SUMMARY
Resistance to chemotherapy plays a significant role in cancer mortality. To identify genetic units affecting sensitivity to cytarabine, the mainstay of treatment for AML, we developed a comprehensive and integrated genome wide platform based on a Dual protein-coding and noncoding Integrated CRISPRa Screening (DICaS). Putative resistance genes were initially identified using pharmacogenetic data from 517 human pan-cancer cell lines. Subsequently, genome scale functional characterization of both coding and lncRNA genes by CRISPR activation was performed. For lncRNA functional assessment we developed a CRISPR activation of lncRNA (CaLR) strategy, targeting 14,701 lncRNA genes. Computational and functional analysis identified novel cell cycle regulation, survival/apoptosis, and cancer signaling genes. Furthermore, transcriptional activation of the GAS6-AS2 lncRNA, identified in our analysis, leads to hyperactivation of the GAS6/TAM pathway, a resistance mechanism in multiple cancers, including AML. Thus, DICaS represents a novel and powerful approach to identify integrated coding and non-coding pathways of therapeutic relevance.
ETOC
A CRISPR activation screen identifies both coding and noncoding pathways involved in resistance to chemotherapy
INTRODUCTION
Although precision medicine and targeted therapies offer new hope for treating cancer, chemotherapy still remains the first, and last, line of defense for most patients. Cytarabine (1-p- d-arabinofuranosylcytosine, Ara-C) is a deoxycytidine analogue that is used as part of a standard chemotherapeutic regimen for the treatment of AML (Ramos et al., 2015). However, approximately 30% to 50% of patients relapse with chemotherapy-resistant disease. Thus, there is an ever-present need to better understand the genetic and molecular mechanisms that contribute to chemotherapy resistance.
To date, studies on mechanisms leading to therapy resistance have focused on proteincoding genes, yet cancer development and progression cannot be fully explained by the coding genome (Huarte, 2015; Imielinski et al., 2012). The recent explosion in research and understanding related to the non-coding RNA (ncRNA) transcriptome has highlighted the importance of ncRNAs in biology (Hon et al., 2017; Iyer et al., 2015). Functional validation of various ncRNA species highlights the fact that these RNAs may play important roles in the pathogenesis of diseases including cancer (Schmitt and Chang, 2016).
One large group of ncRNAs is represented by long non-coding RNAs (lncRNA). LncRNAs can be either nuclear or cytoplasmic in localization and play roles in a diverse array of biological processes. As many nuclear lncRNAs behave in a cis-acting manner (Quinn and Chang, 2016), their study requires their expression from endogenous loci, and CRISPR technologies now facilitate the modulation of gene expression directly from the endogenous promoter (Joung et al., 2017a; Konermann et al., 2014). This approach has already been compellingly demonstrated using CRISPR interference (CRISPRi) to silence the expression of lncRNAs genome-wide (Liu et al., 2017).
Although we now have a wealth of high-throughput data delineating expression of coding and non-coding genes across hundreds of cancer cell lines (Barretina et al., 2012; Garnett et al., 2012), there remains a critical lack of integrated high-throughput functional characterization and validation of these data in a disease context. We therefore sought to develop an integrative and comprehensive CRISPR activation (CRISPRa) framework that would complement these publicly available ‘Big Data’ databases to enable the discovery of functional human protein coding and lncRNA genes contributing to chemotherapy resistance. In doing so, we developed a dual coding and non-coding Integrated CRISPRa Screening (DICaS) platform and applied this integrative approach to identify genetic units and pathways that promote resistance to Ara-C treatment.
RESULTS
Pan-Cancer Cell Line Analysis of IncRNAs Affecting Drug Response
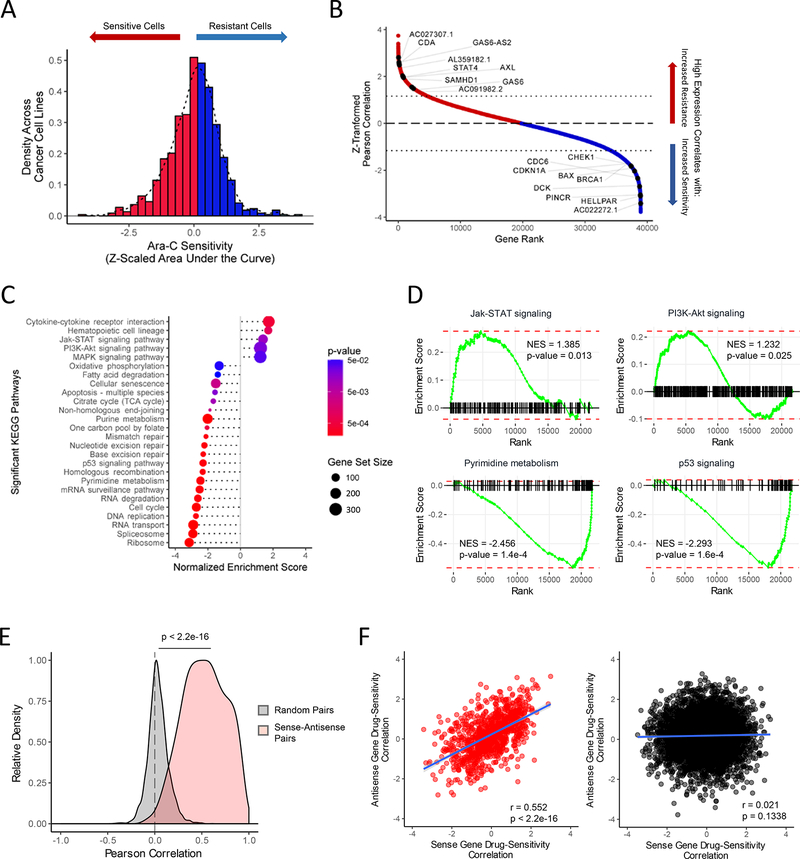
In order to comprehensively define resistance mechanisms to chemotherapy, we chose to examine cellular responses to Ara-C. We developed a computational strategy to identify genes that correlate with sensitivity or resistance to Ara-C by correlating pharmacological profiles from the Cancer Target Discovery and Development (CTD2) database (Basu et al., 2013; Rees et al., 2016) with the transcriptomes of 760 corresponding cell lines from the Cancer Cell Line Encyclopedia (CCLE) (Barretina et al., 2012) (Figure S1A). To identify high confidence gene targets it is imperative to integrate analysis of as many cell lines as possible (Rees et al., 2016); however, we found that the cell line drug sensitivities formed a skewed distribution (Figure S1B), likely conferred by tissue of origin and histological subtype. Indeed, cancer cell type annotations explained a substantial amount of the variation in drug sensitivities (adjusted R2 = 0.5123, ANOVA p < 2.2e-16) (Figure S1A), which were subsequently corrected (Figure S1C). Thus, using a linear regression model to remove these effects we established a normalized distribution of Ara-C sensitivity for the 760 cell lines analyzed (Figure 1A).
Figure 1. Identification of Protein-Coding and Noncoding Gene Biomarkers Correlated with Differential Ara-C Response.
(A) Distribution of Ara-C drug sensitivities across 760 pan-cancer cell lines profiled by both CCLE and CTD2 studies, quantified by their Z-scaled area under the dose response curve values after regressing out lineage-specific effects. See also Table S1.
(B) Distribution of Z-scaled drug resistance-gene expression Pearson correlation values of all analyzed genes. Representative protein-coding and non-coding gene symbols enriched beyond a Z-score threshold of ± 1.16 are demarcated. See also Table S1.
(C) Summary of gene set enrichment analysis (GSEA) of protein-coding genes ranked by drug resistance-gene expression correlation values using annotated KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. See also Table S3.
(D) Representative KEGG pathways from GSEA of protein-coding genes ranked by drug sensitivity-gene expression correlation values as shown in Figures 1B–1C. See also Table S3.
(E) Pearson correlation distributions of gene pair expression levels in the cancer cell line panel across 997 sense-antisense cognate gene pairs and 5,000 random protein coding- lncRNA gene pairs. Wilcoxon rank-sum test: p < 2.2e-16.
(F) Relationship of drug sensitivity-gene expression correlation values between protein coding-lncRNA gene pairs across 997 sense-antisense cognate gene pairs (left panel: Pearson’s R = 0.552, p < 2.2e-16) and 5,000 random gene pairs (right panel: Pearson’s R = 0.021, p = 0.1338).
We subsequently performed a correlation analysis between drug sensitivities and gene expression levels across the 760 cell lines (Figure 1B, Table S1) and determined appropriate Z-score thresholds (Figure S1D-S1E). Interestingly, genes involved in the metabolism of Ara-C were highly enriched, illustrating the applicability of such an approach to identifying chemotherapy resistance mechanisms. Low expression of deoxycytidine kinase (DCK) and equilibrative nucleoside transporter 1 (ENT1) correlated with increased resistance to Ara-C (Z = −2.51 and −1.61, respectively), whereas high expression of cytidine deaminase (CDA) and SAM Domain and HD Domain 1 (SAMHD1) correlated with increased resistance (Herold et al., 2017; Schneider et al., 2016) (Z = 2.54 and 2.03, respectively) (Figure 1B). Interestingly, we also observed a number of cell-cycle and DNA damage regulators previously implicated in modulation of AraC sensitivity (Figure 1B).
To define biological pathways predictive of Ara-C resistance we performed gene set enrichment analysis (GSEA) on the drug sensitivity-gene expression correlations using Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotations (Figure 1C and Table S3)(Kanehisa et al., 2014; Subramanian et al., 2005). We identified positive enrichment of cell survival signaling pathways, including the Jak-STAT (NES = 1.385, p = 0.013), PI3K-Akt (NES = 1.232, p = 0.025), and MAPK (NES = 1.222, p = 0.042) pathways and negative enrichment of the pyrimidine metabolic pathway (NES = −2.456, p = 0.00016), mechanisms related to DNA damage (e.g. p53 signaling pathway: NES = −2.293, p = 0.00016) and RNA regulatory mechanisms (e.g. RNA degradation: NES = −2.613, p = 1.6e-4) (Figure 1C-1D and Figure S1F). To confirm their relevance in human AML, we correlated pre-treatment AML transcriptome profiles with corresponding disease-free survival data from 121 patients treated with Ara-C from The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas Research Network, 2013) and identified a large number of enriched pathways shared with our cell line predictions, including oxidative phosphorylation (NES = −1.994, p = 1.1e-4) and RNA regulatory mechanisms (e.g., RNA degradation: NES = −1.702, p = 0.0011) (Figure S1G-S1H).
As many non-coding genes act in a proximal and localized manner (Schmitt and Chang, 2016), we evaluated coding and non-coding cognate gene pairs for correlation with either resistance or sensitivity to Ara-C and compiled a genome wide set of 997 coding/con-coding sense/antisense gene pairs. Indeed, we observed a significant positive correlation between sense-antisense gene expression levels across the cell line panel (Pearson correlation, median R = 0.5312; Wilcoxon rank-sum test, p < 2.2e-16) (Figure 1E). Furthermore, cognate gene pairs demonstrated significant positive correlation in drug sensitivity (Pearson correlation, R = 0.5636, p < 2.2e-16) (Figure 1F). Importantly, analysis of these same cognate gene pairs among the TCGA AML patient cohort identified a similarly significant positive correlation (Figure S2A-S2C). Interestingly, cognate sense genes were found to be positively enriched in PI3K-Akt (NES = 1.426, p = 0.0764) and MAPK signaling pathways (NES = 1.787, p = 0.0040) (Figure S2D), implicating these sense-antisense gene pairs in a number of the previously identified enriched pathways.
A CRISPRa approach to study AML resistance to Ara-C
To functionally validate our predictive analysis in a high-throughput manner we established a CRISPRa-based system in AML cell lines to provide a comprehensive and integrative genomewide study of both the coding and non-coding genes contributing to Ara-C resistance.
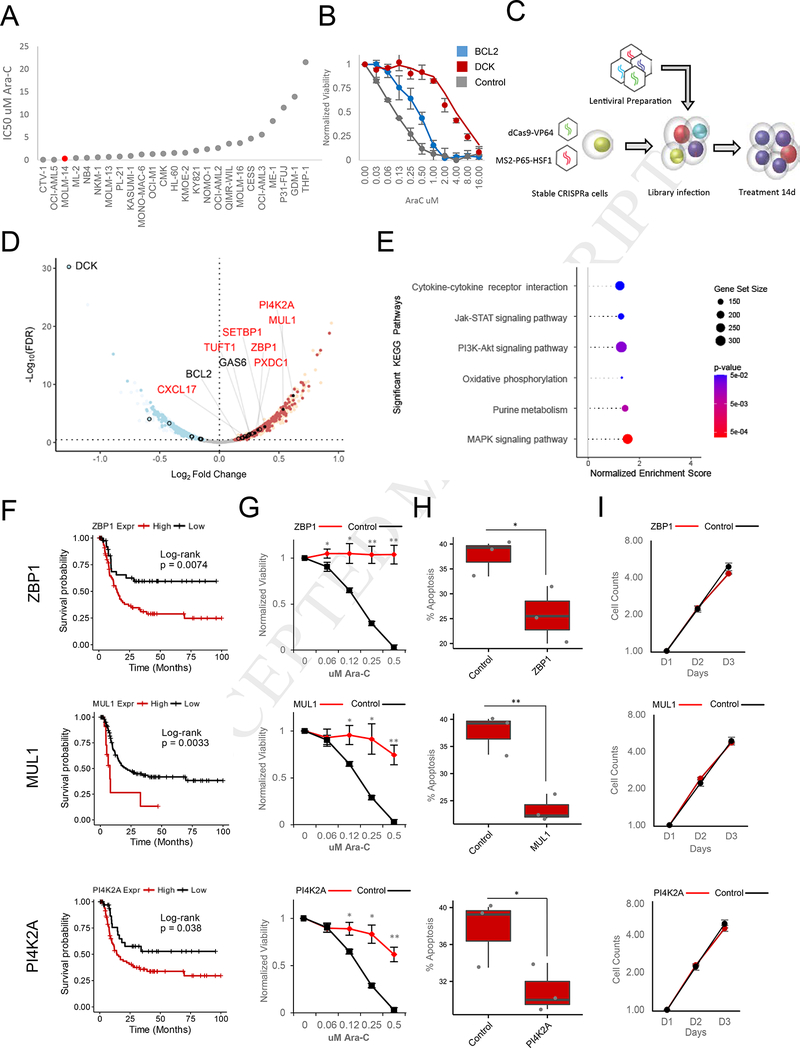
We identified the MOLM14 AML cell line to be the most appropriate model for our screening, as its IC50 (~0.13^M) ranks it amongst the most sensitive AML cell lines (Yang et al., 2013) (Figure 2A). Overexpression of the anti-apoptotic B-cell lymphoma 2 (BCL2) gene increased the IC50 of MOLM14 for Ara-C, while shRNA-mediated knockdown of DCK provided an even more significant protection, increasing its IC50 almost 300-fold (Figure 2B), confirming that sensitivity to Ara-C can be readily manipulated.
Figure 2. CRISPRa Functional Screening of Coding Genes Modulating Ara-C Response.
(A) Distribution of Ara-C IC50 values across a panel of AML cell lines.
(B) Effect of BCL2 overexpression (Blue) or DCK knockdown on sensitivity to Ara-C in MOLM14 cells. Data are represented as mean ± SD, n = 3.
(C) Schematic of CRISPRa pooled screening for the identification of genes whose activation modulate sensitivity to Ara-C in MOLM14 cells.
(D) Volcano plot summarizing the global changes in sgRNA representation of protein-coding genes before and after 14 days of treatment with Ara-C. A subset of genes validated herein (red text) or previously annotated (black text) to modulate Ara-C sensitivity are labeled. A false discovery rate threshold of 0.339 was determined by receiver operating characteristic analysis (Figure S3F). Red - enrichment in the CRISPRa screening; blue depletion in the CRISPRa screening; open black circles - genes previously associated with differential Ara-C sensitivity and above the significance threshold; filled black points genes validated herein. See also Figure S3C-F, S3H, and Table S4.
(E) Summary of gene set enrichment analysis (GSEA) of protein-coding genes ranked by CRISPRa screening using annotated KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. See Table S3.
(F) Disease-free survival association with expression levels of ZBP1, MUL1, and PI4K2A, genes enriched in both protein-coding CRISPRa screening and drug sensitivity-gene expression correlation analyses among patients treated with Ara-C therapy within the TCGA-LAML patient cohort. ZBP1: VST expression level cutoff = 6.13 (low, n = 42; high, n = 79), log-rank test: p-value = 0.0074. MUL1: VST expression level cutoff = 9.64 (low, n = 108; high, n = 13), log-rank test: p-value = 0.0033. PI4K2A: VST expression level cutoff = 7.23 (low, 36; high, n = 85), log-rank test: p-value = 0.038.
(G) Ara-C efficacy measurements in MOLM14 cells expressing sgRNAs targeting ZBP1, MUL1, or PI4K2A based on normalized MTS reads following 48 hours of treatment. Data are represented as mean ± SD, n = 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p<0.001
(H) Modulation of apoptotic response upon stable expression of sgRNAs targeting ZBP1, MUL1, or PI4K2A in MOLM14 cells. The percentage of apoptosis is determined by annexin V and propidium iodide (PI) staining of cells treated with 0.25 pM Ara-C for 72 hours. Data are represented as mean ± SD, n = 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001
(I) Proliferation of unchallenged MOLM14 cells expressing sgRNAs targeting ZBP1, MUL1, or PI4K2A. Proliferation is quantified over four days (D1-D4). Data are represented as mean ± SD, n = 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001
We also tested synergistic activation mediator (SAM) mediated CRISPRa (Konermann et al., 2014) in MOLM14 cells as compared with two additional leukemia cell lines, K562 and HL60, and the previously validated HEK293T. Using a panel of validated sgRNAs targeting the promoters of both coding (TTN, RHOXF, ASCL1, HBG1) and non-coding (MIAT, TUNA) genes (Chavez et al., 2015, 2016), we established that the majority of sgRNAs gave the highest activation in MOLM14 among the leukemia cell lines (Figure S3A).
Genome wide CRISPRa Screening of Protein-Coding Genes in AML
We next applied our CRISPRa platform to screen for protein-coding genes using a genome wide sgRNA library (Konermann et al., 2014) (Figure 2C). For library screening cells were treated for 14 days with 0.25 pM Ara-C, and cell viability monitored over the treatment period (Figure S3B). Following treatment sgRNA abundances were quantified and analyzed for quality control (Figure S3C-S3E). Transcript-level representation between T0 and T14 identified a host of genes enriched and depleted in Ara-C treated cells (Figure 2D and Figure S3F). Interestingly, both the correlation analysis and our forward genetic screen revealed DCK to be the most significantly depleted gene, thereby indicating that strong transcriptional activation of DCK by CRISPRa confers high sensitivity to Ara-C (Figure 1B and 2D). Indeed, this was confirmed by overexpressing the top-scoring DCK targeting sgRNA (Figure S3G). Furthermore, multiple genes suspected to modulate sensitivity to Ara-C were also identified (Table S4).
Gene set enrichment analysis identified a number of pathways congruent with our cell line analysis (Figure 2E and Table S3). Importantly, we identified a large overlap of 2,411 genes significantly enriched/depleted in both our cell line and protein-coding CRISPRa screening (Figure S3H). We subsequently validated a subset of these genes, including ZBP1, MUL1 and PI4K2A, whose expression was associated with poor prognosis and decreased disease-free survival (Figure 2F). Cells expressing the relevant sgRNAs demonstrated increased survival upon treatment with Ara-C (Figure 2G and Figure S3I), and a decrease in apoptosis (Figure 2H and Figure S3J), thereby validating our findings. Importantly, the proliferative capacity of cells was not affected by the overexpression of these sgRNAs (Figure 2I).
Functional Genome Wide Screening of IncRNAs in AML
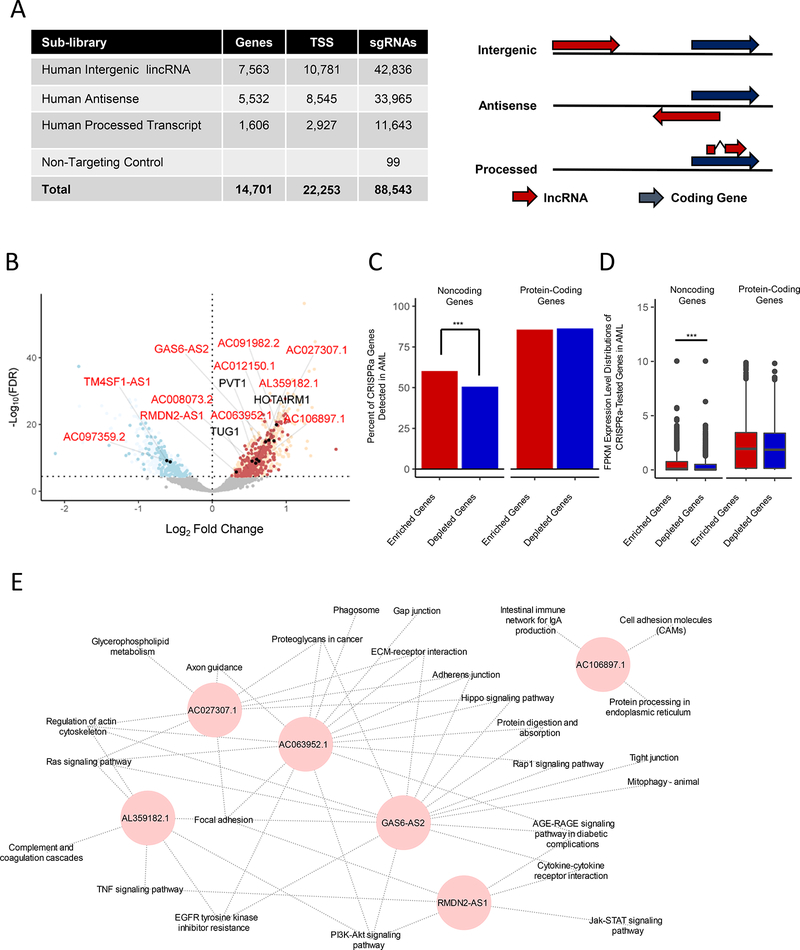
To study the functional roles of IncRNA genes in Ara-C resistance, we designed an sgRNA library using a comprehensive set of 14,701 lncRNA genes, covering all major classifications of lncRNAs (Figure 3A and Table S5). We designed at least 4 sgRNAs per lncRNA, accounting for 22,253 transcriptional start sites (TSSs), covering multiple TSSs per individual lncRNA. This resulted in a library with 88,444 targeting guides (Figure 3A). We termed this CRISPRa SAM- mediated approach "CRISPR activation of lncRNA” (CaLR).
Figure 3. CRISPRa Functional Screening of Noncoding Genes Modulating Ara-C Response.
(A) Left panel: summary of the CaLR library design specifications, including lncRNA gene numbers, transcriptional start sites (TSS), and total sgRNA numbers. Right panel: relationships between coding genes and lncRNA genes for corresponding lncRNA classifications. See also Table S5.
(B) Volcano plot summarizing the global changes in sgRNA representation of noncoding genes before and after 14 days of treatment with Ara-C. A subset of genes either validated herein to modulate Ara-C sensitivity (red text) or previously annotated in various cancer-related pathways (black text) are labeled. A false discovery rate threshold of 3.51e-5 was determined by analysis of nontargeting sgRNA negative controls at the transcript level (Figure S4H). Red points - enrichment in the CRISPRa screening; blue points - depletion in the CRISPRa screening; filled black points - genes validated herein. See also Figure S4E-I and Table S6.
(C) Percentages of significantly enriched or depleted protein-coding or noncoding genes from CRISPRa screens detected in the TCGA-LAML patient samples. Chi-squared test: ***, p = 6.92e-3,
(D) Gene expression level distributions of significantly enriched or depleted protein-coding or noncoding genes from CRISPRa screens detected in the TCGA-LAML patient samples. Wilcoxon rank-sum test: ***, p = 5.4e-7.
(E) Guilt-by-association pathway annotation of enriched genes identified in the CaLR screen. KEGG pathway gene sets were used for this analysis.
To test our library, we picked sgRNAs targeting the TUNA lncRNA gene (n=4 sgRNAs) (Figure S4A) and two alternative TSSs for the MIAT lncRNA gene (MIAT-01, n=5 sgRNAs, MIAT-06, n=4 sgRNAs), and we confirmed activation of each TSS using at least two sgRNAs (see Figure S3A, and Figure S4A-S4B). An additional set of randomized sgRNAs was also tested on HEK293T and MOLM14, revealing that the majority of sgRNAs demonstrated transcriptional activation in at least one of these cell lines (Figure S4D). Next, we carried out screening using our CaLR library similar to that for the protein-coding library above.
After quantifying sgRNA abundance, library preparations were analyzed as above for potential technical bias (Figure S4E-S4G). In order to estimate the false positive rate within our non-coding RNA screening, we included 99 non-targeting sgRNAs (Figure 3A). These 99 nontargeting sgRNAs behaved as expected (Figure S4G) and were utilized to determine an appropriate FDR cut-off to control for the false positive rate (Figure S4H). Interestingly, several cancer-associated lncRNA genes were identified among enriched sgRNAs, including Taurine Up-Regulated 1 (TUG1), HOXA Transcript Antisense RNA, Myeloid-Specific 1 (HOTAIRM1) and Plasmacytoma variant translocation 1 (PVT1) (Figure 3B, Figure S4I, and Table S6).
Interestingly, expression analysis of lncRNAs and coding genes from the TCGA AML patient cohort revealed that enriched lncRNAs from our screen tended to be both detected at a higher rate (p = 6.92e-3) and expressed more highly than depleted lncRNAs (p = 5.4e-7), whereas a similar pattern was not observed among the enriched/depleted protein-coding mRNAs (Figure 3C-3D).
Furthermore, guilt by association analysis of the enriched lncRNAs identified two distinct gene set networks: (1) oxidative phosphorylation and fatty acid metabolism and (2) leukemia development and progression (Figure S4J). Enrichment of these pathways in the first network is reflective of the role of the mitochondria in regulating nucleotide metabolism, while specific pathways enriched in the latter network include leukemia associated pro-survival pathways (e.g. Interferon response, IL6/JAK/STAT3 signaling, TNFa/NF«B signaling) (Steelman et al., 2004) (Stavropoulou et al., 2016).
We compiled a short list of novel annotated lncRNAs to characterize further, which were significantly enriched in both our functional screening and our cell line analysis (Figure 3B). Coexpression analysis to associate individual lncRNA transcript levels with their most highly correlated protein coding genes from our CCLE cell line panel identified many of the pathways uncovered in our global analysis, suggesting that these lncRNAs play roles in survival pathways know to affect leukemia and drug resistance (Figure 3E and Table S3).
Validation of Top lncRNA Candidates
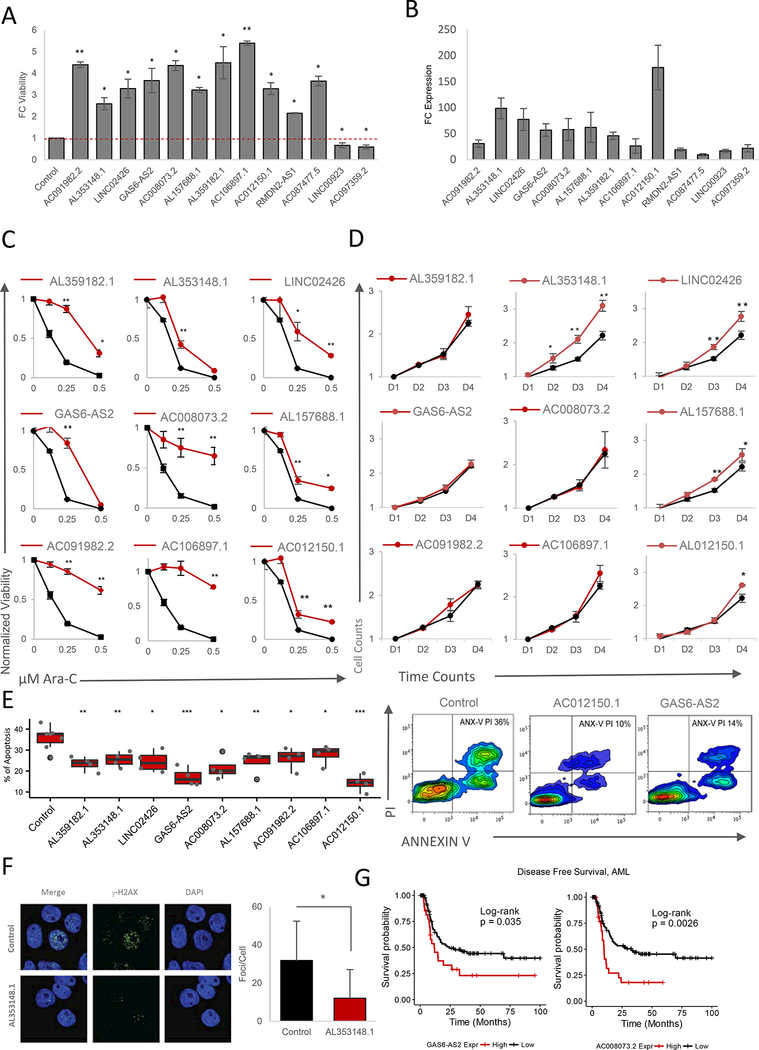
To validate the findings from our screening experimentally, we chose eleven genes significantly enriched and two genes significantly depleted in our screening for further characterization (Figure 3B and 3E). Of these 13 genes selected from our screening, 10 were also found to be candidate genes predicted to influence Ara-C response in our cell line analysis. The enriched sgRNAs resulted in a significant protection over control cells, while the two depleted genes resulted in decreased viability in response to Ara-C (Figure 4A). Indeed, we confirmed increased lncRNA expression across the different sgRNAs examined (Figure 4B). To further characterize the ability of induced lncRNA expression to resist Ara-C cytotoxicity, we treated cells expressing the relevant sgRNAs with Ara-C (Figure 4C and Figure S5A-S5B). Expression of each enriched sgRNA resulted in decreased Ara-C sensitivity (Figure 4C and Figure S5A), correlating with the protective effect observed in Figure 4A, while the depleted lncRNA genes also behaved as expected (Figure S5B).
Figure 4. Validation of CaLR Screening Results.
(A) Fold change (FC) of MOLM14 cell viability treated with 0.25 pM Ara-C for 48 hours. Data are represented as mean ± SD, n = 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01,***, p < 0.001.
(B) Fold change (FC) of expression levels of targeted lncRNAs upon overexpression of enriched sgRNAs versus endogenous levels. Data are represented as mean ± SD, n = 3.
(C) Ara-C efficacy measurements in MOLM14 cells expressing sgRNAs targeting indicating genes based on normalized MTS reads following 48 hours of treatment with the indicated concentrations of Ara-C. Data are represented as mean ± SD, n = 3, Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001
(D) Proliferation of unchallenged MOLM14 cells expressing sgRNAs targeting indicating genes. Proliferation is quantified over four days (D1-D4). Data are represented as mean ± SD, n = 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001.
(E) Left panel: modulation of apoptotic response upon stable expression of sgRNAs targeting a panel of significantly enriched sgRNAs as determined through CaLR screening in MOLM14 cells. The percentage of apoptosis is determined by annexin V and propidium iodide (PI) staining of MOLM14 cells stably infected with individual sgRNAs and treated with 0.25 pM Ara-C for 72 hours. Data are represented as mean ± SD, n = 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001. Right panel: representative flow cytometry plots of annexin V/PI staining intensities corresponding to two sgRNAs promoting survival versus nontargeting control.
(F) Immunofluroscence images (left panel) for DAPI and phospho-YH2A.X staining in MOLM14 cells stably infected with sgRNAs targeting the lncRNA genes shown, and treated with 25 pM Ara-C for 24 hours. Staining is quantified in the right panel. Data are represented as mean ± SD, n = 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001.
(G) Disease-free survival association with expression levels of GAS6-AS2 and AC008073.2, genes enriched in both noncoding CRISPRa screening and drug resistance-gene expression correlation analyses among patients treated with Ara-C therapy within the TCGA-LAML patient cohort. GAS6-AS2: VST expression level cutoff = 3.38 (low, n = 92; high, n = 29), log-rank test: p-value = 0.035. AC008073.2: VST expression level cutoff = 4.39 (low, n = 93; high, n = 28), log-rank test: p-value = 0.0026.
To address how these lncRNAs may be promoting cell viability, we examined lncRNA ability to promote either increased proliferation or increased survival. Out of our candidate lncRNAs, only three appeared to promote proliferation in the absence of Ara-C (AL353148.1, LINC02426; AL157688.1) (Figure 4D and Figure S5C-S5D), suggesting that their enrichment might be facilitated by increased proliferation. On the other hand, while all sgRNAs were able to promote increased survival to some extent (Figure 4E, Figure S5E), both AC012150.1 and GAS6-AS2 demonstrated a significant ability to attenuate apoptosis (Figure 4E, right panel). These results were further confirmed in an independent HL-60 hematopoietic cell line (Figure S5F-S5H).
Given that Ara-C promotes extensive genotoxic stress we tested if our IncRNAs may affect the DNA damage response (DDR). Indeed, we found that the lncRNA AL353148.1 affected DDR response following Ara-C treatment (Figure 4F).
For two of our candidate lncRNAs (GAS6-AS2 and AC008073.2), not only were they identified as candidates in both our cell line analysis and functional CaLR screening, but higher expression levels of these lncRNA genes were also associated with poor prognosis and decreased disease-free survival in AML patients treated with Ara-C (Figure 4G).
Taken together these data reify our screening process as a platform to identify clinically relevant lncRNAs that may modulate Ara-C cytotoxicity through targeting a number of cellular processes.
lncRNA GAS6-AS2 Regulates the GAS/AXL Signaling Axis
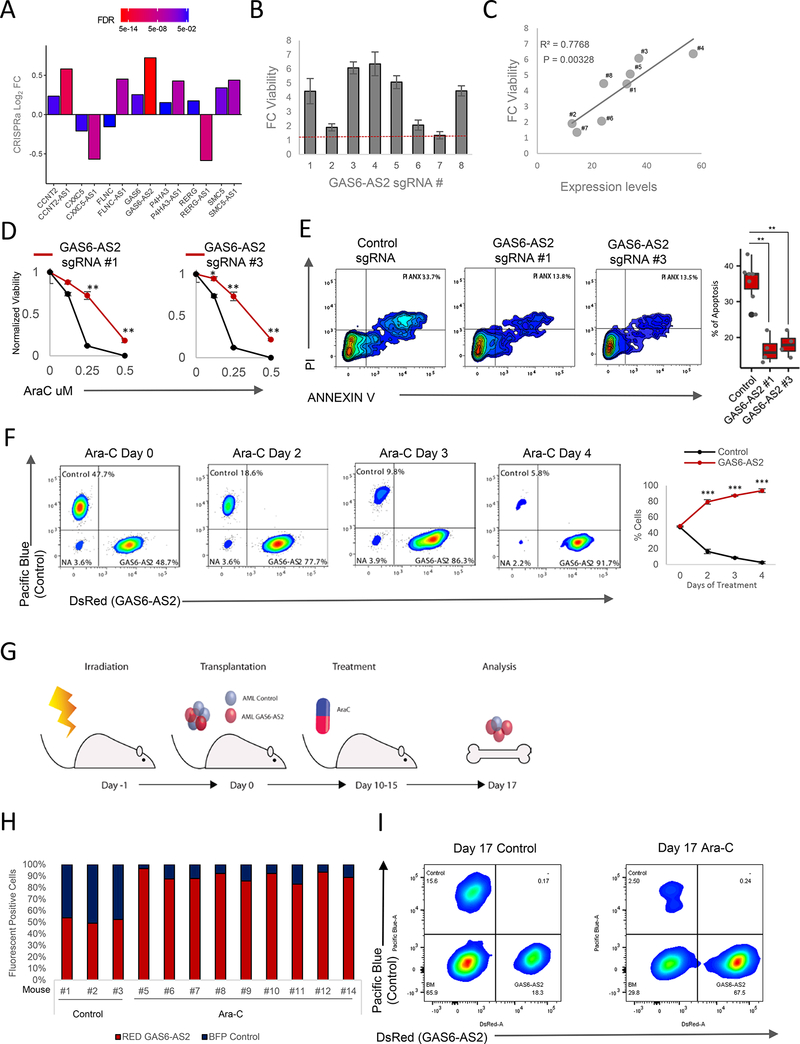
We next integrated our computational analysis with both the coding and non-coding functional screens. Statistical analysis demonstrated significant enrichment of 7 sense-antisense gene pairs (Chi-Squared Test, p < 2.2e-16) (Figure 5A and Figure S2B). Of the 7 cognate pairs identified, GAS6/GAS6-AS2 appeared to be one of the best candidate pairs for further analysis as both were highly enriched, GAS6 is already known to play an important role in drug resistance in cancer, including AML (Figure S6A), while the role and function of GAS6-AS2 remains unknown.
Figure 5. GAS6-AS2 Promotes Drug Resistance In Vitro and In Vivo.
(A) Integration of drug resistance-gene expression correlative analysis and forward genetic screenings identifies seven sense-antisense gene pairs which pass all significance thresholds, a higher number than expected by chance alone (Chi-squared test: p = 9.85e-7).
(B) Fold change (FC) of MOLM14 cell viability treated with 0.25 pM Ara-C for 48 hours. Cells expressing individual sgRNAs targeting GAS6-AS2. Data are represented as mean ± SD, n = 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001.
(C) Pearson correlation between cell viability versus GAS6-AS2 expression level for each of the 8 sgRNAs targeting GAS6-AS2.
(D) Ara-C efficacy measurements in MOLM14 cells expressing sgRNAs #1 and #3 targeting GAS6-AS2 based on normalized MTS reads following 48 hours of treatment. Data are represented as mean ± SD, n = 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001.
(E) Left panel: representative flow cytometry data of MOLM14 cells expressing either control or GAS6-AS2-targeting sgRNAs, treated with 25 pM Ara-C for 24 hours and labeled with viability (propidium iodide (PI)) and apoptotic (annexin V) markers. Right panel percentage of apoptosis determined from quantification of staining results. Data are represented as mean ± SD, n > 3, Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001.
(F) Competition assay between populations of MOLM14 control-Blue and MOLM14 GAS6- AS2-Red following 25 pM Ara-C treatment. Left panels: representative flow cytometry plots. Right panel: ratios between red and blue cells over time. Data are represented as mean ± SD, n > 3. Welch two sample t-test: *, p < 0.05. **, p < 0.01, ***, p < 0.001.
(G) Schematic of an orthotopic xenograft competition assay between control (blue) and GAS6-AS2 (Red) MOLM14 cells with Ara-C treatment.
(H) Ratios of control (blue) versus GAS6-AS2 (Red) MOLM14 cells from bone marrow of mice treated and analyzed at day 17 as outlined in Figure 5G.
(I) Representative flow cytometry results of cells harvested from mouse bone marrow 17 days following transplantation and treatment with vehicle or Ara-C for 5 days.
To confirm the on-target effect of our CRISPRa sgRNA, we overexpressed 8 different GAS6-AS2-targeting sgRNAs (Figure S6B). As expected, the majority of these sgRNAs led to a significant increase in cell survival (Figure 5B). Importantly, we found a strong correlation between the levels of GAS6-AS2 activation and the resistance to Ara-C, indicating a dose dependent specific effect of GAS6-AS2 (Figure 5B and 5C). Similarly, expression of the strong inducer sgRNAs #1 and #3 promoted decreased sensitivity to Ara-C (Figure 5C and 5D) and a potent ability to reduce apoptosis (Figure 5E). Thus, the GAS6-AS2 lncRNA appears to be a bona fide promoter of Ara-C resistance.
AML is known to develop as a multi-clonal disease, and resistant clones are frequently observed in early stages of the disease. The selective pressure of treatment leads to rapid clonal evolution and the emergence of resistant clones. Indeed, following Ara-C treatment of a mixed population of two MOLM14 cells, one expressing high GAS6-AS2 (labeled with red florescent protein) and the other expressing a non-targeting sgRNA (labeled with blue florescent protein), the GAS6-AS2 expressing clone emerged as dominant and was significantly enriched post-treatment (Figure 5F). These results were also confirmed in vivo as outlined in Figure 5G, with NSG mice engrafted with equal numbers of GAS6-AS2 overexpressing and control MOLM14 cells. Analysis of the bone marrow found significant enrichment (p = 0.002) of the GAS6-AS2-Red cells (Red/Blue cells=10.9±6.4) (Figure 5H-5I, and Figure S6D-S6E). Importantly, within a non-treated cohort, both populations of cells were present in an equal ratio, demonstrating that GAS6-AS2 did not exert a proliferative advantage (Figure 5H and 5I). Furthermore, mice transplanted with GAS6-AS2 overexpressing cells alone had a greater tumor burden post-Ara-C treatment as compared with control cells (Figure S6F).
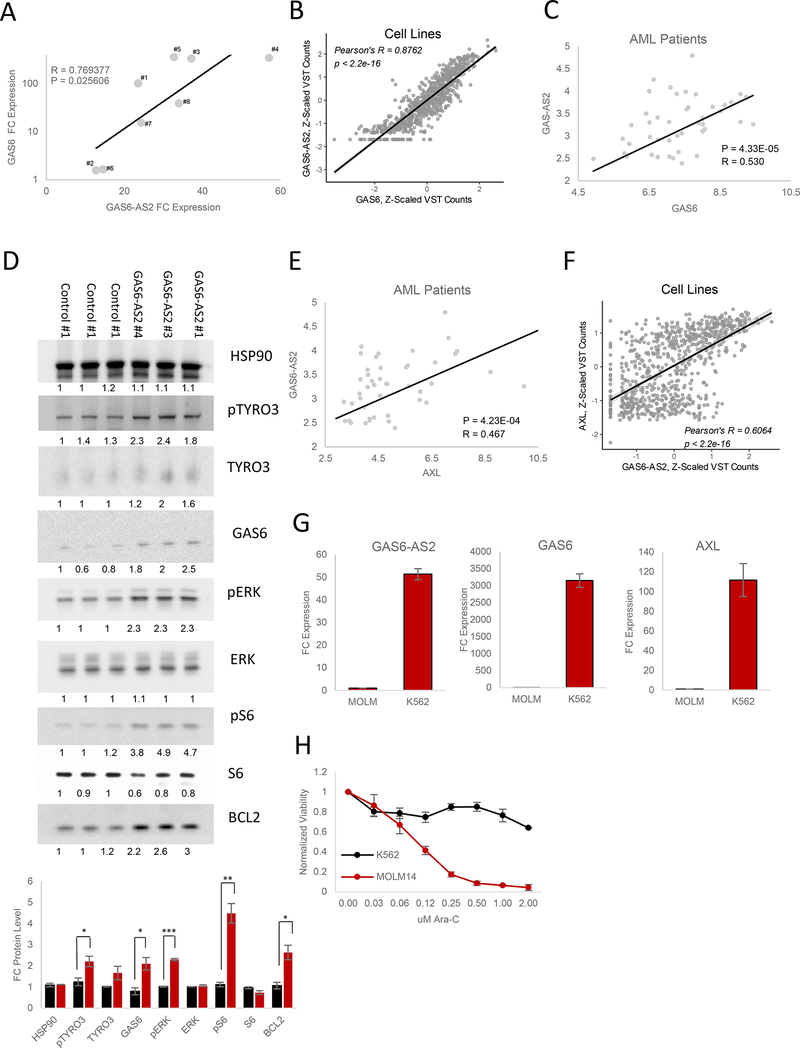
Several lncRNAs have been shown to exert their functional role by cis-regulation of neighboring genes (Rinn and Chang, 2012), and this is further supported by our genome wide analysis of sense-antisense cognate gene pairs (Figure 1E-1F and Figure 5A). As GAS6-AS2 lies in an antisense head-to-head manner with GAS6 (Figure S6G), we hypothesized that the GAS6/GAS6-AS2 cognate gene pair may function in this manner. Importantly, GAS6-AS2 displayed nuclear (and cytoplasmic) localization (Figure S6H), suggesting that it may have the potential to regulate the GAS6 locus. GAS6 is an important ligand for the TYRO3-AXL-MER2K (TAM) receptor tyrosine kinase signaling axis, controlling known pro-survival signals in AML (Wu et al., 2017). Indeed, upregulation of GAS6/TAM signaling strongly correlates with resistance to chemotherapy and is a predictor of poor survival (Hong et al., 2008). In line with our hypothesis, GAS6 expression levels were found to be strongly correlated with GAS6-AS2 expression upon CRISPRa modulation (Figure 6A). In addition, we also observed a striking correlation between these cognate gene pairs across the diverse 760 CCLE cell line panel (Pearson’s R = 0.8762, p < 2.2e-16) (Figure 6B), as well as for a diverse set of primary human cancer types including AML (Figure 6C and Figure S7A).
Figure 6. GAS6-AS2 Activates GAS6/TAM Signaling.
(A) Pearson correlation between GAS6-AS2 and GAS6 expression levels following GAS6- AS2 activation. Data are represented as mean of triplicate measurements.
(B) Pearson correlation between GAS6-AS2 and GAS6 expression levels across the 760 cancer cell lines analyzed (Figure 1A-B).
(C) Pearson correlation between GAS6-AS2 and GAS6 expression levels in AML patient samples.
(D) Western blot analysis of differential GAS6/TAM signaling activation in response to individual control or GAS6-AS2 sgRNA overexpression.
(E) Pearson correlation between GAS6-AS2 and AXL expression levels in AML patient samples.
(F) Pearson correlation between GAS6-AS2 and AXL expression levels across the 760 cancer cell lines analyzed (Figure 1A-B).
(G) Expression levels of GAS6-AS2, GAS6, and AXL in MOLM14 and K562 cell lines.
(H) Ara-C efficacy measurements in MOLM14 and K562 cell lines, based on normalized MTS reads following 48 hours of treatment with the indicated concentrations of Ara-C. Data are represented as mean ± SD, n = 3.
Activation of the GAS6/TAM pathway has been reported to promote MAPK, JAK/STAT, and NFkB signaling (Schoumacher and Burbridge, 2017), with both MEK-ERK and S6K-RPS6 signaling axes being known downstream targets of TAM signaling (Xu et al., 2017). Western blot analysis of lysates from cells expressing three distinct GAS6-AS2 targeting sgRNAs confirmed activation of the GAS6/TAM pathway (Figure 6D). Importantly, both pERK and pRPS6 were strongly phosphorylated in response to GAS6-AS2 activation (Figure 6D).
Surprisingly in a variety of cancer subtypes, including AML, GAS6-AS2 expression levels share strong correlations not only with GAS6 but also to its target receptor AXL (Figure 6E and S6B). AXL correlation was also mirrored in our 760 CCLE cell line panel (Pearson’s R = 0.6064, p < 2.2e-16) (Figure 6F) and by overexpression of GAS6-AS2 in both MOLM14 and HEK293 cells in vitro (Figure S7C). These data suggested that GAS6-AS2 may be able to regulate the TAM receptor signaling axis at a number of levels.
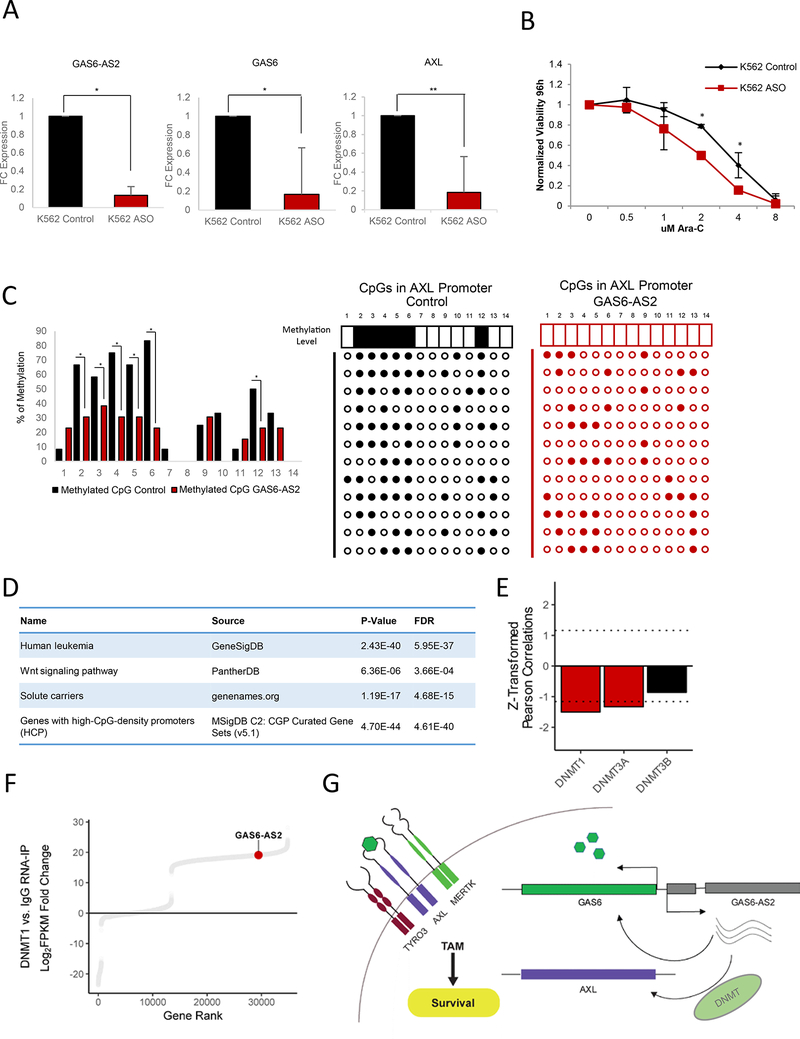
To further investigate the role of GAS6-AS2 in regulating GAS6 and AXL, we took advantage of the K562 leukemia cell line, which we found to express high levels of GAS6-AS2, GAS6, and AXL relative to MOLM14 (Figure 6G) and which we demonstrate to be highly resistant to Ara-C treatment (Figure 6G). Interestingly, knockdown of GAS6-AS2 using two specific locked nucleic acid (LNA)-enhanced anti-sense oligonucleotides (ASOs) led to a significant decrease in both GAS6 and AXL mRNA levels (Figure 7A and Figure S7I-S7J) as well as an increased sensitivity of K562 cells to the activity of Ara-C (Figure 6B).
Figure 7. GAS6-AS2 Demonstrates Trans-Regulation of AXL.
(A) Fold change (FC) of GAS6-AS2, GAS6, and AXL in response to GAS6-AS2 knockdown via ASO in K562 cells. Data are represented as mean ± SD, n = 3. Welch two sample t- test: *, p < 0.05. **, p < 0.01, ***, p < 0.001.
(B) Modulation of Ara-C response upon GAS6-AS2 knockdown via ASO in K562 cells. Data are represented as mean ± SD, n = 3, Welch two sample t-test: *, p < 0.05. **, p < 0.01,***, p < 0.001.
(C) Methylation of CpG islands in the HEK293T AXL promoter following modulation of GAS6-AS2 expression. n = 12, Chi-square test: *, p < 0.05. **, p < 0.01, ***, p < 0.001.
(D) Gene ontology analysis of coding genes clustered with GAS6-AS2 as determined by k- means clustering (cluster #6 in Figure S7D).
(E) Drug sensitivity-gene expression Pearson correlation values of DNA methyltransferases. Genes enriched beyond a Z-score threshold of ± 1.16 are colored in red. See also Figure 1B.
(F) Distribution of FPKM-normalized transcript abundances associated with DNMT1 versus IgG.
(G) Model summarizing the mechanism by which GAS6-AS2 regulates GAS6/TAM signaling.
Previous studies found that AXL transcription is regulated by methylation of CpGs upstream of its TSS (Mudduluru and Allgayer, 2008). Direct methylation analysis using a bisulfite assay identified 6 highly methylated sites in the AXL promoter. Correspondingly, GAS6-AS2 overexpressing cells show significant decreases in methylation of these CpG sites (Figure 7C), suggesting that GAS6-AS has the potential to act in both a cis- and trans-acting manner.
To characterize the global function of GAS6-AS2 in cancer we performed an unbiased K- means clustering based on coding and non-coding gene expression across 53 AML patients (Garzon et al., 2014) (Figure S7D). A large number of genes known to be regulated by promoter methylation were clustered together with GAS6-AS2 (Figure 7D), supporting our hypothesis that GAS-AS2 mediates CpG modification.
Based on these results, we hypothesized that GAS6-AS2 trans-activity may function through a DNA methyltransferase. An unbiased screening of our CCLE panel for candidate DNA methyltransferases that would correlate with Ara-C sensitivity identified decreased expression of DNMT1 and DNMT3A (Figure 7E). Importantly, we observed GAS6-AS2 to be significantly enriched in RNAs bound to DNMT1 using a DNMT1 RNA-IP sequencing (RIP-Seq) dataset (Figure 7F) (Di Ruscio et al., 2013). This suggests that GAS6-AS2 mediates trans-regulation of AXL by coordinating activity of DNMT proteins at the AXL promoter.
Thus, our data support a model whereby increased transcription and expression of GAS6- AS2 promotes upregulation of both the GAS6 ligand and its TAM receptors to promote cellular survival and resistance to Ara-C treatment in AML (Figure 7G).
DISCUSSION
Although thousands of IncRNAs have now been detected and annotated in the human genome, the need to characterize their functions remains a critical challenge. Here, we developed a global approach to integrate computational analysis of cell line pharmacogenomic data sets with functional CRISPRa screens targeting coding and noncoding genes. This approach aimed to uncover integrated mechanisms regulating normal cellular homeostasis and disease and was applied to identifying functional lncRNAs modulating the cytotoxic effect of Ara-C, a front-line chemotherapy agent frequently used in the treatment of AML patients.
Because lncRNAs are poorly annotated, we developed a bioinformatic framework to facilitate the prioritization of candidate genes by their functional and physiological relevance. Using pharmacogenomic and transcriptomic data, we obtained a list of coding and non-coding genes whose expression levels are associated with cellular response to Ara-C. Our list identified many of the coding genes and pathways previously shown to regulate the response to Ara-C treatment. In addition, not only did this analysis reveal a large number of lncRNAs to influence response to Ara-C, but it also implicated a pattern of cis-regulation by lncRNAs on their adjacent cognate coding genes. Thus, this analysis provides us with a unique resource that can both deliver a wealth of novel predictive biomarkers for response to therapy and prioritize functionally relevant genes identified through functional screening.
For our purposes, functional screening was carried out with CRISPRa based technologies using both an established protein-coding sgRNA library (Konermann et al., 2014) and a new genome wide non-coding sgRNA (CaLR) library. In adapting the CRISPRa technology, we found that appropriate cell models and optimization are critical. For our new CaLR library we chose to generate sgRNAs targeting lncRNA genes that are well annotated. This in turn enabled extensive promoter coverage upstream of TSSs and targeting of multiple TSSs for individual lncRNA genes. In addition, this screening approach offers the advantage of driving lncRNA overexpression from the endogenous genomic locus, enabling us to capture c/s-acting and nuclear lncRNA functions, which cannot be readily studied by traditional overexpression approaches (Shechner et al., 2015). It should be noted however, that as some genes harbor other small noncoding RNAs including microRNAs and snoRNAs within intronic regions, driving expression from the endogenous promoter may also result in their expression.
Indeed, our novel CaLR approach identified lncRNAs that facilitate resistance to Ara-C treatment. These data demonstrate that many lncRNA genes are functionally relevant for cancer and modulate distinct cellular programs. Integrating coding and noncoding screening approaches also allows us to categorize lncRNA genes by function, and although we have applied these libraries to the identification of novel genes involved in chemotherapeutic resistance, this platform alone can be applied to the functionalization of lncRNAs across a wide range of biological questions.
Thus, complete integration of computational cell line analysis, coding/non-coding CRISPRa screens, and patient outcome data resulted in the discovery of a distinct set of 7 cognate sense- antisense gene pairs. Of these seven pairs, we pursued the GAS6/GAS6-AS2 cognate gene pair as drivers of resistance to Ara-C in AML. We found that GAS6-AS2 functions through cis- regulation of its adjacent cognate gene, coding for the GAS6 ligand, as well as the transregulation of its receptor AXL to drive aberrant downstream signaling of this pathway.
While each of these approaches as individual modules (computational and screening) has been shown to be useful to identify genes regulating specific cellular processes, each harbors inherent limitations and bias requiring extensive validation of hits. However, our integrated approach described here serves as a more powerful framework for the screening and discovery of protein-coding and non-coding networks regulating biological processes, thereby providing a resource to facilitate improvements towards the annotation and functionalization of non-coding RNAs at large. Our analysis suggests that there are a substantial number of coding and noncoding genes that at a minimum serve as predictive biomarkers that correlate with differential Ara-C responses and may serve as therapeutic targets for the tuning of Ara-C response through modulation of their expression levels. Indeed, this approach may facilitate the identification of novel high confidence and clinically relevant therapeutic opportunities across a broad spectrum of human diseases.
Supplementary Material
Acknowledgments
We thank P.P.P. laboratory members for critical discussions. This work was supported by EMBO Long-Term Fellowship (ALTF 318–2013) and Fulbright awards to A.C.B. A.C. was funded by NCI grant 5T32CA009216–34, and a Burroughs Wellcome Fund Career Award for Medical Scientists. G.M.C was supported by NIH grants RM1 HG008525 and P50 HG005550. P.P.P. is supported by an NIH NCI R35 grant CA197529 and through support from the Ludwig Center at Harvard. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the NIH. The data used for the analyses described in this manuscript were obtained from the GTEx Portal. The results published here are fully or partially based upon data generated by the Cancer Target Discovery and Development (CTD2) Network established by the NCI Office of Cancer Genomics. The authors would like to thank Rory Kirchner of the Harvard Chan Bioinformatics Core, Harvard T.H. Chan School of Public Health, Boston, MA for assistance with computational analysis. The authors would like to thank Davide Cora and Claudio Isella of the Dept. of Oncology, and GenoBiToUS, Genomics and Bioinformatics Service, University of Turin, Italy for assistance with computational analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
A.C.B. conceived the project, designed and performed experiments, interpreted results, and cowrote the manuscript. A.C. designed and created the CaLR library, designed and performed experiments, and interpreted results. Y.R.L., D.N., and J.V. performed experiments. S.V. and M.S. designed the CaLR library. A.C.B, J.D.L. and P.P. carried out computational analysis. J.L.R. and G.M.C. supervised experimental designs. J.G.C. designed and supervised experiments, interpreted results and co-wrote the manuscript. P.P.P. conceived the project, supervised experimental designs, interpreted results, and co-wrote the manuscript. A.C.B., J.G.C., and P.P.P. provided overall project leadership.
Declaration of Interest
G.M.C. is a founder and advisor for Editas Medicine. G.M.C. has equity in Editas and Caribou Biosciences (for full disclosure list, please see: http://arep.med.harvard.edu/gmc/tech.html). All other authors declare no competing interests.
References
- Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet, J., Billis K, Garda Giron, C., Hourlier T, et al. (2016). The Ensembl gene annotation system. Database (Oxford). 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin A. a., Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. (2012). The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Bodycombe NE, Cheah JH, Price EV, Liu K, Schaefer GI, Ebright RY, Stewart ML, Ito D, Wang S, et al. (2013). An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 154, 1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benekli M (2002). Constitutive activity of signal transducer and activator of transcription 3 protein in acute myeloid leukemia blasts is associated with short disease-free survival. Blood 99, 252-257. [DOI] [PubMed] [Google Scholar]
- Cabili M, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, and Rinn JL (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2013). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med 368, 2059-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ, et al. (2015). Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Tuttle M, Pruitt BW, Ben E-C, Chari R, Dmitry T-O, Haque SJ, Cecchi RJ, Kowal EJK, Buchthal J, et al. (2016). Comparison of Cas9 activators in multiple species. Nat Methods 13, 563-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Xu H, Aronow BJ, and Jegga AG (2007). Improved human disease candidate gene prioritization using mouse phenotype. BMC Bioinformatics 8, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Gong Y, Ma Y, Lu K, Lu X, Pierce LA, Thompson RC, Muller S, Knapp S, and Wang J (2013). Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin. Cancer Res 19, 1748-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. (2016). TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 44, e71-e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, and Huber W (2005). BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439-3440. [DOI] [PubMed] [Google Scholar]
- Farge T, Saland E, de Toni F, Aroua N, Hosseini M, Perry R, Bosc C, Sugita M, Stuani L, Fraisse M, et al. (2017). Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov 7, 716-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. (2012). Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 483, 570-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Volinia S, Papaioannou D, Nicolet D, Kohlschmidt J, Yan PS, Powell BL, Kolitz JE, Moore JO, Eisfeld A, et al. (2014). Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc. Natl. Acad. Sci. U. S. A. 1, 1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, and Ploegh HL (2014). Resource Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold N, Rudd SG, Ljungblad L, Sanjiv K, Myrberg IH, Paulin CBJ, Heshmati Y, Hagenkort A, Kutzner J, Page BDG, et al. (2017). Targeting SAMHD1 with the Vpx protein to improve cytarabine therapy for hematological malignancies. Nat. Med 23, 256-263. [DOI] [PubMed] [Google Scholar]
- Hon C, Ramilowski J, Harshbarger J, Bertin N, Rackham O, Gough J, Denisenko E, Schmeier S, Poulsen T, Severin J, et al. (2017). An atlas of human long non-coding RNAs with accurate 5‘ ends. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CC, Lay JD, Huang JS, Cheng AL, Tang JL, Lin MT, Lai GM, and Chuang SE (2008). Receptor tyrosine kinase AXL is induced by chemotherapy drugs and overexpression of AXL confers drug resistance in acute myeloid leukemia. Cancer Lett 268, 314-324. [DOI] [PubMed] [Google Scholar]
- Hu X, Feng Y, Zhang D, Zhao SD, Hu Z, Greshock J, Zhang Y, Yang L, Zhong X, Wang L-P, et al. (2014). A Functional Genomic Approach Identifies FAL1 as an Oncogenic Long Noncoding RNA that Associates with BMI1 and Represses p21 Expression in Cancer. Cancer Cell 26, 344-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M (2015). The emerging role of lncRNAs in cancer. Nat. Med 21, 1253-1261. [DOI] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. (2012). Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 150, 1107-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet 47, 199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayavelu K, and Srivastava VM (2015). Pharmacogenomics RNA expression of genes involved in cytarabine metabolism and transport. 16, 877-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, et al. (2017a). Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung J, Konermann S, Gootenberg JS, Abudayyeh OO, Platt RJ, Brigham MD, Sanjana NE, and Zhang F (2017. b). Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc 12, 828-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaimal V, Bardes EE, Tabar SC, Jegga AG, and Aronow BJ (2010). ToppCluster: a multiple gene list feature analyzer for comparative enrichment clustering and network-based dissection of biological systems. Nucleic Acids Res 38, W96-W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, and Goto S (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, and Tanabe M (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42, D199-D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. (2014). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba JK (2009). Genetic factors influencing cytarabine therapy. Pharmacogenomics 10, 1657-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Sherick a B., Eisenman KM, Sather S, McGranahan A, Armistead PM, McGary CS, Hunsucker S. a, Schlegel J, Martinson H, Cannon C, et al. (2013). Aberrant Mer receptor tyrosine kinase expression contributes to leukemogenesis in acute myeloid leukemia. Oncogene 32, 1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Han L, Roebuck P, Diao L, Liu L, Yuan Y, Weinstein JN, and Liang H (2015). TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 75, 3728-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, and Liu XS (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923-930. [DOI] [PubMed] [Google Scholar]
- Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y, et al. (2017). CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science (80-. ). 355, eaah7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal; Vol 17, No 1 Next Gener. Seq. Data Anal. [Google Scholar]
- Mudduluru G, and Allgayer H (2008). The human receptor tyrosine kinase Axl gene - promoter characterization and regulation of constitutive expression by Sp1, Sp3, and CpG methylation. Biosci. Rep 28, 161-176. [DOI] [PubMed] [Google Scholar]
- Perron U, Provero P, and Molineris I (2017). In silico prediction of lncRNA function using tissue specific and evolutionary conserved expression. BMC Bioinformatics 18, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, and Chang HY (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet 17, 47-62. [DOI] [PubMed] [Google Scholar]
- Ramos N, Mo C, Karp J, and Hourigan C (2015). Current Approaches in the Treatment of Relapsed and Refractory Acute Myeloid Leukemia. J. Clin. Med 4, 665-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees MG, Seashore-Ludlow B, Cheah JH, Adams DJ, Price EV, Gill S, Javaid S, Coletti ME, Jones VL, Bodycombe NE, et al. (2016). Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat. Chem. Biol 12, 109-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, and Vilo J (2016). g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res 44, W83-W89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, and Chang HY (2012). Genome Regulation by Long Noncoding RNAs. Annu. Rev. Biochem 81, 145-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, et al. (2013). DNMT1- interacting RNAs block gene-specific DNA methylation. Nature 503, 371-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saland E, Boutzen H, Castellano R, Pouyet L, Griessinger E, Larrue C, de Toni F, Scotland S, David M, Danet-Desnoyers G, et al. (2015). A robust and rapid xenograft model to assess efficacy of chemotherapeutic agents for human acute myeloid leukemia. Blood Cancer J. 5, e297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AM, and Chang HY (2016). Long Noncoding RNAs in Cancer Pathways. Cancer Cell 29, 452-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Oellerich T, Baldauf H-M, Schwarz S-M, Thomas D, Flick R, Bohnenberger H, Kaderali L, Stegmann L, Cremer A, et al. (2016). SAMHD1 is a biomarker for cytarabine response and a therapeutic target in acute myeloid leukemia. Nat. Med 23, 250-255. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M, and Burbridge M (2017). Key Roles of AXL and MER Receptor Tyrosine Kinases in Resistance to Multiple Anticancer Therapies. Curr. Oncol. Rep 19, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, and Rinn JL (2015). Multiplexable, locus- specific targeting of long RNAs with CRISPR-Display. Nat. Methods 12, 664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, and Ideker T (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavropoulou V, Kaspar S, Brault L, Sanders MA, Juge S, Morettini S, Tzankov A, Iacovino M, Lau I-J, Milne TA, et al. (2016). MLL-AF9 Expression in Hematopoietic Stem Cells Drives a Highly Invasive AML Expressing EMT-Related Genes Linked to Poor Outcome. Cancer Cell 30, 43-58. [DOI] [PubMed] [Google Scholar]
- Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, and McCubrey JA (2004). JAK/STAT, Raf/MEK/ERK, PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia 18, 189-218. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A 102, 15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Ma Z, Hu W, Wang D, Gong B, Fan C, Jiang S, Li T, Gao J, and Yang Y (2017). Molecular insights of Gas6/TAM in cancer development and therapy. Cell Death Dis 8, e2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Chen Y, Dutra-Clarke M, Mayakonda A, Hazawa M, Savinoff SE, Doan N, Said JW, Yong WH, Watkins A, et al. (2017). BCL6 promotes glioma and serves as a therapeutic target. Proc. Natl. Acad. Sci. U. S. A 114, 3981-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, Bindal N, Beare D, Smith J. a., Thompson IR, et al. (2013). Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 41, 955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.