Abstract
The bacterium Bacillus thuringiensis produces several insecticidal proteins, such as the crystal proteins (Cry) and the vegetative insecticidal proteins (Vip). In this work, we report that a specific interaction between two B. thuringiensis toxins creates insecticidal synergism and unravel the molecular basis of this interaction. When applied together, the three-domain Cry toxin Cry9Aa and the Vip Vip3Aa exhibited high insecticidal activity against an important insect pest, the Asiatic rice borer (Chilo suppressalis). We found that these two proteins bind specifically to brush border membrane vesicles of C. suppressalis and that they do not share binding sites because no binding competition was observed between them. Binding assays revealed that the Cry9Aa and Vip3Aa proteins interacted with high affinity. We mapped their specific interacting regions by analyzing binding of Cry9Aa to overlapping fragments of Vip3Aa and by analyzing binding of Vip3Aa to individual domains of Cry9Aa. Binding to peptide arrays helped narrow the binding sites to domain II loop-3 of Cry9Aa and to 428TKKMKTL434 in Vip3Aa. Site-directed mutagenesis confirmed that these binding regions participate in binding that directly correlates with the synergism between the two proteins. In summary, we show that the B. thuringiensis Cry9Aa and Vip3Aa toxins display potent synergy based on a specific interaction between them. Our results further our understanding of the complex synergistic activities among B. thuringiensis toxins and are highly relevant to the development of toxin combinations for effective insect control and for delaying development of insect resistance.
Keywords: protein-protein interaction, toxin, Bacillus, insect, epitope mapping, pesticide, synergism, synergistic activity
Introduction
Bacillus thuringiensis produces different insecticidal proteins, such as the crystal proteins (Cry)3 during the sporulation phase of growth and the vegetative insecticidal proteins (Vip) during the vegetative phase. Both Cry and Vip proteins have been expressed in genetically modified crops, resulting in extraordinary activity to control lepidopteran insect pests. However, evolution of insect resistance to Cry toxins in the field has been reported, representing a major threat for the use of these toxins in insect control (1–3).
Different Cry protein families produced by B. thuringiensis, not related phylogenetically, have been described, such as the three-domain Cry (3d-Cry) family, the Mtx-like Cry family, and Bin-like Cry family (4). Among these, the 3d-Cry family represents the bigger family of proteins produced by B. thuringiensis (composed of more than 54 subgroups); they are globular molecules containing three distinct domains connected by single linkers (4). Cry proteins receive their mnemonic Cry name and four hierarchical ranks depending on their primary sequence identity. The first rank is a different number that is given if the protein shares less than 45% identity with all the other Cry proteins. The second rank is a capital letter that is given if the protein shares less than 78% but more than 45% identity with other Cry proteins. The third rank gives a lowercase letter to distinguish proteins that share more than 78% identity and less than 95% with other Cry proteins (4). Cry9Aa is a member of this 3d-Cry family (4) and shows insecticidal activity against economically important Lepidoptera such as Plutella xylostella, Exorista larvarum, and Spodoptera exigua (5–7). The activated form of the 3d-Cry family is composed of three domains: an α-helical domain (domain I) and two β-sheet domains (domains II and III). In particular, the exposed loop regions of domain II are important in binding larva midgut proteins to trigger toxicity (8). The 3d-Cry toxins bind to specific protein receptors located in midgut brush border membrane, inducing toxin oligomerization and insertion into the membrane forming lytic pores, causing cell swelling and lysis (8). Cry9 toxins have been proposed as promising proteins because they showed a wide spectrum of action against lepidopteran pests (9–12). Binding assays showed that Cry9 toxins do not share receptors with Cry1Ab in different insects, suggesting that Cry9 toxins could be useful for resistance management (13, 14).
The Vip3A toxins display a different mechanism of action and do not share receptors with Cry1A, Cry1F, and Cry2A toxins in brush border membranes isolated from various insect species (15–20). They are toxic to a large number of lepidopteran species, such as Helicoverpa armigera, Agrotis ipsilon, Spodoptera frugiperda, Spodoptera littoralis, and S. exigua (21–23). The insecticidal mechanism of Vip3 proteins is still unclear. It has been reported that Vip3A toxin is activated by midgut proteases, resulting in major proteolytic products of 65 and 20 kDa (16, 24, 25). It was proposed that both fragments are necessary for toxicity and that the 20-kDa N-terminal fragment stabilizes the 65-kDa C-terminal domain (26). Transmission EM studies and analytical ultracentrifugation sedimentation velocity assays showed that trypsin-activated Vip3A formed a complex composed of four subunits (27, 28). The identity of Vip3Aa receptors remains unknown (15, 22). Finally, Vip3A inserts into the membrane, resulting in pore formation activity that lyses midgut cells (18, 22). Recently, it was shown that Vip3A induces apoptosis in the Sf9 cell line (30). The activation of apoptosis was also found in S. exigua larvae when treated with Vip3Aa or Vip3Ca toxins as determined by terminal deoxynucleotidyltransferase–mediated dUTP nick end labeling staining (31). The expression level of Caspase-4, proposed as an initiator Caspase, was up-regulated after 3–12 h of toxin treatment (31), whereas the cobatoxin B gene and immune signaling pathway JAK-STAT gene were found to be slightly down-regulated (31). The JAK-STAT pathway has been shown to be involved in the activation of midgut renewal by the proliferation and differentiation of stem cells (32). Thus, the negative regulation of this pathway may affect the renewal of midgut tissue damaged by Vip3Aa toxin.
Here, we show that Cry9Aa and Vip3Aa have a strong synergistic toxicity when tested together in bioassays against Chilo suppressalis larvae. C. suppressalis is an important rice pest in Asia, especially in China (33). This insect is susceptible to several Cry proteins, such as Cry1Ab, Cry1Ac, Cry1Ca, Cry1Fa, and Cry2Aa toxins (34). The Cry1Ab and Cry1Ac toxins have been expressed in genetically modified rice, showing excellent control of C. suppressalis larvae (35).
The Vip toxins have gained increased interest because of their broad toxicity against lepidopteran pests and their different mode of action (15–20, 22, 24, 25, 30). Transgenic crops expressing Vip3Aa toxin along with other Cry toxins, such as Cry1Ac, have prompted different groups to determine the potential synergism of Vip3 with other Cry toxins that can be used as pyramid toxins in transgenic crops (36, 37). The molecular mechanism responsible for the synergism between Cry and Vip toxins has not been studied before.
We found that Cry9Aa and Vip3Aa toxins do not share binding sites on brush border membrane vesicles (BBMVs) from C. suppressalis, but these two proteins showed a specific interaction between them that directly correlates with their synergistic activity in C. suppressalis. We identified the binding regions in Cry9Aa and Vip3Aa and show that their binding interaction is the basis for their synergistic activity. The finding that the synergism between Vip3Aa and Cry9Aa relies on their specific binding could result in the future development of improved toxin combinations for effective insect control.
Results
Toxicity of Cry9Aa and Vip3Aa against C. suppressalis
In a screening for B. thuringiensis toxins useful for the control of C. suppressalis, four Cry proteins (Cry1Ai, Cry1Ie, Cry9Aa, and Cry9Ee) and Vip3Aa were analyzed individually and in combination against this pest. These analyses showed that Cry9Aa and Vip3Aa had a synergistic effect against this pest, whereas other combination did not (data not shown). For the quantitative analysis of toxicity, these toxins were expressed in Escherichia coli and purified by nickel-affinity chromatography as described under “Experimental procedures” (Fig. S1A). Bioassays against C. suppressalis larvae showed that both proteins have a moderate toxicity and that the toxicity of Cry9Aa was 53-fold higher than the Vip3Aa activity (Table 1). To determine the effect on toxicity of a mixture of Cry9Aa and Vip3A, toxicity assays were performed with a mixture of Cry9Aa and Vip3Aa (1:1 ratio). Table 1 shows that Cry9Aa and Vip3Aa display a significant synergistic interaction with a synergism factor (SF) value of 10.6-fold. We found that this combination of toxins induces synergism in other insect pests because the same combination of proteins at a 1:1 ratio induced synergism against Ostrinia furnacalis larvae, showing an SF value of 4.5-fold (Table S1).
Table 1.
Bioassays of insecticidal activity of Cry9Aa and Vip3Aa toxins against C. suppressalis larvae
| Toxins | Observed LC50 in μg/g diet (95% confidence limits) | Expected LC50 in μg/g diet | SF |
|---|---|---|---|
| Cry9Aa | 0.70 (0.38–1.24) | ||
| Vip3Aa | 37.41 (17.11–79.31) | ||
| Cry9Aa + Vip3Aa | 0.13 (0.07–0.24) | 1.37 | 10.6 |
| Cry9Aa-R495A/R496A/S497A | 4.02 (0.85–71.50) | ||
| Vip3Aa-N364A/D365A/S366A | 163.66 (34.12–1975.17) | ||
| Vip3Aa-K432A/T433A/L434A | 29.59 (13.68–47.99) | ||
| Cry9Aa-R495A/R496A/S497A + Vip3Aa | 1.69 (0.37–35.06) | 7.26 | 4.3 |
| Cry9Aa + Vip3Aa-N364A/D365A/S366A | 0.12 (0.06–0.31) | 1.39 | 11.6 |
| Cry9Aa + Vip3Aa-K432A/T433A/L434A | 1.07 (0.48–2.19) | 1.37 | 1.3 |
Analysis of Cry9Aa and Vip3Aa binding with BBMVs from C. suppressalis
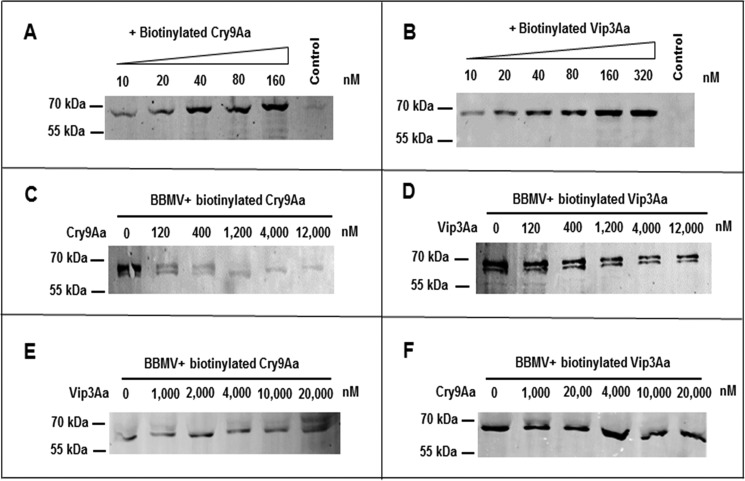
Saturation binding assays using increasing concentrations of biotinylated activated Cry9Aa or Vip3Aa toxins showed that the binding of these proteins to BBMVs from C. suppressalis larvae was saturable (Fig. 1, A and B). The controls in Fig. 1, A and B, are the highest concentration of biotinylated Cry9Aa (160 nm) or Vip3Aa (320 nm) proteins without BBMVs under the same conditions, showing no signal, suggesting that there was no precipitation of the two toxins and that the signals detected in the lanes with BBMVs are due to toxin binding to the BBMVs. Homologous binding competition assays using 40 nm biotinylated Cry9Aa and Vip3Aa proteins showed that both toxins were displaced by the corresponding unlabeled toxin, indicating that they bind specifically to the BBMVs from C. suppressalis (Fig. 1, C and D). The heterologous binding competition assays were also performed with 40 nm biotinylated Cry9Aa and Vip3Aa proteins. These assays showed that these toxins do not share binding sites because unlabeled Vip3Aa did not compete the binding of biotinylated Cry9Aa, and unlabeled Cry9Aa did not compete the binding of labeled Vip3Aa to C. suppressalis BBMVs (Fig. 1, E and F).
Figure 1.
Binding assay of Cry9Aa and Vip3Aa toxins to BBMVs from C. suppressalis. A and B, saturation binding assays were done using increasing concentrations of Cry9Aa and Vip3Aa to BBMVs, respectively, showing that binding of these toxins to the BBMVs is saturable. A control of the highest concentration of biotinylated toxins without BBMVs was included in this figure, showing that the biotinylated toxins did not precipitate in the absence of membranes. C and D, homologous binding competition assays of 40 nm biotinylated Cry9Aa or biotinylated Vip3Aa to BBMVs in the presence of increasing concentrations of the same unlabeled toxin were performed, showing that both toxins were competed by their corresponding unlabeled toxin. E and F, heterologous binding competition assays of 40 nm biotinylated Cry9Aa or Vip3Aa to BBMVs performed in the presence of increasing concentrations of the other unlabeled toxin, showing that binding of biotinylated toxins could not be competed by the other unlabeled toxin.
Binding interaction between Cry9Aa and Vip3Aa proteins
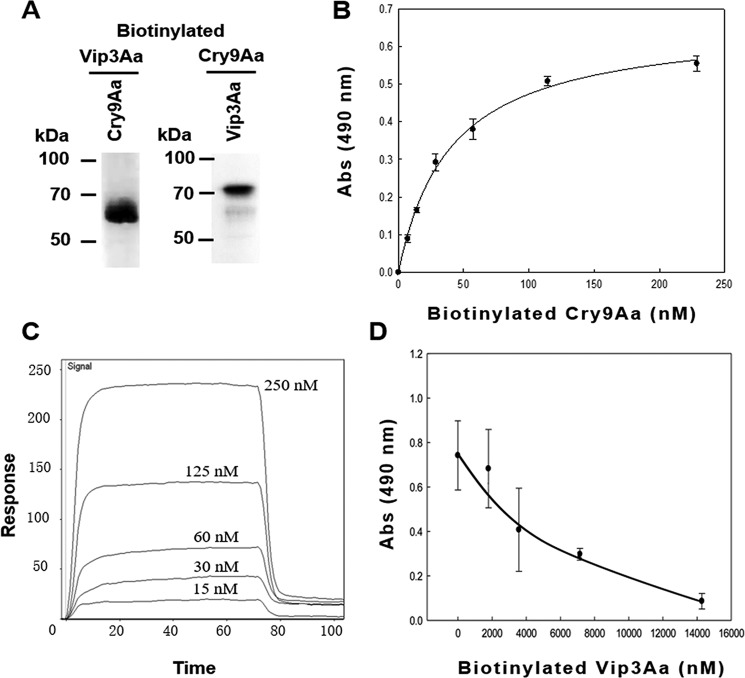
To determine whether the synergistic activity relies on the interaction between Cry9Aa and Vip3Aa toxins, we analyzed the binding between these toxins by ligand blotting (Fig. 2A). In these assays, an excess of Cry9Aa or Vip3Aa toxins was loaded for SDS-PAGE, and after transfer to the membrane and blocking, the other biotinylated toxin was incubated with the membrane. The results showed a clear binding interaction between Cry9Aa and Vip3Aa toxins. In addition, the binding of Cry9Aa to Vip3Aa toxin was confirmed by enzyme-linked immunosorbent assay (ELISA), showing that the binding of biotinylated Cry9Aa with the Vip3Aa toxin fixed in the ELISA well plate is saturable with an apparent dissociation constant (Kd) of 79.8 ± 21.1 nm (Fig. 2B). Surface plasmon resonance assays confirmed that these two toxins display a high-affinity interaction with a Kd value of 222 nm (Fig. 2C). Finally, the specific binding between Cry9Aa and Vip3Aa was verified by homologous competition binding assays, showing that excess unlabeled Vip3Aa competed the binding of biotinylated Vip3Aa to Cry9Aa (Fig. 2D).
Figure 2.
Binding assay between Cry9Aa and Vip3Aa toxins. A, ligand-blot binding assays of 5 nm biotinylated Vip3Aa or Cry9Aa toxins bound to Cry9Aa or Vip3Aa blotted on the PVDF membrane (0.5 μg of each protein), showing that they can interact with each other. B, ELISA binding of increasing concentrations of biotinylated Cry9Aa to 1 μg of Vip3Aa protein bound to the ELISA plate, showing saturable binding. Error bars represent S.D. C, SPR binding analyses of Vip3Aa were performed by immobilizing Vip3Aa toxin by conventional amine coupling. Sensorgrams of serial doubling dilutions of Cry9Aa are shown. D, competition ELISA binding assay of 28 nm biotinylated Vip3Aa to 0.5 μg of Cry9Aa protein bound to the ELISA plate performed in the presence of different excesses of unlabeled Vip3Aa as competitor, showing that their interaction is specific. Error bars represent S.D. Abs, absorbance.
Identification of the Cry9Aa-binding regions
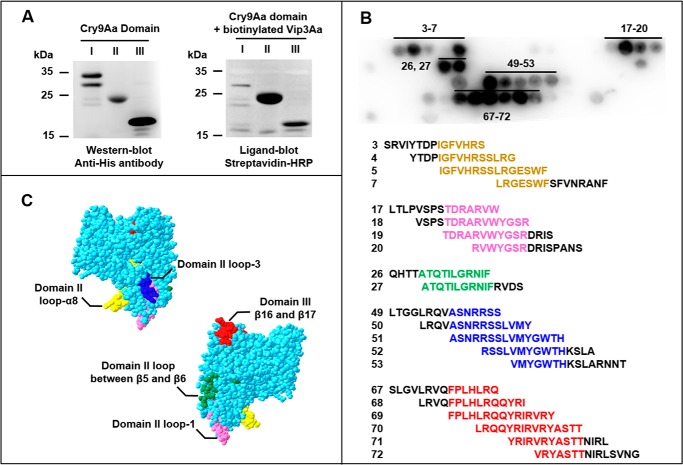
To identify the specific regions involved in the interaction between Cry9Aa and Vip3Aa toxins, we expressed each domain of Cry9Aa separately in E. coli cells. Fig. 3A shows that all three domains were expressed as revealed by Western blotting using Penta-His-horseradish peroxidase (HRP) antibody. Ligand-blot assays showed that biotinylated Vip3Aa bound to domains II and III of Cry9Aa. To further narrow the binding regions in Cry9Aa, a library of overlapping peptides derived from domain II and III amino acid sequences was synthesized and bound to a nitrocellulose membrane. A total of 84 peptides of 15 amino acids each (where 11 residues overlap with the next peptides) were screened with biotinylated Vip3Aa. Fig. 3B shows the interacting signals with five overlapping amino acid sequences: 308IGFVHRSSLRGESWF322 (spots 3–7), 364TDRARVWYGSR374 (spots 17–20), 396ATQTILGRNIF407 (spots 26, 27), 492ASNRRSSLVMYGWTH506 (spots 49–53), and 564FPLHLRQQYRIRVRYASTT582 (spots 67–72). These five regions were localized on a space-filled model of Cry9Aa toxin, which was obtained by comparison to the Cry8Ea structure (38) that shows 33.7% identity with Cry9Aa (Fig. 3C). Residues 308–322 are located in domain II loop-α8, residues 364–374 are in domain II loop-1, residues 396–407 are between β5 and β6 of domain II, residues 492–506 are in domain II loop-3, and residues 564–582 are between β16 and β17 of domain III.
Figure 3.
Identification of Cry9Aa regions involved in the interaction with Vip3Aa. A, expression of each individual Cry9Aa domain detected by anti-His antibody and analysis of binding of biotinylated Vip3Aa (5 nm) to these Cry9Aa domains, showing that Vip3Aa binds to domains II and III of Cry9Aa. B, binding of biotinylated Vip3Aa (5 nm) to the cellulose membrane–bound peptides corresponding to the sequence of domains II and III from Cry9Aa, showing five regions of interaction. C, localization of the identified sequences in a model of Cry9Aa constructed by SWISS-MODEL.
Identification of the Vip3Aa-binding regions
To identify Vip3Aa regions involved in the interaction with Cry9Aa, 10 overlapping fragments of 150–200 amino acids were cloned separately and expressed in E. coli. All the Vip3Aa protein fragments showed similar expression as revealed by Western blotting (Fig. 4A). Ligand-blot binding analysis of biotinylated Cry9Aa to these Vip3Aa fragments showed that Cry9Aa binds to Vip3Aa fragments F5 and F6 (Fig. 4A) that showed an overlapping region of 100 amino acids corresponding to the Vip3Aa region Lys352–Asp451. Further binding analysis of biotinylated Cry9Aa to 23 cellulose membrane–bound peptides of 15 amino acids each (where 11 residues overlap with the next peptides) that correspond to the Vip3Aa amino acid sequence Lys352–Asp451 allowed us to identify two overlapping regions of Vip3Aa (Fig. 4B) corresponding to fragments 356ALIGFEISNDS366 and 428TKKMKTL434 (Fig. 4B).
Figure 4.
Identification of Vip3Aa regions involved in the interaction with Cry9Aa. A, expression of Vip3Aa-overlapping fragments detected by anti-His antibody and analysis of binding of 5 nm biotinylated Cry9Aa to these Vip3Aa fragments detected with streptavidin-HRP conjugate followed with SuperSignal chemiluminescence substrate as described under “Experimental procedures,” showing that Cry9Aa binds to fragments F5 and F6. The overlapping region of these fragments (Lys352–Asp451) is shown in black. B, binding of 5 nm biotinylated Cry9Aa to the cellulose membrane–bound peptides corresponding to the amino acid sequence of Vip3Aa-overlapping fragment, showing two regions of interaction. Binding of biotinylated Cry9Aa was detected also with streptavidin-HRP conjugate and SuperSignal chemiluminescence substrate.
Isolation of Cry9Aa and Vip3Aa mutants affected in binding
To determine whether the binding regions identified are involved in synergism between these toxins, we isolated Cry9Aa and Vip3Aa triple mutants in the identified regions and analyzed their interaction by ligand blotting and ELISAs. Ten Cry9Aa triple mutants with substitutions by alanine were constructed in the loops of domain II because these regions were identified in the binding assay to the cellulose membrane–bound peptides and have been shown to be important for Cry toxin binding to receptors (8). The isolated Cry9Aa mutants are as follows: loop-α8, Cry9Aa-P307A/I308A/G309A, Cry9Aa-L316A/R317A/H318A, and Cry9Aa-V325A/N326A/R327A; loop-1, Cry9Aa-P359A/V360A/S361A and Cry9Aa-T364A/D365A/R366A; loop between β5 and β6, Cry9Aa-H393A/T394A/T395A; loop-2, Cry9Aa-N416A/D417A/T418A and Cry9Aa-V422A/N423A/R424A; loop-3, Cry9Aa-L488A/R489A/G490A and Cry9Aa-R495A/R496A/S497A. Nine of these mutants showed similar expression as the Cry9Aa toxin in E. coli cells. Cry9Aa-V325A/N326A/R327A mutant was severely affected in its expression in E. coli cells and was not further analyzed (Fig. 5A). Ligand-blot assays performed with the Cry9Aa loop mutants incubated with biotinylated Vip3Aa showed that mutant Cry9Aa-P307A/I308A/G309A located in loop-α8 and mutant Cry9Aa-R495A/R496A/S497A located in loop-3 were affected in binding with Vip3Aa (Fig. 5A). The protein stability of these mutants after treatment with trypsin was assayed. The results show a similar activation as the wildtype (WT), suggesting no major structural modifications of these mutants (Fig. S1).
Figure 5.
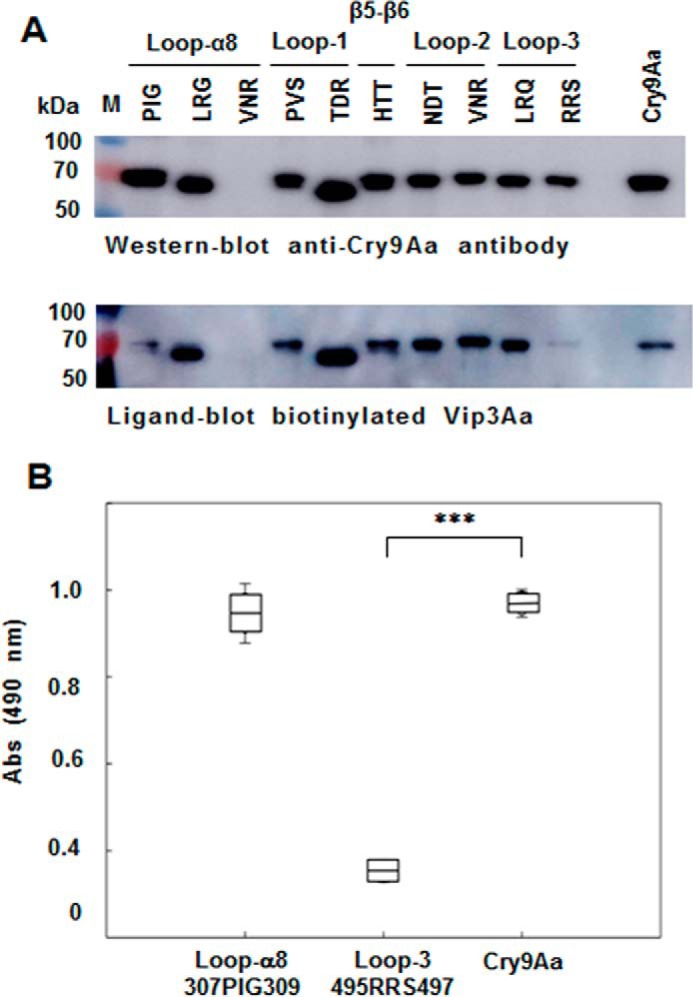
Analysis of binding of Cry9Aa to Vip3Aa WT or mutants. A, expression of different Cry9Aa mutants located in different regions of domain II. In each mutant, three selected amino acids were substituted by alanine. The expression of each Cry9Aa triple mutant was detected by Western blotting with anti-Cry9Aa antibody, and ligand-blot analysis showed the binding of 5 nm biotinylated Vip3Aa to these Cry9Aa mutants. Binding of biotinylated Vip3Aa was detected with streptavidin-HRP conjugate and SuperSignal chemiluminescence substrate as described under “Experimental procedures.” Prestained molecular markers (Thermo Scientific) were used in this figure. B, ELISA binding assays of 56 nm biotinylated Cry9Aa mutants to Vip3Aa (1 μg) fixed on the plate, showing that the binding was severely affected to the loop-3 R495A/R496A/S497A mutant with significant differences (***, p < 0.005) analyzed by t test. Error bars represent S.D. M, molecular mass markers; Abs, absorbance.
ELISA binding assays of immobilized Vip3Aa in the ELISA plate incubated with Cry9Aa mutants showed that the interaction of mutant Cry9Aa-R495A/R496A/S497A and Vip3Aa was significantly decreased in contrast to Cry9Aa-P307A/I308A/G309A that was not affected in binding to Vip3Aa (t test p < 0.005) (Fig. 5B). Surface plasmon resonance assays confirmed that Cry9Aa-R495A/R496A/S497A and Vip3Aa toxins were affected in their interaction, showing a 3-fold higher Kd value, an indication of lower binding affinity between these proteins (data not shown).
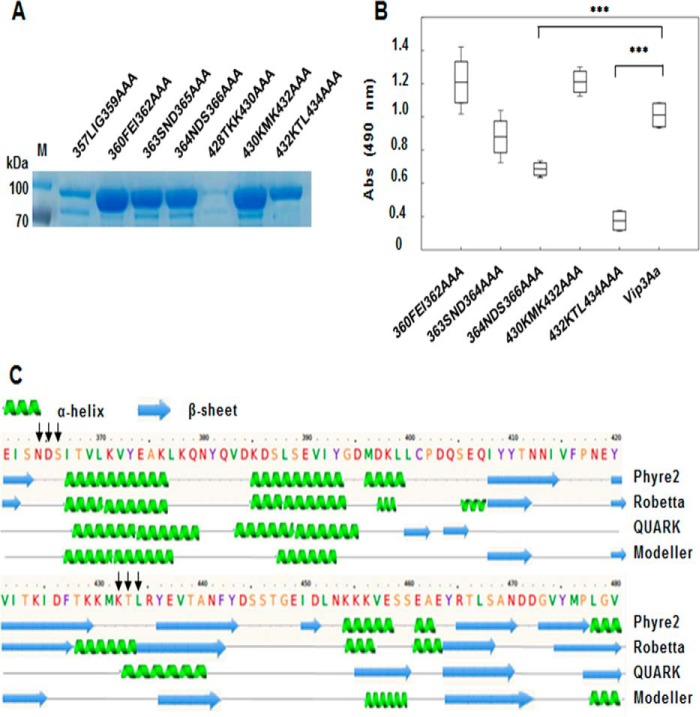
For the identification of Vip3Aa residues involved in Cry9Aa binding, seven triple mutants in two regions of Vip3Aa (Vip3A 356ALIGFEISNDS366 and Vip3A 428TKKMKTL434) were constructed and expressed in E. coli (Fig. 6A). Vip3Aa-T428A/K429A/K430A mutant was not expressed in E. coli and was not further analyzed (Fig. 6A). The protein stability of these mutants after treatment with trypsin was also analyzed, showing that Vip3A-L357A/I358A/G359A was more sensitive to protease digestion, whereas the rest of the proteins were stable and showed similar activation patterns as the WT (Fig. S1). ELISA binding assays showed that Vip3Aa-N364A/D365A/S366A and Vip3Aa-K432A/T433A/L434A showed significantly decreased binding with Cry9Aa (Fig. 6B). ELISA binding assays revealed that the Vip3Aa-K432A/T433A/L434A mutant showed a 6-fold higher Kd value than Vip3A, indicating lower binding affinity (data not shown). Finally, prediction of the secondary structure of Vip3Aa by using four different strategies (Fig. 6C) indicated that Asn364–Ser366 and Lys432–Leu434 residues are likely located in potential exposed loop regions with the first predicted to be found between β-sheet-5 and α-helix-15 and the second predicted to be between β-sheet-7 and β-sheet-8 of Vip3Aa.
Figure 6.
Analysis of binding of Cry9Aa to different Vip3Aa mutants. A, expression of different Vip3Aa mutants located in the identified regions. In each mutant, three selected amino acids were substituted by alanine. The expression of each Vip3Aa triple mutant was analyzed by SDS-PAGE. B, ELISA binding assays of 28 nm biotinylated Vip3Aa mutants to Cry9Aa (0.5 μg) fixed on the plate, showing that the binding was affected to Vip3Aa-N364A/D365A/S366A and Vip3Aa-K432A/T433A/L434A mutants with significant differences (***, p < 0.005) analyzed by t test. Error bars represent S.D. C, secondary structure of Vip3Aa predicted by different programs (Phyre2 (53), Robetta, QUARK (54, 55), and Modeller (56, 57)). The black arrows indicate the mutagenized sites of Vip3Aa, which are probably located in loop regions. M, molecular mass markers; Abs, absorbance.
Analysis of the synergistic activity of Cry9Aa and Vip3Aa mutants against C. suppressalis
The effects of the different Cry9Aa and Vip3Aa mutations that were affected in their binding interaction were evaluated in bioassays against C. suppressalis larvae. Table 1 shows that the observed 50% lethal concentration (LC50) value of the 1:1 mixture of mutant Cry9Aa-R495A/R496A/S497A with Vip3Aa is 13-fold lower than the LC50 value of a 1:1 mixture of both Cry9Aa and Vip3Aa WT toxins; these LC50 values are statistically different because their 95% confidence limits did not overlap. However, it is important to notice that the individual LC50 value of Cry9Aa-R495A/R496A/S497A is 5-fold higher than that of Cry9Aa WT, although their 95% confidence limits overlap. The synergistic activity with Vip3Aa proteins was further affected because the synergy factor was decreased from 10.6 to 4.3 after considering the effect of single activity of Cry9Aa-R495A/R496A/S497A according to Tabashnik's equation as described previously (39). All these data indicate that Cry9Aa domain II loop-3 region Arg495–Ser497 is a key binding region for the synergistic activity with Vip3Aa against C. suppressalis.
Analysis of the insecticidal activity of Vip3Aa-N364A/D365A/S366A mutant showed a 4-fold lower toxicity than Vip3Aa WT, although their 95% confidence limits overlap. However, the analysis of the synergism with Cry9Aa indicated that Vip3Aa-N364A/D365A/S366A was not affected in its synergistic activity with Cry9Aa because the SF value is slightly higher than that of the 1:1 mixture of Cry9Aa and Vip3Aa WT toxins. In contrast, analysis of Vip3Aa-K432A/T433A/L434A mutant showed that this mutant is not affected in its insecticidal activity, but it did show a drastic effect in its synergism with Cry9Aa (8.2-fold lower), indicating that the Vip3Aa Lys432–Leu434 region is implicated in binding and synergism with Cry9Aa.
Discussion
B. thuringiensis Cry and Vip proteins are highly effective in controlling agricultural pests and could be used in pyramided transgenic plants because they have different mechanisms of action (22). Synergistic or antagonistic effects between Vip3Aa and other Cry toxins have been reported previously, such as the synergistic effect of Vip3A with Cry1Ia in some Spodoptera insects, but they resulted in an antagonistic effect in Spodoptera eridania, and it was reported that antagonism correlated with competition for similar binding sites between Cry1Ia and Vip3Aa proteins in S. eridania BBMVs (36). Also, synergism or antagonism between Vip3Aa and Cry1C in different lepidopteran insects has been reported (37). However, the molecular mechanism involved in Vip–Cry synergistic interaction has never been studied at the molecular level. Here, we show that Vip3Aa and Cry9Aa can induce a synergistic response in C. suppressalis and O. furnacalis. In addition, the Cry9A-R495A/R496A/S497A mutant that is affected in synergism with Vip3Aa in C. suppressalis also showed a severe effect in its synergism with Vip3Aa in O. furnacalis (Table S1). The Cry9Aa and Vip3Aa proteins do not share binding sites in BBMVs from C. suppressalis, suggesting that they bind to different receptors. The interaction of Cry9Aa and Vip3Aa with BBMVs from C. suppressalis was saturable and specific (Fig. 1, C and D), and the fact that they do not share binding sites (Fig. 1, E and F) indicates that these two toxins can be good candidates for generating pyramided transgenic rice for the control of C. suppressalis. As mentioned above, it was shown that Vip3Aa and Cry1Ia have an antagonism activity when the two toxins share binding sites in BBMVs (36). In the case of Vip3Aa and Cry9Aa, these toxins do not share receptors, correlating with the lack of antagonistic activity in the insects analyzed.
More importantly, we show that these proteins synergize with each other, resulting in an improved toxicity when both toxins act together against C. suppressalis. In this work, we demonstrated for the first time that Cry9Aa and Vip3Aa toxins interact with each other in a highly specific interaction, and we were able to identify the regions of each toxin that participate in this interaction. Moreover, we showed that the interaction of these two toxins directly correlated with their synergistic activity because mutants affected in the binding interaction are also affected in synergism. The binding regions of Cry9Aa that participate in the interaction with Vip3Aa were narrowed to domains II and III by expressing the isolated Cry9Aa domains in E. coli (Fig. 3), and site-directed mutagenesis revealed that amino acid residues Arg495–Ser497 in loop-3 of domain II are involved in the interaction with Vip3Aa (Fig. 5). Domain II loop-3 and domain III β-16 have been described previously to participate in the interaction of different Cry toxins with their receptors in different insect species (40, 41). Loop-3 is an important binding region of Cry toxins involved in the binding interaction with cadherin and aminopeptidase N (APN) receptors, whereas strand β-16 of domain III participates in interaction with alkaline phosphatase (40–42). The potential role of Cry9Aa domain III amino acid regions identified in toxicity and synergism remains to be analyzed by site-directed mutagenesis.
The binding regions of Vip3Aa with Cry9Aa were also identified in this work; they are located in Vip3Aa Asn364–Ser366 and Lys432–Leu434 (Fig. 6B). The secondary structure of Vip3Aa was predicted using four different strategies (Fig. 6C), which suggested that residues Asn364–Ser366 and Lys432–Leu434 from Vip3Aa are most probably localized in exposed loop regions (Fig. 6C). A predicted model of the three-dimensional structure of Vip3Aa confirmed that these two regions are likely exposed to the solvent (Fig. S2).
Site-directed mutagenesis of the Cry9Aa- and Vip3Aa-binding regions allowed us to demonstrate that a binding interaction between these toxins is crucial for their synergism because mutants that affected their binding interaction, such as Cry9Aa-R495A/R496A/S497A and Vip3Aa-K432A/T433A/L434A, significantly affected their synergism, although both retained toxicity to C. suppressalis. The loss of synergism is especially important in the mutant Vip3Aa-K432A/T433A/L434A, which was not affected in insecticidal activity but was affected in Cry9Aa binding and completely lost the synergism with this toxin. These data strongly support the hypothesis that interaction between these toxins is a requirement for their synergism.
Both Cry9Aa and Vip3Aa toxins are pore-forming toxins that bind to specific receptors located in the BBMVs of C. suppressalis; oligomerization and pore insertion must follow these interactions. We show here that the interaction of Cry9Aa and Vip3Aa is a primary and fundamental step for their synergistic activity. Mapping the specific binding sites involved in the interaction between these toxins gives insights on possible targets for the future modification of these binding sites to improve the toxicity and synergism of other Vip3–Cry toxins.
In the case of the synergistic activity of Cry11A and Cyt1A against the dipteran Aedes aegypti larvae, it was shown that these two toxins bind each other with high affinity by specific regions, and mutations in such regions also resulted in a severe defect in their synergism (43). Interestingly, the Cry11Aa domain II loop region was involved in binding to Cyt1Aa and to the mosquito BBMVs (43). It was proposed that Cyt1Aa synergizes Cry11Aa toxin by functioning as a receptor because it was able to facilitate the oligomerization of Cry11Aa toxin (43, 44). It was notable that the regions of Cry11Aa that were described to be highly important for interaction with its alkaline phosphatase receptor also participate in its interaction with Cyt1Aa (45). The Vip toxins have no structural or sequence relation with Cyt1A toxin; thus, interaction sites of Vip proteins that participate in binding to 3d-Cry toxins were not known until now. In the case of the synergism between Cry9Aa and Vip3Aa, we also found that the Cry9Aa regions that interact with Vip3Aa are exposed loops that were previously identified as important regions for interaction with toxin receptors of other Cry toxins. Cry9Aa domain II exposed loops are likely to be involved in receptor binding, explaining why the Cry9Aa-R495A/R496A/S497A mutations located in loop 3 affected toxicity. The fact that Cry9Aa domain II exposed loop-3 is a region widely recognized as a receptor binding site also involved in binding to Vip3Aa suggests that Vip3Aa may be functioning as a receptor for Cry9Aa, explaining its synergism. The role of this interaction in further steps of the mechanism of action of these toxins remains to be studied. It could be hypothesized that the mechanism of synergism involves oligomerization of Cry9Aa toxin induced by Vip3Aa or that these proteins together could form heteroligomers; an additional hypothesis is that other effects due to their distinct mode of action could contribute to their synergism. It is also possible that apoptosis activation induced by Vip3A could participate in the synergistic effect with Cry9Aa. The molecular mechanism underlying the synergism between Vip3Aa and Cry9Aa toxins remains to be determined.
Our findings highlight that Cry9Aa and Vip3Aa toxins could be used for insect control, especially for construction of novel pyramided rice plants that would help to increase insecticidal activity and delay the resistance in C. suppressalis against insecticidal toxins. Our results help to clarify the binding sites between them, and the fact that this specific binding interaction is involved in synergism indicates that this knowledge could be useful for future rational design of toxins that would improve their toxicity and provide tools for resistance management against other insect pests.
Experimental procedures
Strains and materials
The E. coli BL21 (DE3) strain harboring the recombinant plasmid pEB-cry9Aa655, containing a truncated cry9Aa3 gene (GenBankTM accession number ADE60735) encoding the first 655 amino acids from the N terminus (46) and E. coli BL21 (DE3) harboring the recombinant plasmid pET28a-vip3Aa11, containing full-length vip3Aa11 gene (GenBank accession number AAR36859) were used for Cry9Aa and Vip3Aa production.
Cloning of the different Cry9Aa and Vip3Aa fragments
Cry9Aa or Vip3Aa fragments were amplified by PCR using pEB-cry9Aa655 or pET28a-vip3Aa11 as templates with primers containing a BamHI or XhoI restriction site in the 5′-end of the forward or reverse primers, respectively (Table S2). The three separate domains of Cry9Aa toxin are domain I (Met1–Trp321), domain II (Pro292–Arg511), and domain III (Asn512–Val655). The 10 overlapping fragments of Vip3Aa are F1 (Met1–Gly200), F2 (Asp102–Gly245), F3 (Asp151–Asp300), F4 (Arg246–Leu399), F5 (Ile301–Asp451), F6 (Lys352–Arg500), F7 (Leu400–Glu550), F8 (Leu502–Ser651), F9 (Lys602–Ala749), and F10 (Gln652–Lys789). The PCRs were performed with Phusion High-Fidelity PCR Master Mix (New England Biolabs, Beverly, MA). The PCR products were purified with a QIAquick PCR Purification kit (Qiagen, Valencia, CA), digested with BamHI and XhoI restriction enzymes (New England Biolabs), and ligated into the previously digested vector pET28b (Novagen, EMD Biosciences Inc.).
Site-directed mutagenesis of Cry9Aa and Vip3Aa toxins
Mutagenesis of Cry9Aa and Vip3Aa was performed using a QuikChange XL kit (Stratagene, La Jolla, CA). Appropriate oligonucleotides were synthesized (Table S3) for each mutant construction. Mutants were sequenced and transformed into E. coli BL21 (DE3) strain.
Purification of Cry9Aa and Vip3Aa toxins and fragments
E. coli strains containing cry9Aa and vip3Aa genes, their fragments, or their mutants were grown in 300 ml of LB medium, and the expression of the proteins was induced with 0.5 mm isopropyl β-d-thiogalactopyranoside at 18 °C overnight. Cells were collected by centrifugation at 10,000 × g for 8 min, and the cell pellet was suspended in 30 ml of binding buffer (20 mm Tris-HCl with 0.5 m NaCl and 50 mm imidazole, pH 8.0). Cells were lysed by sonication (Ningbo Scientz Biotechnology Co., Ltd., China) for 5 min (70% power, 3-s pulse on, 5-s pulse off) and centrifuged at 13,000 × g for 15 min at 4 °C.
Proteins were purified by nickel-affinity chromatography (GE Healthcare) pre-equilibrated with binding buffer. Proteins bound to the column were eluted with elution buffer (20 mm Tris-HCl, 500 mm NaCl, 250 mm imidazole, pH 8.0). Fig. S1A shows the soluble Vip3Aa protoxin and Cry9Aa toxin fragment.
Cry9Aa and Vip3Aa proteins were activated with trypsin (Amresco, Solon, OH) in a 20:1 mass ratio (protoxin:trypsin), and activated toxins were recovered in 20 mm Tris-HCl buffer, pH 8.0, with Sephadex G50 (GE Healthcare). The fractions included in 280-nm absorbance peaks were collected. Fig. S1 shows the trypsin-activated Vip3Aa and Cry9Aa WT toxins. Purified Cry9Aa and Vip3Aa mutants were also activated with trypsin as described above (Fig. S1).
Insect bioassays
The insecticidal activity of the Cry9Aa and Vip3Aa proteins was tested against C. suppressalis or O. furnacalis neonates. C. suppressalis larvae were obtained from Beijing Genralpest Biotech Research Co., Ltd. O. furnacalis was obtained from the Corn Pest Group from the Institute of Plant Protection, Chinese Agricultural Academy of Sciences. Bioassays were performed with different toxin concentrations applied to the diet using 25 insects per toxin dose, and the percentage of mortality was recorded after 7 days at 28 °C with a 16-h light and 8-h dark cycle and 60–80% relative humidity. The LC50 was calculated by Probit analysis (47). The expected toxicity of each combination of toxins (ratio of 1:1) was evaluated according to Tabashnik's equation (37). The expected LC50 value is the harmonic mean of the intrinsic LC50 values of each component weighted by the ratio used in the mixture as follows,
| (Eq. 1) |
where rCry9Aa and rVip3Aa are the Cry9Aa and Vip3Aa protein proportions used in the final ratio of the mixture. LC50 Cry9Aa and LC50 Vip3Aa are the LC50 values for each individual toxin. The SF was calculated with the following equation: SF = LC50exp(Cry9Aa + Vip3Aa)/LC50obs(Cry9Aa + Vip3Aa) where LC50exp is the expected LC50 calculated above and LC50obs is the observed LC50. SF values >1 indicate synergism.
Preparation of BBMVs
Third instar larvae of C. suppressalis were obtained from Beijing Genralpest Biotech Research Co., Ltd. The midgut tissue was dissected and stored immediately at −70 °C. BBMVs were prepared by the magnesium precipitation method (48) and stored at −70 °C until used. The purity of BBMV preparations was determined by estimating the enrichment of APN-specific activity in the BBMVs compared with the initial midgut tissue homogenate (49). Table S4 shows the APN activity enrichment in the BBMV preparation, which was 4.25-fold higher compared with the initial midgut homogenate, indicating that BBMVs were correctly prepared.
Toxin labeling
Purified Cry9Aa or Vip3Aa activated toxins and protoxins were labeled with biotin by incubating with maleimide-PEG2-biotin (Thermo Scientific, Rockford, IL) or (+)-biotin N-hydroxysuccinimide ester (Sigma-Aldrich) in phosphate-buffered saline (PBS) buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.4) at room temperature for 2 h. Free biotin was removed by using a desalting column (GE Healthcare) with 20 mm Tris-HCl, pH 8.0, with 1 mm DTT. Purified biotinylated Cry9Aa and Vip3Aa proteins were quantified by the method of Bradford (50) (Bio-Rad). Fig. S1C shows the biotinylated toxins detected with streptavidin–Alexa Fluor 488 conjugate (Thermo Scientific) as explained below.
Saturation binding assays of Cry9Aa and Vip3Aa toxins to BBMVs
Twenty or 10 μg of BBMVs from C. suppressalis were incubated with increasing concentrations of biotinylated Cry9Aa or Vip3Aa (10–160 or 10–320 nm), respectively, in a final volume of 100 μl of binding buffer (PBS with 0.1% Tween 20 and 0.1% BSA, pH 7.4) for 1 h at room temperature. Binding reactions were stopped by centrifugation for 10 min at 18,000 × g after which the BBMV pellet containing the bound toxin was washed with 0.1 ml of ice-cold binding buffer twice. A control of the highest concentration of biotinylated toxins (160 nm biotinylated Cry9Aa or 320 nm biotinylated Vip3Aa) without BBMVs was also included to monitor toxin precipitation. Pellets were solubilized in 10 μl of sample buffer (51), heat-denatured for 10 min at 100 °C, and loaded for SDS-PAGE. Proteins were electrotransferred to Hybond low-fluorescence polyvinylidene difluoride (PVDF) blotting filters (Merck Millipore Ltd., Tullagreen, Ireland). Filters were blocked for 40 min at room temperature in PBS containing 2% Tween 20 and probed with streptavidin–Alexa Fluor 488 conjugate at 1:10,000 dilution in PBST (PBS with 0.1% Tween 20) for 1 h at room temperature. Filters were washed with PBST twice and PBS twice (10 min per wash) and air-dried on filter paper. The dried filters were scanned for blue fluorescence (488 nm) in a Typhoon Trio scanner (GE Healthcare).
Homologous and heterologous competition binding assays of Cry9Aa and Vip3Aa toxins to BBMVs
In homologous competition binding assay, C. suppressalis BBMVs were incubated with 40 nm biotinylated Cry9Aa or Vip3Aa in the presence of 120–12,000 nm unlabeled corresponding toxin for 1 h at room temperature. The following steps were continued as described above for saturation binding assay.
For heterologous competition binding assays, BBMVs were incubated with 40 nm biotinylated Cry9Aa or Vip3Aa in the presence of a 1000–20,000 nm concentration of the other unlabeled toxin for 1 h at room temperature. The following steps were done as described above for saturation binding assay.
Ligand-blot assays
The Cry9Aa or Vip3Aa proteins or their fragments (0.5 μg) were separated by SDS-PAGE and electrotransferred to PVDF membranes. After blocking with PBS containing 2% Tween 20 for 40 min, the membranes were incubated for 1 h with 5 nm biotinylated Vip3Aa or Cry9Aa toxins, and the bound protein was revealed with streptavidin-HRP conjugate (Thermo Scientific) at 1:50,000 dilution in PBST and detected with SuperSignal chemiluminescence substrate (Thermo Scientific) in an Amersham Biosciences Imager 600 (GE Healthcare).
ELISA
96-well ELISA plates were incubated 12 h at 4 °C with 1 μg of Vip3Aa in PBS, pH 7.4, followed by five washes with PBST. The plates were incubated with PBST with 2% BSA for 2 h at 37 °C. The ELISA plates were subsequently incubated with different concentrations of biotinylated Cry9Aa (0, 7, 14, 28, 56, 112, and 224 nm) for 1 h at 37 °C, and after washing with PBST five times, the biotinylated proteins were detected with streptavidin-HRP (1:25,000 dilution) for 1 h at 37 °C. The HRP enzymatic activity was revealed with 40 mg of o-phenylenediamine and 18 ml of H2O2 in 100 ml of 100 mm NaH2PO4, pH 5.0. The enzymatic reaction was stopped with 1 m HCl, and the absorbance was read at 490 nm with an LKB Ultraspec II spectrophotometer (Amersham Biosciences). Binding data were analyzed and plotted with SigmaPlot v.13.0 software (Systat Software, San Jose, CA). Scatchard plot analysis of the data from the saturation binding curve was used to determine the apparent dissociation constants (Kd) of Cry9Aa and Vip3Aa. For binding competition assays, 0.5 μg of Cry9Aa was immobilized on the plate, and 28 nm biotinylated Vip3Aa was incubated in the presence of different concentrations (0, 1750, 3500, 7000, and 14,000 nm) of unlabeled Vip3Aa.
For binding analysis between Cry9Aa and Vip3Aa and their mutants, 0.5 μg of Cry9Aa or 1 μg of Vip3Aa was fixed on the plate, and after blocking (2% no protein blocking solution (Sangon Biotech, Shanghai, China) for Cry9Aa and 2% BSA for Vip3Aa) they were incubated with the different Vip3Aa mutants at 28 nm or the different Cry9Aa mutants at 56 nm for 1 h at 37 °C. After washing with PBST five times, the plates were incubated with anti-Vip3Aa or anti-Cry9Aa antibody (the antibody preparation is described in the supporting methods) (1:10,000 dilution) at 37 °C for 1 h, washed again, and incubated with the secondary goat anti-rabbit-HRP antibody (Santa Cruz Biotechnology Inc., Dallas, TX) (1:10,000 dilution) at 37 °C for 1 h. The detection was done as described above.
Biosensor (surface plasmon resonance (SPR)) analysis of affinity between Cry9Aa and Vip3Aa
A SensiQ instrument (Oklahoma City, OK) was used for performing SPR measurements. Running buffer (HEPES-buffered saline, pH 7.4, containing 10 mm HEPES, 150 mm NaCl, and 0.005% (v/v) Tween 20) was freshly prepared, filtered (pore size, 0.22 mm), and degassed. Vip3Aa toxin was immobilized onto a COOH-functionalized sensor chip (ICx Nomadics) by conventional amine coupling at densities less than 1500 response units. 1 m ethanolamine at a flow rate of 10 ml/min for 5 min was injected to block flow cells. The analyte, Cry9Aa, was injected at a flow rate of 25 ml/min. Serial doubling dilutions of Cry9Aa were analyzed, and the surface was regenerated with a 1-min injection of 20 mm NaOH. Injections were performed three times for each toxin concentration. The data were analyzed using the SensiQ software Qdat version B.02. This software uses nonlinear regression and the Levenberg–Marquadt algorithm to fit experimental data to a binding interaction model that defines the interaction.
Western blot analysis of the expression of Cry9Aa and Vip3Aa fragments and corresponding mutants
The Cry9Aa fragments, Vip3Aa fragments, or their mutants (0.5 μg) were loaded for SDS-PAGE and electrotransferred to PVDF filters. After blocking for 40 min at room temperature in PBS containing 2% Tween 20, filters were probed with 1:20,000 Penta-His-HRP (Qiagen), anti-Cry9Aa, or anti-Vip3Aa antibody at a 1:50,000 dilution in PBST for 1 h at room temperature followed by 1-h incubation with the HRP-labeled goat anti-rabbit secondary antibody at 1:20,000 dilution at room temperature in PBST. Membranes were washed with PBST three times (10 min per wash) and developed with SuperSignal chemiluminescence substrate.
Cellulose membrane–bound peptides
Eighty-four cellulose-bound peptides corresponding to the amino acid sequence of domains II and III of Cry9Aa and 23 cellulose-bound peptides corresponding to the amino acid sequence of Vip3Aa Lys352–Asp451 fragment were prepared by automated spot synthesis and bound to nitrocellulose membrane (JPT Peptide Technologies, Berlin, Germany) (52). Each peptide contained 15 amino acids and overlapped 11 residues with the following peptide. These membranes containing the Cry9Aa- or Vip3Aa-bound peptides were washed with methanol and then with PBS before blocking for 40 min with PBS containing 2% Tween 20. Each membrane was incubated with 5 nm biotinylated Vip3Aa or Cry9Aa in PBST for 1 h, after three wash steps detected with streptavidin-HRP conjugate (1:50,000 dilution) at room temperature, and finally visualized with SuperSignal chemiluminescence substrate.
Prediction of secondary structure of Vip3Aa and Cry9Aa model construction
The secondary structure of Vip3Aa was predicted using different programs (Phyre2 (53), Robetta, QUARK (54, 55), and Modeller (56, 57)). A model of the tridimensional structure of Cry9Aa toxin was constructed using SWISS-MODEL (29) (https://swissmodel.expasy.org),4 performing structure alignments using the crystallographic structure of Cry8Ea (Protein Data Bank code 3EB7) as template. The model of the Vip3Aa toxin was constructed using QUARK protein structure prediction because this is suitable for proteins considered without homologous template (54, 55).
Author contributions
Z. W. and A. B. data curation; Z. W., Z. Z., and A. B. software; Z. W. formal analysis; Z. W. and L. F. validation; Z. W., L. F., and I. G. investigation; Z. W. and A. B. visualization; Z. W., L. F., Z. Z., S. P., and I. G. methodology; Z. W. writing-original draft; Z. Z., S. P., I. G., M. S., J. Z., and A. B. conceptualization; S. P., I. G., F. S., M. S., J. Z., and A. B. supervision; F. S. and J. Z. resources; F. S., M. S., J. Z., and A. B. funding acquisition; F. S., M. S., J. Z., and A. B. project administration; M. S., J. Z., and A. B. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Kanglai He for supplying O. furnacalis neonates and Changhui Li, Yuxiao Liu, and Jorge Sanchez for technical assistance.
This work was supported by National Key Research and Development Program of China Grant 2017YFD0200400 and the Consejo Nacional de Ciencia y Tecnología (Fronteras de la Ciencia 008). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2, Tables S1–S4, and supporting methods.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- Cry
- crystal proteins
- Vip
- vegetative insecticidal protein
- 3d-Cry
- three-domain Cry
- BBMV
- brush border membrane vesicle
- SF
- synergism factor
- SPR
- surface plasmon resonance
- HRP
- horseradish peroxidase
- JAK
- Janus kinase
- STAT
- signal transducers and activators of transcription
- APN
- aminopeptidase N
- LC50
- 50% lethal concentration.
References
- 1. Carrière Y., Crickmore N., and Tabashnik B. E. (2015) Optimizing pyramided transgenic Bt crops for sustainable pest management. Nat. Biotechnol. 33, 161–168 10.1038/nbt.3099 [DOI] [PubMed] [Google Scholar]
- 2. Tabashnik B. E. (2015) ABCs of insect resistance to Bt. PLoS Genet. 11, e1005646 10.1371/journal.pgen.1005646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carrière Y., Fabrick J. A., and Tabashnik B. E. (2016) Can pyramids and seed mixtures delay resistance to Bt crops? Trends Biotechnol. 34, 291–302 10.1016/j.tibtech.2015.12.011 [DOI] [PubMed] [Google Scholar]
- 4. Crickmore N., Baum J., Bravo A., Lereclus D., Narva K., Sampson K., Schnepf E., Sun M., and Zeigler D. R. (2018) Bacillus thuringiensis toxin nomenclature. http://www.btnomenclature.info/ (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.)
- 5. Kuvshinov V., Koivu K., Kanerva A., and Pehu E. (2001) Transgenic crop plants expressing synthetic cry9Aa gene are protected against insect damage. Plant Sci. 160, 341–353 10.1016/S0168-9452(00)00398-8 [DOI] [PubMed] [Google Scholar]
- 6. Marchetti E., Alberghini S. B., Andrea Squartini A., Baronio P., and Dindo M. L. (2009) Effects of conventional and transgenic Bacillus thuringiensis galleriae toxin on Exorista larvarum (Diptera: Tachinidae), a parasitoid of forest defoliating Lepidoptera. Biocontrol Sci. Technol. 19, 463–473 10.1080/09583150902807535 [DOI] [Google Scholar]
- 7. Naimov S., Nedyalkova R., Staykov N., Weemen-Hendriks M., Minkov I., and de Maagd R. A. (2014) A novel Cry9Aa with increased toxicity for Spodoptera exigua (Hübner). J. Invertebr. Pathol. 115, 99–101 10.1016/j.jip.2013.11.003 [DOI] [PubMed] [Google Scholar]
- 8. Bravo A., Likitvivatanavong S., Gill S. S., and Soberón M. (2011) Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 41, 423–431 10.1016/j.ibmb.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lambert B., Buysse L., Decock C., Jansens S., Piens C., Saey B., Seurinck J., Van Audenhove K., Van Rie J., Van Vliet A., and Peferoen M. (1996) A Bacillus thuringiensis insecticidal crystal protein with a high activity against members of the family Noctuidae. Appl. Environ. Microbiol. 62, 80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smulevitch S. V., Osterman A. L., Shevelev A. B., Kaluger S. V., Karasin A. I., Kadyrov R. M., Zagnitko O. P., Chestukhina G. G., and Stepanov V. M. (1991) Nucleotide sequence of a novel δ-endotoxin gene cryIg of Bacillus thuringiensis ssp. galleriae. FEBS Lett. 293, 25–28 10.1016/0014-5793(91)81144-W [DOI] [PubMed] [Google Scholar]
- 11. Van Frankenhuyzen K., Gringorten L., and Gauthier D. (1997) Cry9Ca1 toxin, a Bacillus thuringiensis insecticidal crystal protein with high activity against the Spruce Budworm (Choristoneura fumiferana). Appl. Environ. Microbiol. 63, 4132–4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jansens S., vanVliet A., Dickburt C., Buysse L., Piens C., Saey B., DeWulf A., Gossele V., Paez A., Gobel E., and Peferoen M. (1997) Transgenic corn expressing a Cry9C insecticidal protein from Bacillus thuringiensis protected from European corn borer damage. Crop Sci. 37, 1616–1624 10.2135/cropsci1997.0011183X003700050035x [DOI] [Google Scholar]
- 13. Hua G., Masson L., Jurat-Fuentes J. L., Schwab G., and Adang M. J. (2001) Binding analyses of Bacillus thuringiensis Cry δ-endotoxins using brush border membrane vesicles of Ostrinia nubilalis. Appl. Environ. Microbiol. 67, 872–879 10.1128/AEM.67.2.872-879.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ruiz de Escudero I., Estela A., Escriche B., and Caballero P. (2007) Potential of the Bacillus thuringiensis toxin reservoir for the control of Lobesia botrana (Lepidoptera: Tortricidae), a major pest of grape plants. Appl. Environ. Microbiol. 73, 337–340 10.1128/AEM.01511-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ben Hamadou-Charfi D., Boukedi H., Abdelkefi-Mesrati L., Tounsi S., and Jaoua S. (2013) Agrotis segetum midgut putative receptor of Bacillus thuringiensis vegetative insecticidal protein Vip3Aa16 differs from that of Cry1Ac toxin. J. Invertebr. Pathol. 114, 139–143 10.1016/j.jip.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 16. Chakroun M., and Ferré J. (2014) In vivo and in vitro binding of Vip3Aa to Spodoptera frugiperda midgut and characterization of binding sites by 125I radiolabeling. Appl. Environ. Microbiol. 80, 6258–6265 10.1128/AEM.01521-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee M. K., Miles P., and Chen J. S. (2006) Brush border membrane binding properties of Bacillus thuringiensis Vip3A toxin to Heliothis virescens and Helicoverpa zea midguts. Biochem. Biophys. Res. Commun. 339, 1043–1047 10.1016/j.bbrc.2005.11.112 [DOI] [PubMed] [Google Scholar]
- 18. Liu J. G., Yang A. Z., Shen X. H., Hua B. G., and Shi G. L. (2011) Specific binding of activated Vip3Aa10 to Helicoverpa armigera brush border membrane vesicles results in pore formation. J. Invertebr. Pathol. 108, 92–97 10.1016/j.jip.2011.07.007 [DOI] [PubMed] [Google Scholar]
- 19. Sena J. A., Hernández-Rodríguez C. S., and Ferré J. (2009) Interaction of Bacillus thuringiensis Cry1 and Vip3A proteins with Spodoptera frugiperda midgut binding sites. Appl. Environ. Microbiol. 75, 2236–2237 10.1128/AEM.02342-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gouffon C., Van Vliet A., Van Rie J., Jansens S., and Jurat-Fuentes J. L. (2011) Binding sites for Bacillus thuringiensis Cry2Ae toxin on heliothine brush border membrane vesicles are not shared with Cry1A, Cry1F, or Vip3A toxin. Appl. Environ. Microbiol. 77, 3182–3188 10.1128/AEM.02791-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gayen S., Hossain M. A., and Sen S. K. (2012) Identification of the bioactive core component of the insecticidal Vip3A toxin peptide of Bacillus thuringiensis. J. Plant Biochem. Biotechnol. 21, Suppl. 1, 128–135 10.1007/s13562-012-0148-8 [DOI] [Google Scholar]
- 22. Lee M. K., Walters F. S., Hart H., Palekar N., and Chen J. S. (2003) The mode of action of the Bacillus thuringiensis vegetative insecticidal protein Vip3A differs from that of Cry1Ab-endotoxin. Appl. Environ. Microbiol. 69, 4648–4657 10.1128/AEM.69.8.4648-4657.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Song R., Peng D., Yu Z., and Sun M. (2008) Carboxy-terminal half of Cry1C can help vegetative insecticidal protein to form inclusion bodies in the mother cell of Bacillus thuringiensis. Appl. Microbiol. Biotechnol. 80, 647–654 10.1007/s00253-008-1613-0 [DOI] [PubMed] [Google Scholar]
- 24. Chakroun M., Banyuls N., Bel Y., Escriche B., and Ferré J. (2016) Bacterial vegetative insecticidal proteins (Vip) from entomopathogenic bacteria. Microbiol. Mol. Biol. Rev. 80, 329–350 10.1128/MMBR.00060-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bel Y., Banyuls N., Chakroun M., Escriche B., and Ferré J. (2017) Insights into the structure of the Vip3Aa insecticidal protein by protease digestion analysis. Toxins 9, E131 10.3390/toxins9040131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zack M. D., Sopko M. S., Frey M. L., Wang X., Tan S. Y., Arruda J. M., Letherer T. T., and Narva K. E. (2017) Functional characterization of Vip3Ab1 and Vip3Bc1: two novel insecticidal proteins with differential activity against lepidopteran pests. Sci. Rep. 7, 11112 10.1038/s41598-017-11702-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Palma L., Scott D. J., Harris G., Din S. U., Williams T. L., Roberts O. J., Young M. T., Caballero P., and Berry C. (2017) The Vip3Ag4 insecticidal protoxin from Bacillus thuringiensis adopts a tetrameric configuration that is maintained on proteolysis. Toxins 9, E165 10.3390/toxins9050165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kunthic T., Surya W., Promdonkoy B., Torres J., and Boonserm P. (2017) Conditions for homogeneous preparation of stable monomeric and oligomeric forms of activated Vip3A toxin from Bacillus thuringiensis. Eur. Biophys. J. 46, 257–264 10.1007/s00249-016-1162-x [DOI] [PubMed] [Google Scholar]
- 29. Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42, W252–W258 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang K., Mei S. Q., Wang T. T., Pan J. H., Chen Y. H., and Cai J. (2016) Vip3Aa induces apoptosis in cultured Spodoptera frugiperda (Sf9) cells. Toxicon 120, 49–56 10.1016/j.toxicon.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 31. Hernández-Martínez P., Gomis-Cebolla J., Ferré J., and Escriche B. (2017) Changes in gene expression and apoptotic response in Spodoptera exigua larvae exposed to sublethal concentrations of Vip3 insecticidal proteins. Sci. Rep. 7, 16245 10.1038/s41598-017-16406-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang H., Patel P. H., Kohlmaier A., Grenley M. O., McEwen D. G., and Edgar B. A. (2009) Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355 10.1016/j.cell.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu M. X., Cao S. S., Du Y. Z., Liu Z. X., Liu P., and Li J. (2013) Diapause, signal and molecular characteristics of overwintering Chilo suppressalis (Insecta: Lepidoptera: Pyralidae). Sci. Rep. 3, 3211 10.1038/srep03211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiao Y., Yang Y., Meissle M., Peng Y., and Li Y. (2016) Comparison of susceptibility of Chilo suppressalis and Bombyx mori to five Bacillus thuringiensis proteins. J. Invertebr. Pathol. 136, 95–99 10.1016/j.jip.2016.03.010 [DOI] [PubMed] [Google Scholar]
- 35. Meng F., Wu K., Gao X., Peng Y., and Guo Y. (2003) Geographic variation in susceptibility of Chilo suppressalis (Lepidoptera: Pyralidae) to Bacillus thuringiensis toxins in China. J. Econ. Entomol. 96, 1838–1842 10.1603/0022-0493-96.6.1838 [DOI] [PubMed] [Google Scholar]
- 36. Guo S., Ye S., Liu Y., Wei L., Xue J., Wu H., Song F., Zhang J., Wu X., Huang D., and Rao Z. (2009) Crystal structure of Bacillus thuringiensis Cry8Ea1: an insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela. J. Struct. Biol. 168, 259–266 10.1016/j.jsb.2009.07.004 [DOI] [PubMed] [Google Scholar]
- 37. Tabashnik B. E. (1992) Evaluation of synergism among Bacillus thuringiensis toxins. Appl. Environ. Microbiol. 58, 3343–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bergamasco V. B., Mendes D. R., Fernandes O. A., Desidério J. A., and Lemos M. V. (2013) Bacillus thuringiensis Cry1Ia10 and Vip3Aa protein interactions and their toxicity in Spodoptera spp. (Lepidoptera). J. Invertebr. Pathol. 112, 152–158 10.1016/j.jip.2012.11.011 [DOI] [PubMed] [Google Scholar]
- 39. Lemes A. R., Davolos C. C., Legori P. C., Fernandes O. A., Ferré J., Lemos M. V., and Desiderio J. A. (2014) Synergism and antagonism between Bacillus thuringiensis Vip3A and Cry1 proteins in Heliothis virescens, Diatraea saccharalis and Spodoptera frugiperda. PLoS One 9, e107196 10.1371/journal.pone.0107196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pacheco S., Gómez I., Arenas I., Saab-Rincon G., Rodríguez-Almazán C., Gill S. S., Bravo A., and Soberón M. (2009) Domain II loop 3 of Bacillus thuringiensis Cry1Ab toxin is involved in a “ping pong” binding mechanism with Manduca sexta aminopeptidase-N and cadherin receptors. J. Biol. Chem. 284, 32750–32757 10.1074/jbc.M109.024968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arenas I., Bravo A., Soberón M., and Gómez I. (2010) Role of alkaline phosphatase from Manduca sexta in the mechanism of action of Bacillus thuringiensis Cry1Ab toxin. J. Biol. Chem. 285, 12497–12503 10.1074/jbc.M109.085266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gómez I., Arenas I., Benitez I., Miranda-Ríos J., Becerril B., Grande R., Almagro J. C., Bravo A., and Soberón M. (2006) Specific epitopes of domains II and III of Bacillus thuringiensis Cry1Ab toxin involved in the sequential interaction with cadherin and aminopeptidase-N receptors in Manduca sexta. J. Biol. Chem. 281, 34032–34039 10.1074/jbc.M604721200 [DOI] [PubMed] [Google Scholar]
- 43. Perez C., Fernandez L. E., Sun J., Folch J. L., Gill S. S., Soberón M., and Bravo A. (2005) Bacillus thuringiensis subsp. israelensis Cyt1Aa synergizes Cry11Aa toxin by functioning as a membrane-bound receptor. Proc. Natl. Acad. Sci. U.S.A. 102, 18303–18308 10.1073/pnas.0505494102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pérez C., Muñoz-Garay C., Portugal L. C., Sánchez J., Gill S. S., Soberón M., and Bravo A. (2007) Bacillus thuringiensis ssp. israelensis Cyt1Aa enhances activity of Cry11Aa toxin by facilitating the formation of a pre-pore oligomeric structure. Cell. Microbiol. 9, 2931–2937 10.1111/j.1462-5822.2007.01007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fernandez L. E., Pérez C., Segovia L., Rodríguez M. H., Gill S. S., Bravo A., and Soberón M. (2005) Cry11Aa toxin from Bacillus thuringiensis binds its receptor in Aedes aegypti mosquito larvae through loop α-8 of domain II. FEBS Lett. 579, 3508–3514 10.1016/j.febslet.2005.05.032 [DOI] [PubMed] [Google Scholar]
- 46. Fang L. F., Wang B., Zhou Z. S., Yang S. J., Shu C. L., Song F. P., Bravo A., Soberon M., and Zhang J. (2016) Oligomerization of Cry9Aa in solution without receptor binding, is not related with insecticidal activity. Electron. J. Biotechnol. 21, 54–57 10.1016/j.ejbt.2016.02.005 [DOI] [Google Scholar]
- 47. Finney D. J. (1971) Probit Analysis, 3rd Ed., p. 333, Cambridge University Press, Cambridge, UK [Google Scholar]
- 48. Wolfersberger M., Luethy P., Maurer A., Parenti P., Sacchi F. V., Giordana B., and Hanozet G. M. (1987) Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Physiol. A Physiol. 86, 301–308 10.1016/0300-9629(87)90334-3 [DOI] [Google Scholar]
- 49. Lorence A., Darszon A., and Bravo A. (1997) Aminopeptidase dependent pore formation of Bacillus thuringiensis Cry1Ac toxin on Trichoplusia ni membranes. FEBS Lett. 414, 303–307 10.1016/S0014-5793(97)01014-4 [DOI] [PubMed] [Google Scholar]
- 50. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 51. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 52. Kramer A., and Schneider-Mergener J. (1998) Synthesis and screening of peptide libraries on continuous cellulose membrane supports. Methods Mol. Biol. 87, 25–39 [DOI] [PubMed] [Google Scholar]
- 53. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu D., and Zhang Y. (2012) Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins 80, 1715–1735 10.1002/prot.24065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu D., and Zhang Y. (2013) Toward optimal fragment generations for ab initio protein structure assembly. Proteins 81, 229–239 10.1002/prot.24179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Webb B., and Sali A. (2016) Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 10.1002/cpps.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Martí-Renom M. A., Stuart A. C., Fiser A., Sánchez R., Melo F., and Sali A. (2000) Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 10.1146/annurev.biophys.29.1.291 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.