Abstract
Neonatal vocalization is structurally altered in mouse models of autism spectrum disorder. Our published data showed that pup vocalization, under conditions of maternal separation, contains sequences whose alterations in a genetic mouse model of ASD impair social communication between pups and mothers (Takahashi et al., 2016). We describe details of a method which reveals the statistical structure of call sequences that are functionally critical for optimal maternal care. Entropy analysis determines the degree of non-random call sequencing. A Markov model determines the actual call sequences used by pups. Sparse partial least squares discriminant analysis (sPLS-DA) identifies call sequences that differentiate groups and reveals the degrees of individual variability in call sequences between groups. These three sets of analyses can be used to identify the otherwise hidden call structure that is altered in mouse models of developmental neuropsychiatric disorders, including not only autism, but also schizophrenia.
Keywords: USV, mouse models, Markov model, entropy, sPLS-DA, autism spectrum disorder, schizophrenia
INTRODUCTION
It is possible to diagnose autism spectrum disorder (ASD) by two years of age in humans. However, much earlier identification of signs of ASD is critical, because early intervention is highly effective(Green et al., 2015; Green et al., 2017; Rogers et al., 2014; Wallace & Rogers, 2010; Wetherby et al., 2014). Decreased eye contact, decreased visual attention to social scenes, atypical preverbal vocalizations and atypical development of other neonatal behaviors are indicative of later development of ASD(Constantino et al., 2017; Esposito, Hiroi, & Scattoni, 2017; Jones & Klin, 2013; Ozonoff et al., 2010; Zwaigenbaum, Bryson, & Garon, 2013).
Neonatal vocalization is a very early, primary means of social communication with the mother in that its expression in newborns signals the need for care(Soltis, 2004; Zeifman, 2001). Cries of infants with incipient ASD are characterized by higher-pitch, lower waveform modulation and rhythm and more dysphonation, compared to infants with intellectual disability or typically developing infants(Esposito et al., 2017; Esposito & Venuti, 2008, 2009).
It is, however, difficult to establish the functional role of atypical vocalizations as a component of social communication in humans, as many atypical features in the cognitive, motor, and social domains cannot be isolated to test functionality of each atypical feature in social communication and interaction. Moreover, it is difficult to study genetic and epigenetic mechanisms underlying the role of atypical neonatal social communication as an integral component of ASD. Such mechanisms cannot be easily assessed using human brains.
Although ASD is a human disorder and the symptomatology of ASD cannot be entirely recapitulated in mice, genetic mouse models satisfy construct validity in that they recapitulate ASD-associated human genetic variants in mice (N Hiroi, Hiramoto, Harper, Suzuki, & Boku, 2012; N. Hiroi et al., 2013; Silverman, Yang, Lord, & Crawley, 2010). Moreover, mouse models of such genetic variants exhibit ASD-related behaviors. Mouse call types have been assessed based on number, duration and proportions (Lai et al., 2014; Michetti, Ricceri, & Scattoni, 2012; Nishi & Hiroi, 2016), as well as call types and sequences(Burkett, Day, Penagarikano, Geschwind, & White, 2015; Chabout et al., 2016; Ey et al., 2013; Hiramoto et al., 2011; Scattoni, Gandhy, Ricceri, & Crawley, 2008; Van Segbroeck, Knoll, Levitt, & Narayanan, 2017) in genetic mouse models of ASD. Pup vocal calls clearly influence maternal care in mice (Okabe et al., 2013).
The scientific premise of our neonatal vocalization analytical methods is based on the demonstration that atypical call sequences of a genetic mouse model of ASD are less effective in inducing maternal care (Kikusui & Hiroi, 2017; Takahashi et al., 2016). Our analysis further revealed altered sequences and a lack of individual variability in call sequences among pups, as early as postnatal day 7–8, with a genetic variant implicated in ASD (Takahashi et al., 2016). There is no other standardized protocol to determine individual variability in sequence structure of various call types of genetic mouse models of ASD, in which sequence structures have been validated as functional in neonatal social communication.
Here we describe a set of computational methods to characterize pup call sequences. The techniques described here are designed to record calls (see Basic Protocol 1), identify sequences (see Basic Protocol 2), determine the non-random nature of call sequences (see Basic Protocol 3), determine connections and directionality between call types in sequences (see Basic Protocol 4), and identify individual variability of call sequences among pups (see Basic Protocol 5). These analytical tools can be applied to any genetic mouse model of developmental neuropsychiatric disorders.
BASIC PROTOCOL 1
RECORDING OF PUP VOCALIZATION
Our protocol is based on an UltraSoundGate condenser microphone capsule (CM16, Avisoft, Germany) and its analytical software (Avisoft-Recorder software). Pup vocalization is recorded under a maternal separation condition.
Materials
Mouse pups at postnatal days (P) 4, 8, and 12 (±12 hr).
UltraSoundGate (Avisoft) microphone
Computer equipped with the Avisoft-RECORDER software
Plastic tray (Inner, 18 cm L × 15 cm W × 3 cm H)
Styrofoam box (Outer, 34cm L × 28cm W × 27cm H; Inner, 22cm L × 17.5cm W × 16cm H)
A sound-attenuating box (Outer, 63.5cm L × 39cm W × 60cm H; Inner, 59cm L × 35cm W × 56cm H)
Transport a mouse cage containing pups and their dam from the animal room to the laboratory at least 1 hr before the experiment.
- Place each pup in the plastic tray within in the Styrofoam box. The room temperature is maintained at 26°C.The microphone is positioned, through a hole, in the center of the Styrofoam box coverlid. The tip of the microphone is placed about 10 cm above the plastic tray. The Styrofoam box is placed in a sound-attenuating box.
- Record the pup vocalization for 5 minutes.Conduct recording at a sampling rate of 300 kHz with 16-bit format. The resulting frequency resolution should be 300000/256 FFT =1172Hz. A lower cutoff frequency of 15 kHz should be used to reduce background noise outside the relevant frequency band. The frequency window for analysis ranges from 20 kHz to 200 kHz. Call detection is provided by an automatic threshold-based algorithm and a hold time mechanism.The Nyquist Sampling Theorem dictates that sampling frequencies are a minimum of twice the maximal frequency of the signal of interest. Therefore, considering that vocalizations can occur at fundamental frequencies of approximately 100 kHz, a minimum recording rate of 200 kS/s is required (Zeskind et al., 2011).
- Return the pup to the mouse cage with the dam. Mark each pup with tattoo ink after the first recording to differentiate them in the litter.Collect tail samples for genotyping from each pup after completing vocal recording for all neonatal ages.
Save an audio (.wav) file, using the “save as” command: use the commands “Analyze > Create Spectrogram” to create a spectrogram for viewing the audio file.
- Create an automated measurement report showing calculated timestamps and durations of individual vocal calls.The Avisoftware generates a report (REPORT) of detected sound signals which are defined by a series of parametric criteria, including maximum change, minimum duration and hold time.“Max change” is the maximum difference in frequency between two waves one pixel apart that can still be considered a continuous call. It is the tolerated maximum change of the peak frequency between two consecutive time bins on the spectrogram, expressed as the number of pixels on the spectrogram (frequency bins)."Minimum duration" is the time duration below which a signal is rejected as a short single pulse.“Hold time” is the time duration for which no new call is recognized even if the amplitude of the spectrogram goes below the threshold. A short pause within a hold time is ignored and any sound before and after the silence is considered one call.We applied a wide range of max change/minimum duration/hold time values to generate a REPORT and found that minimum duration at 1 msec and hold time 2.5 msec generates the smallest numbers of false negative cases. In a standard setting, max change 10 pixels (11718 Hz) is recommended. In some mouse lines, some calls (e.g., frequency steps and two syllable; see Basic Protocol 2, Step 7 for call type definitions) might contain a frequency change, within a call, of as high as 40kHz without any temporal gap (as defined by the hold time). However, we found that a higher max change level increased to 20kHz or 40kHz doubled the number of false positive cases at each step, making the file unmanageably large; the number of false negatives, in which one call is erroneously counted as more than one call, was reduced by ~20% at each step, but the number of simple false negative cases, in which a call is not detected, was not changed. Thus, we recommend using max change 10 pixels and correcting false negative calls manually.
Save the audio records and all reports using a cloud-based service (e.g., DropBox) and an external hard drive as a back-up.
BASIC PROTOCOL 2
GENERATION OF DATA FILE
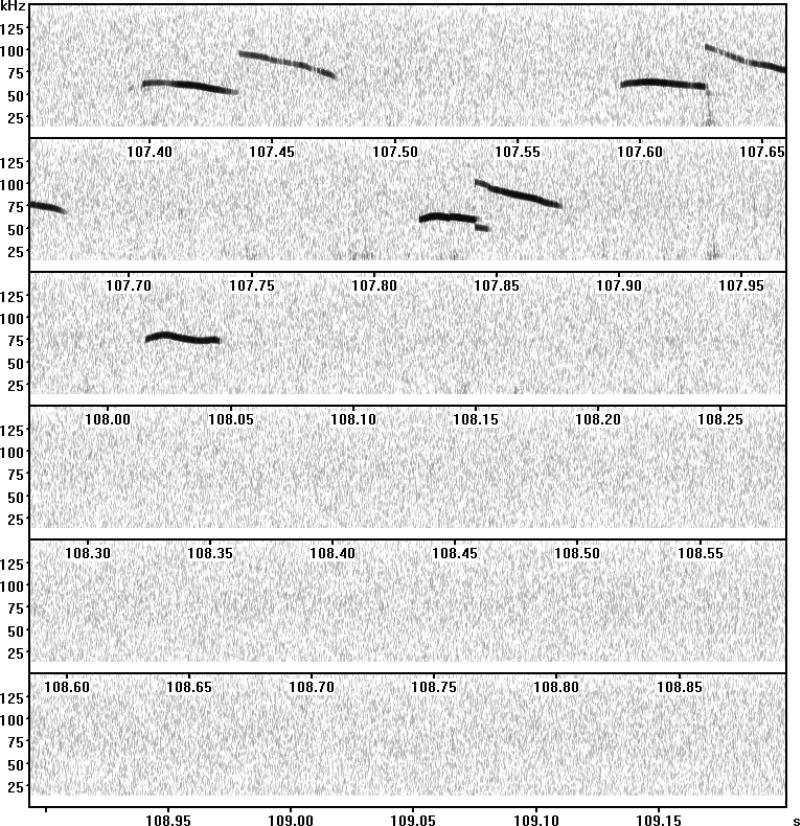
Pup calls are not emitted evenly along the time scale; calls are often uttered in a cluster, creating bursts of calls separated by pauses (see Fig. 1). A challenge is how to objectively judge a series of calls as a sequence and a pause as a functional pause. We used a Poisson model to separate calls into different clusters.
Figure 1.
A representative spectrogram. Taken from Takahashi et al., (2016).
Materials
Computer equipped with the Avisoft-RECORDER software
Audacity software (http://www.audacityteam.org/download/)
Excel
- Use Audacity (http://www.audacityteam.org/download/) software to open the audio file.We do not recommend reliance solely on a sonogram printout for classification of call types, as printouts are not sufficiently sensitive enough for visualization of low amplitude calls.
Click on the filename in the audio file window with the little black triangle pointing down.
Select “Spectrogram” to view the file in spectrogram view.
Click on the black triangle again and select “Spectrogram settings”.
- Set minimum frequency to 0 Hz and Maximum frequency to 150000 Hz.For improved visualization of calls, select the box that says “Show the spectrum using grayscale colors”.
- Use the magnifying glass icon with the + sign to zoom in until calls are visible in the view screen.The REPORT automatically generated by Avisoft-RECORDER software might include false positive cases in which noise is detected as calls and false negative cases in which genuine calls are not detected. These cases should be corrected when comparing the REPORT and images on Audacity.
- Manually determine call types using images viewed on the Audacity screen.Use the classifications by Scattoni (Scattoni et al., 2008) which include complex, harmonics, two syllable, upward, downward, flat, hump, short, composite and frequency steps. We used a correlation plot of PLS-DA analysis to validate classification of calls into ten call types in our genetic mouse model of ASD (Takahashi et al., 2016).Complex: one continuous sound wave with two or more directional pitch changes, each equal to or more than 6.25 kHz.Harmonics: one main sound wave with additional waves at different frequencies surrounding the main wave.Two syllable: two sound waves sequentially emitted with a sudden shift to a higher frequency without a temporal gap.Upward: a sound wave with a continuous increase in pitch that is equal to or larger than 12.5 kHz, with a terminal dominant frequency at least 6.25 kHz higher than the beginning of the sound wave.Downward: a sound wave with a continuous decrease in pitch that is equal to or larger than 12.5 kHz, with a terminal dominant frequency at least 6.25 kHz lower than the beginning of the sound wave.Flat: a sound wave that does not fluctuate in frequency more than 3 kHz.Hump (Chevron): an inverted-U shaped sound wave, composed of a continuous frequency increase equal to or larger than 12.5 kHz, followed by a decrease equal to or larger than 6.25 kHz.Short: a sound wave that lasts for less than 5 ms.Composite: two sound waves emitted simultaneously at different frequencies.Frequency steps: a call that changes into two separate waves simultaneously emitted at different frequencies in the middle with no interruption in time.Atypical: call types that cannot be categorized as any of the above.The method by which differences are detected between mutant and wild-type mice should be empirically determined. There are many classification schemes based on acoustic parameters, including frequencies at the start, end, and peak of calls and frequency modulation and duration. Various schemes categorize calls into three(Young, Schenk, Yang, Jan, & Jan, 2010), five (Branchi, Santucci, Vitale, & Alleva, 1998; Panksepp et al., 2007), nine (Portfors, 2007; Sugimoto et al., 2011), or ten or more distinct call types (Grimsley, Monaghan, & Wenstrup, 2011; Scattoni et al., 2008). We use the call classification scheme developed by Scattoni and colleagues, (Scattoni et al., 2008) for three reasons. First, grouping of different call types potentially masks otherwise detectable differences; the more call types that are categorized, the more sensitive the classification becomes in detecting subtle, type-specific differences. Second, our partial least-square discriminant analysis validated that the ten call types in this scheme are well separated (Takahashi et al., 2016). Third, this call classification and its variations reliably detect call type-dependent atypicalities in pups of genetic models of ASD (Burkett et al., 2015; Ey et al., 2013; Fraley et al., 2016; Hiramoto et al., 2011; Scattoni et al., 2008; Yang, Mahrt, et al., 2015); other classification schemes have not been widely used to detect atypical pup vocalization in genetic mouse models of ASD.Annotation of call types should be performed by at least two raters independently; an initial inter-rater agreement of >97% is desirable. Cases in which the initial annotations are different for the two raters should be re-evaluated by both raters to reach consensus.
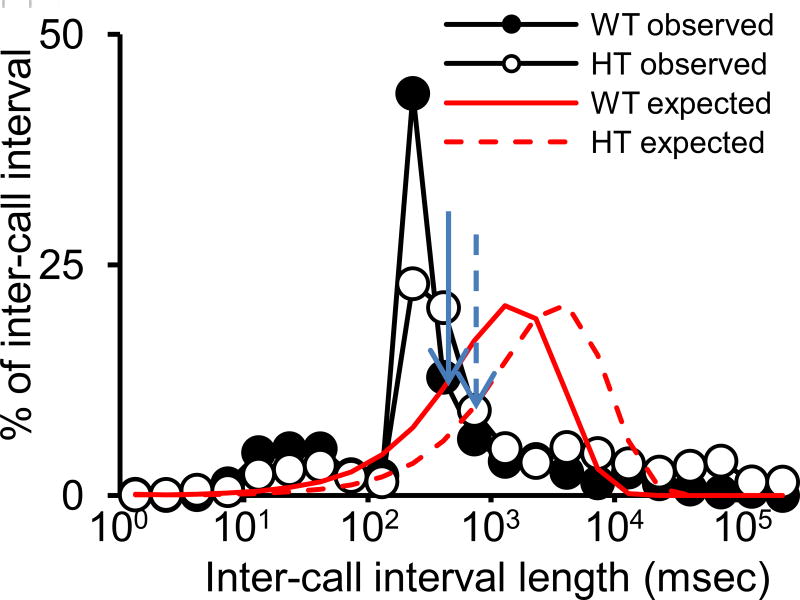
- Construct the theoretically expected distribution of inter-call intervals with a given number of calls within the 5-min recording time for each experimental group (see Fig. 2).The Avisoftware defines an inter-call interval as the distance from the beginning of one call to the beginning of the next. We define an inter-call interval as the distance from the end of one call to the beginning of the next. We add one column in Excel that lists so-defined inter-call intervals. This can be automatically done on Excel using the start and end addresses of each call that are generated by Avisoftware.
Construct the observed distribution of inter-call intervals based on the average of each experimental group.
Compare the theoretical curve to the observed distribution and determine the cross point between the two curves.
- Define sequences within which calls are emitted with inter-call intervals below the cross point value. Tally sequences in an Excel table (see Table 1).As the precise length of a functional unit of call sequences is not known, we do not consider our method to be final or definitive. Instead, we see this method as a tentative way to reduce instances in which the last call of a sequence is erroneously considered to be connected with the first call of the next sequence. This data set is the basis of analyses, described in Protocols 3–5 below.
Figure 2.
An example to determine the cut-off interval beyond which to judge two calls which are not parts of a sequence. Blue solid and broken arrows indicate the cross points of wild-type and heterozygous groups, respectively (Takahashi et al., 2016).
Table 1.
Call sequences.
| Cx | H | Cx | ||||||||||||
| Sh | ||||||||||||||
| H | H | |||||||||||||
| H | ||||||||||||||
| Cx | Cx | Cx | Cx | Cx | Cx | Cx | Cx | |||||||
| D | H | |||||||||||||
| F | ||||||||||||||
| Cx | Cx | Cx | Cx | Cx | D | Cx | D | U | ||||||
| U | ||||||||||||||
| Cx | ||||||||||||||
| Ts | H | Sh | D | D | Sh | H | U | Cx | Cx | Cx | Cx | D | D | Cx |
| H | H | H | H | |||||||||||
| H | ||||||||||||||
| Sh | D | Sh | H | Cx | F | H | D | H | H | D |
Call sequences from an individual pup are shown. Each row represents a sequence defined by Basic Protocol 2, step 11. Cx, complex; H, hump; Sh, short; D, downward; F, flat; U, upward; Ts, two syllable.
BASIC PROTOCOL 3
ENTROPY ANALYSIS
This protocol determines the presence of any structure inherent in the call sequences as well as the entropy level to be used as the basis for supervised classification. We assume that the continuous vocal calls are separated into discrete call sequences (see Table 1). Entropy measures are calculated based on a Markov model of call sequences.
In the zero-order model, entropy is based only on m, the number of call types used out of the ten call types, and is calculated as log2 (m). In the first-order model, each call-type is assumed to be independent and entropy is calculated based on individual call probabilities according to the following equation: Σ pi(log2(pi)), in which pi is the probability of observing call-type i. By extending this equation to higher-order Markov models, we can determine if different length combinations of call-type might be used to discriminate between genotypes.
In the second part of the protocol, we determine the most informative entropy level at which to perform the classification analysis (see BASIC PROTOCOL 5) - this will be the level at which the largest statistical difference is observed in entropy scores between our two genotypes. Here, we use a linear mixed model approach in the R statistical programming environment via the 'lmer' function from the 'lmerTest' package. It is also possible to perform this step using a two-way repeated measures ANOVA with the 'aov' function from the base 'stats' package in R.
Materials
Perl (version ≥ 5.20)
Author-supplied Perl script (‘entropy.pl’)
R (version ≥ 3.0.0) with installed packages: ‘ggplot2’ and ‘lmerTest’
Calculate entropy scores
- Open a terminal window in the folder containing both of your data files and the entropy.pl script, then type:
- perl entropy.pl file.csvThis script expects as input, one comma-separated value (csv) file per mouse, in which each line is a call sequence emitted by that mouse (see example file provided in Table1. The script’s default outputs are the input file name and entropy scores from entropy levels zero to four. Note that this script can easily be applied to each call sequence (csv) file in the folder by using a bash for-loop:
- for f in *.csv; do perl entropy.pl $f; done
Click on the R logo to launch the application, or, from a terminal window, type 'R'.
- Read in the data.
- mydata <- read.csv("entropy_scores.csv", sep=",", header=T)Here we assume a csv input file, although it is also possible to read in Microsoft Excel files in .xlsx format using the 'xlsx' package from R. Note that the data from Step 1 has been re-shaped so that the columns now contain: mouse ID, entropy level, genotype, and entropy score respectively (see example provided in Table2). The exact details of this file will depend on the number of samples of each type in your experimental design.
- Create entropy plot (see Fig. 3).
- library(ggplot2)
- ggplot(mydata, aes(x=level, y=entropy, fill=genotype)) + geom_boxplot()Make sure that the column names in the ‘entropy_scores.csv’ file match the arguments provided to the ggplot command.
- Fit a linear model to determine statistically significant differences in entropy scores at different levels.
- library(lmerTest)
- mod1 <- lmer (entropy ~ genotype*level + (1|mouse), data=mydata)
- summary(mod1)Here, we treat genotype and entropy level as fixed effects and assume that each mouse has a different baseline entropy. The summary command displays information on the fitted model, including the statistical significance associated with differences in entropy scores between mice from different genotypes at each entropy level; choose the most significant in terms of p-value and effect size as the basis for the classification analysis.
Table 2.
Entropy scores
| mouse | level | genotype | entropy |
|---|---|---|---|
| 1 | H0 | HT | 2.807355 |
| 2 | H0 | HT | 3.321928 |
| 3 | H0 | HT | 3 |
| 4 | H0 | HT | 2.321928 |
| 5 | H0 | HT | 2 |
| 6 | H0 | HT | 2.807355 |
| 7 | H0 | HT | 2.584963 |
| 8 | H0 | HT | 2.584963 |
| 9 | H0 | HT | 2 |
| 10 | H0 | HT | 3.169925 |
| 1 | H1 | HT | 1.89364 |
| 2 | H1 | HT | 2.449691 |
| 3 | H1 | HT | 2.235275 |
| 4 | H1 | HT | 1.603024 |
| 5 | H1 | HT | 0.843866 |
| 6 | H1 | HT | 2.011808 |
| 7 | H1 | HT | 1.504393 |
| 8 | H1 | HT | 1.899953 |
| 9 | H1 | HT | 1.509969 |
| 10 | H1 | HT | 2.381264 |
Entries from an example entropy scores file. Each row contains information on the entropy analysis for an individual mouse at a specific entropy level. The columns show mouse ID, entropy level, genotype, and entropy score, respectively. Only a portion of the entire file is shown; subsequent entries would contain information from additional entropy levels and wild-type mice. HT, heterozygous.
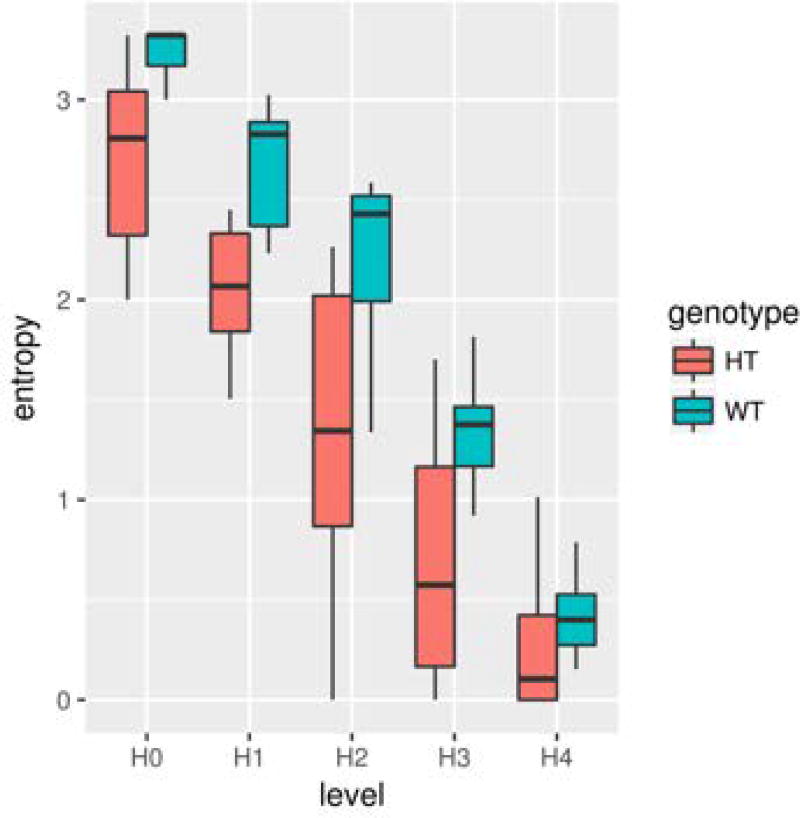
Figure 3.
Entropy plot. Red boxplots show an example of the range of entropy scores for Tbx1 heterozygous (HT) mice at each level in the entropy analysis; blue boxplots show the corresponding scores for wild-type (WT) mice.
BASIC PROTOCOL 4
IDENTIFICATION OF CALL CONNECTIONS
Markov model analysis
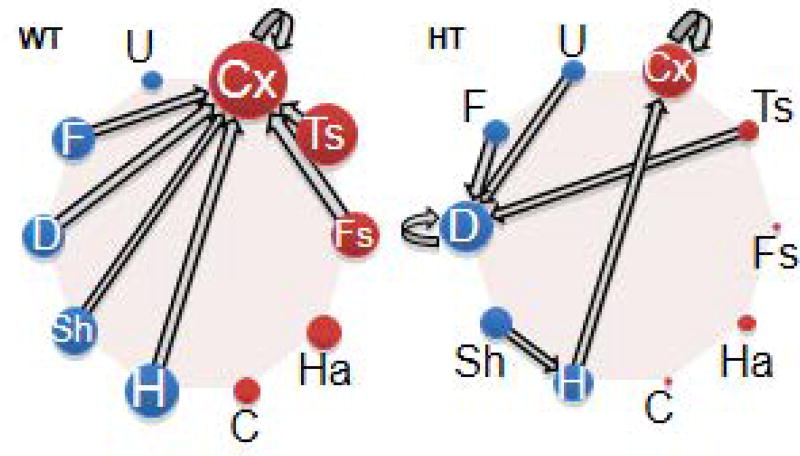
We use Markov chains (Takahashi et al., 2016) to identify the actual call types used in sequences (see Fig. 4). We assume that a pup's emitted call type is in an explicit state (i.e., S1). The probability that a mouse will transition to another state (S2) is calculated. An additional assumption is that the probability of choosing S2 depends solely on the call type of S1. We use custom-written Matlab routines (Mathworks) to calculate the probability for each transition (ST). The number of occurrences of each ST is tallied and divided by the total number of times the S1 call type is made. Connections and directions between call types are identified.
Figure 4.
Connections and their directions between two calls identified using the Markov model. Thickness of arrows and size of call circles represent the relative proportion of a connection and call numbers (Takahashi et al., 2016).
Materials
Download MatLab at https://www.mathworks.com/programs/trials/trial_request.html?prodcode=ML to install a free trial. See if your institution already has a MatLab campus-wide license.
Prepare MatLab
-
1.
Install MatLab.
-
2.
Download “call_probability.m” (see Script 1 in Supporting Information) and “call_probability_averaging.m” scripts (see Script 2 in Supporting Information).
-
3.Make sure “call_probability.m” (Script 1 in Supporting Information) and “call_probability_averaging.m” (Script 2 in Supporting Information) are somewhere in your paths. To test this, open MatLab and try “edit call_probability”.If it does not open in editor, navigate to the folder it is located in and add it using: addpath(‘C:\path\to\script’)
Calculate the Markov Probabilities
-
4.
Create a new folder containing all of your vocalization spreadsheets. For ease of use make sure nothing else is in this folder.
-
5.
Open MatLab.
-
6.Set your working directory as the folder containing the spreadsheets:
- e.g. cd(‘C:\ yourfoldername \’)
-
7.Run the “call probability” program by executing (see Script 1):
- [call_prob] = call probability(200);The input variable is the inter-call interval (ms) which defines the call string; change this value as needed.
-
8.
Save work using: save(‘markovprobs’).
Determine the relative frequencies and directions of call connections
-
9.Run “call_probability_averaging” (see Script 2) and select the saved data file from step 8
- [markovprobabilities] = call_probability_averaging;This will automatically run the calculation for all genotypes.This is your final output. Each cell is a genotype. Rows in the matrix are the first call and columns are the second for each pair.
-
10.
Save your work and figures.
BASIC PROTOCOL 5
CLASSIFICATION ANALYSIS OF CALL SEQUENCES
Supervised classification refers to the process of building a statistical model which uses a matrix, X, of predictor variables (call-type frequencies) to predict a vector Y, of class labels (genotypes). Each row in the n x m matrix, X, corresponds to a single sample (mouse), whereas each column corresponds to an individual call-type. There are many possible approaches to this problem, drawn primarily from the fields of statistics and machine learning; popular techniques include: Linear Discriminant Analysis (LDA), Decision Trees, Random Forest (RF), and Support Vector Machines (SVM). Here, we use sparse Partial Least Squares-Discriminant Analysis (sPLS-DA) approach (Le Cao, Boitard, & Besse, 2011) as implemented in the R 'mixOmics' package. This approach has the additional advantage of including variable selection during classification analysis so that only a subset of call-types are used to predict a class label (genotype) for each sample.
Materials
R (version ≥ 3.0.0) with installed package ‘mixOmics 6.3.0’
Load the data
- Open R and load the 'mixomics' library. Click on the R logo to launch the application, or from a terminal window, type: 'R'.
- library(mixOmics)
- Read in the data and create a class vector specifying the label (genotype) for each sample.
- data <- read.csv("call_frequencies.csv", sep=",", header=T)
- data.mat<-as.matrix(data[,−1])
- clvec <- c(rep(“HT”,20), rep(“WT”,8))Here, we assume a csv file, consisting of a matrix of call-type frequencies (see example provided in Table 3). Each row in the matrix is an individual mouse and each column is a specific call-type combination. The call-type combinations are based on the entropy level selected as part of BASIC PROTOCOL 3. Note that, with some minor modification, call frequency matrices can be automatically generated as part of the ‘normalize’ function within the Perl script provided. In this example, we conduct the classification on a matrix of bigram (two-call) frequencies (with 100 possible call-type combinations based on our alphabet size of ten). The clvec vector specifies the class labels (genotype) associated with each sample (row) in the call frequency matrix. In this example, we have 20 mice in our HT sample group and 8 mice in our WT sample group.
- Choose the number of variables and components using cross validation.
- keepX <- c(1:10, seq(20, 50, 10))
- tuned.model <- tune.splsda(data.mat, clvec, ncomp = 6, validation = 'Mfold', folds = 5, progressBar = TRUE, dist = 'max.dist', measure = "BER", test.keepX = keepX, nrepeat = 50, cpus = 2)In order to construct the final sPLS-DA model, two key parameters must first be tuned: ncomp, the number of components in the model, and keepX, the number of variables (call frequencies) to choose from the matrix X in order to perform the classification. The optimal combination of these parameters (measured by the balanced error rate, BER) is chosen through cross validation (either Mfold of leave-one-out). We recommend choosing from three to ten components for the model and (for small datasets with less than 100 predictor variables) up to 50% of the call-type combinations. Note that depending on the size of the dataset, these choices may significantly affect the time taken for the algorithm to complete. For further information on the various parameter choices, see either the help documentation in R, or visit mixomics.org. Final values for the tuned parameters can then be accessed via tuned.model$choice.ncomp$ncomp and tuned.model$choice.keepX.
- Build final model and plot results (Figs. 5 and 6).
- final <- splsda(data.mat, clvec, ncomp = 2, keepX = c(40,4))
- plotIndiv(final, comp = c(1,2), group = clvec, ind.names = FALSE, ellipse = TRUE, legend = TRUE, title="sPLS-DA Scores Plot Comp 1&2")
- plotLoadings(final, comp = 1, title = 'Loadings on Comp 1', contrib = 'max', method = 'mean')Alternative visualizations of the sPLS-DA results including stability plots, arrow plots, correlation circles, and clustered image maps are also available (see the mixOmics package documentation or website for further details).
Table 3.
Call-type frequency matrix.
| Cx_C | Cx_Cx | Cx_D | Cx_F | Cx_Fs | Cx_H | Cx_Ha | |
|---|---|---|---|---|---|---|---|
| Mouse1 | 0 | 0 | 0.021739 | 0 | 0 | 0 | 0.021739 |
| Mouse2 | 0.002212 | 0.245575 | 0.04646 | 0.042035 | 0.002212 | 0.026549 | 0.006637 |
| Mouse3 | 0 | 0.17033 | 0.090659 | 0.002747 | 0.002747 | 0.043956 | 0 |
| Mouse4 | 0 | 0.111111 | 0 | 0 | 0 | 0 | 0.111111 |
| Mouse5 | 0 | 0.027778 | 0 | 0 | 0 | 0 | 0 |
| Mouse6 | 0 | 0.391304 | 0 | 0 | 0 | 0.130435 | 0 |
| Mouse7 | 0 | 0.567164 | 0.029851 | 0 | 0.044776 | 0.014925 | 0 |
| Mouse8 | 0 | 0.15625 | 0 | 0 | 0 | 0.09375 | 0 |
| Mouse9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mouse10 | 0 | 0.239583 | 0.078125 | 0.020833 | 0 | 0.036458 | 0.005208 |
A matrix of call-type frequencies is shown. Each row in the matrix is an individual mouse and each column is a specific call-type combination.
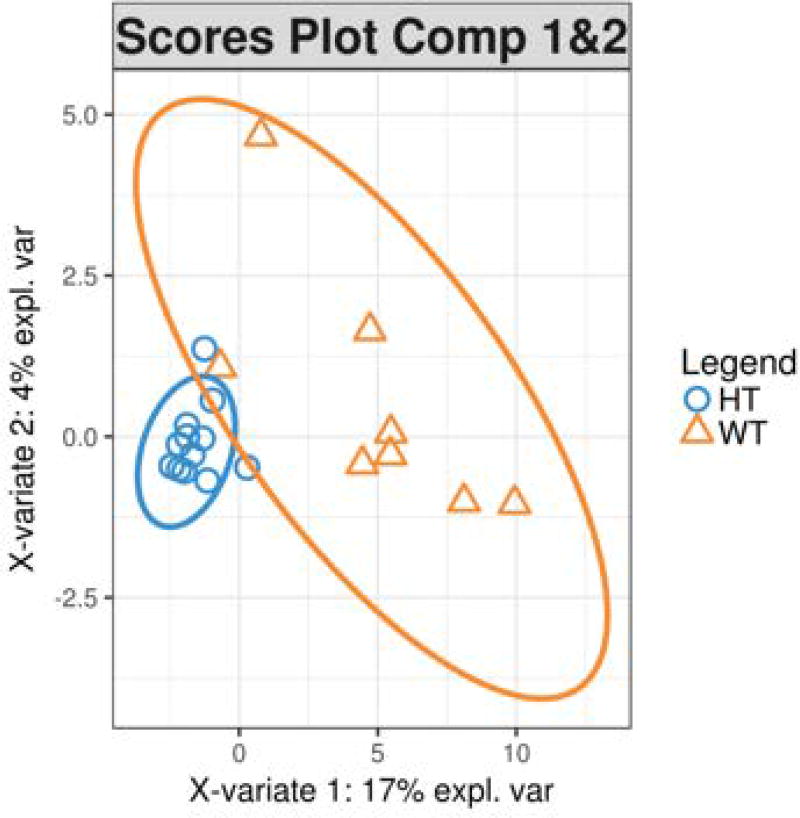
Figure 5.
Scores plot. The scores plot depicts the samples, colored according to class (genotype), projected into the space defined by the first two sPLS-DA model components. Axis labels show the level of variance explained (expl. var) by each component. sPLS-DA, sparse partial least squares discriminant analysis.
Figure 6.
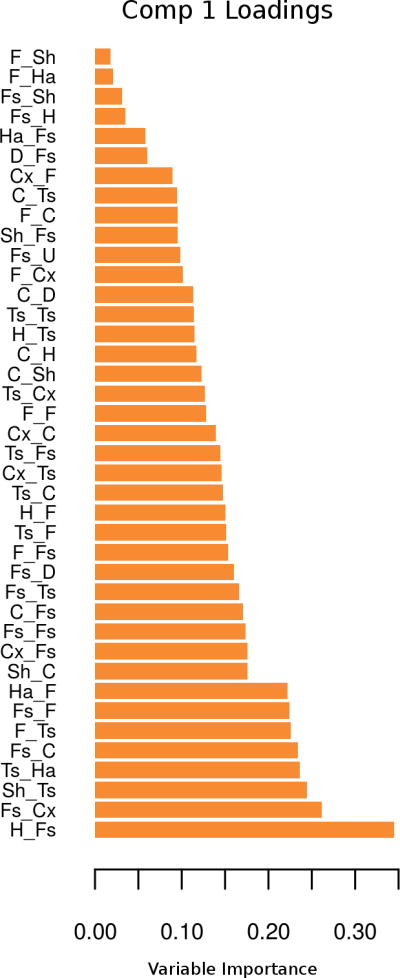
Loading plot. The loading plot shows the strength of the association for each of the selected variables on a particular component (Comp) with the class labels (genotypes).
COMMENTARY
Background Information
There are several other published software packages to analyze mouse vocal call types (Burkett et al., 2015; Chabout et al., 2016; Van Segbroeck et al., 2017). We recommend that interested investigators compare results obtained from this and other methods to evaluate relative power to detect structural differences between mutant and wild-type littermates. The optimal analytical software might depend on the type of gene mutations and the level of noise and false positive and false negative cases.
Critical Parameters and Troubleshooting
Sex
Both sexes should be included in analysis.
Age
Pup vocal calls generally emerge at P4, peak at P8 and decline at P12 (Michetti et al., 2012). These time points should be analyzed because male and female mice reach developmental milestones at different ages (Bronson, Dagg, & Snell, 2007), and the age at which calls peak might vary in mutant mice (Lai et al., 2014; Michetti et al., 2012; Scattoni, Crawley, & Ricceri, 2009; Sungur, Schwarting, & Wohr, 2016). Recording of pups at P4 might be too stressful for some mutant mouse lines and result in high mortality afterwards. If this is the case, such a recording time point should be avoided.
Genetic background
Any genetic mouse model can be used for this analysis, but one technical aspect requires close attention (N Hiroi, 2018). Inbred mouse lines widely differ in ultrasonic vocalization (Faure et al., 2017; Scattoni et al., 2008; Scattoni, Ricceri, & Crawley, 2011). Mouse models are maintained using various breeding methods. In many cases, mutations are generated using embryonic stem (ES) cells of one inbred mouse (e.g., 129sv). A developed mouse is then crossed with a mouse line with high fecundity (e.g., C57BL/6J). Offspring carry the genetic backgrounds of alleles of ES cells and alleles of a breeder strain. Chromosomal regions linked to the target gene are expected to contain more genomic materials from ES cells in mutant mice. In contrast, t chromosomal regions linked to the gene are expected to contain more genomic materials of a breeder line in wild-type littermates. The systematic bias in genetic backgrounds between mutant and wild-type littermates poses an interpretation difficulty for behavioral phenotypes (Crusio, 2004; Flaherty & Bolivar, 2007; Gerlai, 2001; N Hiroi, 2018; Kelly et al., 1998; Wolfer, Crusio, & Lipp, 2002). The best strategies to circumvent this interpretative obstacle are to use congenic mice, co-isogenic mice or F1 inbred hybrid mouse lines. In these mouse lines, the genetic background is identical between mutant and wild-type littermates (i.e., co-isogenic or F1 hybrid) or the genetic background is maximally saturated with the breeder’s genetic background (i.e., congenic) in both mutant and wild-type littermates. The phenotypic difference between non-congenic wild-type and mutant littermates cannot be unambiguously ascribed to the gene mutation in question. Data derived from non-congenic mice should not be viewed as valid evidence to conclude the impact of gene mutation on ultrasonic vocalization or any behavioral or neuronal phenotype(N Hiroi, 2018).
Breeder
To control for the impact of male breeders on dams, male partners should be removed from the breeder cage immediately after a vaginal plug is noted. One critical parameter is the sex of a mutant breeder. Male and female heterozygous mice need to be paired as breeders if homozygous mice are to be examined. If an over-expressing transgenic mouse or hemizygous mouse is to be examined, breeders are a mouse with one mutant copy and a wild-type mouse. The issue is whether the mutant breeder should be a mother or a father. The maternal behaviors of mutant and wild-type mothers might differ, depending on the gene mutated. Ideally, this too should be examined, but if it is beyond the aim of the project, we recommended that a wild-type mother be used to minimize the impact of atypical maternal behaviors of mutant mothers.
Housing
Many phenotypic differences are noted in an experiment in which mice are housed in a litter with the same genotype or a different genotype (Kalbassi, Bachmann, Cross, Roberton, & Baudouin, 2017; Yang, Lewis, Foley, & Crawley, 2015). The manner in which pups are maintained in a litter is an important determinant for many ASD-related phenotypes. This should be analyzed as a factor or a co-variant. Variations in maternal care among strains and those induced by maternal separation/deprivation, cross-fostering, and communal nesting affect the behavioral profiles of offspring and the epigenetic profiles of many genes involved in the hypothalamic pituitary adrenal (HPA) axis (Cierpial, Murphy, & McCarty, 1990; Curley et al., 2010; Kember et al., 2012; Kundakovic & Champagne, 2015; Kundakovic, Lim, Gudsnuk, & Champagne, 2013; Ressler, 1963, 1966; Umemura, Imai, Mimura, Fujiwara, & Ebihara, 2015; Weller et al., 2003). Thus, we do not recommend culling a litter to the same size and genotype composition, because a reduction of the litter size and introduction of a pup from another litter might impact maternal behaviors toward pups. However, the litter size should be recorded as a co-variant.
Changing of cage bedding should be avoided once recording starts at P4; also avoid a brief separation of the dam and pups other than the time of testing (Own & Patel, 2013; Tsuda & Ogawa, 2012).
Number of recordings
Neonatal maternal separation and brief handling of pups influences a wide range of ASD-related behaviors in mice(Kikusui, Isaka, & Mori, 2005; Macri & Laviola, 2004; Tsuda & Ogawa, 2012; Tsuda, Yamaguchi, & Ogawa, 2011; Venerosi, Cirulli, Capone, & Alleva, 2003), although their effects depend on the strain, behavior, and experimental parameter(s) (Cirulli, Berry, & Alleva, 2003; Cirulli et al., 2009; Millstein & Holmes, 2007). Thus, recording at more than three postnatal ages in the same pup is not recommended. Recording a second session on the same day is not recommended for two reasons. First, we noticed the number of calls is drastically reduced for a second session. Second, the mortality rate of pups increases when they are repeatedly tested on the same day.
Anticipated Results and Interpretations
Basic Protocols 1 and 2 are preparatory steps. Data are obtained in Basic Protocols 3–5.
Basic Protocol 3. The outcome of this protocol shows the non-random nature of call selection for sequences of two, three, and four calls and the difference in the degree of non-random call sequences between the genotypes. The zero-order level (H[0]) reflects how many call types each genotype uses. The lower the entropy scores at H[0], the narrower the call type repertoire. H[1] indicates how variably the call types are used within their call repertoires; the lower the score, the more skewed the use of specific call types is within the repertoire. H[2] to H[4] levels indicate the degree of non-random sequencing of various call types within two, three, and four successive calls; the lower the score, the more selective the call types in forming call sequences.
Basic Protocol 4. The outcome of this protocol shows the relative frequencies of call connections and directions (see Fig. 4). Data are interpreted in terms of differences in the most frequently used call connections between groups.
Basic Protocol 5. The outcome of this protocol shows the extent to which individual pups differ in formation of call sequencing (see Fig. 5). The more scattered the data points are, the more variable call sequences are among individual pups.
Time Considerations
Basic Protocol 1: For each pup, we estimate the following times. Steps 1, 1.5 hrs; Steps 2–4, 7 min; Step 5, 3 min; Step 6, 2 min; Step 7, 5 min.
Basic Protocol 2: For each pup we estimate the following times. Steps 1–6, 1 min; Step 7, ~ 1–2 hrs per 100 calls; Steps 8–11, ~1 hr per 100 call data.
Basic Protocol 3: Steps 1 to 5, a few minutes to an hour depending on user.
Basic Protocol 4: Step 1, minutes to hours, depending on computer speed and number of toolboxes to be installed with MatLab. Step 2– 3, a few minutes. Step 4, 5 minutes to run through all following steps. Step 5–11, a few minutes, depending on computer speed
Basic Protocol 5: Step 1 to 3, minutes to hours to tune the classification model; highly dependent on tuning parameters and data size; step 4 to 5, varies, as above.
Supplementary Material
Significance Statement.
Although the number of neonatal ultrasonic vocal calls and different vocal call types and their sequences have been used as indexes of neonatal social communication in mice, our analytical protocol provides a computational means to determine the call sequence of various call types, the degree of non-randomness of various call types used in sequences, and individual variability of call sequences that have been demonstrated to functionally affect maternal behaviors.
Acknowledgments
Research reported in this publication was supported in part by the National Institutes of Health under Award Numbers R01MH099660, R01DC015776, R21HD053114 and U54HD090260 to NH and R01DC007690 to JLP. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest Statement
The authors declare no conflict of interest.
LITERATURE CITED
- Branchi I, Santucci D, Vitale A, Alleva E. Ultrasonic vocalizations by infant laboratory mice: a preliminary spectrographic characterization under different conditions. Dev. Psychobiol. 1998;33(3):249–256. doi: 10.1002/(sici)1098-2302(199811)33:3<249::aid-dev5>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Bronson FH, Dagg CP, Snell GD. Reproduction. In: Green EL, editor. Biology of the Laboratory Mouse. Second. New York: Dover Publications, Inc.; 2007. Oneline publication). (Reprinted from: Not in File) [Google Scholar]
- Burkett ZD, Day NF, Penagarikano O, Geschwind DH, White SA. VoICE: A semi-automated pipeline for standardizing vocal analysis across models. Sci. Rep. 2015;5:10237. doi: 10.1038/srep10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabout J, Sarkar A, Patel SR, Radden T, Dunson DB, Fisher SE, Jarvis ED. A Foxp2 Mutation Implicated in Human Speech Deficits Alters Sequencing of Ultrasonic Vocalizations in Adult Male Mice. Front Behav Neurosci. 2016;10:197. doi: 10.3389/fnbeh.2016.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cierpial MA, Murphy CA, McCarty R. Maternal behavior of spontaneously hypertensive and Wistar-Kyoto normotensive rats: effects of reciprocal cross-fostering of litters. Behav. Neural Biol. 1990;54(1):90–96. doi: 10.1016/0163-1047(90)91271-c. [DOI] [PubMed] [Google Scholar]
- Cirulli F, Berry A, Alleva E. Early disruption of the mother-infant relationship: effects on brain plasticity and implications for psychopathology. Neurosci. Biobehav. Rev. 2003;27(1–2):73–82. doi: 10.1016/s0149-7634(03)00010-1. [DOI] [PubMed] [Google Scholar]
- Cirulli F, Francia N, Berry A, Aloe L, Alleva E, Suomi SJ. Early life stress as a risk factor for mental health: role of neurotrophins from rodents to non-human primates. Neurosci. Biobehav. Rev. 2009;33(4):573–585. doi: 10.1016/j.neubiorev.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino JN, Kennon-McGill S, Weichselbaum C, Marrus N, Haider A, Glowinski AL, Jones W. Infant viewing of social scenes is under genetic control and is atypical in autism. Nature. 2017;547(7663):340–344. doi: 10.1038/nature22999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crusio WE. Flanking gene and genetic background problems in genetically manipulated mice. Biol. Psychiatry. 2004;56(6):381–385. doi: 10.1016/j.biopsych.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Curley JP, Rock V, Moynihan AM, Bateson P, Keverne EB, Champagne FA. Developmental shifts in the behavioral phenotypes of inbred mice: the role of postnatal and juvenile social experiences. Behav. Genet. 2010;40(2):220–232. doi: 10.1007/s10519-010-9334-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Hiroi N, Scattoni ML. Cry, baby, cry: Expression of Distress as a Biomarker and Modulator in Autism Spectrum Disorder. Int J Neuropsychopharmacol. 2017 doi: 10.1093/ijnp/pyx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Venuti P. How is crying perceived in children with Autistic Spectrum Disorder. Research in Autism Spectrum Disorders. 2008;2:371–384. [Google Scholar]
- Esposito G, Venuti P. Comparative analysis of crying in children with autism, developmental delays, and typical development. Focus on Autism and Other Developmental Disabilities. 2009;24(4):240–247. [Google Scholar]
- Ey E, Torquet N, Le Sourd AM, Leblond CS, Boeckers TM, Faure P, Bourgeron T. The Autism ProSAP1/Shank2 mouse model displays quantitative and structural abnormalities in ultrasonic vocalisations. Behav. Brain Res. 2013;256:677–689. doi: 10.1016/j.bbr.2013.08.031. [DOI] [PubMed] [Google Scholar]
- Faure A, Pittaras E, Nosjean A, Chabout J, Cressant A, Granon S. Social behaviors and acoustic vocalizations in different strains of mice. Behav Brain Res. 2017;320:383–390. doi: 10.1016/j.bbr.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Flaherty L, Bolivar V. Congenic and consomic strains. In: Jones BC, Mormede P, editors. Neurobehavioral Genetics. 2. New York: Taylor & Francis; 2007. pp. 115–127. (Reprinted from: In File) [Google Scholar]
- Fraley ER, Burkett ZD, Day NF, Schwartz BA, Phelps PE, White SA. Mice with Dab1 or Vldlr insufficiency exhibit abnormal neonatal vocalization patterns. Sci. Rep. 2016;6:25807. doi: 10.1038/srep25807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlai R. Gene targeting: technical confounds and potential solutions in behavioral brain research. Behav. Brain Res. 2001;125(1–2):13–21. doi: 10.1016/s0166-4328(01)00282-0. [DOI] [PubMed] [Google Scholar]
- Green J, Charman T, Pickles A, Wan MW, Elsabbagh M, Slonims V, Johnson MH. Parent-mediated intervention versus no intervention for infants at high risk of autism: a parallel, single-blind, randomised trial. Lancet Psychiatry. 2015;2(2):133–140. doi: 10.1016/S2215-0366(14)00091-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J, Pickles A, Pasco G, Bedford R, Wan MW, Elsabbagh M, British Autism Study of Infant Siblings, T Randomised trial of a parent-mediated intervention for infants at high risk for autism: longitudinal outcomes to age 3 years. J Child Psychol Psychiatry. 2017;58(12):1330–1340. doi: 10.1111/jcpp.12728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimsley JM, Monaghan JJ, Wenstrup JJ. Development of social vocalizations in mice. PLoS. One. 2011;6(3):e17460. doi: 10.1371/journal.pone.0017460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramoto T, Kang G, Suzuki G, Satoh Y, Kucherlapati R, Watanabe Y, Hiroi N. Tbx1: identification of a 22q11.2 gene as a risk factor for autism spectrum disorder in a mouse model. Hum. Mol. Genet. 2011;20(24):4775–4785. doi: 10.1093/hmg/ddr404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N. Critical Reappraisal of Mechanistic Links of Copy Number Variants to Dimensional Constructs of Neuropsychiatric Disorders in Mouse Models. Psychiatry and Clinical Neuroscience. 2018 Jan 25; doi: 10.1111/pcn.12641. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N, Hiramoto T, Harper KM, Suzuki G, Boku S. Mouse models of 22q11.2-associated autism spectrum disorder. Autism. 2012;S1(001):1–9. doi: 10.4172/2165-7890.S1-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroi N, Takahashi T, Hishimoto A, Izumi T, Boku S, Hiramoto T. Copy Number Variation at 22q11.2: from rare variants to common mechanisms of developmental neuropsychiatric disorders. Mol. Psychiatry. 2013;18:1153–1165. doi: 10.1038/mp.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W, Klin A. Attention to eyes is present but in decline in 2–6-month-old infants later diagnosed with autism. Nature. 2013;504(7480):427–431. doi: 10.1038/nature12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbassi S, Bachmann SO, Cross E, Roberton VH, Baudouin SJ. Male and Female Mice Lacking Neuroligin-3 Modify the Behavior of Their Wild-Type Littermates. eNeuro. 2017;4(4) doi: 10.1523/ENEURO.0145-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MA, Rubinstein M, Phillips TJ, Lessov CN, Burkhart-Kasch S, Zhang G, Low MJ. Locomotor activity in D2 dopamine receptor-deficient mice is determined by gene dosage, genetic background, and developmental adaptations. J Neurosci. 1998;18(9):3470–3479. doi: 10.1523/JNEUROSCI.18-09-03470.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kember RL, Dempster EL, Lee TH, Schalkwyk LC, Mill J, Fernandes C. Maternal separation is associated with strain-specific responses to stress and epigenetic alterations to Nr3c1, Avp, and Nr4a1 in mouse. Brain Behav. 2012;2(4):455–467. doi: 10.1002/brb3.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikusui T, Hiroi N. A Self-Generated Environmental Factor as a Potential Contributor to Atypical Early Social Communication in Autism. Neuropsychopharmacology. 2017;42(1):378. doi: 10.1038/npp.2016.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikusui T, Isaka Y, Mori Y. Early weaning deprives mouse pups of maternal care and decreases their maternal behavior in adulthood. Behav. Brain Res. 2005;162(2):200–206. doi: 10.1016/j.bbr.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Kundakovic M, Champagne FA. Early-life experience, epigenetics, and the developing brain. Neuropsychopharmacology. 2015;40(1):141–153. doi: 10.1038/npp.2014.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundakovic M, Lim S, Gudsnuk K, Champagne FA. Sex-specific and strain-dependent effects of early life adversity on behavioral and epigenetic outcomes. Front Psychiatry. 2013;4(78):1–13. doi: 10.3389/fpsyt.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JK, Sobala-Drozdowski M, Zhou L, Doering LC, Faure PA, Foster JA. Temporal and spectral differences in the ultrasonic vocalizations of fragile X knock out mice during postnatal development. Behav. Brain Res. 2014;259:119–130. doi: 10.1016/j.bbr.2013.10.049. [DOI] [PubMed] [Google Scholar]
- Le Cao KA, Boitard S, Besse P. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinformatics. 2011;12:253. doi: 10.1186/1471-2105-12-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macri S, Laviola G. Single episode of maternal deprivation and adult depressive profile in mice: interaction with cannabinoid exposure during adolescence. Behav. Brain Res. 2004;154(1):231–238. doi: 10.1016/j.bbr.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Michetti C, Ricceri L, Scattoni ML. Modeling social communication deficits in mouse models of autism. Autism Open Access. 2012;S1(007):1–8. [Google Scholar]
- Millstein RA, Holmes A. Effects of repeated maternal separation on anxiety- and depression-related phenotypes in different mouse strains. Neurosci. Biobehav. Rev. 2007;31(1):3–17. doi: 10.1016/j.neubiorev.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Nishi A, Hiroi N. Genetic Mechanisms Emerging from Mouse Models of CNV-Associated Neuropsychiatric Disorders. In: Abel T, Nickl-Jockschat T, editors. The Neurobiology of Schizophrenia. New York: Academic Press/Elsevier; 2016. pp. 397–417. (Reprinted from: Not in File) [Google Scholar]
- Okabe S, Nagasawa M, Kihara T, Kato M, Harada T, Koshida N, Kikusui T. Pup odor and ultrasonic vocalizations synergistically stimulate maternal attention in mice. Behav. Neurosci. 2013;127(3):432–438. doi: 10.1037/a0032395. [DOI] [PubMed] [Google Scholar]
- Own LS, Patel PD. Maternal behavior and offspring resiliency to maternal separation in C57Bl/6 mice. Horm. Behav. 2013;63(3):411–417. doi: 10.1016/j.yhbeh.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Ozonoff S, Iosif AM, Baguio F, Cook IC, Hill MM, Hutman T, Young GS. A prospective study of the emergence of early behavioral signs of autism. J. Am. Acad. Child Adolesc. Psychiatry. 2010;49(3):256–266. [PMC free article] [PubMed] [Google Scholar]
- Panksepp JB, Jochman KA, Kim JU, Koy JJ, Wilson ED, Chen Q, Lahvis GP. Affiliative behavior, ultrasonic communication and social reward are influenced by genetic variation in adolescent mice. PLoS. One. 2007;2(4):e351. doi: 10.1371/journal.pone.0000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portfors CV. Types and functions of ultrasonic vocalizations in laboratory rats and mice. J. Am. Assoc. Lab Anim Sci. 2007;46(1):28–34. [PubMed] [Google Scholar]
- Ressler RH. GENOTYPE-CORRELATED PARENTAL INFLUENCES IN TWO STRAINS OF MICE. J. Comp Physiol Psychol. 1963;56:882–886. doi: 10.1037/h0043542. [DOI] [PubMed] [Google Scholar]
- Ressler RH. Inherited environmental influences on the operant behavior of mice. J. Comp Physiol Psychol. 1966;61(2):264–267. doi: 10.1037/h0023140. [DOI] [PubMed] [Google Scholar]
- Rogers SJ, Vismara L, Wagner AL, McCormick C, Young G, Ozonoff S. Autism treatment in the first year of life: a pilot study of infant start, a parent-implemented intervention for symptomatic infants. J. Autism Dev. Disord. 2014;44(12):2981–2995. doi: 10.1007/s10803-014-2202-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, Crawley J, Ricceri L. Ultrasonic vocalizations: a tool for behavioural phenotyping of mouse models of neurodevelopmental disorders. Neurosci. Biobehav. Rev. 2009;33(4):508–515. doi: 10.1016/j.neubiorev.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, Gandhy SU, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in the BTBR T+tf/J mouse model of autism. PLoS. One. 2008;3(8):e3067. doi: 10.1371/journal.pone.0003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scattoni ML, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav. 2011;10(1):44–56. doi: 10.1111/j.1601-183X.2010.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat. Rev. Neurosci. 2010;11(7):490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis J. The signal functions of early infant crying. Behav. Brain Sci. 2004;27(4):443–458. [PubMed] [Google Scholar]
- Sugimoto H, Okabe S, Kato M, Koshida N, Shiroishi T, Mogi K, Koide T. A role for strain differences in waveforms of ultrasonic vocalizations during male-female interaction. PLoS. One. 2011;6(7):e22093. doi: 10.1371/journal.pone.0022093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungur AO, Schwarting RK, Wohr M. Early communication deficits in the Shank1 knockout mouse model for autism spectrum disorder: Developmental aspects and effects of social context. Autism Res. 2016;9(6):696–709. doi: 10.1002/aur.1564. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Okabe S, O'Broin P, Nishi A, Ye K, Beckert M, Hiroi N. Structure and function of neonatal social communication in a genetic mouse model of autism. Mol. Psychiatry. 2016;21(9):1208–1214. doi: 10.1038/mp.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda MC, Ogawa S. Long-lasting consequences of neonatal maternal separation on social behaviors in ovariectomized female mice. PLoS. One. 2012;7(3):e33028. doi: 10.1371/journal.pone.0033028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda MC, Yamaguchi N, Ogawa S. Early life stress disrupts peripubertal development of aggression in male mice. Neuroreport. 2011;22(6):259–263. doi: 10.1097/WNR.0b013e328344495a. [DOI] [PubMed] [Google Scholar]
- Umemura S, Imai S, Mimura A, Fujiwara M, Ebihara S. Impaired Maternal Behavior in Usp46 Mutant Mice: A Model for Trans-Generational Transmission of Maternal Care. PLoS. One. 2015;10(8):e0136016. doi: 10.1371/journal.pone.0136016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Segbroeck M, Knoll AT, Levitt P, Narayanan S. MUPET-Mouse Ultrasonic Profile ExTraction: A Signal Processing Tool for Rapid and Unsupervised Analysis of Ultrasonic Vocalizations. Neuron. 2017;94(3):465–485 e465. doi: 10.1016/j.neuron.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venerosi A, Cirulli F, Capone F, Alleva E. Prolonged perinatal AZT administration and early maternal separation: effects on social and emotional behaviour of periadolescent mice. Pharmacol. Biochem. Behav. 2003;74(3):671–681. doi: 10.1016/s0091-3057(02)01068-7. [DOI] [PubMed] [Google Scholar]
- Wallace KS, Rogers SJ. Intervening in infancy: implications for autism spectrum disorders. J. Child Psychol. Psychiatry. 2010;51(12):1300–1320. doi: 10.1111/j.1469-7610.2010.02308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller A, Leguisamo AC, Towns L, Ramboz S, Bagiella E, Hofer M, Brunner D. Maternal effects in infant and adult phenotypes of 5HT1A and 5HT1B receptor knockout mice. Dev. Psychobiol. 2003;42(2):194–205. doi: 10.1002/dev.10079. [DOI] [PubMed] [Google Scholar]
- Wetherby AM, Guthrie W, Woods J, Schatschneider C, Holland RD, Morgan L, Lord C. Parent-implemented social intervention for toddlers with autism: an RCT. Pediatrics. 2014;134(6):1084–1093. doi: 10.1542/peds.2014-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer DP, Crusio WE, Lipp HP. Knockout mice: simple solutions to the problems of genetic background and flanking genes. Trends Neurosci. 2002;25(7):336–340. doi: 10.1016/s0166-2236(02)02192-6. [DOI] [PubMed] [Google Scholar]
- Yang M, Lewis F, Foley G, Crawley JN. In Tribute to Bob Blanchard: Divergent Behavioral Phenotypes of 16p11.2 Deletion Mice Reared in Same-Genotype Versus Mixed-Genotype Cages. Physiol Behav. 2015 doi: 10.1016/j.physbeh.2015.04.023. [DOI] [PubMed] [Google Scholar]
- Yang M, Mahrt EJ, Lewis F, Foley G, Portmann T, Dolmetsch RE, Crawley JN. 16p11.2 Deletion Syndrome Mice Display Sensory and Ultrasonic Vocalization Deficits During Social Interactions. Autism Res. 2015 doi: 10.1002/aur.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DM, Schenk AK, Yang SB, Jan YN, Jan LY. Altered ultrasonic vocalizations in a tuberous sclerosis mouse model of autism. Proc. Natl. Acad. Sci. U. S. A. 2010;107(24):11074–11079. doi: 10.1073/pnas.1005620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeifman DM. An ethological analysis of human infant crying: answering Tinbergen's four questions. Dev. Psychobiol. 2001;39(4):265–285. doi: 10.1002/dev.1005. [DOI] [PubMed] [Google Scholar]
- Zeskind PS, McMurray MS, Garber KA, Neuspiel JM, Cox ET, Grewen KM, Johns JM. Development of translational methods in spectral analysis of human infant crying and rat pup ultrasonic vocalizations for early neurobehavioral assessment. Front Psychiatry. 2011;2:56. doi: 10.3389/fpsyt.2011.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaigenbaum L, Bryson S, Garon N. Early identification of autism spectrum disorders. Behav. Brain Res. 2013;251:133–146. doi: 10.1016/j.bbr.2013.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.