Abstract
Thiol-dependent enzymes, including the thioredoxin (Trx) and glutathione (GSH) systems, have recently been found as promising bactericidal targets in multidrug-resistant (MDR) bacteria. We previously discovered that silver acted synergistically with ebselen in the inhibition of the Trx system and also resulted in a fast depletion of GSH in Gram-negative bacteria. Silver has been found by others to improve the sensitivity of bacteria to certain conventional antibiotics. Here, we found that the synergistic antibacterial effects of silver with four conventional antibiotics was correlated with the blockage of bacterial Trx system by silver. The synergistic antibacterial effect came along with the production of reactive oxygen species. All these results suggested that silver primarily enhanced the bactericidal activities of conventional antibiotics towards Gram-negative strains through the upregulation of ROS production.
Introduction
The worldwide ever-increasing of bacterial resistance to the conventional medical antimicrobial agents is currently one of the most serious health crisis for modern medicine1–5. This has serious negative impacts such as decreases of the effectiveness of existing treatments, and causes higher morbidity and mortality rates in patients with infections caused by multidrug-resistant (MDR) bacteria1,2,6. European Antimicrobial Resistance Surveillance Network (EARS-Net) latest data showed high and increasing resistance of Gram-negative bacteria, which are combined with resistance to third-generation cephalosporins, fluoroquinolones, and aminoglycosides for both Escherichia coli and Klebsiella pneumoniae7. Given the fact that even education for the public, controlled use in food animals, and prudent antibiotic prescribing8 cannot stop the growth and spread of bacterial resistance and can only slow and delay it2,8,9. An option to overcome bacterial resistance is the combination of selected antibiotics with each other and with other drugs to improve antibacterial efficacy1,8,10.
We and other groups previously found that 2-phenyl-1,2 benzisoselenazol-3(2H)-one (ebselen), is an effective compound against Gram-positive pathogens (including MDR mycobacteria tuberculosis) by targeting thioredoxin (Trx), which could transfer electrons from NADPH to their substrates via thioredoxin reductase (TrxR) that is critical for bacterial survival4,11,12. Further, the addition of silver acted synergistically with ebselen in treating infections caused by MDR Gram-negative pathogens, including five clinically most difficult-to-treat Gram-negative bacteria: Acinetobacter baumannii, Enterobacter cloacae, Escherichia coli, Klebsiella pneumonia and Pseudomonas aeruginosa. The combination directly inhibited Trx, deplete glutathione (GSH), and induced striking ROS production13. Moreover, the possibility of selection of resistant mutants is very rare14. Thus, thiol-dependent antioxidant systems are promising targets for the development of antibiotics against MDR strains.
Dwyer et al.15 found that antibiotics (ampicillin, gentamicin, and norfloxacin) could alter bacterial redox physiology, and the lethality was accompanied by ROS generation, and Belenky et al.16 profiled the E. coli metabolome and showed that antibiotics (ampicillin, kanamycin, and norfloxacin)-induced metabolic alterations elevated redox state, nucleotide oxidation, and increased oxidative stress by tightly regulated glutathione pools. Kohanski et al.17 stated that all three classes of bactericidal drugs (aminoglycoside, quinolone, and beta-lactam) mediated hydroxyl radical damage. All these above reports pointed out that antibiotics can influence the redox balance in bacteria, but the exact mechanism is still not clear. Meanwhile, Collins and coworkers17,18 proved that ROS production is one of the lethal factors of bactericidal antibiotics, and silver can enhance their antibacterial efficacy.
Antibiotic-mediated cell death is a complex, multi-faceted process that cannot be fully accounted for by the direct interactions of antibiotics with specific cellular targets. Here, we try to figure out whether antibiotics representing five different functional categories (beta-lactams, aminoglycosides, synthesis, tetracycline, and macrolides) can work synergistically with silver against Gram-negative bacteria through targeting redox system as ebselen and silver do13. Our results showed that silver could enhance certain antibiotics antibacterial effects against Gram-negative bacteria which are correlated to reactive oxygen species (ROS) production.
Results
Silver and certain antibiotics in combination exhibited synergistic antibacterial effects against E. coli
The antibacterial effects of silver nitrate (AgNO3) and nine antibiotics representing five different functional categories (beta-lactams, aminoglycosides, synthesis, tetracycline, and macrolides) on the growth of a model Gram-negative bacterium, E. coli, which were detected in the 96 wells microplates. Overnight cultured E. coli DHB4 cells were diluted 1:1000 times in Luria Bertani (LB) medium, and incubated with serial dilutions of ionic silver (Ag+) as a nitrate salt and 9 antibiotics in combinations, separately, for 24 h. Ebselen was used as the positive control, which acted synergistically with silver against Gram-negative bacteria in our recent report13. The results here showed that 4 (gentamicin, kanamycin, geneticin, tetracycline) out of 9 antibiotics had synergistic activity on E. coli DHB4 growth under the conditions we tested (Table S1). Further, the Bliss model was used to determine the nature of the therapeutic effects exhibited by the Ag+ and antibiotics in combinations. We quantified the degree of synergy at 1 h and 4 h between Ag+ and 4 antibiotics (gentamicin, kanamycin, geneticin, and tetracycline) in combinations, and the results showed that Ag+ and 4 antibiotics indeed had synergistic antibacterial effects against E. coli (Fig. 1). All the results pointed out that Ag+ could enhance the antibacterial effects of certain antibiotics against Gram-negative bacteria. In the following experiments, we used these 4 antibiotics for further studies.
Figure 1.
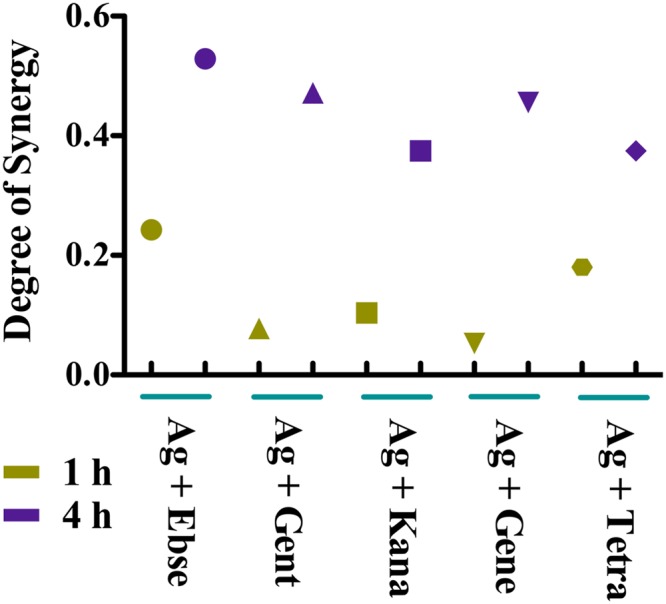
The Bliss model for synergy confirms the synergistic effects, between Ag+ and 4 antibiotics, against a model Gram-negative bacteria, E. coli. The degree of synergy was quantified, using the Bliss Model for Synergy, after 1 and 4 h of treatment with 5 µM AgNO3 in combination with the following antibiotics: 80 µM gentamicin, 80 µM kanamycin, 80 µM geneticin, 80 µM tetracycline, and 80 µM Ebselen was used as the positive control.
ROS was one of the lethal factors for synergistic bactericidal effects of Ag+ and antibiotics
Ag+ and ebselen in combination could induce a high level of ROS production13, and the effects of Ag+ and antibiotics in combinations need further studies. We, therefore, detected the production levels of ROS in Ag+ and 4 antibiotics combinations treated cells, and Ag+ and ebselen in combination was used as the positive control. The results showed that treatment with 5 μM Ag+ alone could not cause a ROS response, and 5 μM Ag+ and 80 μM antibiotics in combinations resulted in the upregulation of ROS (p < 0.0001) when compare with either antibiotics (Fig. 2A) or silver (Fig. S1A). Furthermore, the increased levels of H2O2 caused by the treatment with 5 μM Ag+ and 80 μM antibiotics in combinations were also notarized by Amplex Red assay15 (p < 0.0001) (Figs 2B and S1B). All the results demonstrated that ROS might be one of the determining factors for synergistic bactericidal effects of Ag+ and antibiotics in combinations against E. coli.
Figure 2.
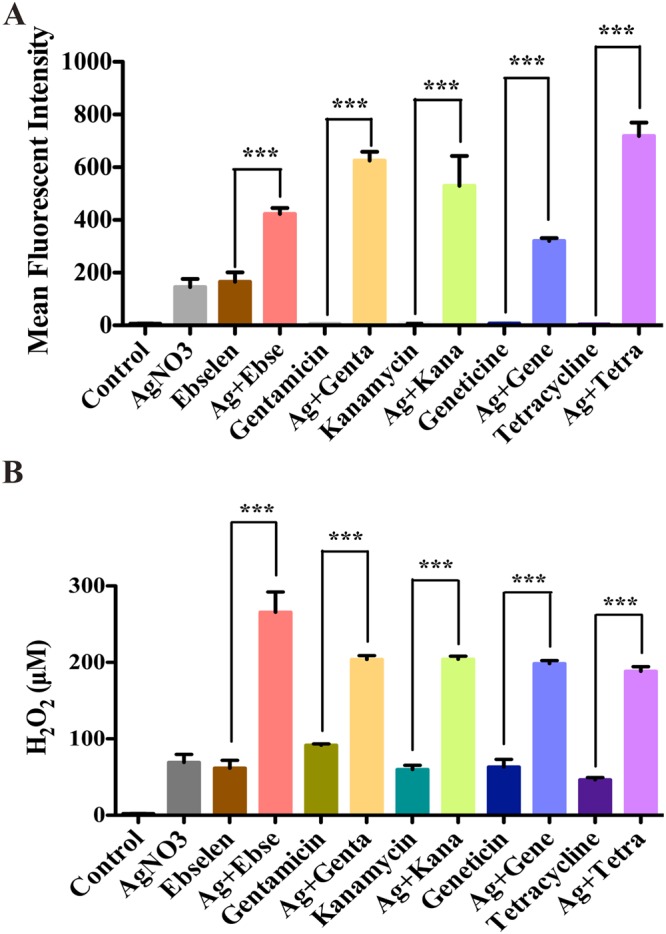
ROS was one of the lethal factors for antibacterial toxicity of silver and antibiotics in combinations. E. coli DHB4 cells grown to OD600 nm of 0.4 were incubated with 80 µM antibiotics and 5 µM Ag+ in combinations, and 80 µM ebselen and 5 µM Ag+ in combination was used as the positive control. (A) The producion level of ROS was detected by flow cytometry (CyAnadp, Beckman coulter), and mean fluorescent intensity (MFI) ± .s. d. of H2DCF-DA-stained E. coli was detected. (B) The production level of H2O2 was detected by the Amplex® Red Hydrogen Peroxide/Peroxidase method (Invitrogen). Reaction buffer contains 50 μM Amplex® Red reagent, 0.1 U/mL HRP, and the indicated amount of H2O2 in 50 mM sodium phosphate buffer (pH 7.4), which was incubated for 30 minutes at 25 °C and further verified with absorbance at 560 nm. The background obsorbance was detected by a non-H2O2 control reaction, which has been subtracted from each value. Data that are presented here as means ± s. d. of 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t-test).
Ag+ could disrupt bacterial Trx system
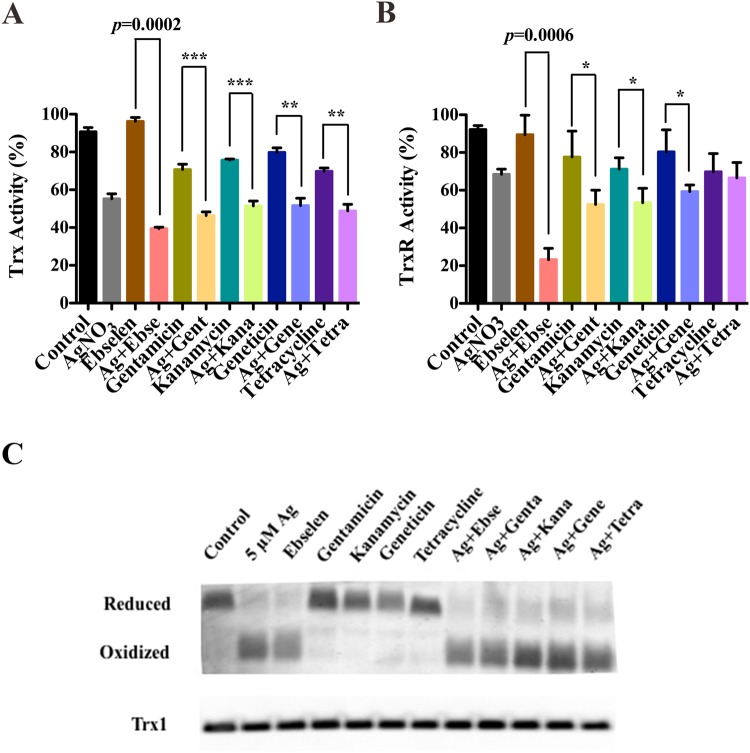
One of the main functions of thiol-dependent antioxidant systems is to scavenge ROS to keep cellular redox homeostasis and protect against oxidative stress. The disruption of the Trx and GSH systems may responsible for the upregulation of ROS. Ag+ and ebselen in combination has been proven to target both bacterial Trx and GSH systems13, while the effects of Ag+ and antibiotics in combinations need further studies. E. coli DHB4 cells grown to OD600 nm of 0.4 were incubated with 5 µM Ag+ and 80 µM antibiotics in combinations, and Ag+ and ebselen in combination was used as the positive control. Results here showed that after 10 min treatment, the Trx activities in cell extracts treated by Ag+ and antibiotics in combinations were dramatically inhibited compared with antibiotics or control group (Fig. 3A, p < 0.001); meanwhile, the TrxR activities in cell extracts treated by Ag+ and antibiotics in combinations were also statistically lowered when compared with antibiotics or control group (Fig. 3B, p < 0.05). We also gained the same results when we prolonged the treatment time to 60 min (Fig. S2). But, there were no statistic difference between Ag+ and antibiotics in combinations and silver (Fig. 3, p > 0.05). These results suggested that silver could disrupt bacterial Trx system; meanwhile, silver and antibiotics in combinations have direct influences on Trx1 when compared with antibiotics alone.
Figure 3.
Silver could directly disrupt bacterial Trx system. E. coli DHB4 cells grown to OD600nm of 0.4 were incubated with antibiotics and Ag+ in combinations for 10 min, and ebselen and Ag+ in combination was used as the positive control. (A) Trx and (B) TrxR activities were detected by using DTNB reduction assay in the presence of TrxR or Trx in E. coli DHB4 cells extracts. (C) E. coli DHB4 cells grown to OD600 nm of 0.4 were incubated with antibiotics and Ag+ in combinations for 60 min, and ebselen and Ag+ in combination was used as the positive control. E. coli DHB4 cells extracts were precipitated by 5% TCA, and further alkylated with 15 mM AMS and the redox state of Trx1 was analyzed by redox Western blot. The mean ± s. d. of 3 independent experiments was calculated. The t-test significances were calculated between control and rest groups, and *p < 0.05, **p < 0.01, ***p < 0.001.
Bring into correspondence with above observation, the redox state of Trx1 verified by Redox Western Blot was also affected by Ag+ and antibiotics in combinations after 60 min treatment when compared to antibiotics. Trx1 was in reduced form in untreated cells (negative control), and became oxidized upon the treatment by Ag+ and antibiotics in combinations (Fig. 3C). In contrast, only 10 min treatment by Ag+ and antibiotics in combinations could not cause Trx1 oxidization (Fig. S3). At the same time, the total protein levels of Trx1were not affected following a 10 min or 60 min treatment by Ag+ and antibiotics in combinations (Figs 3C and S3). These results showed that when targeting Trx system, silver and antibiotics in combinations were not acting as fast as silver and ebselen do, which could effect Trx system in 10 min13.
Ag+ and antibiotics could not directly disrupt bacterial GSH system
In our previous work, 5 µM Ag+ and 80 μM ebselen in combination has been proven to deplete the GSH after 10 min treatment13. In our study, only 5 µM Ag+ and 80 μM gentamicin or kanamycin in combinations could slightly deplete the total GSH amount in cell extracts when compared with antibiotics themselves (p < 0.05) (Fig. 4A), meanwhile other combinations showed no differences (Fig. 4A) (p > 0.05). Further, the protein S-glutathionylation was decreased only in Ag+ and ebselen in combination incubated bacteria13, but not in those bacteria cells treated with 5 µM Ag+ and antibiotics in combinations for 10 min (Fig. 4B) or 60 min treatments (Fig. 5B).
Figure 4.
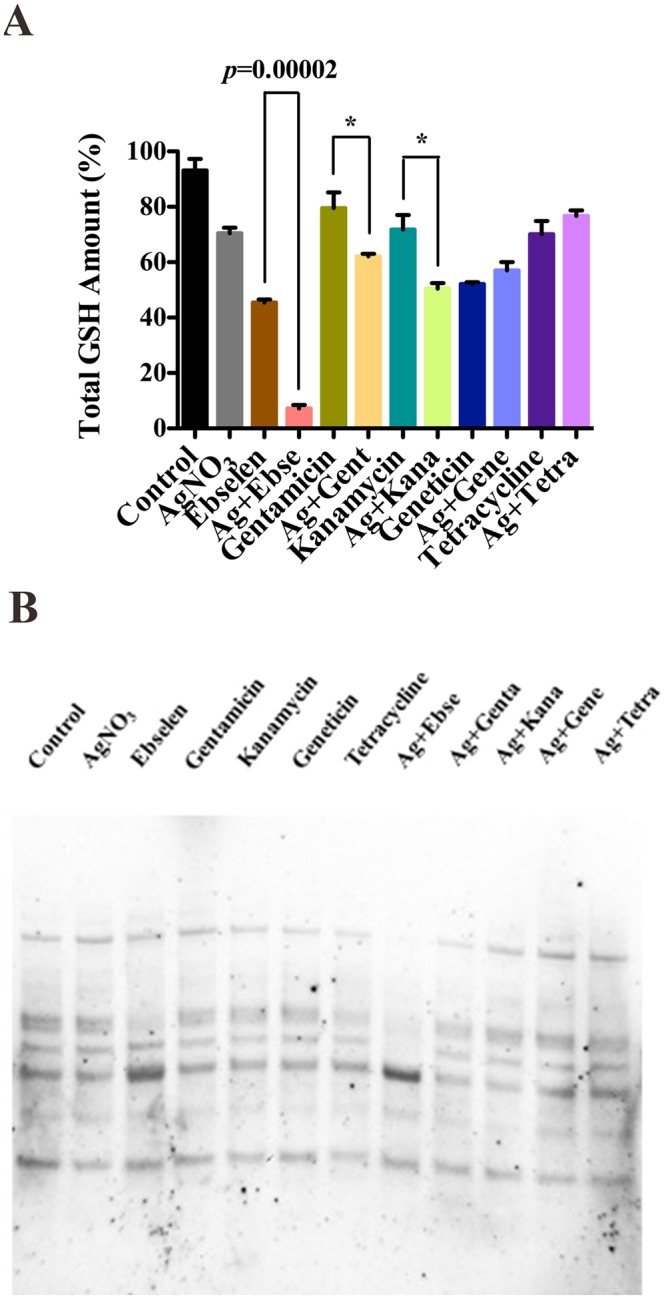
Silver and antibiotics in combinations could not directly disrupt the bacterial GSH system. E. coli DHB4 cells grown to OD600nm of 0.4 were incubated with antibiotics and Ag+ in combinations for 10 min, and ebselen and Ag+ in combination was used as a positive control. (A) Total amounts of GSH were detected by GR-coupled DTNB reduction assay in E. coli DHB4 cells extracts. (B) Changes in protein S-glutathionylation in E. coli. The mean ± s. d. of 3 independent experiments is calculated. The t-test significances were calculated between control and test groups, and *p < 0.05, **p < 0.01, ***p < 0.001.
Figure 5.
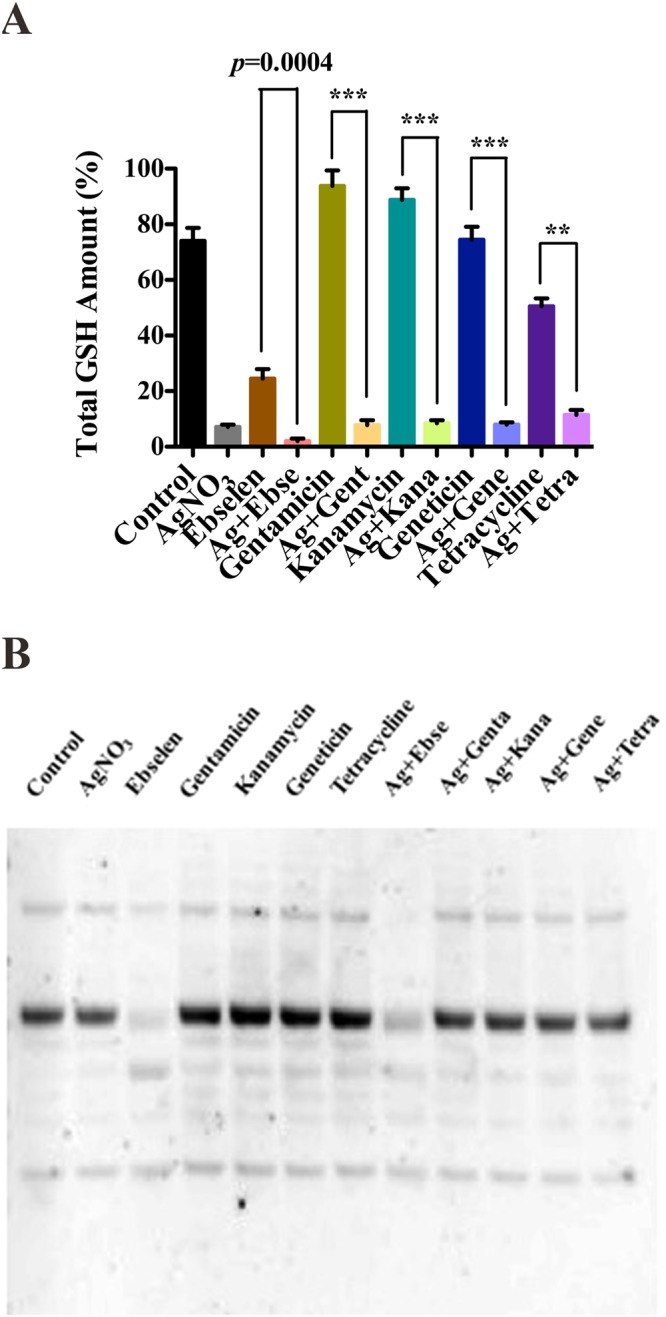
Silver and antibiotics in combinations could not directly disrupt the bacterial GSH system. E. coli DHB4 cells grown to OD600 nm of 0.4 were incubated with antibiotics and Ag+ in combinations for 60 min, and ebselen and Ag+ in combination was used as the positive control. (A) Total amounts of GSH were detected by GR-coupled DTNB reduction assay in E. coli DHB4 cells extracts. (B) Changes in protein S-glutathionylation in E. coli. The mean ± s. d. of 3 independent experiments was depicted. The t-test significances were calculated between control and rest groups, and *p < 0.05, **p < 0.01, ***p < 0.001.
All the results above suggested that silver and antibiotics had no direct effects on bacterial GSH system when acting against Gram-negative bacteria.
Five most difficult-to-treat MDR Gram-negative clinical isolates were highly sensitive to Ag+ and ebselen in combination
We have isolated five clinically most difficult-to-treat MDR Gram-negative pathogen strains: Acinetobacter baumannii, Enterobacter cloacae, Escherichia coli, Klebsiella pneumonia and Pseudomonas aeruginosa. Overnight cultures of five isolates were diluted 1:1000 times in LB medium, and incubated with serial concentrations of Ag+ and antibiotics in combinations for 24 h (Table 1). The results showed that Ag+ and antibiotics in combinations exhibited weak antibacterial effects on these MDR Gram-negative bacteria. Meanwhile, Ag+ and ebselen in combination might be the only effective antibiotic against a range of resistant bacteria. The results might be explained by the fact that the synergistic antibacterial effect of silver and antibiotics only dependent on the upregulation the ROS production.
Table 1.
MIC of silver (μM) in the presence of different antibiotics against MDR Gram-negative bacteria.
| Antibiotics (4 μM) | MIC of silver (μM) | ||||
|---|---|---|---|---|---|
| KP-2 | AB-1 | PA-1 | ECL-2 | ECO-1 | |
| Genta | 160 | 80 | 40 | 160 | 40 |
| Kana | 160 | 80 | 80 | 160 | 40 |
| Gene | 160 | 80 | 80 | 160 | 40 |
| Tetra | 160 | 80 | 80 | 160 | 80 |
| Ebse | 40 | 10 | 20 | 10 | 5 |
Gentamycin: Genta; Kanamycin: Kana; Geneticin: Gene; Tetracycline: Tetra; Ebse: Ebselen.
Discussion
Recently a first new antibiotic in decades called Teixobactin was described inhibiting the peptidoglycan cell wall synthesis3. However, Teixobactin only inhibits Gram-positive bacteria like Staphylococcus aureus. Gram-positive bacteria in general have several differences to Gram-negative one which have a cell wall with an extra membrane. Most of Gram-positive bacteria lack GSH and associated enzymes like glutaredoxins19.
The current antibiotics principles are mainly based on disruption of protein synthesis, cell wall or cell membrane assembly, DNA or RNA replication16. We recently showed that targeting bacterial thiol-dependent redox enzymes is a distinct antibacterial mechanism compared with traditional antibiotics4,13.
There are two prime thiol-dependent enzyme systems, namely, thioredoxin (Trx) system, and glutathione (GSH)/glutaredoxin (Grx) system19. For the Trx system, electrons transferoccurs from NADPH to their substrates via thioredoxin reductases (TrxR)20–23; meanwhile, for GSH/Grx system, electron transfer from NADPH to their substratesvia glutathione reductases (GR)21,24,25. Thiol-dependent enzymes are critical for ribonucleotide reductase (RNR) and DNA replication and repair, ordefense against oxidative stress via peroxiredoxins (Prxs) and methionine sulfoxide reductases (MSRs)21,24,25. Therefore, these two systems are critical for cell viability and proliferation.
Trx systems are ubiquitous in all bacteria, whereas the GSH system, as mentioned above, has been found to be absent in many pathogenic bacteria, including nearly all Gram-positive species19,22,26. We previously reported that ebselen, which is a substrate of mammalian TrxR but a competitive inhibitor of bacterial TrxR, executs selective antibacterial effect toward Gram-positive bacteria4,12,27,28. Further, we recently presented that silver and ebselen in synergistic combination had a strong selective bactericidal effect against Gram-negative bacterial infections. The combination directly inhibited both bacterial Trx and TrxR, and depleted intracellular GSH13. Based on these new findings, and combined with other published reports about antibiotics having influence on redox balance15–17, and that silver could sensitize Gram-negative bacteria to conventional antibiotics, thus we asked whether they also targeted the redox systems.
Drug-drug interactions can be classified as synergistic, antagonistic, or additive (no interaction). Nine antibiotics used in this work represent five different functional categories (beta-lactams, aminoglycosides, synthesis, tetracycline, and macrolides). Among them, total synthesis and tetracycline are broad-spectrum, aminoglycosides are particularly effective against Gram-negative bacteria, and beta-lactams and macrolides are particularly effective against Gram-positive bacteria. In the work reported here, 4 out of 9 antibiotics acted synergistically with silver against E. coli, a model Gram-negative bacterium (Fig. 1), which might occur through inducing ROS production as one of the lethal strategies (Figs 2 and 3).
E. coli has one TrxR, two Trxs (Trx1, and Trx2), and three major thiol peroxidases (bacterioferrit in comigratory protein (BCP), thiol peroxidase (Tpx), and alkyl hydroperoxide peroxidase subunit C (AhpC)29,30. Trx1 in E. coli is involved in protein repair by providing the electrons to E. coli methionine sulfoxide reductases (Msr)20, which participates in the protection of E. coli against oxidative damage from reactive nitrogen intermediates31. Trx1 in E. coli also acts as a specific reductase for homodimeric Tpx to scavenge ROS32. Thus, the decrease of Trx1 activity via its oxidization is at least partially responsible for the ROS production to cause E. coli cell death. Also the oxidative folding of disulfide containing membrane proteins is dependent on Trx and is possibly targeted33.
Meanwhile, although silve could inhibit Trx and TrxR (Fig. 3), yet the effect targeting GSH system is not universal (Figs 4 and 5). The presence of the GSH-Grx system in E. coli may be regarded as a backup for the Trx system. GSH/Grxs in E. coli participate in the antioxidant process by deglutathionylation and transfer electrons to ribonucleotide reductase. Silver and antibiotics in combinations showed no effects on GSH amount, and the S-glutathionylated proteins are not much different from that of in control group (Figs 4 and 5). Thus, this might be one of the reason to explain the anti-MDR-Gram-negative bacteria activities of silver and antibiotics in combinations were much weaker than silver and ebselen in combination (Table 1 and Fig. 6).
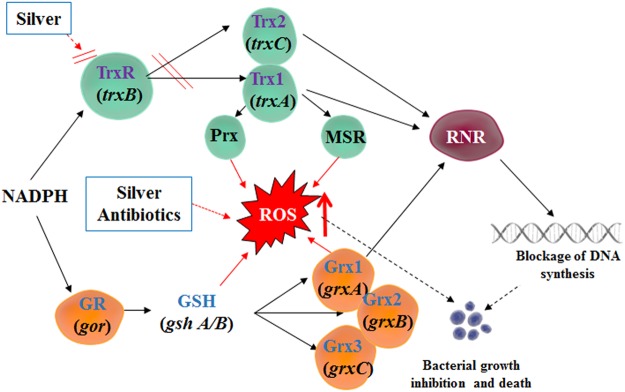
Figure 6.
A graphic illustration that showed mechanisms for the synergistic antibacterial activity of Ag+ with antibiotics in combination. Antibiotics and Ag+ could result in the upregulation of ROS production, and Ag+ could directly inhibit E. coli TrxR and Trx, both of which determine cell death. (A) Ag+ was a strong inhibitor of both E. coli thioredoxin (Trx) and thioredoxin reductase (TrxR). (B) Antibiotics and Ag+ could result in upregulation of ROS production to determine cell death. (C) It is time-dependent. The inhibition is stronger in 60 min treatment than 10 treatment. Information: Trx, thioredoxin; TrxR, thioredoxin reductase; GSH, glutathione; GR, glutathione reductase; Grx: glutaredoxin; Prx, peroxiredoxins; MSR, methionine sulfoxide reductase; RNR, ribonucleotide reductase; ROS, reactive oxygen species.
All in all, silver enhanced the antibacterial effects of certain antibiotics against Gram-negative bacteria through upregulate the ROS production. In order to be more effective, the Trx and GSH systems must be targeted, as well, since both systems are particularly important for ribonucleotide reductase in bacteria which is essential for DNA replication and repair.
Materials and Methods
Bacterial strains
Escherichia coli (E. coli) DHB4 cells and multidrug-resistance (MDR) Gram-negative clinical isolatess (Tables 2 and 3) were used in in vitro experiments. Five Clinical MDR Gram-negative isolates were obtained from clinical patients in People’s Hospital of China Three Gorges University in Hubei Province, PRC, with all approvals and informed consents. All experiments were performed in accordance with relevant guidelines and regulations, and were subject to protocols approved by the Medical Care and Welfare Committee of China Three Gorges University.
Table 2.
Clinically isolated multidrug-resistant Gram-negative strains used in this work.
| Strain | Description | Source |
|---|---|---|
| KP-2 | K. pneumoniasubsp. pneumonia 0322# | This work |
| AB-1 | Acinetobacter baumannii (A. baumannii) H# | This work |
| PA-1 | Pseudomonas aeruginosa (P. aeruginose) 1298# | This work |
| ECL-2 | E. cloacae 2301# | This work |
| ECO-1 | Escherichia coli (E. coli) 1139# | This work |
Table 3.
Drug sensitivity of clinical isolated multidrug-resistant Gram-negative bacteria.
| Antibiotic | KP-2 | AB-1 | PA-1 | ECL-2 | ECO-1 |
|---|---|---|---|---|---|
| Amikacin | S | R | S | S | S |
| Ampicillin | R | R | R/S | / | S |
| Aztreonam | R | R | R | R | R |
| Cefazolin | R | R | R | / | R |
| Cefepime | R | R | R | R | R |
| Cefotaxime | R | R | R | R | R |
| Ceftazidime | R | R | R | R | R/S |
| Chloramphenicol | R | R | / | R | S |
| Ciprofloxacin | R | R | R | R | R |
| Gentamicin | R | R | S | R | S |
| Imipenem | S | R | S | R | S |
| Levofloxacin | R | R | R | R | R |
| Meropenem | S | R | R | R | S |
| Piperacillin | R | R | R/S | R | R |
| Polymyxin | / | S | S | S | / |
| Sulbactam | R | R | / | / | R/S |
| Sulfanilamide | R | / | R | R | R |
| Tazobactam | R | R | R/S | R | S |
| Tetracycline | R | R | R | R | R |
*R: resistant; S: sensitive.
Antibiotics and chemicals
Bacteria cells were cultured in Luria Bertani (LB) medium (EMD millipore). 2-Phenyl-1, 2-benzisoselenazol-3(2H)-one (ebselen) (Daiichi), 9 antibiotics (Table 4): ampicillin, carbenicillin, gentamicin, streptomycin, geneticin, kanamycin, chloramphenicol, tetracycline, erythromycin, silver nitrate (Sigma-Aldrich), Methoxypolyethylene glycol maleimide (MeO-PEG-Mal) (Sigma-Aldrich), Iodoacetamide (IAM) (Sigma-Aldrich), protease inhibitor cocktails (Roche), N-acelytcysteine (NAC) (Sigma-Aldrich), DCTM protein assay (Bio-RAD), E. coli DHB4 TrxR, sheep anti-E. coli Trx1 antibodywas from IMCO Corp. (Stockholm, Sweden; http://www.imcocorp.se), Rabbit anti-sheep IgG-HRP (Santa cruz), IgG2a mouse monoclonal antibody for glutathione-protein complexes (VIROGEN), 4–12% bolt Bis-Tris gel (VWR), all the other reagents were from Sigma-Aldrich.
Table 4.
Antibiotics used in the study with their dosages and primary targets.
| Antibiotic | Abbreviation | Dose (µM) used | Primary target |
|---|---|---|---|
| Ampicillin | Amp | 0/1/2/4 | Cell wall formation |
| Carbenicillin | Car | 0/1/2/4 | Cell wall formation |
| Chloramphenicol | Chl | 0/1/2/4 | Protein synthesis, 50S ribosomal subunit |
| Erythromycin | Ery | 0/1/2/4 | Protein synthesis, 50S ribosomal subunit |
| Gentamicin | Gent | 0/1/2/4/80 | Protein synthesis, 30S ribosomal subunit |
| Geneticin | Gene | 0/1/2/4/80 | Protein synthesis, 30S ribosomal subunit |
| Kanamycin | Kan | 0/1/2/4/80 | Protein synthesis, 30S ribosomal subunit |
| Streptomycin | Str | 0/1/2/4 | Protein synthesis, 30S ribosomal subunit |
| Tetracycline | Tet | 0/1/2/4/80 | Protein synthesis, 30S ribosomal subunit |
Synergistic antibacterial activity of silver and antibiotics in combinations on the E. coli DHB4 growth
E. coli DHB4 cells from frozen stock were grown overnight at 37 °C, 400 rpm. The overnight culture was diluted 1:100 with 5 ml of LB medium in 15 ml tubes and incubated at 37 °C at 400 rpm. Overnight cultured DHB4 cells were grown until an OD600 nm of 0.4 and were used for antibiotic treatment. Briefly, DHB4 cells were diluted 1:1,000 into 100 µl of LB medium. Serial dilutions of antibiotics 100 µl (0, 1, 2, 4 µM) and silver nitrate (AgNO3, 0, 1.25, 2.5, 5, 10, 20, 40, 80 µM) were added. The minimum inhibitory concentration (MIC) was verified as the lowest concentration of drugs that inhibited 90% of growth compared to the untreated cells after 24 hours culture at 37 °C. The cultures treated with the same serial dilutions of 100 µlebselen and silver nitratewere used as the positive control.
Quantifying synergy of ebselenand silver using the bliss model
Drug synergism was determined using the Bliss Independence Model, which calculates a degree of synergy using the following formula: S = (fX0/f00)(f0Y/f00) − (fXY/f00), where fXY refers to the wild-type growth rate in the presence of the combined drugs at a concentration X, for one of the drugs, and Y for the other; fX0 and f0Y refer to the wild-type growth rates in the presence of the individual drugs at a concentration of X and Y, respectively; f00 refers to the wild-type growth rate in the absence of drugs; and S corresponds to the degree of synergy, a parameter that determines a synergistic interaction for positive values and an antagonistic interaction for negative ones. Growth rates at different time points are determined by calculating the slope of the growth or kill curve being analyzed18.
Detection the production of ROS
The E. coli DHB4 were grown until the OD600 nm of 0.4 in 5 ml LB medium, and the DHB4 cells were incubated with silver and antibiotics in combinations for 10 min. To measure the production level of ROS in the treated bacteria, DHB4 cells were harvested by centrifugation at 6,000 rpm for 5 min and thoroughly washed by PBS, which were further stained by 5 µM H2DCF-DA that protected from light for 20 min. After the incubation, treated cells were spun down and re-suspended in PBS, and the amount of ROS was detected by flow cytometry (CyAnadp, Beckman coulter).
Measurement of H2O2 Production
The E. coli DHB4 were grown until the OD600 nm of 0.4 in 5 ml LB medium, and the DHB4 cells were incubated with silver and antibioticsin combinations for 10 min. Treated cells were harvested by centrifugation at 6,000 rpm for 5 min and thoroughly washed 3 times by PBS, and sonicated for 10 s. 50 μl samples were incubated with 50 μM Amplex® Red reagent, and 0.1 U/mL HRP in 50 mM sodium phosphate buffer (pH 7.4) for 30 minutes at 25 °C that protected from light and measured with absorbance at 560 nm (Molecular Probes, Eugene, OR).
Detection of Trx/TrxR activities and GSH amount in antibiotics and silver treated E. coli cell lysates
E. coli DHB4 were grown until the OD600 nm of 0.4 in LB medium, and the DHB4 cells were incubated with 80 µM antibiotics and 5 µM AgNO3 for 10 and 60 min, respectively. The cultures that treated with 5 µM AgNO3 and 80 µM ebselen were used as the positive control. Treated cells were harvested by centrifugation at 6,000 rpm for 5 min and thoroughly washed 3 times by PBS, and cells were further re-suspended in lysis buffer (100 mM NaCl, 20 mM NaF, 2.5 mM EDTA, 1 mM Na3VO4, 2.5 mM EGTA, 20 mM sodium ß-glycerophosphate, 0.5% Triton X-100, 10 mM sodium pyrophosphate, 25 mM Tris·HCl (pH 7.5)), which contains protease inhibitor cocktail, and further lysed by sonication. The treated cell lysates were collected by centrifugation at 13,000 rpm for 20 min and the protein concentrations were measured with the Lowry protein kit.
The TrxR activity in E. coli DHB4 cell extracts was detected by the DTNB reduction activity assay23. These experiments were performed in the 96 microwell plates that contains 5 µM E. coli Trx1, 200 µM NADPH, 1 mM DTNB and 50 mM Tris·HCl (pH 7.5). The absorbance at 412 nm was detected for 5 min with a VERSA microwell plate reader and the slope of initial 2 min was set to verify TrxR activity in cell extracts. The Trx activity was detected by the same method which coupled with 100 nM E. coli TrxR instead of 5 µM E. coli Trx in the above discribed reaction solution. To measure the amount of GSH in cell extracts, 25 µg cell lysates were added into the reaction mixture containing 50 nM GR, 200 µM NADPH, 50 mM Tris·HCl (pH 7.5), 1 mM DTNB and 1 mM EDTA. The absorbance at 412 nm was detected for 5 min.
Redox state of Trx1 in E. coli treated by silver and antibiotics in combinations
E. coli DHB4 were grown until the OD600 nm of 0.4 in LB medium, and the DHB4 cells were incubated with 80 µM antibiotics and 5 µM AgNO3 for 10 and 60 min, respectively. The cultures treated with 5 µM AgNO3 and 80 µM ebselen were used as the positive control. Redox Western blotting was performed to detect the redox state of Trx1 in the DHB4 cells. The treated cells werefurther harvested by centrifugation at 6,000 rpm for 5 min and thoroughly washed 3 times by PBS, and the protein was precipitated by 5% TCA in 1.0 ml. The precipitates were washed throughly 3 times with 1 ml pre-ice-cold acetone and dissolved in 50 mM Tris·HCl (pH 8.5) with 0.5% SDS containing 15 mM MeO-PEG-Mal at 37 °C for 2 hours. Proteins were collected by centrifugation at 13,000 rpm for 20 min and the protein concentration was verified with the Lowry protein kit. Proteins were incubated with SDS-loading buffer at 90 °C for 10 min, and then separated on the 4–12% bolt Bis-Tris gel with MES running buffer (150 V, 40 min). The redox state of Trx1was detected withsheep anti-E. coli Trx1 antibody at 1:1000 dilution, followed by the detection of Chemiluminescence Reagent Plus.
Proteins S-glutathionylation in E. coli cells treated by silver and antibiotics in combinations
Total protein S-glutathionylation of antibiotics and AgNO3 in combinations treated E. coli DHB4 cells was verified by Western blot. E. coli DHB4 were grown until the OD600 nm of 0.4 in LB medium, and the DHB4 cells were incubated with 80 µM antibiotics and 5 µM AgNO3 for 10 and 60 min, respectively. The cells treated with 5 µM AgNO3 and 80 µM ebselen were used as the positive control. Cells were washed 3 times, and re-suspended in lysis buffer (10 mM sodium pyrophosphate, 100 mM NaCl, 2.5 mM EDTA, 25 mM Tris·HCl (pH 7.5), 2.5 mM EGTA, 1 mM Na3VO4, 20 mM sodium ß-glycerophosphate, 0.5% Triton X-100, 20 mM NaF and 50 mM IAM) containing protease inhibitor cocktail. Afte sonication, the treated cell lysates were collected by centrifugation at 13,000 rpm for 20 min. Protein concentration was measured with Lowry protein kit, and Western blot was performed as described above with IgG2a mouse monoclonal antibody (VIROGEN, 101-A/D8) for S-glutathione-protein complexes.
Antibacterial activities of antibiotics and silver on the growth of MDR Gram-negative clinical isolates
Five MDR Gram-negative clinical isolates were grown till an OD600 nm of 0.4, and were diluted 1:1,000 into 100 µl LB medium. Serial dilutions of 100 µl antibiotics (0, 1, 2, 4 µM) and AgNO3 (0, 1.25, 2.5, 5, 10, 20, 40, 80, 160 µM) in combinations were added to the individual wells. The MBC was verified after 16 hours culture at 37 °C. The cells treated with the same serial dilutions of 100 µl ebselen and silver nitratewere used as the positive control.
Statistical analysis
Mean, Standard Deviation (SD) and t-test (two tails, unpaired) significances were calculated in GrapPad Prism Software. *p < 0.05, **p < 0.01, ***p < 0.001.
Data availability
The data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This work was supported by the Swedish Cancer Society (961), the Swedish Research Council Medicine (13X-3529), the K&A Wallenberg Foundation, and grants from Karolinska Institutet, Nature foundation of China No. 81550028 to J.W.
Author Contributions
L.Z., J.L. and A.H. conceived the study. L.Z., J.W., J.L., A.H. designed experiments. L.Z., J.W., Y.G. performed experiments. L.Z., J.L., A.H. wrote the paper. All authors analyzed the results and approved the final version of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Lili Zou and Jun Wang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29313-w.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jun Lu, Email: junlu@swu.edu.cn.
Arne Holmgren, Email: arne.holmgren@ki.se.
References
- 1.Allahverdiyev AM, Kon KV, Abamor ES, Bagirova M, Rafailovich M. Coping with antibiotic resistance: combining nanoparticles with antibiotics and other antimicrobial agents. Expert Rev Anti-Infe. 2011;9:1035–1052. doi: 10.1586/eri.11.121. [DOI] [PubMed] [Google Scholar]
- 2.Panacek, A. et al. Strong and Nonspecific Synergistic Antibacterial Efficiency of Antibiotics Combined with Silver Nanoparticles at Very Low Concentrations Showing No Cytotoxic Effect. Molecules21, 10.3390/molecules21010026 (2016). [DOI] [PMC free article] [PubMed]
- 3.Ling, L. L. et al. A new antibiotic kills pathogens without detectable resistance. Nature517, 455–+, 10.1038/nature14098 (2015). [DOI] [PMC free article] [PubMed]
- 4.Lu J, et al. Inhibition of bacterial thioredoxin reductase: an antibiotic mechanism targeting bacteria lacking glutathione. Faseb J. 2013;27:1394–1403. doi: 10.1096/fj.12-223305. [DOI] [PubMed] [Google Scholar]
- 5.Favrot, L. et al. Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen. Nat Commun4, 10.1038/ncomms3748 (2013). [DOI] [PMC free article] [PubMed]
- 6.Thangamani, S., Younis, W. & Seleem, M. N. Repurposing Clinical Molecule Ebselen to Combat Drug Resistant Pathogens. Plos One10, 10.1371/journal.pone.0133877 (2015). [DOI] [PMC free article] [PubMed]
- 7.Weist K, Hogberg LD. ECDC publishes 2015 surveillance data on antimicrobial resistance and antimicrobial consumption in Europe. Eurosurveillance. 2016;21:39–39. doi: 10.2807/1560-7917.ES.2016.21.46.30399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee CR, Cho IH, Jeong BC, Lee SH. Strategies to Minimize Antibiotic Resistance. Int J Env Res Pub He. 2013;10:4274–4305. doi: 10.3390/ijerph10094274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Kraker MEA, et al. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infec. 2013;19:860–868. doi: 10.1111/1469-0691.12028. [DOI] [PubMed] [Google Scholar]
- 10.Wright, G. D. Q & A: Antibiotic resistance: where does it come from and what can we do about it? Bmc Biol8, 10.1186/1741-7007-8-123 (2010). [DOI] [PMC free article] [PubMed]
- 11.Thangamani, S., Younis, W. & Seleem, M. N. Repurposing ebselen for treatment of multidrug-resistant staphylococcal infections. Sci Rep-Uk5, 10.1038/srep11596 (2015). [DOI] [PMC free article] [PubMed]
- 12.Holmgren A, Masayasu H, Zhao R. Ebselen: A drug targeting thioredoxin and thioredoxin reductase. Free Radical Bio Med. 2002;33:S93–S93. doi: 10.1016/S0891-5849(02)00934-6. [DOI] [Google Scholar]
- 13.Zou L, et al. Synergistic antibacterial effect of silver and ebselen against multidrug-resistant Gram-negative bacterial infections. EMBO Mol Med. 2017;9:1165–1178. doi: 10.15252/emmm.201707661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gustafsson TN, et al. Ebselen and analogs as inhibitors of Bacillus anthracis thioredoxin reductase and bactericidal antibacterials targeting Bacillus species, Staphylococcus aureus and Mycobacterium tuberculosis. Bba-Gen Subjects. 2016;1860:1265–1271. doi: 10.1016/j.bbagen.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Dwyer DJ, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. P Natl Acad Sci USA. 2014;111:E2100–E2109. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belenky P, et al. Bactericidal Antibiotics Induce Toxic Metabolic Perturbations that Lead to Cellular Damage. Cell Rep. 2015;13:968–980. doi: 10.1016/j.celrep.2015.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 18.Morones-Ramirez, J. R., Winkler, J. A., Spina, C. S. & Collins, J. J. Silver Enhances Antibiotic Activity Against Gram-Negative Bacteria. Science translational medicine5, 10.1126/scitranslmed.3006276 (2013). [DOI] [PMC free article] [PubMed]
- 19.Lu J, Holmgren A. The thioredoxin antioxidant system. Free radical biology & medicine. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 20.Holmgren A. Thioredoxin. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 21.Holmgren A. Thioredoxin and glutaredoxin systems. The Journal of biological chemistry. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 22.Martin JL. Thioredoxin–a fold for all reasons. Structure. 1995;3:245–250. doi: 10.1016/S0969-2126(01)00154-X. [DOI] [PubMed] [Google Scholar]
- 23.Holmgren A, Bjornstedt M. Thioredoxin and thioredoxin reductase. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]
- 24.Ritz D, Beckwith J. Roles of thiol-redox pathways in bacteria. Annual review of microbiology. 2001;55:21–48. doi: 10.1146/annurev.micro.55.1.21. [DOI] [PubMed] [Google Scholar]
- 25.Lillig CH, Holmgren A. Thioredoxin and related molecules–from biology to health and disease. Antioxid Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson LJ, White RJ, Chipman JK. Silver and nanoparticles of silver in wound dressings: a review of efficacy and safety. Journal of wound care. 2011;20:543–549. doi: 10.12968/jowc.2011.20.11.543. [DOI] [PubMed] [Google Scholar]
- 27.Zhao R, Holmgren A. A novel antioxidant mechanism of ebselen involving ebselen diselenide, a substrate of mammalian thioredoxin and thioredoxin reductase. Journal of Biological Chemistry. 2002;277:39456–39462. doi: 10.1074/jbc.M206452200. [DOI] [PubMed] [Google Scholar]
- 28.Zhao R, Masayasu H, Holmgren A. Ebselen: A substrate for human thioredoxin reductase strongly stimulating its hydroperoxide reductase activity and a superfast thioredoxin oxidant. P Natl Acad Sci USA. 2002;99:8579–8584. doi: 10.1073/pnas.122061399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potamitou A, Holmgren A, Vlamis-Gardikas A. Protein levels of Escherichia coli thioredoxins and glutaredoxins and their relation to null mutants, growth phase, and function. Journal of Biological Chemistry. 2002;277:18561–18567. doi: 10.1074/jbc.M201225200. [DOI] [PubMed] [Google Scholar]
- 30.Poole LB, Ellis HR. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium .1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry-Us. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 31.St John G, et al. Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. P Natl Acad Sci USA. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baker LMS, Poole LB. Catalytic mechanism of thiol peroxidase from Escherichia coli - Sulfenic acid formation and overoxidation of essential CYS61. Journal of Biological Chemistry. 2003;278:9203–9211. doi: 10.1074/jbc.M209888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J, Holmgren A. The thioredoxin superfamily in oxidative protein folding. Antioxid Redox Signal. 2014;21:457–470. doi: 10.1089/ars.2014.5849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.