Abstract
Hydrogen peroxide (H2O2) is a prime member of the reactive oxygen species (ROS) family of molecules produced during normal cell function and in response to various stimuli, but if left unchecked, it can inflict oxidative damage on all types of biological macromolecules and lead to cell death. In this context, a major source of H2O2 for redox signaling purposes is the NADPH oxidase (Nox) family of enzymes, which were classically studied for their roles in phagocytic immune response but have now been found to exist in virtually all mammalian cell types in various isoforms with distinct tissue and subcellular localizations. Downstream of this tightly regulated ROS generation, site-specific, reversible covalent modification of proteins, particularly oxidation of cysteine thiols to sulfenic acids, represents a prominent posttranslational modification akin to phosphorylation as an emerging molecular mechanism for transforming an oxidant signal into a dynamic biological response. We review two complementary types of chemical tools that enable (a) specific detection of H2O2 generated at its sources and (b) mapping of sulfenic acid post-translational modification targets that mediate its signaling functions, which can be used to study this important chemical signal in biological systems.
Keywords: reactive oxygen species, oxidative stress, molecular imaging, fluorescent probes, posttranslational modifications, bioorthogonal chemistry
INTRODUCTION
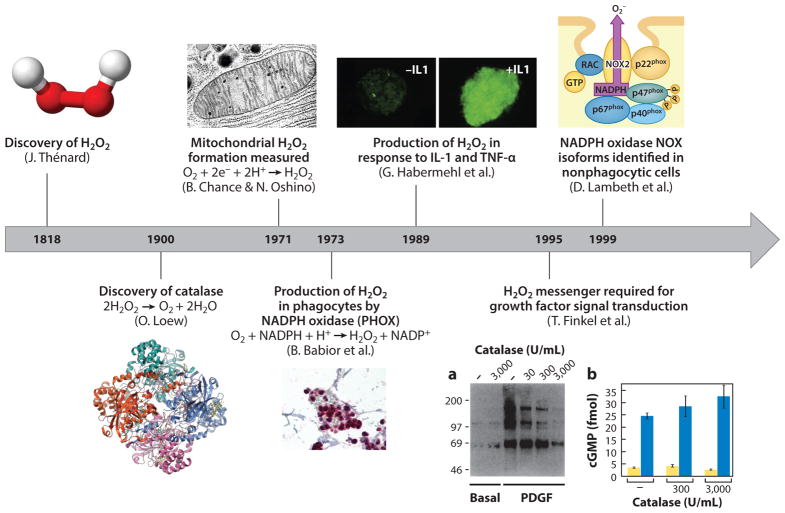
Hydrogen peroxide (H2O2) is widely perceived as a reactive and toxic chemical inimical to the processes of life. It was discovered by Thénard (1) in 1818, after which it was used as a bleaching agent and disinfectant. In 1900, Loew’s (2) discovery of catalase, which efficiently decomposes and thus eliminates this molecule, reinforced the perception of H2O2 as an unwanted toxin. The 1971 observation by Chance & Oshino (3) that the body itself generates H2O2 in the mitochondria was interpreted as an inefficiency of aerobic metabolism, wherein H2O2 was an accidental by-product (4). Even Babior et al.’s (5) discovery of the phagocytic oxidase in 1973, which deliberately produces superoxide (O2−) and its dismutation product H2O2, did not alter this view. The enzyme seemed to be confined to specialized immune cells, in which it produced H2O2 in the interior of the phagosome specifically for its toxic properties in order to destroy engulfed pathogens (5, 6). Indeed, subsequent documentation by Habermehl and colleagues (7) of the production of H2O2 in response to the proinflammatory cytokines interleukin-1 and tumor necrosis factor α reinforced the idea that the biological role of H2O2 was restricted to the immune system. More recently, however, H2O2 has become widely appreciated as a beneficial molecule with more subtle functions than bacterial killing. The Finkel group’s (8) 1995 discovery of deliberate H2O2 production by vascular smooth muscle cells, followed by the detection of widespread enzymatic production of reactive oxygen species (ROS, a term used to describe diverse small oxygen-containing molecules such as O2−, H2O2, hypochlorite, and hydroxyl radical) in 1999 by Lambeth and colleagues (9), has supported the concept that H2O2 is a cellular signaling agent and challenged the idea that its presence is indicative solely of oxidative stress (10). From that point forward, the perceived role of H2O2 in biology has thus progressed from a toxin to an agent of the immune system to a key component of such diverse biological phenomena as the circadian rhythm (11, 12), cell migration (13–15), and stem cell proliferation (Figure 1) (16).
Figure 1.
The perceived role of H2O2 in biology has evolved from toxin to agent of the immune system to a key component of signaling pathways. Abbreviations: cGMP, cyclic guanosine monophosphate; IL, interleukin; PDGF, platelet-derived growth factor; TNF, tumor necrosis factor.
Elucidating the signaling roles of H2O2 in normal physiology and disease has been hampered by the difficulty of detecting H2O2 itself, as well as H2O2-induced modifications, with chemical specificity in complex biological systems. After a brief introduction reprising the major ROS produced by cells and mechanisms of thiol oxidation, we focus on recent progress in chemical approaches for specific detection of H2O2 and different oxidative posttranslational modifications (oxPTMs) of protein cysteine thiols; we emphasize those chemical properties that differentiate one ROS or cysteine modification from another. In keeping with this general theme, we restrict our attention to reagents that have enabled the discovery and understanding of new redox biology within native cellular environments. Along the way, we complement this discussion with examples from the literature that highlight ways in which redox signaling via cysteine oxidation can be used to control protein function and signal transduction pathways.
HYDROGEN PEROXIDE AND REACTIVE OXYGEN SPECIES
Among the diverse collection of cellular ROS, H2O2 stands out as a molecule with ideal signaling properties. It possesses sufficient reactivity to modify potential signaling targets, yet it is also stable enough to diffuse appreciable distances within the cell, reacting with select targets (17). Furthermore, it can be rapidly produced and quickly eliminated, making it ideal for signaling bursts. Average H2O2 concentrations in healthy mammalian cells can range from 1 to 700 nM; higher levels are associated with oxidative stress and cell death (18). However, local H2O2 concentrations can be considerably higher than the average value. The floodgate model of oxidative signaling proposes that rapid increases in local H2O2 can temporarily overwhelm antioxidant defenses, allowing reaction with targets for signaling purposes (19). Thus, whereas high global concentrations of H2O2 cause oxidative stress, brief bursts can be beneficial to cellular physiology. Cellular sources of H2O2 include primarily the mitochondria and the endoplasmic reticulum (ER) (20), as well as various oxidases, including the NADPH oxidase (Nox) proteins (4), amine oxidases (21), acyl-CoA oxidases (22), and cytochrome P450 enzymes (23). Among these sources, the Nox family of proteins appears to be critical for the production of H2O2 for signaling purposes. Unlike other oxidases, the Nox proteins generate O2− as their primary product, which can then disproportionate to H2O2. The founding member of the Nox family is the phagocytic oxidase, which was characterized in the context of immune defense before the discovery of H2O2 as a signaling agent. This enzyme generates ROS within phagocytes in order to damage and eliminate engulfed pathogens. More recently, however, Nox family members were found to be ubiquitously expressed in mammalian tissue types (10). Thus, the widespread expression and deliberate H2O2 production of these enzymes are incompatible with the idea that H2O2 is solely a toxic by-product. Indeed, these facts suggest that H2O2 serves a beneficial function. Combined with the ideal chemical properties of H2O2 as a messenger molecule, the aforementioned observations have motivated the study of Nox-derived H2O2 signaling.
CHEMICAL TOOLS FOR DETECTION OF HYDROGEN PEROXIDE SOURCES
Criteria for Hydrogen Peroxide Probes
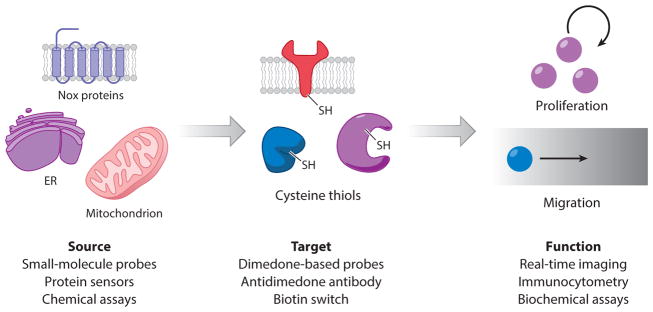
To gain a complete understanding of the function of H2O2, we must have the ability to detect its production at the source, the effects on its immediate target, and the downstream consequences (Figure 2). Novel probes must meet a high bar in terms of their selectivity, kinetics, and photo-physical properties. An ideal chemical probe for H2O2 must be inert to the conditions present in the cell and should not require the addition of other reagents. Furthermore, the probe must be selective against similar analytes, and it should exhibit a turn-on, rather than turn-off, response in order to minimize background. Ideally, a family of probes with varying wavelengths should be available so that their use can be tailored to the specific application and made compatible with probes for different species.
Figure 2.
Strategies for detection of the sources, targets, and downstream consequences of H2O2 signaling. Abbreviations: ER, endoplasmic reticulum; Nox, NADPH oxidase.
Types of Hydrogen Peroxide Detection Technologies
Numerous strategies have been implemented to address the issue of detecting H2O2, including approaches based on proteins (24–27), nanotubes (28), hyperpolarization (29), ultrasound (30), mass spectrometry (MS) (31), and chemiluminescence (32). Of particular interest is a recent innovative enzyme-based detection method utilizing a variant of ascorbate peroxidase (APX). APX expressed intracellularly catalyzes the H2O2-dependent conversion of Amplex Red into a detectable fluorescent product, and this method has been used successfully to detect H2O2 produced as a result of antibiotic treatment in Escherichia coli (27). The potential of the chemiluminescence modality is best demonstrated by the firefly luciferin–based boronate probe PCL1 (Figure 3b), which can be used in a luciferase-expressing mouse to measure H2O2 bursts in live mice (Figure 4a) (33). The next-generation probe PCL2 contains a second reactive trigger to report on simultaneous H2O2 generation and caspase activity in a model of acute inflammation (34). The most promising class of probes consists of small-molecule fluorescent sensors. These sensors do not require additives or particular enzymes, and a wide variety of analogs are accessible to synthetic chemists, permitting the development of a flexible array of tools with differing optical, photophysical, and chemical properties.
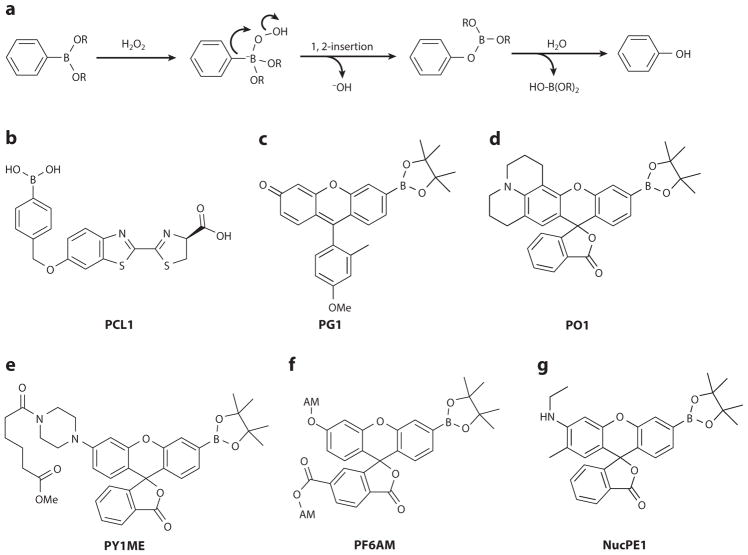
Figure 3.
Aryl boronates are a selective platform for H2O2 detection. (a) General oxidation of an aryl boronate by H2O2. (b–g) Selected small-molecule boronate-based probes for H2O2. Abbreviations: PG1, Peroxy Green 1; PO1, Peroxy Orange 1; NucPE1, Nuclear Peroxy Emerald 1; PF6AM, Peroxyfluor-6 acetoxymethyl ester; PY1ME, Peroxy Yellow 1 Methyl Ester.
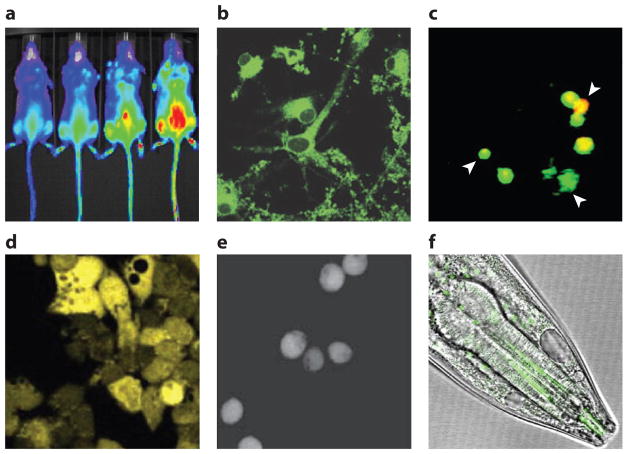
Figure 4.
Applications of small-molecule boronate probes. (a) Response of PCL1 to increasing amounts of H2O2 in live mice (33). (b) PG1 detects H2O2 production by neurons stimulated with epidermal growth factor (51). (c) PO1 and APF detect three distinct types of phagosomes that produce primarily H2O2 (orange), primarily hROS (green), and both (yellow) (43). (d) PF6AM detects H2O2 production by adult hippocampal progenitor cells stimulated with fibroblast growth factor 2 (55). (e) PY1ME detects increased H2O2 uptake by cells expressing aquaporin-3 (16). (f) NucPE1 detects nuclear H2O2 fluxes in Caenorhabditis elegans (64). Abbreviations: APF, aminophenyl fluorescein; hROS, highly reactive oxygen species; NucPE1, Nuclear Peroxy Emerald 1; PF6AM, Peroxyfluor-6 acetoxymethyl ester; PG1, Peroxy Green 1; PO1, Peroxy Orange 1; PY1ME, Peroxy Yellow 1 Methyl Ester. Modified from References 33, 51, 43, 55, 16, and 64.
Early Small-Molecule Fluorescent Probes
The fluorescent probes dihydrodichlorofluorescein (H2DCF) and dihydrodichlorofluorescein di-acetate (H2DCFDA) have been used extensively to detect ROS (35). These probes have been fruitfully employed in biological studies, but their lack of ROS specificity and photoactivation of the unoxidized molecule can confound results (36). Another ROS detection technology of historical interest is the Amplex Red assay (37). This assay is sensitive, but the requirement of the enzyme additive makes it difficult to use within many cell types. In this context, the Chang laboratory (38–41) introduced a useful class of biocompatible small-molecule fluorescent probes that rely on the oxidation of an aryl boronate by H2O2 to produce a phenol (Figure 3a). The boronate moiety masks a phenol, causing the unactivated probe to exhibit significantly lower fluorescence than the oxidized molecule. By appending this chemical modification to various fluorescent scaffolds, one can produce various probe molecules. The boronate oxidation takes advantage of the chemical characteristics of H2O2—specifically, its enhanced, α-effect nucleophilicity and its weak O–O bond (42). The electrophilic boronate undergoes a nucleophilic addition by H2O2, forming a charged tetrahedral boronate complex. The complex then undergoes a 1,2 insertion in which the C–B bond migrates onto a peroxide oxygen molecule, breaking the weak O–O bond and releasing water. Hydrolysis of this intermediate produces boric acid, concomitantly freeing the activated probe.
This oxidation is highly selective for H2O2 over other biologically relevant species, including O2−, hypochlorite, hydroxyl radical, alkyl hydroperoxides, and nitric oxide (43). Although more recent findings have shown that certain boronates also react toward highly reactive species such as peroxynitrite, these molecules are generally produced in much lower quantities and exhibit short biological half-lives compared with that of H2O2 (44–46). In experimental settings in which peroxynitrite or similar highly reactive species are a concern, boronate-based probes must be used with caution in concert with appropriate controls, generally either reducing H2O2 levels with antioxidants or inhibiting its enzymatic production. The first-generation probe Peroxyfluor 1 (PF1) featured dual aryl boronates, which produced a large turn-on response. However, the turn-on kinetics were relatively slow because the probe required two equivalents of H2O2 to produce the maximal turn-on (40). Nevertheless, this probe marked the innovation of a water- and cell-compatible chemistry that was subsequently improved and has allowed the discovery of novel H2O2 signaling processes relating to neuronal, stem, and immune cells (41). In this context, note that this H2O2-sensitive aryl boronate chemistry has been applied toward other goals, such as the use of masked metal chelators (47, 48) and enzyme inhibitors (49) as prodrugs.
Monoboronate Probes for Detecting Endogenous Hydrogen Peroxide Production
The monoboronate probe Peroxy Green 1 (PG1) (Figure 3c) marked a key advance over the first-generation PF1. This asymmetric probe, based on a Tokyo Green platform (50), features a single reactive boronate handle, allowing for much more rapid signal accumulation (51). This probe has been used to directly image the endogenous H2O2 burst of A431 cells upon epidermal growth factor (EGF) stimulation. PG1 has also been validated in primary neuronal culture, a more technically challenging model system, enabling the first direct fluorescence imaging of the neuronal H2O2 burst in response to EGF (Figure 4b). PG1 demonstrates the utility of real-time fluorescence imaging in mapping out cellular signaling pathways. Specifically, owing to the rapid and reliable response of this probe, investigators determined that the neuronal H2O2 burst in response to EGF depends on the EGF receptor (EGFR) kinase domain, phosphatidylinositol 3-kinase (PI3K), Rac1, and Nox. Furthermore, H2O2 production is independent of NO synthase and, by extension, peroxynitrite, as shown by inhibitor controls. This study (51) established these conclusions by sequentially applying pharmacological inhibitors of these various enzymes and monitoring the response of the probe to the H2O2 produced, if any. Thus, it is evident that the boronate probe platform opens up a host of possibilities for elucidating fundamental biological processes, particularly in the realm of cellular signal propagation.
Further derivatives exploit the modularity of the monoboronate platform to expand the available color palette. Access to probes with sensitivity to varying wavelengths of light allows users to monitor two or more analytes concurrently, taking advantage of the power of multichannel imaging experiments. Additional green monoboronate probes include Peroxyfluor 2 (PF2) and Peroxyfluor 3 (PF3), based on the fluorescein scaffold, and the slightly redshifted Peroxy Emerald 1 (PE1), a rhodol derivative. Peroxy Yellow 1 (PY1) is an additional rhodol-based probe with an excitation maximum at 514 nm, and Peroxy Orange 1 (PO1), a rhodol constructed from a julolidine building block (Figure 3d), can be excited at 540 nm (43). The latter probe was used successfully in a dual-channel imaging experiment in RAW264.7 macrophages in combination with aminophenyl fluorescein (APF), a green probe that reacts with highly reactive oxygen species (hROS), which include a subset of ROS, such as peroxynitrite, hypochlorite, and hydroxyl radical, that are more oxidizing relative to H2O2 (52). PO1 exhibits sufficient spectral separation from APF to allow simultaneous imaging of both H2O2 and hROS. With the ability to visualize these two ROS fluxes in the same space at the same time, investigators found that RAW264.7 macrophages contain three types of phagosomes when activated: phagosomes that produce primarily H2O2, those that produce primarily hROS, and those that produce both (Figure 4c) (43). Perhaps more importantly, direct treatment with HOCl did not generate a fluorescence response from the PO1 boronate, demonstrating that these types of probes show selectivity for H2O2 over this ROS in cellular contexts.
Trappable Hydrogen Peroxide Probes
The boronate-based scaffold is also flexible enough to accommodate trapping and targeting groups, allowing for the modulation of the spatial localization of these chemical tools. A trappable probe is one that can freely diffuse across the cellular membrane to enter the cell but then tends to remain inside the cell, enhancing the sensitivity of the probe by increasing its intracellular concentration (53). In practice, this goal can be achieved by masking a carboxylic acid or phenol as an ester. Trappable probes can be removed from the extracellular media, allowing confirmation that the observed signal corresponds to an increase in H2O2 flux within the cell rather than a result of extracellular oxidation.
A first-generation trappable yellow H2O2 probe, Peroxy Yellow 1 Methyl Ester (PY1ME) (Figure 3e), has been used to address an apparent paradox in Nox-derived H2O2 signaling. Nox proteins produce extracellular H2O2; therefore, to play an intracellular signaling role, this Nox-derived H2O2 must cross the plasma membrane. PY1ME was used to test the hypothesis that H2O2 can travel through mammalian cell aquaporins (54). PY1ME features good cellular retention upon washing, and importantly, washing guarantees that the probe signal originates only from within the cells. H2O2 treatment of probe-loaded human embryonic kidney (HEK) 293T cells overexpressing aquaporin-1 (AQP1), AQP3, AQP8, or a control plasmid demonstrates that H2O2 flux into the cell is enhanced over control by AQP3 and AQP8, but not AQP1 (Figure 4d) (55). This result is reasonable given that AQP1 is a classical aquaporin known to transport only water, whereas AQP3 and AQP8 are exemplars of aquaglyceroporins and unorthodox aquaporins, respectively, which are known to transport small solutes in addition to water (54).
The green boronate probe Peroxyfluor-6 acetoxymethyl ester (PF6AM) features two acetoxymethyl (AM) groups, one masking a carboxylic acid and the other masking a phenol (Figure 3f), which are well-known trapping groups (53). These AM groups greatly enhance the accumulation of PF6AM in cells, as well as its sensitivity. This probe has been used to elucidate a beneficial role of H2O2 in the brain. Specifically, exposing adult hippocampal progenitor cells to fibroblast growth factor 2 (FGF-2), which is known to regulate proliferation, results in H2O2 production that is detectable by PF6AM (Figure 4e) (16). The H2O2 produced oxidizes the active-site Cys124 of the protein phosphatase PTEN, converting it into a disulfide with Cys71. The oxidized PTEN is unable to catalyze dephosphorylation (56), most importantly of the kinase Akt, which propagates the FGF-2 signal (57). This H2O2-dependent cascade ultimately maintains normal neuronal stem cell growth and proliferation. Use of PF6AM has also demonstrated that the H2O2 burst in neural stem cells is Nox dependent, as the Nox inhibitor diphenyleneiodonium chloride (DPI) abrogates both the probe’s fluorescence turn-on and the downstream dephosphorylation of Akt. Furthermore, RNA interference knockdown of the specific isoform Nox2 also abolishes the signaling phenotype, which conclusively implicates this protein in the pathway (16). PF6AM has also been fruitfully employed to detect endogenously generated H2O2 in the developing chick lens (58), Nox-dependent H2O2 in stimulated neutrophils (59), and H-RASV12-induced H2O2 production in human fibroblasts (60).
Targetable Hydrogen Peroxide Probes
In addition to trapping strategies, targeting H2O2 probes to specific subcellular regions can enhance their sensitivity and spatial resolution. The mitochondrion is a natural organelle to target with such a strategy, owing to its known O2− and H2O2 production profile. The boronate-based probe MitoPY1 features a triphenylphosphonium moiety for mitochondrial targeting (61). This lipophilic, cationic group allows MitoPY1 to travel down the cellular charge gradient into the mitochondrion (62). MitoPY1 is a yellow probe, and it is sensitive enough to detect paraquat-induced H2O2 in the mitochondria, which presages its utility in elucidating oxidative stress in models of neurodegeneration, such as in Parkinson’s disease. MitoPY1, in conjunction with PF6AM, has also been used to detect Na+-dependent H2O2 increases in mitochondria-rich epithelial cells of the renal medullary thick ascending limb (63). The nucleus is another chemically targetable organelle of interest with respect to H2O2 signaling, and the H2O2-responsive probe Nuclear Peroxy Emerald 1 (NucPE1) (Figure 3g) was determined to be localized to the nucleus. This probe has been used to demonstrate a decrease in nuclear H2O2 flux in Caenorhabditis elegans overexpressing the Sir-2.1 gene, which is implicated in extending the life span of model organisms (Figure 4f) (64). One can target further subcellular locales with H2O2-selective probes by taking advantage of SNAP labeling technology (65). In this paradigm, the enzyme O6-alkylguanine-DNA alkyl-transferase (AGT) is fused to a protein of interest or a localizing sequence and expressed in a relevant biological system. The fluorescent probes SNAP–Peroxy Green 1 (SPG1) and SNAP–Peroxy Green 2 (SPG2) can then be introduced into the cells to tag the arbitrarily localized AGT protein. This strategy has been demonstrated for localization to the plasma membrane, nucleus, mitochondria, and ER (66).
CHEMICAL TOOLS FOR ELUCIDATING HYDROGEN PEROXIDE OXIDATIVE POSTTRANSLATIONAL MODIFICATION TARGETS
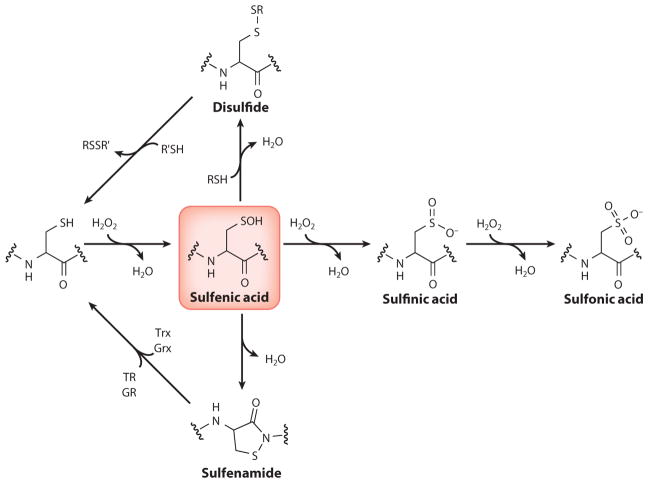
Cysteine is the most sensitive protein side chain to H2O2-mediated oxidation. Cysteine is oxidized via a two-electron nucleophilic substitution that yields a sulfenic acid (a process known as sulfenylation), and this oxidation is implicated in several important biochemical transformations (Figure 5). The propensity of cysteine residues toward oxidation is influenced mainly by three general factors: (a) thiol nucleophilicity, (b) protein microenvironment, and (c) proximity to ROS (67). Second-order rate constants for cysteine oxidation can vary dramatically within proteins, ranging from 20 M/s for protein tyrosine phosphatases (PTPs) to 107 M/s for peroxiredoxin (Prx), and suggest that proximity to their Nox oxidant source may be an important determinant of protein oxidation (68). Oxidized cysteine residues can exist as stable sulfenic acids if they are inaccessible to low-molecular-weight thiols, such as glutathione (GSH), and such stable modifications have been observed in more than 40 proteins (69–71). Depending on the surrounding protein microenvironment and redox conditions, the sulfenic acid may also undergo secondary reactions to yield more stable cysteine oxoforms, such as disulfides, cyclic sulfenamides (72–75), sulfinic acids (RSO2H), and sulfonic acids (RSO3H) (76–78). The condensation of a sulfenic acid to a disulfide or sulfenamide protects against irreversible overoxidation (73, 74), as S–S and S–N bonds can be reduced through the activity of thioredoxin/thioredoxin reductase (Trx/TR) or glutaredoxin/glutaredoxin reductase (Grx/GR) systems (68, 79).
Figure 5.
Oxidative modifications of cysteine residues by H2O2. Abbreviations: GR, glutaredoxin; Grx, glutaredoxin reductase; TR, thioredoxin; Trx, thioredoxin reductase.
Cysteine sulfenylation has emerged as an important oxPTM in the spatial and temporal regulation of protein activity during redox signaling events (80). Sulfenic acids have been identified in the catalytic cycle of multiple enzymes, and the formation of sulfenic acids has also been linked to oxidative stress–induced transcriptional changes in bacteria (81, 82). Less is known about the mechanisms that underlie sulfenic acid–mediated regulation of mammalian protein function and signaling pathways; however, the oxidation of cysteine residues has been associated with an expanding range of biological activities and has been implicated in the regulation of metabolism, immune responses, stem cell biology, pathogenesis of cancers, neurodegeneration, and growth factor signaling pathways (67, 83, 84).
Indirect Methods to Detect Sulfenic Acids
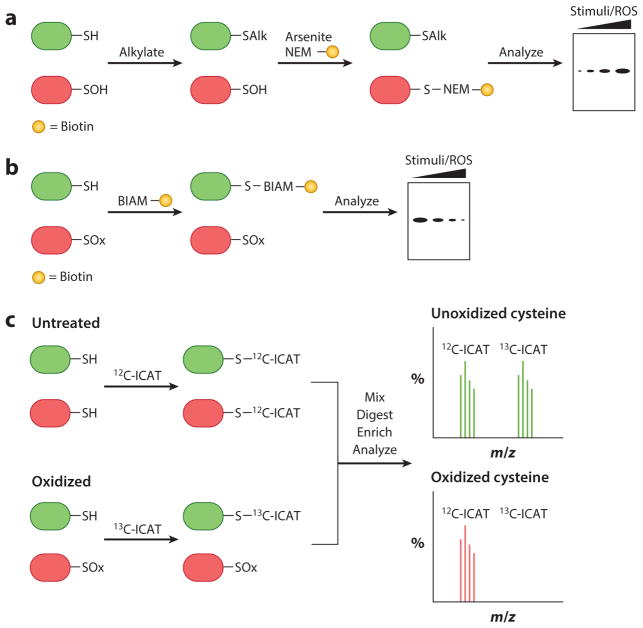
Indirect approaches monitor the reactivity of unoxidized cysteines as a proxy for changes in redox homeostasis. Oxidized cysteines are identified by either the loss of labeling by thiol-modifying reagents or the restoration of labeling after reduction. These indirect approaches block all free thiols with alkylating agents and therefore are typically restricted to analyses of cell lysates or purified proteins. The main indirect chemical method is derived from the biotin switch assay and involves (a) alkylation of free thiols, (b) reduction of sulfenylated cysteines with arsenite, and (c) labeling of nascent thiols with biotinylated N-ethylmaleimide (NEM) or other tagged alkylating agents (Figure 6a). The biotinylated proteins can be enriched and digested for proteomic analysis or imaged by Western blot analysis to assess changes in cysteine oxidation (85, 86). Alternatively, unoxidized cysteines can be alkylated using biotinylated iodoacetamide (BIAM), in which loss of labeling correlates with sulfenylation (Figure 6b). A variation of this approach led to the development of an acid-cleavable BIAM-based isotope-coded affinity tag (ICAT) that allows for the simultaneous identification and quantification of redox-sensitive cysteine thiols (Figure 6c) (87, 88).
Figure 6.
Indirect approaches to detect protein sulfenic acids. (a) Modified biotin switch technique adapted to indirectly detect sulfenic acid–modified proteins (85, 86). (b) Loss of reactivity with thiol-modifying reagents, such as BIAM, indirectly monitors cysteine oxidation. (c) ICAT reagents determine the ratio of oxidized cysteine residues (87, 88). Abbreviations: BIAM, biotinylated iodoacetamide; ICAT, isotope-coded affinity tag; NEM, N-ethylmaleimide; ROS, reactive oxygen species.
Nucleophilic Methods to Directly Detect Sulfenic Acids
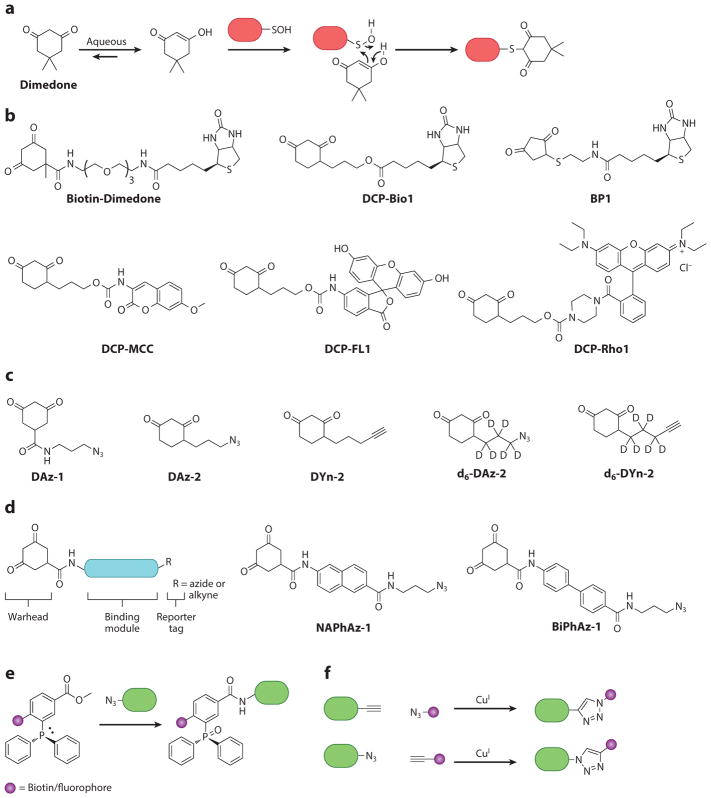
Several methods that take advantage of the electrophilic nature of sulfenic acids have been developed for their direct detection. The selective reaction between dimedone and protein sulfenic acids was first reported in 1974 by Benitez & Allison (89) (Figure 7a). Under aqueous conditions, cyclic 1,3-diketones do not react with thiols, sulfinic acids, or other functional groups commonly found in biomolecules (90). Indeed, dimedone itself can be a powerful tool to identify sulfenic acid modifications through whole-proteome shotgun proteomics (91), and the chemically selective reaction of dimedone has been exploited to detect sulfenylated proteins by MS, fluorescence, and avidin blotting approaches (92).
Figure 7.
Dimedone-based probes for chemoselective detection of protein sulfenic acids. (a) Chemoselective reaction to yield a stable thioether adduct. (b) Dimedone-based probes directly conjugated with biotin or fluorescent tags (90, 93, 94, 100, 101). (c) Dimedone-based probes with bioorthogonal handles for subsequent enrichment or detection of sulfenic acid–modified proteins (71, 101, 108, 114, 116, 118). (d) Redox-based probes that target the redox-sensitive catalytic cysteine in protein tyrosine phosphatases through incorporation of a chemical scaffold with high affinity for the enzyme’s active site (107, 124). (e,f) Bioorthogonal reactions for appending tags to dimedone-based probes with handles (109, 110). Abbreviations: BP1, 4-(ethylthio)cyclopentane-1,3-dione; DCP-FL1, fluoresceinamine-5′-N-[3-(2,4-dioxocyclohexyl)propyl)]carbamate; DCP-MCC, 3-(2,4-dioxocyclohexyl)propyl 7-methoxy-2-oxo-2H-chromen-3-ylcarbamate; DCP-Rho1, rhodamine B [4-[3-(2,4-dioxocyclohexyl)propyl]carbamate]piperazine amide.
Dimedone lacks spectral or affinity tags to aid in subsequent analysis of sulfenylated proteins. The first dimedone-based probes consisted of 1,3-cyclohexadione nucleophiles directly conjugated to fluorophores such as isatoic acid and 7-methoxycoumarin (DCP-MCC) (Figure 7b) (90). A stable protein sulfenic acid derived from H2O2 treatment of a mutant form of the bacterial peroxidase AhpC (C165S) selectively reacted with the fluorophore-functionalized 1,3-cyclohexadione derivatives. Additionally, the compounds did not react toward protein thiols, disulfides, nitrosothiols, sulfinic acids, or sulfonic acids.
Probes that incorporate biotin handles (Biotin-Dimedone and DCP-Bio1) for affinity tag isolation, or fluorophore tags such as fluorescein (DCP-FL1) and rhodamine (DCP-Rho1) for visualization, have also been synthesized (Figure 7b) (93, 94). Uses of these probes to study sulfenylated proteins in cells and in cell lysates include a proteomic analysis of isolated H2O2-perfused rat hearts to understand oxidative stress in cardiac tissues (94); detection of vascular endothelial growth factor (VEGF)-stimulated oxidation of proteins to elucidate VEGF receptor signaling and angiogenesis (95, 96); and detection of sulfenylated PTPs and protein tyrosine kinases (PTKs) during cell signaling (97–99). Furthermore, 1,3-diketone nucleophiles that deviate from the quintessential dimedone scaffold, such as 4-(ethylthio)cylopentane-1,3-dione (BP1) (100) and β-ketoesters (101), have also been reported to act as chemoselective nucleophiles toward sulfenylated proteins (Figure 7b).
Direct conjugation of a probe to biotin or a fluorophore can have detrimental effects on the efficiency of labeling. Commonly, tagged derivatives suffer from diminished cell uptake and trafficking properties (102–105). As a consequence, sulfenylated proteins often need to be labeled from cell lysate. However, the redox balance of the cell is disrupted by lysis (106), leading to artificial oxidation, which can both increase the probability of observing false positives and overoxidize sulfenylated proteins. Additionally, biotin- or fluorophore-tagged probes display decreased binding efficiency and biased protein labeling due to the increased steric bulk of the probe (104, 107).
An alternative approach for the detection of sulfenylated proteins within cells utilizes small, membrane-permeable dimedone derivatives bearing either an alkyne or azide handle (Figure 7c). These smaller probes alleviate issues surrounding diffusion across the cell membrane (Figure 7c). DAz-1 consists of a 1,3-cyclohexanedione nucleophile functionalized with an azide handle through an amide bond at the 5 position (108). Following covalent modification of sulfenylated proteins, biotin or fluorophore tags can be conjugated onto the azide via Staudinger ligation (Figure 7e) or Huisgen [3 + 2] cycloaddition (click chemistry) (Figure 7f) (109, 110). DAz-1 is selective for sulfenylated proteins in their purified form and in living cells (71). Initial studies with DAz-1 revealed differences in protein labeling patterns between cell lysates and live cells, highlighting the importance of probing intact cells to investigate the redox regulation of proteins. DAz-1 was also used to study the importance of sulfenylation of a resolving cysteine in the thiol peroxidase (Gpx3), which senses oxidative stress in yeast (111), and to identify a unique reducing system in the bacterial periplasm that protects single cysteine residues from oxidation (112).
Subsequent dimedone-based probes, DAz-2 and DYn-2, directly incorporate an azide or alkyne handle at the 4 position of the warhead and show enhanced nucleophilicity over DAz-1 (Figure 7c) (113). DAz-2 was used to profile the global sulfenome in human tumor cell lines and uncovered more than 175 new sulfenylated proteins, which are distributed throughout the cell and function in a diverse array of biological processes (114). DYn-2 incorporates an alkyne handle because several studies indicate that alkynyl-chemical reporters, in combination with azido-detection tags, offer superior sensitivity relative to the reverse combination (105, 115). Recently, DYn-2 was used in global profiling studies to reveal dynamic protein sulfenylation during EGF signaling in human epidermal A431 cells and determined that a key residue (Cys797) of the EGF receptor kinase is susceptible to sulfenylation and that it regulates receptor kinase activity. Additionally, several PTPs were found to be sulfenylated in response to EGF stimulation, and the sensitivity of each protein toward oxidation correlated with its proximity to the oxidant source (Nox). EGFR is the most sensitive to oxidation because it forms a complex with Nox2 (116). Therefore, it is surmised that the extent of sulfenylation of redox-sensitive proteins depends on their proximity to the Nox oxidant source and is likely to be distinct for varying cell signaling pathways and cell type–specific phenomena.
Immunological Methods to Detect Sulfenic Acids
Immunological methodologies may offer enhanced selectivity and sensitivity for detecting sulfenylated proteins from a complex proteome. To this end, an antibody that is specific for the dimedone–cysteine thioether adduct epitope has been developed and applied to a protein microarray to monitor differences in sulfenic acid modification across various cancer cell lines (103). Additionally, the dimedone antibody has been used to demonstrate cysteine sulfenylation during EGF signaling and colocalization of the oxidized proteins with Nox2 (116). A similar antibody has been employed to detect dimedone-modified glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and to study GAPDH sulfenylation in cardiac myocytes exposed to exogenous H2O2 (117).
Methods to Quantify Sulfenic Acids
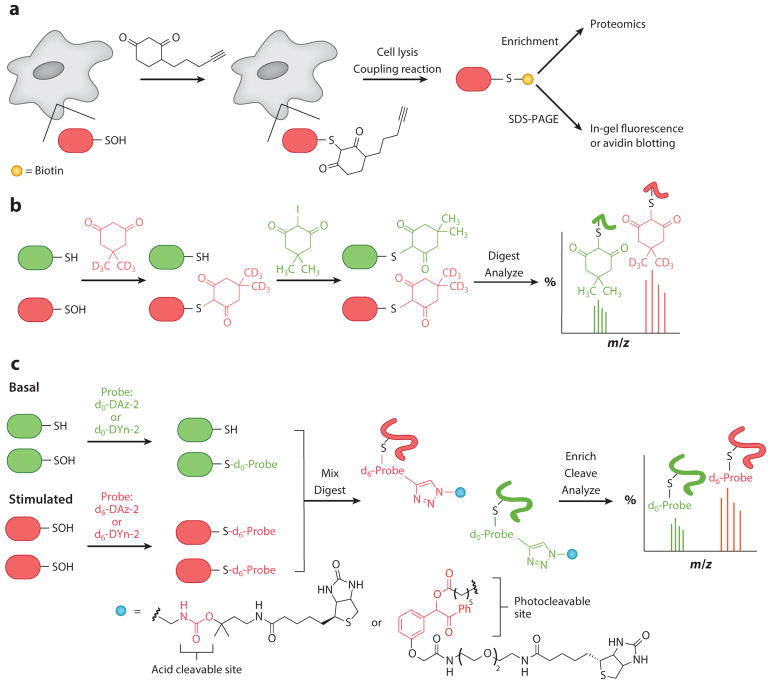
In addition to detection, identification, and mapping of sulfenylated proteins, dimedone-based probes can be used to quantify redox-dependent changes associated with physiological and pathological processes. The isotope-coded d6-dimedone and 2-iododimedone (ICDID) strategy was developed for this purpose. The key feature of the ICDID strategy is that the pair of probes generate nearly chemically identical proteins that differ only in a 6-deuterium label. The ICDID method consists of the following steps: (a) Sulfenylated proteins are derivatized with d6-dimedone, (b) excess reagent is removed and free thiols are labeled with iododimedone, (c) protein samples are digested, and (d) the resulting peptides are separated and analyzed by liquid chromatography–mass spectrometry (LC-MS) (Figure 8b). The extent of sulfenylation of a particular cysteine residue can be determined by dividing the heavy-isotope-labeled peak by the sum of the heavy-and light-isotope-labeled peaks (118).
Figure 8.
Methods to directly detect protein sulfenic acids. (a) Direct in situ labeling of sulfenylated proteins using cell-permeable chemoselective reagents. (b) Isotope-coded dimedone 2-iododimedone allows for quantification of sulfenylated proteins (118). (c) Use of ACLs and isotopically labeled DAz-2 to facilitate enrichment and quantification of sulfenylated proteins (119). Abbreviations: ACL, acid-cleavable linker; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Alternatively, a set of isotopically light and heavy versions of DAz-2 were developed to monitor relative changes in protein sulfenylation (119). A key feature of the isotopically labeled DAz-2 is that it facilitates mapping and quantifying the specific site of cysteine oxidation. The use of biotin tags for enrichment and purification of tagged protein sulfenic acids can give rise to complicated tandem mass spectrometry (MS/MS) spectra due to fragmentation of the biotin handle, making database searching challenging. Therefore, it would be ideal to remove the biotin tag after enrichment of the labeled proteins or peptides, and various cleavable linkers have been reported for this purpose. The isotopically labeled DAz-2 derivatives can be used in combination with a trifluoroacetic acid–cleavable linker to facilitate enrichment, elution, detection, and quantification of sulfenylated proteins (Figure 8c) (119). Additionally, β-ketoesters functionalized with an alkyne handle have been reported and evaluated as NH2OH-cleavable probes for the detection of sulfenylated proteins (101), although we note that linear diketones have been reported to cross-react with amines such as lysine (120). Recently, a highly efficient chemoproteomics work flow that utilizes isotopically light and heavy versions of DYn-2 (Figure 7c) was used to identify ~1,000 sulfenylated sites on more than 700 different proteins in cells (121). Moreover, the work flow allowed for quantification of changes in cellular sulfenylation during growth factor stimulation and identified a novel site of oxidation on SIRT6, a histone deacetylase, revealing an unexpected role of this protein as a redox sensor and transducer.
Protein-Specific Methods to Detect Sulfenic Acids
Although dimedone-based probes are broadly useful in investigations of protein cysteine oxidation, the low abundance of specific signaling proteins has hindered their detection in traditional dimedone-based approaches. Reversible PTP oxidation is an important cellular regulatory mechanism (122), and due to the lack of sensitive and robust methods for detecting oxidized PTPs (123), investigators have developed a direct method to detect oxidized phosphatases in cells to facilitate investigations of PTP redox cycling during cell signaling events. These redox-based probes (RBPs) consist of three components: (a) a cyclic 1,3-diketone nucleophile that reacts with oxidized active-site cysteine; (b) a module that directs binding to the PTP target; and (c) a reporter tag used for the identification, purification, or direct visualization of the probe-labeled proteins (Figure 7d) (107). The RBPs exhibited enhanced binding and detection of the sulfenylated PTPs compared with the parent compounds lacking the binding module. More recently, RBPs bearing an alkyne handle were used to investigate insulin-induced H2O2 production and ROS-mediated oxidative inactivation of PTP1B (124), revealing a physiological role of redox regulation during insulin signaling. It seems likely that the RBP methodology can be applied to other phosphatases to facilitate a better understanding of their role in redox biology.
Immunochemical approaches have also been developed to detect specific proteins prone to oxidation. One approach utilizes an antibody that recognizes the conserved catalytic motif found in all PTPs in conjunction with an antibody that recognizes the terminally hyperoxidized catalytic cysteine (125). This indirect method has been used for the global proteomic assessment of the PTP “redoxome” (126). Importantly, this method relies on an alkylate–reduce–oxidize approach, similar to the alkylate–reduce–alkylate modified biotin switch approach mentioned above (Figure 6a), so it cannot differentiate phosphatases regulated by reversible oxidation from those that are inherently hyperoxidized. Another approach utilizes an antibody consisting of a single-chain variable fragment (ScFv) that has been generated to directly and selectively detect the unique structural conformation adopted by the oxidized PTP1B once the oxidized active-site cysteine condenses to the cyclic sulfenamide (127). These conformation-sensing antibodies trapped PTP1B in the inactive conformation, permitting sustained insulin signaling in HEK cells.
As phosphatases are viable therapeutic targets, in the broader sense the development of protein-targeted probes has unveiled the possibility of developing a new class of lead compounds for the development of alternative therapeutics to ameliorate diseases associated with aberrant phosphatase activity. Indeed, trapping of an oxidized phosphatase by way of small molecules bearing a nucleophilic center may be a practical means to inhibit further catalytic activity and offers an out-of-the-box approach for the design and development of a new class of therapeutics. This concept can be further adapted to other classes of proteins that contain a redox-sensitive cysteine, such as EGFR (128–130).
Electrophilic Probes for Detecting Sulfenic Acids
The sulfur atom of the sulfenic acid acts as both an electrophile and a weak nucleophile. Due to this duality of cysteine sulfenic acids, electrophilic compounds can react with the oxidized cysteine residue. 7-Chloro-4-nitrobenzo-2-oxa-1,3-dizole (NBD-Cl) (131) has been used to detect sulfenylation of several proteins, including AhpC peroxidase, NADH peroxidase, OhrR repressor, PTPs, recombinant human α1-antitrypsin, and human serum albumin (HSA) (70, 131–134). Strained cycloalkynes such as bicyclo[6.1.0]nonyne (BCN) can detect sulfenylated proteins in vitro and may be used to detect sulfenylated proteins in vivo, although alternative reaction pathways may interfere because cysteine thiols are highly susceptible to thiol–yne addition and, in the presence of ROS, may lead to formation of a doubly conjugated by-product (135–139). Additionally, arylboronic acids and cyclic benzoxaboroles have been implemented for the reversible detection of sulfenylated proteins (140). Because boronic acids are susceptible to oxidation and can react with H2O2, these probes may not be suitable for the detection of sulfenylated proteins during redox signaling events.
Direct Methods to Detect Sulfinic and Sulfonic Acids
Sulfenic acids can be oxidized to sulfinic acids in the presence of high concentrations of ROS, a process termed sulfinylation. Recent studies have implicated sulfinylation as a regulator of biological function. The formation and function of the cysteine sulfinic acid have been extensively characterized in Prx and have been proposed to regulate signaling events (19). The discovery of the sulfinic acid reductase sulfiredoxin (Srx), an ATP-dependent protein that specifically reduces the sulfinic acid in Prx, led to a paradigm shift in the understanding of protein regulation by H2O2, whereby sulfinic acids can serve a regulatory function analogous to that of a disulfide or sulfenic acid (141). Sulfinic acid has also been observed in several other proteins, including nitrile hydratase, matrilysin, and the Parkinson’s disease protein DJ-1 (142–144). Current methods to detect protein sulfinic acids are relatively few, but include (a) a mass increase of 32 Da, (b) acidic electrophoretic gel shifts, and (c) antibodies that recognize a sulfinic or sulfonic acid peptide from a specific protein (145–147). Such approaches facilitate studies of sulfinic acids in individual proteins but have limited utility in global analyses.
An alternative approach is to design a probe in which the product of the reaction with the sulfinic acid is uniquely stable. Along these lines, researchers have studied aryl nitroso compounds for their unique reaction with sulfinic acids (148). The initial reaction to form a sulfinic acid–derived N-sulfonyl hydroxylamine product is reversible, but the intermediate can be trapped by ester-functionalized aryl nitroso compounds to yield an irreversible N-sulfonylbenziosoxazolone adduct with excellent first-order reaction rates (7.86 min−1) in the formation of cyclic product (Figure 9a). The aryl nitroso compounds are chemoselective because reaction with a thiol yields a sulfonamide species that can be cleaved with nucleophiles. Additionally, the aryl nitroso compounds do not cross-react with other sulfur- and non-sulfur-containing biological functional groups. This novel selective ligation reaction may enable the detection of protein sulfinylation in biological systems.
Figure 9.
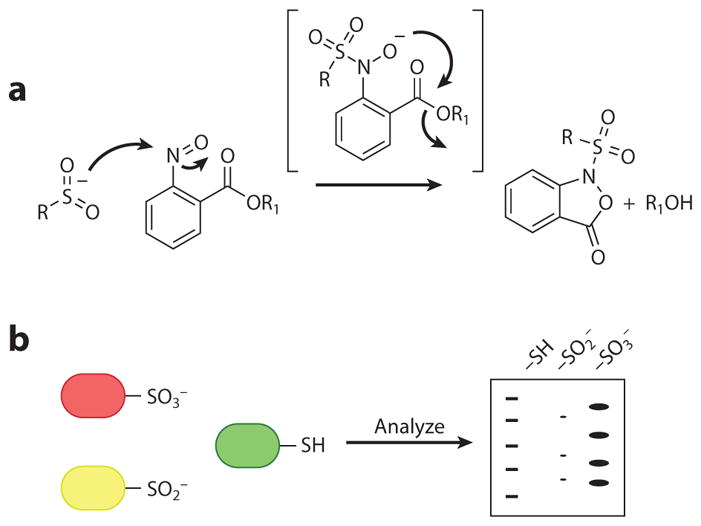
Detection of sulfinic acid– and sulfonic acid–modified proteins. (a) Reaction of a sulfinic acid with an aryl nitroso compound to yield a stable cyclic sulfonamide analog (148). (b) Antibodies that recognize the sulfonic acid–modified proteins (146).
In the presence of strong oxidizing reagents, sulfinic acids can be further oxidized to sulfonic acids, which represent the highest oxidation state of the cysteine sulfur atom. The sulfonic acid modification has been characterized in a small group of proteins, including mammalian copper/zinc superoxide dismutase (Cu/Zn-SOD), in which damage resulting from hyperoxidation may play a role in diseases such as familial amyotrophic lateral sclerosis (149). Also, the sulfonic acid modification may be used to target proteins for degradation (150). Another study proposed that sulfonic acid modification of Prx can enhance its proposed chaperone activity (151). Elucidation of the sulfonic acid modification in biological and pathological states has been hindered by a lack of tools that can trap and tag the sulfonic acid modification on proteins. The use of antibodies to detect sulfonic acid is the most common technique for detection in specific proteins (Figure 9b) (146). However, the antibodies cannot be used to detect sulfonic acids in a whole proteome, and they cannot clearly differentiate the sulfinic from the sulfonic acids (152). Polyarginine (PA)-coated nanodiamonds have been used to selectively enrich sulfonic acid–modified peptides. BSA, used as a model system in this study, was oxidized with performic acid and digested; then sulfonic acid–containing peptides were enriched and eluted from PA-coated nanodiamonds with phosphoric acid, enabling the identification of oxidized peptides by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) (152).
OUTLOOK
As the perceived role of H2O2 in biology has evolved from toxin to immune agent to signaling agent, so too has the chemical toolbox for studying ROS expanded in breadth and depth. Fluorescence and bioluminescence imaging offer attractive modalities for visualizing the sources and generation of H2O2 and for exploring the role of this potentially harmful molecule in both pathology and physiology. Dimedone-based probes have shown great utility in the identification of the cysteine targets of H2O2-derived oxPTMs. However, the understanding of the importance of cysteine oxidation and of the roles it plays in redox biology continues to expand, and so must the chemical toolbox for elucidation of oxidative modifications in biological settings. The cellular lifetime of sulfenic acids has been hypothesized to be relatively short, and recent studies from the Carroll group (113) have shown that the reaction between dimedone and a sulfenic acid is relatively slow. Thus, it is likely that many sulfenylated proteins remain undetected due to incomplete trapping of this oxPTM by dimedone-based probes. We need to expand our search toward novel nucleophiles that show enhanced reactivity and selectivity toward sulfenic acid. Moreover, the emergence of protein sulfinylation as a cell signaling mechanism motivates the development of robust tools and new approaches that can identify protein targets that are susceptible to sulfinylation. The widespread ability of different tissues to produce H2O2, and its relevance to numerous biological pathways and systems, presages further integration of this enigmatic ROS in the processes of life.
SUMMARY POINTS.
Nox-derived H2O2 is an important signaling agent in biological systems, and H2O2-mediated oxidation of cysteines to cysteine sulfenic acids appears to play a key role in many signaling pathways.
The selective reaction between H2O2 and aryl boronates has allowed the design of many aryl boronate–based probes, which offer a means to visualize H2O2 fluxes with high spatial and temporal resolution in living systems when used with appropriate controls.
Based on the specific application, aryl boronate probes with differing excitation/emission wavelengths and subcellular localization can be selected for H2O2 visualization.
Dimedone-based probes offer a way to visualize and identify the cysteine targets of H2O2 oxidation. Many variants allow cysteine sulfenic acid detection by fluorescence or Western blot.
Dimedone-based probes have been developed for proteomics applications, in which sulfenic acid–containing proteins can be identified and the extent of sulfenylation can be quantified.
FUTURE ISSUES.
Whereas the sensitivity of current aryl boronate–based probes can detect endogenous H2O2, it is likely that many physiological H2O2 fluxes remain below their detection limit. Detecting very small amounts of H2O2 is an important goal for future methodologies.
Quantification of H2O2 fluxes remains a difficult task with currently available small-molecule methodologies. This challenge will require the development of reversible, ratiometric sensors based on different chemical reactions than the ones currently employed.
Although dimedone-based probes have demonstrated broad utility in identifying sulfenylated proteins, their relatively slow reactivity may hinder the detection of low-abundance or transiently sulfenylated proteins. The development of new probes with faster reaction kinetics will be an important goal for the future.
Current methodologies for the detection of cysteine sulfinic acids and sulfonic acids have significant limitations. The development of new, broadly applicable detection methodologies will enhance our understanding of the scope of these oxPTMs, as well as the role(s) they play.
Acknowledgments
This work was supported by the National Institutes of Health (NIH) (GM79465 to C.J.C. and R01-GM102187 and R01-CA174864 to K.S.C.). C.J.C. is an Investigator with the Howard Hughes Medical Institute. T.F.B. and C.S.O. were partially supported by a Chemical Biology Interface Training Grant from the NIH (T32-GM066698), and C.S.O. thanks the National Science Foundation for a graduate fellowship.
Glossary
- ROS
reactive oxygen species
- oxPTM
oxidative posttranslational modification
- Nox
NADPH oxidase
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- PTP
protein tyrosine phosphatase
- Prx
peroxiredoxin
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Thénard LJ. Observations sur des combinaisons nouvelles entre l’oxigène et divers acides. Ann Chim Phys. 1818;8:306–13. [Google Scholar]
- 2.Loew O. A new enzyme of general occurrence in organisms. Science. 1900;11:701–2. doi: 10.1126/science.11.279.701. [DOI] [PubMed] [Google Scholar]
- 3.Chance B, Oshino N. Kinetics and mechanisms of catalase in peroxisomes of the mitochondrial fraction. Biochem J. 1971;122:225–33. doi: 10.1042/bj1220225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 5.Babior BM, Kipnes RS, Curnutte JT. Biological defense mechanisms—production by leukocytes of superoxide, a potential bactericidal agent. J Clin Investig. 1973;52:741–44. doi: 10.1172/JCI107236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossi F, Zatti M. Biochemical aspects of phagocytosis in polymorphonuclear leucocytes. NADH and NADPH oxidation by the granules of resting and phagocytizing cells. Cell Mol Life Sci. 1964;20:21–23. doi: 10.1007/BF02146019. [DOI] [PubMed] [Google Scholar]
- 7.Radeke HH, Meier B, Topley N, Flöge J, Habermehl GG, Resch K. Interleukin 1α and tumor necrosis factor α induce oxygen radical production in mesangial cells. Kidney Int. 1990;37:767–75. doi: 10.1038/ki.1990.44. [DOI] [PubMed] [Google Scholar]
- 8.Sundaresan M, Yu Z-X, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–99. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 9.Suh Y-A, Arnold RS, Lassegue B, Shi J, Xu X, et al. Cell transformation by the superoxide-generating oxidase Mox1. Nature. 1999;401:79–82. doi: 10.1038/43459. [DOI] [PubMed] [Google Scholar]
- 10.Bedard K, Krause K-H. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87:245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 11.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, et al. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–58. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J-S, Huang TY, Bokoch GM. Reactive oxygen species regulate a slingshot-cofilin activation pathway. Mol Biol Cell. 2009;20:2650–60. doi: 10.1091/mbc.E09-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gianni D, Taulet N, DerMardirossian C, Bokoch GM. c-Src-mediated phosphorylation of NoxA1 and Tks4 induces the reactive oxygen species (ROS)–dependent formation of functional invadopodia in human colon cancer cells. Mol Biol Cell. 2010;21:4287–98. doi: 10.1091/mbc.E10-08-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–99. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickinson BC, Peltier J, Stone D, Schaffer DV, Chang CJ. Nox2 redox signaling maintains essential cell populations in the brain. Nat Chem Biol. 2011;7:106–12. doi: 10.1038/nchembio.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–24. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 18.Stone JR, Yang S. Hydrogen peroxide: a signaling messenger. Antioxid Redox Signal. 2006;8:243–70. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 19.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–53. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 20.Araki K, Inaba K. Structure, mechanism, and evolution of Ero1 family enzymes. Antioxid Redox Signal. 2012;16:790–99. doi: 10.1089/ars.2011.4418. [DOI] [PubMed] [Google Scholar]
- 21.Dawkes HC, Phillips SEV. Copper amine oxidase: cunning cofactor and controversial copper. Curr Opin Struct Biol. 2001;11:666–73. doi: 10.1016/s0959-440x(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 22.Kim J-JP, Miura R. Acyl-CoA dehydrogenases and acyl-CoA oxidases. Eur J Biochem. 2004;271:483–93. doi: 10.1046/j.1432-1033.2003.03948.x. [DOI] [PubMed] [Google Scholar]
- 23.Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford, UK: Oxford Univ. Press; 1999. [Google Scholar]
- 24.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, et al. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–86. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 25.Markvicheva KN, Bilan DS, Mishina NM, Gorokhovatsky AY, Vinokurov LM, et al. A genetically encoded sensor for H2O2 with expanded dynamic range. Bioorg Med Chem. 2011;19:1079–84. doi: 10.1016/j.bmc.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 26.Bilan DS, Pase L, Joosen L, Gorokhovatsky AY, Ermakova YG, et al. HyPer-3: a genetically encoded H2O2 probe with improved performance for ratiometric and fluorescence lifetime imaging. Am Chem Soc Chem Biol. 2012;8:535–42. doi: 10.1021/cb300625g. [DOI] [PubMed] [Google Scholar]
- 27.Dwyer DJ, Belenky PA, Yang JH, MacDonald IC, Martell JD, et al. Antiobiotics induce redox-related physiological alterations as part of their lethality. PNAS. 2014;111:e2100–9. doi: 10.1073/pnas.1401876111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Huang Y, Gu A, Wang G, Fang B, Wu H. Hydrogen peroxide sensor based on carbon nanotubes/β-Ni(OH)2 nanocomposites. Chin J Chem. 2012;30:501–6. [Google Scholar]
- 29.Lippert AR, Keshari KR, Kurhanewicz J, Chang CJ. A hydrogen peroxide–responsive hyperpolarized 13C MRI contrast agent. J Am Chem Soc. 2011;133:3776–79. doi: 10.1021/ja111589a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson ES, Orozco J, Wu Z, Malone CD, Yi B, et al. Toward in vivo detection of hydrogen peroxide with ultrasound molecular imaging. Biomaterials. 2013;34:8918–24. doi: 10.1016/j.biomaterials.2013.06.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cochemé HM, Quin C, McQuaker SJ, Cabreiro F, Logan A, et al. Measurement of H2O2 within living Drosophila during aging using a ratiometric mass spectrometry probe targeted to the mitochondrial matrix. Cell Metab. 2011;13:340–50. doi: 10.1016/j.cmet.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee D, Khaja S, Velasquez-Castano JC, Dasari M, Sun C, et al. In vivo imaging of hydrogen peroxide with chemiluminescent nanoparticles. Nat Mater. 2007;6:765–69. doi: 10.1038/nmat1983. [DOI] [PubMed] [Google Scholar]
- 33.Van de Bittner GC, Dubikovskaya EA, Bertozzi CR, Chang CJ. In vivo imaging of hydrogen peroxide production in a murine tumor model with a chemoselective bioluminescent reporter. PNAS. 2010;107:21316–21. doi: 10.1073/pnas.1012864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van de Bittner GC, Bertozzi CR, Chang CJ. Strategy for dual-analyte luciferin imaging: in vivo bioluminescence detection of hydrogen peroxide and caspase activity in a murine model of acute inflammation. J Am Chem Soc. 2013;135:1783–95. doi: 10.1021/ja309078t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cathcart R, Schwiers E, Ames BN. Detection of picomole levels of hydroperoxides using a fluorescent dichlorofluorescein assay. Anal Biochem. 1983;134:111–16. doi: 10.1016/0003-2697(83)90270-1. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Zhong Z, Xu Z, Chen L, Wang Y. 2′,7′,-Dichlorodihydrofluorescein as a fluorescent probe for reactive oxygen species measurement: forty years of application and controversy. Free Radic Res. 2010;44:587–604. doi: 10.3109/10715761003709802. [DOI] [PubMed] [Google Scholar]
- 37.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal Biochem. 1997;253:162–68. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 38.Kuivila HG, Wiles RA. Electrophilic displacement reactions. VII Catalysis by chelating agents in the reaction between hydrogen peroxide and benzeneboronic acid. J Am Chem Soc. 1955;77:4830–34. [Google Scholar]
- 39.Kuivila HG, Armour AG. Electrophilic displacement reactions. IX Effects of substituents on rates of reactions between hydrogen peroxide and benzeneboronic acid. J Am Chem Soc. 1957;79:5659–62. [Google Scholar]
- 40.Chang MC, Pralle A, Isacoff EY, Chang CJ. A selective, cell-permeable optical probe for hydrogen peroxide in living cells. J Am Chem Soc. 2004;126:15392–93. doi: 10.1021/ja0441716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lippert AR, Van de Bittner GC, Chang CJ. Boronate oxidation as a bioorthogonal reaction approach for studying the chemistry of hydrogen peroxide in living systems. Acc Chem Res. 2011;44:793–804. doi: 10.1021/ar200126t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jencks WP, Carriuolo J. Reactivity of nucleophilic reagents toward esters. J Am Chem Soc. 1960;82:1778–86. [Google Scholar]
- 43.Dickinson BC, Huynh C, Chang CJ. A palette of fluorescent probes with varying emission colors for imaging hydrogen peroxide signaling in living cells. J Am Chem Soc. 2010;132:5906–15. doi: 10.1021/ja1014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sikora A, Zielonka J, Lopez M, Joseph J, Kalyanaraman B. Direct oxidation of boronates by peroxynitrite: mechanism and implications in fluorescence imaging of peroxynitrite. Free Radic Biol Med. 2009;47:1401–7. doi: 10.1016/j.freeradbiomed.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zielonka J, Sikora A, Hardy M, Joseph J, Dranka BP, Kalyanaraman B. Boronate probes as diagnostic tools for real time monitoring of peroxynitrite and hydroperoxides. Chem Res Toxicol. 2012;25:1793–99. doi: 10.1021/tx300164j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun X, Xu Q, Kim G, Flower SE, Lowe JP, et al. A water-soluble boronate-based fluorescent probe for the selective detection of peroxynitrite and imaging in living cells. Chem Sci. 2014;5:3368–73. [Google Scholar]
- 47.Charkoudian LK, Pham DM, Franz KJ. A prochelator triggered by hydrogen peroxide inhibits iron-promoted hydroxyl radical formation. J Am Chem Soc. 2006;128:12424–25. doi: 10.1021/ja064806w. [DOI] [PubMed] [Google Scholar]
- 48.Wei Y, Guo M. Hydrogen peroxide triggered prochelator activation, subsequent metal chelation, and attenuation of the Fenton reaction. Angew Chem Int Ed. 2007;46:4722–25. doi: 10.1002/anie.200604859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Major Jourden JL, Cohen SM. Hydrogen peroxide activated matrix metalloproteinase inhibitors: a prodrug approach. Angew Chem Int Ed. 2010;49:6795–97. doi: 10.1002/anie.201003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urano Y, Kamiya M, Kanda K, Ueno T, Hirose K, Nagano T. Evolution of fluorescein as a platform for finely tunable fluorescence probes. J Am Chem Soc. 2005;127:4888–94. doi: 10.1021/ja043919h. [DOI] [PubMed] [Google Scholar]
- 51.Miller EW, Tulyathan O, Isacoff EY, Chang CJ. Molecular imaging of hydrogen peroxide produced for cell signaling. Nat Chem Biol. 2007;3:263–67. doi: 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- 52.Setsukinai K-I, Urano Y, Kakinuma K, Majima HJ, Nagano T. Development of novel fluorescence probes that can reliably detect reactive oxygen species and distinguish specific species. J Biol Chem. 2003;278:3170–75. doi: 10.1074/jbc.M209264200. [DOI] [PubMed] [Google Scholar]
- 53.Tsien RY. A non-disruptive technique for loading calcium buffers and indicators into cells. Nature. 1981;290:527–28. doi: 10.1038/290527a0. [DOI] [PubMed] [Google Scholar]
- 54.Rojek A, Praetorius J, Frøkiaer J, Nielsen S, Fenton RA. A current view of the mammalian aquaglyceroporins. Annu Rev Physiol. 2008;70:301–27. doi: 10.1146/annurev.physiol.70.113006.100452. [DOI] [PubMed] [Google Scholar]
- 55.Miller EW, Dickinson BC, Chang CJ. Aquaporin-3 mediates hydrogen peroxide uptake to regulate downstream intracellular signaling. PNAS. 2010;107:15681–86. doi: 10.1073/pnas.1005776107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kwon J, Lee S-R, Yang K-S, Ahn Y, Kim YJ, et al. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. PNAS. 2004;101:16419–24. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peltier J, O’Neill A, Schaffer DV. PI3K/Akt and CREB regulate adult neural hippocampal progenitor proliferation and differentiation. Dev Neurobiol. 2007;67:1348–61. doi: 10.1002/dneu.20506. [DOI] [PubMed] [Google Scholar]
- 58.Basu S, Rajakaruna S, Dickinson BC, Chang CJ, Menko AS. Endogenous hydrogen peroxide production in the epithelium of the developing embryonic lens. Mol Vis. 2014;20:458–67. [PMC free article] [PubMed] [Google Scholar]
- 59.Sakai J, Li J, Subramanian KK, Mondal S, Bajrami B, et al. Reactive oxygen species–induced actin glutathionylation controls actin dynamics in neutrophils. Immunity. 2012;37:1037–49. doi: 10.1016/j.immuni.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang M, Haase AD, Huang F-K, Coulis G, Rivera KD, et al. Dephosphorylation of tyrosine 393 in argonaute 2 by protein tyrosine phosphatase 1B regulates gene silencing in oncogenic RAS-induced senescence. Mol Cell. 2014;55:782–90. doi: 10.1016/j.molcel.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dickinson BC, Chang CJ. A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J Am Chem Soc. 2008;130:9638–39. doi: 10.1021/ja802355u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ross MF, Kelso GF, Blaikie FH, James AM, Cochemé HM, et al. Lipophilic triphenylphosphonium cations as tools in mitochondrial bioenergetics and free radical biology. Biochemistry. 2005;70:222–30. doi: 10.1007/s10541-005-0104-5. [DOI] [PubMed] [Google Scholar]
- 63.Ohsaki Y, O’Connor P, Mori T, Ryan RP, Dickinson BC, et al. Increase of sodium delivery stimulates the mitochondrial respiratory chain H2O2 production in rat renal medullary thick ascending limb. Am J Physiol Ren Physiol. 2011;302:95–102. doi: 10.1152/ajprenal.00469.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dickinson BC, Tang Y, Chang Z, Chang CJ. A nuclear-localized fluorescent hydrogen peroxide probe for monitoring sirtuin-mediated oxidative stress responses in vivo. Chem Biol. 2011;18:943–48. doi: 10.1016/j.chembiol.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juillerat A, Gronemeyer T, Keppler A, Gendreizig S, Pick H, et al. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem Biol. 2003;10:313–17. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 66.Srikun D, Albers AE, Nam CI, Iavarone AT, Chang CJ. Organelle-targetable fluorescent probes for imaging hydrogen peroxide in living cells via SNAP-tag protein labeling. J Am Chem Soc. 2010;132:4455–65. doi: 10.1021/ja100117u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lo Conte M, Carroll KS. The redox biochemistry of protein sulfenylation and sulfinylation. J Biol Chem. 2013;288:26480–88. doi: 10.1074/jbc.R113.467738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–61. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 69.Salsbury FR, Jr, Knutson ST, Poole LB, Fetrow JS. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carballal S, Alvarez B, Turell L, Botti H, Freeman BA, Radi R. Sulfenic acid in human serum albumin. Amino Acids. 2007;32:543–51. doi: 10.1007/s00726-006-0430-y. [DOI] [PubMed] [Google Scholar]
- 71.Reddie KG, Seo YH, Muse WB, III, Leonard SE, Carroll KS. A chemical approach for detecting sulfenic acid–modified proteins in living cells. Mol Biosyst. 2008;4:521–31. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. PNAS. 2007;104:8743–48. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–73. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 74.van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–77. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 75.Yang J, Groen A, Lemeer S, Jans A, Slijper M, et al. Reversible oxidation of the membrane distal domain of receptor PTPα is mediated by a cyclic sulfenamide. Biochemistry. 2007;46:709–19. doi: 10.1021/bi061546m. [DOI] [PubMed] [Google Scholar]
- 76.Hugo M, Turell L, Manta B, Botti H, Monteiro G, et al. Thiol and sulfenic acid oxidation of AhpE, the one-cysteine peroxiredoxin from Mycobacterium tuberculosis: kinetics, acidity constants, and conformational dynamics. Biochemistry. 2009;48:9416–26. doi: 10.1021/bi901221s. [DOI] [PubMed] [Google Scholar]
- 77.Sohn J, Rudolph J. Catalytic and chemical competence of regulation of Cdc25 phosphatase by oxidation/reduction. Biochemistry. 2003;42:10060–70. doi: 10.1021/bi0345081. [DOI] [PubMed] [Google Scholar]
- 78.Turell L, Botti H, Carballal S, Ferrer-Sueta G, Souza JM, et al. Reactivity of sulfenic acid in human serum albumin. Biochemistry. 2008;47:358–67. doi: 10.1021/bi701520y. [DOI] [PubMed] [Google Scholar]
- 79.Berndt C, Lillig CH, Holmgren A. Thiol-based mechanisms of the thioredoxin and glutare-doxin systems: implications for diseases in the cardiovascular system. Am J Physiol Heart Circ Physiol. 2007;292:1227–36. doi: 10.1152/ajpheart.01162.2006. [DOI] [PubMed] [Google Scholar]
- 80.Finkel T. Signal transduction by reactive oxygen species. J Cell Biol. 2011;194:7–15. doi: 10.1083/jcb.201102095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paulsen CE, Carroll KS. Cysteine-mediated redox signaling: chemistry, biology, and tools for discovery. Chem Rev. 2013;113:4633–79. doi: 10.1021/cr300163e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paulsen CE, Carroll KS. Orchestrating redox signaling networks through regulatory cysteine switches. Am Chem Soc Chem Biol. 2010;5:47–62. doi: 10.1021/cb900258z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–21. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 84.Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Curr Biol. 2014;24:R453–62. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marino SM, Li Y, Fomenko DE, Agisheva N, Cerny RL, Gladyshev VN. Characterization of surface-exposed reactive cysteine residues in Saccharomyces cerevisiae. Biochemistry. 2010;49:7709–21. doi: 10.1021/bi100677a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boivin B, Tonks NK. Analysis of the redox regulation of protein tyrosine phosphatase superfamily members utilizing a cysteinyl-labeling assay. Methods Enzymol. 2010;474:35–50. doi: 10.1016/S0076-6879(10)74003-9. [DOI] [PubMed] [Google Scholar]
- 87.Sethuraman M, McComb ME, Huang H, Huang S, Heibeck T, et al. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J Proteome Res. 2004;3:1228–33. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- 88.Sethuraman M, Clavreul N, Huang H, McComb ME, Costello CE, Cohen RA. Quantification of oxidative posttranslational modifications of cysteine thiols of p21ras associated with redox modulation of activity using isotope-coded affinity tags and mass spectrometry. Free Radic Biol Med. 2007;42:823–29. doi: 10.1016/j.freeradbiomed.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Benitez LV, Allison WS. The inactivation of the acyl phosphatase activity catalyzed by the sulfenic acid form of glyceraldehyde 3-phosphate dehydrogenase by dimedone and olefins. J Biol Chem. 1974;249:6234–43. [PubMed] [Google Scholar]
- 90.Poole LB, Zeng BB, Knaggs SA, Yakubu M, King SB. Synthesis of chemical probes to map sulfenic acid modifications on proteins. Bioconjug Chem. 2005;16:1624–28. doi: 10.1021/bc050257s. [DOI] [PubMed] [Google Scholar]
- 91.Martinez-Acedo P, Gupta V, Carroll KS. Proteomic analysis of peptides tagged with dimedone and related probes. J Mass Spectrom. 2014;49:257–65. doi: 10.1002/jms.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leonard SE, Carroll KS. Chemical ‘omics’ approaches for understanding protein cysteine oxidation in biology. Curr Opin Chem Biol. 2011;15:88–102. doi: 10.1016/j.cbpa.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 93.Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, et al. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjug Chem. 2007;18:2004–17. doi: 10.1021/bc700257a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Charles RL, Schröder E, May G, Free P, Gaffney PR, et al. Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics. 2007;6:1473–84. doi: 10.1074/mcp.M700065-MCP200. [DOI] [PubMed] [Google Scholar]
- 95.Oshikawa J, Urao N, Kim HW, Kaplan N, Razvi M, et al. Extracellular SOD-derived H2O2 promotes VEGF signaling in caveolae/lipid rafts and post-ischemic angiogenesis in mice. PLOS ONE. 2010;5:e10189. doi: 10.1371/journal.pone.0010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kaplan N, Urao N, Furuta E, Kim SJ, Razvi M, et al. Localized cysteine sulfenic acid formation by vascular endothelial growth factor: role in endothelial cell migration and angiogenesis. Free Radic Res. 2011;45:1124–35. doi: 10.3109/10715762.2011.602073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Michalek RD, Nelson KJ, Holbrook BC, Yi JS, Stridiron D, et al. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J Immunol. 2007;179:6456–67. doi: 10.4049/jimmunol.179.10.6456. [DOI] [PubMed] [Google Scholar]
- 98.Wani R, Qian J, Yin L, Bechtold E, King SB, et al. Isoform-specific regulation of Akt by PDGF-induced reactive oxygen species. PNAS. 2011;108:10550–55. doi: 10.1073/pnas.1011665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klomsiri C, Rogers LC, Soito L, McCauley AK, King SB, et al. Endosomal H2O2 production leads to localized cysteine sulfenic acid formation on proteins during lysophosphatidic acid–mediated cell signaling. Free Radic Biol Med. 2014;71:49–60. doi: 10.1016/j.freeradbiomed.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qian J, Klomsiri C, Wright MW, King SB, Tsang AW, et al. Simple synthesis of 1,3-cyclopentanedione derived probes for labeling sulfenic acid proteins. Chem Commun. 2011;47:9203–5. doi: 10.1039/c1cc12127h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Qian J, Wani R, Klomsiri C, Poole LB, Tsang AW, Furdui CM. A simple and effective strategy for labeling cysteine sulfenic acid in proteins by utilization of β-ketoesters as cleavable probes. Chem Commun. 2012;48:4091–93. doi: 10.1039/c2cc17868k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cohen MS, Hadjivassiliou H, Taunton J. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat Chem Biol. 2007;3:156–60. doi: 10.1038/nchembio859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Seo YH, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid–specific antibodies. PNAS. 2009;106:16163–68. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hang HC, Loureiro J, Spooner E, van der Velden AW, Kim YM, et al. Mechanism-based probe for the analysis of cathepsin cysteine proteases in living cells. Am Chem Soc Chem Biol. 2006;1:713–23. doi: 10.1021/cb600431a. [DOI] [PubMed] [Google Scholar]
- 105.Speers AE, Cravatt BF. Profiling enzyme activities in vivo using click chemistry methods. Chem Biol. 2004;11:535–46. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 106.Go YM, Jones DP. Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta. 2008;1780:1273–90. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Leonard SE, Garcia FJ, Goodsell DS, Carroll KS. Redox-based probes for protein tyrosine phosphatases. Angew Chem. 2011;50:4423–27. doi: 10.1002/anie.201007871. [DOI] [PubMed] [Google Scholar]
- 108.Seo YH, Carroll KS. Facile synthesis and biological evaluation of a cell-permeable probe to detect redox-regulated proteins. Bioorg Med Chem Lett. 2009;19:356–59. doi: 10.1016/j.bmcl.2008.11.073. [DOI] [PubMed] [Google Scholar]
- 109.Patterson DM, Nazarova LA, Prescher JA. Finding the right (bioorthogonal) chemistry. Am Chem Soc Chem Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]
- 110.Truong TH, Carroll KS. Bioorthogonal chemical reporters for analyzing protein sulfenylation in cells. Curr Protoc Chem Biol. 2012;4:101–22. [Google Scholar]
- 111.Paulsen CE, Carroll KS. Chemical dissection of an essential redox switch in yeast. Chem Biol. 2009;16:217–25. doi: 10.1016/j.chembiol.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 112.Depuydt M, Leonard SE, Vertommen D, Denoncin K, Morsomme P, et al. A periplasmic reducing system protects single cysteine residues from oxidation. Science. 2009;326:1109–11. doi: 10.1126/science.1179557. [DOI] [PubMed] [Google Scholar]
- 113.Gupta V, Carroll KS. Sulfenic acid chemistry, detection and cellular lifetime. Biochim Biophys Acta. 2014;1840:847–75. doi: 10.1016/j.bbagen.2013.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. Am Chem Soc Chem Biol. 2009;4:783–99. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 115.Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, et al. Robust fluorescent detection of protein fatty-acylation with chemical reporters. J Am Chem Soc. 2009;131:4967–75. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- 116.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, et al. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nat Chem Biol. 2012;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Maller C, Schröder E, Eaton P. Glyceraldehyde 3-phosphate dehydrogenase is unlikely to mediate hydrogen peroxide signaling: studies with a novel anti-dimedone sulfenic acid antibody. Antioxid Redox Signal. 2011;14:49–60. doi: 10.1089/ars.2010.3149. [DOI] [PubMed] [Google Scholar]
- 118.Seo YH, Carroll KS. Quantification of protein sulfenic acid modifications using isotope-coded dimedone and iododimedone. Angew Chem. 2011;50:1342–45. doi: 10.1002/anie.201007175. [DOI] [PubMed] [Google Scholar]
- 119.Truong TH, Garcia FJ, Seo YH, Carroll KS. Isotope-coded chemical reporter and acid-cleavable affinity reagents for monitoring protein sulfenic acids. Bioorg Med Chem Lett. 2011;21:5015–20. doi: 10.1016/j.bmcl.2011.04.115. [DOI] [PubMed] [Google Scholar]
- 120.Zhu X, Tanaka F, Lerner RA, Barbas CF, 3rd, Wilson IA. Direct observation of an enamine intermediate in amine catalysis. J Am Chem Soc. 2009;131:18206–7. doi: 10.1021/ja907271a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yang J, Gupta V, Carroll KS, Liebler DC. Site-specific mapping and quantification of protein S-sulphenylation in cells. Nat Commun. 2014;5:4776. doi: 10.1038/ncomms5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tanner JJ, Parsons ZD, Cummings AH, Zhou H, Gates KS. Redox regulation of protein tyrosine phosphatases: structural and chemical aspects. Antioxid Redox Signal. 2011;15:77–97. doi: 10.1089/ars.2010.3611. [DOI] [PubMed] [Google Scholar]
- 123.Karisch R, Neel BG. Methods to monitor classical protein tyrosine phosphatase oxidation. FEBS J. 2013;280:459–75. doi: 10.1111/j.1742-4658.2012.08626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Garcia FJ, Carroll KS. Redox-based probes as tools to monitor oxidized protein tyrosine phosphatases in living cells. Eur J Med Chem. 2014;88:28–33. doi: 10.1016/j.ejmech.2014.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Persson C, Sjöblom T, Groen A, Kappert K, Engström U, et al. Preferential oxidation of the second phosphatase domain of receptor-like PTP-α revealed by an antibody against oxidized protein tyrosine phosphatases. PNAS. 2004;101:1886–91. doi: 10.1073/pnas.0304403101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Karisch R, Fernandez M, Taylor P, Virtanen C, St-Germain JR, et al. Global proteomic assessment of the classical protein tyrosine phosphatome and “redoxome”. Cell. 2011;146:826–40. doi: 10.1016/j.cell.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Haque A, Andersen JN, Salmeen A, Barford D, Tonks NK. Conformation-sensing antibodies stabilize the oxidized form of PTP1B and inhibit its phosphatase activity. Cell. 2011;147:185–98. doi: 10.1016/j.cell.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Truong TH, Carroll KS. Redox regulation of epidermal growth factor receptor signaling through cysteine oxidation. Biochemistry. 2012;51:9954–65. doi: 10.1021/bi301441e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Truong TH, Carroll KS. Redox regulation of protein kinases. Crit Rev Biochem Mol Biol. 2013;48:332–56. doi: 10.3109/10409238.2013.790873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schwartz PA, Kuzmic P, Solowiej J, Bergqvist S, Bolanos B, et al. Covalent EGFR inhibitor analysis reveals importance of reversible interactions to potency and mechanisms of drug resistance. PNAS. 2014;111:173–78. doi: 10.1073/pnas.1313733111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ellis HR, Poole LB. Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry. 1997;36:15013–18. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
- 132.Denu JM, Tanner KG. Specific and reversible inactivation of protein tyrosine phosphatases by hydrogen peroxide: evidence for a sulfenic acid intermediate and implications for redox regulation. Biochemistry. 1998;37:5633–42. doi: 10.1021/bi973035t. [DOI] [PubMed] [Google Scholar]
- 133.Fuangthong M, Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine–sulfenic acid derivative. PNAS. 2002;99:6690–95. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Griffiths SW, King J, Cooney CL. The reactivity and oxidation pathway of cysteine 232 in recombinant human α1-antitrypsin. J Biol Chem. 2002;277:25486–92. doi: 10.1074/jbc.M203089200. [DOI] [PubMed] [Google Scholar]
- 135.Poole TH, Reisz JA, Zhao W, Poole LB, Furdui CM, King SB. Strained cycloalkynes as new protein sulfenic acid traps. J Am Chem Soc. 2014;136:6167–70. doi: 10.1021/ja500364r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.van Geel R, Pruijn GJ, van Delft FL, Boelens WC. Preventing thiol–yne addition improves the specificity of strain-promoted azide–alkyne cycloaddition. Bioconjug Chem. 2012;23:392–98. doi: 10.1021/bc200365k. [DOI] [PubMed] [Google Scholar]
- 137.Beatty KE, Fisk JD, Smart BP, Lu YY, Szychowski J, et al. Live-cell imaging of cellular proteins by a strain-promoted azide–alkyne cycloaddition. ChemBioChem. 2010;11:2092–95. doi: 10.1002/cbic.201000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lo Conte M, Staderini S, Marra A, Sanchez-Navarro M, Davis BG, Dondoni A. Multi-molecule reaction of serum albumin can occur through thiol–yne coupling. Chem Commun. 2011;47:11086–88. doi: 10.1039/c1cc14402b. [DOI] [PubMed] [Google Scholar]
- 139.Kim EJ, Kang DW, Leucke HF, Bond MR, Ghosh S, et al. Optimizing the selectivity of DIFO-based reagents for intracellular bioorthogonal applications. Carbohydr Res. 2013;377:18–27. doi: 10.1016/j.carres.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu CT, Benkovic SJ. Capturing a sulfenic acid with arylboronic acids and benzoxaborole. J Am Chem Soc. 2013;135:14544–47. doi: 10.1021/ja407628a. [DOI] [PubMed] [Google Scholar]
- 141.Jonsson TJ, Johnson LC, Lowther WT. Structure of the sulphiredoxin–peroxiredoxin complex reveals an essential repair embrace. Nature. 2008;451:98–101. doi: 10.1038/nature06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Murakami T, Nojiri M, Nakayama H, Odaka M, Yohda M, et al. Post-translational modification is essential for catalytic activity of nitrile hydratase. Protein Sci. 2000;9:1024–30. doi: 10.1110/ps.9.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fu X, Kassim SY, Parks WC, Heinecke JW. Hypochlorous acid oxygenates the cysteine switch domain of promatrilysin (MMP-7). A mechanism for matrix metalloproteinase activation and atherosclerotic plaque rupture by myeloperoxidase. J Biol Chem. 2001;276:41279–87. doi: 10.1074/jbc.M106958200. [DOI] [PubMed] [Google Scholar]
- 144.Blackinton J, Lakshminarasimhan M, Thomas KJ, Ahmad R, Greggio E, et al. Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1. J Biol Chem. 2009;284:6476–85. doi: 10.1074/jbc.M806599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 146.Woo HA, Kang SW, Kim HK, Yang KS, Chae HZ, Rhee SG. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J Biol Chem. 2003;278:47361–64. doi: 10.1074/jbc.C300428200. [DOI] [PubMed] [Google Scholar]
- 147.Hamann M, Zhang T, Hendrich S, Thomas JA. Quantitation of protein sulfinic and sulfonic acid, irreversibly oxidized protein cysteine sites in cellular proteins. Methods Enzymol. 2002;348:146–56. doi: 10.1016/s0076-6879(02)48634-x. [DOI] [PubMed] [Google Scholar]
- 148.Lo Conte M, Carroll KS. Chemoselective ligation of sulfinic acids with aryl-nitroso compounds. Angew Chem. 2012;51:6502–5. doi: 10.1002/anie.201201812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fujiwara N, Nakano M, Kato S, Yoshihara D, Ookawara T, et al. Oxidative modification to cysteine sulfonic acid of Cys111 in human copper–zinc superoxide dismutase. J Biol Chem. 2007;282:35933–44. doi: 10.1074/jbc.M702941200. [DOI] [PubMed] [Google Scholar]
- 150.Tasaki T, Kwon YT. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci. 2007;32:520–28. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 151.Lim JC, Choi HI, Park YS, Nam HW, Woo HA, et al. Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J Biol Chem. 2008;283:28873–80. doi: 10.1074/jbc.M804087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chang YC, Huang CN, Lin CH, Chang HC, Wu CC. Mapping protein cysteine sulfonic acid modifications with specific enrichment and mass spectrometry: an integrated approach to explore the cysteine oxidation. Proteomics. 2010;10:2961–71. doi: 10.1002/pmic.200900850. [DOI] [PubMed] [Google Scholar]