Abstract
Mutations of the CYP21A2 gene encoding adrenal 21-hydroxylase cause congenital adrenal hyperplasia (CAH). The CYP21A2 gene is partially overlapped by the TNXB gene, which encodes an extracellular matrix protein called Tenascin-X (TNX). Mutations affecting both alleles of TNXB cause a severe, autosomal recessive form of Ehlers-Danlos Syndrome (EDS). Rarely, patients with severe, salt-wasting CAH have deletions of CYP21A2 that extend into TNXB, resulting in a ‘contiguous gene syndrome’ consisting of CAH and EDS. Heterozygosity for TNXB mutations causing haploinsufficiency of TNX may be associated with the mild ‘hypermobility form’ of EDS, which principally affects small and large joints. Studies of patients with salt-wasting CAH found that up to 10% had clinical features of EDS, associated joint hypermobility, haploinsufficiency of TNX and heterozygosity for TNXB mutations, now called ‘CAH-X’. These patients have joint hypermobility and a spectrum of other co-morbidities associated with their connective tissue disorder, including chronic arthralgia, joint subluxations, hernias and cardiac defects. Other disorders are beginning to be associated with TNX deficiency, including familial vesicoureteral reflux and neurologic disorders. Further work is needed to delineate the full spectrum of TNX-deficient disorders, with and without associated CAH.
Introduction
Congenital adrenal hyperplasia (CAH) caused by 21-hydroxylase deficiency is one of the most common disorders of the adrenal gland and is familiar to most endocrinologists. In ‘classic CAH’ the adrenal cannot produce cortisol adequately and overproduces androgens and androgen precursors resulting in prenatal virilization of female fetuses, whereas in ‘non-classic CAH’ cortisol production is essentially normal and adrenal hyperandrogenism is minimal or mild. Classic CAH is further subdivided into ‘salt-wasting CAH’, a severe, potentially life-threatening disorder in which the synthesis of both cortisol and aldosterone are impaired, and ‘simple virilizing CAH’ in which there is no salt loss, but there is hyperandrogenism and impaired cortisol synthesis. All CAH is caused by mutations in the single functional adrenal 21-hydroxylase gene, CYP21A2; the three ‘forms’ of CAH simply reflect the degree to which the CYP21A2 gene is impaired, and represent convenient clinical pictures in a continuous spectrum of disease severity. CAH is a complicated disorder that is fairly well understood and requires intensive management [1]; this review focusses on a newly described variant called ‘CAH-X’ in which CAH is accompanied by defects in the overlapping gene for Tenascin-X (TNX).
21-hydroxylase genes
Although small amounts of 21-hydroxylase activity (especially in the synthesis of aldosterone) can be catalyzed by some hepatic enzymes [2], only deficiency of the adrenal 21-hydroxylase causes CAH. Adrenal 21-hydroxylase activity is catalyzed by an enzyme called P450c21 or CYP21. The gene encoding CYP21 lies in the middle of the human leukocyte antigen (HLA) locus on chromosome 6p21.33, hence CAH is closely linked to specific HLA types [3]. The region containing the CYP21 genes is duplicated and contains several other very closely linked (sometimes overlapping) genes, thus this locus is among the most complex in the human genome [4,5]. The duplicated 30 kb units contain the functional CYP21A2 gene that encodes adrenal 21-hydroxylase and the non-functional CYP21A1P pseudogene duplicated in tandem with the C4A and C4B genes that encode the fourth component of serum complement [6–8]. In some literature, the functional, centromeric, CYP21A2 gene is termed 21B (because it is in the ‘B unit’) and the nonfunctional, telomeric CYP21A1P gene is termed 21A. Somewhat surprisingly, CYP21A1P pseudogene is transcribed, but the resultant RNAs do not encode protein [9,10]. The CYP21A2 and CYP21A1P genes are about 3.4 kb long, are divided into 10 exons, and differ in only 87 or 88 bases [11–13]. This high degree of sequence similarity indicates that these two genes are evolving in tandem through intergenic exchange of DNA. This happens because the HLA locus has a very high rate of genetic recombination. Thus, 99% of mutations causing CAH derive either from gene conversion events where some or all of the CYP21A1P pseudogene replaces the corresponding area of the CYP21A2 gene, of from gene deletoins where homologous recombination excises the functional gene. This high rate of genetic recombination explains why 21-hydroxylase deficiency is so much more common than deficiencies of other steroidogenic enzymes. The CYP21 genes of most, but not all mammals are duplicated similarly, albeit with different duplication boundaries; however, while only the CYP21A2 gene functions in human beings, only the cyp21a1 gene corresponding to CYP21A1P functions in mice [14,15] and both genes function in cattle [16,17]. Sequencing of the gene duplication boundaries show that the human locus duplicated after mammalian speciation [18], consistent with data that indicate that other mammals have single copies of this locus [19].
Other genes in the 21-hydroxylase locus
Several other genes lie in the CYP21 gene locus, and several adrenal-specific transcripts have been identified that do not have known functions (Fig. 1). The best-studied of these neighboring genes are the tandemly duplicated C4A and C4B genes that encode the two isoforms of C4, the fourth component of serum complement. The C4B protein has >99% amino acid sequence identity with C4A but has more hemolytic activity [20]. The C4A and C4B genes are ~22 kb long, although C4B may be only 16 kb long on some alleles due to a deletion within one intron [21]. Whereas in most regions of the genome where genes are separated by long segments of intergenic DNA, the 3' ends of the C4A and C4B genes are only 2466 base pairs upstream from the transcriptional start sites of the CYP21A1P and CYP21A2 genes, respectively. The C4 and CYP21 genes have remained in close physical association throughout evolution because the promoter sequences responsible for adrenal-specific expression of the mouse [22] and human [23] CYP21 genes lie far upstream within intron 35 of the C4 genes. Just upstream from C4A is a gene now called STK19; it was originally called G11 [24] or RP [25], but the name STK19 is is now accepted because it encodes a Serine/Threonine Kinase [26]. The STK19 protein is localized to the nucleus, suggesting a role in gene transcription, but its function remains unknown. There is no homologue of STK19 immediately upstream from the C4B gene because much of the coding DNA in this region was lost during the duplication of the C4/CYP21/TNX locus, so that only 914 bases of the 3’ end of the gene lie upstream from C4B [24,25].
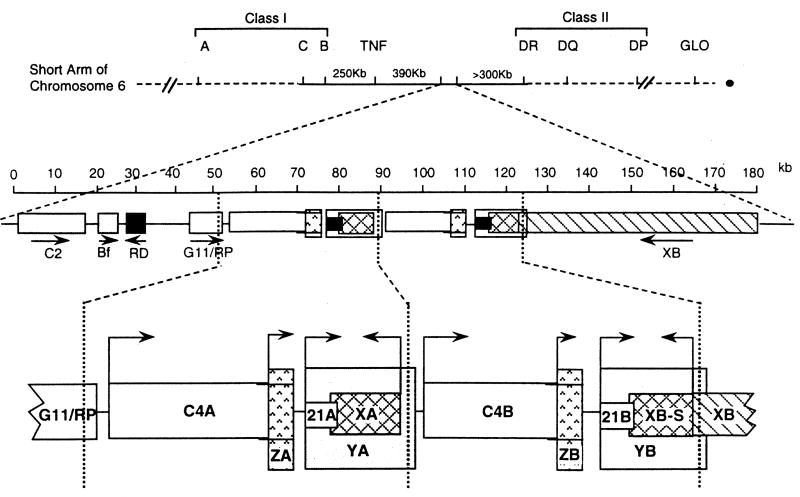
Fig. 1.
The 21-hydroxylase gene locus. Top: diagram of the short arm of chromosome 6, showing the HLA Class I and Class II regions; the telomere is to the left and the centromere to the right. Middle: enlarged view of ~180 kb showing the Class III region on chromosome 6p21.33. C2, complement factor C2; Bf, properedin factor Bf; RD (now known as NEFLE), negative elongation factor subunit E; G11/RP (now known as STK19), serine/threonine kinase 19. The arrows indicate transcriptional orientation. Bottom: the duplicated 30 kb ‘A’ and ‘B’ units: C4A and C4B, genes for complement component 4; 21A, the inactive CYP21A1P pseudogene; 21B the active CYP21A2 gene; XA, YA, and YB, adrenal transcripts that lack open reading frames; XB, the TNXB gene for TNX; XB-S a short, adrenal-specific form of TNX of unknown function; ZA and ZB, adrenal-specific transcripts with open reading frames arising from promoters within the C4 genes; the ZA and ZB promoters are essential components of the CYP21A1P and CYP21A2 promoters. The vertical dotted lines designate the boundaries of the genetic duplication event that resulted in the A and B regions. © WL Miller
The TNXA and TNXB genes lie on the opposite strand of DNA from the C4 and CYP21 genes and hence have the opposite transcriptional orientation. These genes partially overlap the 3' ends of the CYP21genes: the last exon of TNXA and TNXB lies within the 3' untranslated region of exon 10 in CYP21A1P and CYP21A2, respectively [27], and contain fibronectin type III repeats [28]. The TNXA gene was truncated during the duplication of the ancestral C4/CYP21/TNX genetic unit, but nevertheless it is transcribed in the adrenal [18]. Immediately upstream of TNXB lies a gene called CREB-RP, which encodes a protein related to the CREB (cyclic AMP response element binding protein) transcription factor [29]. Transcription of TNXB is initiated from multiple start sites in or near CREB-RP [30,31]. Thus TNXB us unique in having both ends overlapping other genes. The TNXB gene is very large, spanning 68.2 kb of DNA and includes 43 exons encoding a 12kb mRNA that encodes the extracellular matrix protein, Tenascin-X (TNX) [30,32]. The TNXB gene also encodes a truncated 74 kDa form of Tenascin-X, called XB-S (XB-Short), which is identical to the carboxy-terminal 673 amino acids of TNX, arises from an intergenic promoter and is expressed uniquely in the adrenal [33]. Expression of XB-S is induced by hypoxia [34], and proteomics studies indicate it associates with mitotic motor kinesin Eg5, but the precise function of XB-S remains unclear [35]. In addition, RNA transcripts termed YA and YB arise from the CYP21A1P and CYP21A2 promoters, respectively, but do not encode protein [9]. Transcripts having an open reading frame, termed ZA and ZB, arise from a promoter element within intron 35 of the C4 genes, but it is not clear whether or not they encode protein [36].
Tenascin-X
The tenascins are a widely-expressed family of extracellular matrix proteins. Their functions typically oppose those of fibronectin and are largely associated with anti-adhesive effects but extend beyond cellular architecture [37]. There are four mammalian tenascins: according to the nomenclature first proposed by Bristow et al [32], these are now called tenascin-C (TNC, formerly called cytotactin) [38,39], tenascin-R (TNR, formerly called ‘restrictin’ or ‘janusin’) [40,41], tenascin-X (TNX), and tenascin-W (TNW, also tenascin-N) [42,43]. The tenascin proteins are characterized structurally from N-terminus to C-terminus by: i) an N-terminus with 7-amino acid repeats flanked by cysteine residues (TNX has 4 such repeats); ii) a series of repeats that resemble epidermal growth factor (TNX has 18.5); iii) a stretch of fibronectin type III repeats that vary in number as a result of alternative RNA splicing (TNX has 33); iv) a large C-terminal domain structurally related to fibrinogen [32,44]. The N-terminal heptad repeats mediate oligomerization; TNX [45] and TNR [46] form homo-trimers; TNC [44] and TNW form homo-hexamers. Each TNX monomer is 4267 amino acids long with a mass of about 450 kDa; TNX is variably glycosylated and forms trimers whose masses approach 1.4 million daltons; TNX is expressed in most tissues, especially connective tissues [47].
Identification of a CAH patient with a 'contiguous gene syndrome' comprising a deletion of both the CYP21A2 and TNXB genes, demonstrated that Tenascin X deficiency results in Ehlers-Danlos syndrome (EDS) [48]. The causative role of TNX deficiency in connective tissue disorders is confirmed in tnx-knockout mice [49]; it is noteworthy that mouse knockouts of other tenascins do not yield identifiable phenotypes [50–52]. EDS is typically caused by autosomal dominant mutations in collagen, but recessive forms can be caused by mutations in genes for collagen-modifying factors, such as TNX, which is associated with and stabilizes collagen fibrils [45,53]. Tenascin-X deficiency causes a clinically distinct, more severe form of EDS, either with or without associated CAH [54–56]. Heterozygosity for severe TNXB mutations causing haploinsufficiency for TNX may cause ‘hypermobility type EDS’, characterized by joint hypermobility, recurring joint dislocations and joint pain. Among 20 obligate heterozygotes for a severely defective TNXB allele, 9 of 14 females but no males had hypermobility EDS [57]. The diagnosis of TNX-deficient EDS or TNX haploinsufficiency is facilitated by the existence of a 140 kDa proteolytic fragment of TNX in normal serum, but not in the sera of TNX-deficient patients; this 140 kDa fragment of TNX can be measured easily [54, 58].
Beyond classical EDS, TNX is important in development as it promotes epithelial-mysenchymal transitions via TGF-β [59], and may be associated with tumor invasion [60,61]. TNX deficiency has been associated with primary myopathy [62,63], recurrent gastrointestinal perforation [64], and missense mutations in TNX have been found in vesicoureteral reflux [65,66]. TNX, which is expressed in the leptomeninges and choroid plexus [67,68], may play a role in brain and behavior. Single nucleotide polymorphisms in TNXB are associated with schizophrenia [69,70], and tnxb knockout mice have increased anxiety, improved memory and higher sensorimotor coordination than control animals [71].
CAH-X
The initial report of TNX deficiency was in a single patient with CAH and EDS [48]. To evaluate whether isolated TNX deficiency was associated with EDS, Schalkwijk et al [54] evaluated 151 patients with EDS of unknown etiology and found that 5 patients had TNX deficiency, one with CAH. These initial studies of TNX deficiency were consistent with an autosomal recessive pattern of inheritance and the two patients with concomitant CAH had 30 kb deletions involving both the CYP21A2 and TNXB genes. Relatives who were heterozygous for TNXB mutations were either asymptomatic or had mild hypermobility-type EDS of unknown clinical importance [57].
The first evaluation of the potential clinical implications of TNXB heterozygosity in CAH patients was performed in an ongoing observational study of CAH at the National Institutes of Health Clinical Center. In a prospective study, 193 consecutive unrelated patients with CAH were evaluated clinically for manifestations of EDS and genetically for TNXB mutations. Heterozygosity for a TNXB deletion was present in 7% of CAH patients; these CAH patients were more likely than age-and sex-matched CAH patients with normal TNXB to have joint hypermobility, chronic joint pain, multiple joint dislocations and a structural cardiac valve abnormality by echocardiography [72]. Six of 13 probands had a cardiac abnormality including the rare finding of a quadricuspid aortic valve [73], a left ventricular diverticulum and an elongated anterior mitral valve leaflet [72]. Six parents were also evaluated representing relatives who did not have CAH but were heterozygous for a TNXB deletion. Similar to the earlier findings of relatives of the autosomal recessive TNX deficient EDS patients, parents displayed variable symptomatology ranging from no EDS manifestations to hypermobility-type EDS, two with cardiac findings (dilated aortic root and redundant anterior mitral leaflet) [72]. In general, carrying a contiguous deletion of the CYP21A2 and TNXB genes resulted in a more severe EDS phenotype in CAH patients than in their CAH unaffected relatives. As a result of this study, the term CAH-X was coined to describe the subset of CAH patients who display an EDS phenotype due to the monoallelic presence of a CYP21A2 deletion extending into the TNXB gene.
The study of CAH-X has provided insight into the recombination events that occur in the class III region of the MHC locus. This region of the genome is predisposed to genetic recombination due to the presence of duplicated genes; both the CYP21A2 and TNXB genes have corresponding homologous pseudogenes (CYP21A1P and TNXA) giving rise to misalignment during meosis. Approximately 20% of deleterious CYP21A2 mutations are 30 kb gene deletions that result in CYP21A1P/CYP21A2 chimeric genes [74–76]. Nine types of CYP21A1P/CYP21A2 chimera have been identified and named chronologically CH-1 to CH-9 following determination of the junction sites [77]. Similarly, chimeric recombination between TNXB and TNXA occurs (Fig. 2A). Such a recombination event deletes CYP21A2, and therefore also represents a CAH disease-causing allele. The contiguous gene deletion described in the first studies of TNXB in EDS [48, 54] and CAH populations [72] was a TNXA/TNXB chimeric gene [48] which was later termed TNXA/TNXB CH-1 or CAH-X CH-1, in keeping with CYP21A1P/CYP21A2 terminology [78]. CAH-X CH-1 retains a TNXA pseudogene derived 120-bp deletion and renders the gene nonfunctional, resulting in reduced dermal and serum TNX expression supporting a haploinsufficent mechanism [54, 72].
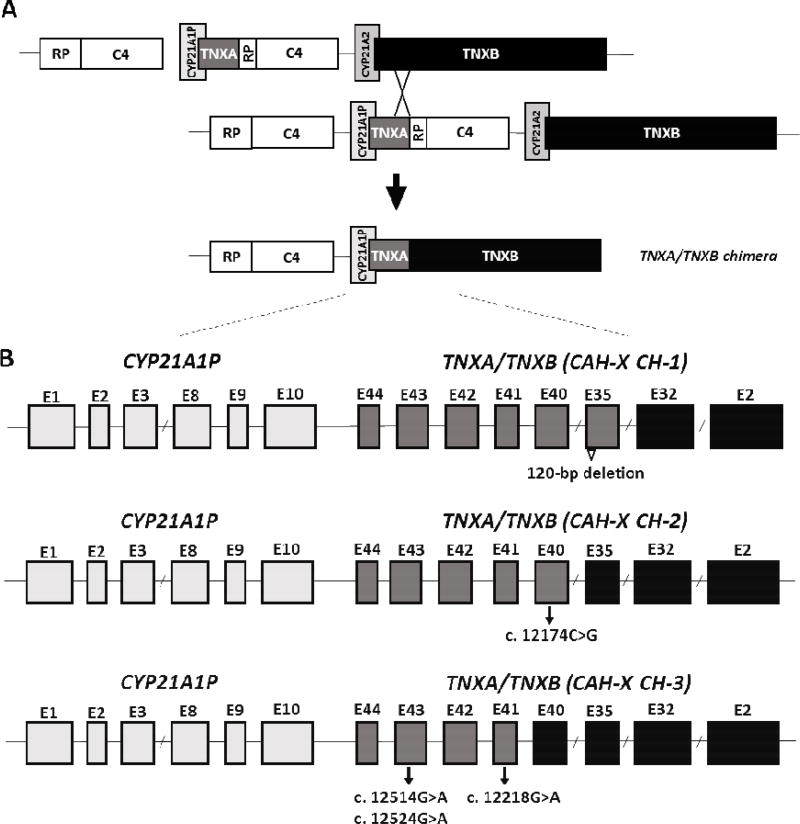
Fig. 2.
Schematic diagram of the TNXA/TNXB chimeric genes. Most common is a bimodular RP-C4-CYP21-TNX region. TNXB encodes the active tenascin-X gene (black) and TNXA encodes the corresponding pseudogene (dark grey). CYP21A2 encodes the active 21-hydroxylase gene (medium grey) and CYP21A1P encodes the corresponding pseudogene (light grey). Panel A shows formation of a TNXA/TNXB chimeric gene due to misalignment during meiosis resulting in deletion of the CYP21A2 gene. Panel B is a schematic of exons (rectangles) of representative TNXA/TNXB chimera. TNXA/TNXB chimeric genes have been classifed into 3 types (CH-1 to CH-3) based on the junction site location. The 120-bp deletion at the boundary of exon 35 and intron 35 of TNXB is shown by an open triangle and present in CAH-X CH-1. The c.12174C>G pseudogene variant in Exon 40 identifies CAHX CH-2, and CAH-X CH-3 is characterized by a cluster of 3 pseudogene variants: c.12218G>A in exon 41, and c.12514 G>A, and c.12524 G>A in exon 43. Adapted from reference [78]; with permission.
After the initial description of CAH-X in a large cohort of patients with CAH, novel TNXA/TNXB chimeras, named CAH-X CH-2 and CAH-X CH-3 were identified [78, 79]. CAH-X CH-2 is characterized by the variant c.12174C>G (p.C4058W) derived from the TNXA pseudogene [78] and CAH-X CH-3 is characterized by a cluster of 3 closely linked mutations (p.R4073H, p.D4172N, and p.S4175N), also derived from the TNXA pseudogene [79] (Fig. 2B). Unlike CAH-X CH-1, both CAH-X CH-2 and CAH-X CH-3 cause structural changes in the TNX protein. CH-2 (p.C4058W) deletes a cysteine that forms a disulfide bond, and computational studies show that it alters protein structure [78]. CH-3 changes three residues (p.R4073H, p.D4172N, and p.S4175N). The p.R4073H change is predicted to reduce protein folding energy by interfering with a cation-pi interaction between p.R4073 and p.F4080 [79]. The changes p.D4172N, and p.S4175N are not predicted to significantly affect the folding energies in the models, but computational analysis is imperfect and future experimental verification is warranted. Because CAH-X CH-2 and CH-3 produce altered proteins, rather than reducing TNX expression, we no longer use the term ‘haploinsufficiency’ in describing the monoallelic presence of a TNXA/TNXB chimera. Similarly, the term ‘autosomal recessive’ is not used to describe patients with CAH-X who have TNXB disease causing mutations on both alleles because ‘autosomal recessive’ implies that having one affected allele does not result in a clinical phenotype. This is not the case with CAH-X.
To date, 24 patients (19 families) with monoallelic CAH-X [72, 78] and 5 patients (5 families) with biallelic CAH-X [48, 54, 79] have been described. Approximately 10 percent of patients with CAH due to 21-hydroxylase deficiency are now estimated to be affected by CAH-X [78]. Overall, CAH-X patients have generalized joint hypermobility, subluxations, chronic arthralgia and about 25% have cardiac abnormalities (Table 1, Figure 3). CAH-X CH-2 causes a more severe phenotype than CAH-X CH-1, characterized by greater skin and joint involvement [78] (Table 1). Patients heterozygous for CAH-X CH-1 have normal skin, while 40% of CAH-X CH-2 patients have skin laxity. Gastrointestinal disorders, such as chronic gastroesophageal reflux and irritable bowel syndrome, and hernia or rectal prolapse are more commonly found in patients heterozygous for CAH-X CH-2 than CAH-X CH-1 (Table 1). Other clinical findings in monoallelic CAH-X include bifid uvula (n=4), scoliosis (n=3), pectus excavatum (n=1) and early onset osteoarthritis (n=1). CAH-X CH-3 has only been described in biallelic CAH-X, so the CAH phenotype associated with heterozygosity for CAH-X CH-3 is unknown. Once again, relatives of CAH-X patients who carry a CAH-X CH-2 or CAH-X CH-3 allele but do not have CAH had a milder phenotype than CAH-X patients, although the majority of relatives had hypermobile joints and two had cardiac findings (atrial septum aneurysm with patent foramen ovale, and chamber enlargement) [78, 79].
Table 1.
Clinical Characteristics of Patients with CAH-X
| Monoallelic CAH-X CH-1a (n=14) |
Monoallelic CAH-X CH-2b (n=10) |
Biallelic CAH-Xc (n=5) |
|
|---|---|---|---|
| Age (years, mean ± s.d., range) | 18.1 ± 8.3 (8–32) | 20.8 ± 16.4 (2–45) | 24.0 ± 7.4 (14–32) |
| Females [no, (%)] | 8 (57.1) | 6 (60.0) | 1 (20.0) |
| Musculoskelatal (no, (%) | |||
| Generalized hypermobilityd | 7 (50.0) | 10 (100.0) | 5 (100.0) |
| Subluxations | 5 (35.7) | 4 (40.0) | 2 (40.0) |
| Chronic arthralgia | 5 (35.7) | 4 (40.0) | 2 (40.0) |
| Chronic tendonitis, bursitis or fasciitis | 2 (11.8) | 3 (30.0) | 2 (40.0) |
| Dermatologic | |||
| Hyperextensible skin' | 0 | 4 (40.0) | 5 (100.0) |
| Wide scars | 3 (21.4) | 2 (20.0) | 2 (40.0) |
| Easy bruising | 5 (35.7) | 4 (40.0) | 5 (100.0) |
| Poor wound healing | 0 | 0 | 2 (40.0) |
| Gastrointestinal | |||
| Chronic disordere | 1 (7.1) | 4 (40.0) | 1 (20.0) |
| Hernia or rectal prolapse | 0 | 3 (30.0) | 2 (40.0) |
| Cardiac | |||
| Congenital defectf | 3 (21.4) | 3 (30.0) | 0 |
| Chamber enlargement | 2 (11.8) | 0 | 2 40.0) |
| Enlarged aortic root | 0 | 2(20.0) | 0 |
In Merke et al 2013 [72].
In Morissette et al 2015 [78].
Generalized hypermobility defined as a Beighton score 5 of 9 or greater for children and 4 of 9 or greater for postpubertal adolescents and adults.
Chronic disorder includes gastroesphageal reflux or irritable bowel syndrome.
Congenital heart defect includes structural valve abnormality, left ventricular diverticulum and patent foramen ovale.
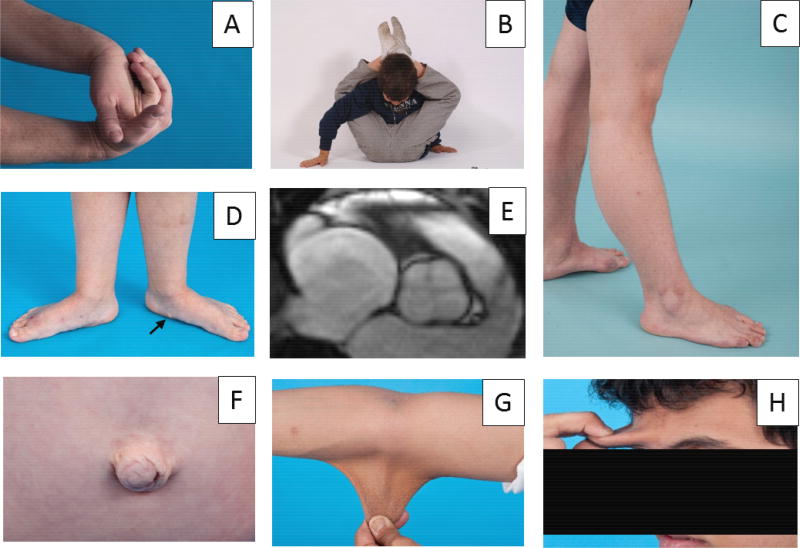
Fig. 3.
Clinical manifestations of CAH-X. Patients with CAH-X due to heterozygosity of a TNXA/TNXB chimera commonly have hypermobility of small joints, shown in Panel A, and large joints, shown in Panels B and C. Pes planus and piezogenic papules (arrow), shown in Panel D, are frequently observed. Approximately 25 percent of patients have a congenital cardiac defect. Panel E shows a quadricuspic aortic valve in a patient heterozygous for CAH-X CH-1 [73]. Hernias are most often observed in patients heterozygous for CAH-X CH-2, shown in Panel F, or biallelic for CAH-X. Hyperextensible skin is observed with CAH-X CH-2, and most severe with biallelic CAH-X, shown in Panels G and H. Panels C and F from reference [78]; with permission.
All patients with biallelic CAH-X show severe skin hyperextensibility (Fig 3G,H), and significant joint hypermobility, and two (19 yo male, 26 yo male) had delayed wound healing [48, 79]. Other EDS manifestations in CAH-X biallelic patients include multiple joint dislocations, chronic arthralgia, chronic tendonitis and/or bursitis, rectal prolapse, severe gastroesphagelal reflux and cardiac abnormalities (ventricular enlargement in two). Biallelic CAH-X patients appear to have a more severe phenotype than biallelic TNX deficient type EDS patients without CAH who have normal wound healing [54, 56, 80]. TNXA/TNXB chimeras may lead to a more severe phenotype than TNXB missense or nonsense mutations, but the hormonal factors characteristic of CAH including chronic glucocorticoid treatment may also influence phenotype.
Most previously reported TNXB mutations causing EDS are located in the region encoding the EGF-like repeats or the fibronectin type III domain of the tenascin protein, while TNXA/TNXB chimeras either interfere with TNX production (CAH-X CH-1) or affect the fibrinogen-like domain (CAH-X CH-2, CAH-X CH-3). The junction site of a TNXA/TNXB chimera could be anywhere between exons 32 and 44, the homologous region between TNXA and TNXB. Unlike CYP21A2 and CYP21A1P, variations in the sequences of TNXB and TNXA are not well characterized and novel chimeras may yet exist. Chimeric junction sites can be clinically meaningful. Two rare CYP21A1P/CYP21A2 chimera (CH-4 and CH-9) result in a milder CAH phenotype than other CYP21A1P/CYP21A2 chimeras [77, 81]. Mutation specific effects on TNX protein expression have been described between CAH-X chimeras and might occur across the spectrum of TNXB mutations. Further studies are needed as variations within the TNXA and TNXB genes have not been investigated in detail.
Extensive studies of the CYP21A2 gene led to the discovery of TNXB and have expanded our understanding of the spectrum of TNX-related disorders. The study of CAH-X led to the discovery of different TNXA/TNXB chimera; 3 have been identified to date. The TNX matrix protein interacts with TGF-β [59] but the precise molecular mechanisms have yet to be determined. The identification of CAH-X syndrome and additional TNXB-related diseases promise to expand our understanding of this widely expressed extracellular matrix protein in various disease processes.
Acknowledgments
Dr. Miller’s research was supported in the past by grants from the NIH and the March of Dimes; Dr. Merke’s research was supported in part by the Intramural Research Program of the NIH.
Footnotes
The authors have no conflicts to declare.
References
- 1.Speiser PW, Azziz R, Baskin LS, Ghizzoni L, Hensle TW, Merke DP, Meyer-Bahlburg HFL, Miller WL, Montori VM, Oberfield SE, Ritzen EM, White PC. Congenital adrenal hyperplasia due to steroid 21-hydroxylase deficiency: An Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2010;95:4133–4160. doi: 10.1210/jc.2009-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomes LG, Huang N, Agrawal V, Mendonca BB, Bachega TASS, Miller WL. Extra-adrenal 21-hydroxylation by CYP2C19 and CYP3A4: Effect on 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2009;94:89–95. doi: 10.1210/jc.2008-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dupont B, Oberfield SE, Smithwick ER, Lee TD, Levine LS. Close genetic linkage between HLA and congenital adrenal hyperplasia (21-hydroxylase deficiency) Lancet. 1977;ii:1309–1312. doi: 10.1016/s0140-6736(77)90362-2. [DOI] [PubMed] [Google Scholar]
- 4.Morel Y, Miller WL. Clinical and molecular genetics of congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Adv Hum Genet. 1991;20:1–68. doi: 10.1007/978-1-4684-5958-6_1. [DOI] [PubMed] [Google Scholar]
- 5.Miller WL, Auchus RJ. The molecular biology, biochemistry and physiology of human steroidogenesis and its disorders. Endocr Rev. 2011;32:81–151. doi: 10.1210/er.2010-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amor M, Tosi M, Duponchel C, Steinmetz M, Meo T. Liver cDNA probes disclose two cytochrome P450 genes duplicated in tandem with the complement C4 loci of the mouse H-2S region. Proc Natl Acad Sci USA. 1985;82:4453–4457. doi: 10.1073/pnas.82.13.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carroll MC, Campbell RD, Porter RR. Mapping of steroid 21-hydroxylase genes to complement component C4 genes in HLA, the major histocompatibility locus in man. Proc Natl Acad Sci USA. 1985;82:521–525. doi: 10.1073/pnas.82.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White PC, Grossberger D, Onufer BJ, Chaplin DD, New MI, Dupont B, Strominger JL. Two genes encoding steroid 21-hydroxylase are located near the genes encoding the fourth component of complement in man. Proc Natl Acad Sci USA. 1985;82:1089–1093. doi: 10.1073/pnas.82.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bristow J, Gitelman SE, Tee MK, Staels B, Miller WL. Abundant adrenal-specific transcription of the human P450c21A "pseudogene". J Biol Chem. 1993;268:12919–12924. [PubMed] [Google Scholar]
- 10.Chang SF, Chung BC. Difference in transcriptional activity of two homologous CYP21A genes. Mol Endocrinol. 1995;9:1330–1336. doi: 10.1210/mend.9.10.8544841. [DOI] [PubMed] [Google Scholar]
- 11.Higashi Y, Yoshioka H, Yamane M, Gotoh O, Fujii-Kuriyama Y. Complete nucleotide sequence of two steroid 21-hydroxylase genes tandemly arranged in human chromosome: A pseudogene and genuine gene. Proc Natl Acad Sci USA. 1986;83:2841–2845. doi: 10.1073/pnas.83.9.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.White PC, New MI, Dupont B. Structure of the human steroid 21-hydroxylase genes. Proc Natl Acad Sci USA. 1986;83:5111–5115. doi: 10.1073/pnas.83.14.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodrigues NR, Dunham I, Yu CY, Carroll MC, Porter RR, Campbell RD. Molecular characterization of the HLA-linked steroid 21-hydroxylase B gene from an individual with congenital adrenal hyperplasia. EMBO J. 1987;6:1653–1661. doi: 10.1002/j.1460-2075.1987.tb02414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker KL, Chaplin DD, Wong M, Seidman JG, Smith JA, Schimmer BP. Expression of murine 21-hydroxylase in mouse adrenal glands and in transfected Y1 adrenocortical tumor cells. Proc Natl Acad Sci USA. 1985;82:7860–7864. doi: 10.1073/pnas.82.23.7860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaplin DD, Galbraith LJ, Seidman JG, White PC, Parker KL. Nucleotide sequence analysis of murine 21-hydroxylase genes: mutations affecting gene expression. Proc Natl Acad Sci USA. 1986;83:9601–9605. doi: 10.1073/pnas.83.24.9601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung B, Matteson KJ, Miller WL. Structure of the bovine gene for P450c21 (steroid 21-hydroxylase) defines a novel cytochrome P450 gene family. Proc Natl Acad Sci USA. 1986;83:4243–4247. doi: 10.1073/pnas.83.12.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John ME, Okamura T, Dee A, Adler B, John MC, White PC, Simpson ER, Waterman MR. Bovine steroid 21-hydroxylase: Regulation of biosynthesis. Biochemistry. 1986;25:2846–2853. doi: 10.1021/bi00358a016. [DOI] [PubMed] [Google Scholar]
- 18.Gitelman SE, Bristow J, Miller WL. Mechanism and consequences of the duplication of the human C4/P450c21/gene × locus. Mol Cell Biol. 1992;12:2124–2134. doi: 10.1128/mcb.12.5.2124. Correction: Mol Cell Biol 12:3313–3314, 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geffrotin C, Chardon P, DeAndres-Cara DR, Feil R, Renard C, Vaiman M. The swine steroid 21-hydroxylase gene (CYP21): Cloning and mapping within the swine leukocyte antigen locus. Animal Genet. 1990;21:1–13. doi: 10.1111/j.1365-2052.1990.tb03202.x. [DOI] [PubMed] [Google Scholar]
- 20.Law SKA, Dodds AW, Porter RR. A comparison of the properties of two classes, C4A and C4B, of the human complement component C4. EMBO J. 1984;3:1819–1823. doi: 10.1002/j.1460-2075.1984.tb02052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu CY, Belt KT, Giles CM, Campbell RD, Porter RR. Structural basis of the polymorphism of the human complement components C4A and C4B: gene size, reactivity and antigenicity. EMBO J. 1986;5:2873–2881. doi: 10.1002/j.1460-2075.1986.tb04582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milstone DS, Shaw SK, Parker KL, Szyf M, Seidman JG. An element regulating adrenal-specific steroid 21-hydroxylase expression is located within the slp gene. J Biol Chem. 1992;267:21924–21927. [PubMed] [Google Scholar]
- 23.Wijesuriya SD, Zhang G, Dardis A, Miller WL. Transcriptional regulatory elements of the human gene for cytochrome P450c21 (steroid 21-hydroxylase) lie within intron 35 of the linked C4B gene. J Biol Chem. 1999;274:38097–38106. doi: 10.1074/jbc.274.53.38097. [DOI] [PubMed] [Google Scholar]
- 24.Sargent CA, Anderson MJ, Hsieh SL, Kendall E, Gomez-Escobar N, Campbell RD. Characterisation of the novel gene G11 lying adjacent to the complement C4A gene in the human major histocompatibility complex. Hum Mol Genet. 1994;3:481–488. doi: 10.1093/hmg/3.3.481. [DOI] [PubMed] [Google Scholar]
- 25.Shen L, Wu LC, Sanlioglu S, Chen R, Mendoza AR, Dangel AW, Carroll MC, Zipf WB, Yu CY. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon-intron structure, composite retroposon, and breakpoint of gene duplication. J Biol Chem. 1994;269:8466–8476. [PubMed] [Google Scholar]
- 26.Gomez-Escobar N, Chou CF, Lin WW, Hsieh SL, Campbell RD. The G11 gene located in the major histocompatibility complex encodes a novel nuclear serine/threonine protein kinase. J Biol Chem. 1998;273:30954–30960. doi: 10.1074/jbc.273.47.30954. [DOI] [PubMed] [Google Scholar]
- 27.Morel Y, Bristow J, Gitelman SE, Miller WL. Transcript encoded on the opposite strand of the human steroid 2l-hydroxylase/complement component C4 gene locus. Proc Natl Acad Sci USA. 1989;86:6582–6586. doi: 10.1073/pnas.86.17.6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto K, Arai M, Ishihara N, Ando A, Inoko H, Ikemura T. Cluster of fibronectin type III repeats found in the human major histocompatibility complex class III region shows the highest homology with the repeats in an extracellular matrix protein, tenascin. Genomics. 1992;12:485–491. doi: 10.1016/0888-7543(92)90438-x. [DOI] [PubMed] [Google Scholar]
- 29.Min J, Shukla H, Kozono H, Bronson SK, Weissman SM, Chaplin DD. A novel Creb family gene telomeric of HLA-DRA in the HLA complex. Genomics. 1995;30:149–156. doi: 10.1006/geno.1995.9891. [DOI] [PubMed] [Google Scholar]
- 30.Speek M, Barry F, Miller WL. Alternate promoters and alternate splicing of human Tenascin-X, a gene with 5' and 3' ends buried in other genes. Hum Mol Genet. 1996;5:1749–1758. doi: 10.1093/hmg/5.11.1749. [DOI] [PubMed] [Google Scholar]
- 31.Wijesuriya SD, Bristow J, Miller WL. Localization and analysis of the principal promoter for human Tenascin-X. Genomics. 2002;80:443–452. doi: 10.1006/geno.2002.6852. [DOI] [PubMed] [Google Scholar]
- 32.Bristow J, Tee MK, Gitelman SE, Mellon SH, Miller WL. Tenascin-X: A novel extracellular matrix protein encoded by the human XB gene overlapping P450c21B. J Cell Biol. 1993;122:265–278. doi: 10.1083/jcb.122.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tee MK, Thomson AA, Bristow J, Miller WL. Sequences promoting the transcription of the human XA gene overlapping P450c21A correctly predict the presence of a novel, adrenal-specific, truncated form of Tenascin-X. Genomics. 1995;28:171–178. doi: 10.1006/geno.1995.1128. [DOI] [PubMed] [Google Scholar]
- 34.Kato A, Endo T, Abiko S, Ariga H, Matsumoto K. Induction of truncated form of Tenascin-X (XB-S) through dissociation of HDAC1 from SP-1/HDAC1 complex in response to hypoxic conditions. Exp Cell Res. 2008;314:2661–2673. doi: 10.1016/j.yexcr.2008.05.019. [DOI] [PubMed] [Google Scholar]
- 35.Endo T, Ariga H, Matsumoto K. Truncated form of Tenascin-X,XB-S, interacts with mitotic motor kinesin Eg5. Mol Cell Biochem. 2009;320:53–66. doi: 10.1007/s11010-008-9898-y. [DOI] [PubMed] [Google Scholar]
- 36.Tee MK, Babalola GO, Aza-Blanc P, Speek M, Gitelman SE, Miller WL. A promoter within intron 35 of the human C4A gene initiates adrenal-specific transcription of a 1kb RNA: location of a cryptic CYP21 promoter element? Hum Mol Genet. 1995;4:2109–2116. doi: 10.1093/hmg/4.11.2109. [DOI] [PubMed] [Google Scholar]
- 37.Valcourt U, Alcaraz LB, Exposito JY, Lethias C, Bartholin L. Tenascin-X: beyond the architectural function. Cell Adhesion & Migration. 2015;9(1–2):154–165. doi: 10.4161/19336918.2014.994893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grumet M, Hoffman S, Crossin KL, Edelman GM. Cytotactin, an extracellular matrix protein of neural and non-neural tissues that mediates glia-neuron interaction. Proc Natl Acad Sci USA. 1985;82:8075–8079. doi: 10.1073/pnas.82.23.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearson CA, Pearson D, Shibahara S, Hofsteenge J, Chiquet-Ehrismann R. Tenascin: cDNA cloning and induction by TGF-β. EMBO J. 1988;7:2977–2982. doi: 10.1002/j.1460-2075.1988.tb03160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rathjen FG, Wolff JM, Chiquet-Ehrismann R. Restrictin: A chick neural extracellular matrix protein involved in cell attachment co-purifies with the cell recognition molecule F11. Development. 1991;113:151–164. doi: 10.1242/dev.113.1.151. [DOI] [PubMed] [Google Scholar]
- 41.Schachner M, Taylor J, Bartsch U, Pesheva P. The perplexing multifunctionality of janusin, a tenascin-related molecule. Perspect Dev Neurobiol. 1994;2:33–41. [PubMed] [Google Scholar]
- 42.Neidhardt J, Fehr S, Kutsche M, Lohler J, Schachner M. Tenascin-N: Characterization of a novel member of the tenascin family that mediates neurite repulsion from hippocampal explants. Mol Cell Neurosci. 2003;23:193–209. doi: 10.1016/s1044-7431(03)00012-5. [DOI] [PubMed] [Google Scholar]
- 43.Scherberich A, Tucker RP, Samandari E, Brown-Luedi M, Martin D, Chiquet-Ehrismann R. Murine tenascin-W: A novel mammalian tenascin expressed in kidney and at sites of bone and smooth muscle development. J Cell Sci. 2004;117(Pt 4):571–581. doi: 10.1242/jcs.00867. [DOI] [PubMed] [Google Scholar]
- 44.Spring J, Beck K, Chiquet-Ehrismann R. Two contrary functions of tenascin: dissection of the active sites by recombinant tenascin fragments. Cell. 1989;59:325–334. doi: 10.1016/0092-8674(89)90294-8. [DOI] [PubMed] [Google Scholar]
- 45.Lethias C, Carisey A, Comte J, Cluzel C, Exposito J-Y. A model of tenascin-X integration within the collagenous network. FEBS Lett. 2006;580:6281–6285. doi: 10.1016/j.febslet.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 46.Norenberg U, Wille H, Wolff JM, Frank R, Rathjen FG. The chicken neural extracellular matrix molecule restrictin: similarity with EGF-, fibronectin type III-, and fibrinogen-like motifs. Neuron. 1992;8:849–863. doi: 10.1016/0896-6273(92)90199-n. [DOI] [PubMed] [Google Scholar]
- 47.Burch GH, Bedolli MA, McDonough S, Rosenthal SM, Bristow J. Embryonic expression of tenascin-X suggests a role in limb, muscle, and heart development. Dev Dynamics. 1995;203:491–504. doi: 10.1002/aja.1002030411. [DOI] [PubMed] [Google Scholar]
- 48.Burch GH, Gong Y, Liu W, Dettman RW, Curry CJ, Smith L, Miller WL, Bristow J. Tenascin-X deficiency is associated with Ehlers-Danlos syndrome. Nature Genet. 1997;17:104–108. doi: 10.1038/ng0997-104. [DOI] [PubMed] [Google Scholar]
- 49.Mao JR, Taylor G, Dean WB, Wagner DR, Afzal V, Lotz JC, Rubin EM, Bristow J. Tenascin-X deficiency mimics Ehlers–Danlos syndrome in mice through alteration of collagen deposition. Nat Genet. 2002;30:421–425. doi: 10.1038/ng850. [DOI] [PubMed] [Google Scholar]
- 50.Saga Y, Yagi T, Ikawa Y, Sakakura T, Aizawa S. Mice develop normally without tenascin. Genes Dev. 1992;6:1821–1831. doi: 10.1101/gad.6.10.1821. [DOI] [PubMed] [Google Scholar]
- 51.Forsberg E, Hirsch E, Frohlich L, Meyer M, Ekblom P, Aszodi A, Werner S, Fassler R. Skin wounds and severed nerves heal normally in mice lacking tenascin-C. Proc Natl Acad Sci USA. 1996;93:6594–6599. doi: 10.1073/pnas.93.13.6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weber P, Bartsch U, Rasband MN, Czaniera R, Lang Y, Bluethmann H, Margolis RU, Levinson SR, Shrager P, Montag D, Schachner M. Mice deficient for tenascin-R display alterations of the extracellular matrix and decreased axonal conduction velocities in the CNS. J Neurosci. 1999;19:4245–4262. doi: 10.1523/JNEUROSCI.19-11-04245.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elefteriou F, Exposito JY, Garrone R, Lethias C. Characterization of the bovine Tenascin-X. J Biol Chem. 1997;272:22866–22874. doi: 10.1074/jbc.272.36.22866. [DOI] [PubMed] [Google Scholar]
- 54.Schalkwijk J, Zweers MC, Steijlen PM, Dean WB, Taylor G, van Vlijmen IM, van Haren B, Miller WL, Bristow J. A recessive form of the Ehlers-Danlos syndrome caused by tenascin-X deficiency. New England J Med. 2001;345:1167–1175. doi: 10.1056/NEJMoa002939. [DOI] [PubMed] [Google Scholar]
- 55.Lindor NM, Bristow J. Tenascin-X deficiency in autosomal recessive Ehlers-Danlos syndrome. Am J Med Genet. 2005;135A:75–80. doi: 10.1002/ajmg.a.30671. [DOI] [PubMed] [Google Scholar]
- 56.Demirdas S, Dulfer E, Robert L, Kempers M, van Beek D, Micha D, van Engelen BG, Hamel B, Schalkwijk J, Loeys B, Maugeri A, Voermans NC. Recognizing the tenascin-X deficient type of Ehlers–Danlos syndrome: a cross-sectional study in 17 patients. Clin Genet. 2017;91:411–425. doi: 10.1111/cge.12853. [DOI] [PubMed] [Google Scholar]
- 57.Zweers MC, Bristow J, Steijlen PM, Dean WB, Hamel BC, Otero M, Kucharekova M, Boezman JB, Schalkwijk J. Haploinsufficiency of TNXB is associated with hypermobility type of Ehlers-Danlos Syndrome. Am J Hum Genet. 2003;73:214–217. doi: 10.1086/376564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamada K, Watanabe A, Takeshita H, Matsumoto K. A method for quantification of serum tenascin-X by nano-LC/MS/MS. Clin Chim Acta. 2016;459:94–100. doi: 10.1016/j.cca.2016.05.022. [DOI] [PubMed] [Google Scholar]
- 59.Alcaraz LB, Exposito JY, Chuvin N, Pommier RM, Cluzel C, Martel S, Sentis S, Bartholin L, Lethias C, Valcourt U. Tenascin-X promotes epithelial-to-mesenchymal transition by activating latent TGF-β. J Cell Biol. 2014;205:409–428. doi: 10.1083/jcb.201308031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsumoto K, Takayama N, Ohnishi J, Ohnishi E, Shirayoshi Y, Nakatsuji N, Ariga H. Tumour invasion and metastasis are promoted in mice deficient in tenascin-X. Genes Cells. 2001;6:1101–1111. doi: 10.1046/j.1365-2443.2001.00482.x. [DOI] [PubMed] [Google Scholar]
- 61.Levy P, Ripoche H, Laurendeau I, Lazar V, Ortonne N, Parfait B, Leroy K, Wechsler J, Salmon I, Wolkenstein P, Dessen P, Vidaud M, Vidaud D, Bieche I. Microarray-based identification of Tenascin C and Tenascin XB, genes possibly involved in tumorigenesis associated with neurofibromatosis type 1. Clin Cancer Res. 2007;13:398–407. doi: 10.1158/1078-0432.CCR-06-0182. [DOI] [PubMed] [Google Scholar]
- 62.Voermans NC, van Alfen N, Pillen S, Lammens M, Schalkwijk J, Zwarts MJ, van Rooij IA, Hamel BCJ, van Engelen BG. Neuromuscular involvement in various types of Ehlers-Danlos syndrome. Ann Neurol. 2009;65:687–697. doi: 10.1002/ana.21643. [DOI] [PubMed] [Google Scholar]
- 63.Penisson-Besnier I, Allamand V, Beurrier P, Martin L, Schalkwijk J, van Vlijmen-Willems I, Gartioux C, Malfait F, Syx D, Macchi L, Marcorelles P, Arbeille B, Croue A, De Paepe A, Dubas F. Compound heterozygous mutations of the TNXB gene cause primary myopathy. Neuromuscular Disorders. 2013;23:664–669. doi: 10.1016/j.nmd.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 64.Sakiyama T, Kubo A, Sasaki T, Yamada T, Yabe N, Matsumoto K, Futei Y. Recurrent gastrointestinal perforation in a patient with Ehlers–Danlos syndrome due to tenascin-X deficiency. J Dermatol. 2015;42:511–514. doi: 10.1111/1346-8138.12829. [DOI] [PubMed] [Google Scholar]
- 65.Gbadegesin RA, Brophy PD, Adeyemo A, Hall G, Gupta IR, Hains D, Bartkowiak B, Rabinovich CE, Chandrasekharappa S, Homstad A, Westreich K, Wu G, Liu Y, Holanda D, Clarke J, Lavin P, Selim A, Miller S, Wiener JS, Ross SS, Foreman J, Rotimi C, Winn MP. TNXB mutations can cause vesicoureteral reflux. J Am Soc Nephrol. 2013;24:1313–1322. doi: 10.1681/ASN.2012121148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tokhmafshan F, Brophy PD, Gbadegesin RA, Gupta IR. Vesicoureteral reflux and the extracellular matrix connection. Pediatr Nephrol. 2017;32:565–576. doi: 10.1007/s00467-016-3386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Geffrotin C, Garrido JJ, Tremet L, Vaiman M. Distinct tissue distribution in pigs of Tenascin-X and Tenascin-C transcripts. Eur J Biochem. 1995;231:83–92. doi: 10.1111/j.1432-1033.1995.tb20673.x. [DOI] [PubMed] [Google Scholar]
- 68.Imura K, Sato I. Identification of the novel localization of tenascin X in the monkey choroid plexus and comparison with the mouse. Euro J Histochem. 2009;53:225–231. doi: 10.4081/ejh.2009.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wei J, Hemmings GP. TNXB locus may be a candidate gene predisposing to schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2004;125:43–49. doi: 10.1002/ajmg.b.20093. [DOI] [PubMed] [Google Scholar]
- 70.Tochigi M, Zhang X, Ohashi J, Hibino H, Otowa T, Rogers M, Kato T, Okazaki Y, Kato N, Tokunaga K, Sasaki T. Association study between the TNXB locus and schizophrenia in a Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:305–309. doi: 10.1002/ajmg.b.30441. [DOI] [PubMed] [Google Scholar]
- 71.Kawakami K, Matsumoto K. Behavioral alterations in mice lacking the gene for Tenascin-X. Biol Pharm Bull. 2011;34:590–593. doi: 10.1248/bpb.34.590. [DOI] [PubMed] [Google Scholar]
- 72.Merke DP, Chen W, Morissette R, Xu Z, Van Ryzin C, Sachdev V, Hannoush H, Shanbhag SM, Acevedo AT, Nishitani M, Arai AE, McDonnell NB. Tenascin-X haploinsufficiency associated with Ehlers-Danlos syndrome in patients with congenital adrenal hyperplasia. J Clin Endocrinol Metab. 2013;98:E379–387. doi: 10.1210/jc.2012-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen W, Kim MS, Shanbhag S, Arai A, VanRyzin C, McDonnell NB, Merke DP. The phenotypic spectrum of contiguous deletion of CYP21A2 and tenascin XB. quadricuspid aortic valve and other midline defects. Am J Med Genet. 2009;149A:2803–2808. doi: 10.1002/ajmg.a.33092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vrzalova Z, Hruba Z, Hrabincova ES, Vrabelova S, Votava F, Kolouskova S, Fajkusova L. Chimeric CYP21A1P/CYP21A2 genes identified in Czech patients with congenital adrenal hyperplasia. Eur J Med Genet. 2011;54:112–117. doi: 10.1016/j.ejmg.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 75.Finkielstain GP, Chen W, Mehta SP, Fujimura FK, Hanna RM, Van Ryzin C, McDonnell NB, Merke DP. Comprehensive genetic analysis of 182 unrelated families with congenital adrenal hyperplasia due to 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2011;96:E161–172. doi: 10.1210/jc.2010-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krone N, Braun A, Roscher AA, Knorr D, Schwarz HP. Predicting phenotype in steroid 21-hydroxylase deficiency? Comprehensive genotyping in 155 unrelated, well defined patients from southern Germany. J Clin Endocrinol Metab. 2000;85:1059–1065. doi: 10.1210/jcem.85.3.6441. [DOI] [PubMed] [Google Scholar]
- 77.Chen W, Xu Z, Sullivan A, Finkielstain GP, Van Ryzin C, Merke DP, McDonnell NB. Junction site analysis of chimeric CYP21A1P/CYP21A2 genes in 21-hydroxylase deficiency. Clin Chem. 2012;58:421–430. doi: 10.1373/clinchem.2011.174037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Morissette R, Chen W, Perritt AF, Dreiling JL, Arai AE, Sachdev V, Hannoush H, Mallappa A, Xu Z, McDonnell NB, Quezado M, Merke DP. Broadening the Spectrum of Ehlers Danlos Syndrome in Patients With Congenital Adrenal Hyperplasia. J Clin Endocrinol Metab. 2015;100:E1143–1152. doi: 10.1210/jc.2015-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen W, Perritt AF, Morissette R, Dreiling JL, Bohn MF, Mallappa A, Xu Z, Quezado M, Merke DP. Ehlers-Danlos Syndrome Caused by Biallelic TNXB Variants in Patients with Congenital Adrenal Hyperplasia. Hum Mutat. 2016;37:893–897. doi: 10.1002/humu.23028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hendriks AG, Voermans NC, Schalkwijk J, Hamel BC, van Rossum MM. Well-defined clinical presentation of Ehlers-Danlos syndrome in patients with tenascin-X deficiency: a report of four cases. Clin Dysmorphol. 2012;21:15–18. doi: 10.1097/MCD.0b013e32834c4bb7. [DOI] [PubMed] [Google Scholar]
- 81.L'Allemand D, Tardy V, Gruters A, Schnabel D, Krude H, Morel Y. How a patient homozygous for a 30-kb deletion of the C4-CYP 21 genomic region can have a nonclassic form of 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2000;85:4562–4567. doi: 10.1210/jcem.85.12.7018. [DOI] [PubMed] [Google Scholar]