Abstract
Group VII ethylene response factors (ERF-VIIs) regulate transcriptional adaptation to flooding-induced hypoxia in plants. ERF-VII stability is controlled in an O2-dependent manner by the Cys/Arg branch of the N-end rule pathway whereby oxidation of a conserved N-terminal cysteine residue initiates target degradation. This oxidation is catalyzed by plant cysteine oxidases (PCOs), which use O2 as cosubstrate to generate Cys-sulfinic acid. The PCOs directly link O2 availability to ERF-VII stability and anaerobic adaptation, leading to the suggestion that they act as plant O2 sensors. However, their ability to respond to fluctuations in O2 concentration has not been established. Here, we investigated the steady-state kinetics of Arabidopsis thaliana PCOs 1–5 to ascertain whether their activities are sensitive to O2 levels. We found that the most catalytically competent isoform is AtPCO4, both in terms of responding to O2 and oxidizing AtRAP2.2/2,12 (two of the most prominent ERF-VIIs responsible for promoting the hypoxic response), which suggests that AtPCO4 plays a central role in ERF-VII regulation. Furthermore, we found that AtPCO activity is susceptible to decreases in pH and that the hypoxia-inducible AtPCOs 1/2 and the noninducible AtPCOs 4/5 have discrete AtERF-VII substrate preferences. Pertinently, the AtPCOs had Km(O2)app values in a physiologically relevant range, which should enable them to sensitively react to changes in O2 availability. This work validates an O2-sensing role for the PCOs and suggests that differences in expression pattern, ERF-VII selectivity, and catalytic capability may enable the different isoforms to have distinct biological functions. Individual PCOs could therefore be targeted to manipulate ERF-VII levels and improve stress tolerance in plants.
Keywords: hypoxia, enzyme kinetics, protein degradation, Arabidopsis, plant biochemistry, post-translational modification (PTM), ERF-VII, N-end rule, oxygen-sensing, plant cysteine oxidase
Introduction
Plants are sessile organisms that require sufficient mechanisms to effectively respond to low-oxygen conditions (hypoxia), such as those experienced during submergence or waterlogging, to survive. Molecular adaptation to hypoxia is mediated by group VII ethylene response factors (ERF-VIIs),5 transcriptional activators that up-regulate the expression of core anaerobic genes during periods of low oxygen (1, 2). At a physiological level, this can result in an escape strategy whereby petioles/shoots elongate above the surface to improve O2 supply (mediated by the ERF-VIIs SNORKEL 1 and SNORKEL 2 in deep-water rice varieties (3)) or in a quiescence response whereby plants enter a temporary anaerobic state to conserve O2 reserves until floodwaters subside (mediated by the ERF-VIIs HRE1/2 and RAP2.2 in Arabidopsis (4, 5)).
Five members of the ERF-VII family have been identified in Arabidopsis thaliana (RAP2.2, RAP2.3, RAP2.12, HRE1, and HRE2), all of which have the conserved N-terminal motif CGGA(I/V)ISD(F/Y) following cotranslational methionine excision. ERF-VII stability is controlled in an O2-dependent manner by the Cys/Arg branch of the N-end rule pathway (6, 7). During hypoxia, the ERF-VIIs can translocate to the nucleus to promote the expression of anaerobic genes. Under normoxic conditions, their N-terminal cysteine residues are oxidized, promoting subsequent arginylation (8), ubiquitination (9), and degradation (2, 10) of the target by arginyltransferases, the E3 ligase proteolysis 6, and the 26S proteasome, respectively. Furthermore, as well as regulating the plant hypoxic response, N-end rule–mediated ERF-VII stability has been linked to other abiotic stress responses (2, 11, 12).
ERF-VII N-terminal cysteine oxidation has been shown to be enzymatically regulated in vitro and in Arabidopsis by the plant cysteine oxidases (8, 13). These monomeric, nonheme iron (Fe2+)-dependent dioxygenases use both atoms of molecular O2 to promote the formation of Cys-sulfinic acid, which is a suitable and sufficient substrate for arginyltransferase 1–catalyzed arginylation, at the N terminus of their ERF-VII targets (8). The direct use of molecular O2 to promote the proteasomal degradation of primary anaerobic response elements has led to the suggestion that the PCOs function as plant O2 sensors (13). In animals, the hypoxic response is coordinated by hypoxia-inducible transcription factor (HIF) levels, which are regulated by Fe2+, 2-oxoglutarate, and O2-depedendent HIF prolyl hydroxylase enzymes (PHDs) in an analogous manner to the ERF-VIIs and PCOs (14). Hydroxylation of specific prolyl residues in HIF-α (coupled with 2-oxoglutarate decarboxylation to succinate and CO2) targets HIF-α for proteasomal degradation (14). These enzymes are termed O2 sensors due to the dependence of their rate of activity on O2 availability. This is biochemically defined by high Km(O2)app values, particularly for the primary O2 sensor, PHD2 (240–1700 μm (15–18)). For the PCOs to effectively function in the same capacity, their rate of activity must also be sensitive to O2 availability in a physiologically relevant manner.
In this study, we sought to determine whether the PCOs have the biochemical potential to act as O2 sensors via a steady-state kinetic investigation. We have optimized assay conditions for analyzing PCO activity and determined the kinetic parameters of each A. thaliana PCO isoform (AtPCO) in terms of pH, a peptide representing the N termini of the AtERF-VIIs AtRAP2.2/2.12, and O2. Importantly, we report that the AtPCOs have Km(O2)app values that fall within a physiologically relevant range, implicating their biochemical capacity to act as O2 sensors in plants. We also found that each isoform possesses distinct catalytic capability and substrate specificity, which may correlate with expression behavior. Consequently, our work provides biochemical evidence supporting the assertion that the AtPCOs act as plant O2 sensors and that suggests that different isoforms could have divergent biological functions.
Results
Recombinant production of AtPCOs 1–5
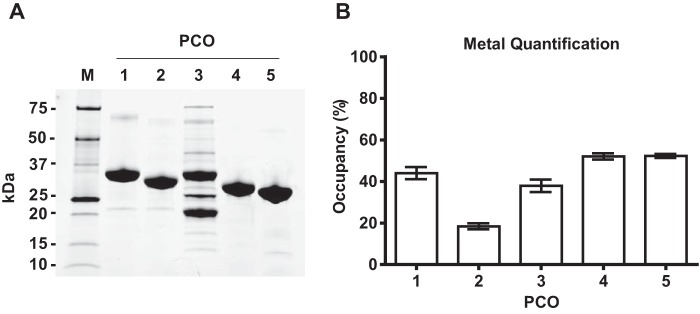
We have previously reported the recombinant production of A. thaliana AtPCOs 1 and 4 to over 90% purity by immobilized nickel-affinity chromatography (8). In the current study, all five AtPCO isoforms were subcloned into the expression vector pET28a and purified with an additional size-exclusion chromatography step to improve protein quality. Each AtPCO was obtained at over 95% purity (Fig. 1A) except for AtPCO3, which appeared to be ∼50% pure by SDS-PAGE despite generating a size-exclusion elution profile similar to the other isoforms. Proteomic analysis suggests that the additional bands correspond to degraded fragments of AtPCO3 and contaminating Escherichia coli proteins (data not shown). Consistent with previous findings (8), iron quantification using a ferrozine-like assay revealed that each AtPCO isoform binds substoichiometric amounts of Fe2+ (∼20–50%; Fig. 1B) with AtPCO2 possessing the lowest measure of metal.
Figure 1.
Purification and iron content of AtPCOs 1–5. A, SDS-PAGE analysis showing purified AtPCO isoforms 1–5 following sequential steps of immobilized nickel-affinity and size-exclusion chromatography with M indicating the molecular mass marker. Each AtPCO protein migrated to the expected mass, demonstrating over 95% purity, except for AtPCO3, which was estimated to be ∼50% pure. This was accounted for in all activity measurements. B, the iron content of each AtPCO was quantified using a ferrozine-like assay, allowing metal occupancy to be estimated from known protein concentrations. All samples possessed substoichiometric amounts of cofactor with AtPCO2 containing the least iron. Error bars display S.D. (n = 3).
AtPCOs 1–5 catalyze oxidation of AtRAP2(2–15)
AtPCOs 1 and 4 can catalyze the dioxygenation of cysteines situated at the N terminus of 10-mer peptides representing AtERF-VII substrates (8). For this study, we wished to confirm that all five AtPCO isoforms can catalyze the oxidation of cysteines positioned at the N terminus of a 14-amino acid–long peptide corresponding to the methionine excised N terminus of AtRAP2.2 and AtRAP2.12 (CGGAIISDFIPPPR), hereby referred to as AtRAP2(2–15) (14-mer peptides allowed distinction between AtRAP2(2–15) and other AtERF-VII peptides; see below). AtRAP2.2 and AtRAP2.12 have been reported as two of the most prominent activators of anaerobic genes in Arabidopsis; therefore, understanding their regulation will provide valuable information on molecular adaptation to hypoxia in plants (5, 19, 20).
AtRAP2.2(2–15) was incubated with AtPCOs 1–5 at 25 °C for 30 min before being quenched with 1% formic acid and analyzed by liquid chromatography–coupled mass spectrometry (LC-MS). As expected, AtRAP2(2–15) was a suitable substrate for all AtPCO isoforms with complete conversion observed in each sample (Fig. S1).
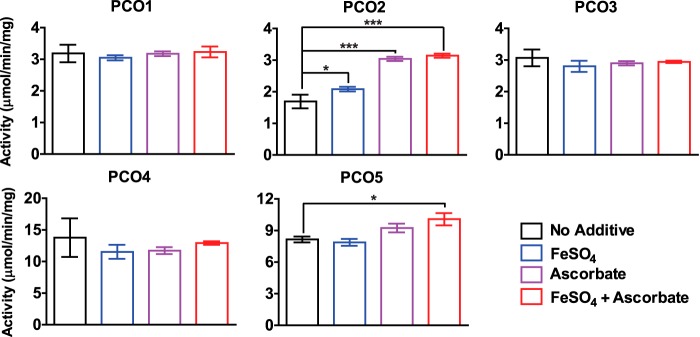
We subsequently determined whether the addition of exogenous Fe2+ and/or ascorbate would enhance the activity of the AtPCOs, analogous to other nonheme iron–dependent oxygenases (21, 22), by comparing the specific activities of AtPCOs 1–5 with and without FeSO4, ascorbate, or both. Stopped assays were conducted at 0-, 30-, 60-, and 90-s intervals to monitor the initial rate of AtRAP2(2–15) oxidation by LC-MS. The addition of both Fe2+ and ascorbate significantly increased the specific activity of AtPCO2 and to a lesser extent AtPCO5 but had little or no effect on the activity of the other isoforms (Fig. 2). In the case of AtPCO2, this could be attributed to enhanced iron occupancy, compensating for the low quantities estimated in Fig. 1B. In light of these results, both additives were retained in all subsequent reactions to facilitate optimal AtPCO activity, and turnover rates are reported per mg of total enzyme present. It is important to note that the specific activities of each isoform varied significantly under equivalent conditions with AtPCOs 4 and 5 catalyzing AtRAP2(2–15) oxidation at ∼13 and 10 μmol/min/mg, respectively, compared with AtPCOs 1, 2, and 3, which processed AtRAP2(2–15) at around 3 μmol/min/mg (taking into account the 50% purity of AtPCO3).
Figure 2.
Analyzing the influence of exogenous iron and ascorbate on AtPCO activity. The specific activities of AtPCOs 1–5 were determined with and without FeSO4 (100-fold in excess of enzyme concentration) and sodium ascorbate (1 mm) by measuring the rate of AtRAP2(2–15) cysteine oxidation at regular time intervals (0, 30, 60, and 90 s) by LC-MS. The addition of exogenous additives, particularly ascorbate, significantly increased the specific activities of AtPCO2 and, to a lesser extent, AtPCO5 but had little effect on the other isoforms. AtPCOs 4 and 5 had the greatest specific activity. Reactions were conducted in 50 mm HEPES, 50 mm NaCl, and 1 mm TCEP, pH 7.5, at 25 °C. Statistical analysis was completed using a one-way analysis of variance, post hoc Dunnett test using the “no additive” sample as the control reference with * and *** denoting p ≤ 0.05 and p ≤ 0.001, respectively. Error bars display S.D. (n = 3).
The activities of AtPCOs 1–5 are pH-dependent
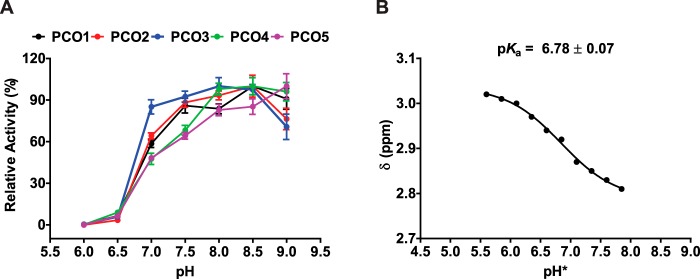
The specific activities of AtPCOs 1–5 were tested between pH 6.0 and 9.0 to determine the optimum buffering conditions for onward kinetic investigation. Bis tris propane was selected for analysis because of its broad buffering capacity, preventing any variability in activity caused by different buffer molecules. Initial rates of AtPCO activity were measured at 0.5 pH intervals using the LC-MS–based assay described above.
All isoforms were observed to exhibit a similar pH profile, showing preference for slightly basic conditions (Fig. 3A). Maximum activity was achieved at pH 8.0 for AtPCO3; pH 8.5 for AtPCOs 1, 2, and 4; and pH 9.0 for AtPCO5. Interestingly, a significant reduction in AtPCO activity was seen for all AtPCO isoforms below pH 7.0 with almost all enzyme activity eliminated at pH 6.0. 1H NMR analysis of the AtRAP2(2–15) peptide revealed that the thiol group pKa of the N-terminal cysteine is 6.78 (Fig. 3B and Fig. S2), at least 1.0 pH unit lower than the free amino acid, suggesting that the thiol is deprotonated for binding and/or turnover.
Figure 3.
The activity of AtPCOs 1–5 depends on pH. A, pH profile showing the relative activities of AtPCOs 1–5 with 200 μm RAP2(2–15) at 25 °C and different pH values using 50 mm Bis tris propane, 50 mm NaCl, and 5 mm TCEP as buffer. A significant reduction in AtPCO activity is observed below pH 7.0 for all isoforms. This correlates with the side-group pKa for the N-terminal cysteine of RAP2(2–15) (B), which was determined by monitoring the chemical shift of cysteine β-protons in D2O (>95%) using 1H NMR. This suggests that the thiol needs to be deprotonated for binding and/or turnover. pH* denotes the pH reading given by a pH meter calibrated for solutions in H2O for D2O samples. pKa was corrected to H2O using the formula pKH = 0.929pKH* + 0.42 derived in Ref. 41. Error bars display S.E. (n = 3).
Collectively, the highest level of activity for all AtPCO isoforms was observed at pH 8.0 and 8.5. The former was selected for all subsequent kinetic analysis to combine optimal activity with the most physiologically relevant conditions.
AtPCOs 1–5 have different catalytic capacities toward AtRAP2(2–15) under aerobic conditions
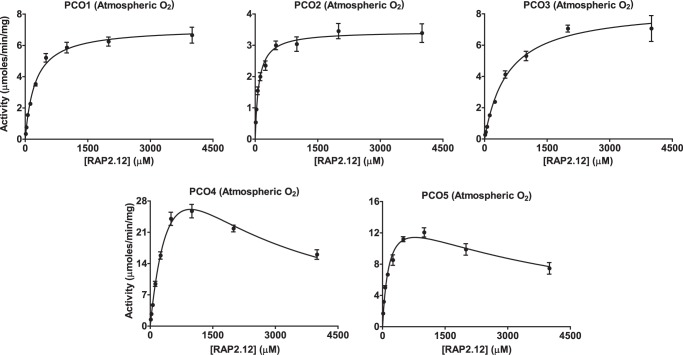
The kinetic parameters of AtPCOs 1–5 for AtRAP2(2–15) were determined under atmospheric conditions as a prelude to establishing the O2 sensitivities of each isoform. As well as identifying the peptide concentrations needed to generate saturating conditions for Km(O2) determination, a comparison of kinetic values for AtRAP2.2 and -2.12, two of the most prominent activators of anaerobic adaptation (19, 20), may contribute to a greater understanding of the physiological role of each AtPCO isoform during the plant hypoxic response. Initial rates of AtPCO activity (Fig. S3) were determined at varying AtRAP2(2–15) concentrations using the assay described above before fitting to the Michaelis–Menten model to estimate catalytic parameters.
The Michaelis–Menten plots (Fig. 4) and derived kinetic constants (Table 1) indicate striking differences between each isoform. AtPCO4 had the highest turnover number at 31.0 s−1, whereas AtPCOs 1, 3, and 5 had turnover values of 4.18, 4.70, and 7.90 s−1, respectively. AtPCO2 was the least active with a kcat of 1.98−1. AtPCOs 4 and 5 exhibited a decline in rate at AtRAP2(2–15) concentrations above ∼1 mm, which could be fitted to a standard equation for substrate inhibition, resulting in an inhibition constant of 1.4 mm for AtPCO4 and 3.8 mm for AtPCO5. Km values for AtRAP2(2–15) ranged from ∼88 μm for AtPCO2 to 690 μm for AtPCO4 with AtPCOs 1, 3, and 5 generating Michaelis constants of 238, 560, and 161 μm, respectively.
Figure 4.
Analyzing the dependence of AtPCOs 1–5 activity on AtRAP2(2–15) availability under atmospheric O2. Michaelis–Menten kinetic plots for AtPCOs 1–5 with respect to AtRAP2(2–15) concentration (calculated from initial rates presented in Fig. S3) are shown. Assays were conducted under aerobic conditions at 25 °C using 50 mm Bis tris propane, 50 mm NaCl, and 5 mm TCEP, pH 8.0, as buffer. Data collected for AtPCO4 and AtPCO5 were fitted to an equation for substrate inhibition to address the decline in rate at high RAP2(2–15) concentrations. Error bars display S.E. (n = 3).
Table 1.
Steady-state kinetic parameters of AtPCOs 1–5 toward AtRAP2(2–15) at atmospheric O2
The turnover numbers (kcat), Michaelis constants (Km), and related maximum velocities (Vmax) of AtPCOs 1–5 for RAP2(2–15) were calculated by analyzing the initial rate of AtPCO-catalyzed cysteine oxidation (Fig. S3) as a subject of AtRAP2(2–15) concentration using the Michaelis–Menten model of enzyme kinetics (Fig. 4). Substrate inhibition was observed for AtPCOs 4 and 5 with inhibition constants calculated as 1.35 ± 0.24 and 3.78 ± 1.03 mm, respectively.
| AtPCO | kcat | Km | Vmax |
|---|---|---|---|
| s−1 | mm | μmol min−1 mg−1 | |
| 1 | 4.18 ± 0.09 | 0.24 ± 0.02 | 7.14 ± 0.16 |
| 2 | 1.98 ± 0.04 | 0.09 ± 0.01 | 3.45 ± 0.08 |
| 3 | 4.70 ± 0.17 | 0.56 ± 0.06 | 8.43 ± 0.31 |
| 4 | 31.0 ± 3.49 | 0.69 ± 0.11 | 63.4 ± 7.13 |
| 5 | 7.90 ± 0.74 | 0.16 ± 0.03 | 16.1 ± 1.51 |
Overall the kinetic data strongly indicated that AtPCO4 is the most catalytically potent isoform despite possessing a relatively high Km for AtRAP2(2–15) as the large turnover number of AtPCO4 overcomes the weak substrate association. Furthermore, the substrate inhibition exhibited by AtPCO4 is only observed at high AtRAP2(2–15) concentrations, which may not be biologically relevant.
The Km(O2)app values for AtPCOs 1–5 are consistent with an O2-sensing role
For enzymes to act as O2 sensors, their activity must be rate-limited by O2 availability in a physiologically relevant range. This can be predicted by comparing biochemically determined Km(O2) values with reported (sub)cellular O2 concentrations when available. To ascertain whether the AtPCOs have the biochemical potential to act as O2 sensors, we determined the Kmapp of each isoform for O2 using steady-state kinetic assays.
To ensure that AtPCO dependence on O2 was not limited by AtRAP2(2–15) availability, peptide concentrations of 5 times the respective Km(AtRAP2(2–15)) value were used for analysis (Table 1). A higher AtRAP2(2–15) concentration (for example 10 times the Km) was not selected both to conserve substrate stocks and to avoid substrate inhibition of AtPCO5 (Fig. 4). Unfortunately, this criterion could not avoid the more significant substrate inhibition region of AtPCO4; therefore, the O2 dependence of this isoform was tested using a concentration of peptide that provided a maximum rate of AtPCO4 activity under aerobic conditions (1 mm).
Steady-state kinetic assays were conducted at different O2 concentrations using a method established previously in our laboratory (17). AtRAP2(2–15) solutions were prepared in gas-tight MS vials and equilibrated with different ratios of O2 and N2 before adding enzyme to initiate the reaction. Single time points within the linear rate ranges established above (Fig. S3) were taken by quenching the reaction with formic acid, allowing AtRAP2(2–15) oxidation to be quantified by LC-MS analysis.
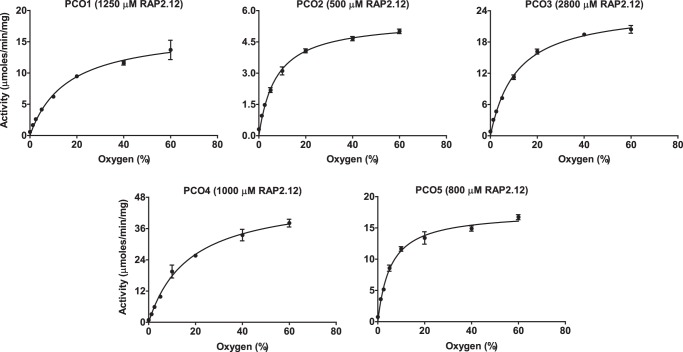
All of the AtPCOs demonstrated a clear dependence on O2 availability with respect to AtRAP2(2–15) oxidation rate. Michaelis–Menten kinetic plots (Fig. 5) allowed determination of Km(O2)app values, which varied from 5.45% O2 for AtPCO5 to 17.3% O2 for AtPCO4 with AtPCOs 1, 2, and 3 generating constants of 15.7, 7.37, and 11.1% O2, respectively (Table 2). These quantities fall within the linear range of concentrations that can be measured by an optic fiber oxygen monitor, allowing headspace gas percentages to be converted into solution partial pressures. Consequently, corresponding Km(O2)app values of 15.2, 6.57, 10.6, 16.5, and 4.83 kPa were determined for AtPCOs 1–5, respectively. The turnover numbers of AtPCOs 1 and 3 were higher than those obtained when measuring AtRAP2(2–15) dependence, indicating that their activity is submaximal at atmospheric O2 concentrations. This reflects their relatively high Km(O2)app values, which are significantly greater than AtPCOs 2 and 5, isoforms that produced similar kcat values in both experiments. In contrast, the turnover number of AtPCO4 was greater during AtRAP2(2–15) dependence assays as the influence of substrate inhibition is considered during the calculation of these kinetic parameters.
Figure 5.
Dependence of AtPCOs 1–5 activity on O2 availability. Michaelis–Menten kinetic plots display the activities of AtPCOs 1–5 with respect to O2 concentration (using single time points within the linear rate range identified in Fig. S3). Assays were conducted at 25 °C using 50 mm Bis tris propane, 50 mm NaCl, and 5 mm TCEP, pH 8.0, as buffer. Peptide concentrations of 5 times the relative Km for AtRAP2(2–15) were used for analysis (given in the graph titles) to ensure that AtPCO turnover was not limited by peptide availability with the exception of AtPCO4, which was analyzed at 1 mm AtRAP2(2–15) to avoid the substrate inhibition region of this isoform (Fig. 4). Error bars display S.E. (n = 3).
Table 2.
Steady-state kinetic parameters of AtPCOs 1–5 toward O2
The turnover numbers (kcat), Michaelis constants (Km), and related maximum velocities (Vmax) of AtPCOs 1–5 for O2 were calculated by analyzing single time points of AtPCO-catalyzed cysteine oxidation within the linear rate range established in Fig. S3 as a subject of O2 concentration using the Michaelis–Menten model of enzyme kinetics (Fig. 5). Gas headspace ratios (percent headspace) were converted into solution partial pressures of O2 (kPa solution) by directly monitoring concentration using an O2 probe.
| AtPCO | kcat | Km | Km | Vmax |
|---|---|---|---|---|
| s−1 | % | kPa solution | μmol min−1 mg−1 | |
| 1 | 9.84 ± 0.45 | 15.7 ± 1.90 | 15.2 ± 1.84 | 16.8 ± 0.76 |
| 2 | 3.05 ± 0.09 | 7.37 ± 0.74 | 6.57 ± 0.66 | 5.56 ± 0.17 |
| 3 | 13.7 ± 0.40 | 11.1 ± 0.97 | 10.6 ± 0.92 | 24.6 ± 0.72 |
| 4 | 23.8 ± 0.90 | 17.3 ± 1.69 | 16.5 ± 1.61 | 48.6 ± 1.83 |
| 5 | 8.59 ± 0.25 | 5.45 ± 0.57 | 4.83 ± 0.51 | 17.5 ± 0.51 |
Significantly, the Km(O2)app values determined are above or comparable with the O2 concentrations typically measured in vascular tissue and root structures (5–10% (23, 24)), indicating that the AtPCOs have the potential to act as physiological O2-sensing enzymes in plants.
AtPCOs 1/2 and 4/5 show different AtERF-VII substrate preferences
Degradation of all five MetCys-initiating ERF-VII transcription factors from Arabidopsis (AtHRE1, AtHRE2, AtRAP2.2, AtRAP2.12, and AtRAP2.3) has been reported to be O2-dependent. Because of the sequence conservation at their N termini, it is likely that destabilization of all the AtERF-VIIs arises due to AtPCO-catalyzed N-terminal cysteine oxidation and subsequent N-end rule–mediated proteasomal degradation in normoxia. However, it is possible that different AtPCO isoforms preferentially target different AtERF-VII substrates; therefore, we sought to ascertain whether there was any substrate selectivity among the enzymes.
We first performed assays to confirm that 14-mer peptides corresponding to the methionine-excised N termini of AtHRE1, AtHRE2, and AtRAP2.3 (hereby referred to as AtHRE1(2–15), AtHRE2(2–15), and AtRAP2.3(2–15), respectively) were substrates for AtPCOs 1–5. Each peptide was incubated with AtPCOs 1–5 for 30 min at 25 °C before being analyzed by LC-MS similar to that described above. As expected, all AtPCO isoforms were capable of oxidizing each peptide (Figs. S4–S6), implicating AtPCO activity in the O2-dependent destabilization of all AtERF-VIIs in vivo.
To gain insight into the physiological function of each AtPCO, we next sought to investigate whether AtPCOs 1–5 have any preferences toward the AtERF-VII substrates. To this end, we used a competition assay in which the AtERF-VII peptides were pooled together, incubated with each AtPCO, and monitored for oxidation at regular time intervals. Equal concentrations of each peptide were used at a concentration equivalent to the respective Km for AtRAP2(2–15) for each AtPCO isoform. Rates of AtERF-VII oxidation were normalized to the optimal substrate for each enzyme such that substrate preferences could be easily compared between enzymes (Fig. 6A). Absolute rates of substrate oxidation are presented in Table S1.
Figure 6.
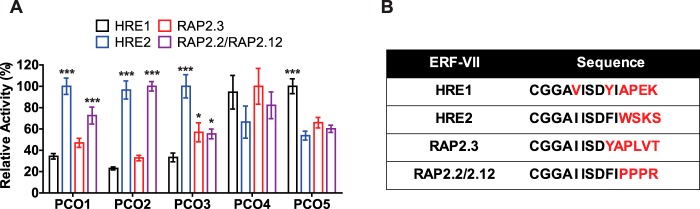
AtERF-VII substrate preferences of AtPCOs 1–5. A, the relative activities of AtPCOs 1–5 for each AtERF-VII substrate were determined by pooling the four peptides at a concentration equal to the Km for RAP2(2–15) and normalizing the rate of N-terminal cysteine oxidation to that of the best substrate. Specific activities are given in Table S1. Significant AtERF-VII preferences (relative to the poorest substrate) could be observed for most isoforms despite high conservation in the peptide sequences (B, where black indicates conservation with and red indicates deviation from RAP2.2/2.12). Statistical analysis was completed using a one-way analysis of variance, post hoc Dunnett test using the substrate that generated the lowest activity as the control reference, with * and *** denoting p ≤ 0.05 and p ≤ 0.001, respectively. Error bars display S.D. (n = 3).
We observed that each AtPCO isoform displayed different substrate preferences toward the AtERF-VII peptides despite high sequence conservation in the first nine amino acids of the peptide substrate (Fig. 6B). AtPCOs 1, 2, and 3 demonstrated selectivity for AtHRE2(2–15), whereas AtPCO2 and, to a lesser but significant extent, AtPCO1 also showed selectivity for AtRAP2(2–15). In contrast, AtPCO5 demonstrated selectivity toward AtHRE1(2–15), whereas AtPCO4 showed no significant preference for any AtERF-VII. The differences in substrate preference are likely due to the subtle differences in AtERF-VII sequence at the C terminus of the peptides (for example a Glu in position 14 of AtHRE1 and a Trp in position 12 of AtHRE2). Assuming these residues do not mechanistically contribute to N-terminal cysteine oxidation, this suggests that extensive interactions between the AtERF-VII N termini and the AtPCOs are important for substrate binding. Although these data were determined under isolated in vitro conditions, the results suggest that the AtPCO isoforms could have different biological roles.
Discussion
ERF-VIIs enable transient survival in hypoxic conditions, such as those encountered during submergence, by up-regulating anaerobic response genes (1). ERF-VII stability is regulated in an O2-dependent manner by the PCOs, which use their cosubstrate to signal for ERF-VII degradation through the Cys/Arg branch of the N-end rule pathway (6–8, 13). This direct link among O2 availability, ERF-VII stability, and the hypoxic response has led to the suggestion that the PCOs act as O2 sensors, but their sensitivity to different substrates (including O2) has not been kinetically determined. Work described here defines the dependence of each PCO from A. thaliana on pH, AtRAP2.2/2.12, and O2 before comparing their activity with alternative AtERF-VII peptide substrates in an attempt to understand enzyme function.
While optimizing assay conditions, we found that AtPCO activity is susceptible to changes in pH with enzyme activity rapidly decreasing below pH 7.0 (Fig. 3A). This correlated with a side-group pKa of 6.78 for the N-terminal cysteine of RAP2(2–15) (which is low relative to the free amino acid), suggesting that this thiol needs to be deprotonated for substrate binding and/or turnover. Physiologically, PCO dependence on slightly basic conditions might act to enhance the plant hypoxic response as low-O2 conditions can reduce cytosolic pH by accumulating weak acids during anaerobic metabolism (25) and lowering ATP-driven proton pump activity at the plasma membrane, causing a buildup of H+ (26, 27). Accordingly, the cellular environment might influence PCO activity in addition to (co)substrate availability.
Kinetic constants were derived for AtPCOs 1–5 following steady-state analysis with respect to AtRAP2(2–15) and O2. The turnover numbers and Michaelis constants for each substrate varied considerably between different isoforms, but AtPCO4 retained the highest catalytic power in both instances. Its significantly greater kcat appears to overcome the relatively weak Km for peptide and O2, allowing AtPCO4 to process large quantities of substrate even at low AtRAP2(2–15) and O2 concentrations. The expression of AtPCO4 is not up-regulated in hypoxia (in contrast to AtPCOs 1 and 2) (13), suggesting that it plays a constitutive role in RAP2.2/2.12 N-terminal cysteine oxidation. The high catalytic efficiency of AtPCO4 may provide plant cells with an underlying ability to target ERF-VIIs for removal both under normoxia and upon reoxygenation following hypoxia, offering continuous and general transcription factor maintenance. This is supported by the relatively broad substrate specificity of this isoform (Fig. 6), which hints that AtPCO4 can competently process all members of the AtERF-VII family.
AtPCO5 may also play a significant role in ERF-VII regulation in vivo. Although the turnover numbers calculated for this isoform are weaker than for AtPCO4, strong substrate association enables it to process large quantities of RAP2(2–15) at relatively low concentrations of peptide and O2, which may more accurately reflect physiological conditions.
It should be noted that the activity of both AtPCO4 and AtPCO5 was observed to decline at high AtRAP2(2–15) concentrations, indicating susceptibility to substrate inhibition. This could limit their ability to process AtERF-VII destabilization at high substrate levels. It is difficult to know if this apparent inhibition is a real property of AtPCO4/5 catalysis or the consequence of using a representative peptide as substrate. In either case, these negative effects were only detected above 1 mm, which might not be applicable in planta.
In contrast to AtPCOs 4 and 5, AtPCOs 1 and 2 (which are hypoxically induced (13)) demonstrated relatively low catalytic capability toward AtRAP2(2–15) and O2 but a clear substrate preference for AtHRE2 and AtRAP2.2/2.12. This raises the possibility that their up-regulation in hypoxia could help “turn off” the plant hypoxic response when O2 concentrations increase, particularly by targeting two of the most prominent anaerobic gene activators, AtRAP2.2/2.12, for degradation.
AtPCO3 showed preference for AtHRE2 but overall had relatively low kinetic capability (at least toward AtRAP2(2–15)) and is not hypoxically up-regulated (13). Although it is clear that it can contribute to AtERF-VII regulation, it must be considered for AtPCO3, as well as other isoforms, that there are over 200 MC-initiating proteins identified in Arabidopsis (28). It may be that there are alternative substrates for some or all of the AtPCOs that are more kinetically relevant than AtERF-VII oxidation.
We cannot rule out that additional interactions between the PCOs and full-length ERF-VIIs could influence their kinetic parameters. However, the peptides used represent N termini of transcription factors (often intrinsically disordered (29)) and as such are unlikely to be flanked by extensively structured regions. AtPCO kinetic parameters determined with the peptide substrates used in this study are therefore deemed likely to reflect their relative properties in planta; the use of peptide substrates is a strategy that has been widely used in the study of oxygen-sensing enzymes in animals (16, 30, 31).
A clear relationship between O2 concentration and AtPCO activity was observed for each isoform, highlighting their dependence on O2 for catalysis (Fig. 5 and Table 2). AtPCOs 2 and 5 have Km(O2)app values of 7.37 and 5.45% (6.57 and 4.83 kPa), respectively, whereas AtPCOs 1 and 4 have higher Km(O2)app values of 15.7 and 17.3% (15.2 and 16.5 kPa), respectively. AtPCO Km(O2)app values are lower than those reported for mammalian O2-sensing enzymes, such as PHD2, which has a reported Km(O2)app value of over 450 μm (equivalent to 40%, and at least twice the molar concentration calculated for the AtPCOs, as estimated by Henry's law) determined using the same technique (17). They also fall below standard atmospheric O2 concentrations in aqueous solution. However, all the isoforms showed a gradient of activity as oxygen concentrations depleted from normal levels.
Accurate measurement of O2 concentrations in plants is challenging and requires further technological optimization (32). Nevertheless, the lack of a circulatory system in plants dictates that O2 levels will be variable. Microprobes have been used to measure O2 in a number of different tissues (for a review, see Ref. 33), and steep gradients have been reported, ranging from near atmospheric levels in surface cells to mildly hypoxic (5–10% O2) in vascular tissues of the castor bean plant (24) and maize root meristems (23). O2 levels in seed embryos vary depending on coating and chlorophyll content (34) but can be near anoxic; e.g. sunflower seed embryos have O2 partial pressures of 0.08 kPa/<1 μm (35). O2 concentrations fall in potato tubers as they grow (to ∼5% (36)), and large fruit can also have O2 gradients from surface to core (e.g. 20–12 kPa in apples (37)). The Km(O2)app values we report for the AtPCOs range from 5.45 to 17.3% (4.83–16.5 kPa; Table 2). Although these values were obtained under nonphysiological conditions, they nevertheless suggest that the activity of these enzymes is likely to be sensitive to physiologically relevant O2 gradients, thus enabling them to act as plant O2 sensors. It will be interesting to establish whether the expression of different PCO isoforms varies between different tissues depending on the O2 environment and enzyme sensitivity so that ERF-VII stability can be regulated appropriately.
Collectively, our results suggest that the AtPCOs are likely to be effective O2-sensing enzymes, which are capable of efficiently regulating the stability of ERF-VII transcription factors in response to hypoxia, potentially in a targeted manner. The relatively high activity of the constitutively expressed AtPCOs 4 and 5 could allow residual ERF-VIIs to be rapidly degraded under normoxic conditions, preventing the adverse consequences of anaerobic gene induction at regular oxygen concentrations. AtPCOs 1 and 2, whose expression is induced in hypoxia (13), could be responsible for terminating anaerobic processes when plants withdraw from hypoxia by targeting the primary activators RAP2.2/2.12 for degradation through the N-end rule pathway as soon as normal oxygen levels start to return. This would help alleviate the strain of prolonged fermentative metabolism, giving the organism the best chance of surviving transient periods of hypoxia, such as those experienced during flooding.
Our results reveal the potential for distinct roles for AtPCO isoforms. This raises the possibility that individual isoforms could be targeted for specific biological outcomes. Overexpression of AtRAP2.12 in Arabidopsis and stabilization of ERF-VII homologs in barley and rice have led to increased plant recovery following submergence (7, 38, 39). ERF-VIIs are also connected to tolerance of other forms of abiotic and biotic stress (12, 40). It is conceivable that a detailed understanding of PCO ERF-VII functionality, beginning with this kinetic study, could enable a rational approach to enhancing crop tolerance to a range of stresses through directed chemical or genetic inhibition methods.
Experimental procedures
Cloning and transformation of Atpco1–5 for heterologous expression in E. coli
The five pco genes from A. thaliana were amplified from initial constructs donated by Dr. Daan A. Weits and Professor Francesco Licausi using the Phusion polymerase system (New England Biolabs) and cloned into the NdeI and XhoI restriction sites of pET28a (Novagen) using ligation-independent methods (In-Fusion, Clontech). In-Fusion products were transformed into E. coli NEB5α competent cells (New England Biolabs) for insert validation by Sanger sequencing (Source Bioscience) before being transformed into E. coli BL21 (DE3) competent cells (New England Biolabs) for protein production.
Heterologous expression of AtPCOs 1–5 in E. coli
E. coli transformants were grown in 600 ml of 2× Yeast extract Tryptone medium supplemented with 50 μm FeSO4 and 40 μg/ml kanamycin sulfate at 37 °C and 180 rpm until an A600 nm was reached. Cultures were induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside and grown overnight at 20 °C and 180 rpm before being harvested by centrifugation at 10,000 × g for 10 min.
Purification of AtPCOs 1–5
Cell pellets were resuspended in 50 mm Tris and 400 mm NaCl, pH 7.5, supplemented with DNase and a Complete EDTA-free protease inhibitor mixture tablet and lysed by sonication. Cellular debris was removed by centrifugation at 48,000 × g for 30 min and filtration through a 0.45-μm membrane. The soluble supernatant was loaded onto an equilibrated HisTrap HP column (GE Healthcare) and washed with resuspension buffer containing increasing concentrations of imidazole until a definitive A280 nm peak was collected. Contaminating proteins were removed using a HiLoad 26/600 Superdex 75 prep grade size-exclusion column (GE Healthcare) equilibrated with resuspension buffer. Protein purity was verified by SDS-PAGE, and protein concentrations were estimated using A280 nm measurements obtained by a Cary UV-visible spectrometer.
Metal quantification of AtPCOs 1–5
Enzyme iron concentrations were estimated using a bathophenanthroline disulfonic acid–based assay, which produces an absorbance reading at 535 nm in the presence of metal. Iron was extracted from protein samples by acid and heat denaturation, allowing absorbance comparison with a standard curve of known iron concentrations. Each sample contained a final concentration of 20% saturated ammonium acetate, 5 mm sodium metabisulfite, and 1 mm bathophenanthroline disulfonic acid as well as 0–250 μm Fe(II).
Analyzing the activity of AtPCOs 1–5 with AtRAP2.2/2.12 at atmospheric O2
The activities of AtPCOs 1–5 were examined by incubating synthesized peptide (GL Biochem) corresponding to the first 14 amino acids of the methionine-excised N terminus of AtRAP2.2/2.12 (AtRAP2(2–15)), AtRAP2.3 (AtRAP3(2–15)), AtHRE1 (AtHRE1(2–15)), or AtHRE2 (AtHRE2(2–15)) with 0.1–1.0 μm enzyme in a benchtop thermocycler (Eppendorf) at 25 °C under aerobic conditions. 5 mm TCEP, 20 μm FeSO4, and 1 mm ascorbate were added to all samples to help maintain a reductive environment unless otherwise specified. 50 mm Bis tris propane and 50 mm NaCl were used to buffer most solutions with pH and peptide concentrations given in the text or figures. Time points were taken at regular intervals by quenching the reaction 1:10 with 1% formic acid for rate analysis. Oxidation was monitored by ultrahigh-performance LC (UPLC)-MS, and turnover was quantified by comparing the areas underneath the product and substrate ions extracted from the total ion current chromatogram. All figures and kinetic parameters were generated using Prism (GraphPad).
UPLC-MS measurements were obtained using an Acquity UPLC system coupled to a Xevo G2-S Q-ToF mass spectrometer (Waters) operated in positive electrospray mode. Instrument parameters, data acquisition, and data processing were controlled by MassLynx 4.1 with source conditions adjusted to maximize sensitivity and minimize fragmentation. Samples were injected onto a Chromolith Performance RP-18e 100 2-mm column (Merck) heated to 40 °C and eluted at 0.3 ml/min using a gradient of 95% deionized water supplement with 0.1% (v/v) formic acid to 95% acetonitrile.
Determining the oxygen sensitivity of AtPCOs 1–5 with AtRAP2.2/2.12
The activities of AtPCOs 1–5 were examined at different O2 concentrations using peptide at 5 times the relevant Km for AtRAP2(2–15) unless otherwise specified. 100-μl aliquots of AtRAP2(2–15) were prepared as above, transferred into silicone-sealed MS vials, and equilibrated with different ratios of nitrogen and oxygen gas for 10 min using a mass flow controller (Brooks Instruments). Reactions were initiated by injecting 1 μl of enzyme using a gas-tight syringe (Hamilton) and allowed to proceed for 1 min (within the linear range of activity). Reactions were terminated by injecting 10 μl of 10% formic acid and analyzed as above, diluting the sample in 1% formic acid to avoid saturation of the detector if necessary. UPLC-MS analysis was carried out as described above.
The partial pressure of O2 in solution was estimated for each Km(O2)app using an Oxylite probe (Oxford Optronix), which measures concentrations in the physiological range. Buffer samples equilibrated with the relevant gas ratios were prepared as above and tested for O2 concentration by inserting the probe through a needle. Multiple readings were taken to ensure accuracy, giving a standard error of 0.37 kPa for PCO1, 0.07 kPa for PCO2, 0.33 kPa for PCO3, 0.23 kPa for PCO4, and 0.03 kPa for PCO5.
Comparing the activity of AtPCOs 1–5 with different ERF-VII substrates at atmospheric oxygen
The activities of AtPCOs 1–5 with AtRAP2.2/2.12 (AtRAP2(2-15)), AtRAP2.3 (AtRAP3(2–15)), AtHRE1 (AtHRE1(2–15)), and AtHRE2 (AtHRE2(2–15)) were compared using a competition assay where all substrates were pooled at a concentration equivalent to the relevant Km for AtRAP2(2–15) and analyzed for turnover rate at 25 °C as described above under aerobic conditions.
Determining the thiol pKa of AtRAP2.2/2.12 using NMR
A selection of 10 buffer solutions of NaH2PO4 (50 mm) in D2O were prepared at a set range of pH values ranging from 5.60 to 7.85 with 0.25 increments (pH* 5.60, 5.85, 6.10, 6.35, 6.60, 6.85, 7.10, 7.35, 7.60, and 7.85). The peptide (1.03 mg) was dissolved in D2O (160 μl). A stock solution of 1,4-dioxane (12.9 μl) was prepared by dissolving in D2O (1.5 ml) and used as a reference. A total of 10 samples were prepared at the different pH* values by mixing buffer (152 μl), peptide solution (8 μl) and 1,4-dioxane stock (1 μl). Samples were subsequently analyzed by 1H NMR at 298 K.
NMR experiments were carried out on a Bruker Avance III 700 MHz equipped with a TCI inverse cryoprobe with sample temperature regulated at 298 K. Data were analyzed using Bruker Topspin 3.5. Processing of spectra was done with a Lorentzian line broadening of 0.3 Hz. Chemical shifts (δ) are given as ppm relative to 1,4-dioxane (δH 3.68 ppm for 1H NMR). The pKa value for H2O was established using the formula derived previously (41).
Author contributions
M. D. W. formal analysis; M. D. W. and E. F. supervision; M. D. W. and E. F. validation; M. D. W., J. J. A. G. K., S. E., L. J. T. K., and E. F. investigation; M. D. W. and E. F. visualization; M. D. W., J. J. A. G. K., S. E., L. J. T. K., and E. F. methodology; M. D. W. and E. F. writing-original draft; M. D. W. and E. F. project administration; M. D. W., J. J. A. G. K., S. E., L. J. T. K., and E. F. writing-review and editing; E. F. conceptualization; E. F. funding acquisition.
Supplementary Material
Acknowledgments
We thank Dr. Daan Weits and Prof. Francesco Licausi for both plasmid donation and useful discussions about the manuscript. We thank Dr. James Wickens and Elisabete Pires of the Mass Spectrometry Research Facility in the Department of Chemistry at the University of Oxford for advice and machine maintenance. We acknowledge Profs. Christopher J. Schofield, Timothy D. W. Claridge, and Rosalind Rickaby for supervision of Jos J. A. G. Kamps and Leah J. Taylor Kearney, respectively.
This work was supported in part by Biotechnology and Biological Sciences Research Council (UK) New Investigator Grant BB/M024458/1 (to E. F.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6 and Table S1.
- ERF-VII
- group VII ethylene response factor
- PCO
- plant cysteine oxidase
- At
- A. thaliana
- kPa
- kilopascal(s)
- HIF
- hypoxia-inducible transcription factor
- PHD
- prolyl hydroxylase enzyme
- Bis tris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- TCEP
- tris(2-carboxyethyl)phosphine
- UPLC
- ultrahigh-performance LC.
References
- 1. Bailey-Serres J., Fukao T., Gibbs D. J., Holdsworth M. J., Lee S. C., Licausi F., Perata P., Voesenek L. A., and van Dongen J. T. (2012) Making sense of low oxygen sensing. Trends Plant Sci. 17, 129–138 10.1016/j.tplants.2011.12.004 [DOI] [PubMed] [Google Scholar]
- 2. Giuntoli B., and Perata P. (2018) Group VII ethylene response factors in Arabidopsis: regulation and physiological roles. Plant Physiol. 176, 1143–1155 10.1104/pp.17.01225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hattori Y., Nagai K., Furukawa S., Song X. J., Kawano R., Sakakibara H., Wu J., Matsumoto T., Yoshimura A., Kitano H., Matsuoka M., Mori H., and Ashikari M. (2009) The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030 10.1038/nature08258 [DOI] [PubMed] [Google Scholar]
- 4. Licausi F., van Dongen J. T., Giuntoli B., Novi G., Santaniello A., Geigenberger P., and Perata P. (2010) HRE1 and HRE2, two hypoxia-inducible ethylene response factors, affect anaerobic responses in Arabidopsis thaliana. Plant J. 62, 302–315 10.1111/j.1365-313X.2010.04149.x [DOI] [PubMed] [Google Scholar]
- 5. Hinz M., Wilson I. W., Yang J., Buerstenbinder K., Llewellyn D., Dennis E. S., Sauter M., and Dolferus R. (2010) Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153, 757–772 10.1104/pp.110.155077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gibbs D. J., Lee S. C., Isa N. M., Gramuglia S., Fukao T., Bassel G. W., Correia C. S., Corbineau F., Theodoulou F. L., Bailey-Serres J., and Holdsworth M. J. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418 10.1038/nature10534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Licausi F., Kosmacz M., Weits D. A., Giuntoli B., Giorgi F. M., Voesenek L. A., Perata P., and van Dongen J. T. (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422 10.1038/nature10536 [DOI] [PubMed] [Google Scholar]
- 8. White M. D., Klecker M., Hopkinson R. J., Weits D. A., Mueller C., Naumann C., O'Neill R., Wickens J., Yang J., Brooks-Bartlett J. C., Garman E. F., Grossmann T. N., Dissmeyer N., and Flashman E. (2017) Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat. Commun. 8, 14690 10.1038/ncomms14690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garzón M., Eifler K., Faust A., Scheel H., Hofmann K., Koncz C., Yephremov A., and Bachmair A. (2007) PRT6/At5g02310 encodes an Arabidopsis ubiquitin ligase of the N-end rule pathway with arginine specificity and is not the CER3 locus. FEBS Lett. 581, 3189–3196 10.1016/j.febslet.2007.06.005 [DOI] [PubMed] [Google Scholar]
- 10. Gibbs D. J., Bacardit J., Bachmair A., and Holdsworth M. J. (2014) The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol. 24, 603–611 10.1016/j.tcb.2014.05.001 [DOI] [PubMed] [Google Scholar]
- 11. Gibbs D. J., Conde J. V., Berckhan S., Mendiondo G. M., Prasad G., and Holdsworth M. J. (2015) Group VII ethylene response factors co-ordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 169, 23–31 10.1104/pp.15.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vicente J., Mendiondo G. M., Movahedi M., Peirats-Llobet M., Juan Y.-T., Shen Y.-Y., Dambire C., Smart K., Rodriguez P. L., Charng Y.-Y., Gray J. E., and Holdsworth M. J. (2017) The Cys-Arg/N-end rule pathway is a general sensor of abiotic stress in flowering plants. Curr. Biol. 27, 3183–3190.e4 10.1016/j.cub.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weits D. A., Giuntoli B., Kosmacz M., Parlanti S., Hubberten H. M., Riegler H., Hoefgen R., Perata P., van Dongen J. T., and Licausi F. (2014) Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat. Commun. 5, 3425 10.1038/ncomms4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Myllyharju J. (2013) Prolyl 4-hydroxylases, master regulators of the hypoxia response. Acta Physiol. 208, 148–165 10.1111/apha.12096 [DOI] [PubMed] [Google Scholar]
- 15. Ehrismann D., Flashman E., Genn D. N., Mathioudakis N., Hewitson K. S., Ratcliffe P. J., and Schofield C. J. (2007) Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay. Biochem. J. 401, 227–234 10.1042/BJ20061151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirsilä M., Koivunen P., Günzler V., Kivirikko K. I., and Myllyharju J. (2003) Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 278, 30772–30780 10.1074/jbc.M304982200 [DOI] [PubMed] [Google Scholar]
- 17. Tarhonskaya H., Chowdhury R., Leung I. K., Loik N. D., McCullagh J. S., Claridge T. D., Schofield C. J., and Flashman E. (2014) Investigating the contribution of the active site environment to the slow reaction of hypoxia-inducible factor prolyl hydroxylase domain 2 with oxygen. Biochem. J. 463, 363–372 10.1042/BJ20140779 [DOI] [PubMed] [Google Scholar]
- 18. Dao J. H., Kurzeja R. J., Morachis J. M., Veith H., Lewis J., Yu V., Tegley C. M., and Tagari P. (2009) Kinetic characterization and identification of a novel inhibitor of hypoxia-inducible factor prolyl hydroxylase 2 using a time-resolved fluorescence resonance energy transfer-based assay technology. Anal. Biochem. 384, 213–223 10.1016/j.ab.2008.09.052 [DOI] [PubMed] [Google Scholar]
- 19. Bui L. T., Giuntoli B., Kosmacz M., Parlanti S., and Licausi F. (2015) Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci 236, 37–43 10.1016/j.plantsci.2015.03.008 [DOI] [PubMed] [Google Scholar]
- 20. Gasch P., Fundinger M., Müller J. T., Lee T., Bailey-Serres J., and Mustroph A. (2016) Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28, 160–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Imsand E. M., Njeri C. W., and Ellis H. R. (2012) Addition of an external electron donor to in vitro assays of cysteine dioxygenase precludes the need for exogenous iron. Arch. Biochem. Biophys. 521, 10–17 10.1016/j.abb.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 22. Flashman E., Davies S. L., Yeoh K. K., and Schofield C. J. (2010) Investigating the dependence of the hypoxia-inducible factor hydroxylases (factor inhibiting HIF and prolyl hydroxylase domain 2) on ascorbate and other reducing agents. Biochem. J. 427, 135–142 10.1042/BJ20091609 [DOI] [PubMed] [Google Scholar]
- 23. Ober E. S., and Sharp R. E. (1996) A microsensor for direct measurement of O2 partial pressure within plant tissues3. J. Exp. Bot 47, 447–454 10.1093/jxb/47.3.447 [DOI] [Google Scholar]
- 24. van Dongen J. T., Schurr U., Pfister M., and Geigenberger P. (2003) Phloem metabolism and function have to cope with low internal oxygen. Plant Physiol. 131, 1529–1543 10.1104/pp.102.017202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vartapetian B. B., and Jackson M. B. (1997) Plant adaptations to anaerobic stress. Ann. Bot. 79, 3–20 10.1093/oxfordjournals.aob.a010303 [DOI] [Google Scholar]
- 26. Wang F., Chen Z.-H., and Shabala S. (2017) Hypoxia sensing in plants: on a quest for ion channels as putative oxygen sensors. Plant Cell Physiol. 58, 1126–1142 10.1093/pcp/pcx079 [DOI] [PubMed] [Google Scholar]
- 27. Gout E., Boisson A.-M., Aubert S., Douce R., and Bligny R. (2001) Origin of the cytoplasmic pH changes during anaerobic stress in higher plant cells. Carbon-13 and phosphorous-31 nuclear magnetic resonance studies. Plant Physiol. 125, 912–925 10.1104/pp.125.2.912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gibbs D. J., Bailey M., Tedds H. M., and Holdsworth M. J. (2016) From start to finish: amino-terminal protein modifications as degradation signals in plants. New Phytol. 211, 1188–1194 10.1111/nph.14105 [DOI] [PubMed] [Google Scholar]
- 29. Staby L., O'Shea C., Willemoës M., Theisen F., Kragelund B. B., and Skriver K. (2017) Eukaryotic transcription factors: paradigms of protein intrinsic disorder. Biochem. J. 474, 2509–2532 10.1042/BCJ20160631 [DOI] [PubMed] [Google Scholar]
- 30. Hancock R. L., Masson N., Dunne K., Flashman E., and Kawamura A. (2017) The activity of JmjC histone lysine demethylase KDM4A is highly sensitive to oxygen concentrations. ACS Chem. Biol. 12, 1011–1019 10.1021/acschembio.6b00958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tarhonskaya H., Hardy A. P., Howe E. A., Loik N. D., Kramer H. B., McCullagh J. S., Schofield C. J., and Flashman E. (2015) Kinetic investigations of the role of factor inhibiting hypoxia-inducible factor (FIH) as an oxygen sensor. J. Biol. Chem. 290, 19726–19742 10.1074/jbc.M115.653014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmidt R. R., Weits D. A., Feulner C. F. J., and van Dongen J. T. (2018) Oxygen sensing and integrative stress signaling in plants. Plant Physiol. 176, 1131–1142 10.1104/pp.17.01394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. van Dongen J. T., and Licausi F. (2015) Oxygen sensing and signaling. Annu. Rev. Plant Biol. 66, 345–367 10.1146/annurev-arplant-043014-114813 [DOI] [PubMed] [Google Scholar]
- 34. Borisjuk L., and Rolletschek H. (2009) The oxygen status of the developing seed. New Phytol. 182, 17–30 10.1111/j.1469-8137.2008.02752.x [DOI] [PubMed] [Google Scholar]
- 35. Rolletschek H., Borisjuk L., Sánchez-García A., Gotor C., Romero L. C., Martínez-Rivas J. M., and Mancha M. (2007) Temperature-dependent endogenous oxygen concentration regulates microsomal oleate desaturase in developing sunflower seeds. J. Exp. Bot. 58, 3171–3181 10.1093/jxb/erm154 [DOI] [PubMed] [Google Scholar]
- 36. Geigenberger P., Fernie A. R., Gibon Y., Christ M., and Stitt M. (2000) Metabolic activity decreases as an adaptive response to low internal oxygen in growing potato tubers. Biol. Chem. 381, 723–740 10.1515/BC.2000.093 [DOI] [PubMed] [Google Scholar]
- 37. Ho Q. T., Verboven P., Verlinden B. E., Schenk A., Delele M. A., Rolletschek H., Vercammen J., and Nicolaï B. M. (2010) Genotype effects on internal gas gradients in apple fruit. J. Exp. Bot. 61, 2745–2755 10.1093/jxb/erq108 [DOI] [PubMed] [Google Scholar]
- 38. Mendiondo G. M., Gibbs D. J., Szurman-Zubrzycka M., Korn A., Marquez J., Szarejko I., Maluszynski M., King J., Axcell B., Smart K., Corbineau F., and Holdsworth M. J. (2016) Enhanced waterlogging tolerance in barley by manipulation of expression of the N-end rule pathway E3 ligase PROTEOLYSIS6. Plant Biotechnol. J. 14, 40–50 10.1111/pbi.12334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xu K., Xu X., Fukao T., Canlas P., Maghirang-Rodriguez R., Heuer S., Ismail A. M., Bailey-Serres J., Ronald P. C., and Mackill D. J. (2006) Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442, 705–708 10.1038/nature04920 [DOI] [PubMed] [Google Scholar]
- 40. Zhao Y., Wei T., Yin K. Q., Chen Z., Gu H., Qu L. J., and Qin G. (2012) Arabidopsis RAP2.2 plays an important role in plant resistance to Botrytis cinerea and ethylene responses. New Phytol. 195, 450–460 10.1111/j.1469-8137.2012.04160.x [DOI] [PubMed] [Google Scholar]
- 41. Krezel A., and Bal W. (2004) A formula for correlating pKa values determined in D2O and H2O. J. Inorg. Biochem. 98, 161–166 10.1016/j.jinorgbio.2003.10.001 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.