Abstract
In the chicken genome there are two closely-linked genes, U4B and U4X, that code for different sequence variants of U4 small nuclear RNA (snRNA). Both genes are expressed with nearly equal efficiency in the early embryo, but U4X gene expression is specifically down-regulated relative to U4B as development proceeds. At the present time, little is known about the mechanisms that regulate differential expression of snRNA genes. We have now identified a novel chicken factor, PPBF, that binds sequence-specifically in vitro to the proximal regulatory region of the U4X gene, but not to the proximal region of the U4B gene. PPBF is itself regulated during development and may therefore be a key factor involved in differentially regulating U4X gene transcription relative to U4B. The U4X and U4B enhancers contain distinct sequence variants of two essential motifs (octamer and SPH). The Oct-1 transcription factor binds with similar affinities to both the U4X and U4B octamer motifs. However, a second essential snRNA enhancer-binding protein, SBF, has a 20- to 30-fold lower affinity for the SPH motif in the U4X enhancer than for the homologous SPH motif in the U4B enhancer. A potential role therefore exists for SBF, as well as PPBF, in the preferential down-regulation of the U4X RNA gene during chicken development.
The small nuclear RNAs (snRNAs) U1, U2, U4, U5, and U6 are involved in the splicing of messenger RNA precursors in eucaryotic cells (Steitz et al., 1988). In general, the snRNAs are encoded by families of genes present in multiple copies in the genomes of higher organisms (Dahlberg and Lund, 1988). In the chicken, there are two (and only two) genes that code for U4 small nuclear RNA (Hoffman et al., 1986; McNamara and Stumph, 1989). These genes, designated the U4X gene and the U4B gene, are closely linked within 500 bp of each other in the chicken genome (see Fig. 1A). Moreover, these two genes encode distinct sequence variants of U4 RNA that differ at seven nucleotide positions (Hoffman et al., 1986; Korf et al., 1988).
Figure 1.
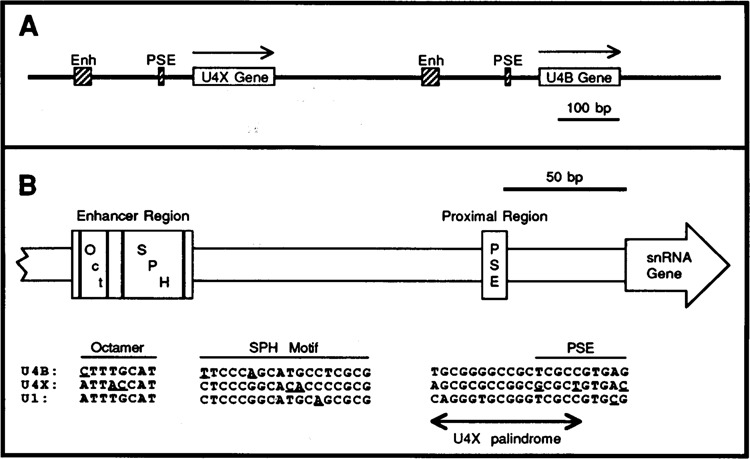
A. Genomic organization of the chicken U4X and U4B snRNA genes. The genes are transcribed left-to-right, as indicated by the arrows. Cross-hatched boxes indicate the locations of the enhancer (Enh) and proximal sequence elements (PSE) conserved in the 5′ flanking DNA of vertebrate snRNA genes. B. Similarities and differences in the promoters of the chicken U4B, U4X, and U1 snRNA genes. The schematic diagram shows the locations of the PSE and of the octamer and SPH motifs that are conserved at nearly identical positions upstream of the U4B, U4X, and U1 genes. The sequence comparisons at the bottom of the figure show that among these three genes the regulatory motifs are quite similar but not identical in sequence. Underlines point out nucleotides in the octamer, SPH, and PSE motifs that are different in one sequence but conserved in the other two. The double-ended arrow indicates the location of a palindromic sequence unique to the U4X gene that partially overlaps the U4X PSE. The U1 sequences shown are from the Ul-52a gene (Earley et al., 1984). The U1 gene octamer and SPH motifs occur naturally in the opposite orientation compared to those in the U4 genes; therefore the U1 octamer and SPH motif sequences shown in the figure are from the template strand, whereas all other sequences are from the non-template strands.
The U4B gene codes for the chicken homologue of mammalian U4B RNA, a major U4 RNA characterized in mammals and other vertebrates (Reddy and Busch, 1988). The U4X gene encodes a sequence variant of U4 RNA that was unknown prior to the cloning of the U4X gene. Interestingly, the relative accumulation of the products of the U4B and U4X RNA genes is differentially regulated during chicken development in a tissue-specific manner (Korf et al., 1988). The data indicate that the U4X and U4B genes are expressed at nearly equal levels during early embryogenesis, but that U4X gene expression is specifically down-regulated relative to U4B as development proceeds. For example, the ratio of U4X:U4B RNA in gizzard (the tissue with the most extreme variation) was found to decrease more than 7-fold during development from the 10-day embryo to the adult (Korf et al., 1988). In other tissues, the U4X gene was similarly down-regulated relative to U4B as a function of development, but the timing of the down-regulation and the final ratio of U4X:U4B RNA in the adult varied from tissue to tissue. Developmental or tissue-specific expression of variant snRNA genes has also been observed in other organisms ranging from fruit flies to mammals (Forbes et al., 1984; Lund et al., 1985; Howard et al., 1986; Lund and Dahlberg, 1987; Lund et al., 1987; Lund, 1988; Santiago and Marzluff, 1989; Lo and Mount, 1990). Despite this, the mechanisms responsible for the differential expression of variant snRNA genes remain unknown.
With the exception of U6, the snRNAs are synthesized by RNA polymerase II (Dahlberg and Lund, 1988; Parry et al., 1989a). However, the genes that code for the snRNAs are atypical RNA polymerase II transcription units. In vertebrates, for example, the site of snRNA transcription initiation is specified not by a TATA box, but rather by a unique cis-acting signal termed the proximal sequence element (PSE) located about 50–60 base pairs upstream of snRNA genes (Dahlberg and Lund, 1988; Parry et al., 1989a). The PSE also plays an essential role in 3′ end formation of the snRNAs (Neuman de Vegvar et al., 1986; Hernandez and Lucito, 1988; Parry et al., 1989b; Neuman de Vegvar and Dahlberg, 1989). Additional control of vertebrate snRNA gene expression is accomplished by a transcriptional enhancer located approximately 180–250 bp upstream of the transcription start site. Like mRNA enhancers, snRNA enhancers serve to elevate the level of gene expression and are composed of multiple functional motifs; furthermore, they contribute to the formation of a stable transcription complex (Mattaj et al., 1985; McNamara et al., 1987; Murphy et al., 1987; Roebuck et al., 1987; Kazmaier et al., 1987; Weller et al., 1988; Tebb and Mattaj, 1989; Roebuck et al., 1990; Janson and Pettersson, 1990).
Although there is no direct evidence linking the differential expression of the chicken U4X and U4B RNA genes to transcriptional control, the structure of their promoter regions is consistent with such a mechanism. A comparison of the chicken U4X, U4B, and U1 gene promoters is shown in Figure 1B. Outside of the PSE and enhancer regions, there is no obvious sequence similarity between the 5′ flanking DNA of these three genes. Notably, however, the U4X gene enhancer and PSE regions exhibit certain features suggesting that U4X transcription may be regulated differently from U4B (and U1). For example, just 5′ of the U4X gene PSE there exists a dyad symmetry element (U4X palindrome, Fig. 1B) not found near the U4B or U1 PSEs. This represents a potential cis-acting element that may be uniquely involved in U4X gene expression.
In the distal region, at least two motifs (octamer and SPH, Fig. 1B) are functionally important for U4B (and Ul) gene enhancer activity. The octamer motif (ATTTGCAT and its variants) is recognized by the transcription factor Oct-1 and seems to be universally present in the enhancers of vertebrate snRNA genes. Although the U4B octamer sequence (CTTTGCAT) is known to be a functional variant of the octamer, the U4X sequence (ATTACCAT) contains two base transversions that could potentially interfere with octamer activity. The second enhancer motif, SPH, is essential for chicken U4B (and Ul) enhancer function and is recognized by a protein that we have termed the SPH motif Binding Factor, or SBF (Roebuck et al., 1990; Zamrod and Stumph, 1990). It is not yet known how general a role SBF plays in mediating vertebrate snRNA gene enhancer activity.
To investigate the molecular basis for the developmental regulation of the U4X and U4B RNA genes, we have now systematically studied the interactions of cellular factors with the known regulatory regions in the 5′ flanking DNA. As a result, we have identified a novel factor (termed PPBF) that interacts specifically with the dyad symmetry element adjacent to the PSE of the U4X gene. PPBF may be an important factor involved in the differential expression of the U4X and U4B snRNA genes. Our data further indicate that Oct-1 protein binds to the U4X and U4B enhancers with similar efficiencies; in contrast, SBF has a significantly lower affinity for the U4X enhancer than for the U4B enhancer. This suggests that the U4X and U4B enhancers, under appropriate circumstances, may also contribute to the differential expression of the chicken U4 RNA genes.
Materials and methods
Identification and purification of PPBF
Highly purified nuclei were prepared from 8- to 10-day chick embryos by ultracentrifugation through two consecutive sucrose gradients, as previously described (Roebuck et al., 1990). Liver, heart, and kidney nuclei were prepared in the same way using tissues taken from chicks 3 weeks post-hatching. Nuclei were lysed and proteins extracted as described previously (Roebuck et al., 1990). By using the electrophoretic mobility shift assay (EMSA), an activity was originally detected in the embryo extracts that specifically recognized the U4XPal/PSE oligo-nucleotide (sequence shown in Fig. 2). Embryo nuclear extract (35–40 mg total protein) was fractionated on a 5 ml heparin agarose column equilibrated with TM100 buffer (50 mM Tris-HC1 [pH 7.5], 12.5 mM MgCl2, 1 mM EDTA, 20% glycerol, 1 mM dithiothreitol, 100 mM KC1). Bound protein was step-eluted in the same buffer containing increasing amounts of KC1 (250, 325, 450, and 1000 mM). These stepelutions are referred to as the HA-250, HA-325, HA-450, and HA-1000 fractions respectively. SBF activity eluted exclusively in the HA-250 fraction and was subsequently further purified by sequence-specific DNA affinity chromatography as previously described (Kadonaga and Tjian, 1986; Roebuck et al., 1990).
Figure 2.
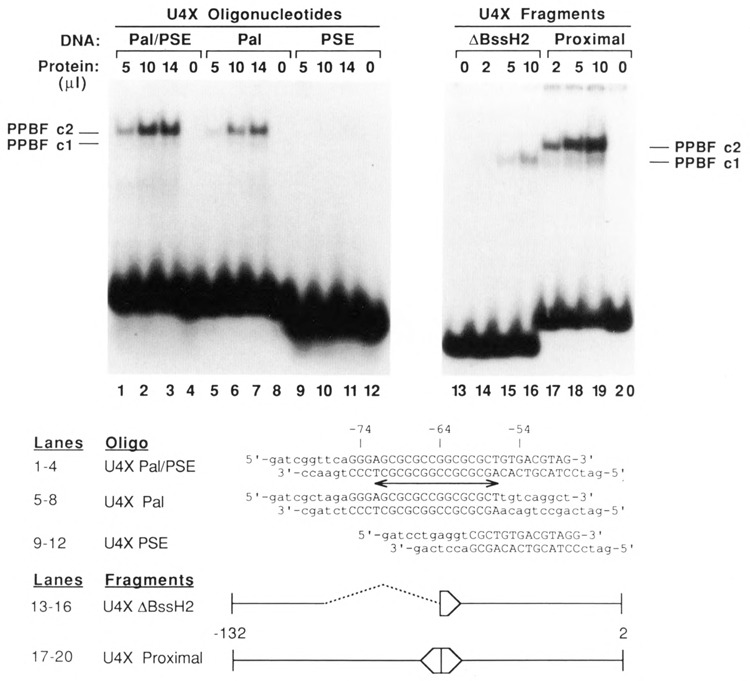
Identification of PPBF, a factor that binds specifically to a palindromic sequence in the proximal regulatory region of the U4X gene. EMSAs were performed by incubating increasing amounts of affinity-purified factor, PPBF, with various synthetic oligonucleotides and restriction fragments that contained DNA sequences from the U4X gene proximal region. The sequences of the synthetic DNA oligonucleotides are shown below the autoradiograms. The U4X Pal/PSE oligonucleotide contained the complete PSE and overlapping palindromic sequence, whereas the U4X Pal and U4X PSE oligonucleotides contained complete sequences only for the palindrome or the PSE respectively. Wild-type nucleotides are shown in upper case, and the double-ended arrow indicates the position of the palindrome. The restriction fragment designated U4X proximal contained U4X sequences from position −132 in the 5′ flanking DNA to position 2 in the coding region of the gene. The U4X ΔBssH II fragment was similar, except that nucleotides from positions −105 to −64 had been deleted by BssH II digestion and re-ligation. Only DNA oligonucleotides or fragments that contained the intact palindromic sequence were able to form the upper complex, c2.
EMSAs indicated the activity that recognized the U4X Pal/PSE sequence (PPBF) eluted entirely in the HA-450 fraction, which contained approximately 2% of the protein applied to the heparin agarose column. PPBF was further purified by sequence-specific DNA affinity chromatography of the HA-450 fraction in a manner similar to that described for SBF (Roebuck et al., 1990), except that the affinity ligand coupled to the Sepharose resin was the concatenated U4XPal/PSE oligonucleotide shown in Figure 2, and poly(dA-dT)poly(dA-dT) and was used as the nonspecific competitor at a concentration of 10 μg/ml. After elution of the affinity-purified PPBF fraction, nuclease- and protease-free bovine serum albumin (Bethesda Research Laboratories) was added to a final concentration of 0.25 mg/ml. This was dialyzed against TM100 containing 0.1 % NP40 and concentrated using Centricon-10 microconcentrators (Amicon Corp.).
Protein–DNA binding assays
End-labeled DNA restriction fragments and double-stranded DNA oligonucleotides used for EMSAs and DNase I footprinting assays are in most cases shown in the figures and described in the legends. The octamer and SPH motif probes used in Figure 8A and B contained the U4B gene octamer and the U1 gene SPH motif sequences respectively, and consisted of the following annealed oligonucleotides: 5′-CACCT-ACTTTGCATAGCGCT-3′ and 3′-GTGGATGAA-ACGTATCGCGA-5′ (for octamer) and 5′-GAT-CAAACCGCGCGCTGCATGCCGGGAGCAC-CAC-3′ and 3’TTTGGCGCGCGACGTACGGCC CTCGTGGTGCTAG-5′ (for SPH motif). All assays of the activities of SBF and Oct-1 included poly(dI-dC)poly(dI-dC) as a non-specific competitor, whereas it was necessary to use poly(dA-dT)poly(dA-dT) to detect PPBF activity with a reasonable efficiency.
Figure 8.
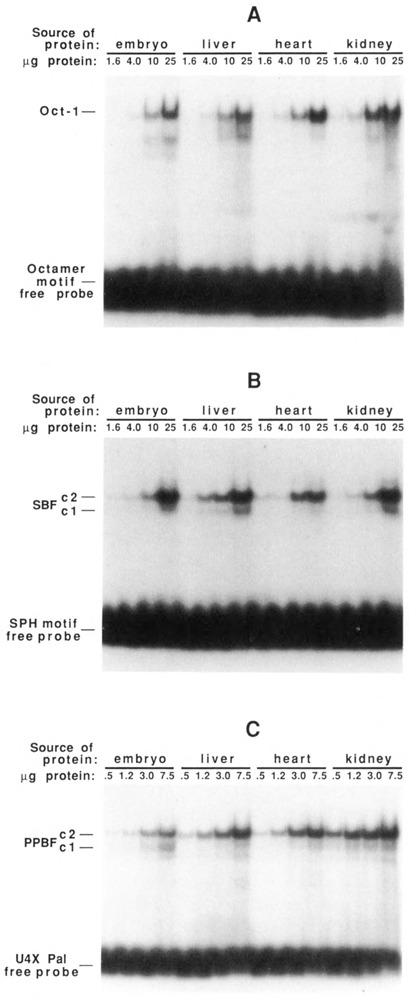
DNA-binding activities of Oct1, SBF, and PPBF as a function of development and tissue-type. Nuclear protein extracts were prepared from 8-day embryos and from liver, heart, and kidneys of chicks 3 weeks post-hatching. Increasing amounts of each unfractionated extract were incubated with 32P-labeled double-stranded oligonucleotides containing specific binding sites for Oct1 (A), SBF (B), or PPBF (C) and analyzed on EMSA gels. Oct1 and SBF DNA-binding activities appear relatively constant in the various tissues; the specific DNA-binding activity of PPBF is lowest in embryo, intermediate in liver and heart, and highest in kidney.
EMSAs were carried out as previously described (Roebuck et al., 1987) when measuring Oct-1 activity or when using affinity-purified factors. When specific competitor DNA fragments were included in the EMSA reactions (as in Figures 6 and 7), the labeled and cold competitor DNAs were added prior to protein addition. For EMSAs measuring SBF and PPBF activities in unfractionated nuclear extracts (Fig. 8), the sensitivities of the assays were increased by using the following buffer in the binding reactions: 25 mM HEPES (pH 7.5), 0.1 M KC1, 0.01 mM ZnSO4, 0.1% NP40, 20% glycerol, and 10 mM dithiothreitol. EMSA gels were run using recirculating buffer (6.7 mM Tris-HCl [pH 7.5], 3.3 mM sodium acetate, 1 mM EDTA) or non-recirculating buffer (25 mM Tris-HCl [pH 83], 190 mM glycine, 1 mM EDTA) with comparable results.
Figure 6.
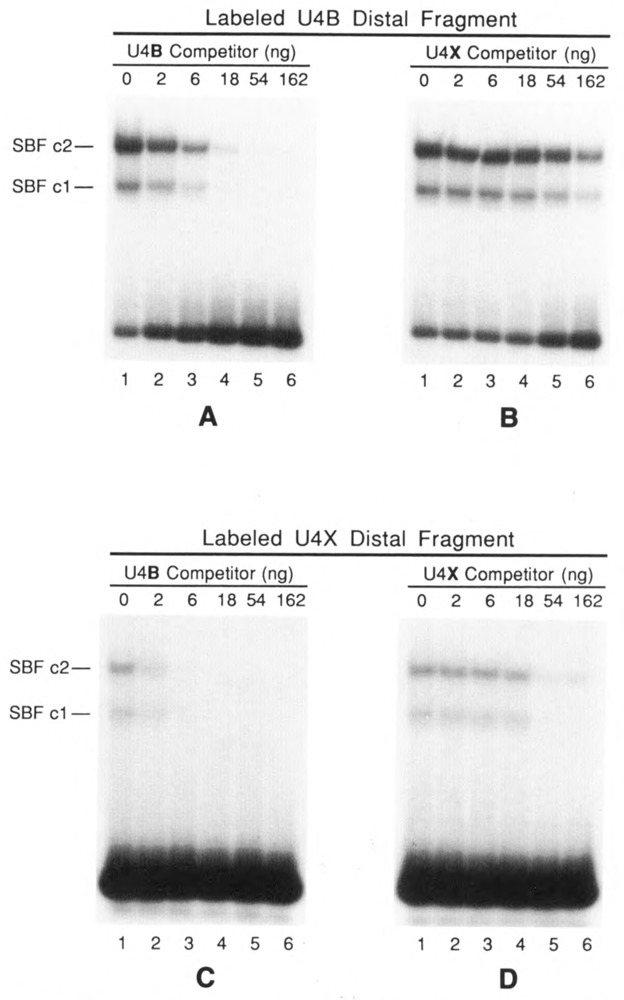
EMSAs demonstrating the differential interaction of SBF with the U4B and U4X enhancers. Assays were performed by incubating 6 μl of affinity-purified SBF with a 32P-labeled DNA fragment encompassing either the U4B enhancer (A and B) or the U4X enhancer (C and D). Incubation mixtures also contained as specific competitor DNA increasing amounts of either the unlabeled U4B fragment (A and C) or the unlabeled U4X fragment (B and D). In each case, the U4B fragment was a 20- to 30-fold more effective competitor than the U4X fragment. The autoradiograms in C and D are 3-fold longer exposures than those shown in A and B. The U4B and U4X distal DNA fragments are diagrammed in Fig. 4C.
Figure 7.
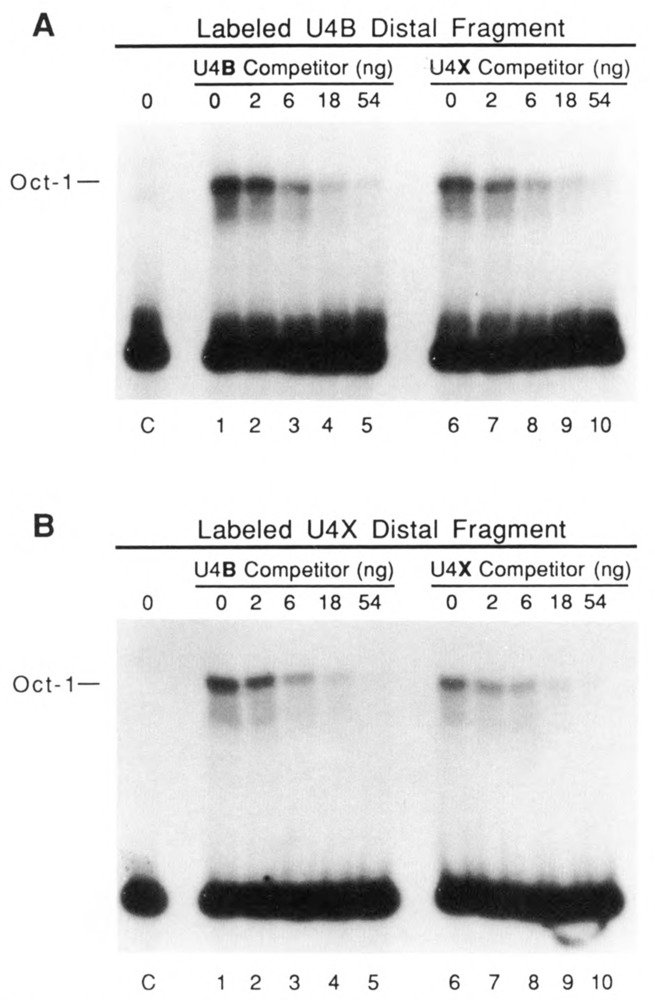
Oct1 factor has similar affinities for the U4B and U4X enhancers. EMSAs were performed similar to those in Figure 6, except that in vitro translated Oct1 factor was used in place of SBF. A and B show results obtained by using labeled DNA fragments from the U4B and U4X distal regions, respectively. Reactions loaded in lanes 1-5 (both panels) contained increasing amounts of the unlabeled U4B distal fragment as competitor, and reactions loaded in lanes 6–10 (both panels) contained increasing amounts of the U4X distal fragment as competitor. Lanes labeled C contained control reactions lacking competitor but incubated in translation extracts from which in vitro synthesized Oct1 mRNA had been omitted.
DNase I footprinting of PPBF on the U4X proximal region was carried out under conditions previously used to map the binding site of SBF in the U1 enhancer (Roebuck et al., 1990), except that poly(dA-dT)poly(dA-dT) was used as the non-specific competitor. The same protocol, however, was unsuccessful at generating a detectable footprint of SBF on the U4X enhancer, presumably because of the lower affinity of the interaction. Therefore, footprinting of SBF on the U4X enhancer employed preparative EMSA as an intermediate step as previously described (Roebuck et al., 1987). Each footprinting mixture (195 μl final volume) contained 10 mM Tris-HCl (pH 7.5), 5 mM NaCl, 11 mM dithiothreitol, 5% glycerol, 1 mM EDTA, 64 μg poly(dI-dC)poly(dI-dC), 200,000 cpm of end-labeled DNA, and 120 ul of affinity-purified SBF. After incubation for 30 minutes at 20°C, 5 μl of 0.1 M MgCl2 was added, followed by addition of DNase I. After a 1.5-minute digestion, EDTA was added to a final concentration of 5 mM. The reactions were electrophoresed on a 4% non-denaturing polyacrylamide preparative mobility shift gel. DNA bands corresponding to the bound and unbound fragments were excised and eluted by shaking in 0.5 mM sodium acetate (pH 7.5), 0.1% SDS, and 1 mM EDTA at room temperature overnight. The supernatant was extracted sequentially with phenol-chloroformisoamyl alcohol (25:24:1), and chloroformisoamyl alcohol (24:1), and the DNA was precipitated by ethanol with carrier RNA. After a second precipitation by ethanol, the products were analyzed on a denaturing polyacrylamide gel.
In vitro synthesis of Oct-1 protein
Plasmid pBSoct-l+ (generously provided by W. Herr, Cold Spring Harbor Laboratory) contains coding sequences for the human ubiquitous octamer binding protein Oct-1 (Sturm et al., 1988). The human and chicken Oct-1 proteins have identical amino acid sequences in the POU-specific and homeobox domains (Petryniak et al., 1990). The plasmid was linearized with Hind III, and 3 μg were used as a template for RNA synthesis by T7 RNA polymerase. Then 1 Hg of RNA transcript was used for in vitro translation in the presence of [35S]methionine. Translation was carried out in a final volume of 50 μl in a rabbit reticulocyte lysate according to the manufacturer’s instructions (Promega). For the EMSAs shown in Figure 7, 3 μl of the translation reaction were used per lane.
Results
Identification of a factor that specifically interacts with the proximal regulatory region of the U4X gene
To search for a factor that specifically recognizes the U4X gene proximal regulatory region, a synthetic double-stranded DNA oligonucleotide probe, corresponding to positions −75 to −48 upstream of the U4X gene, was prepared and labeled with 32P (U4XPal/PSE oligo; sequence shown in Fig. 2). With this probe in an electrophoretic mobility shift assay (EMSA), a sequence-specific DNA-binding activity was detected in unfractionated nuclear extracts prepared from 8- to 10-day old chicken embryos (data not shown, but also see Fig. 8). This DNA-binding activity eluted from a heparin agarose column between 325 and 450 mM KC1 and was further purified by DNA affinity chromatography (Kadonaga and Tjian, 1986) using an affinity resin containing the U4XPal/PSE oligonucleotide covalently linked to Sepharose.
Results of EMSAs using the affinity-purified fraction and the U4XPal/PSE oligo are shown in Figure 2, lanes 1–4. A major band corresponding to a slowly migrating protein-DNA complex (designated c2) was clearly evident. A very faint band of slightly faster mobility (labeled cl) was also visible on the original autoradiogram.
Since the U4XPal/PSE oligo contained DNA sequences extending from position −75 to −48 upstream of the U4X gene, two additional synthetic oligonucleotides were used in EMSAs to further localize the DNA sequences required for the formation of the specific protein-DNA complex. These oligonucleotides, shown in Figure 2, contained different subsets of the U4XPal/PSE sequence. The results (Fig. 2, lanes 5–12) reveal that the palindromic sequence, unique to the U4X gene and just upstream of the PSE, is the target site of the interaction. The sequence-specific DNA-binding activity was therefore named proximal palindrome binding factor (PPBF).
EMSAs were also performed using affinity-purified PPBF and a DNA fragment containing the first 132 bp of U4X gene 5′ flanking DNA (Fig. 2, lanes 17–20). The predominant shifted band corresponded to the c2 complex, and a faint c1 complex was observed. When a fragment lacking sequences between −105 and −64 was used in the assay, only a complex with the mobility of cl was formed (lanes 13–16). These results indicate that affinity-purified PPBF interacts with the palindromic region to form the c2 complex. The nature of the cl complex remains ambiguous from the present data, and it has not been further investigated.
A DNase I protection assay was also used to examine the binding of PPBF to DNA (Fig. 3). Using affinity-purified PPBF, we observed a region of strong protection over the palindromic sequence on both the non-template (upper) and template (lower) strands. A summary of the PPBF footprint at the nucleotide sequence level is shown at the bottom of the figure. The strongest protection occurred directly over the palindrome. Substantially weaker protection extended for some distance toward the gene (in the region corresponding to the PSE). It is possible that this weak protection is due to a PSE-binding protein that co-purifies with PPBF, but this has not yet been studied. Several hypersensitive sites, denoted by black dots, were also observed flanking the protected region.
Figure 3.
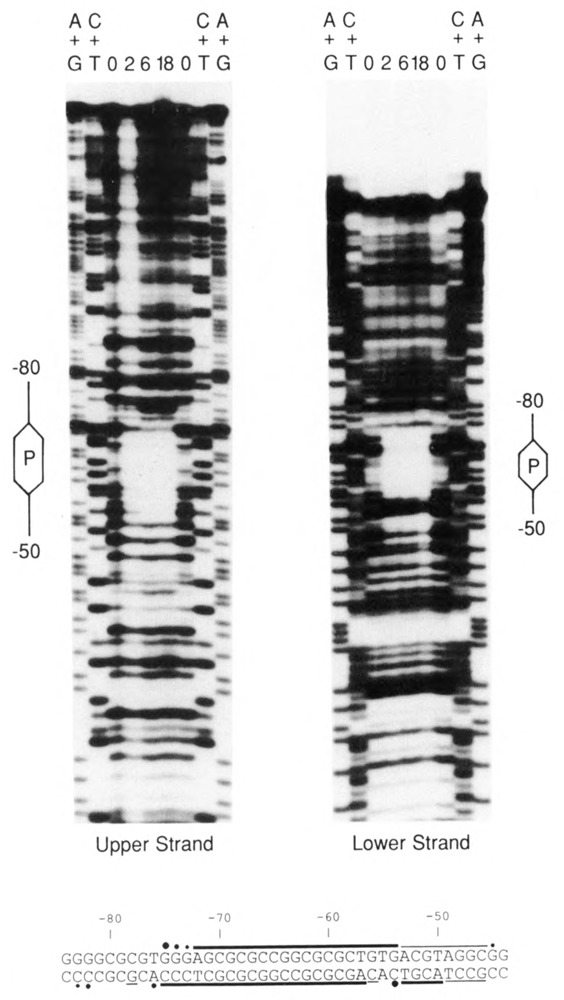
Footprint of PPBF on the U4X proximal region. DNA fragments end-labeled on either the upper or lower strand were incubated with 0, 2, 6, or 18 μl of affinity-purified PPBF (as indicated above the individual lanes), subjected to partial DNase I digestions, and analyzed in denaturing gels. A + G and C + T chemical sequencing reactions were run alongside as markers. The location of the palindromic sequence in each ladder is indicated by the hexagons labeled “P,” and the extent of DNA sequences from −50 to −80 (relative to the transcription initiation site) is delineated by the lines extending from the hexagons. (In the fourth lane of the left panel [2 μl PPBF], the bands are light due to over-digestion and partial loss of sample.) At the bottom of the figure, the nucleotide sequence of the U4X proximal regulatory region is shown, and the horizontal lines indicate the protected regions on the upper and lower strands. Thick lines denote strong protection, and thinner lines weaker protection. Dots indicate sites of enhanced DNase I cleavage induced by the binding of PPBF.
PPBF is a distinct activity from SBF
Earlier work from this laboratory identified the factor SBF, which binds to functional SPH motifs present in the chicken U1 and U4B enhancers (Roebuck et al., 1990; Zamrod and Stumph, 1990). SBF elutes from heparin agarose between 100 and 250 mM KC1, whereas PPBF elutes between 325 and 450 mM KC1. This indicates that SBF and PPBF are very likely distinct factors. However, the sequences recognized by these factors share some similarity:

To compare the binding specificities of PPBF and SBF functionally, we isolated distal and proximal restriction fragments from the 5′flanking DNA of the U4X and U4B genes. These fragments, diagrammed at the bottom of Figure 4, were then incubated with either affinity-purified PPBF or affinity-purified SBF (Roebuck et al., 1990), and the interactions analyzed by EMSA. As expected, PPBF bound strongly to the U4X proximal restriction fragment, forming complexes cl and c2 (Fig. 4A, lanes 4–6) but, importantly, did not detectably bind to the U4B proximal region (lanes 10–12). There was also no detectable binding of PPBF to the distal region of the U4X gene. Interestingly, the affinity-purified PPBF fraction interacted with the U4B distal region fragment, but in a distinctive way. In this case the mobility of the shifted band corresponded to the cl complex only. This qualitatively distinct interaction appears similar to that observed using the U4XABssH2 fragment that lacks half of the PPBF recognition site (Fig. 2, lanes 15 and 16).
Figure 4.
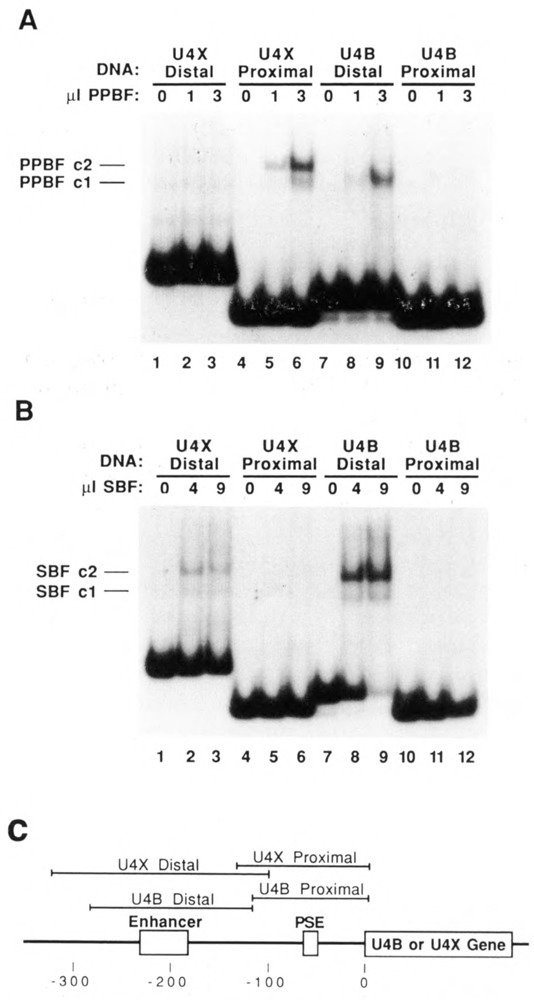
EMSAs demonstrating that PPBF and SBF have distinct DNA-binding specificities. A. Restriction fragments encompassing the U4X enhancer, U4X PSE, U4B enhancer, or U4B PSE were end-labeled and incubated with 0, 1, or 3 μl of affinity-purified PPBF (as indicated above each lane); the formation of protein-DNA complexes was analyzed by electrophoresis in a native polyacrylamide gel. B. The same four DNA fragments were incubated with 0, 4, or 9 μl of affinity-purified SBF, an snRNA gene enhancer-binding protein; complex formation was analyzed as in A. C. Diagram showing the regions encompassed by the U4X distal, U4X proximal, U4B distal, and U4B proximal restriction fragments used in the EMSA analyses presented in A and B.
When SBF was incubated with the same four DNA fragments, a very different EMSA pattern was obtained (Fig. 4B). SBF did not bind detect-ably to the proximal region of either the U4X or the U4B gene. On the other hand, SBF bound strongly to the U4B distal fragment; this was expected, since the U4B enhancer contains an SPH motif recognized by SBF (Zamrod and Stumph, 1990). Importantly, SBF also bound to the distal region of the U4X gene. Although the interaction was apparently weak in comparison to U4B, this result suggests that SBF plays a role in mediating U4X as well as U4B and U1 gene enhancer activity.
In summary, the results shown in Figure 4 confirm that PPBF and SBF are distinct DNA-binding activities. Since PPBF binds to the U4X-but not U4B- proximal region, PPBF may be a key factor involved in the developmental regulation of U4X gene expression.
SBF binds specifically to an SPH-like motif in the U4X enhancer, but the interaction is weaker than with the U4B SPH motif
The results presented in Figure 4B show that SBF binds to a DNA fragment containing the U4X gene enhancer. The site of binding was further localized by a footprinting assay. Following limited DNase I digestion, DNA fragments bound by SBF were separated from unbound DNA fragments by preparative EMSA, eluted from the native gel, and analyzed on a sequencing gel (Fig. 5). Although the footprint was weak (due to the comparatively low affinity of the interaction; see below), the area of protection corresponded to a region of sequence that exhibits homology to the U4B SPH motif. As in the U4B (and Ul) enhancers, this U4X SPH motif is located just downstream of the conserved octamer motif.
Figure 5.
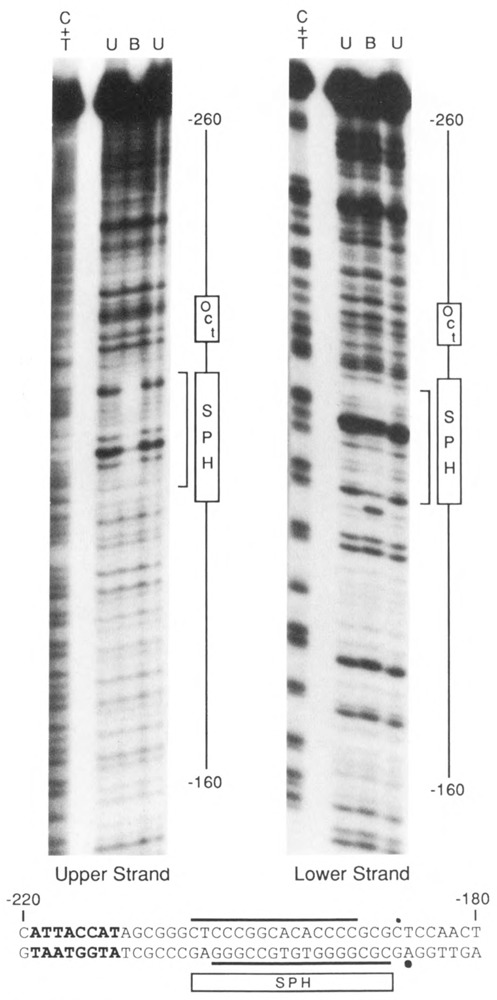
Footprint of SBF on the U4X enhancer region. A restriction fragment encompassing the U4X enhancer region was labeled on either the upper or lower strand at approximately position −105, incubated with affinity-purified SBF, and partially digested with DNase I. The sample was loaded on a preparative EMSA gell and bands corresponding to bound (B) and unbound (U) DNA fragments were eluted and run in separate lanes of a sequencing gel. The lanes labeled C + T contained chemical sequencing reactions as markers. The region protected from DNase I digestion by bound SBF is indicated by brackets alongside the autoradiograms. The protected region is also denoted at the bottom of the figure by horizontal lines above and below the DNA sequence of the U4X enhancer region. Dots indicate sites of enhanced DNase I cleavage induced by the binding of SBF. The rectangular box labeled SPH points out the 18 bp of sequence similarity shared with the U4B and Ul SPH motifs. The nearby octamer sequence variant is shown in bold type.
It was apparent from the data presented above that the interaction of SBF with the U4X enhancer was considerably weaker than its interaction with the U4B enhancer. To better quantitate this difference in affinity, 32P-labeled DNA fragments containing the U4B or U4X enhancer were separately incubated with a constant amount of SBF and increasing amounts of the same DNA fragments as unlabeled competitors. The samples were then analyzed by EMSA. Figure 6 (A and B) shows the results using labeled U4B DNA competed by unlabeled U4B or U4X DNA. The results indicate that the U4B fragment competed 20- to 30-fold better than the U4X fragment for binding to SBF. In the complementary experiment (Fig. 6C and D), binding to the labeled U4X DNA was not as efficient, and the U4B fragment was again a 20- to 30-fold stronger competitor than the homologous U4X fragment.
Oct-1 protein binds to the U4B and U4X octamer motifs with similar affinities
The octamer motifs present in the U4B and U4X enhancers are noticeably different in sequence (Fig. 1). To determine whether these are recognized by the Oct-1 factor with significantly different affinities, EMSAs similar to those in Figure 6 were carried out using in vitro-synthesized Oct-1 protein in place of SBF. The results (Fig. 7A and B) revealed that Oct-1 bound both the U4B and U4X distal region fragments with approximately equal efficiency. Moreover, the ability of the U4B and U4X fragments to compete for Oct-1 binding was indistinguishable. Thus Oct-1 apparently recognizes the U4B and U4X enhancers with similar affinities. From this result, it seems unlikely that Oct-1 factor would be directly responsible for differences in the relative expression levels of the U4B and U4X RNA genes.
Analysis of Oct-1, SBF, and PPBF DNA-binding activities during development
We wished to ascertain whether the differential expression of the U4B and U4X RNA genes could be explained by developmental differences in the specific DNA-binding activities of the proteins described above. Therefore, EMSAs were used to examine the DNA-binding activities of Oct-1, SBF, and PPBF as a function of chicken development and tissue type. Nuclear extracts were prepared from 8-day chick embryos and from the liver, heart, and kidneys of chicks 3 weeks post-hatching. The unfractionated nuclear extracts were analyzed by EMSA using double-stranded labeled oligonucleotides containing specific binding sites for Oct-1, SBF, or PPBF (Fig. 8). The results indicated that the specific DNA-binding activities of Oct-1 and SBF were more or less constant in all the tissues examined (Fig. 8A and B). A greater variation was exhibited by PPBF (Fig. 8C). The specific DNA-binding activity of PPBF appeared to be about 2.5-fold higher in liver and heart, and about 6-fold higher in kidney, than in the 8-day embryo nuclear extracts.
Discussion
In the chicken, expression of the U4X snRNA gene is down-regulated relative to the U4B gene as development proceeds from the early embryo to the adult (Korf et al., 1988). Our data provide evidence for differential protein-DNA interactions occurring at both the enhancer and proximal regulatory regions of these two snRNA genes. These differences may contribute to the observed developmental regulation of U4 gene expression.
Differential protein-DNA interactions at the proximal region
A structural feature unique to the U4X proximal region is the GCrich palindromic sequence that is located immediately upstream of and partially overlapping the U4X PSE. This site is recognized by a newly identified factor, PPBF, that binds sequence-specifically to the U4X proximal region, but not to the U4B proximal region. It is not yet known if PPBF is a positive or negative regulator of U4X transcription. The data presented in Figure 8 favor a role for PPBF as a repressor, since the DNA-binding activity of PPBF is elevated in tissues that exhibit a lower U4X:U4B RNA ratio. For example, from previous data (Korf et al., 1988) we can estimate that U4X RNA gene expression is down-regulated by a factor of about 1.5 in adult heart and liver, and by a factor of about 3 in adult kidney, compared to its level in 8-day whole embryos. Thus, U4X gene expression correlates inversely with the observed levels of PPBF DNA-binding activity. Conclusive evidence for the role of PPBF in regulating U4X gene expression will require the cloning of the gene for PPBF and its use in cotransfection studies and/or the development of a highly efficient in vitro transcription system for chicken snRNA genes.
Recently Burgess’s laboratory reported the characterization and purification of the factor PSE1, which footprints over sequences located 44 to 64 bp upstream of the human U1 gene (Knuth et al., 1990). They also showed that this factor activates transcription from the U1 promoter in vitro (Gunderson et al., 1990). Because the sequences recognized by PSE1 and PPBF are quite different, it seems unlikely that they are the same factor. However, it is possible that they could have similar (or antagonistic) functions.
Role of the U4B and U4X enhancers in differential expression
The U4B and U4X enhancers contain at least two motifs in common: octamer and SPH. Although the sequences in the U4X and U4B enhancers are similar, they are not identical (Fig. 1). Presumably the nucleotide differences could affect the efficiency with which each motif is utilized. Thus, we examined the relative affinities of Oct-1 and SBF for their respective binding sites in the two enhancers.
We found that SBF binds 20- to 30-fold more strongly to the U4B SPH motif than to the U4X SPH motif (Fig. 6). From these data, it is possible to envision an enhancer-based mechanism for the specific down-regulation of U4X transcription–namely, that SBF is limiting in activity in adult tissues, but not a limiting factor in the early embryo. Under such a scenario, high SBF activity in the early embryo would support comparable levels of U4X and U4B transcription, whereas limiting amounts of SBF in adult tissues would efficiently activate only the U4B gene (which contains the high-affinity SBF binding site). However, the data presented in Figure 8B provide no direct support for this hypothesis, since SBF DNA-binding activity seems to be present in roughly equal amounts in all tissues examined. Nevertheless, we would not want to rule out the possibility that such a mechanism may be at work on a subtle scale or in tissues other than those examined.
The Oct-1 factor bound with similar affinities to the U4B and U4X enhancers (Fig. 7). Also, its DNA-binding activity was similar in different tissues and at different developmental stages (Fig. 8A). Together these two results make it seem unlikely that Oct-1 is directly involved in controlling developmental differences in the expression of the U4X and U4B genes relative to each other. It is conceivable that the Oct-1 factor may play other roles in modulating snRNA gene expression (e.g., cell cycle regulation). Also, we cannot exclude the possibility that other octamer binding factors (Rosner et al., 1990; Okamoto et al., 1990; Scholer et al., 1990; Meijer et al., 1990; Smith and Old, 1991), some of which are developmentally regulated, have preferential specificity for one of the octamer variants and thereby regulate the differential expression of the U4B and U4X genes. However, in a recent study Lea et al. (1991) concluded that differential expression of the mouse Ula and Ulb snRNA genes is not dependent on sequence differences in the octamer motif.
Mechanisms controlling variant snRNA gene expression
Lund et al. (1987) determined that the preferential expression of X. laevis U1b2 RNA genes in oocytes was mediated by cis-acting sequences in the proximal region, yet preferential expression of Ulbl genes in activated eggs depended upon sequences in the enhancer region. Thus, at different developmental stages, different mechanisms can apparently account for the differential activity of variant snRNA genes. The studies reported here provide insight into the specific protein-DNA interactions that may contribute to the differential expression of the U4B and U4X RNA genes during chicken development.
Acknowledgments
We thank Winship Herr for pBSoct-l+, and we thank Kathleen McNamara, Bryan Maltby, and Douglas Allen for expert technical assistance. We also thank Sanford Bernstein, Michael Breindl, and Kenneth Roebuck for critically reading the manuscript prior to submission.
This work was supported by Public Health Service grant GM-33512 and National Science Foundation grant DCB-8615964 to W E. Stumph, and in part by the California Metabolic Research Foundation. J. H. Miyake is a predoctoral student in the San Diego State University Department of Biology.
The costs of publishing this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC Section 1734 solely to indicate this fact.
Ihab W. Botros is currently in the Department of Molecular and Cellular Biology, University of Arizona, Tucson, AZ 85721.
References
- Dahlberg J. E. and Lund E. (1988), in Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles (Birnstiel M. L., ed.), Springer Verlag KG, Heidelberg, pp. 38–70. [Google Scholar]
- Earley J. M. III, Roebuck K. A., and Stumph W. E. (1984), Nucl Acids Res 12, 7411–7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes D. J., Kirschner M. W., Caput D., Dahlberg J. E., and Lund E. (1984), Cell 38, 681–689. [DOI] [PubMed] [Google Scholar]
- Gunderson S. I., Knuth M. W., and Burgess R. R. (1990), Genes Dev 4, 2048–2060. [DOI] [PubMed] [Google Scholar]
- Hernandez N. and Lucito R. (1988), EMBO J 7, 3125–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman M. L., Korf G. M., McNamara K. J., and Stumph W. E. (1986), Mol Cell Biol 6, 39l0–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard E. F., Michael S. K., Dahlberg J. E., and Lund E. (1986), Nucl Acids Res 14, 9811–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson L. and Pettersson U. (1990), Proc Natl Acad Sci USA 87, 4732–4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T. and Tjian R. (1986), Proc Natl Acad Sci USA 83, 5889–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmaier M., Tebb G., and Mattaj I. W (1987), EMBO J 6, 3071–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuth M. W, Gunderson S. I., Thompson N. E., Strasheim L. A., and Burgess R. R. (1990), J Biol Chem 265, 17911–17920. [PubMed] [Google Scholar]
- Korf G. M., Botros I. W., and Stumph W. E. (1988), Mol Cell Biol 8, 5566–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea I., Moore H. D. M., and Latchman D. S. (1991), Biochem J 277, 719–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo P. C. H. and Mount S. M. (1990), Nucl Acids Res 18, 6971–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E., Kahan B., and Dahlberg J. E. (1985), Science 229, 1271–1274. [DOI] [PubMed] [Google Scholar]
- Lund E., Bostock C. J., and Dahlberg J. E. (1987), Genes Dev 1, 47–56. [DOI] [PubMed] [Google Scholar]
- Lund E. and Dahlberg J. E. (1987), Genes Dev 1, 39–46. [DOI] [PubMed] [Google Scholar]
- Lund E. (1988), Nucl Acids Res 16, 5813–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W., Lienhard S., Jiricny J., and De Robertis E. M. (1985), Nature 316, 163–167. [DOI] [PubMed] [Google Scholar]
- McNamara K. J., Walker R. J., Roebuck K. A., and Stumph W. E. (1987), Nucl Acids Res 15, 9239–9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara K. J. and Stumph W. E. (1989), Nucl Acids Res 17, 6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer D., Graus A., Kraay R., Langeveld A., Mulder M. P., and Grosveld G. (1990), Nucl Acids Res 18, 7357–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J. T., Skuzeski J. M., Lund E., Steinberg T. H., Burgess R. R., and Dahlberg J. E. (1987), J Biol Chem 262, 1795–1803. [PubMed] [Google Scholar]
- Neuman de Vegvar H. E., Lund E., and Dahlberg J. E. (1986), Cell 47, 259–266. [DOI] [PubMed] [Google Scholar]
- Neuman de Vegvar H. E. and Dahlberg J. E. (1989), Nucl Acids Res 17, 9305–9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Okazawa H., Okuda A., Sakai M., Muramatsu M., and Hamada H. (1990), Cell 60, 461–472. [DOI] [PubMed] [Google Scholar]
- Parry H. D., Scherly D., and Mattaj I. W. (1989a), Trends Biochem Sci 14, 15–19. [Google Scholar]
- Parry H. D., Tebb G., and Mattaj I. W. (1989b), Nucl Acids Res 17, 3633–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryniak B., Staudt L. M., Postema C. E., McCormack W. T., and Thompson C. B. (1990), Proc Natl Acad Sci USA 87, 1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R. and Busch H. (1988), in Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles (Birnstiel M. L., ed.), Springer Verlag KG, Heidelberg, pp. 1–37. [Google Scholar]
- Roebuck K. A., Walker R. J., and Stumph W. E. (1987), Mol Cell Biol 7, 4185–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebuck K. A., Szeto D. P., Green K. P., Fan Q. N., and Stumph W. E. (1990), Mol Cell Biol 10, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner M. H., Vigano M. A., Ozato K., Timmons P. M., Poirier F., Rigby P. W. J., and Staudt L. M. (1990), Nature 345, 686–692. [DOI] [PubMed] [Google Scholar]
- Santiago C. and Marzluff W. F. (1989), Proc Natl Acad Sci USA 86, 2572–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer H. R., Ruppert S., Suzuki N., Chowdhury K., and Gruss P. (1990), Nature 344, 435–439. [DOI] [PubMed] [Google Scholar]
- Smith D. P. and Old R. W. (1991), Nucl Acids Res 19, 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A., Black D. L., Gerke V., Parker K. A., Kramer A., Frendewey D., and Keller W. (1988), in Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles (Birnstiel M. L., ed.), Springer Verlag KG, Heidelberg, pp. 115–154. [Google Scholar]
- Sturm R. A., Das G., and Herr W. (1988), Genes Dev 2, 1582–1599. [DOI] [PubMed] [Google Scholar]
- Tebb G. and Mattaj I. W. (1989), Mol Cell Biol 9, 1682–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller P., Bark C., Janson L., and Pettersson U. (1988), Genes Dev 2, 1389–1399. [DOI] [PubMed] [Google Scholar]
- Zamrod Z. and Stumph W. E. (1990), Nucl Acids Res 18, 7323–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]


