Abstract
FLT3 internal tandem duplication (FLT3ITD) are common mutations in acute myeloid leukemia (AML) associated with poor patient prognosis. Although new generation FLT3 tyrosine kinase inhibitors (TKI) have shown promising results, the outcome of FLT3ITD AML patients remains poor and demands the identification of novel, specific and validated therapeutic targets for this highly aggressive AML subtype. Utilizing an unbiased genome-wide CRISPR/Cas9 screen, we identify GLS, the first enzyme in glutamine metabolism, as synthetically lethal with FLT3-TKI treatment. Using complementary metabolomic and gene-expression analysis, we demonstrate that glutamine metabolism, through its ability to support both mitochondrial function and cellular redox metabolism, becomes a metabolic dependency of FLT3ITD AML, specifically unmasked by FLT3-TKI treatment. We extend these findings to AML subtypes driven by other tyrosine kinase (TK) activating mutations, and validate the role of GLS as a clinically actionable therapeutic target in both primary AML and in vivo models. Our work highlights the role of metabolic adaptations as a resistance mechanism to several TKI, and suggests glutaminolysis as a therapeutically targetable vulnerability when combined with specific TKI in FLT3ITD and other TK activating mutation driven leukemias.
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous disease at both the molecular and clinical level. Recent sequencing efforts have helped to categorize different subtypes based on their mutation profile and its putative effect on AML pathogenesis. Common subgroups include those carrying mutations in transcription factors and epigenetic regulators, cases carrying mutations in genes encoding for components of the spliceosome machinery and cohesin complexes, and those carrying mutations in signaling genes1,2. Within the last group, activating mutations of tyrosine kinases (TK) are the most frequent and generally predict for a poor outcome3. In particular, mutations in the type-III receptor TK FLT3 are present in about 30% of AML patients, are mostly secondary to an internal tandem duplication (FLT3ITD) of the juxtamembrane domain and predict for an increased relapse rate following standard therapies and a poor prognosis4. Although FLT3ITD mutations are acquired relatively late in leukemia evolution1,5 and are unable to produce an AML phenotype in animal models without collaborating mutations6, they are capable of conferring a state of oncogene addiction by activating survival pathways7. Their importance for the maintenance of the leukemic phenotype and as a relevant therapeutic target has also been confirmed by the results of a recent phase 3 randomized study (RATIFY), where a survival benefit for patients treated with FLT3 TK inhibitor (TKI) was demonstrated for the first time8, leading to recent FDA approval of the FLT3 inhibitor Midostaurin. However, despite our understanding of the role played by FLT3ITD mutations in AML and the rational design of targeted inhibitors of their TK activity, the overall outcome of AML patients carrying FLT3ITD mutations remains poor, suggesting that resistance mechanisms to targeted inhibitors might hinder the efficacy of these therapies9. Indeed mutations in the FLT3 TK domain have already been described as a frequent mechanism of resistance7. However, more recently, mutational analysis of patient samples obtained following relapse after FLT3-TKI treatment and a handful of preclinical studies have suggested that cellular adaptive mechanism might also play a role in FLT3-TKI resistance10–13 although these remain overall poorly defined.
FLT3ITD mutations are known to activate survival/proliferation signaling pathways, including the PI3-kinase/AKT, Ras/MAP kinase and JAK/STAT pathways14–17 that are also known to directly or indirectly alter cell metabolism18–20. As a result, leukemias harboring FLT3ITD mutations are often associated with a very proliferative and aggressive phenotype, high tumor bulk, and are accompanied by alterations in cellular metabolism to sustain this proliferative phenotype4,21.
Metabolic reprogramming has emerged as a hallmark of transformed cells22 and several reports have recently highlighted the role of specific metabolic enzymes and metabolites in normal hematopoietic stem cell homeostasis and leukemogenesis through both direct effects on energy production, macromolecule biosynthesis, and their ability to modulate redox balance, epigenetic regulation, and signaling pathways23–29. Moreover, metabolism is able to rapidly respond to changing conditions within a cell, and it has already been shown, in both solid cancers and hematological malignancies, that metabolic adaptations, under therapeutic selective pressure, can act as key resistance mechanisms to standard therapeutics30,31.
In this work, we aimed to identify novel cellular adaptive resistance mechanisms to FLT3-TKI treatment in FLT3ITD AML. Using several unbiased complementary approaches, we identify glutamine metabolism as a protective and adaptive response to FLT3-TKI, and describe the mechanisms underlying this phenotype. Finally, we validate glutaminolysis as a clinically actionable therapeutic vulnerability in both FLT3ITD and other AML subtypes carrying TK activating mutations, following TKI treatment.
Methods
An extended methods section is available in the online supplemental Data.
Cell culture
MV411, MOLM13, THP1, K562 were cultured in RPMI1640 (Sigma) supplemented with 10% dialyzed fetal bovine serum (FBS) (Sigma) and 1% penicillin/streptomycin/glutamine.
Lineage depleted bone marrow cells from Rosa26Cas9/+, FLT3ITD/+ mice were transduced with retrovirus constructs pMSCV-MLL-AF9-IRES-YFP, pMSCV-MLL-AF4-PGK-puro and pMSCV-MLL-ENL-IRES-Neo and cultured in X-VIVO 20 (Lonza) supplemented with 10ng ml-1 IL3, 10ng ml-1 IL6 and 50ng ml-1 of SCF (Peprotech).
Generation of genome-wide mutant libraries, CRISPR screening and gRNA competition assays
CRISPR screens were performed using the previously reported WT Sanger genome-wide CRISPR library32. gRNA competition assays were performed using single and dual gRNA vectors as described previously32. The gRNA sequences are listed in supplemental Methods. Details are provided in supplemental Methods.
Liquid chromatography coupled to mass spectrometry (LC-MS) for metabolomics analysis
MV411 cells were plated at 0.5 x 106 cells per mL in media supplemented with uniformly-labelled-13Carbon (U-13C6) glucose (11 mM) or uniformly-labelled-13Carbon, 15Nitrogen (U-13C5,15N2) glutamine (2 mM) (Cambridge Isotope laboratories) for 48 h before sampling. Details of metabolite extraction and LC-MS analysis are provided in supplemental Methods.
Adult Primary Leukaemia and Cord Blood Samples Drug and Proliferation Assays
Human AML MNC were obtained from bone marrow or peripheral blood of patients. Normal CD34 samples were obtained from leukapheresis products of myeloma/lymphoma patients in bone marrow remission. Informed consent was obtained in accordance with the Declaration of Helsinki and the study was conducted under local ethical approval (REC 07-MRE05-44). Culture condition for methylcellulose and liquid culture assay of primary samples are as previously described33.
In vivo experiments
MV411 cells transduced with control “Scramble” shRNA or GLS shRNA were transplanted (3x106) into sublethally irradiated (2 Gy) 8-12 weeks old NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) male mice, via tail vein injection. Three days following transplant, mice were fed a doxycycline diet (1g/kg) to induce the shRNA and following disease dissemination treatment was started. Mice were treated by gavage either with vehicle (22% hydroxypropyl-β-cyclodextrin/0.3% DMSO) or AC220 at 1 mg/kg daily for 8 days and then 0.1mg/kg till they succumbed to disease. Survival was measured as the time from transplantation until the point at which mice had to be humanely culled due to overt clinical symptoms typical of the MV411 xenotransplant model34.
Results
A genome-wide CRISPR/Cas9 screen identifies GLS as a synthetic lethal gene in TKI treated FLT3ITD cells
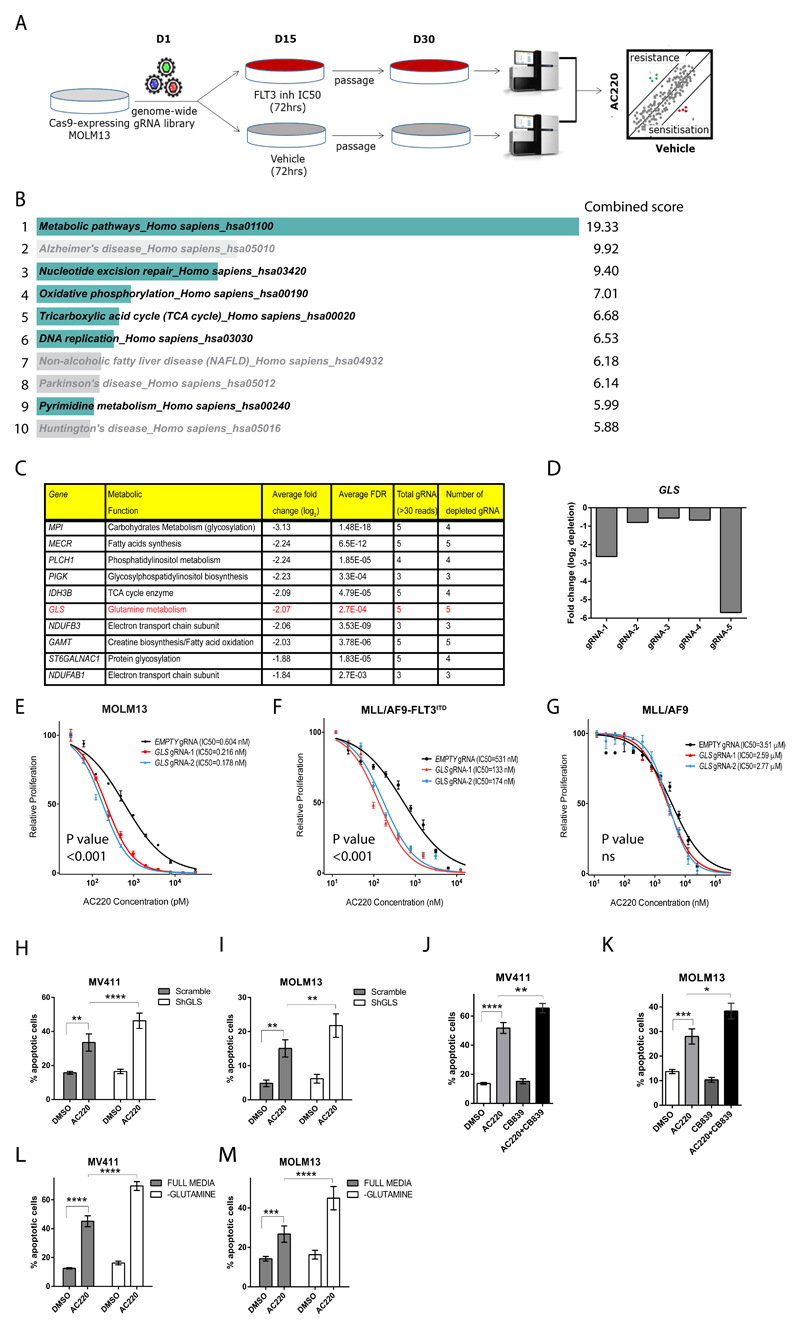
In order to identify genes and pathways that would sensitize FLT3ITD AML to FLT3-TKI treatment in an unbiased manner, we performed a genome-wide CRISPR/Cas9 synthetic lethality screen in the FLT3ITD cell line MOLM13 during treatment with the highly potent and specific FLT3ITD inhibitor AC220 (quizartinib), that is currently being assessed in phase 3 clinical trials34 or vehicle control (Figure 1A). A total of 304 genes dropped out following AC220 treatment (defined as genes showing a drop out of ≤ 0.5 log2 fold change in at least 80% of gRNA at false discovery rate <0.01) (Supplemental Table 1). KEGG gene set enrichment analysis, using Enrichr software35,36, demonstrated significant enrichment for genes involved in several pathways, including some obviously relevant in the AML biology (highlighted in the figure). Amongst these, metabolic pathways, including mostly genes involved in oxidative phosphorylation and the tricarboxylic acid (TCA) cycle were significantly enriched (Figure 1B). Amongst the top metabolic genes depleted following AC220 treatment, glutaminase (GLS) demonstrated the highest number of significantly depleted gRNA (5 out of all 5 gRNA targeting the gene), indicating a strong synthetic lethal interaction with AC220 (Figure 1C-D). GLS is the first enzyme in glutamine catabolism, a metabolic pathway with well-established anaplerotic and biosynthetic roles in cancer cells37 which also regulates the availability of substrates for both the TCA cycle and oxidative phosphorylation, two of the most affected pathways in our screen. Importantly, a potent and selective GLS inhibitor CB839 is currently being investigated in clinical trials38. Given the strong synthetic lethal interaction in the screen, its central role in regulating metabolic pathways shown to be affected in our screen and the availability of a clinical grade inhibitor, we decided to further investigate the role of GLS as a clinically relevant synthetic lethal pair in AC220-treated FLT3ITD cells.
Figure 1. GLS gene deletion and chemical inhibition are synthetically lethal with FLT3 tyrosine kinase inhibitors.
(A) Schematic of the genome-wide CRISPR/Cas9 synthetic lethality screen in the FLT3ITD mutant cell line MOLM13. (B) KEGG pathways enrichment analysis of drop-out genes from CRISPR/Cas9 screen sorted by combined score calculated using Enrichr software35,36. Pathways relevant to AML biology are highlighted while the remaining are grayed out. (C) List of top ten genes from the “metabolic pathways” gene list sorted according to average fold depletion from gRNAs. (D) Bar graph depicting individual fold depletion for each gRNA targeting GLS. (E-G) Growth inhibition curves to AC220 of MOLM13 (E) and murine bone marrow cells expressing MLL/AF9-FLT3ITD (F) and MLL/AF9 (G) transduced respectively with “empty” gRNA control or 2 different gRNA targeting GLS (mean ± s.e.m., n=3, P<0.001 for treatment effect comparing control and both Gls knockout for E and F, ns, not significant for G, two-way ANOVA). (H-I) Apoptosis in the FLT3ITD mutant cell lines MV411 (H) and MOLM13 (I) transduced with control “Scramble” shRNA and GLS shRNA following treatment with AC220 0.5nM for 48hrs (for MV411 mean ± s.e.m., n=7, **** P<0.0001, ** P=0.0086, for MOLM13 mean ± s.e.m., n=3, ** P=0.0014 for AC220 treatments comparison between Scramble and GLS shRNA, ** P=0.0050 for DMSO versus AC220 comparison in the scramble shRNA, two-way ANOVA with Bonferroni’s multiple comparisons). (J-K) Apoptosis in MV411 (J) and MOLM13 (K) following treatment with AC220 1nM, CB839 100nM or their combination (for MV411 mean ± s.e.m., n=14, ** P=0.0033, **** P<0.0001, for MOLM13 mean ± s.e.m., n=16, * P=0.0126,*** P=0.0003, ANOVA with Tukey’s multiple comparisons). (L-M) Apoptosis in MV411 (L) and MOLM13 (M) grown in the presence (full media) or absence of glutamine following treatment with AC220 1nM for 48hrs (for MV411 mean ± s.e.m., n=24, for MOLM13 mean ± s.e.m., n=11, **** P<0.0001, *** P=0.0007, two-way ANOVA with Bonferroni’s multiple comparisons). (s.e.m., standard error of mean).
In single targeting experiments, GLS was validated as synthetically lethal with AC220 in human and murine FLT3ITD mutant, but not in wild-type FLT3 (FLT3wt) cells (Figure 1E-G and Supplemental Figure 1A-C). Moreover, the genetic ablation of GLS was non-toxic in untreated FLT3ITD cells (Supplemental Figure 1D-F). We then confirmed that the silencing of GLS by short hairpin RNA (shRNA) and its chemical inhibition using the specific clinical grade inhibitor CB839 at concentrations shown to be inhibiting GLS enzymatic activity in a specific fashion38, produced similar effects on cell proliferation when combined with AC220 in FLT3ITD-mutant cells, and the combination treatment induced higher levels of apoptosis compared to AC220 treatment alone in FLT3ITD, but not FLT3wt cells (Figure 1H-K and Supplemental Figure 1G-K). In line with these findings, glutamine starvation sensitized FLT3ITD cells to AC220, while having negligible effects in untreated cells (Figure 1L-M). Taken together, these data demonstrate that glutamine metabolism represents a metabolic dependency in FLT3ITD cells that is only unmasked by FLT3-TKI, making genetic and chemical inhibition of GLS a feasible strategy to sensitize these cells to AC220.
FLT3 tyrosine kinase inhibition markedly reduces glycolysis without affecting glutamine uptake in FLT3ITD cells
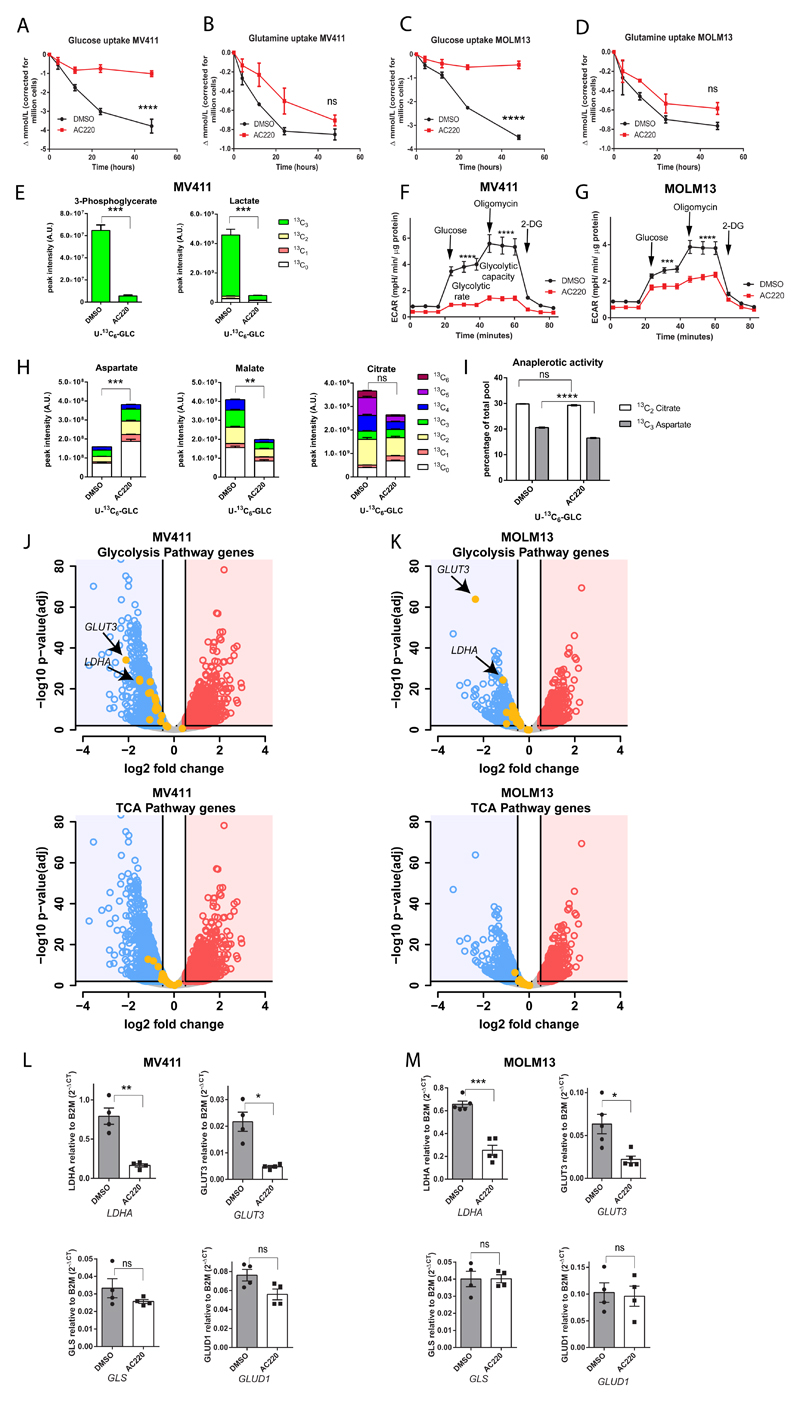
Previous studies have demonstrated that cells carrying FLT3ITD display a highly glycolytic phenotype and enhanced central carbon metabolism21. Indeed, gene set enrichment analysis (GSEA)39,40 of published gene expression datasets of untreated AML patients at diagnosis1,41,42, demonstrate that signatures involving glucose metabolism, TCA cycle, and electron transport chain (ETC) are consistently upregulated in FLT3ITD compared to FLT3wt samples across all datasets analyzed (Supplemental Figure 2A-D). Furthermore, murine bone marrow (BM) cells carrying FLT3ITD demonstrate both increased glycolytic activity/capacity and oxygen consumption compared to their FLT3wt counterpart (Supplemental Figure 2E-F). Considering that glucose and glutamine are the main fuels for central carbon metabolism in cultured cells37,43, we investigated the effects of FLT3-TKI on the utilization of these nutrients and on central carbon metabolism. Dynamic measurement of the concentration of glucose and glutamine in FLT3ITD cell-conditioned medium confirmed that, whilst glucose uptake was almost completely blocked during treatment with AC220, glutamine uptake was only modestly reduced and by 48 hours it was not significantly different between treated and untreated cells (Figure 2A-D). Liquid chromatography-mass spectrometry (LC-MS) analysis using U-13C6-glucose confirmed a marked reduction in glucose labelling of glycolytic intermediates/products upon FLT3 TK inhibition in FLT3ITD mutant cells (Figure 2E and Supplemental Table 2). The effects of AC220 on glycolysis were further confirmed by time-resolved metabolic profiling (Figure 2F-G). As might be expected based on the profound antiproliferative effects of AC220 in the same cells (Supplemental Figure 3A-B), a reduction in total levels of most TCA cycle intermediates was also observed but this was less pronounced and the presence of 13C2 citrate and 13C3 aspartate isotopologues – products, respectively, of the activity of the anaplerotic enzymes pyruvate dehydrogenase (PDH) and pyruvate carboxylase (PC) - suggests that anaplerotic oxidative metabolism is still active in these cells (Figure 2H-I and Supplemental Table 2). Moreover, the preservation of the unlabeled (13C0) and partially labelled (13C2) fractions of TCA cycle intermediates also suggests that alternative carbon sources, such as glutamine, were utilized by the cells to support the production of TCA cycle metabolites following AC220 treatment (Figure 2H and Supplemental Table 2). Gene-expression studies, performed prior to induction of significant levels of apoptosis by AC220, confirmed the more pronounced effects of FLT3 inhibition on glycolytic enzymes compared to TCA cycle and anaplerotic genes including glutaminolytic enzymes, GLS and glutamate dehydrogenase 1 (GLUD1) (Figure 2J-M, Supplemental Figure 3C-D and Supplemental Table 3). Overall these data highlight that FLT3 inhibition significantly impairs the utilization of glucose as a carbon source and particularly glycolysis in FLT3ITD cells, whereas glutamine utilization and anaplerotic oxidative metabolism via the TCA cycle were not equally affected.
Figure 2. FLT3 tyrosine kinase inhibition reduces glucose uptake and central carbon metabolism without affecting glutamine uptake.
(A-D) Time-course analysis of glucose and glutamine uptake from media by MV411 (A-B) and MOLM13 (C-D) cells treated with AC220 1 nM or vehicle control (mean ± s.e.m., n=3, **** P<0.0001, two-way ANOVA with Bonferroni’s multiple comparisons). (E) Total and isotopologue abundance of selected glycolytic intermediates and products measured by liquid chromatography-mass spectrometry (LC-MS) analysis in MV411 cell extracts treated with AC220 1nM or vehicle control and grown in media containing uniformly-labelled-13Carbon [U-13C6] glucose (GLC) (mean ± s.e.m., n=5, *** P=0.0004 for total 3-Phosphoglycerate and P=0.0007 for total lactate, two-tailed paired t-test). (F-G) Extracellular acidification rate (ECAR) of MV411 (F) and MOLM13 (G) cells treated with AC220 1nM or vehicle control (mean ± s.e.m., n=3, **** P<0.0001, *** P=0.0003, two-way ANOVA with Bonferroni’s multiple comparisons). (H) Total and isotopologue levels of selected TCA cycle intermediates in MV411 cells treated as in (E) (mean ± s.e.m., n=5, *** P=0.0009,**P=0.0011, ns, not significant, two-tailed paired t-test). (I) Percentage of total levels of citrate and aspartate provided respectively by 13C2 and 13C3 fraction following AC220 treatment as in (E) (mean ± s.e.m., n=5, **** P<0.0001, ns, not significant, two-way ANOVA with Bonferroni’s multiple comparisons). (J-K) Volcano plot for gene expression changes by RNA-sequencing (n=2 for each cell line) of MV411 and MOLM13 cells treated with AC220 1 nM compared to vehicle control highlighting reduced expression of glycolysis genes (top panels) and the minimal effects on TCA cycle genes (bottom panels). (L-M) q-PCR validation in MV411 (L) and MOLM13 (M) for the reduced expression of lactate dehydrogenase, LDHA and glucose transporter, GLUT3 following treatment with AC220 1nM (top panels) (mean ± s.e.m., MV411 n=4, MOLM13 n=5, for LDHA ** P=0.0053, *** P=0.0005, for GLUT3, MV411 * P=0.0150, MOLM13, * P=0.0387, two-tailed paired t-test) and for the lack of changes in expression in glutamine metabolism genes GLS and glutamate dehydrogenase, GLUD1 (bottom panels) (mean ± s.e.m., n=4, ns, not significant by two-tailed paired t-test).
Glutamine supports the TCA cycle and glutathione production following FLT3 inhibition
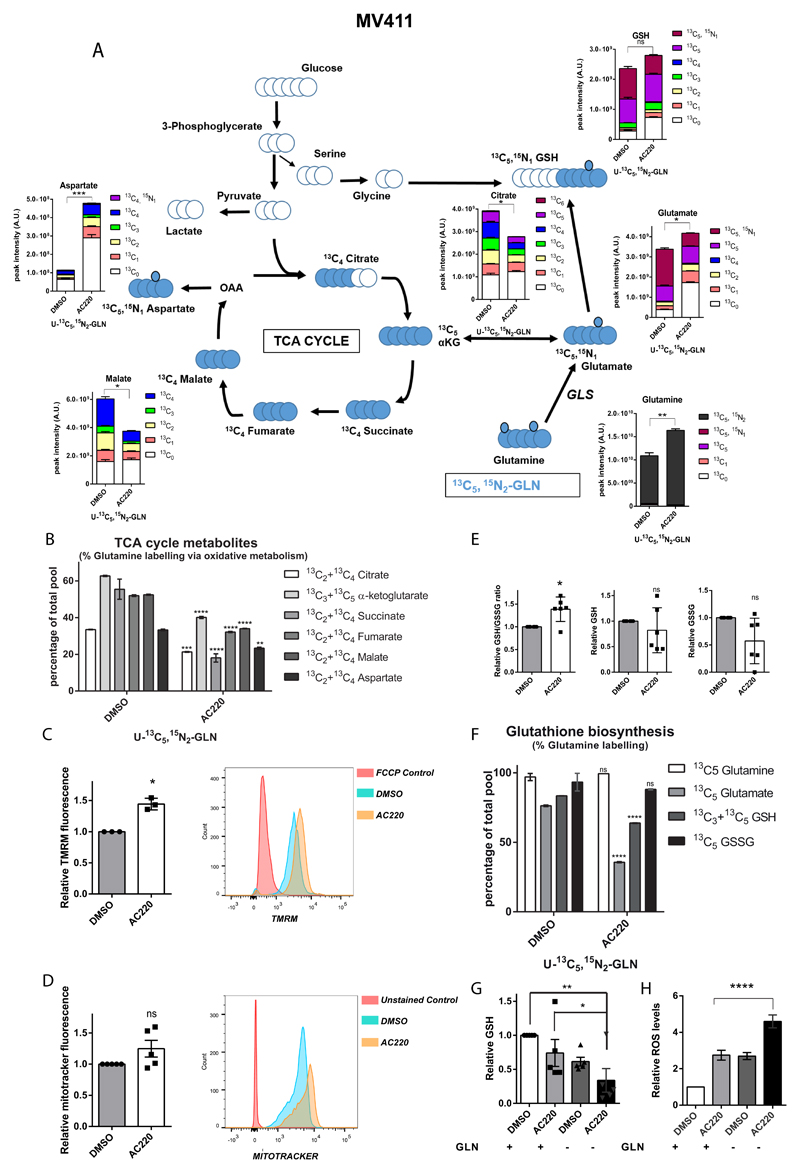
In order to understand the fate of glutamine metabolism in FLT3ITD cells following AC220 treatment, we performed LC-MS analysis upon incubation with stable isotope labelled glutamine (U-13C5,15N2-glutamine). Intracellular levels of labelled glutamine were increased in treated cells, confirming that glutamine uptake was not impaired in these cells but, as expected, given the antiproliferative effects of AC220, incorporation of labelled glutamine in TCA cycle intermediates was reduced and overall the level of most TCA cycle intermediates was decreased compared to vehicle treated cells. However, between 20-40% of the total pool of TCA cycle intermediates was still labelled from glutamine oxidative metabolism in AC220-treated cells compared to 30-60% in vehicle treated cells, suggesting that despite a significant reduction in overall TCA cycle activity, glutamine is still a major anaplerotic substrate in FLT3-TKI treated cells (Figure 3A-B and Supplemental Table 2). Of note, AC220 treated cells were still able to produce aspartate (Figure 3A), a readout of ETC activity44,45, indicating that their respiratory function was not compromised by FLT3-TKI treatment (total levels of aspartate were actually increased, possibly reflecting lack of utilization due to FLT3-TKI antiproliferative effects). Consistent with this hypothesis, AC220-treated cells increased their mitochondrial membrane potential and showed a trend towards increased mitochondrial mass (Figure 3C-D and Supplemental Figure 4A). We did not observe any significant contribution from glutamine reductive metabolism in FLT3ITD mutant cells and this did not change following AC220 treatment (Supplemental Figure 4B)
Figure 3. Glutamine supports both mitochondrial function and glutathione production following FLT3 tyrosine kinase inhibition.
(A) Total and isotopologue levels of TCA cycle intermediates, glutamate and reduced glutathione (GSH) measured by LC-MS analysis in MV411 cells treated with AC220 1 nM or vehicle control and grown in media containing U-13C5 and 15Nitrogen [15N2] glutamine (GLN) (mean ± s.e.m., n=5, *** P=0.0005, ** P=0.0017, * P=0.0195 for citrate, P=0.0228 for malate, P=0.0216 for glutamate, ns, not significant, two-tailed paired t-test). A schematic representation of glutamine metabolism and labelling pattern of metabolites by is also provided. (B) Percentage of total levels of TCA cycle metabolites labelled by U-13C5,15N2-GLN through its oxidative metabolism measured by LC-MS analysis in MV411 cells treated with AC220 1 nM or vehicle control (mean ± s.e.m., n=5, **** P<0.0001, *** P=0.0008, ** P=0.0067, two-way ANOVA with Bonferroni’s multiple comparisons). (C) Relative mitochondrial membrane potential of MV411 cells treated with AC220 1 nM or vehicle control (left panel) with representative flow-cytometry histogram (right panel) (mean ± s.e.m., n=3, * P=0.0145, two-tailed paired t-test). (D) Relative mitochondrial mass of MV411 cells treated with AC220 1 nM or vehicle control (left panel) with representative flow-cytometry histogram (right panel) (mean ± s.e.m., n=3, ns, not significant, P=0.13, two-tailed paired t-test). (E) Relative GSH/GSSG ratio, GSH and GSSG of MV411 cells treated with AC220 1 nM or vehicle control (mean ± s.e.m., n=6, * P=0.0173, ns, not significant, two-tailed paired t-test). (F) Percentage of total levels of glutamine, glutamate, GSH and GSSG labelled by U-13C5,15N2-GLN measured by LC-MS analysis in MV411 cells treated as in (A) (mean ± s.e.m., n=5, **** P<0.0001, ns, not significant, two-way ANOVA with Bonferroni’s multiple comparisons. (G) Relative GSH levels of MV411 cells treated with AC220 1 nM or vehicle control in the presence or absence of glutamine (mean ± s.e.m., n=5, ** P=0.0017, * P=0.0471, q=4.248, DF=12, ANOVA with Tukey’s multiple comparisons). (H) Relative cytoplasmic ROS levels of MV411 cells treated with AC220 1 nM or vehicle control in the presence or absence of glutamine (mean ± s.e.m., n=14, **** P<0.0001, ANOVA with Tukey’s multiple comparisons).
In addition to supporting TCA cycle activity, glutamine, via glutamate, is also a precursor of glutathione, the major cellular anti-oxidant46. Of note, the reduced/oxidized glutathione ratio (GSH/GSSG) was generally preserved in AC220 treated cells (Figure 3E), and glutathione metabolism genes, including the master regulator of antioxidant response NFE2L2, were not affected by AC220 treatment in FLT3ITD mutant cells (Supplemental Figure 4C-D and Supplemental Table 3). As our labelling experiments showed that glutamine largely contributes to glutamate and GSH generation (Figure 3A, F), we hypothesized that glutamine metabolism might play a role in maintaining redox homeostasis in AC220 treated cells. Consistent with this hypothesis, glutamine starvation markedly reduced GSH levels in AC220 treated cells and these effects correlated with a significant increase in intracellular reactive oxygen species (ROS) levels (Figure 3G-H and Supplemental Figure 4E). Overall these data suggest a role for glutamine metabolism in supporting both mitochondrial function and redox homeostasis in FLT3ITD cells under the cellular stress of TK inhibition.
The effects of combined FLT3-TKI and GLS inhibitor treatment can be rescued by the glutamine downstream product α-ketoglutarate
In order to clarify the relative importance of the metabolic pathways supported by glutamine in the survival of FLT3ITD cells upon TKI treatment, we specifically targeted both glutathione metabolism and respiratory function using respectively buthionine sulfoximine (BSO), an inhibitor of GSH synthesis47, or phenformin, an ETC (Complex I) inhibitor 44 in addition to AC220. However, neither of the two combinations was able to fully phenocopy the effects of glutamine starvation or GLS inhibition (Supplemental figure 5A-B). We also failed to completely rescue the effects of glutamine starvation or GLS inhibition using the antioxidant N-acetylcysteine (NAC) or anaplerotic substrates such as pyruvate or aspartate (Supplemental Figure 5C-H). These data support a model whereby both branches of glutamine metabolism, supporting TCA cycle/mitochondrial function and GSH synthesis, are important for continued cell survival and blocking only one of these branches is insufficient to recapitulate the effects of glutamine starvation or GLS inhibition.
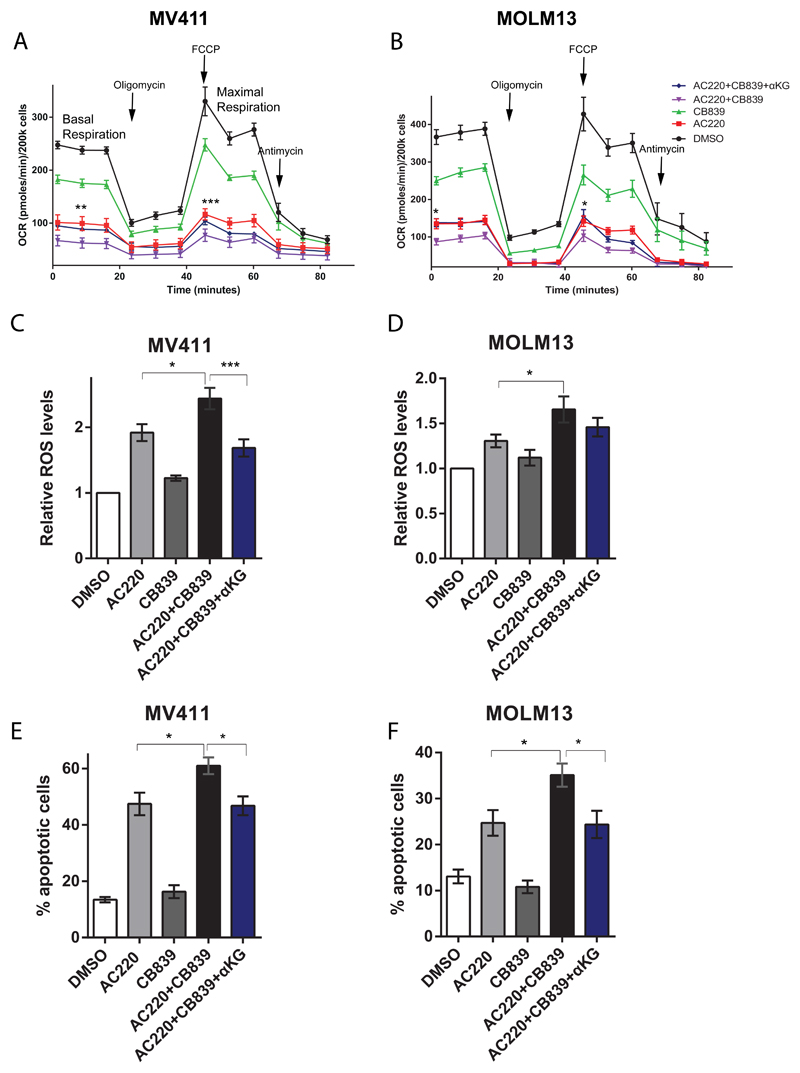
To confirm this hypothesis, we used a cell permeable form of α-ketoglutarate (αKG), a downstream metabolic product of glutamine metabolism, to rescue the effects of combined AC220 and CB839 treatment in FLT3ITD cells. Amongst other functions48, αKG supports both the TCA cycle and glutamate production and can therefore rescue both branches of glutamine metabolism. Moreover, αKG is known to regulate redox homeostasis in cancer cells49. Treatment of FLT3ITD cells with combined AC220 and CB839 resulted in reduced oxygen consumption, during both basal and maximal respiration, and increased intracellular ROS production compared to single agent alone. However, these effects were rescued by concomitant treatment with αKG, in keeping with its anaplerotic and antioxidant properties (Figure 4A-D). The salvage of the metabolic phenotype correlated with a complete rescue of the additional cell death related to the combination treatment by αKG (Figure 4E-F). Overall, these data further confirm the importance of glutamine metabolism in supporting both TCA cycle and redox metabolism in FLT3ITD cells treated with AC220.
Figure 4. Rescue of combined effects of AC220 and CB839 by α-ketoglutarate.
(A-B) Oxygen consumption rate (OCR) in MV411 (A) and MOLM13 (B) cells treated with vehicle control, AC220 1 nM, CB839 100 nM, AC220 and CB839 combination and AC220/CB839 combination + 4 mM of dimethyl-α-ketoglutarate (αKG) measured using a Seahorse analyzer (for MV411 mean ± s.e.m., n=3, for basal respiration ** P=0.0019 between AC220 versus AC220+CB839, and P= 0.018 between AC220+CB839 versus AC220+CB839+αKG, for maximal respiration *** P=0.0002 between AC220 versus AC220+CB839, and P=0.0224 between AC220+CB839 versus AC220+CB839+αKG, two-way ANOVA with Bonferroni’s multiple comparisons; for MOLM13 cells mean ± s.e.m., n=3, for basal respiration * P=0.0298 between AC220 versus AC220+CB839, and P= 0.0182 between AC220+CB839 versus AC220+CB839+αKG, for maximal respiration between * P=0.02 AC220 versus AC220+CB839, and P=0.0148 between AC220+CB839 versus AC220+CB839+αKG, two-way ANOVA with Bonferroni’s multiple comparisons). (C-D) Relative cytoplasmic ROS levels of MV411 (C) and MOLM13 (D) cells treated as in (A-B) (for MV411 mean ± s.e.m., n=5, * P=0.0195, *** P=0.0008; for MOLM13 mean ± s.e.m., n=6, * P=0.0345, ANOVA with Tukey’s multiple comparisons). (E-F) Apoptosis in MV411 (E) and MOLM13 (F) cells treated as in (A-B) (for MV411 mean ± s.e.m., n=10, * P=0.0158 between AC220 versus AC220+CB839 and * P=0.0102 between AC220+CB839 versus AC220+CB839+αKG; for MOLM13 mean ± s.e.m., n=9, * P=0.0105 between AC220 versus AC220+CB839, and * P=0.0120 between AC220+CB839 versus AC220+CB839+αKG, ANOVA with Tukey’s multiple comparisons).
The effects of combined GLS and TKI extend to other TK activating mutations, primary AML samples and in vivo models
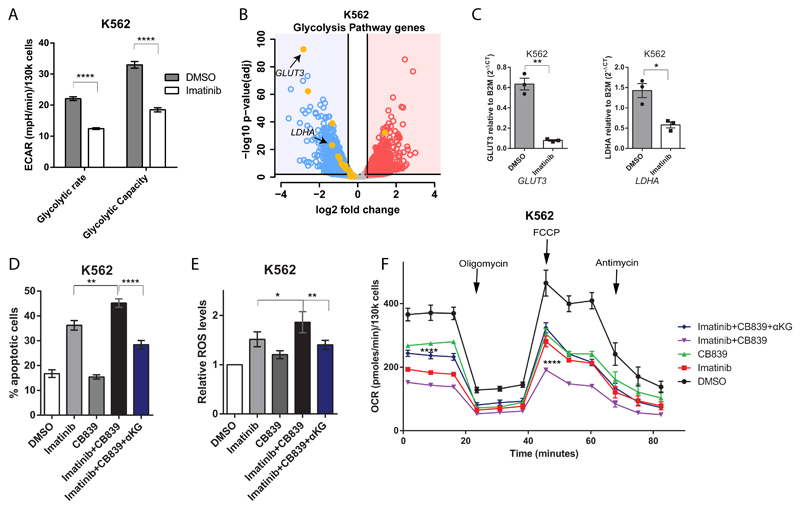
To determine if a similar rewiring of metabolism occurs in leukemia driven by other activated TK that are amenable to targeted inhibition, we analyzed the metabolic consequences of inhibiting the chimeric BCR-ABL tyrosine kinase, which is central to the pathogenesis of chronic myeloid leukemia (CML) and Philadelphia chromosome positive acute lymphoblastic leukaemia 50, using its specific inhibitor imatinib51. Indeed, in a BCR-ABL positive cell line, imatinib treatment resulted in a reduction of glycolytic activity that also correlated with a decrease in gene expression levels of glycolytic enzymes (Figure 5A-C). Conversely, the effects on TCA cycle and glutathione metabolism genes were much less pronounced and although a significant reduction in both GLS and GLUD1 gene expression levels were noted following imatinib treatment, this was less than 50% and much smaller than those observed on glycolytic genes (Supplemental Figure 6A-C and Supplemental table 3). As observed in FLT3ITD cells, combining imatinib with CB839 led to increased apoptosis of BCR-ABL positive cells, which correlated with enhanced intracellular ROS production and reduced oxygen consumption. Moreover, as with FLT3ITD AML, these effects could also be fully rescued by αKG (Figure 5D-F).
Figure 5. Combined effects of tyrosine kinase inhibitors and CB839 in BCR-ABL positive leukemia.
(A) Glycolytic rate and capacity of BCR-ABL mutated K562 cells treated with vehicle control or Imatinib 2 µM (mean ± s.e.m., n=3, P<0.0001 for both glycolytic rate and capacity, respectively, two-way ANOVA with Bonferroni’s multiple comparison) (B) Volcano plot for gene expression changes by RNA-sequencing of K562 cells treated with Imatinib 2 µM or vehicle control highlighting reduced expression of genes involved in glycolysis in treated cells (n=2). (C) q-PCR validation in K562 cells for the reduction in expression levels of GLUT3 (mean ± s.e.m., n=3, ** P=0.0081, two-tailed paired t-test) and LDHA (mean ± s.e.m., n=3, * P=0.0132, two-tailed paired t-test) following treatment as in (B). (D) Apoptosis in K562 cells treated with vehicle control, Imatinib 2 µM, CB839 100 nM, Imatinib and CB839 combination and Imatinib/CB839 combination + 4 mM αKG (mean ± s.e.m., n=9, ** P=0.0019 between Imatinib versus Imatinib+CB839, and **** P<0.0001 between Imatinib+CB839 versus Imatinib+CB839+αKG, ANOVA with Tukey’s multiple comparisons) (E) Relative cytoplasmic ROS levels in BCR-ABL mutated K562 cells treated as in (D) (mean ± s.e.m., n=6, * P=0.0495 between Imatinib versus Imatinib+CB839, and ** P=0.0089 between Imatinib+CB839 versus Imatinib+CB839+αKG, ANOVA with Sidak’s multiple comparisons). (F) Oxygen consumption rate in K562 cells treated as in (D) (mean ± s.e.m., n=3, for basal respiration P=0.0606 between Imatinib versus Imatinib+CB839, and P<0.0001 between Imatinib+CB839 versus Imatinib+CB839+aKG, for maximal respiration between **** P<0.0001 between both Imatinib versus Imatinib+CB839 and Imatinib+CB839 versus Imatinib+CB839+αKG, two-way ANOVA with Bonferroni’s multiple comparisons).
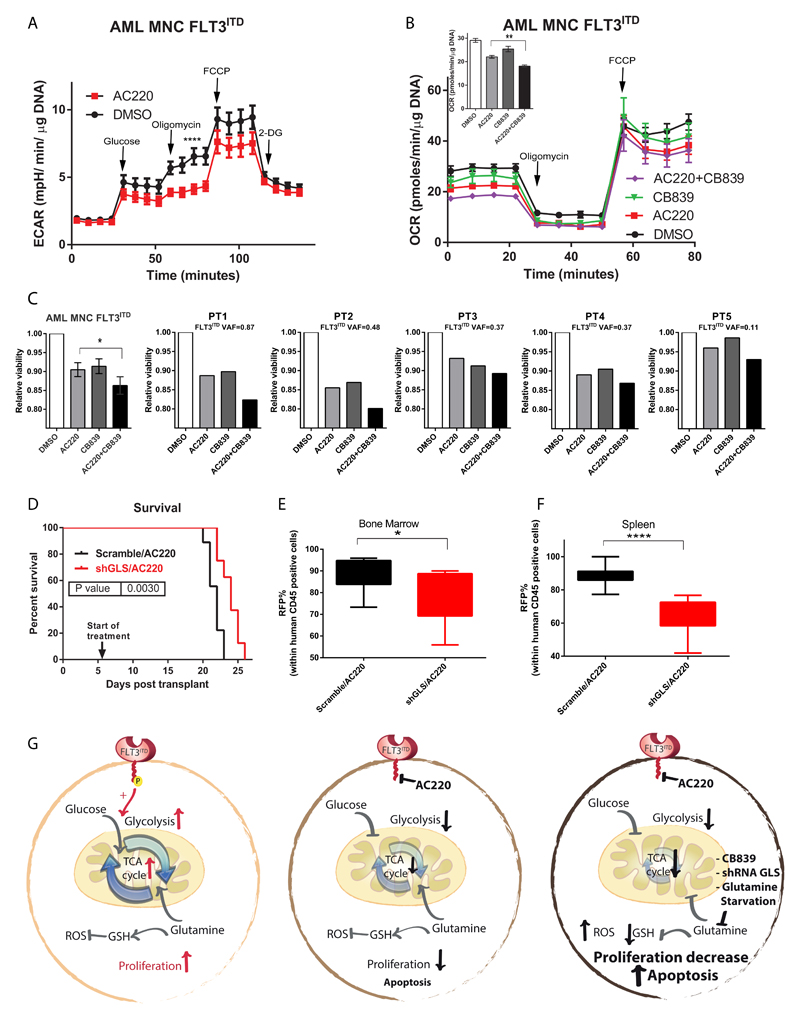
Finally, we sought to confirm our findings in more physiological and clinically relevant models. Using primary AML samples from patients carrying a FLT3ITD mutation, we found that AC220 treatment led to a reduction in glycolytic capacity, and combined treatment with AC220 and CB839 led to a reduction in basal oxygen consumption (Figure 6A-B). These effects correlated with a further reduction in the viability of FLT3ITD primary samples following combined treatment that appeared proportional to the levels of FLT3 mutation, as measured by variant allele frequency (VAF), in each sample while similar effects were not observed in FLT3wt samples (Figure 6C and Supplemental Figure 7A-B). The combined treatment also led to reduced colony-forming-cell (CFC) output in BM murine cells and AML primary samples carrying FLT3ITD mutations while similar effects were not observed in normal CD34+ samples or FLT3wt AML patient samples, suggesting that these effects are specific to FLT3ITD cells (Supplemental Figure 7C-F). Finally, we tested the effects of combined GLS and FLT3 TK inhibition in vivo using FLT3ITD MV411 cells stably expressing a doxycycline inducible GLS or scrambled shRNA alongside a red fluorescent protein (RFP) reporter to allow tracking of shRNA expression. We transplanted shRNA GLS or scrambled cells into recipient immunocompromised mice, allowed leukemia to develop, at which point we fed mice with a doxycycline containing diet and also initiated AC220 treatment by oral gavage. MV411 generated leukemia are extremely aggressive but also highly sensitive to FLT3 inhibitor treatment34 (data not shown). Using a low dose of AC220 in combination with a doxycycline containing diet all mice succumbed to disease while on treatment. However, despite the very aggressive nature of this leukemia, mice transplanted with cells carrying shRNA targeting GLS showed a modest but statistically significant increase in survival compared to mice transplanted with control cells (Figure 6D). We also observed that the mice transplanted with shRNA targeting GLS had lower levels of disease burden in bone marrow and spleen (as measured by percentage of RFP positive cell within human CD45 cells) and a trend towards smaller spleen size (Figure 6E-F and Supplemental Fig 7G). Finally GLS depletion was measured in vivo from human cells isolated from mice organs. Interestingly, in the shRNA GLS transduced cells, AC220 treatment resulted in a 50% reduction in GLS knockdown efficiency suggesting preferential killing of cells having lower levels of GLS expression (Supplemental Figure 7H).
Figure 6. Combined effects of AC220 and CB839 in FLT3ITD primary samples and in vivo.
(A) Extracellular acidification rate of primary FLT3ITD mutated AML samples treated with vehicle control or AC220 2.5 nM measured using a Seahorse analyzer (mean ± s.e.m., n=4, maximal glycolytic capacity **** P<0.0001, two-way ANOVA with Bonferroni’s multiple comparisons). (B) Oxygen consumption rate of primary FLT3ITD mutated AML samples treated with vehicle control, AC220 2.5nM, CB839 100 nM or their combination measured using a Seahorse analyzer. Real-time basal and maximal respiration are shown and in the inset a bar charts for the basal respiration in the 4 different conditions is shown (mean ± s.e.m., n=4, ** P=0.0034 between AC220 versus AC220+CB839, ANOVA with Tukey’s multiple comparisons). (C) Relative viability in primary FLT3ITD mutated AML samples treated with vehicle control, AC220 2.5 nM, CB839 100 nM or their combination. Far left panel shows a summary plot for all 5 patients (mean ± s.e.m., n=5, ** P=0.0176 between AC220 versus AC220+CB839, ANOVA with Tukey’s multiple comparisons). The other panels show data for each individual patient with VAF (variant allele frequency) for FLT3ITD. Note in PT5 AC220 was used at 5 nM given low VAF for FLT3ITD. (D) Survival curve of mice transplanted respectively with MV411 transduced with control “Scramble” shRNA (n=9) and GLS shRNA (n=8) following treatment with AC220 (P=0.0030 by Log-rank test). (E-F) Percentage of RFP positive cells, measured by flow-cytometry, within 45 positive human cells from the bone marrow (E) and spleen (F) of mice transplanted respectively with MV411 transduced with control “Scramble” shRNA (n=9) and GLS shRNA (n=8) following treatment with AC220 (box and whiskers showing minimum to maximum range * for bone marrow, P=0.0201; for spleen, **** P<0.0001, unpaired t test). (G) Schematic model showing the action mechanism of combined GLS and FLT3 TK inhibition. FLT3ITD mutant cells utilize both glucose and glutamine to support their metabolism (left panel). FLT3-TKI treatment (AC220) blocks glucose uptake and mostly glycolysis rendering the cells dependent on glutamine metabolism (middle panel). GLS gene silencing, chemical inhibition (CB839) or glutamine starvation enhance the efficacy of FLT3-TKI by blocking glutamine metabolism and its ability to support both TCA cycle/mitochondrial function and GSH synthesis/redox metabolism (right panel).
Discussion
In this study we used orthogonal unbiased approaches including CRISPR/Cas9 synthetic lethality screen, metabolomics and gene expression analysis, to reveal that FLT3ITD cells develop a metabolic dependency on glutamine metabolism following FLT3 TK inhibition. Targeted inhibition of FLT3 TK activity appears to suppress the enhanced central carbon metabolism typical of FLT3ITD cells by mostly hindering glucose uptake and utilization thus predominantly reversing the glycolytic phenotype. However, TCA cycle activity and respiratory function, although reduced, are less affected and are supported by continuous uptake of glutamine, the other main fuel for central carbon metabolism. Combined suppression of FLT3 TK activity and glutamine metabolism using both GLS chemical inhibition and gene silencing, leads to an increased cell death in FLT3ITD cells, including models previously shown to be already highly sensitive to FLT3 TK inhibition. We also extend these findings to a model of leukemia carrying BCR-ABL TK activating mutations, primary AML samples and in vivo models. We demonstrate that glutamine metabolism supports both the TCA cycle and redox metabolism upon FLT3 TK inhibition and, through rescue experiments, we further validate the role of all these branches of glutamine metabolism in cellular survival (Figure 6G). Our data expand the findings of a recent report on the activity of combined GLS and FLT3 TK inhibition in FLT3ITD AML by providing in depth mechanistic explanation for these findings and extending their validity to primary AML samples and other TK activating mutated leukemias52. Moreover they also explain mechanistically previous published observations suggesting that combined targeting of FLT3 TK activity and redox metabolism or ETC might enhance toxicity in FLT3 mutated AML53,54. However, it is entirely plausible that another consequence of glutamine metabolism might also underlie, at least in part, the effects of targeting it. For instance, branched chain amino acids, produced by the transamination of glutamine derived glutamate, have recently been shown to support the maintenance and progression of myeloid leukemias27.
GLS has recently emerged as a therapeutic target in both solid and hematological malignancies and potent GLS inhibitors, including the one used in this study, have now entered clinical trials in several malignancies [NCT02071862 and NCT02071927]. GLS is the most abundant isoform present in hematopoietic cells and has already been suggested as a potential therapeutic target in AML55. However, our data show that specifically in FLT3ITD mutated AML, GLS inhibition, on its own, produces only mild antiproliferative effects and only becomes a metabolic vulnerability following FLT3 TK inhibition, with similar effects not observed in normal cells or leukemic cells that lack TK activating mutations. Our results therefore suggest a therapeutic window for this combination therapy and confirm its specificity and potential utility in several TK mutated leukemias.
Of note, the best effects in combination with FLT3 TK inhibition were observed when FLT3ITD cells were starved of glutamine rather than following GLS inhibition. This suggests that FLT3ITD mutated cells might also rely on ancillary pathways of glutamine metabolism releasing its γ-nitrogen and producing glutamate. Moreover, glutamine γ-nitrogen is a central substrate for the biosynthesis of nucleotides, NAD, amino acids and glucosamine-6-phosphate56, and given that this function is not targeted by GLS inhibition, it is also plausible that these glutamine dependent metabolic pathways support cell survival following AC220 and combination treatment.
The ability of FLT3-TKI to predominantly revert the glycolytic phenotype, while having a less pronounced effect on TCA cycle activity and oxidative metabolism is another important observation stemming from this work. With regards to this it is noteworthy that, consistent with our findings, two recent reports have suggested that both AML and CML therapy-resistant cells display increased mitochondrial mass and a high oxidative phosphorylation status which is therapeutically actionable57,58. However, the exact mechanisms whereby some metabolic phenotypes are particularly dependent on FLT3 TK activity and how the described metabolic adaptations are established remains unknown and these fundamental questions merit further studies. Speculatively the ability of FLT3 TK to control glycolysis could be explained by its activation of AKT17 which can modulate transcription factors, such as FOXO, known to regulate glycolysis59 or directly control the activity of several glycolytic enzymes21,60,61. However an improved understanding of the molecular mechanisms leading to this metabolic phenotype and whether other anaplerotic substrates, such as fatty acids, also contribute to oxidative phosphorylation in therapy resistant cells might help to clarify the basis of resistance and help target it more effectively.
In conclusion, our results highlight the importance of FLT3 mutations and downstream signaling in the control of leukemia cell metabolism, extend our understanding of the role of metabolic adaptations in the resistance to treatment with FLT3 and other TK inhibitors and provide an example of a complementary unbiased approach to study the role of metabolism in leukemia and as a tool for the design of novel and specific therapeutic strategies targeting cell metabolism in AML.
Supplementary Material
Keypoints.
-
1)
FLT3ITD tyrosine kinase (TK) inhibition impairs glycolysis and glucose utilization without equally affecting glutamine metabolism
-
2)
Combined targeting of FLT3 TK activity and glutamine metabolism weakens FLT3ITD mutant cells leukemogenic potential in vitro and in vivo
Acknowledgments
P.G. is funded by the Wellcome Trust (109967/Z/15/Z) and was previously supported by the Academy of medical Sciences and Lady Tata Memorial Trust. The Huntly lab is funded by European Research Council, MRC, Bloodwise, the Kay Kendall Leukaemia Fund, the Cambridge NIHR Biomedical Research Centre, and core support grants to the Wellcome Trust - Medical Research Council Cambridge Stem Cell Institute. C.F. and A.S.H.C are funded by the Medical Research Council, Core Grant to the Cancer Unit. P.M-P. is supported by a grant from Cancer Research UK (C56179/A21617). D.S. is a Postdoctoral Fellow of the Mildred-Scheel Organisation, German Cancer Aid. This research was supported by the CIMR Flow Cytometry Core Facility. We would like to thank the Welcome Trust Sanger Institute facility for the MiSeq run.
Footnotes
Author Contributions
P.G., C.F. and B.J.P.H. conceived the study; P.G., G.G., K.T., A.S.H.C., G.V., C.F., B.J.P.H. designed and/or conducted experiments, performed data analysis, interpretation and informed study direction. P.M-P., F.B., L.M., L.D.L., H.Y., D.S., S.J.H. helped with experimental work. S.V. and J.M.L.D. performed bioinformatics analyses. P.G., C.F. and B.J.P.H. drafted the manuscript. All authors discussed the results and commented on the manuscript.
Disclosure of Conflicts of Interest
The authors declare that they have no conflicts of interest to disclose
References
- 1.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Dohner H, Campbell PJ. Genomic Classification in Acute Myeloid Leukemia. N Engl J Med. 2016;375(9):900–901. doi: 10.1056/NEJMc1608739. [DOI] [PubMed] [Google Scholar]
- 3.Kiyoi H, Naoe T, Nakano Y, et al. Prognostic implication of FLT3 and N-RAS gene mutations in acute myeloid leukemia. Blood. 1999;93(9):3074–3080. [PubMed] [Google Scholar]
- 4.Levis M. FLT3 mutations in acute myeloid leukemia: what is the best approach in 2013? Hematology Am Soc Hematol Educ Program. 2013;2013:220–226. doi: 10.1182/asheducation-2013.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly LM, Liu Q, Kutok JL, Williams IR, Boulton CL, Gilliland DG. FLT3 internal tandem duplication mutations associated with human acute myeloid leukemias induce myeloproliferative disease in a murine bone marrow transplant model. Blood. 2002;99(1):310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 7.Smith CC, Wang Q, Chin CS, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485(7397):260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone RM, Mandrekar SJ, Sanford BL, et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N Engl J Med. 2017;377(5):454–464. doi: 10.1056/NEJMoa1614359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wander SA, Levis MJ, Fathi AT. The evolving role of FLT3 inhibitors in acute myeloid leukemia: quizartinib and beyond. Ther Adv Hematol. 2014;5(3):65–77. doi: 10.1177/2040620714532123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Osdal T, Ho Y, et al. SIRT1 activation by a c-MYC oncogenic network promotes the maintenance and drug resistance of human FLT3-ITD acute myeloid leukemia stem cells. Cell Stem Cell. 2014;15(4):431–446. doi: 10.1016/j.stem.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park IK, Mundy-Bosse B, Whitman SP, et al. Receptor tyrosine kinase Axl is required for resistance of leukemic cells to FLT3-targeted therapy in acute myeloid leukemia. Leukemia. 2015;29(12):2382–2389. doi: 10.1038/leu.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith CC, Paguirigan A, Jeschke GR, et al. Heterogeneous resistance to quizartinib in acute myeloid leukemia revealed by single-cell analysis. Blood. 2017;130(1):48–58. doi: 10.1182/blood-2016-04-711820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piloto O, Wright M, Brown P, Kim KT, Levis M, Small D. Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood. 2007;109(4):1643–1652. doi: 10.1182/blood-2006-05-023804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Fukuda S, Lee Y, et al. Essential role of signal transducer and activator of transcription (Stat)5a but not Stat5b for Flt3-dependent signaling. J Exp Med. 2000;192(5):719–728. doi: 10.1084/jem.192.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tse KF, Mukherjee G, Small D. Constitutive activation of FLT3 stimulates multiple intracellular signal transducers and results in transformation. Leukemia. 2000;14(10):1766–1776. doi: 10.1038/sj.leu.2401905. [DOI] [PubMed] [Google Scholar]
- 16.Mizuki M, Fenski R, Halfter H, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96(12):3907–3914. [PubMed] [Google Scholar]
- 17.Brandts CH, Sargin B, Rode M, et al. Constitutive activation of Akt by Flt3 internal tandem duplications is necessary for increased survival, proliferation, and myeloid transformation. Cancer Res. 2005;65(21):9643–9650. doi: 10.1158/0008-5472.CAN-05-0422. [DOI] [PubMed] [Google Scholar]
- 18.Elstrom RL, Bauer DE, Buzzai M, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64(11):3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 19.Kerr EM, Gaude E, Turrell FK, Frezza C, Martins CP. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531(7592):110–113. doi: 10.1038/nature16967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang HB, Fan J, Lin R, et al. Metabolic Rewiring by Oncogenic BRAF V600E Links Ketogenesis Pathway to BRAF-MEK1 Signaling. Mol Cell. 2015;59(3):345–358. doi: 10.1016/j.molcel.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ju HQ, Zhan G, Huang A, et al. ITD mutation in FLT3 tyrosine kinase promotes Warburg effect and renders therapeutic sensitivity to glycolytic inhibition. Leukemia. 2017 doi: 10.1038/leu.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Guitart AV, Panagopoulou TI, Villacreces A, et al. Fumarate hydratase is a critical metabolic regulator of hematopoietic stem cell functions. J Exp Med. 2017;214(3):719–735. doi: 10.1084/jem.20161087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taya Y, Ota Y, Wilkinson AC, et al. Depleting dietary valine permits nonmyeloablative mouse hematopoietic stem cell transplantation. Science. 2016;354(6316):1152–1155. doi: 10.1126/science.aag3145. [DOI] [PubMed] [Google Scholar]
- 25.Agathocleous M, Meacham CE, Burgess RJ, et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature. 2017 doi: 10.1038/nature23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang YH, Israelsen WJ, Lee D, et al. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell. 2014;158(6):1309–1323. doi: 10.1016/j.cell.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hattori A, Tsunoda M, Konuma T, et al. Cancer progression by reprogrammed BCAA metabolism in myeloid leukaemia. Nature. 2017;545(7655):500–504. doi: 10.1038/nature22314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herranz D, Ambesi-Impiombato A, Sudderth J, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat Med. 2015;21(10):1182–1189. doi: 10.1038/nm.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanaka K, Sasayama T, Irino Y, et al. Compensatory glutamine metabolism promotes glioblastoma resistance to mTOR inhibitor treatment. J Clin Invest. 2015;125(4):1591–1602. doi: 10.1172/JCI78239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzelepis K, Koike-Yusa H, De Braekeleer E, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 2016;17(4):1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giotopoulos G, Chan WI, Horton SJ, et al. The epigenetic regulators CBP and p300 facilitate leukemogenesis and represent therapeutic targets in acute myeloid leukemia. Oncogene. 2016;35(3):279–289. doi: 10.1038/onc.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarrinkar PP, Gunawardane RN, Cramer MD, et al. AC220 is a uniquely potent and selective inhibitor of FLT3 for the treatment of acute myeloid leukemia (AML) Blood. 2009;114(14):2984–2992. doi: 10.1182/blood-2009-05-222034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuleshov MV, Jones MR, Rouillard AD, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gross MI, Demo SD, Dennison JB, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014;13(4):890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 41.Verhaak RG, Wouters BJ, Erpelinck CA, et al. Prediction of molecular subtypes in acute myeloid leukemia based on gene expression profiling. Haematologica. 2009;94(1):131–134. doi: 10.3324/haematol.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whitman SP, Maharry K, Radmacher MD, et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116(18):3622–3626. doi: 10.1182/blood-2010-05-283648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105(48):18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162(3):540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162(3):552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffith OW. Mechanism of action, metabolism, and toxicity of buthionine sulfoximine and its higher homologs, potent inhibitors of glutathione synthesis. J Biol Chem. 1982;257(22):13704–13712. [PubMed] [Google Scholar]
- 48.Zdzisinska B, Zurek A, Kandefer-Szerszen M. Alpha-Ketoglutarate as a Molecule with Pleiotropic Activity: Well-Known and Novel Possibilities of Therapeutic Use. Arch Immunol Ther Exp (Warsz) 2017;65(1):21–36. doi: 10.1007/s00005-016-0406-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin L, Li D, Alesi GN, et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell. 2015;27(2):257–270. doi: 10.1016/j.ccell.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren R. Mechanisms of BCR-ABL in the pathogenesis of chronic myelogenous leukaemia. Nat Rev Cancer. 2005;5(3):172–183. doi: 10.1038/nrc1567. [DOI] [PubMed] [Google Scholar]
- 51.O'Brien SG, Guilhot F, Larson RA, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348(11):994–1004. doi: 10.1056/NEJMoa022457. [DOI] [PubMed] [Google Scholar]
- 52.Gregory MA, Nemkov T, Reisz JA, et al. Glutaminase inhibition improves FLT3 inhibitor therapy for acute myeloid leukemia. Exp Hematol. 2017 doi: 10.1016/j.exphem.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alvarez-Calderon F, Gregory MA, Pham-Danis C, et al. Tyrosine kinase inhibition in leukemia induces an altered metabolic state sensitive to mitochondrial perturbations. Clin Cancer Res. 2015;21(6):1360–1372. doi: 10.1158/1078-0432.CCR-14-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregory MA, D'Alessandro A, Alvarez-Calderon F, et al. ATM/G6PD-driven redox metabolism promotes FLT3 inhibitor resistance in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2016;113(43):E6669–E6678. doi: 10.1073/pnas.1603876113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacque N, Ronchetti AM, Larrue C, et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126(11):1346–1356. doi: 10.1182/blood-2015-01-621870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang J, Pavlova NN, Thompson CB. Cancer cell metabolism: the essential role of the nonessential amino acid, glutamine. EMBO J. 2017;36(10):1302–1315. doi: 10.15252/embj.201696151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuntz EM, Baquero P, Michie AM, et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med. 2017 doi: 10.1038/nm.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farge T, Saland E, de Toni F, et al. Chemotherapy-Resistant Human Acute Myeloid Leukemia Cells Are Not Enriched for Leukemic Stem Cells but Require Oxidative Metabolism. Cancer Discov. 2017;7(7):716–735. doi: 10.1158/2159-8290.CD-16-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gross DN, van den Heuvel AP, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene. 2008;27(16):2320–2336. doi: 10.1038/onc.2008.25. [DOI] [PubMed] [Google Scholar]
- 60.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452(7184):181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 61.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.