Abstract
Despite the utility of CRISPR-Cas9 nucleases for genome editing, the potential for off-target activity limits their application, especially for therapeutic purposes1,2. We developed a yeast-based assay to identify optimized Streptococcus pyogenes Cas9 (SpCas9) variants that enables simultaneous evaluation of on- and off-target activity. We screened a library of SpCas9 variants carrying random mutations in the REC3 domain and identified mutations that increased editing accuracy whilst maintaining editing efficiency. We combined four beneficial mutations to generate evoCas9, a variant that has fidelity exceeding both wild-type (79-fold improvement) and rationally designed Cas9 variants3,4 (4-fold average improvement), while maintaining near wild-type on-target editing efficiency (90% median residual activity). Evaluating evoCas9 on endogenous genomic loci, we demonstrated a substantial improved specificity and observed no off-target sites for 4 of the 8 sgRNAs tested. Finally, we showed that following long-term expression (40 days), evoCas9 strongly limited the unspecific cleavage of a difficult-to-discriminate off-target site and fully abrogated the cleavage of two additional off-targets.
RNA-guided endonucleases are currently considered the state-of-the-art tool for genome editing and are widely adopted in research. However, the clinical application of Cas9 is potentially limited by its unspecific activity that strongly depends on the amount and duration of nuclease expression5, as well as by the genomic target site6. Three main strategies were developed to reduce unwanted cleavages: 1) control of the activity of SpCas9 through direct delivery (ribonucleoprotein complexes [RNP]5,7, mRNA8,9 and self-limiting circuits10) or tunable systems (reconstitution from inactive split fragments and inducible activation2), 2) modification of SpCas9 (paired nickases11–13, SpCas9-FokI chimeric nucleases14,15 and fusion of SpCas9 with specific DNA-binding domains16) and 3) engineering of the sgRNAs (tru-sgRNAs17 and ggX20 sgRNAs18). The improvements offered by these strategies are counterbalanced by significant loss of on-target activity, restricted number of targetable sites and increased technical complexity. In search for better SpCas9-mediated editing, two studies have recently reported novel more specific nuclease variants obtained through structure-guided rational engineering3,4. And while our manuscript was under review a third highly-specific variant obtained through a rational approach has been reported19. These reports have demonstrated that tailored substitutions abolishing unspecific contacts between SpCas9 and the DNA substrate generate more precise RNA-guided endonucleases (eSpCas9(1.1)3 and SpCas9-HF14) with increased dependency to sgRNA:target DNA pairing. Even though these variants offer improved specificity, for some sites off-target cleavage remains a problem3,4,19.
As opposed to rational approaches, a randomized screening allows the exploration of a wide mutational library and the isolation of suitable residues directly in vivo, potentially enabling the identification of SpCas9 variants with higher fidelity. Similarly to previous studies on zinc-finger nucleases20,21, we approached SpCas9 optimization through a single round of directed evolution screening performed in Saccharomyces cerevisiae by designing a reporter yeast platform to isolate highly specific SpCas9 variants from a library of random mutants of the REC3 domain (aa. 308-718). The assay exploits two genomic reporter cassettes that allow monitoring the editing status of selected on-target and off-target loci by auxotrophic yeast selection (see Supplementary Note). The REC3 domain was chosen for its extensive interactions with sgRNA: target DNA heteroduplex (Supplementary Fig. 1a).
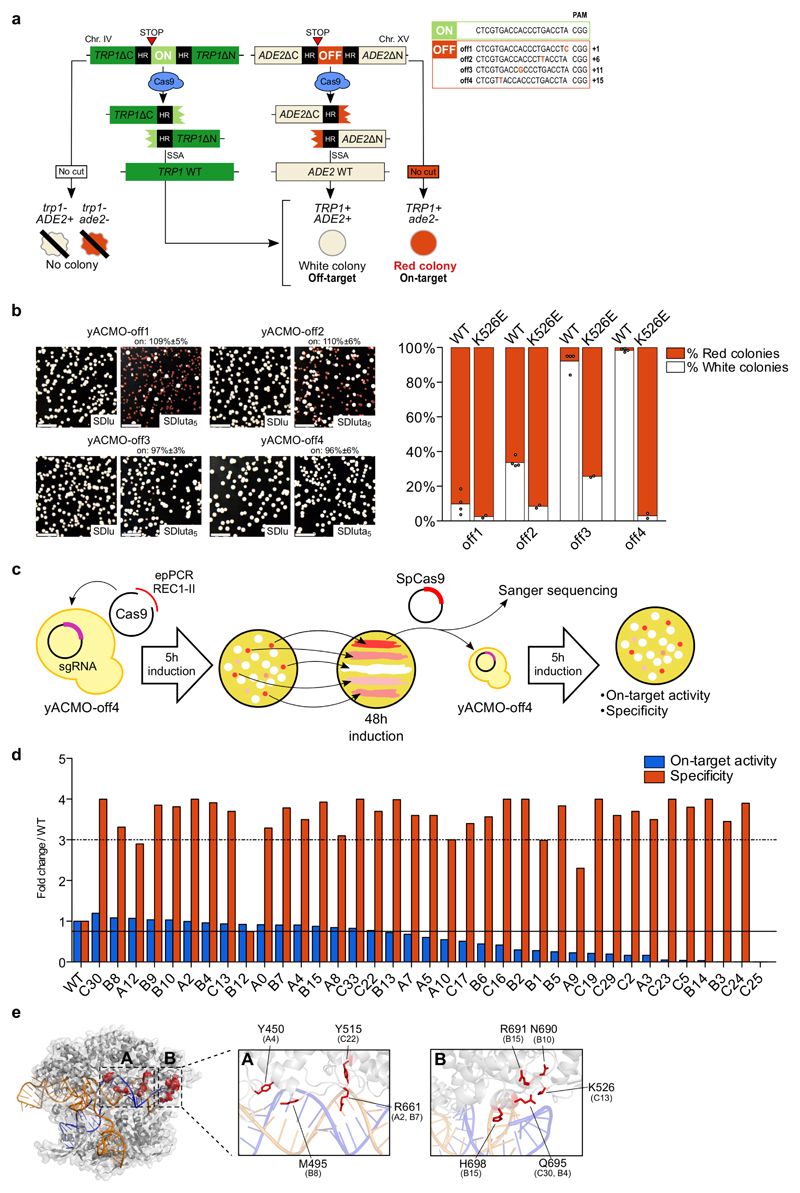
We first generated four reporter yeast strains (yACMO-off1/4) carrying a common genomic on-target site together with an off-target site with a single mismatch positioned more (off4) or less (off1) distal from the PAM trinucleotide (Fig. 1a). The yACMO strains were then transformed with wild-type SpCas9 and the sgRNA to validate the screening system (Fig. 1b). A single round of screening was performed into the yACMO-off4 strain to select the randomly generated variants in the most stringent conditions where unspecific cleavages can be produced with the highest probability (Fig. 1b,c).
Figure 1. Isolation of high-fidelity SpCas9 variants through a yeast-based screening.
(a) Design of yeast reporter strains. The survival of a colony indicates TRP1 on-target cleavage, while the cleavage of ADE2 is revealed by the pigmentation: white and red indicate presence and absence of off-target activity, respectively. See Supplementary Notes for details. (b) Validation of the yACMO strains. Representative plates obtained after transformation of wild-type SpCas9 into the yACMO-off1/off4 reporter strains stably expressing a sgRNA matching the on-target sequence in the TRP1 locus. Plates shown in the left panels (SDlu) select for the total transformants, whereas reporter plates are shown in the right panels (SDluta5). The percentages (± s.e.m. for n=4 biologically independent samples) reported above the SDluta5 plate panels indicate the mean on-target cleavage efficiency. Percentages above 100% are caused by slight deviations in automated counting and should be interpreted as complete on-target cleavage. Scale bar: 1 cm. The graph on the right reports the mean specificity of wild-type SpCas9 (n=4 biologically independent samples) and the C13 mutant (n=2 biologically independent samples), individual samples are represented overlaid as white circles. (c) Schematic representation of the yeast screening workflow (see Supplementary Note). (d) Evaluation of the on-target activity (blue bars) and the specificity (red bars) of SpCas9 variants obtained from the yeast screening through re-challenging of the yACMO-off4 reporter strain. The continuous line indicates an on-target activity of at least 75% of wild-type Cas9; the dashed line indicates an increase in specificity of at least a 3-fold. (e) Identification of target substitutions for specificity optimization. Representation of SpCas9 crystal structure (PDB ID 4UN3) in which selected mutations from the best performing yeast-isolated variants are indicated in red. A conformational cluster of mutations can be identified at one end of the cavity accommodating the heteroduplex (see inset B). In brackets, codes of the variants in (d) containing the corresponding mutation.
The library of REC3 mutants was generated by error-prone PCR and was assembled directly in yeast cells by homologous recombination with a plasmid encoding a REC3-deleted SpCas9 (Fig. 1c). Selected variants were isolated from the red colonies, whose pigmentation results from the absence of SpCas9 off-target activity yACMO-off4 (Fig.1a and Supplementary Note), and were re-analyzed in the same strain to quantify the on/off-target activity with respect to wild-type SpCas9 (Fig. 1c,d and Supplementary Note). From this screen, we identified several substitutions, some of which were present more than once in the mutants’ pool (Supplementary Table 1 and Supplementary Fig. 1b). Notably, we found one SpCas9 variant characterized by a single mutation that has–to our knowledge–not been reported before (K526E), with a substantially reduced off-target activity and a well preserved on-target activity (Fig. 1b, right panel). Of note, the K526E mutation was part of a conformational cluster of substitutions located at one end of the REC3 domain which is in contact with the more PAM-distal part of the target DNA sequence (nt. 17-20, Fig. 1e).
We next reasoned that further increased fidelity could be obtained by the combination of the best performing variants (listed in Table 1). To this aim, we selected a set of most promising mutations positioned in close proximity with the sgRNA:DNA duplex (Fig. 1e) and combined them with the K526E substitution.
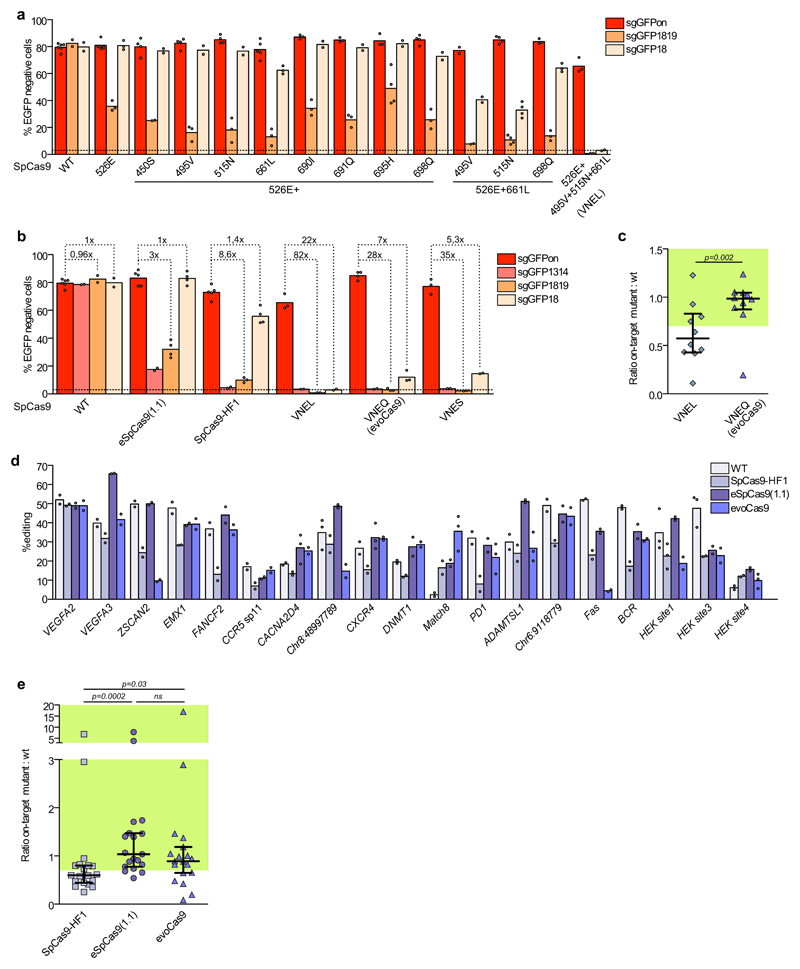
These new variants were tested in an EGFP disruption assay (293multiEGFP cells) using either a fully matching or mismatched sgRNAs (Fig. 2a). The mismatched sgRNAs used to measure the off-target activity contained one or two non-pairing nucleotides in distal positions from the PAM sequence (sgGFP18 and sgGFP1819, mismatched nucleotides in position 18 and 18-19, respectively). Whereas wild-type SpCas9 cut the target sequence with equal efficiency irrespectively of the presence of mismatches in the sgRNA, our best mutant, which contained the M495V/Y515N/K526E/R661L (VNEL) substitutions, induced little or no loss of EGFP fluorescence when tested in combination with both mismatched sgRNAs (sgGFP18 and sgGFP1819) (Fig. 2a). Nevertheless, we observed that this strong increase in specificity came at the cost of a small but measurable loss of on-target activity (~20% drop, Fig. 2b).
Figure 2. Selection of highly specific SpCas9 variants in mammalian cells.
(a) Combination of selected mutations obtained from the yeast screening. Mean EGFP knockout levels in 293multiEGFP following transfection with single, double and triple mutants generated by the combination of mutations obtained from the best performing yeast-isolated variants together with the on-target sgRNA (sgGFPon) or each of the indicated mismatched guides (sgGFP1819 and sgGFP18). (b) Side-by-side comparison of the best-generated variants with previously published mutants. Wild-type SpCas9 (WT) and the indicated variants were tested in 293multiEGFP cells to evaluate their mean on-target efficiency (sgGFPon) and specificity (sgGFP1314, sgGFP1819 and sgGFP18 with mismatches in position 13-14, 18-19 and 18, respectively, from the PAM). Dotted lines indicate on/off target ratios calculated for the indicated on/off couples.(c) Ratio of the on-target activity of evoCas9 (VNEQ) and the VNEL variant to wild-type SpCas9 calculated on EGFP targets and endogenous loci in 293T cells (knockout levels are reported in Supplementary Fig. 2b,c). The median and interquartile range are shown. On-target activity above 70% of the wild-type protein is indicated in green. (d) evoCas9 activity on endogenous loci. Wild-type SpCas9, evoCas9, SpCas9-HF1 and eSpCas9(1.1) were transfected in 293T cells to measure the mean indel formation towards endogenous loci 7 days post-transfection using the TIDE tool. (e) Ratio of the mean on-target activity of evoCas9, SpCas9-HF1 and eSpCas9(1.1) over wild-type SpCas9 relative to endogenous loci in (d). The median and interquartile range are shown. On-target activity above 70% of the wild-type protein is indicated in green. In panels (a-b) loss of EGFP fluorescence was measured by FACS analysis 7 days post-transfection; dashed lines indicate the background loss of EGFP fluorescence. In panels (c) and (e) statistical significance was assessed using the two-sided paired Wilcoxon signed-rank test, sample size n=10 in (c) and n=19 in (e). Summary of data distributions and statistical details are reported in Supplementary Table 5; ns, not significant.
To address this issue, based on the available crystal structures (PDB ID: 4OO8 and 4UN3), we rationally modeled substitutions alternative to the R661L mutation that was predicted to produce steric clashes with the sgRNA molecule in all its rotamers. We selected the R661Q and R661S mutations since they may preserve favorable contacts with the sgRNA backbone without perturbing neighboring residues, generating the VNEQ and VNES variants, respectively (Supplementary Fig. 2a). Their analysis in 293multiEGFP cells showed complete restoration of the on-target cleavage efficiency coupled with a small loss of specificity, which yet remained considerably better than the original nuclease as well as the other analyzed variants (Fig. 2b). These results suggest that the optimization of SpCas9 may result from a set of mutations generating a balanced compromise between increased specificity and preservation of the on-target activity.
The VNEL, VNEQ and VNES mutants were compared with two recently rationally designed high-fidelity variants, eSpCas9(1.1)3 and SpCas9-HF14, revealing a marked increase in fidelity (Fig. 2b). Notably, this was also true when the three variants were tested with the most stringent surrogate off-target (sgGFP18), showing a 5- to 22-fold and a 4- to 16-fold improvement in the on/off-ratio relatively to eSpCas9(1.1) and SpCas9-HF1, respectively (Fig. 2b).
The two more promising variants, VNEL and VNEQ, were then tested using different targets within the EGFP coding sequence, as well as against a selection of 19 endogenous loci previously reported in literature, revealing that VNEQ editing activity was similar to wild-type SpCas9, whereas the VNEL mutant was less active at some of the sites, with a substantial drop in activity for one EGFP locus and several of the genomic sites (Fig. 2c and Supplementary Fig. 2b,c). We excluded potential cleavage alterations related to protein levels of our variants by testing their expression and their activity in titration experiments (Supplementary Fig. 3a,b). The best performing mutant, VNEQ, was named evoCas9 (evolved Cas9) and was further tested against a panel of endogenous genomic sites comparing it side-by-side with wild-type SpCas9, SpCas-HF1 and eSpCas9(1.1) (Fig. 2d). For the majority of the tested loci, evoCas9, similarly to eSpCas9(1.1), showed a near wild-type targeting efficiency (Fig. 2d,e). By contrast, in all but three loci (ZSCAN2, Fas and Chr8:48997789) evoCas9 was significantly (p=0.03) more active than the SpCas9-HF1 variant, which in turn showed lower cleavage efficiency (90% and 60% of median residual activity, respectively, Fig. 2d,e).
The investigation of the sgRNA requirements of evoCas9 and of the other high-specificity variants so far reported demonstrated their incompatibility with sgRNAs having either truncated or longer spacers and with sgRNAs carrying an additional 5’ mismatched guanine, commonly introduced at the beginning of the guide RNA to favor transcription from the U6 promoter (Supplementary Fig. 4a-e)3,4. This feature reduces the total number of targetable sites; nevertheless, this limitation can be circumvented by exploiting alternative methods to synthesize functional guide RNAs (see Supplementary Note). Finally, evoCas9 worked efficiently with sgRNAs with optimized scaffolds22 while preserving its improved specificity. This sgRNA modification consists in the elongation of the stem and a base flip to interrupt a polyT stretch to allow increased cleavage activity (Supplementary Fig. 4f).
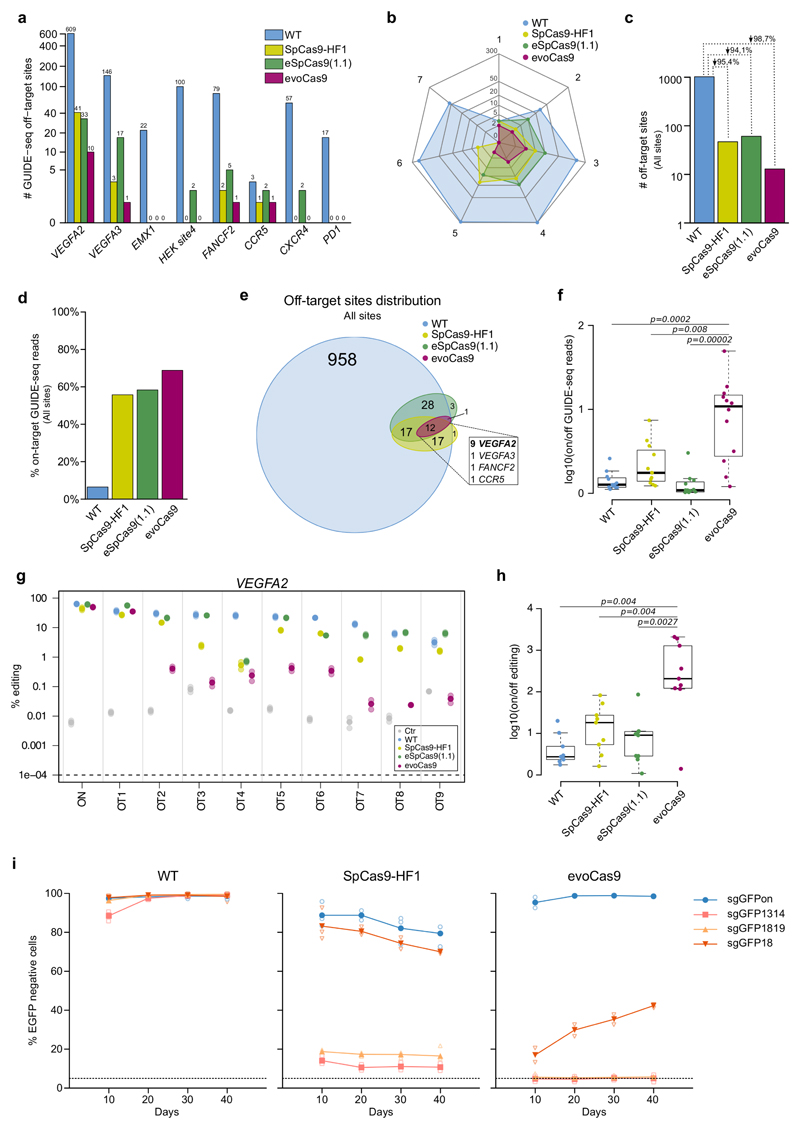
To evaluate evoCas9 genome-wide off-target activity, we performed head-to-head GUIDE-seq experiments with the high-fidelity variants and the wild-type nuclease, in combination with sgRNAs targeting eight different genomic loci (Fig. 3a and Supplementary Fig. 5-6). Among the sites chosen for the analysis we included three loci of potential clinical relevance: CCR5, CXCR4 and PD1. All the tested sgRNAs in combination with wild-type SpCas9 induced cleavage at numerous off-targets differing from the original target site for up to seven mismatches (Fig. 3a,b and Supplementary Fig. 5-6); the highest number of detected off-target sites corresponded to the two repetitive VEGFA2 and VEGFA3 target loci, which were reported to be highly vulnerable to associated non-specific cleavages4,6 (Fig. 3a and Supplementary Fig. 6). EvoCas9 was able to abolish the majority of the off-targets generated by the wild-type nuclease in combination with the analyzed sgRNAs (Fig. 3a,c), with the few residual sites mainly associated with repetitive target sequences (VEGFA2 and VEGFA3). These refractory sites are often characterized by a high degree of similarity with the intended target (Fig. 3b and Supplementary Fig. 5-6). The comparison of the specificity profiles between evoCas9 and the other high-fidelity variants across the eight different sgRNAs revealed that, although in combination with certain guides all three mutants completely avoided unspecific cleavages (EMX1 and PD1, see Fig. 3a), overall evoCas9 showed the highest reduction in the total number of detected off-targets (98.7% versus 95.4% for SpCas9-HF1 and 94.1% for eSpCas9(1.1)) (Fig. 3a,c and Supplementary Fig. 5-6), outperforming the two rationally designed mutants.(Fig. 3b). To further compare the performance of the different nucleases we measured the global on-target specificity by calculating the overall percentage of GUIDE-seq reads corresponding to the intended targets (Fig. 3d and Supplementary Fig. 7). Even though both rationally designed mutants showed a marked increase in the on-target specificity with respect to wild-type SpCas9 (Fig. 3d), evoCas9 showed the highest improvement, with approximately 70% of all GUIDE-seq reads being captured by the on-target sites, followed by eSpCas9(1.1) and SpCas9-HF1 with 60% and 55% total on-target GUIDE-seq reads, respectively. In addition, the few off-target sites that were still cleaved by evoCas9 were also detected among the ones generated by SpCas-HF1 (12 out of 13 sites, Fig. 3e) and eSpCas9(1.1) (13 out of 13 sites, Fig. 3e). To further evaluate the performance of the different high-fidelity variants we calculated the on-/off-target ratio relative to the 12 off-sites shared by wild-type SpCas9 and the three variants (Fig. 3f). Consistently with previous results, evoCas9 showed a significant increase in the ratios when compared to wild-type SpCas9 (p=0.0002), as well as with the two previously published mutants (p=0.00002 for eSpCas9(1.1) and p=0.008 for SpCas9-HF1), indicating superior specificity on difficult-to-discriminate off-target sites (Fig. 3f).
Figure 3. evoCas9 shows unprecedented specificity.
(a-f) GUIDE-seq based comparative analyses of evoCas9, wild-type SpCas9 (WT), SpCas9-HF1, and eSpCas9(1.1) for eight sgRNAs. (a) Total number of off-target sites for each of the tested sgRNA (see Supplementary Fig. 5-6 and Supplementary Data 1-2). (b) Radar plot showing the distribution of the number of mismatches (from 1 to 7) for the off-targets detected by GUIDE-seq. (c) Total number of off-target sites for each type of SpCas9 captured by GUIDE-seq with all the tested sgRNAs. (d) Global on-target cleavage specificity expressed by the percentages of the on-target reads captured by GUIDE-seq with tested sgRNAs. The on-target specificities for each site are reported in Supplementary Fig. 7. (e) Venn diagram showing the intersection among all the off-targets identified by GUIDE-seq. The inset reports the distribution of the off-target sites shared by wild-type SpCas9, SpCas9-HF1, eSpCas9(1.1) and evoCas9. (f) On- versus off-target ratios of GUIDE-seq reads relative to the 12 shared off-targets (see inset of (e)). (g-h) Indel analysis by targeted deep-sequencing of nine VEGFA2 off-targets (OT1-9) identified by GUIDE-seq and shared by wild-type SpCas9 (WT), SpCas9-HF1, eSpCas9(1.1) and evoCas9. Cells not expressing SpCas9 were sequenced to determine background indel levels. Two independent experiments were performed by mixing genomic DNA from three biological replicates before library preparation. For each off-target in (g), the solid circle corresponds to the mean of the editing percentage, while the shaded circles represent each experiment. (h) For each type of SpCas9, on- versus off-target ratios calculated from the mean editing levels measured by targeted deep-sequencing of the nine common VEGFA2 off-target sites. See Supplementary Fig. 8a for indel percentages.(i) Long-term specificity of evoCas9. Stable expression of wild-type SpCas9, evoCas9 and SpCas9-HF1 in 293blastEGFP cells transduced with lentiviral vectors. Each lentiviral vector also expressed the on-target sgRNA (sgGFPon) or a mismatched sgRNAs (sgGFP1314, sgGFP1819 or sgGFP18). EGFP knockout was evaluated by FACS analysis at the time points indicated in the graphs. Cells were kept under puromycin selection for the whole duration of the experiments. n=3 biologically independent samples except n=2 for WT and SpCas9-HF1 sg1819 (10 days) and evoCas9 sg1314 and sg1819 (all time points). Individual samples are reported as empty shaded symbols, while the overlaid solid symbol indicates the mean EGFP knockout level. In panels (f) and (h) statistical significance was assessed using the two-sided paired Wilcoxon signed-rank test, sample size n=12 in (f) and n=9 in (h). Summary of data distributions and statistical details are reported in Supplementary Table 5.
To confirm these findings, we directly assessed indel formation induced by wild-type SpCas9, evoCas9, SpCas9-HF1 and eSpCas9 (1.1) at the nine common off-target sites associated with the editing of the VEGFA2 locus (Fig. 3e) by using targeted deep-sequencing. Consistently with GUIDE-seq data, the two rationally designed variants generated indels clearly above background for all the tested sites (Fig. 3g). In contrast, we measured near-background levels of unspecific editing in for evoCas9 at approximately half of the loci (OT3,7,8,9, Fig. 3g) and at least 10 times less off-target activity than the other variants for three other loci (OT2, 5, 6, Fig. 3g); only at two sites (OT1,4) the three different mutants produced comparable levels of unspecific cleavages (Fig. 3g). The increased specificity of evoCas9 is further evident from the on-/off-target ratios calculated from the targeted deep-sequencing data (Fig. 3h and Supplementary Fig. 8a).
Additional targeted deep-seq analyses were performed on the EMX1 and VEGFA3 loci (Supplementary Fig. 8b,c). Consistent with the GUIDE-seq results, we did not measure any unspecific cleavage for most of the tested sites except for the previously detected VEGFA3 off-target site (OT1), where evoCas9 showed the highest specificity, and three additional sites associated with the same locus (OT4,8,10) where only eSpCas9(1.1) showed measurable unspecific indels (Supplementary Fig. 8c). Finally, evoCas9 high specificity was further confirmed through the evaluation of indel formation at difficult-to-discriminate off-target sites associated with the CCR5 and FANCF sgRNAs by Tracking of Indels by Decomposition (TIDE)23 analysis (Supplementary Fig. 9).
We next investigated the off-target activity associated with long-term evoCas9 expression by using a cellular EGFP-knockout model (293blastEGFP cells) and the set of mismatched sgRNAs described above (sgGFP1314, sgGFP1819, sgGFP18). Evaluation of the loss of EGFP fluorescence at different time points after stable transduction revealed that, whereas wild-type SpCas9 induced rapid accumulation of unspecific cleavages with mismatched sgRNAs, evoCas9 completely abolished the off-target activity generated by two out of three tested sgRNAs (sgGFP1314 and sgGFP1819). Notably, even at early time points (10 days), both wild-type SpCas9 and SpCas9-HF1 were not able to discriminate the most stringent surrogate off-target (sgGFP18) from the specific site, whereas evoCas9 generated approximately half unspecific GFP knockouts in comparison to the other SpCas9 variants at 40 days post-transduction (Fig. 3i). These differences in specificity did not correlate with the variability of the intracellular levels of the different variants (Supplementary Fig. 3c), as observed in transient transfection experiments (Supplementary Fig. 3a).
This study reports the discovery of evoCas9, a SpCas9 variant with very high specificity compared to the wild-type nuclease. A comparison with the best characterized rational high-fidelity variants revealed that evoCas9 is more specific and preserves full catalytic activity across the majority of the tested sites.
In contrast to entirely rational approaches based on loss of function alanine substitution, our strategy consisted in the in vivo selection of mutations within the REC3 domain suitable to generate high-fidelity SpCas9 nucleases followed by rational optimization to isolate the best performing combination. The advantage offered by this approach is particularly evident when considering that SpCas9-HF1 contains three out of four mutations in the same REC3 domain and shares a substituted residue with evoCas9 (R661), but is nevertheless less specific than the in vivo selected evoCas9.
Similarly, our screening approach correctly predicted the importance of a conformational cluster of mutations (Fig. 1e, panel B) that increased the specificity of a new SpCas9 variant, HypaCas9, reported while our manuscript was under review19. The comparison of evoCas9 with HypaCas9 through meta-analysis on three genomic loci (VEGFA2, VEGFA2 and FANCF2) suggested that, overall, evoCas9 is more specific, generating less off-target sites (12 vs. 20 total off-target loci for evoCas9 and HypaCas9, respectively) and showing a better on-/off-target ratio for 7 out of 9 commonly detected off-targets (Supplementary Fig.10). In agreement with this recent report the increased specificity of evoCas9 does not depend on decreased affinity for the DNA substrate (Supplementary Note and Supplementary Fig. 11).
Comparing the ratio between on- and off-target cleavage rates for our mutant, as well as for SpCas9-HF1 and eSpCas9(1.1), demonstrates the existence of a trade-off between specificity and cleavage activity: increased specificity comes along with decreased editing efficiency. We attempted to overcome this limitation by optimizing the interactions between SpCas9 and the sgRNA through rationally designed alternative substitutions as occurred with the VNEL, VNES and evoCas9 variants (see also Supplementary Fig. 2c). It is likely that further improvement in targeting specificity could be obtained by either screening domains other than the REC3 or by combining the evoCas9 substitutions with other not yet investigated mutations derived from our screening. Furthermore, our yeast platform can be easily adapted to improve the specificity of emerging homologous and non-homologous RNA-guided endonucleases24–27. On the same line, our screening platform can be applied for the development of SpCas9 variants tailored on specific loci with therapeutic potential poorly discriminated from non-specific sites or variants differentiating closely related alleles.
Reduced off-target activity is considered an essential step towards a safe clinical use of SpCas9. Accordingly, evoCas9 eliminates 98.7% of the detectable off-targets associated with both standard and clinically relevant genomic loci, while retaining near-wild-type levels of on-target activity. The advancement of the CRISPR toolbox towards a safe and efficient in vivo use with our variant is further evident from the limited accumulation of off-target cleavages observed after its long-term expression through a viral-based delivery. So far, off-target accumulation associated with SpCas9 long-term expression was limited by mRNA8,9 or RNP complexes5,7 delivery techniques that are nevertheless limiting in terms of efficiency and in vivo use. These delivery techniques could also be combined with evoCas9 if further increases in specificity are desired.
Overall, this study provides evidence that the unbiased screening for SpCas9 mutants generates optimized genomic surgery tools towards more controlled biotechnological and clinical applications.
Methods
Additional information on experimental design, software, materials and reagents can be found in the Life Sciences Reporting Summary.
Plasmids
The plasmid p415-GalL-Cas9-CYC1t was used to express Cas9 in yeast (Addgene #43804)28. To allow the precise removal of the REC3 domain by restriction digest, synonymous mutations were generated by PCR to introduce two restriction sites, NcoI and NheI, upstream and downstream of the REC3 domain, respectively (for primers, see Supplementary Table 2). The expression cassette for the sgRNA was obtained from the p426-SNR52p-gRNA.CAN1.Y-SUP4t plasmid (Addgene #43803)28. In order to swap the original spacer sequence with the desired target, an assembly-PCR based strategy was adopted. The resulting fragment was blunt-end cloned into pRS316, a low copy number centromeric plasmid carrying an URA3 yeast selectable marker, pre-digested with SacII/XhoI and blunted, generating the pRS316-SNR52p-gRNA.ON-SUP4t plasmid.
For the expression of SpCas9 in mammalian cells we employed a pX330 (Addgene #42230)29 derived plasmid, where the sgRNA coding cassette has been removed by NdeI digestion, pX-Cas910. The plasmids coding for improved Cas9 variants were obtained by sequential site-directed mutagenesis starting from the pX-Cas9 plasmid. For the expression of previously published enhanced SpCas9 mutants the VP12 (Addgene #72247)4 and the eSpCas9(1.1) (Addgene #71814)3 plasmids were used. Desired spacer sequences were cloned as annealed oligonucleotides with appropriate overhangs into a double BbsI site located upstream the guide RNA constant portion in a pUC19 plasmid containing a U6 promoter-driven expression cassette10. The same pUC19 plasmid containing an optimized guide RNA constant region (stem extension and base flip)22 was used for the preparation of optimized sgRNAs. For the experiments involving lentiviral vectors, the lentiCRISPRv1 transfer vector (Addgene #49535)30 was employed together with the pCMV-delta8.91 packaging vector and pMD2.G, coding for the vesicular stomatitis virus glycoprotein (VSVG), to produce viral particles. Annealed oligonucleotides corresponding to the desired spacers were cloned into the guide RNA expression cassette using a double BsmBI site. The lentiCRISPRv1-based vectors coding for enhanced SpCas9 variants were generated by swapping part of the SpCas9 coding sequence with a PCR fragment corresponding to the region of the CDS containing the mutations (for primers, see Supplementary Table 2). The pEGFP-IRES-Puro plasmid was generated by subcloning the EGFP ORF from pEGFP-N1 (Clontech) to the pIRESpuro3 plasmid (Clontech). dCas9-VP64 was expressed from pcDNA-dCas9-VP6431. evo-dCas9-VP64 was generated by sequential mutagenesis of the original plasmid. The pTRE-GFP plasmid was obtained by subcloning the EGFP coding sequence from the pEGFP-N1 (Clontech) plasmid into the pTRE-Tight cloning vector (Clontech). A complete list of the sgRNA target sites is available in Supplementary Table 3.
Yeast culture
The yLFM-ICORE yeast strain32,33 was used to generate the reporter yeast strains used in this study. Synthetic minimal media (SD) were employed in all yeast experiments. Single amino acids were omitted according to the experimental setup, when selective medium was required. For the induction of Cas9 expression, 20 g/L D-(+)-galactose and 10 g/L D-(+)-raffinose were used instead of dextrose. Specific medium for ade2 mutants colour screening was prepared using low adenine concentrations (5 mg/L). When non-selective medium was required, YPDA rich medium was employed. All chemicals to prepare yeast media were obtained from Sigma-Aldrich. Yeasts were transformed using the lithium acetate/single strand carrier DNA/PEG method34. After transformation, cells were resuspended in the appropriate SD selective medium or directly plated on selective SD agarose plates and incubated at 30°C. For spontaneous reversion frequency evaluation, after transformation with p415-GalL-Cas9-CYC1t, cells were grown in selective medium for 24 hours. The concentration of cells was then evaluated by measuring the OD600 and 1000 cells were plated on selective plates depleted of leucine or 106 cells were spread on plates further depleted of adenine of tryptophan, to evaluate the number of revertants for each locus.
Colony PCRs were performed after cell-wall digestion with 10 U of lyticase (Sigma-Aldrich) for 30 minutes at 30 °C using the Phusion High-Fidelity DNA Polymerase (Thermo Scientific).
Yeast screening for SpCas9 mutants
Yeast reporter strains were generated using the delitto perfetto approach to edit the ADE2 and TRP1 genomic loci, according to published protocols35. Briefly, the coding sequence of each of the two genes was substituted with an in vitro assembled reporter cassette containing the coding sequence split in half by the insertion of the desired target sequence flanked by a 100bp duplicated region (Fig. 1a). The primers used to amplify the genomic fragments necessary for cassette assembly are reported in Supplementary Table 2. The newly generated yeast strains were called yACMO-off1, yACMO-off2, yACMO-off3 and yACMO-off4, and were characterized by a selected on-target sequence in the TRP1 locus and four different off-target sequences in the ADE2 locus, each containing a single mismatch with respect to the on-target sequence in a position that is more PAM-proximal for off1 and more PAM-distal for off4 (see Supplementary Table 2).
The mutants’ library was generated by error-prone PCR (epPCR) using the GeneMorph II kit (Agilent). Following the manufacturer's instructions, the initial amount of template DNA (p415-GalL-Cas9-CYC1t) and the number of cycles were set to obtain an average of 4-5 mutations per kilobase. 50 bp-long primers were selected to anneal 150 bp upstream and downstream of the REC3 coding sequence (see Supplementary Table 2). The PCR library was directly assembled in vivo by co-transformation of the mutagenized amplicon pool with the p415-GalL-Cas9-CYC1t plasmid, previously digested with NcoI and NheI to remove the REC3 domain, with an insert/plasmid ratio of 3:1. The mutagenic library was screened concomitantly to its assembly by co-transformation of the fragments in the yACMO-off4 yeast strain stably expressing a sgRNA towards the on-target sequence located in the TRP1 locus. After transformation, the culture was grown overnight in SD medium lacking uracil and leucine, for selecting cells carrying both the sgRNA- and Cas9-expressing plasmids, to allow recovery and correct recombination. The next day, Cas9 expression was induced by growing the culture in galactose-containing medium for 5 hours prior to plating on several selective plates lacking tryptophan and containing low concentrations of adenine, to discriminate colonies according to the editing status of the TRP1 and ADE2 loci. After 48 hours, TRP1+/ade2- (red) colonies were streaked on selective plates with low adenine and no tryptophan containing galactose and raffinose to keep Cas9 expression constitutively induced and force the generation of off-target cleavages. After further 48 hours of incubation, Cas9-expressing plasmids were extracted from the most red pigmented streaks, corresponding to colonies in which Cas9 cleaved only the on-target site, and the mutations were characterized by Sanger sequencing. To isolate the mutant Cas9 plasmids from yeast, single colonies were grown overnight in SD medium without leucine, to select for the presence of the p415-GalL-Cas9-CYC1t plasmid, while relaxing the selection on the sgRNA-expressing plasmid to induce its dilution and loss. Cells were then mechanically lysed using acid-washed glass beads (Sigma-Aldrich) and plasmid DNA was recovered using standard miniprep silica columns (Macherey-Nagel), treated with NcoI and NheI (New England Biolabs) to digest residual sgRNA-expressing vectors and transformed using chemically competent E. coli. The theoretical complexity of the REC3 mutagenic library, based on an estimation of the yeast transformants obtained, is close to 105. The acquired plate images were analyzed with OpenCFU36. For all images an inverted threshold (value = 2) was used with a radius between 8 and 50 pixels. Discrimination between white and red colonies was obtained by computing the average signal in the RGB channels and setting a manual threshold that accurately discriminates between red and white colonies in each experiment.
Mammalian cells and transfections
293T/17 cells were obtained from the American Type Culture Collection (ATCC) and were cultured in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% fetal calf serum (Life Technologies) and antibiotics (Life Technologies). 293multiEGFP cells were generated by stable transfection of 293T/17 with pEGFP-IRES-Puromycin and selected with 1 μg/ml of puromycin. 293blastEGFP, used in lentiCRISPR transduction experiments to allow selection with puromycin, were obtained by low MOI infection of 293T/17 cells with an EGFP-expressing lentiviral vector carrying a blasticidin resistance gene followed by clonal selection with 5 μg/ml of blasticidin. All cell lines were verified mycoplasma-free (PlasmoTest, Invivogen). For transfection, 1×105 293multiGFP or 293T/17 cells/well were seeded in 24-well plates and transfected the next day using TransIT-LT1 (Mirus Bio) according to manufacturer’s protocol with 400-750 ng of Cas9-expressing plasmids and 200-250 ng of sgRNA-expressing plasmids. For transient transcriptional activation experiments 100 ng of the pTRE-EGFP plasmid were used. To determine the level of EGFP downregulation by Cas9 after transfection into 293multiGFP, cells were collected 7 days post-transfection and were analysed by flow cytometry using a FACSCanto (BD Biosciences). In transcriptional activation experiments, cells were collected and analysed at 2 days post-transfection.
Lentiviral vector production and transductions
Lentiviral particles were produced by transfecting approximately 8x106 293T/17 cells with 10 μg of each lentiCRISPR-based transfer vector together with 6.5 μg of pCMV-deltaR8.91 packaging vector and 3.5 μg of pMD2.G using the polyethylenimine (PEI) method. After 48 hours the supernatants containing the viral particles was collected and filtered through a 0.45 μm PES filter. Quantification of the vector titers was performed using the SG-PERT method37. Vectors stocks were conserved at -80°C for future use.
For transductions, 105 293blastGFP cells were seeded in a 24-well plate and the next day were transduced with 0.4 Reverse Transcriptase Units (RTU)/well of each vector by centrifuging at 1600xg 16°C for 2 hours. Cells were kept in culture for a total of 48 hours before adding 0.5 μg/ml puromycin selection that was maintained throughout the experiment. To determine the level of EGFP downregulation by Cas9 after infection, 293blastGFP cells were collected at the indicated time-points after transduction and were analysed by flow cytometry using a FACSCanto (BD Biosciences).
Detection of Cas9-induced genomic mutations
Genomic DNA was obtained at 7 days post-transfection, using the QuickExtract DNA extraction solution (Epicentre). PCR reactions to amplify genomic loci were performed using the Phusion High-Fidelity DNA polymerase (Thermo Fisher). Samples were amplified using the oligos listed in Supplementary Table 4. Purified PCR products were analyzed by sequencing and applying the TIDE tool38. To quantify the CCR2-CCR5 chromosomal deletion, a semi-quantitative PCR approach was set-up using primers flanking the CCR5 on-target site and the CCR2 off-target locus (Supplementary Table 4). Quantifications were obtained by densitometric analyses using the ImageJ software and exploiting the FANCF genomic locus as an internal normalizer.
Western blots
Cells were lysed in NEHN buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 0.5% NP40, NaCl, 1 mM EDTA, 20% glycerol supplemented with 1% of protease inhibitor cocktail (Pierce). Cell extracts were separated by SDS-PAGE using the PageRuler Plus Protein Standards as the standard molecular mass markers (Thermo Fisher Scientific). After electrophoresis, samples were transferred to 0.22 μm PVDF membranes (GE Healthcare). The membranes were incubated with primary antibodies mouse anti-FLAG® M2 (Sigma) for detecting SpCas9 and the different high-fidelity variants, and mouse anti-α-tubulin (Sigma) or mouse anti-actin (Sigma) for a loading control and with HRP conjugated goat anti-mouse (KPL) secondary antibodies for ECL detection. Images were acquired using the UVItec Alliance detection system.
GUIDE-seq
2x105 293T/17 cells were transfected with 750 ng of each Cas9 expressing plasmid, together with 250 ng of each sgRNA-coding plasmid or an empty pUC19 plasmid, 10 pmol of the bait dsODN containing phosphorothioate bonds at both ends (designed according to the original GUIDE-seq protocol6) and 50 ng of a pEGFP-IRES-Puro plasmid, expressing both EGFP and the puromycin resistance gene. The day after transfection cells were detached and selected with 2 μg/ml of puromycin for 48 hours to eliminate non-transfected cells. Cells were then collected and genomic DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen) following the manufacturer’s instructions and sheared to an average length of 500bp with the Bioruptor Pico sonication device (Diagenode). Library preparations were performed with the original adapters and primers according to previous work6. Libraries were quantified with the Qubit dsDNA High Sensitivity Assay kit (Invitrogen) and sequenced with the MiSeq sequencing system (Illumina) using an Illumina Miseq Reagent kit V2 - 300 cycles (2x150bp paired-end).
Raw sequencing data (FASTQ files) were analyzed using the GUIDE-seq computational pipeline39. After demultiplexing, putative PCR duplicates were consolidated into single reads. Consolidated reads were mapped to the human reference genome GrCh37 using BWA-MEM40; reads with mapping quality lower than 50 were filtered out. Upon the identification of the genomic regions integrating double-stranded oligodeoxynucleotide (dsODNs) in aligned data, off-target sites were retained if at most seven mismatches against the target were present and if absent in the background controls. Visualization of aligned off-target sites is available as a color-coded sequence grid.
To demonstrate the reproducibility of the GUIDE-seq method a replicate experiment on the VEGFA3 sgRNA was performed for wild-type SpCas9. In addition, our datasets were compared with the ones available in literature (Supplementary Fig. 12).
Targeted deep-sequencing
Selected off-target sites41 for the VEGFA2, VEGFA3 and EMX1 genomic loci, together with their relative on-target, were amplified using the Phusion high-fidelity polymerase (Thermo Scientific) or the EuroTaq polymerase (Euroclone) from 293T genomic DNA extracted 7 days after transfection with wild-type SpCas9 or evoCas9 together with sgRNAs targeting the EMX1, the VEGFA2 or VEGFA3 loci, or a pUC empty vector. Off-target amplicons were pooled in near-equimolar concentrations before purification and indexing. Libraries were indexed by PCR using Nextera indexes (Illumina), quantified with the Qubit dsDNA High Sensitivity Assay kit (Invitrogen), pooled according to the number of targets and sequenced on an Illumina Miseq system using an Illumina Miseq Reagent kit V3 - 150 cycles (150bp single read). The complete primer list used to generate the amplicons is reported in Supplementary Table 4.
A reference genome was built using Picard (http://broadinstitute.github.io/picard) and samtools42 from DNA sequences of the considered on-/off-target regions. Raw sequencing data (FASTQ files) were mapped against the created reference genome using BWA-MEM40 with standard parameters and resulting alignment files were sorted using samtools. Only reads with mapping quality above or equal to 30 were retained. The presence of indels in each read for each considered region was determined by searching indels of size 1bp directly adjacent to the predicted cleavage site or indels of size >=2bp overlapping flanking regions of size 5bp around the predicted cleavage site.
Supplementary Material
Acknowledgements
The authors wish to thank the LaBSSAH - CIBIO Next Generation Sequencing Facility of the University of Trento for sequencing samples. We are grateful to Daniele Arosio for helpful discussion throughout the development of this study. This work was supported by intramural funding from the University of Trento and by the European Research Council grant ERCCoG648670 (F.D.).
Footnotes
Code availability. Indels identification in targeted deep-seq data analysis was performed implementing a script in the R language that is available upon request.
Data availability. GUIDE-seq and targeted deep-sequencing data have been deposited at BioProject (https://www.ncbi.nlm.nih.gov/bioproject/) under the accession number PRJNA423000. All other relevant data are available from the authors upon request.
Author contributions
A.Ca., M.O., C.M. and G.R. designed and performed the experiments; A.Ca., M.O., G.R., C.M., G.M. and G.P. collected and analyzed the data; F.L., D.P., A.R. and F.D. contributed with GUIDE-seq experiments and targeted deep-sequencing analysis; A.I. contributed with the yeast assay design and setup. A.Ca., M.O., G.P. and A.C. conceived and designed the study, wrote and edited the paper; A.C. was responsible for the coordination of the study. All authors read, corrected, and approved the final manuscript.
Additional information
Competing financial interests: The authors declare competing financial interests. A patent has been filed for the high-specificity Cas9 variants.
References
- 1.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nuñez JK, Harrington LB, Doudna JA. Chemical and Biophysical Modulation of Cas9 for Tunable Genome Engineering. ACS Chem Biol. 2016;11:681–688. doi: 10.1021/acschembio.5b01019. [DOI] [PubMed] [Google Scholar]
- 3.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleinstiver BP, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S, Kim D, Cho SW, Kim J, Kim JS. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Research. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2014;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramakrishna S, et al. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Research. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang X, et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. Journal of Biotechnology. 2015;208:44–53. doi: 10.1016/j.jbiotec.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 9.Yin H, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Petris G, et al. Hit and go CAS9 delivered through a lentiviral based self-limiting circuit. Nat Comms. 2017;8 doi: 10.1038/ncomms15334. 15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ran FA, et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen B, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 14.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolukbasi MF, et al. DNA-binding-domain fusions enhance the targeting range and precision of Cas9. Nat Methods. 2015;12:1150–1156. doi: 10.1038/nmeth.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim D, Kim S, Kim S, Park J, Kim J-S. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Research. 2016;26:406–415. doi: 10.1101/gr.199588.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen JS, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017 doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyon Y, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proceedings of the National Academy of Sciences. 2010;107:12028–12033. doi: 10.1073/pnas.0914991107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen B, et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HK, et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat Methods. 2017;14:153–159. doi: 10.1038/nmeth.4104. [DOI] [PubMed] [Google Scholar]
- 26.Burstein D, et al. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542:237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shmakov S, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Micro. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiCarlo JE, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Research. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jegga AG, Inga A, Menendez D, Aronow BJ, Resnick MA. Functional evolution of the p53 regulatory network through its target response elements. Proc Natl Acad Sci U S A. 2008;105:944–949. doi: 10.1073/pnas.0704694105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomso DJ, et al. Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proceedings of the National Academy of Sciences. 2005;102:6431–6436. doi: 10.1073/pnas.0501721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 35.Stuckey S, Storici F. Gene knockouts, in vivo site-directed mutagenesis and other modifications using the delitto perfetto system in Saccharomyces cerevisiae. Meth Enzymol. 2013;533:103–131. doi: 10.1016/B978-0-12-420067-8.00008-8. [DOI] [PubMed] [Google Scholar]
- 36.Geissmann Q. OpenCFU, a New Free and Open-Source Software to Count Cell Colonies and Other Circular Objects. PLoS ONE. 2013;8:e54072–10. doi: 10.1371/journal.pone.0054072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casini A, Olivieri M, Vecchi L, Burrone OR, Cereseto A. Reduction of HIV-1 infectivity through endoplasmic reticulum-associated degradation-mediated Env depletion. Journal of Virology. 2015;89:2966–2971. doi: 10.1128/JVI.02634-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Research. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai SQ, Topkar VV, Joung JK, Aryee MJ. Open-source guideseq software for analysis of GUIDE-seq data. Nat Biotechnol. 2016;34:483. doi: 10.1038/nbt.3534. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.