Abstract
Streptomyces actuosus ATCC 25421 was famous for producing thiopeptide nosiheptide, which has widely been used as a feed additive for the promotion of animal growth. Herein, we report the complete genome sequence of S. actuosus ATCC 25421, which consists of an 8,145,579‐bp circular chromosome with a G+C content of 72.53 % containing 7 536 protein‐coding genes. The antiSMASH 3.0 program was used to identify 49 biosynthetic gene clusters for putative secondary metabolites, including a putative lantipeptide gene cluster that showed 85 % similarity to the reported informatipeptin biosynthetic gene cluster, indicating that the putative lantipeptide gene cluster has the ability to generate the informatipeptin analogue. Compared with avermipeptin, the lantipeptide precursor peptide (termed avermipeptin B) from S. actuosus ATCC 25421 contains a 14‐aa leader peptide and a 24‐aa core peptide, in which Ile15 was different from Val15 in avermipeptin. We also deduced the structure and the biosynthetic mechanism of avermipeptin B. Heterologous expression of the avermipeptin B biosynthetic gene cluster in S. lividans TK24 was characterized by high‐resolution mass spectrometry (ESI‐MS/MS). Finally, we found that avermipeptin B displayed strong activity against Gram‐positive strains. The genome sequence reported here can encourage us to mine novel secondary metabolites and investigate their biosynthetic mechanism in the future.
Keywords: avermipeptin B, genome sequencing, lantipeptides, secondary metabolites, Streptomyces actuosus
Streptomyces actuosus ATCC 25 421 (also designated as 40037 or NRRL 2954), first isolated from a soil sample in Argentina, has been characterized to produce a typical thiopeptide antibiotic, nosiheptide, which has widely been used as a feed additive for the promotion of animal growth.1 Nosiheptide, one of the parent compounds in the e series of the thiopeptide family, was the first compound identified from S. actuosus ATCC 25421.2 The biosynthesis of nosiheptide has been elaborated previously.3 To explore other secondary metabolites produced by S. actuosus ATCC 25421, whole‐genome sequencing is an efficient method to discover novel natural products in microorganisms.4 Therefore, to further discover novel secondary metabolites, it is necessary to obtain the complete genome sequence of S. actuosus ATCC 25421.
The genome of S. actuosus ATCC 25421 was obtained by using a PacBio RS II platform and an Illumina HiSeq 4000 platform at the Beijing Genomics Institute (BGI, Shenzhen, China), and about 1 030‐Mb Hiseq clean data and 1 009‐Mb PacBio subreads were produced. Four SMRT cells Zero‐Mode Waveguide arrays of sequencing were used by the PacBio platform to generate the set of subreads.5 The average depth of the genome coverage was 120‐fold. The complete genome of S. actuosus ATCC 25421 was composed of a circular chromosome of 8 145 579 bp with an average G+C content of 72.53 %. The chromosome harbored 7 536 protein‐coding genes, 18 rRNA genes, 70 tRNA genes, and 16 sRNA genes (Figure 1 and Table S1). Based on the clusters of the orthologous genes of proteins (COG), gene ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) designation, 6 411, 13 833, and 3 877 categories were classified (Figures S1–S3; note that genes might be classified into more than one category).
Figure 1.
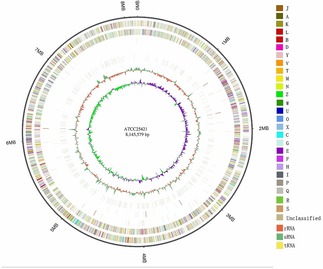
Circular genome map representation of the S. actuosus ATCC 25421 complete chromosome. The outer scale indicates the location in intervals of 1.0 Mbp. From the outer to the inner circle: genome size, forward strand gene (colored according to COG classification), reverse strand gene (colored according to COG classification), forward strand ncRNA, reverse strand ncRNA, repeat, GC content and GC‐SKEW(G‐C/G+C).
Then, the biosynthetic gene clusters were identified for putative secondary metabolites, which were verified by manual inspection by using the antiSMASH 3.0 program.6 A total of 49 putative biosynthetic gene clusters were observed, including two polyketides (PKS), two nonribosomal peptides (NRPS), four saccharides, three terpenes, one siderophore, three fatty acids, one lantipeptide, one ectoine, one indole, one lassopeptide, one PKS‐NRPS, one β‐lactam, one melanin, one saccharide‐melanin‐butyrolactone‐fatty acid‐PKS, one fatty acid‐ butyrolactone, one arylpolyene, one thiopeptide, one bacteriocin, one terpene‐butyrolactone, one bacteriocin‐PKS‐siderophore, and 20 other metabolites (Figure S4 and Table S2). Eight putative gene clusters displayed high homology (>70 % of genes showed homology) to known antimycin, ectoine, desferrioxamine B, spore pigment, nosiheptide, albaflavenone, γ‐butyrolactone, and informatipeptin biosynthetic gene clusters. Five putative gene clusters displayed moderate homology (30–70 % of genes showed homology) to known melanin, carotenoid, macrotetrolide, rabelomycin, and RK‐682 gene clusters. In addition, 35 putative gene clusters displayed low homology (<30 % of genes showed similarity) or no homology to known biosynthetic gene clusters.
The existence of these highly similar gene clusters suggests that S. actuosus ATCC 25421 has tremendous potential to generate these secondary metabolites or their analogues. To date, nosiheptide is the only known secondary metabolite produced by S. actuosus ATCC 25421.1b Surprisingly, one putative lantipeptide gene cluster that showed 85 % similarity to the reported informatipeptin biosynthetic gene cluster was found, indicating that the putative lantipeptide gene cluster has the ability to generate informatipeptin analogues. Informatipeptin was a newly characterized class III lantipeptide from Streptomyces viridochromogenes DSM 40736.7 Lantipeptide are a class of short RiPPs formed through the post ribosomal peptide synthesis pathway and are a prominent group of peptides with pharmaceutical and food industrial applications.3a, 8 To further evaluate whether S. actuosus ATCC 25421 has the ability to generate novel lantipeptide, a number of known type III lantipeptide biosynthetic gene clusters were used to compare with the putative lantipeptide gene cluster in S. actuosus ATCC 25421. The results showed that the putative lantipeptide gene cluster in S. actuosus ATCC 25421 has high homology to the known lantipeptide biosynthetic gene clusters (Figure 2 A). Type III lantipeptide gene clusters have four common open reading frames (ORFs; S, T, B, and A), which encode the precursor peptide, lantipeptide synthetase, and two ABC transporter ATP‐binding proteins.9 The lantipeptide precursor peptide from S. actuosus ATCC 25421 showed 97 % similarity to a known avermipeptin precursor peptide, which contains a 14‐aa leader peptide and a 24‐aa core peptide. The only difference is that Ile15 was replaced by Val15 in avermipeptin (Figure 2 B), indicating that a novel type III lantipeptide may be generated. As it has not been reported previously, we termed the new peptide avermipeptin B, and the avermipeptin B putative biosynthetic gene cluster contained the following four ORFs: aveT encoding lantipeptide synthetase, aveS encoding the precursor peptide, and aveB and A encoding the ABC transporter ATP‐binding proteins (Table S3). The avermipeptin B putative biosynthetic gene cluster showed high homology to the avermipeptin biosynthetic gene cluster, indicating that the structure and the biosynthetic mechanism may also be highly similar. Analysis of other class III lanthipeptides such as avermipeptin, informatipeptin, erythreapeptin, SapB, and griseopeptin showed that the Ser7/Ser10/Cys14‐motif and the Ser16/Ser29/Cys23‐motif might be converted into either labionin (lab) or lan (Figure 2 C and D), which may provide four structural candidates of two lanthionines, two labionins, or one lanthionine and one labionin in the peptide (Figure 2 D). The common structural feature of class III lantipeptides contained the unusual thioether amino acid lanthionine (Lan) and/or methyllanthionine (MeLan, Figure 2 C), which were generated through the modification of the Ser/Thr and Cys amino acids after post‐translation.10 Moreover, dehydration of Thr8 and Ser13 in the avermipeptin B core peptide might convert it into 2,3‐didehydrobutyrine (Dhb) and Dha, which were observed in avermipeptin and informatipeptin (Figure 2 D). Similar to other class III lantipeptides, avermipeptin B biosynthesis was also speculated to occur through the RiPP system. AveT, a lantipeptide synthetase encoded by aveT, was responsible for the conversion of the Ser7/Ser10/Cys14‐motif and the Ser16/Ser29/Cys23‐motif into either lab or lan, as well as Dhb and Dha. Finally, AveB and AveA, two ABC transporter ATP‐binding proteins, respectively encoded by aveB and aveA, may be responsible for the transport and processing to form mature avermipeptin B (Figure 3). To further evaluate the ability of producing avermipeptin B, the complete avermipeptin B gene cluster (Figure 3 A) was cloned into the plasmid pSET152 to construct a new vector pSET152‐ave, which was transformed into the Streptomyces lividans TK24 by conjugation. Then, the extracts from S. lividans TK24/pSET152‐ave were further characterized by high‐resolution HPLC–ESI‐MS/MS according to the method previously reported.9 HPLC analysis results showed that a new peak was observed in the extracts from S. lividans TK24/pSET152‐ave (Figure S5). Fragmentation of the triply charged ion ([M+3 H]3+=691.61) and double charged ion ([M+2 H]2+=1036.92) of avermipeptin B, as well as a complex of daughter ions that consisted of doubly and singly charged ions, were observed from the MS/MS spectra (Figure 4), indicating that heterologous expression of the speculative avermipeptin B gene cluster could produce avermipeptin B in S. lividans TK24. Then, the antimicrobial activity of avermipeptin B was detected and the results showed that avermipeptin B has strong activity against Staphylococcus aureus subsp. aureus ATCC43300, Enterococcus faecalis ATCC29212, and Bacillus subtilis ATCC6633 (Table S4).
Figure 2.

A new class III lantipeptide avermipeptin B biosynthetic cluster in S. actuosus ATCC 25421. A) Comparison of the biosynthetic gene clusters of avermipeptin B and other class III lantibiotics. B) Alignment of type III lantipeptide precursor peptides. C) Structures of lanthionine (Lan) and labionin (Lab), characteristic AAs of type I–III lantibiotics. D) Speculated structure of avermipeptin B and other known class II lantipeptides, including avermipeptin, informatipeptin, erythreapeptin, SapB, and griseopeptin. Dha: 2,3‐didehydroalanine; Dhb: 2,3‐didehydrobutyrine.
Figure 3.
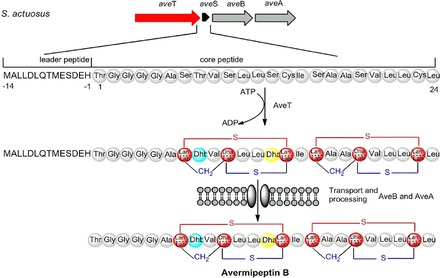
Proposed biosynthetic mechanism of avermipeptin B from the precursor.
Figure 4.
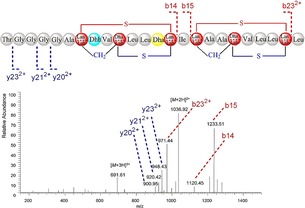
ESI‐MS/MS spectra of avermipeptin B from S. lividans TK24/pSET152‐ave.
In summary, by analyzing the complete genome sequence of S. actuosus ATCC 25421, we found the novel class III lantipeptide avermipeptin B biosynthetic gene cluster in this strain, which could be heterologously expressed in S. lividans TK24, and was characterized by high‐resolution ESI‐MS/MS. The biosynthetic mechanism of avermipeptin B would be further investigated by genetic engineering in our subsequent studies.
Experimental Section
Details of the experimental procedures and materials used in this study, as well as other tables and figures, are given in the Supporting Information. The complete genome sequence of S. actuosus ATCC 25421 was deposited at GenBank under accession number CP029788.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
This work was supported by National Natural Science Foundation of China (nos: 81602512 and 81702905)
W. Liu, F. Sun, Y. Hu, ChemistryOpen 2018, 7, 558.
References
- 1.
- 1a. Shirling E. B., Int. J. Syst. Bacteriol. 1969, 19, 391–512; [Google Scholar]
- 1b. Benazet F., Cartier M., Florent J., Godard C., Jung G., Lunel J., Mancy D., Pascal C., Renaut J., Tarridec P., Experientia 1980, 36, 414–416; [DOI] [PubMed] [Google Scholar]
- 1c. Pascal C., Gaillard C., Moreau M. O., J. Assoc. Off. Anal. Chem. 1979, 62, 976–981. [PubMed] [Google Scholar]
- 2. Prange T., Ducruix A., Pascard C., Lunel J., Nature 1977, 265, 189–190. [DOI] [PubMed] [Google Scholar]
- 3.
- 3a. Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., Camarero J. A., Campopiano D. J., Challis G. L., Clardy J., Nat. Prod. Rep. 2012, 30, 108–160; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3b. Yu Y., Duan L., Zhang Q., Liao R., Ding Y., Pan H., Wendtpienkowski E., Tang G., Shen B., Liu W., ACS Chem. Biol. 2009, 4, 855–864; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3c. Liu W., Ma M., Xue Y., Liu N., Wang S., Chen Y., ChemBioChem 2013, 14, 573–576; [DOI] [PubMed] [Google Scholar]
- 3d. Liu W., Xue Y., Ma M., Wang S., Liu N., Chen Y., ChemBioChem 2013, 14, 1544–1547. [DOI] [PubMed] [Google Scholar]
- 4.
- 4a. Winter J. M., Behnken S., Hertweck C., Curr. Opin. Chem. Biol. 2011, 15, 22–31; [DOI] [PubMed] [Google Scholar]
- 4b. Sun F., Xu S., Jiang F., Liu W., Appl. Microbiol. Biotechnol. 2018, 102, 2225–2234. [DOI] [PubMed] [Google Scholar]
- 5. Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E. E., Nat. Methods 2013, 10, 563–569. [DOI] [PubMed] [Google Scholar]
- 6. Weber T., Kai B., Duddela S., Krug D., Kim H. U., Bruccoleri R., Sang Y. L., Fischbach M. A., Müller R., Wohlleben W., Nucleic Acids Res. 2015, 43, W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mohimani H., Kersten R. D., Liu W. T., Wang M., Purvine S. O., Wu S., Brewer H. M., Pasatolic L., Bandeira N., Moore B. S., ACS Chem. Biol. 2014, 9, 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Willey J. M., Wa V. D. D., Annu. Rev. Microbiol. 2007, 61, 477–501. [DOI] [PubMed] [Google Scholar]
- 9. Völler G. H., Krawczyk J. M., Pesic A., Krawczyk B., Nachtigall J., Süssmuth R. D., ChemBioChem 2012, 13, 1174–1183. [DOI] [PubMed] [Google Scholar]
- 10. Jung G., Angew. Chem. Int. Ed. 1991, 30, 1051–1068. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


