Abstract
Genetic variation and susceptibility to disease are shaped by human demographic history and adaptation. We can now study the genomes of extant Africans and uncover traces of population migration, admixture, assimilation and selection by applying sophisticated computational algorithms. There are four major ethnolinguistic divisions among present day Africans: Hunter-gatherer populations in southern and central Africa; Nilo-Saharan speakers from north and northeast Africa; Afro-Asiatic speakers from north and east Africa; and Niger-Congo speakers who are the predominant ethnolinguistic group spread across most of sub-Saharan Africa. The enormous ethnolinguistic diversity in sub-Saharan African populations is largely paralleled by extensive genetic diversity and until a decade ago, little was known about detailed origins and divergence of these groups. Results from large-scale population genetic studies, and more recently whole genome sequence data, are unravelling the critical role of events like migration and admixture and environmental factors including diet, infectious diseases and climatic conditions in shaping current population diversity. It is now possible to start providing quantitative estimates of divergence times, population size and dynamic processes that have affected populations and their genetic risk for disease. Finally, the availability of ancient genomes from Africa provides historical insights of unprecedented depth. In this review, we highlight some key interpretations that have emerged from recent African genome studies.
Introduction
African populations are known to harbour the greatest genetic diversity. However, most large-scale population genetic studies, until a few years ago, had minimal representation of these populations and our understanding of African genetic diversity and demographic histories was largely based on a limited set of genetic markers. Although African genetic data remain under-represented, the scenario is gradually changing and several recent studies have used dense genotyping arrays and whole genome sequences to provide a less biased view of genetic diversity on the continent (1–4). These studies are challenging some previous estimates of the chronological sequence and dating of major events and providing a more comprehensive view of African demographic history.
The migration of agro-pastoralist Bantu-speakers from their homeland in central-west Africa to large parts of the continent in the last few thousand years changed African history (5,6). The migration into southern and eastern Africa is likely to have involved many levels of interactions ranging from complete replacement to large-scale admixture with the hunter-gatherer and forager populations that inhabited these regions. Although some of these hunter-gatherer populations have survived to the present, their geographic spread and population sizes have been reduced significantly (1,4,7) and varying degrees of gene flow from Bantu-speakers have been observed (8,9). In studying bi-directional gene flow, a key focus of some recent studies is the genetic diversity of extant hunter-gatherer populations and to identify their genetic contribution to the present day Bantu-speakers (8,10,11). Furthermore, ancient genomes are playing an important role in providing snapshots of populations that inhabited various parts of Africa in the past few thousand years and more specimens could shed light on aspects of the Bantu-migration (9,12).
In addition to the intrinsic diversity within the continent due to population structure and isolation, back migration of Eurasian populations into Africa has emerged as a critical contributor to the genetic diversity (1,3,13). These migrations involved the influx of different Eurasian populations at different times and to different parts of Africa. Comprehensive characterization of the details of these migrations through genetic studies on existing populations could help to explain the strong genetic differences between some geographically neighbouring populations. Slave trade in the last few millennia, on the other hand, carried a significant amount of African genetic variation to other parts of the world. Defining the geographic and genetic sources of these African diaspora populations is gaining much interest (2,14–17).
Adaptations to climate-change, diet and a strong burden of pathogens have also played a critical role in shaping genomic diversity (17,18). With the availability of large-scale genomic data from various African populations several novel loci under adaptive selection have been identified in the recent years (2,10,19,20). Moreover, admixture with non-African populations is emerging as an important source of variants in African populations (2,19). The strong correlation between selected loci and geographic distribution of associated diseases are further emphasizing the role of natural selection as an important contributor to the genetic diversity in African populations (1,17,21,22).
In this review we have highlighted some of the key recent insights into African demographic history, migrations within the continent, Eurasian back migration, African diaspora genetics and adaptive evolution. We discuss some ways in which new African data are contributing to population and disease genomics research.
Early Population Divergence in Africa
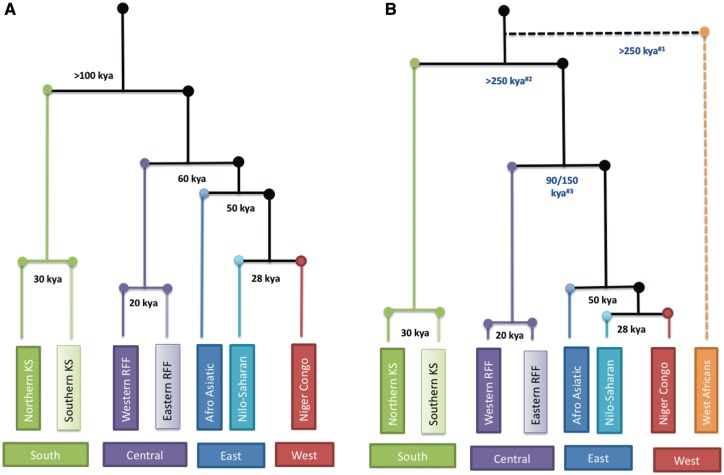
The genomic diversity in Africa has largely been shaped by deep population structure maintained by relatively long periods of geographic and cultural isolation. Fig.1A summarizes some of the major divergence events that have been critical to the origin of the major ethnolinguistic divisions. The oldest split in the human population has been suggested to be between Khoesan (KS) hunter–gatherers and other human populations. Although the estimated date of this event varies across studies, there is general consensus that the split occurred over 100 thousand years ago (kya) (8,23). The next major split was between the rain-forest foragers (RFF) and other human lineages, and is estimated to have occurred about 60–70 kya (23–26). Studies suggest that around 30 kya the KS population differentiated into the northern KS (e.g. Ju/'hoansi) and southern KS (e.g. ≠Khomani) groups (8,27). Similarly, it is estimated that around 20 kya the western RFF (e.g. Baka) diverged from the eastern RFF (e.g. Mbuti) (23,24,26). The separation of the Afro-Asiatic speakers currently living in north and east Africa from other sub-Saharan African (SSA) populations has been dated to about 50 kya (28). The final major separation is suggested to be between the Nilo-Saharan and Niger-Congo speakers dated to ∼28 kya (28). Despite being restricted to a rather small geographic area, genetic data suggest a significant differentiation within both Afro-Asiatics and Nilo-Saharans in east Africa (29–31). Similarly the Bantu and non-Bantu Niger-Congo speakers (from west Africa) also demonstrate observable genetic differentiation (1,3).
Figure 1.
Major early African population splits showing our understanding prior to and after the availability of WGS data and novel analysis approaches. The events of the past ∼5000 years, prior to the Bantu expansion are not shown and therefore the African regions (South, Central, East and West) reflect the groups that predominated in these regions at ∼5000 years ago (kya). Both trees are rooted to the most recent common ancestor (MRCA) and the estimated major splits are shown in kya. (A) shows our understanding prior to ∼2016 when the MRCA was estimated to be ∼150 kya and (B) following further analyses that place the MRCA at ∼300 kya, with revised estimates of major splits shown in blue. The length of the branches are not to scale. The dotted line shows the recently proposed deep split of a western African ancestry population ∼250 kya. #1Skoglund et al. (12); #2Schlebusch et al. (9); #3Hsieh et al. (10). KS: Khoesan; RFF: Rain Forest Foragers.
Notably, most of the divergence time estimates described above were based on uniparental markers or a limited set of autosomal markers. Computational analyses based on whole genome sequence (WGS) data from extant and ancient individuals are providing alternative timelines for some of these events. A recent ancient genome study (9) supports a much earlier date (>260 kya) for KS divergence from other human lineages as proposed by Scally and Durbin in 2012 (32). Similarly, the RFF split has been proposed to be up to 150 kya old (10). Both of these estimates could be about ∼100 thousand years older than previous estimates hinting at deep population structure on the continent (Fig. 1B). Coalescence analyses of WGS data are providing better estimates of demographic history, for example, population size estimates using Markovian coalescent simulations have suggested large population sizes of KS for the majority of human history (4,7). These populations appear not to have been affected by the events ∼30–120 kya that significantly reduced the size of all other human populations (7). The observation of possible ancient KS admixture in western Africa (1) and widespread distribution of KS-related ancestry in eastern and southern Africa (9,12) further indicate a much wider geographic spread of this group compared to their present distribution.
Recently, based on an analysis of 16 ancient African genomes, Skoglund and colleagues proposed a model that suggests an early separation of a west African branch from other human lineages that might predate the split of KS hunter–gatherers (12) (Fig. 1B). Though this dating requires further investigation, the existence of at least two ancient and semi-independent populations, one in east and one in west Africa, in addition to the ancestors of the present day hunter–gatherer and forager populations seems a strong possibility. This could also help to explain the differentiation of the Bantu and non-Bantu genetic components that have been observed in several studies of west African populations (1,3). In addition, there is also strong evidence that these populations were not isolated, and that there was bidirectional gene flow between east and west Africa for a significant proportion of history. For example, a study based on Y chromosome markers in thousands of individuals showed considerable population movement and demographic expansion across the present day Sahara during the recent ‘green Sahara’ period that ended ∼5 kya (33). Similarly, computational models also predict multiple ancient gene flow events between RFF and Niger-Congo speakers (10,28). Although the evidence for ancient introgression in Africa is still not adequate, it is highly plausible that such events have contributed to a fraction of the current genetic diversity (11,34).
Studies based on populations from a wide range of geographic locations are uncovering evidence of other early migration events. One such migration was of east African pastoralists to southern Africa, at a time that predates the Bantu migration by more than a thousand years (12,35,36). This migration event is thought to be the source of lactase persistence variants in southern Africa (37–39). Similarly, evidence for a relatively old Nilo-Saharan migration to western Africa, across the Sahel region, has been identified in some of the populations from this region (3). The availability of more deep-sequenced genomes, including ancient genomes, along with the development of improved algorithms would lead to more robust models of African demographic history, ancient migrations and gene flow.
The Bantu Expansion ∼5000 Years Ago
One of the most recent events shaping the genomic landscape in Africa is the movement of Bantu-speaking populations from the central western region of Nigeria and Cameroon into the rest of SSA. Archaeological evidence supports the spread of Bantu languages together with the advent of agricultural practices from western central Africa to eastern, western and southern Africa between ∼4 and 5 kya (40). The exact route and extent of the Bantu migration across SSA remains an area of active research although several recent genetic studies have provided evidence of regional admixture events that support specific routes. It is likely that multiple waves of migration, coupled with exposure to new environments and interactions with indigenous hunter–gatherer populations, are obscuring some historical events (5).
There are currently two proposed routes to explain the migration of Bantu speakers into SSA (3,6,41). The early-split model suggests that the western and eastern branches split early within the region of the Nigeria/Cameroon frontier around 3–2.5 kya and migrated directly east to the Great Lakes Region of east Africa. The late-split model suggests that populations first moved into the equatorial rainforest region near Gabon/Angola with a split to the east ∼2 kya. Although both routes are supported by several genetic studies, more recently there is greater support for the late-split hypothesis (2) based on data showing that eastern Bantu speakers and southeastern Bantu (SEB) speakers are genetically closer to western Bantu speakers from the southern, rather than the northern region. Furthermore, haplotype analyses showed that eastern Bantu speakers arose from two consecutive admixture events between western Bantu speakers and an Afro-Asiatic speaking population from Ethiopia, the first occurring 1000−1500 years ago and the second a mere 150−400 years ago. The subsequent best-matched western Bantu-speaking parental population for both admixture events was identified from Angola. Furthermore, SEB speakers show a unique admixture event ∼700 years ago between a parental Bantu-speaking population, most likely located in Angola, and the Jo/’hoansi San from Namibia. Together with further evidence of rain-forest hunter–gatherer admixture dated at ∼800 years ago in western Bantu speakers, this presents strong support that the Bantu migration first travelled south through the central rainforests before moving to the eastern and southern regions of the continent (2).
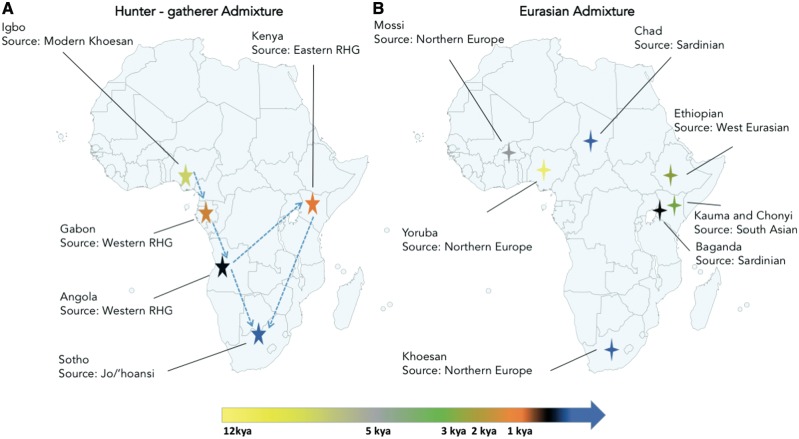
The late-split hypothesis further supports two main migration paths into southern Africa, one along the east of Africa and another along the west, giving rise to the SEB speakers and southwestern Bantu (SWB) speakers, respectively (40). The Bantu migration into southern Africa resulted in interactions with several established groups already present in the region, including various KS groups (Fig. 2A) and this contributed to the extensive genetic diversity observed in the current Bantu-speaking populations in South Africa (6,42).
Figure 2.
Hunter-gatherer (HG) and Eurasian admixture in African populations showing possible sources and timing of events. (A) shows HG admixture events while (B) shows Eurasian admixture events. Blue dotted arrows indicate the route of the Bantu expansion (reflecting the late split), while the colours of the stars and crosses correspond to the suggested timing of admixture events (kya) as shown on the scale at the bottom of the diagram. Populations are labelled as identified in the literature.
Eurasian Gene Flow Back to Africa
Genome-wide data have revealed significant Eurasian ancestry in African populations assimilated across a long time scale. Initial studies limited this Eurasian migration to a single event in east Africa (43), but subsequent studies revealed a more complex series of distinct admixture events occurring in western and eastern Africa (Fig. 2B). The African Genome Variation Project (AGVP) used genome data from 18 ethnolinguistic groups in eight African countries to identify Eurasian admixture ranging from 0% to 50% in and across central, west and east African populations (1). This distinctive Eurasian admixture appears to have occurred over at least three time periods with ancient admixture in central west Africa (e.g. Yoruba from Nigeria) occurring between ∼7.5 and 10.5 kya (1), older admixture in east Africa (e.g. Ethiopia) occurring between ∼2.4 and 3.2 kya (1,36,43) and more recent admixture between ∼0.15 and 1.5 kya in some east African (e.g. Kenyan) populations (1).
Subsequent studies based on LD decay and haplotype sharing in an extensive set of African and Eurasian populations confirmed the presence of Eurasian signatures in west, east and southern Africans. In the west, in addition to Niger-Congo speakers from The Gambia and Mali, the Mossi from Burkina Faso showed the oldest Eurasian admixture event ∼7 kya (3). In the east, these analyses inferred Eurasian admixture within the last 4000 years in Kenya, with the Chonyi and Kauma showing possible south Asian admixture that may have been facilitated by Medieval trade across the Indian Ocean. Eurasian admixture in the Afro-Asiatic speaking populations of east Africa appears to have occurred in two waves, one ∼2 kya and one in the last 200 years with ancestry best matched to the Tuscans from Italy. In the south, more recent direct Eurasian admixture from northern European populations has been observed in KS-speaking populations in South Africa (Fig. 2B). These include the Khomani and Karretjie, with events dating to ∼225 years ago, with an historical link to the European colonial period in South Africa (3).
More Recent Gene Flow and the African Diaspora
The second millennium CE saw significant population exchanges between Africa and other continents. Well before the age of European exploration, there were well-established trade routes between Africa and centres in Asia. Brucato et al. (44) recently surveyed these economic connections and compared them to finger prints of population exchange that can be seen in the genomes of current populations. Based on historical records, trade patterns are divided into four phases. In the first two phases, the trade routes where mainly land-based through the Sinai Peninsula or via the Bab al-Mandab, reaching deep into southern Africa. During the third phase, starting in the eleventh century CE, there was significant sea-borne trade from the Arabian Peninsula and Indonesia to east Africa and to Madagascar. Lastly, trade extended further into central Africa through to the Gulf of Guinea. Brucato et al. (44) show that this trade came with gene flow, and these events were highly correlated with peaks and troughs in trade routes.
The biggest drivers at a population level, out of and into Africa, were the slave trade and colonization that led to millions of Africans forcibly leaving Africa for the Americas and Asia, and, to a lesser extent European and Asian populations establishing themselves in Africa, with some admixture into African populations. Slavery and forced migration of people is a human practice dating millennia. Over the last thousand years, millions of Africans were enslaved and sent to new homes thousands of kilometres away. Although there are many detailed records of where people boarded boats that took them to captivity, less is known about where they actually came from. Recent research, including genetic studies, on the African diaspora and groups who have oral traditions of African ancestry have shed new light on these histories.
The Arab slave trade from at least the eighth century to the late nineteenth century had two distinct routes. The first was the trade either into the Mahgreb and Egypt or through these places into Asia. A second Indian Ocean trade route from the east coast, particularly from the sixteenth century onwards enslaved a further four million people (14). In the first phase mainly Nilotic or Afro-Asiatic speaking people from the Horn of Africa were involved and from the nineteenth century, mainly Bantu-speaking people, probably via regions controlled by the Omani Empire from their base in Zanzibar. There is no good method of accurately estimating the numbers, but several million people have conservatively been estimated to have been enslaved (45). At this scale, there is expected to be significant evidence of gene flow, but relatively little research has been done on the origins of these people or what became of them.
Laso-Jadart et al. (14) studied people from the Makrani group in Pakistan who have traditions suggesting descent from African slaves. Genotype data from 102 individuals showed an average 25% Africa ancestry, with significant diversity, and suggested that the likely source is SEB-speaking people. GLOBEtrotter analysis suggests a single wave of admixture dating to the late eighteenth century, although other models are possible.
The European slave trade from Africa took place over a shorter period of time, from the sixteenth to nineteenth centuries, but at a much more intense scale. Numbers are difficult to estimate, especially as many people who were enslaved died en route, but approximately 12 million people were enslaved and taken to the Americas from the sixteenth to nineteenth centuries (15) of whom 7 million went to South America (16). Several recent studies have investigated their African origins (2,17,46). Patin et al. (2) used data from 2500 Africans and 5200 African-Americans to explore the range of African ancestry in African-Americans. They estimate that ∼50% of the African ancestry of African-Americans originates from the Bight of Benin, ∼30% from western central Africa (current day Angola and DRC), ∼13% from Senegambia and ∼7% from the Windward Coast (present day Cote d'Ivoire). Mathias et al. (46) detected similar patterns and showed considerable variability in admixture patterns. Sex-biased admixture patterns are evident, with significantly higher proportions of European male-biased ancestry (46,47), contrasting with historical records showing that more African men were brought to the Americas than women (48). Complementing the broad-brushstroke papers, several recent studies have explored admixture in particular communities. These studies are bringing out the complexity of history, as well as the impact of admixture on health today (16,49–52).
Admixture into Africa in the Last Two Millennia
The most significant admixture in Africa in the last two thousands years is likely due to invasions from Arabia after the rise of Islam. Recent work has shown very significant impact on north African and north-eastern populations (13,31,53,54). This impact is greatest in Egypt and Mahgreb, and strong signals can be found elsewhere. However, there is strong intriguing evidence for admixture down the east coast of Africa into southern Africa (44,55).
The peopling of Madagascar has been of great interest for decades. It has been long known from archaeological, linguistic, mt and Y DNA data that Madagascar was settled by people from Africa and Austronesia in the first millennium but the dates and sequence of events has been unclear. Madagascar comprised a set of independent kingdoms until being unified in the early nineteenth century and subsequently colonized by the French in the late nineteenth century. Recent genetic evidence demonstrates a complex pattern of settlement and interaction resulting in a heterogeneous population (56). The genetic evidence suggests that two major settlements occurred: Austronesian populations settling in the east of the country roughly 1.5–2.5 kya, and Bantu-speaking populations settling roughly around 1.5 kya, with admixture occurring over the last 1500 years. On an average the relative contributions are ∼37% Austronesian and ∼59% African, although there is significant geographically clustered variation.
European colonization brought many people from elsewhere into Africa, particularly in southern Africa. The most remarkable story is that of the admixed Coloured community in South Africa. The Dutch brought slaves from India and Indonesia, who mixed with Europeans and Khoe, San, and Bantu-speaking people. This community came into being from the end of the seventeenth to the start of the nineteenth century. The latest evidence from genotyping and sequencing studies illustrates this complex admixture (42,57,58).
Contributions from African Whole Genome Sequence Data
Population-level deep-sequencing whole genome data are enhancing our understanding of diversity and LD architecture in African populations (1,59,60). On an average, Africans have ∼20–30% more SNVs compared to non-Africans (1,42,59,60) illustrating their tremendous potential for novel variant discovery. For example, the AGVP study of 320 African genomes identified ∼9.5 million novel SNVs (1). Similarly, the Southern African Human Genome Programme study identified ∼0.8 million novel SNVs from only 24 African genomes (42). These and other African WGS data have been used to develop genotyping platforms such at the Human Heredity and Health in Africa Consortium (H3Africa) SNP array that has better coverage of African genetic diversity and a more comprehensive representation of African LD blocks to enhance African genome-wide association studies (GWASs) (https://commonfund.nih.gov/globalhealth/h3aresources; date last accessed April 2018).
The statistical technique of imputation, providing genotypes for variants not present on a genotyping array, has emerged as a widely used technique to improve the effectiveness of GWASs (61,62). However, the quality and accuracy of imputation depends largely on the size of reference panels and the representation of haplotypes from the population to be studied (1,63,64). African haplotypes are still underrepresented in most reference panels (65,66) and therefore the inclusion of more African genomes into current reference panels is expected to significantly improve the quality of imputation in African studies. An African-centric reference panel of about 5000 genomes (about half of which are novel African genomes) is available at the Sanger Imputation server (https://imputation.sanger.ac.uk/; date last accessed April 2018) and others are under development. Better representation of African ethnolinguistic groups will provide a more comprehensive catalogue of genetic diversity and LD architecture in Africa and such studies are underway within the H3Africa Consortium (67,68).
Natural Selection Has Also Shaped African Genetic Diversity
Sub-Saharan Africa has a high burden of life-threatening non-communicable diseases, especially in regions around the tropical rain forests and such environments are suggested to have contributed to a number of local adaptations (1,17). Consistent with the current spread and prevalence of malaria in Africa, many of the first loci identified to be under strong selection in African populations have been associated with adaptation to malaria. The most studied example is the sickle cell mutation in the HBB gene (69–71), with haplotype analyses suggesting that the mutation arose independently at least four times in Africa (72). Allele frequencies are highest in west Africa and some east African populations where Plasmodium falciparum malaria is endemic. The CD36 malaria associated allele is most common in Nigerian populations with lower frequencies elsewhere. Selection of variants in the DARC gene are known to be adaptive to Plasmodium vivax malaria, with some variants having reached fixation in selected west and east African populations (Table 1) (2,73,74). Recent studies showed that copy number variants and complex structural rearrangements in the GYPA, GYPB and GYPC genes, which reduce the risk of severe malaria by up to 40%, have also been selected in some east African populations (21). As another example, adaptive selection has been observed for APOL1, LARGE and IL21 variants that protect against African trypanosomiasis and Lassa fever (1,18,22,75,76).
Table 1.
Allele frequencies in AGVP and KGP African populations of variants previously detected to be under selection (west African in blue, southern African in green, east African in pink and non-African in yellow)
| Gene | SNP ID | DA | GWD | MSL | ESN | YRI | ZUL | LWK | BAG | ETH | CEU | CHB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lactase Persistence | ||||||||||||
| MCM6 | rs145946881 | G | 0.000 | 0.000 | 0.000 | 0.000 | 0.070 | 0.086 | 0.075 | 0.000 | 0.000 | 0.000 |
| MCM6 | rs41380347 | C | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.017 | 0.000 | 0.000 |
| MCM6 | rs41525747 | C | NA | NA | NA | NA | 0.000 | NA | 0.000 | 0.050 | NA | NA |
| MCM6 | rs4954490 | A | 0.128 | 0.088 | 0.116 | 0.056 | 0.105 | 0.126 | 0.110 | 0.258 | 0.798 | 0.340 |
| MCM6 | rs4988235 | A | 0.009 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.058 | 0.737 | 0.000 |
| MCM6 | rs869051967 | C | NA | NA | NA | NA | 0.000 | NA | 0.000 | 0.008 | NA | NA |
| Malaria | ||||||||||||
| CD36 | rs3211938 | G | 0.009 | 0.006 | 0.242 | 0.292 | 0.005 | 0.076 | 0.050 | 0.000 | 0.000 | 0.000 |
| DARC | rs12075 | G | 0.000 | 0.000 | 0.000 | 0.000 | 0.070 | 0.000 | 0.000 | 0.113 | 0.429 | 0.932 |
| DARC | rs2814778 | C | 1.000 | 1.000 | 1.000 | 0.995 | 0.800 | 1.000 | 1.000 | 0.738 | 0.000 | 0.000 |
| DARC | rs3027011 | G | 0.080 | 0.123 | 0.081 | 0.102 | 0.185 | 0.121 | 0.185 | 0.058 | 0.000 | 0.000 |
| DARC | rs55872368 | T | 0.925 | 0.882 | 0.929 | 0.898 | 0.730 | 0.879 | 0.825 | 0.700 | 0.131 | 0.000 |
| DARC | rs7550207 | C | 0.080 | 0.123 | 0.081 | 0.097 | 0.190 | 0.121 | 0.185 | 0.125 | 0.197 | 0.000 |
| DARC | rs863004 | T | 0.925 | 0.882 | 0.929 | 0.903 | 0.760 | 0.874 | 0.825 | 0.771 | 0.404 | 0.034 |
| G6PD | rs1050828 | T | 0.040 | 0.070 | 0.160 | 0.210 | NA | 0.180 | NA | NA | 0.000 | 0.000 |
| G6PD | rs1050829 | C | 0.360 | 0.280 | 0.350 | 0.380 | NA | 0.340 | NA | NA | 0.000 | 0.000 |
| HBB (HbS) | rs334 | A | 0.115 | 0.123 | 0.121 | 0.139 | 0.005 | 0.101 | 0.070 | 0.000 | 0.000 | 0.000 |
| HBB (HbC) | rs33930165 | T | 0.004 | 0.006 | 0.000 | 0.028 | 0.005 | 0.000 | 0.020 | 0.000 | 0.000 | 0.000 |
| Skin pigmentation | ||||||||||||
| DDB1 | rs11230664 | T | 0.208 | 0.229 | 0.455 | 0.310 | 0.195 | 0.253 | 0.255 | 0.296 | 0.995 | 0.971 |
| HERC2 | rs4932620 | T | 0.071 | 0.076 | 0.096 | 0.083 | 0.030 | 0.131 | 0.100 | 0.242 | 0.010 | 0.019 |
| HERC2 | rs6497271 | G | 0.549 | 0.559 | 0.722 | 0.699 | 0.475 | 0.712 | 0.410 | 0.467 | 0.015 | 0.029 |
| KITLG | rs12821256 | C | 0.000 | 0.000 | 0.000 | 0.000 | 0.005 | 0.000 | 0.000 | 0.004 | 0.131 | 0.000 |
| MFSD12 | rs10424065 | T | 0.389 | 0.394 | 0.273 | 0.269 | 0.130 | 0.253 | 0.235 | 0.304 | 0.005 | 0.000 |
| MFSD12 | rs6510760 | A | 0.907 | 0.853 | 0.849 | 0.898 | 0.675 | 0.818 | 0.815 | 0.529 | 0.051 | 0.078 |
| OCA2 | rs1800404 | T | 0.150 | 0.082 | 0.136 | 0.069 | 0.145 | 0.086 | 0.045 | 0.258 | 0.823 | 0.393 |
| OCA2 | rs1800417 | G | 0.027 | 0.059 | 0.086 | 0.042 | 0.160 | 0.076 | 0.040 | 0.013 | 0.000 | 0.000 |
| SLC24A5 | rs1426654 | A | 0.075 | 0.088 | 0.025 | 0.014 | 0.075 | 0.076 | 0.025 | 0.371 | 1.000 | 0.029 |
| SLC45A2 | rs16891982 | G | 0.004 | 0.012 | 0.000 | 0.000 | 0.000 | 0.005 | 0.005 | 0.021 | 0.980 | 0.015 |
| TMEM138 | rs7948623 | T | 0.270 | 0.241 | 0.177 | 0.264 | 0.230 | 0.348 | 0.295 | 0.425 | 0.000 | 0.000 |
| TYR | rs1042602 | A | 0.000 | 0.006 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.013 | 0.399 | 0.000 |
| Trypanosomiasis | ||||||||||||
| APOL1 | rs60910145 | G | 0.243 | 0.123 | 0.495 | 0.375 | 0.120 | 0.056 | 0.055 | 0.000 | 0.000 | 0.000 |
| APOL1 | rs73885319 | G | 0.243 | 0.123 | 0.495 | 0.375 | 0.125 | 0.056 | 0.055 | 0.000 | 0.000 | 0.000 |
| APOL1 | rs143830837 | – | 0.195 | 0.182 | 0.116 | 0.079 | NA | 0.091 | NA | NA | 0.000 | 0.000 |
DA, derived allele; NA, missing data; Frequency of 0.000, not observed in that population.
Population codes: Esan in Nigeria (ESN); Yoruba in Ibadan, Nigeria (YRI); Gambian in Western Divisions in the Gambia (GWD); Mende in Sierra Leone (MSL); Luhya in Webuye, Kenya (LWK); Han Chinese in Beijing, China (CHB), Utah Residents with Northern and Western European Ancestry (CEU) from KGP Phase 3 and Baganda (BAG), Zulu (ZUL), Ethiopian (ETH) populations from AGVP. Both KGP Phase 3 and AGVP dataset have ∼100 individuals per population.
A major selective sweep around the LCT and MCMC6 genes has been associated with dietary adaptation in pastoralist populations around the world. Variants in this genomic region confer a lactase persistence phenotype (the ability to digest fresh milk as an adult) and have been found to have higher allele frequencies in east and southern African populations (not well represented in the populations shown in Table 1) in parallel with the geographic spread of pastoralism (38,77,78). Similarly, variants in several genes, including VAV3, ARNT2 and THRB, are suggested to be involved in high-altitude adaptation and were identified to be more common in populations native to the Ethiopian highlands (18). Selective sweeps have also been detected for variants in loci associated with short stature (POU1F1, HESX1, DOCK3, CISH and STAT5) in both east central and west central RFF populations (34,79). It is hypothesized that the short stature phenotype may confer some benefits in response to food scarcity, high humidity and heat or as a trade-off between cessation of growth and early reproduction (80).
With the availability of genome-scale data, there has been a surge in studies aimed at identifying signatures of selection in a wide range of African populations and this has led to the discovery of many new adaptive loci (2,12,19,20,81). Some studies have explored novel approaches to detect regions under selection, in addition to the traditionally used neutrality tests. Skoglund et al. (12) for example, used the comparison of ancient southern African genomes to genomes of modern-day San, and identified taste-receptor gene clusters to be under selection in the present day populations. Crawford et al. (19) employed a GWAS-based approach to identify loci associated with skin pigmentation and then studied selective sweeps around the signals (including MFSD12, HERC2/OCA2, SLC24A5, TYR, DDB1/TMEM). Sugden et al. (20) used a novel method to identify selection in KS populations and detected selective sweeps around various metabolism-related genes.
Computational analyses are also generating insights into the evolutionary history of some of the positively selected variants. For example, some of the variants that show signatures of selection in present-day populations have likely been introduced by admixture (gene flow), a process referred to as adaptive introgression. The HLA loci in western Bantu speakers and LCT loci in eastern Bantu-speakers were found to show an excess of local ancestry from rainforest hunter-gatherers and east Africans (Nilo-Saharan/Afro-Asiatic), respectively, hinting at possible adaptive introgression (2). Similarly, the positively selected variant (rs1426654) in the SLC24A5 gene was estimated to have evolved roughly 30 thousand years ago in Eurasian lineages and is proposed to have been re-introduced in the east African populations in the last 3–9 thousand years (13,19,82). Interestingly, another study showed the SLC24A5 derived allele to be more common in the lighter pigmented KS populations hinting at either convergent evolution or Eurasian gene flow thousands of years ago (83). These studies suggest that the use of a wider variety of African populations along with a more comprehensive toolkit for detecting selective sweeps could generate a better understanding of the variation due to adaptive selection across the continent.
Conclusions and Future Perspectives
Our inherent optimism in the power of scientific discovery has not diminished the challenges of understanding complex population relationships. These include the interpretation of genetic diversity to reconstruct human history and our ability to understand gene-to-gene, gene-to-environment interactions and inter-organismal relationships in shaping health and susceptibility to disease. Many unresolved questions related to the genetic variation observed in African populations remain. For example, when and where did key events during the Bantu migration take place? What processes during these events essentially shaped the emergence of the Bantu speakers? How did cultural practices such as sex-biased behaviour influence admixture events during migrations?
African genome data remain scarce and many African populations are yet to be studied. It is now evident that hunter-gatherer groups historically populated large regions of SSA and left traces of their genomes in almost all the populations currently living on the continent. It is important that we engage with extant hunter-gatherer groups about participating in genome studies to ensure a better understanding of their demographic history and the high genetic diversity among their members. The San communities remain poorly studied and to address potential exploitation of this vulnerable group, The San Council of South Africa launched a code of ethics for research in 2017 where they document their views and terms for promoting equitable and fair research in their communities, with respect and honesty (http://trust-project.eu/san-council-launches-san-code-of-ethics/; date last accessed April 2018).
There is great opportunity in studying ancient African genomes. The race for accessing specimens is evident (84) and therefore this intensifies the need to share responsibly and to preserve specimens for a time when improved techniques could provide more insights.
Engrained in the genomes of Africans is the deep and rich history of our species and evidence of migrations that reach the furthest corners of our planet. Over the past decade novel approaches to interpreting DNA sequence divergence have generated hypotheses that have yet to stand the test of time. African populations continue to provide fascinating insights on how diversity was shaped by historical events triggered by changing environments, including climatic and dietary adaptation and encounters with infectious pathogens, and sometimes by chance.
Acknowledgements
M.R. is a South African Research Chair in Genomics and Bioinformatics of African populations hosted by the University of the Witwatersrand, funded by the Department of Science and Technology and administered by National Research Foundation of South Africa (NRF). A.C. and D.S. are funded by the National Institutes of Health (National Human Genome Research Institute, NHGRI) AWI-Gen Collaborative Centre under award number U54HG006938, as part of the H3Africa Consortium. S.H. is partially funded and S.A. is funded by the National Institutes of Health, through the Pan-African Bioinformatics Network for H3Africa (H3ABioNet) award number U41HG006941. SH also acknowledges funding from the NRF IRF160214158079.
Conflict of Interest statement. None declared.
References
- 1. Gurdasani D., Carstensen T., Tekola-Ayele F., Pagani L., Tachmazidou I., Hatzikotoulas K., Karthikeyan S., Iles L., Pollard M.O., Choudhury A.. et al. (2015) The African Genome Variation Project shapes medical genetics in Africa. Nature, 10.1038/nature13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Patin E., Lopez M., Grollemund R., Verdu P., Harmant C., Quach H., Laval G., Perry G.H., Barreiro L.B., Froment A.. et al. (2017) Dispersals and genetic adaptation of Bantu-speaking populations in Africa and North America. Science, 10.1126/science.aal1988. [DOI] [PubMed] [Google Scholar]
- 3. Busby G.B., Band G., Si Le Q., Jallow M., Bougama E., Mangano V.D., Amenga-Etego L.N., Enimil A., Apinjoh T., Ndila C.M.. et al. (2016) Admixture into and within sub-Saharan Africa. Elife, 5, 10.7554/eLife.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A.. et al. (2016) The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature, 538, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tishkoff S.A., Reed F.A., Friedlaender F.R., Ehret C., Ranciaro A., Froment A., Hirbo J.B., Awomoyi A.A., Bodo J.M., Doumbo O.. et al. (2009) The genetic structure and history of Africans and African Americans. Science, 324, 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li S., Schlebusch C., Jakobsson M. (2014) Genetic variation reveals large-scale population expansion and migration during the expansion of Bantu-speaking peoples. Proc. R. Soc. B Biol. Sci., 10.1098/rspb.2014.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim H.L., Ratan A., Perry G.H., Montenegro A., Miller W., Schuster S.C. (2014) Khoisan hunter-gatherers have been the largest population throughout most of modern-human demographic history. Nat. Commun., 5, 5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schlebusch C.M., Skoglund P., Sjödin P., Gattepaille L.M., Hernandez D., Jay F., Li S., De Jongh M., Singleton A., Blum M.G.B.. et al. (2012) Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science, 338, 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schlebusch C.M., Malmström H., Günther T., Sjödin P., Coutinho A., Edlund H., Munters A.R., Vicente M., Steyn M., Soodyall H.. et al. (2017) Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science, 358, 652–655. [DOI] [PubMed] [Google Scholar]
- 10. Hsieh P.H., Veeramah K.R., Lachance J., Tishkoff S.A., Wall J.D., Hammer M.F., Gutenkunst R.N. (2016) Whole-genome sequence analyses of Western Central African Pygmy hunter-gatherers reveal a complex demographic history and identify candidate genes under positive natural selection. Genome Res., 26, 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hsieh P.H., Woerner A.E., Wall J.D., Lachance J., Tishkoff S.A., Gutenkunst R.N., Hammer M.F. (2016) Model-based analyses of whole-genome data reveal a complex evolutionary history involving archaic introgression in Central African Pygmies. Genome Res., 26, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skoglund P., Thompson J.C., Prendergast M.E., Mittnik A., Sirak K., Hajdinjak M., Salie T., Rohland N., Mallick S., Peltzer A.. et al. (2017) Reconstructing prehistoric African population structure. Cell, 171, 59–71.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pagani L., Schiffels S., Gurdasani D., Danecek P., Scally A., Chen Y., Xue Y., Haber M., Ekong R., Oljira T.. et al. (2015) Tracing the route of modern humans out of Africa by using 225 human genome sequences from Ethiopians and Egyptians. Am. J. Hum. Genet., 96, 986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laso-Jadart R., Harmant C., Quach H., Zidane N., Tyler-Smith C., Mehdi Q., Ayub Q., Quintana-Murci L., Patin E. (2017) The genetic legacy of the Indian Ocean slave trade: recent admixture and post-admixture selection in the Makranis of Pakistan. Am. J. Hum. Genet., 101, 977–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schroeder H., Ávila-Arcos M.C., Malaspinas A.-S., Poznik G.D., Sandoval-Velasco M., Carpenter M.L., Moreno-Mayar J.V., Sikora M., Johnson P.L.F., Allentoft M.E.. et al. (2015) Genome-wide ancestry of 17th-century enslaved Africans from the Caribbean. Proc. Natl. Acad. Sci. U.S.A., 112, 201421784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fortes-Lima C., Gessain A., Ruiz-Linares A., Bortolini M.C., Migot-Nabias F., Bellis G., Moreno-Mayar J.V., Restrepo B.N., Rojas W., Avendaño-Tamayo E.. et al. (2017) Genome-wide ancestry and demographic history of African-descendant Maroon Communities from French Guiana and Suriname. Am. J. Hum. Genet., 101, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rotimi C.N., Tekola-Ayele F., Baker J.L., Shriner D. (2016) The African diaspora: history, adaptation and health. Curr. Opin. Genet. Dev., 41, 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vitti J.J., Grossman S.R., Sabeti P.C. (2013) Detecting natural selection in genomic data. Annu. Rev. Genet., 47, 97–120. [DOI] [PubMed] [Google Scholar]
- 19. Crawford N.G., Kelly D.E., Hansen M.E.B., Beltrame M.H., Fan S., Bowman S.L., Jewett E., Ranciaro A., Thompson S., Lo Y.. et al. (2017) Loci associated with skin pigmentation identified in African populations. Science, 358, eaan8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sugden L.A., Atkinson E.G., Fischer A.P., Rong S., Henn B.M., Ramachandran S. (2018) Localization of adaptive variants in human genomes using averaged one-dependence estimation. Nat. Commun., 9, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leffler E.M., Band G., Busby G.B.J., Kivinen K., Le Q.S., Clarke G.M., Bojang K.A., Conway D.J., Jallow M., Sisay-Joof F.. et al. (2017) Resistance to malaria through structural variation of red blood cell invasion receptors. Science, 356, eaam6393.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Andersen K.G., Shylakhter I., Tabrizi S., Grossman S.R., Happi C.T., Sabeti P.C. (2012) Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Philos. Trans. R. Soc. Lond. B. Biol. Sci., 367, 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Veeramah K.R., Wegmann D., Woerner A., Mendez F.L., Watkins J.C., Destro-Bisol G., Soodyall H., Louie L., Hammer M.F. (2012) An early divergence of Khoesan ancestors from those of other modern humans is supported by an ABC-based analysis of autosomal resequencing data. Mol. Biol. Evol., 29, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patin E., Laval G., Barreiro L.B., Salas A., Semino O., Santachiara-Benerecetti S., Kidd K.K., Kidd J.R., Der Veen L., Van Hombert J.M.. et al. (2009) Inferring the demographic history of African farmers and Pygmy hunter-gatherers using a multilocus resequencing data set. PLoS Genet., 5, e1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Verdu P., Austerlitz F., Estoup A., Vitalis R., Georges M., Théry S., Froment A., Le Bomin S., Gessain A., Hombert J.M.. et al. (2009) Origins and genetic diversity of pygmy hunter-gatherers from western central Africa. Curr. Biol., 19, 312–318. [DOI] [PubMed] [Google Scholar]
- 26. Batini C., Lopes J., Behar D.M., Calafell F., Jorde L.B., Van Der Veen L., Quintana-Murci L., Spedini G., Destro-Bisol G., Comas D. (2011) Insights into the demographic history of African pygmies from complete mitochondrial genomes. Mol. Biol. Evol., 28, 1099–1110. [DOI] [PubMed] [Google Scholar]
- 27. Pickrell J.K., Patterson N., Barbieri C., Berthold F., Gerlach L., Güldemann T., Kure B., Mpoloka S.W., Nakagawa H., Naumann C.. et al. (2012) The genetic prehistory of southern Africa. Nat. Commun., 3, 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shriner D., Tekola-Ayele F., Adeyemo A., Rotimi C.N. (2014) Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry. Sci. Rep., 4, 6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gebremeskel E.I., Ibrahim M.E. (2014) Y-chromosome E haplogroups: their distribution and implication to the origin of Afro-Asiatic languages and pastoralism. Eur. J. Hum. Genet., 22, 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dobon B., Hassan H.Y., Laayouni H., Luisi P., Ricaño-Ponce I., Zhernakova A., Wijmenga C., Tahir H., Comas D., Netea M.G.. et al. (2015) The genetics of East African populations: a Nilo-Saharan component in the African genetic landscape. Sci. Rep., 5, 9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hollfelder N., Schlebusch C.M., Günther T., Babiker H., Hassan H.Y., Jakobsson M. (2017) Northeast African genomic variation shaped by the continuity of indigenous groups and Eurasian migrations. PLoS Genet., 13, e1006976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scally A., Durbin R. (2012) Revising the human mutation rate: implications for understanding human evolution. Nat. Rev. Genet., 13, 745–753. [DOI] [PubMed] [Google Scholar]
- 33. D’Atanasio E., Trombetta B., Bonito M., Finocchio A., Di Vito G., Seghizzi M., Romano R., Russo G., Paganotti G.M., Watson E.. et al. (2018) The peopling of the last Green Sahara revealed by high-coverage resequencing of trans-Saharan patrilineages. Genome Biol., 19, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lachance J., Vernot B., Elbers C.C., Ferwerda B., Froment A., Bodo J.M., Lema G., Fu W., Nyambo T.B., Rebbeck T.R.. et al. (2012) Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell, 150, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gomez F., Hirbo J., Tishkoff S.A. (2014) Genetic variation and adaptation in Africa: implications for human evolution and disease. Cold Spring Harb. Perspect. Biol., 6, a008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pickrell J.K., Patterson N., Loh P.-R., Lipson M., Berger B., Stoneking M., Pakendorf B., Reich D. (2014) Ancient west Eurasian ancestry in southern and eastern Africa. Proc. Natl. Acad. Sci. U.S.A., 111, 2632–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Breton G., Schlebusch C.M., Lombard M., Sjödin P., Soodyall H., Jakobsson M. (2014) Lactase persistence alleles reveal partial east African ancestry of southern African Khoe pastoralists. Curr. Biol., 24, 852–858. [DOI] [PubMed] [Google Scholar]
- 38. Macholdt E., Slatkin M., Pakendorf B., Stoneking M. (2015) New insights into the history of the C-14010 lactase persistence variant in Eastern and Southern Africa. Am. J. Phys. Anthropol., 156, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Macholdt E., Lede V., Barbieri C., Mpoloka S.W., Chen H., Slatkin M., Pakendorf B., Stoneking M. (2014) Tracing pastoralist migrations to southern Africa with lactase persistence alleles. Curr. Biol., 24, 875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Robertshaw P.T., Phillipson D.W. (1985) African Archaeology, 3rd edn. Cambridge University Press, Cambridge. [Google Scholar]
- 41. de Filippo C., Bostoen K., Stoneking M., Pakendorf B. (2012) Bringing together linguistic and genetic evidence to test the Bantu expansion. Proc. R. Soc. B Biol. Sci., 279, 3256–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Choudhury A., Ramsay M., Hazelhurst S., Aron S., Bardien S., Botha G., Chimusa E.R., Christoffels A., Gamieldien J., Sefid-Dashti M.J.. et al. (2017) Whole-genome sequencing for an enhanced understanding of genetic variation among South Africans. Nat. Commun., 8, 2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pagani L., Kivisild T., Tarekegn A., Ekong R., Plaster C., Gallego Romero I., Ayub Q., Mehdi S.Q., Thomas M.G., Luiselli D.. et al. (2012) Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. Am. J. Hum. Genet., 91, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brucato N., Kusuma P., Beaujard P., Sudoyo H., Cox M.P., Ricaut F.-X. (2017) Genomic admixture tracks pulses of economic activity over 2,000 years in the Indian Ocean trading network. 7, 5–10. [DOI] [PMC free article] [PubMed]
- 45. Austen R.A. (1992) The Mediterranean Islamic slave trade out of Africa: a tentative census. Slavery Abol., 13, 214–248. [Google Scholar]
- 46. Mathias R.A., Taub M.A., Gignoux C.R., Fu W., Musharoff S., Connor T.D.O., Vergara C., Torgerson D.G., Pino-yanes M., Shringarpure S.S.. et al. (2016) A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat. Commun., 7, 12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Adhikari K., Mendoza-Revilla J., Chacón-Duque J.C., Fuentes-Guajardo M., Ruiz-Linares A. (2016) Admixture in Latin America. Curr. Opin. Genet. Dev., 41, 106–114. [DOI] [PubMed] [Google Scholar]
- 48. Eltis D., Engerman S.L. (1993) Fluctuations in sex and age ratios in the transatlantic slave trade, 1663–1864. Econ. Hist. Rev., 46, 308–323. [Google Scholar]
- 49. Conley A.B., Rishishwar L., Norris E.T., Valderrama-Aguirre A., Mariño-Ramírez L., Medina-Rivas M.A., Jordan I.K. (2017) A comparative analysis of genetic ancestry and admixture in the Colombian populations of Chocó and Medellín. G3 Genes Genomes Genet., 7, 3435–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fuller H., Torres J.B. (2018) Investigating the “Taíno” ancestry of the Jamaican Maroons: a new genetic (DNA), historical, and multidisciplinary analysis and case study of the Accompong Town Maroons. Can. J. Lat. Am. Caribb. Stud., 43, 47–78. [Google Scholar]
- 51. Pardo-Seco J., Heinz T., Taboada-Echalar P., Martinón-Torres F., Salas A. (2016) Mapping the genomic mosaic of two ‘Afro-Bolivians’ from the isolated Yungas valleys. BMC Genomics, 17, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Santangelo R., González-Andrade F., Børsting C., Torroni A., Pereira V., Morling N. (2017) Analysis of ancestry informative markers in three main ethnic groups from Ecuador supports a trihybrid origin of Ecuadorians. Forensic Sci. Int. Genet., 31, 29–33. [DOI] [PubMed] [Google Scholar]
- 53. Arauna L.R., Mendoza-Revilla J., Mas-Sandoval A., Izaabel H., Bekada A., Benhamamouch S., Fadhlaoui-Zid K., Zalloua P., Hellenthal G., Comas D. (2017) Recent historical migrations have shaped the gene pool of Arabs and Berbers in North Africa. Mol. Biol. Evol., 34, 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Font-Porterias N., Solé-Morata N., Serra-Vidal G., Bekada A., Fadhlaoui-Zid K., Zalloua P., Calafell F., Comas D. (2018) The genetic landscape of Mediterranean North African populations through complete mtDNA sequences. Ann. Hum. Biol., 45, 98–104. [DOI] [PubMed] [Google Scholar]
- 55. Soodyall H. (2013) Lemba origins revisited : tracing the ancestry of Y chromosomes in South African and Zimbabwean Lemba. South African Med. J., 103, 1009–1013. [DOI] [PubMed] [Google Scholar]
- 56. Pierron D., Heiske M., Razafindrazaka H., Rakoto I., Rabetokotany N., Ravololomanga B., Rakotozafy L.M.-A., Rakotomalala M.M., Razafiarivony M., Rasoarifetra B.. et al. (2017) Genomic landscape of human diversity across Madagascar. Proc. Natl. Acad. Sci. U.S.A., 114, E6498–E6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chimusa E.R., Daya M., Möller M., Ramesar R., Henn B.M., van Helden P.D., Mulder N.J., Hoal E.G. (2013) Determining ancestry proportions in complex admixture scenarios in South Africa using a novel proxy ancestry selection method. PLoS One, 8, e73971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Uren C., Möller M., van Helden P.D., Henn B.M., Hoal E.G. (2017) Population structure and infectious disease risk in southern Africa. Mol. Genet. Genomics, 292, 499–411. [DOI] [PubMed] [Google Scholar]
- 59. Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., Flicek P., Gabriel S.B.. et al. (2012) An integrated map of genetic variation from 1, 092 human genomes. Nature, 6, 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E., Flicek P.. et al. (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Howie B., Fuchsberger C., Stephens M., Marchini J., Abecasis G.R. (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet., 44, 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J. (2017) 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet., 101, 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang P., Zhan X., Rosenberg N.A., Zöllner S. (2013) Genotype imputation reference panel selection using maximal phylogenetic diversity. Genetics, 195, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Roshyara N.R., Horn K., Kirsten H., Ahnert P., Scholz M. (2016) Comparing performance of modern genotype imputation methods in different ethnicities. Sci. Rep., 6, 34386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chou W.C., Zheng H.F., Cheng C.H., Yan H., Wang L., Han F., Richards J.B., Karasik D., Kiel D.P., Hsu Y.H. (2016) A combined reference panel from the 1000 Genomes and UK10K projects improved rare variant imputation in European and Chinese samples. Sci. Rep., 6, 39313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McCarthy S., Das S., Kretzschmar W., Delaneau O., Wood A.R., Teumer A., Kang H.M., Fuchsberger C., Danecek P., Sharp K.. et al. (2016) A reference panel of 64, 976 haplotypes for genotype imputation. Nat. Genet., 48, 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hindorff L.A., Bonham V.L., Brody L.C., Ginoza M.E.C., Hutter C.M., Manolio T.A., Green E.D. (2018) Prioritizing diversity in human genomics research. Nat. Rev. Genet., 19, 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rotimi C., Abayomi A., Abimiku A., Adabayeri V.M., Adebamowo C., Adebiyi E., Ademola A.D., Adeyemo A., Adu D., Affolabi D.. et al. (2014) Research capacity. Enabling the genomic revolution in Africa. Science, 344, 1346–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Allison A.C. (1954) Protection afforded by sickle-cell trait against subtertian malareal infection. Br. Med. J., 1, 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pasvol G., Weatherall D.J., Wilson R.J.M. (1978) Cellular mechanism for the protective effect of haemoglobin S against P. falciparum malaria. Nature, 274, 701–703. [DOI] [PubMed] [Google Scholar]
- 71. Aidoo M., Terlouw D.J., Kolczak M.S., McElroy P.D., ter Kuile F.O., Kariuki S., Nahlen B.L., Lal A.A., Udhayakumar V. (2002) Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet, 359, 1311–1312. [DOI] [PubMed] [Google Scholar]
- 72. Bitoungui V.J.N., Pule G.D., Hanchard N., Ngogang J., Wonkam A. (2015) Beta-globin gene haplotypes among Cameroonians and review of the global distribution: is there a case for a single sickle mutation origin in Africa? OMICS, 19, 171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hamblin M.T., Di Rienzo A. (2000) Detection of the signature of natural selection in humans: evidence from the Duffy Blood Group Locus. Am. J. Hum. Genet., 66, 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Howes R.E., Patil A.P., Piel F.B., Nyangiri O.A., Kabaria C.W., Gething P.W., Zimmerman P.A., Barnadas C., Beall C.M., Gebremedhin A.. et al. (2011) The global distribution of the Duffy blood group. Nat. Commun., 2, 266.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ko W.-Y., Rajan P., Gomez F., Scheinfeldt L., An P., Winkler C.A., Froment A., Nyambo T.B., Omar S.A., Wambebe C.. et al. (2013) Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am. J. Hum. Genet., 93, 54–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Grossman S.R., Shylakhter I., Karlsson E.K., Byrne E.H., Morales S., Frieden G., Hostetter E., Angelino E., Garber M., Zuk O.. et al. (2010) A composite of multiple signals distinguishes causal variants in regions of positive selection. Science, 327, 883–886. [DOI] [PubMed] [Google Scholar]
- 77. Ranciaro A., Campbell M.C., Hirbo J.B., Ko W.Y., Froment A., Anagnostou P., Kotze M.J., Ibrahim M., Nyambo T., Omar S.A.. et al. (2014) Genetic origins of lactase persistence and the spread of pastoralism in africa. Am. J. Hum. Genet., 94, 496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liebert A., López S., Jones B.L., Montalva N., Gerbault P., Lau W., Thomas M.G., Bradman N., Maniatis N., Swallow D.M. (2017) World-wide distributions of lactase persistence alleles and the complex effects of recombination and selection. Hum. Genet., 136, 1445–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jarvis J.P., Scheinfeldt L.B., Soi S., Lambert C., Omberg L., Ferwerda B., Froment A., Bodo J.M., Beggs W., Hoffman G.. et al. (2012) Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African pygmies. PLoS Genet., 8, e1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Perry G.H., Foll M., Grenier J.-C., Patin E., Nédélec Y., Pacis A., Barakatt M., Gravel S., Zhou X., Nsobya S.L.. et al. (2014) Adaptive, convergent origins of the pygmy phenotype in African rainforest hunter-gatherers. Proc. Natl. Acad. Sci. U.S.A., 111, E3596–E3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Busby G., Christ R., Band G., Leffler E., Le Q.S., Rockett K., Kwiatkowski D., Spencer C. (2017) Inferring adaptive gene-flow in recent African history. bioRxiv, 10.1101/205252.
- 82. Basu Mallick C., Iliescu F.M., Möls M., Hill S., Tamang R., Chaubey G., Goto R., Ho S.Y.W., Gallego Romero I., Crivellaro F.. et al. (2013) The light skin allele of SLC24A5 in South Asians and Europeans shares identity by descent. PLoS Genet., 9, e1003912.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Martin A.R., Lin M., Granka J.M., Myrick J.W., Liu X., Sockell A., Atkinson E.G., Werely C.J., Möller M., Sandhu M.S.. et al. (2017) An unexpectedly complex architecture for skin pigmentation in Africans. Cell, 171, 1340–1353.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Morris A.G. (2017) Ancient DNA comes of age, but still has some teenage problems. S. Afr. J. Sci., 113, 9–10. [Google Scholar]