Significance
Stacking traits in microalgae is limited by a lack of robust genome modification tools and selectable marker availability. This presents a key hurdle in developing strains for renewable products including biofuels. Here, we overcome these limitations by combining inducible Cre recombinase with constitutive Cas9 nuclease expression in the industrial strain, Nannochloropsis gaditana. With this system, we demonstrate marker- and reporter-free recapitulation of an important lipid productivity trait. In addition, we generate a strain harboring seven-gene knockouts within the photosystem antennae encoding genes. The combined use of relatively mature (Cre) and emerging (CAS9) genome modification technologies can thus accelerate the pace of industrial strain development and facilitate basic research into functionally redundant gene families.
Keywords: genome editing, algal biotechnology, Cas9
Abstract
Robust molecular tool kits in model and industrial microalgae are key to efficient targeted manipulation of endogenous and foreign genes in the nuclear genome for basic research and, as importantly, for the development of algal strains to produce renewable products such as biofuels. While Cas9-mediated gene knockout has been demonstrated in a small number of algal species with varying efficiency, the ability to stack traits or generate knockout mutations in two or more loci are often severely limited by selectable agent availability. This poses a critical hurdle in developing production strains, which require stacking of multiple traits, or in probing functionally redundant gene families. Here, we combine Cas9 genome editing with an inducible Cre recombinase in the industrial alga Nannochloropsis gaditana to generate a strain, NgCas9+Cre+, in which the potentially unlimited stacking of knockouts and addition of new genes is readily achievable. Cre-mediated marker recycling is first demonstrated in the removal of the selectable marker and GFP reporter transgenes associated with the Cas9/Cre construct in NgCas9+Cre+. Next, we show the proof-of-concept generation of a markerless knockout in a gene encoding an acyl-CoA oxidase (Aco1), as well as the markerless recapitulation of a 2-kb insert in the ZnCys gene 5′-UTR, which results in a doubling of wild-type lipid productivity. Finally, through an industrially oriented process, we generate mutants that exhibit up to ∼50% reduction in photosynthetic antennae size by markerless knockout of seven genes in the large light-harvesting complex gene family.
Microalgae have long served as important model organisms in the study of various biological phenomena—including photosynthesis, flagellar function, and cell cycle control (1, 2). In the past decade, a resurgence in efforts to develop microalgae as industrial chassis for biofuels, commodity and high-value chemicals, food/feed, and nutraceutical products has been seen in both nonprofit and the private sectors (3–7). Previous efforts beginning in the 1950s have revealed that, in the case of microalgal biofuels, substantial improvements are needed in photosynthetic efficiency and partitioning to lipid to meet the economics at scale (8, 9). As in other industrial microbe engineering fields (10), programs to develop algae into various products are increasingly benefiting from transformative progress in next-generation sequencing technologies. This has fostered a paradigm shift in which establishing a high-quality draft genome is an early-stage development allowing for genomics-informed approaches in strain engineering. The growing number of microalgal genomes, from both model organisms and industrially relevant strains, have revealed vast diversity in genome complexity, potential for genetic redundancy, and regulation of gene expression (11). This is not surprising given the polyphyletic distribution of microalgae (1, 12), but progress in microalgal genomics and systems-level analyses has outpaced the ability to test hypotheses generated from these types of studies through reverse genetics. This includes the ability to rapidly modify gene target(s) to improve strain performance in an applied setting—or, as importantly, the ability to address basic research hypotheses through molecular/genetic dissection of potentially redundant regulatory or metabolic processes. As such, the development of robust tools for targeted genome modification is seen within the algal research community as a key prerequisite to engineering strain improvement traits and/or gaining deep mechanistic insight into biological processes with complex genetic underpinnings (13, 14).
Progress in exploiting targetable nucleases, such as zinc finger nucleases, transcription activator-like effector nucleases (TALENs), and Cas9, to modify genomes at desired loci has been most recently seen in the rapid enablement of Cas9-mediated gene modification in many organisms (15, 16). Cas9 mutagenesis has been demonstrated in a growing number of algae, where most published reports have served as important proof-of-concept studies targeting genes with known phenotypic outcomes in model organisms; the low efficiency of generating mutants remains a critical hurdle (17–21). We recently developed a Cas9-mediated genome-targeting platform (NgCas9+) in Nannochloropsis gaditana, an industrially relevant marine microalga that is inherently capable of high levels of lipid accumulation (22). The NgCas9+ Cas9 expression line, into which in vitro-synthesized guide RNA (gRNA) can be coelectroporated with an antibiotic selection cassette, allows for efficient targeted integration with up to ∼80% of colonies harboring an insert at the desired locus. In NgCas9+, 18 candidate transcriptional regulators were tested by Cas9-mediated knockout—identifying one, ZnCys, whose expression could be fine-tuned to result in a strain with double the wild-type lipid productivity and a minimal biomass productivity deficit. However, stacking additional traits to improve biomass productivity or further partition carbon to lipids in such a strain is limited because of having only one remaining selectable marker (Ble, conferring Zeocin resistance; Methods), as blasticidin deaminase (BSD) and hygromycin phosphotransferase (HygR), used in generating the NgCas9+ Cas9 expression parental line and the primary Cas9-mediated ZnCys insertion mutant, respectively. A Ble-dependent trait addition would thus render a “dead-end” strain in which no further selectable markers are available for additional strain development.
This limitation could, in principle, be ameliorated by multiplexing gRNAs to target multiple genes, but a method allowing for marker recycling and iterative stacking of traits in the same line would be highly advantaged. While a number of approaches to achieve marker recycling have been developed, among the most widely adapted is the use of Cre recombinase with vectors in which selectable marker cassettes are flanked by lox recombination sites (19, 23–25). This technology has been widely adapted in animals and plants but has only recently been demonstrated in algae (24). In the latter context, the removal of selectable markers with only minimal remaining “lox scars” is an important means of generating “marker-free” genetically modified GM crops to meet acceptability in various markets (26).
Here, we describe the development of an improved genome modification platform in Nannochloropsis gaditana in which constitutive Cas9 expression is combined with an inducible Cre construct. Through several examples, we show (i) the new Cas9/Cre expression line exhibits a similar Cas9-mediated insertional mutagenesis efficiency compared with our previously reported Cas9 expression line (NgCas9+), (ii) markerless versions of important lipid productivity traits [i.e., ZnCys attenuation (22)] can readily be generated, and (iii) the ability to generate lines in which up to seven genes encoding light-harvesting complexes of the photosynthetic apparatus are knocked out through an industrial biotechnology-oriented process combining iterative gRNA multiplexing and Cre-mediated marker removal. These vignettes demonstrate the power of combining Cas9 gene editing with the well-established Cre-mediated recombination technology to generate a host strain where the stacking of industrially relevant traits is not restrained by marker availability, and in which genetically redundant biological processes can be investigated.
Results
Generation of a Marker- and Reporter-Free Cas9/Cre Expression Line.
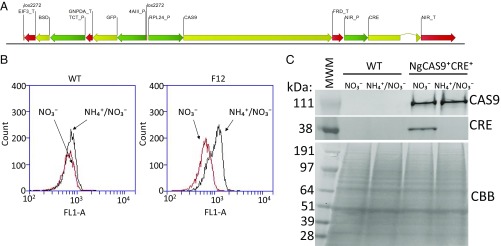
To generate a marker-reporter–free Cre/CAS9 system in Nannochloropsis gaditana, we designed a plasmid (pSliceN’Excise) for constitutive expression of the BSD marker, GFP reporter, and SpCas9 gene using endogenous constitutive promoters and terminators (Fig. 1A). In addition, we included an inducible expression cassette for Cre recombinase controlled by the promoter and terminator of the endogenous nitrite reductase, a gene that is commonly transcriptionally repressed by ammonium (NH4+) in microalgae (27–29). The section of the plasmid harboring BSD and GFP expression cassettes was flanked by lox2272 sites (or, “floxed”) in a direct orientation to enable excision by Cre after expression is induced by absence of NH4+ in the medium (Fig. 1A). Six lines expressing in the entire cell population were split into two cultures of nonselective Cre-repressive media (CRM) (NH4+/NO3−) or Cre-inducing media (CIM) (NO3−), which act to repress or induce Cre expression, respectively. Line F12 lost GFP expression in CIM after 5 d, presumably due to Cre-mediated excision of the region expressing BSD and GFP, and was carried forward for further characterization (Fig. 1B).
Fig. 1.
Generation of a Nannochloropsis line with Cas9 and ammonium-repressible Cre expression (NgCas9+Cre+). (A) Diagram of the construct “pSliceN’Excise” transformed into wild-type (WT) Nannochloropsis containing the blasticidin resistance (BSD) and GFP reporter expression cassettes flanked by lox sites (floxed) adjacent to Cas9 and Cre expression cassettes. Promoters are depicted in green, coding sequences in yellow, and terminator sequences in red. (B) Histograms of GFP fluorescence assessed by flow cytometry of WT and a transformant (F12) selected for comparative growth on CRM (NH4+/NO3−) and CIM (NO3−). (C) Western blot analysis of WT and NgCas9+Cre+, an isolate of the transformant F12 that arose from the NO3−-grown culture depicted in B, under CRM (NH4+/NO3−) and CIM (NO3−) with commercially available antibodies recognizing Cas9 and Cre. A Coomassie Brilliant Blue (CBB)-stained gel is shown for assessment of equal protein loading.
Diagnostic PCR (SI Appendix, Fig. S1 A and B and Table S3) was conducted on genomic DNA from line F12 7 d after the parallel split into Cre inducing or repressing medium. Five primer sets were designed to amplify various areas of the floxed region, one set for the Cas9 and Cre expression cassette region, and one set for the expected extrachromosomal circular Cre-mediated recombination product of the integrated construct. These results indicated that an apparent loss of the floxed region of the construct was accelerated in the F12 CIM culture, although faint PCR products were still detected for the floxed region (SI Appendix, Fig. S1B). Additional PCR analysis using primer set “H” (SI Appendix, Table S1) designed to yield a 4.9-kb band for nonfloxed and 1.3-kb band for floxed Bsd/GFP region revealed that, while both products were observed from gDNA extracted from Cre-repressed F12 culture, only the 1.3-kb product was detectable in Cre-induced F12 culture (SI Appendix, Fig. S1C). Isolated colonies from the CIM-NO3− F12 culture were found to completely lack detectable Bsd and GFP regions, whereas the Cre region remained detectable (SI Appendix, Fig. S1D). Isolate F12-2 was carried forward as a new clonal line, hereafter referred to as NgCas9+Cre+. NgCas9+Cre+ was first analyzed for protein expression of Cas9 and Cre from cultures grown in both CRM and CIM media, revealing that Cas9 expression was constitutive in both media, while Cre was only detected when grown in CIM (Fig. 1C)—consistent with Cre transcript levels measured by qRT-PCR under these conditions (SI Appendix, Fig. S2A).
The genome of NgCas9+Cre+ was sequenced using single-molecule real-time sequencing technology from Pacific Biosciences (PacBio) to characterize integration and subsequent marker/reporter excision of the original pSliceN’Excise vector fragment in the Nannochloropsis genome. A single integration site was observed near the end of chromosome 16, where local alterations to the chromosome region were revealed. The NheI–AscI transgenic region originating from the pSliceN’Excise vector was confirmed to have the floxed BSD and GFP cassettes excised, resulting in an 8.9-kb segment that still harbored Cas9 and Cre expression cassettes in a direct orientation and a single lox2272 site (SI Appendix, Fig. S3B). Immediately upstream of this 8.9-kb segment, a 3.7-kb fragment corresponding to a region of the BAC plasmid backbone was inserted in an inverted orientation. This represents a cointegrated fragment arising from the digestion of pSliceN’Excise with NheI and AscI (Methods). Genome sequencing also revealed a 17-kb deletion near the end of chromosome 16 during integration. This resulted in the disruption of an ABC transporter gene in the middle of the sixth intron by the inverted 3.7-kb NheI–NheI fragment, followed by the 8.9-kb segment harboring Cas9 and Cre. In addition to the rest of the disrupted ABC transporter gene, five other genes were entirely lost, leaving the 8.9-kb Cas9–Cre fragment adjacent to the telomere at one end of chromosome 16 (SI Appendix, Fig. S3B). The NgCas9+Cre+ was found to be indistinguishable from wild-type Nannochloropsis in terms of lipid or biomass productivity when cultured under semicontinuous conditions, which result in optimal wild-type biomass productivity (SI Appendix, Fig. S2B). The impact of the deletion in NgCas9+Cre+ on other aspects of Nannochloropsis physiology is not known, and this should be considered when using the NgCas9+Cre+ strain for molecular/genetic dissection of gene function.
Cas9-Mediated Knockout of Aco1 and Subsequent Cre-Mediated Marker Removal.
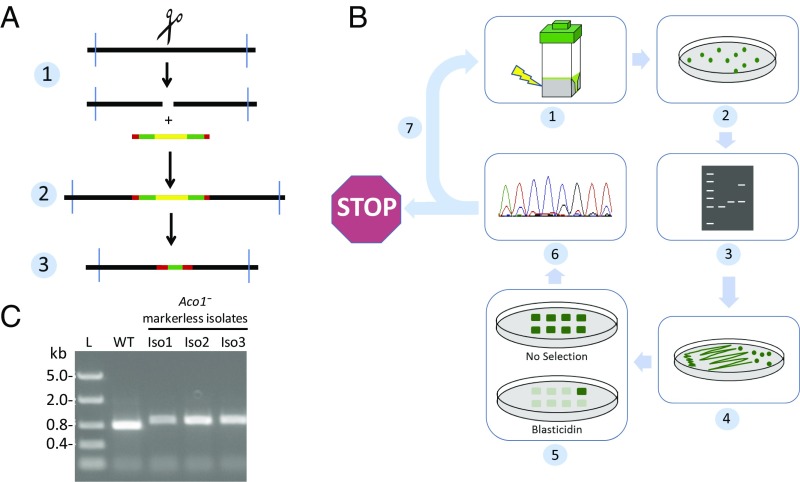
We next tested the relative efficiency in generating Cas9-mediated insertional mutants in a control gene, Aco1, which is one of three genes encoding peroxisomal acyl-CoA oxidases in N. gaditana. Aco1 was selected based on our findings in the previous Cas9 expression host, NgCas9+, that knockouts in this gene had no distinguishing phenotypes from control lines (e.g., significant impact on lipid content; SI Appendix, Fig. S4) and that the particular gRNA used (SI Appendix, Table S2) to knockout Aco1 resulted in a high frequency of generating on-target insertional mutants (up to ∼50%; SI Appendix, Fig. S4). In NgCas9+Cre+, the Aco1 gene was targeted with the same gRNA coelectroporated with a linear marker/reporter gene construct harboring expression cassettes for Bsd and GFP flanked by lox sites in the same orientation (Fig. 2A). Stop codons in all three translational frames were also included on the distal end of each lox site to ensure a stop to translation after marker excision regardless of the orientation of the fragment (Fig. 2A). Markerless insertional mutants were isolated through a process summarized in Fig. 2B, which briefly included (i) selecting blasticidin-resistant colonies on agar plates containing CRM, (ii) colony PCR scoring for lines harboring the expected insertion at the targeted locus, (iii) growth of colonies in CIM liquid media for 7 d followed by plating on CIM agar medium to single colonies, (iv) analysis of the isolated colonies for reversion to a blasticidin-sensitive state, and (v) DNA sequence confirmation of the postrecombination lox “scar.” Of 19 transformants analyzed by PCR for the insert, 9 indicated targeted insertion of the floxed marker/reporter. One line was advanced to Cre induction conditions in CIM media for 7 d, followed by plating to single colonies. Repatched clonal isolates sensitive to blasticidin, and PCR analysis using primers flanking the targeted locus in Aco1 indicated that these clonal isolates all had a small increase in mass over the wild-type band, indicating that the expected lox scar of 134 bp was present (Fig. 2C). Sequencing of the PCR products revealed that the Aco1 gene was disrupted at the expected location in the second exon, exactly 3 bp upstream of the PAM sequence, by a small residual transgenic insert in an inverted orientation, including a single lox site and an in-frame stop codon, which terminates translation of the coding sequence (SI Appendix, Fig. S5). No insertions or deletions were observed to the native DNA flanking the Cas9 targeted cut, while the inverted cassette lost 1 bp at the 5′ junction and 3 bp at the 3′ junction—resulting in a 130-bp scar.
Fig. 2.
Generation of a markerless knockout of acyl-CoA oxidase 1 (Aco1) in NgCAS9+Cre+. (A) Scheme depicting stages for generating a markerless Aco1 gene knockout, where a floxed selectable marker insert integrates into a Cas9-mediated double-stranded DNA break, followed by inducible Cre-mediated excision of the selectable marker region leaving a markerless lox scar. (B) Scheme depicting the protocol for generating single markerless knockouts in NgCAS9+Cre+. After transformation of NgCAS9+Cre+ with an in vitro-synthesized chimeric gRNA and floxed marker DNA, transformants selected by plating on solid media containing blasticidin and NH4+ are screened for insertion of the floxed DNA by colony PCR using locus-specific primers. PCR-positive lines are streaked to isolation on solid media with no selection and medium with NO3− as the sole N source to induce Cre recombination and marker excision. Isolates are then assessed for sensitivity to the selectable agent and remaining lox scar confirmed by PCR sequencing. (C) Isolates of an Aco1 knockout line (Iso1-3) assessed by PCR amplified with locus-specific primers, compared with wild type (WT). A DNA ladder “L” was included with the masses of each fragment indicated on the Left.
Generation of Markerless Attenuation ZnCys Lines with Induced Lipid Productivity by Insertion of a 2-kb Fragment in the 5′-UTR.
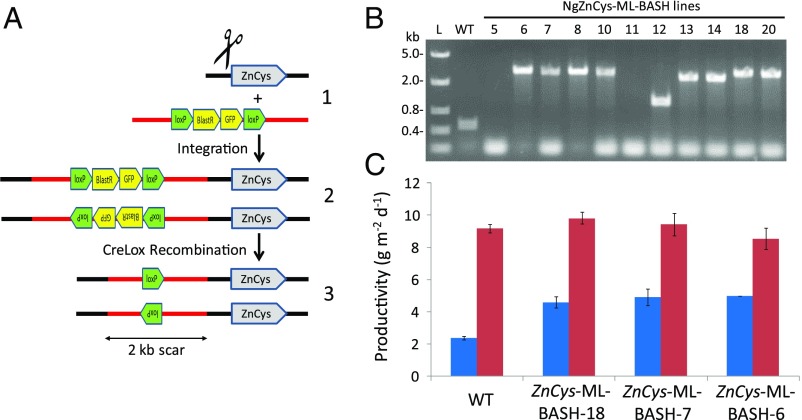
Next, we employed the same methodology described above to instead target the 5′-UTR of the ZnCys gene as a means of markerless recapitulation of the optimized attenuation of ZnCys expression, which leads to a ∼100% increase in lipid productivity (22). This approach, which we refer to as in vivo “promoter bashing,” was first demonstrated by targeting the ZnCys gene 5′-UTR in N. gaditana resulting in stable knockin of a ∼2-kb selectable marker cassette (22). Here, before coelectroporation with gRNA targeting a site 66 bp upstream of the ZnCys start codon, the vector containing the floxed BSD-GFP expression cassettes was digested in a manner that leaves 1.5 and 0.7 kb of backbone DNA outside of the lox sites on the 5′ and 3′ ends of the linearized vector, respectively (Methods). After Cre-mediated marker removal, a ∼2.2-kb markerless insert is expected (Fig. 3A). Twenty-four transformants were scored for visual phenotypes in liquid CIM containing NO3− as the sole N source (SI Appendix, Fig. S6), and 11 lines with a “pale green” appearance similar to the ZnCys-RNAi-7 line (22) were streaked to isolation on CIM agar and analyzed further by colony PCR. Several putative ZnCys markerless 5′-UTR insertion lines (referred to as ZnCys-ML-BASH) showed the intended residual scar size of 2.2 kb (Fig. 3B), indicating that the floxed region was excised, and PCR sequencing further confirmed this finding. Similar to the Aco1 markerless knockouts above, ZnCys-ML-BASH isolates regained sensitivity to blasticidin. Three ZnCys-ML-BASH lines were assessed for biomass and lipid productivity phenotypes under semicontinuous growth conditions identical to those used to assess the original ZnCys attenuation lines, where an incident irradiance mimicking a typical summer day in southern California is provided (22). As seen in Fig. 3C and SI Appendix, Fig. S7, all three ZnCys-ML-BASH lines exhibited total organic carbon (TOC) (a biomass proxy) productivity equivalent to wild type with ∼100% increases in lipid productivity [measured as fatty acid methyl esters (FAMEs)] and increased 16:0 and 16:1 in the total fatty acid profile—as seen previously in ZnCys attenuation lines generated by RNAi and/or stable integration of a selection marker in the 5′-UTR.
Fig. 3.
Generation of markerless promoter insertions in the ZnCys promoter region. (A) Scheme for developing markerless ZynCys promoter insertion mutants (ZnCys-ML-BASH lines). In the first step, a Cas9-mediated double-stranded break is generated in the target locus by coelectroporation of an in vitro-synthesized gRNA along with a lox site flanked marker/reporter cassette (Blast-GFP), which contains additional vector backbone sequence (∼2.2 kb in total, shown in red) outside of the lox sites. NHEJ-mediated repair at the Cas9 target site results in the insertion of the coelectroporated cassette in either orientation. After selection of transgenic lines, Cre is induced, resulting in the recombinatorial removal of the marker/reporter cassette. The additional backbone sequence is stably integrated into the intended locus. (B) Cell patch PCR analysis of wild-type (WT) and ZnCys-ML-BASH lines after Cre recombination using primers (5′-Bash-ZnCys-forward and 5′-Bash-ZnCys-reverse; SI Appendix, Table S1) that flank the Cas9 target site. “L” is Thermo Fisher FastRuler Middle Range DNA Ladder (100–5,000 bp; catalog #SM1113). (C) Areal FAME (blue) and TOC productivity (red) for ZnCys-ML-BASH lines 7, 6, and 18 under semicontinuous growth mode. Shown are the average and SD of biological triplicate cultures where daily FAME and TOC measurements were taken for 7 d. See SI Appendix, Fig. S7 for individually plotted FAME and TOC data for this experiment.
Stacking Multiple Knockouts in the Light-Harvesting Complex Gene Family.
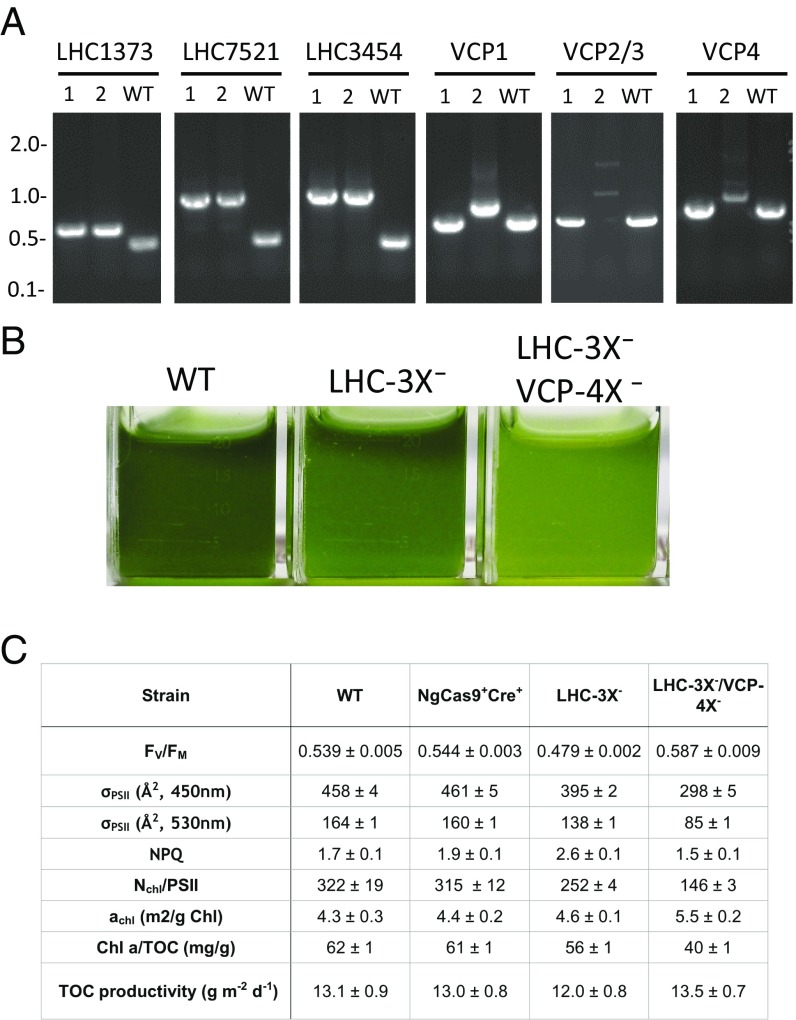
Engineering strains with reduced photosynthetic antennae is widely viewed as a critical component of improving biomass productivity in mass outdoor cultivation (30). Strategies in eukaryotic algae have mainly focused on generating mutations in genes of the chloroplast signal-recognition particle pathway or attenuation of the chlorophyll-b synthase (CAO) gene (31–33)—in addition to reduced antenna mutants from random mutagenesis and screening where the underlying causative genetic lesions are unknown (34, 35). RNAi-mediated silencing of LHCII genes was also demonstrated in the model Chlamydomonas reinhardtii (36). The inability to stack targeted mutations has, however, prevented generation of eukaryotic algal strains with varying degrees of reduced antennae harboring multiple stable knockouts in light-harvesting antennae genes. While stepwise iterative stacking of desired knockout traits is, perhaps, the most straightforward approach, it takes over a month to generate a single markerless knockout at a desired locus when employing Cas9 and Cre as described above. Hence, we tested whether an industrial research-oriented approach combining iterative multiplexing of gRNAs and three selectable marker cassettes (Ble, BSD, and Hyg) in NgCas9+Cre+ could generate a strain with many stacked traits in a substantially reduced time frame (SI Appendix, Fig. S8). For this, we targeted the photosynthetic antenna complex genes in N. gaditana, which is composed of up to 26 light-harvesting complexes (LHCs) including 4 violaxanthin/chlorophyll-a protein (VCP) genes that we identified in the genome of N. gaditana (37). We first targeted the five most highly expressed LHC (non-VCP) genes, identified from RNA-seq on RNA from low light-adapted N. gaditana (SI Appendix, Fig. S9) by simultaneously targeting the genes with unique gRNAs in three successive rounds of transformation and selection, using one of three available floxed selectable markers for each round. Here, the process was modified to employ liquid selection and culturing after the first two rounds of transformation, with plating onto selective agar media after the third round of transformation. Of ∼100 colonies analyzed, 32 with noticeable losses in pigmentation were screened by PCR for insertions at the five LHC loci. While no lines had all five genes knocked out, 12 lines with apparent mutations in three of the targeted LHC genes were carried forward and streaked for isolation on CIM agar plates to induce Cre and complete marker excision. After testing for loss of sensitivity to all three selectable markers and visual scoring cell patches for reduced pigment appearance, one strain, LHC-3X−, was confirmed to harbor knockouts in LHC1373, LHC7521, and LHC3454 (Fig. 4A). LHC-3X− was carried forward as a markerless triple-knockout mutant strain for photophysiological studies and for additional stacking described below (SI Appendix, Fig. S8, first “STOP” point).
Fig. 4.
Genetic and photophysiological characterization of LHC-3X− and LHC-3X−/VCP-4X−. (A) PCR genotyping presence/absence of insertions in LHC-3X− (lane 1) and LHC-3X−/VCP-4X− (lane 2). Wild type (WT) is included as a control. (B) Image of low-light–adapted cultures normalized to equal cell number. (C) TOC productivity, chlorophyll content, and FIRe-based photophysiological measurements for the functional absorption cross-section of PSII (σPSII) assessed at 450 and 530 nm, quantum yield of photochemistry in photosystem II (Fv/Fm), number of chlorophyll per PSII (Nchl/PSII), and the optical absorption cross-section (achl). Shown are the average and SD of technical triplicates of cell culture triplicates (n = 9) for FIRe measurements, and technical duplicates of cell culture triplicates (n = 6) for chlorophyll content. Volumetric TOC data used to calculate steady-state areal TOC productivities are shown in SI Appendix, Fig. S11, along with exemplary FIRe fluorescence traces for the four strains assessed.
Next, the markerless LHC-3X− knockout strain was transformed with a single gRNA against a conserved target found across the nearly identical four Vcp genes (Vcp1-4) and a floxed Bsd-GFP fragment (SI Appendix, Fig. S10). Of 24 colonies analyzed, 10 with a colony appearance visibly paler than LHC-3X− were screened for insertion at all four loci by colony PCR. Six were found to have multiple Vcp genes knocked out, and streaked out to isolation on agar CIM media as above. Markerless isolates were again characterized by PCR, and one isolate, LHC-3X−/VCP-4X−, was shown to have residual scars at all four loci (Fig. 4A); PCR sequencing confirmed that all four copies of the Vcp genes were successfully knocked out (Dataset S1). This resulted in a markerless septuple-knockout mutant strain, LHC-3X−/VCP-4X− (SI Appendix, Fig. S9, second STOP point).
A visible reduction in pigment was observed in liquid culture for LHC-3X− strain compared with wild-type N. gaditana; LHC-3X−/VCP-4X− showed an even greater reduction in pigmentation when grown under ∼50 µE (Fig. 4B). When grown under high-irradiance semicontinuous conditions as described above (Fig. 2), assessment of chlorophyll content revealed ∼10% and ∼35% decrease in the amount of chlorophyll a to TOC, for LHC-3X− and LHC-3X−/VCP-4X− respectively, compared with wild type (Fig. 4C). To further confirm that reduction in chlorophyll a originated from the loss of light-harvesting proteins, we measured the functional absorption cross-section of photosystem II (σPSII) using fluorescence induction and relaxation (FIRe) technique. The observed decrease in σPSII followed the same trend seen in chlorophyll measurements, with a ∼16% reduction of functional cross-section for LHC-3X− and ∼48% reduction for LHC-3X−/VCP-4X− (Fig. 4C). These results are consistent with photophysiological measurements taken under ∼50-µE irradiance conditions (SI Appendix, Table S4), confirming that these marker-free LHC knockout strains exhibit similar reductions in photosynthetic antenna under different growth conditions. TOC productivities were assessed under diel semicontinuous conditions conferring an optimal wild-type productivity, and no significant increase in either of the LHC knockout strains was observed (Fig. 4C).
Discussion
Engineering microalgae for industrial applications faces many challenges often exacerbated by a limited availability of effective selectable markers and robust genome-editing tools. Here, we demonstrate advances made by combining CAS9 editing with Cre-mediated marker recycling in N. gaditana, resulting in a robust platform in which iterative strain improvement is not restrained by marker availability. Induction (as well as strong repression) of Cre expression allows transformants to grow under selective pressure while Cre is repressed in NH4+-containing medium, and enables the marker to be excised and recycled after Cre is induced in medium containing NO3−. This was first demonstrated by the removal of the floxed BSD selectable marker and GFP reporter expression region on the construct used to integrate Cas9 and Cre expression cassettes into the genome, generating the strain NgCas9+Cre+ (Fig. 1). Although not demonstrated here, the same approach can be used in NgCas9+Cre+ to overexpress other transgenes or refactored endogenous genes for various applications. Genome sequencing of NgCas9+Cre+ revealed that nonhomologous end joining (NHEJ)-mediated integration of the Cas9/Cre expression construct resulted in the loss of a ∼17-kb region of chromosome 16, and a consequent loss of function of six genes of unknown function, and similar transgenic events have been observed in other eukaryotic algae in which NHEJ-mediated random insertion is employed (38). Nonetheless, NgCas9+Cre+ was found to be equivalent to wild type in biomass/lipid productivity and in multiple photophysiology parameters (Fig. 4 and SI Appendix, Fig. S2 B and C).
As a first proof of concept in generating a markerless knockout in a gene of interest in NgCas9+Cre+, we targeted an acyl-CoA oxidase encoding gene, Aco1. In our previously reported N. gaditana Cas9 expression strain, NgCas9+, Aco1 has served as a routine control for assessing overall efficiency in generating Cas9-mediated knockouts. Compared with the previous strain, NgCas9+Cre+ exhibited a near-equivalent rate of targeted integration in resistant colonies tested by PCR (∼50%, SI Appendix, Fig. S4B). After Cre-mediated marker removal in one of the Aco1− lines, molecular analysis of three isolates revealed an identical lox scar, which includes a single lox site, no deletion of endogenous DNA adjacent to the Cas9 cut site, deletion of only 1–3 bp on either end of the inserted floxed marker, and introduction of an in-frame translational stop codon in the gene, thereby rendering it nonfunctional.
Optimization of complex traits in some cases requires fine-tuning of endogenous gene expression rather than a strong loss-of-function allele. We previously demonstrated this with the N. gaditana ZnCys transcription factor through stable Cas9-mediated attenuation of gene expression by targeting insertions into the 5′-UTR, resulting in a strain with double the wild-type lipid productivity without significantly impacting TOC productivity (22). Here, we have generated three markerless strains (ZnCys-ML lines) in which the double lipid productivity trait is recapitulated by modifying the floxed construct to leave ∼2 kb of inserted DNA in the 5′-UTR of ZnCys after Cre-mediated excision (Fig. 2). It should be noted that a similar strategy could also be used to exchange or modify endogenous gene promoters to alter expression as desired.
Finally, we demonstrate the potential of the NgCas9+Cre+ strain in rapidly generating strains with multiple markerless knockouts by targeting genes encoding components of the photosynthetic light-harvesting antennae. Although an iterative single-gene targeting approach may be preferred in addressing basic research questions, the industrially oriented process described above (SI Appendix, Fig. S8) was able to rapidly generate the markerless strains LHC-3X− and LHC-3X−/VCP-4X−, which harbor three and seven gene knockouts, respectively. LHC-3X− and LHC-3X−/VCP-4X− showed ∼16% and ∼48% reduction in σPSII compared with the wild type, and similar reduction in chlorophyll a to TOC (∼10% and ∼35%, respectively) under high-irradiance semicontinuous dilution growth conditions. These results show that it is possible to achieve a substantial reduction in σPSII by stacking multiple stable knockouts in genes encoding the structural components of the light-harvesting antenna. While the reduced antenna strains did not exhibit increases in TOC productivity, this may be due to a compromised photosynthetic capacity from knocking out one or more of the LHCs in generating the LHC-3X− knockout strain, which exhibited a reduction in quantum yield of photochemistry in photosystem II (Fv/Fm) and an ∼8% decrease in TOC productivity (Fig. 4C). Selection of LHCs to target based on abundance (relative transcript abundance in this case) may need to be paired with a better understanding of photosynthetic antennae superstructure to reap the maximal benefit from this overall approach.
Combining well-established marker recycling tools like Cre recombinase with the recently emerging CRISPR-Cas genome-editing technologies offers multiple advantages to addressing basic research questions and industrial applications where iterative stacking of genome modifications is required. To date, examples have only been reported in the combined use of Cas9 and Cre in generating marker-free transgenic pigs and markerless genome editing of Caenorhabditis elegans (39, 40)—and the analogous combination of TALENS and/or Cas9 with piggyBac transposase in the seamless correction/modification of genes in human cell lines (41, 42). One consequence of continued Cas9-mediated integration of floxed markers followed by Cre-mediated marker removal is the continued increase in number of lox sites accumulating in the genome. While this can potentially lead to genomic instability with continued expression of Cre (e.g., ref. 43), the use of incompatible mutant lox sites or generation of inert double-mutant lox sites (44), and/or the repression or self-excision of the Cre transgene, can largely prevent this issue.
Recently published work (45) demonstrated the generation of markerless nitrate reductase knockout (NR-KO) mutants in Nannochloropsis oceanica via delivery of a nonintegrated episomal plasmid expressing Cas9 and gRNA. The resultant NR-KO lines were found to lack transgenic DNA after a selection process to remove the episomal DNA was employed. While trait stacking was not demonstrated by Poliner et al., this approach could be employed to generate markerless strains with stacked Cas9-mediated mutations by successive rounds of episome delivery and removal. As such, it is difficult to compare the efficiencies of the Cas9-Cre–mediated approach we describe here compared with the episomal approach. Differences in the outcomes of gene editing are readily apparent, with a residual lox scar present in NgCas9+Cre+ versus an NHEJ-mediated random insertion/deletion event in the episomal approach. Importantly, we have demonstrated the ability to stably add transgenic DNA to the endogenous chromosome followed by marker removal as seen in the generation of NgCas9+Cre+ (Fig. 1) and the ZnCys-ML lines (Fig. 3). Use of episomal Cas9 editing to stably integrate transgenic DNA (e.g., foreign metabolic pathways) into Nannochloropsis, to our knowledge, has not been reported. The development of the dual Cas9-Cre expression strain, NgCas9+Cre+, in N. gaditana places this strain in unrivaled territory among industrially relevant microalgae—not only with the ability to efficiently stack markerless knockouts in multiple genes, but also with the potential for adding foreign metabolic pathways or modifying regulation of endogenous genes in a markerless iterative fashion. As such, this work represents a significant advancement in developing robust tools that will hasten the pace of multitrait industrial strain improvement for algae-based biofuels and other product applications.
Methods
Media Formulations and Strains.
CIM (CIM-NO3−; also referred to standard medium SM-NO3− in ref. 22) consisted of 35 g/L Instant Ocean, 10× F/2 trace metals and vitamins, and 0.361 mM NaH2PO4. The N source was 8.8 mM NaNO3 for the semicontinuous assay and 15 mM NaNO3 for the batch growth screen, both described below. CRM (CRM-NH4+/NO3−; also referred to as SM-NH4+/NO3−) consists of the same ingredients as CIM, but further supplemented with 10 mM NH4Cl and buffered with 15 mM Hepes, pH 8.0. N. gaditana strain CCMP1894 was obtained from CCMP, now known as Provasoli-Guillard National Center for Marine Algae and Microbiota. All strains described in this manuscript were derived from this parental background.
Batch and Semicontinuous Growth Assessment.
Batch culturing was performed in 175-mL culture volume in 75-cm2 rectangular tissue culture flasks, and productivity measurements under semicontinuous daily dilutions were performed in 550-mL culture volume in 225-cm2 flasks exactly as described by Ajjawi et al. (22). In general, 3 d of acclimation to semicontinuous dilution precede days in which TOC and FAME data are collected for determining productivity. Due to necessary upgrades to maintain function for high-irradiance LED growth, different LED sources were used in productivity assessments presented in Figs. 2 and 4; while both provide similar total irradiance under a diel cycle, the upgraded LED system used for Fig. 4 provides a higher amount of blue light, and thus an increased photosynthetically usable radiation (46), explaining the differences observed in wild-type TOC productivity between experiments. The two emission profiles of the LED sources compared with wild-type absorbance spectra are shown in SI Appendix, Fig. S12.
RNA Extractions and qRT-PCR.
RNA extraction was performed as in Ajjawi et al. (22). cDNAs were prepared with the iScript Reverse Transcription Supermix kit (Bio-Rad) and used as templates for qRT-PCR with the Ssofast EvaGreen Supermix (Bio-Rad). The primer sequences for Cre were as follows: forward, 5′-CCAGAGGATTGGCAGTGAAA-3′; reverse, 5′-CACCAGAGACACGGCATTAG-3′. qRT-PCR primers were evaluated for efficiency and the 2−ΔΔCT method was used to estimate gene expression normalized against a control gene (Naga_100004g25; with primer sequences: forward, 5′-CTCTCCTATTGCTTTCCCTCG-3′; reverse, 5′-CTACCAACACCTCTACACTTCC-3′) that was empirically determined to possess a low coefficient of variation across different conditions.
Generation of the NgCas9+Cre+ Markerless Editor Line.
The markerless N. gaditana Cas9/Cre editor line (Ng-Cas9+Cre+) was generated by random integration of an AscI/NheI restriction enzyme-digested vector, pSliceN’Excise, harboring expression cassettes for the selectable marker BSD, Cas9, Cre, and TurboGFP (Evrogen) (Fig. 1). The BSD, Cas9, and Cre genes were optimized for N. gaditana codon usage, whereas TurboGFP was directly amplified from pTurboGFP-C purchased from Evrogen. The Cas9 coding sequence contained an N-terminal nuclear localization signal (NLS) and a FLAG epitope followed by a linker region as described in ref. 22, while the Cre coding sequence contained an N-terminal NLS and was split in half with a native intron from Nannochloropsis gaditana to prevent any basal expression of a functional Cre protein in Escherichia coli; previous constructs without the intron were found to self-excise the floxed marker and reporter genes in E. coli, while introduction of the intron prevented this phenomenon (SI Appendix, Fig. S1A), as reported in ref. 47. The expression of all four genes were driven by endogenous promoters and terminators, which are described in SI Appendix, Table S5. The Cas9/Cre expression construct was assembled with the Gibson Assembly HiFi 1 Step Kit (Synthetic Genomics) into an altered pCC1 BAC vector backbone; the confirmed DNA sequence of this plasmid is shown in GenBank format in SI Appendix.
This plasmid was digested with NheI and AscI to release the DNA fragment harboring Cas9–Cre–Bsd–GFP. While AscI cuts once, NheI cuts twice, resulting in three restriction digest fragments. This includes the desired 12.4-kb NheI–AscI fragment, intended for stable integration of the Cas9 and Cre transgenes, and two fragments from the BAC plasmid backbone: a 5.2-kb AscI–NheI fragment and a 3.7-kb NheI–NheI fragment, which were all present during transformation of Nannochloropsis cells. Transformation of the digested Cas9/Cre expression construct by electroporation was conducted according to ref. 22, except that 1 × 109 cells were transformed in a 0.2-cm cuvette using a field strength of 7,000 V/cm delivered with the Gene Pulser II (Bio-Rad). After overnight recovery in liquid CRM (CRM-NH4+/NO3−), cells were plated on CRM agar containing 100 mg⋅L−1 blasticidin (Invivogen). Putative transformant colonies were repatched on the same selective medium plates, and after sufficient growth, cells were resuspended in liquid CRM and analyzed on the Accuri C6 cytometer (BD Biosciences) for GFP fluorescence (Fig. 1B). Lines exhibiting GFP expression were split into parallel cultures in nonselective CRM and CIM liquid media and cultured for 5 d. For the generation of NgCas9+Cre+, one line (F12) was carried forward after confirming the loss of GFP expression in CIM.
Generation of Targeted Insertional Mutants in NgCas9+Cre+, Followed by Selectable Marker Excision.
gRNAs were synthesized in vitro according to ref. 48, and coelectroporated with a PCR-amplified floxed (flanked by lox sites) DNA fragment containing the following: (i) a codon-optimized selectable marker gene (BSD, HygR, or Ble—see GenBank files in SI Appendix) driven by the endogenous TCT_P promoter and EIF3_T terminator; (ii) a GFPTurbo reporter gene (Evrogen) driven by the endogenous 4AIII_P promoter and GNPDA_T terminator; (iii) two identical or compatible lox sites (loxP, lox2272, and loxN; see sequences in relevant GenBank files in SI Appendix) in the same orientation, which together flank both ends of the tandem marker and reporter expression cassettes; and (iv) spacer DNA at the distal ends of each lox site to protect lox sites from damage/deletion during NHEJ-mediated integration, as well as to add additional features, such as six-frame stop codon regions, to the scar that remains postexcision by Cre. Approximately 5 µg of gRNA and 1 µg of floxed DNA fragment were added to the cuvette, electroporated, and plated onto CRM agar plates containing the appropriate antibiotic (100 mg⋅L−1 blasticidin, 500 mg⋅L−1 hygromycin, or 10 mg⋅L−1 zeocin). Colonies were patched onto the same selective CRM media and analyzed by colony PCR using locus-specific primers that flank the target, screening for integration of the fragment at the target site, as indicated by an expected increase in PCR amplicon size over the wild-type control. A large band indicating the original intact floxed form, as well as a small band indicating the postexcised form with a single lox site and scar, was sometimes observed and interpreted as the presence of a subpopulation of cells in the colony PCR in which Cre had excised the floxed region. Secondary confirmation of the loss of GFP expression in the induced liquid cultures was assessed by flow cytometry, which was conducted in parallel to PCR genotyping, as described above. Lines that lost GFP expression after Cre induction were carried forward to plate for isolation. Three clonal isolates from each induced line were repatched onto CIM agar media with and without appropriate antibiotic selection, and lines sensitive to the selectable agent were carried forward for final PCR sequencing validation. Colony PCR was done as described above using locus-specific primers, and PCR products with an apparent mass indicative of the intended scar with a single lox site were analyzed by Sanger sequencing. This allowed for the determination of insert orientation and potential loss of chromosomal and/or insert DNA, or the gain of small insertions generated during the NHEJ-mediated dsDNA break repair process. In general, loss-of-function “knockout” mutants were generated by selecting a gRNA target locus in the first half of exonic coding sequence, whereas Cas9-mediated insertional attenuation mutants were created by targeting insertion to promoter or 5′-UTRs of a given gene, as described in the main text (see SI Appendix, Table S2, for gRNA sequences). The confirmed DNA sequence of loxP-Bsd-GFP-loxP fragment is shown in GenBank format in SI Appendix.
FAME and TOC Analysis.
FAME and TOC analyses were determined as described in ref. 22.
Western Blots.
Strains were grown to an OD730 of 2.0 in either CIM or CRM media, and 5 mL of culture were pelleted by centrifugation. Pellets were washed once with TBS buffer (50 mM Tris-Cl, pH 7.6, 150 mM NaCl), and then resuspended in 300 µL of SDS/PAGE extraction buffer consisting of 125 mM Tris, pH 8.8, 10% glycerol, and 2% SDS. One hundred microliters of zirconium beads were added to the cell slurry, and cells were vortexed for 30 s before a 10-min incubation at 85 °C. Lysates were vortexed for 30 s three more times throughout the incubation at 85 °C, and then centrifuged, and the supernatant was collected. Supernatant was mixed with NuPAGE LDS Sample Buffer (Thermo Fisher) at a 3:1 ratio, and incubated for 10 min at 85 °C. Twenty-five microliters of the mixture were loaded into each well of the gels. For CRE detection, a 4–12% Bis-Tris gel was used, and electrophoresis was performed using Mops running buffer. For CAS9 detection, a 3–8% Tris-acetate gel was used, and electrophoresis was performed using Tris-acetate running buffer. iBind Western blotting devices (Thermo Fisher Scientific) were used to incubate the blots with primary and secondary antibodies. CRE blots were incubated with primary antibody (rabbit anti-CRE; Millipore at 1:1,000 dilution) and secondary antibody (goat anti-rabbit AP; Novex at 1.5:1,000 dilution), while CAS9 blots were incubated with primary antibody (mouse anti-CAS9; Diagenode at a 2.5:10,000 dilution) and secondary antibody (goat anti-mouse AP; Novex at a 1.5:1,000 dilution). Immunosignals were detected using the Novex AP Chromogeneic Substrate BCIP/NBT kit (Invitrogen).
Chlorophyll Quantitation and FIRe-Based Photophysiological Measurements.
Cells were grown in culture flasks with vent caps for 5 d on an orbital shaker in a growth chamber (25 °C) supplied with 1% CO2. The cultures were illuminated in continuous light (50 μmol photons⋅m−2⋅s−1) from cool-white fluorescent lamps, and cell densities were maintained around 5 × 107 cells per mL by daily dilution. Chlorophyll in the harvested cell pellets was extracted with DMSO/acetone mixed solvent [1:1 (vol/vol)] by vigorous vertex for 3 min. Total chlorophyll a was determined according to ref. 49 by using a Cary Series UV–VIS spectrophotometer (Agilent Technologies).
The functional absorbance cross-section of PSII, , was measured using FIRe kinetics with a multicolor mini-FIRe fluorometer (50). Presented values for were calculated as an average of six measurements (three measurements of each of the two biological replicates)—typical errors for these parameters did not exceed 5%.
Absorption spectra were measured using Perkin-Elmer Lambda 650 spectrophotometer equipped with an integrating sphere. Optical absorption cross-section, (averaged over emission spectrum of a light source), was estimated using the following equation:
| [1] |
where is the chlorophyll concentration in the sample, is the measured OD of the sample at wavelength , is the measuring beam pathlength in the cuvette (1 cm), and is the intensity of the light source used to grow algae at wavelength . The apparent absorption cross-section of individual chlorophyll molecule (averaged over spectrum of white LED), , was calculated assuming mass of chlorophyll molecule of 1.49 × 10−21 g. Using cross-sections of PSII determined with blue/green excitation and optical absorption cross-sections of individual chlorophyll molecule averaged over FIRe LED emission spectra, we estimated the number of chlorophyll molecules in photosystem 2 (Nchl/PSII) using the following relationship:
| [2] |
where , the quantum yield of photochemical reactions in PSII, was assumed to be 0.8.
Supplementary Material
Acknowledgments
We thank the members of the bioinformatic and analytical teams at Synthetic Genomics, Inc., for contributing to this work. This work was funded by ExxonMobil and Synthetic Genomics, Inc.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718193115/-/DCSupplemental.
References
- 1.Cock JM, Coelho SM. Algal models in plant biology. J Exp Bot. 2011;62:2425–2430. doi: 10.1093/jxb/err117. [DOI] [PubMed] [Google Scholar]
- 2.Harris EH. Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- 3.Gangl D, et al. Biotechnological exploitation of microalgae. J Exp Bot. 2015;66:6975–6990. doi: 10.1093/jxb/erv426. [DOI] [PubMed] [Google Scholar]
- 4.Gomaa MA, Al-Haj L, Abed RM. Metabolic engineering of Cyanobacteria and microalgae for enhanced production of biofuels and high-value products. J Appl Microbiol. 2016;121:919–931. doi: 10.1111/jam.13232. [DOI] [PubMed] [Google Scholar]
- 5.Hu Q, et al. Microalgal triacylglycerols as feedstocks for biofuel production: Perspectives and advances. Plant J. 2008;54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- 6.Trentacoste EM, Martinez AM, Zenk T. The place of algae in agriculture: Policies for algal biomass production. Photosynth Res. 2015;123:305–315. doi: 10.1007/s11120-014-9985-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wijffels RH, Barbosa MJ. An outlook on microalgal biofuels. Science. 2010;329:796–799. doi: 10.1126/science.1189003. [DOI] [PubMed] [Google Scholar]
- 8.Berlew JS. Algal Culture: From Laboratory to Pilot Plant. Carnegie Institution of Washington; Washington, DC: 1953. [Google Scholar]
- 9.Sheehan J, Dunahay T, Benemann J, Roessler P. A Look Back at the U.S. Department of Energy’s Aquatic Species Program: Biodiesel from Algae. National Renewable Energy Laboratory; Golden, CO: 1998. [Google Scholar]
- 10.Kuipers OP. Genomics for food biotechnology: Prospects of the use of high-throughput technologies for the improvement of food microorganisms. Curr Opin Biotechnol. 1999;10:511–516. doi: 10.1016/s0958-1669(99)00019-1. [DOI] [PubMed] [Google Scholar]
- 11.Gimpel JA, Henríquez V, Mayfield SP. In metabolic engineering of eukaryotic microalgae: Potential and challenges come with great diversity. Front Microbiol. 2015;6:1376. doi: 10.3389/fmicb.2015.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Archibald JM, Keeling PJ. Recycled plastids: A “green movement” in eukaryotic evolution. Trends Genet. 2002;18:577–584. doi: 10.1016/s0168-9525(02)02777-4. [DOI] [PubMed] [Google Scholar]
- 13.US Department of Energy 2016 Algal Biology Toolbox Workshop Summary Report (US Department of Energy, Washington, DC). Available at https://energy.gov/sites/prod/files/2016/09/f33/algal_biology_toolbox_workshop_summary_report.pdf. Accessed July 15, 2017.
- 14.Doron L, Segal N, Shapira M. Transgene expression in microalgae-from tools to applications. Front Plant Sci. 2016;7:505. doi: 10.3389/fpls.2016.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh V, Braddick D, Dhar PK. Exploring the potential of genome editing CRISPR-Cas9 technology. Gene. 2017;599:1–18. doi: 10.1016/j.gene.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Komor AC, Badran AH, Liu DR. CRISPR-based technologies for the manipulation of eukaryotic genomes. Cell. 2017;168:20–36. doi: 10.1016/j.cell.2016.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baek K, et al. DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci Rep. 2016;6:30620. doi: 10.1038/srep30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W, Brueggeman AJ, Horken KM, Plucinak TM, Weeks DP. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot Cell. 2014;13:1465–1469. doi: 10.1128/EC.00213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nymark M, Sharma AK, Sparstad T, Bones AM, Winge P. A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci Rep. 2016;6:24951. doi: 10.1038/srep24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shin SE, et al. CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci Rep. 2016;6:27810. doi: 10.1038/srep27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, et al. Genome editing of model oleaginous microalgae Nannochloropsis spp. by CRISPR/Cas9. Plant J. 2016;88:1071–1081. doi: 10.1111/tpj.13307. [DOI] [PubMed] [Google Scholar]
- 22.Ajjawi I, et al. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat Biotechnol. 2017;35:647–652. doi: 10.1038/nbt.3865. [DOI] [PubMed] [Google Scholar]
- 23.Abuin A, Bradley A. Recycling selectable markers in mouse embryonic stem cells. Mol Cell Biol. 1996;16:1851–1856. doi: 10.1128/mcb.16.4.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasai Y, Harayama S. Construction of marker-free transgenic strains of Chlamydomonas reinhardtii using a Cre/loxP-mediated recombinase system. PLoS One. 2016;11:e0161733. doi: 10.1371/journal.pone.0161733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zuo J, Niu QW, Møller SG, Chua NH. Chemical-regulated, site-specific DNA excision in transgenic plants. Nat Biotechnol. 2001;19:157–161. doi: 10.1038/84428. [DOI] [PubMed] [Google Scholar]
- 26.Yau YY, Stewart CN., Jr Less is more: Strategies to remove marker genes from transgenic plants. BMC Biotechnol. 2013;13:36. doi: 10.1186/1472-6750-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujiwara T, et al. A nitrogen source-dependent inducible and repressible gene expression system in the red alga Cyanidioschyzon merolae. Front Plant Sci. 2015;6:657. doi: 10.3389/fpls.2015.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koblenz B, Lechtreck KF. The NIT1 promoter allows inducible and reversible silencing of centrin in Chlamydomonas reinhardtii. Eukaryot Cell. 2005;4:1959–1962. doi: 10.1128/EC.4.11.1959-1962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poulsen N, Kröger N. A new molecular tool for transgenic diatoms: Control of mRNA and protein biosynthesis by an inducible promoter-terminator cassette. FEBS J. 2005;272:3413–3423. doi: 10.1111/j.1742-4658.2005.04760.x. [DOI] [PubMed] [Google Scholar]
- 30.de Mooij T, et al. Antenna size reduction as a strategy to increase biomass productivity: A great potential not yet realized. J Appl Phycol. 2015;27:1063–1077. [Google Scholar]
- 31.Mussgnug JH, et al. NAB1 is an RNA binding protein involved in the light-regulated differential expression of the light-harvesting antenna of Chlamydomonas reinhardtii. Plant Cell. 2005;17:3409–3421. doi: 10.1105/tpc.105.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perrine Z, Negi S, Sayre RT. Optimization of photosynthetic light energy utilization by microalgae. Algal Res. 2012;1:134–142. [Google Scholar]
- 33.Tetali SD, Mitra M, Melis A. Development of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii is regulated by the novel Tla1 gene. Planta. 2007;225:813–829. doi: 10.1007/s00425-006-0392-z. [DOI] [PubMed] [Google Scholar]
- 34.Cazzaniga S, et al. Domestication of the green alga Chlorella sorokiniana: Reduction of antenna size improves light-use efficiency in a photobioreactor. Biotechnol Biofuels. 2014;7:157. doi: 10.1186/s13068-014-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakajima Y, Tsuzuki M, Ueda R. Improved productivity by reduction of the content of light-harvesting pigment in Chlamydomonas perigranulata. J Appl Phycol. 2001;13:95–101. [Google Scholar]
- 36.Oey M, et al. RNAi knock-down of LHCBM1, 2 and 3 increases photosynthetic H2 production efficiency of the green alga Chlamydomonas reinhardtii. PLoS One. 2013;8:e61375. doi: 10.1371/journal.pone.0061375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schwartz AS, et al. Complete genome sequence of the model oleaginous alga Nannochloropsis gaditana CCMP1894. Genome Announc. 2018;6:e01448-17. doi: 10.1128/genomeA.01448-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 2005;137:545–556. doi: 10.1104/pp.104.055244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi Y, et al. Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci Rep. 2016;6:31729. doi: 10.1038/srep31729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dickinson DJ, Pani AM, Heppert JK, Higgins CD, Goldstein B. Streamlined genome engineering with a self-excising drug selection cassette. Genetics. 2015;200:1035–1049. doi: 10.1534/genetics.115.178335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie F, et al. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye L, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA. 2014;111:9591–9596. doi: 10.1073/pnas.1407473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campo N, Daveran-Mingot ML, Leenhouts K, Ritzenthaler P, Le Bourgeois P. Cre-loxP recombination system for large genome rearrangements in Lactococcus lactis. Appl Environ Microbiol. 2002;68:2359–2367. doi: 10.1128/AEM.68.5.2359-2367.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Missirlis PI, Smailus DE, Holt RA. A high-throughput screen identifying sequence and promiscuity characteristics of the loxP spacer region in Cre-mediated recombination. BMC Genomics. 2006;7:73. doi: 10.1186/1471-2164-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poliner E, Takeuchi T, Du ZY, Benning C, Farré EM. Nontransgenic marker-free gene disruption by an episomal CRISPR system in the oleaginous microalga, Nannochloropsis oceanica CCMP1779. ACS Synth Biol. 2018;7:962–968. doi: 10.1111/tpj.14314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morel A. Available, usable, and stored radiant energy in relation to marine photosynthesis. Deep Sea Res. 1978;25:673–688. [Google Scholar]
- 47.Agaphonov M, Alexandrov A. Self-excising integrative yeast plasmid vectors containing an intronated recombinase gene. FEMS Yeast Res. 2014;14:1048–1054. doi: 10.1111/1567-1364.12197. [DOI] [PubMed] [Google Scholar]
- 48.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 49.Inskeep WP, Bloom PR. Extinction coefficients of chlorophyll a and b in N,N-dimethylformamide and 80% acetone. Plant Physiol. 1985;77:483–485. doi: 10.1104/pp.77.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorbunov MY, Falkowski PG. Fluorescence induction and relaxation (FIRe) technique and instrumentation for monitoring photosynthetic processes and primary production in aquatic ecosystems. In: van der Est A, Bruce D, editors. Photosynthesis: Fundamental Aspects to Global Perspectives. Alliance Communications Group; Lawrence, KS: 2004. pp. 1029–1031. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.