Abstract
Dynamin-related protein 1 (Drp1) constricts mitochondria as a mechanochemical GTPase during mitochondrial division. The Drp1 gene contains several alternative exons and produces multiple isoforms through RNA splicing. Here we performed a systematic analysis of Drp1 transcripts in different mouse tissues and identified a previously uncharacterized isoform that is highly enriched in the brain. This Drp1 isoform is termed Drp1ABCD because it contains four alterative exons: A, B, C, and D. Remarkably, Drp1ABCD is located at lysosomes, late endosomes, and the plasma membrane in addition to mitochondria. Furthermore, Drp1ABCD is concentrated at the interorganelle interface between mitochondria and lysosomes/late endosomes. The localizations of Drp1ABCD at lysosomes, late endosomes, and the plasma membrane require two exons, A and B, that are present in the GTPase domain. Drp1ABCD assembles onto these membranes in a manner that is regulated by its oligomerization and GTP hydrolysis. Experiments using lysosomal inhibitors show that the association of Drp1ABCD with lysosomes/late endosomes depends on lysosomal pH but not their protease activities. Thus, Drp1 may connect mitochondria to endosomal–lysosomal pathways in addition to mitochondrial division.
Keywords: mitochondria, subcellular organelle, GTPase, brain, lysosome
Introduction
Drp1 is a dynamin-related GTPase that mediates the constriction of mitochondria during mitochondrial division (1–4). Drp1 is a soluble protein in the cytosol and recruited to mitochondria through interactions with the mitochondrial outer membrane proteins such as Mff, Mid49/51, and Fis1 (5, 6). On mitochondria, Drp1 oligomerizes into spiral filaments that circle mitochondria. Oligomerization-stimulated GTP hydrolysis by Drp1 regulates constriction of the oligomers and initiates mitochondrial division.
Drp1-driven mitochondrial division is critical for controlling the number and size of mitochondria through a dynamic balance in collaboration with the opposite activity, mitochondrial fusion mediated by mitofusins and Opa1 (4). Demonstrating the physiological importance of Drp1, complete knockout of Drp1 leads to embryonic lethality in mice (7, 8). Brain-specific knockout of Drp1 causes neurodegeneration with accumulation of enlarged mitochondria, which results from unopposed mitochondrial fusion in the absence of division (8–12). These enlarged mitochondria become ineffective in turnover and transport in axons and dendrites because of increases in their size. Similar to neurons, loss of Drp1 in cardiomyocytes in the heart results in mitophagy defects and leads to lethal heart failure (10, 13, 14). When Drp1 is deleted in oocytes, enlarged mitochondria entangle with other organelles, such as the endoplasmic reticulum, leading to female infertility (15). In addition to these physiological roles, Drp1-mediated mitochondrial division regulates mitochondrial shape during oncogenic transformation, and this dynamic shape change is critical for tumorigenesis (16, 17).
Previous studies have identified Drp1 isoforms that are enriched in particular organs and cell types. For example, two Drp1 isoforms (termed Drp1–101 and Drp1–001 in the original study (18) and Drp1BD and Drp1D here (Fig. 1, A and D)) are highly expressed in immune cells and associate with microtubules in addition to mitochondria (18). This unique localization is determined by the two alternative exons: the combination of the absence of exon C and the presence of exon D in the variable domain (also known as the B insert) (18). Another example is the isoform of Drp1 that contains exons B, C, and D (Drp1BCD; Fig. 1, A and C), and this isoform is mainly expressed in the brain (18–20). Exon B is located in the GTPase domain. Drp1BCD has a relatively lower propensity of oligomerization at the basal level but is strongly activated by the major Drp1 receptor/effector protein Mff (19). This property of Drp1BCD may enable robust control of mitochondrial division in neurons, in which mitochondrial division and fusion are highly regulated.
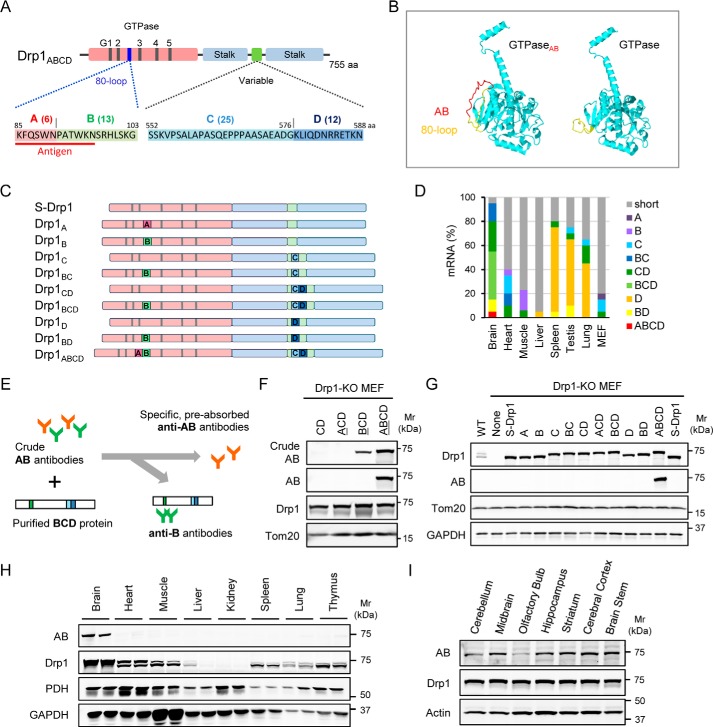
Figure 1.
Drp1ABCD is a brain-enriched Drp1 isoform. A, the positions of the four alternative exons, A, B, C and D, in Drp1. Exons A and B are present in the GTPase domain, whereas exons C and D are located in the variable domain. The amino acid (aa) sequences of these exons are shown. The number of amino acids comprising each exon is shown in parentheses. B, the location of exons A and B (red) in the 3D structure of the GTPase domain of Drp1ABCD (1–362 amino acid residues) and S-Drp1 (1–343) are predicted using I-TASSER (35) and visualized using Jmol (www.jmol.org; please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site). A short protein domain unique to the mitochondrial division dynamin, termed the 80-loop, is shown in yellow (21, 27). C and D, expression analysis of Drp1 transcripts. mRNAs isolated from the indicated mouse organs and MEFs are used as templates to PCR-amplify Drp1 cDNAs. ∼100 Drp1 cDNAs were analyzed by DNA sequencing for each organ. 10 isoforms were identified (C). Different tissues contain different combinations of these 10 isoforms (D). E, production of antibodies specific to the junction of exons A and B. A peptide derived from 12 amino acids (underlined in red in A) was used as an antigen to raise antibodies in rabbits. Antibodies were affinity-purified using the original peptide. To remove antibodies that recognize exon B alone, antibodies were preincubated with purified recombinant His6-Drp1BCD overnight. We also incubated the antibodies with nitrilotriacetic acid–agarose beads coupled to His6-Drp1BCD and removed antibodies that bind exon B alone by centrifugation. F, Drp1-KO MEFs individually expressing each of Drp1CD, Drp1ACD, Drp1BCD, or Drp1ABCD were subjected to Western blotting using anti-exon AB antibodies in the presence (AB) or absence (crude AB) of overnight preincubation with His6-Drp1BCD. We also used antibodies to Drp1 (pan-Drp1) and the mitochondrial outer membrane protein Tom20. G, Drp1-KO MEFs individually expressing all Drp1 isoforms were analyzed by Western blotting using antibodies against AB, Drp1 (pan-DRP1), Tom20, and GAPDH. H and I, immunoblotting of mouse organs (H) and subregions of the brain (I) using antibodies to AB, Drp1 (pan-Drp1), the mitochondrial protein pyruvate dehydrogenase (PDH), GAPDH, and actin. Equivalent amounts of total proteins were loaded in each lane.
In this study, we tested whether there are additional Drp1 isoforms. We extensively examined Drp1 transcripts in multiple mouse organs and identified an isoform that is enriched in the brain and associates with lysosomes and late endosomes in addition to mitochondria. Interestingly, this new isoform appeared to be concentrated at contact sites between mitochondria and lysosomes/late endosomes. We also characterized mechanisms that target this Drp1 isoform to the lysosomal, endosomal, and plasma membranes.
Results
Identification of a novel Drp1 isoform that is enriched in the brain
We systematically analyzed the expression profile of Drp1 transcripts using mRNAs isolated from different mouse organs and cells, including the brain, heart, skeletal muscle, liver, spleen, testis, lung, and embryonic fibroblasts. We identified 10 different Drp1 isoforms with four alternative exons (exons A, B, C, and D; Fig. 1, A–C). Exons A and B are located in the GTPase domain, whereas exons C and D are present in the variable domain. We found that different tissues contain different combinations of these 10 isoforms (Fig. 1D). Of these, short-Drp1 (S-Drp1), which lacks all of the four alternative exons (Fig. 1, A and C), is the most ubiquitously expressed (Fig. 1D), consistent with previous reports (18–20). In contrast to the common isoform S-Drp1, two long isoforms, Drp1BCD, which contains three alternative exons, and a novel isoform, Drp1ABCD, which contains all of the four alternative exons, were only detected in the brain (Fig. 1D). The brain also expresses four other Drp1 isoforms, S-Drp1, Drp1BC, Drp1CD, and Drp1D. Drp1BCD and Drp1CD are major isoforms in the brain and account for ∼65% of total Drp1 mRNAs (Fig. 1D). Because Drp1ABCD has not been characterized previously, we focused on this isoform in the rest of the study.
To examine the expression of Drp1ABCD at the protein level, we generated rabbit polyclonal antibodies that specifically recognize Drp1ABCD. As an antigen, we used a peptide that corresponds to the junction of exons A and B (six amino acids from each exon) (Fig. 1A) because Drp1ABCD is the only isoform that carries this sequence (Fig. 1, C and D). Antibodies were affinity-purified using the same peptide (crude exon AB antibodies) (Fig. 1E). To determine the specificity of the crude exon AB antibodies, we transduced Drp1-KO MEFs2 with lentiviruses carrying each isoform and used their whole-cell lysates for Western blotting. Results showed that both Drp1BCD and Drp1ABCD were detected by the crude exon AB antibodies (Fig. 1F). To neutralize antibodies that recognize exon B alone, we preincubated the crude exon AB antibodies with recombinant His6-Drp1BCD purified from Escherichia coli (Fig. 1E). Western blotting showed that the preabsorbed anti-exon AB antibodies only detected Drp1ABCD but not other isoforms (Fig. 1, F and G). As a control, the expression of these isoforms in Drp1-KO MEFs was confirmed using anti-pan-Drp1 antibodies that recognize all Drp1 isoforms (Fig. 1, F and G).
Using these preabsorbed anti-exon AB antibodies, we examined the expression of Drp1ABCD in mouse organs. Consistent with the transcript analysis (Fig. 1D), Western blotting showed that Drp1ABCD is primarily expressed in the brain (Fig. 1H). Anti-pan-Drp1 antibodies showed that the expression level of total Drp1 varies in different organs, with the highest levels in the brain, heart, skeletal muscle, and thymus (Fig. 1H). Further analysis showed that Drp1ABCD is expressed in most of the subregions of the brain, including the cerebellum, midbrain, hippocampus, striatum, cerebral cortex, and brain stem (Fig. 1I). A relatively low level of Drp1ABCD was observed in the olfactory bulb, whereas anti-pan-Drp1 antibodies showed similar expression levels of total Drp1 in these brain subregions (Fig. 1I).
Drp1ABCD functions in mitochondrial and peroxisomal division
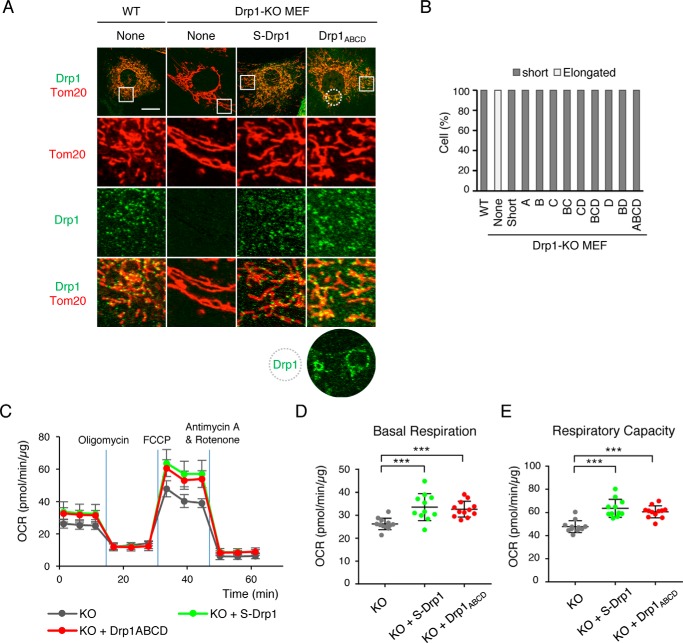
To test the function of Drp1ABCD in mitochondrial division, we individually expressed Drp1ABCD and other isoforms in Drp1-KO MEFs using the lentivirus transduction system (Fig. 1G). The lentivirus system provided almost 100% transduction efficiency and yielded similar expression levels for each Drp1 isoform in Drp1-KO MEFs (Fig. 1G) (10). Using these cells, we performed confocal immunofluorescence microscopy with antibodies to Drp1 (pan-Drp1) and the mitochondrial protein Tom20. Drp1-KO MEFs showed an elongated mitochondrial morphology because of defects in mitochondrial division compared with WT MEFs (Fig. 2, A and B). All of the isoforms, including Drp1ABCD, restored short tubular mitochondria in Drp1-KO MEFs, which look similar to those seen in WT MEFs (Fig. 2, A and B). Consistent with its function in mitochondrial division, Drp1ABCD formed punctate structures on mitochondria, similar to other Drp1 isoforms (Fig. 2A). In addition to mitochondrial division, Drp1ABCD also functions in mitochondrial respiration. We measured oxygen consumption rates in Drp1-KO MEFs and Drp1-KO MEFs expressing S-Drp1 or Drp1ABCD. Results showed that the expression of S-Drp1 and Drp1ABCD similarly increased basal respiration and respiratory capacity (Fig. 2, C–E).
Figure 2.
Drp1ABCD locates at mitochondria and functions in mitochondrial division. A, we examined MEFs and Drp1-KO MEFs that individually express Drp1 isoforms using confocal immunofluorescence microscopy with antibodies to Drp1 and the mitochondrial outer membrane protein Tom20. Images of MEFs and Drp1-KO MEFs carrying no Drp1, S-Drp1, or Drp1ABCD are shown. Boxed and circled regions are enlarged. Scale bar = 20 μm. B, quantification of mitochondrial morphology. 30 cells were analyzed for each isoform. C–E, the oxygen consumption rate (OCR) was measured in Drp1-KO MEFs and Drp1-KO MEFs that express S-Drp1 or Drp1ABCD. Oligomycin (2 μm), carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP, 1 μm), and antimycin A (0.5 μm)/rotenone (0.5 μm) were added at the times indicated by the blue lines. Values are means ± S.D. (n ≥ 11) in C. Basal respiration (D) and respiratory capacity (E) are shown. Error bars are means ± S.D. in D and E. Statistical analysis was performed using Student's t test: ***, p < 0.001.
Previous studies have shown that Drp1 mediates peroxisomal division (8). We therefore tested whether Drp1ABCD promotes peroxisomal division using immunofluorescence microscopy with antibodies against Drp1 and a peroxisomal protein, Pex14. Consistent with defects in peroxisomal division, Drp1-KO MEFs showed elongated peroxisomes (Fig. S1). In contrast, Drp1-KO MEFs expressing S-Drp1 or Drp1ABCD displayed normal oval and round peroxisomes. Both S-Drp1 and Drp1ABCD localized to peroxisomes (Fig. S1).
Drp1ABCD associates with lysosomes and late endosomes
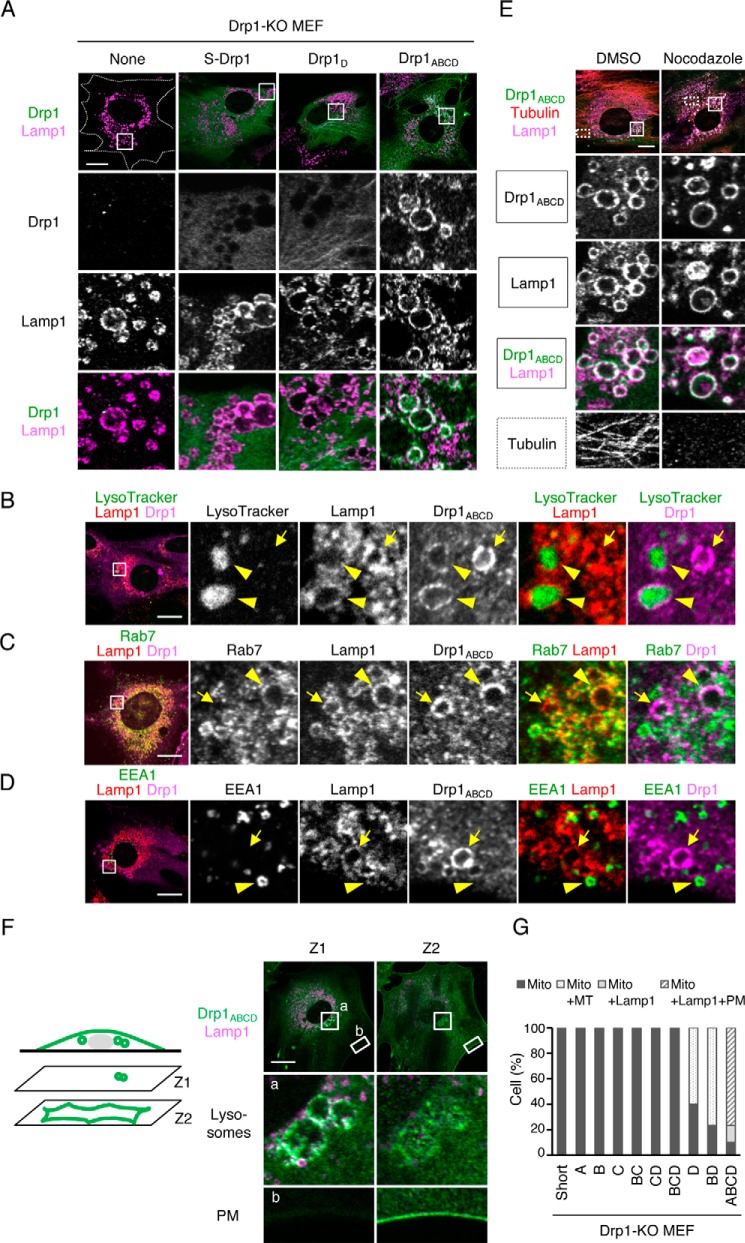
Interestingly, we noticed that Drp1ABCD, but not the other nine isoforms, is also located at vesicular structures in the cytosol in addition to mitochondria when we changed focal planes (Fig. 2A, circled area). To characterize these vesicles, we performed confocal immunofluorescence microscopy with antibodies against Drp1 and Lamp1, a membrane protein associated with lysosomes and late endosomes. We found that Drp1ABCD-positive vesicles were co-labeled with the anti-Lamp1 antibody (Fig. 3A). To further test whether Drp1ABCD is associated with lysosomes and/or late endosomes, we stained Drp1-KO MEFs expressing Drp1ABCD with LysoTracker and then performed immunofluorescence microscopy with Drp1 and Lamp1 antibodies. We found that a subset of Drp1ABCD-positive vesicles co-labeled with LysoTracker (Fig. 3B). We also analyzed Drp1-KO MEFs expressing Drp1ABCD using antibodies against Drp1, Lamp1, and a late endosome marker, Rab7. The results showed that some of Drp1ABCD-positive vesicles co-localized with Rab7 (Fig. 3C). In contrast, when we used antibodies against EEA1, an early endosomal marker, we found no co-localization with Drp1ABCD (Fig. 3D). These data suggest that Drp1ABCD is associated with both lysosomes and late endosomes.
Figure 3.
Drp1ABCD is associated with lysosomes, late endosomes, and the plasma membrane. A, we analyzed Drp1-KO MEFs that carry each Drp1 isoform using immunofluorescence microscopy with antibodies to Drp1 and Lamp1, a membrane protein that is associated with lysosomes and late endosomes. Images of Drp1-KO MEFs carrying no Drp1, S-Drp1, Drp1D, or Drp1ABCD are shown. Boxed regions are enlarged. Scale bar = 20 μm. B, Drp1-KO MEFs expressing Drp1ABCD were stained with 75 nm LysoTracker (Invitrogen, L7528) for 30 min and then subjected to immunofluorescence microscopy with antibodies against Lamp1 and Drp1. The boxed region is enlarged. Scale bar = 20 μm. C and D, immunofluorescence microscopy of Drp1-KO MEFs expressing Drp1ABCD was performed using antibodies against Rab7, Lamp1, and Drp1 (C) and EEA1, Lamp1, and Drp1 (D). Scale bars = 20 μm. E, Drp1-KO MEFs expressing Drp1ABCD were treated with 20 μm nocodazole for 4 h and subjected to immunofluorescence microscopy with antibodies to tubulin, Drp1, and Lamp1. Scale bar = 20 μm. F, confocal immunofluorescence microscopy images of Drp1-KO MEFs expressing Drp1ABCD with antibodies against Lamp1 and Drp1. Images were taken at two different focal planes, Z1 and Z2. The Z1 and Z2 focal planes are 0.9 μm apart. Boxed regions are enlarged. Scale bar = 20 μm. G, quantification of Drp1 localization. 30 cells were analyzed for each isoform. Mito, mitochondria; MT, microtubules; Lamp1, Lamp1-positive vesicles; PM, plasma membrane.
Because lysosomes/late endosomes are associated with microtubules, we tested whether depolymerizing microtubules changes the localization of Drp1ABCD at these membrane vesicles. We found that a microtubule-depolymerizing drug, nocodazole, does not affect the localization of Drp1ABCD on lysosomes/late endosomes, indicating that association of Drp1ABCD with these vesicles is independent of microtubules (Fig. 3E).
By moving the focal plane close to the bottom of cells, we found that Drp1ABCD was associated with the plasma membrane (Fig. 3F). We often found fewer Lamp1-positive vesicles in the focal planes, where we observed localization of Drp1ABCD at the plasma membrane (Fig. 3, F and G). To confirm this plasma membrane localization, we examined Drp1-KO MEFs co-expressing Drp1ABCD and a plasma membrane marker, Lyn-mCherry (22). We found that Drp1ABCD co-localized with Lyn-mCherry (Fig. S2)
Because Drp1 has the ability to control the size of mitochondria through mitochondrial division, we tested whether Drp1ABCD also changes the size of lysosomes/late endosomes. We stained WT MEFs, Drp1-KO MEFs, and Drp1-KO MEFs expressing either S-Drp1 or Drp1ABCD using LysoTracker and analyzed the size of LysoTracker-stained vesicles using confocal microscopy. We found that the size distribution of LysoTracker-stained vesicles was similar in these cells (Fig. 4A). Mean and median areas of LysoTracker-stained vesicles were also indistinguishable (Fig. 4B).
Figure 4.
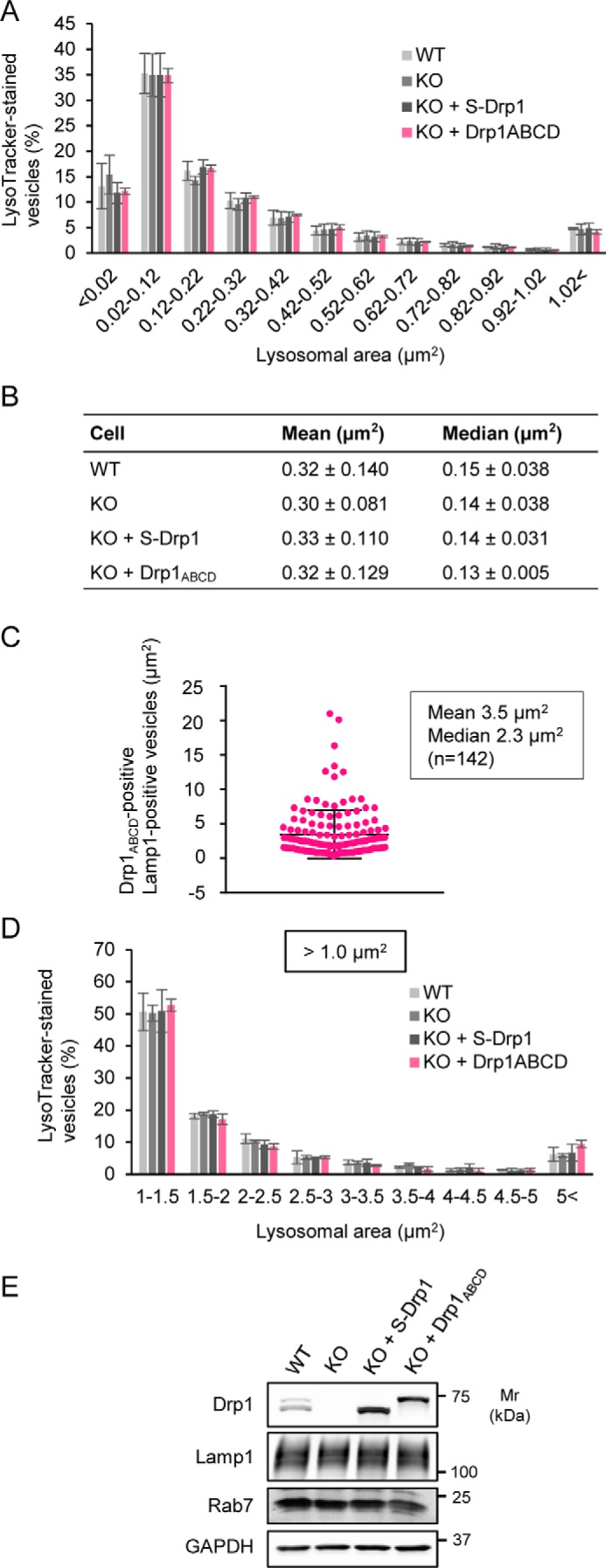
Drp1ABCD does not affect the size of lysosomes/late endosomes. A, WT MEFs, Drp1-KO MEFs, and Drp1-KO MEFs expressing either S-Drp1 or Drp1ABCD were stained with LysoTracker (75 nm for 30 min). The size distribution of the areas of LysoTracker-stained vesicles is shown. Thirty cells were analyzed. Values represent means ± S.D. (n = 3). B, the mean and median of the area of LysoTracker-stained vesicles (n = 3, 30 cells were analyzed for each cell type in each experiment). C, Drp1-KO MEFs expressing Drp1ABCD were analyzed by immunofluorescence microscopy with antibodies against Drp1 and Lamp1. The size distribution of Lamp1-positive vesicles associated with Drp1ABCD is shown. Error bars represent means ± S.D., and 142 vesicles were analyzed. D, the size distribution of LysoTracker-stained vesicles of an area larger than 1 μm2. The same set of MEFs as described in A was analyzed. Values represent means ± S.D. (n = 3). E, Western blotting of whole-cell lysates from MEFs, Drp1-KO MEFs, and Drp1-KO MEFs expressing either S-Drp1 or Drp1ABCD.
To examine vesicles associated with Drp1ABCD, we performed immunofluorescence microscopy with antibodies against Drp1 and Lamp1 in Drp1-KO MEFs expressing Drp1ABCD. Although the areas of individual LysoTracker-stained vesicles were ∼0.3 μm2 (mean) and 0.15 μm2 (median) in Drp1-KO MEFs expressing Drp1ABCD (Fig. 4B), the areas of those associated with Drp1ABCD were 3.5 μm2 (mean) and 2.3 μm2 (median) (Fig. 4C).
We then tested whether Drp1ABCD affects the size of larger LysoTracker-stained vesicles by reanalyzing the size distribution for LysoTracker-stained vesicles that have larger area (>1 μm2). Again, we found similar sizes of LysoTracker-stained vesicles in WT MEFs, Drp1-KO MEFs, and Drp1-KO MEFs expressing either S-Drp1 or Drp1ABCD (Fig. 4D). Thus, Drp1ABCD does not affect the size of lysosomes/late endosomes. Western blotting of whole-cell lysates showed that the expression levels of S-Drp1 and Drp1ABCD are comparable (Fig. 4E).
Drp1ABCD is located at interfaces between mitochondria and lysosomes/late endosomes
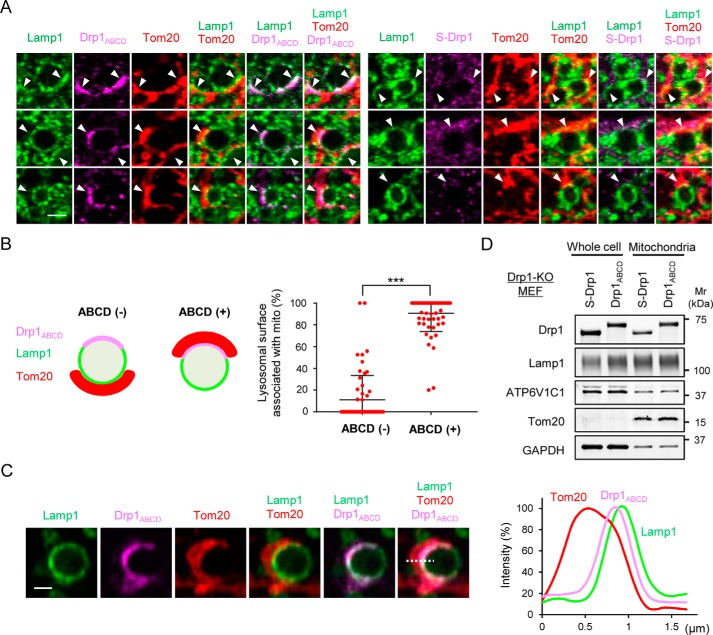
We noticed that the distribution of Drp1ABCD along the Lamp1-positive membrane is not uniform; even on the same vesicles, there are regions that are associated with Drp1ABCD whereas other parts are not (see gallery of Drp1ABCD-associated Lamp1-positive vesicles in Fig. 5A). Interestingly, parts of Lamp1-positive membranes that associated with Drp1ABCD appear to be close contact with mitochondria labeled with antibodies to the mitochondrial outer membrane protein Tom20 (Fig. 5A, arrowheads). Quantification of these images showed that Lamp1-positive vesicles are significantly more closely located to mitochondria where Drp1ABCD is present (Fig. 5B). To further determine the spatial relationship between Lamp1, Drp1ABCD, and the mitochondrial outer membrane, we performed a line scan analysis of the fluorescent signal of Lamp1, Drp1, and Tom20. As shown in Fig. 5C, the peak of the Drp1ABCD signal is located between the peak of Lamp1 and Tom20 signals. It appears that Drp1ABCD is present between the Lamp1-positive membranes and mitochondria.
Figure 5.
Drp1ABCD is located at interfaces between mitochondria and lysosomes/late endosomes. A, Drp1-KO MEFs carrying Drp1ABCD and S-Drp1 were analyzed by immunofluorescence microscopy with antibodies to Lamp1, Drp1, and the mitochondrial protein Tom20. A gallery of Lamp1-positive vesicles is shown. Three examples for each Drp1 construct are shown. Scale bar = 2 μm. B, quantification of Lamp1-positive vesicles that are in close contact with mitochondria in Drp1-KO MEFs expressing Drp1ABCD. We measured the frequency of Lamp1-positive membranes that are closely faced to mitochondria when Drp1ABCD is present (+) or not (−), as depicted in the schematic. Error bars represent the average ± S.D. (n = 61). Statistical analysis was performed using Student's t test: ***, p < 0.001. C, line scan analysis of the fluorescent signal of Lamp1, Drp1ABCD, and Tom20 in immunofluorescence microscopy of Drp1-KO MEFs expressing Drp1ABCD. Scale bar = 1 μm. The fluorescent intensity was quantified along the dotted line. The peak position of Drp1 is located between those of Lamp1 and Tom20. D, mitochondria were isolated from Drp1-KO MEFs expressing S-Drp1 or Drp1ABCD using a mitochondrial isolation kit (Thermo, 89874). Whole-cell lysates and mitochondrial fractions were analyzed by Western blotting with antibodies against Drp1, Lamp1, ATP6V1C1, Tom20, and GAPDH.
To determine whether contacts between mitochondria and Lamp1-positive vesicles increase when Drp1ABCD is ectopically expressed in Drp1-KO MEFs, we biochemically fractionated mitochondria and examined how many lysosomes/late endosomes co-fractionate using Western blotting with antibodies against Lamp1 and V-type proton ATPase subunit C. The results showed that similar amounts of these two proteins associated with the mitochondrial fraction derived from Drp1-KO MEFs expressing S-Drp1 or Drp1ABCD (Fig. 5D). These data suggest that Drp1ABCD is recruited to existing interorganelle interfaces between mitochondria and lysosomes/late endosomes.
Exons A and B target Drp1ABCD to lysosomes/late endosomes and the plasma membrane
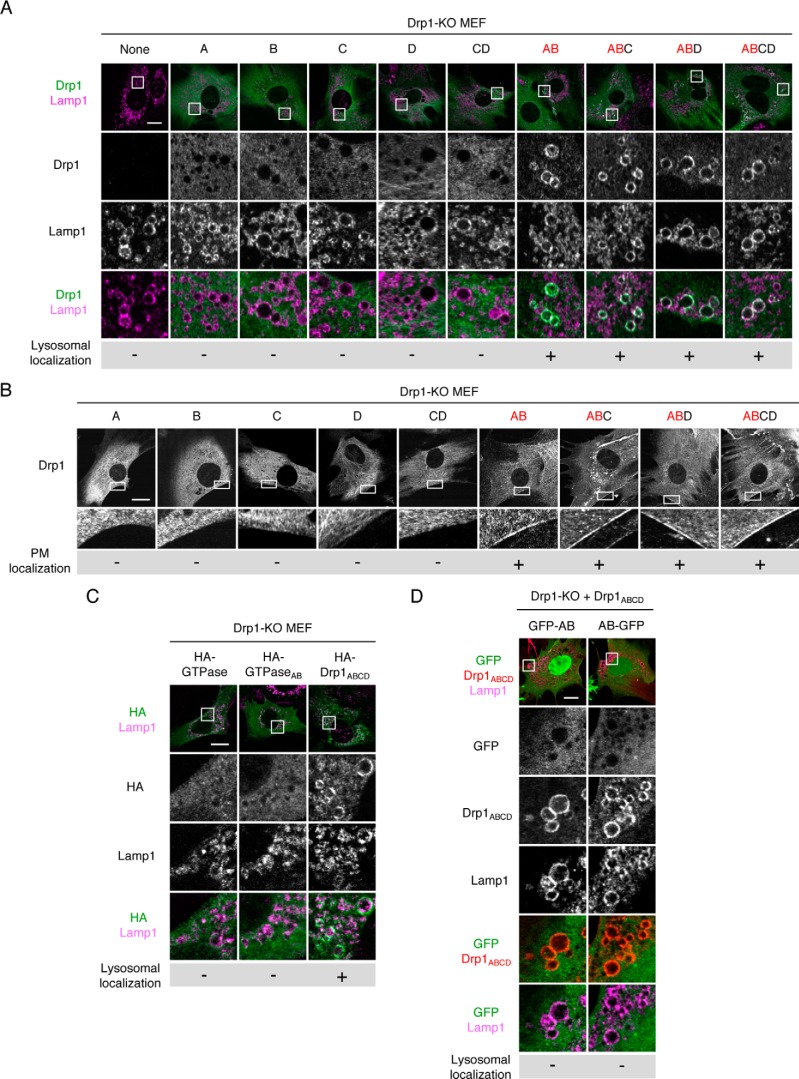
To determine the mechanisms by which Drp1ABCD is associated with lysosomes/late endosomes and the plasma membrane, we tested which alternative exons are required for these localizations. We transduced Drp1-KO MEFs with lentiviruses expressing Drp1 carrying different combinations of the four alternative exons: A, B, C, D, CD, AB, ABC, ABD, and ABCD. Immunofluorescence microscopy of these MEFs using antibodies to Drp1 and Lamp1 showed that Drp1A, Drp1B, Drp1C, Drp1D, and Drp1CD do not associate with Lamp1-positive vesicles (Fig. 6A), consistent with the earlier data (Fig. 3A). In contrast, all of the Drp1 isoforms that contained both exon A and B (e.g. Drp1AB, Drp1ABC, Drp1ABD, and Drp1ABCD) are localized to Lamp1-positive vesicles (Fig. 6A). Therefore, the presence of both exons A and B determines the localization of Drp1ABCD at lysosomes and late endosomes.
Figure 6.
Exons A and B are important for the localization of Drp1ABCD at lysosomes/late endosomes and the plasma membrane. A, Drp1-KO MEFs individually expressing the indicated Drp1 isoforms were analyzed by confocal immunofluorescence microscopy with antibodies to Drp1 and Lamp1. Scale bar = 20 μm. B, the indicated MEFs with specific Drp1 isoforms were analyzed by immunofluorescence microscopy using anti-Drp1 antibodies. Boxed regions are enlarged. Scale bar = 20 μm. C, Drp1-KO MEFs expressing the HA-tagged GTPase domain carrying no exon, AB exons, or full-length Drp1ABCD were analyzed by immunofluorescence microscopy using antibodies to the HA epitope and Lamp1. D, Drp1-KO MEFs expressing Drp1ABCD along with GFP-tagged AB exons (GFP-AB or AB-GFP) were analyzed by immunofluorescence microscopy using antibodies to Drp1 and Lamp1. Scale bar = 20 μm.
Using the same set of cells, we also examined the localization of these Drp1 proteins at the plasma membrane. Similar to lysosomal/late endosomal localization, we found that exons A and B are also necessary for the plasma membrane localization of Drp1 (Fig. 6B).
To test whether the GTPase domain with the AB insertion or the AB insertion itself is sufficient for localization to Lamp1-positive vesicles and the plasma membrane, we performed immunofluorescence microscopy using antibodies against the HA epitope and Drp1 in Drp1-KO MEFs expressing either the HA-GTPase domain or the HA-GTPaseAB domain. We found that both of the GTPase domains were present in the cytosol (Fig. 6C). Similarly, GFP fused to the AB insertion (GFP-AB and AB-GFP) localized to the cytosol when expressed in Drp1-KO MEFs (Fig. 6D). Therefore, in addition to the GTPase domain, other regions of Drp1ABCD are also necessary for targeting Drp1ABCD to Lamp1-positive vesicles and the plasma membrane.
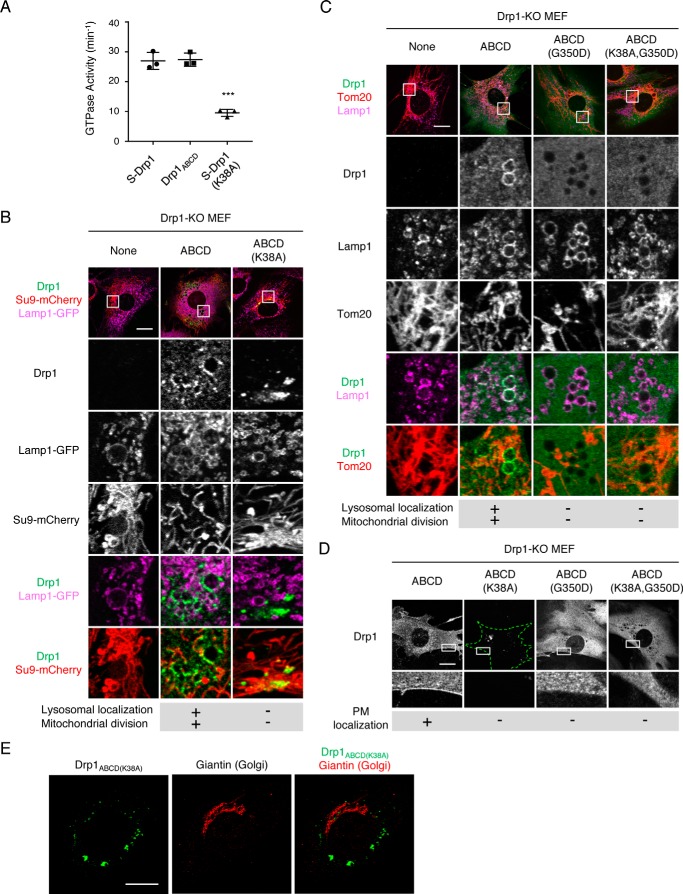
GTP hydrolysis and oligomerization of Drp1ABCD is required for its localization to lysosomes/late endosomes and the plasma membrane
Localization of Drp1 to mitochondria is regulated by its oligomerization and disassembly of oligomers, which is triggered by GTP hydrolysis (5). To further define the mechanism underlying the subcellular localization of Drp1ABCD, we tested the role of the GTPase activity of Drp1ABCD and its oligomerization in localization at lysosomes/late endosomes and the plasma membrane. We first determined whether Drp1ABCD has GTPase activity because Drp1ABCD has an extra amino acid insertion derived from exons A and B in the GTPase domain. We purified recombinant His6-Drp1ABCD proteins from E. coli and incubated them with GTP (Fig. 7A). GTP hydrolysis was analyzed by measuring the release of phosphate using a malachite green assay as we performed previously (23). The results showed that His6-Drp1ABCD hydrolyzes GTP similar to a positive control, His6-S-Drp1 (Fig. 7A). As a negative control, we used His6-S-Drp1 (K38A), which carries a mutation in the GTPase domain that blocks GTP hydrolysis (24) (Fig. 7A). To assess the role of the GTPase activity of Drp1ABCD in its localization in cells, we expressed Drp1ABCD (K38A) in Drp1-KO MEFs and performed confocal immunofluorescence microscopy. We found that Drp1ABCD (K38A) does not localize to Lamp1-positive vesicles, the plasma membrane, or mitochondria, instead forming aggregate in the cytosol (Fig. 7, B and D). Although we sometimes observed Drp1ABCD (K38A) aggregates in perinuclear regions, these aggregates were not associated with the Golgi complex (Fig. 7E). Similar aggregation has been observed for S-Drp1 (K38A) and likely results from the defect of disassembly of Drp1 oligomers because GTP hydrolysis is required for this disassembly (5).
Figure 7.
Localizations of Drp1ABCD at lysosomes/late endosomes and the plasma membrane depends on GTP hydrolysis and oligomerization. A, the GTPase activity of purified, recombinant His6-S-Drp1, His6-Drp1ABCD, and His6-S-Drp1 (K38A) was measured in a malachite green assay. Error bars are mean ± S.D. (n = 3). Statistical analysis was performed using Student's t test: ***, p < 0.001. B, Drp1-KO MEFs expressing mitochondrion-targeted Su9-mCherry and Lamp1-GFP along with mutant Drp1ABCD proteins were subjected to immunofluorescence microscopy with anti-Drp1 antibodies. Scale bar = 20 μm. C and D, Drp1-KO MEFs expressing mutant Drp1ABCD proteins were subjected to immunofluorescence microscopy with antibodies to Drp1, Lamp1, and Tom20 (C) and Drp1 (D). Scale bars = 20 μm. E, Drp1-KO MEFs expressing Drp1ABCD proteins were subjected to immunofluorescence microscopy with antibodies to Drp1 and a Golgi protein, giantin. Scale bar = 20 μm.
Second, we introduced the G350D mutation, which inhibits the oligomerization of Drp1 (25), and expressed Drp1ABCD (G350D) in Drp1-KO MEFs. Confocal immunofluorescence microscopy showed that Drp1ABCD (G350D) diffuses in the cytosol and does not associate with Lamp1-positive vesicles, the plasma membrane, or mitochondria (Fig. 7, C and D). Furthermore, combination of the K38A and G350D mutations showed effects on Drp1ABCD localization similar to the G350D mutation alone, suggesting that oligomerization defects are epistatic to GTP hydrolysis defects (Fig. 7, C and D). Taken together, these data suggest that the localizations of Drp1ABCD at lysosomes/late endosomes and the plasma membrane require its ability to hydrolyze GTP and homo-oligomerize, suggesting that Drp1ABCD is associated with these membranes as assembled structures.
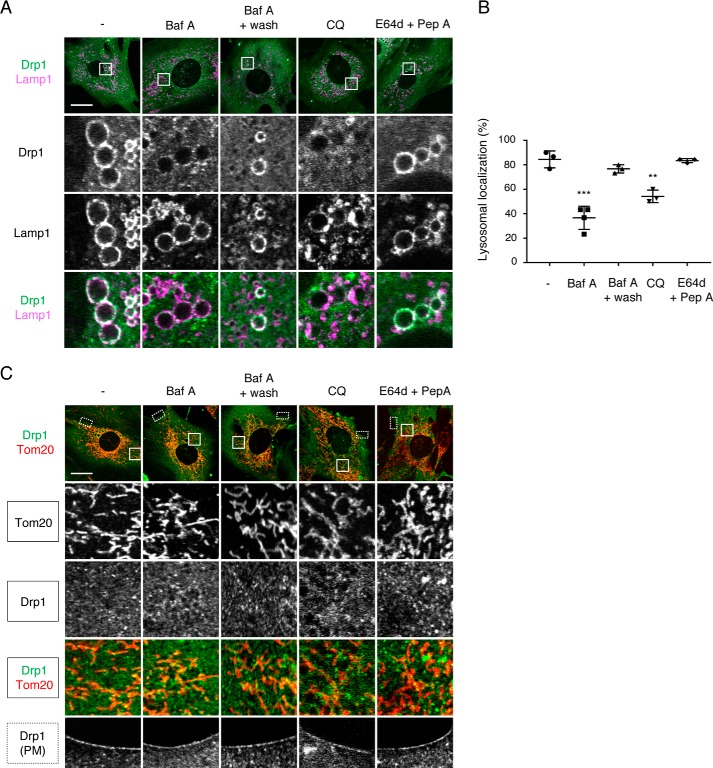
The localization of Drp1ABCD at lysosomes/late endosomes is sensitive to bafilomycin A and chloroquine
To determine whether the localization of Drp1ABCD at lysosomes/late endosomes depends on the acidity or function of these membrane compartments, we first treated Drp1-KO MEFs expressing Drp1ABCD with two inhibitors, bafilomycin A and chloroquine, both of which block acidification of lysosomes/endosomes through distinct mechanisms (26). Although bafilomycin A blocks vacuolar H+-ATPase, mild base chloroquine neutralizes the lysosomal/endosomal acidic pH in these organelles. We found that these inhibitors significantly decreased the association of Drp1ABCD with Lamp1-positive vesicles (Fig. 8, A and B). This inhibition was reversible because the removal of bafilomycin A from the culture medium restored the localization of Drp1ABCD at Lamp1-positive vesicles in 2 h (Fig. 8, A and B). These inhibitory effects are specific to Lamp1-positive vesicles; localization of Drp1ABCD at mitochondria and the plasma membrane was not affected by bafilomycin A or chloroquine (Fig. 8C).
Figure 8.
Bafilomycin A and chloroquine decrease the association of Drp1ABCD with lysosomes/late endosomes. A and C, Drp1-KO MEFs carrying Drp1ABCD were treated with 20 nm bafilomycin A for 1 h, or 50 μm chloroquine (CQ) for 1 h, or a combination of 10 μg/ml E-64d and 10 μg/ml pepstatin A. We also washed out bafilomycin A and incubated further for 2 h. These cells were subjected confocal immunofluorescence microscopy with antibodies to Drp1 and Lamp1 (A) or antibodies to Drp1 and Tom20 (C). Scale bars = 20 μm. B, quantification of cells that show association of Drp1ABCD with Lamp1-positive vesicles. Error bars are mean ± S.D. (n = 3, 30 cells are analyzed for each treatment in each experiment). Statistical analysis was performed using Student's t test: **, p < 0.01; ***, p < 0.001.
Increased pH could decrease the activity of acid hydrolyases and compromise the function of lysosomes. To find out whether the function of lysosomes is required for the localization of Drp1ABCD at lysosomes/late endosomes, we treated Drp1-KO MEFs expressing Drp1ABCD with inhibitors to cysteine proteases (E-64d) and to aspartic acid protease (pepstatin A), both of which inhibit lysosomal proteases (26). We found that these protease inhibitors do not change the localization of Drp1ABCD at lysosomes (Fig. 8A and B). These data suggest that the acidity of lysosomes, but not their function, is required for the association of Drp1ABCD with lysosomes/late endosomes.
Discussion
In this work, we identified a new Drp1 isoform, Drp1ABCD, which predominantly expresses in the brain and locates at lysosomes, late endosomes and the plasma membrane in addition to mitochondria. The identification of Drp1ABCD expands a repertoire of Drp1 isoforms that are differentially expressed in tissues and organs. Our data also suggest that neuronal Drp1ABCD may function at lysosome/endosome–mitochondria contact sites in endosome–lysosome pathways in addition to mitochondrial division. We currently do not know the exact function of Drp1ABCD and are planning to address this by specifically knocking down Drp1ABCD in neurons in our future studies.
The unique localization of Drp1ABCD to lysosomes, late endosomes, and the plasma membrane depends on the combination of exons A and B. Based on the crystal structure of Drp1, amino acid sequences encoded in these two exons are predicted to be exposed on the surface of this protein (Fig. 1B). These amino acid sequence are necessary, but likely not sufficient, to localize Drp1ABCD to lysosomes, late endosomes, and the plasma membrane because oligomerization of Drp1ABCD, which is mediated by the stalk domain, is also essential for these localizations. It appears that Drp1ABCD assembles into oligomers that are recruited to these membranes. Exons A and B likely help recruit Drp1ABCD oligomers to lysosomes, late endosomes, and the plasma membrane because we do not find association of Drp1ABCD (G350D) with these membrane structures. However, it is also possible that Drp1ABCD oligomerizes onto the membrane structures and helps recruit more Drp1ABCD molecules in a synergistic manner. The localization signal created by exons A and B is not exclusive, as we observed that a fraction of Drp1ABCD proteins is associated with mitochondria and peroxisomes.
The variable domain, which is a predicted unstructured loop with ∼100 amino acids, has been proposed to mediate several types of Drp1 regulation (27). For example, this domain undergoes phosphorylation to regulate the interaction of Drp1 with its receptor/effector proteins such as MFF, Fis1, and Mid49/51 (28, 29). Such regulations modulate the recruitment of Drp1 to mitochondria and its assembly on mitochondria. In addition, this domain also participates in interactions with two mitochondrial phospholipids, cardiolipin and phosphatidic acid, that regulate Drp1 function at distinct steps. Cardiolipin promotes the oligomerization of Drp1, whereas phosphatidic acid suppresses oligomerized Drp1 to initiate mitochondrial division (4, 23, 30–33). Alternative splicing of exons C and D would regulate these protein–protein and protein–phospholipid interactions that fine-tune mitochondrial division. The alternative splicing in the variable domain also changes the association of Drp1 with microtubules and regulates microtubule stability. In addition to the variable domain, our data show that alternative splicing of exons A and B in the GTPase domain also plays a critical role in the regulation of Drp1 beyond its GTP hydrolysis activity. The amino acid sequence derived from exons A and B is located on the surface of this domain, and these two exons are necessary and sufficient alternative axons for lysosomal/endosomal and plasma membrane localization. We therefore speculate that this region likely contributes to a unique protein–protein or protein–lipid interaction for the association of Drp1ABCD with specific molecules in the endosomal–lysosomal system.
Experimental procedures
Mice and MEFs
All animal work was done according to guidelines established by the Johns Hopkins University Committee on Animal Care. Drp1-KO MEFs were cultured in Iscove's modified Dulbecco's medium supplemented with 10% fetal bovine serum as described previously (10).
Drp1 transcript analysis
Total RNAs were isolated from mouse brains and MEFs using the RNeasy kit (Qiagen). cDNAs of Drp1 isoforms were prepared from total RNAs using the primers 5-BamH1 Drp1 (AAA GGATCC GCCACC ATGGAGGCGCTGATCCCGGT) and 3-Not1 Drp1 (AAA GCGGCCGC TCACCAAAGATGAGTCTCTC) using the OneStep-RT-PCR kit (Qiagen). Drp1 transcripts in the heart, skeletal muscle, liver, lung, spleen, and testis were PCR-amplified from a mouse cDNA library (Mouse MTCTM Panel I, Clontech). We subcloned ∼100 Drp1 transcripts from each organ and from MEFs into the pHR-SIN plasmid at BamHI and Not1 sites. We individually sequenced all of these plasmids and identified isoforms. The relative abundance of each isoform in each organ (percent) was calculated by dividing the number of plasmids carrying a specific isoform by the total number of plasmids sequenced.
Expression and purification of recombinant Drp1 proteins
Recombinant His6-S-Drp1, His6-Drp1BCD, His6-Drp1ABCD, and His6-S-Drp1 (K38A) were purified from E. coli as reported previously (23).
Production of anti-exon AB antibodies
Anti-exon AB antibodies that specifically recognize Drp1ABCDwere produced using a peptide (amino acid sequence KFQSWNPATWKN) as the antigen in rabbits at NeoScientific and affinity-purified using the same peptide. To remove the residual activity of the exon AB antibodies to bind exon B alone, they were preincubated with purified recombinant His6-Drp1BCD overnight prior to Western blotting. In some experiments, we incubated the exon AB antibodies with nitrilotriacetic acid–agarose beads coupled to His6-Drp1BCD (molar ratio of anti-AB IgG His6-Drp1BCD = 1:2.5) and used the supernatant after centrifugation. The specificity of the antibodies was tested using Western blotting of Drp1-KO MEFs individually expressing Drp1ACD, Drp1BCD, or Drp1ABCD.
Western blotting
Proteins were separated using SDS-PAGE and transferred onto PVDF membranes. The antibodies used were exon AB, Drp1 (BD Biosciences, 611113), Tom20 (BD Biosciences, 612278), PDH subunit E2/E3bp (Abcam, ab110333), GAPDH (Thermo, MA5-15738), actin (Santa Cruz Biotechnology, sc-1615), Lamp1 (BD Biosciences, 553792), ATP6V1C1 (ProteinTech, 16054-1-AP), and Rab7 (Cell Signaling Technology, 9367). Immunocomplexes were visualized using fluorescently labeled secondary antibodies and detected using a PharosFX Plus molecular imager (Bio-Rad).
Lentiviruses and plasmids
Lentiviruses were produced as described previously (10). The pHR-SIN plasmid carrying Drp1 isoforms was cotransfected into HEK293T cells with two other plasmids, pHR-CMV8.2ΔR and pCMV-VSVG, using Lipofectamine 2000 (Invitrogen). Two days after transfection, the supernatant of transfected cells containing released viruses was collected. The viruses were quick-frozen in liquid nitrogen and stored at −80 °C.
To generate HA-Drp1ABCD, full-length Drp1ABCD was cloned into BamHI and NotI sites of the pEGFP-N1 vector (Clontech) to replace enhanced GFP with Drp1ABCD. Next, 1× HA was cloned into EcoRI and KpnI sites. To make HA-GTPase, Drp1ABCD was replaced with the GTPase domain of S-Drp1 (1–343 amino acids) or Drp1ABCD (1–362 amino acids) at BamHI and NotI sites. For GFP-AB and AB-GFP, AB exons were cloned into the pEGFP-N1 and pEGFP-C1 vectors at XhoI and BamHI sites. The plasmid carrying Lyn-mCherry was a gift from Dr. Hideki Nakamura (Johns Hopkins University).
Immunofluorescence microscopy
MEFs were fixed using PBS containing 4% paraformaldehyde, washed in PBS, permeabilized with 0.1% Triton X-100/PBS, and blocked in 0.5% BSA/PBS, except for MEFs expressing Lyn-mCherry, which were fixed using methanol at −20 °C for 20 min. The cells were incubated with antibodies to Drp1, Tom20, Lamp1, Rab7, EEA1 (Cell Signaling Technology, 3288S), Pex14 (8), giantin (Covance, PRB-114C), and tubulin (Cell Signaling Technology, 2125), followed by the appropriate secondary antibodies. Samples were viewed using Zeiss LSM510-Meta and LSM800 GaAsPlaser scanning confocal microscopes (9).
To quantify the size of Lamp1-positive vesicles, cells were stained with 75 nm LysoTracker Red DND-99 (Invitrogen, L7528) at 37 °C for 30 min in the culture medium. Images were converted to binary images using ImageJ, and the area of each vesicle was quantified.
Mitochondrial respiration
Mitochondrial oxygen consumption rates were measured in an XF96 Extracellular Flux Analyzer (Seahorse Bioscience) as described previously described (10, 34). Briefly, cells were plated at a density of 5,000 cells/well in an XF 96-well culture microplate and incubated for 24 h. To record oxygen consumption rates, the culture medium was replaced with Seahorse XF base medium supplemented with 25 mm glucose, 1 mm sodium pyruvate, and 2 mm GlutaMAX-I, and then the cells were incubated at 37 °C in a CO2-free incubator for 1 h. Oxygen consumption rate analysis was initiated with a 13.5-min equilibration step, followed by three cycles of 2 min of mixing and 3 min of measurement. All measurement cycles were performed at 37 °C. The basal oxygen consumption rate was determined, followed by sequential injections of 2 μm oligomycin, 1 μm carbonyl cyanide p-trifluoromethoxyphenylhydrazone, and 0.5 μm rotenone plus 0.5 μm antimycin A. Oxygen consumption rates were normalized relative to the amount of protein in each well.
GTP hydrolysis by purified Drp1 proteins
The GTPase activity of His6-S-Drp1, His6-Drp1ABCD, and His6-S-Drp1 (K38A) was measured as we described previously (23). 0.1 μm Drp1 was incubated with 1 mm GTP in 150 mm KCl, 2 mm MgCl2, 1 mm DTT, and 20 mm MES (pH 7.0). 25-μl samples were mixed with 100 μl of malachite green solution (Echelon, K-1500) at room temperature for 20 min, and the absorbance at A620 was measured.
Author contributions
K. I., M. I., and H. S. conceptualization; K. I. and Y. A. formal analysis; K. I., M. I., and H. S. funding acquisition; K. I., Y. A., T. Yamada, T. L. S., T. O., M. I., and H. S. investigation; K. I., Y. A., T. Yamada, T. L. S., and T. O. methodology; K. I., M. I., and H. S. writing-original draft; H. M. M., T. Yoshimori, M. I., and H. S. supervision; M. I. and H. S. resources; M. I. and H. S. project administration.
Supplementary Material
Acknowledgments
We thank the members of the M. I. and H. S. laboratories for helpful discussions and technical assistance.
This work was supported by NIGMS, National Institutes of Health Grants GM084015 (to M. I.) and GM123266 (to H. S.), the Allegheny Health Network–Sidney Kimmel Comprehensive Cancer Center, and the Johns Hopkins University–University of Maryland Diabetes Research Center). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
- MEF
- mouse embryonic fibroblast
- KO
- knockout
- cDNA
- complementary DNA
- HA
- hemagglutinin
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- PDH
- pyruvate dehydrogenase.
References
- 1. Pernas L., and Scorrano L. (2016) Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 78, 505–531 10.1146/annurev-physiol-021115-105011 [DOI] [PubMed] [Google Scholar]
- 2. Friedman J. R., and Nunnari J. (2014) Mitochondrial form and function. Nature 505, 335–343 10.1038/nature12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shutt T. E., and McBride H. M. (2013) Staying cool in difficult times: mitochondrial dynamics, quality control and the stress response. Biochim. Biophys. Acta 1833, 417–424 10.1016/j.bbamcr.2012.05.024 [DOI] [PubMed] [Google Scholar]
- 4. Kameoka S., Adachi Y., Okamoto K., Iijima M., and Sesaki H. (2018) Phosphatidic acid and cardiolipin coordinate mitochondrial dynamics. Trends Cell Biol. 28, 67–76 10.1016/j.tcb.2017.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roy M., Reddy P. H., Iijima M., and Sesaki H. (2015) Mitochondrial division and fusion in metabolism. Curr. Opin. Cell Biol. 33, 111–118 10.1016/j.ceb.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tamura Y., Itoh K., and Sesaki H. (2011) SnapShot: mitochondrial dynamics. Cell 145, 1158.e1 10.1016/j.cell.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S. O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y., Taguchi N., Morinaga H., Maeda M., Takayanagi R., Yokota S., and Mihara K. (2009) Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 11, 958–966 10.1038/ncb1907 [DOI] [PubMed] [Google Scholar]
- 8. Wakabayashi J., Zhang Z., Wakabayashi N., Tamura Y., Fukaya M., Kensler T. W., Iijima M., and Sesaki H. (2009) The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J. Cell Biol. 186, 805–816 10.1083/jcb.200903065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kageyama Y., Zhang Z., Roda R., Fukaya M., Wakabayashi J., Wakabayashi N., Kensler T. W., Reddy P. H., Iijima M., and Sesaki H. (2012) Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J. Cell Biol. 197, 535–551 10.1083/jcb.201110034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kageyama Y., Hoshijima M., Seo K., Bedja D., Sysa-Shah P., Andrabi S. A., Chen W., Höke A., Dawson V. L., Dawson T. M., Gabrielson K., Kass D. A., Iijima M., and Sesaki H. (2014) Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 33, 2798–2813 10.15252/embj.201488658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamada T., Adachi Y., Fukaya M., Iijima M., and Sesaki H. (2016) Dynamin-related protein 1 deficiency leads to receptor-interacting protein kinase 3-mediated necroptotic neurodegeneration. Am. J. Pathol. 186, 2798–2802 10.1016/j.ajpath.2016.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shields L. Y., Kim H., Zhu L., Haddad D., Berthet A., Pathak D., Lam M., Ponnusamy R., Diaz-Ramirez L. G., Gill T. M., Sesaki H., Mucke L., and Nakamura K. (2015) Dynamin-related protein 1 is required for normal mitochondrial bioenergetic and synaptic function in CA1 hippocampal neurons. Cell Death Dis. 6, e1725 10.1038/cddis.2015.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Song M., Mihara K., Chen Y., Scorrano L., and Dorn G. W. 2nd. (2015) Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 21, 273–286 10.1016/j.cmet.2014.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ikeda Y., Shirakabe A., Maejima Y., Zhai P., Sciarretta S., Toli J., Nomura M., Mihara K., Egashira K., Ohishi M., Abdellatif M., and Sadoshima J. (2015) Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ. Res. 116, 264–278 10.1161/CIRCRESAHA.116.303356 [DOI] [PubMed] [Google Scholar]
- 15. Udagawa O., Ishihara T., Maeda M., Matsunaga Y., Tsukamoto S., Kawano N., Miyado K., Shitara H., Yokota S., Nomura M., Mihara K., Mizushima N., and Ishihara N. (2014) Mitochondrial fission factor Drp1 maintains oocyte quality via dynamic rearrangement of multiple organelles. Curr. Biol. 24, 2451–2458 10.1016/j.cub.2014.08.060 [DOI] [PubMed] [Google Scholar]
- 16. Serasinghe M. N., Wieder S. Y., Renault T. T., Elkholi R., Asciolla J. J., Yao J. L., Jabado O., Hoehn K., Kageyama Y., Sesaki H., and Chipuk J. E. (2015) Mitochondrial division is requisite to RAS-induced transformation and targeted by oncogenic MAPK pathway inhibitors. Mol. Cell 57, 521–536 10.1016/j.molcel.2015.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kashatus J. A., Nascimento A., Myers L. J., Sher A., Byrne F. L., Hoehn K. L., Counter C. M., and Kashatus D. F. (2015) Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell 57, 537–551 10.1016/j.molcel.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Strack S., Wilson T. J., and Cribbs J. T. (2013) Cyclin-dependent kinases regulate splice-specific targeting of dynamin-related protein 1 to microtubules. J. Cell Biol. 201, 1037–1051 10.1083/jcb.201210045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Macdonald P. J., Francy C. A., Stepanyants N., Lehman L., Baglio A., Mears J. A., Qi X., and Ramachandran R. (2016) Distinct splice variants of dynamin-related protein 1 differentially utilize mitochondrial fission factor as an effector of cooperative GTPase activity. J. Biol. Chem. 291, 493–507 10.1074/jbc.M115.680181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoon Y., Pitts K. R., Dahan S., and McNiven M. A. (1998) A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J. Cell Biol. 140, 779–793 10.1083/jcb.140.4.779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wenger J., Klinglmayr E., Fröhlich C., Eibl C., Gimeno A., Hessenberger M., Puehringer S., Daumke O., and Goettig P. (2013) Functional mapping of human Dynamin-1-like GTPase domain based on X-ray structure analyses. PLoS ONE 8, e71835 10.1371/journal.pone.0071835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inoue T., Heo W. D., Grimley J. S., Wandless T. J., Meyer T. (2005) An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat. Methods. 2, 415–418 10.1038/nmeth763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adachi Y., Itoh K., Yamada T., Cerveny K. L., Suzuki T. L., Macdonald P., Frohman M. A., Ramachandran R., Iijima M., and Sesaki H. (2016) Coincident phosphatidic acid interaction restrains Drp1 in mitochondrial division. Mol. Cell 63, 1034–1043 10.1016/j.molcel.2016.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kageyama Y., Zhang Z., and Sesaki H. (2011) Mitochondrial division: molecular machinery and physiological functions. Curr. Opin. Cell Biol. 23, 427–434 10.1016/j.ceb.2011.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chang C. R., Manlandro C. M., Arnoult D., Stadler J., Posey A. E., Hill R. B., and Blackstone C. (2010) A lethal de novo mutation in the middle domain of the dynamin-related GTPase Drp1 impairs higher order assembly and mitochondrial division. J. Biol. Chem. 285, 32494–32503 10.1074/jbc.M110.142430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li M., Khambu B., Zhang H., Kang J. H., Chen X., Chen D., Vollmer L., Liu P. Q., Vogt A., and Yin X. M. (2013) Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J. Biol. Chem. 288, 35769–35780 10.1074/jbc.M113.511212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fröhlich C., Grabiger S., Schwefel D., Faelber K., Rosenbaum E., Mears J., Rocks O., and Daumke O. (2013) Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 32, 1280–1292 10.1038/emboj.2013.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Merrill R. A., and Strack S. (2014) Mitochondria: a kinase anchoring protein 1, a signaling platform for mitochondrial form and function. Int. J. Biochem. Cell Biol. 48, 92–96 10.1016/j.biocel.2013.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang C. R., and Blackstone C. (2010) Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N.Y. Acad. Sci. 1201, 34–39 10.1111/j.1749-6632.2010.05629.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stepanyants N., Macdonald P. J., Francy C. A., Mears J. A., Qi X., and Ramachandran R. (2015) Cardiolipin's propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol. Biol. Cell 26, 3104–3116 10.1091/mbc.e15-06-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Adachi Y., Iijima M., and Sesaki H. (2017) An unstructured loop that is critical for interactions of the stalk domain of Drp1 with saturated phosphatidic acid. Small GTPases 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Francy C. A., Alvarez F. J., Zhou L., Ramachandran R., and Mears J. A. (2015) The mechanoenzymatic core of Dynamin-related protein 1 comprises the minimal machinery required for membrane constriction. J. Biol. Chem. 290, 11692–11703 10.1074/jbc.M114.610881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bustillo-Zabalbeitia I., Montessuit S., Raemy E., Basañez G., Terrones O., and Martinou J. C. (2014) Specific interaction with cardiolipin triggers functional activation of Dynamin-related protein 1. PLoS ONE 9, e102738 10.1371/journal.pone.0102738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gajendiran P., Vega L. I., Itoh K., Sesaki H., Vakili M. R., Lavasanifar A., Hong K., Mezey E., and Ganapathy-Kanniappan S. (2018) Elevated mitochondrial activity distinguishes fibrogenic hepatic stellate cells and sensitizes for selective inhibition by mitotropic doxorubicin. J. Cell Mol. Med. 22, 2210–2219 10.1111/jcmm.13501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y. (2009) I-TASSER: fully automated protein structure prediction in CASP8. Proteins 77, 100–113 10.1002/prot.22588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.