Abstract
Analysing transcriptomes of cell populations is a standard molecular biology approach to understand how cells function. Recent methodological development has allowed performing similar experiments on single cells. This has opened up the possibility to examine samples with limited cell number, such as cells of the early embryo, and to obtain an understanding of heterogeneity within populations such as blood cell types or neurons. There are two major approaches for single-cell transcriptome analysis: quantitative reverse transcription PCR (RT-qPCR) on a limited number of genes of interest, or more global approaches targeting entire transcriptomes using RNA sequencing. RT-qPCR is sensitive, fast and arguably more straightforward, while whole-transcriptome approaches offer an unbiased perspective on a cell’s expression status.
Keywords: single-cell, heterogeneity, transcriptomics, RNA-seq, RNA sequencing, microarrays, RT-qPCR
Why is single-cell transcriptomics useful?
Transcriptomics, defined as high-throughput quantitative study of the total complement of cellular RNA (or more narrowly mRNA) molecules, is a powerful and widely used approach for describing states of cellular activity. This includes dynamic changes in cell state during development and differentiation, and responses to environmental or experimental perturbations. While the quantity of mRNA is not the only determinant of expression and activity of the encoded protein, it provides a highly usable proxy. Therefore, transcriptomics often represents the most efficient means for defining cellular states and studying phenotypic changes and the underlying signalling networks.
Transcriptomics techniques, such as microarrays and massively parallel sequencing, are typically applied on samples consisting of thousands or millions of cells. This implies an assumption that phenotypically similar cells in a population are similar also in terms of molecular composition, and are thus represented with reasonable accuracy by the average values of the population. However, a growing body of data contradicts this assumption [1–5]. In fact, the emerging view strongly suggests that transcriptomes of even closely related cells exhibit considerable heterogeneity. Conceptually, the biological heterogeneity can be divided into (1) heterogeneity originating from stochastic nature of biochemical processes including gene expression, (2) heterogeneity originating from slightly different molecular microenvironments and different signalling histories of each cell and (3) population heterogeneity, which is deterministic and ‘hard-wired’ causing subsets of cells intrinsically to express different properties (Figure 1A) [6].
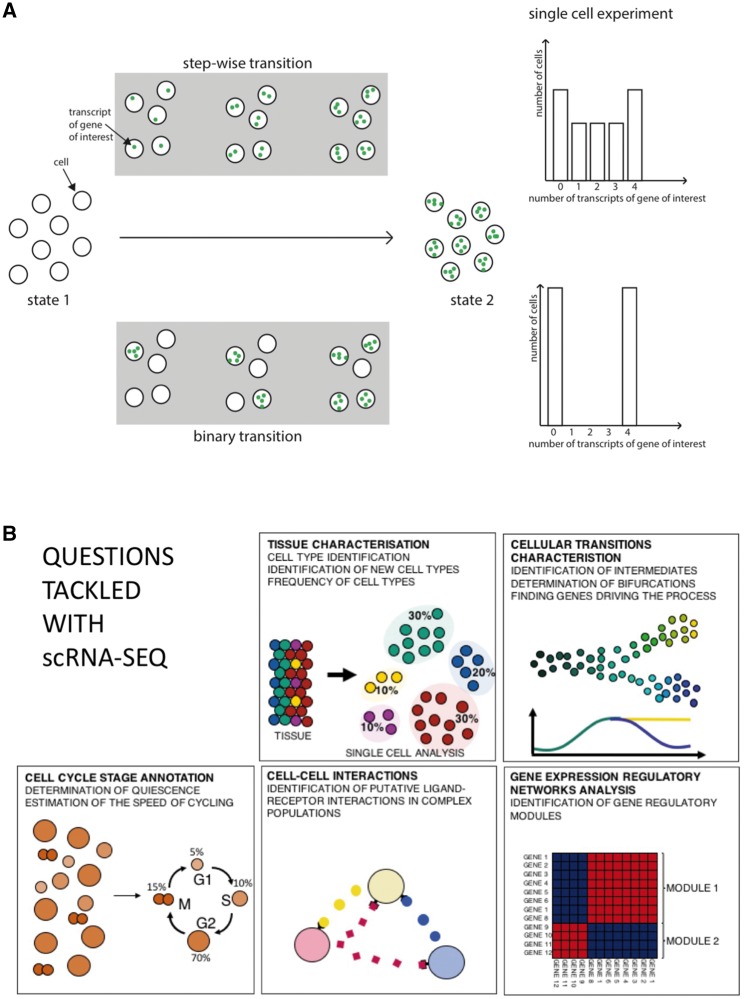
Figure 1.
Single-cell methods provide insight into the nature of a population, its subpopulation structure and heterogeneity. (A) A conceptual example is the switch of cells from State 1 to State 2 in this schematic diagram. This process could be either a binary or gradual switch in transcriptomic state. While population methods cannot distinguish between the two states, single-cell methods can discriminate between these two transitions. (B) Examples of biological questions addressed with single-cell RNA sequencing (scRNA-seq).
Cell-intrinsic and environmental factors contributing to this heterogeneity are incompletely understood, as are its full biological consequences. In an experimental setting, such variation may arise from asynchronous stages of cell cycle [7–11], uneven partitioning of molecules during cell divisions and differences in cellular signalling histories or epigenetic modifications before the experiment in question. Moreover, transcription of both prokaryotic and eukaryotic genes has been documented to frequently follow stochastic burst-like kinetic patterns, with relatively short but intense bursts of transcription being followed by longer inactive periods during which mRNA levels decay [4, 12–14]. Recent studies suggest that such bursting is widespread, although the duration of the bursts and intervals can vary considerably [15]. Mechanistically, expression bursts are dependent on the stochastic processes of transcription factors and RNA polymerase binding [16]. In line with this intrinsic stochasticity, single-cell gene expression data typically follows negative binomial distribution [17]. An important implication of this is that the ubiquitously used population-wide average values are not accurate representations of the typical single cell. In terms of understanding the basic biology of gene expression and the structure of a cell population, these reasons make a strong argument for performing transcriptomics analyses at the single-cell level, and highlight the need for developing robust system-wide methods.
Single-cell efforts have also been further motivated by the innumerable potential applications involved (Figure 1B). An obvious benefit is the possibility to study rare types of cells, either too limited in number or too sparsely distributed for conventional bulk transcriptomics [18]. Important examples include early stages of embryonic development [19] and circulating cancer cells [20, 21]. Another important issue that can be potentially addressed by single-cell analysis is tissue heterogeneity. Many biological systems of high medical significance, such as hematopoietic lineages [22] and neural cells [23], are composed of intermixed differentiated cell types acting in coordination but using different molecular pathways. The response of such a population to a perturbation is likely to be profoundly mixed, and thus data obtained from bulk methods most certainly blend true single-cell transcriptomes and hence will be challenging to interpret [24, 25]. For example, only selected subsets of blood cells are likely to react to a vaccine, or cells of heterogeneous tumours can display widely different responses to a drug. With single-cell transcriptomics, such complex population structures can be dissected and cells of interest can be studied without the confounding effects of population-level averaging. Importantly, resolving heterogeneous populations potentially provides valuable information about transitions between distinct developmental or activation states. By identification of cells in transitional intermediate states one can infer order of regulatory events leading to cellular state transitions. Thus, while single-cell transcriptomics is still in many ways a relatively immature field of research in a state of rapid development, it is already proving its potential and a multitude of research and diagnostic applications are likely to follow.
RT-qPCR is a sensitive method for targeted analysis of genes of interest
The initial and still widely used way of studying gene expression in single cells is by quantitative reverse transcription PCR (RT-qPCR). Its sensitivity, precision, reproducibility and wide dynamic range has made it a tool of choice for studying mRNA expression and validating findings from high-throughput studies, in particular microarrays. In addition to widespread use in research, numerous diagnostic applications of RT-qPCR have been developed [26]. The mainstream RT-qPCR strategies are based on real-time optical monitoring of complementary DNA (cDNA) amplification using either intercalating dyes [27] or fluorescing hydrolysis probes [28]. As the PCR reaction is intrinsically scalable, in suitable conditions it allows amplification from even single-cell quantities, which was demonstrated early for both DNA and cDNA templates [29, 30]. Accordingly, the standard RT-qPCR workflow is conceptually applicable to single-cell material without profound modifications.
A key consideration in these single-cell applications is prevention of loss of RNA, leading to requirement for so-called single-tube protocols where cell lysis, reverse transcription and PCR are performed without intervening purification steps. This is made possible by low final concentrations of sample-derived RNase and potential inhibitory molecules such as salts, urea, heparin or immunoglobulins [31–33], which in bulk studies typically require depletion by a dedicated purification, precipitation or extraction process. The buffers of the subsequent reaction steps, including lysis, are designed to be compatible and enzymes used in the previous step are inactivated by heat treatment. The unforgivingly low amount of starting material also sets high demands on RT efficiency, although absolute efficiency is also gene-dependent [34, 35]. Overall, many of the technical considerations and pitfalls are in common with bulk RT-qPCR assays and include template quality, standardization of the RT reaction and assay design [31, 36]. The recently proposed the minimum information for publication of quantitative real-time PCR experiments (MIQE) guidelines serve to draw attention to these critical and often neglected issues and should also be taken into account in single-cell studies [37].
Unlike microarrays or RNA sequencing (RNA-seq), single-cell RT-qPCR has the potential for detecting transcripts without a preamplification step (Table 1). Theoretically, single molecules can be detected, although reproducible quantification has been reported to require ∼≥20 copies per cell, thus limiting the analysis to intermediate to high copy number mRNAs [32]. Furthermore, without preamplification only a few (∼≤10) genes can be measured simultaneously, as parallel assays require aliquoting of the sample [33]. Taniguchi et al. [35] have proposed a bead-based strategy for immobilizing and reusing cDNA molecules, thus overcoming the need of aliquoting the sample. However, the number of possible sequential assays from a single cDNA library remains relatively small, theoretical output also being limited by instrument time.
Table 1.
Comparison of approaches for single-cell transcriptome characterization
| Property | RT-qPCR | Microarrays | RNA-seq |
|---|---|---|---|
| Genes analysed | • Hundreds | • Thousands | • All |
| • All genes | • Only genes with poly(A) | • Only genes with poly(A) | |
| • Knowledge-driven choice of genes | • Knowledge-driven choice of genes | • Unbiased | |
| Alternative splicing information | • Yes, but challenging | • Yes, knowledge driven | • Yes, unbiased |
| Biases | • Biases from preamplification (if used) | • Biases from preamplification | • Biases from preamplification |
| • Biases caused by detection method (dye, Taqman probes, etc.) | |||
| • False negatives for probes at 5′ end of long genes | |||
| Sensitivity | • High | • No detection of low copy number transcripts | • No detection of low copy number transcripts |
| Multiplexing | • 96 cells×96 genes with the BiomarkTM | • No | • From 96 cells (C1TM) to thousands of cells (ChromiumTM) |
| Quantification | • Relative (absolute possible with spike-ins) | • Relative | • Absolute |
| Data obtained | • Cq values | • Intensities | • Read counts, RPKM/FPKM/TPM |
| Data analysis | • Standard univariate or multivariate statistics | • Standard microarray data analysis pipelines | • Standard RNA-seq data analysis pipelines |
| + Bespoke methods |
FPKM, fragments per kilobase per million; RPKM, reads per kilobase per million; TPM, transcripts per million.
The possibilities of single-cell RT-qPCR can be significantly extended by methods allowing quantitative preamplification of mRNAs independent of gene sequence or transcript size. These protocols are typically based on the use of poly-dT primers and exploit either exponential PCR amplification or in vitro transcription (IVT)-based linear amplification [38]. Thus, a single cell can provide a virtually infinite supply of cDNA, making the availability of suitable RT-qPCR assays and relatively high running costs the limiting factors for sample throughput. However, amplification also leads to increased noise and can introduce biases and should therefore not be used without appropriate quality control. Allowing more extensive multiplexing and thus more powerful experimental designs, preamplification has become a widely used routine step in single-cell RT-qPCR studies [39–41]. Nevertheless, multiplexing approaches are ultimately limited by the amount of manual work involved as well as assay costs. To overcome these limitations, microfluidics-based multiplex assay platforms have been developed. These include the BiomarkTM Dynamic Arrays (Fluidigm), using which 96 samples can be interrogated with 96 parallel primer–probe assays [42]. A key promise of such tools is the potential to uncover novel regulatory relationships between the genes under investigation [43, 44].
A common pitfall in RT-qPCR workflows is presented by data processing and in particular normalization. The purpose of normalization is to eliminate bias resulting from differences in cDNA amounts between samples, associated with unequal loading of starting material, or unequal losses during sample processing. In single-cell experiments, differences in cell size present an important additional consideration. The functional activity of mRNAs is ultimately determined by their intracellular concentration rather than absolute copy number [45]. Thus, including a normalization step for cell size might improve the biological value of the analysis, especially if the analysed cells are particularly heterogeneous in size. On the other hand, inappropriate choice of normalization strategy, based on subjective or otherwise incorrect assumptions, can lead to biased or downright erroneous results. These considerations are therefore extremely important in single-cell analysis.
The primary output of an RT-qPCR assay is the number of PCR cycles required to reach a predefined level of signal, herein referred as quantification cycle (Cq), other commonly used synonyms, coined by various instrument manufacturers, being threshold cycle (Ct), crossing point (Cp) and take-off point (TOP). In bulk RT-qPCR studies, normalization is most commonly performed by comparing the measured Cq values with the corresponding values from so-called reference genes, the expression level of which is assumed to be constant within the particular experimental model. The selection of such genes should thus be well justified and preferentially validated by statistical measures. If possible, multiple reference genes should be used. However, at the single-cell level, the usability of the reference gene approach is limited by the ubiquitous cell-to-cell variability in gene expression, extending to traditional reference genes such as Actb [46], Gapdh [45] and Tbp [35]. Nevertheless, in both yeast and mice, many housekeeping genes have been found to be constitutively expressed at a high level with a less than average degree of variability [47–49].
Of note, single-cell experiments provide an intrinsic means for normalization, as the number of cells is constant, i.e. one. While this strategy does not take into account the variability related to differences in cell size, it theoretically allows the measured Cq values to be transformed into mRNA copy numbers per cell. However, as this is based on the assumption of 100% efficiency in reverse transcription and PCR reactions, in practice, the Cq data represent the lowest estimate of the possible true copy number in the cell. Importantly, if the limit of detection for a given experiment is known, for any assay with Cq values exceeding that limit, the copy number can be confidently determined as zero. This is a significant conceptual difference to bulk RT-qPCR studies, wherein such measurements are commonly dismissed as missing values. The limit of detection can be determined by addition of external RNA or cDNA standards to each sample during the lysis step. As such, spike-in standards do not control for pre-lysis variability, and even more rigorous normalization could potentially be achieved by use of standards directly injected into the cells.
With the possibility to measure absence of mRNA species, and in keeping with the model of stochastic burst-like gene expression, multiplexed single-cell RT-qPCR data frequently contain a high proportion of cells with no mRNAs detected [50]. Importantly, the detection frequency of an mRNA correlates with the overall population abundance of the transcript, and hence in such cases can be used as a measure for population-level average expression [33]. Another consideration following from the stochastic nature of gene expression is that at the single-cell level, biological variability (noise) is significantly greater than the technical variability of the RT-qPCR methods. Thus, unlike with bulk RNA-seq, resources will in general be better used by maximizing the number of analysed cells instead of performing technical replicates. Altogether, single-cell RT-qPCR data processing can, in general, still be considered straightforward compared with the other single-cell transcriptomics tools. The processed data can often be further analysed by either univariate methods (with necessary corrections for multiple testing), or multivariate analyses, such as hierarchical clustering or principal component analysis. In addition, more specialized probabilistic methods have been proposed[51].
Global measurements of gene expression in single cells
RT-qPCR has several advantages but is limited to relatively small numbers of genes and is impractical to scale above a certain level, even with advanced microfluidic devices. To perform single-cell transcriptome analysis on a global scale, one can use microarray or RNA-seq technologies (Table 1). So far, these methods have mostly been used to screen for candidate genes that are subsequently validated with other methods such as RT-qPCR, flow cytometry or single-molecule fluorescence in situ hybridization (FISH) [45, 47, 52–54]. Each single-cell transcriptomic assay experiment, regardless whether is using microarrays or sequencing, can be divided into the following steps: (1) isolation of single cells, (2) cell lysis, (3) reverse transcription, (4) amplification of cDNA, (5) preparation of sequencing libraries and (6) eventually detection.
Single-cell isolation
The first and sometimes underappreciated step is to isolate single cells. Whereas many immune cell types naturally exist as single-cell suspensions, other cells have to be dissociated from the tissue. Such treatment is far from trivial, as it requires enzymatic or mechanical approaches that may affect not only the intactness and viability of cells but also their transcriptomes.
Historically, in the first single-cell mRNA experiments, single cells were manually selected and picked from the early embryo using micropipetting [17, 55, 56]. The advantage of this approach is that particular cells of interest can be selected and cell losses can be minimized in the process. Suspended single cells can be sorted into wells of a microtitre plate using fluorescence activated cell sorting (FACS) [57], they can be separated using microfluidic devices such as the Fluidigm C1TM [23, 47, 58–61] or they can be encapsulated in nanolitre droplets (Table 2) [11, 62].
Table 2.
Comparison of scRNA-seq platforms
| Characteristic | FACS | Microfluidics (e.g. Fluidigm C1TM) | Droplets (DropSeq, InDrop, ChromiumTM, etc.) |
|---|---|---|---|
| Reaction volume | Microliter | Nanoliter | Nanoliter |
| Throughput | Low to medium (depends on level of automatization of RT, amplification and library preparation processes) | Low to medium (depends on chip design) | High (limited by number of distinct cell barcodes) |
| Flexibility | Great flexibility to choose methods for RT, amplification and library preparation | Some flexibility to choose methods for RT, amplification and library preparation | Only molecule counting (no full-length transcript coverage) |
| Additional measurements | Additional data from index sorting (size, granularity, expression of surface markers, DNA content, etc.) | Imaging of the captured cells before lysis | None |
The key advantage of FACS is the possibility to sort for particular subpopulations using molecular markers. In addition, the intensity of the fluorescence of several fluorescent markers along with values of forward and side scatter can be recorded for each cell. This provides useful phenotypic information about protein abundance, cell size and granularity on top of the single-cell transcriptomes [63]. When studying known, rare cell types (e.g. blood stem cells), FACS can capture essentially all cells from the population of interest. The main disadvantage of using FACS to sort single cells into microtitre plates are the microlitre reagent volumes involved, which can be prohibitively expensive in large-scale experiments as compared with nanolitre volumes involved in microfluidics and droplet-based methods [64]. The Fluidigm C1TM is a microfluidic platform that captures single cells (96 or 800 cells per chip) and performs reverse transcription and amplification of cDNA by PCR on chip. As all these reactions are carried out in nanolitre volumes, this leads to lower reagent costs. Importantly, this platform enables microscopic inspection of each cell on capture, which allows identification of positions where multiple cells or debris were captured. A drawback of the C1TM workflow is the relatively low capture efficiency. To capture 96 cells on C1 TM, one typically requires a starting population of at least 1000 cells, making the method impractical for rare populations. Another important limitation of this method is that cells being captured have to be homogeneous in size and compatible with one of the available capture site sizes (5–10, 10–17 and 17–25 µm in diameter). Nonspherical or sticky cells also do not capture well, but at the same time, this capture method is much more gentle than FACS, and hence is suited to delicate cell types such as neurons, megakaryocytes, etc.
Recently, droplet-based microfluidics methods have been published, namely, inDrop [62], Drop-Seq [11] followed by launching of similar commercial protocols such as the ChromiumTM from 10X Genomics [65]. These protocols encapsulate single cells, or single cells and beads bearing barcodes, in aqueous droplets within a surrounding oil phase. The droplets can be subsequently fused with other droplets to deliver reagents to perform lysis, reverse transcription and PCR. Reagent can also be delivered into droplets using picoinjection [66]. These methods will likely prove especially useful for surveying cells from different tissues to identify new cell types and cell functions, as they allow analysis of several thousands of cells in one experiment.
Less frequently used methods include laser capture microdissection, which is useful for picking cells from a particular position in a tissue. It is low throughput and does not necessarily guarantee that a single cell, rather than small group of cells is captured [67, 68]. Finally, nanolitre plates can be used for capturing single cells. Simply by adjusting the concentration of the cells in suspension, cells can be deposited and virtually every well will receive zero or one cell [69, 70].
Cell lysis
Captured cells are lysed by addition of lysis buffer containing detergent to disrupt the cell membrane. For plant or fungi cells, protoplasts must first be obtained by enzymatic or mechanical removal of the cell wall. Efficient cell lysis is crucial for efficient release of RNAs to the reaction and for the efficiency of subsequent reactions.
Reverse transcription
In the next step, RNAs are reverse transcribed, and this is a key step for achieving high sensitivity. A major goal of this stage is to avoid reverse transcribing ribosomal RNAs (rRNAs), which are high-abundance and would dominate any signal from the much lower abundance mRNAs. Owing to the low abundance of mRNAs, common mRNA purification methods cannot be used. Most protocols for reverse transcription (SmartSeq [53], STRT-Seq [54], QuartzSeq [71]) use poly(T) primers that bind to the poly(A) tail of mRNAs. This way only polyadenylated RNA species are reverse transcribed.
Alternatively, primers that are specifically designed not to bind to rRNAs can be used
[72]. The disadvantage of this approach is that it may lead to amplification biases against some mRNAs. Finally, it was shown recently that random hexamer primers can be used [73, 74], provided reverse transcription is performed at low temperature. In such conditions, most rRNAs are within folded ribosomes and are not transcribed. Moving beyond poly(A) priming would be useful for analyses of non-coding RNAs (ncRNAs), such as circular RNAs [74], and also bacterial RNAs, which are not polyadenylated [75].
Second-strand cDNA synthesis can be done using the template-switching properties of the reverse transcriptase to minimize detection of partially transcribed species: this approach is used in SmartSeq [53]. Alternatively, poly(A) tailing and subsequent second-strand synthesis priming from the polyA sequence can be used, but this leads to stronger 3′ bias of read coverage over transcripts, meaning that there are more reads mapping to the 3′ end of the transcript. This originates from incomplete reverse transcription, as in the first single-cell sequencing protocol by Tang and colleagues and the QuartzSeq protocol [55, 71].
It is estimated that a single cell contains around 10 pg of mRNA [53], which will not produce sufficient cDNA for sequencing library preparation alone; thus, the cDNA must be amplified. There are two main methods of amplification: linear amplification using IVT and exponential amplification using PCR. Most protocols use PCR for amplification: SmartSeq [53], SmartSeq2 [76], STRT [54], the Tang protocol [55] and SC3-seq [77]. The main caveat of PCR is the fact that the exponential amplification that occurs may distort the relative amounts mRNA molecules. The alternative approach of IVT was incorporated into the CEL-Seq [78], CEL-seq2 [79] and MARS-Seq [64] protocols. Amplification via IVT is linear, but it was shown that subsequent IVT causes significant shortening of amplified RNAs and thus only the 3′ ends of mRNAs are amplified [80].
The number of molecules in each cell is limited and it is estimated that only 10% of them are transcribed to cDNA with current technologies [81]. The molecules that are transcribed are selected stochastically. Owing to Poisson sampling, the expression-level estimation may not represent the original set of molecules from the cell, especially for lowly abundant mRNA species leading to so-called ‘drop outs’. Computational approaches are being used to alleviate their effects [82, 83].
Library preparation and detection
Microarrays were initially used for detection of amplified cDNA [71, 84–90], but as they have lower robustness, low sensitivity, limited dynamic range and require large amount of cDNA for hybridization, they are now completely replaced by sequencing for the single-cell transcriptomic applications [91, 92].
Sequencing libraries are prepared from amplified cDNA using the same protocols as for conventional bulk mRNA sequencing experiments and can be sequenced on any sequencing platform. Both SOLID and standard Illumina library preparation protocols, involving Covaris shearing, ligation of adapters and library amplification, were used, but the most common is the NexteraTM kit from Illumina that uses enzymatic Tn5-mediated tagmentation as well as home-brew version of this kit [53].
All RNA-seq methods allow multiplexing with barcoded adapters at the stage of library preparation. This means that barcoded adaptors can be ligated to the cDNA that results from preamplification. Both the standard library preparation kit and the NexteraTM kit from Illumina and library preparation kits for SOLIDTM system have barcoding options. Barcoding before the stage of library preparation allows pooling samples to cut down costs of reagents and dramatically reduces sample handling. The STRT method, as well as currently used droplet methods depend on primers containing cell-specific barcodes that are introduced at the reverse transcription step of the protocol [78]. Similarly in CEL-Seq and CEL-Seq2 barcodes are introduced during IVT stage [79].
Single-cell experiments require internal controls
Single-cell RNA-seq presents challenges that are absent in conventional population-level approaches. Distinguishing biological from technical variation in situation where technical replications are difficult to perform, as there are no two identical cells, is challenging. Furthermore, the sensitivity of the protocols is limited, which leads to so-called ‘drop-outs’, i.e. false-negative values for mRNAs that are present in low amounts but are not being detected. Thus, it is important to measure technical variation to understand which genes can be quantified accurately, and to minimize the rate of false positives in differential expression analysis.
Technical noise can arise at each step of the protocol, including differences in lysis efficiency and reverse transcription and the tendency of some species of mRNA to be preferentially amplified due to their sequence and length of their poly(A) tails. These biases have not yet been sufficiently systematically investigated. Secondly, there is variation in the measurement from batch to batch. This may be because of differences between operators, batches of reagents or other factors. Thirdly, single-cell RNA-seq data have the same biases as conventional RNA-seq, such as PCR amplification bias, sequence bias during fragmentation and coverage biases. Importantly, more rounds of amplification are required than in bulk RNA-seq providing more opportunities for the introduction of base substitutions. If amplification is performed using PCR, then PCR amplification biases are also present. It was also reported that reverse transcription with poly-dT priming leads to 3′ bias in read coverage [53, 93]. This is also the case in bulk-level experiment that uses poly-dT priming.
Technical variation between cells can be estimated using mRNA spike-ins that undergo all the steps of the protocol together with the sample. In early microarray experiments, a set of four Bacillus subtilis mRNAs (Lys, Dap, Phe and Thr) spiked-in at different copy numbers has been used to measure detection limits. ERCC (External RNA Control Consortium) spike-in is the most commonly used, commercially available set of control molecules and consists of 92 synthetic polyadenylated mRNA species of different known concentrations [94]. These were designed so as to lack sequence similarity to any known eukaryotic genome. It allows one to measure the sensitivity and accuracy of each experiment, as well as perform correction of some batch effects. It is also used for estimation of the extent of technical noise [95].
ERCC spike-ins can be used to produce a calibration curve to estimate the absolute number of molecules in each cell [85, 86, 96]. It should be noted that ERCC molecules do not go through cell lysis and are not associated with proteins, thus are not subjected to all the processes that cellular mRNAs are. Furthermore, they are not capped, they have short poly(A) tails in comparison with endogenous mRNAs and because of their easy degradation during normal handling, it is difficult to ensure accurate input concentrations [97]. SIRVs (Spike-In RNA Variant Mixes, Lexogen) are an alternative or complementary to ERCCs spike-in mix. They are designed to in addition to abundance control splicing patterns of RNAs, i.e. the spike-in consists of 69 different transcripts that mimic splice variants of 7 genes.
Interestingly, one can also use minute amounts of total RNA coming from a species alien to the species of interest as a spike-in. This approach provides thousands of technical data points across the whole dynamic range of expression, thereby ensuring that technical noise levels can be well quantified across the whole dynamic range [95]. The drawback of this approach is that a substantial number of reads goes to technical noise control, entailing significant costs. Technical variability within an experiment can be also estimated by performing pool and split experiments [98, 99].
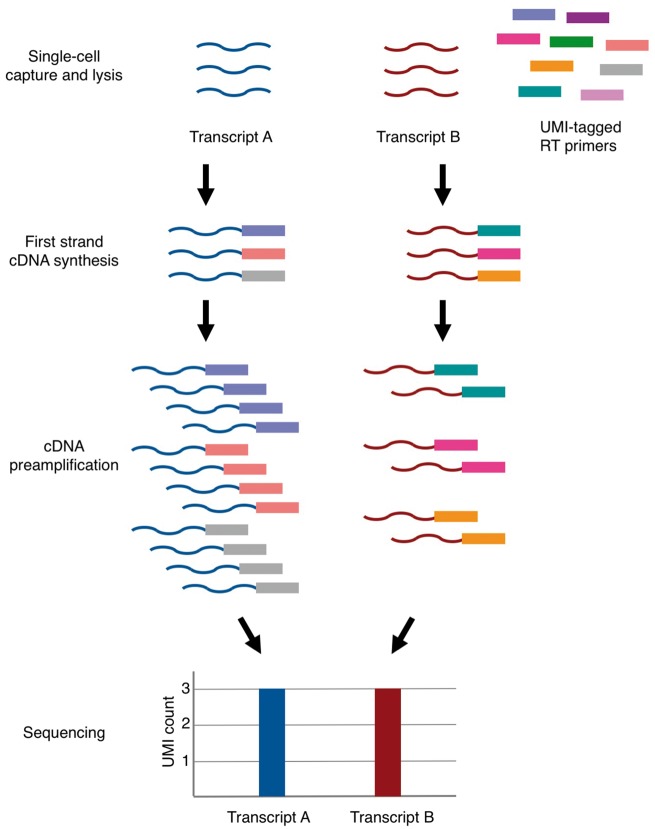
While the use of spike-in RNA is relatively commonplace with protocols based on cell sorting or microfluidic cell capture devices, this strategy is less frequently used in droplet-based workflows. One limitation is that the spike-in molecules will be deposited also in partitions that do not contain cells, and therefore unnecessarily consume sequencing capacity. In addition, reads from such empty droplets might hinder detection of true single cells from the data. Instead of spike-in RNA, droplet-based workflows typically incorporate unique molecular identifiers (UMIs), which are highly diverse, random, unique barcodes for tagging each cDNA molecule generated during reverse transcription [54, 81, 100, 101]. They enable one to count molecules by counting the number of unique UMI sequences associated with each transcript instead of counting the number of sequencing reads that map to a particular transcript (Figure 2). This can ameliorate PCR biases [96]. The main disadvantage of UMIs is that until now they have only been used for methods that count the 3′ end of molecules. In addition, to estimate the number of molecules, one has to sequence deeply.
Figure 2.
Molecular counting with UMIs. UMIs are random n-mer oligonucleotide sequences included in the reverse transcription primers. As the number of different UMI sequences exceeds the number of copies for any single-transcript species, the UMI sequences can be used for quantifying the number of molecules that were successfully captured and amplified, and thus control for amplification biases associated with PCR-based sample preparation.
Choosing the right protocol depends on the biological system under investigation
The optimal single-cell RNA-seq application depends on the desired application. Each of the single-cell methods described above has advantages and disadvantages as summarized in Table 1. Factors to be considered include throughput, sensitivity and robustness, transcript coverage, cost and handling (comprehensive sensitivity and robustness comparison of methods were performed by [97]). For example, for discovery of new cell types, tag-counting droplet methods with high throughput are most advisable, while for analysis of allelic expression or splicing, one must use a protocol that provides sequencing coverage of the entire length of mRNA molecules.
Future outlook
Single-cell transcriptomics brings both new opportunities and new challenges. Measuring gene expression at the single-cell level provides a huge amount of information, which requires adequate data analysis methods. In past couple of years, several different approaches for analysis of single-cell sequencing data emerged and still new computational methods are being developed to access even more information from single-cell data [102, 103].
The current efforts of many groups are focused on approaches for ordering cells along a process in so-called ‘pseudotime’ to describe transitions between cell states and cellular decision points, where cells commit on one of available states [104–110]. As single-cell data have and even with improvement of technology will inevitably suffer from false negatives because of dropout effect, computational approaches to include this effect into models and analysis are crucial [83].
Furthermore, there is room for improvement of experimental side of single-cell methods. One issue that should be addressed is sensitivity and robustness, with more efficient chemistry and RNAse-free reagents we may be able to detect more genes and limit the dropouts. Secondly, sample size, i.e. the number of single cells sequenced is crucial to obtain statistical power and to observe rare cell types. Further developments are needed to increase throughput [111, 112], simultaneously allowing multiplexing different biological samples in one run [113]. Thirdly, further developments are needed to streamline single-cell sequencing of non-polyadenylated RNA species, to detect bacterial RNA as well as eukaryotic ncRNAs and combine it with other measurements in the same cell, such as imaging, genome and epigenome analysis or protein abundance quantification [114].
Finally, it is important to bear in mind that single-cell experiments, though informative and can help elucidate many crucial biological problems, are only part of the equation. Localization of mRNA is as important as its abundance; hence, there is a lot of effort to develop protocols that retain spatial information about the transcripts (TIVA [115], FISSEQ [116, 117] or padlock probe-based methods [118]. Cells are part of complex tissues and interact with each other both physically and by using different chemical signals; thus, understanding single cells in the context of complex tissues will be the next challenge for single-cell research.
Key Points
Studies of cell population heterogeneity and cell transitions benefit from single-cell approaches.
Single-cell quantitative PCR allows study of gene expression heterogeneity in population of cells.
Single-cell RNA-seq experimental approach can be chosen depending on needed number of cell, gene detection efficiency, transcript coverage, cost, etc.
Technical biases and artefacts are determined using spike-ins and UMIs.
Spatial transcriptomics and combination of RNA quantification with other measurements from a single cell are next steps in the field.
Funding
Tapio Lönnberg is supported by the Academy of Finland (Decision 311081).
Aleksandra A. Kolodziejczyk studies single cells using RNA sequencing since 2012, when she started her PhD in the laboratory of Sarah Teichmann Wellcome Trust Sanger Institute and EMBL European Bioinformatics Institute.
Tapio Lönnberg was familiarized with single-cell transcriptomics during his postdoctoral work with Sarah Teichmann at the EMBL European Bioinformatics Institute and the Wellcome Trust Sanger Institute. He is currently an Academy of Finland postdoctoral fellow at the University of Turku, Finland.
References
- 1. Elowitz MB, Levine AJ, Siggia ED, et al. Stochastic gene expression in a single cell. Science 2002;297:1183–6. [DOI] [PubMed] [Google Scholar]
- 2. Levsky JM, Singer RH.. Gene expression and the myth of the average cell. Trends Cell Biol 2003;13:4–6. [DOI] [PubMed] [Google Scholar]
- 3. Raser JM, O'Shea EK.. Noise in gene expression: origins, consequences, and control. Science 2005;309:2010–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raj A, Peskin CS, Tranchina D, et al. Stochastic mRNA synthesis in mammalian cells. PLoS Biol 2006;4:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blake WJ, KAErn M, Cantor CR, et al. Noise in eukaryotic gene expression. Nature 2003;422:633–7. [DOI] [PubMed] [Google Scholar]
- 6. Huang S. Non-genetic heterogeneity of cells in development: more than just noise. Development 2009;136:3853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsang JC, Yu Y, Burke S, et al. Single-cell transcriptomic reconstruction reveals cell cycle and multi-lineage differentiation defects in Bcl11a-deficient hematopoietic stem cells. Genome Biol 2015;16:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kowalczyk MS, Tirosh I, Heckl D, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Res 2015;25:1860–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Buettner F, Natarajan KN, Casale FP, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol 2015;33:155–60. [DOI] [PubMed] [Google Scholar]
- 10. Singh AM, Chappell J, Trost R, et al. Cell-cycle control of developmentally regulated transcription factors accounts for heterogeneity in human pluripotent cells. Stem Cell Reports 2013;1:532–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raj A, van Oudenaarden A.. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 2008;135:216–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chubb JR, Trcek T, Shenoy SM, et al. Transcriptional pulsing of a developmental gene. Curr Biol 2006;16:1018–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suter DM, Molina N, Gatfield D, et al. Mammalian genes are transcribed with widely different bursting kinetics. Science 2011;332:472–4. [DOI] [PubMed] [Google Scholar]
- 15. Lionnet T, Singer RH.. Transcription goes digital. EMBO Rep 2012;13:313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sanchez A, Golding I.. Genetic determinants and cellular constraints in noisy gene expression. Science 2013;342:1188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grün D, Kester L, van Oudenaarden A.. Validation of noise models for single-cell transcriptomics. Nat Methods 2014;11:637–40. [DOI] [PubMed] [Google Scholar]
- 18. Proserpio V, Lönnberg T.. Single-cell technologies are revolutionizing the approach to rare cells. Immunol Cell Biol 2016;94:225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xue Z, Huang K, Cai C, et al. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013;500:593–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miyamoto DT, Zheng Y, Wittner BS, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science 2015;349:1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jordan NV, Bardia A, Wittner BS, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 2016;537:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Velten L, Haas SF, Raffel S, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol 2017;19:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zeisel A, Muñoz-Manchado AB, Codeluppi S, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015;347:1138–42. [DOI] [PubMed] [Google Scholar]
- 24. Shen-Orr SS, Tibshirani R, Khatri P, et al. Cell type-specific gene expression differences in complex tissues. Nat Methods 2010;7:287–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abbas AR, Wolslegel K, Seshasayee D, et al. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One 2009;4:e6098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kubista M, Andrade JM, Bengtsson M, et al. The real-time polymerase chain reaction. Mol Aspects Med 2006;27:95–125. [DOI] [PubMed] [Google Scholar]
- 27. Higuchi R, Dollinger G, Walsh PS, et al. Simultaneous amplification and detection of specific DNA sequences. Biotechnology 1992;10:413–17. [DOI] [PubMed] [Google Scholar]
- 28. Heid CA, Stevens J, Livak KJ, et al. Real time quantitative PCR. Genome Res 1996;6:986–94. [DOI] [PubMed] [Google Scholar]
- 29. Li HH, Gyllensten UB, Cui XF, et al. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature 1988;335:414–17. [DOI] [PubMed] [Google Scholar]
- 30. Rappolee DA, Wang A, Mark D, et al. Novel method for studying mRNA phenotypes in single or small numbers of cells. J Cell Biochem 1989;39:1–11. [DOI] [PubMed] [Google Scholar]
- 31. Nolan T, Hands RE, Bustin SA.. Quantification of mRNA using real-time RT-PCR. Nat Protoc 2006;1:1559–82. [DOI] [PubMed] [Google Scholar]
- 32. Bengtsson M, Hemberg M, Rorsman P, et al. Quantification of mRNA in single cells and modelling of RT-qPCR induced noise. BMC Mol Biol 2008;9:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ståhlberg A, Bengtsson M.. Single-cell gene expression profiling using reverse transcription quantitative real-time PCR. Methods 2010;50:282–8. [DOI] [PubMed] [Google Scholar]
- 34. Ståhlberg A, Håkansson J, Xian X, et al. Properties of the reverse transcription reaction in mRNA quantification. Clin Chem 2004;50:509–15. [DOI] [PubMed] [Google Scholar]
- 35. Taniguchi K, Kajiyama T, Kambara H.. Quantitative analysis of gene expression in a single cell by qPCR. Nat Methods 2009;6:503–6. [DOI] [PubMed] [Google Scholar]
- 36. Ståhlberg A, Kubista M, Aman P.. Single-cell gene-expression profiling and its potential diagnostic applications. Expert Rev Mol Diagn 2011;11:735–40. [DOI] [PubMed] [Google Scholar]
- 37. Bustin SA, Benes V, Garson JA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009;55:611–22. [DOI] [PubMed] [Google Scholar]
- 38. Eberwine J, Yeh H, Miyashiro K, et al. Analysis of gene expression in single live neurons. Proc Natl Acad Sci USA 1992;89:3010–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peixoto A, Monteiro M, Rocha B, et al. Quantification of multiple gene expression in individual cells. Genome Res 2004;14:1938–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warren LA, Rossi DJ, Schiebinger GR, et al. Transcriptional instability is not a universal attribute of aging. Aging Cell 2007;6:775–82. [DOI] [PubMed] [Google Scholar]
- 41. Hayashi K, Lopes SM, Tang F, et al. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell 2008;3:391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Warren L, Bryder D, Weissman IL, et al. Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR. Proc Natl Acad Sci USA 2006;103:17807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guo G, Huss M, Tong GQ, et al. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 2010;18:675–85. [DOI] [PubMed] [Google Scholar]
- 44. Moignard V, Macaulay IC, Swiers G, et al. Characterization of transcriptional networks in blood stem and progenitor cells using high-throughput single-cell gene expression analysis. Nat Cell Biol 2013;15:363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tang F, Lao K, Surani MA.. Development and applications of single-cell transcriptome analysis. Nat Methods 2011;8:S6–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bengtsson M, Ståhlberg A, Rorsman P, et al. Gene expression profiling in single cells from the pancreatic islets of Langerhans reveals lognormal distribution of mRNA levels. Genome Res 2005;15:1388–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shalek AK, Satija R, Adiconis X, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013;498:236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lionnet T, Wu B, Grünwald D, et al. Nuclear physics: quantitative single-cell approaches to nuclear organization and gene expression. Cold Spring Harb Symp Quant Biol 2010;75:113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zenklusen D, Larson DR, Singer RH.. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 2008;15:1263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McDavid A, Finak G, Chattopadyay PK, et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 2013;29:461–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Buettner F, Theis FJ.. A novel approach for resolving differences in single-cell gene expression patterns from zygote to blastocyst. Bioinformatics 2012;28:i626–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tischler J, Surani MA.. Investigating transcriptional states at single-cell-resolution. Curr Opin Biotechnol 2013;24:69–78. [DOI] [PubMed] [Google Scholar]
- 53. Ramsköld D, Luo S, Wang YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 2012;30:777–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Islam S, Kjällquist U, Moliner A, et al. Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 2011;21:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tang F, Barbacioru C, Wang Y, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 2009;6:377–82. [DOI] [PubMed] [Google Scholar]
- 56. Tang F, Barbacioru C, Bao S, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell 2010;6:468–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Macaulay IC, Svensson V, Labalette C, et al. Single-Cell RNA-sequencing reveals a continuous spectrum of differentiation in hematopoietic cells. Cell Rep 2016;14:966–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Treutlein B, Brownfield DG, Wu AR, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 2014;509:371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mahata B, Zhang X, Kolodziejczyk AA, et al. Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep 2014;7:1130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kolodziejczyk AA, Kim JK, Tsang JC, et al. Single Cell RNA-sequencing of pluripotent states unlocks modular transcriptional variation. Cell Stem Cell 2015;17:471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stubbington MJ, Lönnberg T, Proserpio V, et al. T cell fate and clonality inference from single-cell transcriptomes. Nat Methods 2016;13:329–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015;161:1187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hayashi T, Shibata N, Okumura R, et al. Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its “index sorting” function for stem cell research. Dev Growth Differ 2010;52:131–44. [DOI] [PubMed] [Google Scholar]
- 64. Jaitin DA, Kenigsberg E, Keren-Shaul H, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 2014;343:776–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee M, Collins JW, Aubrecht DM, et al. Synchronized reinjection and coalescence of droplets in microfluidics. Lab Chip 2014;14:509–13. [DOI] [PubMed] [Google Scholar]
- 67. Frumkin D, Wasserstrom A, Itzkovitz S, et al. Amplification of multiple genomic loci from single cells isolated by laser micro-dissection of tissues. BMC Biotechnol 2008;8:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Keays KM, Owens GP, Ritchie AM, et al. Laser capture microdissection and single-cell RT-PCR without RNA purification. J Immunol Methods 2005;302:90–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bose S, Wan Z, Carr A, et al. Scalable microfluidics for single-cell RNA printing and sequencing. Genome Biol 2015;16:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Fan HC, Fu GK, Fodor SP.. Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science 2015;347:1258367.. [DOI] [PubMed] [Google Scholar]
- 71. Sasagawa Y, Nikaido I, Hayashi T, et al. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome Biol 2013;14:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bhargava V, Ko P, Willems E, et al. Quantitative transcriptomics using designed primer-based amplification. Sci Rep 2013;3:1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Armour CD, Castle JC, Chen R, et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods 2009;6:647–9. [DOI] [PubMed] [Google Scholar]
- 74. Fan X, Zhang X, Wu X, et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol 2015;16:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kang Y, Norris MH, Zarzycki-Siek J, et al. Transcript amplification from single bacterium for transcriptome analysis. Genome Res 2011;21:925–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Picelli S, Björklund Å, Faridani OR, et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 2013;10:1096–8. [DOI] [PubMed] [Google Scholar]
- 77. Nakamura T, Yabuta Y, Okamoto I, et al. SC3-seq: a method for highly parallel and quantitative measurement of single-cell gene expression. Nucleic Acids Res 2015;43:e60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hashimshony T, Wagner F, Sher N, et al. CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep 2012;2:666–73. [DOI] [PubMed] [Google Scholar]
- 79. Hashimshony T, Senderovich N, Avital G, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol 2016;17:77.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Esumi S, Wu SX, Yanagawa Y, et al. Method for single-cell microarray analysis and application to gene-expression profiling of GABAergic neuron progenitors. Neurosci Res 2008;60:439–51. [DOI] [PubMed] [Google Scholar]
- 81. Islam S, Zeisel A, Joost S, et al. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 2014;11:163–6. [DOI] [PubMed] [Google Scholar]
- 82. Lun AT, Bach K, Marioni JC.. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol 2016;17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Pierson E, Yau C.. ZIFA: dimensionality reduction for zero-inflated single-cell gene expression analysis. Genome Biol 2015;16:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Iscove NN, Barbara M, Gu M, et al. Representation is faithfully preserved in global cDNA amplified exponentially from sub-picogram quantities of mRNA. Nat Biotechnol 2002;20:940–3. [DOI] [PubMed] [Google Scholar]
- 85. Tietjen I, Rihel JM, Cao Y, et al. Single-cell transcriptional analysis of neuronal progenitors. Neuron 2003;38:161–75. [DOI] [PubMed] [Google Scholar]
- 86. Jensen KB, Collins CA, Nascimento E, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 2009;4:427–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kurimoto K, Yabuta Y, Ohinata Y, et al. An improved single-cell cDNA amplification method for efficient high-density oligonucleotide microarray analysis. Nucleic Acids Res 2006;34:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kurimoto K, Yabuta Y, Ohinata Y, et al. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat Protoc 2007;2:739–52. [DOI] [PubMed] [Google Scholar]
- 89. Klein CA, Seidl S, Petat-Dutter K, et al. Combined transcriptome and genome analysis of single micrometastatic cells. Nat Biotechnol 2002;20:387–92. [DOI] [PubMed] [Google Scholar]
- 90. Hartmann CH, Klein CA.. Gene expression profiling of single cells on large-scale oligonucleotide arrays. Nucleic Acids Res 2006;34:e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wang D, Bodovitz S.. Single cell analysis: the new frontier in ‘omics’. Trends Biotechnol 2010;28:281–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nookaew I, Papini M, Pornputtapong N, et al. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nucleic Acids Res 2012;40:10084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mortazavi A, Williams BA, McCue K, et al. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008;5:621–8. [DOI] [PubMed] [Google Scholar]
- 94. Jiang L, Schlesinger F, Davis CA, et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res 2011;21:1543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brennecke P, Anders S, Kim JK, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods 2013;10:1093–5. [DOI] [PubMed] [Google Scholar]
- 96. Kivioja T, Vähärautio A, Karlsson K, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 2011;9:72–4. [DOI] [PubMed] [Google Scholar]
- 97. Svensson V, Natarajan KN, Ly LH, et al. Power analysis of single cell RNA‐sequencing experiments. Nat Methods 2016;14:381–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Deng Q, Ramsköld D, Reinius B, et al. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 2014;343:193–6. [DOI] [PubMed] [Google Scholar]
- 99. Marinov GK, Williams BA, McCue K, et al. From single-cell to cell-pool transcriptomes: stochasticity in gene expression and RNA splicing. Genome Res 2014;24:496–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Fu GK, Hu J, Wang PH, et al. Counting individual DNA molecules by the stochastic attachment of diverse labels. Proc Natl Acad Sci USA 2011;108:9026–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shiroguchi K, Jia TZ, Sims PA, et al. Digital RNA sequencing minimizes sequence-dependent bias and amplification noise with optimized single-molecule barcodes. Proc Natl Acad Sci USA 2012;109:1347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bacher R, Kendziorski C.. Design and computational analysis of single-cell RNA-sequencing experiments. Genome Biol 2016;17:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Rostom R, Svensson V, Teichmann SA, et al. Computational approaches for interpreting scRNA-seq data. FEBS Lett 2017;591:2213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 2014;32:381–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Lönnberg T, Svensson V, James KR, et al. Single-cell RNA-seq and computational analysis using temporal mixture modelling resolves Th1/Tfh fate bifurcation in malaria. Sci Immunol 2017;2:eaal2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Marco E, Karp RL, Guo G, et al. Bifurcation analysis of single-cell gene expression data reveals epigenetic landscape. Proc Natl Acad Sci USA 2014;111:E5643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Reid JE, Wernisch L.. Pseudotime estimation: deconfounding single cell time series. Bioinformatics 2016;32:2973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bendall SC, Davis KL, Amir el-AD, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell 2014;157:714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shin J, Berg DA, Zhu Y, et al. Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 2015;17:360–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Setty M, Tadmor MD, Reich-Zeliger S, et al. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat Biotechnol 2016;34:637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cao J, Packer JS, Ramani V, et al. Comprehensive single cell transcriptional profiling of a multicellular organism by combinatorial indexing. BioRxiv 2017, doi: 10.1101/104844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Rosenberg AB, Roco C, Muscat RA, et al. Scaling single cell transcriptomics through split pool barcoding. BioRxiv 2017, doi: 10.1101/105163. [Google Scholar]
- 113. Kang HM, Subramaniam M, Targ S, et al. Multiplexing droplet-based single cell RNA-sequencing using natural genetic barcodes. BioRxiv 2017, doi: 10.1101/118778. [Google Scholar]
- 114. Macaulay IC, Ponting CP, Voet T.. Single-cell multiomics: multiple measurements from single cells. Trends Genet 2017;33:155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lovatt D, Ruble BK, Lee J, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat Methods 2014;11:190–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lee JH, Daugharthy ER, Scheiman J, et al. Highly multiplexed subcellular RNA sequencing in situ. Science 2014;343:1360–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Mitra RD, Shendure J, Olejnik J, et al. Fluorescent in situ sequencing on polymerase colonies. Anal Biochem 2003;320:55–65. [DOI] [PubMed] [Google Scholar]
- 118. Ke R, Mignardi M, Pacureanu A, et al. In situ sequencing for RNA analysis in preserved tissue and cells. Nat Methods 2013;10:857–60. [DOI] [PubMed] [Google Scholar]