Abstract
Allergic diseases, such as respiratory, cutaneous, and food allergy, have dramatically increased in prevalence over the last few decades. Recent research points to a central role of the microbiome, which is highly influenced by multiple environmental and dietary factors. It is well established that the microbiome can modulate the immune response, from cellular development to organ and tissue formation exerting its effects through multiple interactions with both the innate and acquired branches of the immune system. It has been described at some extent changes in environment and nutrition produce dysbiosis in the gut but also in the skin, and lung microbiome, inducing qualitative and quantitative changes in composition and metabolic activity. Here, we review the potential role of the skin, respiratory, and gastrointestinal tract (GIT) microbiomes in allergic diseases. In the GIT, the microbiome has been proven to be important in developing either effector or tolerant responses to different antigens by balancing the activities of Th1 and Th2 cells. In the lung, the microbiome may play a role in driving asthma endotype polarization, by adjusting the balance between Th2 and Th17 patterns. Bacterial dysbiosis is associated with chronic inflammatory disorders of the skin, such as atopic dermatitis and psoriasis. Thus, the microbiome can be considered a therapeutical target for treating inflammatory diseases, such as allergy. Despite some limitations, interventions with probiotics, prebiotics, and/or synbiotics seem promising for the development of a preventive therapy by restoring altered microbiome functionality, or as an adjuvant in specific immunotherapy.
Keywords: microbiome, microbiota, allergy, allergic diseases, prebiotics, probiotics, synbiotics
Introduction
Allergic diseases, include heterogeneous inflammatory pathologies such as respiratory and food allergies (FA), which are characterized by an immunological response with T lymphocytes producing IL-4, IL-5, and IL-13 and low production of IFN-γ (Th2) (1) and others producing IL-9 and IL-10 (Th9) (2) as the main effector T cells. They promote the induction of other effector cells involved in allergic inflammation, such as mast cells, basophils, and eosinophils (1). These diseases have dramatically increased in prevalence over the last few decades (3–6) and recent research points to a central role of the microbiota (7, 8). It is well established that the microbiome can modulate the immune response, from cellular development to organ and tissue formation (9) exerting its effects through multiple interactions with both the innate and acquired branches of the immune system. In the late 80s, Dr. Strachan proposed what is now referred to as the “hygiene-hypothesis” (10), in which changes in environment and nutrition produce a dysbiosis in the skin, gut, or lung microbiome inducing qualitative and quantitative changes in composition and metabolic activity (11, 12). Furthermore, it was proposed that a lower incidence of infection in early childhood, which may be associated with low microbiota diversity, could explain the increase in prevalence of atopic diseases (13). It should be pointed out that the hygiene hypothesis has not been found to apply to individual hygiene [no relation between personal or home cleanliness and increased risk of asthma or allergy has been found (14)], but to independent host factors such as number of older siblings, contact with pets and rural versus urban living, all of which have been shown to affect microbiome composition and the development of immunologic tolerance (15). Today, the use of bacterial culture-independent tools such as next-generation sequencing to identify different microbes has permitted the investigation of complex populations and their roles in health and disease. Here, we review the potential role of the skin, respiratory, and gastrointestinal tract (GIT) microbiomes in allergic diseases.
Microbiome
The term “microbiome” refers to the microorganisms that live on or inside another organism. They interact with each other and with their host and can be classified as beneficial (symbiotic) or dangerous (pathogenic) (16). Microbiome in humans can account for 90% of the cells by a ratio of 10:1 (17). New studies point out that the number of bacteria in the body is of the same order as the number of human cells (18). Most of these microorganisms inhabit the gut. The microbiome effectively adds a huge amount of genes to the human genome, potentially increasing it up to 200 times (19). As a result, the composition of the human microbiome could be important in the context of health or disease.
Human Gut Microbiome and Implications in Food Allergy
The GIT has a very important immune function in developing either effector or tolerant responses to different antigens by balancing the activities of Th1 and Th2 cells as well as regulating Th17 and T regulatory (Treg) cells in the lamina propria (20–23). Immune dysfunction in allergic diseases such as asthma and atopy seems to be related to differences in the function and composition of the gut microbiome (24).
The gut microbiome constitutes a highly complex ecosystem which includes eukaryotic fungi, viruses, and some archaea, although bacteria are the most prominent components (25). Its composition is generally formed during the first 3 years of life (26); however, recent work has suggested that its colonization may begin in utero (27), contrary to the widely held dogma of the fetus as a sterile environment. Despite its early formation, its composition is highly dynamic and dependent on host-associated factors such as age, diet, and environmental conditions (26, 28–31) with the major phyla being Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria. The gut microbiome is not homogeneous throughout the GIT, showing higher diversity in the oral cavity and intestine, and lower diversity in the stomach, mainly because of the acid environment (32). Aerobic species are mainly located in the upper small intestine and anaerobic species in the colon (33).
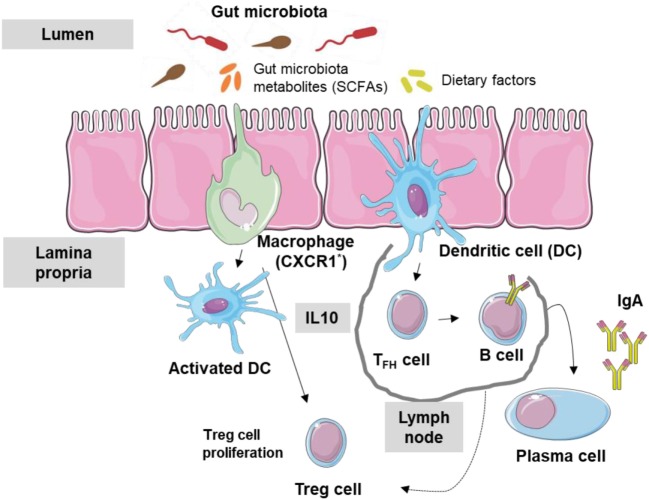
Most antigens in the GIT come from dietary factors and gut microbiota, both of which can affect immune tolerance being the promotion of Treg cells to these dietary factors crucial to avoid an immune response to dietary antigens (34). Alterations in GIT bacterial levels or diversity (dysbiosis) can disrupt mucosal immunological tolerance, leading to allergic diseases including FA (35) and even asthma (36–38). Moreover, low IgA levels at the intestinal surface barrier can also contribute to FA. In fact, low microbiota levels and IgA appear to be related: gut microbiota can stimulate dendritic cells (DCs) in the Peyer’s patches (digestive type of mucosa lymphoid-associated tissue) to activate B cells, leading to specific IgA antibodies production through class switching (39). This stimulation may occur through the production by members of the microbiome of metabolites, such as short chain fatty acids (SCFAs). Thus, the immune tolerance network in the intestinal lumen can be considered to include the gut microbiota, their metabolic products, dietary factors, epithelial cells, DCs, IgA antibodies, and regulatory T cells (Figure 1).
Figure 1.
Interaction between gut microbiota and immune system. Gut microbiota metabolites and dietary factors constitute the main antigen load of the gastrointestinal tract. Macrophages (CXCR1+) and dendritic cells (DCs) are stimulated and T regulatory (Treg) cells are activated by metabolic products such as short chain fatty acid (SCFA). Follicular T cells activate B cells inducing the production of IgA antibodies.
Several factors associated with dysbiosis may influence FA, such as cesarean versus vaginal delivery (40), low versus rich fiber diet (41), breastfeeding (42), and/or early-life-antibiotic exposure, all of which affect bacterial load and diversity.
Once thought to be almost sterile, the esophagus has been shown to comprise around 300 bacteria species. Significant differences in the microbial composition of children with active esophageal inflammation caused by eosinophilic esophagitis compared with controls have been reported (43). Importantly, both the degree of inflammation and the treatment regimen seem to impact the esophageal microbiota (43).
Human Lung Microbiome and Implications in Respiratory Allergy
As with the esophagus and fetus, the lung has long been thought of as sterile; however, recent evidence has shown it to harbor various bacteria phyla, including Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria, even in healthy subjects (44). Similar to the gut, the lung microbiome changes rapidly in the first years of life, before beginning to stabilize (45, 46). Colonization occurs gradually in healthy children, starting with Staphylococcus or Corynebacterium, followed by Moraxella or Alloiococcus (46). A breakdown in the development of the commensal population can lead to dysregulation of the IgE–basophil axis, with elevated serum IgE concentrations and increased of circulating basophil populations as has been described in murine models of allergic airway disease (47). Importantly, this link was found to be B-cell intrinsic and dependent on the MYD88 pathway. Moreover, the lung microbiome may also play a role in driving asthma endotype polarization, by adjusting the balance between Th2 and Th17 patterns. Enterococcus faecalis can suppress Th17 immunity and symptoms of allergic airway disease, and thus it has even been considered a potential therapeutic agent for both asthma and Th17 immunity (48).
Differences in levels and diversity of the lung microbiome have been found between healthy people and patients with asthma and allergic diseases, with an increase of Proteobacteria in the latter; moreover, their presence has been linked to increased severity of asthma probably through the upregulation of Th17-related genes (49, 50).
Early colonization with Haemophilus influenzae, Moraxella catarrhalis, and Streptococcus pneumoniae has been associated with recurrent wheezing and asthma (45, 46, 51, 52). Importantly, as well as bacteria, viruses will also influence asthma development, as has been demonstrated with human rhinovirus infections of the nasopharynx in early-life (46). In addition, other associations such as helminths may be protective for asthma, as helminth infections have been shown to increase the microbiota diversity (53). Associations have been found between the composition of the lung and gut microbiome and the risk of respiratory allergic disease development (54) indicating that both gut and lung mucosa may function as a single organ, sharing immunological functions (44).
Skin Microbiome and Cutaneous Allergic Diseases
Bacterial dysbiosis is associated with chronic inflammatory disorders of the skin, such as atopic dermatitis (AD) and psoriasis (55). The composition of the skin microbiota depends on the body site samples (56). The relevance of AD, often associated with other allergic diseases, has significantly increased in the last few decades. Outgrowths of Staphylococcus and reductions of other communities like Streptococcus or Propionibacterium species correlate with AD flares (57). On the other hand, skin commensal Acinetobacter species have been reported to protect against allergic sensitization and inflammation, playing an important role in tuning the balance of Th1, Th2, and anti-inflammatory responses to environmental allergens (58). Interestingly, studies of cutaneous allergic diseases have found an association with gut microbiome dysbiosis (59), although the underlying mechanisms are still unclear. An initial study of 90 patients with established AD found enrichment for Faecalibacterium prausnitzii and decreased levels of SCFAs in the gut (60).
Therefore, we can summarize that changes in environment and diet produce dysbiosis in gut, skin, and/or lung microbiome inducing qualitative and quantitative changes in the microbiota which directly affect the immunological mechanisms implicated in the prevention of allergic diseases (Figure 2).
Figure 2.
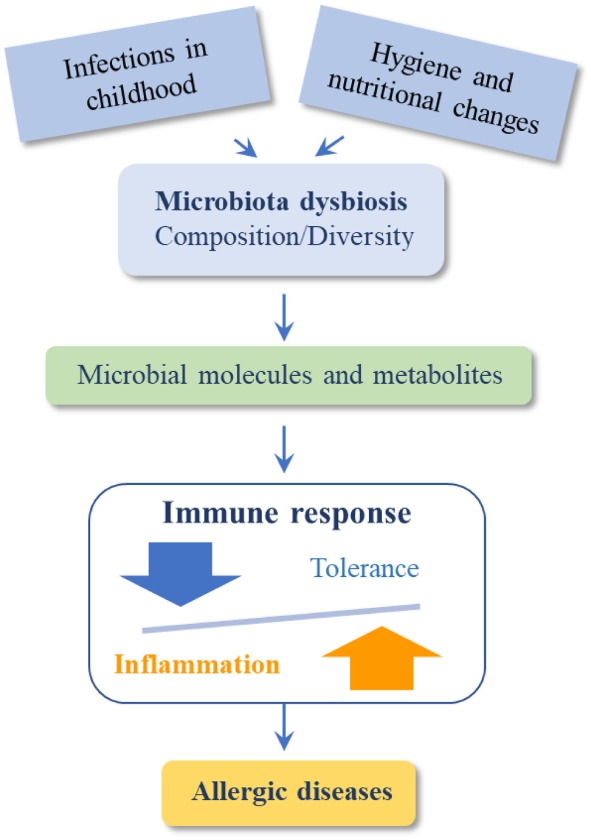
Dysbiosis induce qualitative and quantitative changes in the microbiota that directly affect immunological mechanisms leading to allergic diseases.
Factors Affecting Microbiome Diversity
Childbirth
The mode of delivery in childbirth can produce profound differences in the infant gut microbiome, with lower level of Escherichia coli, Bifidobacterium, and Bacteroides species in children born through cesarean section compared with those delivered vaginally (28, 61, 62). Cesarean-born infants typically have a microbiome enriched with Staphylococcus and Streptococcus, comparable with the maternal skin microbiome (63). These differences appear to be associated with higher risk of allergic diseases and asthma (64–66). Transfer of maternal vaginal microbes at birth may mitigate these effects (67). Time of gestation may also be a factor: premature births are associated with alterations of the gut microbiome, but not atopic sensitization (68).
Importance of Early-Life Microbiome
There is mounting evidence that early-life exposure is critical for the microbiome and that gut microbial dysbiosis heavily influences immune system development (53). Potential factors include perinatal exposure to maternal or infant diet, antibiotic use, and contact with older siblings (16). Data from different populations show that the highest interindividual microbial variability occurs during the first 3 years of age (26). Noteworthy, contact with the microbiome can start before birth, since a low-abundance microbiota in the placenta (69) and meconium (70, 71) have been found.
Microbial exposure during the first months of life induces the activation of the innate immune system in different ways, with consequences for FA. Early inoculation with spore-forming Clostridium class IV and XIV species (72) and other bacteria (53) leads to decreased levels of circulating IgE in adulthood. Conversely, 3-week-old neonates with a higher fecal burden of Clostridium difficile and a higher ratio of C. difficile to Bifidobacterium showed increased numbers of skin test positive results to food and aero-allergens (73). Similarly, high levels of fecal E. coli in infants during their first month are associated with IgE-mediated eczema (74, 75).
Remarkably, the same colonization pattern can have different consequences at different ages. For example, colonization of S. pneumoniae, H. influenzae, or M. catarrhalis within the first month of life increases the risk of asthma, leading to high counts of atopic markers such as eosinophils and serum IgE, but not when colonization occurs at 12 months (45).
Furthermore, respiratory tract infections during early-life are associated with asthma development (76, 77). This may be because viral infections favor other opportunistic respiratory pathogens such as M. catarrhalis and S. pneumoniae, increasing the risk of asthma exacerbations (78). Other possible mechanisms may involve respiratory rhinovirus interacting with airway epithelial cells, increasing IL-25 and IL-33 production and contributing to Th2 immune responses (79). This is in line with the higher levels of house dust mite-specific IgE found in children infected with rhinovirus (80). Moreover, rhinovirus infection can also induce mucus hypersecretion and airway hyperresponsiveness in neonatal mice compared with adults (81).
Diet and Microbiome Metabolic Products
Another key factor influencing gut microbiome diversity is infant feeding, and especially breastfeeding, which has been shown to increase colonization by Lactobacilli and Bifidobacteria (82). Breast milk contains oligosaccharides and a wide range of fatty acids, which will affect the gut microbiome and its capacity to produce metabolites that protect against allergies and asthma (83) through the development of Treg cells (84). This effect is also produced by the intake of unprocessed milk during the first year of life, probably related to higher levels of peptides in the serum fraction and unsaturated omega-3 fatty acids (85). Other dietary components such as polyphenols and fish oils are also important for microbiome diversity (86–88).
Some noteworthy bacteria, such as Lachnospiraceae and Ruminococcaceae, can also influence the gut microbiome by producing SCFAs—including propionate, butyrate, and acetate—through fermentation of complex dietary carbohydrates. Importantly, besides acting as an essential energy source for gastrointestinal colonocytes, these acids exert various anti-inflammatory effects on the immune system that can modulate FA and respiratory diseases (89, 90), by increasing epithelial barrier function (91), and inducing Treg cells (colonic CD103+FoxP3+ cells), DCs precursors, and IL-10 production (8, 90).
Importance of Exposure to Antibiotics
The introduction of antibiotics in the 1950s is associated with an increasing incidence of allergy. This is thought to be causes by antibiotics inducing dysbiosis which has been shown to directly impact the development of AD (92) and asthma (48). The age of initial exposure could be important since maternal intake of antibiotic during pregnancy increases the risk of allergy in children (93), and antibiotic use in the first month of life has been associated with cow’s milk allergy (94). Intrapartum antibiotics have been shown to lead to a modified microbiome in children at 3 and 12 months (95). Other studies showed that antibiotics affect the microbiome in older subjects (96, 97). Antibiotic administration is associated with severe allergic airway inflammation in neonates, but not in adults (98).
Even low doses of antibiotics can affect microbiome composition (99); however, the associations between antibiotic consumption and allergic diseases increase with the number of antibiotics prescribed, and variable effects have been found for different antibiotic families. Some studies have indicated that betalactam antibiotics are the most common triggers when FA is diagnosed before 2 years of age, while macrolides are associated with FA when it is diagnosed later (100). For asthma, further studies are needed to clarify whether it is the infection rather than the antibiotics themselves that increase susceptibility (101).
Interventions
The microbiota can be considered a therapeutical target for treating allergy; moreover, certain species can be used to enhance tolerance response induction. Different approaches for restoring the microbiome involve probiotics, prebiotics, and synbiotics.
Probiotics
According to the Food and Agriculture Organization of the United Nations and the World Health Organization, probiotics are defined as “live microorganisms which, when administered in adequate amounts, confer a health benefit to the host” (102). They do so by promoting the appropriate balance of gut microbiota. The health benefits attributed to one probiotic strain are not necessarily applicable to another one even within one given species (103). Furthermore, the effectiveness may depend on the time of intervention and aspects of the current microbiota composition. In fact, different studies have shown that timing is crucial (104).
In the case of FA, co-administration of bacterial adjuvants with oral immunotherapy (OIT) has been suggested as a potential treatment. Probiotic therapy with Lactobacillus rhamnosus increases efficacy when co-administered with peanut OIT—producing desensitization in 82% of treated patients (105)—or with hydrolyzed casein in milk allergic patients, in which an increase of fecal butyrate levels were found (106, 107). However, other strains of Lactobacilli and/or Bifidobacteria did not demonstrate any effect in preventing allergic diseases (106, 107). Some investigations have shown that the oral administration of probiotics may benefit allergic rhinitis patients (108–110); similarly, local nasal administration of Lactococcus lactis NZ9000 can affect local and systemic immune responses against S. pneumoniae (111). However, Ivory et al. reported that even oral delivery of Lactobacillus casei Shirota modified the immune system of allergic individuals (110), these modifications did not have a significant impact on the allergic status (112), highlighting the fact that analysis of immune parameters per se is not a real indicator of the therapeutical properties of the probiotics.
It has been suggested that probiotics can help preventing eczema and they also show some beneficial effects for other allergic diseases including asthma (113–117); furthermore, another approach based on the intranasal application of bacterial products (endotoxin or flagellin) has demonstrated immunomodulatory ability, mimicking the effect of probiotics, for the lung in different animal models, reducing experimental asthma by either re-establishing the expression of the ubiquitin-modifying enzyme A20 at the endothelial barrier or inducing Tregs (118, 119).
Therefore, it seems that the optimal time periods to apply probiotic intervention are before, during, and just after birth represents. Nevertheless, more studies, using clinical trial methodologies when possible, should be carried out to confirm these findings and determine the optimal probiotics to use.
Prebiotics
Prebiotics are non-digestible food components that benefit the host by selectively stimulating the growth and activity of microorganisms. Studies have shown that fibers and oligosaccharides can improve immunity and metabolism (8) and that the treatment of pregnant and lactating mice increases the proportions of Lactobacillus and Clostridium leptum and promotes a long-term protective effect against FA in the offspring (120).
Studies evaluating the effect of fiber/oligosaccharide intake in modulating asthma (121–123) have shown heterogeneous results, with one study reporting a reduction of wheezing (121) but others reporting no effect (122, 123). A recent Cochrane review has shown that although the addition of prebiotics to infant food may reduce the risk of eczema, it is not clear whether their use may affect other allergic diseases including asthma (124).
Synbiotics
When the use of a combination of prebiotics and probiotics produce synergistic health benefits it is described as a symbiotic. In FA mice models, both the microbiome and diet can affect the development of food tolerance by the induction of Treg cells (34). In cow’s milk allergy, it has been demonstrated that treatment with extensively hydrolyzed casein formula plus L. rhamnosus GG promotes tolerance through changes in the infant gut microbiome (89).
A recent meta-analysis has shown their beneficial effects for eczema treatment (125). However, further well-conducted, randomized, placebo-controlled longitudinal studies are still needed in this area (126).
Conclusion
The microbiota is a highly dynamic environment influenced by multiple environmental and dietary factors, with a complex role in allergic diseases. Further studies with larger number of well-characterized patients and controls are needed to dissect the role of microbiome in allergic diseases are the performance. Despite some limitations, interventions with probiotics, prebiotics, and/or synbiotics show promise for the development of a preventive therapy, either by restoring altered microbiome functionality due to dysbiosis or as a boosting of immunological system in specific immunotherapy. However, the field is still relatively new and we expect many key findings to be made in the next few years. Detailed prospective, randomized, placebo-controlled studies will be essential for this purpose.
Author’s Note
All authors belongs to the Immunology Committee of the Spanish Society of Allergy and Clinic Immunology (SEAIC).
Author Contributions
CM, MP and MP-G conceived and designed this manuscript and were involved in manuscript production contributing equally to this work. TC, MME, MNLL, OL, LM, VM, ES, MZ and ML have read, revised and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by ISCIII (project number PI17/01087) cofounded by FEDER for the thematic network and cooperative research centers ARADyAL (RD16/0006/0007, RD16/0006/0015, and RD16/0006/0001). All the authors acknowledge Tomás Barker for language revision.
Footnotes
Funding. This article have been paid by the Sociedad Española de Alergologia e Inmunologia Clínica (SEAIC) Foundation.
References
- 1.Akdis CA. Allergy and hypersensitivity: mechanisms of allergic disease. Curr Opin Immunol (2006) 18(6):718–26. 10.1016/j.coi.2006.09.016 [DOI] [PubMed] [Google Scholar]
- 2.Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol (2008) 9(12):1341–6. 10.1038/ni.1659 [DOI] [PubMed] [Google Scholar]
- 3.Akinbami LJ, Simon AE, Rossen LM. Changing trends in asthma prevalence among children. Pediatrics (2016) 137(1). 10.1542/peds.2015-2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prescott SL, Pawankar R, Allen KJ, Campbell DE, Sinn J, Fiocchi A, et al. A global survey of changing patterns of food allergy burden in children. World Allergy Organ J (2013) 6(1):21. 10.1186/1939-4551-6-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage J, Johns CB. Food allergy: epidemiology and natural history. Immunol Allergy Clin North Am (2015) 35(1):45–59. 10.1016/j.iac.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sicherer SH, Sampson HA. Food allergy: epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol (2014) 133(2):291–307; quiz 308. 10.1016/j.jaci.2013.11.020 [DOI] [PubMed] [Google Scholar]
- 7.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol (2009) 7(12):887–94. 10.1038/nrmicro2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med (2014) 20(2):159–66. 10.1038/nm.3444 [DOI] [PubMed] [Google Scholar]
- 9.Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol (2015) 159(2):122–7. 10.1016/j.clim.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strachan DP. Hay fever, hygiene, and household size. BMJ (1989) 299(6710):1259–60. 10.1136/bmj.299.6710.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daley D. The evolution of the hygiene hypothesis: the role of early-life exposures to viruses and microbes and their relationship to asthma and allergic diseases. Curr Opin Allergy Clin Immunol (2014) 14(5):390–6. 10.1097/ACI.0000000000000101 [DOI] [PubMed] [Google Scholar]
- 12.Platts-Mills TA. The allergy epidemics: 1870-2010. J Allergy Clin Immunol (2015) 136(1):3–13. 10.1016/j.jaci.2015.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrlander C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med (2011) 364(8):701–9. 10.1056/NEJMoa1007302 [DOI] [PubMed] [Google Scholar]
- 14.Marsland BJ, Salami O. Microbiome influences on allergy in mice and humans. Curr Opin Immunol (2015) 36:94–100. 10.1016/j.coi.2015.07.005 [DOI] [PubMed] [Google Scholar]
- 15.Penders J, Gerhold K, Stobberingh EE, Thijs C, Zimmermann K, Lau S, et al. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J Allergy Clin Immunol (2013) 132(3):601–607.e8. 10.1016/j.jaci.2013.05.043 [DOI] [PubMed] [Google Scholar]
- 16.Riiser A. The human microbiome, asthma, and allergy. Allergy Asthma Clin Immunol (2015) 11:35. 10.1186/s13223-015-0102-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell (2006) 124(4):837–48. 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- 18.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol (2016) 14(8):e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature (2007) 449(7164):804–10. 10.1038/nature06244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Mahony C, Scully P, O’Mahony D, Murphy S, O’Brien F, Lyons A, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog (2008) 4(8):e1000112. 10.1371/journal.ppat.1000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostman S, Rask C, Wold AE, Hultkrantz S, Telemo E. Impaired regulatory T cell function in germ-free mice. Eur J Immunol (2006) 36(9):2336–46. 10.1002/eji.200535244 [DOI] [PubMed] [Google Scholar]
- 22.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell (2009) 139(3):485–98. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov II, Frutos Rde L, Manel N, Yoshinaga K, Rifkin DB, Sartor RB, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe (2008) 4(4):337–49. 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muir AB, Benitez AJ, Dods K, Spergel JM, Fillon SA. Microbiome and its impact on gastrointestinal atopy. Allergy (2016) 71(9):1256–63. 10.1111/all.12943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Power SE, O’Toole PW, Stanton C, Ross RP, Fitzgerald GF. Intestinal microbiota, diet and health. Br J Nutr (2014) 111(3):387–402. 10.1017/S0007114513002560 [DOI] [PubMed] [Google Scholar]
- 26.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature (2012) 486(7402):222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moles L, Gomez M, Heilig H, Bustos G, Fuentes S, de Vos W, et al. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One (2013) 8(6):e66986. 10.1371/journal.pone.0066986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe (2015) 17(5):690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 29.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, et al. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr (2000) 30(1):61–7. 10.1097/00005176-200001000-00019 [DOI] [PubMed] [Google Scholar]
- 30.Li M, Wang M, Donovan SM. Early development of the gut microbiome and immune-mediated childhood disorders. Semin Reprod Med (2014) 32(1):74–86. 10.1055/s-0033-1361825 [DOI] [PubMed] [Google Scholar]
- 31.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature (2010) 464(7285):59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ursell LK, Clemente JC, Rideout JR, Gevers D, Caporaso JG, Knight R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol (2012) 129(5):1204–8. 10.1016/j.jaci.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol (2014) 14(10):667–85. 10.1038/nri3738 [DOI] [PubMed] [Google Scholar]
- 34.Kim KS, Hong SW, Han D, Yi J, Jung J, Yang BG, et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science (2016) 351(6275):858–63. 10.1126/science.aac5560 [DOI] [PubMed] [Google Scholar]
- 35.Aitoro R, Paparo L, Amoroso A, Di Costanzo M, Cosenza L, Granata V, et al. Gut microbiota as a target for preventive and therapeutic intervention against food allergy. Nutrients (2017) 9(7):E672. 10.3390/nu9070672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol (2012) 129(2):e431–2. 10.1016/j.jaci.2011.10.025 [DOI] [PubMed] [Google Scholar]
- 37.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy (2014) 44(6):842–50. 10.1111/cea.12253 [DOI] [PubMed] [Google Scholar]
- 38.Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol (2011) 128(3):e641–5. 10.1016/j.jaci.2011.04.060 [DOI] [PubMed] [Google Scholar]
- 39.Suzuki K, Kawamoto S, Maruya M, Fagarasan S. GALT: organization and dynamics leading to IgA synthesis. Adv Immunol (2010) 107:153–85. 10.1016/B978-0-12-381300-8.00006-X [DOI] [PubMed] [Google Scholar]
- 40.Papathoma E, Triga M, Fouzas S, Dimitriou G. Cesarean section delivery and development of food allergy and atopic dermatitis in early childhood. Pediatr Allergy Immunol (2016) 27(4):419–24. 10.1111/pai.12552 [DOI] [PubMed] [Google Scholar]
- 41.Grimshaw KE, Maskell J, Oliver EM, Morris RC, Foote KD, Mills EN, et al. Diet and food allergy development during infancy: birth cohort study findings using prospective food diary data. J Allergy Clin Immunol (2014) 133(2):511–9. 10.1016/j.jaci.2013.05.035 [DOI] [PubMed] [Google Scholar]
- 42.Muraro A, Halken S, Arshad SH, Beyer K, Dubois AE, Du Toit G, et al. EAACI food allergy and anaphylaxis guidelines. Primary prevention of food allergy. Allergy (2014) 69(5):590–601. 10.1111/all.12398 [DOI] [PubMed] [Google Scholar]
- 43.Benitez AJ, Hoffmann C, Muir AB, Dods KK, Spergel JM, Bushman FD, et al. Inflammation-associated microbiota in pediatric eosinophilic esophagitis. Microbiome (2015) 3:23. 10.1186/s40168-015-0085-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Segal LN, Blaser MJ. A brave new world: the lung microbiota in an era of change. Ann Am Thorac Soc (2014) 11(Suppl 1):S21–7. 10.1513/AnnalsATS.201306-189MG [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bonnelykke K, et al. Childhood asthma after bacterial colonization of the airway in neonates. N Engl J Med (2007) 357(15):1487–95. 10.1056/NEJMoa052632 [DOI] [PubMed] [Google Scholar]
- 46.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe (2015) 17(5):704–15. 10.1016/j.chom.2015.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med (2012) 18(4):538–46. 10.1038/nm.2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adami AJ, Bracken SJ. Breathing better through bugs: asthma and the microbiome. Yale J Biol Med (2016) 89(3):309–24. [PMC free article] [PubMed] [Google Scholar]
- 49.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One (2010) 5(1):e8578. 10.1371/journal.pone.0008578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol (2011) 127(2):e371–3. 10.1016/j.jaci.2010.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korppi M. Bacterial infections and pediatric asthma. Immunol Allergy Clin North Am (2010) 30(4):565–574, vii. 10.1016/j.iac.2010.09.007 [DOI] [PubMed] [Google Scholar]
- 52.Davis MF, Peng RD, McCormack MC, Matsui EC. Staphylococcus aureus colonization is associated with wheeze and asthma among US children and young adults. J Allergy Clin Immunol (2015) 135(3):811–813.e5. 10.1016/j.jaci.2014.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe (2013) 14(5):559–70. 10.1016/j.chom.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell (2012) 148(6):1258–70. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanford JA, Gallo RL. Functions of the skin microbiota in health and disease. Semin Immunol (2013) 25(5):370–7. 10.1016/j.smim.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science (2009) 324(5931):1190–2. 10.1126/science.1171700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res (2012) 22(5):850–9. 10.1101/gr.131029.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fyhrquist N, Ruokolainen L, Suomalainen A, Lehtimaki S, Veckman V, Vendelin J, et al. Acinetobacter species in the skin microbiota protect against allergic sensitization and inflammation. J Allergy Clin Immunol (2014) 134(6):1301–1309.e11. 10.1016/j.jaci.2014.07.059 [DOI] [PubMed] [Google Scholar]
- 59.Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy (2015) 70(2):241–4. 10.1111/all.12549 [DOI] [PubMed] [Google Scholar]
- 60.Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol (2016) 137(3):852–60. 10.1016/j.jaci.2015.08.021 [DOI] [PubMed] [Google Scholar]
- 61.Adlerberth I, Strachan DP, Matricardi PM, Ahrne S, Orfei L, Aberg N, et al. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol (2007) 120(2):343–50. 10.1016/j.jaci.2007.05.018 [DOI] [PubMed] [Google Scholar]
- 62.West CE, Ryden P, Lundin D, Engstrand L, Tulic MK, Prescott SL. Gut microbiome and innate immune response patterns in IgE-associated eczema. Clin Exp Allergy (2015) 45(9):1419–29. 10.1111/cea.12566 [DOI] [PubMed] [Google Scholar]
- 63.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A (2010) 107(26):11971–5. 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kristensen K, Henriksen L. Cesarean section and disease associated with immune function. J Allergy Clin Immunol (2016) 137(2):587–90. 10.1016/j.jaci.2015.07.040 [DOI] [PubMed] [Google Scholar]
- 65.Kolokotroni O, Middleton N, Gavatha M, Lamnisos D, Priftis KN, Yiallouros PK. Asthma and atopy in children born by caesarean section: effect modification by family history of allergies – a population based cross-sectional study. BMC Pediatr (2012) 12:179. 10.1186/1471-2431-12-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karlstrom A, Lindgren H, Hildingsson I. Maternal and infant outcome after caesarean section without recorded medical indication: findings from a Swedish case-control study. BJOG (2013) 120(4):479–86; discussion 486. 10.1111/1471-0528.12129 [DOI] [PubMed] [Google Scholar]
- 67.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, et al. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med (2016) 22(3):250–3. 10.1038/nm.4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Storro O, Oien T, Langsrud O, Rudi K, Dotterud C, Johnsen R. Temporal variations in early gut microbial colonization are associated with allergen-specific immunoglobulin E but not atopic eczema at 2 years of age. Clin Exp Allergy (2011) 41(11):1545–54. 10.1111/j.1365-2222.2011.03817.x [DOI] [PubMed] [Google Scholar]
- 69.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med (2014) 6(237):237ra265. 10.1126/scitranslmed.3008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gosalbes MJ, Llop S, Valles Y, Moya A, Ballester F, Francino MP. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy (2013) 43(2):198–211. 10.1111/cea.12063 [DOI] [PubMed] [Google Scholar]
- 71.Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology (2011) 140(6):1713–9. 10.1053/j.gastro.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science (2011) 331(6015):337–41. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol (2001) 107(1):129–34. 10.1067/mai.2001.111237 [DOI] [PubMed] [Google Scholar]
- 74.Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, van Ree R, et al. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy (2006) 36(12):1602–8. 10.1111/j.1365-2222.2006.02599.x [DOI] [PubMed] [Google Scholar]
- 75.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut (2007) 56(5):661–7. 10.1136/gut.2006.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holt PG. The mechanism or mechanisms driving atopic asthma initiation: the infant respiratory microbiome moves to center stage. J Allergy Clin Immunol (2015) 136(1):15–22. 10.1016/j.jaci.2015.05.011 [DOI] [PubMed] [Google Scholar]
- 77.Smits HH, Hiemstra PS, Prazeres da Costa C, Ege M, Edwards M, Garn H, et al. Microbes and asthma: opportunities for intervention. J Allergy Clin Immunol (2016) 137(3):690–7. 10.1016/j.jaci.2016.01.004 [DOI] [PubMed] [Google Scholar]
- 78.Kloepfer KM, Lee WM, Pappas TE, Kang TJ, Vrtis RF, Evans MD, et al. Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J Allergy Clin Immunol (2014) 133(5):e1301–3. 10.1016/j.jaci.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beale J, Jayaraman A, Jackson DJ, Macintyre JD, Edwards MR, Walton RP, et al. Rhinovirus-induced IL-25 in asthma exacerbation drives type 2 immunity and allergic pulmonary inflammation. Sci Transl Med (2014) 6(256):256ra134. 10.1126/scitranslmed.3009124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soto-Quiros M, Avila L, Platts-Mills TA, Hunt JF, Erdman DD, Carper H, et al. High titers of IgE antibody to dust mite allergen and risk for wheezing among asthmatic children infected with rhinovirus. J Allergy Clin Immunol (2012) 129(6):1499–505.e5. 10.1016/j.jaci.2012.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hong JY, Bentley JK, Chung Y, Lei J, Steenrod JM, Chen Q, et al. Neonatal rhinovirus induces mucous metaplasia and airways hyperresponsiveness through IL-25 and type 2 innate lymphoid cells. J Allergy Clin Immunol (2014) 134(2):429–39. 10.1016/j.jaci.2014.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vangay P, Ward T, Gerber JS, Knights D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe (2015) 17(5):553–64. 10.1016/j.chom.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Loss G, Apprich S, Waser M, Kneifel W, Genuneit J, Buchele G, et al. The protective effect of farm milk consumption on childhood asthma and atopy: the GABRIELA study. J Allergy Clin Immunol (2011) 128(4):766–773.e4. 10.1016/j.jaci.2011.07.048 [DOI] [PubMed] [Google Scholar]
- 84.Lluis A, Depner M, Gaugler B, Saas P, Casaca VI, Raedler D, et al. Increased regulatory T-cell numbers are associated with farm milk exposure and lower atopic sensitization and asthma in childhood. J Allergy Clin Immunol (2014) 133(2):551–9. 10.1016/j.jaci.2013.06.034 [DOI] [PubMed] [Google Scholar]
- 85.Brick T, Schober Y, Bocking C, Pekkanen J, Genuneit J, Loss G, et al. omega-3 fatty acids contribute to the asthma-protective effect of unprocessed cow’s milk. J Allergy Clin Immunol (2016) 137(6):1699–706.e3. 10.1016/j.jaci.2015.10.042 [DOI] [PubMed] [Google Scholar]
- 86.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature (2016) 529(7585):212–5. 10.1038/nature16504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Backhed F. Crosstalk between Gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab (2015) 22(4):658–68. 10.1016/j.cmet.2015.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaliannan K, Wang B, Li XY, Kim KJ, Kang JX. A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia. Sci Rep (2015) 5:11276. 10.1038/srep11276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R, et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J (2016) 10(3):742–50. 10.1038/ismej.2015.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature (2013) 504(7480):451–5. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamada T, Takahashi D, Hase K. The diet-microbiota-metabolite axis regulates the host physiology. J Biochem (2016) 160(1):1–10. 10.1093/jb/mvw022 [DOI] [PubMed] [Google Scholar]
- 92.Lee SY, Yu J, Ahn KM, Kim KW, Shin YH, Lee KS, et al. Additive effect between IL-13 polymorphism and cesarean section delivery/prenatal antibiotics use on atopic dermatitis: a birth cohort study (COCOA). PLoS One (2014) 9(5):e96603. 10.1371/journal.pone.0096603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Metsala J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Prenatal and post-natal exposure to antibiotics and risk of asthma in childhood. Clin Exp Allergy (2015) 45(1):137–45. 10.1111/cea.12356 [DOI] [PubMed] [Google Scholar]
- 94.Metsala J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM. Mother’s and offspring’s use of antibiotics and infant allergy to cow’s milk. Epidemiology (2013) 24(2):303–9. 10.1097/EDE.0b013e31827f520f [DOI] [PubMed] [Google Scholar]
- 95.Azad MB, Konya T, Guttman DS, Field CJ, Sears MR, HayGlass KT, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy (2015) 45(3):632–43. 10.1111/cea.12487 [DOI] [PubMed] [Google Scholar]
- 96.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol (2008) 6(11):e280. 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One (2010) 5(3):e9836. 10.1371/journal.pone.0009836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol (2012) 30:759–95. 10.1146/annurev-immunol-020711-074937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell (2014) 158(4):705–21. 10.1016/j.cell.2014.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hirsch AG, Pollak J, Glass TA, Poulsen MN, Bailey-Davis L, Mowery J, et al. Early-life antibiotic use and subsequent diagnosis of food allergy and allergic diseases. Clin Exp Allergy (2017) 47(2):236–44. 10.1111/cea.12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lapin B, Piorkowski J, Ownby D, Wagner-Cassanova C, Freels S, Chavez N, et al. The relationship of early-life antibiotic use with asthma in at-risk children. J Allergy Clin Immunol (2014) 134(3):728–9. 10.1016/j.jaci.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sanders ME. Probiotics: definition, sources, selection, and uses. Clin Infect Dis (2008) 46(Suppl 2):S58–61. 10.1086/523341 [DOI] [PubMed] [Google Scholar]
- 103.Williams NT. Probiotics. Am J Health Syst Pharm (2010) 67(6):449–58. 10.2146/ajhp090168 [DOI] [PubMed] [Google Scholar]
- 104.Duchmann R, Kaiser I, Hermann E, Mayet W, Ewe K, Meyer zum Buschenfelde KH. Tolerance exists towards resident intestinal flora but is broken in active inflammatory bowel disease (IBD). Clin Exp Immunol (1995) 102(3):448–55. 10.1111/j.1365-2249.1995.tb03836.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang ML, Ponsonby AL, Orsini F, Tey D, Robinson M, Su EL, et al. Administration of a probiotic with peanut oral immunotherapy: a randomized trial. J Allergy Clin Immunol (2015) 135(3):737–744.e8. 10.1016/j.jaci.2014.11.034 [DOI] [PubMed] [Google Scholar]
- 106.Berni Canani R, Nocerino R, Terrin G, Frediani T, Lucarelli S, Cosenza L, et al. Formula selection for management of children with cow’s milk allergy influences the rate of acquisition of tolerance: a prospective multicenter study. J Pediatr (2013) 163(3):771–777.e1. 10.1016/j.jpeds.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 107.Hol J, van Leer EH, Elink Schuurman BE, de Ruiter LF, Samsom JN, Hop W, et al. The acquisition of tolerance toward cow’s milk through probiotic supplementation: a randomized, controlled trial. J Allergy Clin Immunol (2008) 121(6):1448–54. 10.1016/j.jaci.2008.03.018 [DOI] [PubMed] [Google Scholar]
- 108.Costa DJ, Marteau P, Amouyal M, Poulsen LK, Hamelmann E, Cazaubiel M, et al. Efficacy and safety of the probiotic Lactobacillus paracasei LP-33 in allergic rhinitis: a double-blind, randomized, placebo-controlled trial (GA2LEN Study). Eur J Clin Nutr (2014) 68(5):602–7. 10.1038/ejcn.2014.13 [DOI] [PubMed] [Google Scholar]
- 109.Ouwehand AC, Nermes M, Collado MC, Rautonen N, Salminen S, Isolauri E. Specific probiotics alleviate allergic rhinitis during the birch pollen season. World J Gastroenterol (2009) 15(26):3261–8. 10.3748/wjg.15.3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ivory K, Chambers SJ, Pin C, Prieto E, Arques JL, Nicoletti C. Oral delivery of Lactobacillus casei Shirota modifies allergen-induced immune responses in allergic rhinitis. Clin Exp Allergy (2008) 38(8):1282–9. 10.1111/j.1365-2222.2008.03025.x [DOI] [PubMed] [Google Scholar]
- 111.Medina M, Villena J, Salva S, Vintini E, Langella P, Alvarez S. Nasal administration of Lactococcus lactis improves local and systemic immune responses against Streptococcus pneumoniae. Microbiol Immunol (2008) 52(8):399–409. 10.1111/j.1348-0421.2008.00050.x [DOI] [PubMed] [Google Scholar]
- 112.Ivory K, Wilson AM, Sankaran P, Westwood M, McCarville J, Brockwell C, et al. Oral delivery of a probiotic induced changes at the nasal mucosa of seasonal allergic rhinitis subjects after local allergen challenge: a randomised clinical trial. PLoS One (2013) 8(11):e78650. 10.1371/journal.pone.0078650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cuello-Garcia CA, Brozek JL, Fiocchi A, Pawankar R, Yepes-Nunez JJ, Terracciano L, et al. Probiotics for the prevention of allergy: a systematic review and meta-analysis of randomized controlled trials. J Allergy Clin Immunol (2015) 136(4):952–61. 10.1016/j.jaci.2015.04.031 [DOI] [PubMed] [Google Scholar]
- 114.Zuccotti G, Meneghin F, Aceti A, Barone G, Callegari ML, Di Mauro A, et al. Probiotics for prevention of atopic diseases in infants: systematic review and meta-analysis. Allergy (2015) 70(11):1356–71. 10.1111/all.12700 [DOI] [PubMed] [Google Scholar]
- 115.Tokunaga T, Ninomiya T, Osawa Y, Imoto Y, Ito Y, Takabayashi T, et al. Factors associated with the development and remission of allergic diseases in an epidemiological survey of high school students in Japan. Am J Rhinol Allergy (2015) 29(2):94–9. 10.2500/ajra.2015.29.4135 [DOI] [PubMed] [Google Scholar]
- 116.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet (2001) 357(9262):1076–9. 10.1016/S0140-6736(00)04259-8 [DOI] [PubMed] [Google Scholar]
- 117.Elazab N, Mendy A, Gasana J, Vieira ER, Quizon A, Forno E. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics (2013) 132(3):e666–76. 10.1542/peds.2013-0246 [DOI] [PubMed] [Google Scholar]
- 118.Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science (2015) 349(6252):1106–10. 10.1126/science.aac6623 [DOI] [PubMed] [Google Scholar]
- 119.Shim JU, Lee SE, Hwang W, Lee C, Park JW, Sohn JH, et al. Flagellin suppresses experimental asthma by generating regulatory dendritic cells and T cells. J Allergy Clin Immunol (2016) 137(2):426–35. 10.1016/j.jaci.2015.07.010 [DOI] [PubMed] [Google Scholar]
- 120.Bouchaud G, Castan L, Chesne J, Braza F, Aubert P, Neunlist M, et al. Maternal exposure to GOS/inulin mixture prevents food allergies and promotes tolerance in offspring in mice. Allergy (2016) 71(1):68–76. 10.1111/all.12777 [DOI] [PubMed] [Google Scholar]
- 121.Arslanoglu S, Moro GE, Schmitt J, Tandoi L, Rizzardi S, Boehm G. Early dietary intervention with a mixture of prebiotic oligosaccharides reduces the incidence of allergic manifestations and infections during the first two years of life. J Nutr (2008) 138(6):1091–5. 10.1093/jn/138.6.1091 [DOI] [PubMed] [Google Scholar]
- 122.Kukkonen AK, Kuitunen M, Savilahti E, Pelkonen A, Malmberg P, Makela M. Airway inflammation in probiotic-treated children at 5 years. Pediatr Allergy Immunol (2011) 22(2):249–51. 10.1111/j.1399-3038.2010.01079.x [DOI] [PubMed] [Google Scholar]
- 123.Arslanoglu S, Moro GE, Boehm G, Wienz F, Stahl B, Bertino E. Early neutral prebiotic oligosaccharide supplementation reduces the incidence of some allergic manifestations in the first 5 years of life. J Biol Regul Homeost Agents (2012) 26(3 Suppl):49–59. [PubMed] [Google Scholar]
- 124.Osborn DA, Sinn JK. Prebiotics in infants for prevention of allergy. Cochrane Database Syst Rev (2013) (3):CD006474. 10.1002/14651858.CD006474.pub3 [DOI] [PubMed] [Google Scholar]
- 125.Chang YS, Trivedi MK, Jha A, Lin YF, Dimaano L, Garcia-Romero MT. Synbiotics for prevention and treatment of atopic dermatitis: a meta-analysis of randomized clinical trials. JAMA Pediatr (2016) 170(3):236–42. 10.1001/jamapediatrics.2015.3943 [DOI] [PubMed] [Google Scholar]
- 126.Tang ML, Lodge CJ. Examining the evidence for using synbiotics to treat or prevent atopic dermatitis. JAMA Pediatr (2016) 170(3):201–3. 10.1001/jamapediatrics.2015.4406 [DOI] [PubMed] [Google Scholar]