Abstract
Mitochondrial DNA (mtDNA) mutations become more prevalent with age and are postulated to contribute to the ageing process. Point mutations of mtDNA have been suggested to originate from two main sources, i.e. replicative errors and oxidative damage, but the contribution of each of these processes is much discussed. To elucidate the origin of mtDNA mutations, we measured point mutation load in mice with deficient mitochondrial base-excision repair (BER) caused by knockout alleles preventing mitochondrial import of the DNA repair glycosylases OGG1 and MUTYH (Ogg1 dMTS, Mutyh dMTS). Surprisingly, we detected no increase in the mtDNA mutation load in old Ogg1 dMTS mice. As DNA repair is especially important in the germ line, we bred the BER deficient mice for five consecutive generations but found no increase in the mtDNA mutation load in these maternal lineages. To increase reactive oxygen species (ROS) levels and oxidative damage, we bred the Ogg1 dMTS mice with tissue specific Sod2 knockout mice. Although increased superoxide levels caused a plethora of changes in mitochondrial function, we did not detect any changes in the mutation load of mtDNA or mtRNA. Our results show that the importance of oxidative damage as a contributor of mtDNA mutations should be re-evaluated.
INTRODUCTION
The mitochondrion is a subcellular organelle required for essential cellular processes such as iron-sulfur cluster synthesis, lipid metabolism and energy conversion by oxidative phosphorylation (OXPHOS). Most mitochondrial proteins are encoded by the nuclear DNA, synthesized in the cytosol and post-translationally transported into mitochondria. However, mitochondria also contain their own genetic information in the circular double-stranded mtDNA, which, in mammals, encodes two rRNAs, 22 tRNAs and 11 mRNAs containing 13 protein-coding open reading frames. The mitochondrial genetic system is required to produce the 13 mitochondrial proteins that are core components of the OXPHOS system. Mammalian mtDNA is present in multiple copies per cell and because of this polyploidy, mutations are often only present in a fraction of all mtDNA molecules, a condition referred to as heteroplasmy. Pathogenic mutations in mtDNA can have very serious consequences and lead to diseases with onset in the neonatal period or adult life. The disease manifestations are typically pleiotropic and can involve symptoms such as severe progressive neurodegeneration (e.g. Leigh syndrome), infantile multisystem disorders (e.g. Pearson′s syndrome), stroke, hearing loss, myoclonic epilepsy, myopathy, optic atrophy, cardiomyopathy and blindness (1,2). Mutations of mtDNA are also heavily implicated in the mammalian ageing process as mutations expand clonally in old individuals to cause a mosaic respiratory chain deficiency affecting only a proportion of all cells in a variety of tissues, such as brain, heart, skeletal muscle and colonic crypts (3–10). The origin of these point mutations is still under debate, although computational modeling (11) and sequence analysis (6,7,12–14), support the hypothesis that they are created by replication errors, mainly during embryonic life.
In 1956, Harman proposed the free radical theory of ageing, which postulates that ageing is driven by oxidative damage to macromolecules (15). Later he refined the theory and focused it on mitochondria as a main producer of reactive oxygen species (ROS) and the target of oxidative damage (16). These ideas subsequently developed into the so-called mitochondrial theory of ageing (17). Mitochondria, and especially mtDNA, have been thought to be the main targets of oxidative damage because the OXPHOS system produces superoxide as a natural side product of cellular respiration. It was previously proposed that ∼1-2% of all oxygen used during respiration was converted into H2O2 (18), but this is likely an overestimation and the levels of H2O2 production during OXPHOS are probably at least an order of magnitude lower (19). In cell culture, exposure to extracellular H2O2 or rotenone has been reported to lead to mtDNA damage (20,21). However, even extended exposure was reported not to increase mtDNA mutation load (21).
When mtDNA was discovered, it was thought to be naked and unprotected (22,23). However, work during the last decades has shown that this is not the case. It is now well established that mammalian mtDNA is fully protein coated, mainly through the action of a high-mobility group box domain protein called mitochondrial transcription factor A (TFAM) (24,25), and condensed into mitochondrial nucleoids with a slightly elongated shape of ∼80 × 80 × 100 nm (25–27).
Another reason for the high mutation load of mtDNA has been attributed to the scarcity of intramitochondrial DNA repair pathways. At the moment, there is reasonably solid evidence showing that the short and long patch base-excision repair (BER) pathways exist in mitochondria (28–31). However, the existence of other repair pathways such as nucleotide-excision repair (NER) (29,32), mismatch repair and double-strand break repair remain poorly documented and is subject to considerable debate.
In this context, it is also important to consider the chemistry of ROS-mediated DNA damage. Importantly, superoxide and its derivative hydrogen peroxide do not directly react with DNA (33). However, superoxide typically reacts with [4Fe–4S] clusters resulting in the release of iron (34), which, in turn, reacts with hydrogen peroxide through Fenton chemistry resulting in the production of the highly reactive hydroxyl radical (35). This radical is able to react with any cellular macromolecule, including mtDNA, inducing a large spectrum of DNA modifications, such as oxidized deoxyguanosine, 8-oxo-dG (36). This oxidized base is mutagenic as it can mispair with adenosine during DNA replication to induce G>T transversion mutations (37). There are three DNA glycosylases reported to be present in both the cytosol/nucleus and mitochondria, i.e. MutT Human Homolog 1 (MTH1), 8-oxoguanine DNA glycosylase 1 (OGG1) and mutY homolog (MUTYH), that recognize and remove 8-oxo-dG related damage in various forms. MTH1 removes 8-oxo-dGTP from the nucleotide pool (38,39), OGG1 removes 8-oxo-dG from double-stranded DNA (40,41) whereas MUTYH removes an adenosine that has been erroneously incorporated opposite to 8-oxo-dG (42,43). After OGG1 and MUTYH have processed the damaged DNA, it is repaired by gap-tailoring, gap-filling, and DNA ligation. The amount of 8-oxo-dG, assessed by DNA digestion assays (44), was not increased in intact mtDNA in double knockout mice with impaired nuclear and mitochondrial BER (Ogg1) and NER (Csb) whereas 8-oxo-dG, assessed by electrochemical detection (45), was increased in Ogg1 knockout mice.
Studies of naturally occurring somatic mtDNA point mutations in aged flies (13), mice (46,47), and humans (7) have documented that most of the detected mutations are transitions rather than transversions. Furthermore, the abundant mtDNA mutations in human tissues (48), tumor cell lines (49,50) and tumors (51,52) are similarly mainly transition mutations. Typically, a mutational spectrum dominated by transition mutations is consistent with replication errors and/or spontaneous deaminations (14,53) rather than with oxidative damage. Studies of mice expressing an error-prone mtDNA polymerase creating abundant replication errors (54,55) have shown that these mice transmit mtDNA mutations through their germ line in a pattern that is very similar to the patterns observed in different wild mouse strains (56), which lend further support to the hypothesis that replication errors are an important factor shaping mtDNA sequence variation.
Recent studies have reported that flies with decreased mitochondrial matrix localized superoxide dismutase (SOD2) activity combined with a loss-of-function mutation in Ogg1 had no increase in mtDNA mutation load as measured by random mutation capture assay (RMC) (13). However, it has been much debated whether this has relevance for mammals. To this end, we have experimentally assessed whether decreased mitochondrial BER in combination with increased mitochondrial ROS production can induce more mtDNA point mutations in mice. We generated mice lacking the mitochondrial targeting sequence (MTS) of the OGG1 (Ogg1 dMTS) and MUTYH (Mutyh dMTS) DNA glycosylases. These mutant mice were designed to study the importance of BER in mitochondria without inducing adverse effects in the nucleus. Surprisingly, Ogg1 dMTS mice had no increase in the mtDNA mutation load at the age of 100 weeks. Based on the hypothesis that DNA repair is especially important in the germ line, we bred mice with impaired mitochondrial BER for five generations but found no increase in the mtDNA mutation load in maternal lineages homozygous for both the Ogg1 dMTS and Mutyh dMTS alleles. As a final intervention, we generated mice with increased ROS levels in heart by disrupting the gene encoding the mitochondrial matrix localized superoxide dismutase (Sod2). Mice lacking SOD2 in the heart developed a severe dilated cardiomyopathy and showed a molecular phenotype consistent with drastically increased oxidative stress, but there was no increase in the mutation load. In addition, when we combined the homozygous knockout of Sod2 in the heart with homozygous expression of the Ogg1 dMTS allele, we found no increase in the mtDNA and mtRNA mutation load. Our findings call for a reassessment on the contribution of oxidative stress to the creation of mtDNA mutations during ageing.
MATERIALS AND METHODS
Mouse lines
To exclude OGG1 and MUTYH from mitochondria, the endogenous genes were modified to lack the genomic region encoding the predicted mitochondrial targeting sequence (MTS). The predictions of mitochondrial targeting sequence were made with Mitoprot II (57) and Target P1.1 (58,59) using AAB94512.1 protein sequence for OGG1 and NP_001153053.1 for MUTYH. To remove the predicted MTS, a targeting vector (BAC C57BL/6J RPCIB-731) was designed to lack the region encoding L2 to W23 from OGG1 or K2 to P33 from MUTYH. The vectors also contained a positive selection marker (Neomycin resistance) flanked by FRT sites. The targeting vectors were transfected into the TaconicArtemis C57BL/6N Tac ES cell line. After successful homologous recombination, the neomycin cassette was removed by Flp recombination to obtain the constitutive knockout alleles of Mutyh or Ogg1 lacking the genomic region encoding the MTS (Ogg1 dMTS and Mutyh dMTS). The mice were generated at TaconicArtemis and they were maintained on the C57Bl/6NCrl background (Charles River Laboratories, Germany strain code 027). Superoxide dismutase loxP mice (60) were received from Prof. Dr Karin Scharffetter-Kochanek from Universitätklinikum Ulm and bred with a Ckmm cre transgenic line (61) to establish mice with tissue specific lack of the SOD2 protein in heart and skeletal muscle. The Sod2 loxP × Ckmm cre mice were maintained on C57Bl/6NCrl background (Charles River Laboratories, Germany strain code 027).
To generate homozygous Ogg1 dMTS or Mutyh dMTS knockout mice, heterozygous Ogg1 dMTS or Mutyh dMTS mice were mated. In these breedings, offspring of wild-type females and homozygous males was used to minimize accumulation of mtDNA mutations. Tissue-specific SOD2 knockout mice were generated by first crossing homozygous Sod2 loxP mice with heterozygous Ckmm cre mice. In the subsequent cross, mice that were homozygous for Sod2 loxP were crossed with mice heterozygous for Sod2 loxP and Ckmm cre.
Mouse husbandry
Mice were maintained at 21°C in a 12-h light/dark cycle and fed ad libitum on a standard diet (ssniff M-H Low-Phytoestrogen). Enhanced diet was used during breeding or with newly weaned mice (ssniff M-Z Low-Phytoestrogen) by Ssniff Spezialdiaeten GmbH. All experiments were approved and permitted by the Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen (LANUV) in accordance with German and European Union regulations. All animal work was performed in accordance to recommendations and guidelines of the Federation of European Laboratory Animal Science Associations (FELASA).
Constructs encoding OGG1 and MUTYH for immunocytochemistry
A cDNA encoding mouse OGG1 (NM_010957.4) was inserted into the multiple cloning site of pCMV-tag4 (Agilent) to create a construct expressing the wild-type form of OGG1. To remove the region encoding for the MTS (4T-69G) a new restriction site and start codon was added by PCR and the resulting oligonucleotide was inserted into pCMV-tag4a. A construct encoding mouse Myc-DDK-tagged MUTYH (pCMV6-Entry) was purchased at Origene (MR208268, NM_133250.1) and modified to lack the MTS encoding sequence (A4-T99) and include the N-terminal extension of human alpha variant (A1-C42). A construct encoding human Myc-DDK-tagged MUTYH alpha variant (pCMV6-Entry) was bought from Origene (NM_001048171.1) and modified to lack the N-terminal extension of alpha3 variant (A1-C42) and the MTS encoding sequence (A4-G162).
Immunocytochemistry
HeLa cells were grown at 37°C, 5% CO2 on coverslips, transfected with plasmids encoding OGG1 or MUTYH using Lipofectamine2000 (ThermoFisher), and subsequently fixed with 4% paraformaldehyde (EMS). Cells were thereafter incubated with antibodies against the FLAG peptide (Sigma-Aldrich, F1804) and TOM20 (Santa Cruz, sc-11415). The primary antibodies were detected with secondary antibodies (goat anti-mouse Alexa Fluor 488, A11001 and goat anti-rabbit Alexa Fluor 594 A11012 (ThermoFisher)). After immunolabeling, the samples were stained with 1 μg/ml DAPI (AppliChem) and mounted in Prolong Gold (ThermoFisher). The image acquisition was performed with a Leica TCS SP8-X inverted confocal microscope (Leica Microsystems) using a 100×/1.4 oil objective.
8-oxo-dG glycosylase/AP lyase activity
8-oxo-dG incision activity was measured with DNA digestion assay essentially as previously described (45). Fresh mouse liver (∼200 mg) was minced into small pieces, rinsed with PBS and homogenized in 10 ml of mitobuffer (320 mM sucrose, 20 ml Tris–HCl, 1 mM EGTA, 0.2% BSA, pH 7.2) with a glass homogenizer with tight Teflon pestle with five strokes at 200 rpm. An aliquot (total) was taken and thereafter the mitochondrial fraction was purified with differential centrifugation and Percoll gradient as described in the mitochondria purification section. Total and mitochondrial fractions, 300 μg, were lysed in lysis buffer (10μl, 20 mM HEPES–KOH (pH 7.6), 1 mM EDTA, 2 mM DTT, 300 mM KCl, 5% glycerol, 0.05% Triton X-100) and resuspended in dilution buffer (20 μl, 20 mM HEPES–KOH (pH 7.6), 1 mM EDTA, 2 mM DTT, 5% glycerol) to bring the KCl concentration to 100 mM and the final protein concentration to 10 μg/μl. Next, the 8-oxo-dG incision activity was measured by incubating 100 μg of protein with 100 fmol of [α-32P]-5′ labeled 8-oxo-dG dsDNA in reaction buffer (60 mM HEPES–KOH (pH 7.6), 10 mM EDTA, 2 mM DTT, 50 mM KCl, 15% glycerol, final volume 20 μl) for 16 h at 32°C. The reaction was stopped by addition of stop solution (final concentration 0.2 mg/ml Protease K, 0.4% SDS), and incubation at 55°C for 15 min. Finally, the DNA was ethanol precipitated and resolved on a 15% Novex TBE–urea polyacrylamide gel (7 M urea, ThermoFisher Scientific). Oligos used 5′-GAACGACTGT[8-oxo-dG]ACTTGACTGCTACTGA-3′, 5′-ATCAGTAGCAGTCAAGTCACAGTCGTTC-3′ (62).
H&E staining from paraffin embedded sections
Mouse hearts were fixed in buffered formalin and embedded in paraffin (ThermoFisher). Tissue sections of 5 μm were cut with a microtome (HM340E, Thermo). Sections were deparaffinized in xylene (AppliChem), rehydrated with an alcohol series (100–70% ethanol, AppliChem) and stained with Haematoxylin (AppliChem) and Eosin Y (Carl Roth), followed by dehydration through an alcohol series (70–100% ethanol, AppliChem). Sections were mounted (Cytoseal, ThermoFisher) and imaged with a brightfield microscope (Nikon Eclipse Ci).
RNA extraction, cDNA synthesis and PCR amplification to verify the expression of Ogg1 dMTS and Mutyh dMTS transcripts
RNA was extracted from snap-frozen heart or liver tissue with TRIzol (Ambion) following manufacturer's recommendations with an overnight isopropanol precipitation at −20°C. 2 μg of total RNA was converted to cDNA using High Capacity cDNA reverse transcription kit (Applied Biosystems) following manufacturer's recommendations. To verify the correct length of the produced transcript, cDNA from Ogg1 dMTS mice was amplified with primers binding to exon 1 and exon 3 (5′-CGTAATGGGCTGGGGCTG-3′, 5′-CAGCACGCCACTCCAGTGAG-3′). cDNA from Mutyh dMTS mice was amplified with primers binding to exon 1 and exon 7 (5′-TCGGAGACTGCGCAGGAG-3′, 5′-GGGAAGCGCTGGCCAGGT-3′) (63). The PCR products were analyzed on an agarose gel.
DNA extraction
Total DNA was extracted from snap-frozen heart or liver tissue with Gentra Puregene Tissue Kit (QIAGEN) following the manufacturer's recommendations.
Copy number analysis with quantitative real-time PCR and Southern blotting
Total DNA was extracted as described above with Gentra Puregene Tissue Kit (QIAGEN) including RNase treatment. The quality of DNA was verified with NanoDrop (2000C) and quantified with a fluorometric method (Qubit, ThermoFisher). qPCR was carried out in a 7900HT qPCR machine (Applied Biosystems) with the Taqman method using the Taqman Universal PCR Master Mix (Applied Biosystems). Reactions were made in triplicates on a 384-well plate using 5 ng of total DNA per reaction (final volume 10 μl). Specific probes were used to detect the 18S gene (Hs99999901_s1, also detecting mouse 18S) for nuclear DNA and the ND1 (Mm04225274_s1), CytB (AIS062S) and ATP6 (Mm03649417-g1) genes for mtDNA (Applied Biosystems). For each experiment, two mtDNA probes were used and results were always consistent between the two different probes. The amplification data was analyzed using a standard-curve method where an artificial standard curve was produced by mixing a small aliquot of all analyzed samples into a standard sample (SDS 2.4). MtDNA copy number (mtDNA/nDNA) was normalized to control samples.
For copy number analysis with Southern blot, total DNA was extracted and quantified as described above. The DNA was digested overnight with SacI-HF restriction enzyme (New England Biolabs) and ethanol precipitated. Around 700–800 ng of digested DNA was separated with agarose gel electrophoresis and the gel was treated with HCl, denaturated, neutralized and set up for a capillary transfer with 20× SSC. After transfer, the membrane (Hybond-N+, GE Healthcare) was cross-linked, blocked with hybridization buffer (Perfect Hyb Plus, Sigma-Aldrich) and incubated with [α-32P]dCTP labeled mtDNA (pAM1) and nuclear DNA (18S) probes and used to expose a phosphorimager screen or for autoradiography with film (Amersham Hyperfilm MP, GE Healthcare).
Post-PCR cloning and Sanger sequencing
The mtDNA mutation load was analyzed with post-PCR cloning and Sanger sequencing of the WANCY-COX1 tRNA-cluster region, as in (64). This ∼1 kb long region has been extensively used in mutation load analysis in our group (64,65). It is expected to allow higher mutation accumulation in comparison with protein encoding regions because of the higher flexibility of tRNA sequences (66). In general, none to three mutations per sample were found in wild-type mice when ∼ 93 000 bp were sequenced, placing the mutation load from <1.07 × 10−5 to 3.21 × 10−5 mutations/base pair (65). Previously, the background error rate of the method was evaluated by clone of a clone experiments (64). One cloned fragment was diluted into total DNA of a Drosophila and amplified, cloned and sequenced. When 295 clones were assayed, only one additional variant from the reference sequence was observed, resulting to error rate of 3.48 × 10−6 mutations/bp.
Total DNA was extracted and quantified as described above. The WANCY-COX1 region of mtDNA was amplified with Phusion DNA polymerase (New England Biolabs) using PCR primers (F 5′-CCTACCCCTAGCCCCCC-3′ R 5′-AGTATAGTAATGCCTGCG-3′) and cloned into TOP10 chemically competent Escherichia coli by using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen). The transformed bacteria were grown on selective plates, the colonies were picked and sent for sequencing (Plateseq service, Eurofins). The obtained sequences were analyzed with SeqScape software, version 2.7 (Applied Biosystems).
mtDNA purification for Illumina sequencing
First, fresh tissue was minced to small pieces (∼100 mg heart, 400–500 mg liver) washed in PBS and mitochondria were purified with differential centrifugation, as described below in the mitochondria purification section with some small modifications. To pellet cellular debris, the homogenate was spun at 800 g for 10 min at 4°C. To collect the mitochondria, the supernatant was spun at 8500g for 10 min at 4°C. The mitochondria pellet was resuspended in 600 μl of Mito-DNase buffer (300 mM sucrose, 10 mM MgCl2, 20 mM Tris–HCl, pH 7.5, 0.15% BSA, 0.03 mg/ml DNase I type IV, 170 ng/μl RNase A) and the liver mitochondria were further divided into 600 μl aliquots each containing ∼100 mg of starting material. The suspensions were incubated at 37°C for 1 h to digest the nuclear DNA and RNA. Then, mitochondria were repelleted at 13 000 g, for 15 min at 4°C and washed twice with 500 μl of mitobuffer (320 mM sucrose, 20 ml Tris–HCl, 1 mM EGTA, 0.2% BSA, pH 7.2). Next, the samples were frozen in N2(l) and stored in −80°C. Later the same day, the mitochondrial pellets were resuspended into 400 μl lysis buffer (20 mM Tris–HCl, 150 mM NaCl, 20 mM EDTA, 1% SDS, pH 8.75, 0.2 mg/ml Proteinase K, 0.2 mg/ml RNase A) and incubated at 56°C overnight. Then, the samples were cooled down to RT and DNA was extracted with chloroform (100 μl of 6M K-acetate, 500 μl chloroform:isoamylalcohol (24:1, Amresco)). 100 μg/200 μg of RNase A was added to each aqueous phase fractions (heart and liver, respectively) and samples were incubated at 37°C for 45 min to digest the remaining RNA. Thereafter, the samples were ethanol precipitated with 15 μg of glycogen (Ambion). The purified DNA pellet was then resuspended to 20–30 μl of 5 mM Tris buffer pH 8.5 (Macherey-Nagel). Prior to sending the samples to Illumina sequencing (Max Planck-Genome-centre Cologne), the purity of the extracted mtDNA was verified using mtDNA and nuclear DNA specific primers for PCR amplification. The yield from each 100 mg fractions were ∼250 ng for heart and ∼350 ng for liver as quantified with a fluorometric method (Qubit, ThermoFisher). The protocol was modified from a previously described protocol (7). For details see (dx.doi.org/10.17504/protocols.io.mycc7sw)
Illumina sequencing
Isolated mtDNA from both the Mutyh dMTS × Ogg1 dMTS livers and Sod2 loxP × Ckmm cre × Ogg1 dMTS hearts were sequenced in two separate sets each, including controls. First, the quality of the samples was verified with Genomic DNA analysis ScreenTape (Agilent) followed by DNA library preparation. The DNA was fragmented with Covaris to 400 bp (50 ng, 50 μl, 5% duty cycle, intensity 5, 200 cycles per burst, treatment time 55 s). First Sod2 loxP × Ckmm cre × Ogg1 dMTS set was fragmented with Covaris to 350 bp with 53 s treatment time. Following DNA fragmentation, the DNA library was prepared with NEBNext Ultra II DNA library prep kit for Illumina (New England Biolabs). Thereafter single-end sequencing was carried out with HiSeq3000, with HiSeq3000/4000 SR Cluster Kit and the corresponding SBS Kit (Illumina) until 1 Gbase of sequence was achieved. The first Mutyh dMTS × Ogg1 dMTS preparation was pre-purified with AMPureBeads (Beckman Coulter) before the library preparation to remove contaminating small DNA/RNA fragments.
Total RNA was extracted from snap-frozen heart tissue with TRIzol (Ambion) following manufacturer's recommendations with an overnight isopropanol precipitation at −20°C. The quality of the RNA was first verified with northern blotting as described below. Then, rRNAs were depleted with RiboZero rRNA Removal Kit (Human/Mouse/Rat) (Epicentre) following manufacturer's recommendations with an input of 1 μg total RNA. Next, the RNA library was prepared with NEBNext Ultra Directional RNA Kit (New England Biolabs) following manufacturer's recommendations. Finally, the library preparation was sequenced with HiSeq3000 by using the HiSeq3000/4000 SR Cluster Kit and the corresponding SBS Kit (Illumina) until 5 Gb of sequence was achieved.
mtDNA and RNA Illumina sequencing experiments, library preparation and sequencing were performed by the Max Planck-Genome-centre Cologne, Germany (http://mpgc.mpipz.mpg.de/home/).
Data analysis and variant calling of Illumina sequencing of mitochondrial RNA and DNA
Demultiplexed sequencing reads were trimmed with Flexbar version 2.5 (67) for TruSeq adapters and quality (default parameters except -q 28 -m 50 -ae ANY -ao 10). RNA reads were aligned to mouse mitochondrial reference genome (GRCm38, release 81) with STAR aligner version 2.4.1d (68) (default parameters except for genome indexing --genomeSAindexNbases 6). Whereas a ’dual alignment’ approach was used for DNA read alignment with BWA version 0.7.12-r1039 (69) invoking mem (default parameters except -T 19 -B 3 -L 5,4). DNA reads were first aligned to the mouse mitochondrial reference genome and then separately to a split reference genome in which the first 8150 bases were transferred to the end of the genome. Such dual alignment approach was applied in order to enable complete alignment and variant detection at the junction region of the circular mitochondrial genome. ∼50% and ∼90% of the reads aligned to mtDNA from heart and liver samples, respectively.
With samtools (70), the aligned reads were converted to bam format and only uniquely aligned reads (parameter -q 1) were kept for downstream analysis. Then the reads were further sorted and indexed. Per base coverage was determined with bedtools version 2.22.1 (71) genomecov (parameters -split -d). Variants were detected with Lofreq* version 2.1.2 (72) using the following command and parameters: lofreq call-parallel --pp-threads 20 -N -B -q 30 -Q 30 --no-default-filter (referred as ‘only quality-filtered data’). Both, RNA and DNA variants were further filtered for quality and strand bias using LoFreq*: lofreq filter --no-defaults --snvqual-thresh 70 -B 60, for minimum number of variant supporting reads using snpSift filter (73) with the expression DP*AF ≥ 15. DNA variants were additionally filtered for minimum of three variant supporting reads on each strand (expression DP4[2] ≥ 3 & DP4[3] ≥ 3). Finally, variant lists were filtered for minimum variant allele frequency (AF value) of 0.5% (quality-filtered and 0.5% minimum variant allele frequency filtered data). Furthermore, known variants in our mouse strain (positions 4891, 9027 and 9461) were removed from all results. In addition, two maternally occurring variants (positions 9993 and 15403) were removed from four Sod2 loxP control siblings (pp). Heavily strand-biased variants (SB Phred score > 100 or 1000 for RNA and DNA variants, respectively) passing the earlier filters were also removed from the minimum allele frequency filtered data. DNA coverage and variant result files originating from the dual alignment approach were combined: results obtained by the alignment to the normal reference genome were kept for genome positions 200–16 099. Results for the rest of the genome positions, i.e. genome junction region, were obtained by the alignment to the split reference genome and the genome positions were corrected to represent the original position numbers.
Final vcf-files were converted to tab-delimited format with SnpEff version 4.2 (74) and mutation loads were calculated as follows: Over the whole mitochondrial genome, a ’unique’ mutation load was calculated by dividing number of detected variants by coverage (total base pairs aligned to mitochondrial genome), whereas total mutation load was calculated by dividing the sum of variant supporting bases (obtained from DP4 values) by coverage. To obtain the corresponding mutation loads per mutation type (e.g. G>T), the variant or total variant read counts were divided by the total coverage on the reference base in question. The variant calls for each mouse are available as a supplementary Excel-file.
Northern blotting
Total RNA was isolated as described above and quantified with a fluorometric method (Qubit, ThermoFisher). Next, 2 μg of total RNA was treated with NorthernMax-Gly sample loading dye (Ambion) and resolved on a formaldehyde-agarose gel followed by incubation in 0.05 M NaOH, DEPC-water and 20× SSC. The gel was set up for a capillary transfer with 20× SSC. After transfer, the membrane (Hybond-NX, GE Healthcare) was cross-linked and incubated with hybridization solution (5× SSC, 20 mM Na2HPO4, 7% SDS, 0.5× RNA secure (Ambion), 100 μg/ml heparin). The transcripts of interest were detected with mouse-specific biotin-labeled oligonucleotides with overnight incubation at 50°C in hybridization solution (CytB, 18S) followed by washing and signal detection with IRDye 800CW dye-labeled streptavidin (LI-COR Biosciences) (dilution 1:5000 in TBS, 0.05% TWEEN-20) in the Odyssey infrared imaging system (LI-COR Biosciences). This method has previously been described (75).
Western blotting
The steady-state levels of proteins of interest were evaluated by western blotting. Differential-centrifugation purified mitochondria (described below) were lyzed and reduced with NuPAGE LDS sample buffer (Invitrogen) and 50 μM DTT. The mitochondrial proteins were resolved on a NuPAGE SDS-PAGE gel system (Invitrogen) and transferred with wet-transfer onto a PVDF membrane (Immobion FL, Millipore or Amesham Hybond, GE Healthcare). Next, the membrane was blocked with 5% milk TBS, 0.05% Tween-20 and proteins of interest were detected with primary antibody (ACO2 ab110321, Abcam; ATP5A ab14748, Abcam; SOD2 06-984, Millipore; SDHA 459200, Invitrogen; SDHC 14575-1-AP, Proteintech; MitoProfile Total OXPHOS Rodent WB Antibody Cocktail MS604, Mitoscience; mtSSB HPA002866, Sigma; POLγ ab128899, Abcam; TFAM ab131607, Abcam; POLRMT selfmade; TFB2M selfmade) and visualized with horseradish peroxidase-linked secondary antibody (Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, HRP, Invitrogen; Amersham ECL Mouse IgG, HRP-linked, GE Healthcare) by using enhanced chemiluminescence (Amersham ECL Western Blotting Detection Reagent, GE Healtcare) and film (Amersham hyperfilm MP, GE Healthcare). After detection, the membrane was stained with staining solution (Coomassie Blu-R, 10% acetic acid, 20% EtOH), destained and scanned either with the Odyssey infrared imaging system (LI-COR Biosciences) or a normal scanner to control for loading.
Mitochondria purification
Mitochondria were purified from fresh tissue with differential centrifugation. First, the tissue (heart ∼100 mg, liver ∼200 mg) was minced into small pieces, rinsed with PBS and homogenized into 10 ml of mitobuffer (320 mM sucrose, 20 ml Tris–HCl, 1 mM EGTA, 0.2% BSA, pH 7.2) with a glass homogenizer. Heart tissue was first homogenized with loose Teflon pestle by hand with 10 strokes. Then, both liver and heart were homogenized with a tight Teflon pestle with 5 (liver) or 10 strokes (heart) at 200 rpm. Next, homogenates were spun 10 min at 1000g to pellet the cell debris, followed by a 10 min spin at 10 000g to pellet the mitochondria. The mitochondria-enriched fractions were thereafter resuspended in mitobuffer lacking BSA. The samples were kept on ice throughout the procedure and centrifugations were carried out at +4°C.
For label-free quantitative proteomics, mitochondria were prepared in the presence of protease inhibitor cocktail (cOmplete, EDTA-free, Roche) and purified further with Percoll gradient to exclude the mitochondria-associated membranes. Percoll gradients, 8 ml, were poured with 20% Percoll solution in mitobuffer with BSA into Ultraclear ultracentrifugation tubes (14 × 89 mm) and the resuspended mitochondria were layered on top of the gradient. The gradient was spun in SW41 swing-out rotor (Beckman) at 40 000g at +4°C for 30 min. Then the brown mitochondrial layer was collected and diluted 10× with mitobuffer lacking BSA to remove the Percoll. Then the diluted mitochondria were re-collected with 6300 g for 10 min at +4°C. The Percoll protocol was a modified from previously published protocol (76).
Mitochondrial respiration analysis
Mitochondrial oxygen consumption was measured with Oxygraph-2k (OROBOROS INSTRUMENTS, Innsbruck, Austria). The measurement was carried out at 37°C using 65–125 μg of fresh differential-centrifugation purified mitochondria that were diluted in 2.1 ml of mitochondrial respiration buffer (120 mM sucrose, 50 mM KCl, 20 mM Tris–HCl, 4 mM KH2PO4, 2 mM MgCl2, 1 mM EGTA, pH 7.2). The oxygen consumption rate was measured in the presence of 10 mM pyruvate, 10 mM glutamate and 5 mM malate (PGM, complex I) or 10 mM succinate and 10 nM rotenone (complex II). The phosphorylating state respiration (state 3) was analyzed in the presence of 1 mM ADP and non-phosphorylating state in the presence of 2.5 μg/ml oligomycin (pseudo state 4). The quality of the mitochondria was verified by measuring respiration control rate (RCR) with 1 mM ADP (state 3) or 1 mM ADP and 2.5 μg/ml oligomycin (pseudo state 4). The RCR values were 10 with PGM and 6 with succinate and rotenone. Finally, the uncoupled respiration was measured by adding carbon cyanide m-chloriphenyl hydrazone (CCCP) up to 3 μM to reach the maximal respiration. The oxygen consumption was normalized to mitochondrial protein content as measured with the protein DC kit (Bio-Rad Laboratories).
Measurement of respiratory chain enzyme activities
Measurements were carried out using differential-centrifugation purified mitochondria stored at −80°C. Mitochondria, 15–50 μg, were diluted in phosphate buffer (50 mM KH2PO4, pH 7.4) and measurements of isolated respiratory chain complex activities were performed with Hitachi UV-3600 spectrophotometer at 37°C. Citric synthase activity (CS) was measured at 412 nm (E = 13 600 M−1 cm−1) in the presence of 0.1 mM acetyl-CoA, 0.5 mM oxaloacetate and 0.1 mM 5,5′-dithiobis-2-nitrobenzoic acid (DTNB). NADH dehydrogenase (Complex I) activity was measured at 340 nm (E = 6,220 M−1 cm−1) in the presence of 0.25 mM NADH, 0.25 mM decylubiquinone and 1 mM KCN, controlling for rotenone sensitivity. Succinate dehydrogenase (Complex II) activity was measured at 600 nm (E = 21 000 M−1 cm−1) after addition of 40 mM succinate, 35 μM dichlorophenolindophenol (DCPIP) and 1 mM KCN. The cytochrome c oxidase (Complex IV, CIV) activity was measured using the classical TMPD/ascorbate assay, explained in detail elsewhere (77). The complex II to complex III (II-III) activity was measured after reduction of cytochrome c at 540 nm (E = 18 000 M−1 cm−1) in the presence of 1 μg/ml decylubiquinol, 80 μg/ml cytochrome c, 1 mM sodium azide, 40 mM succinate and 1 μM antimycin A. The difference in flux before and after addition of antimycin A reflects the complex II to III activity. All chemicals used in the measurements were from Sigma-Aldrich.
Mitochondrial aconitase activity
The aconitase activity was measured from differential-centrifugation purified mitochondria with an aconitase activity kit following manufacturer's recommendations (ab109712, Abcam). Briefly, the conversion of isocitrate to cis-aconitate is measured at 240 nm in the presence of isocitrate and manganese. The increase in absorbance indicates the activity of mitochondrial aconitase.
Topology gel
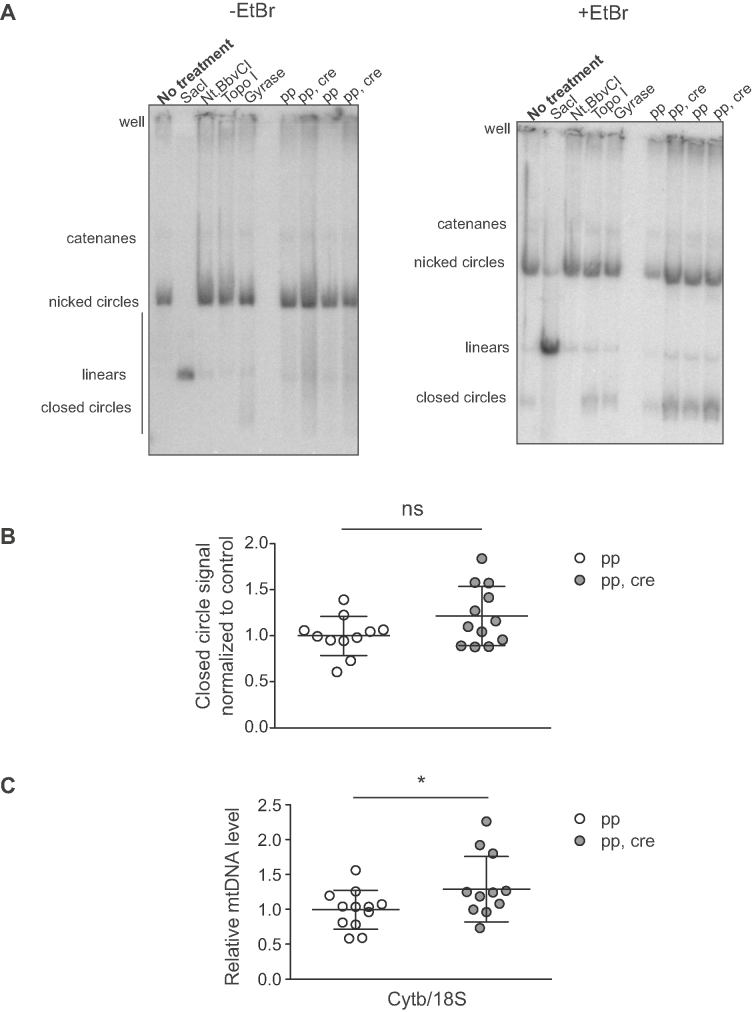
Total DNA was extracted from 50 mg of mouse heart tissue. The minced tissue was first rinsed in PBS and then lyzed in 600 μl of lysis buffer (100 mM Tris–HCl pH 7.5, 100 mM EDTA, 100 mM NaCl, 0.5% SDS, 0.8 mg/ml Proteinase K) at 55°C for 3 h followed by 2-h incubation on ice with premixed LiCl and K-acetate (final concentration 250 mM K-acetate, 760 mM LiCl) to precipitate contaminants. To remove the precipitate, samples were spun for 15 min at 15 000 g at 18°C. DNA was then precipitated from the supernatant with isopropanol with 30-min incubation on ice and collection by centrifugation for 30 min at 16 000g at 18°C. The samples were then washed and resuspended to 10 mM Tris–HCl, 1 mM EDTA pH 8.0 by flicking the tube. To quantify the DNA, a small aliquot of the sample was digested with SacI-HF (New England Biolabs) at 37°C for 30 min and quantified with fluorometric method (Qubit, ThermoFisher). 400 ng of total DNA was then resolved on 0.4% agarose gel (15 × 15 cm) (Seakem Gold agarose, Lonza) with 40 V for 16–20 h with and without EtBr (0.5 mg/ml) to condense the different supercoiling states of the closed circle molecule. After the electrophoresis, the gel was transferred like a Southern blot gel as described above, mtDNA was visualized with a [α-32P]dCTP-labeled probe (pAM1) and the membrane was used to expose a phosphorimager screen or autoradiography film (Amersham hyperfilm MP, GE Healthcare). The quantifications were made from the phosphorimager screen. To visualize different topological isomers of the mtDNA, 400 ng of control total DNA was additionally incubated at 37°C for 30 min with only buffer (no treatment), SacI (linear) (New England Biolabs; 20 U), Nt.BbvCI (nicked circles) (New England Biolabs; 10 U), Topo I (relaxes the closed circles) (New England Biolabs; 5 U), DNA gyrase (compacts the closed circles) (New England Biolabs; 5 U). The method was modified form a previously described protocol (78).
Abasic site analysis
Total DNA was extracted and analysed as in the topology gel method. Prior to loading, a DNA aliquot was digested with EndoIV (New England Biolobs; 10 U) to convert the abasic sites to single-stranded nicks. The amount of abasic sites was quantified by measuring the decrease in closed circle form of mtDNA between the treated and untreated sample after exposure to a phosphorimager screen.
In organello replication
Mitochondria were isolated from ∼100 mg of fresh heart tissue with differential centrifugation as described above. Freshly purified mitochondria, 800 μg, were pelleted at 9000 g, 2 min at 4°C and resuspended to cold incubation buffer (25 mM sucrose, 75 mM sorbitol, 10 mM Tris–HCl, 10 mM K2HPO4, 100 mM KCl, 0.05 mM EDTA, 1 mM ADP, 5 mM MgCl2, 10 mM glutamate, 2.5 mM malate, 1 mg/ml BSA, pH 7.4). The mitochondria were thereafter washed with the incubation buffer twice and resuspended to 500 μl of warm (37°C) incubation buffer with 50 μM of dCPT, dGTP, dTTP and 20 μCi of [α-32P]dATP, and incubated with rotation for 1 h at 37°C to incorporate the radioactivity into mtDNA. Next, the mitochondria were pelleted at 9000 g for 2 min at 4°C and washed twice with cold wash buffer (10% glycerol, 0.15 mM MgCl2, 10 mM Tris–HCl pH 6.8). An aliquot of the mitochondria was collected for loading control. Thereafter, the mtDNA was extracted with Gentra Puregene Tissue Kit (QIAGEN) following the manufacturer's recommendations with some small modifications. The mitochondria were lyzed during 30-min incubation at 55°C and isopropanol precipitation was done overnight in the presence of 30 μg of glycogen (Ambion). The precipitated DNA was resuspended in 20 μl of TE. The labeled DNA (5 μl) was resolved in a 0.9% agarose gel with EtBr (0.5 mg/ml) at 30 V for 14 h. Prior to loading, an aliquot (5 μl) of the purified DNA was heated for 5 min at 95°C to release the 7S DNA. After the run, the gel was transferred by Southern blotting and the membrane (Hybond-N+, GE Healthcare) was used to expose a phosphorimager screen or an autoradiography film (Amersham hyperfilm MP, GE Healthcare). The quantifications were made from exposures to phosphorimager screen. To normalize the loading, the membrane was reprobed to assess steady-state mtDNA levels by using an mtDNA-specific probe (pAM1). Additionally, the previously collected aliquot of mitochondria was resolved on a SDS-PAGE gel and the membrane was stained with Coomassie as described above to normalize the loading for mitochondrial protein per sample. The method was modified form a previously described protocol (79).
In organello transcription
The mitochondria were purified similarly as in in organello replication protocol. In the in organello transcription assay, the washed mitochondria were resuspended to 800 μl of incubation buffer with 50 μCi of [α-32P]UTP and incubated with rotation for 1 hr at 37°C to incorporate the radioactive UTP to mtRNA. The mitochondria were collected with centrifugation at 9000 g for 2 min at 4°C and resuspended into incubation buffer with 80 nM UTP. Half of the samples were incubated for an additional period of 2 h at 37°C to study RNA turnover (chase). The mitochondria were washed twice by pelletting and resuspending them in cold wash buffer (10% glycerol, 0.15 mM MgCl2, 10 mM Tris–HCl pH 6.8). An aliquot of the mitochondria was collected for loading control. Then, the RNA was extracted with 1 ml of TRIzol (Ambion) following manufacturer's recommendations and an overnight isopropanol precipitation at −20°C. The purified RNA was loaded onto a formaldehyde–agarose gel and treated as a northern blot gel as described above. Next, the membrane (Hybond-N+, GE Healthcare) was used to expose a phosphorimager screen or an autoradiography film (Amersham hyperfilm MP, GE Healthcare). The quantifications were made from exposures to phosphorimager screen. To normalize the loading, the membrane was reprobed with an [α-32P]dCTP-labeled probe to assess the steady-state CytB transcripts. Additionally, the previously collected aliquot of mitochondria was resolved on a SDS-PAGE gel and the membrane was stained with Coomassie as described above to normalize the loading for mitochondrial protein per sample. The method was modified form a previously described protocol (80).
Label-free quantitative proteomics
Percoll purified heart mitochondria were prepared for LC MS/MS with guanidinium chloride preparation. The mitochondrial pellets were resuspended in lysis buffer (6 M guanidium chloride, 10 mM TCEP, 40 mM CAA, 100 mM Tris–HCl, 100 μl per 100 mg of heart tissue) and a 20 μl aliquot of the resuspended mitochondria were lyzed with two cycles of heating (95°C, 10 min) and sonication (Bioruptor, 30 s sonication, 30 s break, 10 cycles). The debris was removed by pelleting at 20 000 g for 20 min. A small aliquot of the lyzed sample was diluted 10× with 20 mM Tris–HCl pH 8.3 to dilute the guanidium chloride and 50 μg of the sample was digested with trypsin (1:30 ratio enzyme:protein, Promega Mass spec grade) overnight at 37°C. The digested peptides were cleaned with home-made StageTip (Empore Octadecyl C18; 3M) (81) and eluted to 60% ACN/0.1% formic acid. The elutate was dried with speed-vac and resuspended to 0.1% formic acid.
The peptides were analyzed using an Orbitrap Q Exactive HF mass spectrometer (ThermoFisher Scientific) with a Nano-electrospray ion source, coupled with an EASY-nLC 1000 (ThermoFisher Scientific) UHPLC. A 25 cm long reversed-phase C18 column with 75 μm inner diameter (PicoFrit, LC Packings) was used for separating peptides. The LC runs lasted 130 min with a concentration of 2% solvent B (0.1% formic acid in acetonitrile) increasing to 25% over 120 min and further to 40% over 10 min. The column was subsequently washed and re-equilibrated. The flow rate was 200 nl/min. MS spectra were acquired in a data-dependent manner with a top 10 method. For MS, the mass range was set to 300−1500 m/z and resolution to 60 K at 200 m/z. The AGC target of MS was set to 3e6, and the maximum injection time was 100 ms. Peptides were fragmented with HCD with collision energy of 25. For MSMS, the resolution was set to 30 K. The AGC target was 2e5 and the maximum injection time was 80 ms.
MaxQuant version 1.5.3.8 (82) with integrated Andromeda search engine (83) was used for analyzing the LC/MSMS raw data. The raw data were searched against the mouse proteome from UniProt (knowledgebase 2016_04). The following parameters were used for data analysis: for ‘fixed modification’: cysteine carbamidomethylation, methionine oxidation; for ‘variable modification’: methionine oxidation and protein N-terminal acetylation; for ‘digestion’ specific with Trypsin/P, Max. missed cleavages 2; for label-free quantification, match between runs was selected. Other parameters were set as default. Protein quantification significant analysis was performed with the Perseus statistical framework (84) (http://www.perseus-framework.org/) version 1.5.2.4. After removing the contaminants and reverse identifications, the intensities were transformed to log2. The replicates of each genotype were grouped and filtered with at least three validate values in at least one group. The missing values were replaced from normal distribution with width of 0.3 and down shift of 1.8. Two-sample test was performed to identify the significantly different proteins between knockout and wild-type groups. Proteins with an adjusted P-value (‘BH’ correction) of <0.05 were designated as differentially expressed.
Statistical analysis
All values are expressed as means ± standard deviation, unless differently indicated. Statistical analysis was performed with the Prism software version 5.0f. Statistical significance between two groups was assessed with a two-tailed unpaired Student's t-test, applying Welch correction when required. When more than two groups where compared, 1way ANOVA was performed with either Tukey's or Dunnett's multiple comparison test. Mass spectrometry data were analyzed with the Perseus statistical framework version 1.5.2.4 with two-sample test with Benjamini–Hochberg adjusted P-values. Statistical significance was considered at P < 0.05. (*P < 0.05, **P < 0.005, ***P > 0.001 ****P < 0.0001) Details of statistical analysis applied to each experiment can be found in the figure legends.
RESULTS
OGG1 and MUTYH are predicted to localize to mitochondria in mice
Both MUTYH and OGG1 have been reported to be dually targeted to the nucleus and mitochondria (41,63,85,86). Because loss of BER in the nucleus causes cancer in the mouse (87), studies of BER in mitochondria should ideally involve genetic manipulations that only disrupt the function of MUTYH and OGG1 in mitochondria without affecting their nuclear function. Import of proteins into the mitochondrial matrix is typically dependent on an N-terminal mitochondrial targeting sequence (MTS) and selective ablation of mitochondrial function of MUTYH and OGG1 could thus be achieved by removing gene sequences encoding the MTS. When we analyzed the peptide sequence of mouse OGG1 (AAB94512.1) with subcellular localization prediction tools Mitoprot II (57) and Target P1.1 (58,59), both algorithms identified an N-terminal MTS (M1-W23) with high probability (0.9005 and 0.909, respectively) for mitochondrial localization. Similarly, when we analyzed the peptide sequence of mouse MUTYH (NP_001153053.1) both prediction tools identified an N-terminal MTS (M1-P33) with high probability (0.9184 and 0.871, respectively) for mitochondrial localization.
We tried to verify the predicted mitochondrial localization of the endogenous OGG1 and MUTYH proteins in the mouse by using western blots and cell fractionation. Unfortunately, neither commercially available antibodies nor antibodies generated in our laboratory recognized the endogenous mouse OGG1 or MUTYH proteins in total protein extracts from liver or heart tissue on western blots. It should be pointed out that the antibodies generated several bands on western blots, but this pattern was very similar to the pattern obtained by analysis of liver tissue extracts from double homozygous knockout (Mutyh × Ogg1 null) mice (generous gift from Prof. Lars Eide). We thus conclude that the observed pattern was due to unspecific cross reactivity of the antibodies.
OGG1 is dually targeted to nucleus and mitochondria in the mouse
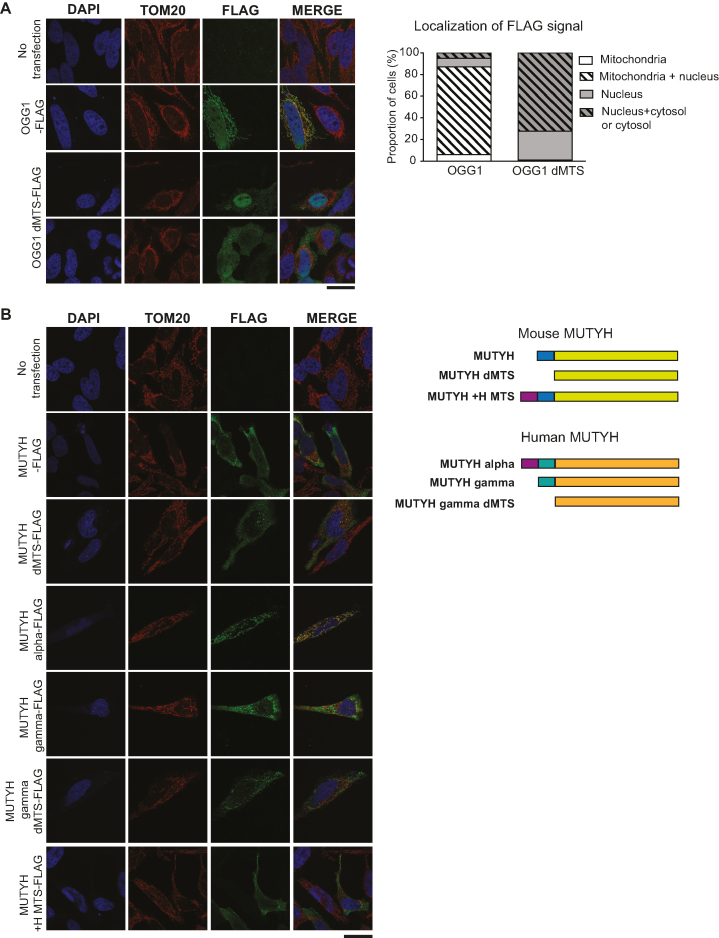
To verify mitochondrial localization, we engineered reporter constructs expressing mouse OGG1 with a C-terminal FLAG-tag. Transient transfection of HeLa cells with constructs expressing tagged OGG1 with and without the predicted N-terminal MTS sequence was performed and analyzed by immunofluorescence microscopy. There was clear co-localization between wild-type OGG1-FLAG and the mitochondrial protein TOM20 as well as with nuclear DAPI staining, thus verifying the known dual localization of this protein (Figure 1A). In contrast, cells that were transfected with the OGG1 dMTS-FLAG construct showed a dispersed signal that did not co-localize with TOM20. In ∼ 30% of the cells the signal was mostly found in the nucleus and in ∼70% in both the nucleus and cytosol (Figure 1A). We thus conclude that OGG1 contains an MTS and that removal of this import signal excludes OGG1 from mitochondria without affecting the nuclear localization. Based on these results we engineered a mouse mutant expressing OGG1 without the MTS.
Figure 1.
Evaluating mitochondrial targeting of OGG1 and MUTYH. (A) Subcellular localization of OGG1 with and without the predicted sequence encoding for the mitochondrial targeting sequence (dMTS). HeLa cells were transiently transfected with Ogg1-FLAG constructs (NM_010957.4, OGG1-FLAG, OGG1 dMTS-FLAG, ΔL2-W23) and target proteins were visualized by immunocytochemistry. Nuclear staining (DAPI, blue), mitochondrial signal (TOM20, red), OGG1 (green, FLAG). Scale bar represents 25 μm. The subcellular localization of FLAG signal was quantified by counting it from 100 cells. (B) Subcellular localization of mouse and human MUTYH with and without the predicted sequence encoding for the mitochondrial targeting sequence (dMTS). HeLa cells were transiently transfected with Mutyh-FLAG constructs and target proteins were visualized by immunocytochemistry. Nuclear staining (blue, DAPI), mitochondrial signal (red, TOM20), MUTYH (green, FLAG). Mouse MUTYH variant b/2 (NM_133250.2). Human MUTYH alpha3 variant (NM_001048171.1) and gamma3 variant (NM_001048173.1). Scale bar represents 25 μm.
MUTYH has no distinct mitochondrial localization in the mouse
We also engineered reporter constructs expressing mouse MUTYH with and without the predicted N-terminal MTS sequence containing a C-terminal FLAG-tag. Unexpectedly, transient transfection of HeLa cells with mouse MUTYH-FLAG or MUTYH dMTS-FLAG showed a dispersed signal that did not co-localize with mitochondrial TOM20 or nuclear DAPI staining (Figure 1B). In humans, the mitochondrial isoform of MUTYH is encoded by transcript variant alpha that has an N-terminal extension (Figure 1B) (85,88). The mouse MUTYH lacks this N-terminal extension and is similar to the human MUTYH transcript variant gamma that encodes an isoform with a predominant nuclear localization (85,88). To study the role of the human N-terminal extension of MUTYH, we generated a chimeric construct expressing mouse MUTYH-FLAG with the human N-terminal extension but obtained no clear mitochondrial localization (Figure 1B). Moreover, cells that were transiently transfected with the human nuclear isoform of MUTYH, showed a dispersed signal that did not co-localize with mitochondrial TOM20 or nuclear DAPI staining, similar to the localization we obtained when transfecting the corresponding mouse variants (Figure 1B). To summarize, attempts to define the subcellular localization of mouse MUTYH with cell fractionation were unsuccessful and transfection with reporter constructs gave inconclusive results. However, given the fact that the human MUTYH has a mitochondrial isoform and that mouse MUTYH is strongly bioinformatically predicted to localize in mitochondria, we decided to generate a mouse mutant expressing MUTYH without the predicted MTS.
Loss of the mitochondrial OGG1 and MUTYH has no obvious phenotype in mice
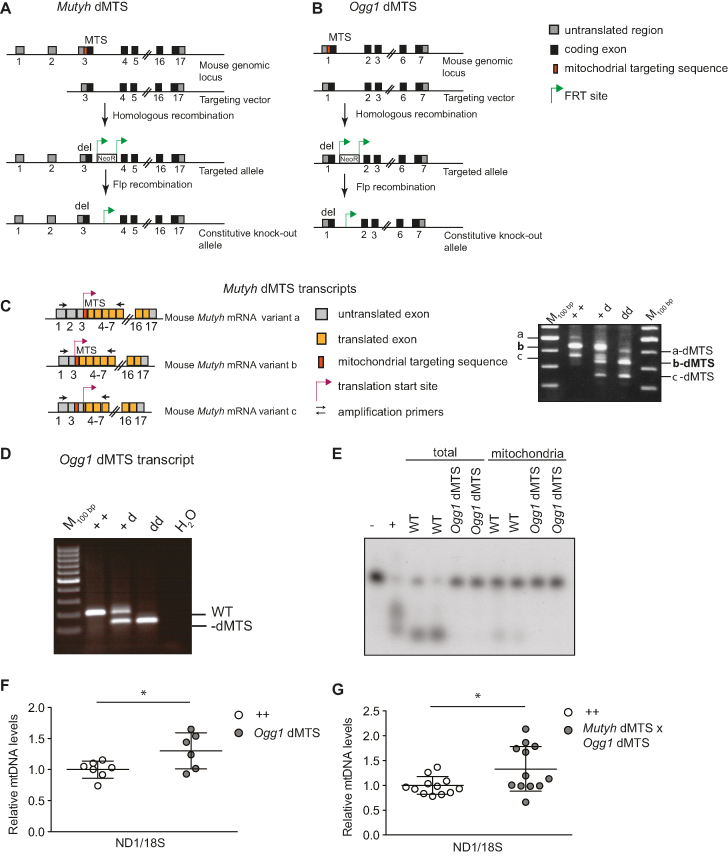
To study the importance of BER in mitochondria, we generated Ogg1 dMTS and Mutyh dMTS knockout mice that express the corresponding glycosylases without the predicted MTS (Figure 2A and B). The lack of nucleotide sequence encoding the MTS was verified by PCR amplification of cDNA from Ogg1 dMTS and Mutyh dMTS mice. As expected, the corresponding transcripts were shorter in the dMTS mice in comparison with wild-type controls (Figure 2C and D). Moreover, the removal of the nucleotide sequence encoding the MTS did not affect the alternative splicing of the Mutyh transcript (Figure 2C), as all the published splice variants were still visible in the PCR analysis (63).
Figure 2.
Excluding OGG1 and MUTYH from mitochondria does not lead to decrease in mtDNA copy number. (A) Targeting strategy of Mutyh dMTS mice to remove sequence encoding the predicted mitochondrial targeting sequence (dMTS) of endogenous MUTYH (ΔK2-P33). (B) Targeting strategy of Ogg1 dMTS mice to remove the sequence encoding the predicted mitochondrial targeting sequence (dMTS) of endogenous OGG1 (ΔL2-W23). (C) Splice variants of Mutyh dMTS and PCR amplification from cDNA to verify the presence of all the splice variants and correct length of the modified transcripts. Transcript variants a, b and c are also known as variants 1, 2 and 3, respectively. (D) PCR amplification of Ogg1 dMTS transcripts from cDNA to verify the correct length of the modified transcript from various genotypes. (E) 8-oxo-dG glycosylase/AP lyase assay to verify that Ogg1 dMTS animals lack OGG1 8-oxo-dG glycosylase activity. Total and mitochondrial lysates were incubated with 8-oxo-dG containing double-stranded oligonucleotide and reaction products were resolved on a denaturing acrylamide gel. WT n = 4, Ogg1 dMTS n = 5 Recombinant OGG1 was used as positive control (+) and in negative control no protein lysate was added. For longer exposure see Supplementary Figure S1A. (F) Relative mtDNA copy number of Ogg1 dMTS mice assessed from liver with qPCR. MtDNA levels were analyzed with a ND1 probe and nuclear DNA with a 18S probe. White circles indicate samples from wild-type controls (++, n = 7, 95–109 week old) and gray circles indicate samples from homozygous Ogg1 dMTS mice (dd, n = 6, 88–107 week old). Horizontal lines represent means, error bars represent SD, *P < 0.05, Student's t-test. For relative copy number analysis with Southern blot see Supplementary Figure S2A. (G) Relative mtDNA copy number of Mutyh dMTS × Ogg1 dMTS mice assessed from liver with qPCR. MtDNA levels were analyzed with a ND1 probe and nuclear DNA with a 18S probe. White circles indicate samples from wild-type controls (++, n = 7, 40–51 week old) and gray circles indicate samples from homozygous Mutyh dMTS × Ogg1 dMTS mice (dd, n = 6, 39–50 week old). Horizontal lines represent means, error bars represent SD, *P < 0.05, Student's t-test, Welch-corrected. For relative copy number analysis with Southern blot see Supplementary Figure S2B.
To verify the absence of 8-oxodG glycosylase/AP lyase activity in Ogg1 dMTS mitochondria, we incubated total and mitochondrial lysates with an 8-oxo-dG containing oligonucleotide and resolved the reaction product on a denaturing acrylamide gel. As expected, the control lysates produced a shorter nicked oligonucleotide whereas the oligonucleotide remained intact when incubated with the Ogg1 dMTS mitochondrial lysate (Figure 2E, Supplementary Figure S1A). This demonstrates that Ogg1 dMTS mitochondria lack 8-oxo-dG glycosylase activity. Unexpectedly, the total lysates from Ogg1 dMTS mice showed no 8-oxo-dG glycosylase activity, suggesting that removal of MTS altered the activity of the enzyme. However, this does not affect our analysis because we are focusing on the mitochondrial phenotype of the Ogg1-dMTS mice.
Next, the overall phenotype of the knockout mice was evaluated. No changes in body weight, heart-to-body weight or spleen-to-body weight ratio were found in the single knockouts (Ogg1 dMTS or Mutyh dMTS) or in the double knockout (Mutyh dMTS × Ogg1 dMTS) mice (Supplementary Figure S1B and C). Consistent with previous studies using single Ogg1 knockout or double Mutyh and Ogg1 knockout mice (89,90), we did not detect alterations in the steady-state levels of OXPHOS proteins in liver mitochondria from Mutyh dMTS × Ogg1 dMTS mice (Supplementary Figure S1D). These studies suggest that the absence of the OGG1 and MUTYH glycosylases in mitochondria does not have an immediate negative impact on mouse physiology or mitochondrial function, in contrast to the clear phenotypes present in mice lacking several other types of proteins involved in mtDNA replication or maintenance (91–93).
Interestingly, the absence of mitochondrial 8-oxo-dG glycosylase activity did not lead to decreased mtDNA levels in Ogg1 dMTS or Mutyh dMTS x Ogg1 dMTS mice (Figure 2F and G). This is surprising because in vitro studies have demonstrated that the mitochondrial DNA polymerase (POLγ) has decreased efficiency in incorporating nucleotides opposite to 8-oxo-dG (94–96). In contrast, a slight increase in mtDNA copy number was seen in the liver of Ogg1 dMTS and Mutyh dMTS x Ogg1 dMTS mice, but there were no changes in mtDNA integrity as evaluated by Southern blot analysis (Supplementary Figure S2A and B).
Mitochondrial BER deficiency does not lead to increase in somatic or maternally transmitted mtDNA mutations
The mtDNA mutation load was analyzed with Sanger sequencing of cloned PCR fragments containing the WANCY-COX1 genomic region of mtDNA. Because recent work has revealed that B[a]P mtDNA adducts do not lead to expected increase in mtDNA mutation load (97) we wanted to verify that post-PCR cloning and Sanger sequencing method has the ability to detect 8-oxo-dG lesions and in vivo G>T mutations. To this end, we used oxidized phenol extraction to isolate total DNA from flies. DNA isolated with this damaging in vitro procedure showed a five-fold increase in the G>T mutation load (2.42 × 10−4 versus control 4.72 × 10−5 mutations per G:C pair) confirming the method's ability to detect G>T mutations, as well as 8-oxo-dG damage to DNA.
The Ogg1 null mice have been reported to have an increase in nuclear DNA mutation load in liver (98,99), and we therefore analyzed the mtDNA mutation load in liver from the Ogg1 dMTS mice. Despite deficient mitochondrial BER, no increase in the mtDNA mutation load was found in Ogg1 dMTS mice (1.82 × 10−5 mutations/bp) in comparison with control mice (1.62 × 10−5 mutations/bp) at 100 weeks of age (Table 1). Although surprising, these results are consistent with the data from Mutyh × Ogg1 null mice, that did not show any increase in the mtDNA mutation load as determined by the random mutation capture assay (RMC) (90).
Table 1.
Mitochondrial BER deficiency does not lead to increase in somatic or maternally transmitted mtDNA mutations. Mutation load analysis from Ogg1 dMTS mice and Mutyh dMTS x Ogg1 dMTS mice after five generations of consecutive breeding. Mutation load was measured with post-PCR cloning and Sanger sequencing from WANCY-COX1 tRNA-cluster region from liver tissue. On average 92 kb were sequenced per sample
| Mouse line | Genotype | Age (w) | # of mutations | Mutation load | Mutations found |
|---|---|---|---|---|---|
| Ogg1 dMTS | ++ | 109 | 2 | 2.16 × 10−5 | G>A, C>G |
| Ogg1 dMTS | ++ | 95 | 0 | <1.08 × 10−5 | |
| Ogg1 dMTS | dd | 107 | 2 | 2.18 × 10−5 | A>C, A>C |
| Ogg1 dMTS | dd | 88 | 0 | <1.10 × 10−5 | |
| Ogg1 dMTS | dd | 107 | 2 | 2.17 × 10−5 | G>A, T>A |
| Mutyh dMTS x Ogg1 dMTS | dd dd | 22 | 0 | <1.07 × 10−5 | |
| Mutyh dMTS x Ogg1 dMTS | dd dd | 22 | 1 | 1.12 × 10−5 | T>C |
| Mutyh dMTS x Ogg1 dMTS | dd dd | 23 | 0 | <1.09 × 10−5 |
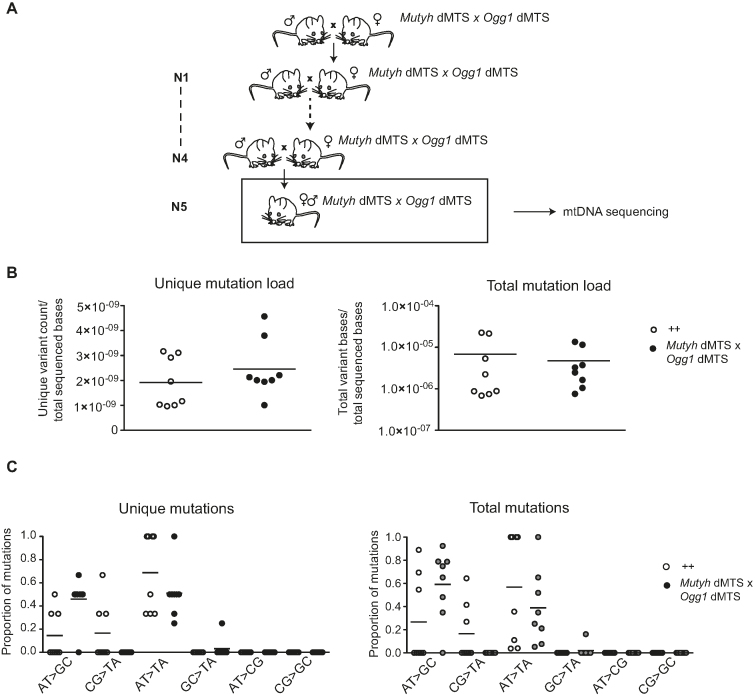
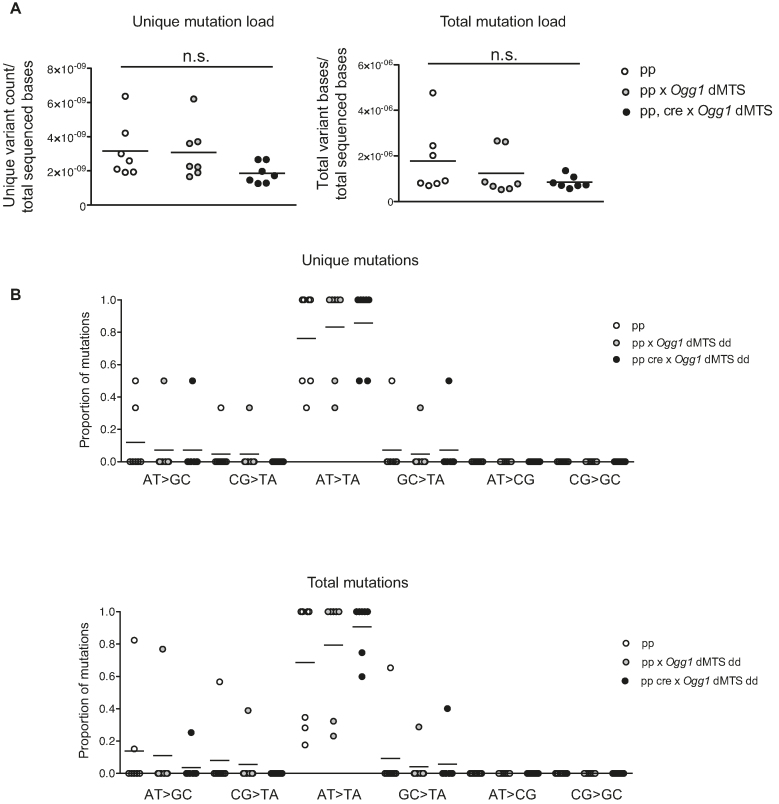
It is possible that the old Ogg1 dMTS mice have no increase or that the increase in mtDNA mutation load is too small to allow detection. We therefore hypothesized that mtDNA mutations passed down the maternal germ line may be easier to detect because of the well-known bottleneck phenomenon that can cause clonal expansion of mtDNA mutations transmitted through the maternal germ line (66). Furthermore, it has also been hypothesized that DNA repair is more stringent in the germ line than in somatic tissues (100). To study possible effects in the maternal germ line, we bred double homozygous Mutyh dMTS × Ogg1 dMTS mice as maternal lineages for five consecutive generations but found no effect on the mtDNA mutation load (1.09 × 10−5 mutations/bp) (Figure 3A, Table 1). This value is close to the minimum mutation load (∼1.1 × 10−5 mutation/bp), that can be detected with the depth of our post-PCR cloning approach. To increase the depth of mutation analysis and to expand the coverage to the entire mtDNA, we used Illumina sequencing of purified mtDNA to assess the mutation load. The variant-called data was quality filtered and the minimum variant allele frequency threshold was set to 0.5%. Based on previous studies we know that mtDNA mutations will clonally expand between generations and reach relative heteroplasmy levels above 1% in the offspring (65), which justifies the applied threshold of 0.5%. Consistent with the results from Sanger sequencing, also the Illumina sequencing showed no increase in unique or total mutation load in Mutyh dMTS × Ogg1 dMTS mice after five generations of consecutive breeding (Figure 3B). To verify that the 0.5% cut off limit was not too stringent, we analyzed data obtained by only applying quality filtering and again found no increase in the mtDNA mutation load (Supplementary Figure S3). In the data sets obtained by only applying quality filtering, we detected high levels of transversion mutations in both control and repair-deficient mouse samples, which argues that the observed G>T mutations were induced during library preparation, as previously described (101–103) (Supplementary Figure S3B).
Figure 3.
Mitochondrial BER deficient mice do not accumulate point mutations to mtDNA after five generations of consecutive breeding. (A) Breeding scheme to accumulate mutations into mtDNA and study germ line mutations. Homozygous Mutyh dMTS × Ogg1 dMTS female mice were bred with homo- or heterozygous Mutyh dMTS × Ogg1 dMTS male mice for five consecutive generations. To minimize the nuclear effects, heterozygote male mice were also used in the breedings. N1–N5 indicates the generations of breeding. (B) Mutation load of mtDNA with Illumina sequencing from Mutyh dMTS × Ogg1 dMTS mice after five generations of consecutive breeding. The sequencing was carried out from purified mtDNA from liver. Data is quality filtered and minimum variant allele frequency is set to 0.5%. In unique mutation load each mutation is counted only once, reflecting how many times a specific mutation has occurred. In total mutation load each mutation is counted as many times as it is seen, reflecting the clonal expansion of mutations. White circles indicate samples from controls (++ n = 6, pp n = 2, 10–13 week old) and gray circles indicate samples from homozygous Mutyh dMTS × Ogg1 dMTS mice (dd dd, n = 8, 10–15 week old). Horizontal lines represent means. C. Mutation profile of mtDNA with Illumina sequencing from Mutyh dMTS × Ogg1 dMTS mice after five generations of consecutive breeding. Samples as in B. Horizontal lines represent means. For only quality filtered data see Supplementary Figure S3.
As we found no increase in total mtDNA mutation load in mice with deficient mitochondrial BER, we proceeded to investigate whether the mutational pattern was different. 8-oxo-dG can induce G:C>T:A mutations when adenosine is incorporated opposite to 8-oxo-dG during replication. However, the prevalence of G:C>T:A transversion mutations showed no increase when we compared Mutyh dMTS × Ogg1 dMTS and control mice. Additionally, A:T>C:G mutations will be formed, when 8-oxo-dGTP is incorporated against adenosine during replication and the 8-oxo-dG:dA mispair is repaired by MUTYH. We did not observe a relative increase in A:T>C:G mutations in the control mice in comparison with Mutyh dMTS × Ogg1 dMTS mice. These results were the same regardless of if we analyzed data obtained by only quality filtering or by quality filtering followed by application of the 0.5% cut-off limit for mutation detection (Figure 3C, Supplementary Figure S3B). Taken together, these results show that 8-oxo-dG driven mutations do not impair mtDNA integrity or increase the mtDNA mutation load when mitochondrial BER is impaired.
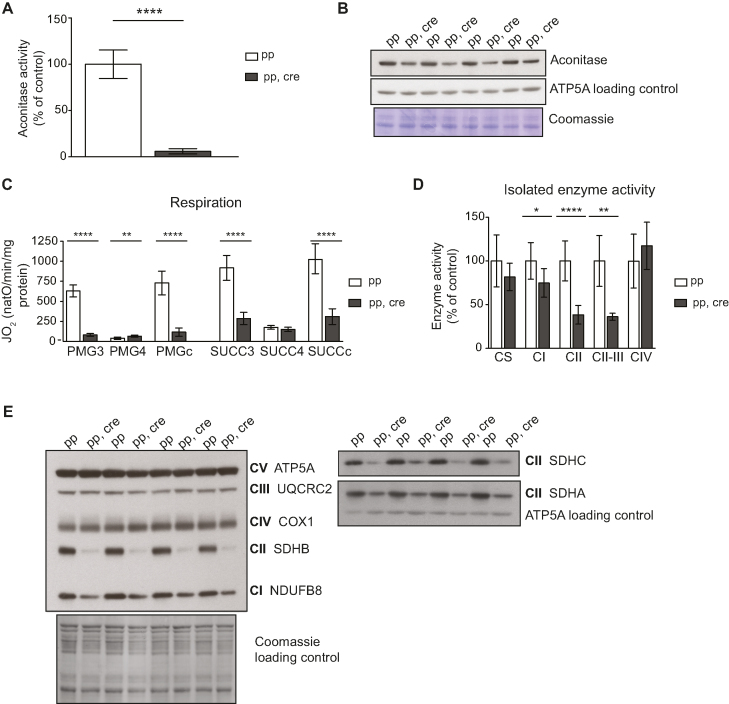
Loss of Sod2 in the heart causes severe oxidative stress and impaired function of [4Fe–4S] cluster proteins
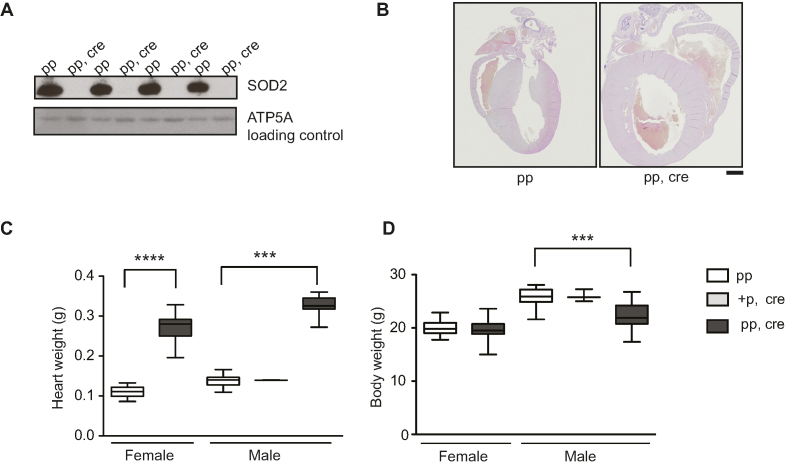
The superoxide dismutases (SODs) convert superoxide into hydrogen peroxide and therefore provide an important ROS defense mechanism (104,105). SOD2 is the sole superoxide dismutase located in the mitochondrial matrix and Sod2 knockout mice have been extensively utilized to study the consequences that increased oxidative stress has on mitochondrial function (60,106–110). We used a conditional knockout allele (60) to disrupt Sod2 in heart (Sod2 loxP × Ckmm cre, Figures 4A and 6) and consistent with previously published observations (108) these mice developed a dilated cardiomyopathy (Figure 4B–D). The absence of the SOD2 protein in heart leads to a number of biochemical aberrations in proteins containing [4Fe–4S] clusters (106,108), including decreased aconitase enzyme activity (Figure 5A and B). Many dehydratases, including aconitase (34,111), harbor a [4Fe–4S] cluster that is highly susceptible to superoxide-induced damage. In aconitase, the [4Fe–4S] cluster is solvent accessible and required for its enzyme activity. Aconitase activity is therefore widely used as a surrogate marker for superoxide levels (112,113). Also, complex I and complex II of the respiratory chain contain [4Fe–4S] clusters, whereas such clusters are absent in complex III and IV (114–117). When analyzing isolated heart mitochondria with polarographic methods, we found a strong decrease in respiration when substrates were used that results in entry of electrons at the level of complex I (pyruvate, glutamate and malate, PGM) or complex II (succinate, SUCC) (Figure 5C). The oxygen consumption was measured in the phosphorylating (state 3: ADP and Pi, PGM3, SUCC3), non-phosphorylating (state 4: oligomycin to inhibit ATP synthase, PMG4, SUCC4) and uncoupled states (CCCP; PMGc, SUCCc). The profound decrease in respiration in state 3 and the uncoupled state was accompanied by a corresponding strong decrease of the enzyme activity of complex I and complex II (Figure 5D). The enzyme activity of complex IV remained unaffected (Figure 5D), consistent with previous COX-SDH enzyme histochemistry results (106,108). Consistent with these results we found no changes in steady-state levels of protein subunits of complex III and complex IV on western blots (Figure 5E) (108). In contrast, the steady-state levels of subunits of complex I and complex II were strongly decreased on western blots (Figure 5E).
Figure 4.
Heart Sod2 knockout mice display severe dilated cardiomyopathy. (A) Western blot analysis of SOD2 protein levels from purified mitochondria of control (pp) and Sod2 loxP × Ckmm cre mice (pp, cre) (9–11 week old). ATP5A was used as a loading control. (B) Vertical sections through the midpoint of paraffin embedded hearts stained with hematoxylin and eosin staining. Control (pp, 11-week old) and Sod2 loxP × Ckmm cre (pp, cre, 10-week old). Scale bar represents 1 mm. (C) Heart weight of control (pp), heterozygous Sod2 loxP x Ckmm cre and homozygous Sod2 loxP x Ckmm cre (pp, cre) mice. White box indicates control mice (pp female n = 22, male n = 35, 9–11 week old), light gray box indicates heterozygous Sod2 loxP x Ckmm cre mice (male +p, cre, n = 2, 9-week old) and dark gray box indicates homozygous Sod2 loxP x Ckmm cre mice (pp, cre, female n = 28, male n = 15, 9–10 week old). D. Body weight of control (pp), heterozygous Sod2 loxP x Ckmm cre and homozygous Sod2 loxP x Ckmm cre (pp, cre) mice. White box indicates control mice (pp female n = 26, male n = 36, 9–11 week old), light gray box indicates heterozygous Sod2 loxP x Ckmm cre mice (male +p, cre, n = 3, 9-week old) and dark gray box indicates homozygous Sod2 loxP x Ckmm cre mice (pp, cre, female n = 30, male n = 19, 9–10 week old). Whiskers represent min and max values, horizontal lines medians; ****P< 0.0001, females Student's t-test, Welch corrected. ***P< 0.001, males one-way ANOVA, Dunnett's multiple comparison test.
Figure 6.
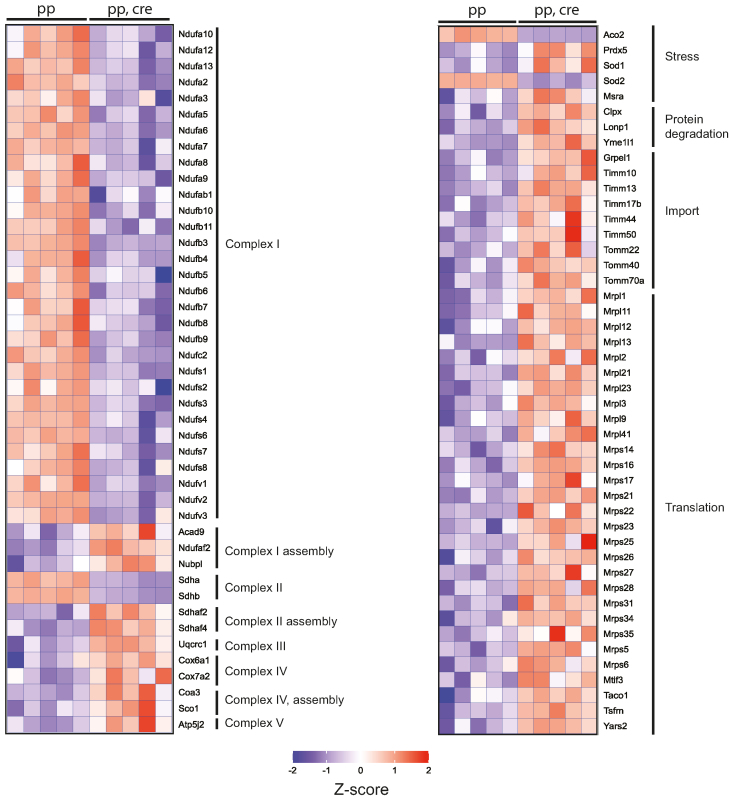
Heart Sod2 knockout mice show global decrease in complex I proteins and indications of general mitochondrial stress in label-free quantitative proteomics. Heat map of selected proteins from label-free quantitative proteomic analysis of Percoll gradient purified heart mitochondria from controls (pp, 8–9 week old) and Sod2 loxP × Ckmm cre mice (pp, cre, 9–10 week old). Changes in the protein steady-state levels are blotted as Z-scores. Blue indicates decreased and red increased steady-state level from the global mean across all samples. Abundances of all the presented proteins were significantly changed, with Benjamini–Hochberg adjusted P-values of <0.05.
Figure 5.
[4Fe–4S] cluster proteins are severely affected in heart Sod2 knockout mice indicating strong increase in superoxide levels. (A) Aconitase activity from purified mitochondria from control (pp) and Sod2 loxP × Ckmm cre mice (pp, cre). White bar indicates activity in control samples (n = 6, 9–10 week old) and gray bar in Sod2 loxP x Ckmm cre samples (n = 6, 9–12 week old). Activity is normalized to control. (B) Western blot analysis of ACO 2 (aconitase) protein levels from purified mitochondria of control (pp) and Sod2 loxP x Ckmm cre mice (pp, cre) (9–10 week old). ATP5A and Coomassie-stained membrane were used as loading controls. (C) Oxygen consumption rate of isolated heart mitochondria from control (pp, white bars, n = 9, 9–11 week old) and Sod2 loxP x Ckmm cre mice (pp, cre, gray bars, n = 9, 9–12 week old). Isolated mitochondria were incubated with complex I (PMG) or complex II (SUCC) substrates. Each set of substrates was successively combined with ADP (to assess the phosphorylating respiration, PMG3, SUCC3), oligomycin (to assess the non-phosphorylating respiration PMG4, SUCC4) and CCCP (to assess uncoupled respiration PMGc, SUCCc). (D) Activity of the respiratory chain complexes I (CI), II (CII), IV (CIV) and the activity from complex II to III (CII-III) of heart mitochondria from control (pp, write bars, n = 3, 11-week old) and Sod2 loxP x Ckmm cre mice (pp, cre, gray bars n = 3, 11–12 week old). Citrate synthase activity (CS) was used as a control. Error bars represent SD. *P< 0.05, **P< 0.005, ****P< 0.0001, Student's t-test, Welch corrected. (E) Western blot analysis of OXPHOS proteins from purified heart mitochondria from control (pp) and Sod2 loxP × Ckmm cre (pp, cre) mice (9–11 week old).
Label-free quantitative proteomics shows profound general mitochondrial stress responses in Sod2 knockout mice
We further analyzed the proteome of Sod2 knockout hearts by using label-free mass spectrometry analysis of purified mitochondria (Figure 6). The steady-state levels of multiple subunits of complex I and II were strongly decreased and there was a concomitant increase of several assembly factors for complexes I, II and IV, indicating a compensatory response to facilitate assembly of OXPHOS complexes (Figure 6). Clear indications of general mitochondrial stress were also observed as the steady-state levels of defense proteins (SOD1, PRDX5, MSRA), mitochondrial proteases (CLPX, LONP1, YME1L1), mitochondrial import machinery components (TIMM, TOM) and proteins involved in mitochondrial translation were increased (Figure 6).
No increase in mtDNA mutation load is observed in Sod2 knockout mice lacking BER
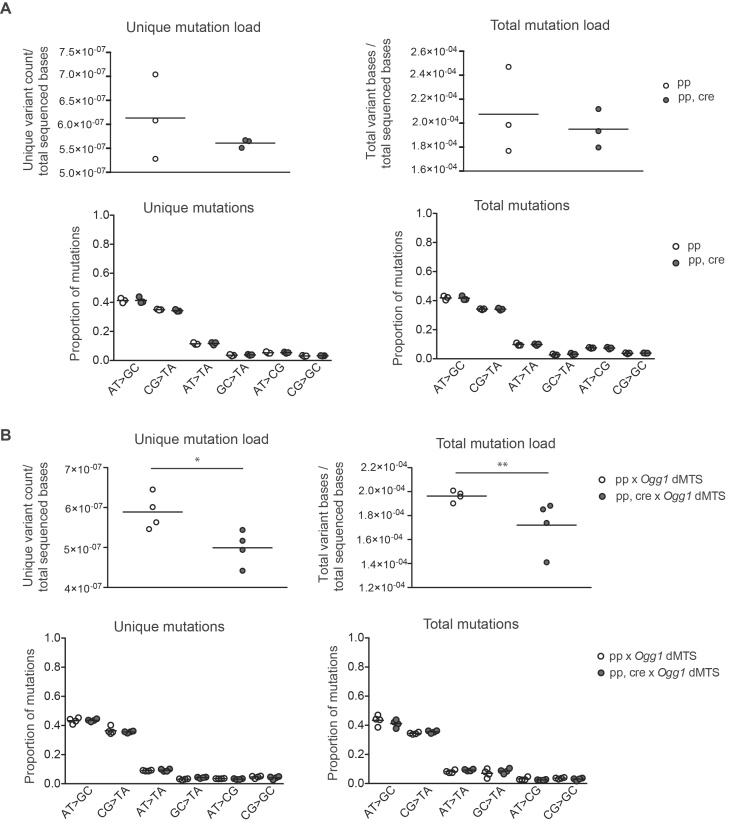
To increase the amount of oxidative stress in the repair-deficient mice, Ogg1 dMTS mice were crossed to generate mice that also had homozygous knockout of Sod2 in heart. Combined increased oxidative stress and absence of OGG1 had no effect on the mtDNA mutation load of 10-week-old mice in comparison with controls (1.30 × 10−5 versus 1.10 × 10−5 mutations/bp) with post-PCR cloning and Sanger sequencing (Table 2). As these values are close to the minimum mutation load (∼1.1 × 10−5 mutations/bp) that can be detected with the depth of our post-PCR cloning approach, they represent the upper limit of the mutation load present in these mice. However, it should be noted that heterozygous mutator mice with one order of magnitude higher mutation load, have normal lifespan and show no obvious phenotype (∼3 × 10−4 mutations/bp (46)). Accordingly, even if the mutation load of heart Sod2 knockout Ogg1 dMTS mice would be increased whilst remaining below our detection limit, this increase in mutation load would not be biologically relevant. Consistent with the lack of mutational increase, it has been previously shown that flies with decreased SOD2 activity combined with a loss-of-function mutation in Ogg1 had no increase in mtDNA mutation load as measured by RMC (13). To exclude the possibility that oxidative damage induced mtDNA mutations are not detected due to being under the threshold of post-PCR cloning and sequencing or positioned outside of the analyzed region, Illumina sequencing was carried out from purified heart mtDNA from mice that were homozygous for the Ogg1 dMTS allele and also had a homozygous knockout for Sod2 in the heart. Again, there was no increase in the mutation load or in the prevalence of G:C>T:A mutations. Furthermore, no increase in MUTYH-induced A:T>C:G mutations were seen in these mice in comparison with controls using the 0.5% minimum variant allele frequency threshold (Figure 7).
Table 2.
No increase in mtDNA mutation load is observed in Sod2 knockout mice lacking mitochondrial BER Mutation load analysis from Sod2 loxP x Ckmm cre x Ogg1 dMTS mice from heart tissue. Mutation load was measured with post-PCR cloning and Sanger sequencing from WANCY-COX1 tRNA-cluster region from heart tissue. On average 92 kb were sequenced per sample
| Mouse line | Genotype | Age (w) | # of mutations | Mutation load | Mutations found |
|---|---|---|---|---|---|
| Sod2 loxp | pp | 10 | 1 | 1.09 × 10−5 | T>C |
| Sod2 loxp x Ckmm cre | pp | 10 | 0 | <1.10 × 10−5 | |
| Sod2 loxp x Ckmm cre x Ogg1 dMTS | pp, cre dd | 10 | 2 | 2.14 × 10−5 | G>T, G>T |
| Sod2 loxp x Ckmm cre x Ogg1 dMTS | pp, cre dd | 9 | 0 | <1.14 × 10−5 | |
| Sod2 loxp x Ckmm cre x Ogg1 dMTS | pp, cre dd | 10 | 1 | 1.09 × 10−5 | G>A |
| Sod2 loxp x Ckmm cre x Ogg1 dMTS | pp, cre dd | 10 | 1 | 1.08 × 10−5 | C>T |
| Sod2 loxp x Ckmm cre x Ogg1 dMTS | pp, cre dd | 10 | 1 | 1.07 × 10−5 | A>T |
Figure 7.
Mitochodrial BER deficient mice do not accumulate point mutations to mtDNA even in the presence of increased oxidative stress. (A) Mutation load of mtDNA from Sod2 loxP x Ckmm cre x Ogg1 dMTS mice with Illumina sequencing. The sequencing was carried out from purified mtDNA from heart. Data is quality filtered and minimum variant allele frequency is set to 0.5%. For the unique mutation load each mutation is counted only once, reflecting how many times a specific mutation has occurred. For the total mutation load each mutation is counted as many times as it is seen, reflecting the clonal expansion of mutations. White circles indicate samples from controls (pp n = 4 or ++ n = 3, 8–12 week old), light gray circles indicate samples from Sod2 loxP x Ogg1 dMTS mice (pp dd n = 4 or +p dd n = 2 or +p cre+ dd n = 1, 8–11 week old) and gray circles indicate samples from Sod2 loxP x Ckmm cre x Ogg1 dMTS mice (pp, cre dd, n = 7, 9–10 week old). Horizontal lines represent means, one-way ANOVA, Tukey's multiple comparison test. (B) Mutation profile of mtDNA from Sod2 loxP x Ckmm cre x Ogg1 dMTS with Illumina sequencing. The sequencing was carried out from purified mtDNA from heart. Samples as in A. Horizontal lines represent mean. For only quality-filtered data see Supplementary Figure S4.
Similar results were obtained using the only quality-filtered data (Supplementary Figure S4).
The mitochondrial RNA mutation load is not increased in heart Sod2 knockouts lacking BER
Based on our results, oxidative stress has limited effects on mtDNA mutagenesis. However, oxidized mtDNA could potentially cause problems during mitochondrial transcription because the mitochondrial RNA polymerase, POLRMT, has been shown to preferentially incorporate adenosine opposite 8-oxoguanosine on a DNA template in vitro (118). To measure whether 8-oxo-dG causes G:C>T:A transversion mutations in mtRNA in either heart Sod2 knockout or heart Sod2 knockouts that also are homozygous for the Ogg1 dMTS allele, variant calling was carried out for reads that mapped to mtDNA from Illumina sequencing of total RNA. There was no increase in mitochondrial RNA mutation load in either heart Sod2 knockout or heart Sod2 knockouts that also are homozygous for the Ogg1 dMTS allele (Figure 8) when only quality-filtered data was used. Similar results were obtained using the quality-filtered and 0.5% cut-off data (Supplementary Figure S5). The prevalence of G:C>T:A mutations was similar between samples. As RNA conversion to DNA is known to be a highly error-prone process, it could induce noise to our data and mask the mutational profile differences between our samples. However, the reverse transcriptase that was used in the RNA library preparation (Moloney murine leukemia virus reverse transcriptase) rarely makes G>T mutations (119), thus justifying our approach to RNA mutation detection. Another possibility explaining the absence of G:C>T:A mutations is that POLRMT has been shown to stall on 8-oxo-dG (118). However, the presence of the mitochondrial transcription elongation factor (TEFM) has been shown to stimulate transcription past 8-oxo-dG in vitro, suggesting that POLRMT should be able to pass 8-oxo-dG in vivo (120).
Figure 8.
Mitochondrial BER deficient mice do not accumulate point mutations of mtRNA even in the presence of increased oxidative stress. (A) Mutation load of mtRNA from Sod2 loxP x Ckmm cre mice from heart. Illumina sequencing was carried out from total RNA considering only the reads that map to mtDNA for variant calling. Data is quality filtered. For the unique mutation load each specific mutation is counted only once, reflecting how many times a mutation has occurred. For the total mutation load each mutation is counted as many times as it is seen, reflecting the clonal expansion of mutations. Mutation profile of mtRNA from Sod2 loxP x Ckmm cre mice from heart. White circles indicate samples from controls (+p n = 1 pp n = 2, 10–11 week old) and gray circles indicate samples from Sod2 loxP x Ckmm cre mice (pp, cre n = 3, 10–11 week old). (B) Mutation load of mtRNA from Sod2 loxP x Ckmm cre x Ogg1 dMTS mice from heart. Illumina sequencing was carried out from total RNA considering only the reads that map to mtDNA for variant calling. Data is quality filtered. Mutation profile of mtRNA from Sod2 loxP x Ckmm cre x Ogg1 dMTS mice from heart. White circles indicate samples from Sod2 loxP x Ogg1 dMTS mice (pp dd n = 4, 9–10 week old) and gray circles indicate samples from Sod2 loxP x Ckmm cre x Ogg1 dMTS mice (pp, cre dd, n = 4, 9–10 week old). *P< 0.05, **P< 0.005, Student's t-test. For quality-filtered data with minimum variant allele frequency set to 0.5% see Supplementary Figure S5.
De novo replication is decreased in heart Sod2 knockout mice whereas de novo transcription is unaffected
Surprised by the lack of mutations in heart Sod2 knockouts and in heart Sod2 knockouts that also are homozygous for the Ogg1 dMTS allele, other forms of DNA damage were considered because these mutant animals have clearly increased oxidative stress. Oxidative stress is known to induce single-stranded breaks as well as abasic sites (21,121). To assess alterations in mtDNA integrity, a topological analysis of mtDNA was carried in heart Sod2 knockout mice. For topological analysis, total DNA was gently extracted from fresh heart tissue and separated onto a low-percent agarose gel to reveal different topological states of mtDNA. The topologies were analyzed in the presence and absence of ethidium bromide (EtBr). As EtBr intercalates with DNA it concentrates different stages of supercoiling of the closed-circle mtDNA into a quantifiable band (Figure 9A). When the proportional distribution of mtDNA was evaluated between different topologies (catenanes, nicked circles, linear and closed circles) no substantial changes were observed between the heart Sod2 knockout and control mice. The proportion of mtDNA that remained as a closed circle was similar between genotypes (Figure 9B). If mtDNA would contain more single-stranded breaks, the proportion of nicked circles would increase at the expense of closed circles. As this was not the case, mtDNA does not seem to accumulate an extensive amount of single-stranded breaks during continuous oxidative stress. The mtDNA topology was analyzed also in heart Sod2 knockout mice that are homozygous for the Ogg1 dMTS allele, but also in these double mutants no change in topology was found (Supplementary Figure S6).
Figure 9.
Heart Sod2 knockout mice show normal mtDNA topology and no decrease in mtDNA copy number. (A) Representative phosphorimager exposure of mtDNA topology analysis of total DNA from heart tissue from 10-week old Sod2 loxP x Ckmm cre mice. MtDNA is visualized using radioactive probes towards mtDNA. Control DNA was treated with various enzymes to reveal the different topologies of mtDNA. SacI cuts both strands of mtDNA once (linear), Nt. BbvCI cuts only one strand of mtDNA (nicked), TopoI relaxes the mtDNA (looser coiling), Gyrase creates coiling to mtDNA (compacted supercoiled DNA). Experimental samples are untreated. First gel does not have ethidium bromide (EtBr), second gel has the same samples and EtBr in the gel to compact the closed circle DNA into a quantifiable band. Phosphorimager images are filtered with averaging to reduce noise. Quantifications were made from the original images. (B) Quantification of the proportion of closed circle form of mtDNA per total mtDNA. Quantification is done from phosphorimager exposure of the topology gels. White circles indicate samples from controls (pp, n = 11, 9–10 week old) and gray circles indicate samples from Sod2 loxP x Ckmm cre mice (pp,cre, n = 12, 10-week old). (C) Relative mtDNA copy number in heart of Sod2 loxP x Ckmm cre mice as assessed with qPCR. MtDNA levels were analyzed with a CytB probe and nuclear DNA with a 18S probe. White circles indicate samples from controls (pp, n = 12, 10–12 week old) and gray circles indicate samples from Sod2 loxP x Ckmm cre mice (pp, cre, n = 11, 10–12 week old). Horizontal lines represent means, error bars represent SD, *P< 0.05, Student's t-test.
The topological analysis does not reveal abasic sites and we therefore treated DNA from heart Sod2 knockout mice with Endonuclease IV, which converts abasic sites to single-stranded breaks. This procedure allows a comparison of the amount of abasic sites by analysis of the proportional change of the closed circle mtDNA in treated and untreated DNA. Heart Sod2 knockout mice did not show an increase in abasic sites with this procedure (Supplementary Figure S7). Taken together, the topological analyses show that mtDNA is either quickly repaired or that the packing mtDNA into nucleoids sufficiently shields mtDNA from extensive damage. However, it needs to be pointed out that the topological analysis detects only considerable changes in mtDNA integrity because even a gentle DNA extraction procedure induces additional nicks to DNA which can mask small changes in integrity. Therefore, we cannot rule out that small changes in mtDNA integrity occurs in the heart Sod2 knockout mice that are below the detection limit of the method.
We also addressed the possibility that damaged mtDNA is rapidly turned over and thus not visible in the analysis. Experiments with mitochondrially targeted restriction enzymes have demonstrated that linear mtDNA fragments are digested rapidly leading to decrease in mtDNA copy number (122,123). To evaluate if rapid loss of the linear form of mtDNA is occurring in heart Sod2 knockout mice, mtDNA copy number was analyzed in heart tissue with qPCR, and a slight increase in copy number was detected (Figure 9C). The mtDNA copy number was also evaluated with Southern blot analyses to verify the absence of deletions (Supplementary Figure S2C).
Other types of damage such as bulky adducts or other modifications that hinder mtDNA replication or transcription (96,118,124) may still prevail and will not be visible on a topology gel because it only reveals nicks and strand breaks. To evaluate mtDNA replication in heart Sod2 knockout mice, in organello replication assays were carried out. In these assays, freshly purified mitochondria were incubated with α-32P-labeled dATP and its incorporation into mtDNA is used as a read out. This de novo replication assay reflects the functionality of the mtDNA replication machinery in intact organelles. A large fraction of mtDNA replication initiating from OH is prematurely terminated in the control region. The resulting abortive replication product is called 7S DNA and it anneals in the control region to form a triple-stranded displacement loop (D-loop) structure (125). To quantify 7S DNA, half of each sample was heated prior to loading on an agarose gel. In heart Sod2 knockout mice, the de novo replication was severely reduced in both of total mtDNA and 7S DNA synthesis (Figure 10A and B). The de novo replication was analyzed also from Sod2 knockout mice that are homozygous for the Ogg1 dMTS but no additional replication effect was induced by the absence of DNA repair (Supplementary Figure S8). Interestingly, the decrease in de novo replication did not lead to an mtDNA copy number decrease in heart Sod2 knockout mice (Figure 9C).
Figure 10.
De novo replication is affected in heart Sod2 knockout mice. (A) Representative experiment of in organello replication assay in heart Sod2 loxP x Ckmm cre mice. The mtDNA was radioactively labeled in isolated mitochondria, purified and half of it was boiled to release newly synthesized 7S DNA. Samples were separated on an agarose gel and transferred to a membrane. Small aliquot representing the input was analyzed with Coomassie staining after the labeling. (B) Quantification of de novo replication, the relative incorporation of radioactivity into mtDNA and newly synthesized 7S DNA in Sod2 loxP x Ckmm cre mice. The incorporation was normalized to steady-state level of mtDNA that was probed from the same membrane after the de novo signal could not be detected anymore. White circles indicate samples from controls (pp, n = 9, 10–11 week old) and gray circles indicate samples from Sod2 loxP x Ckmm cre mice (pp, cre, n = 9, 9–10 week old). (C) Representative experiment of in organello transcription assay from Sod2 loxP x Ckmm cre mice. The mtRNA was radioactively labeled in isolated mitochondria and half of the sample was purified (pulse). Another half was incubated with non-radioactive UTP for 2 h (chase) and purified, and then samples were analyzed with northern blotting. Small aliquot representing the input was analyzed with Coomassie staining after the labeling. (D) Quantification of de novo transcription, the relative incorporation of radioactivity into mtRNA in Sod2 loxP x Ckmm cre mice. The incorporation was normalized to steady-state level of CytB that was probed from the same membrane after the de novo signal could not be detected anymore. See Supplementary Figure S7, which shows that CytB levels do not change in Sod2 loxP x Ckmm cre mice. White circles indicate samples from controls (pp, n = 12, 10–11 week old) and gray circles indicate samples from Sod2 loxP x Ckmm cre mice (pp, cre, n = 12, 9–10 week old). Horizontal lines represent means, error bars represent SD, ****P< 0.0001, Student's t-test.
To decipher whether mtDNA damage or dysfunctional replication machinery were hindering mtDNA replication in heart Sod2 knockout mice, we evaluated the steady-state levels of replication machinery proteins and also performed in organello transcription assays. In organello transcription assays are analogous to replication assays, but UTP is used instead of dATP to assess incorporation into RNA as a measure of de novo transcription. In heart Sod2 knockout mice, de novo transcription was normalized to loading by using the steady-state levels of the CytB mtRNA species (Supplementary Figure S9). Surprisingly, neither de novo transcription (pulse) nor RNA turnover (chase) was altered in heart Sod2 knockout mice (Figure 10C and D), showing that either the decrease in replication was not driven by mtDNA damage or, alternatively, that the replication and transcription machineries do not have the same sensitivity to mtDNA damage. To evaluate the steady-state levels of replication and transcription machinery components, western blot analyses were carried out by using purified mitochondria from heart tissue of Sod2 knockout mice. The analysis revealed that the steady-state levels of DNA polymerase γ were decreased in heart Sod2 knockout mice, whereas the levels of mitochondrial RNA polymerase (POLRMT) were maintained (Figure 11). These findings may explain the observed discrepancy where de novo replication is decreased and de novo transcription is increased. All other proteins involved in mtDNA replication or de novo replication transcription were not strongly affected (TFB2M, TFAM, mtSSB). In line with our findings, the in vitro activity of DNA polymerase γ has been shown to be vulnerable to oxidative stress (126).
Figure 11.
POLγ steady-state levels are decreased in heart Sod2 knockout mice while POLRMT levels are increased. Western blot analysis of proteins involved in replication and transcription from purified heart mitochondria from control (pp) and Sod2 loxP x Ckmm cre (pp, cre) mice (9–10 week old). ATP5A and Coomassie-stained membrane were used as loading controls.
DISCUSSION
Mutations of mtDNA have been long postulated to contribute to ageing. Accordingly, focal OXPHOS dysfunction is more prevalent with age in many tissues and individual cells in these focal aberrations contain mtDNA deletions and/or point mutations (127). In the current study we have focused on elucidating the origin of these point mutations which are currently thought to originate from two, not mutually exclusive, sources: replicative errors or oxidative damage to mtDNA.
In the nuclear genome, it has been estimated that each newborn human carries 50–100 new mutations (128). With age, additional mutations arise and at 60 years of age proliferative cells are estimated to carry 4000 to 40 000 mutations (128). This implies that at around 60 years of age, it is likely that almost every genomic site has acquired a mutation in at least one cell (128). In the cancers lacking a strong environmental component to their development, a clear correlation has been reported between lifetime risk of cancer and lifetime number of stem cell divisions (129). For this subset of cancers, these correlations are not dependent on geographical regions, suggesting that a major driving force behind many cancers are the replication errors rather than exogenous DNA damage sources (129). Although DNA replication is highly accurate process (128,130), mutations slowly accumulate in the nucleus following multiple cell divisions.
Correspondingly, mtDNA replication shows high fidelity (14,131), but point mutations have been reported to be more prevalent with age (6,7,13). The mtDNA goes through an extensive number of replication cycles all the way from when the germ line is set aside during development to old age. In primordial germ cells the mtDNA copy number drops to around 200 copies (132) and these copies will be subsequently amplified to account for all copies of mtDNA present in an adult individual. On the one hand, most mtDNA mutations that are detected with increasing age are transitions and demonstrate a clear strand-specific bias (7,10,13,133–135), which is consistent with the hypothesis that they are likely generated by replication errors (14) or spontaneous deaminations (53), and not by random oxidative damage. On the other hand, oxidative damage has been reported to be more prevalent in old individuals (136,137) and the levels of reactive oxygen species have been reported to increase with age in flies (138), whereas no such increase of H2O2 levels was reported in ageing mice (139).
Many studies that measure oxidative damage of mtDNA interpret an observed increase in damage as a direct causal link that will lead to an increase in the mtDNA mutation load. Interestingly, we detected no increase in the mtDNA mutation load in mitochondrial BER deficient mice that showed drastically increased ROS levels. Consistent with our results, mtDNA has been shown to be less sensitive to exogenous mutagens, such as BPDE (benzo(a)pyrene-7,8-diol- 9,10-epoxide), than nuclear DNA, as concentrations that induce mutations in nuclear DNA are not mutagenic to mtDNA in cells (140). Similarly, Valente and colleagues reported that exposing mice to benzo[a]pyrene (B[a]P) induced adducts in both nuclear and mtDNA. However, these adducts in mtDNA were not converted to mutations and the level of point mutations were only increased in nuclear DNA (97). In cells, only cytotoxic concentrations of N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) produced a reproducible set of point mutations in mtDNA in MNNG-resistant MT1 cells (140).
The nucleus harbors 14 DNA polymerases in total, and a subset of these are translesion polymerases, which allow DNA replication to by-pass various DNA lesions, often at the expense of fidelity (141). In contrast to the nucleus, mitochondria have only one replicative DNA polymerase, which is involved in both repairing and replicating the mtDNA. The multicopy nature of mtDNA might decrease the pressure to bypass various DNA lesions, thus arguing that increased replication termination and/or mtDNA turnover are important reasons why DNA lesions do not commonly lead to mutations in mtDNA.
The mitochondrial theory of ageing suggests that increased oxidative stress with old age induces damage to mtDNA that is readily converted to mutations leading to OXPHOS dysfunction and increase in ROS production in a vicious cycle manner (17). Previously, experiments with the mtDNA mutator mouse expressing proofreading-deficient DNA polymerase gamma have demonstrated that mtDNA mutations do not lead to an increase in superoxide levels or protein carbonylation in mitochondria (142). At a young age, no increase in H2O2 was reported, whereas H2O2 levels increased in heart and kidney, but not in liver and skeletal muscle, as mtDNA mutator mice were at the end stage (139). Additionally, cells that were exposed to H2O2 or rotenone showed mtDNA damage, but no increase in the mtDNA mutation load was detected (21). In the current study, absence of SOD2 induces a profound increase in oxidative stress as exemplified by the loss of aconitase activity. The increased ROS levels cause a plethora of changes in mitochondrial function, however, with the methods used in this study, we did not observe an increase in mtDNA damage or mtDNA mutation load, suggesting that ROS is not a major player in mtDNA mutagenesis. We cannot exclude the possibility that different types of oxidative stress are present during ageing, which might induce demonstrable mtDNA damage and lead to increase in mtDNA mutation load. However, this is unlikely when we consider that the mutational signature from aged tissues is not consistent with ROS driven mutations (7,13). Additionally, de novo mutations occurring at old age would not have enough time to clonally expand to high enough levels needed to impair OXPHOS and are therefore probably not relevant for ageing. Therefore, we conclude that elevated ROS levels do not increase mtDNA mutation load to levels that would be relevant for ageing even in the absence of mitochondrial BER.
In summary, we did not detect any changes in the somatic mtDNA mutation load or in the amount of maternally transmitted mtDNA mutation load in BER-deficient mice. Furthermore, also mice with drastically increased oxidative stress combined with deficient BER showed no increase in the mtDNA or mtRNA mutation load. In support of our findings, sequencing studies of mtDNA in old humans (7) and flies (13) have failed to identify mutation signatures consistent with oxidative damage. Ageing in mammals is typically associated with deficient OXPHOS in a subset of cells in many different tissues (127) but also in these cells the clonally expanded mutations do not have a signature consistent with being induced by oxidative damage (6,10,135). In the light of the experimental results presented here and correlative data from sequencing studies it is now time to re-evaluate the popular hypothesis linking mtDNA point mutations to oxidative stress and instead focus on mtDNA replication errors as a source of somatic mtDNA mutations.
DATA AVAILABILITY
DNA and RNA-seq data have been deposited in the ArrayExpress database (143) at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6532, E-MTAB-6533 and E-MTAB-6534. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (144) partner repository with the dataset identifier PXD008998.
Supplementary Material
ACKNOWLEDGEMENTS
We thank A. Taha, L. Schmitz and N. Hochhard for technical assistance. Additionally, we wish to thank Veronika Georgieva for engineering OGG1-FLAG constructs. Label-free quantitative proteomics was performed at the Proteomics Core Facility, Max Planck Institute for Biology of Ageing from where we would like to specially thank Dr X. Li. Immunocytochemistry, H&E staining of paraffin embedded tissues, and microscopy were performed at the FACS & Imaging Core Facility, Max Planck Institute for Biology of Ageing. DNA and RNA library construction and Illumina sequencing were performed at the Max Plank-Genome-centre Cologne.
Authors Contributions: Conceived and designed the experiments: J.H.K.K., J.B.S. and N.-G.L. Performed the experiments: J.H.K.K, N.A.B., A.M. and A.J. Analyzed the data: J.H.K.K., A.M., M.A.I., N.A.B. and A.J. Wrote the manuscript: J.H.K.K, T.E.S.K. and N.-G.L.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Society [to N.-G.L.]; Swedish Research Council [2015-00418 to N.-G.L.]; Knut and Alice Wallenberg foundation [to N.-G.L.]; Emil Aaltonen foundation [to M.A.I.]. Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Larsson N.G., Clayton D.A.. Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet. 1995; 29:151–178. [DOI] [PubMed] [Google Scholar]
- 2. Lightowlers R.N., Taylor R.W., Turnbull D.M.. Mutations causing mitochondrial disease: What is new and what challenges remain. Science. 2015; 349:1494–1499. [DOI] [PubMed] [Google Scholar]
- 3. Brierley E.J., Johnson M.A., Lightowlers R.N., James O.F., Turnbull D.M.. Role of mitochondrial DNA mutations in human aging: implications for the central nervous system and muscle. Ann. Neurol. 1998; 43:217–223. [DOI] [PubMed] [Google Scholar]
- 4. Bua E., Johnson J., Herbst A., Delong B., McKenzie D., Salamat S., Aiken J.M.. Mitochondrial DNA-deletion mutations accumulate intracellularly to detrimental levels in aged human skeletal muscle fibers. Am. J. Hum. Genet. 2006; 79:469–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cortopassi G.A., Arnheim N.. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990; 18:6927–6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Greaves L.C., Nooteboom M., Elson J.L., Tuppen H.A.L., Taylor G.A., Commane D.M., Arasaradnam R.P., Khrapko K., Taylor R.W., Kirkwood T.B.L. et al. . Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet. 2014; 10:e1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kennedy S.R., Salk J.J., Schmitt M.W., Loeb L.A.. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013; 9:e1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Müller-Höcker J. Cytochrome-c-oxidase deficient cardiomyocytes in the human heart–an age-related phenomenon. A histochemical ultracytochemical study. Am. J. Pathol. 1989; 134:1167–1173. [PMC free article] [PubMed] [Google Scholar]
- 9. Müller-Höcker J. Cytochrome c oxidase deficient fibres in the limb muscle and diaphragm of man without muscular disease: an age-related alteration. J. Neurol. Sci. 1990; 100:14–21. [DOI] [PubMed] [Google Scholar]
- 10. Taylor R.W., Barron M.J., Borthwick G.M., Gospel A., Chinnery P.F., Samuels D.C., Taylor G.A., Plusa S.M., Needham S.J., Greaves L.C. et al. . Mitochondrial DNA mutations in human colonic crypt stem cells. J. Clin. Invest. 2003; 112:1351–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elson J.L., Samuels D.C., Turnbull D.M., Chinnery P.F.. Random intracellular drift explains the clonal expansion of mitochondrial DNA mutations with age. Am. J. Hum. Genet. 2001; 68:802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Baines H.L., Stewart J.B., Stamp C., Zupanic A., Kirkwood T.B.L., Larsson N.-G., Turnbull D.M., Greaves L.C.. Similar patterns of clonally expanded somatic mtDNA mutations in the colon of heterozygous mtDNA mutator mice and ageing humans. Mech. Ageing Dev. 2014; 139:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Itsara L.S., Kennedy S.R., Fox E.J., Yu S., Hewitt J.J., Sanchez-Contreras M., Cardozo-Pelaez F., Pallanck L.J.. Oxidative stress is not a major contributor to somatic mitochondrial DNA mutations. PLoS Genet. 2014; 10:e1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zheng W., Khrapko K., Coller H.A., Thilly W.G., Copeland W.C.. Origins of human mitochondrial point mutations as DNA polymerase γ-mediated errors. Mutat. Res. 2006; 599:11–20. [DOI] [PubMed] [Google Scholar]
- 15. Harman D. Aging: a theory based on free radical and radiation chemistry. J. Gerontol. 1956; 11:298–300. [DOI] [PubMed] [Google Scholar]
- 16. Harman D. The biologic clock: the mitochondria. J Am Geriatr Soc. 1972; 20:145–147. [DOI] [PubMed] [Google Scholar]
- 17. Alexeyev M.F. Is there more to aging than mitochondrial DNA and reactive oxygen species. FEBS J. 2009; 276:5768–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boveris A., Oshino N., Chance B.. The cellular production of hydrogen peroxide. Biochem. J. 1972; 128:617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hansford R.G., Hogue B.A., Mildaziene V.. Dependence of H2O2 formation by rat heart mitochondria on substrate availability and donor age. J. Bioenerg. Biomembr. 1997; 29:89–95. [DOI] [PubMed] [Google Scholar]
- 20. Yakes F.M., VanHouten B.. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:514–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shokolenko I., Venediktova N., Bochkareva A., Wilson G.L., Alexeyev M.F.. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009; 37:2539–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nass M.M., Nass S.. Intramitochondrial fibers with DNA characteristics. I. Fixation and electron staining reactions. J. Cell Biol. 1963; 19:593–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nass M.M., Nass S., Afzelius B.A.. The general occurence of mitochondrial DNA. Exp. Cell Res. 1965; 37:516–539. [DOI] [PubMed] [Google Scholar]
- 24. Kaufman B.A., Durisic N., Mativetsky J.M., Costantino S., Hancock M.A., Grutter P., Shoubridge E.A.. The mitochondrial transcription factor TFAM coordinates the assembly of multiple DNA molecules into nucleoid-like structures. Mol. Biol. Cell. 2007; 18:3225–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kukat C., Davies K.M., Wurm C.A., Spåhr H., Bonekamp N.A., Kühl I., Joos F., Polosa P.L., Park C.B., Posse V. et al. . Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:11288–11293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brown T.A., Tkachuk A.N., Shtengel G., Kopek B.G., Bogenhagen D.F., Hess H.F., Clayton D.A.. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell. Biol. 2011; 31:4994–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kukat C., Wurm C.A., Spåhr H., Falkenberg M., Larsson N.-G., Jakobs S.. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:13534–13539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pettepher C.C., Ledoux S.P., Bohr V.A., Wilson G.L.. Repair of alkali-labile sites within the mitochondrial DNA of RINr 38 cells after exposure to the nitrosourea streptozotocin. J. Biol. Chem. 1991; 266:3113–3117. [PubMed] [Google Scholar]
- 29. Ledoux S.P., Wilson G.L., Beecham E.J., Stevnsner T., Wassermann K., Bohr V.A.. Repair of mitochondrial-DNA after various types of DNA damage in Chinese-hamster ovary cells. Carcinogenesis. 1992; 13:1967–1973. [DOI] [PubMed] [Google Scholar]
- 30. Driggers W.J., Ledoux S.P., Wilson G.L.. Repair of oxidative damage within the mitochondrial DNA of RINr 38 cells. J. Biol. Chem. 1993; 268:22042–22045. [PubMed] [Google Scholar]
- 31. Szczesny B., Tann A.W., Longley M.J., Copeland W.C., Mitra S.. Long patch base excision repair in mammalian mitochondrial genomes. J. Biol. Chem. 2008; 283:26349–26356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clayton D.A., Doda J.N., Friedberg E.C.. Absence of a pyrimidine dimer repair mechanism for mitochondrial DNA in mouse and human cells. Basic Life Sci. 1975; 5B:589–591. [DOI] [PubMed] [Google Scholar]
- 33. Halliwell B., Aruoma O.I.. DNA damage by oxygen-derived species. Its mechanism and measurement in mammalian systems. FEBS Lett. 1991; 281:9–19. [DOI] [PubMed] [Google Scholar]
- 34. Flint D.H., Tuminello J.F., Emptage M.H.. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 1993; 268:22369–22376. [PubMed] [Google Scholar]
- 35. Halliwell B., Gutteridge J.M.. Biologically relevant metal ion-dependent hydroxyl radical generation. An update. FEBS Lett. 1992; 307:108–112. [DOI] [PubMed] [Google Scholar]
- 36. Cadet J., Delatour T., Douki T., Gasparutto D., Pouget J.P., Ravanat J.L., Sauvaigo S.. Hydroxyl radicals and DNA base damage. Mutat. Res. 1999; 424:9–21. [DOI] [PubMed] [Google Scholar]
- 37. Shibutani S., Takeshita M., Grollman A.P.. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991; 349:431–434. [DOI] [PubMed] [Google Scholar]
- 38. Kang D., Nishida J., Iyama A., Nakabeppu Y., Furuichi M., Fujiwara T., Sekiguchi M., Takeshige K.. Intracellular localization of 8-oxo-dGTPase in human cells, with special reference to the role of the enzyme in mitochondria. J. Biol. Chem. 1995; 270:14659–14665. [DOI] [PubMed] [Google Scholar]
- 39. Sakumi K., Furuichi M., Tsuzuki T., Kakuma T., Kawabata S., Maki H., Sekiguchi M.. Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 1993; 268:23524–23530. [PubMed] [Google Scholar]
- 40. Hu J., de Souza-Pinto N.C., Haraguchi K., Hogue B.A., Jaruga P., Greenberg M.M., Dizdaroglu M., Bohr V.A.. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J. Biol. Chem. 2005; 280:40544–40551. [DOI] [PubMed] [Google Scholar]
- 41. Rosenquist T.A., Zharkov D.O., Grollman A.P.. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:7429–7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Slupska M.M., Luther W.M., Chiang J.H., Yang H., Miller J.H.. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J. Bacteriol. 1999; 181:6210–6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guan Y., Manuel R.C., Arvai A.S., Parikh S.S., Mol C.D., Miller J.H., Lloyd S., Tainer J.A.. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol. 1998; 5:1058–1064. [DOI] [PubMed] [Google Scholar]
- 44. Trapp C., McCullough A.K., Epe B.. The basal levels of 8-oxoG and other oxidative modifications in intact mitochondrial DNA are low even in repair-deficient (Ogg1(-/-)/Csb(-/-)) mice. Mutat. Res. 2007; 625:155–163. [DOI] [PubMed] [Google Scholar]
- 45. de Souza-Pinto N.C., Eide L., Hogue B.A., Thybo T., Stevnsner T., Seeberg E., Klungland A., Bohr V.A.. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001; 61:5378–5381. [PubMed] [Google Scholar]
- 46. Vermulst M., Bielas J.H., Kujoth G.C., Ladiges W.C., Rabinovitch P.S., Prolla T.A., Loeb L.A.. Mitochondrial point mutations do not limit the natural lifespan of mice. Nature Genet. 2007; 39:540–543. [DOI] [PubMed] [Google Scholar]
- 47. Ameur A., Stewart J.B., Freyer C., Hagström E., Ingman M., Larsson N.-G., Gyllensten U.. Ultra-deep sequencing of mouse mitochondrial DNA: mutational patterns and their origins. PLoS Genet. 2011; 7:e1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khrapko K., Coller H.A., André P.C., Li X.C., Hanekamp J.S., Thilly W.G.. Mitochondrial mutational spectra in human cells and tissues. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:13798–13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Polyak K., Li Y., Zhu H., Lengauer C., Willson J.K., Markowitz S.D., Trush M.A., Kinzler K.W., Vogelstein B.. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat. Genet. 1998; 20:291–293. [DOI] [PubMed] [Google Scholar]
- 50. Ericson N.G., Kulawiec M., Vermulst M., Sheahan K., O'Sullivan J., Salk J.J., Bielas J.H.. Decreased mitochondrial DNA mutagenesis in human colorectal cancer. PLoS Genet. 2012; 8:e1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ju Y.S., Alexandrov L.B., Gerstung M., Martincorena I., Nik-Zainal S., Ramakrishna M., Davies H.R., Papaemmanuil E., Gundem G., Shlien A. et al. . Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. Elife. 2014; 3:415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stewart J.B., Alaei-Mahabadi B., Sabarinathan R., Samuelsson T., Gorodkin J., Gustafsson C.M., Larsson E.. Simultaneous DNA and RNA mapping of somatic mitochondrial mutations across diverse human cancers. PLoS Genet. 2015; 11:e1005333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. [DOI] [PubMed] [Google Scholar]
- 54. Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J.N., Rovio A.T., Bruder C.E., Bohlooly-Y M., Gidlöf S., Oldfors A., Wibom R. et al. . Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004; 429:417–423. [DOI] [PubMed] [Google Scholar]
- 55. Kujoth G.C., Hiona A., Pugh T.D., Someya S., Panzer K., Wohlgemuth S.E., Hofer T., Seo A.Y., Sullivan R., Jobling W.A. et al. . Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005; 309:481–484. [DOI] [PubMed] [Google Scholar]
- 56. Stewart J.B., Freyer C., Elson J.L., Wredenberg A., Cansu Z., Trifunovic A., Larsson N.-G.. Strong purifying selection in transmission of mammalian mitochondrial DNA. PLoS Biol. 2008; 6:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Claros M.G., Vincens P.. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996; 241:779–786. [DOI] [PubMed] [Google Scholar]
- 58. Emanuelsson O., Nielsen H., Brunak S., Heijne von, G.. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000; 300:1005–1016. [DOI] [PubMed] [Google Scholar]
- 59. Nielsen H., Engelbrecht J., Brunak S., Heijne von, G.. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997; 10:1–6. [DOI] [PubMed] [Google Scholar]
- 60. Strassburger M., Bloch W., Sulyok S., Schüller J., Keist A.F., Schmidt A., Wenk J., Peters T., Wlaschek M., Lenart J. et al. . Heterozygous deficiency of manganese superoxide dismutase results in severe lipid peroxidation and spontaneous apoptosis in murine myocardium in vivo. Free Radic. Biol. Med. 2005; 38:1458–1470. [DOI] [PubMed] [Google Scholar]
- 61. Wang J., Wilhelmsson H., Graff C., Li H., Oldfors A., Rustin P., Brüning J.C., Kahn C.R., Clayton D.A., Barsh G.S. et al. . Dilated cardiomyopathy and atrioventricular conduction blocks induced by heart-specific inactivation of mitochondrial DNA gene expression. Nat. Genet. 1999; 21:133–137. [DOI] [PubMed] [Google Scholar]
- 62. Page M.M., Stuart J.A.. In vitro measurement of DNA base excision repair in isolated mitochondria. Methods Mol. Biol. 2009; 554:213–231. [DOI] [PubMed] [Google Scholar]
- 63. Ichinoe A., Behmanesh M., Tominaga Y., Ushijima Y., Hirano S., Sakai Y., Tsuchimoto D., Sakumi K., Wake N., Nakabeppu Y.. Identification and characterization of two forms of mouse MUTYH proteins encoded by alternatively spliced transcripts. Nucleic Acids Res. 2004; 32:477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wanrooij S., Miralles Fusté J., Stewart J.B., Wanrooij P.H., Samuelsson T., Larsson N.-G., Gustafsson C.M., Falkenberg M.. In vivo mutagenesis reveals that OriL is essential for mitochondrial DNA replication. EMBO Rep. 2012; 13:1130–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ross J.M., Stewart J.B., Hagström E., Brené S., Mourier A., Coppotelli G., Freyer C., Lagouge M., Hoffer B.J., Olson L. et al. . Germline mitochondrial DNA mutations aggravate ageing and can impair brain development. Nature. 2013; 501:412–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stewart J.B., Freyer C., Elson J.L., Larsson N.-G.. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat. Rev. Genet. 2008; 9:657–662. [DOI] [PubMed] [Google Scholar]
- 67. Dodt M., Roehr J.T., Ahmed R., Dieterich C.. FLEXBAR-flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel). 2012; 1:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li H., Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wilm A., Aw P.P.K., Bertrand D., Yeo G.H.T., Ong S.H., Wong C.H., Khor C.C., Petric R., Hibberd M.L., Nagarajan N.. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012; 40:11189–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cingolani P., Patel V.M., Coon M., Nguyen T., Land S.J., Ruden D.M., Lu X.. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012; 3:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Davies S.M.K., Lopez Sanchez M.I.G., Narsai R., Shearwood A.-M.J., Razif M.F.M., Small I.D., Whelan J., Rackham O., Filipovska A.. MRPS27 is a pentatricopeptide repeat domain protein required for the translation of mitochondrially encoded proteins. FEBS Lett. 2012; 586:3555–3561. [DOI] [PubMed] [Google Scholar]
- 76. Wieckowski M.R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P.. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 2009; 4:1582–1590. [DOI] [PubMed] [Google Scholar]
- 77. Mourier A., Motori E., Brandt T., Lagouge M., Atanassov I., Galinier A., Rappl G., Brodesser S., Hultenby K., Dieterich C. et al. . Mitofusin 2 is required to maintain mitochondrial coenzyme Q levels. J. Cell Biol. 2015; 208:429–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kolesar J.E., Wang C.Y., Taguchi Y.V., Chou S.-H., Kaufman B.A.. Two-dimensional intact mitochondrial DNA agarose electrophoresis reveals the structural complexity of the mammalian mitochondrial genome. Nucleic Acids Res. 2013; 41:e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gensler S., Weber K., Schmitt W.E., Pérez-Martos A., Enríquez J.A., Montoya J., Wiesner R.J.. Mechanism of mammalian mitochondrial DNA replication: import of mitochondrial transcription factor A into isolated mitochondria stimulates 7S DNA synthesis. Nucleic Acids Res. 2001; 29:3657–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Enríquez J.A., Pérez-Martos A., López-Pérez M.J., Montoya J.. In organello RNA synthesis system from mammalian liver and brain. Methods Enzymol. 1996; 264:50–57. [DOI] [PubMed] [Google Scholar]
- 81. Rappsilber J., Ishihama Y., Mann M.. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003; 75:663–670. [DOI] [PubMed] [Google Scholar]
- 82. Cox J., Mann M.. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008; 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 83. Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., Mann M.. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011; 10:1794–1805. [DOI] [PubMed] [Google Scholar]
- 84. Cox J., Hein M.Y., Luber C.A., Paron I., Nagaraj N., Mann M.. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteomics. 2014; 13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Takao M., Zhang Q.M., Yonei S., Yasui A.. Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine: 8-oxoguanine DNA glycosylase. Nucleic Acids Res. 1999; 27:3638–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Takao M., Aburatani H., Kobayashi K., Yasui A.. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998; 26:2917–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xie Y.L., Yang H.J., Cunanan C., Okamoto K., Shibata D., Pan J., Barnes D.E., Lindahl T., McIlhatton M., Fishel R. et al. . Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-Ras oncogene in lung tumors. Cancer Res. 2004; 64:3096–3102. [DOI] [PubMed] [Google Scholar]
- 88. Ohtsubo T., Nishioka K., Imaiso Y., Iwai S., Shimokawa H., Oda H., Fujiwara T., Nakabeppu Y.. Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res. 2000; 28:1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stuart J.A., Bourque B.M., de Souza-Pinto N.C., Bohr V.A.. No evidence of mitochondrial respiratory dysfunction in OGG1-null mice deficient in removal of 8-oxodeoxyguanine from mitochondrial DNA. Free Radic. Biol. Med. 2005; 38:737–745. [DOI] [PubMed] [Google Scholar]
- 90. Halsne R., Esbensen Y., Wang W., Scheffler K., Suganthan R., Bjørås M., Eide L.. Lack of the DNA glycosylases MYH and OGG1 in the cancer prone double mutant mouse does not increase mitochondrial DNA mutagenesis. DNA Repair. 2012; 11:278–285. [DOI] [PubMed] [Google Scholar]
- 91. Larsson N.G., Wang J., Wilhelmsson H., Oldfors A., Rustin P., Lewandoski M., Barsh G.S., Clayton D.A.. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 1998; 18:231–236. [DOI] [PubMed] [Google Scholar]
- 92. Milenkovic D., Matic S., Kühl I., Ruzzenente B., Freyer C., Jemt E., Park C.B., Falkenberg M., Larsson N.-G.. TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum. Mol. Gen. 2013; 22:1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kühl I., Miranda M., Posse V., Milenkovic D., Mourier A., Siira S.J., Bonekamp N.A., Neumann U., Filipovska A., Polosa P.L. et al. . POLRMT regulates the switch between replication primer formation and gene expression of mammalian mtDNA. Sci. Adv. 2016; 2:e1600963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Graziewicz M.A., Bienstock R.J., Copeland W.C.. The DNA polymerase gamma Y955C disease variant associated with PEO and parkinsonism mediates the incorporation and translesion synthesis opposite 7,8-dihydro-8-oxo-2′-deoxyguanosine. Hum. Mol. Gen. 2007; 16:2729–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hanes J.W., Thal D.M., Johnson K.A.. Incorporation and replication of 8-oxo-deoxyguanosine by the human mitochondrial DNA polymerase. J. Biol. Chem. 2006; 281:36241–36248. [DOI] [PubMed] [Google Scholar]
- 96. Stojkovič G., Makarova A.V., Wanrooij P.H., Forslund J., Burgers P.M., Wanrooij S.. Oxidative DNA damage stalls the human mitochondrial replisome. Sci. Rep. 2016; 6:28942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Valente W.J., Ericson N.G., Long A.S., White P.A., Marchetti F., Bielas J.H.. Mitochondrial DNA exhibits resistance to induced point and deletion mutations. Nucleic Acids Res. 2016; 44:8513–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D.E.. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Minowa O., Arai T., Hirano M., Monden Y., Nakai S., Fukuda M., Itoh M., Takano H., Hippou Y., Aburatani H. et al. . Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:4156–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kirkwood T.B. Evolution of ageing. Nature. 1977; 270:301–304. [DOI] [PubMed] [Google Scholar]
- 101. Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A.. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Chen L., Liu P., Evans T.C., Ettwiller L.M.. DNA damage is a pervasive cause of sequencing errors, directly confounding variant identification. Science. 2017; 355:752–756. [DOI] [PubMed] [Google Scholar]
- 103. Costello M., Pugh T.J., Fennell T.J., Stewart C., Lichtenstein L., Meldrim J.C., Fostel J.L., Friedrich D.C., Perrin D., Dionne D. et al. . Discovery and characterization of artifactual mutations in deep coverage targeted capture sequencing data due to oxidative DNA damage during sample preparation. Nucleic Acids Res. 2013; 41:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Murphy M.P., Holmgren A., Larsson N.-G., Halliwell B., Chang C.J., Kalyanaraman B., Rhee S.G., Thornalley P.J., Partridge L., Gems D. et al. . Unraveling the biological roles of reactive oxygen species. Cell Metab. 2011; 13:361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009; 417:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Li Y., Huang T.T., Carlson E.J., Melov S., Ursell P.C., Olson J.L., Noble L.J., Yoshimura M.P., Berger C., Chan P.H. et al. . Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 1995; 11:376–381. [DOI] [PubMed] [Google Scholar]
- 107. Lustgarten M.S., Jang Y.C., Liu Y., Muller F.L., Qi W., Steinhelper M., Brooks S.V., Larkin L., Shimizu T., Shirasawa T. et al. . Conditional knockout of Mn-SOD targeted to type IIB skeletal muscle fibers increases oxidative stress and is sufficient to alter aerobic exercise capacity. Am. J. Physiol., Cell Physiol. 2009; 297:C1520–C1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Nojiri H., Shimizu T., Funakoshi M., Yamaguchi O., Zhou H., Kawakami S., Ohta Y., Sami M., Tachibana T., Ishikawa H. et al. . Oxidative stress causes heart failure with impaired mitochondrial respiration. J. Biol. Chem. 2006; 281:33789–33801. [DOI] [PubMed] [Google Scholar]
- 109. Williams M.D., Van Remmen H., Conrad C.C., Huang T.T., Epstein C.J., Richardson A.. Increased oxidative damage is correlated to altered mitochondrial function in heterozygous manganese superoxide dismutase knockout mice. J. Biol. Chem. 1998; 273:28510–28515. [DOI] [PubMed] [Google Scholar]
- 110. Van Remmen H., Ikeno Y., Hamilton M., Pahlavani M., Wolf N., Thorpe S.R., Alderson N.L., Baynes J.W., Epstein C.J., Huang T.-T. et al. . Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol. Genomics. 2003; 16:29–37. [DOI] [PubMed] [Google Scholar]
- 111. Imlay J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006; 59:1073–1082. [DOI] [PubMed] [Google Scholar]
- 112. Gardner P.R., Raineri I., Epstein L.B., White C.W.. Superoxide radical and iron modulate aconitase activity in mammalian cells. J. Biol. Chem. 1995; 270:13399–13405. [DOI] [PubMed] [Google Scholar]
- 113. Tarpey M.M., Wink D.A., Grisham M.B.. Methods for detection of reactive metabolites of oxygen and nitrogen: in vitro and in vivo considerations. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004; 286:R431–R444. [DOI] [PubMed] [Google Scholar]
- 114. Wirth C., Brandt U., Hunte C., Zickermann V.. Structure and function of mitochondrial complex I. Biochim. Biophys. Acta. 2016; 1857:902–914. [DOI] [PubMed] [Google Scholar]
- 115. Sun F., Huo X., Zhai Y., Wang A., Xu J., Su D., Bartlam M., Rao Z.. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell. 2005; 121:1043–1057. [DOI] [PubMed] [Google Scholar]
- 116. Gao X., Wen X., Esser L., Quinn B., Yu L., Yu C.-A., Xia D.. Structural basis for the quinone reduction in the bc1 complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc1 with bound substrate and inhibitors at the Qi site. Biochemistry. 2003; 42:9067–9080. [DOI] [PubMed] [Google Scholar]
- 117. Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S.. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996; 272:1136–1144. [DOI] [PubMed] [Google Scholar]
- 118. Nakanishi N., Fukuoh A., Kang D., Iwai S., Kuraoka I.. Effects of DNA lesions on the transcription reaction of mitochondrial RNA polymerase: implications for bypass RNA synthesis on oxidative DNA lesions. Mutagenesis. 2013; 28:117–123. [DOI] [PubMed] [Google Scholar]
- 119. Ellefson J.W., Gollihar J., Shroff R., Shivram H., Iyer V.R., Ellington A.D.. Synthetic evolutionary origin of a proofreading reverse transcriptase. Science. 2016; 352:1590–1593. [DOI] [PubMed] [Google Scholar]
- 120. Posse V., Shahzad S., Falkenberg M., Hallberg B.M., Gustafsson C.M.. TEFM is a potent stimulator of mitochondrial transcription elongation in vitro. Nucleic Acids Res. 2015; 43:2615–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Pogozelski W.K., Tullius T.D.. Oxidative strand scission of nucleic acids: routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev. 1998; 98:1089–1108. [DOI] [PubMed] [Google Scholar]
- 122. Srivastava S., Moraes C.T.. Double-strand breaks of mouse muscle mtDNA promote large deletions similar to multiple mtDNA deletions in humans. Hum. Mol. Gen. 2005; 14:893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Moretton A., Morel F., Macao B., Lachaume P., Ishak L., Lefebvre M., Garreau-Balandier I., Vernet P., Falkenberg M., Farge G.. Selective mitochondrial DNA degradation following double-strand breaks. PLoS ONE. 2017; 12:e0176795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kasiviswanathan R., Minko I.G., Lloyd R.S., Copeland W.C.. Translesion synthesis past acrolein-derived DNA adducts by human mitochondrial DNA polymerase γ. J. Biol. Chem. 2013; 288:14247–14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Gustafsson C.M., Falkenberg M., Larsson N.-G.. Maintenance and expression of mammalian mitochondrial DNA. Annu. Rev. Biochem. 2016; 85:133–160. [DOI] [PubMed] [Google Scholar]
- 126. Graziewicz M.A., Day B.J., Copeland W.C.. The mitochondrial DNA polymerase as a target of oxidative damage. Nucleic Acids Res. 2002; 30:2817–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Larsson N.-G. Somatic mitochondrial DNA mutations in mammalian aging. Annu. Rev. Biochem. 2010; 79:683–706. [DOI] [PubMed] [Google Scholar]
- 128. Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Tomasetti C., Li L., Vogelstein B.. Stem cell divisions, somatic mutations, cancer etiology, and cancer prevention. Science. 2017; 355:1330–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Keightley P.D. Rates and fitness consequences of new mutations in humans. Genetics. 2012; 190:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Lynch M. The lower bound to the evolution of mutation rates. Genome Biol. Evol. 2011; 3:1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Cree L.M., Samuels D.C., de Sousa Lopes S.C., Rajasimha H.K., Wonnapinij P., Mann J.R., Dahl H.-H.M., Chinnery P.F.. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat. Genet. 2008; 40:249–254. [DOI] [PubMed] [Google Scholar]
- 133. Williams S.L., Mash D.C., Züchner S., Moraes C.T.. Somatic mtDNA mutation spectra in the aging human putamen. PLoS Genet. 2013; 9:e1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Greaves L.C., Elson J.L., Nooteboom M., Grady J.P., Taylor G.A., Taylor R.W., Mathers J.C., Kirkwood T.B.L., Turnbull D.M.. Comparison of mitochondrial mutation spectra in ageing human colonic epithelium and disease: absence of evidence for purifying selection in somatic mitochondrial DNA point mutations. PLoS Genet. 2012; 8:e1003082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Greaves L.C., Barron M.J., Plusa S., Kirkwood T.B., Mathers J.C., Taylor R.W., Turnbull D.M.. Defects in multiple complexes of the respiratory chain are present in ageing human colonic crypts. Exp. Gerontol. 2010; 45:573–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Bokov A., Chaudhuri A., Richardson A.. The role of oxidative damage and stress in aging. Mech. Ageing Dev. 2004; 125:811–826. [DOI] [PubMed] [Google Scholar]
- 137. Stadtman E.R. Protein oxidation and aging. Free Radic. Res. 2006; 40:1250–1258. [DOI] [PubMed] [Google Scholar]
- 138. Cochemé H.M., Quin C., McQuaker S.J., Cabreiro F., Logan A., Prime T.A., Abakumova I., Patel J.V., Fearnley I.M., James A.M. et al. . Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011; 13:340–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Logan A., Shabalina I.G., Prime T.A., Rogatti S., Kalinovich A.V., Hartley R.C., Budd R.C., Cannon B., Murphy M.P.. In vivo levels of mitochondrial hydrogen peroxide increase with age in mtDNA mutator mice. Aging Cell. 2014; 13:765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Marcelino L.A., André P.C., Khrapko K., Coller H.A., Griffith J., Thilly W.G.. Chemically induced mutations in mitochondrial DNA of human cells: mutational spectrum of N-methyl-N'-nitro-N-nitrosoguanidine. Cancer Res. 1998; 58:2857–2862. [PubMed] [Google Scholar]
- 141. Pata J.D. Structural diversity of the Y-family DNA polymerases. Biochim. Biophys. Acta. 2010; 1804:1124–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Trifunovic A., Hansson A., Wredenberg A., Rovio A.T., Dufour E., Khvorostov I., Spelbrink J.N., Wibom R., Jacobs H.T., Larsson N.-G.. Somatic mtDNA mutations cause aging phenotypes without affecting reactive oxygen species production. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17993–17998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kolesnikov N., Hastings E., Keays M., Melnichuk O., Tang Y.A., Williams E., Dylag M., Kurbatova N., Brandizi M., Burdett T. et al. . ArrayExpress update–simplifying data submissions. Nucleic Acids Res. 2015; 43:D1113–D1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. . 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA and RNA-seq data have been deposited in the ArrayExpress database (143) at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6532, E-MTAB-6533 and E-MTAB-6534. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (144) partner repository with the dataset identifier PXD008998.