Abstract
Background
The antitumour efficacy of tyrosine kinase inhibitors (TKIs) in lung cancer patients with compound epidermal growth factor receptor (EGFR) mutations has not been resolved. Our study summarizes a single institutional experience of first-generation TKI therapy for lung cancers with compound EGFR mutations.
Methods
A total of 106 consecutive patients with tumours bearing compound EGFR mutations were identified between January 2012 and May 2016; all patients received first-generation TKI therapy. Deletions in exon 19 and the L858R point mutation in exon 21 were considered common mutations; T790M was considered separately because of its association with TKIs resistances. Any other mutation was defined as a rare mutation. Patients were divided as follows: double common mutations (group A); common plus T790M mutations (group B); common plus rare mutations (group C); double rare mutations (group D); and rare plus T790M mutations (group E). A separate group of 115 consecutive patients with a single common mutation was created for comparative analysis (group F).
Results
The frequency of patients with compound EGFR was 2.9% (114/3925) and their response rate to first-generation TKIs was 50.9%, which was not significantly different from group F (67.0%, P = 0.088). The progression-free survival (PFS) of the 106 patients receiving TKI therapy was worse than that of group F (median, 9.1 vs. 13.0 months, respectively; P < 0.001). The PFS of the compound mutation group was shorter than that of the single common mutation group (median, 10.1 months in group A, P = 0.240; 9.1 months in group B, P < 0.001; 9.6 months in group C, P = 0.010; 6.5 months in group D, P = 0.048; 5.4 months in group E, P = 0.017). Patients with a co-occurring mutation in exon 20 (excluding T790M) exhibited significantly worse PFS than the patients with other compound mutations or with a single common mutation (median, 6.5 vs. 9.1 vs. 13.0 months, respectively, P = 0.002).
Conclusions
There was significant heterogeneity among the compound EGFR mutations and their response to first-generation TKIs. Individualized treatment in clinical practice should be considered for each case.
Keywords: EGFR, TKIs, Compound mutations
Introduction
Lung cancer continues to be the primary cause of cancer death both in China and worldwide. The 5-year survival rate for lung cancer has increased from 12% in the 1970s to 17.7% in 2016 [1], and these sufferers primarily include numerous patients with advanced lung cancer. Many advances have been made in the treatment of advanced lung cancer, especially with regard to targeted therapies, and these treatment strategies have resulted in considerable improvements in survival. Among them, the receptor tyrosine kinases (RTKs) super-family of cell surface receptors serve as mediators of cell signaling by extra-cellular growth factors. Members of the ErbB family of RTKs, such as ErbB1 (also known as EGFR), ErbB2, ErbB3 and ErbB4, have received much attention, given their strong association with malignant proliferation [2, 3].
Over the past decade, three small-molecule ErbB tyrosine kinases inhibitors (TKIs) have been shown to efficiently target tumour cell survival pathways in advanced non-small cell lung cancers (NSCLC) expressing the epidermal growth factor receptor (EGFR): gefitinib (approved by the US Food and Drug Administration in May 2003), erlotinib (approved by the US Food and Drug Administration in November 2004) and icotinib (approved by China’s State Food and Drug Administration in June 2011). The use of these agents has resulted in higher overall response rates (ORRs, up to 60%–70%) and longer progression-free survival (PFS; 9–16 months) and overall survival (OS; exceeding 20 months) than current first-line platinum-based chemotherapies [1–5].
These patients with classical EGFR genetic mutations that are sensitive to TKIs involve in-frame deletions of exon 19 and L858R substitutions of exon 21 and occur in approximately 85%–90% of all EGFR-mutated patients [1, 4–6]. However, compound EGFR mutations with or without classical mutations have been detected within the same tumour tissues in some patients [7–14]. Previous studies reported that only 2%–15% of the population with EGFR mutations exhibits these rare compound mutations [7–14]. The characteristics of this rare population, the efficacy of EGFR TKIs, and the prognostic value of the compound mutations have not been clarified because of the very low rate of these mutations. Furthermore, the lack of adequate evidence-based medical research hinders treatment decisions when these co-existing double-site mutants are detected.
The present cohort analysis examined two major questions: first, we investigated the incidence of different compound EGFR mutation subtypes in a single institution; and second, we investigated this population’s characteristics and the efficacy (ORRs and PFS) of first-generation small-molecule TKI treatment and prognosis compared with patients bearing the classical mutation alone.
Materials and methods
Patient selection
We retrospectively reviewed a molecular diagnostic database of lung cancer in the Sun Yat-sen University Cancer Center (SYSUCC) between January 2012 and May 2016. The database was screened for EGFR-mutated cases, and a cohort of consecutive cases with compound EGFR mutations was identified. The co-existence of two different EGFR mutation sites detected in a single tumour specimen was defined as a compound EGFR mutation. The hospital’s ethics committee approved the research using this micro-database, and all subjects provided written informed consent.
Patients were eligible for enrolment if they had received daily oral EGFR TKIs, such as gefitinib (250 mg, qd), erlotinib (150 mg, qd) or icotinib (125 mg, tid), until disease progression or death; complete follow-up information was obtained from the medical record department. The co-existence of compound mutations with T790M must have been detected prior to initiation of targeted therapy.
Exclusion criteria included second primary neoplasms diagnosed before/after the lung cancer, intolerant levels of toxicity, loss to follow-up, and refusal of treatment.
Sequencing of EGFR mutations in exons 18–21
Details of the genetic sequencing and molecular analysis were described previously [6]. Briefly, tumour cell DNA was extracted from paraffin-embedded specimens that contained at least 50% tumour cells, including surgically resected tumour specimens, fine needle aspiration biopsies of lymph nodes or metastatic lesions, and tumour cells from pleural fluid, using a QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) in strict accordance with the manufacturer’s recommendations. The tumour cell DNA was examined for EGFR mutation(s) in exons 18–21, including 36 TKI-responsive mutation sites and nine TKI-resistant mutation sites, using an amplification refractory mutation system polymerase chain reaction (ARMS-PCR) kit (GP Medical Technology Co. Ltd., Beijing, China) [6]. Mass ARRAY TYPER 4.0 software (Sequenom Inc., San Diego, CA, USA) was used to individually classify EGFR-positive mutation sites when the mutation frequency was higher than 1%.
Follow-up and end-point
A systemic baseline assessment, including chest and abdomen enhanced computed tomography (CT) scanning and brain enhanced magnetic resonance imaging examination, was routinely performed prior to EGFR TKI treatments for locally advanced, recurrent, or metastatic lung cancer. A follow-up assessment was generally performed every 3 months after the 1st day of TKI treatment until February 28, 2017, radiographic progression or death. Two board-certified radiologists independently evaluated the therapeutic effectiveness based on the Response Evaluation Criteria in Solid Tumours (RECIST) and classified the therapeutic effect into four levels: progressive disease (PD), stable disease (SD), partial response (PR), and complete response (CR). Patients exhibiting PD or SD were considered non-responders to targeted treatments, while patients with PR or CR were regarded as effective disease control by antitumour agents. Follow-up information was extracted from the patients’ complete medical and radiological records.
The duration of PFS was calculated from the 1st day of EGFR TKI treatment to the last follow-up, the date of death or when disease progression was first observed. The duration of OS was also evaluated from the date of the 1st day of EGFR TKI treatment until the last follow-up or the date of death from any cause. Patients were censored at their last known progression-free or alive date.
Data analysis
Complete medical, pathological and radiological data and molecular diagnostics were analysed using the Statistical Package for the Social Sciences for Windows version 23.0 (SPSS Inc., Chicago, IL, USA). Categorical variables were compared between the EGFR mutant subgroups (i.e., single common mutation, double common mutations, common plus T790M mutations, common plus rare mutations, rare plus rare mutations, and rare plus T790M mutations) using Chi square (χ2) and Fisher’s exact tests. The Kaplan–Meier method was used to construct the PFS and OS curves of each subgroup, and significant differences between survival curves were examined using the log-rank test. A two-sided P value less than 0.05 was used to confirm significant differences.
Results
Clinical and pathological features of the population with compound EGFR mutations
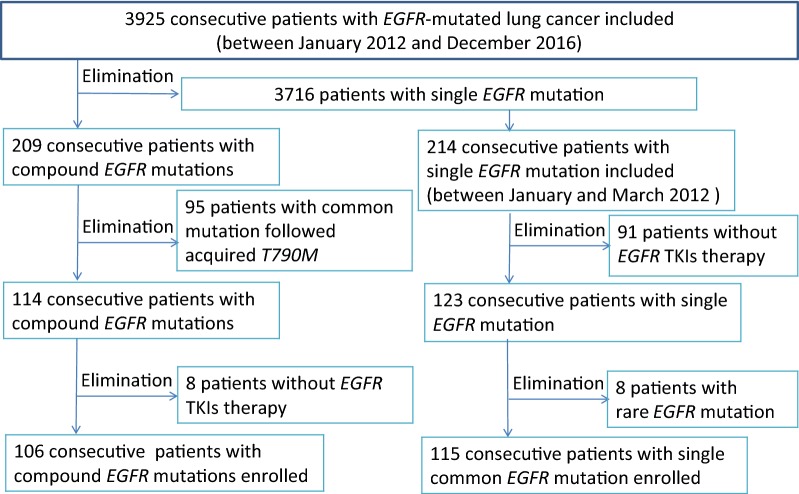
A total of 3925 NSCLC patients with EGFR mutations were identified using ARMS-PCR between January 2012 and December 2016 in SYSUCC. A total of 209 consecutive patients with compound EGFR mutations were identified among the 3925 NSCLC patients. However, 95 cases with the 19Del or L858R mutations that acquired the T790M after TKI therapy and 8 cases that did not receive TKI therapy in our hospital were excluded. Only 106 patients (2.9%) with primary co-existing double EGFR mutations received first-generation TKI therapy and were entered into our analysis (Fig. 1).
Fig. 1.
Screening procedure of the 106 lung cancer patients with compound EGFR mutations
Most of the 106 patients were non-geriatric (63/106, 59.4%; median age at initial diagnosis, 57 ± 10.7 years), non-smokers (69/106, 65.1%) and had advanced lung cancer (86/106, 81.1%) (Table 1). All initial pathological stage I -IIIA patients (20/106, 18.9%) received radical resection. No sex predilection (55 women and 51 men) was present in our cohort. Notably, a compound EGFR mutation was detected in four non-adenocarcinoma patients, including two squamous cell carcinomas (SCC, L858R + L858Q and L858R + T790M), one lymphoepithelioma-like carcinoma (LELC, 19Del + L858R) and one adenosquamous carcinoma (ASC, 19Del + T790M). The first-generation EGFR TKIs were primarily administered as first (55/106, 51.9%) or second line (46/106, 43.4%) treatment.
Table 1.
Clinicopathological characteristics of lung cancer patients with compound EGFR mutations
| Variable | Single common mutation (n = 115) | Compound mutations (n = 106) | P † | Double common mutations (n = 5) | P † | Common + rare mutations (n = 11) | P † | Rare + rare mutations (n = 13) | P † | Rare + T790M mutations (n = 8) | P † | Common + T790M mutations (n = 69) | P † |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex | |||||||||||||
| Female | 56 | 55 | 0.635 | 1 | 0.368 | 6 | 0.711 | 6 | 0.862 | 4 | 0.943 | 38 | 0.402 |
| Male | 59 | 51 | 4 | 5 | 7 | 4 | 31 | ||||||
| Age (years) | |||||||||||||
| < 60 | 57 | 63 | 0.141 | 2 | 0.675 | 5 | 0.794 | 8 | 0.413 | 7 | 0.063 | 41 | 0.195 |
| ≥ 60 | 58 | 43 | 3 | 6 | 5 | 1 | 28 | ||||||
| Smoking status | 0.270 | 0.118 | 0.795 | 0.442 | 0.795 | 0.022 | |||||||
| Non-smoker | 69 | 69 | 1 | 5 | 4 | 5 | 54 | ||||||
| Smoker | 34 | 24 | 3 | 3 | 4 | 3 | 11 | ||||||
| Unknown | 12 | 13 | 1 | 0 | 5 | 0 | 4 | ||||||
| Tumour status | 0.034 | 0.592 | 0.782 | 0.753 | 0.103 | 0.064 | |||||||
| Recurrence | 36 | 20 | 1 | 3 | 3 | 0 | 13 | ||||||
| Initial IIIb–IV | 79 | 86 | 4 | 8 | 10 | 8 | 56 | ||||||
| ECOG PS | |||||||||||||
| 0–1 | 107 | 96 | 0.501 | 4 | 0.327 | 9 | 0.212 | 13 | 0.326 | 6 | 0.071 | 64 | 0.941 |
| 2–4 | 8 | 10 | 1 | 2 | 0 | 2 | 5 | ||||||
| Pathology | 0.529 | 0.141 | 0.672 | 0.899 | 0.948 | 0.534 | |||||||
| ADC | 110 | 102 | 4 | 10 | 13 | 8 | 67 | ||||||
| SCC | 5 | 2 | 0 | 1 | 0 | 0 | 1 | ||||||
| LELC | 0 | 1 | 1 | 0 | 0 | 0 | 0 | ||||||
| ASC | 0 | 1 | 0 | 0 | 0 | 0 | 1 | ||||||
| Timing of TKI | 0.720 | 0.914 | 0.835 | 0.803 | 0.805 | 0.460 | |||||||
| First line | 52 | 55 | 2 | 5 | 6 | 3 | 39 | ||||||
| Second line | 55 | 46 | 3 | 6 | 7 | 5 | 25 | ||||||
| Third line | 7 | 4 | 0 | 0 | 0 | 0 | 4 | ||||||
| Fourth line | 1 | 1 | 0 | 0 | 0 | 0 | 1 | ||||||
| TKI selection | 0.034 | 0.981 | 0.592 | 0.214 | 0.161 | 0.021 | |||||||
| Gefitinib | 64 | 47 | 3 | 5 | 4 | 2 | 33 | ||||||
| Erlotinib | 26 | 41 | 1 | 4 | 4 | 4 | 28 | ||||||
| Icotinib | 25 | 18 | 1 | 2 | 5 | 2 | 8 | ||||||
| Response to TKIs | |||||||||||||
| PD | 10 | 14 | 0.088 | 1 | 0.148 | 2 | 0.652 | 2 | 0.328 | 4 | 0.012 | 5 | 0.293 |
| SD | 21 | 33 | 1 | 3 | 5 | 1 | 22 | ||||||
| PR | 77 | 54 | 3 | 5 | 5 | 3 | 39 | ||||||
| CR | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| NE | 6 | 5 | 0 | 1 | 1 | 0 | 3 | ||||||
ECOG PS Eastern Cooperative Oncology Group performance status, ADC adenocarcinoma, SCC squamous cell carcinoma, LELC lymphoepithelioma-like carcinoma, ASC adenosquamous carcinoma, PD progressive disease, SD stable disease, PR partial response, CR complete response, NE not evaluated, TKI tyrosine kinase inhibitor, PFS progression-free survival, OS overall survival
†All variables of different subgroups were compared with the single common mutation group; P < 0.05 was defined as significantly different
A total of 115 consecutive patients with a single common mutation (19Del or L858R) who received first-generation EGFR TKI therapy between January and March 2012 were selected for comparative analysis (Fig. 1).
Distribution frequency of compound EGFR mutations
We divided the cohort with the compound EGFR mutations into five groups based on the categories of common and rare mutations sites reported in the literature: a double common mutation group (5/106, 4.7%), a common plus rare mutation group (11/106, 10.4%), a common plus T790M mutation group (69/106, 65.1%), a double rare mutation group (13/106, 12.3%), and a rare plus T790M mutation group (8/106, 7.5%) (Table 1) [3]. The most frequent mutation site was T790M (77/106, 72.6%), and the majority of patients harboured 19Del (50/106, 47.2%) or L858R (40/106, 37.7%) as one of the compound mutations (Table 3). Notably, the most frequent compound mutation involved a common mutation co-existing with T790M (69/106, 65.1%), and these common mutations included 19Del or L858R with T790M. The most frequent uncommon mutation was L858Q (13/106, 12.3%), followed by S768I (9/106, 8.5%), G719X (7/106, 6.6%), G719S (5/106, 4.7%), D761Y (2/106, 1.9%), S720P (1/106, 0.9%), K757R (1/106, 0.9%), I744M (1/106, 0.9%), R776C (1/106, 0.9%), L833V (1/106, 0.9%), E709A (1/106, 0.9%), and V774M (1/106, 0.9%).
Table 3.
Frequency, detailed combination patterns, progression-free survival, overall survival and response to first-generation TKIs of compound EGFR mutations
| Subgroup of compound EGFR mutations | Frequency (n, %) | Mutated exons | Response (rate, %) | PFS (range, months) | OS (range, months) |
|---|---|---|---|---|---|
| Double common | 5 (4.7) | 25.0% | 10.1 ± 2.4 | 24.2 ± 8.2 | |
| 19Del + L858R | 5 | 19 and 21 | 3PR, 1SD, 1PD | 4.9–12 | 13.1–25.6 |
| Common + rare | 11 (10.4) | 45.5% | 10.5 ± 3.9 | Not reached | |
| 19Del + L861Q | 2 | 19 and 21 | 1PR, 1SD | 11.9–14.4 | 26.5–41.2 |
| L858R + S720P | 1 | 21 and 18 | PD | 2.1 | 2.1 |
| L858R + K757R | 1 | 21 and 19 | PR | 9.0 | 8.7 |
| L858R + I744 M | 1 | 21 and 19 | PR | 17.6 | 41.2 |
| L858R + S768I | 3 | 21 and 20 | 1PR, 1PD, 1SD | 1.8–6.2 | 4.0–12.5 |
| L858R + R776H | 1 | 21 and 20 | PR | 10.5 | 12.6 |
| L858R + L858Q | 1 | 21 and 21 | NE | 1 | 3 |
| L858R + L833V | 1 | 21 and 21 | SD | 5.0 | 15.9 |
| Common + T790M | 69 (65.1) | 56.5% | 10.3 ± 0.6 | Not reached | |
| 19Del + T790M | 43 | 19 and 20 | 27PR, 12SD, 2 PD, 2NE | 0.6–40.7 | 0.2–88.5 |
| L858R + T790M | 26 | 21 and 20 | 12PR, 10SD, 3PD, 1NE | 0.9–24.1 | 1.2–56.6 |
| Rare + rare | 13 (12.3) | 38.5% | 6.5 ± 1.3 | Not reached | |
| G719C + S768I | 1 | 18 and 20 | PR | 6.5 | 13.2 |
| G719S + S768I | 2 | 18 and 20 | 1PR, 1SD | 1–8.0 | 2.0–8.4 |
| G719S + L858Q | 1 | 18 and 21 | SD | 6.4 | 29.3 |
| G719X + S768I | 3 | 18 and 20 | 2PR, 1SD | 2.0–18 | 2.0–44.0 |
| G719X + L858Q | 3 | 18 and 21 | 1SD, 1PD, 1NE | 0.3–27.3 | 2.3–29.2 |
| G719S + E709A | 1 | 18 and 18 | PR | 4.1 | 4.1 |
| G719S + L858Q | 1 | 18 and 21 | SD | 6.4 | 29.3 |
| S768I + V774M | 1 | 20 and 20 | PD | 2.0 | 13.8 |
| Rare + T790M | 8 (7.5%) | 37.5% | 5.4 ± 2.5 | 23.8 ± 1.5 | |
| G719X + T790M | 1 | 18 and 20 | PR | 11.1 | 55.6 |
| D761Y + T790M | 2 | 19 and 20 | 2PD | 1.1–5.5 | 1.1–8.5 |
| L858Q + T790M | 5 | 21 and 20 | 2PR, 1SD, 2PD | 1.4–20.6 | 18–88.3 |
TKI tyrosine kinase inhibitor, PFS progression-free survival, OS overall survival, PR partial response, SD stable disease, CR complete response, PD progressive disease, NE not evaluated
PFS and patient response after TKI treatment
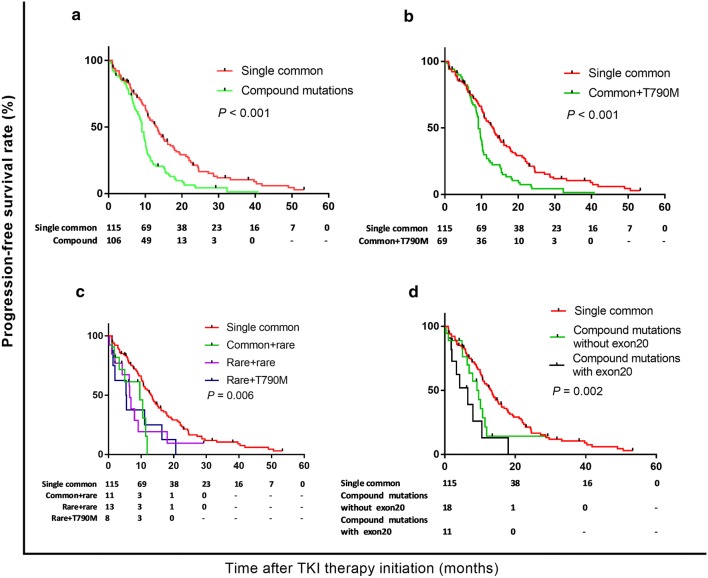
The median follow-up time in the compound EGFR mutation cohort was 29.4 months (range, 1.5–119.5 months), and the 1-, 2-, and 3-year PFS rates after EGFR TKI treatment were 32.7, 4.3, and 1.4%, respectively, which were all significantly lower than the population with a single common mutation (1-, 2-, and 3-year PFS rates were 54.1, 20.1, and 10.5%, respectively, P < 0.001) (Fig. 2a). Twenty-six tumour-related deaths occurred during follow-up, and the median OS was not reached for all patients with compound mutations. Univariate analysis of the total 221 patients who received first-generation TKI therapy revealed that compound mutations were significantly correlated with shorter duration of targeted therapy (HR: 1.883, 95% CI 1.404–2.526, P < 0.001), in addition to initial advanced status, non-adenocarcinoma, and more than second-line treatment (Table 2). Inclusion of these variables in the multivariate analysis revealed that these four factors were also independent significant PFS factors.
Fig. 2.
Progression-free survival by mutation status: a a single common EGFR mutation vs. compound EGFR mutations (median PFS: 9.1 vs. 13.0 months, respectively; P < 0.001), b a single common EGFR mutation vs. common + T790M mutations (median: 9.1 months vs. 13.0 months, P < 0.001), c a single common EGFR mutation vs. common + rare mutations vs. rare + rare mutations vs. rare + T790M mutations (median: 13.0 months vs. 10.5 months vs. 6.5 months vs. 5.4 months, P = 0.006), and d a single EGFR mutation vs. compound mutations without exon 20 vs. compound mutations with exon 20 (median: 13.0 months vs. 9.1 months vs. 6.5 months, P = 0.002)
Table 2.
Univariate and multivariate analysis for progression-free survival to first-generation TKI therapy
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| HR (95% CI) | P † | HR (95% CI) | P † | |
| Sex | ||||
| (Female/male) | 1.061 (0.919–1.224) | 0.420 | ||
| Age | ||||
| (< 60/≥ 60) | 1.319 (0.987–1.763) | 0.061 | ||
| Smoking status | ||||
| (Smoker/nonsmoker/unknown) | 0.726 (0.518–1.018) | 0.063 | ||
| Tumour status | ||||
| (Recurrence/initial IIIb–IV) | 0.721 (0.560–0.926) | 0.011 | 0.706 (0.548–0.909) | 0.007 |
| ECOG PS | ||||
| (0–1/2–4) | 1.438 (0.872–2.372) | 0.154 | ||
| Pathology | ||||
| (Non-adeno/adeno) | 4.175 (2.113–8.250) | 0.001 | 5.472 (2.623–11.417) | 0.001 |
| Timing of TKI | ||||
| (1/≥ 2) | 0.643 (0.480–0.862) | 0.003 | 0.610 (0.452–0.823) | 0.001 |
| EGFR mutation status | ||||
| (Compound/single) | 1.883 (1.404–2.526) | 0.001 | 1.981 (1.466–2.676) | 0.001 |
| TKI selection | ||||
| (Gefitinib/erlotinib/icotinib) | 0.978 (0.819–1.167) | 0.806 | ||
ECOG PS Eastern Cooperative Oncology Group performance status, HR hazard ratio, CI confidence interval
†All variables of different subgroups were compared with the single common mutation group; P < 0.05 was defined as significantly different
Among the five patients with double common EGFR mutations (19Del plus L858R), only one stage IV patient with brain metastases exhibited effective local control, i.e., SD in response to gefitinib, but progression of the primary pulmonary neoplasm was identified after 7.9 months. Three other patients with advanced lung cancer exhibited a PR to oral gefitinib or icotinib therapy, and their prolonged PFS times was longer than 10 months (10.1, 10.7 and 13.5 months, respectively) (Table 3). Notably, the patient with primary pulmonary LELC, whose tumour harboured a 19Del plus L858R mutation, was diagnosed at stage IV because of osseous metastasis. However, no antitumour activity of erlotinib against this rare subtype of lung cancer was observed, i.e., there was PD. There were no significant differences in the response rates (RR, 25.0% vs. 67.0%, P = 0.908, using Chi square tests) or PFS (median, 10.1 months vs. 13.0 months, P = 0.240, using log-rank tests) compared with those of the group with a single common mutation.
The RR to TKI in the patients with common mutations (19Del or L858R) plus T790M, which was not a mutation acquired during oral EGFR TKI, was 56.5% (39/69); this rate was not significantly different than that of patients with a single 19Del or L858R mutation (P = 0.293) (Table 1). These 69 patients exhibited a worse PFS than the 115 patients with a primary T790M (median, 9.1 months vs. 13.0 months, respectively; P < 0.001) (Fig. 2b). Approximately 33% of the patients (23/69) were enrolled in an AZD9291 international multicentre, single-arm phase 2 clinical trial after progression was detected using CT scanning during treatment with first-generation EGFR TKIs.
The compound EGFR-mutated patients with rare site involvement exhibited a lower RR (37.5% vs. 67.8%, P = 0.023) and a shorter median PFS than the single common mutation subgroup (median, 13.0 months vs. 6.5 months, respectively; P < 0.001) (Fig. 2c). However, there was no difference in PFS across the common plus rare mutations subgroup, double rare mutations subgroup, or the rare plus T790M mutations subgroup (median, 10.5 months vs. 6.5 months vs. 5.4 months, respectively; P = 0.984) (Table 1). The co-occurrence of mutations in exon 20 (excluding T790M) had a significant effect on PFS, which was worse than the other compound mutations and the single common mutation patients (median, 13.0 months vs. 9.1 months vs. 6.5 months, respectively; P = 0.002) (Fig. 2d).
Discussion
Our single institution study identified 114 patients with compound EGFR mutations among 3925 patients with EGFR mutations (114/3925, 2.9%). The RR of this rare population to first-generation TKI therapy was 50.9%, which was lower than that of patients with a single common mutation, but the difference was not significant (P = 0.088). Patients with compound mutations exhibited a shorter duration of first-generation TKI therapy in multivariate analysis than patients with a single common mutation (HR, 1.981; 95% confidence interval (CI) 1.466–2.676; P < 0.001). Exclusion of patients with a co-occurrence of mutations in exon 20 (excluding T790M) revealed that the duration of targeted TKI therapy was even shorter for other types of compound mutations (P = 0.002).
The phenomenon of lung cancer cells harbouring multiple EGFR mutations is worth mentioning, and it reportedly has accompanied the clinical use of first-generation small-molecule TKIs since 2004 [7]. Most published studies on multiple mutations were case reports because the techniques for detecting EGFR mutations were used only to detect drug-sensitive mutations in exon 19 and 21, and some patients with compound mutations were likely undetected. Developments in mutational detection and analysis techniques, such as direct sequencing, multiplex PCR systems and next-generation sequencing, increased the number of reported cases with compound mutations between 2004 and 2017 (Table 4). The reported frequency of the rare population with compound mutations ranged from 2.6% to 15% [8–15], which was slightly higher than that observed in our cohort (2.9%).
Table 4.
Literature review of patients harbouring compound EGFR mutations and PFS and response to first-generation TKIs between 2004 and 2017
| Compound mutations | Double common (n, mPFS, response) | Common + rare (n, mPFS, response) | Rare + rare (n, mPFS, response) | Common + T790M (n, mPFS, response) | Rare + T790M (n, mPFS, response) |
|---|---|---|---|---|---|
| Kobayashi et al. [8] | None | 3; 3 monthsa; 2 PR, 1 PD | 4; 8 months; 4 PR | None | None |
| Zhang et al. [9] | 3; 17.5 months; 1 CR, 1 PR, 1 NA | None | None | None | None |
| Hsieh et al. [10] | None | 1; 1.9 months; 1 SR | 6; 11.6 months; 4 PR, 2 PD |
None | None |
| Hata et al. [11] | 8; 12.7 months; 1 CR, 5 PR, 1 SD, 1 NA | 8; 2.5 months; 2 PR, 1 SD, 2 PD, 3 NA | None | None | None |
| Keam et al. [14] | None | 16; 8.1 months; 11 PR, 4 SD, 1 PD | 3; 4.6 months; 1 PR, 1 PD, 1 NA | 5; 8.0 months; 4 PR, 1 PD | None |
| Xu et al. [16] | 14; 9.53 months; 10 PR, 3 SD, 1 PD | 18; 9.8 months; 10 PR, 5 SD, 3 PD | None | 9; 1.9 months; 2 PR, 3 SD, 4 PD | None |
| Wu et al. [17] | None | 7; NA; 2 PR, 1 SD, 4 PD | 3; NA; 2 PR, 1 PD | None | None |
| Chen et al. [18] | None | 10; 8.9 months; 4 PR, 6 NA | None | 3; 6.7 months; 1 PR, 1 SD, 1 NA | 1; 6 months; SD |
| Wu et al. [21] | None | 12; 13.5 months; 10 PR, 1 SD, 1 PD | 7; 4.2 months; 2 PR, 4 SD, 1 PD | 2; NA; 2 PR | None |
| Asahina et al. [26] | None | None | 1; 1.1 months; PD | None | None |
| Zhang et al. [32] | 2; 6.1 months; 2PR | 7; NA; NA | 11; NA; NA | 8; 3.3 months; 1PR, 1SD, 6NA | 3; NA; NA |
| Zhu et al. [33] | None | 3; 5.3 monthsa; 2SD, 1PD | 5; 3.5 monthsa; 2 PR, 2 SD, 1 NA | None | None |
| Wu et al. [34] | None | 9; 8.6 months; 7 PR, 1 SD, 1 PD | 4; 9.2 months; 2 PR, 1 PD, 1 SD | None | None |
| Yang et al. [35] | 1; 2 months; 1 PD | None | None | None | None |
| Svaton et al. [36] | None | None | 1; 8 months; 1 PR | None | None |
| Peng et al. [37] | 2; 11.5 months; 1 PR, 1 SD | 3; 10 months; 3 SD | None | 1; 10 months; 1 SD | None |
| Baek et al. [38] | 12; 7.4 months; 4 CR, 5 PR, 2 SD, 1 PD | None | 11; 5.1 months; 5 CR, 4 PR, 2 SD | None | None |
| Peng et al. [39] | 2; 11.5 months; 1 PR, 1 SD | 4; 8 months; 4 SD | 3; 3 monthsa; 1 CR, 1 SD | 2; 9 months; 2 SD | None |
| Chung et al. [40] | None | 1; 5 monthsa; PR | None | None | None |
| Yang et al. [41] | None | 3; NA; 1 PR, 1 SD, 1 PD | 2; NA; 2 SD | 1; NA; PD | None |
| Ichihara et al. [42] | None | 2; 2.4 months; 2 SD | None | 1; 1.6 months; SD | None |
| Pugh et al. [43] | None | 1; NA; PR | 1; NA; PR | None | None |
| Kimura et al. [44] | None | 1; 5 months; PR | None | None | None |
| Van Zandwijk et al. [45] | None | None | 1; NA; PR | None | None |
| Jackman et al. [46] | None | 1; 14.8 monthsa; SD | None | None | None |
| Pallis et al. [47] | None | 3; NA; 1 PR, 1 SD, 1 PD | 1; NA; PD | None | None |
| Han et al. [48] | None | 1; 13.8 months; 1 SD | 2; 3 months; 2 PR | None | None |
| Kosakaet al. [49] | None | 2; 24.5 months; 2 PR | None | None | None |
| Choong et al. [50] | None | 1; 8 months; PR | None | None | None |
| Oshita et al. [51] | None | 2; 13.2 months; 2 PR | 1; 12 months; SD | None | None |
| Tokumo et al. [52] | None | 1; 2 months; PD | None | None | None |
| Chou et al. [53] | None | None | 2; 4.1 months; 1 PD, 1 PD | None | None |
| Shih et al. [54] | None | 2; NA; 2 PR | 2; NA; 2 PR | None | None |
| Taron et al. [55] | None | 1; 9.4 months; PR | None | None | None |
| Mitsudomi et al. [56] | None | 1, NA,1 PD | None | None | None |
| Takano et al. [57] | None | 2; 12.6 months; 2 PR | None | None | None |
| Pao et al. [58] | None | 1; 13 months; 1 PR | None | None | None |
| Total, n | 44 | 127 | 71 | 32 | 4 |
| ORR, n (%) | 31 (70.5%) | 68 (53.5%) | 34 (47.9%) | 10 (31.2%) | NA |
| mPFS, range (months) | 2–17.5 | 1.9–24.5 | 1.1–12 | 1.6–10 | 6 |
PFS progression-free survival, TKI tyrosine kinase inhibitor, mPFS median progression-free survival, PR partial response, NA not available, SD stable disease, PD progressive disease, CR complete response, SR serological response, ORR overall response rate
amPFS not reached
19Del and L858R mutations are classical sensitizing mutations, and the strong response of these two mutations to TKIs has been demonstrated in many prospective studies [2–5, 9]. However, these two common mutations are frequently detected concomitant with other mutations in the compound EGFR-mutated population. A compound mutation co-existing with a 19Del or L858R mutation was the most common combination in previous reports (203/278, 73.0%) (Table 4) and in our cohort (85/106, 80.2%) (Table 3). Xu et al. [16] reported that tumours with double common EGFR mutations (19Del + L858R, n = 18) exhibited similar antitumour responses to small-molecule TKIs as tumours with single common mutations, and the median PFS and ORR rates were 9.53 months and 71.4% (10/14), respectively, which is consistent with our results (10.1 months and 60%, respectively) and those of Hata Akito (16.5 months and 86%, respectively) [11]. Some case reports also found that first-generation EGFR TKIs may be a desirable therapeutic strategy for patients with advanced lung cancer with synchronous 19Del and L858R mutations [8, 9, 35].
In patients harbouring common plus rare mutation, the L858R mutation was more frequently observed than the 19Del mutation. For example, approximately 10% and 17.3% of NSCLC patients harboured the L858R mutation concomitantly with rare mutations in the two cohorts reported by Wu et al. [17] and Kobayashi et al. [8], respectively. Similarly, in our common plus rare mutation subgroup, L858R, was identified in the majority of the cases (9/11, 81.8%).
The response to TKIs in patients with common plus rare mutations and whether TKI therapy prolonged PFS remains controversial because of the relatively large heterogeneity. Keam [14] reported an RR of 68.8% and median PFS time of 8.1 months in 16 patients, which are similar to our observed RR (45.5%) and median PFS time (10.5 months) in 11 patients. Notably, this above finding was also reported previously [16, 18]. As a whole, this population may benefit from TKIs, but to lesser extent than the population harbouring a single common mutation. We found that the patients with L858R + K757R mutations (exon 21 + exon 19) and L858R + I744M mutations (exon 21 + exon 19) exhibited a partial response to gefitinib and obtained PFS of 9.0- and 9.6-month, respectively. Klughammer et al. [19] and Kempf et al. [20] reported that a single I744M mutation or a single K757R mutation in exon 19 may be TKI-sensitizing mutations, and these mutations were also observed to have PR to oral TKI therapy. Therefore, the above double or single mutation(s) patterns may be candidates for TKI therapy. Patients with exon 20 mutations are considered resistant to TKIs (discussed below), but our study included a patient with L858R plus the R776H mutation (exon 21 + exon 20) showing PR to TKIs for 10.5 months, which is also highly consistent with prior reports [8, 21]. Other patients with the L858R mutation associated with S720P, S768I, L858Q or L833V were classified in the insensitive to TKIs group, and most (5/6, 83.3%) exhibited PD or SD to TKI therapy. To the best of our knowledge, our study is the first to report the combinations of L858R + S720P mutations (exon 21 + exon 18) and L858R + L833V mutations (exon 21 + exon 21). No patient with L833V + H835L mutations (exon 21 + exon 21) was detected in our cohort; however, patients with this combination have been reported to have a good response to gefitinib [22, 23]. One case is especially notable. Leventakos et al. [24] demonstrated that patients with L858R + S768I mutations may be sensitive, or at least not resistant, to TKI therapy, which is in contrast to our results. Our patients with a single L861Q mutation or compound mutations with L861Q exhibited a high RR (66%) and non-inferior PFS (median, 6 months) to TKI therapy. We also detected two cases with 19Del + L861Q mutations, and a good response to TKIs (one PR and one SD) and prolonged PFS (11.4 and 11.9 months) were observed. However, the patient with L858R + L861Q exhibited PD after only 1 month of TKI therapy.
Notably, common mutations concomitant with an initial T790M mutation accounted for 65.1% of all compound mutations in our cohort, which was higher than in previous reports [13]. However, Su et al. [13] verified that pre-treatment of a co-existing EGFR T790M mutation was not a rare event (23/73, 31.5%), and the PFS was significantly shorter than that in patients without T790M (median, 6.7 months vs. 10.2 months, respectively; P = 0.035) [13]. T790M status also affected PFS in our cohort compared with a single-sensitizing EGFR mutation (9.1 months vs. 13.0 months, respectively; P < 0.001). We hypothesize that the scarcity of reports may be due to the bias of excluding patients with T790M [21]. Previous research on compound mutations may have overlooked the fact that—T790M may occur in patients before receiving EGFR TKI treatment [1, 21]. In addition, direct sequencing may be used to detect the classical sensitizing mutations in exons 19 and 21, which leads to a missed opportunity to discover patients with co-occurring T790M. The impact of EGFR TKIs in these patients with 19Del or L858R plus T790M was not clarified because of the scarcity of patients and the varying durations of PFS in the published literature. However, approximately one-third and one-half of patients with concomitant initial T790M as one of the compound mutations in previous studies [13] and our cohort obtained more than 8 months of PFS with the aid of TKI therapy. Therefore, small molecule TKIs may be an optional therapeutic strategy to identify potential beneficiaries after explaining the bias of the therapy to patients in detail to ensure patient understanding and informed consent.
Patients who harbour a single exon 20 mutation in EGFR are reportedly insensitive to small-molecule TKIs [14, 19, 25–27]. However, whether patients with an EGFR exon 20 mutation accompanied by another mutation are candidates for TKI therapy remains unanswered. Marius Lund-Iversen and his colleagues reported seven exon 20-positive patients who received oral TKI, including five patients with single exon 20 mutation and two patients with double mutations. The five patients with single exon 20 mutation were found to have progressive disease at the first post-treatment follow-up, but the two patients with double mutations obtained 11 and 14 months of an ongoing response [28]. Chen et al. [18] also concluded that patients with compound mutations involving mutation in exon 20 benefited from TKIs more than single exon 20 mutations [18]. The duration of response to TKIs in compound EGFR-mutated patients with concomitant exon 20 mutation (excluding T790M) (6.5 months) was still shorter than compound mutated patients without exon 20 mutation (9.1 months) and patients with single common mutations (13.0 months), which is consistent with Keam et al. [14] (< 5 months). The analytical results of Kancha et al. [29] and Wu et al. [17] also support this finding. Together this suggests that first-generation EGFR TKIs may not be suitable for patients with an exon 20 mutation regardless of the presence of other mutations.
Overall, patients with double rare mutations or a rare mutation plus T790M exhibited a lower RR (38.5% and 37.5%, respectively) and worse PFS to TKI therapy (median, 6.5 and 5.4 months, respectively) in our cohort, which is consistent with a previous publication [14]. A patient with a single L861Q point mutation at exon 21 and a G719X point mutation at exon 18 may be classified into the TKI-sensitive mutation group [30]. However, patients with a L861Q or G719X mutation co-occurring with a rare mutation or T790M affected the effectiveness and sensitivity to TKI therapy in our clinical practice (RR, 28.6%; median PFS, 5.1 months). We found five patients with a rare mutation plus T790M, which is more than the overall number of reported cases. One patient with G719X + T790M mutations and one patient with L858Q + T790M mutations exhibited PR to TKI therapy and obtained more than 10 months PFS, which was similar to a case report from Chen et al. [18]. Balak et al. [31] found that the D761Y mutation in exon 19 was a novel secondary resistance mutation to EGFR TKIs [31]. Therefore, the two patients with two resistance mutations (D761Y + T790M) in our study exhibited disease progression very soon after initiating TKI therapy, which was not surprising.
The population in our study was a fairly large cohort to investigate the effectiveness of TKI therapy in patients with compound EGFR-mutated lung cancer. However, this study was a retrospective analysis, which may limit the reliability of the results. Potential selective bias may be unavoidable because of the low incidence of occurrence of these types of mutations. In addition, the RR and PFS of patients with compound EGFR mutations were compared only between patients who received EGFR TKI therapy without inclusion of patients who received chemotherapy. Our data were collected from a single institution, and patients from other areas of China should be examined. All reported cases between 2004 and 2017 were enrolled, but the literature from Asia still accounts for the majority of available data.
Conclusions
Although NSCLC patients with compound mutations exhibited a shorter RFS and lower RR in response to TKI therapy than those with a single common mutation, TKI therapy may still benefit patients with compound mutations. Therefore, after explaining the biases of TKI therapy to patients in detail to ensure their understanding and informed consent, a trial of first-generation small molecule TKIs may be an optional therapeutic strategy to identify potential beneficiaries.
Authors’ contributions
LJZ and YBL designed this study, participated in the interpretation of these results and reviewed the manuscript. XYY, YSW, WDW and YQC were responsible for data collection and assembly, data analysis and interpretation of the results. XWZ, ZCZ and XYY drafted this manuscript together. All authors read and approved the final manuscript.
Acknowledgements
We express our gratitude for the support and suggestions in this study from the staff of the Department of Pathology, Sun Yat-sen University Cancer Center.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used during this study are available from the corresponding author upon reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The ethics committee of Sun Yat-sen University Cancer Center approved our research based on a micro-database. Our research was performed according to the World Medical Association Declaration of Helsinki, and all subjects signed written informed consents.
Funding
The publishing and preparation of this manuscript were funded by the National Key Research and Development Plan (No. 2016YFC0905400), Ministry of Science and Technology of the People’s Republic of China.
Abbreviations
- EGFR
epidermal growth factor receptor
- TKIs
tyrosine kinase inhibitors
- NSCLC
non-small cell lung cancer
- PFS
progression-free survival
- OS
overall survival
- ADC
adenocarcinoma
- SCC
squamous cell carcinoma
- LELC
lymphoepithelioma-like carcinoma
- ASC
adenosquamous carcinoma
- PD
progressive disease
- SD
stable disease
- PR
partial response
- CR
complete response
- NE
not evaluated
Footnotes
Xiangyang Yu, Xuewen Zhang and Zichen Zhang contributed equally to this work
Contributor Information
Xiangyang Yu, Email: yuxy@sysucc.org.cn.
Xuewen Zhang, Email: zhangxuew@sysucc.org.cn.
Zichen Zhang, Email: zhangzch@sysucc.org.cn.
Yongbin Lin, Email: linyb@sysucc.org.cn.
Yingsheng Wen, Email: wenyingsh@sysucc.org.cn.
Yongqiang Chen, Email: chenyongq@sysucc.org.cn.
Weidong Wang, Email: wanggm@sysucc.org.cn.
Lanjun Zhang, Email: zhanglj@sysucc.org.cn.
References
- 1.NCCN clinical practice guidelines in oncology (NCCN guidelines) non-small cell lung cancer, version 4; 2017.
- 2.Gaughan EM, Costa DB. Genotype-driven therapies for non-small cell lung cancer: focus on EGFR, KRAS and ALK gene abnormalities. Ther Adv Med Oncol. 2011;3(3):113–125. doi: 10.1177/1758834010397569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 4.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 5.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 6.Wen YS, Cai L, Zhang XW, Zhu JF, Zhang ZC, Shao JY, et al. Concurrent oncogene mutation profile in chinese patients with stage Ib lung adenocarcinoma. Medicine. 2014;93(29):e296. doi: 10.1097/MD.0000000000000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang SF, Liu HP, Li LH, Ku YC, Fu YN, Tsai HY, et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res. 2004;10(24):8195–8203. doi: 10.1158/1078-0432.CCR-04-1245. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi S, Canepa HM, Bailey AS, Nakayama S, Yamaguchi N, Goldstein MA, et al. Compound EGFR mutations and response to EGFR tyrosine kinase inhibitors. J Thorac Oncol. 2013;8(1):45–51. doi: 10.1097/JTO.0b013e3182781e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang GC, Lin JY, Wang Z, Zhou Q, Xu CR, Zhu JQ, et al. Epidermal growth factor receptor double activating mutations involving both exons 19 and 21 exist in Chinese non-small cell lung cancer patients. Clin Oncol (R Coll Radiol) 2007;19(7):499–506. doi: 10.1016/j.clon.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Hsieh MH, Fang YF, Chang WC, Kuo HP, Lin SY, Liu HP, et al. Complex mutation patterns of epidermal growth factor receptor gene associated with variable responses to gefitinib treatment in patients with non-small cell lung cancer. Lung Cancer. 2006;53(3):311–322. doi: 10.1016/j.lungcan.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Hata A, Yoshioka H, Fujita S, Kunimasa K, Kaji R, Imai Y, et al. Complex mutations in the epidermal growth factor receptor gene in non-small cell lung cancer. J Thorac Oncol. 2010;5(10):1524–1528. doi: 10.1097/JTO.0b013e3181e8b3c5. [DOI] [PubMed] [Google Scholar]
- 12.Yokoyama T, Kondo M, Goto Y, Fukui T, Yoshioka H, Yokoi K, et al. EGFR point mutation in non-small cell lung cancer is occasionally accompanied by a second mutation or amplification. Cancer Sci. 2006;97(8):753–759. doi: 10.1111/j.1349-7006.2006.00233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su KY, Chen HY, Li KC, Kuo ML, Yang JC, Chan WK, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30(4):433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 14.Keam B, Kim DW, Park JH, Lee JO, Kim TM, Lee SH, et al. Rare and complex mutations of epidermal growth factor receptor, and efficacy of tyrosine kinase inhibitor in patients with non-small cell lung cancer. Int J Clin Oncol. 2014;19(4):594–600. doi: 10.1007/s10147-013-0602-1. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Wu BQ, Zhong HH, Hui P, Fang WG. Screening for EGFR and KRAS mutations in non-small cell lung carcinomas using DNA extraction by hydrothermal pressure coupled with PCR-based direct sequencing. Int J Clin Exp Pathol. 2013;6(9):1880–1889. [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J, Jin B, Chu T, Dong X, Yang H, Zhang Y, et al. EGFR tyrosine kinase inhibitor (TKI) in patients with advanced non-small cell lung cancer (NSCLC) harboring uncommon EGFR mutations: a real-world study in China. Lung Cancer. 2016;96:87–92. doi: 10.1016/j.lungcan.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 17.Wu JY, Yu CJ, Chang YC, Yang CH, Shih JY, Yang PC. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin Cancer Res. 2011;17(11):3812–3821. doi: 10.1158/1078-0432.CCR-10-3408. [DOI] [PubMed] [Google Scholar]
- 18.Chen D, Song Z, Cheng G. Clinical efficacy of first-generation EGFR-TKIs in patients with advanced non-small-cell lung cancer harboring EGFR exon 20 mutations. Onco Targets Ther. 2016;9:4181–4186. doi: 10.2147/OTT.S108242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klughammer B, Brugger W, Cappuzzo F, Ciuleanu T, Mok T, Reck M, et al. Examining treatment outcomes with erlotinib in patients with advanced non-small cell lung cancer whose tumors harbor uncommon EGFR mutations. J Thorac Oncol. 2016;11(4):545–555. doi: 10.1016/j.jtho.2015.12.107. [DOI] [PubMed] [Google Scholar]
- 20.Kempf E, Lacroix L, Soria JC. First reported case of unexpected response to an epidermal growth factor receptor tyrosine kinase inhibitor in the I744M uncommon EGFR mutation. Clin Lung Cancer. 2015;16(6):e259–e261. doi: 10.1016/j.cllc.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Wu SG, Chang YL, Hsu YC, Wu JY, Yang CH, Yu CJ, et al. Good response to gefitinib in lung adenocarcinoma of complex epidermal growth factor receptor (EGFR) mutations with the classical mutation pattern. Oncologist. 2008;13(12):1276–1284. doi: 10.1634/theoncologist.2008-0093. [DOI] [PubMed] [Google Scholar]
- 22.Frega S, Conte P, Fassan M, Polo V, Pasello G. A triple rare E709K and L833V/H835L EGFR mutation responsive to an irreversible Pan-HER inhibitor: a case report of lung adenocarcinoma treated with afatinib. J Thorac Oncol. 2016;11(5):e63–e64. doi: 10.1016/j.jtho.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 23.Yang TY, Tsai CR, Chen KC, Hsu KH, Lee HM, Chang GC. Good response to gefitinib in a lung adenocarcinoma harboring a heterozygous complex mutation of L833V and H835L in epidermal growth factor receptor gene. J Clin Oncol. 2011;29(16):e468–e469. doi: 10.1200/JCO.2010.33.5802. [DOI] [PubMed] [Google Scholar]
- 24.Leventakos K, Kipp BR, Rumilla KM, Winters JL, Yi ES, Mansfield AS. S768I mutation in EGFR in patients with lung cancer. J Thorac Oncol. 2016;11(10):1798–1801. doi: 10.1016/j.jtho.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arcila ME, Nafa K, Chaft JE, Rekhtman N, Lau C, Reva BA, et al. EGFR exon 20 insertion mutations in lung adenocarcinomas: prevalence, molecular heterogeneity, and clinicopathologic characteristics. Mol Cancer Ther. 2013;12(2):220–229. doi: 10.1158/1535-7163.MCT-12-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Asahina H, Yamazaki K, Kinoshita I, Yokouchi H, Dosaka-Akita H, Nishimura M. Non-responsiveness to gefitinib in a patient with lung adenocarcinoma having rare EGFR mutations S768I and V769L. Lung Cancer. 2006;54(3):419–422. doi: 10.1016/j.lungcan.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Yasuda H, Kobayashi S, Costa DB. EGFR exon 20 insertion mutations in non-small-cell lung cancer: preclinical data and clinical implications. Lancet Oncol. 2012;13(1):e23–e31. doi: 10.1016/S1470-2045(11)70129-2. [DOI] [PubMed] [Google Scholar]
- 28.Lund-Iversen M, Kleinberg L, Fjellbirkeland L, Helland Å, Brustugun OT. Clinicopathological characteristics of 11 NSCLC patients with EGFR-exon 20 mutations. J Thorac Oncol. 2012;7(9):1471–1473. doi: 10.1097/JTO.0b013e3182614a9d. [DOI] [PubMed] [Google Scholar]
- 29.Kancha RK, von Bubnoff N, Peschel C, Duyster J. Functional analysis of epidermal growth factor receptor (EGFR) mutations and potential implications for EGFR targeted therapy. Clin Cancer Res. 2009;15(2):460–467. doi: 10.1158/1078-0432.CCR-08-1757. [DOI] [PubMed] [Google Scholar]
- 30.Riely GJ, Politi KA, Miller VA, Pao W. Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res. 2006;12(24):7232–7241. doi: 10.1158/1078-0432.CCR-06-0658. [DOI] [PubMed] [Google Scholar]
- 31.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12(21):6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Wang Z, Hao X, Hu X, Wang H, Wang Y, et al. Clinical characteristics and response to tyrosine kinase inhibitors of patients with non-small cell lung cancer harboring uncommon epidermal growth factor receptor mutations. Chin J Cancer Res. 2017;29(1):18–24. doi: 10.21147/j.issn.1000-9604.2017.01.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu X, Bai Q, Lu Y, Qi P, Ding J, Wang J, et al. Response to tyrosine kinase inhibitors in lung adenocarcinoma with the rare epidermal growth factor receptor mutation S768I: a retrospective analysis and literature review. Target Oncol. 2017;12(1):81–88. doi: 10.1007/s11523-016-0455-4. [DOI] [PubMed] [Google Scholar]
- 34.Wu JY, Shih JY. Effectiveness of tyrosine kinase inhibitors on uncommon E709X epidermal growth factor receptor mutations in non-small-cell lung cancer. Onco Targets Ther. 2016;9:6137–6145. doi: 10.2147/OTT.S118071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y, Zhang B, Li R, Liu B, Wang L. EGFR-tyrosine kinase inhibitor treatment in a patient with advanced non-small cell lung cancer and concurrent exon 19 and 21 EGFR mutations: a case report and review of the literature. Oncol Lett. 2016;11(5):3546–3550. doi: 10.3892/ol.2016.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svaton M, Pesek M, Chudacek Z, Vosmiková H. Current two EGFR mutations in lung adenocarcinoma—case report. Klin Onkol. 2015;28(2):134–137. doi: 10.14735/amko2015134. [DOI] [PubMed] [Google Scholar]
- 37.Peng L, Song Z, Jiao S. Comparison of uncommon EGFR exon 21 L858R compound mutations with single mutation. Onco Targets Ther. 2015;8:905–910. doi: 10.2147/OTT.S78984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baek JH, Sun JM, Min YJ, Cho EK, Cho BC, Kim JH, et al. Efficacy of EGFR tyrosine kinase inhibitors in patients with EGFR-mutated non-small cell lung cancer except both exon 19 deletion and exon 21 L858R: a retrospective analysis in Korea. Lung Cancer. 2015;87(2):148–154. doi: 10.1016/j.lungcan.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Peng L, Song ZG, Jiao SC. Efficacy analysis of tyrosine kinase inhibitors on rare non-small cell lung cancer patients harboring complex EGFR mutations. Sci Rep. 2014;4:6104. doi: 10.1038/srep06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chung KP, Shih JY, Yu CJ. Favorable response to gefitinib treatment of lung adenocarcinoma with coexisting germline and somatic epidermal growth factor receptor mutations. J Clin Oncol. 2010;28(34):e701–e703. doi: 10.1200/JCO.2010.28.6260. [DOI] [PubMed] [Google Scholar]
- 41.Yang CH, Yu CJ, Shih JY, Chang YC, Hu FC, Tsai MC, et al. Specific EGFR mutations predict treatment outcome of stage IIIB/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. J Clin Oncol. 2008;26(16):2745–2753. doi: 10.1200/JCO.2007.15.6695. [DOI] [PubMed] [Google Scholar]
- 42.Ichihara S, Toyooka S, Fujiwara Y, Hotta K, Shigematsu H, Tokumo M, et al. The impact of epidermal growth factor receptor gene status on gefitinib-treated Japanese patients with non-small-cell lung cancer. Int J Cancer. 2007;120(6):1239–1247. doi: 10.1002/ijc.22513. [DOI] [PubMed] [Google Scholar]
- 43.Pugh TJ, Bebb G, Barclay L, Sutcliffe M, Fee J, Salski C, et al. Correlations of EGFR mutations and increases in EGFR and HER2 copy number to gefitinib response in a retrospective analysis of lung cancer patients. BMC Cancer. 2007;7:128. doi: 10.1186/1471-2407-7-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kimura H, Suminoe M, Kasahara K, Sone T, Araya T, Tamori S, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA) Br J Cancer. 2007;97(6):778–784. doi: 10.1038/sj.bjc.6603949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Zandwijk N, Mathy A, Boerrigter L, Ruijter H, Tielen I, et al. EGFR and KRAS mutations as criteria for treatment with tyrosine kinase inhibitors: retro- and prospective observations in non-small-cell lung cancer. Ann Oncol. 2007;18(1):99–103. doi: 10.1093/annonc/mdl323. [DOI] [PubMed] [Google Scholar]
- 46.Jackman DM, Yeap BY, Lindeman NI, Fidias P, Rabin MS, Temel J, et al. Phase II clinical trial of chemotherapy-naive patients ≥ 70 years of age treated with erlotinib for advanced non-small-cell lung cancer. J Clin Oncol. 2007;25(7):760–766. doi: 10.1200/JCO.2006.07.5754. [DOI] [PubMed] [Google Scholar]
- 47.Pallis AG, Voutsina A, Kalikaki A, Souglakos J, Briasoulis E, Murray S, et al. ‘Classical’ but not ‘other’ mutations of EGFR kinase domain are associated with clinical outcome in gefitinib-treated patients with non-small cell lung cancer. Br J Cancer. 2007;97(11):1560–1566. doi: 10.1038/sj.bjc.6604068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han SW, Kim TY, Jeon YK, Hwang PG, Im SA, Lee KH, et al. Optimization of patient selection for gefitinib in non-small cell lung cancer by combined analysis of epidermal growth factor receptor mutation, K-ras mutation, and Akt phosphorylation. Clin Cancer Res. 2006;12(8):2538–2544. doi: 10.1158/1078-0432.CCR-05-2845. [DOI] [PubMed] [Google Scholar]
- 49.Kosaka T, Yatabe Y, Endoh H, Yoshida K, Hida T, Tsuboi M, et al. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12(19):5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]
- 50.Choong NW, Dietrich S, Seiwert TY, Tretiakova MS, Nallasura V, Davies GC, et al. Gefitinib response of erlotinib-refractory lung cancer involving meninges–role of EGFR mutation. Nat Clin Pract Oncol. 2006;3(1):50–57. doi: 10.1038/ncponc0400. [DOI] [PubMed] [Google Scholar]
- 51.Oshita F, Matsukuma S, Yoshihara M, Sakuma Y, Ohgane N, Kameda Y, et al. Novel heteroduplex method using small cytology specimens with a remarkably high success rate for analysing EGFR gene mutations with a significant correlation to gefitinib efficacy in non-small-cell lung cancer. Br J Cancer. 2006;95(8):1070–1075. doi: 10.1038/sj.bjc.6603396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tokumo M, Toyooka S, Ichihara S, Ohashi K, Tsukuda K, Ichimura K, et al. Double mutation and gene copy number of EGFR in gefitinib refractory non-small-cell lung cancer. Lung Cancer. 2006;53(1):117–121. doi: 10.1016/j.lungcan.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 53.Chou TY, Chiu CH, Li LH, Hsiao CY, Tzen CY, Chang KT, et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non-small cell lung cancer. Clin Cancer Res. 2005;11(10):3750–3757. doi: 10.1158/1078-0432.CCR-04-1981. [DOI] [PubMed] [Google Scholar]
- 54.Shih JY, Gow CH, Yu CJ, Yang CH, Chang YL, Tsai MF, et al. Epidermal growth factor receptor mutations in needle biopsy/aspiration samples predict response to gefitinib therapy and survival of patients with advanced nonsmall cell lung cancer. Int J Cancer. 2006;118(4):963–969. doi: 10.1002/ijc.21458. [DOI] [PubMed] [Google Scholar]
- 55.Taron M, Ichinose Y, Rosell R, Mok T, Massuti B, Zamora L, et al. Activating mutations in the tyrosine kinase domain of the epidermal growth factor receptor are associated with improved survival in gefitinib-treated chemorefractory lung adenocarcinomas. Clin Cancer Res. 2005;11(16):5878–5885. doi: 10.1158/1078-0432.CCR-04-2618. [DOI] [PubMed] [Google Scholar]
- 56.Mitsudomi T, Kosaka T, Endoh H, Horio Y, Hida T, Mori S, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non–small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23(11):2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 57.Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23(28):6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 58.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101(36):13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during this study are available from the corresponding author upon reasonable request.