A simple physical model enables rapid detection and extraction of linear hierarchies in large networks.
Abstract
We present a physically inspired model and an efficient algorithm to infer hierarchical rankings of nodes in directed networks. It assigns real-valued ranks to nodes rather than simply ordinal ranks, and it formalizes the assumption that interactions are more likely to occur between individuals with similar ranks. It provides a natural statistical significance test for the inferred hierarchy, and it can be used to perform inference tasks such as predicting the existence or direction of edges. The ranking is obtained by solving a linear system of equations, which is sparse if the network is; thus, the resulting algorithm is extremely efficient and scalable. We illustrate these findings by analyzing real and synthetic data, including data sets from animal behavior, faculty hiring, social support networks, and sports tournaments. We show that our method often outperforms a variety of others, in both speed and accuracy, in recovering the underlying ranks and predicting edge directions.
INTRODUCTION
In systems of many individual entities, interactions and their outcomes are often correlated with these entities’ ranks or positions in a hierarchy. While in most cases these rankings are hidden from us, their presence is nevertheless revealed in the asymmetric patterns of interactions that we observe. For example, some social groups of birds, primates, and elephants are organized according to dominance hierarchies, reflected in patterns of repeated interactions in which dominant animals tend to assert themselves over less powerful subordinates (1). Social positions are not directly visible to researchers, but we can infer each animal’s position in the hierarchy by observing the network of pairwise interactions. Similar latent hierarchies have been hypothesized in systems of endorsement in which status is due to prestige, reputation, or social position (2, 3). For example, in academia, universities may be more likely to hire faculty candidates from equally or more prestigious universities (3).
In all these cases, the direction of the interactions is affected by the status, prestige, or social position of the entities involved. But it is often the case that even the existence of an interaction, rather than its direction, contains some information about those entities’ relative prestige. For example, in some species, animals are more likely to interact with others who are close in dominance rank (4–8); human beings tend to claim friendships with others of similar or slightly higher status (9); and sports tournaments and league structures are often designed to match players or teams on the basis of similar skill levels (10, 11). This suggests that we can infer the ranks of individuals in a social hierarchy using both the existence and the direction of their pairwise interactions. It also suggests assigning real-valued ranks to entities rather than simply ordinal rankings, for instance, to infer clusters of entities with roughly equal status with gaps between them.
Here, we introduce a physically inspired model that addresses the problems of hierarchy inference, edge prediction, and significance testing. The model, which we call SpringRank, maps each directed edge to a directed spring between the nodes that it connects and finds real-valued positions of the nodes that minimize the total energy of these springs. Because this optimization problem requires only linear algebra, it can be solved for networks of millions of nodes and edges in seconds.
We also introduce a generative model for hierarchical networks in which the existence and direction of edges depend on the relative ranks of the nodes. This model formalizes the assumption that individuals tend to interact with others of similar rank, and it can be used to create synthetic benchmark networks with tunable levels of hierarchy and noise. It can also predict unobserved edges, allowing us to use cross-validation as a test of accuracy and statistical significance. Moreover, the maximum likelihood estimates of the ranks coincide with SpringRank asymptotically.
We test SpringRank and its generative model version on both synthetic and real data sets, including data from animal behavior, faculty hiring, social support networks, and sports tournaments. We find that it infers accurate rankings, provides a simple significance test for hierarchical structure, and can predict the existence and direction of as-yet unobserved edges. In particular, we find that SpringRank often predicts the direction of unobserved edges more accurately than a variety of existing methods, including popular spectral techniques, minimum violation ranking (MVR), and the Bradley-Terry-Luce (BTL) method.
Related work
Ranking entities in a system from pairwise comparisons or interactions is a fundamental problem in many contexts, and many methods have been proposed. One family consists of spectral methods such as eigenvector centrality (12), PageRank (13), rank centrality (14), and the method of Callaghan et al. (15). These methods propose various types of random walks on the directed network and therefore produce real-valued scores. However, by design, these methods tend to give high ranks to a small number of important nodes, giving us little information about the lower-ranked nodes. In addition, they often require explicit regularization by adding a small term to every element of the adjacency matrix if the graph of comparisons is not strongly connected.
A second family focuses on ordinal rankings, that is, permutations, that minimize various penalty functions. This family includes MVR (16–18), SerialRank (19), and SyncRank (20). MVR imposes a uniform penalty for every violation or “upset,” defined as an edge that has a direction opposite to the one expected by the rank difference between the two nodes. Nonuniform penalties and other generalizations are often referred to as agony methods (21). For common choices of the penalty function, minimization can be computationally difficult (17, 22), forcing us to use simple heuristics that find local minima.
SerialRank constructs a matrix of similarity scores between each pair of nodes by examining whether they produce similar outcomes when compared with the other nodes, thereby relating the ranking problem to a more general ordering problem called seriation. SyncRank is a hybrid method that first solves a spectral problem based on synchronization, embeds node positions on a half-circle in the complex plane, and then chooses among the circular permutations of those ranks by minimizing the number of violations as in MVR.
Random utility models (23), such as the BTL model (24, 25), are designed to infer real-valued ranks from data on pairwise preferences. These models assign a probability to the direction of an edge conditioned on its existence, but they do not assign a probability to the existence of an edge. They are appropriate, for instance, when an experimenter presents subjects with choices between pairs of items and asks them which they prefer.
Methods such as David’s score (26) and the Colley matrix (27) compute rankings from proportions of wins and losses. The latter, which was originally developed by making mathematical adjustments to winning percentages, is equivalent to a particular case of the general method we introduce below. Elo score (28), Go rank (29), and TrueSkill (30) are also widely used win-loss methods, but these schemes update the ranks after each match rather than taking all previous interactions into account. This specialization makes them useful when ranks evolve over sequential matches, but less useful otherwise.
Finally, there are fully generative models such as the probabilistic niche model of ecology (31–33), models of friendship based on social status (9), and, more generally, latent space models (34), which assign probabilities to the existence and direction of edges based on real-valued positions in social space. However, inference of these models tends to be difficult, with many local optima. Our generative model can be viewed as a special case of these models for which inference is especially easy.
In the absence of ground-truth rankings, we can compare the accuracy of these methods using cross-validation, computing the ranks using a subset of the edges in the network, and then using those ranks to predict the direction of the remaining edges. Equivalently, we can ask them to predict unobserved edges, such as which of two sports teams will win a game. However, these methods do not all make the same kinds of predictions, requiring us to use different kinds of cross-validation. Methods such as BTL produce probabilistic predictions about the direction of an edge, that is, they estimate the probability that one item will be preferred to another. Fully generative models also predict the probability that an edge exists, that is, that a given pair of nodes in the network interact. On the other hand, ordinal ranking methods such as MVR do not make probabilistic predictions, but we can interpret their ranking as a coarse prediction that an edge is more likely to point in one direction than another.
RESULTS
The SpringRank model
We represent interactions between N entities as a weighted, directed network, where Aij is the number of interactions i → j suggesting that i is ranked above j. This allows both ordinal and cardinal input, including where pairs interact multiple times. For instance, Aij could be the number of fights between i and j that i has won or the number of times that j has endorsed i.
Given the adjacency matrix A, our goal is to find a ranking of the nodes. To do so, the SpringRank model computes the optimal location of nodes in a hierarchy by imagining the network as a physical system. Specifically, each node i is embedded at a real-valued position or rank si, and each directed edge i → j becomes an oriented spring with a nonzero resting length and displacement si − sj. Since we are free to rescale the latent space and the energy scale, we set the spring constant and the resting length to 1. Thus, the spring corresponding to an edge i → j has energy
| (1) |
which is minimized when si − sj = 1.
This version of the model has no tunable parameters. Alternately, we could allow each edge to have its own rest length or spring constant, based on the strength of each edge. However, this would create a large number of parameters, which we would have to infer from the data or choose a priori. We do not explore this here.
According to this model, the optimal rankings of the nodes are the ranks , which minimize the total energy of the system given by the Hamiltonian
| (2) |
Since this Hamiltonian is convex in s, we can find s* by setting ∇H(s) = 0, yielding the linear system
| (3) |
where 1 is the all-ones vector and Dout and Din are diagonal matrices whose entries are the weighted in- and out-degrees, and . See section S1 for detailed derivations.
The matrix on the left side of Eq. 3 is not invertible. This is because H is translation-invariant: It depends only on the relative ranks si − sj, so that if s* = {si} minimizes H(s), then so does {si + a} for any constant a. One way to break this symmetry is to invert the matrix in the subspace orthogonal to its nullspace by computing a Moore-Penrose pseudoinverse. If the network consists of a single component, then the nullspace is spanned by the eigenvector 1, in which case, this method finds the s* where the average rank (1/N)∑isi = (1/N)s* ⋅ 1 is zero. This is related to the random walk method of (15): If a random walk moves along each directed edge with rate and against each one with rate , then s* is proportional to the perturbation to the stationary distribution to first order in ε.
In practice, it is more efficient and accurate to fix the rank of one of the nodes and solve the resulting equation using a sparse iterative solver (see section S1). Faster still, because this matrix is a Laplacian, recent results (35, 36) allow us to solve Eq. 3 in nearly linear time in M, the number of nonzero edges in A.
Another way to break translation invariance is to introduce an “external field” affecting each node, so that the combined Hamiltonian is
| (4) |
The field H0 corresponds to a spring that attracts every node to the origin. We can think of this as imposing a Gaussian prior on the ranks or as a regularization term that quadratically penalizes ranks with large absolute values. This version of the model has a single tunable parameter, namely, the spring constant α. Since H(s) scales with the total edge weight M = ∑i,jAij while H0(s) scales with N, for a fixed value of α, this regularization becomes less relevant as networks become denser and the average (weighted) degree M/N increases.
For α > 0, there is a unique s* that minimizes Hα, given by
| (5) |
where is the identity matrix. The matrix on the left side is now invertible, since the eigenvector 1 has eigenvalues α instead of 0. In the limit α → 0, we recover Eq. 3; the value α = 2 corresponds to the Colley matrix method (27).
Minimizing H(s), or the regularized version Hα(s), corresponds to finding the “ground state” s* of the model. In the next section, we show that this corresponds to a maximum likelihood estimate of the ranks in a generative model. However, we can use SpringRank not just to maximize the likelihood, but to compute a joint distribution of the ranks as a Boltzmann distribution with Hamiltonian Eq. 4, and thus estimate the uncertainty and correlations between the ranks. In particular, the ranks si are random variables following an N-dimensional Gaussian distribution with mean s* and covariance matrix (section S4)
| (6) |
Here, β is an inverse temperature controlling the amount of noise in the model. In the limit β → ∞, the rankings are sharply peaked around the ground state s*, while for β → 0, they are noisy. As we discuss below, we can estimate β from the observed data in various ways.
The rankings given by SpringRank Eq. 3 and its regularized form Eq. 5 are easily and rapidly computed by standard linear solvers. In particular, iterative solvers that take advantage of the sparsity of the system can find s* for networks with millions of nodes and edges in seconds. However, as defined above, SpringRank is not a fully generative model that assigns probabilities to the data and allows for Bayesian inference. In the next section, we introduce a generative model for hierarchical networks and show that it converges to SpringRank in the limit of strong hierarchy.
A generative model
In this section, we propose a probabilistic generative model that takes as its input a set of node ranks s1, …, sN and produces a weighted, directed network. The model also has a temperature or noise parameter β and a density parameter c. Edges between each pair of nodes i, j are generated independently of other pairs, conditioned on the ranks. The expected number of edges from i to j is proportional to the Boltzmann weight of the corresponding term in the Hamiltonian Eq. 2
where the actual edge weight Aij is drawn from a Poisson distribution with this mean. The parameter c controls the overall density of the network, giving an expected number of edges
while the inverse temperature β controls the extent to which edges respect (or defy) the ranks s. For smaller β, edges are more likely to violate the hierarchy or to connect distant nodes, decreasing the correlation between the ranks and the directions of the interactions: For β = 0, the model generates a directed Erdős-Rényi graph, while in the limit β → ∞, edges only exist between nodes i, j with si − sj = 1, and only in the direction i → j.
The Poisson distribution may generate multiple edges between a pair of nodes, so this model generates directed multigraphs. This is consistent with the interpretation that Aij is the number, or total weight, of edges from i to j. However, in the limit , the Poisson distribution approaches a Bernoulli distribution, generating binary networks with Aij ∈ {0, 1}.
The likelihood of observing a network A given ranks s, inverse temperature β, and density c is
| (7) |
Taking logs, substituting the maximum likelihood value of c, and discarding constants that do not depend on s or β yields a log likelihood (see section S2)
| (8) |
where H(s) is the SpringRank energy defined in Eq. 2. In the limit of large β where the hierarchical structure is strong, the that maximizes Eq. 8 approaches the solution s* of Eq. 3 that minimizes H(s). Thus, the maximum likelihood estimate of the rankings in this model approaches the SpringRank ground state.
As discussed above, we can break translational symmetry by adding a field H0 that attracts the ranks to the origin. This is is equivalent to imposing a prior . The maximum a posteriori estimate then approaches the ground state s* of the Hamiltonian in Eq. 4, which is given by Eq. 5.
This model belongs to a larger family of generative models considered in ecology and network theory (9, 31, 32), and, more generally, the class of latent space models (34), where an edge points from i to j with probability f(si − sj) for some function f. These models typically have complicated posterior distributions with many local optima, requiring Monte Carlo methods [for example, (33)] that do not scale efficiently to large networks. In our case, f(si − sj) is a Gaussian centered at 1, and the posterior converges to the multivariate Gaussian Eq. 6 in the limit of strong structure.
Predicting edge directions
If hierarchical structure plays an important role in a system, then it should allow us to predict the direction of previously unobserved interactions, such as the winner of an upcoming match or which direction social support will flow between two individuals. This is a kind of cross-validation, which lets us test the statistical significance of hierarchical structure. It is also a principled way of comparing the accuracy of various ranking methods for data sets where no ground-truth ranks are known.
We formulate the edge prediction question as follows: Given a set of known interactions, and given that there is an edge between i and j, in which direction does it point? In one sense, any ranking method provides an answer to this question, since we can predict the direction according to whether i or j is ranked higher on the basis of the known interactions. When comparing SpringRank to methods such as SyncRank, SerialRank, and MVR, we use these “bitwise” predictions and define the accuracy σb as the fraction of edges whose direction is consistent with the inferred ranking.
But we want to know the odds on each game, not just the likely winner—that is, we want to estimate the probability that an edge goes in each direction. A priori, a ranking algorithm does not provide these probabilities unless we make further assumptions about how they depend on the relative ranks. These assumptions yield generative models such as the one defined above, where the conditional probability of an edge i → j is
| (9) |
The density parameter c affects the probability that an edge exists, but not its direction. Thus, our probabilistic prediction method has a single tunable parameter, β.
Note that Pij is a logistic curve, is monotonic in the rank difference si − sj, and has width determined by the inverse temperature β. SpringRank has this in common with two other ranking methods: Setting recovers the BTL model (24, 25) for which Pij = γi/(γi + γj), and setting k = 2β recovers the probability that i beats j in the Go rank (29), where . However, SpringRank differs from these methods in how it infers the ranks from observed interactions, so SpringRank and BTL make different probabilistic predictions.
In our experiments below, we test various ranking methods for edge prediction by giving them access to 80% of the edges in the network (the training data) and then asking them to predict the direction of the remaining edges (the test data). We consider two measures of accuracy: σa is the average probability assigned to the correct direction of an edge, and σL is the log likelihood of generating the directed edges given their existence. For simple directed graphs where Aij + Aji ∈ {0, 1}, these are
| (10) |
In the multigraph case, we ask how well Pij approximates the fraction of interactions between i and j that point from i to j (see Eqs. 12 and 13). For a discussion of other performance measures, see section S9.
We perform our probabilistic prediction experiments as follows. Given the training data, we infer the ranks using Eq. 5. We then choose the temperature parameter β by maximizing either σa or σL on the training data while holding the ranks fixed. The resulting values of β, which we denote and , respectively, are generally distinct (table S2 and section S7). This is intuitive, since a single severe mistake where Aij = 1 but Pij ≈ 0 reduces the likelihood by a large amount, while only reducing the accuracy by one edge. As a result, predictions using produce fewer incorrectly oriented edges, achieving a higher σa on the test set, while predictions using will produce fewer markedly incorrect predictions where Pij is very low, and thus achieve higher σL on the test set.
Statistical significance using the ground-state energy
We can measure statistical significance using any test statistic, by asking whether its value on a given data set would be highly improbable in a null model. One such statistic is the accuracy of edge prediction using a method such as the one described above. However, this may become computationally expensive for cross-validation studies with many replicates, since each fold of each replicate requires inference of the parameter . Here, we propose a test statistic that is very easy to compute, inspired by the physical model behind SpringRank: namely, the ground-state energy. For the unregularized version Eq. 2, the energy per edge is (see section S3)
| (11) |
Since the ground-state energy depends on many aspects of the network structure, and since hierarchical structure is statistically significant if it helps us predict edge directions, we focus on the following null model, used previously by de Silva et al. (37): We randomize the direction of each edge while preserving the total number of edges between each pair of vertices. If the real network has a ground-state energy, which is much lower than typical networks drawn from this null model, then we can conclude that the hierarchical structure is statistically significant.
This test correctly concludes that directed Erdős-Rényi graphs have no significant structure. It also finds no significant structure for networks created using the generative model Eq. 7 with β = 0.1, that is, when the temperature or noise level 1/β is sufficiently large, the ranks are no longer relevant to edge existence or direction (fig. S2). However, we see in the next section that it shows statistically significant hierarchy for a variety of real-world data sets, showing that H(s*) is both useful and computationally efficient as a test statistic.
Performance on real and synthetic data
Having introduced SpringRank, an efficient procedure for inferring real-valued ranks, a corresponding generative model, a method for edge prediction, and a test for the statistical significance of hierarchical structure, we now demonstrate it by applying it to both real and synthetic data. For synthetic data sets where the ground-truth ranks are known, our goal is to see to what extent SpringRank and other algorithms can recover the actual ranks. For real-world data sets, in most cases, we have no ground-truth ranking, so we apply the statistical significance test defined above and compare the ability of SpringRank and other algorithms to predict edge directions given a subset of the interactions.
We compare SpringRank to other widely used methods: the spectral methods PageRank (13), eigenvector centrality (12), and rank centrality (14); MVR (16, 17), SerialRank (19), and SyncRank (20), which produce ordinal rankings; David’s score (26); and the BTL random utility model (24, 25) using the algorithm proposed in (38), which, like our generative model, makes probabilistic predictions. We also compare unregularized SpringRank with the regularized version α = 2, corresponding to the Colley matrix method (27). Unfortunately, eigenvector centrality, rank centrality, David’s score, and BTL are undefined when the network is not strongly connected, for example, when there are nodes with zero in- or out-degree. In these cases, we follow the common regularization procedure of adding low-weight edges between every pair of nodes (see section S10).
Performance for synthetic networks
We study two types of synthetic networks, generated by the model described above. Of course, since the log likelihood in this model corresponds to the SpringRank energy in the limit of large β, we expect SpringRank to do well on these networks, and its performance should be viewed largely as a consistency check. But by varying the distribution of ranks and the noise level, we can illustrate types of structure that may exist in real-world data and test each algorithm’s ability to identify them.
In the first type, the ranks are normally distributed with mean zero and variance one (Fig. 1A). In the second type, the ranks are drawn from an equal mixture of three Gaussians with different means and variances, so that nodes cluster into high, middle, and low tiers (Fig. 1C). This second type is intended to focus on the importance of real-valued ranks and to measure the performance of algorithms that (implicitly or explicitly) impose strong priors on the ranks when the data defy their expectations. In both cases, we vary the amount of noise by changing β while keeping the total number of edges constant (see Materials and Methods).
Fig. 1. Performance on synthetic data.
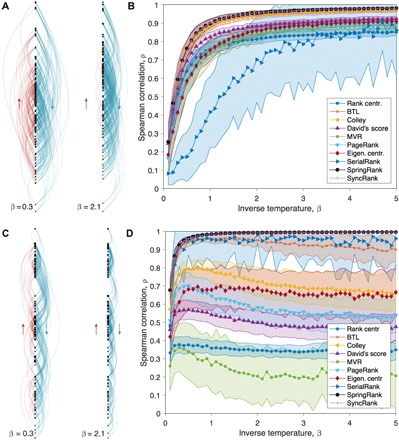
(A) A synthetic network of N = 100 nodes, with ranks drawn from a standard Gaussian and edges drawn via the generative model Eq. 7 for two different values of β and average degree 5. Blue edges point down the hierarchy and red edges point up, indicated by arrows. (B) Accuracy of the inferred ordering defined as the Spearman correlation averaged over 100 independently generated networks. Error bars indicate 1 SD. (C and D) Identical to (A) and (B) but with ranks drawn from a mixture of three Gaussians so that the nodes cluster into three tiers (Materials and Methods). See fig. S1 for performance curves for Pearson correlation r.
Since we wish to compare SpringRank both to methods such as MVR that only produce ordinal rankings and to those like PageRank and David’s score that produce real-valued ranks, we measure the accuracy of each algorithm according to the Spearman correlation ρ between its inferred rank order and the true one. Results for the Pearson correlation, where we measure the algorithms’ ability to infer the real-valued ranks as opposed to just their ordering, are shown in fig. S1.
We find that all the algorithms do well on the first type of synthetic network. As β increases so that the network becomes more structured, with fewer edges (shown in red in Fig. 1A) pointing in the “wrong” direction, all algorithms infer ranks that are more correlated with the ground truth. SpringRank and SyncRank have the highest accuracy, followed closely by the Colley matrix method and BTL (Fig. 1B). Presumably, the Colley matrix works well here because the ranks are drawn from a Gaussian prior, as it implicitly assumes.
Results for the second type of network are more nuanced. The accuracy of SpringRank and SyncRank increases rapidly with β with exact recovery around β = 1. SerialRank also performs quite well on average. The other methods do not improve as β increases, and many of them decrease beyond a certain point (Fig. 1D). This suggests that these algorithms become confused when the nodes are clustered into tiers, even when the noise is small enough that most edges have directions consistent with the hierarchy. SpringRank takes advantage of the fact that edges are more likely between nodes in the same tier (Fig. 1C), so the mere existence of edges helps it cluster the ranks.
These synthetic tests suggest that real-valued ranks capture information that ordinal ranks do not and that many ranking methods perform poorly when there are substructures in the data such as tiered groups. Of course, in most real-world scenarios, the ground-truth ranks are not known, and thus edge prediction and other forms of cross-validation should be used instead. We turn to edge prediction in the next section.
Performance for real-world networks
As discussed above, in most real-world networks, we have no ground truth for the ranks. Thus, we focus on our ability to predict edge directions from a subset of the data and measure the statistical significance of the inferred hierarchy.
We apply our methods to data sets from a diverse set of fields, with sizes ranging up to N = 415 nodes and up to 7000 edges (see table S2): three North American academic hiring networks, where Aij is the number of faculty at university j who received their doctorate from university i, for History (illustrated in Fig. 2, A and B), Business, and Computer Science departments (3); two networks of animal dominance among captive monk parakeets (5) and one among Asian elephants (37), where Aij is the number of dominating acts by animal i toward animal j; and social support networks from two villages in Tamil Nadu referred to (for privacy reasons) by the pseudonyms “Tenpaṭṭi” and “Alakāpuram,” where Aij is the number of distinct social relationships (up to five) through which person i supports person j (2); and 53 networks of National Collegiate Athletic Association (NCAA) Women’s and Men’s college basketball matches during the regular season, spanning 1985–2017 (Men) and 1998–2017 (Women), where Aij = 1 if team i beat team j. Each year’s network comprises a different number of matches, ranging from 747 to 1079 (39).
Fig. 2. Ranking the history faculty hiring network.
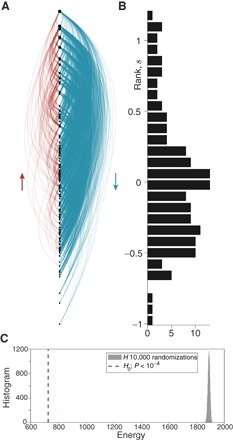
(A) Linear hierarchy diagram with nodes embedded at their inferred SpringRank scores. Blue edges point down the hierarchy and red edges point up. (B) Histogram of the empirical distribution of ranks, with a vertical axis of ranks matched to (A). (C) Histogram of ground-state energies from 10,000 randomizations of the network according to the null model where edge directions are random. The dashed red line shows the ground-state energy of the empirical network depicted in (A) and (B). The fact that the ground-state energy is so far below the tail of the null model is overwhelming evidence that the hierarchical structure is statistically significant, with P < 10−4.
Together, these examples cover prestige, dominance, and social hierarchies. In each of these domains, inferring ranks from interactions is key to further analysis. Prestige hierarchies play an unequivocal role in the dynamics of academic labor markets (40); in behavioral ecology, higher-ranked individuals in dominance hierarchies are believed to have higher fitness (1, 41); and patterns of aggression are believed to reveal animal strategies and cognitive capacities (4–8). Finally, in social support networks, higher-ranked individuals have greater social capital and reputational standing (42, 43), particularly in settings in which social support is a primary way to express and gain respect and prestige (44).
We first applied our ground-state energy test for the presence of statistically significant hierarchy, rejecting the null hypothesis with P < 10− 4 in almost all cases (for example, for history faculty hiring, see Fig. 2C). The one exception is the Asian elephants network for which P > 0.4. This corroborates the original study of this network (37), which found that counting triad motifs shows no significant hierarchy (45). This is despite the fact that one can find an appealing ordering of the elephants using the MVR method, with just a few violating edges (fig. S9). Thus, the hierarchy found by MVR may well be illusory.
As described above, we performed edge prediction experiments using fivefold cross-validation, where 80% of the edges are available to the algorithm as training data, and a test set consisting of 20% of the edges is held out (see Materials and Methods). To test SpringRank’s ability to make probabilistic predictions, we compare it to BTL.
We found that SpringRank outperforms BTL, both in terms of the accuracy σa (Fig. 3A) and, for most networks, the log likelihood σL (Fig. 3B). The accuracy of both methods has a fairly broad distribution over the trials of cross-validation, since in each network some subsets of the edges are harder to predict than others when they are held out. However, as shown in Fig. 4, in most trials, SpringRank was more accurate than BTL. Figure 3A and Table 1 show that SpringRank predicts edge directions more accurately in the majority of trials of cross-validation for all nine real-world networks, where this majority ranges from 62% for the parakeet networks to 100% for the computer science hiring network.
Fig. 3. Edge prediction accuracy over BTL.
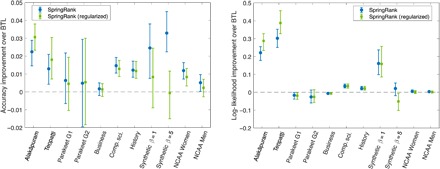
Distribution of differences in performance of edge prediction of SpringRank compared to BTL on real and synthetic networks defined as (A) edge prediction accuracy σa Eq. 12 and (B) the conditional log likelihood σL Eq. 13. Error bars indicate quartiles and markers show medians, corresponding to 50 independent trials of fivefold cross-validation, for a total of 250 test sets for each network. The two synthetic networks are generated with N = 100, average degree 5, and Gaussian-distributed ranks as in Fig. 1A, with inverse temperatures β = 1 and β = 5. For each experiment shown, the fractions of trials in which each method performed equal to or better than BTL are shown in Table 1. These differences correspond to prediction of an additional 1 to 12 more correct edge directions, on average.
Fig. 4. Probabilistic edge prediction accuracy σa of SpringRank versus BTL.
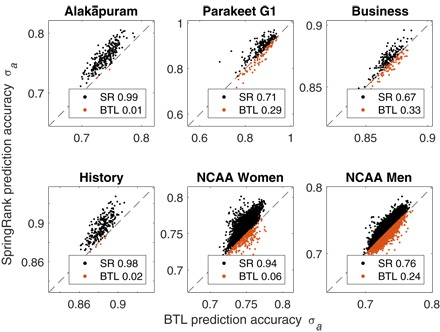
For 50 independent trials of fivefold cross-validation (250 total folds per network), the values of σa for SpringRank (SR) and BTL are shown on the vertical and the horizontal axes, respectively. Points above the diagonal, shown in black, are trials where SpringRank is more accurate than BTL. The fractions for which each method is superior are shown in plot legends, matching Table 1.
Table 1. Edge prediction with BTL as a benchmark.
During 50 independent trials of fivefold cross-validation (250 total folds per network), columns show the percentages of instances in which SpringRank Eq. 3 and regularized SpringRank Eq. 5 with α = 2 produced probabilistic predictions with equal or higher accuracy than BTL. Distributions of accuracy improvements are shown in Fig. 3. Center columns show accuracy σa, and right columns show σL (Materials and Methods). Italics indicate where BTL outperformed SpringRank for more than 50% of tests. NCAA Basketball data sets were analyzed 1 year at a time.
| Data set | Type | % Trials higher σa versus BTL | % Trials higher σL versus BTL | ||
| SpringRank | +Regularization | SpringRank | +Regularization | ||
| Computer science (3) | Faculty hiring | 100.0 | 97.2 | 100.0 | 99.6 |
| Alakāpuram (2) | Social support | 99.2* | 99.6 | 100.0 | 100.0 |
| Synthetic β = 5 | Synthetic | 98.4 | 63.2 | 76.4 | 46.4 |
| History (3) | Faculty hiring | 97.6* | 96.8 | 98.8 | 98.8 |
| NCAA Women (1998–2017) (39) | Basketball | 94.4* | 87.0 | 69.1 | 51.0 |
| Tenpaṭṭi (2) | Social support | 88.8 | 93.6 | 100.0 | 100.0 |
| Synthetic β = 1 | Synthetic | 83.2 | 65.2 | 98.4 | 98.4 |
| NCAA Men (1985–2017) (39) | Basketball | 76.0* | 62.3 | 68.5 | 52.4 |
| Parakeet G1 (5) | Animal dominance | 71.2* | 56.8 | 41.2 | 37.2 |
| Business (3) | Faculty hiring | 66.8* | 59.2 | 39.2 | 36.8 |
| Parakeet G2 (5) | Animal dominance | 62.0 | 51.6 | 47.6 | 47.2 |
*Tests that are shown in detail in Fig. 4.
Table 1 shows that SpringRank also obtained a higher log likelihood σL than BTL for six of the nine real-world networks. Regularizing SpringRank with α = 2 does not appear to significantly improve either measure of accuracy (Fig. 3). We did not attempt to tune the regularization parameter α.
To compare SpringRank with methods that do not make probabilistic predictions, including those that produce ordinal rankings, we measured the accuracy σb of bitwise predictions, that is, the fraction of edges consistent with the inferred ranking. We found that spectral methods perform poorly here, as does SerialRank. BTL does better on the NCAA networks in terms of bitwise prediction than it does for probabilistic predictions, suggesting that it is better at rank-ordering teams than determining their real-valued position.
We found that SyncRank is the strongest of the ordinal methods, matching SpringRank’s accuracy on the parakeet and business school networks, but SpringRank outperforms SyncRank on the social support and NCAA networks (see fig. S4). We show a trial-by-trial comparison of SpringRank and SyncRank in Fig. 5, showing that in most trials of cross-validation, SpringRank makes more accurate predictions for the NCAA networks.
Fig. 5. Bitwise prediction accuracy σb of Spring Rank versus SyncRank.
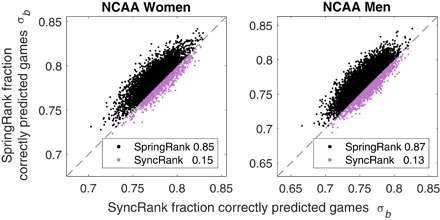
For 50 independent trials of fivefold cross-validation (250 total folds per NCAA season), the fractions of correctly predicted game outcomes σb for SpringRank and SyncRank are shown on the vertical and the horizontal axes, respectively. Points above the equal performance line, shown in black, are trials where SpringRank is more accurate than SyncRank. The fractions for which each method is superior are shown in plot legends.
To check whether our results were dependent on the choice of holding out 20% of the data, we repeated our experiments using twofold cross-validation, that is, using 50% of network edges as training data and trying to predict the other 50%. We show these results in fig. S5. While all algorithms are less accurate in this setting, the comparison between algorithms is similar to that for fivefold cross-validation.
Finally, the real-valued ranks found by SpringRank shed light on the organization and assembly of real-world networks (see figs. S6, S7, S8, S12, and S13). For example, we found that ranks in the faculty hiring networks have a long tail at the top, suggesting that the most prestigious universities are more separated from those below them than an ordinal ranking would reveal. In contrast, ranks in the social support networks have a long tail at the bottom, suggesting a set of people who do not have sufficient social status to provide support to others. SpringRank’s ability to find real-valued ranks makes these distributions amenable to statistical analysis, and we suggest this as a direction for future work.
DISCUSSION
SpringRank is a mathematically principled, physics-inspired model for hierarchical structure in networks of directed interactions. It yields a simple and highly scalable algorithm, requiring only sparse linear algebra, which enables analysis of networks with millions of nodes and edges in seconds. Its ground-state energy provides a natural test statistic for the statistical significance of hierarchical structure.
While the basic SpringRank algorithm is nonparametric, a parameterized regularization term can be included as well, corresponding to a Gaussian prior. While regularization is often required for BTL, eigenvector centrality, and other commonly used methods (section S10), it is not necessary for SpringRank, and our tests indicate that its effects are mixed.
We also presented a generative model that allows one to create synthetic networks with tunable levels of hierarchy and noise, whose posterior coincides with SpringRank in the limit where the effect of the hierarchy is strong. By tuning a single temperature parameter, we can use this model to make probabilistic predictions of edge directions, generalizing from observed to unobserved interactions. Therefore, after confirming its ability to infer ranks in synthetic networks where ground-truth ranks are known, we measured SpringRank’s ability to predict edge directions in real networks. We found that in networks of faculty hiring, animal interactions, social support, and NCAA basketball, SpringRank often makes better probabilistic predictions of edge predictions than the popular BTL model and performs as well or better than SyncRank and a variety of other methods that produce ordinal rankings.
SpringRank is based on springs with quadratic potentials, but other potentials may be of interest. For instance, to make the system more tolerant to outliers while remaining convex, one might consider a piecewise potential that is quadratic for small displacements and linear otherwise. We leave this investigation of alternative potentials to future work. Given its simplicity, speed, and high performance, we believe that SpringRank will be useful in a wide variety of fields where hierarchical structure appears because of dominance, social status, or prestige.
MATERIALS AND METHODS
Synthetic network generation
Networks were generated in three steps. First, node ranks splanted were drawn from a chosen distribution. For test 1, N = 100 ranks were drawn from a standard normal distribution, while for test 2, 34 ranks were drawn from each of three Gaussians, , , and , for a total of N = 102. Second, an average degree < k > and a value of the inverse temperature β were chosen. Third, edges were drawn generated to Eq. 7 with c = < k > N/∑i,j exp [−(β/2)(si − sj − 1)2] so that the expected mean degree is < k > (see section S6).
This procedure resulted in directed networks with the desired hierarchical structure, mean degree, and noise level. Tests were conducted for < k > ∈ [5, 15], β ∈ [0.1, 5], and all performance plots show means and SDs for 100 replicates.
Performance measures for edge prediction
For multigraphs, we defined the accuracy of probabilistic edge prediction as the extent to which Pij is a good estimate of the fraction of interactions between i and j that point from i to j, given that there are any edges to predict at all, that is, assuming . If this prediction were perfect, then we would have . We defined σA as 1 minus the sum of the absolute values of the difference between Aij and this estimate
| (12) |
where M is the number of directed edges in the subset of the network under consideration, for example, the training or test set. Then, σa = 1 if for all i, j, and σa = 0 if for each i, j all the edges go from i to j (for example) but Pij = 0 and Pji = 1.
To measure accuracy via the conditional log likelihood, we asked with what probability we would get the directed network A from the undirected network if each edge between i and j points from i → j with probability Pij and from j → i with probability Pji = 1 − Pij. This gives
| (13) |
where is the binomial coefficient. We disregarded the first term of this sum since it does not depend on P. If we wish to compare networks of different sizes as in Fig. 3, then we can normalize σL by the number of edges. For an extensive discussion of performance metrics, see section S9.
Statistical significance of ranks
We computed a standard left-tailed P value for the statistical significance of the ranks s* by comparing the ground-state energy Eq. 11 of the real network A with the null distribution of ground-state energies of an ensemble of networks drawn from the null model where is kept fixed, but the direction of each edge is randomized.
| (14) |
In practice, this P value is estimated by drawing many samples from the null distribution by randomizing the edge directions of A to produce , computing the ranks from Eq. 3, and then computing the ground-state energy Eq. 11 of each.
Cross-validation tests
We performed edge prediction using fivefold cross-validation. In each realization, we divided the interacting pairs i, j, that is, those with nonzero , into five equal groups. We used four of these groups as a training set, inferring the ranks and setting β to maximize σa or σL (on the left and right of Fig. 3, respectively). We then used the fifth group as a test set, asking the algorithm for Pij for each pair i, j in that group, and reported σa or σL on that test set. By varying which group we used as the test set, we got five trials per realization: For instance, 50 realizations gave us 250 trials of cross-validation. Results for twofold cross-validation are reported in the Supplementary Materials.
Supplementary Material
Acknowledgments
We thank A. Clauset and J. Ugander for helpful comments. Funding: C.D.B. and C.M. were supported by the John Templeton Foundation. C.M. was also supported by the Army Research Office under grant W911NF-12-R-0012. D.B.L. was supported by NSF award SMA-1633747 and the Santa Fe Institute Omidyar Fellowship. Author contributions: All authors derived the model, analyzed results, and wrote the manuscript. C.D.B. wrote Python implementations and D.B.L. wrote MATLAB implementations. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Open-source code in Python, MATLAB, and SAS/IML is available at https://github.com/cdebacco/SpringRank. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaar8260/DC1
Section S1. Deriving the linear system minimizing the Hamiltonian
Section S2. Poisson generative model
Section S3. Rewriting the energy
Section S4. Ranks distributed as a multivariate Gaussian distribution
Section S5. Bayesian SpringRank
Section S6. Fixing c to control for sparsity
Section S7. Comparing optimal β for predicting edge directions
Section S8. Bitwise accuracy σb
Section S9. Performance metrics
Section S10. Parameters used for regularizing ranking methods
Section S11. Supplementary tables
Section S12. Supplementary figures
Table S1. Pearson correlation coefficients between various rankings of faculty hiring networks.
Table S2. Statistics for SpringRank applied to real-world networks.
Fig. S1. Performance (Pearson correlation) on synthetic data.
Fig. S2. Statistical significance testing using the null model distribution of energies.
Fig. S3. Edge prediction accuracy over BTL for NCAA basketball data sets.
Fig. S4. Bitwise edge direction prediction.
Fig. S5. Edge prediction accuracy with twofold cross-validation.
Fig. S6. Summary of SpringRank applied to computer science faculty hiring network.
Fig. S7. Summary of SpringRank applied to history faculty hiring network.
Fig. S8. Summary of SpringRank applied to business faculty hiring network.
Fig. S9. Summary of SpringRank applied to Asian elephants network.
Fig. S10. Summary of SpringRank applied to parakeet G1 network.
Fig. S11. Summary of SpringRank applied to parakeet G2 network.
Fig. S12. Summary of SpringRank applied to Tenpaṭṭi social support network.
Fig. S13. Summary of SpringRank applied to Alakāpuram social support network.
Reference (46)
REFERENCES AND NOTES
- 1.Drews C., The concept and definition of dominance in animal behaviour. Behaviour 125, 283–313 (1993). [Google Scholar]
- 2.Power E. A., Social support networks and religiosity in rural South India. Nat. Hum. Behav. 1, 0057 (2017). [Google Scholar]
- 3.Clauset A., Arbesman S., Larremore D. B., Systematic inequality and hierarchy in faculty hiring networks. Sci. Adv. 1, e1400005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Côté S. D., Festa-Bianchet M., Reproductive success in female mountain goats: The influence of age and social rank. Anim. Behav. 62, 173–181 (2001). [Google Scholar]
- 5.Hobson E. A., DeDeo S., Social feedback and the emergence of rank in animal society. PLOS Comput. Biol. 11, e1004411 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dey C. J., Quinn J. S., Individual attributes and self-organizational processes affect dominance network structure in pukeko. Behav. Ecol. 25, 1402–1408 (2014). [Google Scholar]
- 7.Dey C. J., Reddon A. R., O’Connor C. M., Balshine S., Network structure is related to social conflict in a cooperatively breeding fish. Anim. Behav. 85, 395–402 (2013). [Google Scholar]
- 8.Cant M. A., Llop J. B., Field J., Individual variation in social aggression and the probability of inheritance: Theory and a field test. Am. Nat. 167, 837–852 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Ball B., Newman M. E. J., Friendship networks and social status. Netw. Sci. 1, 16–30 (2013). [Google Scholar]
- 10.Szymanski S., The economic design of sporting contests. J. Econ. Lit. 41, 1137–1187 (2003). [Google Scholar]
- 11.Baumann R., Matheson V. A., Howe C. A., Anomalies in tournament design: The madness of March Madness. J. Quant. Anal. Sports 6 (2010). [Google Scholar]
- 12.Bonacich P., Power and centrality: A family of measures. Am. J. Sociol. 92, 1170–1182 (1987). [Google Scholar]
- 13.L. Page, S. Brin, R. Motwani, T. Winograd, “The PageRank citation ranking: Bringing order to the web” (Technical Report, Stanford InfoLab, 1999).
- 14.Negahban S., Oh S., Shah D., Rank centrality: Ranking from pairwise comparisons. Oper. Res. 65, 266–287 (2016). [Google Scholar]
- 15.Callaghan T., Mucha P. J., Porter M. A., Random walker ranking for NCAA division I-A football. Am. Math. Mon. 114, 761–777 (2007). [Google Scholar]
- 16.Ali I., Cook W. D., Kress M., On the minimum violations ranking of a tournament. Manage. Sci. 32, 660–672 (1986). [Google Scholar]
- 17.Slater P., Inconsistencies in a schedule of paired comparisons. Biometrika 48, 303–312 (1961). [Google Scholar]
- 18.M. Gupte, P. Shankar, J. Li, S. Muthukrishnan, L. Iftode, Finding hierarchy in directed online social networks, in Proceedings of the 20th International Conference on World Wide Web (ACM, 2011), pp. 557–566. [Google Scholar]
- 19.F. Fogel, A. d’Aspremont, M. Vojnovic, Serialrank: Spectral ranking using seriation, in Advances in Neural Information Processing Systems (NIPS, 2014), pp. 900–908. [Google Scholar]
- 20.Cucuringu M., Sync-rank: Robust ranking, constrained ranking and rank aggregation via eigenvector and SDP synchronization. IEEE Trans. Netw. Sci. Eng. 3, 58–79 (2016). [Google Scholar]
- 21.Letizia E., Barucca P., Lillo F., Resolution of ranking hierarchies in directed networks. PLOS ONE 13, e0191604 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatti N., Tiers for peers: A practical algorithm for discovering hierarchy in weighted networks. Data Min. Knowl. Discov. 31, 702–738 (2017). [Google Scholar]
- 23.K. E. Train, Discrete Choice Methods with Simulation (Cambridge Univ. Press, 2009). [Google Scholar]
- 24.Bradley R. A., Terry M. E., Rank analysis of incomplete block designs: The method of paired comparisons. Biometrika 39, 324–345 (1952). [Google Scholar]
- 25.Luce R. D., On the possible psychophysical laws. Psychol. Rev. 66, 81–95 (1959). [DOI] [PubMed] [Google Scholar]
- 26.David H. A., Ranking from unbalanced paired-comparison data. Biometrika 74, 432–436 (1987). [Google Scholar]
- 27.W. N. Colley, Colley’s bias free college football ranking method: The Colley matrix explained (2002); www.colleyrankings.com/matrate.pdf.
- 28.A. E. Elo, The Rating of Chessplayers, Past and Present (Arco Pub., 1978). [Google Scholar]
- 29.R. Coulom, Whole-history rating: A Bayesian rating system for players of time-varying strength, in International Conference on Computers and Games (Springer, 2008), pp. 113–124. [Google Scholar]
- 30.R. Herbrich, T. Minka, T. Graepel, Trueskill: A Bayesian skill rating system, in Advances in Neural Information Processing Systems (NIPS, 2007), pp. 569–576. [Google Scholar]
- 31.Williams R. J., Anandanadesan A., Purves D., The probabilistic niche model reveals the niche structure and role of body size in a complex food web. PLOS ONE 5, e12092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams R. J., Purves D. W., The probabilistic niche model reveals substantial variation in the niche structure of empirical food webs. Ecology 92, 1849–1857 (2011). [DOI] [PubMed] [Google Scholar]
- 33.A. Z. Jacobs, J. A. Dunne, C. Moore, A. Clauset, Untangling the roles of parasites in food webs with generative network models. https://arxiv.org/abs/1505.04741 (2015).
- 34.Hoff P. D., Raftery A. E., Handcock M. S., Latent space approaches to social network analysis. J. Am. Stat. Assoc. 97, 1090–1098 (2002). [Google Scholar]
- 35.Spielman D. A., Teng S.-H., Nearly linear time algorithms for preconditioning and solving symmetric, diagonally dominant linear systems. SIAM J. Matrix Anal. Appl. 35, 835–885 (2014). [Google Scholar]
- 36.I. Koutis, G. L. Miller, R. Peng, A nearly-m log n time solver for SDD linear systems, in Proceedings of the 52nd Foundations of Computer Science (FOCS) (IEEE Press, 2011), pp. 590–598. [Google Scholar]
- 37.de Silva S., Schmid V., Wittemyer G., Fission–fusion processes weaken dominance networks of female Asian elephants in a productive habitat. Behav. Ecol. 28, 243–252 (2017). [Google Scholar]
- 38.Hunter D. R., MM algorithms for generalized Bradley-Terry models. Ann. Stat. 32, 384–406 (2004). [Google Scholar]
- 39.National Collegiate Athletic Association (2018); www.ncaa.org/championships/statistics.
- 40.S. F. Way, D. B. Larremore, A. Clauset, Gender, productivity, and prestige in computer science faculty hiring networks, in Proceedings of the 25th International Conference on World Wide Web (WWW’16), Montréal, Québec, Canada, 11 to 15 April 2016, pp. 1169–1179. [Google Scholar]
- 41.Majolo B., Lehmann J., de Bortoli Vizioli A., Schino G., Fitness-related benefits of dominance in primates. Am. J. Phys. Anthropol. 147, 652–660 (2012). [DOI] [PubMed] [Google Scholar]
- 42.N. Lin, Social Capital: A Theory of Social Structure and Action (Cambridge Univ. Press, 2002), vol. 19. [Google Scholar]
- 43.K. S. Cook, M. Levi, R. Hardin, Whom Can we Trust?: How Groups, Networks, and Institutions Make Trust Possible (Russell Sage Foundation, 2009). [Google Scholar]
- 44.M. Mines, Public Faces, Private Lives: Community and Individuality in South India (University of California Press, 1994). [Google Scholar]
- 45.Shizuka D., McDonald D. B., A social network perspective on measurements of dominance hierarchies. Anim. Behav. 83, 925–934 (2012). [Google Scholar]
- 46.Rosasco L., Vito E. D., Caponnetto A., Piana M., Verri A., Are loss functions all the same? Neural Comput. 16, 1063–1076 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/7/eaar8260/DC1
Section S1. Deriving the linear system minimizing the Hamiltonian
Section S2. Poisson generative model
Section S3. Rewriting the energy
Section S4. Ranks distributed as a multivariate Gaussian distribution
Section S5. Bayesian SpringRank
Section S6. Fixing c to control for sparsity
Section S7. Comparing optimal β for predicting edge directions
Section S8. Bitwise accuracy σb
Section S9. Performance metrics
Section S10. Parameters used for regularizing ranking methods
Section S11. Supplementary tables
Section S12. Supplementary figures
Table S1. Pearson correlation coefficients between various rankings of faculty hiring networks.
Table S2. Statistics for SpringRank applied to real-world networks.
Fig. S1. Performance (Pearson correlation) on synthetic data.
Fig. S2. Statistical significance testing using the null model distribution of energies.
Fig. S3. Edge prediction accuracy over BTL for NCAA basketball data sets.
Fig. S4. Bitwise edge direction prediction.
Fig. S5. Edge prediction accuracy with twofold cross-validation.
Fig. S6. Summary of SpringRank applied to computer science faculty hiring network.
Fig. S7. Summary of SpringRank applied to history faculty hiring network.
Fig. S8. Summary of SpringRank applied to business faculty hiring network.
Fig. S9. Summary of SpringRank applied to Asian elephants network.
Fig. S10. Summary of SpringRank applied to parakeet G1 network.
Fig. S11. Summary of SpringRank applied to parakeet G2 network.
Fig. S12. Summary of SpringRank applied to Tenpaṭṭi social support network.
Fig. S13. Summary of SpringRank applied to Alakāpuram social support network.
Reference (46)


