Significance
Bone marrow failure (BMF) is an inherited life-threatening condition characterized by defective hematopoiesis, developmental abnormalities, and predisposition to cancer. BMF caused by ERCC6L2 mutations is considered to be a genome instability syndrome, because DNA repair is compromised in patient cells. In this study, we report BMF cases with biallelic disease-causing variants and provide evidence from patients’ cells that transcription deficiency can explain the genome instability. Specifically, we demonstrate that ERCC6L2 participates in RNA polymerase II-mediated transcription via interaction with DNA-dependent protein kinase (DNA-PK) and resolves DNA–RNA hybrids (R loops). Collectively, our data point to a causal mechanism in BMF in which patients with ERCC6L2 mutations are defective in the repair of transcription-associated DNA damage.
Keywords: ERCC6L2, DNA-PK, transcription, DNA repair, R loops
Abstract
Biallelic variants in the ERCC excision repair 6 like 2 gene (ERCC6L2) are known to cause bone marrow failure (BMF) due to defects in DNA repair and mitochondrial function. Here, we report on eight cases of BMF from five families harboring biallelic variants in ERCC6L2, two of whom present with myelodysplasia. We confirm that ERCC6L2 patients’ lymphoblastoid cell lines (LCLs) are hypersensitive to DNA-damaging agents that specifically activate the transcription coupled nucleotide excision repair (TCNER) pathway. Interestingly, patients’ LCLs are also hypersensitive to transcription inhibitors that interfere with RNA polymerase II (RNA Pol II) and display an abnormal delay in transcription recovery. Using affinity-based mass spectrometry we found that ERCC6L2 interacts with DNA-dependent protein kinase (DNA-PK), a regulatory component of the RNA Pol II transcription complex. Chromatin immunoprecipitation PCR studies revealed ERCC6L2 occupancy on gene bodies along with RNA Pol II and DNA-PK. Patients’ LCLs fail to terminate transcript elongation accurately upon DNA damage and display a significant increase in nuclear DNA–RNA hybrids (R loops). Collectively, we conclude that ERCC6L2 is involved in regulating RNA Pol II-mediated transcription via its interaction with DNA-PK to resolve R loops and minimize transcription-associated genome instability. The inherited BMF syndrome caused by biallelic variants in ERCC6L2 can be considered as a primary transcription deficiency rather than a DNA repair defect.
Inherited bone marrow failure (IBMF) syndromes are a clinically heterogeneous group of diseases. Affected individuals present with a variety of hematological complications that include myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML). Patients with IBMF often have a variable number of extrahematopoietic features which have historically been the cornerstone for defining inherited BMF subtypes (1). Recognized entities include Fanconi anemia (FA), dyskeratosis congenita, and Shwachman Diamond syndrome, which are associated with primary defects in DNA repair, telomere maintenance, and ribosome biogenesis, respectively (1). Increasingly, through advances in next generation sequencing technologies, new BMF entities are being identified and characterized. This is leading to better management of these complex patients as well as elucidating interesting new biological connections.
We first reported biallelic loss-of-function variants in ERCC6L2 in patients presenting with BMF and microcephaly [Mendelian Inheritance in Man (MIM) 615667; ref. 2]. Recently, several more BMF cases with biallelic variants in ERCC6L2 have been identified (3–6), some of whom presented with MDS (3, 4). We have now characterized five families that add to the inherited BMF entity arising from ERCC6L2 mutations and highlight the link between bone marrow failure and MDS/AML. We also provide data on the biological functions of ERCC6L2, which collectively suggest that this disorder principally arises from a transcription deficiency.
Results
Patient Characteristics.
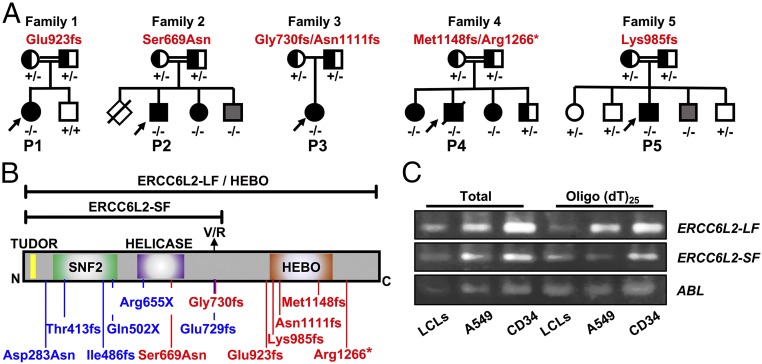
Through a combination of whole exome sequencing and candidate gene sequencing (7) (SI Appendix, Table S1) we have identified eight cases from five families with biallelic variants in ERCC6L2 (NM_020207.4) (Fig. 1 A and B and SI Appendix, Fig. S1 and Table S2). Sanger sequencing of parental DNA confirmed an autosomal recessive pattern of inheritance (Fig. 1A). Two of the seven ERCC6L2 variants identified are reported at low frequency on the Genome Aggregation Database (accessed January 31, 2018) and interestingly, two other variants have recently been reported in patients with BMF (4) (SI Appendix, Table S2). To our knowledge, none of the other variants identified in these families have been described elsewhere.
Fig. 1.
Biallelic mutations in ERCC6L2 are identified in BMF cases. (A) Families identified with ERCC6L2 variants, in autosomal-recessive pattern. Genotypes indicate + for normal and − for mutated alleles. Affected cases are denoted in black. Family members harboring biallelic ERCC6L2 variants but with no disease features are indicated in gray. (B) The variants identified in this paper are indicated in red on a diagram of the long isoform (LF) of ERCC6L2, in which functional domains are annotated. Previously published mutations are shown in blue. The location of the short isoform (SF) is indicated. (C) RT-PCR analysis shows the presence of the long and short isoforms of ERCC6L2 in EBV-transformed LCLs, A549 cells, and CD34+ hematopoietic progenitors.
All probands presented below the age of 20 y with various forms of BMF (SI Appendix, Table S3). Specifically, the index case from family 1 (P1, homozygous Glu923fs) presented with a hypoplastic bone marrow at 8 y of age. Her family history includes a first cousin who was diagnosed with hypoplastic anemia and an unspecified relative with AML (SI Appendix, Table S3). In family 2, the index case (P2, homozygous Ser699Asn) presented at 4 y of age with mild pancytopenia. Ser699 lies in the helicase domain of ERCC6L2, which is highly conserved among vertebrates (SI Appendix, Fig. S2). His elder brother died at 2 y of age, due to neonatal epilepsy, apnea, and organ failure. His younger affected sister had hypocellular bone marrow, while his younger brother, 3 y of age, is also homozygous for Ser699Asn but is asymptomatic. In family 3, the index case (P3, Gly730fs/Asn1111fs) developed microcephaly by the age of 6 y and presented with macrocytosis associated with thrombocytopenia at 17 y of age. The index case from family 4 (P4, Met1148fs/Arg1266*) presented with bone marrow aplasia at age 2 y. He progressed to MDS and AML with a complex karyotype containing a deletion of the long arm of chromosome 5 and loss of one copy of chromosome 18 and a marker chromosome, leading to fatal complications (SI Appendix, Table S3). His two sisters, who are both compound heterozygotes, also presented with bone marrow dysfunction. The index case from family 5 (P5, homozygous Lys985fs) 12 y of age, presented with hypoplastic bone marrow which progressed to MDS with an abnormal karyotype of monosomy 7 and trisomy 20. Somatic features of short stature, café au lait spots, and leukoplakia are also present in this case. One sibling of the index case, 3 y of age, is homozygous for the frameshift variant but is asymptomatic. No indication of abnormal chromosomal breakage was reported in the peripheral blood cells of all affected individuals, when treated with diepoxybutane or mitomycin C, excluding a diagnosis of FA (SI Appendix, Table S3). This strong allelic series, in conjunction with other BMF cohorts studied to date (Fig. 1 A and B), demonstrate that biallelic mutations in ERCC6L2 can cause an inherited BMF syndrome with predisposition to MDS and AML (2–4).
Patients’ Cells Are Hypersensitive to Transcriptional Inhibitors.
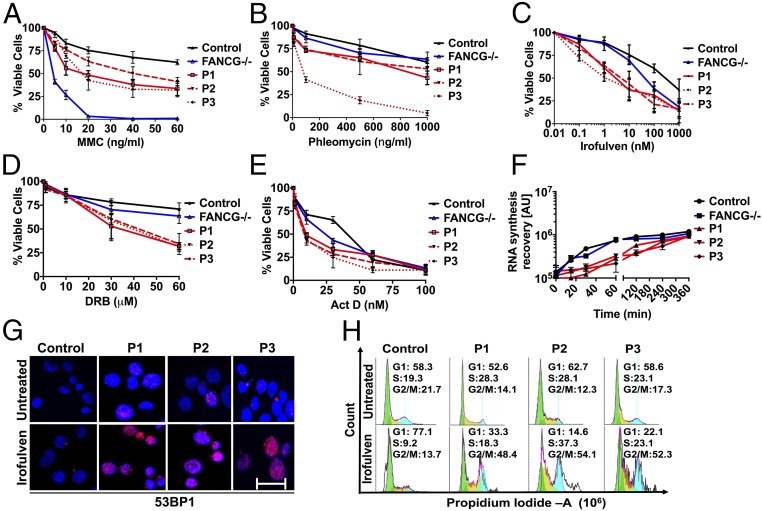
ERCC6L2 belongs to the SWI/SNF family of ATP-dependent chromatin remodellers and is known to participate in the DNA damage response and mitochondrial function (2, 5). It is ubiquitously expressed and has at least two distinct isoforms, a short form and a long form, which has been called a helicase mutated in bone marrow failure (HEBO) (5). The isoforms arise by alternative splicing in exon 14, resulting in replacement of the last Val712 residue of the short form with an 850-aa extension in the long form. Real time-PCR analysis of both total RNA and poly(A)+ mRNA indicated the presence of long and short ERCC6L2 isoform transcripts in both CD34+ hematopoietic stem cell progenitors and differentiated lymphoblastoid cell lines (LCLs) from normal individuals (Fig. 1C). Studies on patient fibroblasts and A549 cells treated with ERCC6L2 siRNA have shown hypersensitivity to DNA damaging agents that induce double stand breaks (DSBs) (2, 5). Patient fibroblasts overexpressing the long form but not the short form of ERCC6L2 show strong resistance to DNA damage induced by phleomycin (5). Notably in A549 cells, knockdown of ERCC6L2 induced sensitivity to irofulven, an RNA polymerase II (RNA Pol II)-interfering agent that triggers the transcription-coupled nucleotide excision repair (TCNER) pathway (2). Irofulven inhibits RNA synthesis and specifically activates TCNER by trapping transcription complexes where RNA Pol II is engaged (8–10). Here, we show that patient-derived LCLs, available from three index cases (P1–P3), show hypersensitivity to irofulven as well as to mitomycin C and phleomycin (Fig. 2 A–C). The sensitivity of the patient’s LCLs to increasing doses of mitomycin C was significantly higher than that of control, although they were by no means comparable to that of LCLs from a patient with Fanconi anemia group G (FANCG) mutant (Fig. 2A). Interestingly patients were also hypersensitive to the RNA Pol II-interfering agents, 5, 6-dichlorobenzimidazole 1-β-d-ribofuranoside (DRB), a transcription elongation inhibitor and actinomycin D (ActD), a general transcription inhibitor (Fig. 2 D and E) compared with both the control and the FANCG patient.
Fig. 2.
ERCC6L2 deficiency causes transcription deficits and arrests patients’ LCLs in G2/M phase. (A–C) Control and patients’ LCLs were plated in the presence of increasing concentrations of indicated compounds for 24 h and assayed for cell viability. (D and E) Compared with control and FANCG LCLs, patient LCLs showed reduced survival after treatment with DRB and ActD in a dose-dependent manner. Error bars represent SEs calculated between octuplets in each individual experiment (n = 2). (F) Control and patient LCLs were incubated with 1 nM irofulven for 1 h and the influence on RNA synthesis after release from irofulven for the indicated time intervals was measured by incorporation of fluorescently labeled 5-EU, that was added to the medium for 15 min. Error bars represent the SEM obtained from three independent experiments. (G) 53BP1 staining in control and patients’ LCLs at 24 h postrelease from irofulven treatment. Panels display 53BP1 (red) and DAPI (blue). (Scale bar: 50 μm.) (H) FACS analysis plots show the cell cycle phases where pre- and 24-h postrelease from irofulven treatment arrest patient cells in G2/M phase.
Irofulven and DRB exert similar effects on transcription, by blocking RNA elongation and inhibiting RNA synthesis (10). Given that patients’ LCLs were hypersensitive to these transcription inhibitors, we next set out to evaluate the effects of defective ERCC6L2 on global RNA synthesis in intact cells. To do this, we measured the incorporation of fluorescently labeled 5-ethynyl uridine (5-EU) into nascent RNA, using a Click-iT RNA Alexa Fluor 488 kit. An initial treatment with irofulven for 3 h inhibited RNA synthesis in both patient and control LCLs, as shown at the zero time point in Fig. 2F. Subsequent labeling of the nascent RNAs, after release from the irofulven insult, revealed a gradual recovery of RNA synthesis, which was notably slower in the patients compared with control and FANCG LCLs (Fig. 2F). The reduced recovery rate, postirofulven treatment, indicates a transcription deficiency in these ERCC6L2 patients. DRB and ActD were used as positive controls to test the fidelity of the assay as they are known to inhibit RNA synthesis by blocking transcription (SI Appendix, Fig. S3). Moreover, irofulven treatment led to a significant increase in DSB markers such as 53BP1 (Fig. 2G) and arrested patients’ LCLs in G2/M phase (Fig. 2H). These results collectively demonstrate that patients are transcription deficient and exhibit increased sensitivity to DNA damaging agents. However, the precise role of ERCC6L2 in regulating transcription and/or DNA repair remains unclear.
The ERCC6L2 Interactome Includes RNA Binding and Processing Factors.
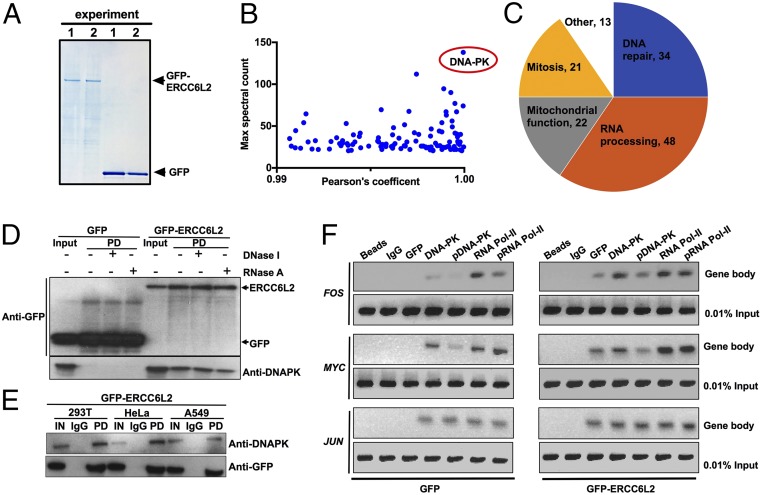
To further elucidate the primary cellular function of the long form of ERCC6L2 (NP_064592.2), we sought to identify its interaction partners. Using an unbiased approach, we performed affinity-based mass spectrometry (MS) analyses (11) after coimmunoprecipitation with GFP TRAP beads in soluble extracts from 293T cells overexpressing GFP-ERCC6L2 (Fig. 3A). Data obtained from MS were analyzed by filtering the interactors based on the best protein MASCOT scores, the spectral counts, and the number of unique peptides (>20) combined with an interaction stringency determined by a Pearson’s correlation coefficient of >0.99 for GFP-ERCC6L2 over GFP alone (Fig. 3B and SI Appendix, Table S4). Pathway analysis and visualization of the resulting proteins (n = 106) using the Uniprot database (12) and Cytoscape (13) revealed an unexpected role for ERCC6L2 in RNA binding (14–16) along with its anticipated role in DNA repair and mitochondrial function (2, 5) (Fig. 3 C and D and SI Appendix, Fig. S4 and Table S5). It had previously been shown that ectopically expressed ERCC6L2 interacts with NBS1 at DNA damage foci (5). In agreement with this, we found that NBS1 bound to ERCC6L2, but only with a MASCOT sum score of 260. This is in stark contrast to the striking interaction observed with the DNA-dependent protein kinase (DNA-PK, also known as PRKDC) which had the highest MASCOT sum score (23,361), number of spectral counts (n = 138), and number of unique peptides (n = 105) of any protein in the dataset (Fig. 3B and SI Appendix, Tables S4 and S6). These MS studies therefore show a close association between ERCC6L2 and DNA-PK and suggest an additional layer of function for ERCC6L2 in RNA processing.
Fig. 3.
ERCC6L2 physically interacts with DNA-PK and occupies gene bodies along with RNA Pol II. (A) Colloidal blue staining of GFP and GFP-ERCC6L2 proteins overexpressed in 293T cells and separated by SDS/PAGE. (B) MS analysis for relative protein abundance that is sorted on the basis of highest spectral counts and ranked by Pearson’s correlation coefficient (Log2 fold change) between GFP-ERCC6L2 and GFP only control. Note the standout signal for DNA-PK. (C) Pie chart overrepresentation of ERCC6L2 interactome. (D and E) Co-IP followed by Western blot analysis confirm the interaction between GFP-ERCC6L2 and DNA-PK in 293T, A549, and HeLa cells. Lysates were treated with DNase I or RNase A where indicated. (F) ChIP-PCR shows occupancy of GFP-ERCC6L2, RNA Pol II, and DNA-PK with the MYC FOS and JUN gene bodies. Note the occupancy of all three genes is seen with GFP-ERCC6L2 and not when GFP alone was expressed. The phosphorylation residues targeted by ChIP antibodies are serine-2056 for DNA-PK and serine-2 for RNA Pol II.
ERCC6L2 Interacts with DNA-PK to Participate in RNA Pol II-Mediated Transcription.
We confirmed the ERCC6L2 interaction with DNA-PK in 293T cells overexpressing GFP-ERCC6L2 by coimmunoprecipitation with GFP TRAP beads and Western blotting. The stringency of ERCC6L2 interaction with DNA-PK was not significantly affected by either DNase I or RNase A (Fig. 3D), indicating only a minor involvement of DNA and/or RNA in mediating this association. Coimmunoprecipitation experiments in HeLa and A549 cells overexpressing GFP-ERCC6L2 confirmed that ERCC6L2 association with DNA-PK is not cell specific (Fig. 3E).
DNA-PK is involved in several vital functions, including a crucial role in signaling nonhomologous end joining (NHEJ) or homologous recombination (HR) in DNA repair (17). However, it has been shown previously that ERCC6L2 is not required for either NHEJ or HR (5). DNA-PK also promotes transcription fidelity by the eviction of RNA polymerase II (RNA Pol II) from active genes at sites of DNA damage (18). Noting the transcription deficiency observed in patients’ LCLs (Fig. 2 D–F) and the multiple RNA binding proteins present in our pull-down experiment (Fig. 3C and SI Appendix, Fig. S4 and Tables S4 and S5), we sought to further investigate the role of ERCC6L2 in transcription, where both DNA-PK and RNA Pol II are involved. To do this, we performed chromatin immunoprecipitation (ChIP)-PCR assays on chromatin extracts prepared from 293T cells expressing GFP alone and GFP-ERCC6L2 using a GFP-specific antibody. These revealed the occupancy of GFP-ERCC6L2 on MYC, FOS, and JUN gene bodies (Fig. 3F), using the same primer pairs that were used previously elsewhere to show occupancy of total and phosphorylated forms of DNA-PK and RNA Pol II on these genes (19). These experiments indicate that ERCC6L2 associates with DNA-PK and occupies the same gene bodies along with RNA Pol II.
ERCC6L2 Is Required for Transcriptional Fidelity.
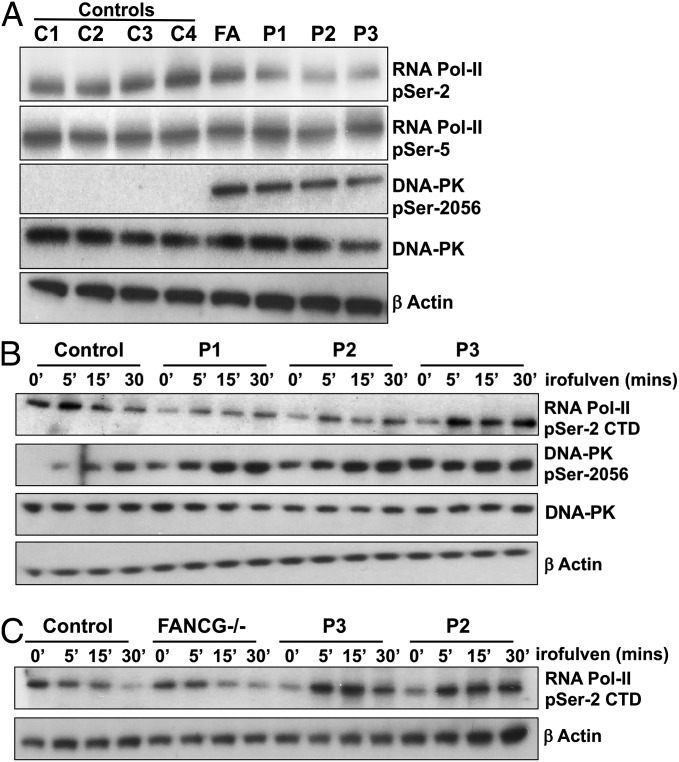
Having shown ERCC6L2 occupancy on gene bodies, we next tested transcriptional fidelity in the available ERCC6L2 patients (P1–P3) by assessing phosphorylation levels of serine-5 and serine-2 residues in the heptapeptide repeat (Tyr-Ser-Pro-Thr-Ser-Pro-Ser) in the carboxyl-terminal domain (CTD) of RNA Pol II. Phosphorylation on serine-5 of the RNA Pol II CTD indicates transcription initiation, whereas phosphorylation of serine-2 is a marker of transcription elongation (20). In the steady state, a reduction in phosphorylation levels of serine-2, but not serine-5 was observed in patients’ LCLs compared with control as well as the FANCG patient (Fig. 4A and SI Appendix, Fig. S5A). This result indicates that ERCC6L2 plays a role in the elongation of RNA Pol II transcripts. We also observed a dramatic increase in DNA-PK catalytic subunit (cs) phosphorylation at serine-2056 in cells from the ERCC6L2 and FA patients (Fig. 4A), indicating a heightened DNA damage response in both diseases. In addition to these defects in the steady state, patients’ LCLs also showed abnormal kinetics of serine-2 phosphorylation on RNA Pol II CTD and serine-2056 phosphorylation of DNA-PKcs when they were treated with irofulven (Fig. 4B). As expected, we observed a reduction in serine-2 phosphorylation in control LCLs within 30 min of irofulven treatment (Fig. 4B). However, in the patients’ LCLs we observed a brief increase in serine-2 phosphorylation on RNA Pol II CTD, indicating a failure of these cells to stall transcription elongation. This increase of serine-2 phosphorylation on RNA Pol II CTD was specific to ERCC6L2 patients, as the transcription termination kinetics in FANCG cells was similar to the normal control (Fig. 4C). At the same time, we saw an increase in the activation of DNA-PKcs in patients (indicated by phosphorylation at serine-2056) over and above the increase that was seen in the control (Fig. 4B and SI Appendix, Fig. S5B). Together, these data indicate that although DNA-PK is hyperactive in patients’ LCLs, it is ineffective in stalling RNA Pol II. This results in an increase in transcription-induced DNA damage and cell cycle arrest (Fig. 2 G and H).
Fig. 4.
Patients’ LCLs are hypersensitive to RNA Pol II-mediated transcription insults. (A) Immunoblotting of patient LCL lysates (P1–P3), controls (C1–C4), and a FANCG line (FA) for total and phosphorylated RNA Pol II CTD at serine-2 (indicating transcription elongation) and serine-5 residues (indicating transcription initiation), total, and phosphorylated DNA-PK at serine-2056 (indicating activation) residue and loading control β-actin. (B) Immunoblotting of control and patient LCL lysates posttreatment with irofulven at indicated time points using antibodies against total and serine-2 phosphorylated RNA Pol II CTD, total, and serine-2056 phosphorylated DNA-PK and the loading control β-actin. (C) Immunoblotting panels show control, FANCG, and patient (P2 and P3) LCL lysates obtained after irofulven treatment for the indicated times, using antibodies against serine-2 phosphorylated RNA Pol II CTD and β-actin loading control.
Patients’ Cells Possess an Increased Number of R Loops.
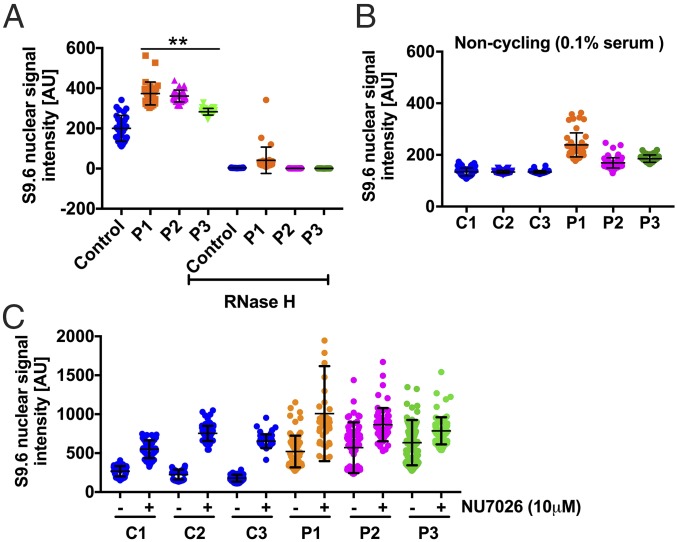
During transcription, nascent RNAs can form hydrogen bonds with one strand of the DNA double helix, leading to the formation of DNA–RNA hybrids, known as R loops. Cells utilize diverse mechanisms to resolve these R loops, either by RNase activity, that specifically degrades the RNA moiety in RNA–DNA hybrids or by helicases, such as Aquarius and Senataxin that unwind the R loops (21). R-loop formation can also be precluded by proteins that bind to nascent RNAs as they emerge from RNA Pol II (22). Besides DNA-PK, ERCC6L2 also interacts with other proteins that are involved in processing of R loops (SI Appendix, Table S7) (23, 24). Noting these interaction partners, we sought to investigate R loops in patients’ LCLs as they possess transcription-associated defects that compromise genome integrity. By immunostaining with S9.6 antibody, which specifically recognizes DNA–RNA hybrids, we did indeed detect R loops in both patients and control (Fig. 5A and SI Appendix, Fig. S6). However, the fluorescence intensity of the DNA–RNA hybrid signal was increased significantly in the nucleoplasm of the patients’ LCLs compared with control. This signal was abolished by pretreatment with RNase H, demonstrating its specificity. Some weak nucleolar S9.6 signal persisted after RNase H treatment, and this could be due to the presence of hybrids that are either resistant to RNase H, or the incomplete action of the nuclease in nucleolus, where DNA–RNA hybrids are abundant. We excluded nucleolar signals from our analysis, as the number of nucleoli varied from cell to cell (SI Appendix, Fig. S6) and concluded that patients have significantly increased numbers of nucleoplasmic R loops.
Fig. 5.
RNA–DNA hybrids (R loops) accumulate in the nucleus of patient LCLs. (A) The graph shows the median of the S9.6 signal intensity, specific for DNA–RNA hybrids, per nucleus after nucleolar signal removal. (B) The graph shows the nucleoplasmic S9.6 signal intensity in noncycling control and patient cells grown in 0.1% FBS. (C) The graph shows the levels of DNA–RNA hybrids in cells grown in the presence or absence of DNA-PK inhibitor NU7026 for 3 h. In all graphs, the S9.6 signal intensity was calculated from at least ∼3,000 cells from different fields of view, indicated by individual points for each sample from two independent experiments. **P < 0.01 (Mann–Whitney U test, two tailed).
To investigate a possible role for replication in this increased R-loop signal in the ERCC6L2 patients’ cells, we compared them to the control cells upon serum starvation. This caused the cells to cease replicating, as demonstrated by the negligible incorporation of bromodeoxyuridine (BrdU) (SI Appendix, Fig. S7). Under these conditions, patient cells still showed significant increase in R-loop signals, compared with controls (Fig. 5B). These data suggest that ERCC6L2 is primarily involved in processing the R loops generated during transcription. Finally, we have established a direct role for DNA-PK in R-loop metabolism. Treating control cells for 3 h with NU7026, a potent inhibitor of DNA-PKcs phosphorylation activity (SI Appendix, Fig. S8), resulted in a dramatic increase of DNA–RNA hybrids (Fig. 5C). Patient cells also showed an increase in signal intensity, above the steady state levels that were already high. This increase in R-loop signal intensity upon inhibition of DNA-PKcs activity indicates that transcription elongation is dependent on the activity of DNA-PK (18). Collectively our results point to a crucial role for the interaction between ERCC6L2 and DNA-PK in the control of transcription and processing R loops.
Discussion
This paper reports on eight cases of bone marrow failure from five families harboring biallelic variants in ERCC6L2 (Fig. 1 A and B). Two of the cases (P4 and P5) had MDS, one of whom progressed to AML (SI Appendix, Table S3). Previous studies have established a DNA repair defect in A549 cells and patient fibroblasts that lack ERCC6L2 (2, 5). Here, we show that patient-derived LCLs also exhibit hypersensitivity to DNA damaging agents (Fig. 2). We also demonstrate that these patients’ cells are hypersensitive to RNA Pol II-interfering agents, particularly irofulven that traps RNA Pol II complexes at DNA lesions and initiates TCNER. Patient cells are compromised in DNA repair capacity, as evidenced by 53BP1 staining, have abnormal delay in RNA synthesis recovery rates, and fail to accurately stall transcription elongation following irofulven treatment (Fig. 2). Extending these studies, we have identified a role for ERCC6L2 in contributing to the DNA damage response through its interaction with DNA-PK (SI Appendix, Table S4). We show that the ERCC6L2 interactome not only contains DNA repair proteins, but is also composed of RNA binding proteins that are involved in processes such as mRNA elongation, maturation, and export (Fig. 3 A–C and SI Appendix, Fig. S4). We also show that ERCC6L2 primarily interacts with DNA-PK and occupies gene bodies alongside RNA Pol II (Fig. 3 D–F). Finally, we have observed an increase in nucleoplasmic R-loop density (Fig. 5A and SI Appendix, Fig. S6), which could be the root cause of the genomic instability in these patients (SI Appendix, Fig. S10). DNA-PK was initially identified as a regulatory component of transcriptionally poised RNA Pol II (17). Given that transcription is perhaps the most fundamental mechanism behind cell fate, the role of DNA-PK in regulating this process, besides DNA repair, may have critical implications in BMF. Our studies suggest the ERCC6L2–DNA-PK interaction could be particularly relevant in the repair of transcription-associated DNA lesions (19). Interestingly, a knockin mouse model harboring defective DNA-PKcs activity has features of congenital BMF causing premature death due to inefficient DNA repair (25). One could argue that DNA-PKcs activity in transcription-associated DNA repair is controlled via its interaction with ERCC6L2 and this is defective in patient cells. Collectively our studies support the notion of a primary transcription defect rather than a DNA repair defect in patients with BMF harboring biallelic ERCC6L2 variants.
To date, the molecular pathology of IBMF has been attributed to defects in DNA repair, telomere maintenance, and ribosome biogenesis (1). Defects in the TCNER pathway have been reported previously in autosomal recessive disorders, including Cockayne syndrome (CS) (MIM 133540) and the functionally related xeroderma pigmentosum (XP) (MIM 278700, 610651, 278720, 278730, 278740, 278760, and 278780) (26). Mutations in CSA and CSB genes cause CS and to date 70% of CS cases are linked to mutations in CSB (27). CS is characterized by growth retardation, retinal abnormalities, progressive neural retardation, and severe photosensitivity (28). However, only five of the 20 ERCC6L2 mutated cases reported to date have neurological defects (2, 5, 6) and conversely, there have been no reports of BMF in cases with CS or XP. Both CSB and ERCC6L2 belong to the family of SWI/SNF proteins that activate transcription by facilitating access of the transcriptional machinery to the chromatin (29). Recent cryo-EM studies have shown a key role for RAD26 (the yeast ortholog of human CSB) in TCNER and RNA Pol II transcription elongation (30). CSB protein is required for processing R loops at the stalled transcription complexes (31). Both CS cells and ERCC6L2 LCLs are defective in RNA synthesis recovery and hypersensitive to clastogens that induce TCNER (32). Failure of the TCNER process also sensitizes CS cells to oxidizing agents that include ionizing radiation (33). ERCC6L2 patients’ LCLs are also hypersensitive to DNA damaging agents that induce oxidative stress (Fig. 2 A–C) (2, 5). Together, these studies indicate that ERCC6L2, like CSB, may participate as a cotranscriptional regulator, facilitating RNA Pol II elongation and processing of R loops. Defects in RNA Pol II-mediated elongation and R-loop accumulation could sensitize patient cells to persistent DNA damage (Fig. 2 A–C) and explain the delay in RNA synthesis resumption after irofulven exposure (Fig. 2F). Another possibility is that when ERCC6L2 is defective, TCNER is not initiated appropriately, resulting in the accumulation of elongating RNA Pol II complexes at sites of irofulven-induced DNA damage. This would also explain the increased sensitivity of patients’ cells to irofulven (Figs. 2C and 4B).
Our MS analysis of proteins that interact with ERCC6L2 (SI Appendix, Fig. S4 and Table S4) indicates that it is a multifunctional protein. In particular, our studies suggest that ERCC6L2 may act primarily in RNA processing, in addition to its role in DNA repair. We observed engagement of multiple RNA binding proteins, including heterogeneous nuclear ribonucleoproteins, as well as splicing and export factors which coordinate mRNA maturation and transport (14–16, 34) (Fig. 3C and SI Appendix, Fig. S4 and Tables S4 and S5). Interestingly, ERCC6L2 also interacts with key proteins (SI Appendix, Fig. S4 and Table S7), that are involved in transcription termination and R-loop metabolism (20, 24, 35). Excessive R loops can exacerbate sensitivity to secondary DNA damaging agents and cause genome instability (SI Appendix, Fig. S10) (36) and we believe this is the case in ERCC6L2 patient cells. R-loop accumulation has been previously reported in FA (37). Cells that lack a functional FA DNA repair pathway possess DNA replication defects due to persistent R loops and show hypersensitivity to DNA replication inhibitors. Our patients’ LCLs also displayed sensitivity to both replication and transcription inhibitors (Fig. 2 A–E), but they do possess a functional FA pathway as evidenced by monoubiquitination of FANCD2 upon mitomycin C treatment (5). We believe that the excessive R-loop accumulation in ERCC6L2 patients may arise due to inefficient transcription stalling. In one possible model, the ERCC6L2–DNA-PK complex, along with other RNA processing factors, may bind nascent mRNAs and prevent them annealing back to DNA. Failure to do so results in persistent R-loop accumulation and defects in transcription (SI Appendix, Fig. S10). Although a full understanding of the mechanistic details of this model awaits further investigation, our work indicates a role for ERCC6L2 in transcription-coupled DNA repair and R-loop biology.
In conclusion, we reveal a molecular function for ERCC6L2 in controlling RNA Pol II-mediated transcript elongation via interaction with DNA-PK to resolve R loops and prevent genome instability. Our genetic and functional data show that pathological recessive variants in ERCC6L2 perturb transcription elongation and impede TCNER. This picture is reminiscent of the cellular defects observed with other chromatin remodellers of the SWI/SNF family, including CSB (31, 32, 38). However, it remains unclear as to why defects in ERCC6L2 function cause bone marrow failure, while defective CSB does not.
Materials and Methods
Exome and candidate gene sequencing was performed on a series of genetically uncharacterized cases in the Dyskeratosis Congenita Registry (held at Barts and The London School of Medicine, London), with written consent under the approval of our local research ethics committee (London, City and East). Transcription assays were performed using a 5-EU labeling kit (Thermo Fisher). Mass spectrometry was performed in an Orbitrap mass spectrometer (Q-Exactive Plus). All statistical analysis was done using GraphPad Prism 7 software.
Supplementary Material
Acknowledgments
We thank the families and clinicians who contributed to this research, Dr. Mike Williams for sending us the patient sample for genotyping, and Prof. Andrew Silver for his critical reading of this manuscript. This work was funded by Bloodwise Program Grant (14032) and the Medical Research Council Research Grant (MR/P018440).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803275115/-/DCSupplemental.
References
- 1.Collins J, Dokal I. Inherited bone marrow failure syndromes. Hematology. 2015;20:433–434. doi: 10.1179/1024533215Z.000000000381. [DOI] [PubMed] [Google Scholar]
- 2.Tummala H, et al. ERCC6L2 mutations link a distinct bone-marrow-failure syndrome to DNA repair and mitochondrial function. Am J Hum Genet. 2014;94:246–256. doi: 10.1016/j.ajhg.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Järviaho T, et al. Bone marrow failure syndrome caused by homozygous frameshift mutation in the ERCC6L2 gene. Clin Genet. 2018;93:392–395. doi: 10.1111/cge.13125. [DOI] [PubMed] [Google Scholar]
- 4.Bluteau O, et al. A landscape of germline mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131:717–732. doi: 10.1182/blood-2017-09-806489. [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, et al. A nonsense mutation in the DNA repair factor Hebo causes mild bone marrow failure and microcephaly. J Exp Med. 2016;213:1011–1028. doi: 10.1084/jem.20151183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shabanova I, et al. ERCC6L2-associated inherited bone marrow failure syndrome. Mol Genet Genomic Med. 2018;6:463–468. doi: 10.1002/mgg3.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walne AJ, et al. Marked overlap of four genetic syndromes with dyskeratosis congenita confounds clinical diagnosis. Haematologica. 2016;101:1180–1189. doi: 10.3324/haematol.2016.147769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaspers NG, et al. Anti-tumour compounds illudin S and Irofulven induce DNA lesions ignored by global repair and exclusively processed by transcription- and replication-coupled repair pathways. DNA Repair (Amst) 2002;1:1027–1038. doi: 10.1016/s1568-7864(02)00166-0. [DOI] [PubMed] [Google Scholar]
- 9.Koeppel F, et al. Irofulven cytotoxicity depends on transcription-coupled nucleotide excision repair and is correlated with XPG expression in solid tumor cells. Clin Cancer Res. 2004;10:5604–5613. doi: 10.1158/1078-0432.CCR-04-0442. [DOI] [PubMed] [Google Scholar]
- 10.Escargueil AE, et al. Influence of irofulven, a transcription-coupled repair-specific antitumor agent, on RNA polymerase activity, stability and dynamics in living mammalian cells. J Cell Sci. 2008;121:1275–1283. doi: 10.1242/jcs.023259. [DOI] [PubMed] [Google Scholar]
- 11.Casado P, et al. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci Signal. 2013;6:rs6. doi: 10.1126/scisignal.2003573. [DOI] [PubMed] [Google Scholar]
- 12.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The UniProt Consortium UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aktaş T, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 15.Leone S, Bär D, Slabber CF, Dalcher D, Santoro R. The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation. EMBO Rep. 2017;18:1248–1262. doi: 10.15252/embr.201744330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okamura M, Inose H, Masuda S. RNA export through the NPC in eukaryotes. Genes (Basel) 2015;6:124–149. doi: 10.3390/genes6010124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodwin JF, Knudsen KE. Beyond DNA repair: DNA-PK function in cancer. Cancer Discov. 2014;4:1126–1139. doi: 10.1158/2159-8290.CD-14-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pankotai T, Bonhomme C, Chen D, Soutoglou E. DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat Struct Mol Biol. 2012;19:276–282. doi: 10.1038/nsmb.2224. [DOI] [PubMed] [Google Scholar]
- 19.Bunch H, et al. Transcriptional elongation requires DNA break-induced signalling. Nat Commun. 2015;6:10191. doi: 10.1038/ncomms10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svejstrup JQ. The RNA polymerase II transcription cycle: Cycling through chromatin. Biochim Biophys Acta. 2004;1677:64–73. doi: 10.1016/j.bbaexp.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 21.Aguilera A, García-Muse T. R loops: From transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Niu T, Manley JL. The RNA binding protein RNPS1 alleviates ASF/SF2 depletion-induced genomic instability. RNA. 2007;13:2108–2115. doi: 10.1261/rna.734407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst) 2011;10:654–665. doi: 10.1016/j.dnarep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 24.Cristini A, Groh M, Kristiansen MS, Gromak N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 2018;23:1891–1905. doi: 10.1016/j.celrep.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S, et al. Congenital bone marrow failure in DNA-PKcs mutant mice associated with deficiencies in DNA repair. J Cell Biol. 2011;193:295–305. doi: 10.1083/jcb.201009074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraemer KH, et al. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: A complex genotype-phenotype relationship. Neuroscience. 2007;145:1388–1396. doi: 10.1016/j.neuroscience.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karikkineth AC, Scheibye-Knudsen M, Fivenson E, Croteau DL, Bohr VA. Cockayne syndrome: Clinical features, model systems and pathways. Ageing Res Rev. 2017;33:3–17. doi: 10.1016/j.arr.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Boer J, Hoeijmakers JH. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 29.Wilson CJ, et al. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 30.Xu J, et al. Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature. 2017;551:653–657. doi: 10.1038/nature24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sollier J, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balajee AS, May A, Dianov GL, Friedberg EC, Bohr VA. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc Natl Acad Sci USA. 1997;94:4306–4311. doi: 10.1073/pnas.94.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cramers P, et al. Impaired repair of ionizing radiation-induced DNA damage in Cockayne syndrome cells. Radiat Res. 2011;175:432–443. doi: 10.1667/RR1972.1. [DOI] [PubMed] [Google Scholar]
- 34.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): Novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 35.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Morales JC, et al. Kub5-Hera, the human Rtt103 homolog, plays dual functional roles in transcription termination and DNA repair. Nucleic Acids Res. 2014;42:4996–5006. doi: 10.1093/nar/gku160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.García-Rubio ML, et al. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015;11:e1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Gool AJ, et al. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16:5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.