Abstract
It is understood that adversity during development has the power to alter behavioral trajectories, and the role of the epigenome in that relationship is currently under intense investigation. Several studies in both non-human animals and humans have established a link between early adversity and epigenetic regulation of genes heavily implicated in the stress response, plasticity and cognition, and psychiatric disorders such as depression and anxiety. Thus, the relatively recent surge of studies centering on the epigenetic outcomes of stress has great potential to inform treatments and interventions for psychiatric disorder precipitated by early adversity. Here we review what we know and what we do not know, and suggest approaches to help further elucidate the relationship between early adversity, epigenetics, and behavior.
Keywords: Epigenetics, DNA methylation, DNA hydroxymethylation, Early-life stress, Early adversity, Cell-type specificity, Sex differences, Non-CG methylation, Peripheral measures
1. Introduction
In the following sections we discuss ongoing considerations in the field of neuroepigenetics. Discussion will begin with current knowledge of each topic, focusing on DNA methylation. We will then narrow the focus to where we are at with that knowledge at the intersection of early adversity, the epigenome, and behavior. Lastly, discussion will turn to what we do not know within these areas, the direction the literature suggests we go next, and some specific steps necessary to address outstanding issues.
1.1 Early Adversity, Epigenetics, & Psychiatric Disorder
Adversity in early life, particularly in relation to the caregiver, is a major predisposing factor in the development of psychiatric disorder.1–3 Investigation into the mechanisms of this relationship reveals epigenetic alterations as promising candidates. Exposure to early adversity elicits changes in the epigenome that can vary by gene, brain region, sex, and across time (see Table 1).4–11 In animal models, these changes are associated with behavioral alterations reflective of psychological dysfunction.7,9,12 In humans, they are present in individuals with a history of early adversity and in those with psychiatric disorder.4,13–17 Taken together, these data present a strong argument for the functional role of epigenetics in adverse psychiatric outcomes induced by disruption of the early environment.
Table 1.
Models of early adversity and their associated epigenetic outcomes, including the brain region and time point at which they were found and any sex differences reported.
| Type of Early Adversity | Epigenetic Outcome | Brain Region/Peripheral Measure | Age | Sex Differences | Reference |
|---|---|---|---|---|---|
| Low Levels of Licking/Grooming | Increased methylation GR | Hippocampus | Adult | Only male brains analyzed | 7 |
| Decreased methylation GR | Hippocampus | Adult | Only female brains analyzed | 69 | |
| Increased methylation ERα | Medial Preoptic Area | Adult | Only female brains analyzed | 11 | |
| Maternal Separation | Decreased methylation Avp | Paraventricular nucleus | Adult | Only male brains analyzed | 9 |
| Increased methylation GR | Hippocampus | Adolescent | Present only in males | 72 | |
| Increased methylation Ntsr1 | Amygdala | Adult | Only male brains analyzed | 12 | |
| Decreased methylation Pomc | Pituitary | Adult | Only male brains analyzed | 79 | |
| Scarcity-Adversity Model Of Low Nesting Resources | Increased methylation Bdnf IX | Whole Prefrontal Cortex | Infant, Adolescent, Adult | Males & Females | 10 |
| Increased methylation Bdnf I | Medial Prefrontal Cortex | Adolescent | Present only in males | 5 | |
| Decreased methylation Bdnf IV | Medial Prefrontal Cortex | Adolescent | Present only in females | 5 | |
| Decreased methylation Bdnf I | Medial Prefrontal Cortex | Adult | Males & Females | 5 | |
| Increased methylation Bdnf IV | Medial Prefrontal Cortex | Adult | Present only in females | 5 | |
| Decreased methylation Bdnf I | Amygdala | Infant | Present only in females | 6 | |
| Increased methylation Bdnf IV | Amygdala | Adolescent | Present only in females | 6 | |
| Decreased methylation Bdnf I | Amygdala | Adult | Present only in males | 6 | |
| Decreased methylation Bdnf IV | Amygdala | Adult | Males & Females | 6 | |
| Decreased methylation Bdnf I, IV | Dorsal Hippocampus | Adult | Present only in males | 6 | |
| Increased methylation Bdnf I | Ventral Hippocampus | Adult | Present only in females | 6 | |
| Increased methylation Bdnf IV | Ventral Hippocampus | Infant, Adolescent | Present only in females | 6 | |
| Childhood Abuse/Neglect (Humans) | Increased methylation NR3C1 | Hippocampus | Adult | Only male brains analyzed | 4 |
| Site-specific differential methylation, BDNF, NR3C1, FKBP5 | Saliva | Child/Adolescent, 5–14 years | Males & Females | 13 | |
| Increased methylation, BDNF, OXTR | Blood | Adult | Males & Females | 29 | |
| Differential methylation Genome-wide | Saliva | Child, Average age 9 | Gene-dependent sex differences | 8 |
1.2 DNA Methylation
Methylation of DNA is an epigenetic modification whereby methyl groups are added to cytosine bases in the DNA sequence. This is generally associated with repression of gene transcription (for review see 18) though it is known in some cases to give rise to enhanced transcriptional states.19 While traditionally discussed as occurring at cytosine-guanine (CG) dinucleotides, non-CG methylation is also known to occur and have considerable implications for gene expression.20–22 Methyl group addition is accomplished by DNA methyltransferases DNMT1, DNMT3a, or DNMT3b depending on whether the mark is new or being maintained during replication.23 DNA demethylation may occur through oxidation of methyl marks24 or via enzymes such as Gadd45b.25 The effects of methylation are partly accomplished via methyl-CpG binding protein 2 (MeCP2), which binds to methylated DNA and facilitates gene silencing or expression via recruitment of corepressors26 or coactivators (see Figure 1 for schematic of epigenetic enzymes and their roles).19 DNA methylation is the most heavily studied epigenetic alteration in the realm of early stress and psychiatric disorder, and is thus the focus of the ensuing discussion.
Figure 1.
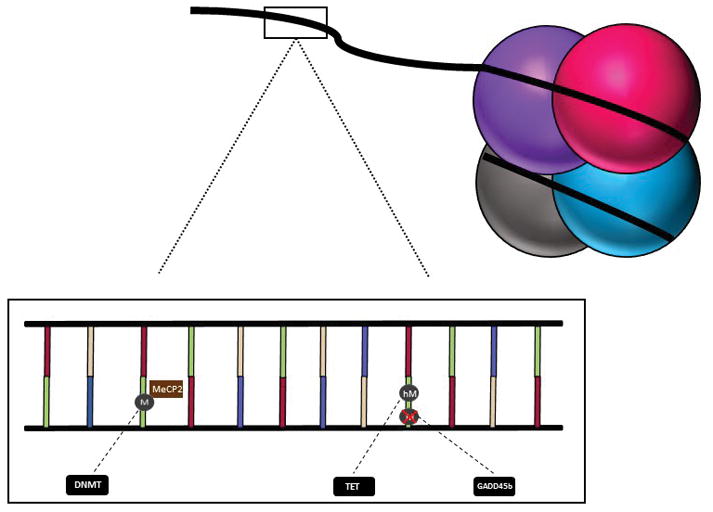
Schematic of DNA wrapped around four core histones. Zoom-in of DNA strand demonstrates the addition of methyl groups (M) to cytosines by DNMT enzymes, association of MeCP2 with methyl groups, oxidation of methyl groups by TET enzymes to create hydroxymethylcytosines (hM), and demethylation activity of GADD45b.
2. Specificity of Experience-induced Methylation Changes
2.1 Tissue Specificity
Though some post-mortem brain studies have been published, human studies of early-stress, psychiatric outcome, and the epigenome generally rely on peripheral measures such as saliva and blood. Use of these peripheral measures has greatly informed the literature on stress-induced methylation statuses of genes critical to mental health such as brain-derived neurotrophic factor (BDNF),27,28 serotonin transporter (SLC6A4),16 and oxytocin receptor (OXTR).29 However, the question remains of whether the epigenetic state of these peripheral tissues accurately reflects what is going in the brain, the target of interest in the study of psychiatric disorder.
Examination of human samples of various tissue types (i.e. brain, blood, saliva) has revealed baseline levels of epigenetic variation between tissues, but has also revealed some promising correlations that prompt further investigation. A 2012 study examining methylated regions of the human genome in multiple brain regions and in whole blood reported high variation between tissues, especially at genes critical to developmental processes. However, a correlation between brain and blood was reported for some of the differences found between individual subjects.30 Importantly, this report shows tissue-specific differences in methylation to occur primarily in intragenic locations, with much less between-tissue variation occurring in promoter regions.30 Given that promoter and intragenic methylation have distinct effects on transcriptional activity (for review see 31), each of these findings bear important implications. Another study comparing methylation between brain and blood in humans found blood to be a poor predictor of brain, and in contrast to the previously mentioned study found that the majority of the few sites exhibiting correlation with brain tissue were located in gene bodies.32 A 2014 study revealed that though overall positively correlated, methylation within CpG islands is highly dissimilar between saliva and blood in humans, with saliva being a closer match to methylation patterns found in the brain.33 Blood is also a poor predictor of human brain BDNF exon I methylation, though muscle tissue methylation at this locus exhibits a high correlation to brain.34 Interestingly, methylation in brain and blood have been found similar in an animal model. A study examining correlations between brain and blood in a mouse model of Cushing’s disease found the two to be positively correlated for methylation of Fkbp5, though methylation changes occurred at different regions in each tissue type.35 However, the disease variable must be taken into account when interpreting these results in terms of tissue specificity as disease could have multiple epigenetic effects between tissues that may not be seen in a normal state. The results of cross-tissue comparison studies in humans should also be interpreted with caution given that, due to complications typically associated with post-mortem tissue collection, it is not often possible to obtain central and peripheral samples from the same subjects (though some of the aforementioned studies were able to do so). Even so, they provide a valuable foundation for exploring the issue of intra-tissue epigenetic variation in relation to the study of psychiatric disorder.
Despite methodological complications, studies of the epigenome following early stress in humans report findings that are promising in terms of the predictive value of peripheral measures. For example, increased methylation of the glucocorticoid receptor (GR) gene following early life stress has been reported in both whole blood14 and saliva.28 This is consistent with a 2009 report of increased GR methylation in post-mortem brain tissue taken from suicide victims with a history of child abuse.4 In an interesting cross-species comparison, researchers examined white blood cell progenitor cells in human and nonhuman primate cord blood alongside brain tissue (prefrontal cortex) from rats, all following exposure to early adversity. They reported similar early stress-induced epigenetic patterns between these species and their respective tissue for a handful of genes, one of which is associated with major depressive disorder in humans.36
2.2 Cell-type Specificity
Specificity of methylation patterns also occurs between cell types. We know that in the central nervous system neurons and glia display distinct patterns of methylation. Compared to glia, neurons exhibit increased levels of both CG and non-CG methylation as well as enriched methylation at CG island shores, enhancers, and intergenic regions.37–40 Methylation patterns also differ by neuron type41,42 in what appears to be a functionally significant manner, especially for developmental processes.20 These cells also exhibit more variability in their epigenetic state than non-neurons,43 suggesting that the neuronal epigenome responds dynamically to experience. This has proven true in response to early stress exposure: neurons in the rodent medial prefrontal cortex (mPFC) exhibit increased levels of Bdnf methylation in response to adverse caregiving when compared to non-neurons.44
Beyond the importance of understanding precisely where in the genome epigenetic modifications are taking place in response to adversity, taking into account cell-type specificity in peripheral samples may be critical for drawing parallels to the brain. For example, saliva samples containing a higher proportion of leukocytes are more positively correlated with blood methylation patterns whereas saliva samples containing a higher proportion of epithelial cells are more positively correlated with brain methylation patterns.33
3. Beyond Methylation
3.1 DNA Hydroxymethylation
Once established, a methyl mark on a cytosine can be oxidized into a hydroxyl group, creating a hydroxymethylated cytosine. This is accomplished by the family of ten-eleven translocation (TET) proteins.45 This recently discovered mark may play multiple roles in the epigenome. Evidence supports a transient role critical to the process of active demethylation24 as well as a stable role in neuronal function46,47 and behavioral outcomes.48 The relationship of this mark to gene expression appears to be quite dynamic49,50 and may interact with methylation, changing gene expression outcomes.51
3.2 Early Adversity and the Hydroxymethylome
Significantly different patterns of hydroxymethylation are seen in non-human primates exposed to early-life adversity (i.e. peer-reared) when compared to their maternally-reared counterparts.51 Our lab has also found significant changes in this mark using a rodent model of adverse caregiving. Specifically, male rats exposed to our model of early adversity (i.e. the scarcity-adversity model of low nesting resources) exhibit decreased levels of hydroxymethylation within amygdala tissue in adolescence.52
The recent discovery of this mark means there has been much less time to address it in studies of early adversity. In fact, methods most commonly used in the field up to this point are unable to distinguish between methylation and hydroxymethylation, though methods do now exist for this purpose53–55 and are certain to become more commonly used. For example, oxidative bisulfite sequencing (oxBS-seq) entails the oxidation of hydroxymethylcytosines followed by conversion to uracil in a bisulfite treatment, allowing for sole detection of methylated cytosines.54 Another option is Tet-assisted bisulfite sequencing (TAB-seq) wherein hydroxymethylcytosines are preserved via glucosylation and methylated cytosines are oxidized to carboxylcytosines via Tet1. The carboxylcytosines are then converted to thymine in a bisulfite treatment, allowing for sole detection of hydroxymethylated cytosines.55
To our knowledge, no studies yet exist that examine hydroxymethylation at any time point following developmental stress exposure in humans. However, this modification has been studied in both the developing and adult human brain56–58 as well as in human peripheral samples (i.e. blood and saliva),59 and we know it is responsive to environmental conditions.60 These factors comprise the necessary foundation to move forward with examination of this mark within the realm of early adversity in humans.
4. Sex Differences
4.1 Animal Models
The epigenome and its many mechanisms are unarguably sexually dimorphic in development and throughout the lifespan. Sex differences have been reported in MeCP2, Gadd45b, and DNMT3a expression in the developing rat amygdala,61–63 in MeCP2 expression in the developing rat ventromedial hypothalamus and preoptic area,62 and in methylation patterns of steroid receptors (e.g. estrogen receptor α) in sexually dimorphic regions of the brain (e.g. mediobasal hypothalamus, preoptic area).64,65 Post development, sex-specific histone modifications have been reported for the bed nucleus of the stria terminalis and preoptic area in adult mice66 and again, in methylation patterns of steroid receptors in sexually dimorphic brain regions.64,65 In humans, baseline (i.e. in non-stress/disease samples) sex differences in methylation have been reported.67,68 Bear in mind that as the axis of this review spins on early adversity, these lists of baseline epigenetic differences are for illustration and thus are not exhaustive.
Though foundational studies of the epigenome in the early stress literature were conducted only in males, a large amount of data have since been produced to support the sex-specific nature of epigenetic modifications following early adversity. One of these foundational studies found that natural variations in maternal care (i.e. high versus low licking and grooming by a dam) resulted in altered methylation patterns and anxiety-like behavior in adult male offspring.7 A study published approximately a decade later examined outcomes in female offspring and found interesting differences (i.e. different in many respects from the male data) in both epigenetic and behavioral responses.69 Though methodological differences between these studies restrict a direct comparison, the collective results nevertheless point to the need for sex-specific examination.
Using a version of the scarcity-adversity model of low nesting resources, wherein pups experience daily exposure to adverse conditions outside the home cage during the first week of life, our lab has observed sex-specific alterations in epigenetic regulators in the mPFC as late as postnatal day 90.70 We have also reported numerous sex differences in adversity-induced Bdnf methylation patterns. We find that females exhibit changes in Bdnf methylation that very rarely match male patterns.5,6,52 We also observe significant, brain-region specific differences in genome-wide DNA methylation and hydroxymethylation in adversity-exposed adolescent males, an effect not observed in females.52 These alterations are accompanied by sex-specific deficits in select behavioral tasks,71 data which support the hypothesis that altered epigenetic patterns are behaviorally functional. Exposure to maternal separation, another commonly used model of early adversity, results in similar sex-specific outcomes. For example, in a mouse model of maternal separation, altered Nr3c1 (GR) methylation is seen in the adolescent male but not female hippocampus of C57BL/6J mice, a change accompanied by various male-specific behavioral alterations.72
4.2 Humans
Interestingly, methylation of NR3C1 in humans exposed to early adversity is not reported to differ between males and females in adulthood.15 The same appears to be true for early stress-induced methylation changes in genes whose methylation status significantly predicts depression.13 However, sex differences in methylation have been reported in children8 and adolescents73 exposed to early stress. Many variables could account for these opposing results. Tissue types less representative of the brain (e.g. blood) may not pick up sex differences that would normally be seen in the brain. Differing types, intensities, and timing of adversity may have more or less of a sex effect. Lastly, gene-specific studies not reporting a sex difference may simply be examining a gene that does not exhibit sex differences in methylation in response to adversity.
5. Methylation across the lifespan
We now know that methylation across the genome is tied to age in both non-human animals9,74 and humans.68,75,76 In addition, data suggest that the effect of early environmental alterations on health at a later age is of an epigenetic nature. For example, prenatal exposure to famine is related to altered methylation of growth- and metabolic-related genes more than half a century after exposure.77,78 Adversity in the context of caregiving in humans and non-humans is known to leave its epigenetic mark early on14,79,80 and/or in adulthood.4,9,79 That is, in some cases these marks are seen persistently across development and into adulthood while in other cases they are dynamic across the lifespan, appearing in some life stages and not in others. These stress-induced changes in methylation have even been reported to override changes typically seen in the aging methylome.9
In non-human primates, serotonin transporter methylation appears to be elevated in both infancy80 and adulthood81 following early stress, though it is important to note that different types of early stress were employed in these two studies. In a mouse model of early stress, Avp hypomethylation was detected at 6 weeks of age and persisted up to the last measurement at 1 year of age.9 In a similar model, Pomc hypomethylation is maintained within the same time frame.79 In contrast to these sustained marks, our lab has found developmental changes in methylation of the Bdnf gene in our adversity-exposed rats. In infancy, we find that adversity-exposed females exhibit a brain region-specific decrease in methylation of Bdnf exon I alongside increased methylation of exon IV, the reverse of which is then found in adulthood.6 In males, the only developmental change detected in Bdnf methylation is at exon I in adolescence.5 Not only is that mark found to be reversed (i.e. opposite direction in methylation) in adulthood, six additional alterations in Bdnf methylation emerge at this time point.5,6 Additionally we find several significant, maltreatment-specific alterations in epigenetic regulators in the mPFC of our adult animals that are not detected in infancy.70 Taken together, these data underline the importance of longitudinal studies of environmental effects upon the epigenome, a type of study that is somewhat lacking in the field at present.
6. What we do not know and how we get there
6.1 Tissue- and Cell-type Specificity
Currently, one of the largest issues in studying epigenetic signatures following adversity in humans is the lack of understanding of the relationship between central and peripheral epigenetic marks. Though we may not necessarily expect similar marks between central and peripheral measures, thorough investigation and comparison of the epigenome between tissue types will aid in understanding if there is a relationship between them, and if there is, how we can use that relationship to understand what is happening in the brain when only peripheral measures are available. An example of the utility of this approach can be seen in work by Walton and colleagues.32 Following initial examination of tissue-specific methylation in samples obtained from epileptic patients during neurosurgery, these researchers isolated a small number of sites in which significant correlations existed between methylation in blood and brain. Using these sites they examined methylation in blood samples of schizophrenia subjects, an approach that led them to discover an association between schizophrenia and methylation of a gene (AVP) and its receptor (AVPR1A) previously associated with this disorder.32
Collection of more biosamples in rodent studies will be incredibly useful in this endeavor. Previous reports of correlation of tissue-specific methylation between-species (i.e. mouse and human)41,82 support the examination of peripheral tissues in animal models to draw parallels in humans, though more studies are needed to know if correlations also exist for adversity-induced methylation patterns. Peripheral and central tissue collection in rodent studies can be easily accomplished within subjects and will greatly contribute to the quest of understanding the peripheral-central relationship of the psychiatric epigenome. However, there are important caveats to consider in making cross-species comparisons. For example, the previously discussed correlation between methylation patterns in mouse and human brain is more highly conserved in inhibitory than excitatory neurons.41
When possible, collecting multiple types of peripheral samples within human subjects will help us better understand intra-tissue epigenetic variation in humans. Given that methylation patterns exhibit brain region-specific changes in response to adversity, it will also be necessary to examine multiple regions individually to understand which of them exhibit a relationship to peripheral measures and what each relationship looks like. Though access to human brain tissue is difficult to obtain (especially when specific conditions, like adversity, are required), the collection of more brain regions from the same subject is warranted.
Currently there are few studies on cell-type specific methylation following early stress. However, existing literature clearly demonstrates stress-induced epigenetic responses and the field would greatly benefit from expanding these studies to understand more precisely where these changes are occurring. Given the known mental health outcomes of developmental stress exposure, this understanding is critical to the intersection of epigenetics and psychopathology.
6.2 Hydroxymethylation and non-CG methylation
Another necessary step in the field will be increased utilization of the techniques referenced in section 3.2 in order to distinguish between methylated and hydroxymethylated cytosines. Considering what is known at this point about the distinct effects of these two modifications on gene expression, it will be critical to parse them apart if we wish to understand epigenetic regulation of behavior following early adversity. Additionally, investigation of hydroxymethylation in human early adversity studies will be essential in moving the field forward.
Another component of the epigenome requiring more attention is non-CG methylation. Examining methylation at CG sites is the dominant practice at present. However, non-CG methylation is not only known to occur (as referenced in section 1.2) but is reportedly the preeminent form of methylation in neurons.38 Furthermore, when occurring in the gene body, it has been reported to be more predictive of neuronal gene expression than both CG methylation and measures of chromatin accessibility.20 Given this powerful relationship to functional outcome, examination of non-CG methylation is paramount to understanding how epigenetic alterations subserve the relationship between early adversity and psychiatric dysfunction.
6.3 Sex Differences
In terms of understanding sex differences in the epigenome following early life stress, female offspring need to be included more consistently in animal research, something that is already occurring based on recently introduced NIH policies on balancing sex in cell and animal studies. Something else to consider is that studies in post-mortem brains suffer from a lack of female tissue, making it difficult to verify sex differences in human studies using peripheral samples. Sex differences are prevalent in psychiatric disorders,83,84 particularly in disorders for which stress is a precipitating factor.85 Given these facts alongside our knowledge of a sexually dimorphic epigenome in individuals exposed to early adversity and in those with psychiatric disorder, it is highly plausible that epigenetic mechanisms are a causal factor in these sex-specific outcomes. Failing to consistently study sex differences will greatly undermine our understanding of and ability to treat psychiatric dysfunction.
6.4 Lifespan Methylation
Concerning aging, we know the human epigenome bears the marks of early stress at individual time points (e.g. adolescence, adulthood) but we have very little information on how methylation changes across the lifespan in these individuals. The literature would greatly benefit from longitudinal studies in early-stress cohorts (in both animal models and humans) that report epigenetic states at various age points following the initial exposure. Understanding the state of the epigenome at a given point in time has great potential to inform the medical community based on the age a given treatment or intervention might be administered.
7. Concluding Remarks
For a young field, great strides have certainly been made. The foundation that exists in the current literature gives us every opportunity to address these challenges and others that will surely arise. Given the nature of these studies, some obstacles will be more difficult to address than others. For example, obtaining post-mortem brain tissue under specific conditions (i.e. from subjects with known histories of early stress) is difficult, as is obtaining peripheral samples from the same subjects due to timing (of death and of the research) as well as the conditions of death. Longitudinal studies also present unique challenges due to issues such as subject attrition. However, the potential of this work to improve the human condition definitively outweighs the cost. The implications are far-reaching, with the potential to shape both policy and medicine in a manner that will meaningfully affect the lives of many in terms of development and/or in the prevention and treatment of disease.
Acknowledgments
Work on this article was supported by a grant from The Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01HD087509) to TLR and a University Dissertation Fellowship from the University of Delaware to TSD.
References
- 1.Vachon DD, Krueger RF, Rogosch FA, Cicchetti D. Assessment of the harmful psychiatric and behavioral effects of different forms of child maltreatment. JAMA psychiatry. 2015;72(11):1135–1142. doi: 10.1001/jamapsychiatry.2015.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carr CP, Martins CMS, Stingel AM, Lemgruber VB, Juruena MF. The Role of Early Life Stress in Adult Psychiatric Disorders: A Systematic Review According to Childhood Trauma Subtypes. The Journal of Nervous and Mental Disease. 2013;201(12):1007–1020. doi: 10.1097/NMD.0000000000000049. [DOI] [PubMed] [Google Scholar]
- 3.Heim C, Nemeroff CB. The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biological Psychiatry. 2001;49(12):1023–1039. doi: 10.1016/s0006-3223(01)01157-x. [DOI] [PubMed] [Google Scholar]
- 4.McGowan PO, Sasaki A, D’Alessio AC, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12(3):342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaze J, Scheuing L, Roth TL. Differential methylation of genes in the medial prefrontal cortex of developing and adult rats following exposure to maltreatment or nurturing care during infancy. Developmental neuroscience. 2013;35(4):306–316. doi: 10.1159/000350716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth TL, Matt S, Chen K, Blaze J. Bdnf DNA methylation modifications in the hippocampus and amygdala of male and female rats exposed to different caregiving environments outside the homecage. Developmental Psychobiology. 2014 doi: 10.1002/dev.21218. n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nature neuroscience. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 8.Cicchetti D, Hetzel S, Rogosch FA, Handley ED, Toth SL. An investigation of child maltreatment and epigenetic mechanisms of mental and physical health risk. Development and Psychopathology. 2016;28(4pt2):1305–1317. doi: 10.1017/S0954579416000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murgatroyd C, Patchev AV, Wu Y, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nature neuroscience. 2009;12(12):1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 10.Roth TL, Lubin F, Funk A, Sweatt J. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biological Psychiatry. 2009;65(9):760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-α1b promoter and estrogen receptor-α expression in the medial preoptic area of female offspring. Endocrinology. 2006;147(6):2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- 12.Toda H, Boku S, Nakagawa S, et al. Maternal separation enhances conditioned fear and decreases the mRNA levels of the neurotensin receptor 1 gene with hypermethylation of this gene in the rat amygdala. PloS one. 2014;9(5):e97421. doi: 10.1371/journal.pone.0097421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weder N, Zhang H, Jensen K, et al. Child abuse, depression, and methylation in genes involved with stress, neural plasticity, and brain circuitry. Journal of the American Academy of Child & Adolescent Psychiatry. 2014;53(4):417–424. e415. doi: 10.1016/j.jaac.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romens SE, McDonald J, Svaren J, Pollak SD. Associations between early life stress and gene methylation in children. Child development. 2015;86(1):303–309. doi: 10.1111/cdev.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perroud N, Paoloni-Giacobino A, Prada P, et al. Increased methylation of glucocorticoid receptor gene (NR3C1) in adults with a history of childhood maltreatment: a link with the severity and type of trauma. Translational Psychiatry. 2011;1(12):e59. doi: 10.1038/tp.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vijayendran M, Beach S, Plume JM, Brody G, Philibert R. Effects of genotype and child abuse on DNA methylation and gene expression at the serotonin transporter. Frontiers in Psychiatry. 2012;3:55. doi: 10.3389/fpsyt.2012.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klengel T, Mehta D, Anacker C, et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nature neuroscience. 2013;16(1):33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bird A. DNA methylation patterns and epigenetic memory. Genes & development. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 19.Chahrour M, Jung SY, Shaw C, et al. MeCP2, a Key Contributor to Neurological Disease, Activates and Represses Transcription. Science. 2008;320(5880):1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mo A, Mukamel Eran A, Davis Fred P, et al. Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron. 2015;86(6):1369–1384. doi: 10.1016/j.neuron.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie W, Barr CL, Kim A, et al. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012;148(4):816–831. doi: 10.1016/j.cell.2011.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bestor TH. The DNA methyltransferases of mammals. Human Molecular Genetics. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 24.Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma DK, Jang M-H, Guo JU, et al. Neuronal activity–induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323(5917):1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nan X, Ng H-H, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 27.Perroud N, Salzmann A, Prada P, et al. Response to psychotherapy in borderline personality disorder and methylation status of the BDNF gene. Translational psychiatry. 2013;3(1):e207. doi: 10.1038/tp.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weder N, Zhang H, Jensen K, et al. Child Abuse, Depression, and Methylation in Genes Involved With Stress, Neural Plasticity, and Brain Circuitry. Journal of the American Academy of Child & Adolescent Psychiatry. 2014;53(4):417–424. e415. doi: 10.1016/j.jaac.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Unternaehrer E, Meyer AH, Burkhardt SC, et al. Childhood maternal care is associated with DNA methylation of the genes for brain-derived neurotrophic factor (BDNF) and oxytocin receptor (OXTR) in peripheral blood cells in adult men and women. Stress. 2015;18(4):451–461. doi: 10.3109/10253890.2015.1038992. [DOI] [PubMed] [Google Scholar]
- 30.Davies MN, Volta M, Pidsley R, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome biology. 2012;13(6):R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13(7):484. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 32.Walton E, Hass J, Liu J, et al. Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophrenia bulletin. 2015;42(2):406–414. doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith AK, Kilaru V, Klengel T, et al. DNA extracted from saliva for methylation studies of psychiatric traits: evidence tissue specificity and relatedness to brain. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2015;168(1):36–44. doi: 10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenz L, Zewdie S, Laforge-Escarra T, et al. BDNF promoter I methylation correlates between post-mortem human peripheral and brain tissues. Neuroscience Research. 2015;91:1–7. doi: 10.1016/j.neures.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Ewald ER, Wand GS, Seifuddin F, et al. Alterations in DNA methylation of Fkbp5 as a determinant of blood–brain correlation of glucocorticoid exposure. Psychoneuroendocrinology. 2014;44:112–122. doi: 10.1016/j.psyneuen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nieratschker V, Massart R, Gilles M, et al. MORC1 exhibits cross-species differential methylation in association with early life stress as well as genome-wide association with MDD. Translational Psychiatry. 2014;4(8):e429. doi: 10.1038/tp.2014.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Baker-Andresen D, Zhao Q, Marshall V, Bredy TW. Methyl CpG Binding Domain Ultra-Sequencing: a novel method for identifying inter-individual and cell-type-specific variation in DNA methylation. Genes, Brain and Behavior. 2014;13(7):721–731. doi: 10.1111/gbb.12150. [DOI] [PubMed] [Google Scholar]
- 38.Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341(6146):1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013;8(3):290–302. doi: 10.4161/epi.23924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kozlenkov A, Roussos P, Timashpolsky A, et al. Differences in DNA methylation between human neuronal and glial cells are concentrated in enhancers and non-CpG sites. Nucleic acids research. 2013;42(1):109–127. doi: 10.1093/nar/gkt838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo C, Keown CL, Kurihara L, et al. Single-cell methylomes identify neuronal subtypes and regulatory elements in mammalian cortex. Science. 2017;357(6351):600–604. doi: 10.1126/science.aan3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozlenkov A, Wang M, Roussos P, et al. Substantial DNA methylation differences between two major neuronal subtypes in human brain. Nucleic acids research. 2015;44(6):2593–2612. doi: 10.1093/nar/gkv1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwamoto K, Bundo M, Ueda J, et al. Neurons show distinctive DNA methylation profile and higher interindividual variations compared with non-neurons. Genome research. 2011;21(5):688–696. doi: 10.1101/gr.112755.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blaze J, Roth TL. Caregiver maltreatment causes altered neuronal DNA methylation in female rodents. Development and Psychopathology. 2017;29(2):477–489. doi: 10.1017/S0954579417000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews Molecular cell biology. 2013;14(6):341. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szulwach KE, Li X, Li Y, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011;14(12):1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kato T, Iwamoto K. Comprehensive DNA methylation and hydroxymethylation analysis in the human brain and its implication in mental disorders. Neuropharmacology. 2014;80:133–139. doi: 10.1016/j.neuropharm.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Wei W, Zhao Q-Y, et al. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proceedings of the National Academy of Sciences. 2014;111(19):7120–7125. doi: 10.1073/pnas.1318906111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robertson J, Robertson AB, Klungland A. The presence of 5-hydroxymethylcytosine at the gene promoter and not in the gene body negatively regulates gene expression. Biochemical and biophysical research communications. 2011;411(1):40–43. doi: 10.1016/j.bbrc.2011.06.077. [DOI] [PubMed] [Google Scholar]
- 50.Valinluck V, Tsai H-H, Rogstad DK, Burdzy A, Bird A, Sowers LC. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic acids research. 2004;32(14):4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Massart R, Suderman M, Provencal N, et al. Hydroxymethylation and DNA methylation profiles in the prefrontal cortex of the non-human primate rhesus macaque and the impact of maternal deprivation on hydroxymethylation. Neuroscience. 2014;268:139–148. doi: 10.1016/j.neuroscience.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Doherty TS, Forster A, Roth TL. Global and gene-specific DNA methylation alterations in the adolescent amygdala and hippocampus in an animal model of caregiver maltreatment. Behavioural Brain Research. 2016;298:55–61. doi: 10.1016/j.bbr.2015.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawasaki Y, Kuroda Y, Suetake I, Tajima S, Ishino F, Kohda T. A Novel method for the simultaneous identification of methylcytosine and hydroxymethylcytosine at a single base resolution. Nucleic acids research. 2017;45(4):e24–e24. doi: 10.1093/nar/gkw994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Booth MJ, Ost TW, Beraldi D, et al. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nature protocols. 2013;8(10):1841. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu M, Hon GC, Szulwach KE, et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nature protocols. 2012;7(12):2159. doi: 10.1038/nprot.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang T, Pan Q, Lin L, et al. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Human molecular genetics. 2012;21(26):5500–5510. doi: 10.1093/hmg/dds394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen L, Li X, Yan L, et al. Whole-genome analysis of 5-hydroxymethylcytosine and 5-methylcytosine at base resolution in the human brain. Genome biology. 2014;15(3):R49. doi: 10.1186/gb-2014-15-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gross JA, Pacis A, Chen GG, Barreiro LB, Ernst C, Turecki G. Characterizing 5-hydroxymethylcytosine in human prefrontal cortex at single base resolution. BMC genomics. 2015;16(1):672. doi: 10.1186/s12864-015-1875-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Godderis L, Schouteden C, Tabish A, et al. Global methylation and hydroxymethylation in DNA from blood and saliva in healthy volunteers. BioMed research international. 2015:2015. doi: 10.1155/2015/845041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tellez-Plaza M, Tang W-Y, Shang Y, et al. Association of global DNA methylation and global DNA hydroxymethylation with metals and other exposures in human blood DNA samples. Environ Health Perspect. 2014;122(9):946–954. doi: 10.1289/ehp.1306674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolodkin MH, Auger AP. Sex Difference in the Expression of DNA Methyltransferase 3a in the Rat Amygdala During Development. Journal of Neuroendocrinology. 2011;23(7):577–583. doi: 10.1111/j.1365-2826.2011.02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurian JR, Forbes-Lorman RM, Auger AP. Sex Difference in Mecp2 Expression During a Critical Period of Rat Brain Development. Epigenetics. 2007;2(3):173–178. doi: 10.4161/epi.2.3.4841. [DOI] [PubMed] [Google Scholar]
- 63.Kigar SL, Chang L, Hayne MR, Karls NT, Auger AP. Sex differences in Gadd45b expression and methylation in the developing rodent amygdala. Brain Research. 2016;1642:461–466. doi: 10.1016/j.brainres.2016.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nugent BM, Schwarz JM, McCarthy MM. Hormonally mediated epigenetic changes to steroid receptors in the developing brain: Implications for sexual differentiation. Hormones and Behavior. 2011;59(3):338–344. doi: 10.1016/j.yhbeh.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwarz JM, Nugent BM, McCarthy MM. Developmental and hormone-induced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinology. 2010;151(10):4871–4881. doi: 10.1210/en.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen EY, Ahern TH, Cheung I, et al. Epigenetics and sex differences in the brain: A genome-wide comparison of histone-3 lysine-4 trimethylation (H3K4me3) in male and female mice. Experimental Neurology. 2015;268:21–29. doi: 10.1016/j.expneurol.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.El-Maarri O, Becker T, Junen J, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Human Genetics. 2007;122(5):505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 68.Boks MP, Derks EM, Weisenberger DJ, et al. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PloS one. 2009;4(8):e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pan P, Fleming AS, Lawson D, Jenkins JM, McGowan PO. Within-and between-litter maternal care alter behavior and gene regulation in female offspring. Behavioral Neuroscience. 2014;128(6):736. doi: 10.1037/bne0000014. [DOI] [PubMed] [Google Scholar]
- 70.Blaze J, Roth T. Exposure to caregiver maltreatment alters expression levels of epigenetic regulators in the medial prefrontal cortex. International Journal of Developmental Neuroscience. 2013;31(8):804–810. doi: 10.1016/j.ijdevneu.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doherty TS, Keller S, Blaze J, Roth TL. Phenotypic outcomes in adolescence and adulthood in the scarcity-adversity model of low nesting resources outside the home cage. Developmental Psychobiology. 2017 doi: 10.1002/dev.21547. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kundakovic M, Lim S, Gudsnuk K, Champagne FA. Sex-specific and strain-dependent effects of early life adversity on behavioral and epigenetic outcomes. Frontiers in Psychiatry. 2013:4. doi: 10.3389/fpsyt.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Essex MJ, Thomas Boyce W, Hertzman C, et al. Epigenetic vestiges of early developmental adversity: childhood stress exposure and DNA methylation in adolescence. Child development. 2013;84(1):58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maegawa S, Hinkal G, Kim HS, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome research. 2010;20(3):332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Horvath S. DNA methylation age of human tissues and cell types. Genome biology. 2013;14(10):3156. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Horvath S, Zhang Y, Langfelder P, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome biology. 2012;13(10):R97. doi: 10.1186/gb-2012-13-10-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proceedings of the National Academy of Sciences. 2008;105(44):17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tobi EW, Lumey L, Talens RP, et al. DNA methylation differences after exposure to prenatal famine are common and timing-and sex-specific. Human molecular genetics. 2009;18(21):4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu Y, Patchev AV, Daniel G, Almeida OF, Spengler D. Early-life stress reduces DNA methylation of the pomc gene in male mice. Endocrinology. 2014;155(5):1751–1762. doi: 10.1210/en.2013-1868. [DOI] [PubMed] [Google Scholar]
- 80.Kinnally EL, Capitanio JP, Leibel R, et al. Epigenetic regulation of serotonin transporter expression and behavior in infant rhesus macaques. Genes, Brain and Behavior. 2010;9(6):575–582. doi: 10.1111/j.1601-183X.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kinnally EL, Feinberg C, Kim D, et al. DNA methylation as a risk factor in the effects of early life stress. Brain, behavior, and immunity. 2011;25(8):1548–1553. doi: 10.1016/j.bbi.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41(2):178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dohrenwend BP, Dohrenwend BS. Sex differences and psychiatric disorders. American journal of sociology. 1976;81(6):1447–1454. doi: 10.1086/226229. [DOI] [PubMed] [Google Scholar]
- 84.Earls F. Sex differences in psychiatric disorders: origins and developmental influences. Psychiatric developments. 1987;5(1):1–23. [PubMed] [Google Scholar]
- 85.Bangasser DA, Valentino RJ. Sex differences in stress-related psychiatric disorders: neurobiological perspectives. Frontiers in neuroendocrinology. 2014;35(3):303–319. doi: 10.1016/j.yfrne.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]


