Abstract
While the peripheral nervous system has regenerative ability, restoration of sufficient function remains a challenge. Vimentin has been shown to be localized in axonal growth fronts and associated with nerve regeneration, including myelination, neuroplasticity, kinase signaling in nerve axoplasm, and cell migration; however, the mechanisms regulating its expression within Schwann cell (SC) remain unexplored. The aim of this study was to profile the spatial and temporal expression profile of microRNA (miRNA) in a regenerating rat sciatic nerve after transection, and explore the potential role of miR-138-5p targeting vimentin in SC proliferation and migration. A rat sciatic nerve transection model, utilizing a polyethylene nerve guide, was used to investigate miRNA expression at 7, 14, 30, 60, and 90 days during nerve regeneration. Relative levels of miRNA expression were determined using microarray analysis and subsequently validated with quantitative real-time polymerase chain reaction. In vitro assays were conducted with cultured Schwann cells transfected with miRNA mimics and assessed for migratory and proliferative potential. The top seven dysregulated miRNAs reported in this study have been implicated in cell migration elsewhere, and GO and KEGG analyses predicted activities essential to wound healing. Transfection of one of these, miRNA-138-5p, into SCs reduced cell migration and proliferation. miR-138-5p has been shown to directly target vimentin in cancer cells, and the luciferase assay performed here in rat Schwann cells confirmed it. These results detail a role of miR-138-5p in rat peripheral nerve regeneration and expand on reports of it as an important regulator in the peripheral nervous system.
Keywords: non-coding RNA, neural regeneration, nerve guide, sciatic nerve transection, peripheral nerve injury, wound healing, Gene Ontology processes, Kyoto Encyclopedia of Genes and Genomes pathways, microarray, luciferase assay
Introduction
Peripheral nerve injury is a major source of morbidity and is responsible for $150 billion in annual health care costs in the United States (Taylor et al., 2008; Thorsin et al., 2012). Although clinical management of these injuries, sometimes requiring nerve grafts or conduits, remains the current standard of care, clinical results are often suboptimal. Yet, there remain issues with donor site morbidity from nerve grafts. Furthermore, in clinical circumstances when surgical intervention is delayed and/or the length of reinervation is great, nerve regeneration is predictably poor and other more complex surgical techniques such as nerve or tendon transfer must be considered (Lee and Wolfe, 2000). Clearly, an ability to manipulate the microenvironment of a regenerating nerve holds the potential to improve functionality after nerve injury.
The general biological processes of peripheral nerve regeneration (PNR) following peripheral nerve injury have been known for some time (Waller, 1850; Brushart, 1993; Costigan et al., 2002; FexSvennigsen and Dahlin, 2013). In brief, following injury macrophages and Schwann cells (SCs) begin to clean debris from the injury site. In addition, the process of Wallerian degeneration begins whereby the axon degenerates distally from the site of injury. In turn, SCs proliferate and migrate from the distal site forming a framework to guide the regenerating nerve. This is followed by early regeneration events involving SC and axon growth from the nerve proximal to the injury site. Finally the regenerating nerve reinervates its target and SCs remyelinate the regenerated axon. This series of coordinated events has been well studied, yet an in depth understanding of the complex processes orchestrating the expression and interaction of multiple factors in the nerve microenvironment remains elusive (Terenghi, 1999; Abe and Cavelli, 2008).
Differential protein expression has been reported in nerve injury (Patodia and Raivich, 2012; Li et al., 2014; Richner et al., 2014) including the spatiotemporal expression profile of key proteins involved in nerve regeneration (Bryan et al., 2012). One such protein, vimentin, has been shown to be localized in axonal growth fronts (Gess et al., 2015). It is an intermediate filament expressed in many cell types, including those associated with nerve regeneration where it has been reported to be involved in myelination (Triolo et al., 2012), neuroplasticity (Qian et al., 2015), and kinase signaling in nerve axoplasm (Perlson et al., 2005). It has also been implicated in cell migration (Chang et al., 2012; Gan et al., 2016; Zhang et al., 2016), but the mechanisms regulating its expression within SCs remain unexplored.
Additionally, earlier studies have revealed > 6000 genes are differently expressed in nerve injury and regeneration (Yao et al., 2013; Akane et al., 2014; Jiang et al., 2014). Likewise, altered expression of microRNA (miRNA) has also been reported in sciatic nerve injury (Viader et al., 2011; Lu et al., 2014; Phay et al., 2015). MiRNAs are small non-coding RNAs that negatively regulate gene expression by targeting messenger RNA post-transcription. They have been implicated in a diverse array of biological processes including epithelial-mesenchymal transition (EMT) in oncogenesis and wound healing, as well as numerous disease pathways. In particular, miRNA have been implicated in several studies of nerve regeneration including axon guidance (Iyer et al., 2014), myelination (He et al., 2012), and SC migration (Yu et al., 2012). Specifically, miR-138-5p has been implicated in several studies of the nervous system including the regulation of vimentin in a model of spinal cord injury (Qian et al., 2015).
In this study, we utilize a rat sciatic nerve injury model to investigate miRNA expression alterations throughout 90 days of PNR. In addition, we investigate the potential role of miR-138-5p in SC migration and proliferation and the regulation of vimentin expression.
Materials and Methods
Surgical procedure
All animal experiments were performed with the approval of the Lahey Hospital & Medical Center Institutional Animal Care and Use Committee (A3377-01). Thirty male Sprague-Dawley rats (Charles River Laboratories, Wilmington, MA, USA), 9–11 weeks old, weighing between 225–250 g served as the model for nerve guide repair. All animals were anesthetized via an intraperitoneal injection of 45 mg/kg sodium pentobarbital. All rats underwent an initial surgery where the right sciatic nerve was exposed through a posterior thigh incision. The dissection was carried down through the gluteal muscle at which point the sciatic nerve was identified. The remainder of the dissection was performed under an operating microscope at 30× magnification. This same approach was carried out during a second harvesting procedure.
A length of at least 20 mm of nerve was exposed. A 10 mm section was removed via sharp transection with straight micro-scissors leaving at least a 5 mm proximal and distal sciatic nerve stump. A 14 mm long polyethylene nerve guide (1.67 mm ID, Becton Dickinson, Franklin Lakes, NJ, USA) was sutured in place with 10-0 nylon suture, using a horizontal mattress stitch. The suture was placed 2 mm from the end of the guide and through the epineurium on each respective nerve stump. The full circumference of each stump was therefore drawn entirely inside the guide with 2 mm of length lying inside each end of the guide, resulting in the proximal and distal stumps positioned 10 mm apart (Figure 1). The 10 mm resected section of nerve served as a per-animal control. The gluteal muscle was then closed with three buried interrupted 3-0 chromic sutures, and the skin was closed with a running 4-0 braided polyglycolic acid suture.
Figure 1.

Schematic of the resected sciatic nerve with placement of the nerve guide.
A 10 mm section of sciatic nerve was removed leaving at least a 5 mm proximal and distal nerve stump. A 14 mm long polyethylene nerve guide was sutured in place. The suture was placed 2 mm from the end of the guide and each stump was therefore drawn entirely inside the guide, resulting in the proximal and distal stumps positioned 10 mm apart.
At each time point (7, 14, 30, 60, and 90 days), six rats underwent a second procedure to harvest the proximal and distal nerve ends and the contents of the guide. All adhesions on the guide were carefully taken down with sharp dissection under the operating microscope (Zeiss, model # OPMI-6-SDFC, Oberkochen, Germany). Sharp transection of the proximal and distal stump was carried out, 5 mm from each end of the guide, with straight micro-scissors. The entire specimen, proximal and distal stumps within the guide, was taken out as one piece. The 10-0 nylon sutures were cut and the remaining tissue was separated into proximal and distal nerve stumps and solid guide contents. Proximal and distal segments included a total of 7 mm of nerve (2 mm of stump within the guide and 5 mm of additional nerve). In addition solid content from within the nerve guide was harvested. Solid guide content was defined as any matrix or tissue that could be macroscopically retrieved from the lumen of the nerve guide. The guide content segment was comprised of only regenerated material and not the nerve stumps themselves. Only tissue from animals with solid guide content was included in the analyses. All tissue was frozen immediately after harvesting, with liquid nitrogen.
Total RNA isolation
Total RNA was isolated from the proximal, distal, solid guide component, and original control sciatic samples for each of the rats with solid guide content at the time of harvest. This was performed using the Ambion mirVana miRNA Isolation Kit (Life Technologies, Foster City, CA, USA) following the manufacturers guidelines for frozen tissue. Tissue disruption was performed using a TissueLyser LT (Qiagen, Hilden, Germany). Total RNA was isolated from cultured rat SC lines using the Ambion mirVana miRNA Isolation Kit following the manufacturer’s guidelines for cultured mammalian cells. RNA quantity and purity was determined using the Epoch spectrophotometer (BioTek, Winooski, VT, USA) by measuring the optical density at 260 and 280 nm (OD260nm and OD280 nm).
Microarray miRNA profiling analysis
Total RNA pools were prepared from each of the four sets of samples (control, proximal, distal and solid guide contents) at 30, 60, and 90 days. The miRNAs were profiled by Exiqon (Vedbaek, Denmark) using the miRCURY™ LNA array, version 11.0 (Exiqon).The quality of the total RNA was verified by an Agilent 2100 Bioanalyzer profile. One µg total RNA from each sample and corresponding control was labeled with Hy3™ and Hy5™ fluorescent label, respectively, using the miRCURY™ LNA Array power labeling kit (Exiqon). The Hy3™-labeled samples and a Hy5™-labeled control RNA sample were mixed pair-wise and hybridized to the LNA array, which contains capture probes targeting 372 rat miRNA. The hybridization was performed according to the miRCURY™ LNA array manual using a Tecan HS4800 hybridization station (Tecan, Grödig, Austria). After hybridization the microarray slides were scanned and stored in an ozone free environment (ozone level below 2.0 ppb) in order to prevent potential bleaching of the fluorescent dyes. The miRCURY™ LNA array microarray slides were scanned using the Agilent G2565BA Microarray Scanner System (Agilent Technologies, Santa Clara, CA, USA) and the image analysis was carried out using the ImaGene 8.0 software (BioDiscovery, El Segundo, CA). The quantified signals were background corrected and normalized using the global LOWESS (LOcally WEighted Scatterplot Smoothing) polynomial regression algorithm.
GO and KEGG functional enrichment analyses
Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were performed using DIANA-mirPath v. 3.0 (Vlachos et al., 2015; University of Thessaly, Volos, Greece). Analyses were performed using the human equivalents of the miRNA with at least a ± five-fold change in expression based on the microarray analysis. Predicted gene targets with significant enrichment for these miRNA, based on TarBase v. 7.0 (University of Thessaly, Volos, Greece), were used. Heat maps depict the clusters with significant pathways or categories union analyses using Fisher’s Exact Test.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Evaluation of mature miRNA expression of the control, proximal, and distal samples at 7, 14, 30, 60, and 90 days was conducted via qRT-PCR using TaqMan miRNA quantification kits following the manufacturer’s protocol (ABI, Foster City, CA, USA). Relative expression levels of miR-138-5p (Cat. # 4427975, Assy ID # 002284, ABI) were normalized using an average of RNU6B (Cat. # 4427975, Assy ID # 001093, ABI) and RNU43 (Cat. # 4427975, Assy ID # 001095, ABI) and compared to corresponding control values using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Likewise, confirmation of transfection of cultured SCs with the miR-138-5p mimic was confirmed via qRT-PCR using TaqMan miRNA quantification kits as described above.
SCs culture
Immortalized rat SCs were established previously (Bryan et al., 2003) and maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, glutamine, hydrocortisone (0.02 µg/mL) insulin (0.25 µg/mL), transferrin (0.12 µg/mL), and glucose (67.5 µg/mL) at 37°C while humidified under 5% CO2.
Cell transfection of pre-miR constructs
SCs (1.3 × 105 cells per well) were transfected with microRNA mimics: 30 nM pre-miR-138-5p (Cat. # PM11727, ABI) or pre-miR-Precursor Negative Control #1 (Cat. # AM17110, ABI) using siPORT NeoFX transfection reagent (ABI) according to the manufacture’s protocol in a CELLSTAR six-well dish (Greiner, Kremsmünster, Austria). Cells were subsequently used in migration or proliferation assays at the time points specified. An additional well of cells was harvested and RNA was analyzed via qRT-PCR to verify overexpression compared to controls (data not shown).
Cell proliferation assay
Three wells of SCs were transfected with either miR-138-5p mimic or the negative control miR. At 24, 48, and 72 hours after cell transfection, the media was aspirated from a well, the cells were washed with 1× PBS, and the cells were detached using 1 mL of 0.1% Trypsin. The cells were brought up in 4 mL of medium and counted using a hemocytometer (American Optical Corporation, Buffalo, NY, USA). Repeated proliferation assays were performed in four separate experiments comparing the number of cells present for transfectants of miR-138-5p vs. negative control miR.
Cell migration assay
Transwell chambers (6.5 mm insert, 8.0 µm pores; Corning Costar, Corning, NY, USA) were prepared for migration by filling the bottom chamber with 600 μL of DMEM supplemented with fibronectin (10 µg/mL). SCs that were transfected 48 hours earlier were resuspended at a concentration of 1 × 106 cells/mL in serum free DMEM. An aliquot (0.1 mL) of the cells was placed in the top chamber of a Transwell and incubated at 37°C for 18 hours. After removal of cells in the upper chamber, the remaining cells were fixed in methanol and stained with 0.2% crystal violet (Sigma, St. Louis, MO, USA). Migration was determined by counting the number of cells that had traversed the membrane. Each experiment was set-up in triplicate, and repeated migration assays were performed in three separate experiments comparing the number of cells migrating for transfectants of miR-138-5p vs. the negative control miR.
Western blot analysis
Cell culture samples grown in six-well dishes to 70–80% confluency, transfected 48 hours earlier were used. After washing with 1× PBS, the cells were lysed in 100 μL of 1× SDS-Laemmli that had been boiled for 5 minutes. Subsequently, the dishes were scraped and the cell lysate was sheared with multiple passes through a 24 gauge needle. Total protein concentration was determined for each sample using the BCA assay (Pierce, Waltham, MA, USA). For gel loading, an equal quantity of total protein per sample was prepared in 1× SDS-Laemmli-DTT, boiled for 5 minutes, and loaded in each lane of a 7.5% polyacrylamide gel (Mini-PROTEAN® TGX™, Bio-Rad, Hercules, CA, USA). Proteins were transferred onto nitrocellulose using the iBlot® gel transfer system (Invitrogen, Carlsbad, CA, USA). Membranes were blocked in 10% milk in TBS with 0.05% Tween (TBST) and placed on primary antibody overnight at 4°C. Antibodies against vimentin (1:500; Cat.# 08-1052, Zymed, San Francisco, CA, USA) and α-tubulin (1:1000; Cat. # 3873, Cell Signaling Technology, Danvers, MA, USA) were used in Western blot analysis. Following incubation with the primary antibodies, blots were washed in TBST, three times for 15 minutes each, and secondary antibody linked to horseradish peroxidase was incubated with the blots for one hour at room temperature. Blots were then washed as described above and developed with an ECL kit (Pierce). Densitometry was performed using the ImageJ software (National Institute of Health, Bethesda, MD, USA) (Schneider et al., 2012), and vimentin values were normalized using α-tubulin. Repeated western blot assays were performed with transfected lysates from six separate experiments.
Luciferase reporter assay
Two separate luciferase assay systems were utilized from different vendors. A single vector prepared by Genecopoeia (Rockville, MD, USA) contained both the rat 3′UTR of vimentin linked to the Firefly luciferase gene as well as a Renilla luciferase gene for assay normalization. Another system used two separate vectors: one vector contained the human 3′UTR of vimentin linked to the Firefly luciferase gene (Origene, Rockville, MD, USA) and the other contained the Renilla luciferase gene (Promega, Madison, WI, USA) for assay normalization.
SCs were plated in a six-well dish at 1.3 × 105 cells/well. Twenty-four hours after plating cells were co-transfected with 0.5 µg/µL of the appropriate vector and either 30 nM miR-138-5p, or the negative control miR mimic, using the Endofectin transfection agent (Genecopoeia). Twenty-four hours after transfection the cells were harvested and the Firefly and Renilla luminescence were measured using the Luc-Pair™ Duo-Luciferase Assay kit (Genecopoeia) or the Dual-Glo® Luciferase Assay kit (Promega) as appropriate, following each manufacturer’s instructions. Transfection efficiency was normalized relative to the control Renilla luminescence. Each experiment was performed in duplicate and repeated assays were performed in three separate experiments.
Statistical analysis
A paired-samples t-test was used to determine if there was a statistically significant difference in traits for cells transfected with the pre-miR negative control or the pre-miR-138-5p. Corresponding plots depict the mean relative response rate for cells transfected with the pre-miR-138-5p relative to the pre-miR negative control. A P-value < 0.05 was considered statistically significant.
Results
Number of animals
Tissue was collected from the proximal and distal ends of the regenerating nerve as well as the solid material within the guide and compared with control material resected during the initial surgery. At 7 and 14 days, rats with any visible amount of solid material within the nerve guide were included in the study. At 30, 60, and 90 days only rats with solid material spanning the entire nerve guide were included in the study. Therefore at each time point there were 5, 5, 4, 3, and 4 rats included in the study, respectively.
Microarray miRNA expression profiles following sciatic nerve injury
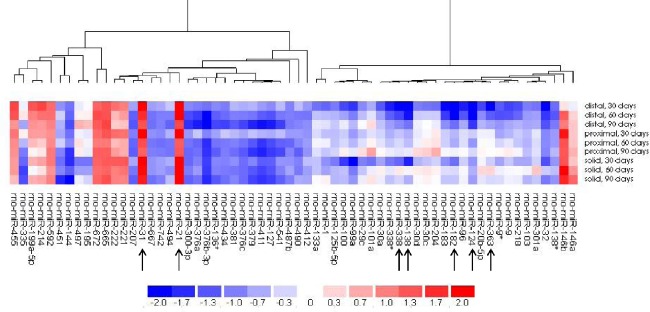
A microarray survey of miRNA expression in the regenerating nerve was performed with the 30-, 60-, and 90-day nerve samples (Additional Table 1 (102.8KB, pdf) ). This encompasses the estimated peak in SC proliferation of ~70 days following rat sciatic nerve transection (Salonen et al., 1988). An insufficient amount of RNA was isolated from the solid component of the seven and 14 day time points for microarray analysis. Expression levels of the proximal, distal, and solid guide components resected during a subsequent surgery were compared relative to the corresponding control nerve harvested during the initial surgery (time = 0). Sixty-two of 372 rat miRNA passed the filtering criteria on variation comparing each sample versus control and were therefore included in the two-way hierarchical clustering analysis (Figure 2). Of these, 40 miRNA were predominately down-regulated across all samples and time points and 12 were up-regulated, compared to control nerve. An evaluation of the array results revealed the top seven miRNA that had at least a ± five-fold change in expression relative to control nerve; all have been implicated in cell migration and proliferation in previous studies (Figure 3) (Xu et al., 2013, 2015; Cai et al., 2015; Chapman et al., 2015; Chen et al., 2015; Li et al., 2015; Lie et al., 2016). Since it is known that SC proliferation and migration are integral to PNR, we investigated this potential association with miRNA-138-5p, the miRNA with greatest change in expression in the microarray analysis.
Figure 2.
The dendogram results of the two-way hierarchical clustering analysis of the 30-, 60-, and 90-day sciatic nerve (experimental) samples relative to control nerve (time 0).
The clustering is performed on log2(Hy3/Hy5) ratios, and the heat map depicts the log Median Ratio (LMR) where a value > 0 indicates up-regulation (red) in experimental samples and a value < 0 indicates down-regulation (blue) compared to control nerve. A LMR > 1.0 is equal to Fold change > 2.0 (i.e. Fold change = 2ΔLMR). The arrows indicate the top seven miRNA that had at least a ± five-fold change in expression relative to control nerve.
Figure 3.
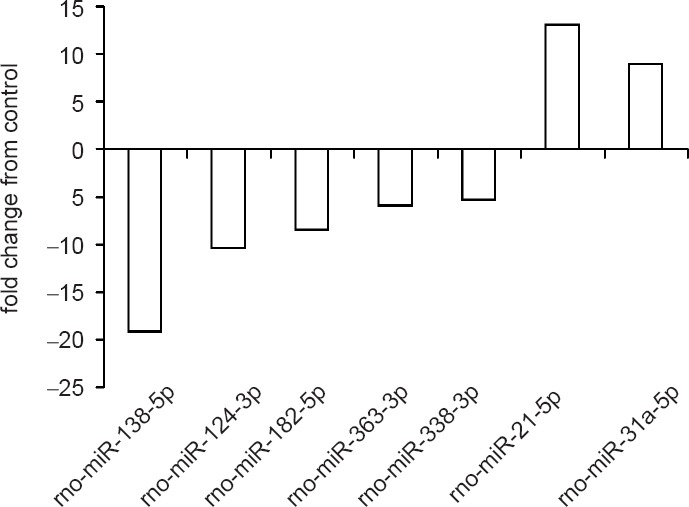
The top seven dysregulated microRNA (miRNA).
Based on the microarray results of the 30-, 60-, and 90-day sciatic nerve samples, these seven miRNA had the greatest change in expression relative to control nerve (fold change of at least ± five). Each has been implicated in cell migration and proliferation in prior studies either in rat, or the equivalent miRNA in human or mouse. A negative fold change indicates that expression levels were higher in the control nerve.
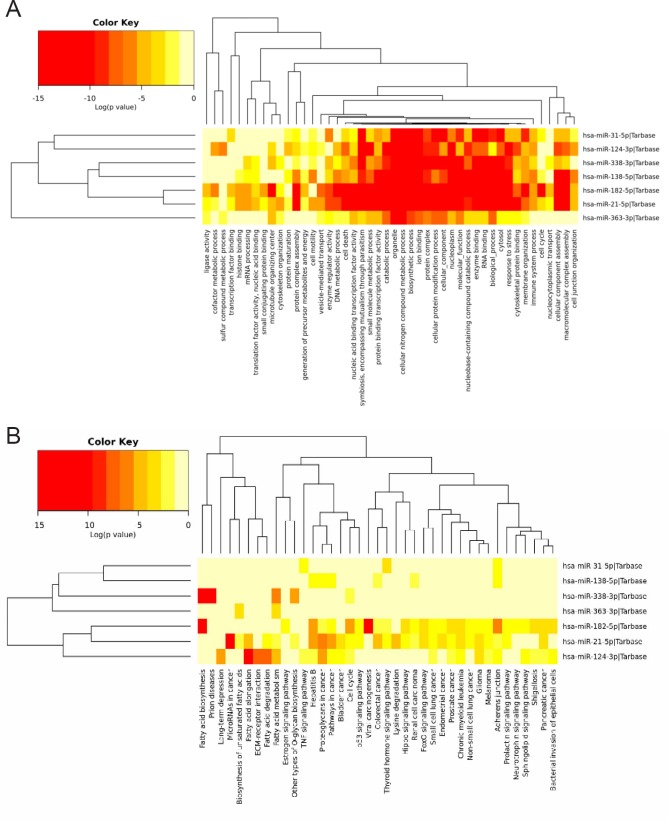
Pathways and processes prediction
For the top seven dysregulated miRNA in this study, the most significant process for the GO term for biological process was cellular nitrogen compound metabolic process with 1726 target genes, for cellular component it was organelle with 3433 genes, and for molecular function it was ion binding with 1858 genes. The most significant KEGG pathway was fatty acid biosynthesis. These predicted activities are primarily involved in biosynthesis and assembly, the cell cycle, the cytoskeletal system, and cell differentiation (Figure 4): all elemental activities of wound healing. With respect to miR138-5p alone, the most significant process for the GO term for biological process was cellular nitrogen compound metabolic process with 223 target genes, for cellular component it was organelle with 408 genes, and for molecular function it was RNA binding with 103 genes. The most significant KEGG pathway was adherens junction.
Figure 4.
The significant major GO processes (A) and KEGG pathways (B) predicted for the gene targets of the seven most altered microRNA (miRNA) observed in this study.
Log of P-values are depicted with greater significance in red. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Confirmation of miR-138-5p expression following sciatic nerve injury
The relative expression level of miR-138-5p was evaluated via qRT-PCR for the control, proximal, and distal samples for each of the 21 rats with adequate solid material within the nerve guide at 7, 14, 30, 60, and 90 days. Expression levels were normalized and reported relative to control nerve (Figure 5). These measures correlated strongly with those observed in the microarray for at 30, 60, and 90 days after neurotomy. For example, the 30 day distal time point was 23 fold lower compared with control. This corresponded well with the 19 fold decrease observed for this sample in the microarray analysis. Expression levels were less than 20% in both nerve stumps at 7 and 14 days. By 30 days the level began to recover in the proximal stump and is near the control baseline between 60-90 days. However, the distal stump lagged behind, and recovery of miR-138-5p to baseline was not achieved within this study interval. The maximal level observed for the distal stump was approximately 50% of control at 90 days.
Figure 5.
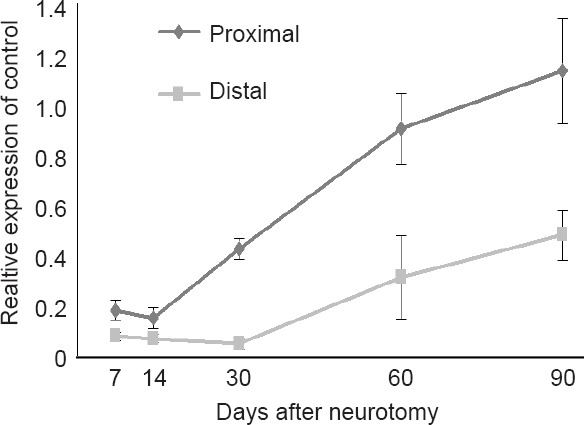
Normalized expression levels of miR-138-5p, reported relative to control nerve, at the five time points throughout this study.
Recovery of miR-138-5p expression level in the distal nerve stump, relative to control nerve, lagged behind the proximal stump which returned to baseline within 60–90 days after neurotomy. Error bars represent standard error of the mean.
Increased expression of miR-138-5p in SCs inhibited cell proliferation and migration
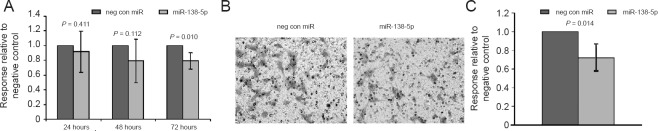
Cell proliferation and migration were measured in rat SCs transfected with the miRNA mimic miR-138-5p or a non-targeting negative control. The rate of proliferation for SCs transfected with miR-138-5p was decreased by ~20% compared to negative control three days following transfection. In addition, the mimic for miR-138-5p decreased the rate of SC migration through Transwell chambers by ~30% compared to negative control after 18 hours (Figure 6).
Figure 6.
The effect of miR-138-5p on Schwann cell (SC) proliferation over 3 days following transfection relative to negative control transfectants.
Overall Schwann cell proliferation decreased by ~20% (A). The effect of miR-138-5p on SC migration through Transwell chambers over 18 hours, following 48 hours of transfection (B). Overall cell migration decreased by ~30% relative to negative control transfectants (C). Error bars represent the 95% confidence interval.
Increased expression of miR-138-5p in SCs inhibited vimentin expression
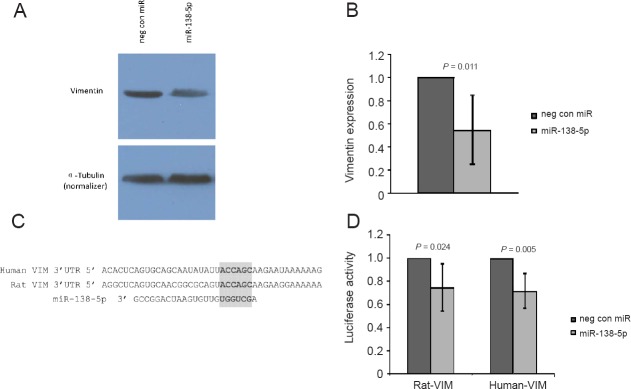
It has been reported, in both rat neuroendocrine tumor cells and human breast cancer cells, that miR-138-5p directly targets vimentin and negatively regulates its expression (Qian et al., 2015; Zhang et al., 2016). This binding is predicted to occur at positions 32-37 of the 3’ UTR of vimentin, in both species (TargetScan, release 7.1). To expand on these findings we investigated the potential effect of miR-138-5p on vimentin expression in rat SCs. Overexpression of miR-138-5p, via transfection of a miRNA mimic, resulted in ~40% decrease in vimentin expression, determined by Western blot analysis, as compared to transfection with a negative control (Figure 7A, B). In addition, Luciferase assays were performed in an effort to verify that vimentin is a direct target of miR-138-5p, utilizing rat SCs. Vectors containing the 3’ UTR of vimentin from rat or human (Additional Figure 1 (132.3KB, pdf) ). were transfected along with miRNA mimics of miR-138-5p or a negative control. Luciferase activity was decreased by ~25%, for both rat and human vimentin vectors, for miR-138-5p transfectants compared with negative controls (Figure 7C, D).
Figure 7.
MicroRNA-138-5p inhibits vimentin expression.
Western blot results for the relative quantification of vimentin in transfected SCs (A). Densitometry analysis revealed that vimentin expression was decreased ~40% in miR-138-5p transfectants as compared to the negative control (B). Positions 10–50 of the human and rat vimentin (VIM) 3’UTR with the predicted binding site of miR-138-5p at positions 32-37 highlighted (C). Transfection into rat SCs of miR-138-5p reduced luciferase activity by ~25%, for both the rat and human vimentin vectors, compared to the negative control miR (D). Error bars represent the 95% confidence interval.
Discussion
SCs are instrumental throughout the process of PNR. They make up the bulk of the cells involved in the process (Salonen et al., 1988), and their activities are multifaceted. Shortly after injury they dedifferentiate and aid macrophages in the process of removing debris from the field of injury and throughout the distal axon; subsequently they form bands of Büngner and produce trophic factors to guide the regenerating axon, and ultimately develop a myelin sheath around the axon (Gaudet et al., 2011). Instrumental to these processes are the ability of SCs to proliferate and migrate. As such, understanding the molecular basis of these processes is fundamental to potentially developing means to enhance nerve regeneration and ultimately improving patient outcomes.
The focus of this work was profiling miRNA expression during PNR and investigating a potential molecular mechanism involved in SC migration and proliferation. As it has been realized that miRNAs are powerful regulators of gene expression, studies of their roles in nerve tissue have broadened the understanding of PNR (Zhou et al., 2016). Recent studies profiling miRNA in PNR after sciatic transection have examined changes in expression within a few hours and up to 14 days after neurotomy (Zhou et al., 2011; Yu et al., 2012). These studies have investigated a diverse range of miRNA involvement including SC migration (Yu et al., 2012; Yi et al., 2016), the impact of hypoxia on SC behavior (Yao et al., 2016), and the survival of dorsal root ganglion neurons (Zhou et al., 2015). The work presented here investigated miRNA expression in PNR over a three month time period, revealing an extended role of miRNA throughout PNR and providing additional basis for further investigation.
The seven miRNA highlighted in the microarray analysis demonstrated at least a ± five-fold change in expression relative to control nerve. The GO processes and KEGG pathways for the predicted target genes of these miRNA center on actions critical to PNR. These include the biosynthesis of the raw materials necessary for cell structure and to promote growth and proliferation. In addition, many of the predicted activities facilitate cell migration, inflammation, and EMT. Indeed, all seven of these miRNA have been implicated in these processes in prior studies, and several are involved in the nervous system. The human miR counterparts of five (-21-5p, 31a-5p, 124-3p, 338-3p, and 363-3p) of these have been associated with proliferation, migration, and/or EMT (Liu et al., 2015; Yan et al., 2016; Ying et al., 2017; Yuan et al., 2017; Zhao et al., 2017) in various other tissues or disease processes, and miR-182-5p has been implicated in rat SC migration specifically (Yu et al., 2012). Furthermore, miR-338-3p has been studied with respect to axonal respiration (Aschrafi et al., 2012), miR-21-5p is considered to promote inflammation in dorsal root ganglion (Simeoli et al., 2017), and miR-124-3p has been examined in several nerve studies where it has been implicated in neurogenesis (Yang et al., 2017).
The microarray results in this study revealed that the most highly dysregulated miRNA at 30, 60, and 90 days of PNR was down-regulation of miR-138-5p; its expression was 19 fold lower than control in the distal segment 30 days after neurotomy. The qRT-PCR results confirmed low expression in both the proximal and distal segments. This analysis also included the 7 and 14 day time points, which also exhibited very low expression relative to control nerve. Due to the need to rapidly freeze the resected nerve tissues to preserve the RNA, these specimens contained a mixture of cells involved in PNR. However, prior studies have indicated that SCs make up the majority of this regenerating tissue (Salonen et al., 1988), and prior work by our group using the same model as our current study, stained the regenerating material with SC specific markers confirming these results(Bryan et al., 2003). Therefore the changes in miRNA expression detailed here are likely primarily reflective of changes in SC miRNA expression.
Previous studies of the nervous system have examined altered expression levels of miR-138-5p and implicated it in the regulation of axon and dendrite growth (Siegel et al., 2009; Liu et al., 2013). It has been implicated in neurologic maladies such as Huntington’s disease (Lee et al., 2011), neural tube defects (Ichi et al., 2010), and panic disorder (Muiños-Gimeno et al., 2011). Taken together, these studies indicate miR-138-5p is an important regulator of gene expression in the nervous system. In addition, the GO processes and KEGG pathways for the predicted target genes of miR-138-5p included components of the cytoskeletal system (GO process organelle) and adherens junctions. Both of these mechanisms are key events in the transition to a mesenchymal phenotype (Lamouille et al., 2014). Furthermore, miR-138-5p has been demonstrated to suppress EMT (Liu et al., 2011; Li et al., 2015), a fundamental process of wound healing, and in keeping with the lower levels we observed following nerve injury. However, additional studies are needed to further detail the role of miR-138-5p with respect to EMT in PNR.
Studies in various cancer cell lines have shown that miR-138-5p inhibits cell migration (Jiang et al., 2010; Tan et al., 2016; Zhang et al., 2016) while another report investigating hypothalamic cells from chicken embryos indicated higher levels promoted migration (Kisliouk et al., 2013). We demonstrated here that forced expression of miR-138-5p resulted in decreased cell proliferation and migration in rat SCs. Cell proliferation and migration are critical attributes for SCs during PNR. In this study, both the proximal and distal nerve stumps exhibited decreased miR-138-5p expression which never fully recovered in the distal stump within 90 days, suggesting a prolonged influence on SCs distally. This correlates with a previous study demonstrating that the number of SCs in the distal segment was elevated throughout 30 weeks of rat sciatic nerve regeneration (Salonen et al., 1988).
Vimentin is one of the most widely expressed intermediate filaments (Satelli and Li, 2011) and functions in cell signaling, attachment, and migration (Ivaska et al. 2007). Within studies of sciatic nerve injury, vimentin has been shown to be upregulated (Perlson et al., 2005) and enhances the ability of SCs to guide and support regenerating axons (Berg et al., 2013). Direct regulation of vimentin expression by miR-138-5p has been previously demonstrated in rat neuroendocrine tumor cells and human breast cancer cells. The work presented in this study noted a decrease in vimentin expression when the level of miR-138-5p was increased in SCs, and luciferase activity assay confirmed the 3’UTR of vimentin was a direct target of miR-138-5p. In conjunction with the low levels of miR-138-5p observed in the regenerating nerve in this study, particularly at the earlier time points corresponding with peak SC migration, this illustrates a potential mechanism of vimentin regulation in PNR: a decrease in miR-138-5p expression leads to increased expression of vimentin which helps facilitate SC migration. In the later stages of PNR axon regeneration and its subsequent myelination proceeds from the proximal segment distally. It has been reported that vimentin negatively regulates myelination (Triolo et al., 2012), and therefore as vimentin decreases it is possible SC migration would decrease as myelination increases. Therefore the trend we observed in the expression of miR-138-5p where the baseline levels returned first in the proximal segment are consistent with prediction of vimentin expression dimunition as the regenerating axon matures.
The results presented here are the first to detail protracted changes in miR-138-5p expression following sciatic nerve transection, as well as its potential role in SC proliferation and migration. This study confirmed previous reports that miR-138-5p targets the 3’UTR of vimentin and therefore may be one of the key regulators of cytoskeletal processes. Due to the critical role of SCs in nerve regeneration, the ability to enhance their migration is an attractive therapeutic strategy (Tomita et al., 2009), and the ultimate goal of the work presented here is to improve outcomes for patients with nerve injury. Therapies utilizing miRNA are in development, and two have proceeded to clinical trial. Targeted methods to modulate miRNA expression using stents, nanoparticles, and viral vectors are being investigated. It is likely that these methods could be adapted into nerve conduit based therapies, and the later method has recently shown the potential for improving nerve regeneration (Wang et al., 2016). Indeed, miRNA based technologies are emerging as promising approaches to delivering safe and effective therapy.
Additional file:
Additional Table 1 (102.8KB, pdf) : An expression matrix for all nine sciatic nerve samples.
Additional Figure 1 (132.3KB, pdf) : Vector map and the sequence for the Genecopoeia vector containing the 3’UTR of vimentin (Rat and Human).
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Financial support: This work was supported in part by the Leisa V. Clayton Foundation. The funder did not participate in data collection and analysis, article writing or submission.
Institutional review board statement: Animal studies were conducted in accordance with Lahey Hosptital & Medical Center Institutional Animal Care and Use Committee (IACUC) guidelines under protocol number A3377-01.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewer: Paula V. Monje, University of Miami, USA.
Funding: This study was supported by the Leisa V. Clayton Foundation.
References
- 1.Abe N, Cavelli V. Nerve injury signaling. Curr Opin Neurobiol. 2008;18:276–283. doi: 10.1016/j.conb.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akane H, Saito F, Shiraki A, Imatanaka N, Akahori Y, Itahashi M, Wang L, Shibutani M. Gene expression profile of brain regions reflecting aberrations in nervous system development targeting the process of neurite extension of rat offspring exposed developmentally to glycidol. J Appl Toxical. 2014;34:1389–1399. doi: 10.1002/jat.2971. [DOI] [PubMed] [Google Scholar]
- 3.Aschrafi A, Kar AN, Natera-Naranjo O, MacGibeny MA, Gioio AE, Kaplan BB. MicroRNA-338 regulates the axonal expression of multiple nuclear-encoded mitochondrial mRNAs encoding subunits of the oxidative phosphorylation machinery. Cell Mol Life Sci. 2012;69:4017–4027. doi: 10.1007/s00018-012-1064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg A, Zelano J, Pekna M, Wilhelmsson U, Pekny M, Cullheim S. Axonal regeneration after sciatic nerve lesion is delayed but complete in GFAP- and vimentin-deficient mice. PLoS One. 2013;8:e79395. doi: 10.1371/journal.pone.0079395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brushart TW. Motor axons preferentially reinnervate motor pathways. J Neurosci. 1993;13:2730–2738. doi: 10.1523/JNEUROSCI.13-06-02730.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryan DJ, Tang JB, Holway AH, Rieger-Christ KM, Trantolo DJ, Wise DL, Summerhayes IC. Enhanced peripheral nerve regeneration elicited by cell-mediated events delivered via a bioresorbable PLGA guide. J Reconstr Microsurg. 2003;19:125–134. doi: 10.1055/s-2003-37820. [DOI] [PubMed] [Google Scholar]
- 7.Bryan DJ, Litchfield CR, Rieger-Christ KM, Manchio J, Logvinenki T, Holway AH, Austin J, Summerhayes IC. Characterization of the spatiotemporal expressions profile of key proteins in rat sciatic nerve regeneration using reverse phase protein arrays. Proteome Sci. 2012;10:9. doi: 10.1186/1477-5956-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai SD, Chen JS, Xi ZW, Zhang LJ, Niu ML, Gao ZY. MicroRNA - 144 inhibits migration and proliferation in rectal cancer by downregulating ROCK 1. Mol Med Rep. 2015;12:7396–7402. doi: 10.3892/mmr.2015.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang IA, Oh MJ, Kim MH, Park SK, Kim BG, Namgung U. Vimentin phosphorylation by Cdc2 in Schwann cell controls axon growth via β1-integrin activation. FASEB J. 2012;26:2401–2413. doi: 10.1096/fj.11-199018. [DOI] [PubMed] [Google Scholar]
- 10.Chapman BV, Wald AI, Akhtar P, Munko AC, Xu J, Gibson SP, Grandis JR, Ferris RL, Khan SA. MicroRNA-363 targets myosin 1B to reduce cellular migration in head and neck cancer. BMC Cancer. 2015;15:861. doi: 10.1186/s12885-015-1888-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JT, Yao KH, Hua L, Zhang LP, Wang CY, Zhang JJ. MiR-338-3p inhibits the proliferation and migration of gastric cancer cells by targeting ADAM17. Int J Clin Exp Pathol. 2015;8:10922–10928. [PMC free article] [PubMed] [Google Scholar]
- 12.Costigan M, Befat K, Karchewski L, Griffin RS, D’Urso D, Allchorne A, Sitarski J, Mannion JW, Pratt RE, Woolf CJ. Replicate high density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. 2002;3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.FexSvennigsen A, Dahlin LB. Repair of the Peripheral Nerve-Remyelination that Works. Brain Sci. 2013;3:1182–1197. doi: 10.3390/brainsci3031182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan Z, Ding L, Burckhardt CJ, Lowery J, Zaritsky A, Sitterley K, Mota A, Costigliola N, Starker CG, Voytas DF, Tytell J, Goldman RD, Danuser G. Vimentinintermediate filaments template microtubule networks to enhance persistence in cell polarity and directed migration. Cell Syst. 2016;3:252–263. doi: 10.1016/j.cels.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaudet AD, Popovich PG, Ramer MS. Wallerian degeneration: gaining perspective on inflammatory events after peripheral nerve injury. J Neuroinflammation. 2011;8:110. doi: 10.1186/1742-2094-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gess B, Rohr D, Lange E, Halfter H, Young P. Desmoplakin is involved in organization of an adhesion complex in peripheral nerve regeneration after injury. Exp Neurol. 2015;264:55–66. doi: 10.1016/j.expneurol.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 17.He X, Yu Y, Awatramani R, Lu QR. Unwrapping myelination by microRNAs. Neuroscientist. 2012;18:45–55. doi: 10.1177/1073858410392382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichi S, Costa FF, Bischof JM, Nakazaki H, Shen YW, Boshnjaku V, Sharma S, Mania-Farnell B, McLone DG, Tomita T, Soares MB, Mayanil CS. Folic acid remodels chromatin on Hes1 and Neurog2 promoters during caudal neural tube development. J Biol Chem. 2010;285:36922–36932. doi: 10.1074/jbc.M110.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivaska J, Pallari HM, Nevo J, Eriksson JE. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp Cell Res. 2007;313:2050–2062. doi: 10.1016/j.yexcr.2007.03.040. [DOI] [PubMed] [Google Scholar]
- 20.Iyer AN, Bellon A, Baudet ML. microRNAs in axon guidance. Front Cell Neurosci. 2014;8:78. doi: 10.3389/fncel.2014.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang L, Liu X, Kolokythas A, Yu J, Wang A, Heidbreder CE, Shi F, Zhou X. Downregulation of the Rho GTPase signaling pathway is involved in the microRNA-138-mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int J Cancer. 2010;127:505–212. doi: 10.1002/ijc.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang N, Li H, Sun Y, Yin D, Zhao Q, Cui S, Yao D. Differential gene expression in proximal and distal nerve segments of rats with sciatic nerve injury during Wallerian degeneration. Neural Regan Res. 2014;9:1186–1194. doi: 10.4103/1673-5374.135325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kisliouk T, Meiri N. MiR-138 promotes the migration of cultured chicken embryonic hypothalamic cells by targeting reelin. Neuroscience. 2013;238:114–124. doi: 10.1016/j.neuroscience.2013.02.020. [DOI] [PubMed] [Google Scholar]
- 24.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SK, Wolfe SW. Peripheral nerve injury and repair. J Am Acad Orthop Surg. 2000;8:243–252. doi: 10.5435/00124635-200007000-00005. [DOI] [PubMed] [Google Scholar]
- 26.Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park JE, Park KH, Jung KH, Lee SK, Kim M, Roh JK. Altered microRNA regulation in Huntington’s disease models. Exp Neurol. 2011;227:172–179. doi: 10.1016/j.expneurol.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Li D, Li X, Wang A, Meisgen F, Pivarcsi A, Sonkoly E, Ståhle M, Landén NX. MicroRNA-31 promotes skin wound healing by enhancing keratinocyte proliferation and migration. J Invest Dermatol. 2015;135:1676–1685. doi: 10.1038/jid.2015.48. [DOI] [PubMed] [Google Scholar]
- 28.Li J, Wang Q, Wen R, Liang J, Zhong X, Yang W, Su D, Tang J. MiR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med. 2015;19:2793–2805. doi: 10.1111/jcmm.12666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, Zhang P, Guo W, Li H, Gu X, Yao D. Protein expression profiling during Wallerian degeneration after rat sciatic nerve injury. Muscle Nerve. 2014;50:73–78. doi: 10.1002/mus.24082. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Wang Z, Wang Y, Gu W. MiR-338 suppresses the growth and metastasis of OSCC cells by targeting NRP1. Mol Cell Biochem. 2015;398:115–122. doi: 10.1007/s11010-014-2211-3. [DOI] [PubMed] [Google Scholar]
- 31.Liu CM, Wang RY, Saijilafu, Jiao ZX, Zhang BY, Zhou FQ. MicroRNA-138 and SIRT1 form a mutual negative feedback loop to regulate mammalian axon regeneration. Genes Dev. 2013;27:1473–1483. doi: 10.1101/gad.209619.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X, Wang C, Chen Z, Jin Y, Wang Y, Kolokythas A, Dai Y, Zhou X. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem J. 2011;440:23–31. doi: 10.1042/BJ20111006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Zhang W, Liu K, Liu S, Ji B, Wang Y. miR-138 suppresses cell proliferation and invasion by inhibiting SOX9 in hepatocellular carcinoma. Am J Transl Res. 2016;8:2159–2168. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Lu A, Huang Z, Zhang C, Zhang X, Zhao J, Zhang H, Zhang Q, Wu S, Yi X. Differential expression of microRNAs in dorsal root ganglia after sciatic nerve injury. Neural Regen Res. 2014;9:1031–1040. doi: 10.4103/1673-5374.133164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muiños-Gimeno M, Espinosa-Parrilla Y, Guidi M, Kagerbauer B, Sipilä T, Maron E, Pettai K, Kananen L, Navinés R, Martín-Santos R, Gratacòs M, Metspalu A, Hovatta I, Estivill X. Human microRNAs miR-22, miR-138-2, miR-148a and miR-488 are associated with panic disorder and regulate several anxiety candidate genes and related pathways. Biol Psychiatry. 2011;69:526–533. doi: 10.1016/j.biopsych.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Patodia S, Raivich G. Downstream effector molecules in successful peripheral nerve regeneration. Cell Tissue Res. 2012;349:15–26. doi: 10.1007/s00441-012-1416-6. [DOI] [PubMed] [Google Scholar]
- 38.Perlson E, Hanz S, Ben-Yaakov K, Segal-Ruder Y, Seger R, Fainzilber M. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45:715–26. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 39.Phay M, Kim HH, Yoo S. Dynamic change and target prediction of axon-specific microRNAs in regenerating sciatic nerve. PLoS One. 2015;10:e0137461. doi: 10.1371/journal.pone.0137461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian BJ, You L, Shang FF, Liu J, Dai P, Lin N, He M, Liu R, Zhang Y, Xu Y, Zhang YH, Wang TH. Vimentin regulates neuroplasticity in transected spinal cord rats associated with micRNA138. Mol Neurobiol. 2015;51:437–447. doi: 10.1007/s12035-014-8745-2. [DOI] [PubMed] [Google Scholar]
- 41.Richner M, Ulrichsen M, Elmegaard SL, Dieu R, Pallesen LT, Vaegter CB. Peripheral nerve injury modulates neurotrophin signaling in the peripheral and central nervous system. Mol Neurobiol. 2014;50:945–970. doi: 10.1007/s12035-014-8706-9. [DOI] [PubMed] [Google Scholar]
- 42.Salonen V, Aho H, Röyttä M, Peltonen J. Quantitation of Schwann cells and endoneurial fibroblast-like cells after experimental nerve trauma. Acta Neuropathol. 1988;75:331–336. doi: 10.1007/BF00687785. [DOI] [PubMed] [Google Scholar]
- 43.Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68:3033–46. doi: 10.1007/s00018-011-0735-1. Taylor CA, Braza D, Rice JB, Dillinghan T (2008) The incidence of peripheral nerve injury in extremity trauma. Am J Phys Med Rehabil 87:381-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hübel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simeoli R, Montague K, Jones HR, Castaldi L, Chambers D, Kelleher JH, Vacca V, Pitcher T, Grist J, Al-Ahdal H, Wong LF, Perretti M, Lai J, Mouritzen P, Heppenstall P, Malcangio M. Exosomal cargo including microRNA regulates sensory neuron to macrophage communication after nerve trauma. Nat Commun. 2017;8:1778. doi: 10.1038/s41467-017-01841-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan Y, Hu H, Tan W, Jin L, Liu J, Zhou H. MicroRNA-138 inhibits migration and invasion of non-small cell lung cancer cells by targeting LIMK1. Mol Med Rep. 2016;14:4422–4428. doi: 10.3892/mmr.2016.5769. [DOI] [PubMed] [Google Scholar]
- 48.Terenghi G. Peripheral nerve regeneration and neurotrophic factors. J Anat. 1999;194:1–14. doi: 10.1046/j.1469-7580.1999.19410001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorsin F, Rosberg HE, Steen CK, Dahlin LB. Digital nerve injuries: epidemiology, results, costs, and impact on daily life. J Plast Surg Hand Surg. 2012;46:184–190. doi: 10.3109/2000656X.2012.676554. [DOI] [PubMed] [Google Scholar]
- 50.Tomita K, Hata Y, Kubo T, Fujiwara T, Yano K, Hosokawa K. Effects of the in vivo predegenerated nerve graft on early Schwann cell migration: quantitative analysis using S100-GFP mice. Neurosci Lett. 2009;461:36–40. doi: 10.1016/j.neulet.2009.05.075. [DOI] [PubMed] [Google Scholar]
- 51.Triolo D, Dina G, Taveggia C, Vaccari I, Porrello E, Rivellini C, Domi T, La Marca R, Cerri F, Bolino A, Quattrini A, Previtali SC. Vimentin regulates peripheral nerve myelination. Development. 2012;139:1359–1367. doi: 10.1242/dev.072371. [DOI] [PubMed] [Google Scholar]
- 52.Viader A, Chang LW, Fahrner T, Nagarajan R, Milbrandt J. MicroRNAs modulate Schwann cell response to nerve injury by reinforcing transcriptional silencing of dedifferentiation-related genes. J Neurosci. 2011;31:17358–17369. doi: 10.1523/JNEUROSCI.3931-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waller A. Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Philos Trans R Soc Lond B Biol Sci. 1850;140:423–429. [Google Scholar]
- 55.Wang J, Muheremu A, Zhang M, Gong K, Huang C, Ji Y, Wei Y, Ao Q. MicroRNA-338 and microRNA-21 co-transfection for the treatment of rat sciatic nerve injury. Neurol Sci. 2016;37:883–890. doi: 10.1007/s10072-016-2500-6. [DOI] [PubMed] [Google Scholar]
- 56.Xu J, Zhang W, Lv Q, Zhu D. Overexpression of miR-21 promotes the proliferation and migration of cervical cancer cells via the inhibition of PTEN. Oncol Rep. 2015;33:3108–3116. doi: 10.3892/or.2015.3931. [DOI] [PubMed] [Google Scholar]
- 57.Xu X, Li S, Lin Y, Chen H, Hu Z, Mao Y, Xu X, Wu J, Zhu Y, Zheng X, Luo J, Xie L. MicroRNA-124-3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J Transl Med. 2013;11:276. doi: 10.1186/1479-5876-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan L, Cao R, Liu Y, Wang L, Pan B, Lv X, Jiao H, Zhuang Q, Sun X, Xiao R. MiR-21-5p links epithelial-mesenchymal transition phenotype with stem-like cell signatures via AKT signaling in keloid keratinocytes. Sci Rep. 2016;6:28281. doi: 10.1038/srep28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J, Zhang X, Chen X, Wang L, Yang G. Exosome mediated delivery of miR-124 promotes neurogenesis after ischemia. Mol Ther Nucleic Acids. 2017;7:278–287. doi: 10.1016/j.omtn.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yao C, Shi X, Zhang Z, Zhou S, Qian T, Wang Y, Ding F, Gu X, Yu B. Hypoxia-induced upregulation of miR-132 promotes Schwann cell migration after sciatic nerve injury by targeting PRKAG3. Mol Neurobiol. 2016;53:5129–5139. doi: 10.1007/s12035-015-9449-y. [DOI] [PubMed] [Google Scholar]
- 61.Yao D, Li M, Shen D, Ding F, Lu S, Zhao Q, Gu X. Expression changes and bioinformatic analysis of Wallerian degeneration after sciatic nerve injury in rat. Neurosci Bull. 2013;29:321–332. doi: 10.1007/s12264-013-1340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yi S, Wang S, Zhao Q, Yao C, Gu Y, Liu J, Gu X, Li S. miR-sc3 a novel microRNA promotes Schwann cell proliferation and migration by targeting Astn1. Cell Transplant. 2016;25:973–982. doi: 10.3727/096368916X690520. [DOI] [PubMed] [Google Scholar]
- 63.Ying J, Yu X, Ma C, Zhang Y, Dong J. MicroRNA-363-3p is downregulated in hepatocellular carcinoma and inhibits tumorigenesis by directly targeting specificity protein 1. Mol Med Rep. 2017;16:1603–1611. doi: 10.3892/mmr.2017.6759. [DOI] [PubMed] [Google Scholar]
- 64.Yu B, Qian T, Wang Y, Zhou S, Ding G, Ding F, Gu X. miR-182 inhibits Schwann cell proliferation and migration by targeting FGF9 and NTM, respectively at an early stage following sciatic nerve injury. Nucleic Acids Res. 2012;40:10356–10365. doi: 10.1093/nar/gks750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuan Q, Sun T, Ye F, Kong W, Jin H. MicroRNA-124-3p affects proliferation, migration and apoptosis of bladder cancer cells through targeting AURKA. Cancer Biomark. 2017;19:93–101. doi: 10.3233/CBM-160427. [DOI] [PubMed] [Google Scholar]
- 66.Zhang J, Liu D, Feng Z, Mao J, Zhang C, Lu Y, Li J, Zhang Q, Li Q, Li L. MicroRNA-138 modulates metastasis and EMT in breast cancer cells by targeting vimentin. Biomed Pharmacother. 2016;77:135–141. doi: 10.1016/j.biopha.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 67.Zhao G, Han C, Zhang Z, Wang L, Xu J. Increased expression of microRNA-31-5p inhibits cell proliferation, migration, and invasion via regulating Sp1 transcription factor in HepG2 hepatocellular carcinoma cell line. Biochem Biophys Res Commun. 2017;490:371–377. doi: 10.1016/j.bbrc.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 68.Zhou S, Yu B, Qian T, Yao D, Wang Y, Ding F, Gu X. Early changes of microRNAs expression in the dorsal root ganglia following rat sciatic nerve transection. Neurosci Lett. 2011;494:89–93. doi: 10.1016/j.neulet.2011.02.064. [DOI] [PubMed] [Google Scholar]
- 69.Zhou S, Zhang S, Wang Y, Yi S, Zhao L, Tang X, Yu B, Gu X, Ding F. MiR-21 and miR-222 inhibit apoptosis of adult dorsal root ganglion neurons by repressing TIMP3 following sciatic nerve injury. Neurosci Lett. 2015;586:43–49. doi: 10.1016/j.neulet.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 70.Zhou S, Ding F, Gu X. Non-coding RNAs as emerging regulators of neural injury responses and regeneration. Neurosci Bull. 2016;32:253–264. doi: 10.1007/s12264-016-0028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.