Dysfunction of OsGPAT3 leads to disrupted anther lipid metabolism and male sterility in rice.
Keywords: Anther development, glycerol-3-phosphate acyltransferase, lipid metabolism, male sterility, microgametophyte, rice, tapetum
Abstract
Lipid molecules are key structural components of plant male reproductive organs, such as the anther and pollen. Although advances have been made in the understanding of acyl lipids in plant reproduction, the metabolic pathways of other lipid compounds, particularly glycerolipids, are not fully understood. Here we report that an endoplasmic reticulum-localized enzyme, Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3), plays an indispensable role in anther development and pollen formation in rice. OsGPAT3 is preferentially expressed in the tapetum and microspores of the anther. Compared with wild-type plants, the osgpat3 mutant displays smaller, pale yellow anthers with defective anther cuticle, degenerated pollen with defective exine, and abnormal tapetum development and degeneration. Anthers of the osgpat3 mutant have dramatic reductions of all aliphatic lipid contents. The defective cuticle and pollen phenotype coincide well with the down-regulation of sets of genes involved in lipid metabolism and regulation of anther development. Taking these findings together, this work reveals the indispensable role of a monocot-specific glycerol-3-phosphate acyltransferase in male reproduction in rice.
Introduction
Male reproductive development in higher plants is a complicated biological process that includes the development of the anther and the generation of pollen (Liu and Qu, 2008; Ma, 2005; Sanders et al., 1999; Zhang et al., 2011; Zhang and Wilson, 2009). The developed anther wall has four somatic layers: the epidermis, the endothecium, the middle layer, and the tapetum (Goldberg et al., 1993). The innermost cell layer of the anther wall, the tapetum, which encompasses the meiotic cells (microsporocytes) at the center, plays a crucial role in regulating programmed anther development and microspore/pollen formation (Li et al., 2006; Parish and Li, 2010; Zhang and Yang, 2014). Tapetal cell differentiation and tapetum development are critical for the early events in male reproduction, including meiosis, while tapetal degeneration is vital for formation of viable pollen during late pollen development (Ma, 2005; Wilson and Zhang, 2009; Zhang and Liang, 2016; Zhang et al., 2011). Tapetal cells are characterized by the presence of abundant organelles and vigorous metabolic activities, secreting various monomers or precursors for the synthesis of the anther cuticle, pollen wall, and pollen intracellular nutrients (Li et al., 2010; Liu and Fan, 2013; Zhang and Li, 2014; Zhang et al., 2011; Zhang and Yang, 2014). The disruption of tapetal function frequently leads to aborted microspore formation and male sterility (Fu et al., 2014; Gómez et al., 2015; Li et al., 2006; Li et al., 2011; Niu et al., 2013; Parish and Li, 2010; Xu et al., 2010; Zhang et al., 2008; Zhang et al., 2010). On the other hand, the mature pollen that is produced within the anther is covered by a specialized pollen wall comprised mainly of three layers: the pollen coat, exine, and intine. Pollen serves as a biological protector of male sperm cells and a communicator between male and female reproductive organs that facilitates pollination and fertilization. The disruption of pollen formation often results in male sterility (Dobritsa et al., 2009; Shi et al., 2015). Recent studies have demonstrated that there is a high level of conservation between anther development and pollen formation in plants, highlighting their joint function in plant reproduction (Gómez et al., 2015; Huang et al., 2015).
Two lipid layers play important protective roles in plant anther development and pollen formation. The first of these is the anther epidermal cuticle and the second is the pollen exine. Both layers share a common lipid metabolic pathway similar to those found in plant cuticular lipids, waxes, and cutin (or suberin) (Jeffree, 1996; Kunst and Samuels, 2003; Li and Zhang, 2010; Li-Beisson et al., 2013; Nawrath, 2006; Zhang et al., 2016), which seems to be conserved at least in land plants (Shi et al., 2015; Wallace et al., 2015). Therefore, cuticular lipid metabolism, including biosynthesis, modification, transport, and transcription regulation, is essential for the normal development of anther and microspores (Shi et al., 2015; Zhang and Li, 2014).
During the past decades, significant advances in the understanding of lipid metabolism in plant reproductive development and male fertility have been achieved on the basis of studies of mutants of various lipid metabolic genes, mainly in Arabidopsis and rice; for example, Male Sterile 2 (MS2) (Aarts et al., 1997; Chen et al., 2011a), CYP703A2 (Morant et al., 2007), CYP704B1 (Dobritsa et al., 2009), ACOS5 (de Azevedo Souza et al., 2009), and WBC27 (Dou et al., 2011) in Arabidopsis, and Defective Pollen Wall (DPW) (Shi et al., 2011), CYP703A3 (Aya et al., 2009; Yang et al., 2014), CYP704B2 (Li et al., 2010), Wax-deficient anther1 (WDA1) (Jung et al., 2006), OsC6 (Zhang et al., 2010), PDA1/OsABCG15 (Qin et al., 2013; Wu et al., 2014; Zhu et al., 2013), DPW2 (Xu et al., 2016), OsDEX1 (Yu et al., 2016), and OsABCG26 (Zhao et al., 2015) in rice. Except WDA1, most of these genes have relatively high expression levels in the tapetum and/or microspores, and their corresponding mutants exhibit defective anther cuticle and/or pollen exine together with the reduction of anther cuticular lipid components and pollen wall lipid constituents, causing complete or partial male sterility (see review by Shi et al., 2015). Transcription factors that govern tapetum development in plants also play important roles in regulating cuticular lipid-mediated anther and pollen development, such as ABORTED MICROSPORES (AMS) (Xu et al., 2014; Xu et al., 2010) in Arabidopsis and Tapetum Degeneration Retardation (TDR) in rice (Li et al., 2006; Zhang et al., 2008). Nevertheless, none of the abovementioned genes and transcription factors is involved in the metabolism of glycerolipids, an important cutin component in plants (Graça et al., 2002; Li-Beisson et al., 2013), let alone the genetic, biochemical, and molecular mechanisms underlying the involvement of glycerolipid metabolism in anther and pollen development.
The glycerolipid triacylglycerol (TAG) and its derivatives are important storage and membrane lipids and indispensable components of biological polymers including cutin and suberin in plants (Pollard et al., 2008). TAG is generated by connecting fatty acids to a glycerol backbone (Coleman and Lee, 2004). Glycerol-3-phosphate acyltransferases (GPATs) catalyze the first step of TAG biosynthesis by acylating glycerol 3-phosphate at the sn-1 or sn-2 hydroxyl with an acyl donor, acyl-CoA or acyl-ACP, and generating lysophosphatidic acids (LPAs) that can act as signaling molecules in regulating cell growth (Moolenaar et al., 1997; Sheng et al., 2015; Takeuchi and Reue, 2009). Because GPAT displays the lowest specific activity toward a very broad group of substrates, it has been considered to be the rate-limiting enzyme (Wendel et al., 2009; Zheng and Zou, 2001). In animals, GPATs usually acylate glycerol-3-phosphate at the sn-1 position, and are required for membrane lipid synthesis and energy storage. In contrast, in land plants, most GPATs are sn-2 GPATs, which catalyze the reaction in which glycerol is an anchor point for the linear or cross link with fatty acids, playing important roles in the assembly of cutin or suberin in plants (Pollard et al., 2008; Yang et al., 2012).
Arabidopsis has eight sn-2 GPATs with different functions (Chen et al., 2011b). GPAT4 and GPAT8 have high sequence similarity and are suggested to be functionally redundant duplicated genes. Neither the gpat4 nor the gpat8 single mutant showed any obvious cuticle defect, whereas the gpat4gpat8 double mutant exhibited a marked decrease in cutin content in leaves and stems (Li et al., 2007). GPAT6 is highly expressed in flowers (Zheng et al., 2003). Its mutant displayed defective nanoridges on petal surfaces and a significant reduction of cutin monomers in flowers (Li-Beisson et al., 2009). Further biochemical analyses demonstrated that GPAT4, GPAT6, and GPAT8 prefer C16:0 and C18:1 ω-oxidized substrates and have additional phosphatase activity, resulting in the conversion of sn-2 LPA to sn-2 MAG, which is also an important intermediate for polyester assembly (Yang et al., 2012). GPAT5 is required for the synthesis of suberin in seed coat and root, and the gpat5 mutant exhibited strong reduction of very long chain (C22–C24) fatty acid monomers and their derivatives (Beisson et al., 2007). GPAT7, which is phylogenetically most closely related to GPAT5, takes part in suberin synthesis in the wounding response (Yang et al., 2012). GPAT5 and GPAT7 accommodate a broad chain length range of both ω-oxidized and unsubstituted substrates, but they do not possess phosphatase activity. GPAT1, which is mainly expressed in flowers and siliques, also has sn-2 acyltransferase activity utilizing both substituted and unsubstituted substrates, but has no phosphatase activity (Zheng et al., 2003). There is no report yet on the function of GPAT2 or GPAT3 (Yang et al., 2012). Notably, GPAT1 and GPAT6 are essential for male plant fertility. Both gpat1 and gpat6 mutants display altered endoplasmic reticulum (ER) profiles in tapetal cells, as well as severely reduced pollen production and decreased pollen pollination (Li et al., 2012; Zheng et al., 2003). The gpat1gpat6 double mutant exhibited short filaments, defective callose degeneration and microspore release, and complete male sterility (Li et al., 2012). Through database searching, we found 17 GPATs in rice. So far, there is only one report of a rice plastidial GPAT on its substrate selectivity and association with chilling tolerance (Zhu et al., 2009). The involvement of GPATs in rice fertility, particularly anther and pollen development, remains unknown.
In this study, we report the functional analysis of an ER-localized GPAT, Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3), which plays a crucial role in rice male fertility. The osgpat3 mutant exhibits abnormal tapetum development and defective anther cuticle and pollen exine formation, which is concomitant with a dramatic reduction in aliphatic contents, as well as the down-regulation of genes involved in lipid metabolism and regulation of anther development. We demonstrate that this monocot GPAT plays different roles in male reproduction from its dicot counterpart, providing new insights into the function of glycerolipid biosynthetic enzymes in male fertility.
Materials and methods
Plant materials and growth conditions
Rice (Oryza sativa L.) plants used in this study were in the 9522 background (japonica) and were grown in the paddy field of Shanghai Jiao Tong University. The F2 mapping population was generated from a cross between osgpat3 (japonica) and GuangLuAi 4 (wild type, indica) for gene mapping. Male sterile plants in the F2 population were chosen for gene mapping.
Characterization of mutant phenotype
Plants or flowers were photographed with a Nikon D90 digital camera and a Leica MZ16FA microscope. Observation of anther development by semi-thin sections and transmission electron microscopy were performed as described by Li et al. (2006). Anther staging was defined as described previously (Zhang et al., 2011; Zhang and Wilson, 2009). Scanning electron microscopy and analyses of anther waxes, cutin, and internal soluble lipids were performed as described by Shi et al. (2011).
Map-based cloning of the OsGPAT3 gene
For fine mapping of the OsGPAT3 locus, bulked segregation analysis was used to identify markers linked to OsGPAT3 as described by Liu et al. (2005). The primer sequences for InDel markers are shown in Supplementary Table S1 at JXB online. The OsGPAT3 locus was first mapped between two InDel molecular markers, CH1132 and RM6094, on chromosome 11. Then, 3000 F2 offspring from the mapping cross were generated, and five InDel markers (ML2, ML4, ML12, AZ11-2, and YUN115.1) were used. OsGPAT3 was finally defined between InDel markers ML4 and ML12, within a 122 kb region. PCR was performed according to Chu et al., (2006). The PCR products were separated on 6% polyacrylamide denaturing gels, and bands were visualized by a silver-staining method (Liu et al., 2005). Sequence data for the genomic DNA and mRNA of OsGPAT3 can be found in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/) and Rice Genome Annotation Project (RGAP; http://rice.plantbiology.msu.edu/) databases under accession numbers Os11g0679700/NM_001074987 and LOC_Os11g45400, respectively.
Complementation of the osgpat3 mutant
For functional complementation, the OsGPAT3 1.632 kb coding DNA sequence (CDS) and the upstream 3 kb promoter were amplified and subcloned into a modified binary vector pCAMBIA1301 with the GUS coding region replaced by an eGFP fragment, which carries a hygromycin resistance marker, using the restriction endonucleases SacI and SpeI. Calluses induced from young panicles of the homozygous osgpat3 plants were used for transformation with Agrobacterium tumefaciens (EHA105) carrying the pCAMBIA1301-OsGPAT3-GFP plasmid or the control plasmid pCAMBIA1301-GFP (Li et al., 2006). For transgenic plants, at least 15 independent lines were obtained for each construct. Transgenic plants were identified by PCR using the primers listed in Supplementary Table S1.
Phylogenetic analysis
Multiple protein sequence alignments were performed using MUSCLE 3.6 (http://www.ebi.ac.uk/Tools/msa/muscle/). A phylogenetic tree was constructed with the alignment of GPAT-like protein sequences of rice, Arabidopsis, and other species. MEGA 4.0 (http://www.megasoftware.net/index.html) and the neighbor-joining method were used, with Poisson correction, pairwise deletion, and bootstrap (1000 replicates; random seed).
Quantitative reverse-transcription real-time PCR assay and in situ hybridization
Total RNA was isolated from various rice tissues, including anthers at different developmental stages, using Trizol reagent (Invitrogen) as described by the supplier. An aliquot of 1 μg RNA per sample was used to synthesize cDNA, using a PrimeScript RT reagent Kit with gDNA eraser (Takara). Quantitative reverse-transcription real-time PCR (qRT-PCR) was performed on a Bio-Rad C1000 machine using Takara SYBR Premix Ex TaqTM GC with a standard two-step protocol, consisting of 95 °C for 30s followed by 40 cycles of 95 °C for 5s and 60 °C for 30s. The expression level of OsACTIN1 was used as an internal control, and a relative quantitation method (Δ cycle threshold) was used to quantify the relative expression level of target genes. Three biological replicates with three technique replicates each were included for statistical analysis and error range analysis. The GenBank accession numbers of the genes used in the qRT-PCR assay are DPW (Os03g0167600), CYP703A3 (Os08g0131100), CYP704B2 (Os03g0168600), TDR (Os02g0120500), TIP2 (Os01g0293100), and MTR1 (Os02g0491300) (see Supplementary Table S1 for primer sequences). RNA in situ hybridizations were performed as described by Li et al. (2006). A 269 bp cDNA fragment of OsGPAT3 was used for making antisense and sense probes (Supplementary Table S1).
Subcellular localization of OsGPAT3
The full-length OsGPAT3 CDS and OsGPAT3ΔN were cloned into the XhoI and SpeI sites of the pA7-35S::GFP plasmid. The resulting plasmids were coupled with gold particles and bombarded into onion epidermal cells, which were observed as previously described by Liu and Mehdy (2007). A laser scanning confocal microscope (Leica TCS SP5) was used for the analysis. GFP fluorescent signals were imaged at the excitation wavelength of 488 nm and the emission wavelength of 505–530 nm.
Results
Phenotypic analysis of osgpat3
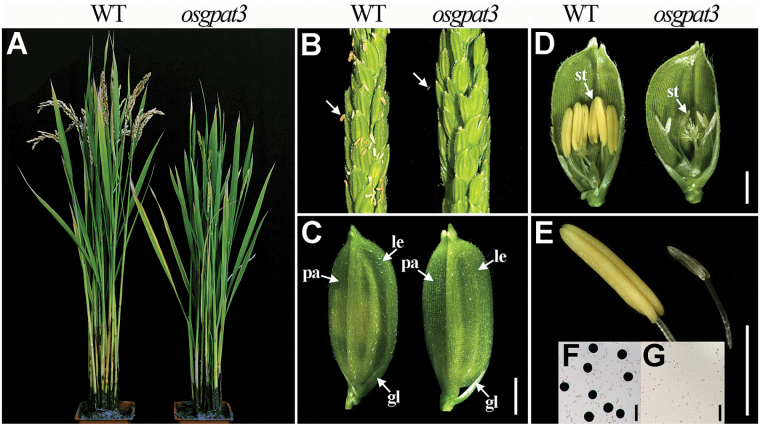
To understand the molecular basis of control of rice male fertility, a completely male-sterile mutant was isolated from our rice mutant library (Chen et al., 2006). This mutant was named osgpat3 because of a deletion of the putative GPAT gene detected in the mutant by a map-based cloning approach (see below). All the F1 progeny from the backcross between the wild type and osgpat3 displayed the wild-type phenotype, and the F2 progeny had an approximate 3:1 segregation ratio of wild-type (fertile) and mutant (sterile) phenotypes (310:105, χ2=0.013, P>0.05), suggesting a monofactorial recessive inheritance of the mutation. The mutant exhibited normal vegetative development and inflorescence morphology (Fig. 1A–C), but had pale yellow to white and much smaller anthers compared with those of the wild type (Fig. 1D, E) and lacked mature pollen grains at the late stages of anther development (Fig. 1F, G); these observations indicated that OsGPAT3 is required for anther development and pollen formation in rice.
Fig. 1.
Phenotypic comparison between wild-type (WT) and osgpat3 mutant rice. (A) A WT plant (left) and an osgpat3 mutant plant (right) after producing seeds. (B) A WT panicle (left) and an osgpat3 mutant panicle (right) at the heading stage. The arrows indicate anthers. (C) A WT spikelet (left) and an osgpat3 mutant spikelet (right). gl, glume; le, lemma; pa, palea. (D) A WT spikelet (left) and an osgpat3 mutant spikelet (right) after removing half of the lemma and palea. st, stamen. (E) A WT anther (left) showing normal yellow color and an osgpat3 mutant anther (right), which is smaller and pale to white in color. (F) WT pollen grains stained with 1% I2-KI solution at stage 12, showing mature pollen grains. (G) Pollen grains of osgpat3 stained with 1% I2-KI solution at stage 12; no mature pollen grains are present. Bars=2 mm in (C–E) and 50 μm in (F) and (G). (This figure is available in colour at JXB online.)
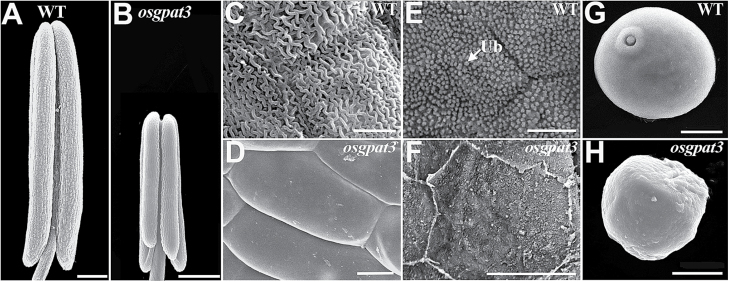
Scanning electron microscopy was used to further observe the phenotypic changes in osgpat3 at various anther developmental stages as defined previously by Zhang and Wilson (2009). At stage 9, there was no observable anther epidermal difference between wild type and osgpat3 (data not shown). At stage 10, dense array of granular Ubisch bodies were observed on the inner locule surface of wild-type anthers but not on that of osgpat3 (Fig. 2E, F). At stage 12, the outer surface of the wild-type anther was covered by well-formed cutinized nanoridges, while the osgpat3 anther surface was quite smooth and cutinized nanoridges were absent (Fig. 2A–D). In addition, normal pollen grains appeared in wild-type anthers at stage 9, while shrunken and irregularly shaped pollen grains, which became completely aborted at the later stages, were observed in osgpat3 (Fig. 2G, H). These results showed that mutation of OsGPAT3 influences the development of the anther cuticle and Ubisch body, and pollen formation.
Fig. 2.
Appearance of the anther and pollen grain in wild-type (WT) and osgpat3 mutant rice under scanning electron microscopy. (A) WT and (B) osgpat3 anthers at stage 12 of development. (C, D) The outermost surface of the epidermis of (C) WT and (D) osgpat3 anthers at stage 12. (E, F) The inner surface of the anther wall layers of (E) WT and (F) osgpat3 anthers at stage 10. Ub, Ubisch bodies. (G, H) Pollen grains of (G) WT and (H) osgpat3 at stage 9. Bars=200 μm in (A) and (B), 5 μm in (C–F), and 10 μm in (G) and (H).
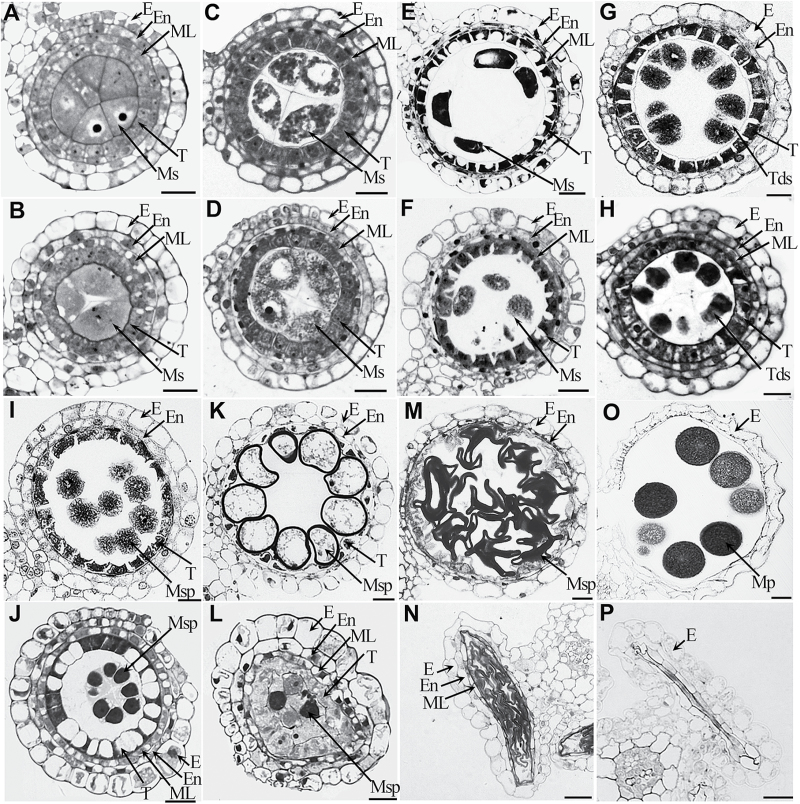
Similarly, light microscopic examination of transverse sections of anthers showed that there were no remarkable morphological differences between the wild type and osgpat3 during the early developmental stages (Fig. 3A–D). At stage 8a, ellipsoidal dyads were formed and tapetal cells became vacuolated and shrunken with darkly stained cytoplasm in the wild type (Fig. 3E). In the osgpat3 mutant, although dyads were formed, the tapetal cells were less vacuolated and less darkly stained (Fig. 3F). At stage 8b, even though osgpat3 formed normal tetrads, the cells of the middle layer did not degenerate as did those in the wild type (Fig. 3G, H). At stage 9, the wild-type anther had condensed and deeply stained tapetal cells, and young microspores were freely released from the tetrads. By contrast, at the same stage, osgpat3 had vacuolated tapetal cells, and microspores were still covered with callose and could not be released from the tetrads (Fig. 3I, J). At stage 10, the wild type displayed degenerating tapetum, as well as vacuolated and round microspores, while the osgpat3 tapetum became swollen and less stained, the microspores started to degrade, and the anther wall started to collapse (Fig. 3K, L). At stage 11, wild-type microspores became falcate in shape and tapetal cells had almost completely degraded into cellular debris, whereas osgpat3 microspores displayed an irregular strip-shaped appearance, and the anther wall collapsed into the locule (Fig. 3M, N). At stage 12, the wild type produced mature pollen grains in the locule, whereas osgpat3 had a flat locule without mature pollen (Fig. 3O, P). These results indicated that OsGPAT3 is essential for the timely differentiation and degradation of the tapetum.
Fig. 3.
Transverse section analysis of anther development in wild-type and osgpat3 mutant rice. Locules from the anther section of wild-type (A, C, E, G, I, K, M, O) and osgpat3 (B, D, F, H, J, L, N, P) from stage 6 to stage 12 of development in sequence (6, 7, 8a, 8b, 9, 10, 11, 12, respectively). E, epidermis; En, endothecium; ML, middle layer; ; Mp, mature pollen Ms, microsporocyte; Msp, microspores; T, tapetum; Tds, tetrads. Bars=15 μm.
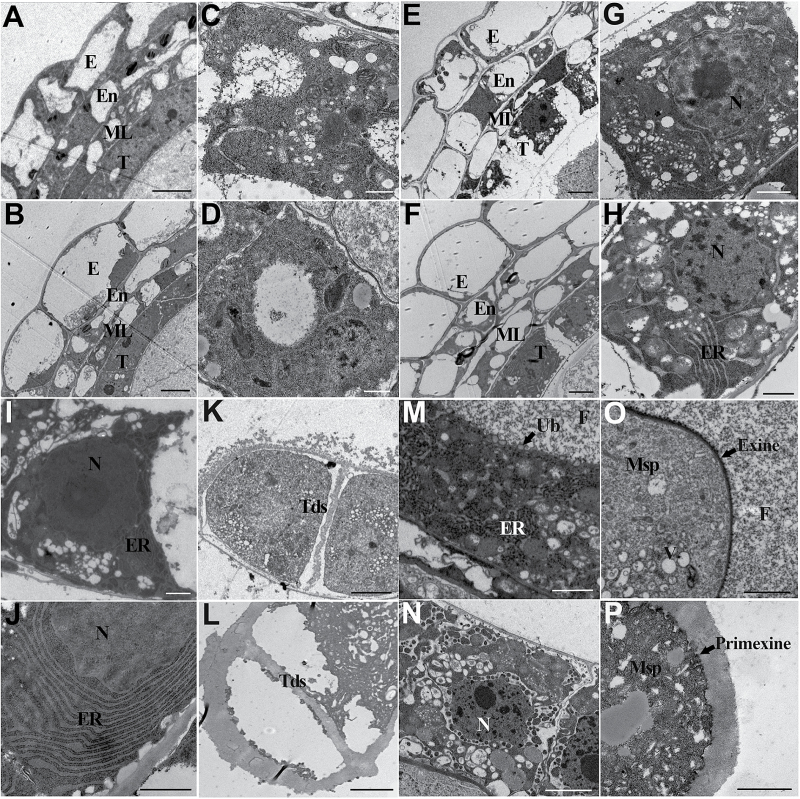
To better understand the developmental abnormalities of anther and pollen in the osgpat3 mutant, transmission electron microscopy was applied. Consistent with the findings of light microscopy of transverse sections, no obvious ultrastructural morphological alteration was observed in osgpat3 at the early stages of anther development (Fig. 4A–D). At stage 8a, the osgpat3 mutant tapetal cells showed less stained cytoplasm, and contained swollen ER and more vacuoles and lipidosomes compared with the wild type (Fig. 4E–H). At stage 8b, osgpat3 tapetal cells displayed markedly increased and expanded ER (Fig. 4I, J). More tapetal cells with increased ER profiles are shown in Supplementary Fig. S3. Consistent with the defective release of microspores from the tetrad, a thicker callose wall surrounding the tetrad was observed in osgpat3 (Fig. 4K, L). At stage 9, the wild-type tapetum became condensed, and proliferated ER appeared in the tapetal cells (Fig. 4M). In contrast, at this stage almost no ER but abundant vacuoles and lipidosomes were observed in osgpat3 tapetal cells (Fig. 4N). Additionally, in wild-type anthers the secretory structures Ubisch bodies were seen on the surface of the tapetal cells, fine fibrillar materials released from the tapetum were distributed uniformly throughout the locule, and pollen exine was formed on the surface of the microspores, whereas in the osgpat3 mutant no Ubisch bodies or fibrillar material, and only coarse primexine, was evident (Fig. 4M–P). These observations confirmed that mutation of OsGPAT3 affects the development of tapetum and Ubisch bodies, pollen exine formation, and also callose wall degradation.
Fig. 4.
Transmission electron microscopy analysis of anthers in wild-type and osgpat3 mutant rice. (A, B) Anthers of (A) the wild type and (B) osgpat3 at stage 6, showing the four layers of the anther wall. (C, D) Tapetal cells of (C) the wild type and (D) osgpat3 at stage 6. (E, F) Anthers of (E) the wild type and (F) osgpat3 at stage 8a. (G, H) Tapetal cells of (G) the wild type and (H) osgpat3 at stage 8a. (I, J) Tapetal cells of (I) the wild type and (J) osgpat3 at stage 8b. Obviously increased and swollen ER can be observed in osgpat3. (K, L) Tetrads in (K) the wild type and (L) osgpat3 at stage 8b. (M, N) Tapetal cells in (M) the wild type and (N) osgpat3 at stage 9. The arrow in (M) indicates an Ubisch body (Ub). (O, P) Young microspores in (O) the wild type and (P) osgpat3 at stage 9. Thick callose surrounds the microspore in osgpat3. No exine, and only a coarse primexine, is formed in osgpat3. E, epidermis; En, endothecium; ML, middle layer; Ms, microsporocyte; Msp, microspores; N, Nucleus; T, tapetum; Tds, tetrads. Bars=5 μm in (A), (B), (E), (F), (K), and (L), 1 μm in (C), (D), (G), (H), (I), and (J), and 2 μm in (M–P).
Changes in aliphatic composition in osgpat3
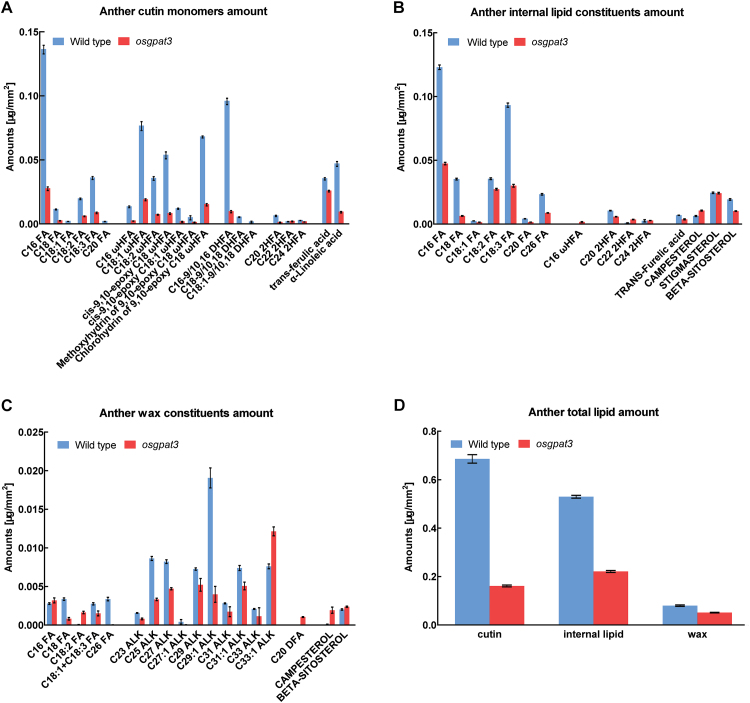
The defective anther cuticle and Ubisch body in osgpat3 suggested that mutation of OsGPAT3 affects anther cuticular lipid profiling. To test this hypothesis, the composition of waxes, cutin, and total soluble lipids in wild-type and osgpat3 anthers was analyzed by gas chromatography-flame ionization detection (GC-FID) combined with gas chromatography-mass spectrometry (GC-MS). We used the approach described by Li et al. (2010) to calculate the surface area of the anthers, in which the calculated values of surface area were plotted against the weight of each sample (Supplementary Fig. S1). Analytical results showed that the total wax, cutin, and total internal lipid levels in osgpat3 anthers were decreased by 37.5%, 76.8%, and 58.5%, respectively, compared with those of the wild type (Fig. 5D), which was attributed to the significant reduction of almost every lipid molecule in osgpat3 mutant anthers (Fig. 5A–C). These chemical analysis data not only confirmed the defective anther cuticle and pollen exine patterning observed by scanning and transmission electron microscopy, but also indicated that the mutation of OsGPAT3 significantly affects the biosynthesis of these lipid compounds, which are required for anther cuticle and pollen exine formation.
Fig. 5.
Analysis of lipid content in wild-type and osgpat3 mutant anthers. Amount per unit surface area (mg mm–2) of (A) cutin monomers, (B) internal lipid constituents, (C) wax constituents, and (D) total anther lipid contents in wild-type and osgpat3 anthers. Error bars indicate SD (n=4). Compound names are abbreviated as follows: C16 FA, palmitic acid; C18 FA, stearic acid; C18:1 FA, oleic acid; C18:2 FA, linoleic acid; C18:3 FA, linolenic acid; C20 FA, eicosanoic acid; C26 FA, cerotic acid; ALK, alkane; C16 ωHFA, 16-hydroxy-hexadecanoic acid; C18:1 ωHFA, 18-hydroxy-oleic acid; C18:2 ωHFA, 18-hydroxy-linoleic acid; cis-9,10-epoxy C18 ωHFA, cis-9,10-epoxy 18-hydroxy-stearic acid; cis-9,10-epoxy C18:1 ωHFA, cis-9,10-epoxy 18-hydroxy-oleic acid; methoxyhydrin of 9,10-epoxy C18 ωHFA, methoxyhydrin of 9,10-epoxy 18-hydroxy-stearic acid; chlorohydrin of 9,10-epoxy C18 ωHFA, chlorohydrin of 9,10-epoxy 18-hydroxy-stearic acid; C16-9/10, 16 DHFA, 9(10), 16-dihydroxy-hexadecanoic acid; C18-9/10, 18 DHFA, 9(10), 18-dihydroxy-stearic acid; C18:1–9/10, 18 DHFA, 9(10), 18-dihydroxy-oleic acid; C20 2HFA, 2-hydroxyeicosanoic acid; C20 DFA, eicosane-1, 20-dioic acid; C22 2HFA, 2-hydroxydocosanoic acid; C24 2HFA, 2-hydroxytetracosanoic acid. (This figure is available in colour at JXB online.)
Map-based cloning and functional complementation of osgpat3
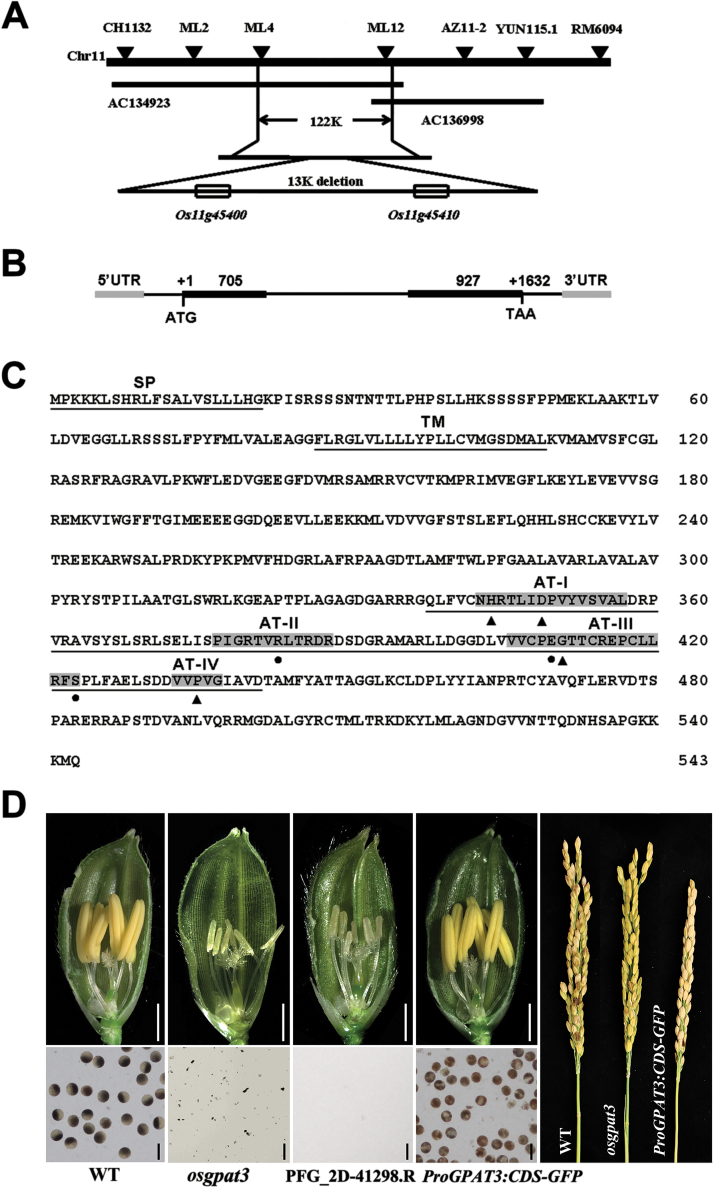
To identify the mutant gene, a map-based cloning approach was used. Through fine mapping, the mutation was located to a 13 kb deletion on chromosome 11. There are two putative genes in the 13 kb deleted region (Fig. 6A). One is LOC_Os11g45410, encoding a pentatricopeptide repeat-containing protein (PPR) without any functional characterization; the other gene is LOC_Os11g45400, which is presumed to encode a GPAT. To clarify which gene is responsible for the mutant phenotype, genetic complementation was performed in the homozygous osgpat3 mutant. Transgenic lines with the coding sequence of LOC_Os11g45400 fused to GFP driven by its own promoter (3 kb) restored male fertility (Fig. 6D). Moreover, a homozygous mutant PFG_2D-41298.R (Jeon et al., 2000) in which the T-DNA fragment was inserted within the second exon of LOC_Os11g45400, showed a similar sterile phenotype to osgpat3 (Fig. 6D). These results confirmed that the deletion of LOC_Os11g45400 is responsible for the developmental defects observed in osgpat3 plants. According to the annotation from RGAP, LOC_Os11g45400 contains two exons and one intron (Fig. 6B), and is predicted to encode a GPAT protein of 543 amino acids, containing a transmembrane domain and an acyltransferase domain, but no phosphatase domain (Fig. 6C; Mañas-Fernández et al., 2010). These results indicated that deletion of the LOC_Os11g45400 gene is responsible for the defective male fertility in osgpat3.
Fig. 6.
Molecular identification and sequence analysis of OsGPAT3. (A) Fine mapping of the osgpat3 mutation on chromosome 11. The names and positions of the molecular markers are indicated. AC134923 and AC136998 are genomic DNA accession numbers. The mutation was mapped to a 122 kb region between two molecular markers (ML4 and ML12). (B) A schematic representation of the exon and intron organization of LOC_Os11g45400. (C) Predicted protein sequence of OsGPAT3. The protein is predicted to contain a 21 amino acid signal peptide (SP), a transmembrane domain (TM), and a conserved acyltransferase domain (AT) with four conserved motifs. Dots indicate key amino acids important for substrate binding and triangles indicate key amino acids important for catalysis. (D) Functional complementation of osgpat3 with the full-length CDS of LOC_Os11g45400 fused to GFP, driven by its own promoter (3 kb). The LOC_Os11g45400 T-DNA insertion homozygous mutant PFG_2D-41298.R also shows a similar male sterility phenotype to that of the osgpat3 mutant. WT, wild type. Bars=2 mm for spikelets and 50 μm for pollen grains. (This figure is available in colour at JXB online.)
OsGPAT3 belongs to a monocot-specific clade of the plant sn-2 GPAT family
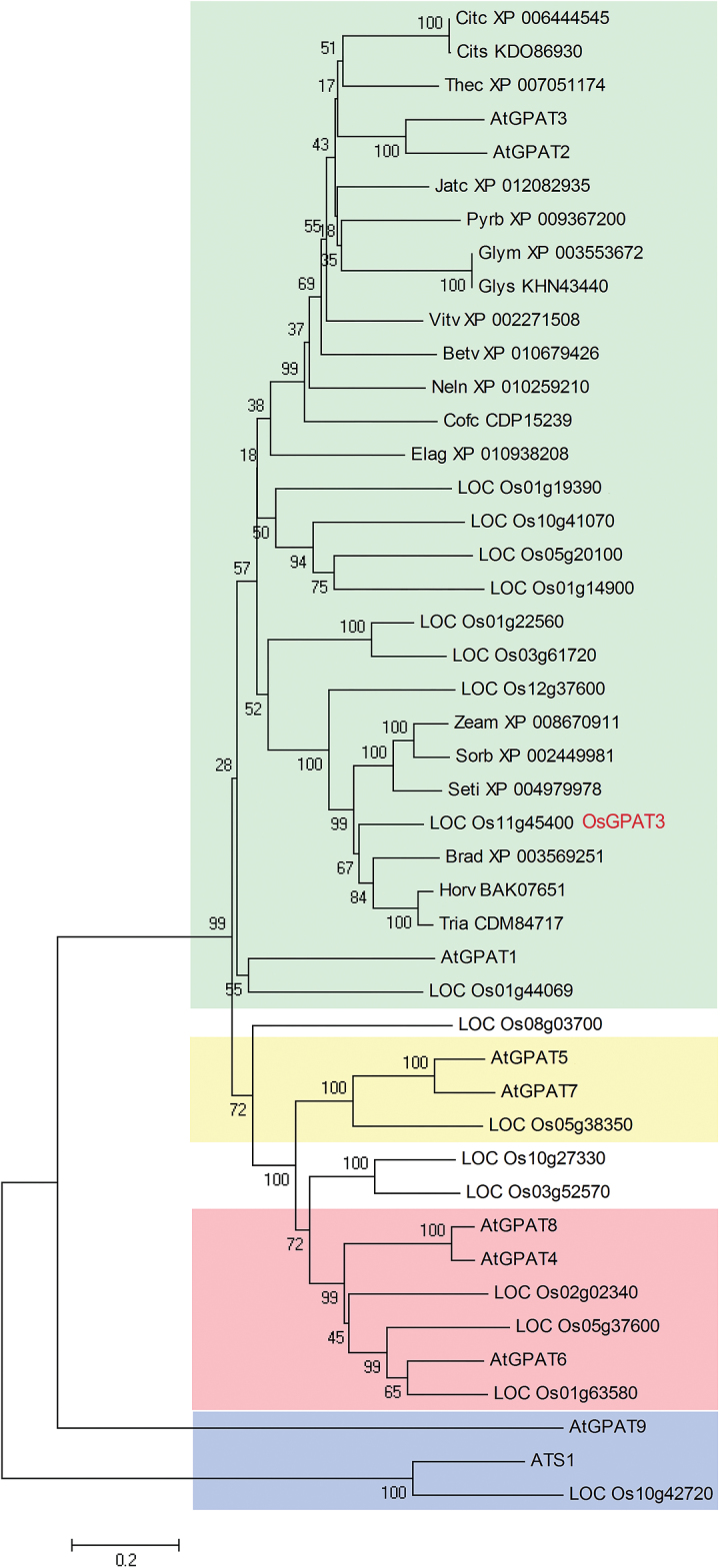
To clarify the evolutionary role and potential function of OsGPAT3, we performed phylogenetic analysis by searching the public databases NCBI, RGAP, and TAIR, using BLASTP with the full length of the OsGPAT3 predicted amino acid sequence as a query. Since the family of GPATs is quite large, we only collected a total of 45 protein sequences of all the GPAT members from Arabidopsis and rice, and the top 18 closely related OsGPAT3 homologs from other 18 plant species. Subsequently, we constructed a neighbor-joining phylogenetic tree of the 45 sequences (Fig. 7), which were grouped into two main clades. Rice plastidial GPAT (LOC_Os10g42720), together with Arabidopsis plastidial ATS1 and ER-bound AtGPAT9, belonged to the sn-1 clade. The sn-2 GPAT clade included all the other plant members, which could be mainly classified into three subclades: the AtGPAT4/6/8-related subclade required for the biosynthesis of cutin, the AtGPAT5/7-related subclade associated with the biosynthesis of suberin, and the AtGPAT1/2/3-related subclade, which was less functionally characterized than the other two subclades. LOC_Os11g45400 was relatively close to AtGPAT3, and therefore was named OsGPAT3. The sn-2 clade is a land-plant-specific lineage associated with the landmark transition from aquatic to terrestrial habitats (Yang et al., 2012). Compared with the first two subclades, the third sn-2 GPAT subclade is more divergent and has more members, and is considered to be the evolutionarily more recent clade. So far, only AtGPAT1 in the third subclade has been found to have in vitro enzyme activity, and is essential for tapetum differentiation and male fertility (Zheng et al., 2003). No enzyme activity was detected for AtGPAT2 or AtGPAT3, and their mutants showed no obvious phenotype in organs such as leaves, flowers, or seeds (Yang et al., 2012). There are nine rice GPATs in the third subclade. Among them, OsGPAT3 is closer to members from Brachypodium distachyon, Hordeum vulgare, Triticum aestivum, Zea mays, Sorghum bicolor, Setaria italica, and three other members from rice. Based on the fact that osgpat3 displayed a remarkably defective phenotype in the anther and pollen, we propose that OsGPAT3 and its homologs in rice may have evolved divergently, leading to diversified functions different from those in dicots. Thus, OsGPAT3 may represent one unique sn-2 GPAT member specific to monocots.
Fig. 7.
Phylogenetic tree of the GPAT family and OsGPAT3-related orthologs. A neighbor-joining phylogenetic tree was constructed with of all the Arabidopsis and rice GPATs and OsGPAT3 orthologs from other 18 species (E-value less than 7e-132) found in the NCBI, RGAP, and TAIR databases. The proteins are named according to their gene names or NCBI accession numbers. Bootstrap values are percentage of 1000 replicates. The length of the branches is proportional to the amino acid variation rates. Different clades are highlighted in different colors, representing the sn-1 GPAT (blue), the GPAT4/6/8 clade (red), the GPAT5/7 clade (yellow), and the GPAT1/2/3 clade (green). At, Arabidopsis thaliana; Os, Oryza sativa; Citc, Citrus clementina; Cits, Citrus sinensis; Thec, Theobroma cacao; Jatc, Jatropha curcas; Pyrb, Pyrus x bretschneideri; Vitv, Vitis vinifera; Glym, Glycine max; Glys, Glycine soja; Betv, Beta vulgaris; Neln, Nelumbo nucifera; Cofc, Coffea canephora; Elag, Elaeis guineensis; Zeam, Zea mays; Sorb, Sorghum bicolor; Seti, Setaria italica; Brad, Brachypodium distachyon; Horv, Hordeum vulgare; Tria, Triticum aestivum. (This figure is available in colour at JXB online.)
OsGPAT3 is mainly expressed in tapetum and microspores
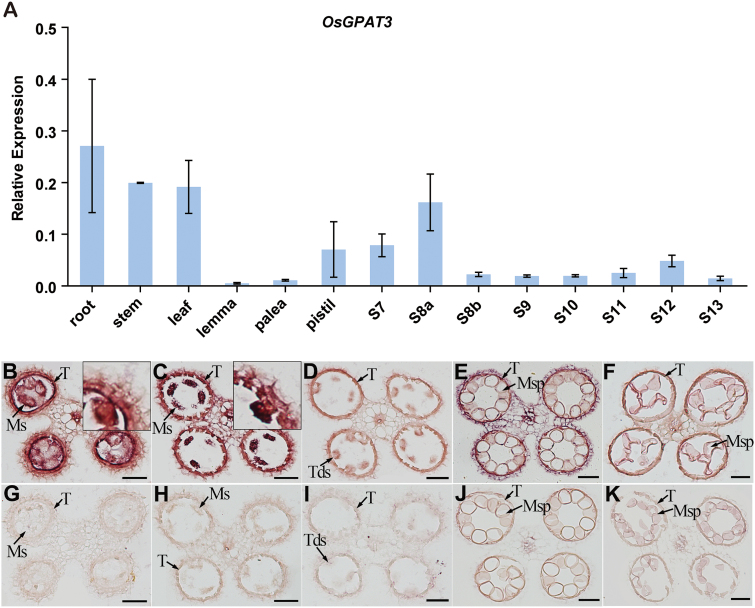
To further understand the function of OsGPAT3 in male sterility, we investigated the expression pattern of OsGPAT3 in anthers at various developmental stages and other tissues in wild-type plants using qRT-PCR. In anther tissues, the expression of OsGPAT3 was detectable as early as stage 7, peaked at stage 8a, declined gradually until stage 10, then increased and peaked again at a lower level at stage 12, and thereafter declined (Fig. 8A). This dynamic expression pattern of OsGPAT3 in anther tissues was also confirmed by GUS staining (Supplementary Fig. S2A), and was consistent with reported data from the Rice Oligonucleotide Array Database (http://www.ricearray.org/; see Supplementary Fig. S2B). Although relatively higher levels of expression of OsGPAT3 were also detected in stem, root, and leaf tissues, and moderate expression was detected in pistil, no obvious morphological abnormalities were observed in these organs in osgpat3 plants, suggesting a possible redundant function of OsGPAT3-related homologs in these tissues.
Fig. 8.
Expression patterns of OsGPAT3 in wild-type anthers and other tissues. (A) qRT-PCR analysis of the expression level of OsGPAT3 in wild-type root, stem, leaf, lemma, palea, pistil, and in anthers at different developmental stages (S7–S13). Rice OsActin1 was used as a normalizer control. Error bars indicate the SE of three biological replicates. (B–K) In situ analysis of the expression of OsGPAT3 in wild-type anthers at stages 7, 8a, 8b, 10, and 11, respectively, with antisense probe (B–F) and sense probe (G–K). The insets in (B) and (C) show an enlarged section. Ms, microsporocyte; Msp, microspores; T, tapetum; Tds, tetrads. Bars=50 μm. (This figure is available in colour at JXB online.)
To determine the spatial and temporal expression patterns of OsGPAT3 more precisely, we performed in situ hybridization with wild-type anther sections. The strong signal of OsGPAT3 transcripts was detected mainly in microspores and tapetum, and weak signals were also detected in the middle layer and the endothecium layer of the anther wall (Fig. 8B–K). The tendency to variation in signal intensity at different stages was also in agreement with the results of qRT-PCR and GUS staining analyses. These results supported that OsGPAT3 is directly involved in tapetal development, anther development, and pollen formation.
OsGPAT3 localizes to endoplasmic reticulum
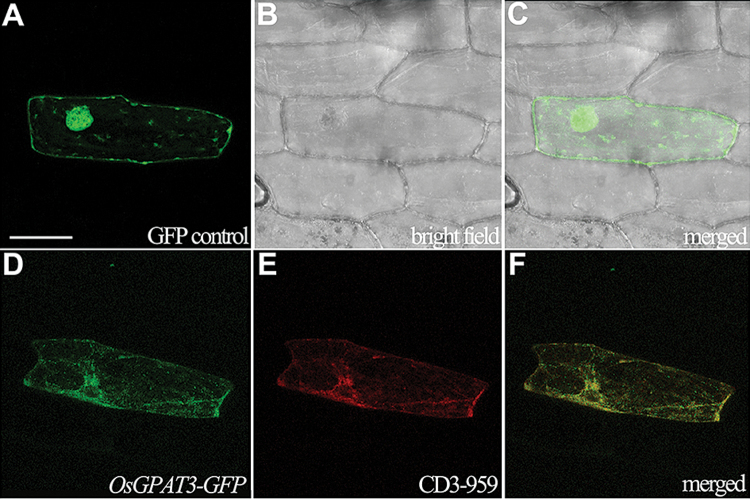
OsGPAT3 has a 21 amino acid putative signal peptide at the N-terminus that was predicted to be localized to the mitochondrion or chloroplast (SignalP and TargetP; http://www.cbs.dtu.dk/services/). To validate this prediction and to further understand the function of OsGPAT3, we made three constructs: OsGPAT3-GFP containing the full-length cDNA of OsGPAT3, OsGPAT3ΔN-GFP without the sequence encoding the 21 amino acid peptide, and a GFP control, all driven by the CaMV35S promoter, to determine the subcellular localization of OsGPAT3. When these individual constructs were transformed into onion epidermal cells by particle bombardment, OsGPAT3-GFP signal was observed mainly at the ER (Fig. 9D), mimicking the signal of an ER-localized marker protein (CD3-959, a combination of the AtWAK2 signal peptide and the ER retention signal His-Asp-Glu-Leu; Fig. 9E, F) (Nelson et al., 2007). Although OsGPAT3 lacks the classic ER retention signal KDEL, it has one KKXX motif at the C-terminus and three internal RXR motifs, all of which are also well-defined ER retrieval motifs (Gao et al., 2014; Shikano and Li, 2003). Furthermore, the co-transformation of OsGPAT3-GFP with a mitochondrial marker gene did not show overlapping of the fluorescent signals (data not shown), which is consistent with previous reports that there is no mitochondrial localization of Arabidopsis AtGPAT2 or AtGPAT3 (Shen et al., 2003; Zheng et al., 2003). In addition, the OsGPAT3ΔN-GFP construct showed the same fluorescent profile as that of OsGPAT3-GFP (data not shown), suggesting that the 21 amino acid sequence at the N-terminus of OsGPAT3 was not a signal peptide. These data showed that OsGPAT3 is an ER-localized GPAT.
Fig. 9.
Confocal microscopic images showing the subcellular localization of OsGPAT3-GFP. (A–C) Localization of GFP control (35S:GFP) in onion epidermal cells. (D) Localization of OsGPAT3-GFP (35S:OsGPAT3-GFP) in onion epidermal cells. (E) Red fluorescence shows the ER localization of an ER marker protein (CD3-959) in the same cell as shown in (D). (F) Merged image of (D) and (E). Bars=100 μm. (This figure is available in colour at JXB online.)
Expression of genes associated with anther development is down-regulated in osgpat3
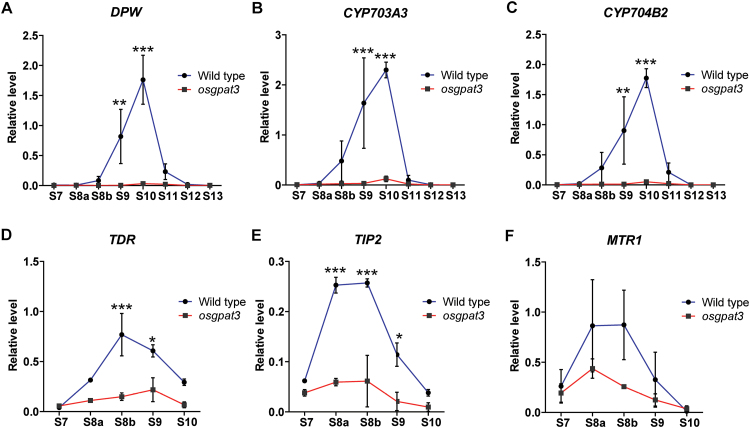
Because osgpat3 showed a dramatic decrease in most kinds of detectable lipid molecules and defects in anther cuticle and pollen exine formation, we hypothesized that osgpat3 may have impacts on the expression of genes involved in the lipid metabolism required for anther cuticle or exine formation. To test this hypothesis, we compared the expression of a set of genes known to be associated with lipid metabolism in wild-type and osgpat3 anthers using qRT-PCR. The results showed that the expression of not only the three lipid biosynthetic genes (DPW, CYB704B2, and CYP703A3) (Fig. 10A–C), but also two transcription factors (TDR and its interaction protein TIP2), which are associated with tapetum degeneration and lipid metabolism (Fig. 10D, E), were significantly down-regulated in the osgpat3 mutant. In addition, the expression level of MTR1 (Fig. 10F), a secretory fasciclin glycoprotein-encoding gene that affects the development of both tapetum and microspores (Tan et al., 2012), was down-regulated in the osgpat3 mutant. These results suggested that OsGPAT3 may play important roles in the early stages of anther development.
Fig. 10.
Analysis of the expression of anther development regulatory genes using qRT-PCR at different developmental stages. (A–C) qRT-PCR expression analysis of the cuticle and pollen wall development-related lipid biosynthetic pathway genes (A) DPW, (B) CYP703A3, and (C) CYP704B2 in stage 7 to 13 anthers from wild-type and osgpat3 rice. (D–F) qRT-PCR expression analysis of anther development-related transcription factor genes (D) TDR and (E) TIP2, and (F) a fasciclin glycoprotein protein-encoding gene, MTR1, in stage 7 to 10 anthers from wild-type and the osgpat3 mutant rice. OsActin1 was used as a normalizer control. Error bars indicate the SE of three biological replicates. *P<0.05, **P<0.01, ***P<0.001. (This figure is available in colour at JXB online.)
Discussion
Lipids, their derivatives, and their pathway-related genes are critical for reproductive development and fertility in higher plants. So far as we know, the tapetal lipid metabolism contributes significantly to anther wall cuticle and pollen exine/tryphine formation during anther development. In tapetal cells, de novo synthesized fatty acids were either reduced to fatty alcohols by MS2/DPW in plastid or translocated to ER for activation via ACOS, elongation through long-chain acyl-CoA synthetase (LACS), and hydroxylation by CYP450s (Shi et al., 2015). Glycerol is an essential backbone of plant polyesters such as suberin (Pollard et al., 2008); however, its presence in plant cutin was discovered only comparatively recently (Graça et al., 2002). The linking of fatty acids to glycerol that is catalyzed by GPAT was suggested to occur after oxidization (Beisson et al., 2012; Yang et al., 2012); therefore, various substituted fatty acids, such as ω-hydroxylated fatty acids and α, ω-dicarboxylic fatty acid, are thought to be the substrates of those GPATs. For example, the Arabidopsis AtGPAT6 prefers C16 and C18 ω-oxidized acyl-CoA substrates, while AtGPAT1 can use both unsubstituted and substituted acyl-CoAs.
To answer the question of whether OsGPAT3 works downstream of the previously reported lipid metabolic enzymes, such as DPW, CYP704B2, and CYP703A3, which are involved in the regulation of rice male fertility, we performed an enzyme activity assay of OsGPAT3 with substrates that are the products of DPW, CYP704B2. and CYP703A3 or the dominant components of plant biopolymers. This assay did not provide any positive results (data not shown); this is consistent with findings in Arabidopsis, in which AtGPAT2 or AtGPAT3 also showed no activity to tested fatty acid-CoA substrates (Yang et al., 2012; Zheng et al., 2003). It is possible that an appropriate substrate has not yet been tested or that unidentified factors are missing from the assay system, and these possibilities deserve further investigation in the future. However, the significantly altered cuticular lipid profile in osgpat3 mutant anthers, together with the high amino acid sequence similarity of OsGPAT3 to other known GPAT family members, suggests that OsGPAT3 is an active enzyme that is required for male fertility in rice. In addition, our comparative gene expression analysis through the Rice Oligonucleotide Array Database found that the expression of OsGPAT3 peaks earlier than that of DPW, CYP704B2, and CYP704A3, consistent with previous studies (Li et al., 2010; Shi et al., 2011; Yang et al., 2014) and our qRT-PCR data (Fig. 8A; Supplementary Fig. S2B). Combining the findings that the expression of DPW, CYP704B2, and CYP703A3, and of tapetum programmed cell death regulatory genes such as TDR and TIP2, were significantly down-regulated in the osgpat3 mutant, we propose that OsGPAT3 may not work directly downstream of DPW, CYP704B2, or CYP703A3. Rather, OsGPAT3 may affect male fertility via its function at earlier developmental stages, with a mechanism that differs from that of these previously reported lipid metabolic genes, which are usually expressed at relatively late anther developmental stages. It is possible that a gene like LOC_Os1g63580, a homolog of AtGPAT6, is likely to work downstream of DPW, CYP704B2, and CYP704A3 (Supplementary Fig. S2B).
In Arabidopsis, AtGPAT1 and AtGPAT6 were reported to be required for male fertility. The atgpat1 mutant showed perturbed tapetum degeneration and massive arrest of pollen development, and the atgpat6 mutant exhibited large-scale abortion of pollen grains and defective pollen wall formation. Both atgpat1 and atgpat6 single mutants were semi-sterile and displayed reduced ER proliferation in the tapetal cells, while the atgpat1atgpat6 double mutant showed a defect of microspore release from tetrads and complete male sterility (Li et al., 2012; Zheng et al., 2003). Our results showed that loss of function of OsGPAT3 expression in tapetum and microspores disrupts tapetum development and metabolism, leading to defective anther cuticle and pollen exine formation (including the absence of Ubisch bodies), and eventual complete abortion of pollen grains. Unlike atgpat1 or atgpat6, the osgpat3 mutant exhibited increased ER expansion and proliferation in tapetal cells at anther developmental stage 8, and almost complete absence of ER but abundant vacuoles and lipidosomes (probably generated from the degradation of ER) at stage 9, indicating that the lesion of OsGPAT3 may lead to abnormal ER development and degradation, which may affect tapetum metabolism and function. This functional difference between OsGPAT3 and AtGPAT1/6 may reflect structural differences in tapetum; the tapeta of rice and other cereals exhibit characteristic orbicules/Ubisch bodies, which have not been observed in members of the Brassicaceae family, including Arabidopsis, which have unique secretory tapeta containing specialized organelles including elaioplasts and tapetosomes (Zhang et al., 2011).
It was reported that the land plant GPATs belong to a sn-2 GPAT family that differs from animal GPATs (Yang et al., 2012). Clade classification of this family reflected their biochemical functions, which are evolutionally associated with the plants’ adaptation during the transition from aquatic to terrestrial habitats. While the first and second clades are conserved among the tracheophytes in the biosynthesis of cutin or suberin, the function of the third clade remains poorly known. Our phylogenetic analysis showed that the monocotyledon subclade to which OsGPAT3 belongs includes three other rice members with unknown function (Fig. 7). While atgpat2 or atgpat3 mutants showed no obvious macroscopic or chemical phenotype (Yang et al., 2012), the osgpat3 mutant showed significant changes in anther cuticular lipid profiling and was male sterile, suggesting that OsGPAT3 plays an indispensable role in male reproduction that is distinct from the role of its dicot counterpart. Further functional characterization of the third land plant sn-2 GPAT clade will facilitate our understanding of the evolutionary and molecular aspects of the grass GPAT proteins in plant evolution and adaptation.
In conclusion, we identified a land plant sn-2 GPAT family member, OsGPAT3, in rice, which encodes an ER-localized GPAT. The dysfunction of OsGPAT3 significantly affects the anther cuticle and pollen exine formation, and leads to eventual male sterility. This work has expanded our understanding of lipid metabolism, particularly the poorly known glycerolipid metabolism, in plant male reproductive development. The conservation and divergence of the sn-2 GPAT family in land plants merit further investigations, which would help us to explore the evolutionary and biochemical functions of lipid metabolism in plant fertility.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The ratio of weight/surface area of the anthers in the wild type and osgpat3 mutant.
Fig. S2. Expression patterns of OsGPAT3 in wild-type anthers.
Fig. S3. Transmission electron microscopy analysis of tapetal cell development in the wild type and osgpat3 mutant at stage 8.
Table S1. Primer sequences used in this study.
Author contributions
DZ and WL conceived the original screening and research plans; DZ and WL supervised the project and experiments; XM performed most of the experiments; QZ and GL participated in primer design, vector construction, and lipid analysis; JS, SQ, LZ, ZL, and MC provided technical assistance to XM; JS and XM conceived and wrote the paper, with contributions from all the authors; DZ supervised and complemented the writing of the paper.
Supplementary Material
Acknowledgements
We thank Prof. John B. Ohlrogge and Drs Weili Yang, Xiuling Cai, and Jiechen Wang for their help in the GPAT activity assay. We thank Mr Changsong Yin for his help in the in situ hybridization. We also thank Ms Na Luo of the University of Hong Kong for editing this paper. This work was supported by funds from the National Key Research and Development Program of China (2016YFD0100804); the National Natural Science Foundation of China (31430009, 31110103915, 32322040, 31271698); China Innovative Research Team, Ministry of Education, the Innovative Research Team in University of the Ministry of Science and Technology of China, and the Program of Introducing Talents of Discipline to Universities (111 Project, B14016); the National Key Basic Research Developments Program, Ministry of Science and Technology, China (2013CB126902); the Science and Technology Commission of Shanghai Municipality (13JC1408200); and the Leading Scientist in Agriculture of Shanghai Municipality.
References
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. 1997. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. The Plant Journal 12, 615–623. [DOI] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M. 2009. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. The Plant Cell 21, 1453–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson F, Li-Beisson Y, Pollard M. 2012. Solving the puzzles of cutin and suberin polymer biosynthesis. Current Opinion in Plant Biology 15, 329–337. [DOI] [PubMed] [Google Scholar]
- Beisson F, Li Y, Bonaventure G, Pollard M, Ohlrogge JB. 2007. The acyltransferase GPAT5 is required for the synthesis of suberin in seed coat and root of Arabidopsis. The Plant Cell 19, 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chu H, Yuan Z, Pan A, Liang W, Huang H. 2006. Isolation and genetic analysis for rice mutants treated with 60 Co γ -Ray. Journal of Xiamen University 45, 82–85. [Google Scholar]
- Chen W, Yu XH, Zhang K, Shi J, De Oliveira S, Schreiber L, Shanklin J, Zhang D. 2011a Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiology 157, 842–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Snyder CL, Truksa M, Shah S, Weselake RJ. 2011b sn-Glycerol-3-phosphate acyltransferases in plants. Plant Signaling & Behavior 6, 1695–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H, Qian Q, Liang W, et al. 2006. The FLORAL ORGAN NUMBER4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiology 142, 1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RA, Lee DP. 2004. Enzymes of triacylglycerol synthesis and their regulation. Progress in Lipid Research 43, 134–176. [DOI] [PubMed] [Google Scholar]
- de Azevedo Souza C, Kim SS, Koch S, Kienow L, Schneider K, McKim SM, Haughn GW, Kombrink E, Douglas CJ. 2009. A novel fatty acyl-CoA synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. The Plant Cell 21, 507–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa AA, Shrestha J, Morant M, Pinot F, Matsuno M, Swanson R, Møller BL, Preuss D. 2009. CYP704B1 is a long-chain fatty acid omega-hydroxylase essential for sporopollenin synthesis in pollen of Arabidopsis. Plant Physiology 151, 574–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou XY Yang KZ Zhang Y Wang W Liu XL Chen LQ Zhang XQ, Ye D. 2011. WBC27, an adenosine tri-phosphate-binding cassette protein, controls pollen wall formation and patterning in Arabidopsis. Journal of Integrative Plant Biology 53, 74–88. [DOI] [PubMed] [Google Scholar]
- Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W. 2014. The rice basic helix-loop-helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. The Plant Cell 26, 1512–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez JF, Talle B, Wilson ZA. 2015. Anther and pollen development: a conserved developmental pathway. Journal of Integrative Plant Biology 57, 876–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Cai Y, Wang Y, Kang BH, Aniento F, Robinson DG, Jiang L. 2014. Retention mechanisms for ER and Golgi membrane proteins. Trends in Plant Science 19, 508–515. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM. 1993. Anther development: basic principles and practical applications. The Plant Cell 5, 1217–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graça J, Schreiber L, Rodrigues J, Pereira H. 2002. Glycerol and glyceryl esters of omega-hydroxyacids in cutins. Phytochemistry 61, 205–215. [DOI] [PubMed] [Google Scholar]
- Huang Q, Dresselhaus T, Gu H, Qu LJ. 2015. Active role of small peptides in Arabidopsis reproduction: expression evidence. Journal of Integrative Plant Biology 57, 518–521. [DOI] [PubMed] [Google Scholar]
- Jeffree C. 1996. Structure and ontogeny of plant cuticles. In: Kersteins G, ed. Plant cuticles: an integrated functional approach. Oxford: BIOS Scientific Publishers, 33–82. [Google Scholar]
- Jeon JS, Lee S, Jung KH, et al. 2000. T-DNA insertional mutagenesis for functional genomics in rice. The Plant Journal 22, 561–570. [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee DY, et al. 2006. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. The Plant Cell 18, 3015–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Samuels AL. 2003. Biosynthesis and secretion of plant cuticular wax. Progress in Lipid Research 42, 51–80. [DOI] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. 2009. Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proceedings of the National Academy of Sciences of the United States of America 106, 22008–22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, et al. 2013. Acyl-lipid metabolism. The Arabidopsis Book 11, e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, et al. 2010. Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. The Plant Cell 22, 173–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson ZA, Zhang D. 2011. PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiology 156, 615–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang D. 2010. Biosynthesis of anther cuticle and pollen exine in rice. Plant Signaling & Behavior 5, 1121–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, et al. 2006. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. The Plant Cell 18, 2999–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XC, Zhu J, Yang J, Zhang GR, Xing WF, Zhang S, Yang ZN. 2012. Glycerol-3-phosphate acyltransferase 6 (GPAT6) is important for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Molecular Plant 5, 131–142. [DOI] [PubMed] [Google Scholar]
- Li Y, Beisson F, Koo AJK, Molina I, Pollard M, Ohlrogge J. 2007. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proceedings of the National Academy of Sciences of the United States of America 104, 18339–18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Mehdy MC. 2007. A nonclassical arabinogalactan protein gene highly expressed in vascular tissues, AGP31, is transcriptionally repressed by methyl jasmonic acid in Arabidopsis. Plant Physiology 145, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Chu H, Li H, et al. 2005. Genetic analysis and mapping of rice (Oryza sativa L.) male-sterile (OsMS-L) mutant. Chinese Science Bulletin 50, 122–125. [Google Scholar]
- Liu J, Qu LJ. 2008. Meiotic and mitotic cell cycle mutants involved in gametophyte development in Arabidopsis. Molecular Plant 1, 564–574. [DOI] [PubMed] [Google Scholar]
- Liu L, Fan XD. 2013. Tapetum: regulation and role in sporopollenin biosynthesis in Arabidopsis. Plant Molecular Biology 83, 165–175. [DOI] [PubMed] [Google Scholar]
- Mañas-Fernández A, Li-Beisson Y, Alonso DL, García-Maroto F. 2010. Cloning and molecular characterization of a glycerol-3-phosphate O-acyltransferase (GPAT) gene from Echium (Boraginaceae) involved in the biosynthesis of cutin polyesters. Planta 232, 987–997. [DOI] [PubMed] [Google Scholar]
- Ma H. 2005. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annual Review of Plant Biology 56, 393–434. [DOI] [PubMed] [Google Scholar]
- Moolenaar WH, Kranenburg O, Postma FR, Zondag GC. 1997. Lysophosphatidic acid: G-protein signalling and cellular responses. Current Opinion in Cell Biology 9, 168–173. [DOI] [PubMed] [Google Scholar]
- Morant M, Jørgensen K, Schaller H, Pinot F, Møller BL, Werck-Reichhart D, Bak S. 2007. CYP703 is an ancient cytochrome P450 in land plants catalyzing in-chain hydroxylation of lauric acid to provide building blocks for sporopollenin synthesis in pollen. The Plant Cell 19, 1473–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C. 2006. Unraveling the complex network of cuticular structure and function. Current Opinion in Plant Biology 9, 281–287. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D. 2013. EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nature Communications 4, 1445. [DOI] [PubMed] [Google Scholar]
- Parish RW, Li SF. 2010. Death of a tapetum: a programme of developmental altruism. Plant Science 178, 73–89. [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. 2008. Building lipid barriers: biosynthesis of cutin and suberin. Trends in Plant Science 13, 236–246. [DOI] [PubMed] [Google Scholar]
- Qin P, Tu B, Wang Y, Deng L, Quilichini TD, Li T, Wang H, Ma B, Li S. 2013. ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant & Cell Physiology 54, 138–154. [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu Y-C, Lee PY, Truong MT, Beals TP, Goldberg RB. 1999. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sexual Plant Reproduction 11, 297–322. [Google Scholar]
- Shen W, Wei Y, Dauk M, Zheng Z, Zou J. 2003. Identification of a mitochondrial glycerol-3-phosphate dehydrogenase from Arabidopsis thaliana: evidence for a mitochondrial glycerol-3-phosphate shuttle in plants. FEBS Letters 536, 92–96. [DOI] [PubMed] [Google Scholar]
- Sheng X, Yung YC, Chen A, Chun J. 2015. Lysophosphatidic acid signalling in development. Development 142, 1390–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Cui M, Yang L, Kim YJ, Zhang D. 2015. Genetic and biochemical mechanisms of pollen wall development. Trends in Plant Science 20, 741–753. [DOI] [PubMed] [Google Scholar]
- Shi J, Tan H, Yu XH, et al. 2011. Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. The Plant Cell 23, 2225–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano S, Li M. 2003. Membrane receptor trafficking: evidence of proximal and distal zones conferred by two independent endoplasmic reticulum localization signals. Proceedings of the National Academy of Sciences of the United States of America 100, 5783–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K, Reue K. 2009. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. American Journal of Physiology. Endocrinology and Metabolism 296, E1195–E1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H, Liang W, Hu J, Zhang D. 2012. MTR1 encodes a secretory fasciclin glycoprotein required for male reproductive development in rice. Developmental Cell 22, 1127–1137. [DOI] [PubMed] [Google Scholar]
- Wallace S, Chater CC, Kamisugi Y, Cuming AC, Wellman CH, Beerling DJ, Fleming AJ. 2015. Conservation of Male Sterility 2 function during spore and pollen wall development supports an evolutionarily early recruitment of a core component in the sporopollenin biosynthetic pathway. The New Phytologist 205, 390–401. [DOI] [PubMed] [Google Scholar]
- Wendel AA, Lewin TM, Coleman RA. 2009. Glycerol-3-phosphate acyltransferases: rate limiting enzymes of triacylglycerol biosynthesis. Biochimica et Biophysica Acta 1791, 501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ZA, Zhang DB. 2009. From Arabidopsis to rice: pathways in pollen development. Journal of Experimental Botany 60, 1479–1492. [DOI] [PubMed] [Google Scholar]
- Wu L, Guan Y, Wu Z, et al. 2014. OsABCG15 encodes a membrane protein that plays an important role in anther cuticle and pollen exine formation in rice. Plant Cell Reports 33, 1881–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Shi J, Rautengarten C, et al. 2016. Defective Pollen Wall 2 (DPW2) encodes an acyl transferase required for rice pollen development. Plant Physiology DOI: 10.1104/pp.16.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Ding Z, Vizcay-Barrena G, et al. 2014. ABORTED MICROSPORES acts as a master regulator of pollen wall formation in Arabidopsis. The Plant Cell 26, 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Yang C, Yuan Z, Zhang D, Gondwe MY, Ding Z, Liang W, Zhang D, Wilson ZA. 2010. The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana. The Plant Cell 22, 91–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Simpson JP, Li-Beisson Y, Beisson F, Pollard M, Ohlrogge JB. 2012. A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference, and evolution. Plant Physiology 160, 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Wu D, Shi J, et al. 2014. Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. Journal of Integrative Plant Biology 56, 979–994. [DOI] [PubMed] [Google Scholar]
- Yu J, Meng Z, Liang W, et al. 2016. A rice Ca2+ binding protein is required for tapetum function and pollen formation. Plant Physiology DOI: 10.1104/pp.16.01261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DS, Liang WQ, Yuan Z, Li N, Shi J, Wang J, Liu YM, Yu WJ, Zhang DB. 2008. Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Molecular Plant 1, 599–610. [DOI] [PubMed] [Google Scholar]
- Zhang D, Li H. 2014. Exine export in pollen. In: Geisler M, ed. Plant ABC transporters. Cham: Springer International Publishing, 49–62. [Google Scholar]
- Zhang D, Liang W. 2016. Improving food security: using male fertility for hybrid seed breeding. In: Sanders S, Hicklin T, eds. Pushing the boundaries of scientific research: 120 years of addressing global issues. Washington, DC: Science/AAAS, 45–48. [Google Scholar]
- Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D. 2010. OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiology 154, 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L. 2011. Cytological analysis and genetic control of rice anther development. Journal of Genetics and Genomics 38, 379–390. [DOI] [PubMed] [Google Scholar]
- Zhang D, Yang L. 2014. Specification of tapetum and microsporocyte cells within the anther. Current Opinion in Plant Biology 17, 49–55. [DOI] [PubMed] [Google Scholar]
- Zhang D, Yang X, Shi J. 2016. Role of lipids in plant pollen development. In: Nakamura Y, Li-Beisson Y, eds. Lipids in plant and algae development. Cham: Springer International Publishing, 315–337. [Google Scholar]
- Zhang DB, Wilson ZA. 2009. Stamen specification and anther development in rice. Chinese Science Bulletin 54, 2342–2353. [Google Scholar]
- Zhao G, Shi J, Liang W, et al. 2015. Two ATP binding cassette G transporters, rice ATP binding cassette G26 and ATP binding cassette G15, collaboratively regulate rice male reproduction. Plant Physiology 169, 2064–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Xia Q, Dauk M, Shen W, Selvaraj G, Zou J. 2003. Arabidopsis AtGPAT1, a member of the membrane-bound glycerol-3-phosphate acyltransferase gene family, is essential for tapetum differentiation and male fertility. The Plant Cell 15, 1872–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Zou J. 2001. The initial step of the glycerolipid pathway. Journal of Biological Chemistry 276, 41710–41716. [DOI] [PubMed] [Google Scholar]
- Zhu L, Shi J, Zhao G, Zhang D, Liang W. 2013. Post-meiotic deficient anther1 (PDA1) encodes an ABC transporter required for the development of anther cuticle and pollen exine in rice. Journal of Plant Biology 56, 59–68. [Google Scholar]
- Zhu SQ, Zhao H, Zhou R, Ji BH, Dan XY. 2009. Substrate selectivity of glycerol-3-phosphate acyl transferase in rice. Journal of Integrative Plant Biology 51, 1040–1049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.