Abstract
PINA is a novel ATPase and DNA helicase highly conserved in Archaea, the third domain of life. The PINA from Sulfolobus islandicus (SisPINA) forms a hexameric ring in crystal and solution. The protein is able to promote Holliday junction (HJ) migration and physically and functionally interacts with Hjc, the HJ specific endonuclease. Here, we show that SisPINA has direct physical interaction with Hjm (Hel308a), a helicase presumably targeting replication forks. In vitro biochemical analysis revealed that Hjm, Hjc, and SisPINA are able to coordinate HJ migration and cleavage in a concerted way. Deletion of the carboxyl 13 amino acid residues impaired the interaction between SisPINA and Hjm. Crystal structure analysis showed that the carboxyl 70 amino acid residues fold into a type II KH domain which, in other proteins, functions in binding RNA or ssDNA. The KH domain not only mediates the interactions of PINA with Hjm and Hjc but also regulates the hexameric assembly of PINA. Our results collectively suggest that SisPINA, Hjm and Hjc work together to function in replication fork regression, HJ formation and HJ cleavage.
INTRODUCTION
DNA replication is a fundamental process of all life forms. In vivo, many replication barriers, such as various DNA lesions, transcribing RNA polymerases, and DNA–RNA loops can slow down the replication fork progression and even make the replication fork stall (1–4). The stalled replication fork needs to be restarted in a timely manner to ensure completion of the DNA replication. Among different pathways to rescue the stalled replication fork, one important pathway is the replication fork regression followed by cleavage of the DNA intermediate and restart of the replication fork (5). In the regression model, when DNA damage occurs, helicases and other motor proteins can promote replication fork regression, leading to the formation of a chicken-foot structure, known as Holliday junction (HJ). HJ is then cleaved by specific endonucleases, or resolvases, generating double stranded DNA breaks (DSBs) which are processed to re-establish the replication fork through homologous recombination. Therefore, in the process of stalled replication fork repair, DNA replication, repair, and recombination are intimately interconnected (4,6–8).
The replication regression and reversal model has been attractive ever since it was proposed 40 years ago (9). This model has been confirmed biochemically and genetically in bacteria (5). Convincingly, Holliday junction structures were experimentally observed in the T4 replication system (10), yeast (11) and cancer cells (12), indicating that the intermediate has physiological relevance. It has now been established that replication fork reversal is an evolutionarily conserved response of cells to various types of DNA replication stress.
So far, proteins that promote HJ formation and migration have been identified in both prokaryotes and eukaryotes. These proteins include the T4 bacteriophage protein UvsW (10), E. coli RecA, RuvAB and RecG (13,14), and eukaryotic recombinase RAD51, RecQ homologues (RECQ1, RECQ5, BLM and WRN), SWI/SNF helicase-like proteins (RAD54, Rad5, HLTF, SMARCAL1 and ZRANB3), and FANCM/Fml1 (5,15,16). Archaea, the third domain of life, resemble Eukarya in many aspects including genetic information processing machinery, cell cycle, cell division mechanism, and membrane remodeling systems (17–22). Therefore, the study of the archaeal biology, including DNA replication and repair, can help understand the evolution of the basic biological processes, and even provide insight into the eukaryotic biological mechanisms that have become complicated during evolution. Although HJ has not been directly observed in archaea, biochemical, structural, and genetic studies have shown that several proteins, mainly Hef (23–25), Hjm/Hel308a (26–31) and Hjc (32–36), have the capacity and potential to process stalled replication fork by HJ formation, migration, or cleavage. Interestingly, as pointed out by Ishino et al., it was the discovery and characterization of the Hef nuclease and helicase that led to the clarification of the enzymatic property of the human homolog FancM (37,38). This is one of many examples that reinforce the significance of investigation on archaeal biology.
Recently, we have identified and characterized a novel ATPase from Sulfolobus islandicus, designated SisPINA (PIN domain-containing ATPase from S. islandicus) (39). The protein is highly conserved among archaeal species; every archaeal strain that has been sequenced possesses a homolog of SisPINA, an indication of its essential role in Archaea. Consistently, we found that the gene is essential for S. islandicus cell survival. We showed that SisPINA is able to drive HJ migration in vitro. The protein interacts physically and functionally with the HJ-specific endonuclease (resolvase), Hjc, and coordinates the HJ processing (39). Structural analysis suggested that ATP binding and hydrolysis cause conformational change of SisPINA which promotes HJ branch migration. However, more biochemical, structural, and functional investigation is needed to fully understand the properties of the protein and reveal the mechanism of homologous recombinational repair at large in Archaea.
Here, we report that SisPINA interacts with Hjm in S. islandicus. SisPINA and Hjm remodel the replication fork in a concerted way. We show that the lysine-rich carboxyl terminal region (residues 493–505) of SisPINA plays a key role in mediating the interaction between SisPINA and Hjm. We solved a novel structure of SisPINA revealing that the last 70 residues form a classical type II KH domain. Based on the results reported here and published previously by our group (39), we propose a model illustrating the mechanisms and roles of PINA in stalled replication fork repair through recombinational repair pathway.
MATERIALS AND METHODS
Construction of the strain encoding a chromosomally-coded N-terminal 6 × His-tagged SisPINA
The SisPINA gene was amplified by PCR using the S. islandicus REY15A genomic DNA as a template and SisPINA-MluI Forward and SisPINA-SalI Reverse as the primers (Supplementary Table S1). The gene fragment was then cloned into pSeSD (40) at the MluI/SalI sites, and the plasmid was named as pSeSD-SisPINA-1. A fragment containing the arabinose promoter, the partial SisPINA gene, and the sequence encoding a 6 × His tag was amplified by PCR using the pSeSD-SisPINA-1 as a template and SisPINA-NcoI Forward and SisPINA-XhoI Reverse as the primers (Supplementary Table S1). Then the fragment was cloned into pMID plasmid at the NcoI/XhoI sites (41), and the plasmid was named as pMID-SisPINA-1. The down-stream fragment of SisPINA was amplified and then cloned into pMID-SisPINA-1 at the SalI/MluI sites, and the resultant plasmid was named as pMID-SisPINA-N-His. pMID-SisPINA-N-His was linearized by digestion with SalI/XhoI. The fragment containing pyrEF-lacs was utilized to transform S. islandicus E233S cells by electroporation as previously described (41). The screening procedure for the strain with chromosomally-coded N-terminal 6 × His-tagged SisPINA (Sis/pMID-SisPINA-in-situ-N-His-T) was the same as for Sis/pMID-SisPINA-T (39).
Expression, purification, and identification of N-terminal 6 × His-tagged SisPINA and its co-eluted proteins in S. islandicus
The cells of Sis/pMID-SisPINA-in-situ-N-His-T were cultured in 2000 ml rich MTSV medium (41) until O.D.600 reached 0.4. The cells were collected by centrifugation at 6300 × g for 10 min, resuspended in 60 ml lysis buffer (25 mM Tris–HCl, pH 8.0, 200 mM NaCl, and 5% glycerol (v/v)), and disrupted by sonication. The sample was centrifuged at 13,500 x g for 40 min. The supernatant was treated with ammonium sulfate at a final concentration of 600 mg/ml. The sample was centrifuged at 13,500 x g for 20 minutes and the precipitant was resuspended in 30 ml lysis buffer and dialyzed against buffer A (25 mM Tris–HCl, pH 8.0 and 200 mM NaCl). The sample was loaded onto the nickel affinity column pre-equilibrated with buffer A. The column was washed with buffer B (25 mM Tris–HCl pH 8.0, 200 mM NaCl and 40 mM imidazole), then SisPINA and its associated proteins were eluted in buffer C (25 mM Tris–HCl, pH 8.0, 200 mM NaCl and 400 mM imidazole). The elution fractions from the nickel affinity column were pooled and analyzed by SDS-PAGE. The proteins of the specific bands were identified by LC-MS/MS (Liquid Chromatography-Mass Spectrometry/Mass Spectrometry) by Hangzhou Jingjie Biotechnology Limited Company (Hangzhou, China).
Gene cloning and construction of plasmid for protein expression in E. coli
Vectors for SisPINA and SisHjc expression were constructed as previously described in Zhai et al. (39). The vector for SisHjm expression was constructed as described in Song et al. (42). The gene encoding SisPINA (1–492) was amplified by PCR using pET15bm-SisPINA as a template and SisPINA-NdeI-Forward and SisPINA (1–492)-SalI-Reverse as primers (Supplementary Table S1). The fragment was digested by NdeI and SalI, and cloned into the NdeI/SalI sites of pET15bm (29). The resultant plasmid was named pET15bm-SisPINA (1–492) and used for the expression of SisPINA (1–492) without any tag (Supplementary Table S3). The plasmid used to express the site-directed mutant SisPINA-R206A was constructed by overlap PCR using the site-specific primers. Unexpectedly, during amplification of the SisPINA-R206A fragment by PCR, two more mutation sites were introduced accidentally: R147K and I199S. This plasmid was named pET22b-SisPINA-R206A/R147K/I199S and used to express the specific mutant without any tag. The fragment encoding KH domain of SisPINA was amplified by PCR using pET15bm-SisPINA as a template and SisPINA-KH-NdeI-Forward and SisPINA-SalI-Reverse as primers. The PCR product was digested by NdeI and SalI, and inserted into the NdeI/SalI sites of pET22b. The plasmid was named pET22b-KH-domain and used for the expression of KH domain of SisPINA without any tag.
Expression and purification of recombinant proteins in E. coli
SisPINA and SisHjc were expressed and purified as described in Zhai et al. (39). SisHjm was expressed as described in Song et al. (42), except the final concentration of IPTG was 0.3 mM. The purification of SisHjm was almost the same as SisPINA, but the ammonium sulfate precipitation saturation was 80%. Similar procedures were used to purify SisPINA (1–492) except the heat-resistant supernatant was precipitated by 60% saturated ammonium sulfate. The procedures for the culture and expression of SisPINA-R206A/R147K/I199S were similar to those for SisPINA while the purification of this mutant was the same as for SisHjc (39). The induction condition for the expression of KH domain of SisPINA was similar to that for SisHjm while the purification procedures were same as those for SisHjc.
Pull-down experiments
The pull-down was performed according to a method previously described in Zhai et al. (39). Briefly, purified SisPINA or SisPINA (1–492) were incubated with SisHjm at 65°C for 45 min, then the proteins were incubated with the pre-equilibrated nickel resin in buffer A (25 mM Tris–HCl, pH 8.0 and 200 mM NaCl) at room temperature for 5 min. The resin was then washed with buffer B (25 mM Tris–HCl, pH 8.0, and 200 mM NaCl, 40 mM imidazole) six times, and protein was then eluted with buffer C (25 mM Tris–HCl, pH 8.0 and 200 mM NaCl, 400 mM imidazole). N-terminal His-tagged SisHjm was used as the positive control, and non-tagged SisPINA or SisPINA (1–492) was used as a negative control. Samples of inputs and elution fractions of SisPINA, SisPINA (1–492), N-His-tagged SisHjm, and N-His-tagged SisHjm/SisPINA and N-His-tagged SisHjm/SisPINA (1–492) mixtures were analyzed by SDS-PAGE with Coomassie blue staining. The same method was used for the analysis of the physical interaction between N-terminal His-tagged SisHjm and non-tagged KH domain of SisPINA.
Gel filtration analysis
The physical interaction between SisPINA and SisHjm were also analyzed by gel filtration chromatography. Purified SisPINA and SisHjm were mixed and incubated at 65°C for 30 min. Samples (500 μl) of SisPINA, SisHjm, or their mixture of the protein mixture was loaded onto a Superdex 200 column (GE Healthcare, UK) pre-equilibrated with a buffer containing 25 mM Tris–HCl, pH 8.0 and 200 mM NaCl. Fractions (1.0 ml each) were collected and analyzed by SDS-PAGE with Coomassie blue staining. The same procedures were used to analyze the interaction between SisHjm and SisPINA (1–492) or SisPINA-R206A/R147K/I99S.
Preparation of DNA substrates and assays for DNA helicase and HJ cleavage activity
Oligonucleotides used in this study are shown in Supplementary Table S2 and labeled at the 5′-end by T4 polynucleotide kinase; the detailed process was described in Zhai et al. (39). The specific DNA substrates were produced by annealing and purified by gel extraction. The DNA helicase activity was carried out as previously described in Zhai et al. with modification as indicated (39). The HJ cleavage assay was performed following the method in Zhai et al. (39).
Crystallization and structure solution of SisPINA-R206A/R147K/I199S
The sitting drop vapor diffusion method was used to screen crystal growth conditions. 1 μl of purified SisPINA-R2016A/R147K/I199S (8 mg/ml) was mixed with an equal volume of stock solution and kept at 18°C. Original crystals of SisPINA-R206A/R147K/I199S were formed in a buffer containing 0.1 M Bis-Tris, pH 6.5 and 20% PEG monomethyl ether 5000. High quality crystals were obtained in the original well and directly used for data collection. The crystals were immersed into reservoir solution containing 20% (v/v) ethylene glycol as a cryoprotectant, plunged into liquid nitrogen, and placed in pucks for transportation to the synchrotron at cryogenic temperature. Diffraction data were collected on Beamline 17U at 100 K at Shanghai Synchrotron Radiation facility (SSRF). The raw data collected were then indexed, integrated, and scaled with HKL-2000 (43). The structure was solved by molecular replacement (MR) with PDB ID 5F4H as a search model with Phaser (44). ARP/wARP was used to build the initial structure (45). Density modification was carried out via DM (46). Coot and Refmac5 were used for manual building and refinement, respectively (47,48). Final structural validation statistics were determined via SFCHECK (49). The statistics of data and structure refinement are detailed in Table 1.
Table 1.
Data collection and refinement statistics
| Native | |
|---|---|
| Data collection | |
| Space group | P 21 2 21 |
| Cell dimensions | |
| a, b, c (Å) | 69.12, 74.77, 107.58 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 19.87–2.33 (2.39–2.33) |
| R pim | 0.046 (0.414) |
| I / σI | 32.38 (2.44) |
| CC1/2 (%) | 0.723 |
| Completeness (%) | 99.4 (97.1) |
| Redundancy | 5.8 (5.7) |
| Refinement | |
| Resolution (Å) | 19.87–2.33 |
| No. reflections | 21020 |
| R work/Rfree | 20.72/23.64 |
| No. atoms | |
| Protein | 3680 |
| Glycerol | 6 |
| Water | 79 |
| B-factors | |
| Mean | 57.82 |
| Protein | 57.88 |
| Ligand | 48.85 |
| Water | 55.65 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.0074 |
| Bond angles (°) | 1.5774 |
| Ramachandran | |
| Core | 92.6% |
| Allowed | 7.1% |
| Gen. Allowed | 0.2% |
| Disallowed | 0.0% |
*Values in parentheses are for the highest-resolution shell.
Diffraction data were collected from one crystal.
Docking Hjm with SisPINA and Hjc
From available models in the PDB, the Pyrococcus furiosus Hjm (PDB ID: 2ZJ8) and Archaeoglobus fulgidus Hjc (PDB ID: 2WCW) models were selected, considering their high resolution and model completeness, and subsequently mutated in Coot (47) to reflect the sequence of S. islandicus Hjm (gene id: SiRe_0250) and Hjc (gene id: SiRe_1431), respectively. Molecular docking of SisPINA and mutated Hjm, along with mutated Hjm and mutated Hjc monomer, was carried out with the Zdock server (50). Resultant docked models were analyzed using the Proteins, Interfaces, Structures, and Assemblies (PISA) server (51). All structural figures were generated with PyMOL (http://www.pymol.org/) and the UCSF Chimera packages (52). Amino acid charge calculations for electrostatic surface images were generated by the PDB2PQR server (53,54), using protonation state prediction by PROPKA (55,56) and PARSE force field parameters (57,58). Poisson–Boltzmann (PB) calculations were carried out by the Adaptive Poisson-Boltzmann Solver (APBS) server (59).
RESULTS
SisPINA physically interacts with SisHjm
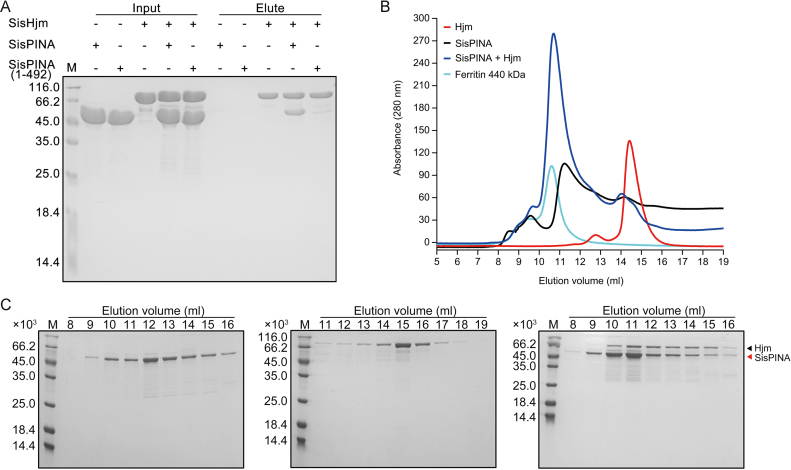
By a method similar in our previous paper (39), a S. islandicus strain encoding a chromosomally-coded N-terminal 6 × His-tagged SisPINA was constructed to search for proteins that interact with SisPINA in vivo (Supplementary Figure S1). Proteins were pulled down with His-tagged SisPINA by Ni-resins. Analysis by LC-MS/MS (Supplementary Figure S2A, B) indicated that Hjm was one of the proteins pulled down with SisPINA. The number of identified unique peptides of SisPINA and Hjm were 60 and 42, and the corresponding sequence coverage of these two proteins were 91% and 64%, respectively (Supplementary Figure S2C). To verify the physical interaction between SisPINA and Hjm, pull-down and gel filtration chromatography experiments were conducted with purified SisPINA and Hjm proteins. As shown in Figure 1A, the N-terminal His-tagged SisHjm was able to pull-down non-tagged SisPINA. Results of the gel filtration chromatography analysis showed that the elution volume of Hjm was around 15 ml (Figure 1B, red and Figure 1C, middle). When SisPINA and Hjm were mixed and incubated at 65°C for 30 min, the elution volume of Hjm (along with PINA) was shifted to around 11 ml together with the majority of SisPINA (Figure 1B, blue and Figure 1C, right), a volume close to that of ferritin (MW 440 kDa, Figure 1B, cyan). The interaction between SisPINA and Hjm was not DNA-mediated, because DNA was not detectable in either protein sample (Supplementary Figure S3). These results indicate that SisPINA interacts directly with Hjm and both proteins form a complex in vitro. The results also confirm that SisPINA forms an oligomer (hexamer) (Figure 1B and C). Estimation based on the intensities of protein bands indicated that about one Hjm monomer is associated with one PINA hexamer (Figure 1C, right and Supplementary Figure S4A).
Figure 1.
SisPINA has physical interaction with SisHjm. Analysis of the interaction between SisPINA and SisHjm by pull-down and gel filtration. (A) Recombinant SisPINA or SisPINA (1–492) (without His-tag) was incubated with N-terminal His-tagged SisHjm. Inputs and elution fractions were analyzed by SDS-PAGE with Coomassie blue staining. M, molecular size markers. ‘–’, no protein, ‘+’, protein added. Molecular masses of standard proteins are indicated at the left of each panel. (B) Gel filtration profile of Hjm (red), SisPINA (black), Hjm/SisPINA mixture (blue), and protein marker ferritin (cyan, 440 kD) (GE Healthcare, UK). The protein sample (500 ul) of SisPINA (430 μg), Hjm (230 μg), or their mixture was loaded onto the Superdex 200 column which was equilibrated with the buffer containing 25 mM Tris–HCl, pH 8.0, 200 mM NaCl, and 5% glycerol. (C) SDS-PAGE analysis of the elute fractions in (B). The fractions (1 ml each) were collected and analyzed.
Interestingly, the interaction between SisPINA (1–492) and SisHjm was dramatically decreased (Figure 1A), and no complex was formed between SisPINA (1–492) and SisHjm (Supplementary Figure S5), similar to the situation between SisPINA and Hjc (39). These results imply that the C-terminal region of SisPINA is important for its interaction and complex formation with both SisHjm and SisHjc.
The C-terminus of SisPINA has a KH domain structure that is involved in protein–protein interaction
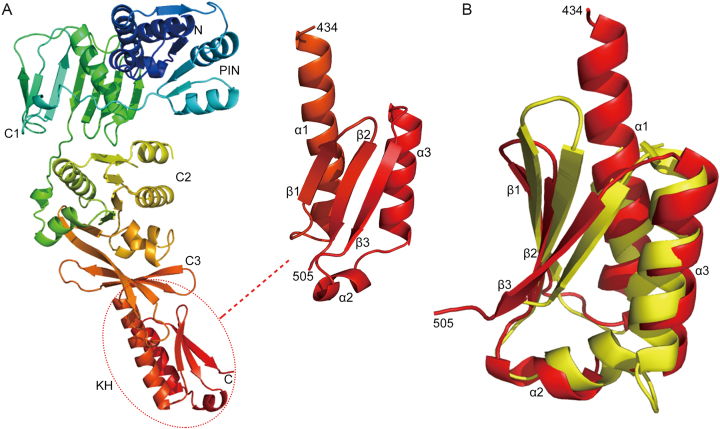
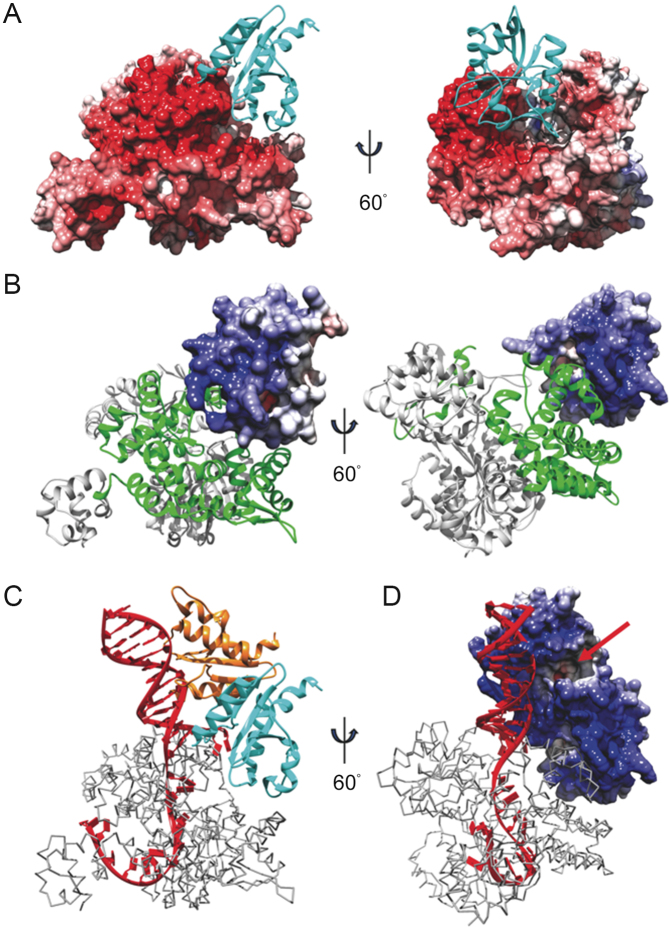
Recently, we determined the structure of SisPINA as a hexamer in crystals (PDB ID: 5F4H) (39). However, due to the lack of electron density, the last 72 amino acids of SisPINA were not defined structurally, although the full length SisPINA was crystallized. To reveal the whole structure of SisPINA, we obtained crystals of an accidental mutant of SisPINA, SisPINA-R206A/R147K/I199S, and determined the crystal structure of the mutant, which defines the full structure of the protein (residues 1-505, Figure 2A). In contrast to the hexameric SisPINA structure, this mutant is a monomer in the crystal and has the PIN and C1 domains in a highly different orientation relative to the C2/ATPase and C3 domains. Residues beyond the C3 domain (434–505) form a novel KH domain (hnRNP K homology domain) (Figure 2A, dashed circle), composed of three α-helices and three anti-parallel β-strands. The SisPINA KH domain lacks the GxxG-motif typical for KH domains (60). Dali analysis (61) identified that this domain shares structural similarity with Aeropyrum pernix NusA homolog (PDB ID: 2CXC), exhibiting a Z-score of 5.6 and an RMSD of 2.9 Å over 138 residues (Figure 2B). NusA in bacteria is a universal regulator of transcript elongation. It contains N-terminal pause-promoting domain and other putative RNA binding domains: an OB-fold S1-like domain, and two KH domains (62). The archaeal NusA homologs are widespread but represent a shortened version of the bacterial factor: it contains two KH domains but lacks the three additional domains that are well conserved in the bacterial factor. The function of archaeal NusA homologs in regulation of transcript elongation is still putative and remains to be investigated (63).
Figure 2.
The C-terminus of SisPINA folds into a KH domain. (A) The structure of SisPINA-R206A/R147K/I199S. The C-terminal domain (dashed circle) was enlarged and the α-helices and β-strands are indicated. (B) Alignment of SisPINA C-terminal KH domain with NusA. The ribbon structure of the C-terminus of SisPINA (residues 434–505) (red) is superimposed on the structure of the NusA (yellow) from Aeropyrum pernix (PDB ID: 2CXC).
Typical KH domain containing proteins are transcriptional factors in bacteria which usually have multiple repeats. Dual KH domain-containing proteins are highly conserved in Archaea (64), however, their function is unclear. We have shown that the interaction between SisPINA(1–492) and Hjm is much weaker than that between SisPINA and Hjm, indicating that the KH domain of SisPINA plays a key role in the interaction between SisPINA and Hjm (Figure 1A and Supplementary Figure S5). Here, for the first time, we show that a single KH domain is a part of the PIN containing ATPase which is involved in protein–protein interaction. We found that the KH domain of SisPINA alone is able to bind ssDNA but not Hjm (Supplementary Figure S6), implying that although the KH domain plays an important role in SisPINA and Hjm interaction, other part(s) of SisPINA also assist the interaction between SisPINA and Hjm.
Monomeric SisPINA-R206A/R147K/I199S forms a complex with Hjm in a 1:1 molar ratio
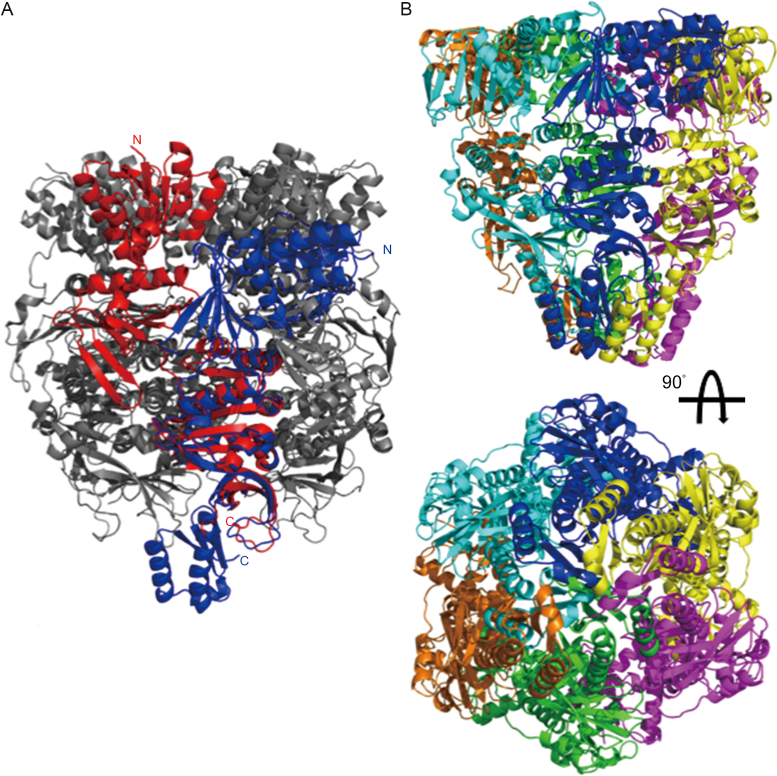
Interestingly, when the KH domain was attached to the monomers of the hexameric PINA by superimposing the mutant monomer over the ATPase domain, there is significant clash between KH domains of neighboring subunits (Figure 3), suggesting that the formation of the PINA hexamer destabilizes the structure of the KH domain as revealed by our previously determined hexameric structure (39).
Figure 3.
Hexameric assembly destabilizes the KH domain of SisPINA. (A) Alignment of the SisPINA-R206A/R147K/I199S structure (blue) with one subunit (red) of the SisPINA-R206A hexamer structure (PDB ID: 5F4H; gray) over the ATPase domain. (B) KH domains clashing among subunits of the hexameric PINA. Six SisPINA-R206A/R147K/I199S monomers (in different colors) are aligned to the SisPINA-R206A hexamer (not shown) over the ATPase domains.
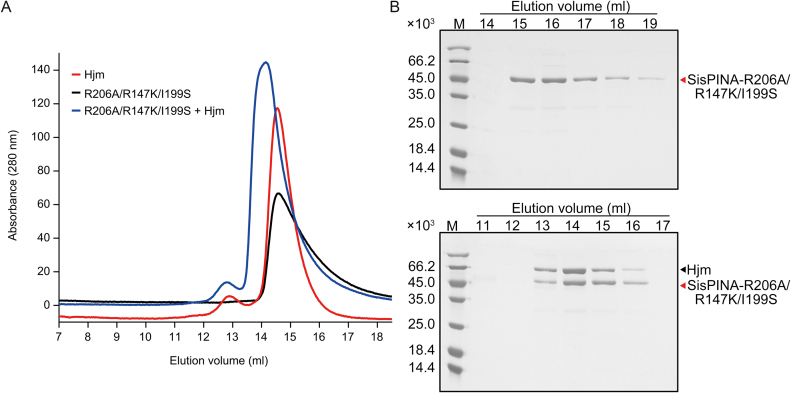
To understand how SisPINA interacts with Hjm, we analyzed the complex formation between SisPINA-R206A/R147K/I199S and Hjm by gel filtration. As shown in Figure 4, SisPINA-R206A/R147K/I199S elution peaked at a volume corresponding to monomeric form (Figure 4A, black), in contrast to the wild type SisPINA which displayed a major hexameric peak (Figure 1B, black). When SisPINA-R206A/R147K/I199S was mixed with Hjm, SisPINA-R206A/R147K/I199S eluted earlier along with Hjm (Figure 4A, blue and Figure 4B, bottom). Interestingly, quantification of band intensity revealed that the molar ratios between Hjm and SisPINA-R206A/R147K/I199S (monomer) in the complex fractions was also approximately 1:1 (Supplementary Figure S4B). It is reasonable to assume that the C-terminus in one subunit of the hexameric SisPINA folds into KH domain which interacts with Hjm, while the KH domains of other subunits could be disordered.
Figure 4.
Analysis of the physical interaction between SisPINA-R206A/R147K/I199S and SisHjm by gel filtration. (A) Gel filtration profile of Hjm (red), SisPINA-R206A/R147K/I199S (black), and the mixture of Hjm and SisPINA-R206A/R147K/I199S (blue). (B) The SDS-PAGE of eluted fractions of SisPINA-R206A/R147KI/199S (top) and its mixture with Hjm (bottom). The procedure for the analysis was same as that for the analysis of the interaction between SisPINA and Hjm.
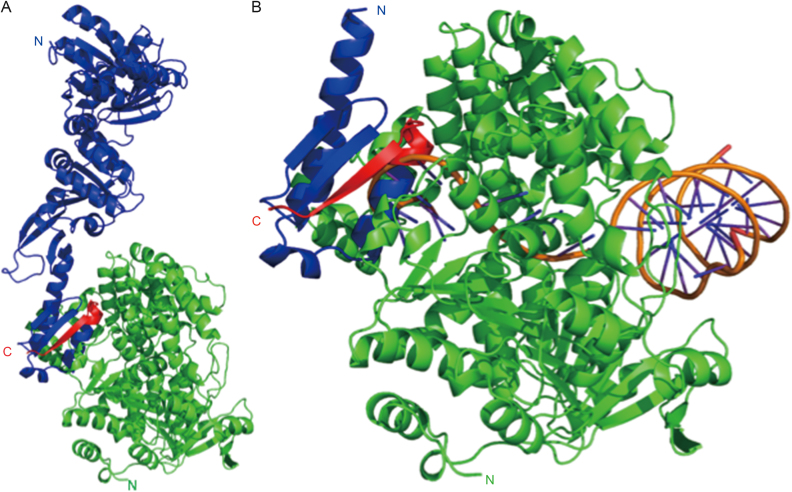
To explore this further, we performed molecular docking via the Zdock server (50) to investigate the potential interaction surfaces of PINA with Hjm (PDB ID: 2ZJ8). Two ‘hot-spots’ within the KH domain of SisPINA were revealed, one of which included the last 13 residues (Figure 5A) which have been shown to facilitate the interaction with SisHjm in vitro (Figure 1A and Supplementary Figure S5). Intriguingly, comparison of the docked Hjm with Pyrococcus furiosus Hjm bound to DNA (PDB ID: 2P6R) reveals the associated SisPINA positions the KH domain directly in the ssDNA exit channel of Hjm, partly occluding its path through the protein (Figure 5B). Such steric hindrance could explain why PINA inhibits the helicase activity observed for SisHjm (Figure 6A).
Figure 5.
Complex model of SisPINA interacting with Hjm. (A) Docking result showing that SisPINA (blue) interacts with Hjm (green) through its C-terminal residues 492–502 (red). (B) SisPINA KH domain (colored as in A) clashing with the ssDNA (orange) bound with Hjm (green). DNA positioned via alignment of PfuHjm:DNA complex (PDB ID: 2P6R) with Hjm.
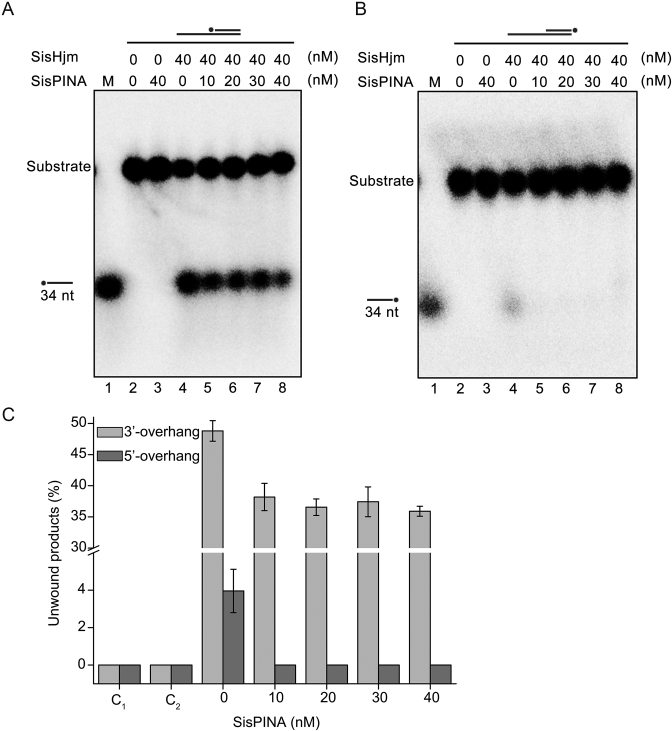
Figure 6.
Effect of SisPINA on the unwinding activity of SisHjm towards 3′-overhang (84 mer/34 mer) (A) and 5′-overhang (84 mer/34 mer) (B). The ‘*’ indicates labeling of the substrates at the 5′ end. Various concentrations of SisPINA (0, 10, 20, 30 and 40 nM, as hexamer) and 40 nM SisHjm (as monomer) were added into the reaction mixture and the reactions were carried out at 45°C for 25 min. The products were separated on 8% native PAGE gels and analyzed using a phosphor imager. (C) Quantification of the results in (A) and (B). The data were obtained with ImageJ software (NIH) and the values were calculated from at least three repeats.
SisPINA inhibits the unwinding activity of SisHjm towards 3′-overhang and 5′-overhang DNA
Previously, the direction of the unwinding activity of Hjm (Hel308a) was characterized in several archaeal species. The unwinding polarity seems to be different among the proteins from different species. It has been reported that, in P. furiosus and Methanothermobacter thermautotrophicus, Hjm proteins unwind DNA strands only in the 3′ to 5′ direction while S. tokodaii Hjm can unwind the DNA strands in both 3′ to 5′ and 5′ to 3′ directions (26,27,29). In S. solfataricus, the most closely related species to S. islandicus of the genus investigated, Hel308a was reported with DNA binding and helicase activities on a 3′-overhang substrate (65). Here, we first determined and confirmed the direction of the unwinding activity of Hjm from S. islandicus (SisHjm). The results show that SisHjm mainly unwinds 3′-overhang DNA but also exhibits very low activity towards 5′-overhang DNA (Figure 6). Subsequently, we analyzed the effect of SisPINA on the unwinding activity of SisHjm. As the concentration of SisPINA increased, the amount of ssDNA unwound from 3′-overhang DNA and 5′-overhang DNA by SisHjm decreased, indicating that SisPINA can inhibit the helicase activity of SisHjm. As described above (Figure 5B), the docking result showed that SisPINA could bind Hjm through a beta sheet of the KH domain to Hjm at the site where the unwound ssDNA is held, leading to the inhibition on Hjm helicase activity.
SisPINA and SisHjm together remodel DNA substrates similar to a replication fork
Until now, the function of Hjm in vivo has remained elusive. More and more studies imply that Hjm is likely associated with stalled replication fork processing rather than driving Holliday junction migration (27,31,37,65). Intriguingly, in our previous study, we found that SisPINA exhibited high affinity to Holliday junction DNA and replication fork. It is able to drive migration of Holliday junction but lacked the ability to unwind the replication fork (39). A possible scenario could be that SisPINA functions together with SisHjm at the stalled replication fork. Here, we characterize the functional interaction between SisPINA and SisHjm on replication fork substrates in vitro. Four substrates were used: replication forks with labels on the lagging strand and leading strand, a 3′-flap, and a 5′-flap (Figure 7). It should be noted that the parental strands of the lagging strand are not complementary, thus it is not a real replication fork. When the reaction mixture contains no protein, the replication fork spontaneously unwound and produced overhang DNA under the reaction conditions (Figure 7A and B, lane 2). In line with previous results, SisPINA did not exhibit ability in unwinding replication forks (Figure 7A and B, lane 3). Instead, SisPINA (40 nM as hexamer) dramatically decreased the amount of the spontaneously unwound products (Figure 7A and B, lane 3). Based on these results, we speculate that SisPINA plays a potential role in stabilizing the replication fork, a function that is presumably important for replication fork regression.
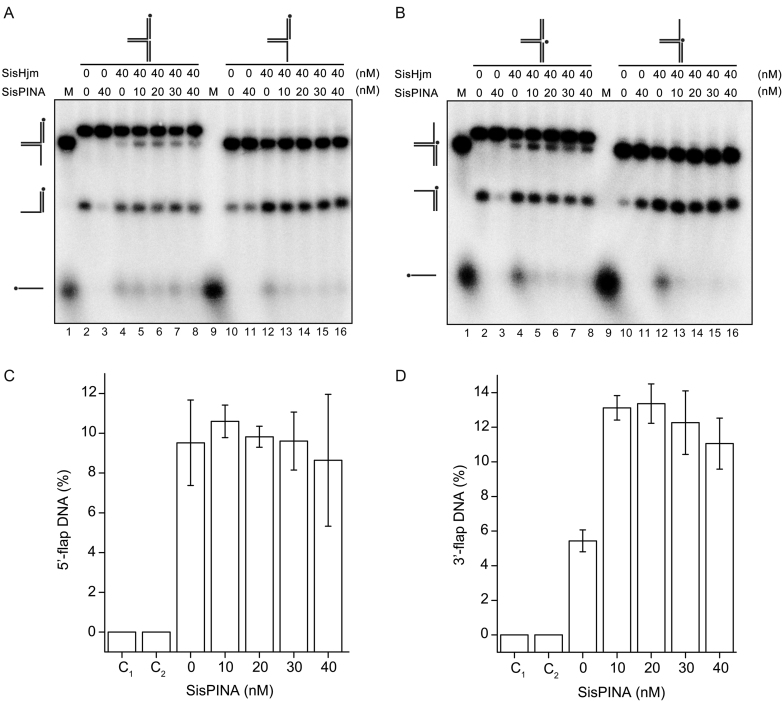
Figure 7.
SisPINA and SisHjm in combination remodel the replication fork. SisPINA and SisHjm were mixed to detect the capability of replication fork DNA processing. The fork substrates were labeled at the leading strand (A) or lagging strand (B) and the representative gels are shown. All the reactions were carried out at 45°C for 30 min. The ‘*’ indicates labeling of the substrates at the 5′ end. (C) Effect of SisPINA on the generation of 5′-flap DNA from a pseudo replication fork by Hjm. (D) Effect of SisPINA on the generation of 3′-flap DNA from a pseudo replication fork by Hjm. The products of 5′-flap DNA (A, lanes 2–8) and 3′-flap DNA (B, lanes 2–8) were quantified by Image J (NIH). The values were based on data of at least three experimental repeats.
On the other hand, SisHjm (40 nM as monomer) turned the replication forks into 5′-flap/3′-flap, overhang, and ssDNA (Figure 7A and B, lane 4), revealing that SisHjm has the ability to unwind replication forks. Notably, we found that during processing replication forks, SisHjm preferred removing the lagging strand (Figure 7A and B, lane 4), which is in agreement with a previous report on Hel308a from halophilic archaea (27). Furthermore, when both SisPINA and SisHjm were added to the reaction mixture, SisPINA only slightly enhanced the activity of SisHjm to produce 3′-flap DNA, but has no obvious effect on the production of 5′-flap DNA (Figure 7C and D). Nevertheless, it inhibited the activity of SisHjm to produce ssDNA (Figure 7A and B, lanes 5–8), consistent with the inhibition of SisPINA on the unwinding of overhang DNA substrates by SisHjm (Figure 6).
SisPINA is able to unwind 3′- flap DNA
We next analyzed the effect of SisPINA on SisHjm's processing of 5′-flap and 3′-flap DNA substrates. SisHjm alone exhibited helicase activities on 5′-flap and 3′-flap DNA substrates, producing overhang DNA and ssDNA, but the amount of overhang DNA was much more than ssDNA (Figure 7A and B, lane 12). When increasing amounts of SisPINA were added to the reaction mixtures, there was no significant change in the amount of overhang DNA generated, but the amount of ssDNA produced by SisHjm decreased (Figure 7A and B, lanes 12–16). These results further confirm that SisPINA inhibits the unwinding activity of SisHjm toward overhang DNA (Figure 6). Notably, SisPINA and SisHjm can unwind 3′-flap DNA, but the amount of 3′-overhang DNA produced by SisHjm or SisPINA and SisHjm (comparing lane 12 and lanes 13–16 in Figure 7B) was almost the same. Currently, the mechanism of this phenomenon is unclear.
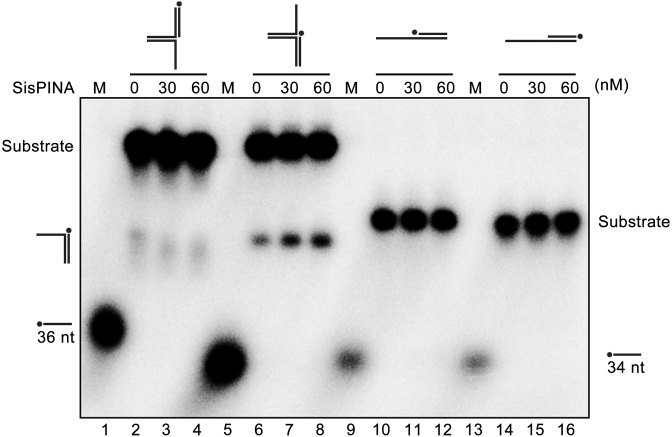
Previously, we reported that SisPINA is able to promote HJ migration and unwind Y DNA (39). To test if SisPINA can unwind 3′-flap or 5′-flap DNA, we analyzed the unwinding ability of SisPINA on four different DNA substrates, including 5′-flap DNA (72 nt with 36 bp matches), 3′-flap DNA (72 nt with 36 bp matches), 5′-overhang DNA (84 nt with 34 bp), and 3′-overhang DNA (84 nt with 34 bp) (Figure 8). Among these substrates, SisPINA can only unwind 3′-flap DNA, producing 3′-overhang DNA, but no unwinding product was observed for the other three substrates. It is worth mentioning that the 36 nt ssDNA marker in lane 5 ran a little faster than that in lane 1, we speculate that this was probably caused by electrophoresis. So far, we have demonstrated that SisPINA possesses activities of mediating HJ migration and unwinding Y-shaped or 3′-flap DNA.
Figure 8.
SisPINA is able to unwind 3′-flap DNA. Lanes 2–4, replication fork without lagging strand; lanes 6–8, replication fork without leading strand; lanes 10–12, 3′-overhang DNA; lanes 14–16, 5′-overhang DNA. Lanes 1 and 5, 32P-labeled 36 nt ssDNA marker; Lanes 9 and 13, 32P-labeled 34 nt ssDNA markers. The ‘*’ indicates labeling of the substrates at the 5′ end. The concentrations of SisPINA (as hexamer) used for the analysis were indicated. All of the reactions were performed at 45°C for 30 min and the products were separated on 8% native page gels and analyzed with a phosphor imager.
SisPINA, SisHjm and SisHjc process HJ together in a concerted way
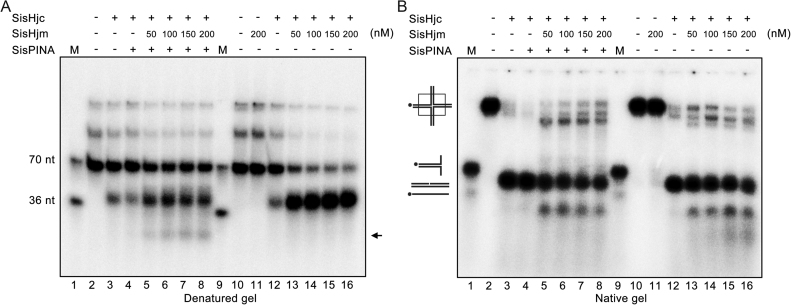
It was reported that Hjm and Hjc interact with each other physically and functionally in vitro in S. tokodaii (29). We also found that SisPINA interacts with SisHjc (39). Considering SisPINA, Hjm, and Hjc have a potential capacity to drive HJ migration or cleavage, we want to know whether these three proteins can process HJ in vitro in a concerted way. Therefore, we examined the processing of a mobile HJ by denatured and native gel electrophoresis. As previously reported, SisHjc has strand preference in HJ cleavage on the unlabeled strand (39) (Figure 9A and B, lanes 2 and 3). When the reaction mixture contained SisPINA and SisHjc, the cleavage pattern of HJ by SisHjc was the same as previously described (39), showing that SisPINA enhanced the preferred strand cleavage of HJ by SisHjc (Figure 9, lanes 3 and 4). In contrast, addition of SisHjm resulted in switching of SisHjc cleavage from the unlabeled strand to the labeled strand (Figure 9A, lanes 12, 13–16). When only SisHjm was added to the SisHjc reaction mixture, additional products were observed (Figure 9B lanes 13–16). As shown in Figure 9, with native gel analysis, as the concentration of Hjm increased, shorter products were observed and the lengths were <70 nt (the length of labelled strand is 70 nt). We speculate that SisHjc would cleave the HJ in the favorite direction (one of the two cleaved sites contained the labelled strand) in the first place and produce two nicked dsDNA which would be unwound by SisHjm.
Figure 9.
Holliday junction processing by SisPINA, SisHjm, and SisHjc. The cleavage of HSL (mobile) HJ substrate by SisHjc in the presence of SisPINA and SisHjm at indicated concentrations was analyzed by denatured (A) and native (B) gel electrophoresis. All of the reactions were carried out at 55°C for 30 min. The black arrow indicates band that is produced by HJ branch migration and cleavage. The ‘*’ indicates labeling of the substrates at the 5′ end.
To gain a better understanding of how this SisHjc:SisHjm complex would act on HJ, we performed molecular docking with the A. fulgidus Hjc (PDB ID: 2WCW) monomer and P. furiosus Hjm (PDB ID: 2ZJ8) mutated to the corresponding S. islandicus homologous sequences. Of the resultant models, a single ‘hot spot’ was found for Hjc to bind to Hjm; this Hjm surface is evidently highly negatively charged, based on electrostatic surface calculations (Figure 10A), and is part of the mapped region of Hjm involved in Hjc binding (30). Conversely, a highly positively charged surface of Hjc (Figure 10B), which was reported to be involved in DNA binding and contains the active site (35), is oriented favorably toward the aforesaid negatively charged Hjm surface and the active site is buried in an inactive orientation. When both the Hjc dimer (PDB ID: 2WCW) and P. furiosus Hjm:DNA complex (PDB ID: 2P6R) were aligned with the complex from the docking result (Figure 10C and D), the resultant model positions the exposed, active Hjc monomer directly adjacent to the Hjm-bound DNA, with the active site oriented for cleavage of the DNA backbone.
Figure 10.
Modeling Hjm:Hjc unwinding and cleavage of DNA. Docking result showing (A) the electrostatic surface of Hjm interacting with Hjc (cyan ribbons), or (B) the electrostatic surface of Hjc interacting with Hjm (ribbons). The Hjc-interaction region of Hjm is highlighted by green. Model of a Hjc dimer (cyan and orange ribbons (C) or electrostatic surface (D)) bound with Hjm (gray trace) and DNA (red) from PfuHjm:DNA complex (PDB ID: 2P6R) based on results in A&B. The Hjc active site is indicated with a red arrow. Red surface - negative charge, white surface - neutral, blue surface - positive charge.
When SisPINA, SisHjm, and SisHjc were all added in the reaction with mobile HJ, another small band (black arrow) appeared in addition to the typical cleavage products (about 70 nt and 36 nt, Figure 9A, lanes 5–8). Under the same reaction conditions, this small band was not produced when SisPINA was omitted or replaced by SisPINA (1–492) in the mixture (Supplementary Figure S7, lanes 5–8). Notably, when the reaction mixture contained SisPINA, SisHjm, and SisHjc, the HJ strand cleavage preference of SisHjc differs from that when the mixture contained SisHjm and SisHjc (labeled strand cleavage in Figure 9, lanes 12–16) or SisPINA and SisHjc (unlabeled strand cleavage in Figure 9, lanes 3–4 (39)). It will be very interesting to know if all these modes of HJ processing occur in vivo. We speculate that different modes of HJ processing are used for different mechanisms in repairing stalled replication forks or double-strand DNA breaks.
DISCUSSION
Repair of stalled replication forks is an essential process for all life forms. Replication fork regression is an attractive model for repair of stalled replication forks and has only been confirmed in Bacteria and Eukarya (5,9,10,13,14). Here we provide further evidence that the conserved archaeal ATPase and DNA helicase PINA participates in the replication fork regression pathway in the thermophilic archaeon S. islandicus. PINA is able to unwind HJ, Y-shaped, and 3′-flapped DNAs, the intermediates of stalled or collapsed replication forks. It interacts with Hjm, an essential protein which was implied to be involved in repair of stalled replication forks in many studies (26–31). We found that PINA inhibits the unwinding activity of Hjm and both proteins work together remodeling replication forks and promote HJ migration (Figures 6,7,9). For further analysis, an authentic replication fork should be utilized as in other reports (66,67).
The inability to knockout of the genes coding for PINA, Hjm, and proteins involved in DNA double strand breaks supported that PINA and these proteins function in the repair pathway of stalled replication forks. We have shown that in S. islandicus, hjm, pina, nurA, herA, mre11, rad50 and radA are all essential (39,68,69). The genes coding for HJ resolvases, Hjc and Hje, can be deleted individually, however, double knockout of both are lethal (70). Since PINA is conserved in all archaea, it will be very interesting to know whether PINA genes are essential or not in other archaea.
Another piece of evidence supporting PINA’s participation in repair of stalled replication forks is that PINA has strong interaction with the small subunit of replication fork protein C (RFCs), the clamp loader in Archaea and Eukarya (71,72). RFCs is among the associated proteins co-eluted with His-tagged PINA (Supplementary Figure S2C) and the physical interaction between SisPINA and SisRFCs has been confirmed by pull down and gel filtration analysis. However, the role of RFCs in the repair of stalled replication fork is still unknown. One possibility might be that RFCs is involved in reassembly of replication fork after D-loop formation. Another more attracting possibility is that RFC comes to the stalled fork to unload PCNA from the DNA strand. Then, RFC recruits PINA through protein–protein interaction to stabilize the fork DNA. The issue is interesting and needs further investigation. Analysis of the physiological role of this interaction is in progress.
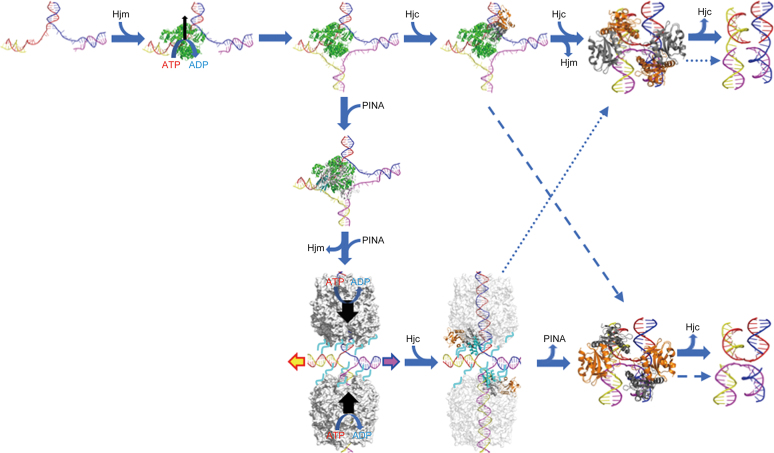
Here, we solved the structure of the SisPINA monomer, which defines the C-terminal KH domain that was not observed previously (Figure 2). We demonstrated that this domain is involved in the interaction with Hjm and Hjc ((39) and this study) and is able to bind ssDNA in vitro not but Hjm (Supplementary Figure S6). The domain alone was unable to bind Hjm, suggesting that other parts of PINA may contain Hjm-interacting site(s), in agreement with the docking results of PINA-Hjm interaction (Figure 5). To our knowledge, this is the first report on the ability of a KH domain to mediate protein–protein interaction. Based on results presented here and in our previous paper (39), we propose the following model on how Hjm, PINA, and Hjc work together in HJ formation, migration, and cleavage (Figure 11). (i) When DNA replication stalls, Hjm is recruited to process (reform) the collapsed replication fork, leading to the formation of HJ or chicken feet DNA. Hjm then directly recruits PINA, via interaction with the KH domain of monomeric PINA of one subunit in the PINA hexermer, which inhibits the unwinding activity of Hjm. Hjm is then released from HJ, with or without the assistance of PINA. Analogous to RuvAB (73), two PINA hexamers assemble on the opposite arms of HJ and drive the HJ branch to migrate until the endonuclease Hjc is recruited through the interaction with the KH domain of one subunit of the PINA hexermer. The interaction of Hjc with PINA stabilizes the KH domain, which destabilizes the PINA hexamer and releases PINA hexamers from HJ, allowing Hjc to finally cleave HJ. (ii) Alternatively, after HJ formation, Hjm recruits Hjc directly to resolve the HJ, bypassing PINA. (iii) During dsDNA break repair and recombination, PINA hexamers directly bind to HJ DNA and drive HJ branch migration until Hjc is recruited to HJ. Hjc disassembles PINA hexamers from HJ and cleaves HJ. Another scenario would be that PINA comes to the stalled replication fork ahead of Hjm by the recruitment of RFC. Then Hjm comes to the fork. Subsequently, both PINA and Hjm work together to process the stalled replication fork through recombination pathway. It is necessary to examine whether Hjm or PINA has higher affinity to stalled replication fork. More investigation is needed to clarify the possibility and the details of the model.
Figure 11.
A proposed model for PINA/Hjm/Hjc to process a stalled DNA replication fork. A stalled DNA replication fork (blue, red, yellow, and magenta strands) is recognized and bound by Hjm (green), which proceeds to regress the replication fork and form a HJ structure. Either Hjc (right) or PINA (down) is recruited to the HJ structure by Hjm. The Hjc dimer (gray and orange) binding to the HJ facilitates DNA handover from Hjm to Hjc, prompting Hjm dismissal and turning the open HJ into an X-HJ structure that is nicked by two Hjc dimers (PDB: 2WJ0). Alternatively, a PINA (white) monomer is recruited by Hjm through the interaction of Hjm and the PINA KH domain (cyan), leading to the assembly of the PINA hexamer on the HJ DNA and the dissociation of Hjm. Two PINA hexamers (white surface) are formed on two opposing HJ arms and mediate HJ migration powered by ATP hydrolysis. Two Hjc dimers then bind to the HJ and interact with the PINA C-terminal domains, destabilizing the PINA hexamers. After PINA hexamers are released, the two Hjc dimers turn the open HJ into the X-HJ structure and mediate two symmetric cuts. HJ migration directionality is indicated by colored arrows. Preferred HJ cleavages by Hjc are indicated with solid arrows while alternative HJ cleavages are indicated with dashed arrows. Unfolded PINA KH domains are indicated by cyan lines.
DATA AVAILABILITY
The atomic coordinates and structural factors for SisPINA-R206A/R147K/I199S are deposited to Protein Data Bank with the accession number of 5YWW. The mass spectrometry proteomics data are deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008644.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Science Foundation of China [31670061, 30930002, 31470184 to Y.S. and 31470200 to J.N.]; State Key Laboratory of Microbial Technology (to Y.S. and J.N.); US NIH [R01GM108893 to L.F.] (in part). Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Branzei D., Foiani M.. Maintaining genome stability at the replication fork. Nat. Rev. 2010; 11:208–219. [DOI] [PubMed] [Google Scholar]
- 2. Kitao H., Iimori M., Kataoka Y., Wakasa T., Tokunaga E., Saeki H., Oki E., Maehara Y.. DNA replication stress and cancer chemotherapy. Cancer Sci. 2018; 109:264–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barlow J.H., Nussenzweig A.. Replication initiation and genome instability: a crossroads for DNA and RNA synthesis. Cell. Mol. Life. Sci. 2014; 71:4545–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friedberg E.C., Walker G.C., Siede W.. DNA Repair and Mutagenesis. 2005; Washington, D.C: ASM Press. [Google Scholar]
- 5. Atkinson J., McGlynn P.. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009; 37:3475–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Polleys E.J., House N.C.M., Freudenreich C.H.. Role of recombination and replication fork restart in repeat instability. DNA Repair. 2017; 56:156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yeeles J.T., Poli J., Marians K.J., Pasero P.. Rescuing stalled or damaged replication forks. Cold Spring Harbor Perspect. Biol. 2013; 5:a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Petermann E., Helleday T.. Pathways of mammalian replication fork restart. Nat. Rev. 2010; 11:683–687. [DOI] [PubMed] [Google Scholar]
- 9. Higgins N.P., Kato K., Strauss B.. A model for replication repair in mammalian cells. J. Mol. Biol. 1976; 101:417–425. [DOI] [PubMed] [Google Scholar]
- 10. Manosas M., Perumal S.K., Croquette V., Benkovic S.J.. Direct observation of stalled fork restart via fork regression in the T4 replication system. Science. 2012; 338:1217–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sogo J.M., Lopes M., Foiani M.. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002; 297:599–602. [DOI] [PubMed] [Google Scholar]
- 12. Neelsen K.J., Zanini I.M., Herrador R., Lopes M.. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J. Cell. Biol. 2013; 200:699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta S., Yeeles J.T., Marians K.J.. Regression of replication forks stalled by leading-strand template damage: I. Both RecG and RuvAB catalyze regression, but RuvC cleaves the Holliday junctions formed by RecG preferentially. J. Biol. Chem. 2014; 289:28376–28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta S., Yeeles J.T., Marians K.J.. Regression of replication forks stalled by leading-strand template damage: II. Regression by RecA is inhibited by SSB. J. Biol. Chem. 2014; 289:28388–28398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Blackford A.N., Schwab R.A., Nieminuszczy J., Deans A.J., West S.C., Niedzwiedz W.. The DNA translocase activity of FANCM protects stalled replication forks. Hum. Mol. Genet. 2012; 21:2005–2016. [DOI] [PubMed] [Google Scholar]
- 16. Deans A.J., West S.C.. FANCM connects the genome instability disorders Bloom's Syndrome and Fanconi Anemia. Mol. Cell. 2009; 36:943–953. [DOI] [PubMed] [Google Scholar]
- 17. Edgell D.R., Doolittle W.F.. Archaea and the origin(s) of DNA replication proteins. Cell. 1997; 89:995–998. [DOI] [PubMed] [Google Scholar]
- 18. Samson R.Y., Bell S.D.. Cell cycles and cell division in the archaea. Curr. Opin. Microbiol. 2011; 14:350–356. [DOI] [PubMed] [Google Scholar]
- 19. Zaremba-Niedzwiedzka K., Caceres E.F., Saw J.H., Backstrom D., Juzokaite L., Vancaester E., Seitz K.W., Anantharaman K., Starnawski P., Kjeldsen K.U. et al. . Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017; 541:353–358. [DOI] [PubMed] [Google Scholar]
- 20. Ling H., Boudsocq F., Woodgate R., Yang W.. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol. Cell. 2004; 13:751–762. [DOI] [PubMed] [Google Scholar]
- 21. Hopfner K.P., Karcher A., Craig L., Woo T.T., Carney J.P., Tainer J.A.. Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell. 2001; 105:473–485. [DOI] [PubMed] [Google Scholar]
- 22. Williams R.S., Moncalian G., Williams J.S., Yamada Y., Limbo O., Shin D.S., Groocock L.M., Cahill D., Hitomi C., Guenther G. et al. . Mre11 dimers coordinate DNA end bridging and nuclease processing in double strand break repair. Cell. 2008; 135:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komori K., Hidaka M., Horiuchi T., Fujikane R., Shinagawa H., Ishino Y.. Cooperation of the N-terminal helicase and C-terminal endonuclease activities of archaeal Hef protein in processing stalled replication forks. J. Biol. Chem. 2004; 279:53175–53185. [DOI] [PubMed] [Google Scholar]
- 24. Lestini R., Laptenok S.P., Kuhn J., Hink M.A., Schanne-Klein M.C., Liebl U., Myllykallio H.. Intracellular dynamics of archaeal FANCM homologue Hef in response to halted DNA replication. Nucleic Acids Res. 2013; 41:10358–10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lestini R., Duan Z., Allers T.. The archaeal Xpf/Mus81/FANCM homolog Hef and the Holliday junction resolvase Hjc define alternative pathways that are essential for cell viability in Haloferax volcanii. DNA Repair. 2010; 9:994–1002. [DOI] [PubMed] [Google Scholar]
- 26. Fujikane R., Shinagawa H., Ishino Y.. The archaeal Hjm helicase has recQ-like functions, and may be involved in repair of stalled replication fork. Genes Cells. 2006; 11:99–110. [DOI] [PubMed] [Google Scholar]
- 27. Guy C.P., Bolt E.L.. Archaeal Hel308 helicase targets replication forks in vivo and in vitro and unwinds lagging strands. Nucleic Acids Res. 2005; 33:3678–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fujikane R., Komori K., Shinagawa H., Ishino Y.. Identification of a novel helicase activity unwinding branched DNAs from the hyperthermophilic archaeon, Pyrococcus furiosus. J. Biol. Chem. 2005; 280:12351–12358. [DOI] [PubMed] [Google Scholar]
- 29. Li Z., Lu S., Hou G., Ma X., Sheng D., Ni J., Shen Y.. Hjm/Hel308A DNA helicase from Sulfolobus tokodaii promotes replication fork regression and interacts with Hjc endonuclease in vitro. J. Bacteriol. 2008; 190:3006–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hong Y., Chu M., Li Y., Ni J., Sheng D., Hou G., She Q., Shen Y.. Dissection of the functional domains of an archaeal Holliday junction helicase. DNA Repair. 2012; 11:102–111. [DOI] [PubMed] [Google Scholar]
- 31. Liew L.P., Lim Z.Y., Cohen M., Kong Z., Marjavaara L., Chabes A., Bell S.D.. Hydroxyurea-mediated cytotoxicity without inhibition of ribonucleotide reductase. Cell Rep. 2016; 17:1657–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Komori K., Sakae S., Shinagawa H., Morikawa K., Ishino Y.. A Holliday junction resolvase from Pyrococcus furiosus: functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in Bacteria, Eukarya, and Archaea. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:8873–8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Komori K., Sakae S., Fujikane R., Morikawa K., Shinagawa H., Ishino Y.. Biochemical characterization of the Hjc Holliday junction resolvase of Pyrococcus furiosus. Nucleic Acids Res. 2000; 28:4544–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Komori K., Sakae S., Daiyasu H., Toh H., Morikawa K., Shinagawa H., Ishino Y.. Mutational analysis of the Pyrococcus furiosus Holliday junction resolvase Hjc revealed functionally important residues for dimer formation, junction DNA binding, and cleavage activities. J. Biol. Chem. 2000; 275:40385–40391. [DOI] [PubMed] [Google Scholar]
- 35. Bond C.S., Kvaratskhelia M., Richard D., White M.F., Hunter W.N.. Structure of Hjc, a Holliday junction resolvase, from Sulfolobus solfataricus. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nishino T., Komori K., Tsuchiya D., Ishino Y., Morikawa K.. Crystal structure of the archaeal Holliday junction resolvase Hjc and implications for DNA recognition. Structure. 2001; 9:197–204. [DOI] [PubMed] [Google Scholar]
- 37. Ishino Y., Nishino T., Morikawa K.. Mechanisms of maintaining genetic stability by homologous recombination. Chem. Rev. 2006; 106:324–339. [DOI] [PubMed] [Google Scholar]
- 38. Meetei A.R., Medhurst A.L., Ling C., Xue Y., Singh T.R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C.G. et al. . A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005; 37:958–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhai B., DuPrez K., Doukov T.I., Li H., Huang M., Shang G., Ni J., Gu L., Shen Y., Fan L.. Structure and function of a novel ATPase that interacts with Holliday junction resolvase Hjc and promotes branch migration. J. Mol. Biol. 2017; 429:1009–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Peng N., Deng L., Mei Y., Jiang D., Hu Y., Awayez M., Liang Y., She Q.. A synthetic arabinose-inducible promoter confers high levels of recombinant protein expression in hyperthermophilic archaeon Sulfolobus islandicus. Appl. Environ. Microbiol. 2012; 78:5630–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deng L., Zhu H., Chen Z., Liang Y., She Q.. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles. 2009; 13:735–746. [DOI] [PubMed] [Google Scholar]
- 42. Song X., Ni J., Shen Y.. Structure-based genetic analysis of Hel308a in the hyperthermophilic archaeon Sulfolobus islandicus. J. Genet. Genomics. 2016; 43:405–413. [DOI] [PubMed] [Google Scholar]
- 43. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 44. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Langer G., Cohen S.X., Lamzin V.S., Perrakis A.. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008; 3:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terwilliger T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 2000; 56:965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P., Cowtan K.. Coot. model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 48. Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vaguine A.A., Richelle J., Wodak S.J.. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 1999; 55:191–205. [DOI] [PubMed] [Google Scholar]
- 50. Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., Weng Z.. ZDOCK server: interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics. 2014; 30:1771–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 52. Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E.. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004; 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 53. Dolinsky T.J., Czodrowski P., Li H., Nielsen J.E., Jensen J.H., Klebe G., Baker N.A.. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007; 35:W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dolinsky T.J., Nielsen J.E., McCammon J.A., Baker N.A.. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004; 32:W665–W667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li H., Robertson A.D., Jensen J.H.. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005; 61:704–721. [DOI] [PubMed] [Google Scholar]
- 56. Sondergaard C.R., Olsson M.H., Rostkowski M., Jensen J.H.. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa Values. J. Chem. Theory Comput. 2011; 7:2284–2295. [DOI] [PubMed] [Google Scholar]
- 57. Sitkoff D., Sharp K.A., Honig B.. Accurate calculation of hydration free energies using macroscopic solvent models. J. Phys. Chem. 1994; 98:1978–1988. [Google Scholar]
- 58. Tang C.L., Alexov E, Pyle A.M., Honig B.. Calculation of pKas in RNA: on the structural origins and functional roles of protonated nucleotides. J. Mol. Biol. 2007; 366:1475–1496. [DOI] [PubMed] [Google Scholar]
- 59. Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A.. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Valverde R., Edwards L., Regan L.. Structure and function of KH domains. FEBS J. 2008; 275:2712–2726. [DOI] [PubMed] [Google Scholar]
- 61. Holm L., Rosenstrom P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Worbs M., Bourenkov G.P., Bartunik H.D., Huber R., Wahl M.C.. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001; 7:1177–1189. [DOI] [PubMed] [Google Scholar]
- 63. Werner F. Molecular mechanisms of transcription elongation in archaea. Chem. Rev. 2013; 113:8331–8349. [DOI] [PubMed] [Google Scholar]
- 64. Shibata R., Bessho Y., Shinkai A., Nishimoto M., Fusatomi E., Terada T., Shirouzu M., Yokoyama S.. Crystal structure and RNA-binding analysis of the archaeal transcription factor NusA. Biochem. Biophys. Res. Commun. 2007; 355:122–128. [DOI] [PubMed] [Google Scholar]
- 65. Richards J.D., Johnson K.A., Liu H., McRobbie A.M., McMahon S., Oke M., Carter L., Naismith J.H., White M.F.. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J. Biol. Chem. 2008; 283:5118–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fugger K., Mistrik M., Neelsen K.J., Yao Q., Zellweger R., Kousholt A.N., Haahr P., Chu W.K., Bartek J., Lopes M. et al. . FBH1 catalyzes regression of stalled replication forks. Cell Rep. 2015; 10:1749–1757. [DOI] [PubMed] [Google Scholar]
- 67. Xue X., Choi K., Bonner J., Chiba T., Kwon Y., Xu Y., Sanchez H., Wyman C., Niu H., Zhao X. et al. . Restriction of replication fork regression activities by a conserved SMC complex. Mol Cell. 2014; 56:436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Huang Q., Liu L., Liu J., Ni J., She Q., Shen Y.. Efficient 5′-3′ DNA end resection by HerA and NurA is essential for cell viability in the crenarchaeon Sulfolobus islandicus. BMC Mol. Biol. 2015; 16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang C., Tian B., Li S., Ao X., Dalgaard K., Gokce S., Liang Y., She Q.. Genetic manipulation in Sulfolobus islandicus and functional analysis of DNA repair genes. Biochem. Soc. Trans. 2013; 41:405–410. [DOI] [PubMed] [Google Scholar]
- 70. Huang Q., Li Y., Zeng C., Song T., Yan Z., Ni J., She Q., Shen Y.. Genetic analysis of the Holliday junction resolvases Hje and Hjc in Sulfolobus islandicus. Extremophiles. 2015; 19:505–514. [DOI] [PubMed] [Google Scholar]
- 71. Yao N.Y., O’Donnell M.. The RFC clamp loader: structure and function. Subcell. Biochem. 2012; 62:259–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hedglin M., Kumar R., Benkovic S.J.. Replication clamps and clamp loaders. Cold Spring Harbor Perspect. Biol. 2013; 5:a010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Eggleston A.K., West S.C.. Cleavage of Holliday junctions by the Escherichia coli RuvABC complex. J. Biol. Chem. 2000; 275:26467–26476. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structural factors for SisPINA-R206A/R147K/I199S are deposited to Protein Data Bank with the accession number of 5YWW. The mass spectrometry proteomics data are deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD008644.