Abstract
The topological state of DNA in vivo is regulated by topoisomerases. Gyrase is a bacterial topoisomerase that introduces negative supercoils into DNA at the expense of ATP hydrolysis. According to the strand-passage mechanism, a double-strand of the DNA substrate is cleaved, and a second double-stranded segment is passed through the gap, converting a positive DNA node into a negative node. The correct orientation of these DNA segments for strand passage is achieved by wrapping of the DNA around gyrase, which involves the C-terminal domains (CTDs) of both GyrA subunits in the A2B2 heterotetramer. Gyrase lacking both CTDs cannot introduce negative supercoils into DNA. Here, we analyze the requirements for the two CTDs in individual steps in the supercoiling reaction. Gyrase that contains a single CTD binds, distorts, and cleaves DNA similarly to wildtype gyrase. It also shows wildtype-like DNA-dependent ATPase activity, and undergoes DNA-induced movement of the CTD as well as N-gate narrowing. Most importantly, the enzyme still introduces negative supercoils into DNA in an ATP-dependent reaction, with a velocity similar to wildtype gyrase, and decreases the linking number of the DNA in steps of two. One CTD is thus sufficient to support DNA supercoiling.
INTRODUCTION
DNA topoisomerases regulate the supercoiling state of DNA in all kingdoms of life. According to mechanistic features, topoisomerases are classified into two types. Type I topoisomerases cleave one strand of their double-stranded DNA substrate and change the linking number of DNA in steps of one. In contrast, type II topoisomerases cleave both strands of the DNA and change the linking number in increments of two (1). Based on their structural features, type II topoisomerases are subdivided into type IIA and type IIB enzymes. Type IIA enzymes form a two-fold symmetric structure with three protein/protein interfaces, termed the N-gate, DNA-gate, and C-gate. Despite their common architecture, members of the type IIA topoisomerase family catalyze different reactions: eukaryotic topoisomerase II (topo II) catalyzes ATP-dependent DNA relaxation (2), bacterial gyrase mediates the ATP-dependent introduction of negative supercoils (3), and topoisomerase IV (topo IV) mediates ATP-dependent DNA decatenation (4). All of these reactions are believed to occur via strand passage (reviewed in (5)), guided by coordinated opening and closing of the three gates: first, a double-stranded DNA-segment, the G-segment (for gate), binds at the DNA-gate, and both strands of the DNA are cleaved. ATP-dependent closing of the N-gate then traps a second double-stranded DNA, the T-segment (for transport). DNA-gate opening allows for passage of the T-segment through the gap in the G-segment (1,6). The G-segment is religated, and the T-segment leaves the enzyme through the C-gate. Topoisomerase VI, a type IIB topoisomerase, lacks the C-gate and catalyzes ATP-dependent DNA relaxation by a hitherto uncharacterized two-gate mechanism (7,8).
The bacterial type IIA topoisomerase gyrase is a heterotetramer of two GyrB and two GyrA subunits (9). GyrB provides the active sites for ATP binding and hydrolysis (10). GyrA contains the catalytic tyrosines for DNA cleavage (11), and the C-terminal domains (CTDs) that play a central role in DNA binding (12–14). DNA supercoiling starts by wrapping of DNA around gyrase in a process that involves the CTDs (15). As a result, a positive node is formed on the enzyme, with a left-handed crossing of the G-segment and the T-segment in the correct orientation for strand passage (1,6). Single-molecule FRET experiments have revealed a series of conformational changes of DNA and gyrase at the beginning of the supercoiling reaction (reviewed in refs. (16,17)). DNA binding at the DNA-gate leads to distortion of the G-segment (18). DNA emanating from the DNA-gate contacts the CTDs and becomes wrapped around their perimeter, which induces an upward movement of the CTDs (19), and a narrowing of the N-gate (20). ATP binding to the DNA/gyrase complex then causes dimerization of the GyrB subunits (10) and closing of the N-gate (20), which leads to trapping of the T-segment above the G-segment (21). Opening of the gyrase DNA- and C-gates, or strand passage itself, have not been visualized up to now, but crystal structures indicate that DNA- and C-gate opening is possible in principle (22).
The topoisomerase core, formed by the N-terminal domains (NTDs) of the GyrA subunits and by the GyrB subunits, is common to all members of the type IIA topoisomerase family. The GyrA CTDs, connected to the NTD by a flexible linker, are the key elements for the negative supercoiling activity of gyrase (19,20,23), and are highly conserved among gyrases. The CTD consists of six β-sheet elements (blades), arranged into a circular β-pinwheel of approx. 50 Å diameter (13). Blades 1 and 6 interact through a loop region with a conserved 7-amino acid sequence, the GyrA-box, which is a hallmark feature of gyrase (24,25). Deletion of the entire CTDs or of the GyrA-box uncouples DNA binding and ATPase activity, and abrogates DNA supercoiling (19,20,23,24,26). The GyrA-box appears to contact the DNA leaving the CTDs towards the upper cavity between the GyrB subunits, ‘at the end of the wrap’ (26,27). The C-terminal tail of the CTDs, following the six blades, has been implicated in species-specific fine-tuning of gyrase activities (28–30). The isolated CTDs bind DNA around their positively charged rim, introducing a sharp bend and a positive writhe into DNA (12,13). In the context of gyrase, the CTDs are involved in DNA wrapping, and help stabilize DNA bound to gyrase in a positive supercoil (15), which positions the T-segment captured by nucleotide-induced N-gate closure above the G-segment bound at the DNA-gate (21). The CTDs prepare gyrase for strand passage in various ways. They are mobile elements that change their position on binding of DNA to gyrase (19), and contribute to the distortion of the DNA bound at the DNA-gate (20). The CTDs functionally link the DNA-gate and the N-gate through the DNA wrapped around their perimeter; they communicate DNA binding at the DNA-gate to the N-gate by mediating DNA-induced N-gate narrowing (19,20,26,28), which requires the GyrA-box (26). They also mediate DNA-stimulation of ATP hydrolysis (20). In a reverse direction of communication, from the N-gate to the DNA-gate, nucleotide-induced N-gate closure alters the interaction of the CTDs with the DNA (31–35), and alleviates distortion of the DNA at the DNA-gate (18). Altogether, the CTDs are thus key elements for coordinating G-segment binding and distortion with N-gate narrowing, ATP hydrolysis, and T-segment capture, and for coordination of the nucleotide cycle with negative supercoiling of DNA.
While the current picture assumes that both CTDs are involved in stabilizing a positive supercoil in DNA by gyrase, only a single T-segment enters the upper cavity and is captured above the G-segment, hence only one of the two CTDs can perform an active role in each catalytic cycle. It has been suggested that the two CTDs take turns in T-segment presentation in a hand-over-hand mechanism, and may guide strand passage by moving from the top to the bottom of the enzyme alongside with the T-segment passing through the gates (36). However, there is no structural evidence of intermediates with only one of the two CTDs facing upwards to deliver a T-segment, or evidence for an active role of the CTDs in strand passage. Previous studies addressing the roles of the CTDs in DNA supercoiling used deletion variants of gyrase lacking both CTDs. Here, we investigate the role of the two CTDs in gyrase by deleting one or both of them. We find that gyrase with a single CTD is still capable of introducing negative supercoils into DNA in an ATP-dependent reaction, with only a small decrease in the rate of supercoiling. Independent of the number of CTDs present, gyrase introduces negative supercoils by decreasing the linking number in steps of two. DNA binding, distortion, and cleavage, DNA-induced N-gate narrowing, and the DNA-stimulated ATPase activity of gyrase with a single CTD are similar to wildtype gyrase. Thus, a single CTD is sufficient for DNA supercoiling by gyrase.
MATERIALS AND METHODS
Cloning, protein production and purification
Bacillus subtilis GyrA, GyrA variants, and GyrB were produced recombinantly in E. coli BL21(DE3) and purified as described (18,19,37). Heterodimeric GyrA containing one subunit with and one subunit lacking a CTD, GyrA·GyrAΔCTD, was generated by co-production of His6-tagged GyrAΔCTD and Strep-tagged GyrA, and purified by tandem-affinity chromatography as described previously (38) (Supplementary Figure S1).
Supercoiling reactions
Supercoiling reactions were performed in 50 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM MgCl2 at 37°C with 20 nM relaxed pUC18, indicated GyrB and GyrA concentrations, and 1.5 mM ATP as described (18,20). All components were pre-incubated for 3 min before starting the reaction by addition of ATP. To determine the change in linking number, a single pUC18 topoisomer was prepared as described (38). Supercoiling reactions were performed with 6 nM single topoisomer, 10 nM GyrA, 20 nM GyrB and 1.5 mM ATP at 37°C for 5 min. Relaxation reactions were performed with 10 nM negatively supercoiled pUC18 for 30 min. Reactions were stopped by adding 0.5% SDS and 25 mM EDTA, and the topoisomer distributions were analyzed on 1.3% agarose gels in TEP buffer (36 mM Tris, 36 mM NaH2PO4, 1 mM EDTA, pH 8.0; 2.6 V/cm, 3 h). To obtain rate constants of supercoiling, the fraction of supercoiled plasmid was quantified by densitometry. Errors (±) represent standard deviations of rate constants from three independent experiments.
Fluorescence equilibrium titrations
Dissociation constants of GyrA- and gyrase/DNA complexes were determined in fluorescence anisotropy titrations of 10 nM of a 60 bp DNA with GyrA in absence or presence of 8 μM GyrB in 50 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM MgCl2 at 37°C using a Fluoromax 3 fluorimeter (Jobin Yvon, Germany). The DNA was labeled with Alexa488 and Alexa546 and contained a preferred gyrase cleavage site in the center (18,39). Alexa546 fluorescence was excited at 555 nm (bandwidth 3 nm), and detected at 571 nm (bandwidth 6 nm). To determine Kd values, binding isotherms were analyzed using a one-to-one binding model as described previously (18). Experiments were performed in duplicate; errors (±) represent the error of the mean.
Topoisomerase I-relaxation of DNA/gyrase complexes
Trapping of positive supercoils by gyrase was assayed by relaxation of gyrase-bound DNA with topoisomerase I as described (34). DNA gyrase (100 nM GyrA/200 nM GyrB, 200 nM GyrA/400 nM GyrB, 300 nM GyrA/600 nM GyrB, 400 nM GyrA/800 nM GyrB, 600 nM GyrA/1200 nM GyrB, 800 nM GyrA/1600 nM GyrB) was incubated with 20 nM relaxed pUC18 (giving a gyrase:DNA ratio of 2.5 to 20) in 50 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM MgCl2 at 37°C. After 15 min, 0.8 units of topoisomerase I were added, the reaction was incubated for 30 min at 37°C, and stopped with 0.5% SDS, 25 mM EDTA. Proteinase K was added to a concentration of 0.2 mg/ml, followed by incubation at 37°C for 10 min. Reaction products were separated by electrophoresis on 1.3% agarose gels in TEP buffer (36 mM Tris, 36 mM NaH2PO4, 1 mM EDTA, pH 8.0; 2.6 V/cm, 3 h).
DNA cleavage
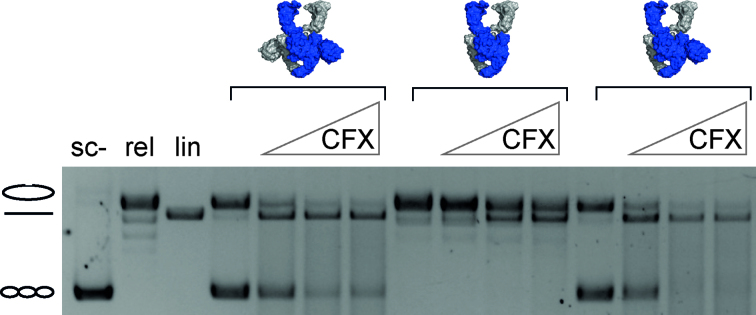
DNA cleavage reactions were performed at 37°C with 20 nM plasmid DNA with 200 nM GyrA and 800 nM GyrB, 1.5 mM ATP and 0–250 μM Ciprofloxacin (CFX) in 50 mM Tris–HCl, pH 7.5, 100 mM KCl, 10 mM MgCl2. Reactions were stopped with 0.5% SDS, 25 mM EDTA after 5 min, and products were separated on 1.3% agarose gels in TEP buffer (36 mM Tris, 36 mM NaH2PO4, 1 mM EDTA, pH 8.0; 2.6 V/cm, 3 h). Samples were pre-incubated at 37°C for 3 min before ATP and CFX were added.
Fluorescent labeling and single-molecule FRET experiments
Single-molecule FRET experiments addressing subunit exchange during the supercoiling reaction were performed with the GyrA variant GyrAD145C (18,38). DNA-induced movement of the CTDs was monitored using a donor/acceptor-labeled GyrAT140C/K594C·GyrA heterodimer (19) containing one or two CTDs (GyrAT140C/K594C·GyrA or GyrAT140C/K594C·GyrAΔCTD, respectively). In all GyrA variants used for smFRET, the intrinsic cysteine had been replaced by leucine (C350L). GyrA dimers were labeled statistically with a mixture of Alexa488 (donor) and Alexa546 (acceptor) maleimides as described previously (18,20,40). This procedure generates a mixture of –/donor-, donor/donor-, donor/acceptor-, acceptor/donor-, acceptor/acceptor- and -/acceptor-labeled species. Single-molecule FRET experiments addressing the conformation of the N-gate were performed with a GyrBE17CGyrA fusion protein (GyrBA) to prevent subunit dissociation (20). The intrinsic cysteines in the GyrA and GyrB parts were replaced by a leucine (GyrAC350L) and by serines (GyrBC58S, GyrBC414S), respectively. (GyrBA)2 was labeled statistically with Alexa488 (donor) and Alexa546 (acceptor) maleimides as described (20). For experiments addressing the conformation of the G-segment, a phosphorothioate-modified 60 bp DNA was used (18). This DNA contains a preferred gyrase sequence (39) in the center, flanked by Alexa488 (donor) and Alexa546 (acceptor), and a 3′-bridging phosphorothioate modification at the cleavage site in one of the strands. Phosphorothioate-modified DNA is cleaved by gyrase, but is re-ligated inefficiently (41), which increases the population of cleavage complexes (18). The sequences used are 5′-GTCTCGCAGTGCCAAAAAAAGCAYGATCATACGXCGACTCTAGAGTCTCGCAGTGTACCG-3′, where Y denotes a T with a 3′-thiolate modification and X a T with Alexa488 attached to an amino-linker at C6, and 5′-CGGTACACTGCGAGACTCTAGAGTCGACGTATGATCATGCTXTTTTTGGCACTGCGAGAC-3′, where X denotes a T with Alexa546 attached to an amino-linker at C6. The preferred gyrase cleavage site is highlighted in bold.
Single-molecule FRET experiments were performed on a home-built confocal microscope, equipped with a frequency-doubled, mode-locked, pulsed titanium-sapphire laser (λex = 475 nm), a 60x water-immersion objective (PlanApo, N.A. 1.2; Olympus), a DM505 dichroic beam splitter (Olympus) and a HQ490LP optical filter (AHF) to reject excitation light, a Q565LP beam splitter (AHF) and emission filters D535/40BP (donor) and E570LP (acceptor), avalanche photodiode detectors (SPCM-14; PerkinElmer), and an SPC-630 counting card (Becker & Hickl) (40). Measurements probing the conformation of a 60 bp DNA bound at the DNA-gate of gyrase were performed with 50 pM double-stranded DNA, 2 μM GyrA and 5 μM GyrB in 50 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM MgCl2 in the absence and presence of 2 mM ADPNP as described previously (16,20). The position of the CTDs was probed in measurements with 150 pM donor/acceptor-labeled GyrA (donor concentration) and 8 μM GyrB in 50 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM MgCl2 as described previously (16,19). Measurements probing the conformation of the N-gate were performed with 150 pM (GyrBA)2(monomer concentration) as described previously (16,20).
Subunit exchange
Subunit exchange of GyrA heterodimers during supercoiling reactions was investigated as described (38). Briefly, 50 nM donor-labeled GyrAD145C·GyrAD145C_ΔCTD and 50 nM acceptor-labeled GyrAD145C·GyrAD145C_ΔCTD (dimer concentration) with 800 nM GyrB, 1.5 mM ATP and 20 nM relaxed pUC18 in 50 mM Tris–HCl, pH 7.5, 100 mM KCl, 10 mM MgCl2 for 5 min, followed by stopping the reaction with 0.5% SDS, 25 mM EDTA. Single-molecule FRET experiments were performed after dilution to 40 pM (donor concentration). Donor/acceptor-labeled GyrAD145C·GyrAD145C_ΔCTD served as a positive control.
Subunit exchange during storage of purified GyrAD145C·GyrAD145C_ΔCTD heterodimers at –80°C was analyzed by size exclusion chromatography on an analytical S200 column in 50 mM Tris–HCl, pH 7.5, 500 mM NaCl, 10 mM MgCl2. Homodimers of GyrA and of GyrAΔCTD were analyzed as a reference.
Steady-state ATPase activity
Steady-state ATPase activity was measured in a coupled enzymatic assay that links ATP hydrolysis to NADH oxidation, which was measured as a decrease in the absorption at 340 nm. Measurements were performed with 0.1 μM GyrB and 0.5 μM GyrA in 50 mM Tris–HCl pH 7.5, 100 mM KCl, 10 mM MgCl2 and 5 mM ATP at 37°C with 0–150 μM relaxed or negatively supercoiled pUC18 in an Ultraspec pro photometer (GE Healthcare, Germany). Data were analyzed with the Michaelis-Menten equation to obtain vmax or kcat and Kapp,DNA values. Errors (±) reflect standard deviations from three independent experiments. The errors σx for the fold-change x in the rate of ATP hydrolysis in the presence of DNA with
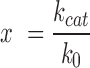 |
were propagated from the experimental errors of k0 (in the absence of DNA) and kcat (in the presence of saturating concentrations of DNA) according to
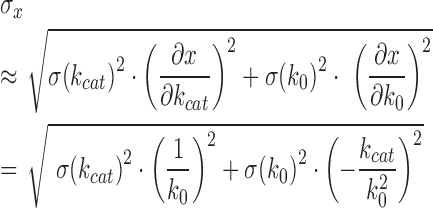 |
RESULTS
DNA gyrase with a single CTD introduces negative supercoils into DNA
To dissect the role of the two CTDs in gyrase, we first tested supercoiling and relaxation activities of gyrase containing two CTDs, one CTD, or no CTDs (Supplementary Figure S2). While wildtype gyrase (B·A·A·B) introduced negative supercoils into relaxed plasmid in the presence of ATP and relaxed negatively supercoiled DNA in the absence of ATP, gyrase lacking both CTDs (B·AΔCTD·AΔCTD·B) failed to catalyze either reaction. Gyrase containing a single CTD (B·A·AΔCTD·B) still catalyzed ATP-dependent negative supercoiling and ATP-independent relaxation. The individual subunits GyrB, GyrA, GyrAΔCTD, or GyrA·GyrAΔCTD heterodimers, did not show supercoiling or relaxation activities (Supplementary Figure S2).
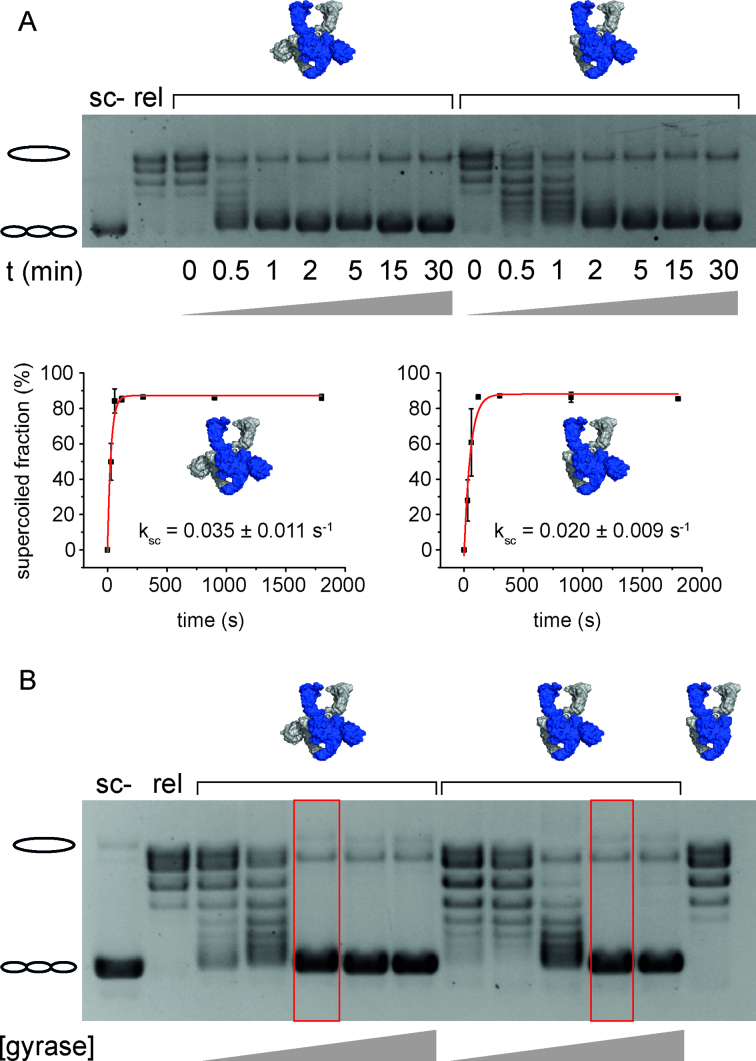
To obtain more insights into the supercoiling efficiencies, we followed the DNA supercoiling reaction as a function of time or gyrase concentration (Figure 1). While gyrase completely supercoils relaxed DNA within 1 min (at a DNA:protein ratio of 1:1), gyrase containing only one CTD needed ∼2 min for complete supercoiling under these conditions. A quantification of the fraction of supercoiled plasmid at each time point gave a rate constant for DNA supercoiling of ksc = 0.035 (±0.011) s-1 for gyrase, and a slightly reduced rate constant of ksc = 0.020 (±0.009) s-1 for gyrase containing a single CTD. Gyrase with only one CTD was required in about twice the concentration compared to wildtype gyrase to achieve complete supercoiling.
Figure 1.
DNA supercoiling by gyrase containing two or one CTD(s). (A) Time trace of ATP-dependent DNA supercoiling by gyrase with two CTDs and gyrase with one CTD (see cartoons of structures). 20 nM relaxed pUC18 was supercoiled by 40 nM GyrA and 160 nM GyrB in presence of 1.5 mM ATP at 37°C, and the reaction was stopped at indicated time points. Error bars depict the standard deviation from three independent experiments. Quantification of the fraction of supercoiled DNA as a function of time gives rate constants of supercoiling of ksc = 0.035 ± 0.011 s−1 for gyrase with both CTDs, and ksc = 0.020 ± 0.009 s−1 for gyrase with a single CTD. Errors are standard deviation from three independent experiments. Gyrase with one CTD thus catalyzes negative supercoiling of DNA only slightly more slowly than gyrase with two CTDs. (B) Concentration dependence of DNA supercoiling for gyrase with two, one, and no CTD(s). 20 nM relaxed pUC18 were incubated with 10, 20, 40, 80, or 160 nM GyrA and 20, 40, 80, 160, or 320 nM GyrB in the presence of 1.5 mM ATP, and reactions were stopped after 5 min. Compared to gyrase with two CTDs, about twice the concentration of gyrase with one CTD is needed to achieve complete supercoiling (red boxes). Gyrase lacking the CTDs (160 nM GyrAΔCTD, 320 nM GyrB) does not supercoil DNA. sc-: negatively supercoiled DNA, rel: relaxed DNA.
To exclude that DNA supercoiling by gyrase with one CTD is caused by subunit exchange and in situ formation of gyrase (B·A·A·B) during the experiment, we performed single-molecule FRET experiments with mixtures of donor-labeled and acceptor-labeled gyrase (containing GyrAD145C) before and after the supercoiling reaction (Supplementary Figure S3) (38). The FRET histogram showed a prominent peak at FRET efficiencies around zero, representing gyrase carrying only the donor dye, and a tiny peak at FRET efficiency of EFRET = 1, caused by direct excitation of gyrase labeled with only the acceptor dye. In a control experiment with donor/acceptor-labeled gyrase, FRET across the GyrA dimer interface gave rise to a peak at EFRET = 0.7. No peak at this FRET efficiency was detected in the histogram of the mixture, neither before nor after the supercoiling reaction. Hence, subunit exchange during the supercoiling reaction is negligible, consistent with previous findings (38).
Finally, we tested for contamination of our heterodimer preparations with homodimeric GyrA, as well as for subunit exchange during purification and storage, by analysis of heterodimeric A·AΔCTD by analytical size-exclusion chromatography (Supplementary Figure S3). GyrA dimers eluted from the column in a single, defined peak at 9.7 ml. The elution volume was increased to 11 ml for GyrA lacking the CTDs (AΔCTD·AΔCTD), consistent with its lower molecular mass. Heterodimeric A·AΔCTD eluted as a broader peak, with a maximum at an elution volume of 10 ml. By describing the chromatogram as a linear combination of the reference chromatogram for GyrA·GyrA and a peak centered at 10.2 ml corresponding to GyrA·GyrAΔCTD, we estimated the fraction of GyrA homodimers in the preparation to approx. 5%. Such a small fraction cannot account for the robust supercoiling activity observed for B·A·AΔCTD·B.
Gyrase with a single CTD decreases the linking number in steps of two
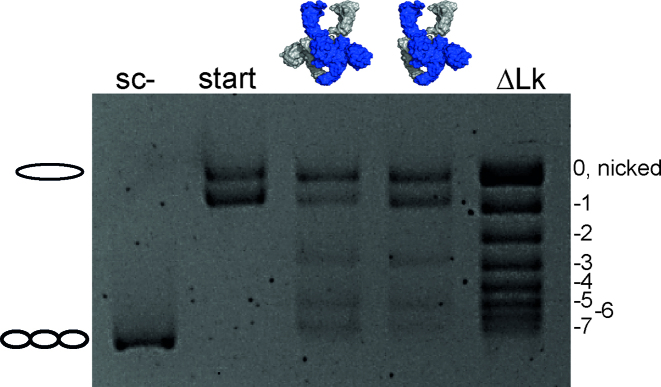
Gyrase wraps a positive supercoil of DNA with the help of the CTDs (15), and thereby stabilizes a positive node that is inverted into a negative node by strand passage, leading to a decrease in the linking number of the DNA by two in each catalytic cycle (1). To test if gyrase containing a single CTD can wrap DNA in a similar manner, we determined the change in linking number in supercoiling reactions with a single topoisomer as a DNA substrate (Figure 2). Gyrase containing both CTDs changed the linking number in steps of two, as observed previously (1,38). Gyrase containing only one of the CTDs also decreased the linking number in steps of two, demonstrating that one CTD is sufficient for DNA wrapping and supercoiling.
Figure 2.
DNA supercoiling of a single topoisomer by gyrase containing two or one CTD(s). Supercoiling reactions were performed with 6 nM single topoisomer (start), 10 nM GyrA, and 20 nM GyrB in the presence of 1.5 mM ATP at 37°C, and reactions were stopped after 5 min. Gyrase with one and two CTDs decreases the linking number of the DNA in steps of two. ΔLk denotes the topoisomer ladder. Gyrase with one and two CTD(s) catalyzes negative DNA supercoiling by decreasing the linking number in steps of two.
The second CTD does not contribute to the stability of the gyrase/DNA complex
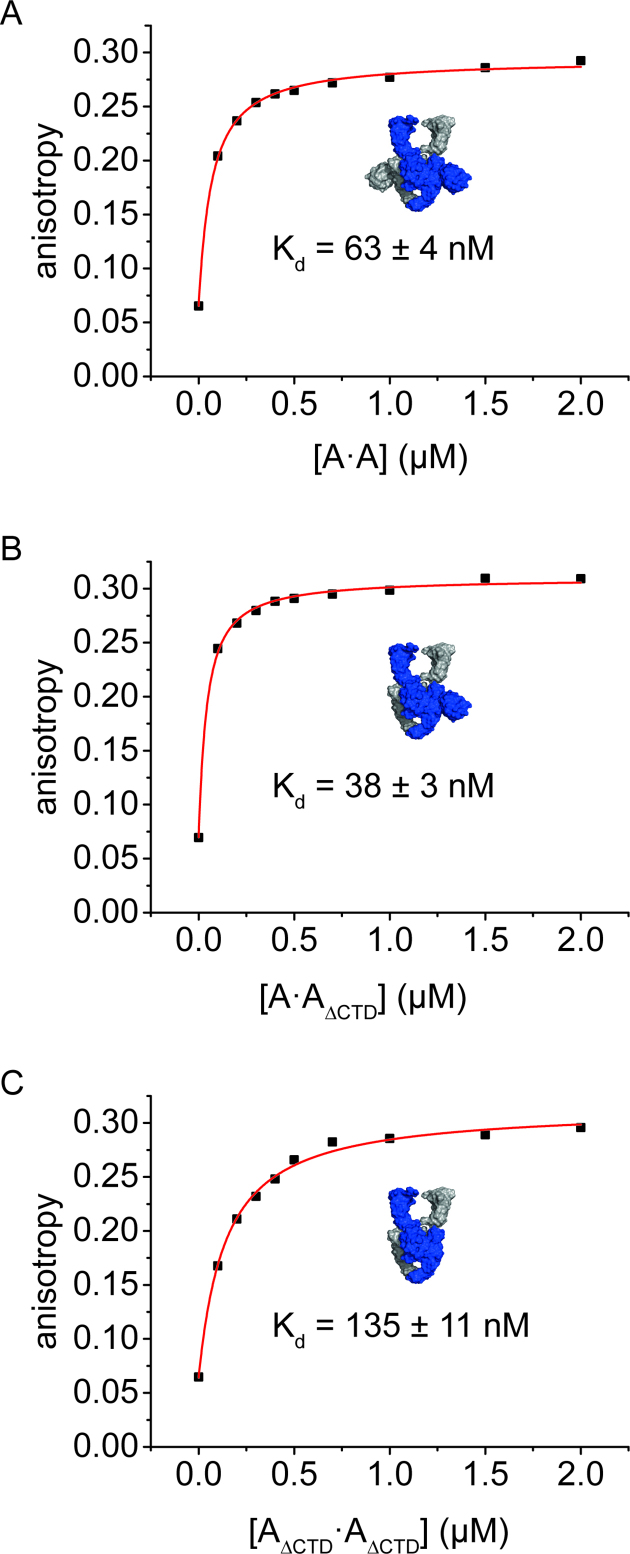
The CTDs bind short linear DNA (12) and are crucial for wrapping of DNA around gyrase (23). Deletion of both CTDs leads to a 5-fold decrease in affinity for a linear 60 bp DNA (18,19), whereas the affinity for shorter DNAs is not affected. These observations are consistent with short DNAs interacting with the DNA-gate, without contacts to the CTD, whereas longer DNAs extend from the DNA-gate towards the CTDs and form interactions with the CTDs (19). The 60 bp DNA is thus a suitable model DNA to probe the effect of the CTDs on DNA binding to regions flanking the G-segment. To test the contribution of each of the CTDs to DNA binding, we measured Kd values of gyrase/DNA complexes in fluorescence anisotropy titrations of the 60 bp DNA (Figure 3). The Kd value for the gyrase/DNA complex was determined as Kd = 63 (±4) nM, as observed previously (51 nM, (18), 63 nM, (19)). Gyrase lacking both CTDs showed a two-fold reduced DNA affinity, with a Kd value of Kd = 135 (±11) nM, similar to previous reports (4.7-fold, 298 nM, (19)). Interestingly, deletion of only one of the two CTDs did not reduce the DNA affinity, and a Kd value of Kd = 38 (±3) nM was determined for gyrase containing only one CTD. For GyrA only, in the absence of GyrB, deletion of a single CTD also had little effect on DNA affinity, whereas deletion of both CTDs reduced the affinity for the 60 bp DNA by 20-fold (Supplementary Figure S4). The Kd values are Kd = 1.05 (±0.11) μM (A·A), Kd = 1.25 (±0.02) μM (A·AΔCTD), and Kd = 22 (±0.4) μM (AΔCTD·AΔCTD). Altogether, these experiments show that a single CTD is sufficient for high-affinity binding of this DNA to gyrase, and demonstrate that the second CTD does not provide additional thermodynamic contributions to DNA binding. To compare binding of gyrase with one or two CTDs in the context of a substrate for the supercoiling reaction, we performed topoisomerase I relaxation assays of gyrase-bound DNA (34) with both enzymes and relaxed plasmid (Supplementary Figure S5). Gyrase trapped positive supercoils in the DNA substrate independent of the number of CTDs present.
Figure 3.
DNA binding to gyrase with two, one, or no CTD(s). Fluorescence anisotropy titrations of an Alexa488/Alexa546-labeled 60 bp DNA with 0.2–2 μM GyrA (monomer concentration) containing two CTDs (A·A, panel A), one CTD (A·AΔCTD, panel B), or no CTDs (AΔCTD·AΔCTD, panel C), in presence of 8 μM GyrB at 37°C. Binding was followed using the fluorescence anisotropy of Alexa546 as a probe. The curves depicted are representatives of duplicate measurements; errors (±) denote the error of the mean. Deletion of one CTD does not reduce the affinity of gyrase for the 60 bp DNA, but deletion of both CTDs leads to a two-fold decrease in DNA affinity.
Gyrase with a single CTD distorts the G-segment
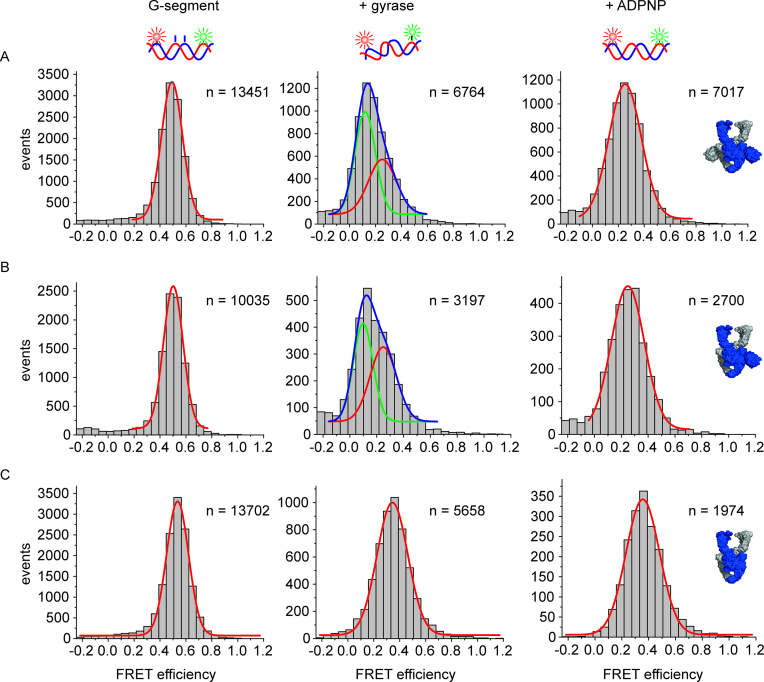
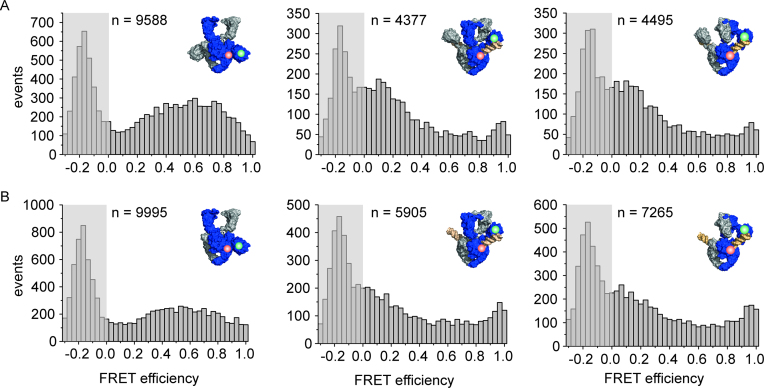
The 60 bp DNA becomes distorted on binding to gyrase (18). This distortion is coupled to DNA cleavage, and is not observed in cleavage-deficient gyrase (18). Gyrase lacking both CTDs also does not distort the DNA, indicating that the CTDs contribute to this process through an unknown mechanism (20). The different effects of gyrase with and without CTDs on DNA conformation raise the question whether one CTD is sufficient to induce this conformational change of the DNA. We therefore performed single-molecule FRET experiments with the 60 bp DNA, labeled with donor and acceptor fluorophores around the gyrase cleavage site (18) (Figure 4). This DNA reports on G-segment conformation: a high-FRET state corresponds to DNA bound to gyrase, while a low-FRET state represents DNA that is bound to gyrase and is severely distorted (18). To increase the population of gyrase complexes with distorted DNA, we used the 60 bp DNA that contained a phosphorothioate modification at the cleavage site (see Material and Methods), which can be cleaved but re-ligation is disfavored (18,41). 49% of DNA bound to gyrase containing a single CTD was observed in a low-FRET state (EFRET = 0.10) corresponding to distorted DNA. In comparison, 57% of the DNA was in the low-FRET state in presence of gyrase containing both CTDs. This value is in the range of previous measurements (40%; (18)). In both cases, the distortion was relieved on ADPNP binding, and only the high-FRET species (EFRET = 0.25) was present. This is in agreement with previous observations (18) and may reflect a change in G-segment conformation caused by the proposed ‘loss of wrap’ on ADPNP binding (35). No low-FRET species was observed with gyrase lacking the CTDs, consistent with previous reports (20). Hence, a single CTD is sufficient to induce distortion in the G-segment.
Figure 4.
Conformation of the G-segment bound to gyrase with two, one, or no CTDs. Single-molecule FRET histograms of an Alexa488/Alexa546-labeled 60 bp DNA (G-segment) containing a phosphorothioate modification at the cleavage site, bound to gyrase with two CTDs (A), with one CTD (B), and without CTDs (C). DNA on its own (left panels) shows a unimodal FRET histogram characteristic of regular B-form DNA. DNA bound to gyrase (center panels) with two or one CTDs exists in a low-FRET state (EFRET = 0.12, green Gaussian; severely distorted from B-form geometry (18)) and a medium-FRET state (EFRET = 0.25, red Gaussian, slightly distorted from B-form geometry (18)); the sum of the two distributions is shown in blue. Addition of ADPNP to the gyrase/DNA complex to induce N-gate closure leads to disappearance of the low-FRET state for both enzymes. Gyrase lacking the CTDs does not distort the G-segment. Addition of ADPNP has no effect on the FRET distribution. The DNA concentration was 50 pM, the concentrations of GyrA and GyrB were 2 and 5 μM, respectively. n denotes the number of events summarized in each histogram.
DNA cleavage
Deletion of the CTDs does not affect the extent of DNA cleavage in the presence of quinolones by E. coli gyrase (42). However, the CTDs are involved in distortion of the DNA bound at the DNA-gate, and DNA distortion is in turn linked to cleavage. As gyrase with a single CTD is still capable of distorting the G-segment, we next investigated if there is a link between the number of CTDs and DNA cleavage in B. subtilis gyrase. Cleavage assays on plasmid DNA with gyrase containing two, one or no CTDs in the presence of increasing concentrations of CFX revealed similar extents of double-strand cleavage at all CFX concentrations tested (Figure 5). Gyrase lacking both CTDs generated relaxed DNA during the cleavage reaction due to its ATP-dependent relaxation activity. Apart from this difference, the cleavage pattern for gyrase with two or one CTD(s) was virtually identical, indicating that, consistent with previous reports (42), the CTDs are not required for double-strand cleavage.
Figure 5.
DNA cleavage by gyrase with two, one, or no CTD(s) in the presence of CFX. DNA cleavage reactions were performed at 37°C with 20 nM plasmid DNA, 200 nM GyrA, 800 nM GyrB, 1.5 mM ATP and increasing concentrations of CFX (0, 10, 100 or 250 μM) for 5 min. All enzymes show similar levels of DNA cleavage at all CFX concentrations tested, irrespective of the number of CTDs present.
DNA-induced CTD movement
The CTDs move away from the GyrA NTD when DNA bound at the DNA-gate is long enough to establish contacts with the CTDs (19). Using single-molecule FRET and gyrase carrying a donor and acceptor fluorophore on the GyrA NTD (T140C) and the CTD (K594C) in one of the GyrA subunits (B·A·AT140C/K594C·B), respectively, we tested whether gyrase that contains only one CTD also undergoes this DNA-induced conformational change (Figure 6). Gyrase alone shows a broad FRET histogram, irrespective of the number of CTDs present. The broad distribution could either point to a substantial flexibility of the CTDs relative to the GyrA body, or it might represent two overlapping distributions, i.e. a low FRET state for gyrase where the labeled CTD is pointing upward, and a high-FRET state where this CTD is pointing downward. When DNA is bound, the FRET efficiency shifts towards lower values, again irrespective of the number of CTDs present, which is indicative of an upward movement of the CTD(s) on DNA binding. Gyrase with a single CTD thus also undergoes DNA-induced movement of its CTD. Addition of ADPNP and closure of the N-gate do not affect the FRET histograms and the position of the CTD(s), in agreement with previous observations for wildtype gyrase (19).
Figure 6.
DNA-induced CTD movement monitored by single-molecule FRET. Single-molecule FRET histograms for gyrase labeled with donor (green) and acceptor (red) in the NTD and CTD of one GyrA subunit. The area at EFRET < 0 contains the peak representing molecules that carry only the donor fluorophore, and is shaded in gray. n denotes the number of events accumulated in each histogram. (A) Gyrase with both CTDs (B·AT140C/K594C·A·B), labeled with donor and acceptor fluorophores in the NTD and CTD of one GyrA subunit, shows a broad FRET histogram (left), in agreement with flexible attachment of the CTDs to the GyrA NTD. Addition of DNA (center) leads to a decrease in FRET efficiencies, indicative of an upward movement of the CTDs. Addition of ADPNP (right) does not further affect the FRET efficiency histogram and thus the position of the CTDs. (B) Gyrase with one CTD (B·AT140C/K594C·AΔCTD·B) shows a similar FRET histogram as gyrase with two CTDs. Addition of DNA (center) also leads to a decrease in FRET efficiency. Addition of ADPNP does not induce any further changes in FRET efficiency and CTD position. Concentrations were 150 pM GyrA (donor concentration), 8 μM GyrB, 20 nM relaxed pUC18, and 2 mM ADPNP.
Gyrase with a single CTD undergoes DNA-induced N-gate narrowing
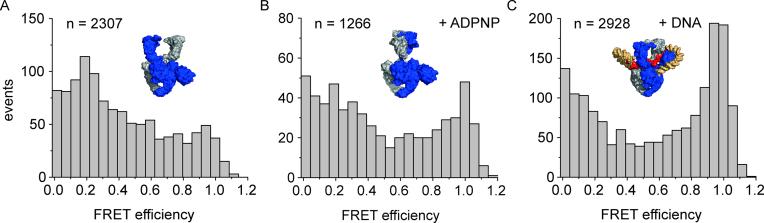
Binding of DNA to the DNA-gate of gyrase and wrapping of the flanking regions around the CTDs triggers a narrowing of the N-gate (20). The CTDs contribute to DNA-induced N-gate narrowing, and gyrase lacking the CTDs does not show N-gate narrowing in response to DNA binding (20). In single-molecule FRET experiments using a dimeric gyrase (GyrBA)2 labeled at the N-gate (E17C), we therefore investigated whether gyrase with a single CTD shows DNA-induced N-gate narrowing (Figure 7). The covalent linkage of GyrB and GyrA in GyrBA prevents dissociation of fluorescently labeled GyrB subunits during single-molecule experiments. Gyrase with only one CTD showed a low FRET efficiency of approx. 0.2, characteristic of an open N-gate. Closing the N-gate by adding ADPNP caused an increase in FRET efficiency to 0.9, as observed previously for wildtype gyrase (20). Addition of DNA also led to the population of a high-FRET state, demonstrating that the N-gate of gyrase with a single CTD narrows in response to DNA binding.
Figure 7.
Gyrase with a single CTD undergoes DNA-induced N-gate narrowing. Single-molecule FRET histograms for gyrase (BE17CA·BE17CAΔCTD) labeled with donor and acceptor fluorophores at the N-gate in the absence (A) and presence of 2 mM ADPNP (B) or 20 nM plasmid DNA (C). In the absence of DNA or ADPNP, the FRET efficiency is low (EFRET = 0.2), consistent with an open N-gate. ADPNP addition leads to N-gate closure, evidenced by a shift of the FRET efficiency to EFRET = 0.95–1. In the presence of DNA, the FRET efficiency is also high (EFRET = 0.95–1), which is consistent with DNA-induced narrowing of the N-gate. n denotes the number of events summarized in each histogram.
DNA-stimulated ATP hydrolysis is unaffected by the deletion of one CTD
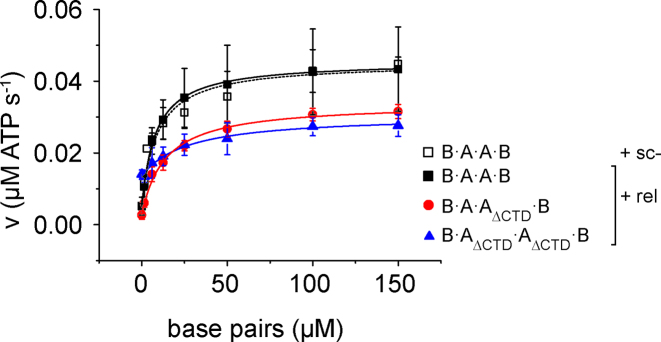
To investigate a possible contribution of the CTDs to coupling of DNA binding and ATP hydrolysis, we measured the DNA-dependent steady-state ATPase activity of gyrase, and gyrase lacking one or both CTDs, in the presence of relaxed plasmid DNA (Figure 8). Gyrase catalyzes ATP hydrolysis with kcat = 0.45 (±0.12) s-1, the apparent KM value for relaxed DNA is Kapp,DNA = 7.7 (±2.4) μM. In the presence of negatively supercoiled DNA, kcat and Kapp,DNA are virtually identical with kcat = 0.45 (±0.05) s−1, and Kapp,DNA = 9.1 (±3.5) μM. These values are in agreement with previous reports (kcat = 0.55 (±0.04) s−1; Kapp,DNA = 6.2 (±0.4) μM; (37)). Gyrase lacking both CTDs hydrolyzed ATP 1.5-fold more slowly (kcat = 0.30 (±0.01) s−1), and had an approximately 3.6-fold increased Kapp,DNA for relaxed DNA of Kapp,DNA = 28 (±13.5) μM. Gyrase with only one CTD catalyzed ATP hydrolysis with kcat = 0.34 (±0.04) s-1 and thus only 1.3-fold more slowly than wildtype gyrase, and had an only 1.7-fold increased Kapp,DNA for relaxed DNA of Kapp,DNA = 13 (±3.7) μM. The kcat and Kapp,DNA values are summarized in Table 1. Strikingly, both gyrase with one and two CTDs show a strong stimulation of their intrinsic ATPase activity by DNA (9.3- and 11.8-fold, respectively), whereas the ATPase activity of gyrase lacking both CTDs shows little (2.3-fold) DNA stimulation (Table 1). Overall, gyrase with a single CTD shows wildtype-like DNA stimulation of ATP hydrolysis.
Figure 8.
Effect of the CTDs on DNA-stimulated ATPase activity of gyrase. Steady-state ATPase activity of gyrase containing two, one, or no CTD(s) at 37°C with 0.1 μM GyrB, 0.5 μM GyrA and 1.5 mM ATP as a function of the concentration of negatively supercoiled DNA (sc-; B·A·A·B; black, open squares) or relaxed DNA (rel; B·A·A·B: black squares, B·A·AΔCTD·B: red circles; B·AΔCTD·AΔCTD·B: blue triangles). Error bars denote standard deviations from three to five independent experiments. The kcat and Kapp,DNA values obtained from analyses with the Michaelis-Menten equation are summarized in Table 1.
Table 1.
DNA-stimulated ATP hydrolysis
| enzyme | A2B2 (2 CTDs) | A2B2 (2 CTDs) | B·AΔCTD·A·B (1 CTD) | B·AΔCTD·AΔCTD·B (no CTDs) |
|---|---|---|---|---|
| DNA | sc- | rel | rel | rel |
| k 0 (s−1) | 0.071 ± 0.02 | 0.048 ± 0.01 | 0.029 ± 0.01 | 0.13 ± 0.01 |
| k cat (s−1) | 0.45 ± 0.05 | 0.45 ± 0.12 | 0.34 ± 0.04 | 0.30 ± 0.01 |
| fold stimulationa | 6.4± 1.9 | 9.3 ± 3.2 | 11.8 ± 4.2 | 2.3 ± 0.2 |
| K app,DNA (μM) | 9.1 ± 3.5 | 7.7 ± 2.4 | 13.4 ± 3.7 | 28 ± 13.5 |
k 0 and kcat denote rate constants of ATP hydrolysis under steady-state conditions in the absence of DNA (k0) and at saturating DNA concentrations (kcat), respectively. sc-: negatively supercoiled pUC18 plasmid, rel: relaxed pUC18 plasmid.
aError propagated from errors of k0 and kcat (see Materials and Methods section).
DISCUSSION
We have previously shown that gyrase containing a single catalytic tyrosine still supercoils DNA in steps of two when one of the two CTDs is deleted (38). This enzyme cannot perform strand passage but catalyzes DNA supercoiling by nicking/closing. Here we show that one of the two CTDs is dispensable for supercoiling in the context of the wildtype enzyme which can cleave both strands of the G-segment and catalyze DNA supercoiling through strand passage. Gyrase with a single CTD supercoils DNA only slightly more slowly than the wildtype enzyme. A single CTD is sufficient for binding and distortion of the DNA bound at the DNA-gate, for DNA cleavage, DNA-induced N-gate narrowing, for coupling of DNA binding and ATP hydrolysis, and for DNA supercoiling. The second CTD, although dispensable for DNA binding and for DNA-stimulated ATP hydrolysis, may provide minor contributions to functional DNA binding, as indicated by a small reduction of the Kapp,DNA when the second CTD is present (Table 1). The observation of robust supercoiling by gyrase with one CTD raises the question whether the two CTDs are functionally independent or cooperate in DNA supercoiling.
If the two CTDs carry out different functions in the catalytic cycle of gyrase, the intrinsic symmetry of the enzyme would have to be broken during the supercoiling reaction. In early publications, cartoons illustrating the current model for DNA supercoiling by gyrase used not to specify the orientation of the CTDs in different intermediates of the catalytic cycle (35,43), or showed gyrase in conformations with both CTDs pointing upward or both pointing downward (44). The first model depicting asymmetric states of gyrase appeared in 2004, and showed one CTD facing upward and the second CTD facing downward. The upward-facing CTD is wrapped around the DNA between the G-segment and the adjacent T-segment to be captured on N-gate closure (13). Since then, publications from many different laboratories have depicted asymmetric conformations of gyrase in their models for DNA supercoiling (19,21,29,36,45–48). Although it seems obvious that only one CTD can present a T-segment, there is no direct structural evidence for the presence of such asymmetric up-and-down structures. The cryo-EM structure of Thermus thermophilus gyrase bound to DNA, ADPNP, and CFX, captured the enzyme in a state where both CTDs wrap the DNA around their perimeter and are facing upwards (21). Small-angle X-ray scattering (27) and single-molecule FRET experiments ((19), this work) are consistent with both CTDs facing upwards when DNA is bound. However, several studies provided indirect evidence for asymmetry in DNA wrapping. Early footprinting studies revealed changes in DNA footprints of gyrase-bound DNA on ADPNP binding and N-gate closure that were asymmetric with respect to the cleavage site (31,33), pointing towards an asymmetry in the gyrase/DNA complex at this stage. In rotor-bead experiments, an intermediate in the catalytic cycle was observed at low ATP concentrations that may wrap the DNA asymmetrically (45). Single-molecule FRET experiments of gyrase/DNA complexes labeled on the GyrA NTD and CTD revealed bimodal FRET histograms for certain labeling geometries, also pointing to a possible asymmetry with respect to CTD position and DNA wrapping (19). This asymmetry may be more subtle, however, such as in the cryo-EM structure, where the CTDs are both oriented upwards, but the planes of their β-pinwheels form different angles with respect to the NTDs (21).
What might be the trigger for breaking the symmetry of gyrase? In recent models for the catalytic cycle, the asymmetric state with different positions of the CTD has been depicted for the state where one of the ATP molecules bound to the GHKL domain of GyrB has been hydrolyzed (47). Recent data indicate that gyrase may indeed hydrolyze ATP sequentially (49), similar to topoisomerase II (50–53), providing support for the presence and functional relevance of an ADP/ATP-bound state. In this state, the N-gate is in the closed conformation (49). The cryo-EM structure of gyrase shows the two GyrB subunits, locked as a dimer and twisted around each other in the ADPNP-bound state, tilted by 145° from the vertical axis of the enzyme (21). The GHKL domains are inclined towards the CTD which is positioned lower, aligned with the DNA-gate. This CTD is in an orientation that brings the exiting DNA into a positive crossover with the G-segment, and might thus represent the ‘active’ CTD (21). Although the captured intermediate does not reflect an ATP/ADP-bound state, the observed tilting of the GyrB subunits may point to a nucleotide-dependent interaction with the CTDs, which may lead to a break in symmetry and to the appearance of asymmetric states.
The observation that gyrase with a single CTD readily supercoils DNA in an ATP-dependent reaction raises the question as to why gyrase contains two CTDs. The only slightly higher rate of supercoiling of gyrase with two CTDs compared to the enzyme carrying only one CTD suggests that a second CTD provides only a small advantage. In general, DNA bound at the DNA-gate could be wrapped around the left-hand or right-hand CTD with equal probability. For gyrase with only one CTD, wrapping can occur only on one side. DNA wrapping requires pliability of the DNA, as it needs to be bent around the CTD(s). The DNA regions to the left and right of the binding site may differ in flexibility due to differences in sequence. Gyrase containing two CTDs can still wrap such DNA, irrespective of which side wraps more easily. In contrast, gyrase with only one CTD would only efficiently wrap the DNA if the more flexible region is located on the side of the CTD. In addition to local differences in DNA flexibility and bendability, DNA in vivo may have different accessibility left and right of the gyrase cleavage side due to binding of other proteins and DNA packing. Furthermore, as long as gyrase contains two CTDs, it would still be able to exert its physiological function when one of the CTDs becomes destabilized or degraded, and is thus less prone to failure. While the second CTD is dispensable for the hallmark reaction of gyrase, we cannot exclude that the second CTD might play an important role in other cellular processes gyrase might be involved in.
From another perspective, the presence of the two CTDs might just be an inevitable consequence of the dimeric nature of GyrA. For a gyrase with only a single CTD to evolve, a duplication of the gyrA gene would be required, followed by mutation of one of the copies such that the CTD is lost. However, in this case organisms would produce two different GyrA subunits, one with and one without CTD, which could statistically assemble into homodimers with two CTDs, homodimers lacking CTDs, and heterodimers with only a single CTD. Assembly of all produced proteins into functional heterodimers would then require an additional level of control. In the absence of selection pressure for a gyrase with only a single CTD, a dimeric, intrinsically symmetric gyrase might just be the most parsimonious solution to a complex biological problem.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jessica Guddorf for excellent technical assistance, and Markus Rudolph for helpful comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [KL5311/5-1, KL5311/9-1]. The open access publication charge for this paper has been waived by Oxford University Press — NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Brown P.O., Cozzarelli N.R.. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979; 206:1081–1083. [DOI] [PubMed] [Google Scholar]
- 2. Goto T., Wang J.C.. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J. Biol. Chem. 1982; 257:5866–5872. [PubMed] [Google Scholar]
- 3. Gellert M., Mizuuchi K., O’Dea M.H., Nash H.A.. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:3872–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peng H., Marians K.J.. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:8571–8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang J.C. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998; 31:107–144. [DOI] [PubMed] [Google Scholar]
- 6. Mizuuchi K., Fisher L.M., O’Dea M.H., Gellert M.. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:1847–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buhler C., Lebbink J.H., Bocs C., Ladenstein R., Forterre P.. DNA topoisomerase VI generates ATP-dependent double-strand breaks with two-nucleotide overhangs. J. Biol. Chem. 2001; 276:37215–37222. [DOI] [PubMed] [Google Scholar]
- 8. Corbett K.D., Benedetti P., Berger J.M.. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat. Struct. Mol. Biol. 2007; 14:611–619. [DOI] [PubMed] [Google Scholar]
- 9. Mizuuchi K., O’Dea M.H., Gellert M.. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:5960–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wigley D.B., Davies G.J., Dodson E.J., Maxwell A., Dodson G.. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991; 351:624–629. [DOI] [PubMed] [Google Scholar]
- 11. Horowitz D.S., Wang J.C.. Mapping the active site tyrosine of Escherichia coli DNA gyrase. J. Biol. Chem. 1987; 262:5339–5344. [PubMed] [Google Scholar]
- 12. Reece R.J., Maxwell A.. The C-terminal domain of the Escherichia coli DNA gyrase A subunit is a DNA-binding protein. Nucleic Acids Res. 1991; 19:1399–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Corbett K.D., Shultzaberger R.K., Berger J.M.. The C-terminal domain of DNA gyrase A adopts a DNA-bending beta-pinwheel fold. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:7293–7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ruthenburg A.J., Graybosch D.M., Huetsch J.C., Verdine G.L.. A superhelical spiral in the Escherichia coli DNA gyrase A C-terminal domain imparts unidirectional supercoiling bias. J. Biol. Chem. 2005; 280:26177–26184. [DOI] [PubMed] [Google Scholar]
- 15. Liu L.F., Wang J.C.. DNA-DNA gyrase complex: the wrapping of the DNA duplex outside the enzyme. Cell. 1978; 15:979–984. [DOI] [PubMed] [Google Scholar]
- 16. Hartmann S., Weidlich D., Klostermeier D.. Single-molecule confocal FRET microscopy to dissect conformational changes in the catalytic cycle of DNA topoisomerases. Methods Enzymol. 2016; 581:317–351. [DOI] [PubMed] [Google Scholar]
- 17. Gubaev A., Klostermeier D.. The mechanism of negative DNA supercoiling: a cascade of DNA-induced conformational changes prepares gyrase for strand passage. DNA Repair (Amst.). 2014; 16:23–34. [DOI] [PubMed] [Google Scholar]
- 18. Gubaev A., Hilbert M., Klostermeier D.. The DNA gate of Bacillus subtilis gyrase is predominantly in the closed conformation during the DNA supercoiling reaction. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:13278–13283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lanz M.A., Klostermeier D.. Guiding strand passage: DNA-induced movement of the gyrase C-terminal domains defines an early step in the supercoiling cycle. Nucleic Acids Res. 2011; 39:9681–9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gubaev A., Klostermeier D.. DNA-induced narrowing of the gyrase N-gate coordinates T-segment capture and strand passage. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:14085–14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papillon J., Menetret J.F., Batisse C., Helye R., Schultz P., Potier N., Lamour V.. Structural insight into negative DNA supercoiling by DNA gyrase, a bacterial type 2A DNA topoisomerase. Nucleic Acids Res. 2013; 41:7815–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rudolph M.G., Klostermeier D.. Mapping the spectrum of conformational states of the DNA- and C-Gates in Bacillus subtilis Gyrase. J. Mol. Biol. 2013; 425:2632–2640. [DOI] [PubMed] [Google Scholar]
- 23. Kampranis S.C., Maxwell A.. Conversion of DNA gyrase into a conventional type II topoisomerase. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:14416–14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kramlinger V.M., Hiasa H.. The “GyrA-box” is required for the ability of DNA gyrase to wrap DNA and catalyze the supercoiling reaction. J. Biol. Chem. 2006; 281:3738–3742. [DOI] [PubMed] [Google Scholar]
- 25. Ward D., Newton A.. Requirement of topoisomerase IV parC and parE genes for cell cycle progression and developmental regulation in Caulobacter crescentus. Mol. Microbiol. 1997; 26:897–910. [DOI] [PubMed] [Google Scholar]
- 26. Lanz M.A., Klostermeier D.. The GyrA-box determines the geometry of DNA bound to gyrase and couples DNA binding to the nucleotide cycle. Nucleic Acids Res. 2012; 40:10893–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Costenaro L., Grossmann J.G., Ebel C., Maxwell A.. Small-angle X-ray scattering reveals the solution structure of the full-length DNA gyrase a subunit. Structure. 2005; 13:287–296. [DOI] [PubMed] [Google Scholar]
- 28. Lanz M.A., Farhat M., Klostermeier D.. The acidic C-terminal tail of the GyrA subunit moderates the DNA supercoiling activity of Bacillus subtilis gyrase. J. Biol. Chem. 2014; 289:12275–12285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tretter E.M., Berger J.M.. Mechanisms for defining the supercoiling setpoint of DNA gyrase orthologs I. A non-conserved acidic C-terminal tail modulates E. coli gyrase activity. J. Biol. Chem. 2012; 287:18636–18644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tretter E.M., Berger J.M.. Mechanisms for defining supercoiling setpoint by DNA gyrase orthologs II. The shape of the GyrA CTD is not a sole determinant for controlling supercoiling efficiency. J. Biol. Chem. 2012; 287:18645–18654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morrison A., Cozzarelli N.R.. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rau D.C., Gellert M., Thoma F., Maxwell A.. Structure of the DNA gyrase-DNA complex as revealed by transient electric dichroism. J. Mol. Biol. 1987; 193:555–569. [DOI] [PubMed] [Google Scholar]
- 33. Orphanides G., Maxwell A.. Evidence for a conformational change in the DNA gyrase-DNA complex from hydroxyl radical footprinting. Nucleic Acids Res. 1994; 22:1567–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kampranis S.C., Bates A.D., Maxwell A.. A model for the mechanism of strand passage by DNA gyrase. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:8414–8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams N.L., Howells A.J., Maxwell A.. Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 2001; 306:969–984. [DOI] [PubMed] [Google Scholar]
- 36. Schoeffler A.J., Berger J.M.. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 2005; 33:1465–1470. [DOI] [PubMed] [Google Scholar]
- 37. Gottler T., Klostermeier D.. Dissection of the nucleotide cycle of B. subtilis DNA gyrase and its modulation by DNA. J. Mol. Biol. 2007; 367:1392–1404. [DOI] [PubMed] [Google Scholar]
- 38. Gubaev A., Weidlich D., Klostermeier D.. DNA gyrase with a single catalytic tyrosine can catalyze DNA supercoiling by a nicking-closing mechanism. Nucleic Acids Res. 2016; 44:10354–10366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bashkirov V.I., Zvingila D.J.. Sequence specificity of Bacillus subtilis DNA gyrase in vivo. Genetica. 1991; 85:3–12. [DOI] [PubMed] [Google Scholar]
- 40. Theissen B., Karow A.R., Kohler J., Gubaev A., Klostermeier D.. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Deweese J.E., Burgin A.B., Osheroff N.. Using 3′-Bridging phosphorothiolates to isolate the forward DNA cleavage reaction of human topoisomerase iialpha. Biochemistry. 2008; 47:4129–4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reece R.J., Maxwell A.. Probing the limits of the DNA breakage-reunion domain of the Escherichia coli DNA gyrase A protein. J. Biol. Chem. 1991; 266:3540–3546. [PubMed] [Google Scholar]
- 43. Williams N.L., Maxwell A.. Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry. 1999; 38:13502–13511. [DOI] [PubMed] [Google Scholar]
- 44. Maxwell A., Costenaro L., Mitelheiser S., Bates A.D.. Coupling ATP hydrolysis to DNA strand passage in type IIA DNA topoisomerases. Biochem. Soc. Trans. 2005; 33:1460–1464. [DOI] [PubMed] [Google Scholar]
- 45. Gore J., Bryant Z., Stone M.D., Nollmann M., Cozzarelli N.R., Bustamante C.. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006; 439:100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nollmann M., Stone M.D., Bryant Z., Gore J., Crisona N.J., Hong S.C., Mitelheiser S., Maxwell A., Bustamante C., Cozzarelli N.R.. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat. Struct. Mol. Biol. 2007; 14:264–271. [DOI] [PubMed] [Google Scholar]
- 47. Basu A., Schoeffler A.J., Berger J.M., Bryant Z.. ATP binding controls distinct structural transitions of Escherichia coli DNA gyrase in complex with DNA. Nat. Struct. Mol. Biol. 2012; 19:538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Basu A., Parente A.C., Bryant Z.. Structural dynamics and mechanochemical coupling in DNA gyrase. J. Mol. Biol. 2016; 428:1833–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hartmann S., Gubaev A., Klostermeier D.. Binding and hydrolysis of a single ATP is sufficient for N-Gate closure and DNA supercoiling by Gyrase. J. Mol. Biol. 2017; 429:3717–3729. [DOI] [PubMed] [Google Scholar]
- 50. Lindsley J.E., Wang J.C.. Study of allosteric communication between protomers by immunotagging. Nature. 1993; 361:749–750. [DOI] [PubMed] [Google Scholar]
- 51. Harkins T.T., Lindsley J.E.. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 1. A DNA-dependent burst in ATP hydrolysis. Biochemistry. 1998; 37:7292–7298. [DOI] [PubMed] [Google Scholar]
- 52. Harkins T.T., Lewis T.J., Lindsley J.E.. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry. 1998; 37:7299–7312. [DOI] [PubMed] [Google Scholar]
- 53. Baird C.L., Harkins T.T., Morris S.K., Lindsley J.E.. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:13685–13690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.