Abstract
Mitochondria are highly dynamic organelles undergoing coordinated cycles of fission and fusion, referred as ‘mitochondrial dynamics’, in order to maintain their shape, distribution and size. Their transient and rapid morphological adaptations are crucial for many cellular processes such as cell cycle, immunity, apoptosis and mitochondrial quality control. Mutations in the core machinery components and defects in mitochondrial dynamics have been associated with numerous human diseases. These dynamic transitions are mainly ensured by large GTPases belonging to the Dynamin family. Mitochondrial fission is a multi-step process allowing the division of one mitochondrion in two daughter mitochondria. It is regulated by the recruitment of the GTPase Dynamin-related protein 1 (Drp1) by adaptors at actin- and endoplasmic reticulum-mediated mitochondrial constriction sites. Drp1 oligomerization followed by mitochondrial constriction leads to the recruitment of Dynamin 2 to terminate membrane scission. Inner mitochondrial membrane constriction has been proposed to be an independent process regulated by calcium influx. Mitochondrial fusion is driven by a two-step process with the outer mitochondrial membrane fusion mediated by mitofusins 1 and 2 followed by inner membrane fusion, mediated by optic atrophy 1. In addition to the role of membrane lipid composition, several members of the machinery can undergo post-translational modifications modulating these processes. Understanding the molecular mechanisms controlling mitochondrial dynamics is crucial to decipher how mitochondrial shape meets the function and to increase the knowledge on the molecular basis of diseases associated with morphology defects. This article will describe an overview of the molecular mechanisms that govern mitochondrial fission and fusion in mammals.
Keywords: Dynamin family, ER-Actin, Mitochondrial dynamics, Molecular Mechanisms, Regulation
Introduction
For a long time, mitochondria have primarily been considered as the ‘powerhouse’ of the cell, producing the energy required for cell metabolism by oxidative phosphorylation (OXPHOS) [1,2]. It is now accepted that mitochondria are also involved in numerous other physiological processes such as programmed cell death, innate immunity, autophagy, redox signalling, calcium homeostasis and stem cells reprogramming [2–4]. Mitochondrial ultrastructure visualized by electron microscopy (EM) is characterized by a double membrane system. The outer mitochondrial membrane (OMM) faces the cytosol, and the inner mitochondrial membrane (IMM) protrudes into the mitochondrial matrix containing mitochondrial DNA (mtDNA). The compartment delimited by the IMM and the OMM is referred as the intermembrane space (IMS). However, the development of live cell imaging over the last 30 years has dramatically changed the concept of mitochondria being static and isolated structures. Indeed, mitochondria can modulate their morphology to create a tubular network coordinated by fission and fusion events. The balance between these two opposite processes regulates mitochondrial number, size and positioning within the cytoplasm and is referred as ‘mitochondrial dynamics’ [5].
Mitochondrial fission is characterized by the division of one mitochondrion into two daughter mitochondria, whereas mitochondrial fusion is the union of two mitochondria resulting in one mitochondrion. The deregulation of these spatio-temporal events results in either a fragmented network characterized by a large number of small round-shape mitochondria or a hyperfused network with elongated and highly connected mitochondria (Figure 1). These balanced dynamic transitions are not only required to ensure mitochondrial function but also to respond to cellular needs by adapting the network to nutrient availability and to the metabolic state of the cell [6]. Moreover, different morphological states are associated with multiple physiological and pathophysiological conditions [7]. Mitochondrial fragmentation is often linked to mitochondrial dysfunction as this morphological state predominates during elevated stress levels and cell death [8]. However, it is also observed in the phase G2/M of the cell cycle and is needed for mitochondrial motility, quality control and mtDNA inheritance [9,10]. Although still under debate, a fused mitochondrial network would allow matrix component distribution and stimulation of OXPHOS activity [11]. Mitochondrial elongation also confers protection against phagophore engulfment during autophagy triggered by nutrient starvation and is mainly associated with cell survival mechanisms [12].
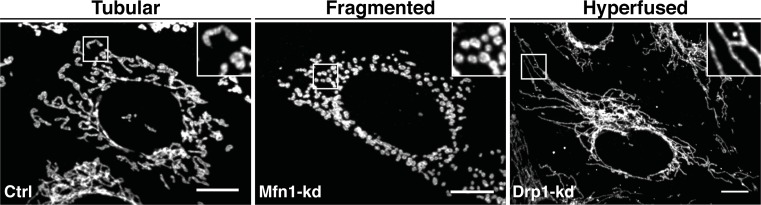
Figure 1. The mitochondrial morphology network.
Representative microscopy confocal images showing the different mitochondrial morphological aspects from control (Ctrl), Mfn1- and Drp1-Knockdown (Kd) mouse embryonic fibroblasts cells. Mitochondria are labelled with an anti-TOM20 antibody (OMM marker). Tubular, fragmented and hyperfused mitochondria are highlighted by zoomed areas (white squares); scale bars: 10 μm. Please note that the bright TOM20-positive structure in the zoom area of the Drp1-Kd is not a mitochondrial fragment but a mitochondria-derived vesicle [192].
The main proteins composing the core machinery are large GTPase proteins belonging to the Dynamin family (Figure 2). These mechanoenzymes can oligomerize and change conformation to drive membrane remodelling, constriction, scission and/or fusion [13]. Mitochondrial constriction and scission are carried out by the Dynamin-related/-like protein 1 (Drp1) and Dynamin2 (Dnm2), respectively [14]. Mitochondrial fusion is ensured by mitofusins 1 and 2 (Mfn1 and Mfn2) and optic atrophy 1 (OPA1), which mediate OMM and IMM fusion, respectively [15]. Knockout (KO) of either of these GTPases is embryonic lethal in mice and embryonic fibroblasts derived from these mice harbour drastic mitochondrial morphology defects [16–19] (except for the Dnm2-KO mouse where mitochondrial morphology has not been investigated). The relevance of mitochondrial dynamics has also been highlighted in humans where pathogenic mutations in genes corresponding to the core fission machinery (Drp1 [20], Dnm2 [21], MFF [22] and Mid49 [23]), fusion (Mfn2 [24] and OPA1 [25,26]), and other factors involved in these events (e.g. MSTO1 [27,28], GDAP1 [29] and SLC25A46 [30,31]) have been reported.
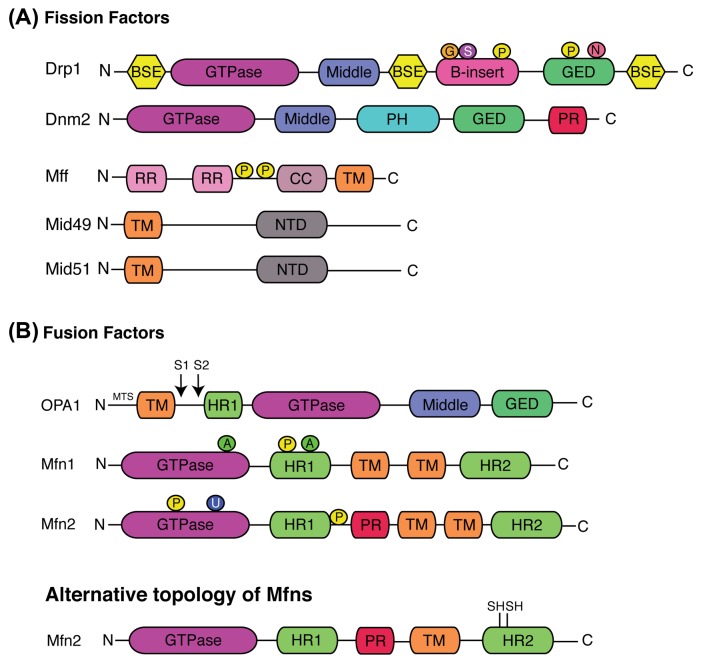
Figure 2. Schematic representation of the structural elements of the fission and fusion proteins, and their associated post-translational modifications.
Illustration of the core machinery proteins involved in (A) mitochondrial fission and (B) fusion. The classical model proposes that Mfns contain two transmembrane (TM) domains in between HR1 and HR2 domains. Alternatively, Mfns have been recently demonstrated to have only one TM that lies between the two HR domains. Cysteine residues, sensitive to oxidative stress are located in the C-terminal located in the IMS (only Mfn2 structural domains are represented but this new topology is also applicable to Mfn1). Domains are depicted in different colours. Identified location of post-translational modifications are indicated by P (Phosphorylation), N (S-nitrosylation), S (SUMOylation), G (O-GLcNAcylation), A (Acetylation) or U (Ubiquitination); BSE, bundle signalling elements; CC, coil-coil; GED, GTPase effector domain; HR, heptad repeat; MTS, mitochondrial targeting sequence; NTD, nucleotidyl transferase domains; PH, Pleckstrin homology; PR, Proline rich; RR, repeat regions; TM, transmembrane.
Together, this highlights the importance of understanding how mitochondrial morphology is regulated, in order to decipher how mitochondrial shape meets the function. In this article, we will present an overview of the recent proposed mechanisms regulating mitochondrial fission and fusion in mammals.
Molecular mechanisms of mitochondrial fusion
Mitofusins and outer mitochondrial membrane fusion
OMM fusion is ensured by the two large GTPases homologues Mfn1 and Mfn2 in mammals, which share approximately 80% sequence similarity in humans. The Mfns orthologue, fuzzy onion (Fzo1), was originally characterized in Drosophila melanogaster [32] and is conserved from yeast [33] to human [34]. Overexpression of either Mfns leads to mitochondrial aggregation around the nucleus [35]. While Mfn1-KO induces mitochondrial fragmentation, Mfn2-KO exhibits swollen spherical mitochondria [16]. This difference can be explained by the fact that Mfn1 has been shown to have a greater guanosine triphosphate (GTP)-dependent membrane tethering activity [36]. In addition, Mfn2 is not only involved in fusion but is also a key regulator of the mitochondria-endoplasmic reticulum (ER) contact sites tethering [37,38]. Nevertheless, overexpression of Mfn1 or Mfn2 in Mfn2-KO or Mfn1-KO MEF cells, respectively, can restore mitochondrial fusion [16]. Both proteins also accumulate at contact areas between two adjacent mitochondria [35] and establish homo or heterotypic complexes leading to mitochondrial fusion [39].
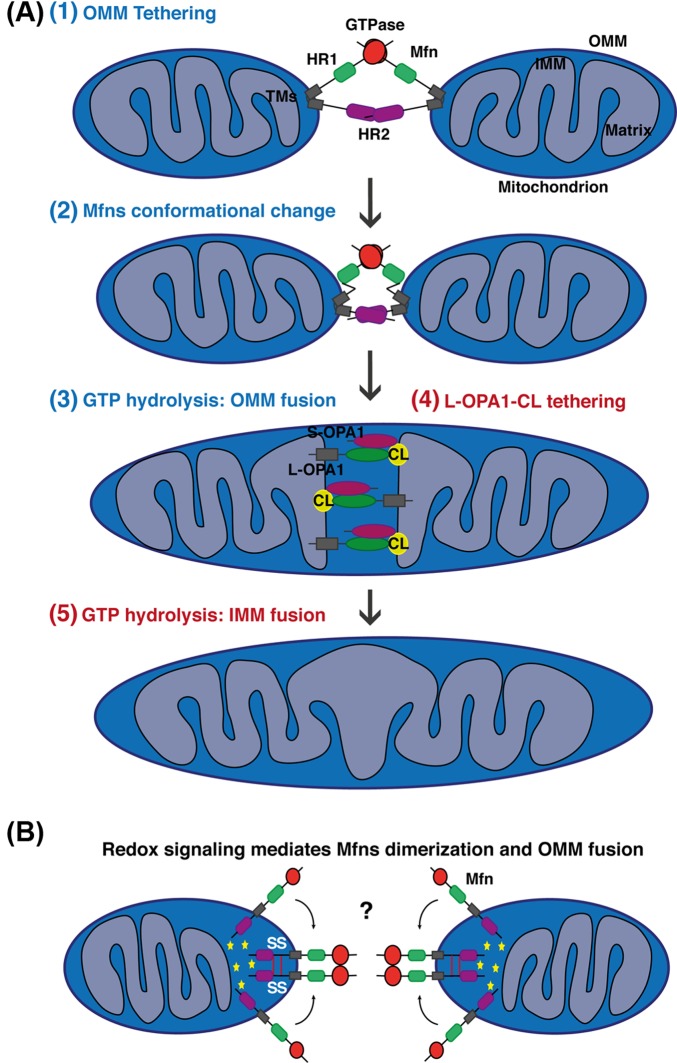
Globally, mitochondrial fusion is characterized by three different steps: the tethering of two mitochondria in trans, the docking of two membranes increasing the contact surface area and decreasing the distance between the two membranes [40], and finally the fusion of the two OMM due to conformational changes induced by GTP hydrolysis [36,41] (Figure 3).
Figure 3. Simplified models for mitochondrial fusion in mammals.
(A) Schematic representations of mitochondrial fusion, based on the Mfns topology suggesting two TM domains with both the HR1 and HR2 domains facing the cytosol. (1) The outer membrane of two opposing mitochondria are tethered by the interaction in trans of the HR2 and/or GTPase domains of Mfns. GTP binding or/and hydrolysis induce Mfns conformational change leading to mitochondrial docking and to an increase of membrane contact sites. For clarity reasons, not all of the recent suggested models leading to Mfns dimerization and conformational change are highlighted in the scheme. (3) Finally, GTPase-dependent power stroke or GTP-dependent oligomerization ensure OMM fusion. The composition of the OMM in phospholipids can also regulate this process. (4) Following OMM fusion, OPA1 and CL drive IMM fusion. The interaction between OPA1 and CL on either side of the membrane tethers the two IMM, which fuse following OPA1-depedent GTP hydrolysis (5). In this model, S-OPA1 has been shown to enhance OPA1–CL interaction and fusion. Please note that after OMM and IMM fusion, Mfn2 and OPA1, as membrane-bound proteins, are still present on the different membranes but are disassembled. (B) Schematic representations of OMM fusion based on the new metazoan Mfns topology suggesting only one TM placing the Mfn C-terminus in the IMS. Oxidized environment in the IMS (ROS production) and increase concentration of GSSG lead to the establishment of two disulphide bonds within the IMS domain. These redox-mediated disulphide modifications induce the dimerization and oligomerization of Mfns molecules which may promote tethering or GTPase activity required for OMM fusion. Interestingly, this redox-regulated Mfns oligomerization is a dynamic and reversible process. Yellow stars indicate an oxidized environment.
Over the last 15 years, the proposed mechanism of mitochondrial fusion by mitofusins has been based on their topology. Like yeast Fzo1 [42], it was accepted that Mfns were inserted in the OMM via two transmembrane (TM) domains separated by a short loop exposing their N-terminal region containing the GTPase and the coil-coil heptad repeat 1 (HR1) domains and their C-terminal harbouring the HR2 domain in the cytosol [34,43–45] (Figures 2 and 3A). Based on this model and some structural insights, the required mechanistic steps of fusion have been proposed (Figure 3A).
For example, it has been proposed that Mfns dimeric antiparallel trans interactions between apposing mitochondria are established via their HR2 domains, followed by GTP hydrolysis resulting in OMM fusion [44].
In contrast to the HR2 trans model, more recent structural studies conducted with a ‘minimal’ recombinant Mfn1 (internal deletion of the HR2 and generation of the predicted TM domains) revealed that the tethering is mediating through the GTPase domains [46,47]. The fusion of the adjacent membranes may then be ensured by a GTPase-dependent power stroke [47] or GTP-dependent oligomerization [46]. While crystal structures clearly reveal the GTPase binding in trans as a primary mechanism of tethering, a peptide that mimics the HR1 helix has also been shown to activate mitochondrial fusion [48]. These peptides, or smaller drugs that alter the conformation of HR1, increase mitochondrial fusion when added to cells. Based on modelling from the structures, the authors propose that these compounds interfere with HR1 binding to HR2, thereby opening the helix and promoting mitochondrial tethering and fusion [48]. These compounds were not tested in a direct mitochondrial fusion assay, so it remains possible that other mechanisms can explain their cellular effects. Finally, it has been recently proposed that the C-terminal tail of Mfn1, harbouring the HR2 domain, contains an amphipathic helix required for Mfn1 insertion and promoting mitochondrial fusion [49]. More recent work has shown that HR1, but not HR2, promotes liposome tethering and lipid mixing in reconstituted assays, hinting that this HR may destabilize the lipids to drive membrane fusion [50]. The idea that the Mfns form a larger fusion pore, visualized as a circular mitochondrial docking complex, has been suggested using cryo-electron microscopy in isolated yeast mitochondria [40]. This study has also revealed that the cycles of GTP hydrolysis are required to assemble this dynamic structure [40].
Importantly, the prediction that HR1 and HR2 both reside on the cytosolic side of the OMM was based on the established topology of the yeast orthologue Fzo1. However, this topology was recently challenged, forcing a reconsideration of the current models [51] (Figures 2 and 3B). Furthermore, phylogenetic analysis has revealed that Mfns from yeast and metazoans are highly divergent, with bioinformatics predicting a single TM domain in metazoan Mfns, with two membrane spanning regions in Mfns of the fungal clade. Classical biochemical experiments confirmed that human Mfns harbour only one TM domain, placing the ∼12 kDa C-terminal and the HR2 domain in the IMS [51]. Interestingly, this new topology has been functionally linked to the control of mitochondrial fusion by redox signalling. Indeed, two cysteines located in the HR2 domains can be oxidized by increased level of oxidized glutathione leading to the formation of disulphide bonds between two Mfns molecules and their oligomerization required for membrane fusion [51]. These results confirmed initial studies describing the role of reactive oxygen species and oxidative stress in promoting mitochondrial fusion [52–54]. This new topology is consistent with the observation that HR2 did not drive liposome tethering or fusion [50]. Given the previous assumptions that the HR2 domain resides in the cytosol, this new topology raises a number of outstanding questions, for example: Are the GTPase domain interactions in trans sufficient to tether two mitochondria with HR1 domain driving bilayer mixing? Do the compounds interfering with HR1 domain affect additional Mfn partners that may participate in fusion?
Together, these data highlight the requirement of a reappraisal of the current acknowledged models and further experiments based on this new model should be performed in the near future to confirm and shed light on the full mechanism of OMM fusion.
OPA1 and inner mitochondrial membrane fusion
IMM fusion occurs downstream of OMM fusion and is mediated by the large GTPase OPA1 and specific IMM lipid components. Indeed, genetic loss of OPA1 leads to mitochondrial fragmentation whereas OPA1 overexpression induces mitochondrial elongation [55]. OPA1, originally described in the yeast model (Mgm1p) [56], is evolutionary conserved and is a complex protein with eight identified splice-variants. Its protein domain organization shares similarities with ‘classical’ dynamins (Figure 2). It is inserted within the IMM via a ∼100 residues N-terminal matrix targeting signal followed by a TM domain, exposing the majority of the protein to the IMS [57]. Despite the role of the GTPase and GTPase effector domain (GED) domains for GTP hydrolysis, the specific roles of the different domains during fusion events are not well understood. OPA1 harbours at least two sites for proteolytic cleavage, which generate shorter and soluble fragments. These cleavages are mediated by two membrane-bound metalloproteases, OMA1 [58,59] and YME1L [60,61], cleaving the protein at S1 and S2 sites, respectively. This results in at least five OPA1 fragments detectable by immunoblot where the two higher molecular weight forms are referred as L-OPA1 and the three shorter as S-OPA1. The abundance of the different OPA1 isoforms is cellular context specific and affects mitochondrial dynamics regulation. Indeed, OMA1-dependent cleavage of OPA1 is a stress response, whereas stimulation of OXPHOS induces YME1L activity. It is interesting to note that a mild mitochondrial stress leads to a stress-induced mitochondrial hyperfusion (SIMH) mechanism regulated by the Stomatin-like protein 2, Mfn1 and OPA1 and acting as a pro-survival response [62].
Initial work has described the requirement of both L- and S-OPA1 isoforms to allow mitochondrial fusion since L-OPA1 and S-OPA1 alone have only little fusion activity [61]. However, recent studies have now shown that the L-OPA1 isoform alone is sufficient to drive fusion. Indeed, L-OPA1 accumulation drives fusion during SIMH [62] and is responsible for the mitochondrial hyperfusion observed in YME1L/OMA1-DKO cells [63]. The balance between OPA1 cleavage by OMA1 and YME1L plays a crucial role in fusion regulation and the precise role of the S-OPA1 generation is not perfectly understood [64]. Indeed, initial work has shown that S-OPA1 isoform is also able to induce membrane tubulation in a liposome assay [65] and stimulation of OXPHOS induces YME1L-dependent S-OPA1 generation leading to mitochondrial fusion [66]. In contrast, mitochondrial stress-induced OPA1 cleavage by OMA1 can lead to mitochondrial fragmentation [58,59].
In mammals, OPA1 localization in only one of the two opposing mitochondria is sufficient to drive the fusion of both membranes [67]. These findings have been recently confirmed and a new model for IMM fusion regulated by OPA1 and a particular phospholipid has been proposed [68] (Figure 3A). As described later in this article, membrane lipid composition and in particular the lipid, cardiolipin (CL), play a crucial role in membrane remodelling and dynamics. CL is a mitochondria specific negatively charged lipid mainly localized in the IMM and required for the assembly and stability of large protein complexes like mitochondrial contact site and cristae organizing system (MICOS) and OXPHOS complexes [69]. Incubation of recombinant L-OPA1 with reconstituted CL-containing liposomes leads to a heterotypic interaction between L-OPA1 and CL driving membrane fusion [68]. In this model, S-OPA1 facilitates OPA1-CL binding and membranes fusion, corroborating studies performed in yeast showing the requirement of both S- and L-OPA1 isoforms as well as cardiolipin for IMM fusion [70,71]. Interestingly, these data have been validated in cellulo using a cell fusion assay, and suggest that the presence of OPA1 and CL on either side of the membrane can promote fusion and represents the minimal IMM fusion machinery [68]. These results also suggest that the OPA1 homotypic interaction is not involved in IMM fusion but in cristae architecture control.
Finally, OPA1-dependent IMM fusion depends on Mfn1 but not Mfn2 [62,72]. This observation raises the possible communication between the two membranes during fusion and suggests a potential interaction of Mfn1 with OPA1, a hypothesis now more plausible based on the new Mfns topology [51].
Overall, because of the complex processing of OPA1 and the lack of 3D structure of the protein, the precise mode of action of OPA1 has remained elusive. Further studies are needed to fully establish the mechanism of IMM fusion.
Molecular mechanisms of mitochondrial fission
Drp1 and adaptors
Mitochondrial fission is a multi-step process where the recruitment of the large GTPase Drp1 plays a crucial role. Drp1 is evolutionary conserved and its role in mitochondrial division was initially described in Caenorhabditis elegans [73] and yeast [74,75] before being extensively studied in mammals [76]. It is mainly a cytosolic protein, which is dynamically recruited to mitochondrial and peroxisomal membranes where it oligomerizes and drives membrane constriction in a GTP-dependent manner [14]. Indeed, genetic loss of Drp1 leads to a drastic elongation of both mitochondria and peroxisomes [77] in multiple cell lines and a variety of animal models [18,19].
Drp1 is composed of four distinct domains, an N-terminal GTPase domain followed by the middle domain, variable domain (or B-insert) and the GED in C-terminal (Figure 2). Like ‘classical dynamins’, Drp1 also contains bundle signalling elements (BSE) and stalk regions, but does not harbour the pleckstrin homology (PH) domain or the proline and arginine rich domain (PRD) at the C-terminal. The BSEs connect the GTPase domain with the stalk domain allowing Drp1 binding to membranes and subsequently its oligomerization [78].
During mitochondrial division, Drp1 is recruited to the OMM where it forms a ring-like structure around mitochondria leading to the narrowing of the membrane [76,78,79] (Figure 4). Then, GTP hydrolysis enhances this membrane constriction [80] which marks a potential future site of mitochondrial scission. Assembly of Drp1 at the OMM is mediated by the central stalk (middle domain) forming Drp1-oligomeric helices starting at two different points of the membrane [78,81]. Interestingly, in contrast with the ‘common pathway’ where cytosolic Drp1 is directly recruited to a constriction site through its membrane-anchored adaptors, a ‘targeted equilibrium’ has been proposed. In this model, dimeric and oligomeric forms of Drp1 are in constant balance between the cytosol and mitochondria [82]. Mitochondria-bound Drp1 puncta can merge into a mature-sized Drp1 complex capable of moving laterally along the mitochondrial tubule, induce constriction and eventually fission [82].
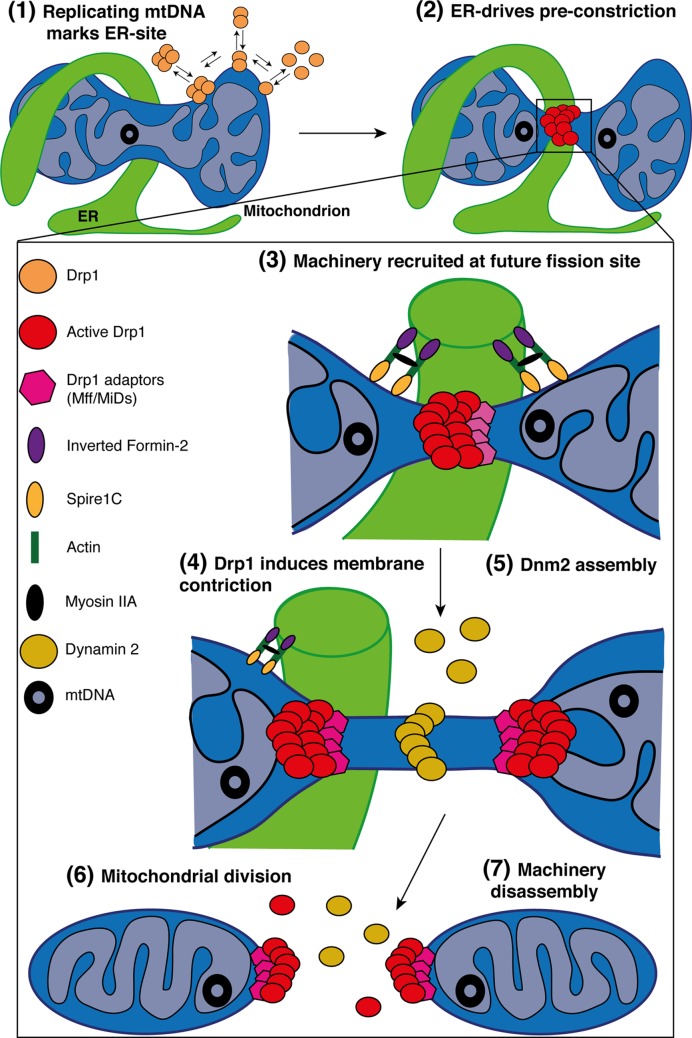
Figure 4. Simplified model for mitochondrial fission in mammals.
Schematic representation of the multi-step processes required for mitochondria division. (1) In the matrix, replication of the mtDNA marks the site for ER-recruitment. In parallel, Drp1 oligomers are in constant balance between the cytosol and mitochondria. In addition, IMM constriction occurs at mitochondria–ER contacts in a Ca2+-dependent process, before Drp1 oligomerization and maturation. (2) Oligomeric forms of Drp1 accumulate at ER-sites where the pre-constriction of the membrane has been initiated. (3) The zoomed area highlights the factors regulating mitochondrial division. The ER-bound INF2 and mitochondrial Spire1C induce actin nucleation and polymerization at mitochondria–ER contact sites. The Myosin IIa may ensure actin cable contraction, providing the mechanical force to drive mitochondria pre-constriction. At these sites, MFF and MiDs recruit Drp1 where it oligomerizes in a ring-like structure and (4) GTP-hydrolysis leads to conformational change, enhancing pre-existing mitochondrial constriction. The composition of the OMM in phospholipids also regulates Drp1 assembly and activity. (5) Then, Dnm2 is recruited to Drp1-mediated mitochondrial constriction neck where it assembles and terminates membrane scission, (6) leading to two daughter mitochondria. (7) The mechanisms of disassembly of the fission machinery following division remain unclear but both adaptors and Drp1 are found at both mitochondrial tips after division.
As Drp1 lacks a PH domain to bind membrane phospholipids directly, its recruitment at the OMM requires adaptors proteins. In the yeast model, Dnm1 (Drp1 orthologue) is recruited to the OMM via the membrane-anchored protein fis1 [83] and two receptors Mdv1 [84] and Caf4 [85]. However, there are no obvious orthologues for Mdv1 and Caf4 in mammals and recent studies suggest that Fis1 is not involved in the fission mechanism in basal conditions [86]. Instead, the tail-anchored proteins mitochondrial fission factor (MFF) [87] and mitochondrial dynamics proteins 49 and 51 (MiD49 and MiD51) [88,89] act as receptors for Drp1 in mammals (Figures 2 and 4). On one hand, overexpression of MFF leads to a fragmented network [90] whereas MFF genetic invalidation induces mitochondrial and peroxisome elongation [87,90], accompanied by a decrease in Drp1 mitochondrial recruitment. Indeed, MFF can specifically recruit high-oligomeric forms of Drp1 in cellulo [91] and stimulate its GTPase [92] activity enhancing membrane constriction in liposome assay. On the other hand, MiDs overexpression leads to mitochondrial elongation due to Drp1 sequestration [88,93], whereas low levels induce mitochondrial fragmentation [94]. MiD49/51-DKO phenocopies MFF-KO, characterized by mitochondrial hyperfusion and Drp1 recruitment defects, showing a potential redundancy between MiD49 and MiD51 [86,95]. MiDs harbour a nucleotidyltransferase domain and MiD51 requires ADP as a cofactor to stimulate Drp1 oligomerization and GTPase activity [96,97]. However, in vitro experiments using MiD51-bound liposomes demonstrated the capacity of MiD51 to inhibit Drp1 GTPase activity [95]. While these receptors colocalize together in discrete foci with Drp1 at ER-constriction sites [86,94], MFF and MiD49/51 can act independently on Drp1 recruitment and activity [98]. Therefore, it is assumed that MFF and MiDs have distinct but complementary roles in mitochondrial fission where MiDs recruit GTP-bound state of Drp1 to facilitate oligomerization whereas MFF selectively recruits oligomeric and active forms of Drp1. These functional differences have been highlighted recently during the cell death programme [86]. Further work would shed light on the precise mechanisms of Drp1 recruitment by these adaptors.
Final step of mitochondrial scission
Although the role Drp1 plays in membrane constriction is well described, its capacity to terminate fission has always been questioned. Recombinant Drp1 expression leads to liposome tubulation but not to their scission [99]. Moreover, cryo-EM imaging in yeast showed that the most representative diameter of Dnm1-lipid tubes constriction upon addition of GTP was 50–60 nm [100], which suggested that final scission required an additional process. Recently, the canonical protein Dnm2, initially involved in endocytic vesicle scission, has been proposed to catalyse this final step [101] (Figure 4). Like Drp1, Dnm2 is a GTPase that assembles in a collar-like structure around the constricting lipid ‘necks’ of budding membrane-bound vesicles [13]. Live-cell imaging experiments have shown that Dnm2 acts downstream of mitochondrial Drp1 activity, is transiently and specifically recruited to ER- and Drp1-induced constriction sites and leads to fission [101]. In addition, silencing Dnm2 induces mitochondrial elongation and the presence of highly narrow and elongated super-constriction sites. This phenotype was not rescued by re-expression of Dnm2 mutants lacking its GTPase, PH or PRD domains suggesting that activity, lipid binding and localization of Dnm2 are required for its role in mitochondrial division [101].
Mitochondrial division occurs at ER contact sites
A groundbreaking discovery in the mitochondrial dynamics field was the discovery that the ER was required for the initial step of mitochondrial division. Indeed, high-resolution and 3D reconstructed images acquired using EM and tomography have shown that not only ER tubules make contact with mitochondria but they can also wrap around them leading to mitochondrial constriction [102] (Figure 4). This pre-constriction step is required to decrease the average mitochondrial diameter from approximately 300–500 nm to approximately 150 nm [102], to allow Drp1-oligomeric ring formation. Therefore, Drp1 and its adaptors MFF and MiD49/51 are also specifically recruited to these mitochondria–ER contact sites prior to mitochondrial division [95,102,103]. With recent evidence implicating the role of phospholipids [104] and calcium transfer [105,106] during the process, it is tempting to suggest that these ER contact sites are not just required for mitochondrial pre-constriction but also represent a signalling platform for metabolite exchange, facilitating membrane remodelling and division.
The ER-bound inverted-formin 2 (INF2) and the mitochondrial anchored formin-binding Spire1C are both actin-nucleating proteins (Figure 4). Silencing either protein leads to mitochondrial elongation and defects in actin polymerization at the mitochondria–ER interface [107,108]. At these contact sites, INF2 cooperates with Spire1C to regulate actin assembly required for mitochondrial constriction before Drp1 recruitment and oligomerization [107,108]. In addition, Myosin IIA may ensure actin cable contraction providing the mechanical force for pre-constriction site formation [109]. Furthermore, transient F-actin bursts have been observed at mitochondria just before Drp1-dependent mitochondrial division [110,111] and other proteins involved in actin cytoskeleton regulation have been shown to regulate mitochondrial fission such as cofilin [110,112], cortactin [110], Arp2/3 [110] and Septin 2 [113]. Finally, Drp1 can bind F-actin in vitro which stimulates its oligomerization and its GTPase activity [114]. Together, the concomitant action of the ER and actin has clearly been identified as a crucial regulator of mitochondrial division and further studies will shed light on new potential regulators and their links with other members of the fission machinery.
Since the discovery of the role of the ER in mitochondrial division and the capacity of the oligomeric form of Drp1 to move along the tubules, it was unclear how the ER identifies and marks the sites for mitochondrial division. It had already been described that mitochondrial nucleoids were localized at mitochondria–ER contact sites in yeast [115] and mammal cells [116]. Recently, using high-resolution microscopy and live cell imaging, replicating mtDNA has specifically been spatially associated at mitochondria–ER contacts and constrictions, marking future mitochondrial fission sites [117] (Figure 4), allowing mtDNA distribution to the two newly generated mitochondria. This new observation describes mtDNA replication as one of the first steps of mitochondrial division raising new questions about the regulating mechanism and how this signal coming from the matrix is transmitted to the ER.
Constriction and division of the inner mitochondrial membrane
While the mechanisms regulating OMM constriction are well documented, the events leading to IMM constriction or division are poorly understood. Until now, no machinery has been associated with IMM division. EM analyses in C. elegans [73] and from rat cardiomyocytes [118] reported the potential presence of IMM constriction or division in the absence of either Drp1 or OMM constriction, which suggested that it could happen early during the mitochondrial division process. However, only very recently, an underlying mechanism for IMM constriction and possibly division has been proposed.
Coupling EM to super-resolution microscopy, two recent studies have suggested that IMM constriction is Ca2+-dependent and occurs at mitochondria–ER contact sites [105,106]. Stimulation of ER-induced calcium release to mitochondria leads to the constriction of the inner membrane compartment and may induce IMM division before Drp1 recruitment, therefore independently of OMM-constriction [105,106]. In addition, this phenomenon is inhibited by the loss of the mitochondrial calcium uniporter (MCU), which also leads to mitochondrial elongation. This is consistent with previous work suggesting a link between mitochondrial Ca2+ influx and mitochondrial fragmentation [119]. While in human Osteosarcoma cells (U2OS), IMM constriction has been attributed to the stimulation of mitochondria–ER contacts and mitochondrial calcium uptake by INF2-mediated actin polymerization [105], in neurons this mechanism is ensured by OPA1 processing [106]. Indeed, calcium entry in mitochondria induces a drop in mitochondrial membrane potential leading to the activation of OMA1 and the processing of OPA1 in S-OPA1. S-OPA1 accumulation disrupts the capacity of the MICOS complex to stabilize OMM–IMM tethering, leading to the IMM untethering and possibly constriction [106]. This proposed mode of action confirms the role previously described for S-OPA1 in fission. Indeed, S-OPA1 can localize at mitochondria–ER contact sites with the OMM fission machinery, but also overexpression enhances mitochondrial fission [120]. However, further studies need to be performed to decipher the players regulating IMM constriction and division and precisely incorporate these events in the global mitochondrial division process.
Additional layers of mitochondrial dynamics regulation
Membrane lipids composition in mitochondrial dynamics
Phospholipids are the major components of mitochondrial membranes and their role in membrane curvature, remodelling and regulation of mitochondrial dynamics has recently emerged. Mitochondrial membranes are mainly composed of phosphatidylcholine and phosphoethanolamine but also contain minor amounts of other phospholipids like phosphatidic acid (PA) and cardiolipin (CL), which play a major role in membrane remodelling. PA, a saturated lipid, is directly transferred from the ER to mitochondria, where it is converted into CL at the IMM [121]. A small amount of CL can be located to the OMM where CL can be converted into PA by the OMM C-anchored member of the phospholipase D family, mitoPLD [104].
Initial work has demonstrated that overexpression of mitoPLD triggered mitochondrial hyperfusion, whereas its ablation inhibited fusion and induced mitochondrial fragmentation [122]. Interestingly, hydrolysis of OMM-localized PA in diacylglycerol (DAG) or lysoPA by cytosolic mitochondrial recruited PA phosphatase (lepin 1b) [123] or phospholipase (PA-PLA1) [124], respectively, inhibits fusion induced by PA accumulation. While PA accumulation enhances Mfn1/2-dependent OMM fusion [122], CL stimulates OPA1 assembly and GTPase activity, subsequently leading to liposomes membrane tubulation [65]. As described earlier, CL plays a major role in the heterotypic interaction with OPA1, which stimulates fusion and represents the minimum machinery to drive inner membranes fusion [68].
Although Drp1 lacks a specific PH domain, it has been shown that it can interact with both CL and PA. Drp1 binding to CL via its B-insert domain drives oligomerization and stimulation of its GTPase activity enhancing constriction and tubulation of liposome membranes [92,125–129], designating CL at the OMM as a pro-fission phospholipid. Interestingly, Drp1 oligomerization and GTP hydrolysis can rearrange liposome membranes containing CL to create a constricted membrane region enriched in CL and favourable to scission [126]. In contrast, PA synthesis by the mitoPLD negatively regulates Drp1-dependent mitochondrial division [130]. In cellulo, Drp1 binds directly PA, via an unstructured loop in its stalk domain, at the OMM constriction sites leading to its oligomerization but to an inhibition of its GTPase activity, which results in mitochondrial hyperfusion [130,131]. Overall, these studies highlight the antagonistic roles of PA and CL microdomain formation in mitochondrial fission and fusion regulation.
Post-translational modifications of the core components
Post-translational modifications of the core protein machinery have been extensively studied in the last 10 years (Figure 2). Drp1 phosphorylation has been the most studied and phosphorylation at serine 616 and serine 637 are considered as pro-fission and pro-fusion forms, respectively. During mitosis, Drp1 is phosphorylated by cdk1/cyclin B kinase dependent on serine 616, stimulating its oligomerization, subsequently inducing mitochondrial fission and ensuring organelle distribution to daughter cells [132]. Drp1 can also be phosphorylated by other kinases at this residue during cell death by protein kinase C (PKC) [133] and the Ca2+-/calmodulin-dependent kinase II (CaMKII) [134,135] and by ERK-1/2 during cancer cell invasion [136,137] and cell reprograming [138]. On the other hand, protein kinase A, recruited to mitochondria through A kinase-anchoring protein 1 (AKAP1), phosphorylates Drp1 on residue 637 inhibiting fission and protecting mitochondria from autophagosomal degradation during nutrient deprivation [139] and cell death [140,141]. Dephosphorylation of this residue is carried out by the calcium-dependent phosphatase calcineurin during cell death [140,142,143] and PGAM5 during necrosis [144]. Finally, other kinases including Rho-associated coiled coil-containing protein kinase 1 (ROCK1) [145] and glycogen synthase kinase 3β (GSK3B) [146,147] can phosphorylate Drp1 and modulate mitochondrial morphology. In addition to phosphorylation, Drp1 can be dynamically SUMOylated/deSUMOylated on multiple non-consensus sites within the B domain controlling its stable association with the membrane, fission activity and cell death [148–153]. Drp1 can also be ubiquitinated by the RING-finger ubiquitin E3 ligase MARCH5/MITOL [154] and Parkin [155]. Finally, Drp1 activity can be controlled by S-nitrolysation [156–158] (but this regulation is still controversial) and O-GluNAcylation [159] modifications in its variable domain.
In addition, Drp1 receptors can also be regulated by post-translational modifications. Indeed, MFF is a substrate of the cellular energy sensor AMP-activated protein kinase (AMPK) upon mitochondrial dysfunction and a decrease in the cytosolic ATP/AMP ratio [160]. Phosphorylation of MFF enhances Drp1 recruitment, mitochondrial fission and damaged mitochondrial degradation [160]. Finally, MiD49 is also ubiquitinated by MARCH5/MITOL leading to its proteasomal degradation [161].
Post-translational modifications of mitochondrial fusion proteins are less documented. Indeed, the IMM OPA1 protein is regulated by proteolytic cleavage as already described, and only few modifications have been associated with Mfn1/Mfn2. The activity and stability of Mfn1 is regulated by ubiquitination and acetylation. MARCH5 ubiquitinates acetylated Mfn1 promoting its proteasomal degradation during mitochondrial stress [162]. During starvation, the protein deacetylase HDAC6 binds to and deacetylates Mfn1 enhancing fusion [163]. Finally, the phosphorylation of Mfn1 in the HR1 domain by the extracellular-signal-regulated kinase (ERK) inhibits mitochondrial fusion and promotes apoptosis [164].
Mfn2 can also be ubiquitinated by the HECT-type E3 ubiquitin-ligase Huwe1 [165], the RING-between RING type E3 ubiquitin-ligase Parkin [166], and the canonical RING-finger ligase MARCH5 [167] to control its activity and stability. Indeed, PINK1-phosphorylated Mfn2 can be ubiquitinated by Parkin leading to mitophagy [166] and JNK-phosphorylated Mfn2 can be ubiquitinated by Huwe1, which leads to its degradation, facilitating fragmentation and apoptosis [165].
Other proteins regulating mitochondrial dynamics
While most work has focused on the core GTPases that govern mitochondrial dynamics, additional factors have been identified that either directly or indirectly modify mitochondrial dynamics.
Ganglioside-induced differentiation associated protein 1 (GDAP1) and SLC25A46 are two proteins which can control mitochondrial fission. GDAP1 has been proposed to participate in fission upstream MFF and Drp1 [29]. SLC25A46 has been suggested to be the mammalian orthologue of the yeast Ugo1, a protein interacting with Fzo1 and Mgm1 to coordinate outer and inner membrane fusion processes in yeast [168,169]. However, loss of SLC25A46 in human cells leads to mitochondrial hyperfusion probably due to a deregulation of mitochondrial membrane phospholipids composition suggesting that the role of SLC25A46 seems to have evolved toward a pro-fission function [30,31]. Finally, additional evidence suggests that inner mitochondrial compartments may drive mitochondrial division. An IMM protein, mitochondrial fission process 1 (MTFP1), also called MTP18, has been involved in early step of mitochondrial division, upstream Drp1 activity [170,171]. Indeed, MTFP1 loss induces mitochondrial hyperfusion and a deregulation of Drp1 phosphorylation in an unknown mechanism [172]. This small, inner membrane protein was recently shown to be a translational target of mTOR, where protein expression was lost upon starvation or inhibition of mTOR, driving mitochondrial hyperfusion which promoted cell survival [172].
MSTO1 (Misato) is a cytoplasmic regulator of the OMM fusion machinery since it depletion leads to impaired fusion [27,28]. Finally, the reactive oxygen species modulator 1 (ROMO1) protein has been identified as a redox-regulated protein required for mitochondrial fusion and normal cristae morphology [53]. ROMO1, or Mgr2 in yeast, has a primary role in regulating the lateral release of membrane proteins transiting through the Tim23 channel during biogenesis [173]. However, under oxidative stress, ROMO1 is required for OPA1 oligomerization and ROMO1 silencing induces mitochondrial fission [53], reinforcing the interplay between redox signalling and the control of mitochondrial fusion. It is unclear whether this function in OPA1 dynamics directly relates to defects in biogenesis, or if it may have a secondary stress-related function in metazoans.
Mitochondrial dynamics: clinical syndromes
Pathogenic mutations in genes encoding the core fission and fusion machinery components have been linked to different severe human disorders, highlighting the physiological role of mitochondrial dynamics in cell homoeostasis (Table 1). These defects are mainly associated with neuromuscular and central nervous system (CNS) clinical syndromes and they are responsible for severe disabilities and progressive clinical course.
Table 1. Clinical syndromes due to mutations in genes encoding fission and fusion machinery components.
| Gene | OMIM | Inheritance | Disease | Symptoms | Refs |
|---|---|---|---|---|---|
| MFN2 | 608507 | AD | Charcot–Marie–Tooth disease type 2A | Distal limb muscle weakness and atrophy, axonal degeneration/regeneration, areflexia, distal sensory loss (pain and temperature more frequent) with or without: (a) CNS involvement (cognitive decline, spasticity, encephalopathy), (b) optic atrophy, (c) hearing loss and (d) vocal cord paresis | [24] |
| AR | Charcot–Marie–Tooth disease type 2A | [174] | |||
| AD | Hereditary motor and sensory neuropathy VIA | [178] | |||
| OPA1 | 605290 | AD | Optic atrophy 1 | Progressive loss of visual acuity, temporal optic nerve pallor, central scotoma with or without: (a) CNS (ataxia, spasticity, hearing loss) and (b) PNS (axonal sensorineural polyneuropathy) symptoms. | [26] |
| AD | Optic atrophy plus syndrome | [25] | |||
| AR | Behr syndrome | Early-onset optic atrophy accompanied by neurologic features, including ataxia, pyramidal signs, spasticity and mental retardation | [185] | ||
| MSTO1 | 617619 | AR/AD | Myopathy and ataxia | Hand and feet muscle weakness, growth impairment, fine tremor, cerebellar hypotrophy with or without: (a) white matter hyperintensities, (b) frontal lobe atrophy and (c) mental retardation | [27] [28] |
| DNM1L | 603850 | AR/AD | Encephalopathy | Abnormal brain development, seizures, hepatic dysfunction, encephalopathy, dysmorphism. | [20] [187] |
| AD | Optic atrophy 5 | Progressive loss of visual acuity, optic nerve atrophy and central scotoma | [188] | ||
| MFF | 614785 | AR | Encephalopathy | Seizures, dysphagia, optic and peripheral neuropathy, developmental delay, microcephaly, cerebellar atrophy and basal ganglia lesions | [22] |
| MIEF2 | 615498 | AR | Mitochondrial myopathy | Progressive muscle weakness, intermittent muscle pain and exercise intolerance | [23] |
| DNM2 | 602378 | AD | Centronuclear myopathy 1 | Slowly progressive muscle weakness. | [21] |
| AD | Charcot–Marie–Tooth disease, axonal type 2M | Distal limb muscle weakness and atrophy and sensory impairment, areflexia +/-neutropenia. | [180] | ||
| AD | Charcot–Marie–Tooth disease, dominant intermediate B | ||||
| AR | Lethal congenital contracture syndrome 5 | Polyhydramnios, decreased foetal movements, intracranial bleeding, retinal haemorrhage, joint contractures and respiratory insufficiency | |||
| SLC25A46 | 610826 | AR | Pontocerebellar hypoplasia type 1 | Early onset of optic atrophy, peripheral axonal sensorimotor neuropathy, ataxia, myoclonus, cerebellar atrophy, hypotonia with variable degree of severity, age at onset and association of symptoms | [189] |
| AR | Hereditary sensory motor neuropathy | [31] | |||
| AR | Optic atrophy spectrum disorders | [30] | |||
| GDAP1 | 606598 | AR | Charcot–Marie–Tooth disease type 4A | Distal limb muscle weakness and atrophy and sensory impairment, areflexia with or without: (a) axonal regeneration and (b) vocal cord paresis | [176] [175] |
| AR/AD | Charcot–Marie–Tooth disease type 2K | ||||
| AR | Charcot–Marie–Tooth disease type A | ||||
| AR | Charcot–Marie–Tooth disease with vocal cord paresis | ||||
| INF2 | 610982 | AD | Charcot–Marie–Tooth disease type E | Distal limb muscle weakness and atrophy and sensory impairment, areflexia, sensorineural hearing loss and foot drop | [177] |
| AD | Focal segmental glomerulosclerosis | Proteinuria and renal failure | [179] |
A non-exhaustive list of the diseases related to the principal identified mutations in genes encoding the core components of mitochondrial dynamics with associated symptoms. Abbreviations: AD, autosomal dominant; AR, autosomal recessive; CNS, central nervous system; OMIM, Online Mendelian Inheritance in Man®; PNS, peripheral nervous system.
Clinical genetics studies have identified pathogenic mutations in MFN2 [24,174], GDAP1 [175,176], INF2 [177] and DNM2 [180] as a cause of different types of Charcot–Marie–Tooth (CMT) disease, a clinically diverse group of inherited peripheral neuropathies. CMT diseases are characterized by degeneration of peripheral sensory and motor axons, causing distal sensory loss, muscle atrophy and weakness. In addition, MFN2 has also been associated with hereditary motor and sensory neuropathy VI, with optic atrophy and vocal cord paresis as potential additional symptoms [178]. Defects in the ER-associated fission protein INF2 can also cause renal focal segmental glomerulosclerosis [177,179] while DNM2 mutations have also been responsible of centronuclear myopathy [21] or lethal congenital contracture syndrome 5 [180].
Dominant mutations have been reported in OPA1, and associated with optic atrophy, the most common hereditary optic neuropathy [181]. This neuropathy is characterized by a loss of retinal ganglion cells in the optic nerve, leading to a gradual and progressive loss of vision [25,26]. It has been challenging to pinpoint the cause of the selective degradation of the optic nerve, because similarly to Mfn2, OPA1 also plays multiple roles, such as in cristae architecture and in apoptosis [182]. A subset of dominant mutations in the GTPase domain of OPA1, which is directly involved in IMM fusion, has been associated with dominant optic atrophy plus syndrome, defined by the development of additional symptoms such as deafness, ataxia and myopathy throughout adulthood [183,184]. Instead, recessive mutations in OPA1 cause Behr syndrome, a complex neurological disorder characterized by early-onset optic atrophy, ataxia, spasticity and mental retardation [185]. These three distinct clinical syndromes due to OPA1 mutations are not clearly explained by the underlying pathophysiology [186].
Mutations in pro-fusion gene MSTO1 cause mitochondrial myopathy and ataxia [27,28] while mutations in DRP1 lead to a severe neurological syndrome with microcephaly, abnormal brain development, optic atrophy and persistent lactic acidemia [20,187,188]. Furthermore, defects involving Drp1 adaptors, MFF [22] and MIEF2 [23], have been linked to human diseases. Indeed, patients harbouring mutations in MFF exhibit seizures, optic neuropathy and microcephaly [22]. Patients carrying mutations in MIEF2 suffer from myopathy, with complex I and complex IV activity deficiency in muscle [23]. Pathogenic mutations have also been associated with other regulators of mitochondrial fission. Indeed, patients carrying mutations in SLC25A46 have been reported to present clinically heterogeneous disorders, ranging from pontocerebellar hypoplasia [189], hereditary sensory motor neuropathy [31] to optic atrophy spectrum disorder [30].
Studies on in vivo and in vitro models with defective fission/fusion machinery components have been fundamental for shedding light on the crucial role of mitochondrial dynamics in the control of cell fate decisions. However, the clinical and genetic complexity of these disorders have not been explained yet and additional studies are required to improve our understanding on the molecular basis of diseases associated with mitochondrial dynamic defects.
Conclusions
Mitochondrial fusion and fission are crucial events and it is evident that these dynamic morphological transitions control cell fate decisions. Elucidating how these events are regulated, from a molecular but also biological point of view, represents a crucial step to the understanding of numerous human diseases. The discovery of new players which regulate these events is in constant evolution, from unexpected organelles [190] to key biological events [191], and that will continue in the following years with the development of novel microscopy technology and genetic tools.
Summary
Mitochondria are highly dynamic organelles that remodel their network in order to maintain their shape, distribution and size.
The balance between fission and fusion events modulates mitochondrial morphology depending on the metabolic needs of the cell.
The components of the core machinery regulating mitochondrial dynamics belong to the Dynamin family and these mechano-GTPases enzymes ensure these dynamic transitions.
Mitochondrial dynamics are controlled by additional layers of regulation including the ER–actin axis, membranes lipid composition and post-translational modifications of the key proteins.
Mitochondrial fission and fusion regulate numerous physiological functions and numerous diseases have reported abnormal mitochondrial morphology.
Acknowledgments
We thank Dr Caterina Garone for constructive and helpful remarks on the mitochondrial dynamics and clinical syndromes section.
Abbreviations
- ADP
adenosine diphosphate
- AMPK
AMP-dependent protein kinase
- BDLP
bacterial dynamin-like protein
- CaMKII
Ca2+-/calmodulin-dependent kinase II
- CL
cardiolipin
- Dnm2
Dynamin 2
- Drp1
Dynamin-related protein 1
- ER
endoplasmic reticulum
- GDAP1
ganglioside-induced differentiation associated protein 1
- GTP
guanosine triphosphate
- HR
heptad repeats
- IMM
inner mitochondrial membrane
- INF-2
inverted formin-2
- MFF
mitochondrial fission factor
- Mfn1
mitofusin 1
- Mfn2
mitofusin 2
- MiD49
mitochondrial dynamic protein 49
- MiD51
mitochondrial dynamic protein 51
- MSTO1
mitochondrial distribution and morphology regulator 1
- mtDNA
mitochondrial DNA
- MTFP1
mitochondrial fission process protein 1
- OMA1
overlapping with the M-AAA protease
- OMM
outer mitochondrial membrane
- OPA1
optic atrophy protein 1
- PA
phosphatidic acid
- PH
pleckstrin homology
- PKC
protein kinase C
- PR
proline rich
- ROMO
reactive oxygen species modulator
- TM
transmembrane
Funding
This work was supported by the Medical Research Council, UK (MC_ UU_00015/7). L.T. is an MRC-funded PhD student. S.N. is a recipient of a Daiichi Sankyo Foundation of Life Science postdoctoral fellowship, Japan. V.P. is supported by a Marie Skłodowska-Curie postdoctoral fellowship.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.McBride H.M., Neuspiel M. and Wasiak S. (2006) Mitochondria: more than just a powerhouse. Curr. Biol. 16, R551–R560 10.1016/j.cub.2006.06.054 [DOI] [PubMed] [Google Scholar]
- 2.Kamer K.J. and Mootha V.K. (2015) The molecular era of the mitochondrial calcium uniporter. Nat. Rev. Mol. Cell Biol. 16, 545–553 10.1038/nrm4039 [DOI] [PubMed] [Google Scholar]
- 3.Nikoletopoulou V., Markaki M., Palikaras K. and Tavernarakis N. (2013) Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta 1833, 3448–3459 10.1016/j.bbamcr.2013.06.001 [DOI] [PubMed] [Google Scholar]
- 4.Rambold A.S. and Pearce E.L. (2018) Mitochondrial dynamics at the interface of immune cell metabolism and function. Trends Immunol. 39, 6–18 10.1016/j.it.2017.08.006 [DOI] [PubMed] [Google Scholar]
- 5.Liesa M., Palacin M. and Zorzano A. (2009) Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 89, 799–845 10.1152/physrev.00030.2008 [DOI] [PubMed] [Google Scholar]
- 6.Wai T. and Langer T. (2016) Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 27, 105–117 10.1016/j.tem.2015.12.001 [DOI] [PubMed] [Google Scholar]
- 7.Nunnari J. and Suomalainen A. (2012) Mitochondria: in sickness and in health. Cell 148, 1145–1159 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zemirli N., Morel E. and Molino D. (2018) Mitochondrial dynamics in basal and stressful conditions. Int. J. Mol. Sci. 19, 10.3390/ijms19020564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otera H., Ishihara N. and Mihara K. (2013) New insights into the function and regulation of mitochondrial fission. Biochim. Biophys. Acta 1833, 1256–1268 10.1016/j.bbamcr.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 10.Pickles S., Vigie P. and Youle R.J. (2018) Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 28, R170–R185 10.1016/j.cub.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra P. and Chan D.C. (2016) Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212, 379–387 10.1083/jcb.201511036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rambold A.S., Kostelecky B., Elia N. and Lippincott-Schwartz J. (2011) Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. U.S.A. 108, 10190–10195 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson S.M. and De Camilli P. (2012) Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 13, 75–88 10.1038/nrm3266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraus F. and Ryan M.T. (2017) The constriction and scission machineries involved in mitochondrial fission. J. Cell Sci. 130, 2953–2960 10.1242/jcs.199562 [DOI] [PubMed] [Google Scholar]
- 15.Pernas L. and Scorrano L. (2016) Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 78, 505–531 10.1146/annurev-physiol-021115-105011 [DOI] [PubMed] [Google Scholar]
- 16.Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E. and Chan D.C. (2003) Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160, 189–200 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies V.J., Hollins A.J., Piechota M.J., Yip W., Davies J.R., White K.E. et al. (2007) Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum. Mol. Genet. 16, 1307–1318 10.1093/hmg/ddm079 [DOI] [PubMed] [Google Scholar]
- 18.Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S.O., Masuda K. et al. (2009) Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958–966 10.1038/ncb1907 [DOI] [PubMed] [Google Scholar]
- 19.Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T.W. et al. (2009) The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186, 805–816 10.1083/jcb.200903065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waterham H.R., Koster J., van Roermund C.W., Mooyer P.A., Wanders R.J. and Leonard J.V. (2007) A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 356, 1736–1741 10.1056/NEJMoa064436 [DOI] [PubMed] [Google Scholar]
- 21.Bitoun M., Maugenre S., Jeannet P.Y., Lacene E., Ferrer X., Laforet P. et al. (2005) Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat. Genet. 37, 1207–1209 10.1038/ng1657 [DOI] [PubMed] [Google Scholar]
- 22.Koch J., Feichtinger R.G., Freisinger P., Pies M., Schrodl F., Iuso A. et al. (2016) Disturbed mitochondrial and peroxisomal dynamics due to loss of MFF causes Leigh-like encephalopathy, optic atrophy and peripheral neuropathy. J. Med. Genet. 53, 270–278 10.1136/jmedgenet-2015-103500 [DOI] [PubMed] [Google Scholar]
- 23.Bartsakoulia M., Pyle A., Troncoso-Chandia D., Vial-Brizzi J., Paz-Fiblas M.V., Duff J. et al. (2018) A novel mechanism causing imbalance of mitochondrial fusion and fission in human myopathies. Hum. Mol. Genet. 27, 1186–1195 10.1093/hmg/ddy033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zuchner S., Mersiyanova I.V., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E.L. et al. (2004) Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 36, 449–451 10.1038/ng1341 [DOI] [PubMed] [Google Scholar]
- 25.Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A. et al. (2000) OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26, 211–215 10.1038/79944 [DOI] [PubMed] [Google Scholar]
- 26.Delettre C., Lenaers G., Griffoin J.M., Gigarel N., Lorenzo C., Belenguer P. et al. (2000) Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26, 207–210 10.1038/79936 [DOI] [PubMed] [Google Scholar]
- 27.Gal A., Balicza P., Weaver D., Naghdi S., Joseph S.K., Varnai P. et al. (2017) MSTO1 is a cytoplasmic pro-mitochondrial fusion protein, whose mutation induces myopathy and ataxia in humans. EMBO Mol. Med. 9, 967–984 10.15252/emmm.201607058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nasca A., Scotton C., Zaharieva I., Neri M., Selvatici R., Magnusson O.T. et al. (2017) Recessive mutations in MSTO1 cause mitochondrial dynamics impairment, leading to myopathy and ataxia. Hum. Mutat. 38, 970–977 10.1002/humu.23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niemann A., Ruegg M., La Padula V., Schenone A. and Suter U. (2005) Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J. Cell Biol. 170, 1067–1078 10.1083/jcb.200507087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abrams A.J., Hufnagel R.B., Rebelo A., Zanna C., Patel N., Gonzalez M.A. et al. (2015) Mutations in SLC25A46, encoding a UGO1-like protein, cause an optic atrophy spectrum disorder. Nat. Genet. 47, 926–932 10.1038/ng.3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Janer A., Prudent J., Paupe V., Fahiminiya S., Majewski J., Sgarioto N. et al. (2016) SLC25A46 is required for mitochondrial lipid homeostasis and cristae maintenance and is responsible for Leigh syndrome. EMBO Mol. Med. 8, 1019–1038 10.15252/emmm.201506159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hales K.G. and Fuller M.T. (1997) Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121–129 10.1016/S0092-8674(00)80319-0 [DOI] [PubMed] [Google Scholar]
- 33.Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J. et al. (1998) Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143, 359–373 10.1083/jcb.143.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santel A. and Fuller M.T. (2001) Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114, 867–874 [DOI] [PubMed] [Google Scholar]
- 35.Eura Y., Ishihara N., Yokota S. and Mihara K. (2003) Two mitofusin proteins, mammalian homologues of FZO, with distinct functions are both required for mitochondrial fusion. J. Biochem. 134, 333–344 10.1093/jb/mvg150 [DOI] [PubMed] [Google Scholar]
- 36.Ishihara N., Eura Y. and Mihara K. (2004) Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 117, 6535–6546 10.1242/jcs.01565 [DOI] [PubMed] [Google Scholar]
- 37.de Brito O.M. and Scorrano L. (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 10.1038/nature07534 [DOI] [PubMed] [Google Scholar]
- 38.Filadi R., Greotti E., Turacchio G., Luini A., Pozzan T. and Pizzo P. (2015) Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proc. Natl. Acad. Sci. U.S.A. 112, E2174–E2181 10.1073/pnas.1504880112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoppins S., Edlich F., Cleland M.M., Banerjee S., McCaffery J.M., Youle R.J. et al. (2011) The soluble form of Bax regulates mitochondrial fusion via MFN2 homotypic complexes. Mol. Cell 41, 150–160 10.1016/j.molcel.2010.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brandt T., Cavellini L., Kuhlbrandt W. and Cohen M.M. (2016) A mitofusin-dependent docking ring complex triggers mitochondrial fusion in vitro. Elife 5, e14618, 10.7554/eLife.14618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Legros F., Lombes A., Frachon P. and Rojo M. (2002) Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol. Biol. Cell 13, 4343–4354 10.1091/mbc.e02-06-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Low H.H., Sachse C., Amos L.A. and Lowe J. (2009) Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139, 1342–1352 10.1016/j.cell.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rojo M., Legros F., Chateau D. and Lombes A. (2002) Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 115, 1663–1674 [DOI] [PubMed] [Google Scholar]
- 44.Koshiba T., Detmer S.A., Kaiser J.T., Chen H., McCaffery J.M. and Chan D.C. (2004) Structural basis of mitochondrial tethering by mitofusin complexes. Science 305, 858–862 10.1126/science.1099793 [DOI] [PubMed] [Google Scholar]
- 45.Huang P., Galloway C.A. and Yoon Y. (2011) Control of mitochondrial morphology through differential interactions of mitochondrial fusion and fission proteins. PLoS One 6, e20655 10.1371/journal.pone.0020655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi Y., Yan L., Yu C., Guo X., Zhou X., Hu X. et al. (2016) Structures of human mitofusin 1 provide insight into mitochondrial tethering. J. Cell Biol. 215, 621–629 10.1083/jcb.201609019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao Y.L., Meng S., Chen Y., Feng J.X., Gu D.D., Yu B. et al. (2017) MFN1 structures reveal nucleotide-triggered dimerization critical for mitochondrial fusion. Nature 542, 372–376 10.1038/nature21077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franco A., Kitsis R.N., Fleischer J.A., Gavathiotis E., Kornfeld O.S., Gong G. et al. (2016) Correcting mitochondrial fusion by manipulating mitofusin conformations. Nature 540, 74–79 10.1038/nature20156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang X., Zhou X., Hu X., Joshi A.S., Guo X., Zhu Y. et al. (2017) Sequences flanking the transmembrane segments facilitate mitochondrial localization and membrane fusion by mitofusin. Proc. Natl. Acad. Sci. U.S.A. 114, E9863–E9872 10.1073/pnas.1708782114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daste F., Sauvanet C., Bavdek A., Baye J., Pierre F., Le Borgne R. et al. (2018) The heptad repeat domain 1 of Mitofusin has membrane destabilization function in mitochondrial fusion, EMBO Rep. 10.15252/embr.201643637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mattie S., Riemer J., Wideman J.G. and McBride H.M. (2018) A new mitofusin topology places the redox-regulated C terminus in the mitochondrial intermembrane space. J. Cell Biol. 217, 507–515 10.1083/jcb.201611194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shutt T., Geoffrion M., Milne R. and McBride H.M. (2012) The intracellular redox state is a core determinant of mitochondrial fusion. EMBO Rep. 13, 909–915 10.1038/embor.2012.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norton M., Ng A.C., Baird S., Dumoulin A., Shutt T., Mah N. et al. (2014) ROMO1 is an essential redox-dependent regulator of mitochondrial dynamics. Sci. Signal. 7, ra10 10.1126/scisignal.2004374 [DOI] [PubMed] [Google Scholar]
- 54.Thaher O., Wolf C., Dey P.N., Pouya A., Wullner V., Tenzer S. et al. (2017) The thiol switch C684 in Mitofusin-2 mediates redox-induced alterations of mitochondrial shape and respiration. Neurochem. Int. 10.1016/j.neuint.2017.05.009 [DOI] [PubMed] [Google Scholar]
- 55.Griparic L., van der Wel N.N., Orozco I.J., Peters P.J. and van der Bliek A.M. (2004) Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. J. Biol. Chem. 279, 18792–18798 10.1074/jbc.M400920200 [DOI] [PubMed] [Google Scholar]
- 56.Sesaki H., Southard S.M., Yaffe M.P. and Jensen R.E. (2003) Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell 14, 2342–2356 10.1091/mbc.e02-12-0788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olichon A., Emorine L.J., Descoins E., Pelloquin L., Brichese L., Gas N. et al. (2002) The human dynamin-related protein OPA1 is anchored to the mitochondrial inner membrane facing the inter-membrane space. FEBS Lett. 523, 171–176 10.1016/S0014-5793(02)02985-X [DOI] [PubMed] [Google Scholar]
- 58.Head B., Griparic L., Amiri M., Gandre-Babbe S. and van der Bliek A.M. (2009) Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 187, 959–966 10.1083/jcb.200906083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ehses S., Raschke I., Mancuso G., Bernacchia A., Geimer S., Tondera D. et al. (2009) Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 187, 1023–1036 10.1083/jcb.200906084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Griparic L., Kanazawa T. and van der Bliek A.M. (2007) Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 178, 757–764 10.1083/jcb.200704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song Z., Chen H., Fiket M., Alexander C. and Chan D.C. (2007) OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178, 749–755 10.1083/jcb.200704110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S. et al. (2009) SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28, 1589–1600 10.1038/emboj.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anand R., Wai T., Baker M.J., Kladt N., Schauss A.C., Rugarli E. et al. (2014) The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 204, 919–929 10.1083/jcb.201308006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacVicar T. and Langer T. (2016) OPA1 processing in cell death and disease - the long and short of it. J. Cell Sci. 129, 2297–2306 10.1242/jcs.159186 [DOI] [PubMed] [Google Scholar]
- 65.Ban T., Heymann J.A., Song Z., Hinshaw J.E. and Chan D.C. (2010) OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Hum. Mol. Genet. 19, 2113–2122 10.1093/hmg/ddq088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mishra P., Carelli V., Manfredi G. and Chan D.C. (2014) Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab. 19, 630–641 10.1016/j.cmet.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Song Z., Ghochani M., McCaffery J.M., Frey T.G. and Chan D.C. (2009) Mitofusins and OPA1 mediate sequential steps in mitochondrial membrane fusion. Mol. Biol. Cell 20, 3525–3532 10.1091/mbc.e09-03-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ban T., Ishihara T., Kohno H., Saita S., Ichimura A., Maenaka K. et al. (2017) Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 19, 856–863 10.1038/ncb3560 [DOI] [PubMed] [Google Scholar]
- 69.Cogliati S., Enriquez J.A. and Scorrano L. (2016) Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem. Sci. 41, 261–273 10.1016/j.tibs.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 70.DeVay R.M., Dominguez-Ramirez L., Lackner L.L., Hoppins S., Stahlberg H. and Nunnari J. (2009) Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186, 793–803 10.1083/jcb.200906098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rujiviphat J., Meglei G., Rubinstein J.L. and McQuibban G.A. (2009) Phospholipid association is essential for dynamin-related protein Mgm1 to function in mitochondrial membrane fusion. J. Biol. Chem. 284, 28682–28686 10.1074/jbc.M109.044933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cipolat S., Martins de Brito O., Dal Zilio B. and Scorrano L. (2004) OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U.S.A. 101, 15927–15932 10.1073/pnas.0407043101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labrousse A.M., Zappaterra M.D., Rube D.A. and van der Bliek A.M. (1999) C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol. Cell 4, 815–826 10.1016/S1097-2765(00)80391-3 [DOI] [PubMed] [Google Scholar]
- 74.Bleazard W., McCaffery J.M., King E.J., Bale S., Mozdy A., Tieu Q. et al. (1999) The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat. Cell Biol. 1, 298–304 10.1038/13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sesaki H. and Jensen R.E. (1999) Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J. Cell Biol. 147, 699–706 10.1083/jcb.147.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smirnova E., Griparic L., Shurland D.L. and van der Bliek A.M. (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell 12, 2245–2256 10.1091/mbc.12.8.2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Koch A., Thiemann M., Grabenbauer M., Yoon Y., McNiven M.A. and Schrader M. (2003) Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278, 8597–8605 10.1074/jbc.M211761200 [DOI] [PubMed] [Google Scholar]
- 78.Frohlich C., Grabiger S., Schwefel D., Faelber K., Rosenbaum E., Mears J. et al. (2013) Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 32, 1280–1292 10.1038/emboj.2013.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ingerman E., Perkins E.M., Marino M., Mears J.A., McCaffery J.M., Hinshaw J.E. et al. (2005) Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 170, 1021–1027 10.1083/jcb.200506078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mears J.A., Lackner L.L., Fang S., Ingerman E., Nunnari J. and Hinshaw J.E. (2011) Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 18, 20–26 10.1038/nsmb.1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koirala S., Guo Q., Kalia R., Bui H.T., Eckert D.M., Frost A. et al. (2013) Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc. Natl. Acad. Sci. U.S.A. 110, E1342–E1351 10.1073/pnas.1300855110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ji W.K., Hatch A.L., Merrill R.A., Strack S. and Higgs H.N. (2015) Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife 4, e11553 10.7554/eLife.11553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mozdy A.D., McCaffery J.M. and Shaw J.M. (2000) Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 151, 367–380 10.1083/jcb.151.2.367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tieu Q. and Nunnari J. (2000) Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J. Cell Biol. 151, 353–366 10.1083/jcb.151.2.353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Griffin E.E., Graumann J. and Chan D.C. (2005) The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J. Cell Biol. 170, 237–248 10.1083/jcb.200503148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Otera H., Miyata N., Kuge O. and Mihara K. (2016) Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J. Cell Biol. 212, 531–544 10.1083/jcb.201508099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gandre-Babbe S. and van der Bliek A.M. (2008) The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell 19, 2402–2412 10.1091/mbc.e07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Palmer C.S., Osellame L.D., Laine D., Koutsopoulos O.S., Frazier A.E. and Ryan M.T. (2011) MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 12, 565–573 10.1038/embor.2011.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Loson O.C., Song Z., Chen H. and Chan D.C. (2013) Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell 24, 659–667 10.1091/mbc.e12-10-0721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J. et al. (2010) Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 191, 1141–1158 10.1083/jcb.201007152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu R. and Chan D.C. (2015) The mitochondrial fission receptor Mff selectively recruits oligomerized Drp1. Mol. Biol. Cell 26, 4466–4477 10.1091/mbc.e15-08-0591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Macdonald P.J., Francy C.A., Stepanyants N., Lehman L., Baglio A., Mears J.A. et al. (2016) Distinct Splice Variants of Dynamin-related Protein 1 Differentially Utilize Mitochondrial Fission Factor as an Effector of Cooperative GTPase Activity. J. Biol. Chem. 291, 493–507 10.1074/jbc.M115.680181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao J., Liu T., Jin S., Wang X., Qu M., Uhlen P. et al. (2011) Human MIEF1 recruits Drp1 to mitochondrial outer membranes and promotes mitochondrial fusion rather than fission. EMBO J. 30, 2762–2778 10.1038/emboj.2011.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Elgass K.D., Smith E.A., LeGros M.A., Larabell C.A. and Ryan M.T. (2015) Analysis of ER-mitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J. Cell Sci. 128, 2795–2804 10.1242/jcs.169136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Osellame L.D., Singh A.P., Stroud D.A., Palmer C.S., Stojanovski D., Ramachandran R. et al. (2016) Cooperative and independent roles of the Drp1 adaptors Mff, MiD49 and MiD51 in mitochondrial fission. J. Cell Sci. 129, 2170–2181 10.1242/jcs.185165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Richter V., Palmer C.S., Osellame L.D., Singh A.P., Elgass K., Stroud D.A. et al. (2014) Structural and functional analysis of MiD51, a dynamin receptor required for mitochondrial fission. J. Cell Biol. 204, 477–486 10.1083/jcb.201311014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Loson O.C., Liu R., Rome M.E., Meng S., Kaiser J.T., Shan S.O. et al. (2014) The mitochondrial fission receptor MiD51 requires ADP as a cofactor. Structure 22, 367–377 10.1016/j.str.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Palmer C.S., Elgass K.D., Parton R.G., Osellame L.D., Stojanovski D. and Ryan M.T. (2013) Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J. Biol. Chem. 288, 27584–27593 10.1074/jbc.M113.479873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoon Y., Pitts K.R. and McNiven M.A. (2001) Mammalian dynamin-like protein DLP1 tubulates membranes. Mol. Biol. Cell 12, 2894–2905 10.1091/mbc.12.9.2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Naylor K., Ingerman E., Okreglak V., Marino M., Hinshaw J.E. and Nunnari J. (2006) Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J. Biol. Chem. 281, 2177–2183 10.1074/jbc.M507943200 [DOI] [PubMed] [Google Scholar]
- 101.Lee J.E., Westrate L.M., Wu H., Page C. and Voeltz G.K. (2016) Multiple dynamin family members collaborate to drive mitochondrial division. Nature 540, 139–143 10.1038/nature20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Friedman J.R., Lackner L.L., West M., DiBenedetto J.R., Nunnari J. and Voeltz G.K. (2011) ER tubules mark sites of mitochondrial division. Science 334, 358–362 10.1126/science.1207385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yamashita S.I., Jin X., Furukawa K., Hamasaki M., Nezu A., Otera H. et al. (2016) Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J. Cell Biol. 215, 649–665 10.1083/jcb.201605093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kameoka S., Adachi Y., Okamoto K., Iijima M. and Sesaki H. (2018) Phosphatidic Acid and Cardiolipin Coordinate Mitochondrial Dynamics. Trends Cell Biol. 28, 67–76 10.1016/j.tcb.2017.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chakrabarti R., Ji W.K., Stan R.V., de Juan Sanz J., Ryan T.A. and Higgs H.N. (2018) INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J. Cell Biol. 217, 251–268 10.1083/jcb.201709111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cho B., Cho H.M., Jo Y., Kim H.D., Song M., Moon C. et al. (2017) Constriction of the mitochondrial inner compartment is a priming event for mitochondrial division. Nat. Commun. 8, 15754 10.1038/ncomms15754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Manor U., Bartholomew S., Golani G., Christenson E., Kozlov M., Higgs H. et al. (2015) A mitochondria-anchored isoform of the actin-nucleating spire protein regulates mitochondrial division. Elife 4, e08828, 10.7554/eLife.08828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Korobova F., Ramabhadran V. and Higgs H.N. (2013) An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467 10.1126/science.1228360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Korobova F., Gauvin T.J. and Higgs H.N. (2014) A role for myosin II in mammalian mitochondrial fission. Curr. Biol. 24, 409–414 10.1016/j.cub.2013.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li S., Xu S., Roelofs B.A., Boyman L., Lederer W.J., Sesaki H. et al. (2015) Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J. Cell Biol. 208, 109–123 10.1083/jcb.201404050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moore A.S., Wong Y.C., Simpson C.L. and Holzbaur E.L. (2016) Dynamic actin cycling through mitochondrial subpopulations locally regulates the fission-fusion balance within mitochondrial networks. Nat. Commun. 7, 12886 10.1038/ncomms12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rehklau K., Hoffmann L., Gurniak C.B., Ott M., Witke W., Scorrano L. et al. (2017) Cofilin1-dependent actin dynamics control DRP1-mediated mitochondrial fission. Cell Death Dis. 8, e3063 10.1038/cddis.2017.448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pagliuso A., Tham T.N., Stevens J.K., Lagache T., Persson R., Salles A. et al. (2016) A role for septin 2 in Drp1-mediated mitochondrial fission. EMBO Rep. 17, 858–873 10.15252/embr.201541612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ji W.K., Chakrabarti R., Fan X., Schoenfeld L., Strack S. and Higgs H.N. (2017) Receptor-mediated Drp1 oligomerization on endoplasmic reticulum. J. Cell Biol. 216, 4123–4139 10.1083/jcb.201610057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Murley A., Lackner L.L., Osman C., West M., Voeltz G.K., Walter P. et al. (2013) ER-associated mitochondrial division links the distribution of mitochondria and mitochondrial DNA in yeast. Elife 2, e00422 10.7554/eLife.00422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ban-Ishihara R., Ishihara T., Sasaki N., Mihara K. and Ishihara N. (2013) Dynamics of nucleoid structure regulated by mitochondrial fission contributes to cristae reformation and release of cytochrome c. Proc. Natl. Acad. Sci. U.S.A. 110, 11863–11868 10.1073/pnas.1301951110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lewis S.C., Uchiyama L.F. and Nunnari J. (2016) ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353, aaf5549 10.1126/science.aaf5549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fujioka H., Tandler B. and Hoppel C.L. (2012) Mitochondrial division in rat cardiomyocytes: an electron microscope study. Anat. Rec. 295, 1455–1461 10.1002/ar.22523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hom J.R., Gewandter J.S., Michael L., Sheu S.S. and Yoon Y. (2007) Thapsigargin induces biphasic fragmentation of mitochondria through calcium-mediated mitochondrial fission and apoptosis. J. Cell. Physiol. 212, 498–508 10.1002/jcp.21051 [DOI] [PubMed] [Google Scholar]
- 120.Baker M.J., Tatsuta T. and Langer T. (2011) Quality control of mitochondrial proteostasis. Cold Spring Harb. Perspect. Biol. 3, a007559, 10.1101/cshperspect.a007559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vance J.E. (2015) Phospholipid synthesis and transport in mammalian cells. Traffic 16, 1–18 10.1111/tra.12230 [DOI] [PubMed] [Google Scholar]
- 122.Choi S.Y., Huang P., Jenkins G.M., Chan D.C., Schiller J. and Frohman M.A. (2006) A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis. Nat. Cell Biol. 8, 1255–1262 10.1038/ncb1487 [DOI] [PubMed] [Google Scholar]
- 123.Huang H., Gao Q., Peng X., Choi S.Y., Sarma K., Ren H. et al. (2011) piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev. Cell 20, 376–387 10.1016/j.devcel.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baba T., Kashiwagi Y., Arimitsu N., Kogure T., Edo A., Maruyama T. et al. (2014) Phosphatidic acid (PA)-preferring phospholipase A1 regulates mitochondrial dynamics. J. Biol. Chem. 289, 11497–11511 10.1074/jbc.M113.531921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bustillo-Zabalbeitia I., Montessuit S., Raemy E., Basanez G., Terrones O. and Martinou J.C. (2014) Specific interaction with cardiolipin triggers functional activation of Dynamin-related protein 1. PLoS One 9, e102738 10.1371/journal.pone.0102738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Stepanyants N., Macdonald P.J., Francy C.A., Mears J.A., Qi X. and Ramachandran R. (2015) Cardiolipin’s propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol. Biol. Cell 26, 3104–3116 10.1091/mbc.e15-06-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Macdonald P.J., Stepanyants N., Mehrotra N., Mears J.A., Qi X., Sesaki H. et al. (2014) A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol. Biol. Cell 25, 1905–1915 10.1091/mbc.e14-02-0728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ugarte-Uribe B., Muller H.M., Otsuki M., Nickel W. and Garcia-Saez A.J. (2014) Dynamin-related protein 1 (Drp1) promotes structural intermediates of membrane division. J. Biol. Chem. 289, 30645–30656 10.1074/jbc.M114.575779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Francy C.A., Alvarez F.J., Zhou L., Ramachandran R. and Mears J.A. (2015) The mechanoenzymatic core of dynamin-related protein 1 comprises the minimal machinery required for membrane constriction. J. Biol. Chem. 290, 11692–11703 10.1074/jbc.M114.610881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Adachi Y., Itoh K., Yamada T., Cerveny K.L., Suzuki T.L., Macdonald P. et al. (2016) Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol. Cell 63, 1034–1043 10.1016/j.molcel.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Adachi Y., Iijima M. and Sesaki H. (2017) An unstructured loop that is critical for interactions of the stalk domain of Drp1 with saturated phosphatidic acid. Small GTPases 1–8 10.1080/21541248.2017.1321614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Taguchi N., Ishihara N., Jofuku A., Oka T. and Mihara K. (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–11529 10.1074/jbc.M607279200 [DOI] [PubMed] [Google Scholar]
- 133.Qi X., Disatnik M.H., Shen N., Sobel R.A. and Mochly-Rosen D. (2011) Aberrant mitochondrial fission in neurons induced by protein kinase C{delta} under oxidative stress conditions in vivo. Mol. Biol. Cell 22, 256–265 10.1091/mbc.e10-06-0551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kim D.I., Lee K.H., Gabr A.A., Choi G.E., Kim J.S., Ko S.H. et al. (2016) Abeta-Induced Drp1 phosphorylation through Akt activation promotes excessive mitochondrial fission leading to neuronal apoptosis. Biochim. Biophys. Acta 1863, 2820–2834 10.1016/j.bbamcr.2016.09.003 [DOI] [PubMed] [Google Scholar]
- 135.Xu S., Wang P., Zhang H., Gong G., Gutierrez Cortes N., Zhu W. et al. (2016) CaMKII induces permeability transition through Drp1 phosphorylation during chronic beta-AR stimulation. Nat. Commun. 7, 13189 10.1038/ncomms13189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Serasinghe M.N., Wieder S.Y., Renault T.T., Elkholi R., Asciolla J.J., Yao J.L. et al. (2015) Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol. Cell 57, 521–536 10.1016/j.molcel.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kashatus J.A., Nascimento A., Myers L.J., Sher A., Byrne F.L., Hoehn K.L. et al. (2015) Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57, 537–551 10.1016/j.molcel.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Prieto J., Leon M., Ponsoda X., Sendra R., Bort R., Ferrer-Lorente R. et al. (2016) Early ERK1/2 activation promotes DRP1-dependent mitochondrial fission necessary for cell reprogramming. Nat. Commun. 7, 11124 10.1038/ncomms11124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gomes L.C., Di Benedetto G. and Scorrano L. (2011) During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13, 589–598 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cribbs J.T. and Strack S. (2007) Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 8, 939–944 10.1038/sj.embor.7401062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chang C.R. and Blackstone C. (2007) Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J. Biol. Chem. 282, 21583–21587 10.1074/jbc.C700083200 [DOI] [PubMed] [Google Scholar]
- 142.Cereghetti G.M., Stangherlin A., Martins de Brito O., Chang C.R., Blackstone C., Bernardi P. et al. (2008) Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc. Natl. Acad. Sci. U.S.A. 105, 15803–15808 10.1073/pnas.0808249105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Slupe A.M., Merrill R.A., Flippo K.H., Lobas M.A., Houtman J.C. and Strack S. (2013) A calcineurin docking motif (LXVP) in dynamin-related protein 1 contributes to mitochondrial fragmentation and ischemic neuronal injury. J. Biol. Chem. 288, 12353–12365 10.1074/jbc.M113.459677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Z., Jiang H., Chen S., Du F. and Wang X. (2012) The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 148, 228–243 10.1016/j.cell.2011.11.030 [DOI] [PubMed] [Google Scholar]
- 145.Wang W., Wang Y., Long J., Wang J., Haudek S.B., Overbeek P. et al. (2012) Mitochondrial fission triggered by hyperglycemia is mediated by ROCK1 activation in podocytes and endothelial cells. Cell Metab. 15, 186–200 10.1016/j.cmet.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chou C.H., Lin C.C., Yang M.C., Wei C.C., Liao H.D., Lin R.C. et al. (2012) GSK3beta-mediated Drp1 phosphorylation induced elongated mitochondrial morphology against oxidative stress. PLoS One 7, e49112 10.1371/journal.pone.0049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Yan J., Liu X.H., Han M.Z., Wang Y.M., Sun X.L., Yu N. et al. (2015) Blockage of GSK3beta-mediated Drp1 phosphorylation provides neuroprotection in neuronal and mouse models of Alzheimer’s disease. Neurobiol. Aging 36, 211–227 10.1016/j.neurobiolaging.2014.08.005 [DOI] [PubMed] [Google Scholar]
- 148.Braschi E., Zunino R. and McBride H.M. (2009) MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 10, 748–754 10.1038/embor.2009.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wasiak S., Zunino R. and McBride H.M. (2007) Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J. Cell Biol. 177, 439–450 10.1083/jcb.200610042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zunino R., Braschi E., Xu L. and McBride H.M. (2009) Translocation of SenP5 from the nucleoli to the mitochondria modulates DRP1-dependent fission during mitosis. J. Biol. Chem. 284, 17783–17795 10.1074/jbc.M901902200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Prudent J., Zunino R., Sugiura A., Mattie S., Shore G.C. and McBride H.M. (2015) MAPL SUMOylation of Drp1 stabilizes an ER/mitochondrial platform required for cell death. Mol. Cell 59, 941–955 10.1016/j.molcel.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 152.Fu J., Yu H.M., Chiu S.Y., Mirando A.J., Maruyama E.O., Cheng J.G. et al. (2014) Disruption of SUMO-specific protease 2 induces mitochondria mediated neurodegeneration. PLos Genet. 10, e1004579 10.1371/journal.pgen.1004579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Guo C., Hildick K.L., Luo J., Dearden L., Wilkinson K.A. and Henley J.M. (2013) SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia. EMBO J. 32, 1514–1528 10.1038/emboj.2013.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Karbowski M., Neutzner A. and Youle R.J. (2007) The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J. Cell Biol. 178, 71–84 10.1083/jcb.200611064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang H., Song P., Du L., Tian W., Yue W., Liu M. et al. (2011) Parkin ubiquitinates Drp1 for proteasome-dependent degradation: implication of dysregulated mitochondrial dynamics in Parkinson disease. J. Biol. Chem. 286, 11649–11658 10.1074/jbc.M110.144238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Cho D.H., Nakamura T., Fang J., Cieplak P., Godzik A., Gu Z. et al. (2009) S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science 324, 102–105 10.1126/science.1171091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Bossy B., Petrilli A., Klinglmayr E., Chen J., Lutz-Meindl U., Knott A.B. et al. (2010) S-Nitrosylation of DRP1 does not affect enzymatic activity and is not specific to Alzheimer’s disease. J. Alzheimers Dis. 20, S513–S526 10.3233/JAD-2010-100552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Haun F., Nakamura T., Shiu A.D., Cho D.H., Tsunemi T., Holland E.A. et al. (2013) S-nitrosylation of dynamin-related protein 1 mediates mutant huntingtin-induced mitochondrial fragmentation and neuronal injury in Huntington’s disease. Antioxid. Redox Signal. 19, 1173–1184 10.1089/ars.2012.4928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Gawlowski T., Suarez J., Scott B., Torres-Gonzalez M., Wang H., Schwappacher R. et al. (2012) Modulation of dynamin-related protein 1 (DRP1) function by increased O-linked-beta-N-acetylglucosamine modification (O-GlcNAc) in cardiac myocytes. J. Biol. Chem. 287, 30024–30034 10.1074/jbc.M112.390682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Toyama E.Q., Herzig S., Courchet J., Lewis T.L. Jr, Loson O.C., Hellberg K. et al. (2016) Metabolism. AMP-activated protein kinase mediates mitochondrial fission in response to energy stress. Science 351, 275–281 10.1126/science.aab4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xu S., Cherok E., Das S., Li S., Roelofs B.A., Ge S.X. et al. (2016) Mitochondrial E3 ubiquitin ligase MARCH5 controls mitochondrial fission and cell sensitivity to stress-induced apoptosis through regulation of MiD49 protein. Mol. Biol. Cell 27, 349–359 10.1091/mbc.e15-09-0678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Park Y.Y., Nguyen O.T., Kang H. and Cho H. (2014) MARCH5-mediated quality control on acetylated Mfn1 facilitates mitochondrial homeostasis and cell survival. Cell Death Dis. 5, e1172 10.1038/cddis.2014.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lee J.Y., Kapur M., Li M., Choi M.C., Choi S., Kim H.J. et al. (2014) MFN1 deacetylation activates adaptive mitochondrial fusion and protects metabolically challenged mitochondria. J. Cell Sci. 127, 4954–4963 10.1242/jcs.157321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Pyakurel A., Savoia C., Hess D. and Scorrano L. (2015) Extracellular regulated kinase phosphorylates mitofusin 1 to control mitochondrial morphology and apoptosis. Mol. Cell 58, 244–254 10.1016/j.molcel.2015.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Leboucher G.P., Tsai Y.C., Yang M., Shaw K.C., Zhou M., Veenstra T.D. et al. (2012) Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol. Cell 47, 547–557 10.1016/j.molcel.2012.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen Y. and Dorn G.W. II (2013) PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 340, 471–475 10.1126/science.1231031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sugiura A., Nagashima S., Tokuyama T., Amo T., Matsuki Y., Ishido S. et al. (2013) MITOL regulates endoplasmic reticulum-mitochondria contacts via Mitofusin2. Mol. Cell 51, 20–34 10.1016/j.molcel.2013.04.023 [DOI] [PubMed] [Google Scholar]
- 168.Sesaki H. and Jensen R.E. (2004) Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J. Biol. Chem. 279, 28298–28303 10.1074/jbc.M401363200 [DOI] [PubMed] [Google Scholar]
- 169.Hoppins S., Horner J., Song C., McCaffery J.M. and Nunnari J. (2009) Mitochondrial outer and inner membrane fusion requires a modified carrier protein. J. Cell Biol. 184, 569–581 10.1083/jcb.200809099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tondera D., Santel A., Schwarzer R., Dames S., Giese K., Klippel A. et al. (2004) Knockdown of MTP18, a novel phosphatidylinositol 3-kinase-dependent protein, affects mitochondrial morphology and induces apoptosis. J. Biol. Chem. 279, 31544–31555 10.1074/jbc.M404704200 [DOI] [PubMed] [Google Scholar]
- 171.Tondera D., Czauderna F., Paulick K., Schwarzer R., Kaufmann J. and Santel A. (2005) The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J. Cell Sci. 118, 3049–3059 10.1242/jcs.02415 [DOI] [PubMed] [Google Scholar]
- 172.Morita M., Prudent J., Basu K., Goyon V., Katsumura S., Hulea L. et al. (2017) mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol. Cell 67, 922e5–935e5 10.1016/j.molcel.2017.08.013 [DOI] [PubMed] [Google Scholar]
- 173.Gebert M., Schrempp S.G., Mehnert C.S., Heisswolf A.K., Oeljeklaus S., Ieva R. et al. (2012) Mgr2 promotes coupling of the mitochondrial presequence translocase to partner complexes. J. Cell Biol. 197, 595–604 10.1083/jcb.201110047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kijima K., Numakura C., Izumino H., Umetsu K., Nezu A., Shiiki T. et al. (2005) Mitochondrial GTPase mitofusin 2 mutation in Charcot-Marie-Tooth neuropathy type 2A. Hum. Genet. 116, 23–27 10.1007/s00439-004-1199-2 [DOI] [PubMed] [Google Scholar]
- 175.Cuesta A., Pedrola L., Sevilla T., Garcia-Planells J., Chumillas M.J., Mayordomo F. et al. (2002) The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat. Genet. 30, 22–25 10.1038/ng798 [DOI] [PubMed] [Google Scholar]
- 176.Baxter R.V., Ben Othmane K., Rochelle J.M., Stajich J.E., Hulette C., Dew-Knight S. et al. (2002) Ganglioside-induced differentiation-associated protein-1 is mutant in Charcot-Marie-Tooth disease type 4A/8q21. Nat. Genet. 30, 21–22 10.1038/ng796 [DOI] [PubMed] [Google Scholar]
- 177.Boyer O., Nevo F., Plaisier E., Funalot B., Gribouval O., Benoit G. et al. (2011) INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N. Engl. J. Med. 365, 2377–2388 10.1056/NEJMoa1109122 [DOI] [PubMed] [Google Scholar]
- 178.Voo I., Allf B.E., Udar N., Silva-Garcia R., Vance J. and Small K.W. (2003) Hereditary motor and sensory neuropathy type VI with optic atrophy. Am. J. Ophthalmol. 136, 670–677 10.1016/S0002-9394(03)00390-8 [DOI] [PubMed] [Google Scholar]
- 179.Boyer O., Benoit G., Gribouval O., Nevo F., Tete M.J., Dantal J. et al. (2011) Mutations in INF2 are a major cause of autosomal dominant focal segmental glomerulosclerosis. J. Am. Soc. Nephrol. 22, 239–245 10.1681/ASN.2010050518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zuchner S., Noureddine M., Kennerson M., Verhoeven K., Claeys K., De Jonghe P. et al. (2005) Mutations in the pleckstrin homology domain of dynamin 2 cause dominant intermediate Charcot-Marie-Tooth disease. Nat. Genet. 37, 289–294 10.1038/ng1514 [DOI] [PubMed] [Google Scholar]
- 181.Ferre M., Amati-Bonneau P., Tourmen Y., Malthiery Y. and Reynier P. (2005) eOPA1: an online database for OPA1 mutations. Hum. Mutat. 25, 423–428 10.1002/humu.20161 [DOI] [PubMed] [Google Scholar]
- 182.Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T. et al. (2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126, 177–189 10.1016/j.cell.2006.06.025 [DOI] [PubMed] [Google Scholar]
- 183.Hudson G., Amati-Bonneau P., Blakely E.L., Stewart J.D., He L., Schaefer A.M. et al. (2008) Mutation of OPA1 causes dominant optic atrophy with external ophthalmoplegia, ataxia, deafness and multiple mitochondrial DNA deletions: a novel disorder of mtDNA maintenance. Brain 131, 329–337 10.1093/brain/awm272 [DOI] [PubMed] [Google Scholar]
- 184.Amati-Bonneau P., Valentino M.L., Reynier P., Gallardo M.E., Bornstein B., Boissiere A. et al. (2008) OPA1 mutations induce mitochondrial DNA instability and optic atrophy ‘plus’ phenotypes. Brain 131, 338–351 10.1093/brain/awm298 [DOI] [PubMed] [Google Scholar]
- 185.Yu-Wai-Man P., Griffiths P.G., Gorman G.S., Lourenco C.M., Wright A.F., Auer-Grumbach M. et al. (2010) Multi-system neurological disease is common in patients with OPA1 mutations. Brain 133, 771–786 10.1093/brain/awq007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Olichon A., Guillou E., Delettre C., Landes T., Arnaune-Pelloquin L., Emorine L.J. et al. (2006) Mitochondrial dynamics and disease, OPA1. Biochim. Biophys. Acta 1763, 500–509 10.1016/j.bbamcr.2006.04.003 [DOI] [PubMed] [Google Scholar]
- 187.Yoon G., Malam Z., Paton T., Marshall C.R., Hyatt E., Ivakine Z. et al. (2016) Lethal Disorder of Mitochondrial Fission Caused by Mutations in DNM1L. J. Pediatr. 171, 313-6 e1-2 10.1016/j.jpeds.2015.12.060 [DOI] [PubMed] [Google Scholar]
- 188.Vanstone J.R., Smith A.M., McBride S., Naas T., Holcik M., Antoun G. et al. (2016) DNM1L-related mitochondrial fission defect presenting as refractory epilepsy. Eur. J. Hum. Genet. 24, 1084–1088 10.1038/ejhg.2015.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Braunisch M.C., Gallwitz H., Abicht A., Diebold I., Holinski-Feder E., Van Maldergem L. et al. (2018) Extension of the phenotype of biallelic loss-of-function mutations in SLC25A46 to the severe form of pontocerebellar hypoplasia type I. Clin. Genet. 93, 255–265 10.1111/cge.13084 [DOI] [PubMed] [Google Scholar]
- 190.Wong Y.C., Ysselstein D. and Krainc D. (2018) Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature 554, 382–386 10.1038/nature25486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Schmitt K., Grimm A., Dallmann R., Oettinghaus B., Restelli L.M., Witzig M. et al. (2018) Circadian control of DRP1 activity regulates mitochondrial dynamics and bioenergetics. Cell Metab. 27, 657e5–666e5 10.1016/j.cmet.2018.01.011 [DOI] [PubMed] [Google Scholar]
- 192.Sugiura A., McLelland G.L., Fon E.A. and McBride H.M. (2014) A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 33, 2142–2156 10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]