Abstract
DNA methylation can contribute to the maintenance of genome integrity and regulation of gene expression. In most situations, DNA methylation patterns are inherited quite stably. However, changes in DNA methylation can occur at some loci as a result of tissue culture resulting in somaclonal variation. To investigate heritable epigenetic changes as a consequence of tissue culture, a sequence-capture bisulfite sequencing approach was implemented to monitor context-specific DNA methylation patterns in ∼15 Mb of the maize genome for a population of plants that had been regenerated from tissue culture. Plants that have been regenerated from tissue culture exhibit gains and losses of DNA methylation at a subset of genomic regions. There was evidence for a high rate of homozygous changes to DNA methylation levels that occur consistently in multiple independent tissue culture lines, suggesting that some loci are either targeted or hotspots for epigenetic variation. The consistent changes inherited following tissue culture include both gains and losses of DNA methylation and can affect CG, CHG, or both contexts within a region. Only a subset of the tissue culture changes observed in callus plants are observed in the primary regenerants, but the majority of DNA methylation changes present in primary regenerants are passed onto offspring. This study provides insights into the susceptibility of some loci and potential mechanisms that could contribute to altered DNA methylation and epigenetic state that occur during tissue culture in plant species.
Keywords: DNA methylation, maize, epigenomics, tissue culture, somaclonal variation, bisulfite sequencing, sequence capture
TISSUE culture is used in many commercially important plant species, both for clonal propagation as well as successful transformation and regeneration of transgenic materials. While it is expected that clonal plants derived from tissue culture will have no changes in genetic information, there are frequent examples of somaclonal variation, manifested as heritable phenotypic differences in plants recovered from tissue culture (Phillips et al. 1994; Kaeppler et al. 2000; Miguel and Marum 2011; Neelakandan and Wang 2012). In some cases, this could be due to activation of transposons during tissue culture (Peschke et al. 1987; Hirochika 1993). However, in other cases there is evidence for epigenetic changes that are directly linked to gene expression variation manifested as phenotypic variation in plants derived from culture (Rhee et al. 2010; Ong-Abdullah et al. 2015). For instance, the Karma epiallele in oil palm is associated with loss of CHG DNA methylation at a transposable element (TE) nested in a gene resulting in production of aberrant transcripts (Ong-Abdullah et al. 2015).
In plants, DNA methylation variation can be associated with heritable differences in gene expression in the absence of DNA sequence changes. Approximately 30% of the cytosines in the maize genome are present as 5-methylcytosine (Papa et al. 2001). DNA methylation is particularly prevalent in CG dinucleotide or CHG (where H is A, C, or T) trinucleotide contexts, with lower levels of DNA methylation at CHH sites (Gent et al. 2013; Regulski et al. 2013; West et al. 2014). There are distinct mechanisms to target and maintain methylation in each of these sequence contexts (Law and Jacobsen 2010; Matzke and Mosher 2014; Springer and Schmitz 2017). Maintenance pathways are required to remethylate daughter strands following DNA replication; otherwise, DNA methylation can be passively lost over rounds of replication. Plants also utilize active mechanisms for demethylation (Zhang and Zhu 2012) and the plant methylome is the result of combined methylation and demethylation activities.
The sources and prevalence of epigenetic variation are of significant interest. There is abundant evidence for natural variation in DNA methylation in plants, as shown in maize and other plant species (Taudt et al. 2016; Springer and Schmitz 2017). There is less evidence for developmental variation in DNA methylation for most vegetative tissues (Eichten et al. 2013a; Schmitz et al. 2013; Li et al. 2015c; Kawakatsu et al. 2016). While several studies have provided examples of DNA methylation changes induced by the environment (Dowen et al. 2012; Jiang et al. 2014; Wibowo et al. 2016), other studies have found quite limited evidence for changes in DNA methylation in response to the environment (Song et al. 2013; Eichten and Springer 2015; Hagmann et al. 2015; Secco et al. 2015; Crisp et al. 2017; Ganguly et al. 2017).
There is evidence that DNA methylation patterns are perturbed by tissue culture. Previous studies have found locus-specific effects on DNA methylation (Kaeppler and Phillips 1993), and several epialleles resulting from tissue culture have been characterized (Krizova et al. 2009; Rhee et al. 2010; Ong-Abdullah et al. 2015). Recent studies have provided genome-wide evidence for changes in DNA methylation following tissue culture in Arabidopsis, rice, and maize (Tanurdzic et al. 2008; Stroud et al. 2013; Stelpflug et al. 2014). Each of these studies have documented hundreds of loci with altered DNA methylation levels following tissue culture, and have found evidence for losses of DNA methylation that are consistent in independent tissue culture derived lines. Given the widespread use of tissue culture for propagation and transformation of plants, the extent and nature of heritable epigenetic changes as a consequence of culture are of great interest. There is evidence for Mendelian inheritance of epialleles following tissue culture in rice and maize (Stroud et al. 2013; Stelpflug et al. 2014). However, some changes are likely more transient, for instance, asymmetric CHH DNA methylation (Stroud et al. 2013), although this has not been specifically examined in maize. Maize transgenic plants are generally seed propagated prior to widespread usage and therefore heritable changes could be maintained while transient changes would be eliminated following reproduction. Therefore, in this study we sought to examine the level and consistency of heritable changes in DNA methylation in the progeny of plants regenerated from tissue culture.
Previous work on maize utilized a quantitative approach to document locus-specific methylation levels by coupling immunoprecipitation of methylated DNA with hybridization to microarrays (Stelpflug et al. 2014). While this method can be useful for documenting regions with quantitative variation in DNA methylation, the procedure lacks the ability to resolve context-specific differences and lacks the resolution for determining the specific boundaries of altered methylation. In this study, we focused on characterizing the heritable changes that could be observed in the progeny of plants regenerated from tissue culture. We utilized a sequence-capture bisulfite sequencing approach to profile context specific DNA methylation levels in 25 R1 plants recovered from tissue culture. We find many regions with altered DNA methylation levels, particularly in the CG and CHG contexts. A subset of the changes in DNA methylation are consistently found in many independent events. The majority of these consistent changes in methylation are also observed to occur in natural populations. Analysis of context-specific changes in methylation allowed us to identify numerous CG-only and CHG-only differentially methylated regions (DMRs). Moreover, we document multiple examples of Karma-like (Ong-Abdullah et al. 2015) epialleles with context-specific changes in DNA methylation. At these loci, heterochromatic DNA methylation is lost at TEs located within genes. This study may shed light on the potential mechanisms that drive epigenome changes during tissue culture.
Materials and Methods
Tissue culture and plant materials
All tissue culture experiments were conducted using the regenerable A188 maize inbred. Methylomes of the third seedling leaf of 2-week-old seedlings from three biological replicates of B73, two replicates of Mo17, and single replicates of W22, MoG, and Ki11 were captured and analyzed for comparison to A188 and regenerated plants. Inbreds, noncultured control plants and regenerated maize plants of independent maize-immature-embryo-derived cell lines were grown under standard greenhouse conditions at the Gortner Ave. Greenhouse (University of Minnesota, St. Paul, MN). The tissue culture and regeneration process of maize immature embryos was as described by Stelpflug et al. (2014).
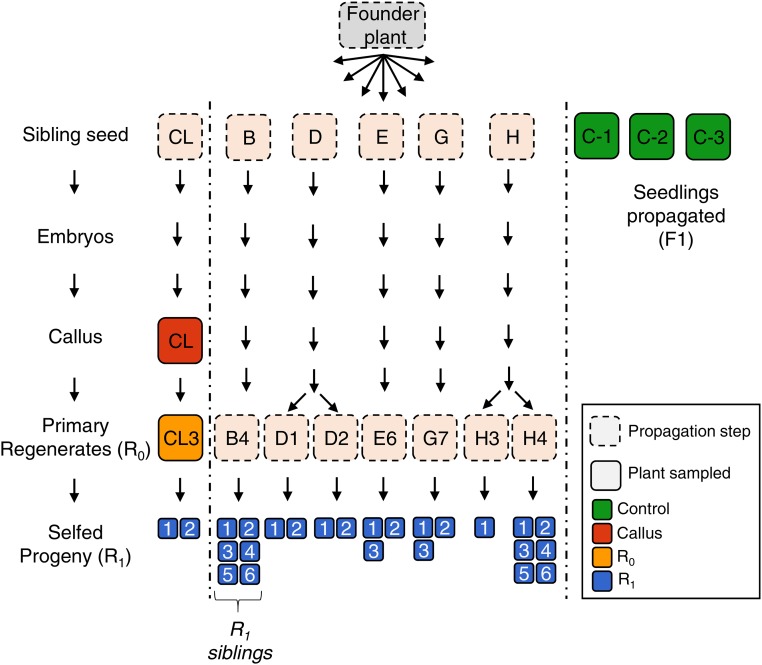
Cell cultures (cell-lines) originated from independent immature embryos named CL, B, D, E, G, and H. R0 plants were regenerated from each of these cell lines and R1 progeny propagated. Each independent R0 and R1 plant was named using their embryo letter followed by an R0 and R1 number (e.g., B4_2 is the second R1 progeny of plant four regenerated from embryo B). One callus sample named CL, an R0 regenerated plant (CL3 - flag leaf), and two R1 progeny (CL3_1 and CL3_2) were selected for comparison of different generations. The cell line “CL” was previously described in Stelpflug et al. (2014); however, sample names were modified for consistency. Sample name descriptions herein correspond to the original sample names as follows: callus sample CL = “CL-3”; R0 CL3 = “3-7”; R1 CL3_1 = “3-7.3”; and R1 CL3_2 = “3-7.7.” Seven R0 regenerated plants (B4, D1, D2, E6, G7, H3, and H4) from five independent maize cell cultures were selfed, and a total of 25 progeny R1 plants were harvested for analysis including one to six replicates from each R0. In addition, the corresponding third leaf was collected from three different control A188 plants, which were sibling plants originating from the same seed source and were not subjected to tissue culture.
Maize bisulfite coupled sequence capture (SeqCap-Epi-v2) probe design
The sequence capture probe set was originally designed based on the B73 RefGen_v2 (AGPv2) assembly of the maize genome and subsequently updated to B73 RefGen_v4 aka AGPv4 (also known as Zm-B73-REFERENCE-GRAMENE-4.0) as detailed in Supplemental Material, File S1. A total of 20,643 nonredundant genomic regions spanning 15,728,511 Mb were used to design probes based on the B73 reference genome (AGPv2). These regions were selected based on various criteria. All regions from the first version of capture probes were included in v2 (Li et al. 2015a); however, the total genome space captured was increased to 15.7 Mb. Additional probes were designed to capture loci satisfying criteria including: DMRs identified between B73, Mo17, and Oh43, and between five tissues of B73 (Li et al. 2015b); tissue culture DMRs (Stelpflug et al. 2014); cryptic promoters (Li et al. 2015b); mCHH islands (Li et al. 2015c); and, various siRNA loci such as phased loci (Zhai et al. 2015). The specific target region of interest was termed “specific region” and the bait region captured by the probe design was termed the “target region.” The target regions often included flanking regions of the specific region, and a single target region can encompass multiple specific regions. A single specific region may satisfy multiple criteria of interest, e.g., a region may be both an mCHH island and a CHH DMR. In total, 23,151 “specific” regions (22,950 nonredundant) were defined, including 201 regions that each was annotated to two classes.
Library construction and sequencing
SeqCap libraries were constructed as described previously (Li et al. 2015a). Briefly, DNA of maize-embryo-derived callus, regenerated plants, and noncultured control plants were isolated using the standard CTAB method. Genomic DNA (500–1000 ng) was used for sequence capture library construction. DNA was sheared to fragments between 180 and 250 bp. The fragments were subject to end repair, dA tailing, ligating to index adapters, and Dual-SPRI Size Selection, followed by bisulfite conversion. The bisulfite converted libraries were then subject to Pre-Capture LM-PCR amplification and purification, and quantified using a PicoGreen dsDNA Assay Kit. Then, the amplified sample library was hybridized to probes designed to target the set of genomic regions selected for analysis. After hybridization, the captured DNA library was bounded to capture beads and the bead-bound DNA was washed. Post-capture LM-PCR amplification was performed, and, following PCR cleanup, the libraries were quantitated using PicoGreen. Libraries were pooled together in several batches and the pools sequenced over multiple lanes at the University of Minnesota Genomics Center on a HiSeq2500, in High-output 2 × 125 bp paired end (PE) mode.
Data analysis
Adapters were trimmed using Trim Galore! Reads were mapped to maize B73 reference genome AGPv4 using BSMAP-2.90 (Xi and Li 2009), allowing five mismatches or less in a read and quality threshold 20 in trimming 3′ end of reads (-v 5 -q 20). Only uniquely mapped reads were kept for subsequent analysis. PCR duplicates were removed using picard-tools-1.102 “MarkDuplicates,” and bamtools was used to remove any improperly paired reads. Overlapping reads were then clipped using “bam clipOverlap” command from bamUtils. Conversion rate was determined using the reads mapped to the cytosines of the unmethylated chloroplast genome. The filtered alignment files were then used to derive methylation ratios (i.e., number of methylated and unmethylated reads) for each cytosine in all three sequence contexts (CG, CHG, and CHH) using BSMAP tools. The methylation level of a specific target region was calculated based on the cytosines within the region using the weighted DNA methylation method [#C/(#C+#T)]. Read coverage per target regions was obtained by counting the number of reads overlapping with the target regions, which was determined using BEDTools. Mapping statistics can be found in Table S1.
Following mapping and preprocessing of the raw sequence data, region level methylation data were filtered and processed as follows. Of the 22,749 nonredundant v4 “specific” regions of interest, we obtained data for 21,725 regions in at least one sample. Per sample, regions with at least three reads were retained. Following 3× read coverage filtering, the mean of the controls per methylation context was calculated for any region that had data for two or more control samples. Per context, the number of regions passing the filter were 15,478 (CG), 15,554 (CHG), and 15,751 (CHH); 17,140 unique regions in total. Regions with significant variance among control samples were removed using a strict filter if control[max – min] was >30% for CG, >30% for CHG, and >10% for CHH. Per context, the numbers of regions retained after this filter were 13,518 (CG); 13,609 (CHG); and 13,344 (CHH); 15,325 unique regions in total. In total, this yielded 15,325 distinct loci for differential methylation analysis. Per-sample DMRs were identified by comparison to the mean of the control samples where there was a >40% difference in CG/CHG context, and for CHH one sample <5% and one sample >25%. The DMR thresholds were determined based on the bimodal distribution of methylation levels in the CG and CHG context (Figure S1D), and based on prior characterization of CHH islands (Li et al. 2015c). For instance, in the CG and CHG context, 91.3% and 88%, respectively, of all regions had methylation levels either >50 or <10%, so a threshold of 40% was determined to predominately identify regions that switched between low and high methylation. In maize, regions of high CHH (“CHH islands”) we considered region that have levels of around >25%. We used a criteria of >25% in one sample and <5% in another to slightly increase the stringency; however, adjusting these threshold had a minor impact on the number of CHH DMRs. In total, 3921 nonredundant regions had differential methylation in at least one context in at least one sample (Table S3). Next, the sequence context and overlap of each DMR per sample was determined and DMRs were categorized as CHH, CG-only, CG-CHG, and CHG-only. For CG- and CHG-only, a DMR is defined as context specific (“-only”) where the change in the other context is <10% compared to the controls (e.g., “CG-only hypo” occurs where CHG does not drop by >10%).
Lastly, all CG and CHG DMRs were aggregated into a nonredundant list and consensus calls across all samples were evaluated as described in Table S3. A consensus direction for each region was determined if there was a >80% consensus among samples regarding the direction of the DMR call. The consistency of each DMR was then determined by calculating the percent support for each DMR among the samples. Finally, a specificity consensus of the DMR was determined, indicating whether the DMR is specific to a single context and occurring without a significant change in the other context. Principal component analysis (PCA) was performed in R using the pcaMethods package, using data summarized to 100 bp tiles for each sample. Gene feature files (GFF) were downloaded from MaizeGDB and the Transposable Element annotation file from Jiao et al. (2017).
RNA-seq analysis
RNA-seq data for R1 plants regenerated from tissue culture were downloaded from SRA SRP040690 (Stelpflug et al. 2014). Quality control was performed with FastQC v.0.11.2. Adapters were removed using scythe v.0.991 with flags -p 0.01 for the prior, and reads were quality trimmed with sickle v.1.33 with flags -q 20 (quality threshold) -l 20 (minimum read length after trimming). The trimmed and quality filtered reads were aligned to the to maize B73 reference genome AGPv4 using the subjunc v.1.6.0 aligner to report only reads with a single unambiguous best mapping location (Liao et al. 2013). Reads were sorted, indexed using samtools v1.3 (Li et al. 2009) and the number of reads mapping per AGPv4 gene loci was summarized using featureCounts v.1.6.0 (Liao et al. 2014) with flags -P and -c to discard read pairs mapping to different chromosomes and the -s flag for strand specificity. Multi-mapping reads and multi-overlapping reads (i.e., reads mapping to overlapping regions of more than one gene locus) were not counted. Reads were summarized to parent gene loci rather than individual splice variants. Statistical testing for relative gene expression changes was performed in R using following the “edgeR-limma-voom” approach (https://www.bioconductor.org/help/workflows/RNAseq123/) using, edgeR v.3.4.2. and voom in the limma package 3.20.1.
Data availability
Supplementary material including Figures S1–S7, File S1, and Tables S1–S4 has been uploaded to Figshare. File S1 contains detailed descriptions of the conversion between B73 RefGen_v2 assembly of the maize genome and B73 RefGen_v4. The annotation of the sequence capture design of SeqCap-Epi-v2 is available under the DOI: https://doi.org/10.13020/D69X0H. Sequence files and metadata are available in the NCBI Short Read Archive SRP141150 and BioProject database PRJNA450979. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6203180.
Results
We employed a strategy that coupled bisulfite modification with sequence capture termed “SeqCap-Epi-v2” to document context-specific DNA methylation patterns of the maize genome (AGPv4 - Jiao et al. 2017). The design captured 22,749 specific target regions covering 15.7 Mb (see https://doi.org/10.13020/D69X0H and methods for detailed description of the sequence capture design). Methylation patterns were profiled in three sibling A188 plants that had not passed through tissue culture as well as one callus sample, one primary R0 regenerant, and 25 R1 plants that resulted from self-pollination of primary regenerants (Figure 1 and Table S1). The level of DNA methylation in the CG, CHG, and CHH context was determined for each region in each sample, and we focused our analysis on 15,325 regions with coverage of at least three reads and consistent methylation levels among the three control siblings (see Materials and Methods). Regions with >30% variation between control samples in the CG or CHG context, or >10% in the CHH context, were excluded. We reasoned that these regions likely represent metastable loci with DNA methylation variation unrelated to tissue culture treatment. In addition, we included profiles for five other maize inbreds (B73, Mo17, W22, Ki11, and MoG) to enable a comparison of the relative effect of tissue culture compared to genetically distinct lines (Table S1).
Figure 1.
Transgenerational tissue culture experimental design. Overview of the tissue-culture experimental design comprised of six cell-lines initiated from independent embryos originating from the same progenitor plant. Samples are represented by the squares, solid outlines indicate samples harvested for DNA methylation profiling.
Changes in DNA methylation following tissue culture often include loss of CG and CHG methylation
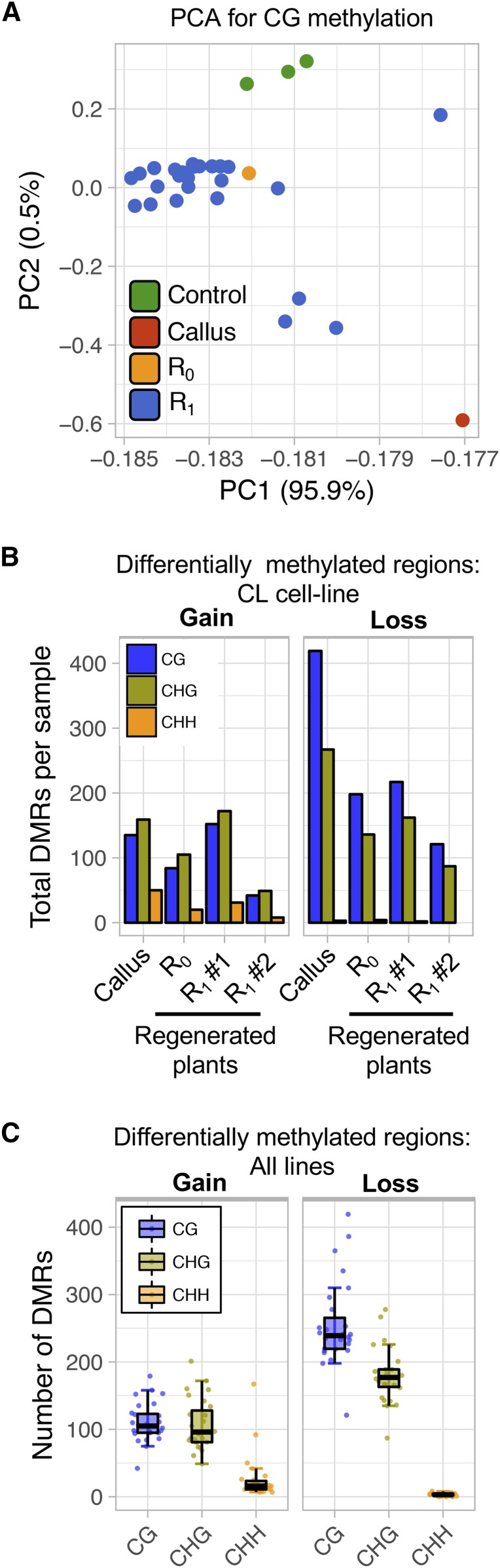
Genome-wide methylation patterns were assessed in A188 plants before, during, and after tissue culture, and were subsequently contrasted with the five maize inbreds from diverse genetic backgrounds. PCA of CG methylation revealed clustering driven largely by genotype (Figure S1, A and B). The tissue culture samples were generally clustered closely with A188 plants that had not experienced tissue culture (Figure S1, A and B) when analyzed together with other genotypes. When PCA was performed only on the tissue culture experiment (A188 background plants), plants that have been passed through tissue culture were differentiated from those that have not (Figure 2A). DMRs were identified (see Materials and Methods for details) for analysis of context-specific DNA methylation levels for each region in each sample relative to the controls (Figure 2, B and C and Figure S1C). The DMR criteria were selected based on the genomic distribution of methylation levels (Figure S1D) to allow for discovery of regions that shift between methylated and unmethylated states. Within the CL cell-line pedigree, we were able to compare a callus sample (“CL”), the R0 plant (“CL3”) and two sibling R1 plants (“CL3-1” and “CL3-2,” Figure 1). There are relatively similar numbers of DMRs per sample throughout this lineage (Figure 2B). The callus sample has a higher number of CHH gains than subsequent materials and has an elevated number of losses in the CG and CHG context. However, there are no consistent differences in the numbers of DMRs between the R0 and R1 plants. Among all regenerated plants that were monitored, in total 3921 distinct regions in the genome were identified as DMRs in one or more plants (Figure 2B). There are many more CG and CHG DMRs than CHH DMRs even though less strict criteria are used for CHH DMR discovery (Figure 2, B and C and Figure S1C). CHH DMRs are more likely to represent gains of methylation if a sample that has experienced tissue culture relative to the controls, while CG and CHG DMRs are more likely to exhibit losses of methylation in the regenerated plants (Figure 2, B and C). In many cases, the change in DNA methylation level is consistent with effects on methylation for both alleles such that the homozygous A188 incurred changes in DNA methylation at both alleles following tissue culture.
Figure 2.
Documenting changes in DNA methylation following tissue culture. (A) PCA of CG methylation levels for A188 samples from the tissue culture experiment (noncultured seedling, callus, R0, and R1 regenerated seedlings). (B) The number of DMRs per context for each sample in the “CL” cell-line lineage, including the callus tissue, an R0 regenerated seedling, and two R1 regenerated seedlings. Bars indicate the total number of DMRs for both gains in methylation compared to the controls (hypermethylated) and loss compared to the controls (hypomethylated). (C) The average number of DMRs per context identified for all samples passed through tissue culture (n = 25). The error bars report the SD and each dot represents an individual sample.
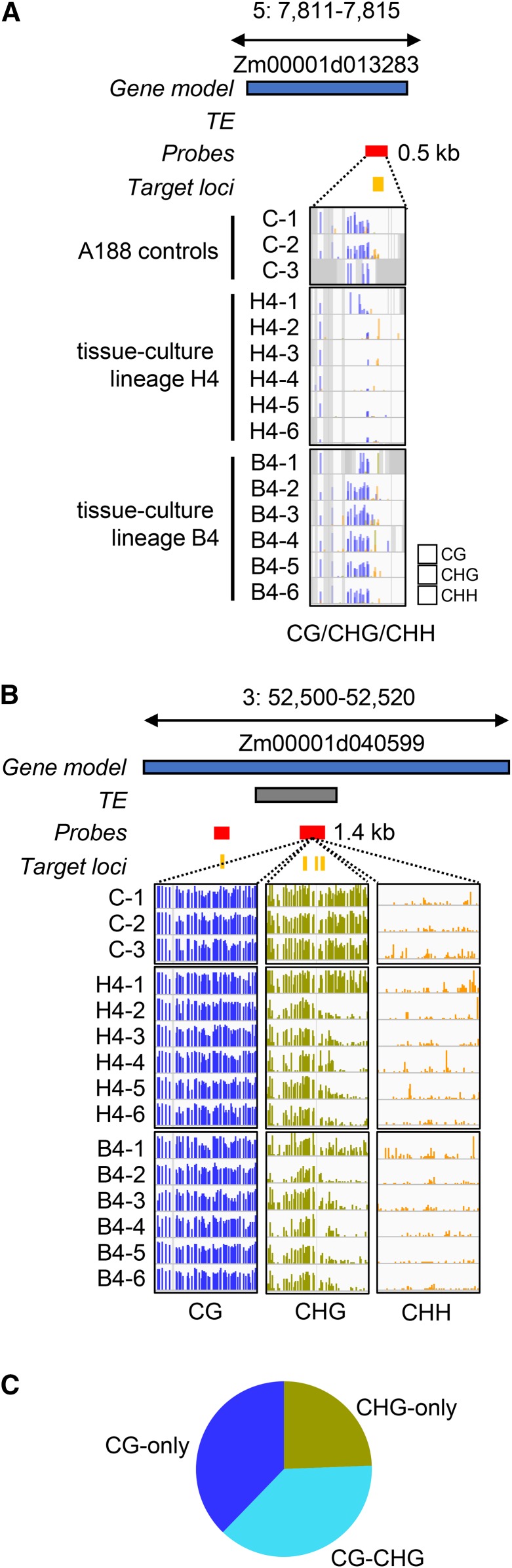
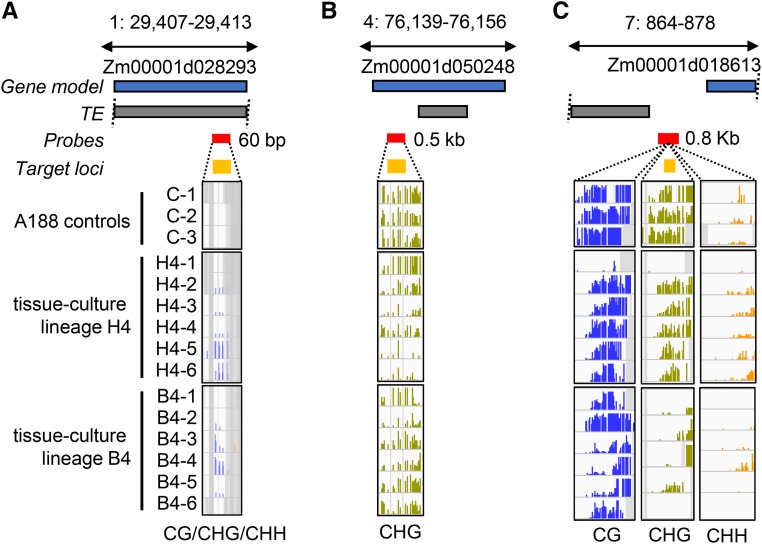
A visual inspection of several of the DMRs revealed evidence for context-specific changes in DNA methylation (Figure 3, A and B). Examples for a CG-only and a CHG-only DMR are shown in Figure 3, A and B. The CG-only DMR in Figure 3A occurs within a gene body and represents a loss of CG methylation. This loss is seen in five of the six H4-family siblings, but one individual has partial CG methylation, potentially suggesting that it has regained methylation. None of the individuals in the B4 lineage exhibit changes in CG methylation at this locus. The CHG-only DMR in Figure 3B occurs in a region that has high levels of CG and CHG methylation in the control plants. CHG methylation is substantially reduced, especially at the 3′ region, in all the B4 individuals and in five of the six H4 individuals. A detailed analysis of the relative changes in different contexts for all DMRs suggests limited overlap of changes in CHH methylation with CG or CHG changes (Figure S2). Changes in CG and CHG contexts often were found in the same region but there were examples of CG-only or CHG-only changes as well (Figure S2). Each of the CG and/or CHG DMRs were classified as CG/CHG, CG-only or CHG-only for further analyses (Figure 3C).
Figure 3.
Identification of context-specific DMRs resulting from tissue culture. (A) Example of a gene body CG-only DMR in the B4 and H4 tissue-culture lineages. The blue and gray boxes represent the relative positions of gene and TE models (v4 genome annotation); the red and yellow boxes indicate the SeqCap-Epi_v2 capture probes locations and target loci, respectively. For each sample track, bar height represents % methylation (0–100%), purple = CG, green = CHG, and yellow = CHH. Gray shading indicates noncapture regions. (B) Example of a karma-like CHG-only hypomethylation at a TE nested in gene Zm00001d040599. (C) The CG and CHG DMRs were classified based on whether they exhibit changes in both contents (CG-CHG), or had CG-only or CHG-only changes in DNA methylation. The proportion of all DMRs (n = 7844) that are classified as CG-only, CHG-only, or CG-CHG DMRs is shown.
The maize B73 genome can be divided into four methylation domains, characterized by distinct methylation profiles (noting that more sophisticated definitions have also been proposed (Gouil and Baulcombe 2016)). A small fraction (3.9%) is unmethylated in all contexts; 0.7% has high CHH and represents RdDM targets, 7.1% has high CG-only and is found in gene bodies, and the largest fraction (47.9%) has high levels of both CG and CHG and represents heterochromatin (Springer and Schmitz 2017). The remaining 40.3% of the maize genome is unclassified due to either a lack of coverage or intermediate methylation levels. We assessed how the changes in methylation induced by tissue culture affected the shifts among these types of domains. For instance, the CG-CHG DMRs largely represented a conversion of heterochromatic region in control plants to unmethylated domains in regenerated plants. The CG-only DMRs had low levels of CHG methylation in both the control and tissue culture samples (Figure S3A). These CG-only DMRs therefore reflect shifts between the unmethylated and CG-only methylation domains. In contrast, CHG-only DMRs frequently occurred where CG methylation was high (Figure S3B). These CHG-only DMRs represent interconversion between CG-CHG heterochromatin and “gene body-like” CG-only methylation.
A subset of methylation changes induced by tissue culture are common to many independent samples
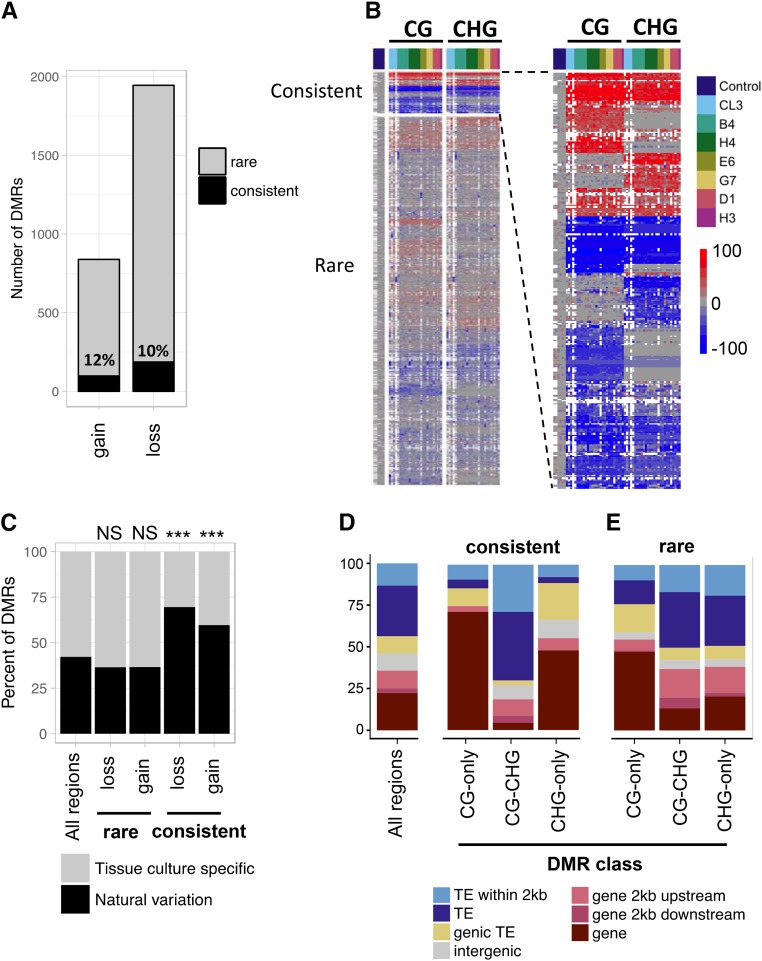
Next, we examined the consistency of changes in methylation among the 25 R1 plants (Figure 4). The overlap of DMRs among multiple individuals far exceeded the frequency predicted by simulations (Figure S4A). For instance, given the total number of regions that lost CG methylation (CG-hypo DMRs) per sample, we would expect by random chance to find less than one DMR to recur in six or more samples (P < < 0.001), yet we observe 303 DMRs common to six or more samples.
Figure 4.
A subset of methylation changes induced by tissue culture are common to many independent samples. (A) The relative numbers of rare and consistent DMRs is shown for CG and CHG gains or losses of DNA methylation. Loci were considered consistent if >50% of samples exhibited the DMR. The number indicates the proportion of DMRs classified as consistent. (B) Hierarchical clustering of the change in methylation levels (sample - control) for all samples at each DMR in the CG and CHG context. White shading denotes missing data. Each column represents a different sample with the controls on the left, and the color coding at the top indicating the sample lineages for the remaining samples. Each row indicates a distinct DMR and there is separate clustering for the consistent (top) and rare (bottom) DMRs. The section showing the consistent DMRs is enlarged on the right to better visualize these regions. (C) The percentage of tissue culture DMRs overlapping natural variation DMRs (determined using a panel of 19 inbreds) is shown. At the top for each set of DMRs the significance is indicated (* P < 0.001; NS, not significant, hypergeometric test). (D and E) The genomic feature(s) overlapping each DMR—categorized as (D) consistent or (E) rare—were determined and the percentage of each feature is shown. The distribution for all regions sampled is shown to the left.
DMRs that were identified in <50% of samples were classified as “rare” while DMRs identified in >50% of samples were classified as “consistent” (Figure S4B, Table 1, and Table S3). The vast majority (99%) of CHH DMRs were classified as rare. A larger subset of CG and CHG DMRs display strong consistency between samples. Consistent DMRs accounted for ∼10% of both the losses (hypo) and gains (hyper) in methylation (Figure 4A). The patterns for rare and consistent DMRs can be visualized through hierarchical clustering of methylation levels for all samples (Figure 4B). In each individual sample approximately half the DMRs identified are consistent and shared with other samples, while half are rare (Figure S4C). An example of the DNA methylation patterns exhibited in a common DMR is shown in Figure S4D. Note that, in this case, the A188 plants have essentially no methylation while the levels of DNA methylation in plants regenerated from tissue culture is quite high, suggesting gain of DNA methylation for both alleles.
Table 1. Characterization of CG, CHG, and CHH DMRs.
| Specificity consensus | Direction consensus | Consistency | Number of regions | Natural variants (%) |
|---|---|---|---|---|
| CG-only | Gain | Consistent | 21 | 76.1 |
| Rare | 152 | 59.6 | ||
| Loss | Consistent | 36 | 85.7 | |
| Rare | 380 | 52.3 | ||
| CG-CHG | Gain | Consistent | 22 | 54.5 |
| Rare | 37 | 51.4 | ||
| Loss | Consistent | 49 | 79.1 | |
| Rare | 165 | 34.5 | ||
| CHG-only | Gain | Consistent | 12 | 100 |
| Rare | 114 | 40.7 | ||
| Loss | Consistent | 16 | 81.3 | |
| Rare | 236 | 40.3 | ||
| unclassified | Gain | Consistent | 43 | 48.3 |
| Rare | 438 | 30.0 | ||
| Loss | Consistent | 86 | 59.3 | |
| Rare | 977 | 31.9 | ||
| CHH | Gain | Consistent | 4 | 1.1 |
| Rare | 372 | 98.9 | ||
| Loss | Consistent | 1 | 2.1 | |
| Rare | 47 | 97.9 |
All CG, CHG and CHH DMRs were categorized based on the methylation context consensus and consistency between samples. CHH DMRs were analyzed separately from CG and CHG. For CG and CHG specificity and consistency a consensus of ≥80% of samples was required or else regions were denoted “unclassified.” If there was ≥50% support for a DMR between samples it was classified as “consistent,” all others were considered “rare.”
Features of consistent DMRs
The features of CG and CHG DMRs were further assessed to better understand why some regions exhibit consistent changes in methylation given that many other DMRs behave more stochastically. The tissue culture induced DMRs were compared to a set of naturally occurring DMRs found among a wider panel of 19 inbreds subject to whole genome bisulfite sequencing (WGBS) (Anderson et al. 2018). DMRs identified by WGBS were filtered to only include those within the sequence capture space of the SeqCap-Epi-v2. Consistent DMRs exhibited a significant overlap with loci that exhibit natural variation (Figure 4C P < 0.001, hypergeometric test). In contrast, the rare DMRs do not exhibit significant enrichment or depletion for overlap with naturally occurring DMRs (P > 0.5, hypergeometric test).
Next, we examined the annotation for the genomic locations of consistent CG and CHG DMRs. Consistent DMRs displayed significant bias in their location in the genome (Figure 4D). Consistent CG-only and CHG-only DMRs were both enriched in genes and depleted in TEs [false discovery rate (FDR) < 0.05, hypergeometric test], while consistent CG-CHG DMRs were depleted at genes (FDR < 0.05) and enriched at TEs (although not statistically significant after FDR correction). Rare CG-only DMRs had a similar profile to consistent CG-only DMRs. By contrast, rare CG-CHG/CHG-only DMRs had a profile more consistent with the background frequency, with some enrichment in promoters (FDR < 0.05) and close to TEs (FDR < 0.05; Figure 4E).
The enrichment of CG-only DMRs in genes was consistent with our expectation that these DMRs have the characteristics of gene-body methylation. It was somewhat unexpected to also find CHG-only DMRs enriched in genes given that these DMRs occur in a high CG and CHG methylation context characteristic of heterochromatin (Figure 3B). In total, 64% (18/28) of the consistent CHG-only DMRs occur at locations of genic heterochromatin. We also observed that many of these genes have nested TEs (for example Figure 3B and Figure 6B) reminiscent of the Karma locus in oil palm (Ong-Abdullah et al. 2015). This prompted us to examine the prevalence of potential Karma-like epialleles in more detail. Indeed, the example shown in Figure 3E of a CHG-only DMR occurs within a TE that is located within a gene. Across the maize genome, >10% of maize genes contain at least one TE insertion over 1 kb within an intron (West et al. 2014; Hirsch et al. 2016). In total, our sequence capture design profiled methylation information at 1447 genic loci that have high CG and CHG methylation in the control A188 plants indicative of heterochromatin (control average >50% CG and >50% CHG). We detected significantly reduced levels of CHG methylation at 200 of these loci (219 DMRs, Table S4). These genes were found to have diverse functions, and gene ontology (GO) analysis revealed that several genes were linked to nucleotide metabolic processes (e.g., GO:0009165 and GO:0055086); although, following FDR correction, no GO categories were statistically enriched relative to the background set of genes profiled. The heterochromatic DMRs were mostly rare (187/219); nevertheless, 37 (20%) were consistently observed in multiple independent plants (Table S5).
Figure 6.
Evidence for segregation of heterozygous changes during culture or stochastic reacquisition of DNA methylation. (A–C) Examples of potentially segregating CG-hyper, CHG-hypo and CG-CHG-hypo DMRs. The blue and gray boxes represent the relative positions of gene and TE models (v4 genome annotation); the red and yellow boxes indicate the SeqCap-Epi_v2 capture probes locations and target loci respectively. For each sample track, bar height represents % methylation (0–100%), purple = CG, green = CHG and yellow = CHH. Gray shading indicates noncapture regions (missing data).
We examined the expression of genes that overlapped DMRs or were within 2 kb upstream or downstream of a DMR using existing RNA-seq data from seedling leaf tissue of regenerated and control plants (Stelpflug et al. 2014). The RNA-seq data included the CL3_1 and CL3_2 samples that were profiled in this study, and an additional four R1 regenerated plants not part of the present study. The expression of genes near or overlapping consistent DMRs was not significantly affected (Figure S5A). At consistent CG-CHG DMRs, a trend (though not statistically significant) toward downregulation at loci that had reduced methylation was observed. A more focused analysis was performed on 25 expressed genes that contained regions of CHG methylation within the gene that exhibited consistent losses following tissue culture (Figure S5B). At seven of these genes, we found evidence for replicable changes in expression (two or more samples, more than twofold change; Figure S5B). These included four examples of increases in expression following tissue culture, two genes with decreases in expression in multiple R1 individuals and one gene that had multiple R1 individuals with either increases or decreases of expression.
Heritability and evidence for targeted remodeling of the methylome
Previous investigations identified variability in the inheritance patterns of DMRs, some of which exhibited stochastic behavior or incomplete penetrance (Stroud et al. 2013; Stelpflug et al. 2014). This could result from heterozygous changes induced during tissue culture, which would segregate in later generations or could result from reacquisition of the original epigenetic state at one or both alleles following tissue culture. We examined the variation within the CL3 cell lineage and among six sibling R1 plants in each of the B4 and H4 lineages to assess conservation or variability of DMRs within related plants and to assess inheritance of DNA methylation levels in R1 plants.
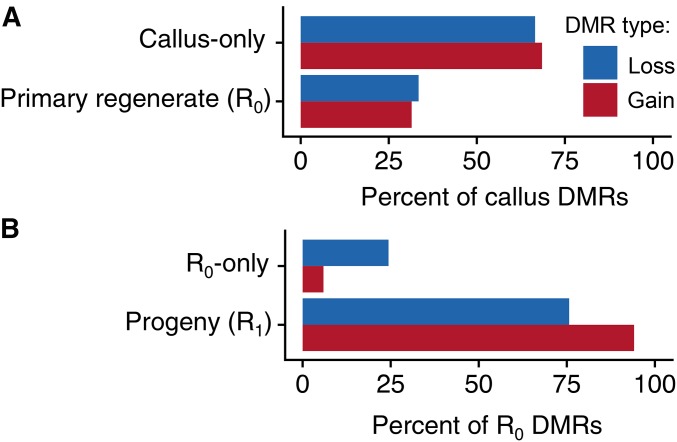
The DMRs identified in the CL callus sample, its R0 (CL3) and R1 plants (CL3-1 and CL3-2) were compared in order to document the origin or loss of DMRs throughout this lineage (Figure 5, A and B and Figure S6, A–F). We determined whether the DMRs that were present in the callus or R0 plant were present in subsequent materials (Figure 5). Only a subset (∼30%) of the DNA methylation gains or losses detected in callus tissue were found in the R0 primary regenerant (Figure 5A). Based on this single lineage, this suggests that some of the DNA methylation changes that are triggered by tissue culture are not passed on to the regenerated plantlets. In contrast, the majority of DNA methylation changes found in the R0 were also detected in the R1 progeny (Figure 5B). Over 90% of the DNA methylation gains that are detected in the R0 and ∼75% of the DNA methylation losses detected in the R0 are also observed in the R1 plants.
Figure 5.
Evidence for inheritance of tissue culture associated methylation changes in the CG and CHG context. (A) The inheritance patterns of callus DMRs. Each CG and CHG callus DMR was defined as either callus-only (not a DMR in the R0) or R0 inherited; then expressed as percent of the total number of callus DMRs. This analysis only included callus CG and CHG DMRs with sufficient sequence data for determination of methylation state in all samples (n = 287). (B) The transmission of R0 DMRs to the R1. Callus DMRs that we present in the R0 were then examined in the R1 (n = 95) and expressed a percent of the total number of R0 DMRs.
Although we did not profile the R0 plant that was the progenitor of the H4 or B4 family we could look at the correspondence of DMRs among R1 sibling individuals within these families (Figure S7). A substantial number of the DMRs within each family are found in at least five of the six individuals (Figure S7, A–D) and show consistent levels of change (Figure S7, E–H). The fact that major changes in the level of DNA methylation are detected (>50%), and that for many DMRs, the majority of siblings exhibit these changes suggests homozygous epiallelic change during culture. However, we were interested in assessing whether there was also evidence for either segregation of heterozygous changes during culture or stochastic reacquisition of DNA methylation. Examples of potentially segregating CG-hyper, CHG-hypo and CG-CHG-hypo DMRs are shown in Figure 6, A–C. In Figure 6A, for both lineages, there are examples of plants with both substantial and partial gain in methylation. In Figure 6B, only the H4 lineage exhibits varying levels of CHG hypomethylation. In Figure 6C, there are varying levels of CG and CHG hypomethylation in the B4 lineage; however, here methylation is only partially lost from the left side of the region, and could either represent segregation or stochastic reacquisition of DNA methylation seeded by the flanking methylation (noting that this is the most parsimonious explanation without knowing for certain the methylation levels in the particular R0).
Discussion
DNA methylation can provide additional heritable information beyond DNA sequence in plant genomes. Many studies have found that a substantial proportion of the methylome is highly heritable and not sensitive to environmental perturbation (Song et al. 2013; Eichten and Springer 2015; Hagmann et al. 2015; Secco et al. 2015). However, locus-specific changes in DNA methylation have been observed in response to environmental conditions in several studies (Jiang et al. 2014; Wibowo et al. 2016). In particular, several studies have found changes in DNA methylation following passage through tissue culture and regeneration (Kaeppler and Phillips 1993; Stroud et al. 2013; Stelpflug et al. 2014; Ong-Abdullah et al. 2015). In this study, we have assessed the context-specific DNA methylation patterns for a portion of the maize genome. In comparison to the significant difference in DNA methylation patterns that can be observed between inbred lines, the plants that had been passed through tissue culture still are quite similar to the control plants of the same genotype. Nevertheless, a PCA reveals evidence for a set of changes that distinguish plants passed through tissue culture.
In this study, we have focused on changes in DNA methylation that are observed in the progeny of plants regenerated from tissue culture (R1). By analyzing the R1 we have focused on the heritable changes rather than transient effects that may be restricted to culture/callus or the R0. For the cell line for which we had callus-R0-R1 data, we found that a lower proportion of callus DMRs were observed in the R0; whereas a very high proportion of the R0 DMRs were present in R1 plants. Unfortunately, due to sample losses we did not have access to profiles of the callus and R0 for the other lineages. A limitation of our approach is that we were not able to profile each callus and R0 for the other R1 plants included in this study. Thus, we have limited sampling of the full spectrum of DMRs that arise during tissue culture that cannot be maintained through sexual reproduction; this likely includes many asymmetric CHH changes. We also cannot rule out the appearance of rare spontaneous epimutations that may occur at unstable loci between the R0 and the R1. However, our sampling did enable the analysis of a large population of R1 plants to characterize the consistent and heritable effects of tissue culture, which may be of most interest to efforts to transform seed-propagated plants such as maize or other plants.
Tissue culture could impact DNA methylation through a variety of mechanisms. It is possible that tissue culture could simply reduce the efficacy of the DNA methylation, or demethylation, machinery, resulting in widespread loss or gain of DNA methylation. There are more hypomethylation events than hypermethylation events, but we do find evidence for both gains and losses of DNA methylation. Additionally, we do not see major changes in overall DNA methylation levels in plants recovered from tissue culture. This suggests that there is not a global failure of DNA methylation or demethylation during tissue culture. Another possibility is that tissue culture simply leads to higher epimutation rates. This would be expected to produce a set of random changes in each event, and would also be expected to result in heterozygous changes to DNA methylation. However, we see evidence for significant overlap of DNA methylation changes, even in independent lineages, and it seems that the many loci with changes in DNA methylation are affected at both alleles. These observations argue for a targeted process (as opposed to random) or for specific loci being particularly sensitive to the effects of tissue culture. In this respect, very unstable loci where maintenance of methylation is easily disrupted may be readily targeted in tissue culture. It may be difficult to restore methylation at these loci as has been observed for particular regions in Arabidopsis that do not recover following epimutagenesis (Ji et al. 2018).
The basis for this potential targeted mechanism remains unclear. We find both homozygous gains, and losses, of DNA methylation to be more consistent among independent events than expected by chance. In addition, we find that these consistent gains and losses of DNA methylation can affect CHG, CG or both contexts. These observations likely rule out simple hypotheses such as the targeting, or failure to target, specific methyltransferases to certain regions. In addition, we do not find particular features of the target loci that are able to explain why these regions have altered DNA methylation. It is possible that changes in expression at these loci at early stages of tissue culture could alter chromatin and impact the DNA methylation state. Further work will be needed to assess the normal chromatin at these loci and the potential changes during initiation and maintenance of callus.
A subset of the changes in DNA methylation are consistently found in many independent events and appear to represent homozygous changes in DNA methylation that occur during tissue culture. The majority of these consistent changes in methylation are also observed as DMRs among diverse inbred lines that have not been subjected to tissue culture (Anderson et al. 2018). The fact that we see differing epigenetic states at these loci in A188 before and after tissue culture suggests that the DNA methylation state at these loci is not determined solely by the underlying genetic sequence, and that these are genuine epialleles (Crisp et al. 2016). In many cases, natural variation DMRs can be associated with cis sequence variation (Eichten et al. 2013b; Schmitz et al. 2013), indicative of a mechanism tied to underlying DNA sequence variation. However, these epialleles that are found following tissue culture, and, in natural populations, may represent a set of pure epialleles (Richards 2006) that exhibit partially unstable behaviors.
The CG-only DMRs often occurred within gene bodies and could reflect the somewhat unstable nature of gene body methylation (Schmitz et al. 2013). There is little evidence that changes in gene body CG methylation influence expression levels (Bewick et al. 2016; Bewick and Schmitz 2017; Picard and Gehring 2017). In contrast, CHG methylation levels are usually quite stable. The Karma locus in oil palm is an example in which a loss of CHG methylation associated with tissue culture results in phenotypic changes (Ong-Abdullah et al. 2015). In this case, specific loss of CHG methylation, with no change in CG methylation, at an intronic TE results in altered transcripts for the DEFICIENS gene and the defective mantled phenotype. Interestingly, we found that many of the CHG-only DMRs identified in this study shared characteristics with the Karma locus. These CHG-only DMRs often occurred at TEs located within genes. Notably, these included genes involved in developmental processes that could possibly impact somatic embryogenesis including, histone H2A (Zm00001d012837), meg4 (Zm00001d019541), and Hox1a (Zm00001d010758). Analysis of RNA-seq data also showed that the expression of both histone H2A and meg4 was altered in many regenerated plants (Figure S5B). This suggests that the ability to maintain heterochromatin within a largely euchromatin environment may be compromised in tissue culture. There is evidence that changes in CHG methylation without loss of CG methylation can result in changes in gene expression in maize (Anderson et al. 2018). Previous studies have suggested that the correct expression and repression of a suite of genes is required for somatic embryogenesis (Salvo et al. 2014). It will be interesting to determine whether the inability to maintain CHG methylation within or near genes is a common cause of somaclonal variation in plant species.
Acknowledgments
We thank Peter Hermanson for sequence-capture library preparation, and technical support; Yaniv Brandvain for statistical advice; and Jackie Noshay, Sarah Anderson, and Peng Zhou for legendary computational assistance. This work was funded by a grant from the National Science Foundation (IOS-1237931) to N.M.S. Illumina sequencing was performed at the University of Minnesota Genomics Center. Computational support was provided by the Minnesota Supercomputing Institute and the Texas Advanced Computing Center.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6203180.
Communicating editor: A. Houben
Literature Cited
- Anderson S. N., Zynda G., Song J., Han Z., Vaughn M., et al. , 2018. Subtle perturbations of the maize methylome reveal genes and transposons silenced by chromomethylase or RNA-directed DNA methylation pathways. G3 (Bethesda) 8: 1921–1932. 10.1534/g3.118.200284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick A. J., Schmitz R. J., 2017. Gene body DNA methylation in plants. Curr. Opin. Plant Biol. 36: 103–110. 10.1016/j.pbi.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewick A. J., Ji L., Niederhuth C. E., Willing E. M., Hofmeister B. T., et al. , 2016. On the origin and evolutionary consequences of gene body DNA methylation. Proc. Natl. Acad. Sci. USA 113: 9111–9116. 10.1073/pnas.1604666113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp P. A., Ganguly D., Eichten S. R., Borevitz J. O., Pogson B. J., 2016. Reconsidering plant memory: intersections between stress recovery, RNA turnover, and epigenetics. Sci. Adv. 2: e1501340 10.1126/sciadv.1501340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp P. A., Ganguly D. R., Smith A. B., Murray K. D., Estavillo G. M., et al. , 2017. Rapid recovery gene downregulation during excess-light stress and recovery in Arabidopsis. Plant Cell 29: 1836–1863. 10.1105/tpc.16.00828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen R. H., Pelizzola M., Schmitz R. J., Lister R., Dowen J. M., et al. , 2012. Widespread dynamic DNA methylation in response to biotic stress. Proc. Natl. Acad. Sci. USA 109: E2183–E2191. 10.1073/pnas.1209329109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S. R., Springer N. M., 2015. Minimal evidence for consistent changes in maize DNA methylation patterns following environmental stress. Front. Plant Sci. 6: 308 10.3389/fpls.2015.00308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichten S. R., Vaughn M. W., Hermanson P. J., Springer N. M., 2013a Variation in DNA methylation patterns is more common among maize inbreds than among tissues. Plant Genome 6 10.3835/plantgenome2012.06.0009. [Google Scholar]
- Eichten S. R., Briskine R., Song J., Li Q., Swanson-Wagner R., et al. , 2013b Epigenetic and genetic influences on DNA methylation variation in maize populations. Plant Cell 25: 2783–2797. 10.1105/tpc.113.114793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly D. R., Crisp P. A., Eichten S. R., Pogson B. J., 2017. The Arabidopsis DNA methylome is stable under transgenerational drought stress. Plant Physiol. 175: 1893–1912. 10.1104/pp.17.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gent J. I., Ellis N. A., Guo L., Harkess A. E., Yao Y., et al. , 2013. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23: 628–637. 10.1101/gr.146985.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouil Q., Baulcombe D. C., 2016. DNA methylation signatures of the plant chromomethyltransferases. PLoS Genet. 12: e1006526 10.1371/journal.pgen.1006526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann J., Becker C., Muller J., Stegle O., Meyer R. C., et al. , 2015. Century-scale methylome stability in a recently diverged Arabidopsis thaliana lineage. PLoS Genet. 11: e1004920 10.1371/journal.pgen.1004920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirochika H., 1993. Activation of tobacco retrotransposons during tissue culture. EMBO J. 12: 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch C. N., Hirsch C. D., Brohammer A. B., Bowman M. J., Soifer I., et al. , 2016. Draft assembly of elite inbred line PH207 provides insights into genomic and transcriptome diversity in maize. Plant Cell 28: 2700–2714. 10.1105/tpc.16.00353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., Jordan W. T., Shi X., Hu L., He C., et al. , 2018. TET-mediated epimutagenesis of the Arabidopsis thaliana methylome. Nat. Commun. 9: 895 10.1038/s41467-018-03289-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Mithani A., Belfield E. J., Mott R., Hurst L. D., et al. , 2014. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 24: 1821–1829. 10.1101/gr.177659.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Peluso P., Shi J., Liang T., Stitzer M. C., et al. , 2017. Improved maize reference genome with single-molecule technologies. Nature 546: 524–527. 10.1038/nature22971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppler S. M., Phillips R. L., 1993. Tissue culture-induced DNA methylation variation in maize. Proc. Natl. Acad. Sci. USA 90: 8773–8776. 10.1073/pnas.90.19.8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeppler S. M., Kaeppler H. F., Rhee Y., 2000. Epigenetic aspects of somaclonal variation in plants. Plant Mol. Biol. 43: 179–188. 10.1023/A:1006423110134 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T., Stuart T., Valdes M., Breakfield N., Schmitz R. J., et al. , 2016. Unique cell-type-specific patterns of DNA methylation in the root meristem. Nat. Plants 2: 16058 10.1038/nplants.2016.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizova K., Fojtova M., Depicker A., Kovarik A., 2009. Cell culture-induced gradual and frequent epigenetic reprogramming of invertedly repeated tobacco transgene epialleles. Plant Physiol. 149: 1493–1504. 10.1104/pp.108.133165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. A., Jacobsen S. E., 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Suzuki M., Wendt J., Patterson N., Eichten S. R., et al. , 2015a Post-conversion targeted capture of modified cytosines in mammalian and plant genomes. Nucleic Acids Res. 43: e81 10.1093/nar/gkv244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Song J., West P. T., Zynda G., Eichten S. R., et al. , 2015b Examining the causes and consequences of context-specific differential DNA methylation in maize. Plant Physiol. 168: 1262–1274. 10.1104/pp.15.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Gent J. I., Zynda G., Song J., Makarevitch I., et al. , 2015c RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. USA 112: 14728–14733. 10.1073/pnas.1514680112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W., 2013. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41: e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K., Shi W., 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Matzke M. A., Mosher R. A., 2014. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15: 394–408 (erratum: Nat. Rev. Genet. 15: 570) 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- Miguel C., Marum L., 2011. An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond. J. Exp. Bot. 62: 3713–3725. 10.1093/jxb/err155 [DOI] [PubMed] [Google Scholar]
- Neelakandan A. K., Wang K., 2012. Recent progress in the understanding of tissue culture-induced genome level changes in plants and potential applications. Plant Cell Rep. 31: 597–620. 10.1007/s00299-011-1202-z [DOI] [PubMed] [Google Scholar]
- Ong-Abdullah M., Ordway J. M., Jiang N., Ooi S. E., Kok S. Y., et al. , 2015. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525: 533–537. 10.1038/nature15365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa C. M., Springer N. M., Muszynski M. G., Meeley R., Kaeppler S. M., 2001. Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell 13: 1919–1928. 10.1105/tpc.13.8.1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschke V. M., Phillips R. L., Gengenbach B. G., 1987. Discovery of transposable element activity among progeny of tissue culture–derived maize plants. Science 238: 804–807. 10.1126/science.238.4828.804 [DOI] [PubMed] [Google Scholar]
- Phillips R. L., Kaeppler S. M., Olhoft P., 1994. Genetic instability of plant tissue cultures: breakdown of normal controls. Proc. Natl. Acad. Sci. USA 91: 5222–5226. 10.1073/pnas.91.12.5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard C. L., Gehring M., 2017. Proximal methylation features associated with nonrandom changes in gene body methylation. Genome Biol. 18: 73 10.1186/s13059-017-1206-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski M., Lu Z., Kendall J., Donoghue M. T., Reinders J., et al. , 2013. The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23: 1651–1662. 10.1101/gr.153510.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee Y., Sekhon R. S., Chopra S., Kaeppler S., 2010. Tissue culture-induced novel epialleles of a Myb transcription factor encoded by pericarp color1 in maize. Genetics 186: 843–855. 10.1534/genetics.110.117929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E. J., 2006. Inherited epigenetic variation — revisiting soft inheritance. Nat. Rev. Genet. 7: 395–401. 10.1038/nrg1834 [DOI] [PubMed] [Google Scholar]
- Salvo S. A. G. D., Hirsch C. N., Buell C. R., Kaeppler S. M., Kaeppler H. F., 2014. Whole transcriptome profiling of maize during early somatic embryogenesis reveals altered expression of stress factors and embryogenesis-related genes. PLoS One 9: e111407 10.1371/journal.pone.0111407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R. J., Schultz M. D., Urich M. A., Nery J. R., Pelizzola M., et al. , 2013. Patterns of population epigenomic diversity. Nature 495: 193–198. 10.1038/nature11968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D., Wang C., Shou H., Schultz M. D., Chiarenza S., et al. , 2015. Stress induced gene expression drives transient DNA methylation changes at adjacent repetitive elements. eLife 4: 09343 10.7554/eLife.09343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q. X., Lu X., Li Q. T., Chen H., Hu X. Y., et al. , 2013. Genome-wide analysis of DNA methylation in soybean. Mol. Plant 6: 1961–1974. 10.1093/mp/sst123 [DOI] [PubMed] [Google Scholar]
- Springer N. M., Schmitz R. J., 2017. Exploiting induced and natural epigenetic variation for crop improvement. Nat. Rev. Genet. 18: 563–575. 10.1038/nrg.2017.45 [DOI] [PubMed] [Google Scholar]
- Stelpflug S. C., Eichten S. R., Hermanson P. J., Springer N. M., Kaeppler S. M., 2014. Consistent and heritable alterations of DNA methylation are induced by tissue culture in maize. Genetics 198: 209–218. 10.1534/genetics.114.165480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud H., Ding B., Simon S. A., Feng S., Bellizzi M., et al. , 2013. Plants regenerated from tissue culture contain stable epigenome changes in rice. eLife 2: e00354 10.7554/eLife.00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanurdzic M., Vaughn M. W., Jiang H., Lee T. J., Slotkin R. K., et al. , 2008. Epigenomic consequences of immortalized plant cell suspension culture. PLoS Biol. 6: 2880–2895. 10.1371/journal.pbio.0060302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taudt A., Colome-Tatche M., Johannes F., 2016. Genetic sources of population epigenomic variation. Nat. Rev. Genet. 17: 319–332. 10.1038/nrg.2016.45 [DOI] [PubMed] [Google Scholar]
- West P. T., Li Q., Ji L., Eichten S. R., Song J., et al. , 2014. Genomic distribution of H3K9me2 and DNA methylation in a maize genome. PLoS One 9: e105267 10.1371/journal.pone.0105267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibowo A., Becker C., Marconi G., Durr J., Price J., et al. , 2016. Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity. eLife 5: e13546 10.7554/eLife.13546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Li W., 2009. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10: 232 10.1186/1471-2105-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Zhang H., Arikit S., Huang K., Nan G.-L., et al. , 2015. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA 112: 3146–3151. 10.1073/pnas.1418918112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhu J. K., 2012. Active DNA demethylation in plants and animals. Cold Spring Harb. Symp. Quant. Biol. 77: 161–173. 10.1101/sqb.2012.77.014936 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplementary material including Figures S1–S7, File S1, and Tables S1–S4 has been uploaded to Figshare. File S1 contains detailed descriptions of the conversion between B73 RefGen_v2 assembly of the maize genome and B73 RefGen_v4. The annotation of the sequence capture design of SeqCap-Epi-v2 is available under the DOI: https://doi.org/10.13020/D69X0H. Sequence files and metadata are available in the NCBI Short Read Archive SRP141150 and BioProject database PRJNA450979. Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6203180.