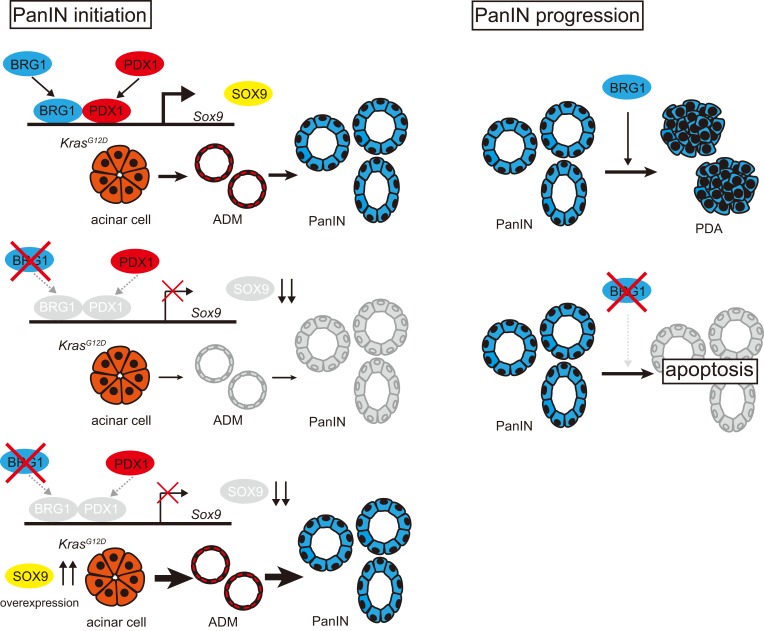
Abstract
Chromatin remodeler Brahma related gene 1 (BRG1) is silenced in approximately 10% of human pancreatic ductal adenocarcinomas (PDAs). We previously showed that BRG1 inhibits the formation of intraductal pancreatic mucinous neoplasm (IPMN) and that IPMN-derived PDA originated from ductal cells. However, the role of BRG1 in pancreatic intraepithelial neoplasia–derived (PanIN-derived) PDA that originated from acinar cells remains elusive. Here, we found that exclusive elimination of Brg1 in acinar cells of Ptf1a-CreER; KrasG12D; Brg1fl/fl mice impaired the formation of acinar-to-ductal metaplasia (ADM) and PanIN independently of p53 mutation, while PDA formation was inhibited in the presence of p53 mutation. BRG1 bound to regions of the Sox9 promoter to regulate its expression and was critical for recruitment of upstream regulators, including PDX1, to the Sox9 promoter and enhancer in acinar cells. SOX9 expression was downregulated in BRG1-depleted ADMs/PanINs. Notably, Sox9 overexpression canceled this PanIN-attenuated phenotype in KBC mice. Furthermore, Brg1 deletion in established PanIN by using a dual recombinase system resulted in regression of the lesions in mice. Finally, BRG1 expression correlated with SOX9 expression in human PDAs. In summary, BRG1 is critical for PanIN initiation and progression through positive regulation of SOX9. Thus, the BRG1/SOX9 axis is a potential target for PanIN-derived PDA.
Keywords: Gastroenterology, Oncology
Keywords: Cancer, Epigenetics, Mouse models
Introduction
Pancreatic ductal adenocarcinoma (PDA) carries one of the most dismal prognoses of all human malignancies (1). To identify strategies for diagnosing and treating PDA during the preinvasive stages, it is essential to improve our understanding of the molecular pathogenesis leading to PDA initiation and progression. PDA is thought to arise from 3 premalignant lesions, including pancreatic intraepithelial neoplasia (PanIN), intraductal pancreatic mucinous neoplasm (IPMN), and mucinous cystic neoplasm (MCN) (2), with the majority of PDAs thought to arise from PanIN (1). The occurrence of oncogenic KRAS mutations is the earliest genetic event in PDA pathogenesis, and these mutations are present in over 90% of all human PanIN1A/B (3). The significance of oncogenic KRAS in PDA initiation and progression has been proven using genetically engineered mouse models (4, 5). Furthermore, lineage-tracing studies using transgenic mice have demonstrated that pancreatic acinar cells possessing the KrasG12D mutation lose their acinar differentiation status and acquire a duct-like phenotype in a process called acinar-to-ductal metaplasia (ADM) (6–9). ADM is thought to evolve into PanIN lesions and eventually progress into invasive PDA. Thus, ADM is considered to be the initial morphological change in PanIN-derived PDA formation.
Recent global genomic studies revealed that human PDAs have mutations in 10 core signaling pathways (10). The SWI/SNF chromatin remodeling complex, which is part of a family of complexes that permit DNA-protein contacts to regulate gene expression, is one of these pathways. Approximately 14% of all human PDAs have inactivating mutations in components of SWI/SNF chromatin remodeling complexes (10). Brahma related gene 1 (BRG1, human SMARCA4) is 1 of 2 catalytic ATPase subunits of the SWI/SNF-chromatin remodeling complexes, and a recent study demonstrated that BRG1 inactivation occurs in approximately 10% of human PDAs (11). Recently, we focused on the role of BRG1 in pancreatic tumorigenesis and showed that the pancreas-specific loss of Brg1 in the presence of oncogenic Kras results in the formation of cystic neoplastic lesions that resemble human IPMN capable of progressing to PDA (12). Moreover, pancreatic duct–specific loss of Brg1 in the presence of oncogenic Kras results in IPMN formation, showing that IPMN is derived from pancreatic ductal cells. In contrast, not only does loss of Brg1 in pancreatic acinar cells in the presence of oncogenic Kras prevent IPMN formation, it also reduces spontaneous PanIN formation. Therefore, BRG1 appears to possess cell type–specific roles in KRAS-driven pancreatic tumorigenesis: inhibition of IPMN formation from ductal cells and promotion of PanIN formation from acinar cells (12). Furthermore, we recently showed that BRG1 suppresses IPMN formation by inhibiting the dedifferentiation of ductal cells, whereas BRG1 promotes tumorigenesis in full-blown IPMN-PDA by supporting a mesenchymal-like transcriptional landscape (13). However, the precise role of BRG1 in the formation of acinar cell–derived PanIN and PanIN-derived PDA is not fully understood.
Here, we investigate the contribution of BRG1 to the formation of PanIN and PanIN-derived PDA. For this purpose, we used genetically engineered mouse models and ex vivo acinar cell culture experiments. We provide evidence that BRG1 plays a critical role in acinar cell–derived Kras-induced ADM and PanIN formation through regulation of Sox9 expression in mice. Furthermore, we showed that BRG1 is critical for maintenance of established PanIN by using an inducible dual recombinase system in mice. In summary, our data highlight cell type–specific, context-dependent roles for BRG1 in the initiation and progression of PDA.
Results
Acinar cell–specific ablation of Brg1 drastically attenuates KrasG12D-driven spontaneous ADM and PanIN formation.
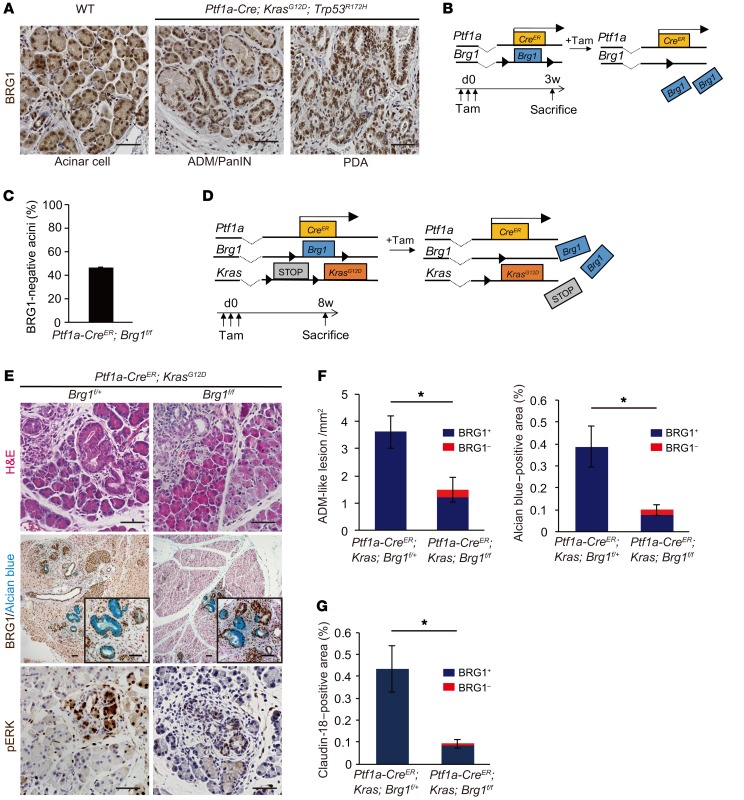
We first ascertained the expression pattern for BRG1 in all the lineages of mouse PanIN-derived PDAs. Immunohistochemical analysis revealed that BRG1 was expressed in adult pancreatic acinar cells in WT mice and in the ADMs, PanINs, and PDAs of Ptf1a-Cre; KrasG12D; Trp53R172H mice, an established model for PDA in which 1 allele of the tumor suppressor p53 is mutated through Cre recombination in pancreatic epithelial cells in parallel with expression of oncogenic KrasG12D (Figure 1A).
Figure 1. Acinar-specific ablation of Brg1 attenuates oncogenic KRAS-driven ADM and PanIN formation.
(A) Immunohistochemistry for BRG1 in adult Ptf1a-Cre; KrasG12D; Trp53R172H mice. Scale bars: 50 μm. (B) The genetic strategy for determining the efficiency of acinar cell–specific Brg1 deletion following tamoxifen (Tam) induction and the experimental design for tamoxifen administration and analysis. (C) Deletion rate of BRG1 in Ptf1a-CreER; Brg1fl/fl mice at 3 weeks after tamoxifen administration. n = 3 mice. Data are shown as mean ± SEM. (D) The genetic strategy used to delete Brg1 and activate oncogenic Kras in adult pancreatic acinar cells and the experimental design for tamoxifen administration and analysis. (E) H&E staining and immunohistochemistry for BRG1 with Alcian blue and phospho-ERK staining in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice with littermate controls. Scale bars: 50 μm. (F) Quantification of Alcian blue–negative ADM-like lesions and Alcian blue–positive late ADMs and PanINs in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice with littermate controls. Red bars show incidence of BRG1-negative late ADMs and PanINs. n = 3–4 mice per genotype. Data are shown as mean ± SEM. *P < 0.05, Student’s t test. (G) Quantification of PanINs in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice with littermate controls. Claudin-18–positive area was counted. Red bars show incidence of BRG1-negative PanINs. n = 4 mice per genotype. Data are shown as mean ± SEM. *P < 0.05, Student’s t test.
A previous study showed that acinar-specific loss of Brg1 in an oncogenic Kras background reduces spontaneous PanIN formation (12). To confirm this and to investigate whether Brg1 deletion affects acinar cell–derived ADM formation, we generated Ptf1a-CreER; KrasG12D; Brg1fl/fl mice. In these animals, Brg1 is deleted and oncogenic Kras is expressed exclusively in adult pancreatic acinar cells upon tamoxifen induction of Cre activity. To determine the efficiency of Brg1 deletion following tamoxifen induction, we quantified the number of BRG1-deficient acinar cells in Ptf1a-CreER; Brg1fl/fl mice 3 weeks after subcutaneous tamoxifen injection (Figure 1B) and found that BRG1 protein expression was lost in 45% of all pancreatic acinar cells via quantification of immunohistochemical analysis (Figure 1C). Next, we assessed ADM and PanIN formation 8 weeks after 3 consecutive subcutaneous tamoxifen injections in 4-week-old Ptf1a-CreER; KrasG12D; Brg1fl/fl mice and littermate control Ptf1a-CreER; KrasG12D; Brg1fl/+ mice (Figure 1D). We used both Brg1fl/+ and Brg1+/+ mice as controls because we previously had not observed any differences in histology and overall survival between Ptf1a-Cre; KrasG12D; Brg1fl/+ and Ptf1a-Cre; KrasG12D; Brg1+/+ mice (12). We examined ADM and PanIN formation by H&E and Alcian blue staining, respectively, to assess intestinal mucin production (Figure 1E). Phosphorylated ERK (phospho-ERK), one of the downstream targets of KRAS, was detected by immunohistochemistry to confirm Kras activation (Figure 1E). The number of Alcian blue–negative ADM-like lesions and the areas of Alcian blue–positive staining of late ADMs and PanINs were significantly reduced in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice compared with those in Ptf1a-CreER; KrasG12D; Brg1fl/+ mice (Figure 1, E and F). Importantly, costaining for BRG1 and Alcian blue revealed that most of the ADMs and PanINs that developed in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice had escaped Brg1 recombination and thus retained BRG1 expression (Figure 1E). Indeed, BRG1-negative ADM-like lesions and Alcian blue–positive areas constituted only 18.5% and 22% of the total ADM-like lesions and Alcian blue–positive areas, respectively (Figure 1F). Furthermore, we performed staining for claudin-18 as a PanIN-specific marker. We found that claudin-18–positive PanINs were significantly reduced in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice compared with those in Ptf1a-CreER; KrasG12D; Brg1fl/+ mice (Figure 1G). Most of the claudin-18–positive PanINs in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice were BRG1 positive, and the grade of all of the few BRG1-negative PanINs was PanIN1. These data indicate that the BRG1-positive ADMs and PanINs that developed in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice arose from acinar cells, which had recombined the KrasG12D allele, but had escaped Brg1 deletion. Therefore, acinar cell–specific ablation of Brg1 drastically attenuated KRAS-driven spontaneous ADM and PanIN formation at both stages, indicating that BRG1 is critical for acinar cell–derived spontaneous KRAS-driven ADM and PanIN formation in mice.
Brg1 is required for PanIN formation after pancreatic injury in the presence of mutant Kras.
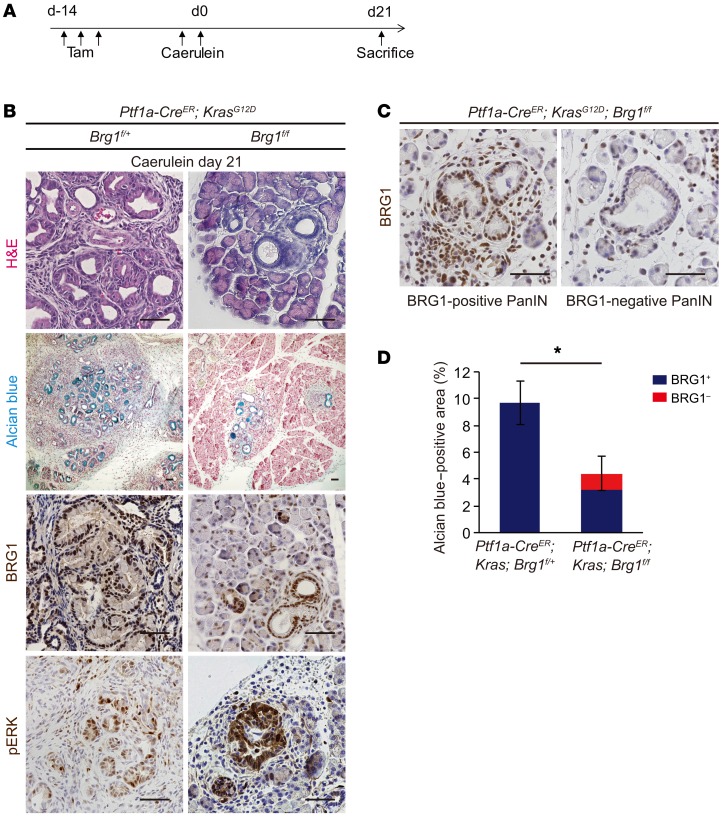
Chronic pancreatitis is known to be a risk factor in humans for developing PDA (1). Previous studies have demonstrated that caerulein-induced acute or chronic pancreatitis significantly accelerates PanIN formation in mutant Kras backgrounds (6, 14). Therefore, we tested to determine whether Brg1 deletion also affects PanIN formation in the context of acute pancreatitis. We induced caerulein-mediated acute pancreatitis 2 weeks after tamoxifen injection in 4-week-old Ptf1a-CreER; KrasG12D; Brg1fl/fl mice with the littermate control Ptf1a-CreER; KrasG12D; Brg1fl/+ mice. PanIN formation was assessed 21 days after caerulein injection (Figure 2A). We identified PanIN formation using H&E and Alcian blue staining (Figure 2B). BRG1 and phospho-ERK immunostainings confirmed BRG1 deletion and Kras activation, respectively (Figure 2B). We found that the areas of Alcian blue–positive late ADMs and PanINs decreased significantly in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice 21 days after caerulein-induced pancreatitis compared with those in Ptf1a-CreER; KrasG12D; Brg1fl/+ mice (Figure 2, B and D). Furthermore, BRG1 staining revealed that most of the Alcian blue–positive areas that developed in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice following acute pancreatitis retained BRG1 expression (Figure 2, C and D). Indeed, BRG1-negative Alcian blue–positive areas constituted only 28% of the total Alcian blue–positive areas present (Figure 2D). These data indicate that, under inflammatory conditions, PanINs develop predominately from cells that escaped Brg1 deletion. Thus, BRG1 function is critical in pancreatitis-induced PanIN formation.
Figure 2. Acinar-specific ablation of Brg1 attenuates acute pancreatitis–induced PanIN formation in an oncogenic KRAS background.
(A) Schematic showing experimental design for tamoxifen administration and caerulein-induced acute pancreatitis in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice. (B) H&E and Alcian blue staining and immunohistochemistry for BRG1 and phospho-ERK in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice and littermate controls at 21 days after caerulein induction. Scale bars: 50 μm. (C) Immunohistochemistry for BRG1 in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice at 21 days after caerulein induction. Representative BRG1-positive and BRG1-negative PanINs. Scale bars: 50 μm. (D) Quantification of Alcian blue–positive area in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice with littermate controls. Red bars show incidence of BRG1-negative late ADMs and PanINs. n = 4 mice per genotype. Data are shown as mean ± SEM. *P < 0.05, Student’s t test.
Brg1 is required for PanIN and PDA formation in the presence of mutant p53.
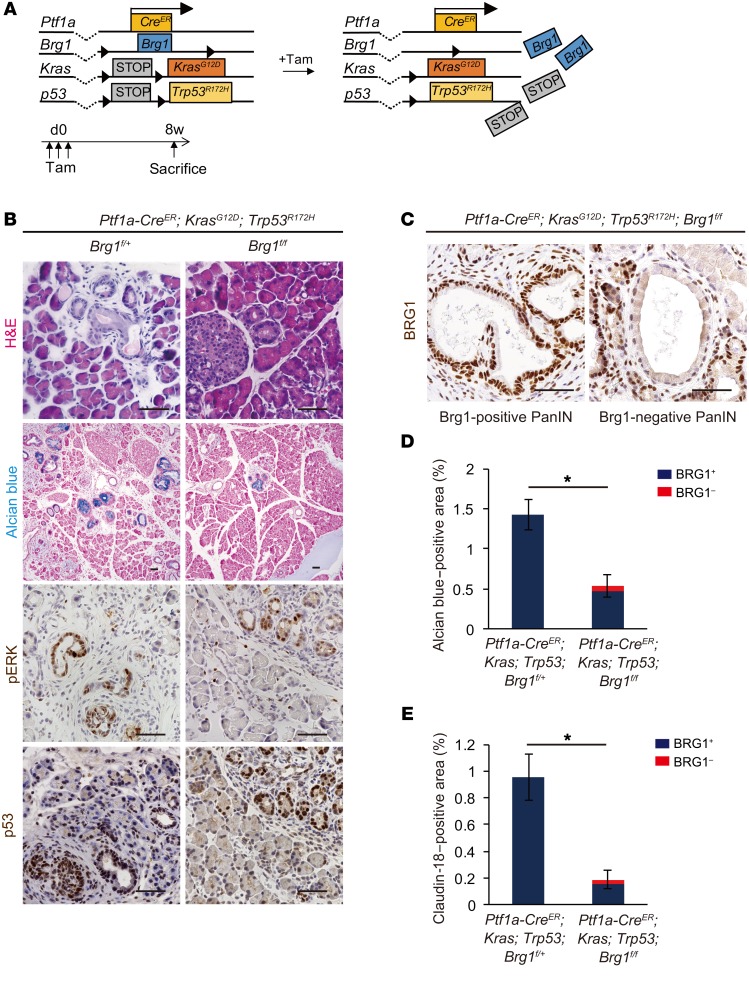
We previously reported that expression levels of tumor-suppressor genes, such as p16 and p53, are altered in Brg1-depleted IPMN lesions (12). Therefore, we examined the expression levels of p16 and p53 in Brg1-depleted PanINs. Immunohistochemical analysis revealed that expression levels of p16 and p53 were reduced in BRG1-depleted PanINs (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI94287DS1). Although these genes were suppressed, loss of BRG1 in acinar cells resulted in decreased PanIN formation. These data suggest that BRG1 plays a critical role in PanIN formation independently of p16 and WT p53. Mutant p53 is known to not only suppress WT p53 function, but also to have additional oncogenic roles (15). Furthermore, p53 mutations are the second most frequent mutations found in human PDAs (16). A previous study has demonstrated that mice expressing oncogenic Kras and mutant p53 during embryogenesis develop PanINs that eventually progress to lethal PDAs within their first 4 to 5 months of life (5). Therefore, we generated Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice carrying a commonly found p53 mutation in human PDA and assessed PanIN formation 8 weeks after tamoxifen injection in 4-week-old animals with littermate Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/+ mice as controls (Figure 3A). We identified PanINs by H&E and Alcian blue staining (Figure 3B). Phospho-ERK and p53 immunostaining confirmed Kras and mutant p53 activity, respectively (Figure 3B). We found that the Alcian blue–positive areas of late ADMs and PanINs, similarly to those of transgenic mice with WT p53, were significantly decreased in Ptf1a-CreER; KrasG12D; p53R172H; Brg1fl/fl mice compared with those in Ptf1a-CreER; KrasG12D; p53R172H; Brg1fl/+ controls (Figure 3, B and D). BRG1 staining likewise revealed that most of the Alcian blue–positive lesions in Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice retained BRG1 expression (Figure 3, C and D). Indeed, the BRG1-negative Alcian blue–positive area accounted for only 12% of the total Alcian blue–positive area (Figure 3D). As observed for the p53 WT mice, we also found that claudin-18–positive PanINs were significantly reduced in Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice compared with those in Ptf1a-CreER; KrasG12D; p53R172H; Brg1fl/+ mice (Figure 3E). Again, most of the claudin-18–positive PanINs in Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice were BRG1 positive, and the grade of all of the few BRG1-negative PanINs was PanIN1. These data support the notion that BRG1-positive PanINs in Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice have escaped Brg1 deletion and that Brg1 deletion markedly attenuates KRAS-driven ADM and PanIN formation in the mutant p53 background.
Figure 3. Acinar-specific ablation of Brg1 attenuates spontaneous PanIN formation in a background of oncogenic KRAS and mutant p53.
(A) The genetic strategy used to delete Brg1 and activate oncogenic Kras and mutant p53 in adult pancreatic acinar cells in addition to experimental design for tamoxifen administration and analysis. (B) H&E and Alcian blue staining and immunohistochemistry for phospho-ERK and p53 in Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice and littermate controls. Scale bars: 50 μm. (C) Immunohistochemistry for BRG1 in Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice. Representative BRG1-positive and BRG1-negative PanINs. Scale bars: 50 μm. (D) Quantification of Alcian blue–positive late ADMs and PanINs in Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice with littermate controls. Red bars show incidence of BRG1-negative late ADMs and PanINs. n = 3 mice per genotype. Data are shown as mean ± SEM. *P < 0.05, Student’s t test. (E) Quantification of claudin-18–positive PanIN area in Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice with littermate controls. Red bars show incidence of BRG1-negative PanINs. n = 3 mice per genotype. Data are shown as mean ± SEM. *P < 0.05, Student’s t test.
Next, to investigate whether BRG1 remains essential for development of acinar-derived PDA, 4-week-old Ptf1aCreER; KrasG12D; TP53R172H; Brg1fl/+ and Ptf1aCreER; KrasG12D; TP53R172H; Brg1fl/fl mice were administered tamoxifen and analyzed 6 months after tamoxifen administration (Supplemental Figure 2A). We found that 1 of 3 Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice developed PDA, whereas 3 of 3 Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/+ mice developed PDA (Supplemental Figure 2B). Notably, the 1 PDA that was formed in 1 of 3 Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice was BRG1 positive, indicating that PDA developed from acinar cells that had escaped Brg1 deletion. Furthermore, we also induced caerulein pancreatitis to Ptf1a-CreER; KrasG12D; Trp53R172H; Brg1fl/fl mice and assessed PDA formation 4 months after caerulein injection. One of three mice carried PDA, and this PDA was likewise BRG1 positive (Supplemental Figure 2C). Therefore, these results suggest that BRG1 is critical for the formation of PanIN-derived PDA that originates from acinar cells.
Brg1 is required for ADM formation ex vivo.
To gain further insights into the underlying molecular mechanisms controlling the role of BRG1 in acinar cell–derived KRAS-driven tumorigenesis, we next performed ex vivo acinar cell culture experiments (17). First, we isolated primary acinar cell explants from Ptf1a-Cre; Brg1fl/fl and WT pancreata. These acinar cell explants were cultured under 3D conditions for 5 days. Then, ADM formation was assessed with or without TGF-α treatment, which is known to induce ADMs and PanINs (17, 18). ADM formation in Ptf1a-Cre; Brg1fl/fl acini was significantly attenuated compared with that in WT acini with or without TGF-α administration ex vivo, indicating that BRG1 is required for ADM formation in this context (Supplemental Figure 3A).
Acinar cell identity is compromised in Brg1-depleted acinar cells.
To examine transcriptional changes in Brg1-depleted versus WT acini during the acinar-to-ductal metaplastic conversion, we next performed reverse transcriptase PCR (RT-PCR) on isolated acini for genes important for pancreatic development and maintenance of pancreatic differentiation. We found that expression levels of multiple genes involved in acinar cell identity, including Cpa1, Amy2a4, Gata6, were significantly downregulated in acini isolated from Ptf1a-Cre; Brg1fl/fl mice (Supplemental Figure 3B). We also found a strong, but not statistically significant, downregulation in the expression levels of Ptf1a and Nr5a2. Ptf1a, Nr5a2, and Cpa1 are expressed in multipotent progenitor cells during pancreatic development and in adult acinar cells (19–22). Gata6 is required for the differentiation and maintenance of acinar cells, and it is expressed in adult acinar cells (23). Notably, expression of Sox9, which is commonly expressed in multipotent progenitors during pancreatic development (24, 25) and transiently expressed during acinar-to-ductal metaplastic transformation (6), was also downregulated in Brg1-depleted dedifferentiated acinar cells. These results suggest that acinar cell identity is compromised in Brg1-depleted acinar cells.
KrasG12D; Brg1fl/fl acini display a transcriptional signature distinct from that of KrasG12D acini during the acinar-to-ductal conversion.
While the results from our ex vivo experiments of acini isolated from Ptf1a-Cre; Brg1fl/fl mice were instructive, such an analysis did harbor some limitations. In particular, Ptf1a expression in Ptf1a-Cre mice was downregulated compared with that in WT mice due to the heterozygosity of the Ptf1a allele, as reported previously (26). Furthermore, acini from Ptf1a-Cre; KrasG12D; Brg1fl/fl mice could not be analyzed because acinar tissues were significantly reduced and replaced by cystic IPMN lesions (12). To solve these problems, we performed cell-culture experiments using acini from Brg1fl/fl, KrasG12D, and KrasG12D; Brg1fl/fl mice infected with Cre recombinase–expressing adenovirus (Ad-Cre), which can initiate genetic recombination of the loxP sites at the Kras and Brg1 loci by infection. We found that ex vivo ADM formation was markedly reduced in acinar cell explants of KrasG12D; Brg1fl/fl mice compared with those in KrasG12D mice (Supplemental Figure 3C). To explore transcriptional changes acutely after Brg1 deletion, microarray analyses were performed on KrasG12D and KrasG12D; Brg1fl/fl acinar cell explants 24 hours after adenovirus infection. At this point, acinar cell explants underwent dedifferentiation (27). Notably, 6,321 genes were differentially expressed using 1.5-fold change. (Supplemental Table 1). Pathway enrichment analyses of Gene Ontology using the software program DAVID 6.8 (https://david.ncifcrf.gov/) identified 32 biological processes that were significantly enriched between Brg1-depleted and Brg1-WT dedifferentiated acinar cell explants with oncogenic Kras (FDR set at 0.05). These processes included transcription, phosphorylation, chromatin modification, cell cycle, apoptotic process, angiogenesis, ubiquitination, cell migration, embryo development, and cell differentiation (Supplemental Table 2). We next focused on the expression of genes important for pancreatic development and maintenance of pancreatic differentiation. We found that the expression of some genes regulating acinar cell identity, including Ptf1a and Xbp1 (28), and some genes important for progenitor potential, including Sox9, Foxo1 (29, 30), and Glis3 (31), were acutely compromised in Brg1-depleted dedifferentiated acinar cells (Supplemental Figure 3D). These data suggest that loss of BRG1 in dedifferentiated acinar cells acutely changes the expression of pancreatic developmental genes, including Sox9.
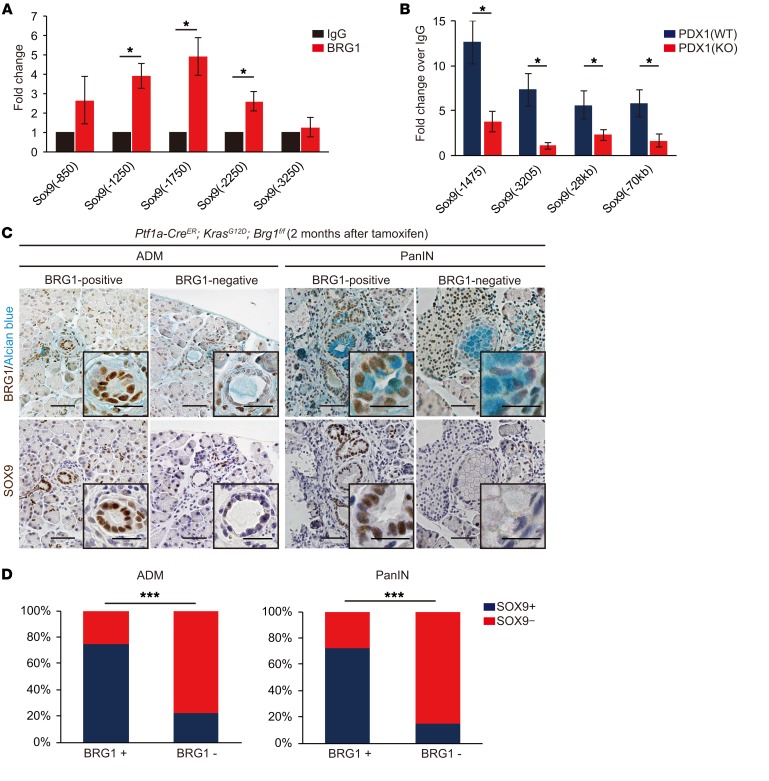
BRG1 directly binds to the Sox9 promoter and is critical for recruitment of PDX1 to the Sox9 promoter and enhancer in acinar cells.
Given that SOX9 has been identified as a critical factor for PanIN formation (32–34), downregulation of Sox9 expression in Brg1-depleted dedifferentiated acinar cells prompted us to examine the possibility that BRG1 directly regulates Sox9 expression in acinar cells during ADM/PanIN formation. To this end, we next performed ChIP experiments. We found that BRG1 binds to evolutionary conserved regions within the promoter of Sox9 (12) in murine isolated acinar cells, as determined by ChIP experiments (Figure 4A). These results were consistent with our previous finding that BRG1 binds to the Sox9 promoter in PanIN-derived mouse PDA cell lines (12). Therefore, these results suggest that BRG1, at least in part, directly regulates Sox9 expression in acinar cells during ADM/PanIN formation.
Figure 4. BRG1 promotes Sox9 expression in murine ADMs and PanINs.
(A) ChIP qPCR of isolated acinar cells from WT mice on the Sox9 promoter regions. n = 3 mice per genotype. Relative fold enrichment of BRG1 over IgG control on the Sox9 promoter regions. Numbers in primer names denote distance from transcription start site (TSS) of the Sox9 gene. Data are shown as mean ± SEM. *P < 0.05, Student’s t test. (B) ChIP qPCR of isolated acinar cells from WT and Ptf1a-CreER; Brg1fl/fl mice that were treated with tamoxifen (KO). n = 3–4 mice per genotype. Relative fold enrichment of PDX1 over IgG control on the Sox9 promoter and enhancer. Numbers in primer names denote distance from TSS of the Sox9 gene. Data are shown as mean ± SEM. *P < 0.05, Student’s t test. (C) Immunohistochemistry of serial sections for BRG1 and SOX9. Left: BRG1-positive and BRG1-negative ADMs in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice at 2 months after tamoxifen administration. Right: BRG1-positive and BRG1-negative PanINs in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice at 2 months after tamoxifen administration. Scale bars: 50 μm. (D) Quantification of the relationship between expression of BRG1 and SOX9 in ADM and PanIN in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice. BRG1-positive ADMs or PanINs and SOX9-positive ADMs or PanINs are defined as the lesions in which more than half of the cells are positive for BRG1 and SOX9, respectively. n = 4 mice. ***P < 0.001, Fisher’s exact test.
To gain further insight into the mechanisms by which BRG1 regulates Sox9 expression, we performed RT-PCR analysis of genes that are known to regulate Sox9 expression in the pancreas. Specifically, Pdx1 and Sox9 generate a feed-forward loop during foregut differentiation (35), Notch signaling regulates Sox9 expression positively in pancreatic development (36), and Onecut1 regulates Sox9 expression in ADM (37). RT-PCR analyses revealed that none of these upstream regulators of Sox9 in the pancreas were significantly changed in their expression in either Brg1-depleted acini (Supplemental Figure 3B and Supplemental Figure 4A). However, loss of BRG1 compromised the binding of PDX1 to the Sox9 promoter and enhancer, as determined by ChIP analysis of isolated acini from WT mice compared with Brg1-depleted acini from Ptf1a-CreER; Brg1fl/fl mice that were treated with tamoxifen (Figure 4B). These data suggest that BRG1 is critical for recruitment of upstream regulators, including PDX1, to the Sox9 promoter and enhancer to promote its expression in acinar cells during ADM/PanIN formation.
BRG1 regulates Sox9 expression during ADM/PanIN formation in vivo.
To determine whether BRG1 promotes Sox9 expression in vivo, we next performed immunohistochemical staining of SOX9 and BRG1 in ADMs and PanINs in serial sections from Ptf1a-CreER; KrasG12D; Brg1fl/fl mice 2 months after tamoxifen injection. Quantification revealed a significant positive correlation between expression of BRG1 and SOX9 in ADM and PanIN (Figure 4, C and D). Of note, SOX9 expression was markedly downregulated in the rarely observed BRG1-negative ADM/PanINs compared with BRG1-positive ADM/PanINs in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice. Therefore, these data suggest that BRG1 positively regulates Sox9 expression during KRAS-driven ADM/PanIN formation in mice.
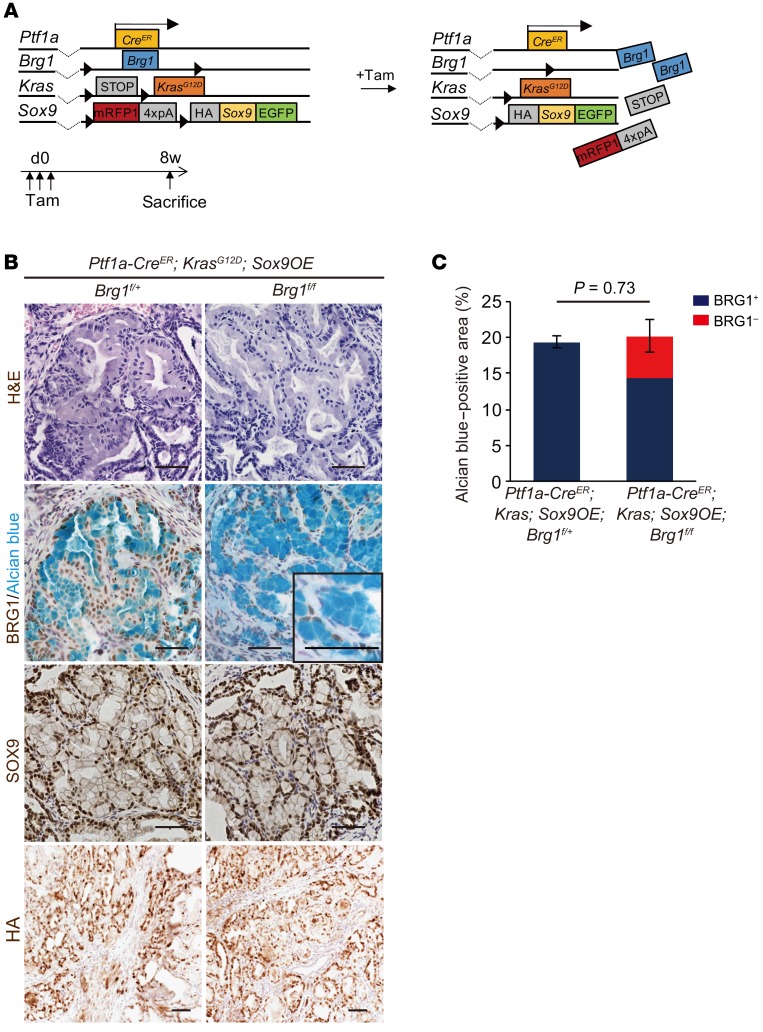
Sox9 overexpression rescues the reduction of KRAS-driven PanIN formation by Brg1 deletion in mice.
Given that SOX9 expression was markedly downregulated in BRG1-depleted acini during KRAS-driven ADM/PanIN formation in mice, we hypothesized that Sox9 overexpression might rescue the reduction of PanIN formation in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice. To test this hypothesis, we generated Ptf1a-CreER; KrasG12D; Brg1fl/fl; Sox9OE mice. In these animals, Sox9 was constitutively overexpressed in pancreatic acinar cells and Brg1 was concomitantly deleted in the presence of mutant Kras upon tamoxifen injection. Four-week-old Ptf1a-CreER; KrasG12D; Brg1fl/fl; Sox9OE mice were administered tamoxifen, analyzed 8 weeks after tamoxifen administration, and compared with control Ptf1a-CreER; KrasG12D; Brg1fl/+; Sox9OE mice (Figure 5A). Strikingly, normal acinar cells were lost and numerous PanINs were observed in both Ptf1a-CreER; KrasG12D; Brg1fl/fl; Sox9OE and control Ptf1a-CreER; KrasG12D; Brg1fl/+; Sox9OE mice (Figure 5B). In these PanINs, SOX9 protein was broadly expressed and broad recombination of the Sox9OE allele was also confirmed by immunostaining for the HA tag. Prevalence of PanINs in Ptf1a-CreER; KrasG12D; Brg1fl/fl; Sox9OE mice was comparable to that in Ptf1a-CreER; KrasG12D; Brg1fl/+; Sox9OE mice (Figure 5C). Notably, 30% of PanINs in Ptf1a-CreER; KrasG12D; Brg1fl/fl; Sox9OE mice formed in the absence of BRG1 (Figure 5C and Supplemental Figure 5A). BRG1-negative PanIN areas in Ptf1a-CreER; KrasG12D; Brg1fl/fl; Sox9OE mice were significantly increased (267-fold change) compared with those in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice 8 weeks after tamoxifen injection. Similarly, BRG1-positive PanIN areas in Ptf1a-CreER; KrasG12D; Brg1fl/fl; Sox9OE mice were increased (185-fold change) compared with those in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice (Supplemental Figure 5B). These results indicate that Sox9 overexpression is capable of promoting formation of KRAS-driven PanINs from acinar cells that have lost BRG1 expression. Therefore, Sox9 overexpression offsets the reduction of PanIN formation by Brg1 deletion. These results provide evidence that Sox9 is a key downstream effecter of Brg1 in acinar cell–derived KRAS-driven PanIN formation in mice.
Figure 5. Sox9 overexpression cancels the decrease in KRAS-driven PanIN formation following Brg1 deletion in mice.
(A) The genetic strategy used to delete Brg1 and activate oncogenic Kras and Sox9 expression in adult pancreatic acinar cells in addition to the experimental design of tamoxifen administration and analysis. (B) H&E staining and immunohistochemistry for BRG1 with Alcian blue staining, SOX9, and the HA-tag in Ptf1a-CreER; KrasG12D; Sox9OE; Brg1fl/fl mice and Ptf1a-CreER; KrasG12D; Sox9OE; Brg1fl/fl mice. Scale bars: 50 μm. (C) Quantification of PanINs in Ptf1a-CreER; KrasG12D; Sox9OE; Brg1fl/+ mice and Ptf1a-CreER; KrasG12D; Sox9OE; Brg1fl/fl mice. Red bars show incidence of BRG1-negative PanINs. n = 3 per genotype. Data are shown as mean ± SEM. Student’s t test.
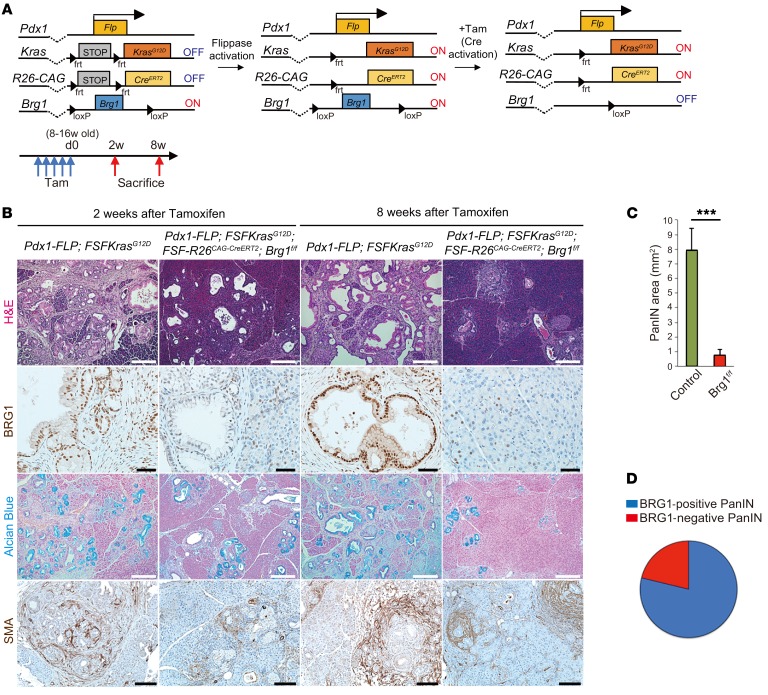
BRG1 ablation in formed PanIN resulted in regression of the lesions.
One caveat of the above-mentioned in vivo experiments is that the activation of oncogenic KRAS and deletion of Brg1 happens simultaneously. Therefore, we decided to test to determine whether BRG1 deletion in an established PanIN has any adverse consequences on the neoplastic lesion. To address this question, we used the recently developed 2-hit mouse model, which permits independent temporal modification of 2 alleles using the Flp and Cre recombinases (38) (Figure 6A). For example, we activated oncogenic KRAS using the Flp recombinase directed by the mouse Pdx1 promoter. To delete BRG1 in Flp-recombined cells via Cre recombinase at a later time point of our choice, we used a tamoxifen-inducible CreERT2 allele silenced by an FRT-STOP-FRT cassette in the Rosa26 locus. Tamoxifen treatment induced the expression of the CreERT2 allele, which in turn prompted recombination of the LoxP site at the Brg1 locus. We aged Pdx1-Flp; FSF-KrasG12D animals up to 8 to 16 weeks to ensure PanIN formation and then deleted Brg1 via tamoxifen administration. Two or eight weeks after tamoxifen administration, we euthanized these animals and analyzed their pancreata. At the 2-week time point, control (Pdx1-Flp; FSF-KrasG12D) animals had developed ADM and PanINs, whereas Brg1-deleted mice exhibited significantly less disease burden (Figure 6B). More pronounced differences were observed at the 8-week time point. Whereas control animals displayed abundant PanIN formation, deletion of Brg1 ablated formation of neoplasia, which was evident in the H&E and Alcian blue stainings. Quantification revealed 8 times less PanIN burden in Brg1-depleted animals (Figure 6C). When assessed for BRG1 expression in these PanINs developed in Brg1-floxed animals, 80% of lesions were BRG1 positive, clearly indicating that PanINs require BRG1 expression for maintenance (Figure 6D). We further interrogated why BRG1 expression is critical for the maintenance of established PanINs. To address this question, 8- to 16-week-old Pdx1-Flp; FSF-KrasG12D; FSF-Rosa26CreER; Brg1fl/fl mice were administered tamoxifen and analyzed 3 days after tamoxifen administration (Supplemental Figure 6A). We found that deletion of Brg1 resulted in widespread apoptosis of PanIN cells (Supplemental Figure 6B). These data indicate that BRG1 is critical for the maintenance and progression of established PanINs and acts by inhibiting apoptosis.
Figure 6. BRG1 is required for PanIN maintenance.
(A) The genetic strategy used to activate oncogenic Kras at the embryonic stage and delete Brg1 at a subsequent adult stage and experimental design for tamoxifen administration and analysis. (B) H&E and Alcian blue staining and immunohistochemistry for BRG1 and SMA in Pdx1-Flp; FSF-KrasG12D and Pdx1-Flp; FSF-KrasG12D; FSF-R26CAG-CreERT2; Brg1fl/fl mice. Scale bars: 200 μm (H&E, Alcian blue, and SMA): 50 μm (BRG1). (C) Quantification of PanINs in Pdx1-Flp; FSF-KrasG12D (green; n = 7) and Pdx1-Flp; FSF-KrasG12D; FSF-R26CAG-CreERT2; Brg1fl/fl (red; n = 6) mice. Data are shown as mean ± SEM. ***P < 0.001, Student’s t test. (D) Quantification of BRG1-positive/negative PanINs in Pdx1-Flp; FSF-KrasG12D; FSF-R26 CAG-CreERT2; Brg1fl/fl mice. n = 22 mice.
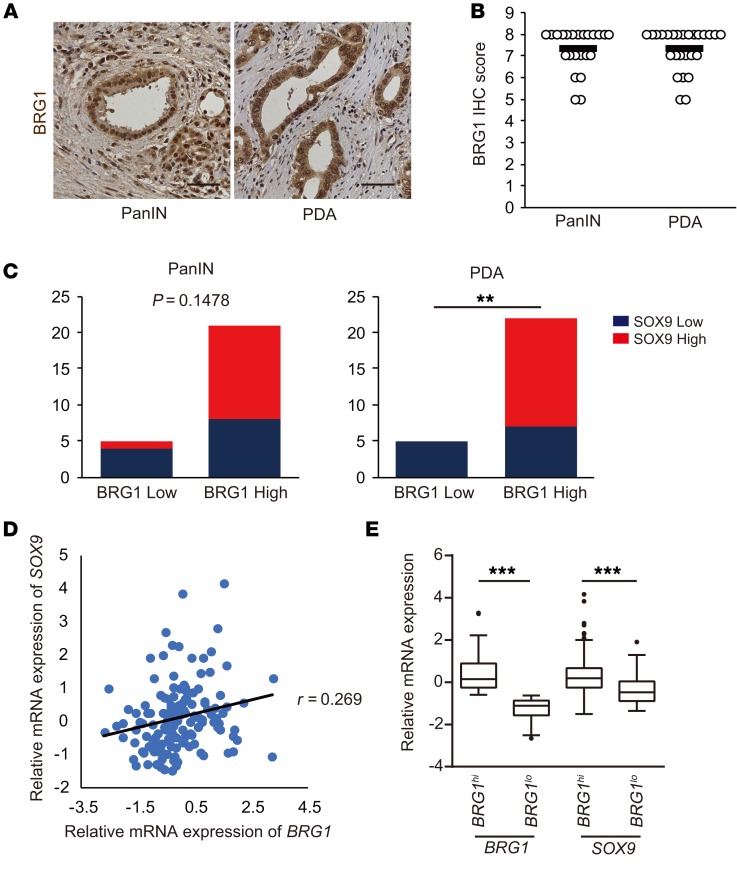
BRG1 expression correlated with SOX9 expression in human PDA.
Finally, to determine whether a BRG1/SOX9 axis is observed in human PanIN-derived PDAs, we performed immunohistochemistry on 27 PanIN-derived PDA samples that had no evidence of IPMN and had surrounding PanINs (Figure 7A). BRG1 was highly expressed in 21 of 26 PanINs and 22 of 27 PDAs (Figure 7B). This finding supported the notion that BRG1 is important for PanIN-derived PDA formation.
Figure 7. BRG1 expression correlates with SOX9 expression in human PDAs.
(A) Immunohistochemistry for BRG1 in human PanINs and PDAs. Scale bars: 50 μm. (B) BRG1 immunohistochemistry score from patient samples with PanINs (n = 26) and PanIN-derived PDAs (n = 27). PDA samples containing PanINs are selected and BRG1 expression is scored using an IHC score ranging from 0 to 8 (low to high). Means are shown. (C) Quantification of SOX9 expression in BRG1lo or BRG1hi human PDA samples. The cut-off IHC score is 0 to 6 for low and 7 to 8 for high expression. Left: PanIN (n = 26); right: PDA (n = 27). **P < 0.01, Fisher’s exact test. (D) Plots of mRNA expression of BRG1 and SOX9 from a cohort of 150 patients in the TCGA data set. (E) The box-and-whisker plot demonstrates the differential expression for SOX9 between the BRG1hi (n = 112, the higher 75%) and BRG1lo groups (n = 38, the lower 25%) from a cohort of 150 patients in the TCGA data set. In the box-and-whisker plots, horizontal bars indicate the medians, boxes indicate 25th to 75th percentiles, and whiskers indicate minimum to maximum without outlier. ***P < 0.001, Student’s t test.
To determine whether genomic alterations were present in human PDA cases, we performed targeted sequencing of 5 BRG1lo samples. In total, 20 somatic mutations were identified, and TP53 and KRAS mutations were detected in all 5 cases. However, no BRG1 somatic mutations were detected. Using copy number analysis, we identified deletions in 19p, the chromosomal location of BRG1, in 2 cases (Supplemental Figure 7, A and B, Supplemental Table 3). Furthermore, analysis of The Cancer Genome Atlas (TCGA, https://cancergenome.nih.gov/) copy number alteration data revealed that BRG1 gene deletion has significant correlation with low BRG1 mRNA expression (Supplemental Figure 7C).
Next, to investigate whether BRG1 and SOX9 expression were correlated in human PanINs and PanIN-derived PDAs, we performed immunohistochemistry for SOX9 on serial sections of these tissues. SOX9 expression was reduced in 4 of 5 PanIN samples with decreased BRG1 expression, but only 8 of 21 PanIN samples with high BRG1 expression showed reductions in SOX9 expression. Furthermore, SOX9 expression was reduced in 5 of 5 PDA samples with decreased BRG1 expression and only in 7 of 22 PDA samples marked by high BRG1 expression (Figure 7C). While we observed a strong, but statistically nonsignificant, trend toward BRG1 and SOX9 coexpression in PanINs, a clear statistically significant (P < 0.01) correlation was observed for PDAs. To confirm this correlation, we analyzed BRG1 and SOX9 expression levels against each other across the TCGA mRNA expression data sets of human PDAs (Figure 7D). To determine whether Sox9 expression was decreased in the samples in which BRG1 expression was low, we next separated the TCGA samples into high (the higher 75% of BRG1 expression) and low (the lower 25% of BRG1 expression) groups. As a result, we found significant correlation between BRG1 and SOX9 expression in human PDAs (Figure 7E). Most of the TCGA samples are considered to be PanIN-derived PDAs, reflecting the general frequency in the patient population (1). We also examined the relationship between BRG1 and SOX9 expression in human PDAs using the Queensland Centre for Medical Genomics (QCMG) mRNA expression data sets (ref. 10) and found the same correlation (Supplemental Figure 8, A and B). These data further support the conclusion that BRG1 positively regulates SOX9 expression in human PanIN-derived PDAs.
Discussion
Increasing evidence supports the role of chromatin-remodeling regulators in the formation and progression of neoplastic lesions in the pancreas. In particular, loss of the SWI/SNF complex protein BRG1 has been implicated in the formation of IPMN lesions (12).
In this study, we demonstrated a critical role for the BRG1/SOX9 axis in acinar cell–derived PanIN-PDA formation. We first showed that loss of BRG1 suppressed KRAS-induced ADM and PanIN formation from pancreatic acinar cells. ADMs and PanINs were significantly (Figure 1, F and G) decreased by Brg1 deletion in acinar cells using Ptf1a-CreER; KrasG12D; Brg1fl/fl mice. To emphasize this finding, most ADMs and PanINs observed in Ptf1a-CreER; KrasG12D; Brg1fl/fl mice retained BRG1 expression, indicating that these ADMs and PanINs were derived from the cells that had escaped Brg1 deletion. This critical role of BRG1 in ADM formation was confirmed by ex vivo primary acinar cell culture. Also, this critical role of BRG1 in PanIN formation was confirmed in the background of pancreatitis and mutant p53 in mice. Furthermore, we also showed that loss of BRG1 suppressed formation of PanIN-derived PDA originating from acinar cells in the background of mutant p53 and that PDA developed from acinar cells that had escaped Brg1 deletion. Taken together with our previous findings that Brg1 inhibits duct cell–derived IPMN and IPMN-derived PDA formation (12), our data presented here highlight the cell type–specific context-dependent function of BRG1 in pancreatic tumorigenesis.
To address the underlying molecular mechanism by which BRG1 regulates aspects of acinar cell–derived ADM and PanIN formation, we used ex vivo acinar cell culture approaches and analyzed transcriptional profiling. We found that Sox9 expression was downregulated in slightly dedifferentiated acini immediately after adenoviral BRG1 deletion as well as in BRG1-deleted acini isolated from Ptf1a-Cre; Brg1fl/fl mice ex vivo. In our acinar cell culture experiment, we found decreased ADM formation in Brg1-deleted acinar cells, although surprisingly, acinar cell regulator genes, including Ptf1a and Nr5a2, which are known to inhibit ADM formation, were also downregulated. It is likely that reduced Sox9 expression in these cells provides an explanation for the lack of ADM formation in cells with reduced expression of acinar identity genes. Prior work has demonstrated that the loss of Ptf1a in acinar cells induces expression of Sox9 (39) and ADM formation (40), likely mediated via upregulation of Sox9, which is critical for ADM formation in vitro (32). Therefore, the lack of Sox9 expression in Brg1-deleted acinar cells impairs efficient ADM formation despite a reduction of acinar regulator genes.
Furthermore, our studies reveal that BRG1 positively regulates Sox9 expression during ADM/PanIN formation in vivo. Sox9 has been identified as a critical factor during PanIN-derived PDA formation (32–34). Sox9 overexpression in pancreatic epithelial cells accelerates KRAS-induced PanIN formation, whereas Sox9 deletion in adult acinar cells prevents spontaneous as well as pancreatitis-induced ADM and PanIN formation. Notably, the Sox9 deletion phenotype in acinar cells appears to phenocopy that of Brg1 deletion in acinar cells. In support of the hypothesis that SOX9 acts downstream of BRG1, we observed that Sox9 overexpression promotes KRAS-driven PanIN formation following Brg1 deletion in mice. Thus, this in vivo evidence strongly suggests that essential aspects of BRG1 function during PanIN formation are mediated through positive regulation of Sox9 expression.
How does BRG1 regulate Sox9 expression? Consistent with our previous report (12), here, we show that BRG1 binds to the Sox9 promoter regions to support its expression in murine acinar cells by ChIP analysis. Furthermore, we show that BRG1 is critical for recruitment of upstream regulators, including PDX1, to the Sox9 promoter and enhancer in acinar cells during ADM/PanIN formation. In support of these data, a recent study suggested that BRG1 is essential for PDX1 transactivation in pancreatic islet cells (41). This notion is further emphasized by our acinar cell experiment data demonstrating that Sox9 expression was decreased immediately after Brg1 deletion in isolated acinar cells. It is possible that other upstream regulators of Sox9 in the pancreas, including Onecut1 and Notch signaling, also contribute to promoting Sox9 expression in acinar cells in collaboration with BRG1 during ADM/PanIN formation.
We also addressed the question of BRG1 function during PanIN maintenance by using the Flp/Cre system to eliminate Brg1 after PanIN formation had been initiated. The finding that the majority of PanINs that were still present 8 weeks after tamoxifen treatment retained BRG1 expression strongly supports the notion that BRG1 activity remains critical in established PanIN. Further studies will be needed to investigate the exact mechanism by which BRG1 prevents apoptosis in established PanINs in more detail.
Furthermore, we observed a correlation between BRG1 and SOX9 expression in human PanIN-derived PDAs, supporting the notion that the BRG1/SOX9 axis is conserved in human PDAs as well. Interestingly, we showed that BRG1 downregulation occurred in approximately 20% of human PanIN-derived PDAs, a fact that might suggest a discrepancy in our data presented here that describe a role for BRG1 in PanIN initiation and maintenance. Of note, a similar discrepancy is also observed in lung cancer (42). Although BRG1 is mutated at a very high percentage in human lung cancer, BRG1 is required for initiation of KRAS-induced lung adenoma. Two different scenarios might explain these divergent observations. One hypothesis is that human PanINs lacking BRG1 expression might be derived from pancreatic ductal cells. While the formation of PanIN lesions from duct cells in human is still a matter of intense debate, our prior work has shown that Brg1 inactivation in ductal cells in the presence of oncogenic Kras leads to neoplastic lesions in mice (12). Recent work by others illustrates a transition from duct cells to PanIN/PDA in organoids, but the role of BRG1 in this process has not been ascertained (43). Another hypothesis is that the BRG1/SOX9 axis might play a tumor-suppressive role in established acinar cell–originated PanIN-derived PDA in a context-dependent manner, similar to what has been seen during IPMN formation and established IPMN-derived PDA (13). In support of this latter hypothesis, it is known that both low BRG1 and low SOX9 expression correlate with poor overall survival rates for human PanIN-derived PDAs (12, 32). Further investigation will be required to fully address the temporal activities of BRG1 during PDA formation and progression.
In this study, using copy number analysis, we identified deletions in 19p, the chromosomal location of BRG1, in 2 out of 5 cases with low BRG1 protein expression. We also found that BRG1 gene deletion has a correlation with low BRG1 mRNA expression by analyzing TCGA data. Furthermore, recent studies showed that loss of BRG1 protein via aberrant splicing and epigenetic regulations is more common than reductions in gene expression via mutations (11), suggesting the possibility for similar mechanisms in our 5 PDA cases with low BRG1 protein expression.
In conclusion, we demonstrate that BRG1 is critical for ADM/PanIN formation and maintenance of established PanIN. BRG1 elicits its function at least in part through regulation of SOX9 expression during ADM/PanIN formation. Importantly, similar interactions between BRG1 and SOX9 expression are observed in human PDAs. Although future studies are needed to clarify the roles of BRG1 in established PanIN-derived PDA, our findings reveal a tumor-promoting role for BRG1 in the initiation and maintenance of PanIN. Our data underscore the cell type–specific, context-dependent roles of BRG1 in PDA initiation and progression and suggest that the BRG1/SOX9 axis is a potential druggable target for prevention of PDA.
Methods
Mice.
The following mouse strains were used: Ptf1a-Cre (gift of Yoshiya Kawaguchi, Kyoto University) (19), Ptf1a-CreER (20), KrasG12D (gift of David Tuveson, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA) (44), Brg1fl (gift of David Reisman, University of Florida, Gainesville, Florida, USA, with permission from Pierre Chambon, University of Strasbourg Institute for Advanced Study, Strasbourg, France) (45), Trp53R172H (gift of Yoshiya Kawaguchi) (46), Pdx1-Flp, FSF-KrasG12D/+, FSF-R26CAG-CreERT2/+ (38), and Sox9OE (47). Mice were crossed in a mixed background, and no selection for a specific sex was done in this study. Tamoxifen (Sigma-Aldrich) was dissolved in corn oil and administered subcutaneously at a concentration of 6 mg/20 g body weight per injection. In an inducible dual-recombinase system, tamoxifen was dissolved in peanut oil and administered intraperitoneally at a concentration of 2 mg/mouse for 5 days. Acute pancreatitis was induced by injecting caerulein (50 mg/kg diluted in saline; Sigma-Aldrich) intraperitoneally on 2 consecutive days once every hour for 8 hours each day.
Clinical samples.
Twenty-seven surgically resected specimens of pancreatic cancer tissues were obtained from patients admitted to Kyoto University Hospital.
RNA isolation and quantitative real-time PCR analysis.
RNA was isolated using the RNeasy kit (QIAGEN). Complementary DNA was synthesized using the ReverTra Ace qPCR RT Kit (Toyobo). Quantitative PCR (qPCR) was performed with a SYBR Green–based gene-expression assay using a LightCycler 480 System (Roche). Expression levels were normalized using Gapdh as a reference gene. Primers were designed using the Massachusetts General Hospital PrimerBank (https://pga.mgh.harvard.edu/primerbank/). Primer sequences are described in Supplemental Table 4. All reactions were performed in duplicate.
Immunohistochemistry.
Mouse tissue was fixed overnight in 4% paraformaldehyde/PBS, dehydrated into 70% ethanol, embedded in paraffin, and cut into 5 μm thick sections. Paraffin-embedded sections were stained with H&E, Alcian blue, and Nuclear Fast Red (Vector Laboratories). For immunohistochemical staining, antigen retrieval was performed by incubating sections in citric acid buffer (pH 6.0) or EDTA buffer (pH 8.0) for 15 minutes at 98°C. Blocking was performed by incubating sections in blocking solution (Dako). For primary antibodies, incubation was performed overnight at 4°C or for 2 hours at room temperature (RT). Secondary antibodies were incubated for 1 hour at RT. For immunohistochemistry, slides were developed using either the ABC Kit (Vector Laboratories) and DAB Kit (Dako) or the EnVision Kit (Dako), followed by counterstaining with hematoxylin. The primary antibodies used in this study were as follows: rabbit anti-BRG1 (Santa Cruz Biotechnology Inc., catalog sc-10768; dilution 1:500–1000 [mouse], 1:200 [human]), rabbit anti-SOX9 (MilliporeSigma, catalog AB5535; dilution 1:10000–100000 [mouse], 1:1000 [human]), rabbit anti-p53 (Vector Laboratories, catalog VP-P956; dilution 1:500), rat anti-HA (Roche, catalog 1867423; dilution 1:200), rabbit anti-p44/p42-MAPK (Cell Signaling Technology, catalog 4370; dilution 1:400), mouse anti-smooth muscle actin (Sigma-Aldrich, catalog A5228; dilution 1:100), rabbit anti–claudin-18 (Invitrogen, catalog 700718; dilution 1:200–2000), mouse anti-Ki67 (BD, catalog 550609; dilution 1:400), rabbit anti–cleaved caspase 3 (Cell Signaling Technology, catalog 9661; dilution 1:100), and rabbit anti-p16 (Santa Cruz Biotechnology Inc., catalog sc-1207; dilution 1:400).
For quantitative analysis of PanIN areas, measurements were performed either by assessing Alcian blue–positive areas or claudin-18–positive areas using ImageJ (NIH). Five complete pancreas sections per mouse were analyzed. ADM-like lesions contained metaplastic units that exhibited open and/or distended lumens containing acinar and/or ductal-like cells, as previously described (48, 49). The entire pancreatic area was also measured using ImageJ. Quantifications of BRG1-positive/negative ADM-like lesions/PanINs were performed on samples stained with Alcian blue and immunostained for BRG1. Three complete pancreas sections per mouse were counted. In the Sox9 overexpression animals, BRG1 staining of PanINs in 5 random high-power microscopic fields for 3 sections per mouse was quantified. We defined the BRG1-positive ADMs or PanINs and SOX9-positive ADMs or PanINs as the lesions in which more than half of the cells of each ADM or PanIN lesion were positive for BRG1 and SOX9, respectively. All quantitative analyses were performed in at least 3 animals per genotype.
Acinar cell isolation and culture.
Mouse pancreata were chopped into small pieces and digested with collagenase P (0.2 mg/ml) for 10 minutes. After 2 washes with HBSS buffer containing 5% FBS, the tissue suspension was filtered through a 100 μm cell strainer. The flowthrough was layered onto an HBSS plus 30% FBS solution and centrifuged. The cell pellet was centrifuged, resuspended in 1:1 acinar cultured media that included a rat-tail collagen type I solution (1 mg/ml final collagen concentration), and added onto a well coated with rat-tail collagen type I. Acinar-cultured media was composed of Waymouth’s Medium (Thermo Fisher Scientific) plus 10% FBS plus 1% penicillin/streptomycin plus 100 μg/ml trypsin inhibitor plus 1 μg/ml dexamethasone (17, 50).
Adenovirus infection.
After acinar cell isolation, cell pellets were resuspended into acinar-cultured media, infected with 300 MOI Ad5-Cre-GFP or Ad5-GFP (Vector Biolabs), and incubated for 2 to 3 hours. Cells were then pelleted and resuspended in acinar-cultured media. For the RT-PCR analysis of adenovirus-infected acini, paired t test was used to compare expression changes among individual mice.
Gene-expression analysis.
Total pooled RNA isolated from 3 individual pools of cultured acinar cell clusters 24 hours after Cre-expressing adenovirus infection was used for gene-expression analysis. Mouse genotypes used for analysis were WT, Brg1fl/fl, KrasG12D, and KrasG12D; Brg1fl/fl. Transcriptome profiling was performed using SurePrint G3 Mouse Gene Expression 8x60K v2 (Agilent Technologies). Raw data were normalized using GeneSpring GX 14.5 (Agilent Technologies). To gain functional insights into the differentially expressed genes, DAVID Bioinformatics Resources 6.8 from the National Institute of Allergy and Infectious Diseases was used.
For the BRG1-associated gene-expression analysis, a cohort of patients with pancreatic cancer was studied (150 patients in the TCGA data set and 96 patients in the QCMG data set). BRG1hi tumors were defined as the higher 75% of tumors with positive z scores in each data set. Conversely, BRG1lo tumors were defined as the lower 25% of tumors with negative z scores in each data set.
ChIP experiments.
ChIP was performed using the iDeal ChIP-seq Kit for Transcription Factors (Diagenode). Briefly, freshly isolated acinar cells were dual crosslinked using ChIP cross-link Gold (Diagenode) for 30 minutes at RT, followed by fixation in 1% formaldehyde (28906, Thermo Fisher) for 10 minutes at RT and quenching with 125 mM glycine. Fixed cells were lysed and subsequently sheared using the Bioruptor Pro (Diagenode) for 2 × 10 cycles of 30 seconds ON/30 seconds OFF at a high-power setting to obtain fragments of 200–500 bp. Sheared chromatin (1/25) was kept aside as input, and the remaining chromatin was incubated overnight with 1 μg rabbit IgG antibody (provided in kit), 2 μg rabbit anti-Brg1 antibody (catalog sc-10768, Santa Cruz Biotechnology Inc.), or 3.45 μg rabbit anti-Pdx1 antibody (catalog 5679, Cell Signaling Technology). Antibody-chromatin complexes were pulled down using protein A–coated magnetic beads and decrosslinked for 4 hours. The DNA was purified using IPure Beads (provided in kit) and qPCR performed using the FastStart Universal SYBR Green Master Mix (Roche). Fold enrichment over IgG was calculated based on percentage of input values: 100 × 2(Ct[adjusted input] – Ct[IP]).
Primer sequences for putative binding sites of BRG1 on Sox9 promoter regions were described previously (12). The following primer sequences were used for putative binding sites of PDX1 on Sox9 promoter regions: Sox9 (–1475 bp): forward: 5′-TAAAGCGAATCGGCCTGTAT-3′, reverse: 5′-ACGAGTTCCTTTCCCCAAAT-3′; Sox9 (–3205 bp): forward: 5′-TAGACGAGGTCCCCACTTTG-3′, reverse: 5′-GGCTCAGTCACTTGCATTTTC-3′; Sox9 (–28 kb): forward: 5′-AGATCCACCCTGCAAACAAG-3′, reverse: 5′-AAGATCCTGGGTGTTTGCTG-3′; Sox9 (–70 kb): forward: 5′-CATCGTGCCTAATATCGCCTA-3′, reverse: 5′-TGATTGTTGCACCACCAACT-3′.
Targeted sequencing.
For DNA extraction, 5 cut sections of 10 μm thickness were prepared from formalin-fixed, paraffin-embedded (FFPE) tumor tissue from each patient, and genomic DNA was extracted using the GeneRead DNA FFPE Kit (QIAGEN). For sample preparation, 200 ng of genomic DNA was used as input for library preparation using the KAPA Hyper Prep Kit (Kapa Biosystems), followed by targeted capture using a SureSelect Custom Kit (Agilent Technologies), for which 67 pancreatic cancer–associated genes, including BRG1, and 1,250 SNPs regularly scattered throughout the genome were selected. Next-generation sequencing was performed using the Illumina HiSeq 2500 platform with a standard 125-bp paired-end read protocol. The average read depth was ×457, and the average percentage of targeted bases covered by at least ×100 was more than 99%. Sequence alignment and mutation calling were performed using our in-house pipelines, as previously described (51, 52), with minor modifications. Mutations were called and filtered by excluding (i) synonymous SNVs; (ii) variant allele frequency (VAF) of less than 0.05; (iii) strand ratio equal to 0 or 1; (iv) EBCall –log P value of less than 4, using sequencing data of 20 FFPE normal pancreatic tissues as controls; (v) known variants listed in the 1000 Genomes Project (http://www.internationalgenome.org/; October 2014 release), National Heart, Lung, and Blood Institute Exome Sequencing Project (ESP) 5400 (http://evs.gs.washington.edu/EVS/), and the Human Genetic Variation Database (HGVD: http://www.hgvd.genome.med.kyoto-u.ac.jp/, April 2016 release). The variants with 0.35 < VAF < 0.65 that were protein-truncating variants or were registered in the Catalogue of Somatic Variations in Cancer (COSMIC) (https://cancer.sanger.ac.uk/cosmic) were considered to be somatic mutations. All variants with VAF less than 0.35 or VAF greater than 0.65 were considered as somatic mutations. In PDA_3, a missense variant in BRG1 was confirmed in tumor and paired normal tissues by amplicon sequencing and found to be a germline variant. Copy number analyses were performed by in-house pipelines, named CNACS, as previously described (53). Briefly, copy number abnormalities were identified using the data of allele frequencies and sequenced depth of SNPs.
Data deposition.
All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE93615).
Statistics.
Data are presented as mean ± SEM. Statistical comparisons between groups were made using the 2-tailed Student’s t test. P values less than 0.05 were considered statistically significant. All statistical analyses were performed using either GraphPad Prism (version 6.0) or Microsoft Excel 2016.
Study approval.
All mouse experiments were approved by the IACUC and the Ethics Committee of Kyoto University. In human experiments, the Ethics Committee of the Kyoto University Hospital approved the protocol before the initiation of the study, and written informed consent was obtained from each patient.
Author contributions
MT and AF conceived and designed the study. MT, NR, YH, LL, NK, Satoshi Ogawa, NG, KI, YK, YM, and GEK performed the experiments and analyzed the data. HA generated Sox9OE mice. CVW generated Ptf1a-CreER mice. DS generated Pdx1-Flp, FSF-KrasG12D/+, and FSF-R26 CAG-CreERT2/+ mice. MT, YT, TY, TM, YY, KT, KH, Seishi Ogawa, and SU contributed reagents, materials, and analysis tools. MT and NR wrote the manuscript, and AF, MH, TC, and HS revised it.
Supplementary Material
Acknowledgments
The authors thank Yoshiya Kawaguchi for sharing Ptf1a-Cre and Trp53R172H mice, David Tuveson for sharing KrasG12D mice, David Reisman for sharing Brg1flox mice, Katsutoshi Kuriyama and Yuta Kawamata for technical support, and Yuzo Kodama and all of the Fukuda/Seno/Kodama laboratory members for helpful discussions. This work was supported in part by Grants-in-Aid KAKENHI (25112707, 26293173, 14J03460, 16K09394, and 16K15427); a research program as part of the Project for Development of Innovative Research on Cancer Therapeutics (P-Direct) from the Ministry of Education, Culture, Sports, Science, and Technology; and the Japan Society for the Promotion of Science. It was also supported by Health Labor Sciences Research grants from the Ministry of Health, Labor, and Welfare (for the development of therapeutic drugs for intractable inflammatory bowel disease); the Kobayashi Foundation for Cancer Research; the Naito Foundation; the Princess Takamatsu Cancer Research Fund (13-24514); the Mochida Foundation; the Mitsubishi Foundation, the Japanese Society of Gastroenterology, and NCI RO1CA172045 (to MH).
Version 1. 07/16/2018
Electronic publication
Version 2. 08/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(8):3475–3489.https://doi.org/10.1172/JCI94287.
Contributor Information
Motoyuki Tsuda, Email: mtsuda@kuhp.kyoto-u.ac.jp.
Akihisa Fukuda, Email: fukuda26@kuhp.kyoto-u.ac.jp.
Nilotpal Roy, Email: nilotpalr@gmail.com.
Yukiko Hiramatsu, Email: hirayuki@kuhp.kyoto-u.ac.jp.
Laura Leonhardt, Email: Laura.Leonhardt@ucsf.edu.
Nobuyuki Kakiuchi, Email: kakiuchi@kuhp.kyoto-u.ac.jp.
Kaja Hoyer, Email: kaja.hoyer@charite.de.
Satoshi Ogawa, Email: sogawa@kuhp.kyoto-u.ac.jp.
Norihiro Goto, Email: norihiro@kuhp.kyoto-u.ac.jp.
Kozo Ikuta, Email: kikuta@kuhp.kyoto-u.ac.jp.
Yoshito Kimura, Email: ykimu80@kuhp.kyoto-u.ac.jp.
Yoshihide Matsumoto, Email: matkyoto@kuhp.kyoto-u.ac.jp.
Yutaka Takada, Email: yutakada@kuhp.kyoto-u.ac.jp.
Takuto Yoshioka, Email: takuto@kuhp.kyoto-u.ac.jp.
Takahisa Maruno, Email: maruno@kuhp.kyoto-u.ac.jp.
Yuichi Yamaga, Email: ymgych49@kuhp.kyoto-u.ac.jp.
Grace E. Kim, Email: Grace.Kim@ucsf.edu.
Haruhiko Akiyama, Email: hakiyama@gifu-u.ac.jp.
Seishi Ogawa, Email: sogawa-tky@umin.ac.jp.
Dieter Saur, Email: dieter.saur@tum.de.
Kyoichi Takaori, Email: takaori@kuhp.kyoto-u.ac.jp.
Shinji Uemoto, Email: uemoto@kuhp.kyoto-u.ac.jp.
Matthias Hebrok, Email: mhebrok@diabetes.ucsf.edu.
Tsutomu Chiba, Email: chiba@kuhp.kyoto-u.ac.jp.
Hiroshi Seno, Email: seno@kuhp.kyoto-u.ac.jp.
References
- 1.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med. 2014;371(11):1039–1049. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 2.Matthaei H, Schulick RD, Hruban RH, Maitra A. Cystic precursors to invasive pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2011;8(3):141–150. doi: 10.1038/nrgastro.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanda M, et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142(4):730–733.e9. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/S1535-6108(03)00309-X. [DOI] [PubMed] [Google Scholar]
- 5.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7(5):469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 6.Morris JP, Cano DA, Sekine S, Wang SC, Hebrok M. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest. 2010;120(2):508–520. doi: 10.1172/JCI40045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De La O JP, et al. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105(48):18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Habbe N, et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105(48):18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi G, et al. Loss of the acinar-restricted transcription factor Mist1 accelerates Kras-induced pancreatic intraepithelial neoplasia. Gastroenterology. 2009;136(4):1368–1378. doi: 10.1053/j.gastro.2008.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 11.Marquez-Vilendrer SB, Thompson K, Lu L, Reisman D. Mechanism of BRG1 silencing in primary cancers. Oncotarget. 2016;7(35):56153–56169. doi: 10.18632/oncotarget.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Figura G, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol. 2014;16(3):255–267. doi: 10.1038/ncb2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy N, et al. Brg1 promotes both tumor-suppressive and oncogenic activities at distinct stages of pancreatic cancer formation. Genes Dev. 2015;29(6):658–671. doi: 10.1101/gad.256628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerra C, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11(3):291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 15.Freed-Pastor WA, Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26(12):1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waddell N, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Means AL, et al. Pancreatic epithelial plasticity mediated by acinar cell transdifferentiation and generation of nestin-positive intermediates. Development. 2005;132(16):3767–3776. doi: 10.1242/dev.01925. [DOI] [PubMed] [Google Scholar]
- 18.Wagner M, Lührs H, Klöppel G, Adler G, Schmid RM. Malignant transformation of duct-like cells originating from acini in transforming growth factor transgenic mice. Gastroenterology. 1998;115(5):1254–1262. doi: 10.1016/S0016-5085(98)70098-8. [DOI] [PubMed] [Google Scholar]
- 19.Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32(1):128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 20.Pan FC, et al. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development. 2013;140(4):751–764. doi: 10.1242/dev.090159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hale MA, et al. The nuclear hormone receptor family member NR5A2 controls aspects of multipotent progenitor cell formation and acinar differentiation during pancreatic organogenesis. Development. 2014;141(16):3123–3133. doi: 10.1242/dev.109405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Q, Law AC, Rajagopal J, Anderson WJ, Gray PA, Melton DA. A multipotent progenitor domain guides pancreatic organogenesis. Dev Cell. 2007;13(1):103–114. doi: 10.1016/j.devcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Martinelli P, Cañamero M, del Pozo N, Madriles F, Zapata A, Real FX. Gata6 is required for complete acinar differentiation and maintenance of the exocrine pancreas in adult mice. Gut. 2013;62(10):1481–1488. doi: 10.1136/gutjnl-2012-303328. [DOI] [PubMed] [Google Scholar]
- 24.Furuyama K, et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43(1):34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 25.Kopp JL, et al. Sox9+ ductal cells are multipotent progenitors throughout development but do not produce new endocrine cells in the normal or injured adult pancreas. Development. 2011;138(4):653–665. doi: 10.1242/dev.056499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukuda A, et al. Reduction of Ptf1a gene dosage causes pancreatic hypoplasia and diabetes in mice. Diabetes. 2008;57(9):2421–2431. doi: 10.2337/db07-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pinho AV, et al. Adult pancreatic acinar cells dedifferentiate to an embryonic progenitor phenotype with concomitant activation of a senescence programme that is present in chronic pancreatitis. Gut. 2011;60(7):958–966. doi: 10.1136/gut.2010.225920. [DOI] [PubMed] [Google Scholar]
- 28.Hess DA, et al. Extensive pancreas regeneration following acinar-specific disruption of Xbp1 in mice. Gastroenterology. 2011;141(4):1463–1472. doi: 10.1053/j.gastro.2011.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talchai SC, Accili D. Legacy effect of foxo1 in pancreatic endocrine progenitors on adult β-cell mass and function. Diabetes. 2015;64(8):2868–2879. doi: 10.2337/db14-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamura T, et al. Regulation of pancreatic juxtaductal endocrine cell formation by FoxO1. Mol Cell Biol. 2009;29(16):4417–4430. doi: 10.1128/MCB.01622-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang HS, et al. Transcription factor Glis3, a novel critical player in the regulation of pancreatic beta-cell development and insulin gene expression. Mol Cell Biol. 2009;29(24):6366–6379. doi: 10.1128/MCB.01259-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grimont A, et al. SOX9 regulates ERBB signalling in pancreatic cancer development. Gut. 2015;64(11):1790–1799. doi: 10.1136/gutjnl-2014-307075. [DOI] [PubMed] [Google Scholar]
- 33.Kopp JL, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22(6):737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fukuda A, Chiba T. Sox9-dependent acinar-to-ductal reprogramming is critical for pancreatic intraepithelial neoplasia formation. Gastroenterology. 2013;145(4):904–907. doi: 10.1053/j.gastro.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 35.Shih HP, et al. A gene regulatory network cooperatively controlled by Pdx1 and Sox9 governs lineage allocation of foregut progenitor cells. Cell Rep. 2015;13(2):326–336. doi: 10.1016/j.celrep.2015.08.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shih HP, et al. A Notch-dependent molecular circuitry initiates pancreatic endocrine and ductal cell differentiation. Development. 2012;139(14):2488–2499. doi: 10.1242/dev.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prévot PP, et al. Role of the ductal transcription factors HNF6 and Sox9 in pancreatic acinar-to-ductal metaplasia. Gut. 2012;61(12):1723–1732. doi: 10.1136/gutjnl-2011-300266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schönhuber N, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med. 2014;20(11):1340–1347. doi: 10.1038/nm.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoang CQ, et al. Transcriptional maintenance of pancreatic acinar identity, differentiation, and homeostasis by PTF1A. Mol Cell Biol. 2016;36(24):3033–3047. doi: 10.1128/MCB.00358-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krah NM, et al. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. Elife. 2015;4:e07125. doi: 10.7554/eLife.07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna B, Guo M, Reynolds A, Hara M, Stein R. Dynamic recruitment of functionally distinct Swi/Snf chromatin remodeling complexes modulates Pdx1 activity in islet β cells. Cell Rep. 2015;10(12):2032–2042. doi: 10.1016/j.celrep.2015.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter DM, et al. Systematic in vivo inactivation of chromatin-regulating enzymes identifies Setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res. 2017;77(7):1719–1729. doi: 10.1158/0008-5472.CAN-16-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boj SF, et al. Organoid models of human and mouse ductal pancreatic cancer. Cell. 2015;160(1-2):324–338. doi: 10.1016/j.cell.2014.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15(24):3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17(10):5976–5986. doi: 10.1128/MCB.17.10.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olive KP, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119(6):847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 47.Kim Y, et al. Generation of transgenic mice for conditional overexpression of Sox9. J Bone Miner Metab. 2011;29(1):123–129. doi: 10.1007/s00774-010-0206-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei D, et al. KLF4 Is essential for induction of cellular identity change and acinar-to-ductal reprogramming during early pancreatic carcinogenesis. Cancer Cell. 2016;29(3):324–338. doi: 10.1016/j.ccell.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu L, Shi G, Schmidt CM, Hruban RH, Konieczny SF. Acinar cells contribute to the molecular heterogeneity of pancreatic intraepithelial neoplasia. Am J Pathol. 2007;171(1):263–273. doi: 10.2353/ajpath.2007.061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fukuda A, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19(4):441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 52.Shiraishi Y, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41(7):e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoshizato T, et al. Genetic abnormalities in myelodysplasia and secondary acute myeloid leukemia: impact on outcome of stem cell transplantation. Blood. 2017;129(17):2347–2358. doi: 10.1182/blood-2016-12-754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.