MR, Masci AM, Lin Y, Hero A, Smith B, He Y. The Ontology of Biological and Clinical Statistics (OBCS) for standardized and reproducible statistical analysis. J Biomed Semantics. 2016;7:53. doi: 10.1186/s13326-016-0100-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He Y, Cowell L, Diehl AD, Mobley HL, Peters B, Ruttenberg A, Scheuermann RH, Brinkman RR, Courtot M, Mungall C, et al. VO: Vaccine Ontology. The 1st International Conference on Biomedical Ontology (ICBO-2009). Nature Precedings; 2009. http://precedings.nature.com/documents/3552/version/1. [Google Scholar]
- 13.Özgür A, Xiang Z, Radev DR, He Y. Mining of vaccine-associated IFN-γ gene interaction networks using the Vaccine Ontology. J Biomed Semantics. 2011;2:S8. doi: 10.1186/2041-1480-2-S2-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y, He Y. Ontology representation and analysis of vaccine formulation and administration and their effects on vaccine immune responses. J Biomed Semantics. 2012;3:17. doi: 10.1186/2041-1480-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beasley DW, McAuley AJ, Bente DA. Yellow fever virus: genetic and phenotypic diversity and implications for detection, prevention and therapy. Antiviral Res. 2015;115:48–70. doi: 10.1016/j.antiviral.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Gardner CL, Ryman KD. Yellow fever: a reemerging threat. Clin Lab Med. 2010;30:237–260. doi: 10.1016/j.cll.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theiler M, Smith HH. The use of yellow fever virus modified by in vitro cultivation for human immunization. J Exp Med. 1937;65:787–800. doi: 10.1084/jem.65.6.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Norrby E. Yellow fever and Max Theiler: the only Nobel Prize for a virus vaccine. J Exp Med. 2007;204:2779–2784. doi: 10.1084/jem.20072290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roukens AH, Visser LG. Yellow fever vaccine: past, present and future. Expert Opin Biol Ther. 2008;8:1787–1795. doi: 10.1517/14712598.8.11.1787. [DOI] [PubMed] [Google Scholar]
- 20.Pulendran B. Learning immunology from the yellow fever vaccine: innate immunity to systems vaccinology. Nat Rev Immunol. 2009;9:741–747. doi: 10.1038/nri2629. [DOI] [PubMed] [Google Scholar]
- 21.Pulendran B, Li S, Nakaya HI. Systems vaccinology. Immunity. 2010;33:516–529. doi: 10.1016/j.immuni.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, Moser JM, Mehta RS, Drake DR, 3rd, Castro E, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med. 2008;205:3119–3131. doi: 10.1084/jem.20082292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scherer CA, Magness CL, Steiger KV, Poitinger ND, Caputo CM, Miner DG, Winokur PL, Klinzman D, McKee J, Pilar C, et al. Distinct gene expression profiles in peripheral blood mononuclear cells from patients infected with vaccinia virus, yellow fever 17D virus, or upper respiratory infections. Vaccine. 2007;25:6458–6473. doi: 10.1016/j.vaccine.2007.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smyth GK, Ritchie M, Thorne N, Wettenhall J. Statistics for Biology and Health. New York: Springer; 2005. Limma: Linear Models for Microarray Data; pp. 397–420. [Google Scholar]
- 25.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang Z, Courtot M, Brinkman RR, Ruttenberg A, He Y. OntoFox: web-based support for ontology reuse. BMC Res Notes. 2010;3:175. doi: 10.1186/1756-0500-3-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arp R, Smith B, Spear AD. Building Ontologies With Basic Formal Ontology. Cambridge: MIT Press; 2015. [Google Scholar]
- 29.Musen MA. The protégé project: a look back and a look forward. AI Matters. 2015;1:4–12. doi: 10.1145/2757001.2757003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The protege ontology editor. http://protege.stanford.edu/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.