 Integrated analysis of DNA modification and gene expression was conducted in mouse hepatoocellular adenomas promoted by phenobarbital.
Integrated analysis of DNA modification and gene expression was conducted in mouse hepatoocellular adenomas promoted by phenobarbital.
Abstract
Phenobarbital (PB) is a nongenotoxic hepatocellular carcinogen in rodents. PB induces hepatocellular tumors by activating the constitutive androstane receptor (CAR). Some previous research has suggested the possible involvement of epigenetic regulation in PB-promoted hepatocellular tumorigenesis, but the details of its molecular mechanism are not fully understood. In the present study, comprehensive analyses of DNA methylation, hydroxymethylation and gene expression using microarrays were performed in mouse hepatocellular adenomas induced by a single 90 mg kg–1 intraperitoneal injection dose of diethylnitrosamine (DEN) followed by 500 ppm PB in the diet for 27 weeks. DNA modification and expression of hundreds of genes are coordinately altered in PB-induced mouse hepatocellular adenomas. Of these, gene network analysis showed alterations of CAR signaling and tumor development-related genes. Pathway enrichment analysis revealed that differentially methylated or hydroxymethylated genes belong mainly to pathways involved in development, immune response and cancer cells in contrast to differentially expressed genes belonging primarily to the cell cycle. Furthermore, overlap was evaluated between the genes with altered expression levels with 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) alterations in mouse hepatocellular adenoma induced by DEN/PB and the genes with altered expression levels in the liver of CD-1 mice or humanized chimeric mice treated with PB for 7 days. With the integration of transcriptomic and epigenetic approaches, we detected candidate genes responsible for early key events of PB-promoted mouse hepatocellular tumorigenesis. Interestingly, these genes did not overlap with genes altered by the PB treatment of humanized chimeric mice, thus suggesting a species difference between the effects of PB in mouse and human hepatocytes.
Introduction
Phenobarbital (PB) is a nongenotoxic agent which has been shown to produce hepatocellular tumors in rats and mice.1 PB is considered to increase hepatocellular tumors by activating the CAR and inducing its nuclear translocation.2,3 In the nucleus, the CAR forms a heterodimer with the retinoid X receptor (RXR) and regulates the expression of downstream genes by binding PB-responsive enhancer modules in their promoters.4–6 Previous studies have shown that none of the Car–/– mice developed liver tumors, whereas all Car+/+ mice developed hepatocellular carcinoma and/or adenoma by PB treatment after tumor initiation with a genotoxic carcinogen, diethylnitrosamine (DEN).7 The potent mouse CAR activator 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) also does not produce liver tumors in mice with or without DEN initiation in Car–/– mice.8
In recent years, epigenetic modifications associated with gene expression regulation have been notable as important indicators of biological processes and diseases including tumorigenesis. Well-defined mechanisms of epigenetic regulation are DNA modification such as changing cytosine (C) to 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC).9–13 The cytosine residues at CpG sites can be methylated to form 5mC in mammalian genomes. Methylated-CpG plays various roles in gene and chromatin regulation, one of which is interaction with other epigenetic modifications and 5mC-specific binding proteins in a coordinated manner which leads to gene repression at the gene promoter. In contrast, 5hmC occurs in DNA demethylation processes. 5mC can be enzymatically oxidized to 5hmC, which can be further oxidized to unmethylated cytosine. The 5hmC levels at gene promoters and enhancers may be partially associated with the transcriptional activation of genes. Several reports have suggested that alterations in 5mC and 5hmC profiles correlate with various diseases including hepatic cancers.14–18
Some previous research has suggested the possible involvement of epigenetic regulation in carcinogenic promotion after administration of PB. Exposure to PB in mice globally altered the patterns of both 5mC and 5hmC over the entire genome including promoter regions in liver tissue.18–23 There were several genes differentially expressed with correlated epigenetic changes and possible associations with carcinogenesis.23 In addition, 5mC and 5hmC alterations also occurred in PB-elicited tumor tissues of the liver.18,24–26 At the promoter regions of some genes, decreased 5hmC and increased 5mC were observed in PB-induced mouse liver tumors.26
Regarding human relevance of the CAR-mediated MOA in hepatocellular tumor production, numerous publications have mentioned that the CAR-mediated MOA is qualitatively not relevant to humans, based on the lack of the key event of increased cell proliferation in PB-induced hepatocellular tumorigenesis.3,27–42 PB does not stimulate replicative DNA synthesis in hepatocytes of humanized chimeric mice39 and in a number of studies does not induce replicative DNA synthesis in cultured human hepatocytes.31,41,43 Epidemiological reports have also supported the conclusion of no increased risk regarding liver tumors in PB-treated humans.1,3,44
In contrast, some studies of PB treatment indicated that PB induces cell cycle transcriptional responses in the humanized CAR8 and humanized CAR/PXR45 mouse liver, and, furthermore, that PB-treatment produced liver tumors in the humanized CAR/PXR mouse similar to wild type mice but at a significantly lesser extent than the wild type mice.46 Schwarz's group indicated that the human relevance of the tumorigenicity of PB through CAR activation remains the subject of an ongoing debate.37,47–50
We should recognize that the human receptors of the hCAR/hPXR mouse model are operational in a mouse hepatocyte environment. Although the mouse CAR/PXR has been replaced by that of human, cell proliferation is likely to occur in the same way with wild type mouse since all other genes are those of the original mouse. To enhance the scientific understanding of the human relevance of CAR-mediated liver tumorigenesis, it is necessary to determine the mechanism of action for CAR-mediated liver tumorigenesis. Thus, it is important to determine key molecule(s) (especially in the early phase of treatment) involved in CAR-mediated hepatocellular tumorigenesis in rodents and to investigate its human relevance.
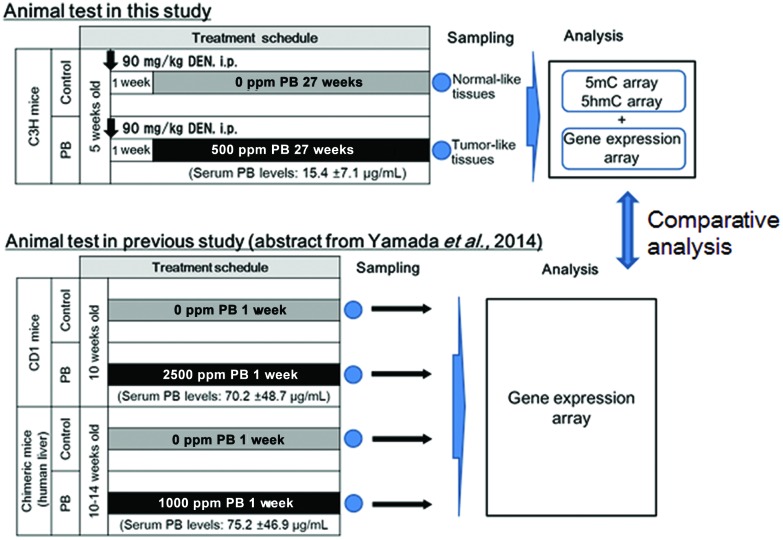
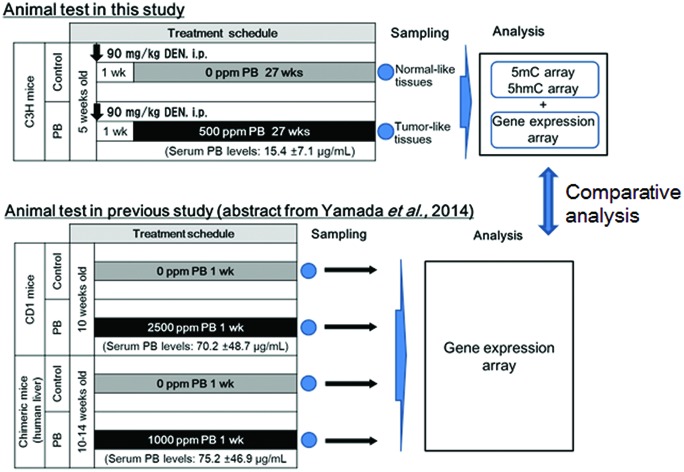
While epigenetics are suggested to be related to PB-mediated liver tumorigenesis,18–20,22,23,25,26,51 as mentioned above, the present study focused on genes with alterations of DNA methylation and hydroxymethylation with concomitant alteration of expression levels. These genes were selected from PB-induced hepatocellular adenomas in mice, by comprehensive analyses of DNA methylation, hydroxymethylation and gene expression using microarrays; we determined the differences in 5mC, 5hmC and transcriptional profiles between hepatocellular adenomas in mice treated with DEN + PB (DEN/500 ppm PB) and control liver samples in age-matched mice treated with DEN alone (DEN/0 ppm PB), and then performed integrated analysis to discover genes with epigenetic and transcriptional correlations (Fig. 1). Using these selected genes, to find the candidates of key genes of CAR-mediated hepatocellular tumorigenesis by transcriptomic and epigenetic approaches in mice, we built gene networks associated with CAR and hepatocellular tumors and investigated pathways enriched for the results. In addition to this, to determine the possible candidates for early key event genes for CAR-mediated MOA of liver tumor formation, the selected genes were evaluated for the overlap of the early altered genes observed in CD-1 mice treated with PB for 7 days.39 Furthermore, to evaluate its human relevance, the selected early altered genes were also evaluated for the overlap of the altered genes observed in humanized chimeric mice treated with PB for 7 days.39
Fig. 1. Summary of study design for this research. The effects of phenobarbital (PB) on DNA methylation (5mC), DNA hydroxymethylation (5hmC) and gene expression were determined in hepatocellular adenomas induced by DEN/500 ppm PB. Then the altered genes detected in the above analysis were compared to those altered at an early phase of PB treatment in the liver of wild-type mice and chimeric mice with human hepatocytes.39.
Materials and methods
Animals and husbandry
All animal experiments were approved by the Institutional Animal Care and Use Committee at the Environmental Health Science Laboratory of the Sumitomo Chemical Co. Ltd (Approval No. 12-shuyouseibutu-02; 13-shuyouseibutu-02) and were performed in accordance with the Guide for Animal Care and Use at Sumitomo Chemical Co. Ltd. Male C3H/HeNCrlCrlj mice aged four weeks were purchased from Charles River Japan, Inc., Hino Breeding Center (Shiga, Japan). Animals were acclimated to laboratory conditions individually for 1 week prior to administration. During the course of the study, the environmental conditions in the animal room were targeted within a temperature range of 22–26 °C and a relative humidity range of 40–70%, with frequent ventilation (more than 10 times per hour) and a 12 h light (8:00–20:00)/12 h dark (20:00–8:00) illumination cycle. One or two animals per cage were housed in suspended aluminum cages with stainless steel wire-mesh fronts and floors (Yamato Scientific Co., Ltd, Tokyo, Japan). A commercially available pulverized diet (CRF-1; Oriental Yeast Co., Ltd, Tokyo) and filtered tap water were provided ad libitum throughout the study.
Study design
The study design is summarized in Fig. 1. Experimental procedures were described previously.52 Twenty mice were treated with a single intraperitoneal injection of 90 mg per kg bw diethylnitrosamine (DEN, Tokyo Chemical Industry Co., Ltd, Tokyo, Japan; dissolved in saline, 10 ml per kg bw) at 6 weeks before randomly allocating into two groups (ten animals per dose). One week after administration with DEN, diet including sodium phenobarbital (NaPB, referred to as PB in this paper, Wako Pure Chemical Industries, Ltd, Osaka, Japan) at 0 (control) and 500 ppm was administered to each group for 27 weeks. Mortality, body weights, food consumption, PB intake, liver weight, plasma concentration of PB, gross pathology, and liver histopathology (light microscopy) were examined. The animals were sacrificed without fasting by blood withdrawal after decapitation. A part of the liver was fixed in buffered formalin, dehydrated, embedded in paraffin, sectioned, stained with hematoxylin and eosin, and examined by light microscopy. In addition, adjacent parts of the liver sampled for histopathology were stored at –80 °C for gene analysis. The samples of non-neoplastic tissue collected from 3 controls (DEN/0 ppm PB) and neoplastic lesions collected from 3 animals in the PB-treated group (DEN/500 ppm PB) were examined. The plasma concentration of PB was determined by the method previously described.39
DNA and RNA isolation
Genomic DNA and total RNA were isolated using the AllPrep DNA/RNA/miRNA Universal kit (QIAGEN). Liver samples were homogenized and purified with spin columns according to the manufacturer's instructions. The extracted genomic DNA and total RNA were stored at 4 °C and –80 °C, respectively, until just before use.
5mC antibody preparation
Mouse monoclonal antibodies to 5mC were generated based on the mouse iliac lymph node method53 carried out by ITM Co., Ltd. Briefly, the mice were immunized by injecting the tail base with the adjuvant synthetic thiolated 5-methylcytidine. Two weeks later, serum and lymph nodes of the mice were obtained and used for cell fusion of lymph node lymphocytes with a myeloma cell line to make a hybridoma. The resulting hybridoma cells were cultured onto 96-well plates. The monoclonal antibodies were purified from the positive clone of hybridoma supernatants.
MeDIP and HmeDIP (5mC and 5hmC immunoprecipitation)
Two μg of genomic DNA from each sample were sonicated to a length between 200–1000 bp using a Q500 Sonicator (Qsonica). Hundred μM Primer-1 and -2 (see ESI Table S1†) were mixed equally, denatured for 5 min at 95 °C and cooled down to room temperature. Sonicated DNA was then end-repaired by incubation for 30 min at 20 °C using the NEBNext End Repair Module (NEB). The reaction mixture was purified using the Agencourt AMPure XP (Beckman Coulter), and subjected to 3′-dA tailing by incubation for 30 min at 37 °C using the NEBNext dA-Tailing Module (NEB). After purification as described above, linker ligation was performed by incubation in a reaction containing 2.5 μM linkers for 1 h at 20 °C using the NEBNext Ultra Ligation Module (NEB). The reaction mixture was purified as described above. Linker-ligated DNA was then denatured for 10 min at 95 °C, followed by cooling rapidly and removing 1% of each sample as input DNA. Denatured DNA (600 ng for MeDIP and 500 ng for HmeDIP) was then immunoprecipitated with each antibody. For MeDIP, 2.1 μg of mouse monoclonal 5mC antibody (as described above) and 10 μl of Magna ChIP™ Protein A + G Magnetic Beads (Millipore) were mixed and incubated overnight at 4 °C in 200 μl of IP buffer (10 mM Tris-HCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.1% Na-deoxycholate and 140 mM NaCl). The beads were washed with IP buffer and incubated with denatured DNA overnight at 4 °C in 500 μl of IP buffer. For HmeDIP, denatured DNA and 0.6 μg of rabbit polyclonal 5hmC antibody (Active Motif) were mixed and incubated for 3 h at 4 °C in 500 μl of IP buffer, followed by incubation with 6.3 μl of magnetic beads for 2 h at 4 °C. After immunoprecipitation, the samples were washed with 4 buffers (low salt wash buffer, high salt wash buffer, LiCl wash buffer and TE buffer) (Millipore). Protein was digested with proteinase K (Millipore) overnight at 62 °C. Proteinase K was inactivated by incubating for 10 min at 95 °C. Immunoprecipitated DNA and input DNA were purified as described above, and amplified using Primer-3 (see ESI Table S1†) for 17 cycles in a reaction containing 400 μM dATP, 400 μM dCTP, 400 μM dGTP, 320 μM dTTP, 80 μM dUTP, PfuTurbo Cx Hotstart DNA Polymerase (Agilent Technologies) and 10× reaction buffer. Amplified DNA was purified as described above.
After verification of the specificity of the in-house 5mC antibody, we validated the 5mC- and 5hmC-enriching specificity of MeDIP and HmeDIP in this study based on quantitative PCR analysis for the promoter region of Cyp2b10 in which the epigenetic alterations by PB exposure up to 13 weeks were reported.21–23 Statistical significance was determined by Student's t-test.
Quantitative PCR analysis for evaluation of MeDIP and HmeDIP
Primer sequences and their amplified regions for MeDIP and HmeDIP-quantitative PCR are shown in ESI Table S1.† For quantifying the exon of the Zc3h13 gene, we used Mouse Positive Control Primer Set Zc3h13 (Active Motif). The identity of these target regions in PCR primers was checked using UCSC In-Silico PCR (; http://genome.ucsc.edu/cgi-bin/hgPcr) and NCBI BLAST (; https://blast.ncbi.nlm.nih.gov/Blast.cgi). Quantitative PCR assays were performed using a StepOnePlus™ Real-Time PCR System (Applied Biosystems). Immunoprecipitated and input DNA (2 μl of 1/10 diluted) was mixed respectively with Power SYBR Green PCR Master Mix (Applied Biosystems) (15 μl), 10 μM of each pair primers (0.9 μl each) and ultrapure water (11.2 μl). The mixture was incubated for 10 min at 95 °C, and then subjected to PCR reaction for 40 cycles: denatured for 15 s at 95 °C, annealed/extended for 30 s at 68 °C.
Global DNA methylation and hydroxymethylation analysis
Microarray analysis was conducted on three samples for each group using GeneChip Mouse Promoter 1.0R Arrays (Affymetrix). These arrays contain over 25-mer probes across –7.5 to +2.5 kb from the transcription start site (TSS) of 25 500 mouse promoter regions. The procedure was basically conducted following the manufacturer's protocol (Affymetrix Chromatin Immunoprecipitation Assay Protocol). The obtained image files were analyzed with Partek Genomics Suite 6.6 (Partek). The derived signal values were normalized by quantile normalization and the Robust Multichip Average (RMA) background correlation.54 These normalized scores were transformed log 2 values and calculated log 2 ratios (MeDIP/input and HmeDIP/input). Signal averages of each group, p-values and t-values in statistical significance testing (Student's t-test) were obtained. The 5mC and 5hmC enriched regions were detected using the Model based Analysis of Tiling-arrays (MAT).55 MAT algorithm calculated a trimmed mean of the t-values of each probe in the window (fragment length = 600 bp, minimum number of probes = 10, top and bottom exclusion = 10%). Using the trimmed mean and the number of probes, MAT scores and the corresponding p-values were calculated. All those showing less than 0.01 for the p-value were regarded as 5mC/5hmC significantly changed regions. The regions that overlapped within –7.5 to +2.5 kb from the TSS of Refseq genes (mm8) were annotated as the promoters of those genes. We defined the ‘promoter’ of each gene as the –7.5 kb upstream to +1.5 kb downstream region from the transcription start site (TSS), because the transcription factor binding sites are located mostly in a region upstream to the regulated gene.56 If 5mC and 5hmC enriched regions by MAT algorithm were overlapped with –7.5 kb to +1.5 kb from the TSS of genes, the regions were annotated with these genes.
Global gene expression analysis
Gene expression analysis was conducted on three total RNA samples for each group using SurePrint G3 Mouse GE 8 × 60K Microarrays (Agilent Technologies). These arrays contain over 60-mer probes to examine 39 000 mouse mRNAs and 16 000 lncRNAs. The procedure was basically conducted following the manufacturer's protocol (version 6.5). The scanned image files were converted to TIFF files containing signal values of each probe using Feature Extraction (version 10.7.3.1). The signal values were log 2-transformed and processed to the 75th percentile normalization.57 Signal averages for each group and the p-value in statistical significance testing (Student's t-test) were obtained. All those showing less than 0.05 for the p-value and “1 (present: gIsPosAndSignif)” for Detection Call in PB-treatment arrays were regarded as up-regulated probe sets. All those showing less than 0.05 for the p-value and “1 (present: gIsPosAndSignif)” for Detection Call in the control arrays were regarded as down-regulated probe sets.
Integrated pathway analysis of 5mC, 5hmC and gene expression
Gene network analyses and pathway enrichment analyses were performed using MetaCore® via; https://portal.genego.com. For gene network analyses, we used known interaction information with direction (incoming/outgoing), effect (activation/inhibition) and mechanism (e.g. bind/influence on expression/transcription regulation) of the CAR and CAR/RXR-alpha complex, followed by using known information of causal associations of genes with liver neoplasms in epigenetic, transcriptional or protein levels. The detailed information and references are shown in the MetaCore website. For pathway analyses, we used known canonical pathways of common functional themes in the MetaCore database and performed the analyses based on the gene list of 5mC, 5hmC and gene expression alterations.
Results
Animal data
Although one animal of the DEN/500 ppm PB group was found dead at day 100, treatment with 500 ppm PB did not affect body weight, body weight gain, or food consumption, demonstrating that treatment with 500 ppm PB (average PB intake was 69.9 mg kg–1 day–1) did not show excess toxicity under the present study conditions. Absolute and relative liver weights were significantly increased to 1.4- and 1.5-fold of control, respectively. Histopathological examination revealed that DEN/500 ppm PB induced hepatocellular adenomas in all animals (19/19, vs. 7/20 in DEN/0 ppm PB) and adenocarcinomas in some (3/19, vs. 0/20 in DEN/0 ppm PB). These data are presented in ESI Tables S2 and S3.† Group mean and standard deviation of plasma concentration of PB in DEN/500 ppm PB was 15.4 ± 7.1 μg mL–1 (N = 19).
Global profiles of 5mC and 5hmC are altered in mouse hepatocellular adenoma induced by DEN/PB
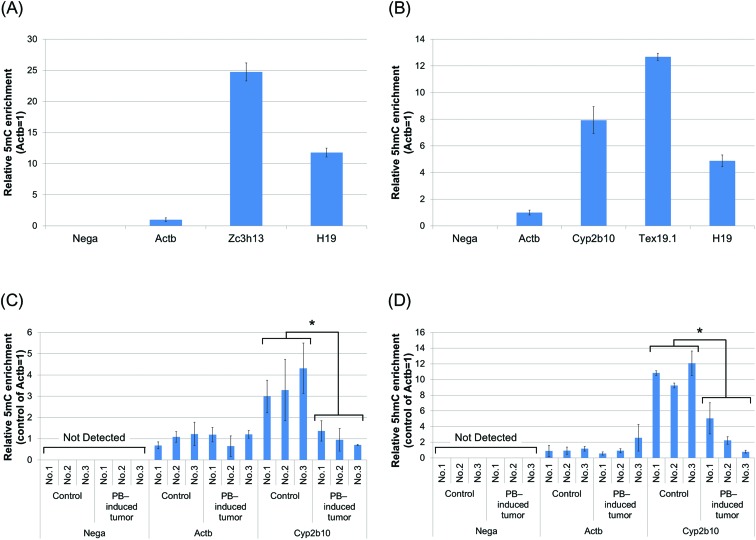
Normal appearing tissues in 3 animals of the DEN/0 ppm PB group and tissues with hepatocellular adenomas in 3 animals of the DEN/500 ppm PB group were used for global analyses (ESI Table S4 and Fig. S1†). First, we verified the specificity of the in-house 5mC antibody (ESI Fig. S2†). Next, we validated the 5mC- and 5hmC-enriching specificity of MeDIP and HmeDIP in this study (Fig. 2). We performed quantitative PCR analysis and confirmed that the region without CpG sites was not detected in immunoprecipitated DNA with either 5mC or 5hmC antibodies. As for the promoter region of Cyp2b10, both the detected 5mC and 5hmC levels differed between immunoprecipitated DNA of PB-induced tumors in the DEN/500 ppm PB group and control tissues in the DEN/0 ppm PB group. These experiments showed that MeDIP and HmeDIP could specifically enrich 5mC- and 5hmC-containing regions, respectively.
Fig. 2. Validation of the 5mC- and 5hmC-enriching specificity of MeDIP and HmeDIP in this study. (A, B) Followed by removing 1% of DNA as the input sample, genomic DNA from liver samples of normal control mice was subjected to MeDIP (A) and HmeDIP (B). Immunoprecipitated and input DNA were analyzed by quantitative PCR with primers for the region without CpG sites (Nega), the promoter region of Actb (low-level 5mC and 5hmC), Tex19.1 (high-level 5hmC), Cyp2b10, the exon of Zc3h13 (high-level 5mC) and the imprinting control region of H19 (high-level 5mC and 5hmC) (ESI Table S1†). (C, D) Likewise, genomic DNA from control tissues (DEN/0 ppm PB) and PB-induced hepatocellular adenoma (DEN/500 ppm PB) was subjected to MeDIP (C) and HmeDIP (D), followed by quantitative PCR with primers as described above for Nega, Actb and Cyp2b10 (known 5mC and 5hmC alteration by PB treatment). Data are mean ± SD from triplicate experiments. *p < 0.01, Student's t-test.
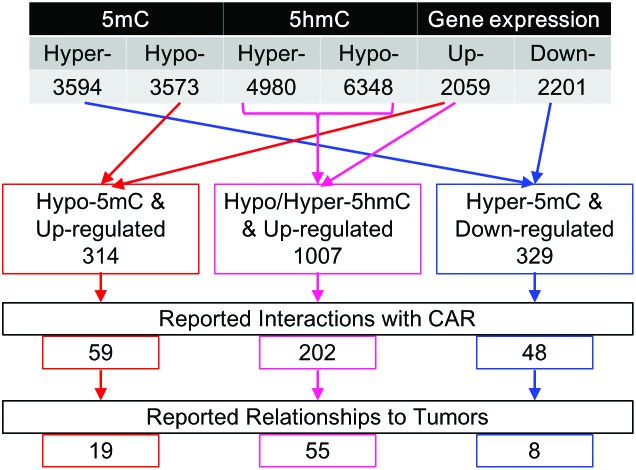
We studied differential epigenetic and expression profiles between tumors of PB-treated mice and normal tissues of control mice by global analyses using microarrays. The 5mC analysis showed significantly hyper- and hypomethylated gene promoters (3594 and 3573) in liver tumors compared to control tissue (Fig. 3). 4980 and 6348 gene promoters were hyper- and hypohydroxymethylated, respectively. Tables 1 and 2 show the top 10 significant differential gene promoters (these regions are arranged in ascending order of p-value and MAT-score). With regard to gene expression, 2059 and 2201 genes were up- and down-regulated, respectively. We also compared the gene expression profile to RNA-sequence data in a previous study focusing on PB-induced liver tumors,26 and confirmed some reproducibility between these datasets (ESI Fig. S3†).
Fig. 3. Summary results of 5mC, 5hmC and gene expression analysis. These genes were significantly altered in hepatocellular adenoma of mice in the DEN/500 ppm PB group compared to normal livers of control mice in the DEN/0 ppm PB group. We used the MetaCore database to search interactions with CAR and relationships to tumor production.
Table 1. Top 10 most hyper- and hypomethylated gene promoters in hepatocellular tumors of PB-treated mice.
| [Hypermethylated] | |||||||
| Gene symbol | Distance to TSS | Chromosome | Region start | Region end | Probes in region | p-Value | MAT-score a |
| Ccnf | –4052 | chr17 | 23983062 | 23984925 | 43 | 8.83 × 10–6 | 7.038 |
| Prr23a | 855 | chr9 | 98652794 | 98653962 | 21 | 8.83 × 10–6 | 6.485 |
| Oxct2a | 0 | chr4 | 122824723 | 122826092 | 38 | 8.83 × 10–6 | 6.241 |
| Calml3 | 0 | chr13 | 3802864 | 3803753 | 25 | 8.83 × 10–6 | 6.217 |
| Zrsr1 | 1249 | chr11 | 22873278 | 22874439 | 28 | 8.83 × 10–6 | 6.171 |
| Pcdh12 | –1023 | chr18 | 38411399 | 38412806 | 35 | 8.83 × 10–6 | 6.109 |
| Slc9a1 | 522 | chr4 | 132642370 | 132643661 | 36 | 8.83 × 10–6 | 6.076 |
| Donson | 0 | chr16 | 91577392 | 91578500 | 29 | 8.83 × 10–6 | 5.792 |
| 5830415F09Rik | 453 | chr4 | 46408754 | 46410071 | 27 | 8.83 × 10–6 | 5.788 |
| Olfr105 | –1015 | chr17 | 36988619 | 36989545 | 26 | 1.77 × 10–5 | 5.705 |
| [Hypomethylated] | |||||||
| Gene symbol | Distance to TSS | Chromosome | Region start | Region end | Probes in region | p-Value | MAT-score a |
| Psen2 | 1221 | chr1 | 182080643 | 182081405 | 15 | 8.83 × 10–6 | –9.763 |
| Pld3 | –3530 | chr7 | 27265403 | 27266148 | 18 | 8.83 × 10–6 | –9.475 |
| 2310022A10Rik | –3088 | chr7 | 27265403 | 27266148 | 18 | 8.83 × 10–6 | –9.475 |
| LOC100041223 | 0 | chrY_random | 12134681 | 12136488 | 83 | 8.83 × 10–6 | –9.082 |
| LOC100039810 | 936 | chrY_random | 12134681 | 12136488 | 83 | 8.83 × 10–6 | –9.082 |
| LOC100041256 | 936 | chrY_random | 12134681 | 12136488 | 83 | 8.83 × 10–6 | –9.082 |
| Pusl1 | –5213 | chr4 | 154740776 | 154741472 | 19 | 8.83 × 10–6 | –8.317 |
| LOC100042428 | 0 | chrY_random | 11615565 | 11617305 | 80 | 8.83 × 10–6 | –8.192 |
| MGC107098 | 1267 | chrY_random | 11615565 | 11617305 | 80 | 8.83 × 10–6 | –8.192 |
| LOC100039753 | 1285 | chrY_random | 11615565 | 11617305 | 80 | 8.83 × 10–6 | –8.192 |
aThe relative methylation level calculated by the MAT (Model based Analysis of Tiling-arrays) algorithm.
Table 2. Top 10 most hyper- and hypohydroxymethylated gene promoters in hepatocellular tumors of PB-treated mice.
| [Hyperhydroxymethylated] | |||||||
| Gene symbol | Distance to TSS | Chromosome | Region start | Region end | Probes in region | p-Value | MAT-score a |
| Mir1982 | –479 | chr10 | 80230053 | 80231447 | 38 | 8.83 × 10–6 | 10.373 |
| Gm9786 | 228 | chr10 | 80230053 | 80231447 | 38 | 8.83 × 10–6 | 10.373 |
| Oaz1 | 268 | chr10 | 80230053 | 80231447 | 38 | 8.83 × 10–6 | 10.373 |
| LOC100041223 | –2138 | chrY_random | 12131429 | 12133005 | 65 | 8.83 × 10–6 | 10.228 |
| LOC100039810 | –740 | chrY_random | 12131429 | 12133005 | 65 | 8.83 × 10–6 | 10.228 |
| LOC100041256 | –740 | chrY_random | 12131429 | 12133005 | 65 | 8.83 × 10–6 | 10.228 |
| Igfbp5 | –320 | chr1 | 72808393 | 72809533 | 29 | 8.83 × 10–6 | 10.170 |
| C030023E24Rik | –4736 | chrX | 57444031 | 57447222 | 82 | 8.83 × 10–6 | 9.465 |
| Cdr1 | 0 | chrX | 57444031 | 57447222 | 82 | 8.83 × 10–6 | 9.465 |
| Trim32 | 0 | chr4 | 65091192 | 65092726 | 40 | 8.83 × 10–6 | 9.444 |
| [Hypohydroxymethylated] | |||||||
| Gene symbol | Distance to TSS | Chromosome | Region start | Region end | Probes in region | p-Value | MAT-score a |
| 2210021J22Rik | –2995 | chr15 | 85642459 | 85645379 | 81 | 8.83 × 10–6 | –13.554 |
| Hspa1a | –4679 | chr17 | 34584892 | 34587462 | 67 | 8.83 × 10–6 | –12.096 |
| Lsm2 | –2724 | chr17 | 34584892 | 34587462 | 67 | 8.83 × 10–6 | –12.096 |
| Gstt3 | –1851 | chr10 | 75226982 | 75227939 | 22 | 8.83 × 10–6 | –12.018 |
| Eid3 | 0 | chr10 | 82296212 | 82297783 | 41 | 8.83 × 10–6 | –11.725 |
| Stk40 | –5130 | chr4 | 125599936 | 125601131 | 34 | 8.83 × 10–6 | –11.623 |
| Lsm10 | 979 | chr4 | 125599936 | 125601131 | 34 | 8.83 × 10–6 | –11.623 |
| Emg1 | –1464 | chr6 | 124679262 | 124682316 | 79 | 8.83 × 10–6 | –11.309 |
| Phb2 | 1354 | chr6 | 124679262 | 124682316 | 79 | 8.83 × 10–6 | –11.309 |
| BC021614 | 542 | chr19 | 4057119 | 4058753 | 41 | 8.83 × 10–6 | –11.135 |
aThe relative methylation level calculated by the MAT (Model based Analysis of Tiling-arrays) algorithm.
In some cases, a certain correlation was found between DNA methylation and gene expression. DNA methylation of gene promoter regions is possibly associated with transcriptional repression of some genes, while hydroxymethylated DNA was associated with transcriptional activation.10–13 To identify the candidates important for PB-mediated hepatocellular tumorigenesis from the thousands of altered genes described above, we integrated 5mC/5hmC and gene expression analyses and focused on these epigenetic and transcriptional correlations. In hepatocellular adenomas developing after PB treatment (DEN/500 ppm PB), 314 genes were up-regulated and hypomethylated, 1007 genes were up-regulated and differentially hydroxymethylated, and 329 genes were down-regulated and hypermethylated compared to control (DEN/0 ppm PB) (Fig. 3).
Gene network analysis shows possible interactions with CAR and relationships to liver tumor production in mouse hepatocellular adenoma induced by DEN/PB
To examine whether differential genes in DNA methylation and gene expression are associated with the CAR, we then investigated gene networks using the MetaCore database. In MetaCore, CAR's downstream transcription factors (HNF4-alpha, ESR1 and HIF1A) are reported to interact with many genes of integrated analysis results. We defined these transcription factors as ‘hub’ genes, and we identified genes that have interaction information of the CAR or its hub genes. In epigenetically and transcriptionally changed genes (up-regulated and hypomethylated; up-regulated and differentially hydroxymethylated; down-regulated and hypermethylated), 59, 202 and 48 genes were reported to have a possible interaction with the CAR or its downstream hub genes, respectively (Fig. 3).
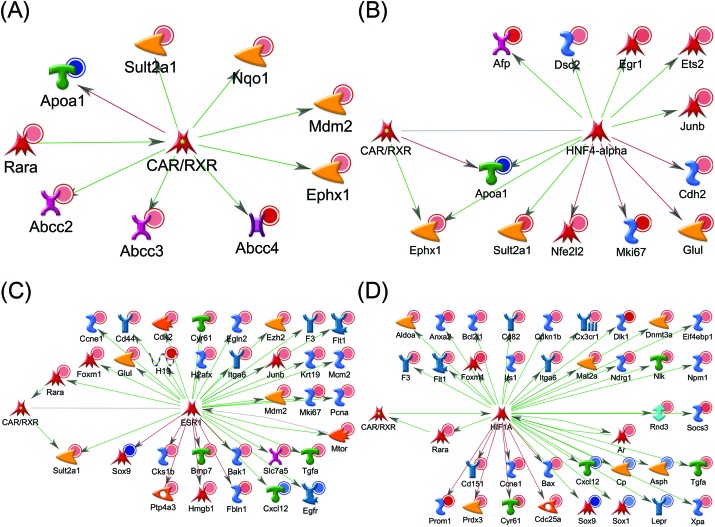
We further investigated associations of our data with liver tumor production-related genes. As a result of analyzing the information on disease, we found that dozens of genes focused above were reported to have causal relationships to liver neoplasms. Fig. 3 shows the number of significantly differential genes that have possible interactions with the CAR and relate to liver tumor production, and Table 3 shows the details of these genes. As for differentially hydroxymethylated and up-regulated genes, both MAT-scores of hyper- and hypohydroxymethylated promoter regions are shown here. Using our data shown in Table 3, we built whole gene networks including the CAR and its downstream hub genes (Fig. 4).
Table 3. Genes reported having interactions with CAR and relationships to tumors.
| [Hypo-5mC & upregulated] | ||||
| Hypo-5mC |
Upregulated |
|||
| Gene symbol | p-Value | MAT-score a | p-Value | Fold-change |
| Abcc2 | 9.90 × 10–3 | –3.069 | 7.55 × 10–5 | 2.234 |
| Afp | 9.68 × 10–3 | –3.080 | 2.80 × 10–2 | 13.655 |
| Anxa2 | 9.34 × 10–3 | –3.097 | 4.43 × 10–2 | 6.270 |
| Bmp7 | 9.59 × 10–3 | –3.085 | 1.39 × 10–3 | 5.299 |
| Ccne1 | 8.77 × 10–3 | –3.126 | 7.11 × 10–4 | 4.063 |
| Cyr61 | 2.05 × 10–3 | –3.749 | 4.09 × 10–2 | 5.303 |
| Dsc2 | 6.04 × 10–3 | –3.292 | 2.31 × 10–2 | 1.293 |
| Eif4ebp1 | 6.29 × 10–3 | –3.277 | 4.47 × 10–3 | 1.250 |
| F3 | 8.20 × 10–3 | –3.157 | 1.39 × 10–3 | 2.831 |
| Glul | 5.16 × 10–3 | –3.362 | 7.97 × 10–4 | 9.049 |
| Irs1 | 9.30 × 10–3 | –3.100 | 3.31 × 10–3 | 2.714 |
| Itga6 | 9.44 × 10–3 | –3.094 | 2.32 × 10–3 | 2.933 |
| Junb | 6.50 × 10–3 | –3.258 | 5.47 × 10–3 | 3.254 |
| Mcm2 | 4.41 × 10–5 | –5.577 | 3.20 × 10–2 | 2.721 |
| Mki67 | 5.71 × 10–3 | –3.315 | 1.06 × 10–3 | 21.397 |
| Nfe2l2 | 6.00 × 10–3 | –3.294 | 3.66 × 10–2 | 2.299 |
| Nlk | 5.68 × 10–3 | –3.317 | 5.06 × 10–3 | 3.764 |
| Nqo1 | 9.25 × 10–3 | –3.101 | 3.39 × 10–4 | 6.660 |
| Ptp4a3 | 7.69 × 10–3 | –3.184 | 3.00 × 10–2 | 2.064 |
| [Hyper or Hypo-5hmC & upregulated] | ||||||
| Hyper-5hmC |
Hypo-5hmC |
Upregulated |
||||
| Gene symbol | p-Value | MAT-score a | p-Value | MAT-score a | p-Value | Fold-change |
| Abcc3 | — | — | 8.83 × 10–6 | –7.846 | 1.28 × 10–2 | 1.537 |
| Abcc4 | 2.33 × 10–3 | 4.113 | — | — | 8.93 × 10–5 | 104.220 |
| Aldoa | 6.31 × 10–3 | 3.604 | — | — | 2.40 × 10–3 | 1.426 |
| Anxa2 | — | — | 1.85 × 10–4 | –5.126 | 4.43 × 10–2 | 6.270 |
| Ar | 2.26 × 10–3 | 4.128 | — | — | 4.87 × 10–2 | 1.637 |
| Bak1 | — | — | 6.49 × 10–3 | –3.634 | 1.33 × 10–2 | 1.861 |
| Bax | — | — | 7.94 × 10–5 | –5.403 | 4.47 × 10–2 | 1.699 |
| Bcl2 l1 | — | — | 1.24 × 10–3 | –4.429 | 1.30 × 10–2 | 1.855 |
| Bmp7 | — | — | 1.59 × 10–4 | –5.204 | 1.39 × 10–3 | 5.299 |
| Cd151 | — | — | 3.53 × 10–5 | –5.622 | 3.28 × 10–3 | 1.255 |
| Cd44 | 2.68 × 10–3 | 4.043 | — | — | 3.57 × 10–2 | 4.191 |
| Cd82 | — | — | 9.21 × 10–3 | –3.452 | 1.08 × 10–3 | 1.395 |
| Cdc25a | — | — | 5.19 × 10–3 | –3.739 | 2.31 × 10–2 | 1.572 |
| Cdh2 | 8.76 × 10–3 | 3.461 | 7.91 × 10–3 | –3.534 | 3.76 × 10–2 | 1.706 |
| Cdk2 | 9.50 × 10–3 | 3.414 | — | — | 2.92 × 10–2 | 1.580 |
| Cdkn1b | — | — | 5.38 × 10–4 | –4.736 | 1.20 × 10–2 | 1.271 |
| Cks1b | 2.82 × 10–4 | 5.052 | — | — | 1.60 × 10–2 | 2.038 |
| Cx3cr1 | 6.36 × 10–3 | 3.597 | — | — | 3.14 × 10–2 | 1.938 |
| Cyr61 | 2.67 × 10–3 | 4.046 | 1.85 × 10–4 | –5.144 | 4.09 × 10–2 | 5.303 |
| Dlk1 | 5.66 × 10–3 | 3.656 | — | — | 1.29 × 10–3 | 24.247 |
| Dnmt3a | 8.55 × 10–3 | 3.472 | 2.96 × 10–3 | –4.005 | 3.11 × 10–3 | 2.316 |
| Egln2 | 2.70 × 10–3 | 4.040 | 8.83 × 10–6 | –7.145 | 3.32 × 10–3 | 1.194 |
| Egr1 | 7.57 × 10–3 | 3.524 | 8.83 × 10–6 | –6.222 | 1.67 × 10–2 | 5.828 |
| Eif4ebp1 | — | — | 6.74 × 10–3 | –3.618 | 4.47 × 10–3 | 1.250 |
| Ephx1 | — | — | 1.65 × 10–3 | –4.273 | 4.95 × 10–4 | 2.227 |
| Ets2 | — | — | 1.47 × 10–3 | –4.326 | 2.01 × 10–2 | 2.590 |
| Ezh2 | 6.31 × 10–3 | 3.604 | — | — | 4.61 × 10–2 | 2.018 |
| F3 | 8.79 × 10–3 | 3.458 | 3.88 × 10–4 | –4.828 | 1.39 × 10–3 | 2.831 |
| Fbln1 | 9.27 × 10–4 | 4.543 | 1.60 × 10–3 | –4.295 | 6.48 × 10–4 | 24.599 |
| Flt1 | — | — | 7.29 × 10–3 | –3.579 | 1.93 × 10–2 | 1.718 |
| Foxm1 | — | — | 7.90 × 10–3 | –3.534 | 6.99 × 10–3 | 8.387 |
| Glul | — | — | 8.83 × 10–6 | –6.114 | 7.97 × 10–4 | 9.049 |
| H19 | 4.93 × 10–3 | 3.724 | — | — | 1.65 × 10–3 | 118.119 |
| H2afx | — | — | 8.83 × 10–6 | –6.041 | 1.41 × 10–2 | 2.360 |
| Hmgb1 | 6.34 × 10–3 | 3.600 | — | — | 1.90 × 10–2 | 1.263 |
| Irs1 | — | — | 2.47 × 10–4 | –5.032 | 3.31 × 10–3 | 2.714 |
| Krt19 | 7.50 × 10–3 | 3.529 | — | — | 1.08 × 10–2 | 7.442 |
| Mat2a | 5.45 × 10–3 | 3.671 | — | — | 2.80 × 10–3 | 3.061 |
| Mcm2 | 1.77 × 10–4 | 5.288 | 9.68 × 10–3 | –3.426 | 3.20 × 10–2 | 2.721 |
| Mdm2 | — | — | 1.47 × 10–3 | –4.322 | 1.09 × 10–2 | 1.511 |
| Mtor | 7.59 × 10–4 | 4.620 | — | — | 2.81 × 10–2 | 1.504 |
| Ndrg1 | — | — | 1.90 × 10–3 | –4.220 | 2.46 × 10–2 | 5.770 |
| Nlk | — | — | 8.83 × 10–6 | –6.351 | 5.06 × 10–3 | 3.764 |
| Npm1 | — | — | 2.31 × 10–3 | –4.127 | 9.32 × 10–3 | 1.961 |
| Pcna | 5.23 × 10–3 | 3.688 | 1.05 × 10–3 | –4.512 | 7.92 × 10–3 | 1.868 |
| Prdx3 | — | — | 4.70 × 10–3 | –3.781 | 3.16 × 10–3 | 2.099 |
| Prom1 | 1.87 × 10–3 | 4.223 | — | — | 4.27 × 10–4 | 24.364 |
| Ptp4a3 | 9.89 × 10–3 | 3.401 | 2.58 × 10–3 | –4.067 | 3.00 × 10–2 | 2.064 |
| Rara | 6.70 × 10–3 | 3.578 | 6.81 × 10–3 | –3.613 | 8.85 × 10–3 | 1.659 |
| Rnd3 | 2.80 × 10–3 | 4.020 | — | — | 2.55 × 10–3 | 2.428 |
| Slc7a5 | 6.81 × 10–3 | 3.573 | — | — | 2.20 × 10–3 | 9.769 |
| Socs3 | — | — | 7.94 × 10–5 | –5.403 | 4.21 × 10–3 | 8.617 |
| Sult2a1 | — | — | 2.56 × 10–3 | –4.069 | 1.44 × 10–2 | 7.438 |
| Tgfa | — | — | 8.83 × 10–6 | –6.089 | 1.08 × 10–2 | 1.848 |
| Xpa | — | — | 8.83 × 10–6 | –6.336 | 7.80 × 10–3 | 1.227 |
| [Hyper-5mC & downregulated] | ||||
| Hyper-5mC |
Downregulated |
|||
| Gene symbol | p-Value | MAT-score a | p-Value | Fold-change |
| Apoa1 | 3.91 × 10–3 | 3.404 | 2.36 × 10–3 | –1.483 |
| Asph | 6.44 × 10–4 | 4.062 | 2.18 × 10–2 | –1.532 |
| Cp | 9.96 × 10–3 | 3.004 | 6.15 × 10–3 | –1.207 |
| Cxcl12 | 2.73 × 10–3 | 3.534 | 3.31 × 10–2 | –2.813 |
| Egfr | 2.65 × 10–4 | 4.492 | 3.18 × 10–2 | –3.202 |
| Lepr | 8.26 × 10–3 | 3.086 | 1.73 × 10–2 | –1.354 |
| Sox1 | 4.21 × 10–3 | 3.380 | 2.14 × 10–2 | –1.672 |
| Sox9 | 9.50 × 10–3 | 3.028 | 1.19 × 10–2 | –6.940 |
aThe relative methylation level calculated by the MAT (Model based Analysis of Tiling-arrays) algorithm.
Fig. 4. Gene networks built using the MetaCore database and our gene data. These genes were reported having interactions with (A) CAR and its downstream hub genes, (B) HNF4-alpha, (C) ESR1 and (D) HIF1A. These genes were also reported having relationships to tumors. Genes with red or blue circles indicate up-regulated or down-regulated genes compared to control, respectively. Green, red or gray arrows indicate positive/activation, negative/inhibition or unspecified effects, respectively.
Some cancer-related pathways enriched for the results of 5mC, 5hmC and gene expression analyses in mouse hepatocellular adenoma induced by DEN/PB
Next, we performed pathway enrichment analysis using MetaCore pathways and the number of detected pathways by PB administration (p < 0.01) for 5mC, 5hmC and gene expression alterations were 288, 366 and 231, respectively (data not shown). Tables 4–6 show the results of the top 20 enriched pathways for genes that were significantly changed in 5mC, 5hmC or gene expression by PB administration, respectively (these pathways are arranged in ascending order by p-value). The differentially methylated genes belonged mainly to pathways involved in the development and immune response, such as oncostatin M signaling, megakaryopoiesis, growth hormone signaling and inflammatory response. The main signaling cascades of ovarian cancer are also enriched. The differentially hydroxymethylated genes belonged mainly to pathways involved in development and cancer cells, such as PIP3 (phosphatidylinositol 3,4,5-trisphosphate) signaling, regulation of EMT (epithelial-to-mesenchymal transition), myeloma cells and colorectal cancer. Both changes in 5mC and 5hmC profiles correlated most closely with TGF, WNT and cytoskeletal remodeling. Meanwhile, the differentially expressed genes were intimately related to pathways involved in the cell cycle or immune response such as G1/S transition, the metaphase checkpoint and IL-4-induced regulators of cell growth.
Table 4. Top 20 enrichment for hyper- and hypomethylated gene promoters by MetaCore pathways.
| Maps | Total | p-Value | FDR a | In data |
| Cytoskeleton remodeling_TGF, WNT and cytoskeletal remodeling | 111 | 4.81 × 10–10 | 2.21 × 10–7 | 50 |
| NF-AT signaling in cardiac hypertrophy | 65 | 5.06 × 10–10 | 2.21 × 10–7 | 35 |
| Neurophysiological process_dynein–dynactin motor complex in axonal transport in neurons | 54 | 3.27 × 10–9 | 9.49 × 10–7 | 30 |
| Immune response_oncostatin M signaling via MAPK in human cells | 37 | 7.95 × 10–8 | 1.43 × 10–5 | 22 |
| Development_cytokine-mediated regulation of megakaryopoiesis | 57 | 8.21 × 10–8 | 1.43 × 10–5 | 29 |
| Cell cycle_regulation of G1/S transition (part 1) | 38 | 1.55 × 10–7 | 2.25 × 10–5 | 22 |
| Cell adhesion_PLAU signaling | 39 | 2.90 × 10–7 | 3.61 × 10–5 | 22 |
| Development_epigenetic and transcriptional regulation of oligodendrocyte precursor cell differentiation and myelination | 34 | 4.00 × 10–7 | 4.36 × 10–5 | 20 |
| Immune response_oncostatin M signaling via MAPK in mouse cells | 35 | 7.64 × 10–7 | 7.40 × 10–5 | 20 |
| Development_BMP signaling | 33 | 1.24 × 10–6 | 1.08 × 10–4 | 19 |
| Translation_regulation of EIF2 activity | 39 | 1.53 × 10–6 | 1.16 × 10–4 | 21 |
| Development_growth hormone signaling via PI3K/AKT and MAPK cascades | 42 | 1.60 × 10–6 | 1.16 × 10–4 | 22 |
| Ovarian cancer (main signaling cascades) | 64 | 1.84 × 10–6 | 1.24 × 10–4 | 29 |
| Cytoskeleton remodeling_cytoskeleton remodeling | 102 | 2.26 × 10–6 | 1.41 × 10–4 | 40 |
| Transcription_role of heterochromatin protein 1 (HP1) family in transcriptional silencing | 40 | 2.64 × 10–6 | 1.53 × 10–4 | 21 |
| Immune response_gastrin in inflammatory response | 69 | 3.54 × 10–6 | 1.93 × 10–4 | 30 |
| Immune response_HMGB1/RAGE signaling pathway | 53 | 3.85 × 10–6 | 1.97 × 10–4 | 25 |
| Development_TGF-beta-dependent induction of EMT via SMADs | 35 | 4.12 × 10–6 | 2.00 × 10–4 | 19 |
| Development_VEGF signaling via VEGFR2 – generic cascades | 84 | 5.66 × 10–6 | 2.50 × 10–4 | 34 |
| Development_role of HDAC and calcium/calmodulin-dependent kinase (CaMK) in control of skeletal myogenesis | 54 | 5.86 × 10–6 | 2.50 × 10–4 | 25 |
aMultiple testing correction based on FDR (False Discovery Rate).
Table 5. Top 20 enrichment for hyper- and hypohydroxymethylated gene promoters by MetaCore pathways.
| Maps | Total | p-Value | FDR a | In data |
| Cytoskeleton remodeling_TGF, WNT and cytoskeletal remodeling | 111 | 2.63 × 10–13 | 2.31 × 10–10 | 69 |
| Signal transduction_AKT signaling | 43 | 1.06 × 10–10 | 4.65 × 10–8 | 33 |
| Development_PIP3 signaling in cardiac myocytes | 47 | 8.36 × 10–10 | 2.45 × 10–7 | 34 |
| Development_regulation of epithelial-to-mesenchymal transition (EMT) | 64 | 5.16 × 10–9 | 1.14 × 10–6 | 41 |
| NF-AT signaling in cardiac hypertrophy | 65 | 1.01 × 10–8 | 1.49 × 10–6 | 41 |
| Development_IGF-1 receptor signaling | 52 | 1.01 × 10–8 | 1.49 × 10–6 | 35 |
| Immune response_IL-1 signaling pathway | 44 | 1.33 × 10–8 | 1.67 × 10–6 | 31 |
| Some pathways of EMT in cancer cells | 51 | 2.39 × 10–8 | 2.63 × 10–6 | 34 |
| Main growth factor signaling cascades in multiple myeloma cells | 41 | 3.42 × 10–8 | 3.34 × 10–6 | 29 |
| Colorectal cancer (general schema) | 30 | 8.08 × 10–8 | 6.34 × 10–6 | 23 |
| Immune response_signaling pathway mediated by IL-6 and IL-1 | 30 | 8.08 × 10–8 | 6.34 × 10–6 | 23 |
| Development_epigenetic and transcriptional regulation of oligodendrocyte precursor cell differentiation and myelination | 34 | 8.96 × 10–8 | 6.34 × 10–6 | 25 |
| Signal transduction_additional pathways of NF-kB activation (in the cytoplasm) | 53 | 1.01 × 10–7 | 6.34 × 10–6 | 34 |
| Development_insulin, IGF-1 and TNF-alpha in brown adipocyte differentiation | 53 | 1.01 × 10–7 | 6.34 × 10–6 | 34 |
| Immune response_gastrin in inflammatory response | 69 | 1.16 × 10–7 | 6.71 × 10–6 | 41 |
| IGF family signaling in colorectal cancer | 60 | 1.25 × 10–7 | 6.71 × 10–6 | 37 |
| Transcription_N-CoR/SMRT complex-mediated epigenetic gene silencing | 49 | 1.30 × 10–7 | 6.71 × 10–6 | 32 |
| Development_regulation of lung epithelial progenitor cell differentiation | 41 | 1.93 × 10–7 | 9.45 × 10–6 | 28 |
| Cell adhesion_PLAU signaling | 39 | 2.08 × 10–7 | 9.61 × 10–6 | 27 |
| Development_TGF-beta-dependent induction of EMT via SMADs | 35 | 2.27 × 10–7 | 1.00 × 10–5 | 25 |
aMultiple testing correction based on FDR (False Discovery Rate).
Table 6. Top 20 enrichment for expression-changed genes by MetaCore pathways.
| Maps | Total | p-Value | FDR a | In data |
| Cytoskeleton remodeling_TGF, WNT and cytoskeletal remodeling | 111 | 3.04 × 10–11 | 2.73 × 10–8 | 47 |
| Immune response_IL-4-induced regulators of cell growth, survival, differentiation and metabolism | 63 | 1.48 × 10–10 | 6.63 × 10–8 | 32 |
| Cell cycle_ESR1 regulation of G1/S transition | 33 | 7.70 × 10–9 | 1.99 × 10–6 | 20 |
| Blood coagulation_blood coagulation | 39 | 8.88 × 10–9 | 1.99 × 10–6 | 22 |
| Immune response_IL-6 signaling pathway via JAK/STAT | 72 | 4.39 × 10–8 | 7.89 × 10–6 | 31 |
| Cell cycle_the metaphase checkpoint | 36 | 6.03 × 10–8 | 9.02 × 10–6 | 20 |
| IGF family signaling in colorectal cancer | 60 | 1.07 × 10–7 | 1.25 × 10–5 | 27 |
| Immune response_oncostatin M signaling via MAPK in human cells | 37 | 1.12 × 10–7 | 1.25 × 10–5 | 20 |
| Cytoskeleton remodeling_cytoskeleton remodeling | 102 | 1.48 × 10–7 | 1.47 × 10–5 | 38 |
| Oxidative phosphorylation | 105 | 3.52 × 10–7 | 3.07 × 10–5 | 38 |
| Protein folding and maturation_POMC processing | 30 | 4.10 × 10–7 | 3.07 × 10–5 | 17 |
| Regulation of lipid metabolism_RXR-dependent regulation of lipid metabolism via PPAR, RAR and VDR | 30 | 4.10 × 10–7 | 3.07 × 10–5 | 17 |
| Development_WNT signaling pathway. Part 2 | 53 | 4.72 × 10–7 | 3.26 × 10–5 | 24 |
| Neurophysiological process_dynein–dynactin motor complex in axonal transport in neurons | 54 | 7.21 × 10–7 | 4.63 × 10–5 | 24 |
| Cell adhesion_chemokines and adhesion | 100 | 8.26 × 10–7 | 4.95 × 10–5 | 36 |
| Immune response_oncostatin M signaling via MAPK in mouse cells | 35 | 1.28 × 10–6 | 7.20 × 10–5 | 18 |
| Cell cycle_role of APC in cell cycle regulation | 32 | 1.40 × 10–6 | 7.41 × 10–5 | 17 |
| Immune response_alternative complementary pathway | 53 | 2.04 × 10–6 | 1.02 × 10–4 | 23 |
| Cell cycle_chromosome condensation in prometaphase | 21 | 2.50 × 10–6 | 1.18 × 10–4 | 13 |
| Cytoskeleton remodeling_role of PKA in cytoskeleton reorganisation | 40 | 3.06 × 10–6 | 1.37 × 10–4 | 19 |
aMultiple testing correction based on FDR (False Discovery Rate).
Evaluation of overlap between the altered genes at expression levels with 5mC and 5hmC alterations in mouse hepatocellular adenoma induced by DEN/PB and altered genes at expression levels in liver of CD-1 mice and humanized chimeric mice treated with PB for 7 days
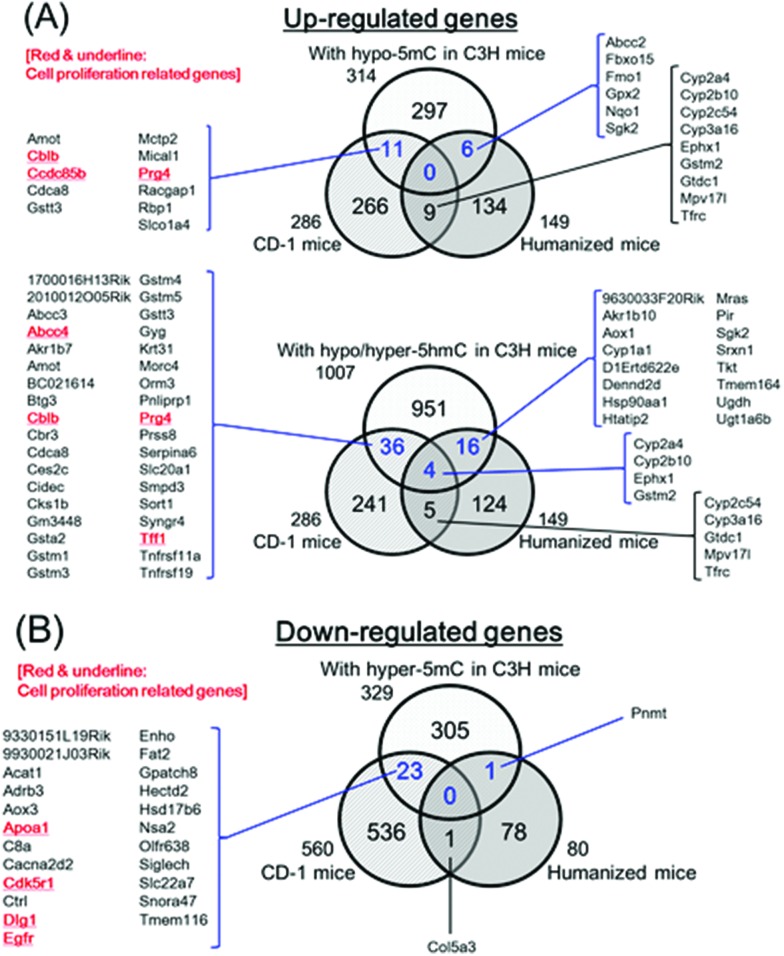
To evaluate the possible candidates of early phase marker genes for liver tumorigenesis by the CAR-mediated MOA, we determined overlap between the genes with altered expression levels with 5mC and 5hmC alterations in mouse hepatocellular adenoma induced by DEN/500 ppm PB and the genes previously determined with altered expression levels in the liver of CD-1 mice treated with 2500 ppm PB for 7 days.39 Several genes with altered expression levels with epigenetic alterations in hepatocellular adenoma induced by DEN/500 ppm PB changed in the same manner (increased or decreased expression levels) in the liver of CD-1 mice treated with 2500 ppm PB for 7 days (Fig. 5, ESI Tables S5–7†). Some of these overlapped genes (Abcc4, Apoa1, Cblb, Ccdc85b, Cdk5r1, Dlg1, Egfr, Prg4 and Tff1) were associated with Gene Ontology (GO) terms related to CAR or beta-catenin and cell proliferation and/or cell growth, and these nine genes previously demonstrated these relationships (Table 7). Furthermore, the overlap between the altered genes at expression levels with 5mC and/or 5hmC alterations in mouse hepatocellular adenoma induced by DEN/500 ppm PB and the altered genes at expression levels of liver in humanized chimeric mice treated with 1000 ppm PB for 7 days, previously determined,39 was also analyzed. While several genes overlapped, none of the genes related to cell proliferation and/or cell growth overlapped (Fig. 5, ESI Tables S5–7†).
Fig. 5. Overlap between the genes with 5mC and 5hmC alterations in mouse hepatocellular adenoma induced by DEN/PB and altered genes of liver in CD-1 and humanized chimeric mice treated with PB for 7 days. (A) and (B) present up-regulated or down-regulated genes compared to control, respectively. “C3H mice” means hepatocellular adenoma induced by DEN/PB. The altered genes of non-tumor liver in CD-1 and humanized chimeric mice treated with PB for 7 days were previously published.39 Information of individual genes overlapping is presented in ESI Tables S5–7.† .
Table 7. Overlapping genes between hepatocellular adenoma induced by DEN/500 ppm PB in C3H mice and livers in CD-1 mice treated with 2500 ppm PB for 7 days: detailed information of genes with gene ontology terms of relationship to CAR or beta-catenin and/or cell proliferation based on published references.
| C3H mice (up-regulated) |
Gene ontology terms of relationship to: |
Ref. | |||
| Gene symbol (mouse) | p-Value | Fold-change | CAR or beta-catenin | Cell proliferation or cell growth | |
| Abcc4 | 8.93 × 10–5 | 104.2 | Activation of CAR induces hepatic up-regulation of Abcc4 in a CAR-dependent manner. | Abcc4 enhances cell growth by progressing cell cycle. | 73 |
| 74 | |||||
| Cblb | 4.21 × 10–2 | 2.2 | — | Cblb regulates growth factor receptor signaling negatively and is involved in the inhibition of cancer cell proliferation. | 75 |
| Ccdc85b | 7.77 × 10–3 | 1.6 | Ccdc85b activates the degradation of beta-catenin. | Ccdc85b inhibits tumor cell growth. | 76 |
| Prg4 | 1.09 × 10–2 | 1.8 | Activation of Wnt/beta-catenin signaling increases Prg4 expression and slow-cell cycle population. | Prg4 suppresses synovial cell proliferation. | 77 |
| 78 | |||||
| Tff1 | 1.97 × 10–4 | 16.3 | Tff1 promotes invasion and chemoresistance in cancer through Wnt/beta-catenin signaling. | Tff1 reduces cell proliferation by delaying G1-S cell cycle transition. | 79 |
| 80 | |||||
| Gene symbol (mouse) | C3H mice (down-regulated) |
Gene ontology terms of relationship to: |
Ref. | ||
| p-Value | Fold-change | CAR or beta-catenin | Cell proliferation or cell growth | ||
| Apoa1 | 2.36 × 10–3 | –1.5 | Activation of CAR decreases plasma HDL level through down-regulation of Apoa1 expression. | Apoa1 promotes proliferation of human endothelial progenitor cells. | 81 |
| 82 | |||||
| Cdk5r1 | 2.43 × 10–2 | –1.2 | — | Cdk5r1 has a role of induction in beta-cell proliferation. | 83 |
| Dlg1 | 9.79 × 10–3 | –1.6 | Beta-catenin contributes to tumorigenesis through inducing DLG degradation (anti-Dlg1 antibody was used in this study). | Dlg1 is involved in inhibition of cell cycle progression by blocking G0/G1 transition. | 84 |
| 85 | |||||
| Egfr | 3.18 × 10–2 | –3.2 | Phenobarbital activates CAR by repressing the Egfr signaling. | Egfr signaling play key roles in hepatic cell proliferation. | 86 |
| 87 | |||||
Discussion
Candidate key genes for a mode of action involving activation of the constitutive androstane receptor (CAR) in mouse hepatocellular tumorigenesis
We analyzed 5mC, 5hmC and gene expression profiles globally in PB-induced hepatocellular adenomas in mice. Since we would like to know gene contributors with epigenetic alterations to PB-induced liver tumorigenesis, we focused on up and down regulated genes with changes in 5mC/5hmC in this paper. These tumors have distinctive alterations of 5mC and 5hmC profiles in thousands of gene promoter regions, suggesting that DNA modifications may characterize liver tumors in PB-treated mice as well as hepatic cancers in humans.16,17 In comparison with the number of differential gene promoters, there are more 5hmC-altered promoters than 5mC. This difference may be partially due to a structural difference between hydroxymethyl and methyl groups. The former group is slightly larger than the latter, which results in a higher titer of 5hmC antibodies and more sensitive detection of 5hmC sites than 5mC. In addition, the correlation between 5hmC alterations and up-regulated genes induced by PB treatment is clearer and more significant than 5mC alterations in mice.9,23
We performed integrated analysis and found that hundreds of genes are coordinately altered in DNA modification and expression in the liver of PB-treated mice. Subsequently, gene network analysis showed that several genes showing relationship between the CAR and tumor development are reported. Phillips et al. reported genes which showed CAR-dependent increases or decreases in liver tumors of mice,19 so we compared our data with their data. We found that some epigenetically and transcriptionally changed genes are reported to be transcriptionally changed in PB-treated wild-type mice but not changed in PB-treated CAR-knockout mice. For example, CAR activation results in direct up-regulation of Mdm2 expression,8 which is purportedly involved in poor survival in patients with hepatocellular carcinoma (HCC).58 Anxa2, whose promoter might contain CAR response elements,19 is expressed more abundantly in HCC tissues than in non-tumorous tissues.59 The CAR opposes the action of the estrogen receptor (Esr1),60 and Esr1 regulates the cell cycle by up-regulating Mki67.61 Some reports show that the high Mki67 expression may be correlated with the cell proliferation of HCC.38,62,63 The CAR probably binds to the PB-responsive enhancer module of the hypoxia inducible factor (HIF)-1A,64 which may contribute to HIF1A-induced activation of Cyr61.65 Cyr61 may relate to invasion and metastasis of HCC.66 These genes illustrated above might play a crucial function in PB-induced liver tumorigenesis in mice.
Another previous publication has reported candidate biomarkers of PB exposure up to 13 weeks of exposure that are differentially expressed correlated with epigenetic changes.23 We also compared our data with those of Laird et al. (2013)9 and Thomson et al. (2013)23 and found that 2 genes (Ndrg1 and Prom1) exhibit the changes in 5hmC levels with induced gene expression in both. Under short-term hypoxia, Ndrg1 expression is induced in a HIF1A-dependent manner.67 Ndrg1 is known to be associated with metastasis, recurrence and poor prognosis in HCC.68 HIF1A increases the promoter activity of Prom1,69 whose expression may indicate high capacity for tumorigenicity in HCC cells.70 These two genes showed continuous change in both 5hmC and gene expression levels from short-term PB-treated liver to long-term PB-induced tumor, suggesting that these genes might also be responsible for a cancer-promoting effect by PB.
Pathway enrichment analysis discovered many pathways enriched significantly, which included our data in about half of the total genes. Interestingly, the major types of top 20 pathways enriched for 5mC- or 5hmC-altered genes show different trends from those for transcriptionally altered genes. Many pathways enriched for gene expression profiles are associated with the cell cycle. This result supports the previous studies that PB administration to B6C3F1 mice or wild-type C57BL/6 mice causes increased expression of cell cycle regulatory genes.20,45 In contrast, both enrichment results for 5mC and 5hmC profiles contain fewer pathways related to cell cycle regulation and more pathways related to development and cancer than gene expression. These differences suggest that the activation of signaling pathways leading to tumorigenesis is more characteristically associated with DNA modification than gene expression.
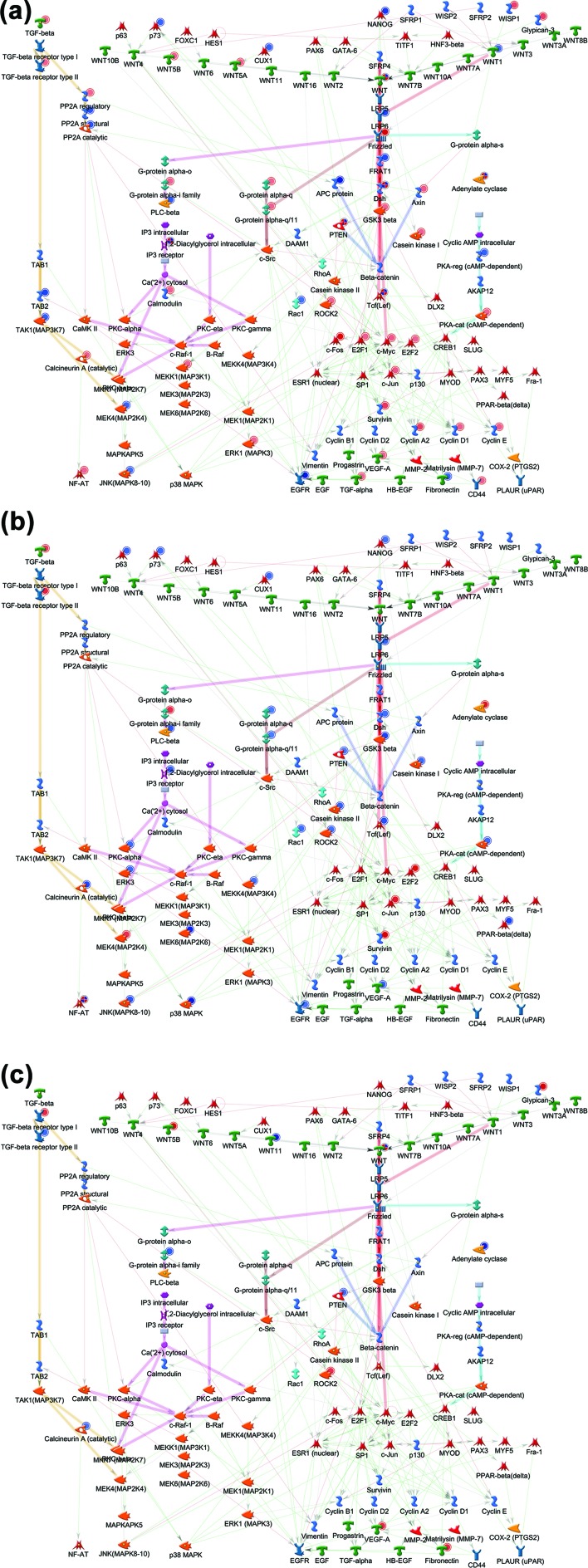
A recent controversy concerning the CAR-mediated MOA for mouse liver tumor formation concerns the phenotype of mouse liver tumors promoted by PB, where beta-catenin gene mutations have been observed.48,49 This gene is often mutated in human liver tumors.71 In our data, the promoter region of the beta-catenin gene was hypermethylated (chr9: 120778584–120779193) and hypohydroxymethylated (chr9: 120780888–120781871), but beta-catenin gene expression was not significantly altered based on the criteria of our study (but close to significant, p = 0.0543, 1.22-fold control). Furthermore, we performed additional gene network analysis of the Wnt/beta-catenin signaling network in MetaCore with our gene expression data of the three animal models (Fig. 6). PB treatment altered the expression of many genes related to the Wnt/beta-catenin signaling network in tumor tissue of C3H mice (51 genes) and in the liver of CD-1 mice (36 genes), however, fewer in chimeric mice with human hepatocytes (12 genes). These findings suggest a species difference between the effects of PB in mouse and human hepatocytes.
Fig. 6. Gene network of Wnt signaling in the MetaCore database and the gene expression data of (A) hepatocellular adenoma in C3H mice, (B) liver in CD-1 mice and (C) chimeric mice with human hepatocytes. Genes with red or blue circles indicate up-regulated or down-regulated genes compared to control, respectively. Green, red or gray arrows indicate positive/activation, negative/inhibition or unspecified effects, respectively.
As shown in Fig. 5 and Table 6, we identified nine genes altered in common in hepatocellular adenoma induced by DEN/PB (both alterations of expression and 5mC/5hmC) and in the liver of mice treated with PB for 1 week (alteration of expression, 5mC/5hmC were not determined). Interestingly, all nine genes were known as cell proliferation/growth-related genes, and seven of the nine genes were also known to relate to CAR or beta-catenin. Thus, taking published information into consideration, the determined nine genes, at least, could provide important information for an evaluation of key genes for CAR-mediated mouse liver tumorigenesis.
The present research may have limitations for reaching the definitive conclusions described above. Dose levels were different between the 27 week and 1 week studies; much higher doses were used in the short duration studies (500 ppm for a 27 week study and 2500 ppm for 1 week). This lack of dose concordance may render conclusions based on similarity between short and long duration as speculative and may not provide strong evidence for inclusion into the mouse MOA for liver tumorigenesis. However, in the 1 week study in CD-1 mice, NaPB equivocally increased hepatic 7-pentoxyresorufin O-depentylase activity (CYP2B marker) at 500, 1000, 1500 and 2500 ppm (4.4, 5.4, 6.4 and 6.4 fold of control).39 These data suggest that CAR activation by PB 500 and 2500 ppm was similar. Thus, we consider that the similarity between short and long duration can provide evidence for inclusion into the mouse MOA. To confirm this, functional analysis of the selected genes remains to be conducted. Another limitation was the confounded study design. We analyzed the tumor/PB vs. non-tumor/control liver from different animals, but this analysis cannot separate PB-specific effects from tumor-specific effects. This means that the identified changed genes carried over into the subsequent analysis may be a result of either the PB treatment or the tumorigenesis process or the interaction of the two. Therefore, when compared with the short duration treatments, any commonality may be due merely to a PB response that is sustained for the duration of treatment. Thus, functional analysis of the selected genes remains to be evaluated in tumorigenesis.
Evaluation of the human relevance of the CAR-mediated MOA for mouse hepatocellular tumorigenesis
In the present study, plasma PB levels in the DEN/500 ppm PB mice were 15 μg ml–1. The plasma levels of PB observed in human subjects given therapeutic doses of 3–6 mg kg–1 ranged from 10 to 25 μg ml–1.72 Therefore, plasma PB levels in the DEN/500 ppm PB mice were equivalent to those reported in humans by Monro (1993).72 Since plasma PB levels in the humanized chimeric mice treated with 1000 ppm (75 μg ml–1) were higher than those in the DEN/500 ppm PB mice (15 μg ml–1) but equivalent to those in CD-1 mice treated with 2500 ppm (70 μg ml–1), we believe that the comparison between the 1000 ppm PB treated humanized chimeric mice and the DEN/500 ppm PB treated mice, and the 2500 ppm PB treated mice is able to provide useful information, at least without underestimation, on species differences in initial key events of hepatocellular tumorigenesis. Non-overlap of the selected nine cell proliferation/growth-related genes described above in humanized chimeric mouse models (namely, nine cell proliferation/growth-related genes observed in the liver of mice given PB for 1 week and in DEN/PB liver tumors of mice given PB for 27 weeks were not increased in the humanized chimeric mice given PB for 1 week) does not conflict with the previous findings that the key species differences are related to the lack of cell proliferation in human hepatocytes.31,39,41,43 Therefore, these findings are consistent with the previous conclusion that CAR-mediated MOA for rodent liver tumorigenesis is not relevant to humans.3,27–42 To confirm this, functional analysis of the selected genes remains to be evaluated in mouse liver tumorigenesis.
Funding
This work was supported by the Sumitomo Chemical Company, Ltd.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We acknowledge Professor Samuel M. Cohen (Department of Pathology and Microbiology, University of Nebraska Medical Center) and Professor Brian G. Lake (Centre for Toxicology, Faculty of Health and Medical Sciences, University of Surrey) for valuable scientific advice. The authors also thank Mr N. Hattori for global DNA methylation, hydroxymethylation and gene expression analyses; Ms N. Takata for preliminary experiments of MeDIP; and Ms H. Kikumoto and K. Tanaka for experimental animal studies in this work.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7tx00163k
References
- IARC, IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Some Thyrotropic Agents: Phenobarbital and its sodium salt, 2001, vol. 79, pp. 161–288. [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T., Sueyoshi T., Zelko I., Moore R., Washburn K., Negishi M. Mol. Cell. Biol. 1999;19:6318–6322. doi: 10.1128/mcb.19.9.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcombe C. R., Peffer R. C., Wolf D. C., Bailey J., Bars R., Bell D., Cattley R. C., Ferguson S. S., Geter D., Goetz A., Goodman J. I., Hester S., Jacobs A., Omiecinski C. J., Schoeny R., Xie W., Lake B. G. Crit. Rev. Toxicol. 2014;44:64–82. doi: 10.3109/10408444.2013.835786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkakoski P., Zelko I., Sueyoshi T., Negishi M. Mol. Cell. Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Sueyoshi T., Inoue K., Moore R., Negishi M. Mol. Pharmacol. 2003;64:1069–1075. doi: 10.1124/mol.64.5.1069. [DOI] [PubMed] [Google Scholar]
- Yoshinari K., Kobayashi K., Moore R., Kawamoto T., Negishi M. FEBS Lett. 2003;548:17–20. doi: 10.1016/s0014-5793(03)00720-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Moore R., Goldsworthy T. L., Negishi M., Maronpot R. R. Cancer Res. 2004;64:7197–7200. doi: 10.1158/0008-5472.CAN-04-1459. [DOI] [PubMed] [Google Scholar]
- Huang W., Zhang J., Washington M., Liu J., Parant J. M., Lozano G., Moore D. D. Mol. Endocrinol. 2005;19:1646–1653. doi: 10.1210/me.2004-0520. [DOI] [PubMed] [Google Scholar]
- Laird A., Thomson J. P., Harrison D. J., Meehan R. R. Epigenomics. 2013;5:655–669. doi: 10.2217/epi.13.69. [DOI] [PubMed] [Google Scholar]
- Moore L. D., Le T., Fan G. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani C. J., Madzo J., Moen E. L., Yesilkanal A., Godley L. A. Cancers. 2013;5:786–814. doi: 10.3390/cancers5030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddington J. P., Pennings S., Meehan R. R. Biochem. J. 2013;451:13–23. doi: 10.1042/BJ20121585. [DOI] [PubMed] [Google Scholar]
- Moen E. L., Mariani C. J., Zullow H., Jeff-Eke M., Litwin E., Nikitas J. N., Godley L. A. Immunol. Rev. 2015;253:36–49. doi: 10.1111/imr.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneto H., Sasaki S., Yamamoto H., Itoh F., Toyota M., Suzuki H., Ozeki I., Iwata N., Ohmura T., Satoh T., Karino Y., Toyota J., Satoh M., Endo T., Omata M., Imai K. Gut. 2001;48:372–377. doi: 10.1136/gut.48.3.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sceusi E. L., Loose D. S., Wray C. J. HPB. 2011;13:369–376. doi: 10.1111/j.1477-2574.2011.00303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.-R., Tian M.-X., Jin L., Yang L.-X., Ding Z.-B., Shen Y.-H., Peng Y.-F., Zhou J., Qiu S.-J., Dai Z., Fan J., Shi Y.-H. J. Exp. Clin. Cancer Res. 2014;33:32. doi: 10.1186/1756-9966-33-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajadian S. O., Ehnert S., Vakilian H., Koutsouraki E., Damm G., Seehofer D., Thasler W., Dooley S., Baharvand H., Sipos B., Nussler A. K. Clin. Epigenet. 2015;7:98. doi: 10.1186/s13148-015-0133-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. P., Meehan R. R. Epigenomics. 2016;9:77–91. doi: 10.2217/epi-2016-0122. [DOI] [PubMed] [Google Scholar]
- Phillips J. M., Burgoon L. D., Goodman J. I. Toxicol. Sci. 2009;110:319–333. doi: 10.1093/toxsci/kfp108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. M., Burgoon L. D., Goodman J. I. Toxicol. Sci. 2009;109:193–205. doi: 10.1093/toxsci/kfp050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempiäinen H., Muller A., Brasa S., Teo S. S., Roloff T. C., Morawiec L., Zamurovic N., Vicart A., Funhoff E., Couttet P., Schubeler D., Grenet O., Marlowe J., Moggs J., Terranova R. PLoS One. 2011;6:e18216. doi: 10.1371/journal.pone.0018216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. P., Lempiainen H., Hackett J. A., Nestor C. E., Muller A., Bolognani F., Oakeley E. J., Schubeler D., Terranova R., Reinhardt D., Moggs J. G., Meehan R. R. Genome Biol. 2012;13:R93. doi: 10.1186/gb-2012-13-10-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. P., Hunter J. M., Lempiainen H., Muller A., Terranova R., Moggs J. G., Meehan R. R. Nucleic Acids Res. 2013;41:5639–5654. doi: 10.1093/nar/gkt232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman A. N., Phillips J. M., Goodman J. I. Toxicol. Sci. 2006;91:393–405. doi: 10.1093/toxsci/kfj155. [DOI] [PubMed] [Google Scholar]
- Phillips J. M., Goodman J. I. Toxicol. Sci. 2009;108:273–289. doi: 10.1093/toxsci/kfp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson J. P., Ottaviano R., Unterberger E. B., Lempiäinen H., Muller A., Terranova R., Illingworth R. S., Webb S., Kerr A. R. W., Lyall M. J., Drake A. J., Wolf C. R., Moggs J. G., Schwarz M., Meehan R. R. Cancer Res. 2016;76:3097–3108. doi: 10.1158/0008-5472.CAN-15-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. M., Arnold L. L. Toxicol. Sci. 2011;120:S76–S92. doi: 10.1093/toxsci/kfq365. [DOI] [PubMed] [Google Scholar]
- Cohen S. M. Toxicol. Pathol. 2010;38:487–501. doi: 10.1177/0192623310363813. [DOI] [PubMed] [Google Scholar]
- Holsapple M. P., Pitot H. C., Cohen S. M., Boobis A. R., Klaunig J. E., Pastoor T., Dellarco V. L., Dragan Y. P. Toxicol. Sci. 2006;89:51–56. doi: 10.1093/toxsci/kfj001. [DOI] [PubMed] [Google Scholar]
- Lake B. G. Xenobiotica. 2009;39:582–596. doi: 10.1080/00498250903098184. [DOI] [PubMed] [Google Scholar]
- Hirose Y., Nagahori H., Yamada T., Deguchi Y., Tomigahara Y., Nishioka K., Uwagawa S., Kawamura S., Isobe N., Lake B. G., Okuno Y. Toxicology. 2009;258:64–69. doi: 10.1016/j.tox.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Yamada T., Uwagawa S., Okuno Y., Cohen S. M., Kaneko H. Toxicol. Sci. 2009;108:59–68. doi: 10.1093/toxsci/kfp007. [DOI] [PubMed] [Google Scholar]
- Osimitz T. G., Lake B. G. Crit. Rev. Toxicol. 2009;39:501–511. doi: 10.1080/10408440902914014. [DOI] [PubMed] [Google Scholar]
- Lake B. G., Price R. J., Osimitz T. G. Pest Manage. Sci. 2015;71:829–834. doi: 10.1002/ps.3854. [DOI] [PubMed] [Google Scholar]
- LeBaron M. J., Gollapudi B. B., Terry C., Billington R., Rasoulpour R. J. Crit. Rev. Toxicol. 2014;44(Suppl. 2):15–24. doi: 10.3109/10408444.2014.910751. [DOI] [PubMed] [Google Scholar]
- LeBaron M. J., Rasoulpour R. J., Gollapudi B. B., Sura R., Kan H. L., Schisler M. R., Pottenger L. H., Papineni S., Eisenbrandt D. L. Toxicol. Sci. 2014;142:74–92. doi: 10.1093/toxsci/kfu155. [DOI] [PubMed] [Google Scholar]
- Marx-Stoelting P., Ganzenberg K., Knebel C., Schmidt F., Rieke S., Hammer H., Schmidt F., Potz O., Schwarz M., Braeuning A. Arch. Toxicol. 2017;91:2895–2907. doi: 10.1007/s00204-016-1925-2. [DOI] [PubMed] [Google Scholar]
- Wood C. E., Hukkanen R. R., Sura R., Jacobson-Kram D., Nolte T., Odin M., Cohen S. M. Toxicol. Pathol. 2015;43:760–775. doi: 10.1177/0192623315576005. [DOI] [PubMed] [Google Scholar]
- Yamada T., Okuda Y., Kushida M., Sumida K., Takeuchi H., Nagahori H., Fukuda T., Lake B. G., Cohen S. M., Kawamura S. Toxicol. Sci. 2014;142:137–157. doi: 10.1093/toxsci/kfu173. [DOI] [PubMed] [Google Scholar]
- Yamada T., Cohen S. M., Lake B. G. Toxicol. Sci. 2015;147:298–299. doi: 10.1093/toxsci/kfv186. [DOI] [PubMed] [Google Scholar]
- Yamada T., Kikumoto H., Lake B. G., Kawamura S. Toxicol. Res. 2015;4:901–913. [Google Scholar]
- Kushida M., Yamada T. and Okuno Y., in Thresholds of Genotoxic Carcinogens, ed. T. Nomi and S. Fukushima, Academic Press, London, 2016, ch. 12, pp. 193–203. [Google Scholar]
- Parzefall W., Erber E., Sedivy R., Schulte-Hermann R. Cancer Res. 1991;51:1143–1147. [PubMed] [Google Scholar]
- La Vecchia C., Negri E. Eur. J. Cancer Prev. 2014;23:1–7. doi: 10.1097/CEJ.0b013e32836014c8. [DOI] [PubMed] [Google Scholar]
- Luisier R., Lempiainen H., Schlerbichler N., Braeuning A., Geissler M., Dubost V., Muller A., Scheer N., Chibout S. D., Hara H., Picard F., Theil D., Couttet P., Vitobello A., Grenet O., Grasl-Kraupp B., Ellinger-Ziegelbauer H., Thomson J. P., Meehan R. R., Elcombe C. R., Henderson C. J., Wolf C. R., Schwarz M., Moulin P., Terranova R., Moggs J. G. Toxicol. Sci. 2014;139:501–511. doi: 10.1093/toxsci/kfu038. [DOI] [PubMed] [Google Scholar]
- Braeuning A., Gavrilov A., Brown S., Wolf C. R., Henderson C. J., Schwarz M. Toxicol. Sci. 2014;140:259–270. doi: 10.1093/toxsci/kfu099. [DOI] [PubMed] [Google Scholar]
- Braeuning A. Arch. Toxicol. 2014;88:1771–1772. doi: 10.1007/s00204-014-1331-6. [DOI] [PubMed] [Google Scholar]
- Braeuning A., Schwarz M. Arch. Toxicol. 2016;90:1525–1526. doi: 10.1007/s00204-016-1712-0. [DOI] [PubMed] [Google Scholar]
- Braeuning A., Henderson C. J., Wolf C. R., Schwarz M. Toxicol. Sci. 2015;147:299–300. [PubMed] [Google Scholar]
- Groll N., Kollotzek F., Goepfert J., Joos T. O., Schwarz M., Braeuning A. Biol. Chem. 2016;397:91–96. doi: 10.1515/hsz-2015-0223. [DOI] [PubMed] [Google Scholar]
- Phillips J. M., Goodman J. I. Toxicol. Sci. 2008;104:86–99. doi: 10.1093/toxsci/kfn063. [DOI] [PubMed] [Google Scholar]
- Sakamoto Y., Inoue K., Takahashi M., Taketa Y., Kodama Y., Nemoto K., Degawa M., Gamou T., Ozawa S., Nishikawa A., Yoshida M. Toxicol. Pathol. 2013;41:1078–1092. doi: 10.1177/0192623313482055. [DOI] [PubMed] [Google Scholar]
- Sado Y., Inoue S., Tomono Y., Omori H. Acta Histochem. Cytochem. 2006;39:89–94. doi: 10.1267/ahc.06001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Bolstad B. M., Collin F., Cope L. M., Hobbs B., Speed T. P. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. E., Li W., Meyer C. A., Gottardo R., Carroll J. S., Brown M., Liu X. S. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal M., Kumar A., Yella V. R. Curr. Opin. Struct. Biol. 2014;25:77–85. doi: 10.1016/j.sbi.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Shippy R., Fulmer-Smentek S., Jensen R. V., Jones W. D., Wolber P. K., Johnson C. D., Pine P. S., Boysen C., Guo X., Chudin E., Sun Y. A., Willey J. C., Thierry-Mieg J., Thierry-Mieg D., Setterquist R. A., Wilson M., Lucas A. B., Novoradovskaya N., Papallo A., Turpaz Y., Baker S. C., Warrington J. A., Shi L., Herman D. Nat. Biotechnol. 2006;24:1123–1131. doi: 10.1038/nbt1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöniger-Hekele M., Hanel S., Wrba F., Muller C. Liver Int. 2005;25:62–69. doi: 10.1111/j.1478-3231.2004.0997.x. [DOI] [PubMed] [Google Scholar]
- Mohammad H. S., Kurokohchi K., Yoneyama H., Tokuda M., Morishita A., Jian G., Shi L., Murota M., Tani J., Kato K., Miyoshi H., Deguchi A., Himoto T., Usuki H., Wakabayashi H., Izuishi K., Suzuki Y., Iwama H., Deguchi K., Uchida N., Sabet E. A., Arafa U. A., Hassan A. T., El-Sayed A. A., Masaki T. Int. J. Oncol. 2008;33:1157–1163. [PubMed] [Google Scholar]
- Min G., Kim H., Bae Y., Petz L., Kemper J. K. J. Biol. Chem. 2002;277:34626–34633. doi: 10.1074/jbc.M205239200. [DOI] [PubMed] [Google Scholar]
- Liao X. H., Lu D. L., Wang N., Liu L. Y., Wang Y., Li Y. Q., Yan T. B., Sun X. G., Hu P., Zhang T. C. FEBS J. 2014;281:927–942. doi: 10.1111/febs.12658. [DOI] [PubMed] [Google Scholar]
- van Dekken H., Verhoef C., Wink J., van Marion R., Vissers K. J., Hop W. C. J., de Man R. A., Ijzermans J. N., van Eijck C. H. J., Zondervan P. E. Acta Histochem. 2005;107:161–171. doi: 10.1016/j.acthis.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Mitselou A., Karapiperides D., Nesseris I., Vougiouklakis T., Agnantis N. J. Anticancer Res. 2010;30:4493–4501. [PubMed] [Google Scholar]
- Shizu R., Shindo S., Yoshida T., Numazawa S. Toxicol. Lett. 2013;219:143–150. doi: 10.1016/j.toxlet.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Kunz M., Moeller S., Koczan D., Lorenz P., Wenger R. H., Glocker M. O., Thiesen H. J., Gross G., Ibrahim S. M. J. Biol. Chem. 2003;278:45651–45660. doi: 10.1074/jbc.M301373200. [DOI] [PubMed] [Google Scholar]
- Zeng Z. J., Yang L. Y., Ding X., Wang W. World J. Gastroenterol. 2004;10:3414–3418. doi: 10.3748/wjg.v10.i23.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangul H. BMC Genet. 2004;5:27. doi: 10.1186/1471-2156-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J., Xie H. Y., Xu X., Wu J., Wei X., Su R., Zhang W., Lv Z., Zheng S., Zhou L. Cancer Lett. 2011;310:35–45. doi: 10.1016/j.canlet.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Ohnishi S., Maehara O., Nakagawa K., Kameya A., Otaki K., Fujita H., Higashi R., Takagi K., Asaka M., Sakamoto N., Kobayashi M., Takeda H. PLoS One. 2013;8:e66255. doi: 10.1371/journal.pone.0066255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin S., Li J., Hu C., Chen X., Yao M., Yan M., Jiang G., Ge C., Xie H., Wan D., Yang S., Zheng S., Gu J. Int. J. Cancer. 2007;120:1444–1450. doi: 10.1002/ijc.22476. [DOI] [PubMed] [Google Scholar]
- Dong B., Lee J. S., Park Y. Y., Yang F., Xu G., Huang W., Finegold M. J., Moore D. D. Nat. Commun. 2015;6:5944. doi: 10.1038/ncomms6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monro A. Regul. Toxicol. Pharmacol. 1993;18:115–135. doi: 10.1006/rtph.1993.1048. [DOI] [PubMed] [Google Scholar]
- Assem M., Schuetz E. G., Leggas M., Sun D., Yasuda K., Reid G., Zelcer N., Adachi M., Strom S., Evans R. M., Moore D. D., Borst P., Schuetz J. D. J. Biol. Chem. 2004;279:22250–22257. doi: 10.1074/jbc.M314111200. [DOI] [PubMed] [Google Scholar]
- Zhao X., Guo Y., Yue W., Zhang L., Gu M., Wang Y. OncoTargets Ther. 2014;7:343–351. doi: 10.2147/OTT.S56029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D., Ma Y., Liu J., Xu L., Zhang Y., Qu J., Liu Y., Qu X. Int. J. Mol. Sci. 2013;14:24399–24411. doi: 10.3390/ijms141224399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai A., Hijikata M., Hishiki T., Isono O., Chiba T., Shimotohno K. Oncogene. 2008;27:1520–1526. doi: 10.1038/sj.onc.1210801. [DOI] [PubMed] [Google Scholar]
- Yasuhara R., Ohta Y., Yuasa T., Kondo N., Hoang T., Addya S., Fortina P., Pacifici M., Iwamoto M., Enomoto-Iwamoto M. Lab. Invest. 2011;91:1739–1752. doi: 10.1038/labinvest.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sharif A., Jamal M., Zhang L. X., Larson K., Schmidt T. A., Jay G. D., Elsaid K. A. Arthritis Rheumatol. 2015;67:1503–1513. doi: 10.1002/art.39087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossenmeyer-Pourie C., Kannan R., Ribieras S., Wendling C., Stoll I., Thim L., Tomasetto C., Rio M. C. J. Cell Biol. 2002;157:761–770. doi: 10.1083/jcb200108056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Ma Y., Huang X. Int. J. Clin. Exp. Pathol. 2015;8:10412–10419. [PMC free article] [PubMed] [Google Scholar]
- Masson D., Qatanani M., Sberna A. L., Xiao R., Pais de Barros J. P., Grober J., Deckert V., Athias A., Gambert P., Lagrost L., Moore D. D., Assem M. J. Lipid Res. 2008;49:1682–1691. doi: 10.1194/jlr.M700374-JLR200. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pecchi V., Valdes S., Pons V., Honorato P., Martinez L. O., Lamperti L., Aguayo C., Radojkovic C. Microvasc. Res. 2015;98:9–15. doi: 10.1016/j.mvr.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Draney C., Hobson A. E., Grover S. G., Jack B. O., Tessem J. S. J. Diabetes Res. 2016;2016:6375804. doi: 10.1155/2016/6375804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiah V. K., Narayan N., Massimi P., Banks L. Int. J. Cancer. 2012;131:2223–2233. doi: 10.1002/ijc.27519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinken M., Decrock E., Leybaert L., Bultynck G., Himpens B., Vanhaecke T., Rogiers V. Biochim. Biophys. Acta. 2012;1818:2002–2008. doi: 10.1016/j.bbamem.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Hashimoto M., Honkakoski P., Negishi M. Arch. Toxicol. 2015;89:1045–1055. doi: 10.1007/s00204-015-1522-9. [DOI] [PubMed] [Google Scholar]
- Komposch K., Sibilia M. Int. J. Mol. Sci. 2016;17:30–60. doi: 10.3390/ijms17010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.