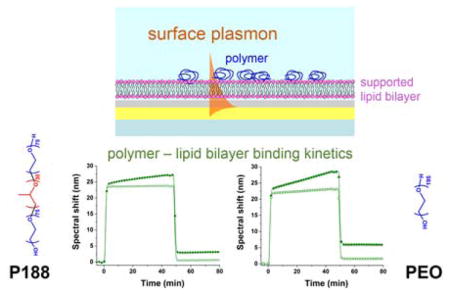
Abstract
Poloxamer 188 (P188), a poly(ethylene oxide)-b-poly(propylene oxide)-b-poly(ethylene oxide) triblock copolymer, protects cell membranes against various external stresses whereas polyethylene oxide (PEO; 8,600 g/mol) homopolymer lacks protection efficacy. As part of a comprehensive effort to elucidate the protection mechanism, we used surface plasmon resonance (SPR) to obtain direct evidence of binding of the polymers onto supported lipid bilayers. Binding kinetics and coverage of the P188 and PEO were examined and compared. Most notably, the PEO exhibited comparable membrane association to that of the P188, evidenced by comparable association rate constants and coverage. This result highlights the need for additional mechanistic understanding beyond simple membrane association to explain the differential efficacy of P188 in therapeutic applications.
Keywords: block copolymer, poly(ethylene oxide), poloxamer, phospholipid membrane, surface plasmon resonance, adsorption
Graphical Abstract
Introduction
Poloxamers are non-ionic, amphiphilic triblock copolymers consisting of two hydrophilic poly(ethylene oxide) (PEO) blocks flanking a central hydrophobic poly(propylene oxide) (PPO) chain. By varying the length of PEO and PPO, a broad spectrum of poloxamers are available1, allowing a wide range of biomedical applications including thermosensitive hydrogels2, drug delivery carriers3,4, and membrane permeation enhancers for chemotherapy5,6. Among these variants, Poloxamer 188 (P188) has gained great attention as a cell membrane stabilizer, due to its ability to protect cell integrity from various cell membrane stresses. P188 has an average molar mass of 8400 g/mol and contains PEO and PPO blocks with approximately 75 and 30 repeat units, respectively (often referred to as PEO75-PPO30-PEO75), which translates to 80 wt% of the more hydrophilic PEO. In vivo administration of P188 to dystrophic mice has been shown to improve recovery from acute cardiac failure7 and protect skeletal muscles from mechanical stress8. Also, P188 improved recovery from myocardial infarction9 and cardiac arrest in pigs10. At the cellular level, P188 significantly attenuated damage from mechanical stress11, high-dose irradiation12, ischemia/hypoxia reperfusion injury13,14 and traumatic brain injury15.
Despite the observed efficacy as a membrane protection agent, the underlying mechanism of the P188 interaction with the membrane and its protection is not yet clear. As an effort to explore the protective mechanism, the effect of copolymer architecture was investigated in a dystrophic mouse model16 and an in vitro cellular assay with model PEO-PPO diblock copolymers17. Increasing the hydrophobicity of the PPO block through incorporation of a polar tert-butyl end group significantly enhanced protection efficacy at fixed PPO and PEO length, implying the possible anchoring of the hydrophobic portion of the polymer to the membrane16. Besides the end group, the hydrophobicity can be modulated by PPO length, which has an optimal range to provide sufficient membrane protection efficacy. No membrane protection was achieved by polymers with PPO length under a threshold value, whereas an excessively long PPO block size led to micelle formation, which limited further enhancement of protection efficacy. Also, when a set of copolymers with fixed PPO length and end group were compared, membrane protection efficacy improved with increasing PEO length17. In contrast, 8600 g/mol PEO homopolymer, a molecular weight comparable to P188 but without a hydrophobic block, exhibited poor protection17. Overall, both PPO and PEO clearly play a role in membrane protection, but the mechanisms involved with the block copolymer-membrane association, including membrane interaction kinetics, remain to be fully elucidated.
Model lipid membranes have been used to investigate the polymer protection mechanism at the molecular level18. P188 prevented POPC/POPG/PLPC liposome degradation from peroxidation stress while a five-fold greater concentration of 8.6 kDa PEO relative to P188 was required to obtain a similar protective effect19. By measuring water hydration dynamics on the surface and intrabilayer of DOPC liposomes, P188 was found to weakly adsorb on the liposome surface, whereas approximately 100 times the concentration of 8.6 kDa PEO was needed to achieve a similar level of interaction20. As measured by a surface force apparatus, 8 kDa PEO did not adsorb to myelin21 or DMPC bilayers,22,23 whereas P18821 and 20 kDa PEO22,24 did bind. When 8 kDa PEO was injected for ~10 min, it did not adsorb to a DOPC bilayer as measured by a quartz crystal microbalance at concentrations of 0.1 – 20 wt%25. When a C18H37 tail was attached to a 5 kDa PEO, binding occured26. For 2.5 kDa PEO, the critical hydrophobic tail length required for binding to phosphatidylcholine supported bilayers and vesicles was determined to be C1227. Similarly, in surface plasmon resonance measurements with spherical vesicles adsorbed to a gold surface, 2.5 kDa PEO-C11 did not appreciably bind, whereas PEO-C12 bound slightly and PEO-C14 bound substantially. Molecular dynamics simulations suggest that the hydrophilic PEO blocks of P188 adsorb at the lipid-water interface while hydrophobic PPO penetrates into the lipid bilayer28. Although the previous studies provide some information, direct evidence as to whether P188 and PEO adhere to the lipid membrane is still lacking. One study used a microcantilever to directly observe P188 adhesion on a supported lipid bilayer29, but PEO homopolymer was not tested.
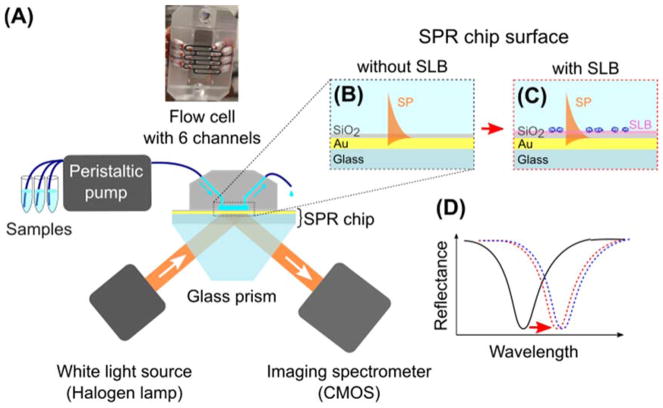
A suitable method to obtain direct evidence of polymer adhesion on a lipid bilayer is surface plasmon resonance (SPR) spectroscopy. As a highly surface sensitive, real-time, and label-free method30,31, SPR has been used to investigate molecular interactions on surfaces including protein-protein32, antimicrobial peptide-lipid bilayer33,34, protein-polymer35–37, and polymer-substrate combinations38–41. In the commonly used prism-coupled (also known as Kretschmann) configuration, the surface plasmon (SP) is excited on an outer surface of a thin metal film by an evanescent wave produced by total internal reflection of p-polarized light on the inner metal-glass interface30 (Fig. 1). At the resonance, the propagation constant of the evanescent wave is matched with that of the SP, resulting in maximum energy transfer between the incident light and the SP. In our case (fixed angle of incidence and polychromatic incident light), this is manifested as a distinct dip in the wavelength spectrum of a reflected wave (Fig. 1D). As the propagation constant of the SP and thus resonance wavelength are dependent on the refractive index of the medium probed by the SP field, real-time monitoring of changes in the bulk medium or molecular transport near the metal surface, including the binding kinetics, is possible.
Figure 1.
(A) Schematic representation of the SPR experiment; (B,C) Pairs of flow channels for the same polymer concentration were used: one with a bare SiO2 surface (B) and the other supporting a lipid bilayer (C); (D) Resonance wavelength shift upon polymer binding (black - bare SiO2 surface, red - with a lipid bilayer, and blue - with bilayer and adhered copolymer).
In this work we used SPR to directly detect polymer binding on a supported lipid bilayer (SLB), which is a simple model system widely used to mimic a cell plasma membrane42. We employed a single-component glycerophospholipid SLB to measure P188 and 8.6 kDa PEO (hereafter referred to as merely PEO) binding kinetics and extent of binding. This direct measurement serves as a valuable technique to elucidate the protection mechanism. Most notably, we find that the PEO homopolymer exhibits membrane association comparable to that exhibited by triblock P188, which highlights the need to consider other mechanisms of interaction beyond simple hydrophilic-hydrophobic interactions to explain the differential efficacy in physiological applications.
Materials and methods
Materials
Poloxamer 188 (Pluronic F68; P188) was generously provided by BASF (Wyandotte, MI). PEO homopolymer with molecular weight 8600 g/mol (PEO) was purchased from EMD Millipore (Billerica, MA). Dispersities of P188 and PEO were 1.07 and 1.06, respectively, based on size exclusion chromatography using tetrahydrofuran as eluent. Salts for buffer solution (sodium chloride, potassium chloride, calcium chloride, magnesium chloride and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES); all bioreagent grade), bovine serum albumin (BSA; purity > 98%), and chloroform (purity > 99%) were purchased from Sigma. 16:0–18:1 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) were purchased from Avanti Polar Lipids (Alabaster, AL). The chemical structures of P188, PEO, and POPC are shown in Figure 2.
Figure 2.
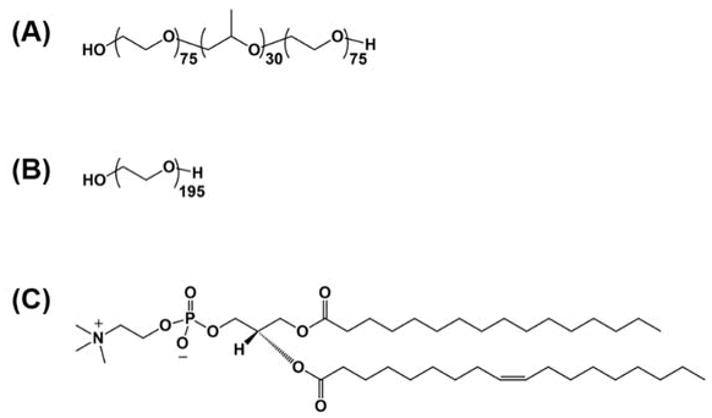
Chemical structure of (A) P188, (B) PEO, and (C) POPC lipid.
SPR chip preparation
Polished BK7 glass substrates (Schott, Malaysia) were coated with 1.5 nm of titanium adhesion layer and 50 nm gold layer using thermal evaporation in vacuum. Then, 10 ± 1 nm of silica film was grown on top of the gold film using atomic layer deposition43 (Savannah ALD system, Cambridge NanoTech, USA) from ozone and tris(dimethylamino)silane precursors at 180 °C. Prior to the SPR experiment, SiO2-coated chips were treated with O2 plasma (100 W) for 2 min or with UV-ozone (UVO-Cleaner 42; Jelight, USA) for 30 min to clean and oxidize the silica surface for reproducible formation of the SLB. The cleaned SPR chips were stored in a vacuum before use. For control SPR experiments, gold-coated SPR chips without top SiO2 layer were used. The gold surface of these chips was also cleaned by UV-ozone treatment for 30 min prior each measurement.
Liposome and polymer solution preparation
Unilamellar vesicles were prepared by the extrusion method. Lipids dissolved in chloroform were first placed under a mild flow of argon to allow evaporation of the chloroform, followed by drying in a desiccator for at least 1 h to remove any residual solvent. The resulting dry lipid film was then hydrated in physiological buffer (140 mM NaCl, 5 mM KCl, 2.5 mM CaCl2, 2 mM MgCl2, and 10 mM HEPES) by stirring overnight at room temperature. The hydrated solution was extruded 49 times at room temperature using an Avanti Mini-Extruder fitted with a 100 nm polycarbonate filter. The unilamellar vesicles were diluted to 1 mg/mL. P188 and PEO were dissolved in the physiological buffer to 0.15, 1.5, and 4.5 mM concentrations and filtered before making the SPR measurements to remove any possible aggregates in the solution. BSA was also dissolved in the buffer at a concentration of 1 mg/mL.
SPR measurements
We employed a spectroscopic SPR sensor based on the Kretschmann configuration of attenuated total reflection developed at the Institute of Photonics and Electronics (Prague, Czech Republic). The sensor was capable of reading the SPR signal from six independent channels addressed separately by the microfluidic system (Fig. 1A). The temperature of the sensing head was kept constant at 25°C or 37°C with ±0.01 °C precision throughout the measurement. The angle of incidence was adjusted to observe the resonance minimum of reflectance at 780 nm when water was in contact with the SiO2-coated chip. Under these conditions, the sensor sensitivity, S, is approximately 5980 nm/refractive index unit (RIU)44. The instrument was turned on at least 30 min before the measurement to stabilize the temperature of the sensor head and the lamp. After installing the cleaned SPR chips to the sensor, degassed distilled water followed by physiological buffer were injected onto the chip surface until a stable baseline was obtained (i.e., spectral shift changes less than 0.5 nm for 15 min). The lipid bilayer was formed by sequentially injecting 1 mg/mL POPC unilamellar vesicles, physiological buffer, water and buffer. After successful lipid bilayer formation, P188 or PEO solution was injected for 45 min and washed out with buffer for 30 min. For certain chips, 1 mg/mL BSA solution was injected for 10 min after lipid bilayer formation to examine the surface coverage. No polymer was injected when the BSA was used. 20 μL/min flow rate was maintained throughout the measurement, except for the injection of POPC vesicles, where the flow rate was 5 μL/min. After each measurement, all fluidic channels were cleaned with 0.1M sodium hydroxide, 0.5% Triton X-100, and distilled water. 5–7 independent measurements were performed with freshly prepared SPR chips.
SPR data analysis
As the polymer molecules are soluble in the buffer and are much smaller than the wavelength of the SP, the change of the effective refractive index of the solution of polymer molecules in buffer can be expressed using effective medium approximation. The Maxwell-Garnett approximation for the effective dielectric constant of the medium εm containing a volume fraction fpolymer of polymer with permittivity εm is45:
| (1) |
In terms of refractive index, this can be in our case (npolymer = 1.465, nbuffer ≈ 1.33 at λ = 750nm) further simplified as Δneff = neff = nbuffer + (npolymer − nbuffer)fpolymer. The validity of this approximation is confirmed by the linear dependence depicted in Figure 4B. npolymer = 1.465 for both P188 and PEO (assumed to be the same as that of Pluronic 123 and Pluronic F127; npolymer was reported by the manufacturer at 589 nm at 20 °C). In the case where no polymer molecules are bound to the sensor surface, the spectral SPR shift R is linearly proportional to the concentration of the polymer and thus neff is given by the following equation:
| (2) |
where S is the sensor sensitivity to bulk refractive index changes. nbuffer was derived from the relationship R = S(nbuffer − nwater) 46, where R is the spectral shift upon water-buffer solution exchange and nwater = 1.33.
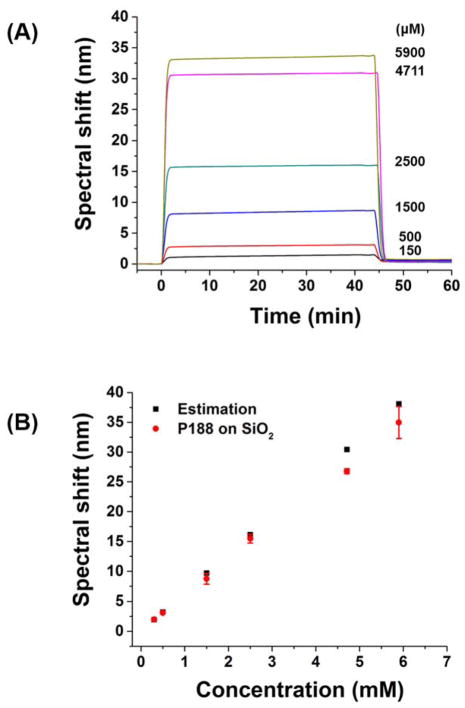
Figure 4.
(A) Spectral shift upon injection of P188 at various concentrations over a bare SiO2 surface. (B) Comparison of spectral shift at 45 min in (A) (red) and estimated spectral shift based on bulk refractive index of P188 solution (black). The measurements were performed at 25°C.
The kinetic curves were fitted using MATLAB according to the Langmuir one-to-one model47 assuming no dissociation (this assumption is discussed in Results and Discussion.):
| (3) |
where Rmax is the maximum binding signal, ka is the association rate constant, and [A]0 is the polymer concentration.
The surface coverage Γ (number of polymers per unit area) was calculated using the following equation40:
| (4) |
where dpolymer is the effective thickness of the polymer and dn/dc of polymer is the differential refractive index increment. dpolymer was determined from the SPR response using npolymer = 1.465 and surface sensitivity (change of resonance wavelength induced by unit change of refractive index in layer with thickness of 1 nm on top of the SLB) calculated by rigorous coupled wave analysis (RCWA)48. dn/dc was measured with a Wyatt differential refractometer (OptiLab rEX) and found to be dn/dc = 0.135 and 0.128 mL/g for P188 and PEO in water, respectively, at 25 °C.
Results and Discussion
Prior to the study of polymer adhesion, a supported lipid bilayer was formed on an SiO2 surface via vesicle rupture. We used POPC, one of the most abundant lipids in mammalian cell membranes, to form a model lipid membrane49. Key processes of the bilayer formation are vesicle adhesion, rupture, and spreading through hydrophobic fusion of bilayer edges42. Vesicle saturation on the surface is often required for successful bilayer formation42. Once the vesicle ruptures, the remaining processes occur rapidly, presumably because the hydrophobic bilayer edge catalyzes cascading liposome rupture and bilayer spreading50,51. Vesicle rupture occurs on a limited set of hydrophilic substrates such as mica52, glass53, or SiO254, which provide enough vesicle-substrate interaction to increase membrane stress near the rim of the contact area55. In contrast, vesicles adhere intact on a bare gold surface56. Hence, we used the bare gold surface as the control and compared the SPR signal induced by the POPC vesicles on top of SiO2-coated vs. bare gold surface. When POPC liposomes were injected onto the gold surface, the surface was rapidly saturated and remained unchanged upon subsequent solution exchanges, indicating that liposomes were strongly adhered without rupturing or detaching (Fig. 3A). Note that the spectral shift change upon solution exchange between buffer and water (on bare gold: 8.9 ± 0.1 nm; on liposome: 11.3 ± 0.2 nm), is due to the change in bulk refractive index, not to changes in the vesicles. In contrast to gold, the SiO2 surface induced noticeable changes throughout the measurement (Fig. 3B). Spectral shifts quickly increased for the first 15 min and gradually rose to 80 nm during liposome injection. Upon buffer wash, the spectral shift slowly decreased to 70 nm caused by the removal of loosely bound liposomes. Solution exchange from buffer to water induced a drop-off in the SPR signal. As the inside of a vesicle is filled with buffer, exchanging the outside environment to water exposes the vesicle to hypo-osmotic stress, which aids vesicle rupture. The system was re-equilibrated with buffer, which causes the change of bulk refractive index while the SLB formed by ruptured vesicles remain adhered to the surface. Experiments at 37 °C yielded similar results (Fig. S1), and the final spectral shift was 18.4 ± 2.8 nm at 37 °C.
Figure 3.
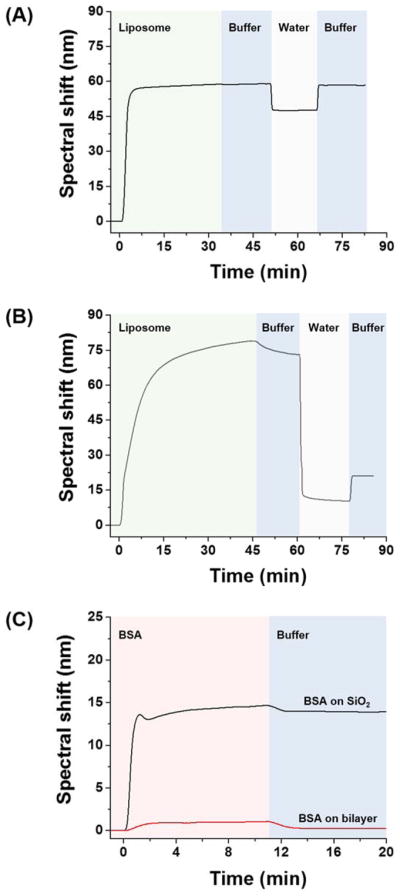
Spectral shift upon liposome injection on (A) gold or (B) SiO2 surface. (C) Spectral shift upon bovine serum albumin injection on lipid bilayer or bare SiO2 surface. Change of solution is indicated by background color change. All measurements were conducted at 25°C.
To verify that a single lipid bilayer was formed, we calculated the associated spectral shift with the RCWA model48 using a refractive index of 1.4557 for the 4 nm thick lipid layer. The estimated spectral shift (17.3 nm) was close to the experimentally observed value (18.4 ± 2.8 nm), thereby supporting that a single lipid bilayer was formed on the SiO2 surface. The 1.1 nm difference between the estimated vs. observed spectral shift translates to a discrepancy of 0.007 RIU (ca. 0.5% error). We also assessed the surface coverage of the lipid bilayer utilizing the fact that BSA does not adhere on this surface58 but does adhere to bare SiO259. At pH 7, the SiO2 surface is negatively charged60 and allows the BSA to bind via electrostatic interaction59. However, a zwitterionic surface such as POPC lipid resists BSA adhesion61. As shown in Figure 3C, when the BSA was injected after bilayer formation, little BSA adhered on the surface after buffer wash (spectral shift: 0.35 ± 0.16 nm), indicating that lipid bilayer covered essentially the entire SiO2 surface. On the other hand, significant BSA binding (spectral shift: 13.9 ± 0.1 nm) was observed on the bare SiO2 surface, in line with previous observations59. In this way we confirmed that a single lipid bilayer with nearly complete coverage was formed.
Because SPR spectroscopy probes subtle variations in refractive index near the surface, polymer concentration-dependent bulk refractive index changes should be taken into account to enable the accurate measurement of binding kinetics. We used a bare SiO2 surface to measure bulk refractive index changes at different P188 concentrations (Fig. 4A). When P188 was injected over a bare SiO2 surface, the spectral shift increased abruptly, within 3 min, and remained constant during subsequent polymer injection. After a buffer wash, the spectral shift dropped to almost zero (0.48 ± 0.19 nm) regardless of the polymer concentration, indicating that the P188 does not adhere to the bare SiO2 surface. The same behavior was observed with PEO. We confirmed that the abrupt spectral shift arises from the bulk refractive index change based on the calculated spectral shift associated with each polymer concentration as described by Eq.2 (Fig. 4B). As mentioned previously, the SiO2 surface is negatively charged (i.e., deprotonated) at pH 7. PEO was reported to adhere onto a protonated SiO2 surface, mediated by hydrogen bonding62,63. Thus, we conclude that P188 and PEO do not adhere onto the SiO2 surface at pH 7 due to the lack of hydrogen bonding sites on the SiO2 surface. These results imply that we can use the SiO2 surface as a reference substrate. We note here that the P188 does not form any micelles over the concentration range used in this study, as confirmed by small-angle neutron scattering and dynamic light scattering measurements (Figs. S2 and S3).
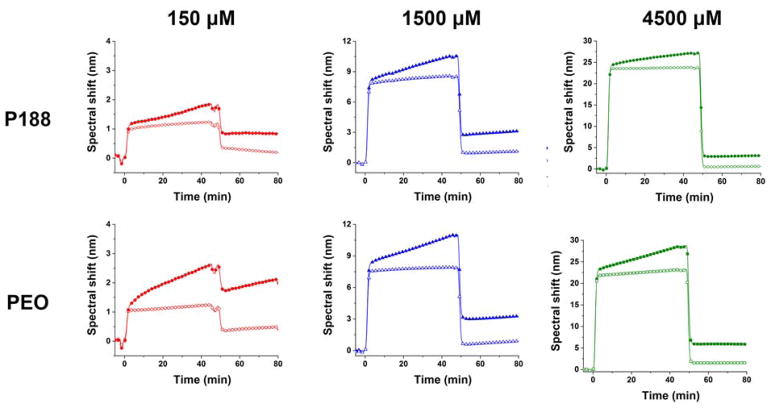
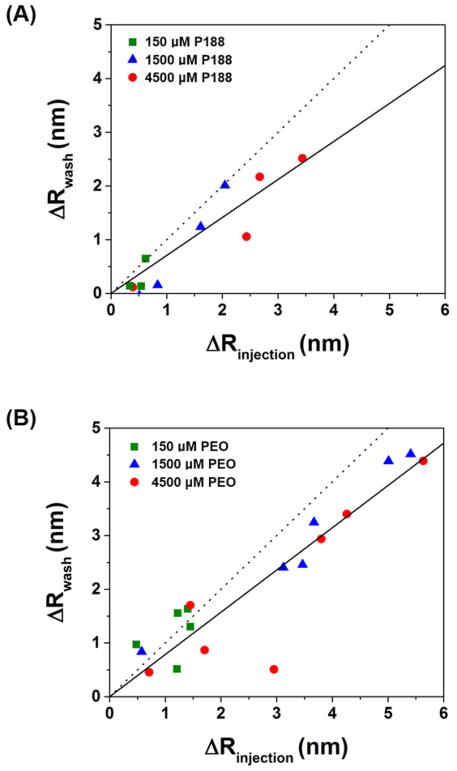
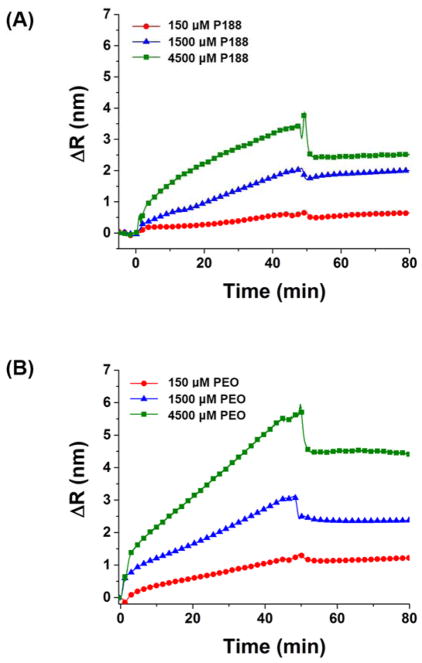
We explored the binding behavior of P188 and PEO by injecting the polymers over the bilayer and monitoring spectral shift changes (Fig. 5). At all test concentrations, the spectral shift with a lipid bilayer was higher than that with the bare SiO2 surface. In other words, there is significant binding of P188 and PEO on the lipid bilayer. Remarkably, the PEO showed comparable adsorption (spectral shift: 2.0 ± 1.6 nm at 4.5 mM after wash) to P188 (spectral shift: 1.5 ± 1.1 nm at 4.5 mM after wash). This finding conflicts with the common assumption that PEO does not bind to lipid bilayers. When the polymers were injected for 45 min and washed with buffer solution for 30 min, an abrupt decrease due to the bulk refractive index change was followed by a spectral shift that remained constant at a value higher than that observed from the reference surface. We further examined the adsorbed amount of polymer before (ΔRinjection) and after (ΔRwash) washing with buffer solution, where ΔR is the difference between the spectral shift from the lipid bilayer and SiO2 surface (Fig. 6). ΔRwash increases linearly with ΔRinjection with a slope near unity, implying that the adsorbed polymers mostly remain after washing. Although the slope is smaller than one, we suspect that this reflects a bulk refractive index mismatch between the SPR channels, which is an experimental bias that is difficult to avoid. The refractive index mismatch is also evident in the plot of ΔR vs. time (Fig. 7). The sudden signal decrease at 45 min nearly matches in magnitude the abrupt increase right after the injection, indicating that the discontinuity likely derives from an insufficient bulk refractive index correction. Thus, we further corrected the data by shifting down the 0–45 min data by the amount of the residual mismatch to obtain the association rate constant.
Figure 5.
Spectral shift upon P188 and PEO adhesion on lipid bilayer at concentrations of 150, 1500, and 4500 μM. Filled symbols indicate polymers on lipid bilayer and open symbols indicate polymers on bare SiO2 surface. The measurements were done at 37°C.
Figure 6.
Adsorbed amount of (A) P188 and (B) PEO before (ΔRinjection) and after (ΔRwash) washing with buffer solution. ΔRinjection and ΔRwash was taken as the spectral shift at 45 min and 95 min in Figure 5, respectively. Solid lines are linear fits with intercept set to zero. Slope of the dotted line is 1.
Figure 7.
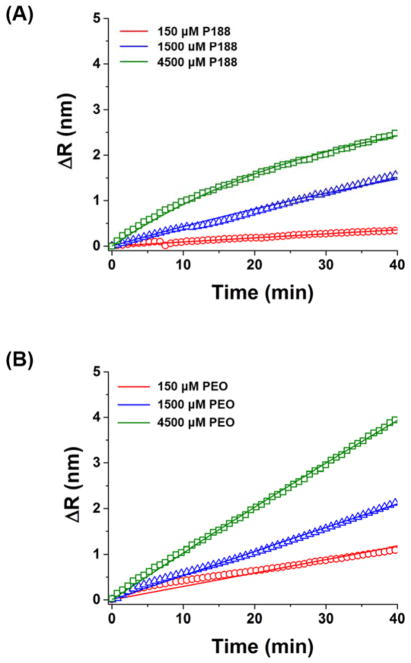
Binding kinetic curves of (A) P188 and (B) PEO. ΔR is the spectral shift after bulk refractive index correction.
The corrected binding kinetic curves were fit with a simple one-to-one binding model (lipid + polymer ⇋ lipid-polymer complex)44 (Fig. 8). The fitting quality was not dramatically improved with a transport-limited model or a model with two bound states (Fig. S4). Since desorption of P188 or PEO was negligible during washing (maximum change of 0.1 nm over during 25 min), the dissociation rate constant was assumed to be zero and only association was considered in the initial binding phase. The association rate constant (ka) of P188 was 0.15 ± 0.12 M−1s−1 and that of PEO was 0.18 ± 0.18 M−1s−1. These ka values for P188 and PEO on the POPC bilayer are much smaller than typical ka values for specific interaction between an antibody and an antigen: ka ~ 105 – 106 M−1s−1 64.
Figure 8.
Binding kinetic curves (symbols) fit to the Langmuir one-to-one binding model (solid curves): (A) P188, (B) PEO.
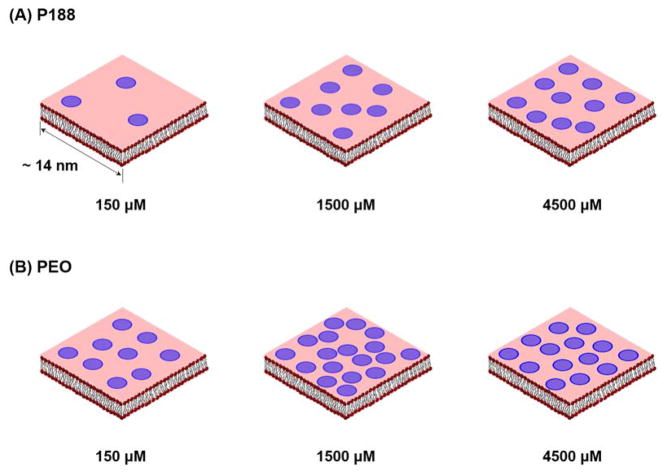
In addition to the binding kinetics, we calculated the coverage of polymers on the lipid bilayer according to Equation 4 based on the values of ΔRwash. The coverage is presented in Table 1 in two ways – the number of polymers per unit area and the number of lipids per polymer molecule – and illustrated schematically in Figure 9. Note that the difference between the surface coverage of P188 and PEO is not statistically significant based on multiple independent experiments (Fig. S5).
Table 1.
Surface coverage of P188 and PEO on lipid bilayer.
| Concentration (μM) | Effective thickness (nm) | Number of polymers per unit area (nm−2) | Number of lipids per polymer | |
|---|---|---|---|---|
| 150 | P188 | 0.08 ± 0.07 | 0.005(4) | 261 |
| PEO | 0.30 ± 0.11 | 0.02(2) | 65 | |
| 1500 | P188 | 0.28 ± 0.23 | 0.02(0) | 71 |
| PEO | 0.73 ± 0.34 | 0.05(4) | 26 | |
| 4500 | P188 | 0.36 ± 0.27 | 0.02(6) | 55 |
| PEO | 0.50 ± 0.38 | 0.03(7) | 38 |
Figure 9.
Schematic representation of adhered amount of (A) P188 and (B) PEO on lipid bilayer. A blue circle denotes single polymer chain with diameter scaled to the radius of gyration of P188 and PEO (ca. 3 nm for both polymers) assuming random coil conformations. The number of the circles is proportional to the number of lipid molecules per polymer listed in Table 1.
Taken together, both P188 and PEO adsorb slowly but extensively on the POPC lipid bilayer. Evidence that PEO binds on the lipid bilayer implies that initial contact of P188 would be mediated by the PEO blocks, which form 80 wt% of this triblock copolymer. Individual P188 molecules will assume a Gaussian coil conformation, with the hydrophilic PEO being exposed to the aqueous environment. Thus, it is reasonable to assume that as a P188 molecule approaches the bilayer the PEO blocks first make contact with lipid head groups. In other words, the P188 and PEO both interact with the lipid bilayer via initial PEO contact, which would explain the comparable binding. The interaction of the PEO blocks in Poloxamers has been reported in several molecular dynamics simulations28,65. After initial contact, the PEO blocks likely remain in close proximity to the lipid head groups positioning the overall P188 molecule for subsequent insertion of the hydrophobic PPO block into the lipid tail region66, as suggested by previous studies using Poloxamers with long PPO chains67. In the case of PEO, which lack hydrophobic repeat units, the molecule most likely remains in a hydrated state near the lipid head groups on the surface of the membrane. In both cases, the binding strength of P188 and PEO was strong enough to resist desorption from the lipid bilayer under a 20 μL/min flow of buffer solution.
PEO may bind to the membrane surface via interaction of ether oxygen atoms with positively charged quaternary ammoniums in the lipid heads68. The oxygen in PEO is known to carry a slight negative charge, which enables the molecule to trap positive metal ions such as lithium, sodium, and potassium69. Also, the oxygen can interact with positively charged lysine, stabilizing the helix structure in PEO-protein conjugates,70 suggesting a similar mode of interaction may be operative with the lipid bilayer. More work, possibly including atomistic molecular simulations, would be needed to support this hypothesis. A recent study by atomistic molecular dynamics simulation suggests that the PEO may weakly adsorb onto lipid membrane via hydrogen bonding of the terminal hydroxyl groups of the PEO to the membrane.71
Another factor governing adsorption is molecular weight. While PEO 8600 g/mol adhered to the lipid bilayer, PEO 2100 g/mol did not exhibit any appreciable binding signal at 4500 μM (Fig. S6); the SPR spectral shift from PEO 2100 g/mol on the lipid bilayer was almost the same as that from PEO 2100 g/mol on the bare SiO2 surface. This result indicates that the binding of PEO 2100 g/mol to the lipid bilayer is minimal, considering that PEO 8600 g/mol exhibited ca. 1 nm of binding signal even at 150 μM (ca. 14% mass concentration of the PEO 2100 g/mol at 4500 μM). The result is in line with previous studies using a surface force apparatus21–24 that reported PEO interaction with lipid bilayer when the PEO molecular weight was higher than 20 kg/mol, although the threshold molecular weight for interaction was higher than what we observed here, i.e., between 2100 and 8600 g/mol (this discrepancy is discussed later in this section). The molecular weight dependence could be explained by configurational entropy: the higher molecular weight PEO suffers less entropic loss than its lower molecular weight counterpart when adhered to the lipid bilayer24.
Excellent surface sensitivity of the SPR method enabled the study of binding of P188 and PEO. To the best of our knowledge, this is the first experimental evidence that PEO binds to a supported lipid bilayer membrane at a comparable level to that of P188. Previous studies reported little or no binding of PEO to bilayer when no alkyl tail was present in the PEO molecule. Adhesion of P188 and PEO on POPC liposomes was negligible from a pulse-field gradient NMR (PFG-NMR) study, when the polymer:lipid ratio was 0.02:172. Based on the SPR results here, the kas of the P188 and PEO obtained from SPR are small. Therefore, a higher polymer to lipid ratio would be needed to observe detectable binding. In this SPR study, the estimated polymer to lipid molar ratio is 6:1 at 150 μM polymer concentration. From equation (3), when the polymer:lipid ratio is 0.02:1, it requires ca. 10 days to obtain comparable SPR signal to the one obtained in the case of 6:1 ratio. Although there would be possibilities of a curvature effect or different sensitivity between the SPR and PFG-NMR, we expect that it would take a long time (i.e., order of several days) to observe detectable binding with PFG-NMR experiments with a low polymer:lipid ratio.
In the case of supported lipid bilayers, a microcantilever was used to probe the P188 binding to a POPC membrane29. The microcantilever signal reached a plateau within ~3 min, whereas the SPR signal in current study did not plateau within the timescale of the experiment (~45 min). Thus, the observed binding kinetics in the microcantilever study were significantly faster than the current case. One difference between the microcantilever and SPR instruments is that the microcantilever surface can bend upon molecule binding on the surface, which might provide more space between the lipid on the surface, resulting in more polymer binding on the surface. PEO did not adhere on the lipid membrane when probed by QCM25 and surface force apparatus21–24. Polymer was injected for 10 min in the QCM experiment in contrast to our SPR experiment, in which the polymer injection was for 45 min. Insufficient polymer injection might result in a weak binding signal. In the surface force apparatus, the two lipid bilayers approach to each other when polymer is sandwiched between the lipid bilayers. Because the apparatus measures force between two approaching surfaces, there is a chance that the bound polymer might be detached and not detected by the force measurement, which might have caused the discrepancy in PEO molecular weight threshold for binding to the lipid bilayer.
Although both P188 and PEO bind to the SLB, only P188 exhibits cell membrane protection; PEO showed poor protection efficacy at the same concentration in an in vitro cellular assay17. This suggests that the differential protection efficacy cannot be solely explained by the extent of polymer binding, implying that the additional insertion of the PPO block plays a key and enabling role. The SLB used in this study is a single component phosphatidylcholine lipid, whereas the cell membrane consists of many types of lipids including phosphatidylserine and phosphatidylinositol, as well as non-lipid components such as cholesterol and proteins73. Another difference between the SLB and cell membrane is the lateral diffusion of the lipid. The lateral diffusion of the SLB (~ 1.8 μm2/sec) is around 6 times slower than that of a free-standing lipid31,74 or lipid in a cell membrane, due to the interaction with the substrate. Moreover, membrane curvature is present in the cellular studies but absent in the current planar SPR assays. In addition, penetration depth differences between P188 and PEO might lead to different biological response or membrane tension/fluidity changes to cope with external membrane injuries. The factors noted here could collectively lead to the differential protection efficacy of the two polymers. In this study, we found that molecular adsorption alone is most likely not the essential mechanism of cell membrane protection.
Conclusion
We have demonstrated direct evidence of P188 and PEO adsorption to supported lipid bilayers using SPR spectroscopy. The superior sensitivity of SPR enabled the quantitative detection of polymer binding to the lipid membrane. Remarkably, PEO showed comparable binding as P188, contrary to a common belief and multiple alternative experimental results that the PEO homopolymer barely interacts with lipid membranes. This study necessitates explanation for differential membrane protection efficacy of P188 and PEO other than molecular adsorption alone.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01 HL122323). to F.S.B.. M.V., C.T.E., and S.-H.O. acknowledge support from the Minnesota Partnership for Biotechnology and Medical Genomics. SPR chips were fabricated in the Minnesota Nano Center (MNC) cleanroom, which receives partial support from the National Science Foundation (NSF) through the National Nanotechnology Coordinated Infrastructure (NNCI).
Footnotes
Supplementary materials and methods, supported bilayer formation dynamics, small-angle neutron scattering of P188, dynamic light scattering of P188 and PEO, analysis of multiple binding models, surface coverage analysis, and surface plasmon resonance dynamics of PEO 2100 g/mol
References
- 1.Alexandridis P, Alan Hatton T. Poly(ethylene Oxide)-Poly(propylene Oxide)-Poly(ethylene Oxide) Block Copolymer Surfactants in Aqueous Solutions and at Interfaces: Thermodynamics, Structure, Dynamics, and Modeling. Colloids Surfaces A Physicochem Eng Asp. 1995;96:1–46. [Google Scholar]
- 2.Yang Z, Nie S, Hsiao WW, Pam W. Thermoreversible Pluronic F127-Based Hydrogel Containing Liposomes for the Controlled Delivery of Paclitaxel: In Vitro Drug Release, Cell Cytotoxicity, and Uptake Studies. Int J Nanomedicine. 2011;6:151. doi: 10.2147/IJN.S15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tagami T, Kubota M, Ozeki T. Effective Remote Loading of Doxorubicin into DPPC/Poloxamer 188 Hybrid Liposome to Retain Thermosensitive Property and the Assessment of Carrier-Based Acute Cytotoxicity for Pulmonary Administration. J Pharm Sci. 2015;104:3824–3832. doi: 10.1002/jps.24593. [DOI] [PubMed] [Google Scholar]
- 4.Kabanov AV, Batrakova EV, Alakhov VY. Pluronic® Block Copolymers for Overcoming Drug Resistance in Cancer. Adv Drug Deliv Rev. 2002;54:759–779. doi: 10.1016/s0169-409x(02)00047-9. [DOI] [PubMed] [Google Scholar]
- 5.Kabanov AV, Batrakova EV, Miller DW. Pluronic® Block Copolymers as Modulators of Drug Efflux Transporter Activity in the Blood-Brain Barrier. Adv Drug Deliv Rev. 2003;55:151–164. doi: 10.1016/s0169-409x(02)00176-x. [DOI] [PubMed] [Google Scholar]
- 6.Alakhova DY, Kabanov AV. Pluronics and MDR Reversal: An Update. Mol Pharm. 2014;11:2566–2578. doi: 10.1021/mp500298q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yasuda S, Townsend D, Michele DE, Favre EG, Day SM, Metzger JM. Dystrophic Heart Failure Blocked by Membrane Sealant Poloxamer. Nature. 2005;436:1025–1029. doi: 10.1038/nature03844. [DOI] [PubMed] [Google Scholar]
- 8.Houang EM, Haman KJ, Filareto A, Perlingeiro RC, Bates FS, Lowe DA, Metzger JM. Membrane-Stabilizing Copolymers Confer Marked Protection to Dystrophic Skeletal Muscle in Vivo. Mol Ther - Methods Clin Dev. 2015;2:15042. doi: 10.1038/mtm.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartos JA, Matsuura TR, Sarraf M, Youngquist ST, McKnite SH, Rees JN, Sloper DT, Bates FS, Segal N, Debaty G, Lurie KG, Neumar RW, Metzger JM, Riess ML, Yannopoulos D. Bundled Postconditioning Therapies Improve Hemodynamics and Neurologic Recovery after 17min of Untreated Cardiac Arrest. Resuscitation. 2015;87:7–13. doi: 10.1016/j.resuscitation.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartos JA, Matsuura TR, Tsangaris A, Olson M, McKnite SH, Rees JN, Haman K, Shekar KC, Riess ML, Bates FS, Metzger JM, Yannopoulos D. Intracoronary Poloxamer 188 Prevents Reperfusion Injury in a Porcine Model of ST-Segment Elevation Myocardial Infarction. JACC Basic to Transl Sci. 2016;1:224–234. doi: 10.1016/j.jacbts.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee RC, River LP, Pan FS, Ji L, Wollmann RL. Surfactant-Induced Sealing of Electropermeabilized Skeletal Muscle Membranes in Vivo. Proc Natl Acad Sci. 1992;89:4524–4528. doi: 10.1073/pnas.89.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenebaum B, Blossfield K, Hannig J, Carrillo CS, Beckett MA, Weichselbaum RR, Lee RC. Poloxamer 188 Prevents Acute Necrosis of Adult Skeletal Muscle Cells Following High-Dose Irradiation. Burns. 2004;30:539–547. doi: 10.1016/j.burns.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Hunter RL, Luo AZ, Zhang R, Kozar RA, Moore FA. Poloxamer 188 Inhibition of Ischemia/reperfusion Injury: Evidence for a Novel Anti-Adhesive Mechanism. Ann Clin Lab Sci. 2010;40:115–125. [PubMed] [Google Scholar]
- 14.Gu JH, Ge JB, Li M, Xu HD, Wu F, Qin ZH. Poloxamer 188 Protects Neurons against Ischemia/Reperfusion Injury through Preserving Integrity of Cell Membranes and Blood Brain Barrier. PLoS One. 2013;8:e61641. doi: 10.1371/journal.pone.0061641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo CL, Chen XP, Li LL, Li QQ, Li BX, Xue AM, Xu HF, Dai DK, Shen YW, Tao LY, Zhao ZQ. Poloxamer 188 Attenuates in Vitro Traumatic Brain Injury-Induced Mitochondrial and Lysosomal Membrane Permeabilization Damage in Cultured Primary Neurons. J Neurotrauma. 2013;30:597–607. doi: 10.1089/neu.2012.2425. [DOI] [PubMed] [Google Scholar]
- 16.Houang EM, Haman KJ, Kim M, Zhang W, Lowe DA, Sham YY, Lodge TP, Hackel BJ, Bates FS, Metzger JM. Chemical End Group Modified Diblock Copolymers Elucidate Anchor and Chain Mechanism of Membrane Stabilization. Mol Pharm. 2017;14:2333–2339. doi: 10.1021/acs.molpharmaceut.7b00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim M, Haman KJ, Houang EM, Zhang W, Yannopoulos D, Metzger JM, Bates FS, Hackel BJ. PEO–PPO Diblock Copolymers Protect Myoblasts from Hypo-Osmotic Stress In Vitro Dependent on Copolymer Size, Composition, and Architecture. Biomacromolecules. 2017;18:2090–2101. doi: 10.1021/acs.biomac.7b00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siontorou C, Nikoleli G, Nikolelis D, Karapetis S. Artificial Lipid Membranes: Past, Present, and Future. Membranes (Basel) 2017;7:38. doi: 10.3390/membranes7030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang JYY, Marks J, Lee KYC. Nature of Interactions between PEO-PPO-PEO Triblock Copolymers and Lipid Membranes: (I) Effect of Polymer Hydrophobicity on Its Ability to Protect Liposomes from Peroxidation. Biomacromolecules. 2012;13:2616–2623. doi: 10.1021/bm300847x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng CYY, Wang JYY, Kausik R, Lee KYC, Han S. Nature of Interactions between PEO-PPO-PEO Triblock Copolymers and Lipid Membranes: (II) Role of Hydration Dynamics Revealed by Dynamic Nuclear Polarization. Biomacromolecules. 2012;13:2624–2633. doi: 10.1021/bm300848c. [DOI] [PubMed] [Google Scholar]
- 21.Banquy X, Lee DW, Kristiansen K, Gebbie MA, Israelachvili JN. Interaction Forces between Supported Lipid Bilayers in the Presence of PEGylated Polymers. Biomacromolecules. 2016;17:88–97. doi: 10.1021/acs.biomac.5b01216. [DOI] [PubMed] [Google Scholar]
- 22.Kuhl T, Guo Y, Alderfer JL, Berman AD, Leckband D, Israelachvili J, Hui SW. Direct Measurement of Polyethylene Glycol Induced Depletion Attraction between Lipid Bilayers. Langmuir. 1996;12:3003–3014. [Google Scholar]
- 23.Kuhl TL, Berman AD, Hui SW, Israelachvili JN. Part 1. Direct Measurement of Depletion Attraction and Thin Film Viscosity between Lipid Bilayers in Aqueous Polyethylene Glycol Solutions. Macromolecules. 1998;31:8250–8257. [Google Scholar]
- 24.Kuhl TL, Berman AD, Hui SW, Israelachvili JN. Part 2. Crossover from Depletion Attraction to Adsorption: Polyethylene Glycol Induced Electrostatic Repulsion between Lipid Bilayers. Macromolecules. 1998;31:8258–8263. [Google Scholar]
- 25.Zhao Z, Ji X, Dimova R, Lipowsky R, Liu Y. Viscoelasticity of Poly(ethylene Glycol) Solutions on Supported Lipid Bilayers via Quartz Crystal Microbalance with Dissipation. Macromolecules. 2015;48:1824–1831. [Google Scholar]
- 26.Liu G, Fu L, Zhang G. Role of Hydrophobic Interactions in the Adsorption of Poly(ethylene Glycol) Chains on Phospholipid Membranes Investigated with a Quartz Crystal Microbalance. J Phys Chem B. 2009;113:3365–3369. doi: 10.1021/jp810304f. [DOI] [PubMed] [Google Scholar]
- 27.Zhao F, Cheng X, Liu G, Zhang G. Interaction of Hydrophobically End-Capped Poly(ethylene Glycol) with Phospholipid Vesicles: The Hydrocarbon End-Chain Length Dependence. J Phys Chem B. 2010;114:1271–1276. doi: 10.1021/jp910024n. [DOI] [PubMed] [Google Scholar]
- 28.Goliaei A, Lau EY, Adhikari U, Schwegler E, Berkowitz ML. Behavior of P85 and P188 Poloxamer Molecules: Computer Simulations Using United-Atom Force-Field. J Phys Chem B. 2016;120:8631–8641. doi: 10.1021/acs.jpcb.6b03030. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Segatori L, Biswal SL. Probing the Association of Triblock Copolymers with Supported Lipid Membranes Using Microcantilevers. Soft Matter. 2014;10:6417–6424. doi: 10.1039/c4sm00928b. [DOI] [PubMed] [Google Scholar]
- 30.Homola J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem Rev. 2008;108:462–493. doi: 10.1021/cr068107d. [DOI] [PubMed] [Google Scholar]
- 31.Wittenberg NJ, Im H, Xu X, Wootla B, Watzlawik J, Warrington AE, Rodriguez M, Oh SH. High-Affinity Binding of Remyelinating Natural Autoantibodies to Myelin-Mimicking Lipid Bilayers Revealed by Nanohole Surface Plasmon Resonance. Anal Chem. 2012;84:6031–6039. doi: 10.1021/ac300819a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, Umana P, Benz J. Unique Carbohydrate-Carbohydrate Interactions Are Required for High Affinity Binding between Fc RIII and Antibodies Lacking Core Fucose. Proc Natl Acad Sci. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mozsolits H, Wirth HJ, Werkmeister J, Aguilar MI. Analysis of Antimicrobial Peptide Interactions with Hybrid Bilayer Membrane Systems Using Surface Plasmon Resonance. Biochim Biophys Acta - Biomembr. 2001;1512:64–76. doi: 10.1016/s0005-2736(01)00303-0. [DOI] [PubMed] [Google Scholar]
- 34.Papo N, Shai Y. Exploring Peptide Membrane Interaction Using Surface Plasmon Resonance: Differentiation between Pore Formation versus Membrane Disruption by Lytic Peptides. Biochemistry. 2003;42:458–466. doi: 10.1021/bi0267846. [DOI] [PubMed] [Google Scholar]
- 35.Wei Q, Becherer T, Angioletti-Uberti S, Dzubiella J, Wischke C, Neffe AT, Lendlein A, Ballauff M, Haag R. Protein Interactions with Polymer Coatings and Biomaterials. Angew Chemie Int Ed. 2014;53:8004–8031. doi: 10.1002/anie.201400546. [DOI] [PubMed] [Google Scholar]
- 36.Holmlin RE, Chen X, Chapman RG, Takayama S, Whitesides GM. Zwitterionic SAMs That Resist Nonspecific Adsorption of Protein from Aqueous Buffer. Langmuir. 2001;17:2841–2850. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 37.Chang Y, Liao SC, Higuchi A, Ruaan RC, Chu CW, Chen WY. A Highly Stable Nonbiofouling Surface with Well-Packed Grafted Zwitterionic Polysulfobetaine for Plasma Protein Repulsion. Langmuir. 2008;24:5453–5458. doi: 10.1021/la800228c. [DOI] [PubMed] [Google Scholar]
- 38.Green RJ, Tasker S, Davies J, Davies MC, Roberts CJ, Tendler SJB. Adsorption of PEO–PPO–PEO Triblock Copolymers at the Solid/Liquid Interface: A Surface Plasmon Resonance Study. Langmuir. 1997;13:6510–6515. [Google Scholar]
- 39.Brandani P, Stroeve P. Kinetics and Equilibria of Adsorption of PEO-PPO-PEO Triblock Copolymers on a Hydrophilic Self-Assembled Monolayer on Gold. Macromolecules. 2004;37:6640–6643. [Google Scholar]
- 40.Brandani P, Stroeve P. Kinetics of Adsorption and Desorption of PEO–PPO–PEO Triblock Copolymers on a Self-Assembled Hydrophobic Surface. Macromolecules. 2003;36:9502–9509. [Google Scholar]
- 41.Liu X, Wu D, Turgman-Cohen S, Genzer J, Theyson TW, Rojas OJ. Adsorption of a Nonionic Symmetric Triblock Copolymer on Surfaces with Different Hydrophobicity. Langmuir. 2010;26:9565–9574. doi: 10.1021/la100156a. [DOI] [PubMed] [Google Scholar]
- 42.Hardy GJ, Nayak R, Zauscher S. Model Cell Membranes: Techniques to Form Complex Biomimetic Supported Lipid Bilayers via Vesicle Fusion. Curr Opin Colloid Interface Sci. 2013;18:448–458. doi: 10.1016/j.cocis.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Im H, Wittenberg NJ, Lesuffleur A, Lindquist NC, Oh SH. Membrane Protein Biosensing with Plasmonic Nanopore Arrays and Pore-Spanning Lipid Membranes. Chem Sci. 2010;1:688. doi: 10.1039/C0SC00365D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Homola J, editor. Springer Series on Chemical Sensors and Biosensors. Vol. 4 Springer Berlin Heidelberg; Berlin, Heidelberg: 2006. Surface Plasmon Resonance Based Sensors. [Google Scholar]
- 45.Liu T, Pang Y, Zhu M, Kobayashi S. Microporous Co@CoO Nanoparticles with Superior Microwave Absorption Properties. Nanoscale. 2014;6:2447. doi: 10.1039/c3nr05238a. [DOI] [PubMed] [Google Scholar]
- 46.Jung LS, Campbell CT, Chinowsky TM, Mar MN, Yee SS. Quantitative Interpretation of the Response of Surface Plasmon Resonance Sensors to Adsorbed Films. Langmuir. 1998;14:5636–5648. [Google Scholar]
- 47.Hu J, Ma L, Wang S, Yang J, Chang K, Hu X, Sun X, Chen R, Jiang M, Zhu J, Zhao Y. Biomolecular Interaction Analysis Using an Optical Surface Plasmon Resonance Biosensor: The Marquardt Algorithm vs Newton Iteration Algorithm. PLoS One. 2015;10:e0132098. doi: 10.1371/journal.pone.0132098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwiecien P, Richter I. Frontiers in Optics 2011/Laser Science. XXVII. OSA; Washington, D.C: 2011. Modeling of Plasmonic Nanostructures Using Efficient Three Dimensional Aperiodic Rigorous Coupled Wave Analysis; p. JWA41. [Google Scholar]
- 49.Harayama T, Riezman H. Understanding the Diversity of Membrane Lipid Composition. Nat Rev Mol Cell Biol. 2018;19:281–296. doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 50.Andrecka J, Spillane KM, Ortega-Arroyo J, Kukura P. Direct Observation and Control of Supported Lipid Bilayer Formation with Interferometric Scattering Microscopy. ACS Nano. 2013;7:10662–10670. doi: 10.1021/nn403367c. [DOI] [PubMed] [Google Scholar]
- 51.Weirich KL, Israelachvili JN, Fygenson DK. Bilayer Edges Catalyze Supported Lipid Bilayer Formation. Biophys J. 2010;98:85–92. doi: 10.1016/j.bpj.2009.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richter RP, Brisson AR. Following the Formation of Supported Lipid Bilayers on Mica: A Study Combining AFM, QCM-D, and Ellipsometry. Biophys J. 2005;88:3422–3433. doi: 10.1529/biophysj.104.053728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schönherr H, Johnson JM, Lenz P, Frank CW, Boxer SG. Vesicle Adsorption and Lipid Bilayer Formation on Glass Studied by Atomic Force Microscopy. Langmuir. 2004;20:11600–11606. doi: 10.1021/la049302v. [DOI] [PubMed] [Google Scholar]
- 54.Reimhult E, Zäch M, Höök F, Kasemo B. A Multitechnique Study of Liposome Adsorption on Au and Lipid Bilayer Formation on SiO2. Langmuir. 2006;22:3313–3319. doi: 10.1021/la0519554. [DOI] [PubMed] [Google Scholar]
- 55.Dacic M, Jackman JA, Yorulmaz S, Zhdanov VP, Kasemo B, Cho NJ. Influence of Divalent Cations on Deformation and Rupture of Adsorbed Lipid Vesicles. Langmuir. 2016;32:6486–6495. doi: 10.1021/acs.langmuir.6b00439. [DOI] [PubMed] [Google Scholar]
- 56.Keller CA, Kasemo B. Surface Specific Kinetics of Lipid Vesicle Adsorption Measured with a Quartz Crystal Microbalance. Biophys J. 1998;75:1397–1402. doi: 10.1016/S0006-3495(98)74057-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salamon Z, Devanathan S, Alves ID, Tollin G. Plasmon-Waveguide Resonance Studies of Lateral Segregation of Lipids and Proteins into Microdomains (Rafts) in Solid-Supported Bilayers. J Biol Chem. 2005;280:11175–11184. doi: 10.1074/jbc.M411197200. [DOI] [PubMed] [Google Scholar]
- 58.Glasmästar K, Larsson C, Höök F, Kasemo B. Protein Adsorption on Supported Phospholipid Bilayers. J Colloid Interface Sci. 2002;246:40–47. doi: 10.1006/jcis.2001.8060. [DOI] [PubMed] [Google Scholar]
- 59.Kubiak-Ossowska K, Tokarczyk K, Jachimska B, Mulheran PA. Bovine Serum Albumin Adsorption at a Silica Surface Explored by Simulation and Experiment. J Phys Chem B. 2017;121:3975–3986. doi: 10.1021/acs.jpcb.7b01637. [DOI] [PubMed] [Google Scholar]
- 60.Kosmulski M. The pH-Dependent Surface Charging and Points of Zero Charge. J Colloid Interface Sci. 2011;353:1–15. doi: 10.1016/j.jcis.2010.08.023. [DOI] [PubMed] [Google Scholar]
- 61.Schlenoff JB. Zwitteration: Coating Surfaces with Zwitterionic Functionality to Reduce Nonspecific Adsorption. Langmuir. 2014;30:9625–9636. doi: 10.1021/la500057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gage RA, Currie EPK, Cohen Stuart MA. Adsorption of Nanocolloidal SiO2 Particles on PEO Brushes. Macromolecules. 2001;34:5078–5080. [Google Scholar]
- 63.Sarkar B, Venugopal V, Tsianou M, Alexandridis P. Adsorption of Pluronic Block Copolymers on Silica Nanoparticles. Colloids Surfaces A Physicochem Eng Asp. 2013;422:155–164. [Google Scholar]
- 64.Schreiber G, Haran G, Zhou HX. Fundamental Aspects of Protein–Protein Association Kinetics. Chem Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hezaveh S, Samanta S, De Nicola A, Milano G, Roccatano D. Understanding the Interaction of Block Copolymers with DMPC Lipid Bilayer Using Coarse-Grained Molecular Dynamics Simulations. J Phys Chem B. 2012;116:14333–14345. doi: 10.1021/jp306565e. [DOI] [PubMed] [Google Scholar]
- 66.Houang EM, Bates FS, Sham YY, Metzger JM. All-Atom Molecular Dynamics-Based Analysis of Membrane-Stabilizing Copolymer Interactions with Lipid Bilayers Probed under Constant Surface Tensions. J Phys Chem B. 2017:10657–10664. doi: 10.1021/acs.jpcb.7b08938. [DOI] [PubMed] [Google Scholar]
- 67.Firestone MA, Wolf AC, Seifert S. Small-Angle X-Ray Scattering Study of the Interaction of Poly(ethylene Oxide)-B-Poly(propylene Oxide)-B-Poly(ethylene Oxide) Triblock Copolymers with Lipid Bilayers. Biomacromolecules. 2003;4:1539–1549. doi: 10.1021/bm034134r. [DOI] [PubMed] [Google Scholar]
- 68.Bortel E, Kochanowski A. No Title. Die Makromol Chemie. 1984;185:1409–1417. [Google Scholar]
- 69.Zhang Z, Ohl M, Diallo SO, Jalarvo NH, Hong K, Han Y, Smith GS, Do C. Dynamics of Water Associated with Lithium Ions Distributed in Polyethylene Oxide. Phys Rev Lett. 2015;115:198301. doi: 10.1103/PhysRevLett.115.198301. [DOI] [PubMed] [Google Scholar]
- 70.Jain A, Ashbaugh HS. Helix Stabilization of Poly(ethylene glycol)–Peptide Conjugates. Biomacromolecules. 2011;12:2729–2734. doi: 10.1021/bm2005017. [DOI] [PubMed] [Google Scholar]
- 71.Liu Y, Agudo-Canalejo J, Grafmüller A, Dimova R, Lipowsky R. Patterns of Flexible Nanotubes Formed by Liquid-Ordered and Liquid-Disordered Membranes. ACS Nano. 2016;10:463–474. doi: 10.1021/acsnano.5b05377. [DOI] [PubMed] [Google Scholar]
- 72.Zhang W, Haman KJ, Metzger JM, Hackel BJ, Bates FS, Lodge TP. Quantifying Binding of Ethylene Oxide-Propylene Oxide Block Copolymers with Lipid Bilayers. Langmuir. 2017;33:12624–12634. doi: 10.1021/acs.langmuir.7b02279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Meer G, Voelker DR, Feigenson GW. Membrane Lipids: Where They Are and How They Behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duro N, Gjika M, Siddiqui A, Scott HL, Varma S. POPC Bilayers Supported on Nanoporous Substrates: Specific Effects of Silica-Type Surface Hydroxylation and Charge Density. Langmuir. 2016;32:6766–6774. doi: 10.1021/acs.langmuir.6b01155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.