Summary
The molecular mechanism underpinning the specification of the ectoderm, a transient germ-layer tissue, during mouse gastrulation was examined here in a stem cell-based model. We captured a self-renewing cell population with enhanced ectoderm potency from mouse epiblast stem cells (EpiSCs) by suppressing Nodal signaling activity. The transcriptome of the Nodal-inhibited EpiSCs resembles that of the anterior epiblast of embryonic day (E)7.0 and E7.5 mouse embryo, which is accompanied by chromatin modifications that reflect the priming of ectoderm lineage-related genes for expression. Nodal-inhibited EpiSCs show enhanced ectoderm differentiation in vitro and contribute to the neuroectoderm and the surface ectoderm in postimplantation chimeras but lose the propensity for mesendoderm differentiation in vitro and in chimeras. Our findings show that specification of the ectoderm progenitors is enhanced by the repression of Nodal signaling activity, and the ectoderm-like stem cells provide an experimental model to investigate the molecular characters of the epiblast-derived ectoderm.
Keywords: nodal signaling, epiblast stem cells, transcriptome, histone modification, ectoderm propensity
Highlights
-
•
Self-renewing epiblast stem cells can be maintained under Nodal inhibition
-
•
Nodal-inhibited epiblast stem cells and the ectoderm display similar transcriptome
-
•
Blocking Nodal changes the epigenome to that associated with ectoderm potency
-
•
Nodal-inhibited epiblast stem cells differentiate preferentially to ectodermal cells
In this article, Liu and colleagues show that Nodal-inhibited epiblast stem cells can self-renew in vitro and display similar transcriptome compared with the ectoderm of late-gastrula embryo. Further analysis shows that inhibition of Nodal signaling predisposes the epigenome to ectoderm-associated status. Moreover, these Nodal-inhibited epiblast stem cells efficiently differentiate to ectodermal cells but not mesendoderm lineages.
Introduction
Mouse gastrulation is a rapid and dynamic process that commences following the formation of the postimplantation embryo, which is made up of the epiblast and visceral endoderm derived from the inner cell mass of the blastocyst, and the extra embryonic ectoderm derived from the trophectoderm. During gastrulation, at embryonic day (E)6.5 to E7.5, epiblast cells are allocated to the primitive streak for the generation of the mesoderm and definitive endoderm (Lu et al., 2001), whereas cells remaining in the epiblast are endowed the ectoderm property (Tam and Loebel, 2007). Fate mapping the E7.5 late-gastrula mouse embryo revealed that the anterior epiblast is predisposed for neural and surface ectoderm cell fates (Tam, 1989). Clonal analysis has further revealed the presence of bipotential progenitors in the anterior epiblast of E7.0 mid-gastrula embryo (Cajal et al., 2012) that can differentiate into both neural and epidermal lineage in vitro (Li et al., 2013). These findings are consistent with the concept that the anterior epiblast of the gastrulating embryo harbors the ectoderm progenitors.
The process of specification and commitment of the ectoderm lineage in developing embryo is less well understood. The paucity of molecular markers that signify the emergence of the lineage and the lack of appropriate experimental model for studying the biology of ectoderm progenitor cells have impeded our understanding of the development of the ectoderm lineage. In particular, there is an unfulfilled requirement for an in vitro cell-based model for studying ectoderm development. Pluripotent stem cells (PSCs) have been isolated from mouse embryos, such as the embryonic stem cells (ESCs) from the epiblast of preimplantation blastocyst (Evans and Kaufman, 1981, Martin, 1981), and the epiblast stem cells (EpiSCs) (Brons et al., 2007, Kojima et al., 2014, Tesar et al., 2007) and “region-selective” EpiSCs (rsEpiSCs) (Wu et al., 2015) from the postimplantation epiblast. None of these stem cell types, however, consistently display predisposed ectoderm lineage potency.
During gastrulation, Nodal and Wnt signaling play pivotal roles on the formation of the primitive streak, progression of gastrulation, and tissue patterning in the anterior-posterior axis of the embryo. The spatial transcriptome study and analysis of gene-expression domain of the gastrula stage mouse embryo revealed that cells in different regions of the epiblast are subject to different levels of Nodal and Wnt signaling (Peng et al., 2016, Pfister et al., 2007). Nodal signaling is active in the posterior epiblast for primitive streak formation and mesendoderm development (Brennan et al., 2001, Conlon et al., 1994). In contrast, the anterior epiblast that is fated for the ectoderm appears to be a “signal-silent” zone for Nodal activity through genome-wide study (Peng et al., 2016). Canonical Wnt-β-catenin signaling is required for axis formation and mesoderm induction in the mouse embryo. In the loss of Wnt3 (Liu et al., 1999) and β-catenin mutants (Huelsken et al., 2000), mesoderm fails to form. The repression of Wnt signaling activity by the antagonist, such as DKK1 emanating from anterior visceral endoderm, is associated with the acquisition of ectoderm potency by the anterior epiblast (Kimura-Yoshida et al., 2005). Loss of Dkk1 function, which creates a gain of Wnt function, leads to the loss of brain and cranial structures (Lewis et al., 2008, Mukhopadhyay et al., 2001), which could be related to an altered ectoderm potency of neural progenitor tissue. These findings imply that a diminished level of Nodal and Wnt signaling activity may underpin the acquisition of the ectoderm lineage potency.
Consistent with the concept that suppressing Nodal signaling enables the acquisition of ectoderm cell fates, blocking Nodal signaling promotes neural ectoderm differentiation of human ESCs in vitro (Li et al., 2011, Patani et al., 2009, Smith et al., 2008, Vallier et al., 2004), and double-mutant embryos of Nodal antagonists Cer1 and Lefty1 result in the loss of neural ectoderm and the ectopic differentiation of mesoderm (Perea-Gomez et al., 2002). Loss of Nodal function leads to precocious neural differentiation and early loss of pluripotency of the epiblast (Camus et al., 2006, Mesnard et al., 2006). In mouse EpiSCs, blocking activin signaling enhances neural differentiation (Brons et al., 2007, Tesar et al., 2007, Vallier et al., 2009). An ectoderm-like state can be induced in the mouse ESC-derived EpiSCs (ESD-EpiSCs) by Nodal inhibition (Li et al., 2015). However, these ectoderm-like cells are unstable and do not self-renew in vitro.
In this study, we tested whether modulation of the Nodal and Wnt signaling activity may impact on the lineage potency of the mouse EpiSCs that were derived and maintained under fibroblast growth factor 2 (FGF2)/activin A conditions. Our findings showed that inhibiting Wnt activity has no discernible effect of the lineage propensity of the established EpiSCs, while inhibiting Nodal activity can enhance the ectoderm lineage propensity. Nodal-inhibited EpiSCs differentiate efficiently to cells of the epidermis lineage in vitro while retaining the neuroectoderm potential, but these cells lose the ability to differentiate into mesendoderm derivatives.
Results
Inhibiting WNT Signaling Activity in Epiblast Stem Cells Has No Impact on Lineage Propensity
EpiSCs were derived from the epiblast of the E6.5 early-primitive-streak-stage mouse embryo and maintained in culture supplemented with FGF2 and activin A. To assess the impact of abrogating WNT activity on the lineage property of the established EpiSC, we added a chemical inhibitor (IWP2), which blocks WNT signaling by inhibiting the function of Porcupine that mediates the trafficking and secretion of WNT ligand (Gao and Hannoush, 2014), to the culture for over ten passages (IW-EpiSC) (Figure S1A). To characterize the differentiation potency of these EpiSCs, we assessed the expression profile of lineage markers over 4 days of in vitro differentiation by microfluidic qPCR. The EpiSC and IW-EpiSC showed similar patterns of lineage differentiation in vitro in terms of the relative number of germ-layer-specific genes that were expressed at days 0 and 4 and the trajectory of lineage differentiation (Figures S1B and S1C). These findings suggest that abrogating WNT activity did not rewire the lineage property of EpiSCs. However, IW-EpiSCs in activin A-containing medium were potentially still subject to transforming growth factor β (TGF-β) signaling activity, such as Nodal, which may counteract any effect elicited by the inhibition of WNT activity on lineage propensity. We therefore proceeded to test the effect of inhibiting Nodal activity on the EpiSCs.
Derivation of EpiSCsS/F from EpiSCs
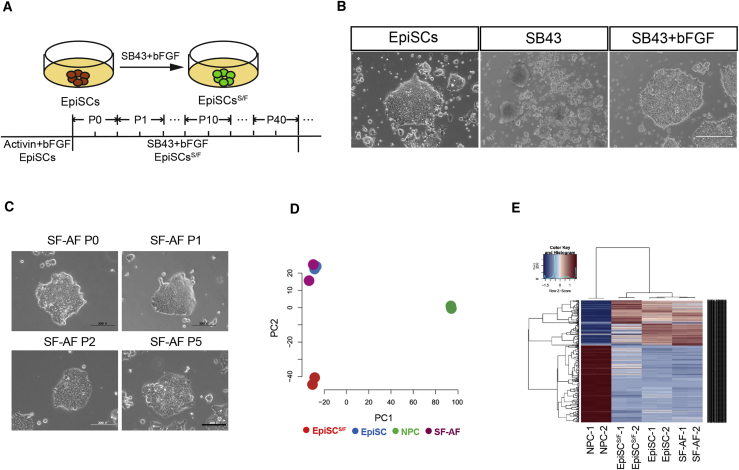
We tested the effect of blocking Nodal pathway using SB431542 (SB43) (Figure 1A), which inhibits the activity of TGF-β receptors. SB43-treated EpiSCs cultured in chemical defined medium (CDM) only (without activin A and FGF2) showed extensive cell death (Figure 1B), whereas those cultured in CDM supplemented with FGF2 formed epithelial colonies (Figure 1B). From these colonies, a stable EpiSC line (denoted as EpiSCsS/F) that self-renewed for over 40 passages was derived and maintained in CDM + FGF2 + SB43. Another EpiSCS/F line was also derived from EpiSCs generated from the epiblast of 129 strain E5.5 mouse embryo (Figures S1D and S1E). EpiSCsS/F can also be derived from single cells from the parental EpiSC line (Figure S1F). EpiSCsS/F could be reverted back to the EpiSC state by withdrawing SB43 and culturing in EpiSCs medium (denoted EpiSCSF-AF). EpiSCsSF-AF showed morphology and transcriptome profiles similar to those of EpiSCs (Figures 1C–1E), indicating that the EpiSCsS/F have not been hard-wired to an SB/F-specific status.
Figure 1.
Derivation of EpiSCS/F from EpiSCs
(A) Strategy for EpiSCsS/F production. P, passage.
(B) EpiSCs on feeder-free culture in three different media supplemented with activin A and bFGF, SB43 only, and SB43 plus bFGF, respectively. Phase-contrast images. Scale bar, 500 μm.
(C) Morphology of EpiSCS/F cultured in activin A and bFGF supplemented medium (SF-AF). Scale bar, 200 μm.
(D) PCA display of RNA-seq data of EpiSCsS/F, EpiSCs, SF-AF, and neural progenitor cells (NPCs).
(E) Hierarchical clustering of EpiSCsS/F, EpiSCs, SF-AF, and NPCs.
Immunocytochemistry revealed that the expression of the pluripotent factors, SOX2 and OCT4, was comparable between EpiSCs and EpiSCsS/F (Figure 2A, upper panels), while single-cell PCR results showed a lower average mRNA level of Oct4 in EpiSCsS/F compared with EpiSCs (Figure 2B). EpiSCsS/F showed weaker alkaline phosphatase activity than ESCs and EpiSCs (Figure S2A), but proliferated at rates as similar to those of EpiSCs (Figure S2B). qRT-PCR analysis revealed that Nodal downstream target genes were suppressed in EpiSCsS/F (Figure S2C). qPCR analysis revealed that treatment with SB43 induced a rapid repression of Fgf5 expression during EpiSCsS/F derivation (Figures S2D and S2E). Analysis of Oct4, Otx2, T, Sox1, and Ck18 at different passages (Figures 2B and S2E), further showed that the EpiSCS/F maintained a stable phenotype on extended passaging.
Figure 2.
Characterization of EpiSCS/F
(A) Expression of SOX2 and OCT4 (upper panels), NANOG, FGF5, and T (middle panels), and OTX2, SIX3, and E-CADHERIN (lower panels) in EpiSCsS/F and EpiSCs. Immunofluorescence and DAPI counterstaining. ∗p < 0.05, ∗∗∗p < 0.001, significant difference in the fraction of cells displaying positive immunofluorescence (% positive/DAPI) by Student's t test, n = 5 samples for each type of EpiSC. n.s., not significant. Scale bars, 50 μm.
(B) qPCR analysis of the expression of Oct4 and Otx2 in EpiSCs and EpiSCsS/F (at passages 23 and 30). Thirty cells per cell type were analyzed by qPCR. Statistical analysis was performed using Student's t test (∗∗∗p < 0.001; n.s., not significant).
Data are means ± SD.
To characterize the EpiSCsS/F, we compared the transcriptome of EpiSCsS/F with that of EpiSCs derived with activin-FGF2 (Kojima et al., 2014) and the ESC-derived EpiSCs. Hierarchical clustering analysis of the transcriptome showed that EpiSCsS/F were grouped separately from EpiSCs and ESC-derived EpiSCs (Zhang et al., 2010) (Figure S3A). The expression of EpiSC markers (Nanog, T, Fgf5, Eomes, and Lefty1) was lower in EpiSCsS/F (Figure 2A, middle panels; Figure S2F). EpiSCs and EpiSCsS/F expressed similar level of junctional gene E-CADHERIN and anterior epiblast marker OTX2, but EpiSCsS/F showed higher expression of another anterior epiblast marker, SIX3 (Figure 2A, lower panels). Both EpiSCs and EpiSCsS/F did not express the markers of ESCs (Rex1, Esrrb, and Klf4), and very low level of markers of mesendoderm cells (Flk1, Gata6, and Sox17), neural progenitor cells (NPCs) (Pax6, Zfp521, and Nestin), and epidermis (Ck18, Ck19, Ck8 and Grhl2) (Figure S2F).
Taken together, stable EpiSCS/F cell lines can be derived from established EpiSCs by inhibiting Nodal signaling, and these self-renewing cells are molecularly distinctive from naive (ESC) and primed pluripotent stem cells (EpiSCs) as well as three tissue progenitor cells.
EpiSCS/F Exhibit Gene-Expression Profiles that Are Similar to the Ectoderm of Late-Mid-Streak-Stage Embryos
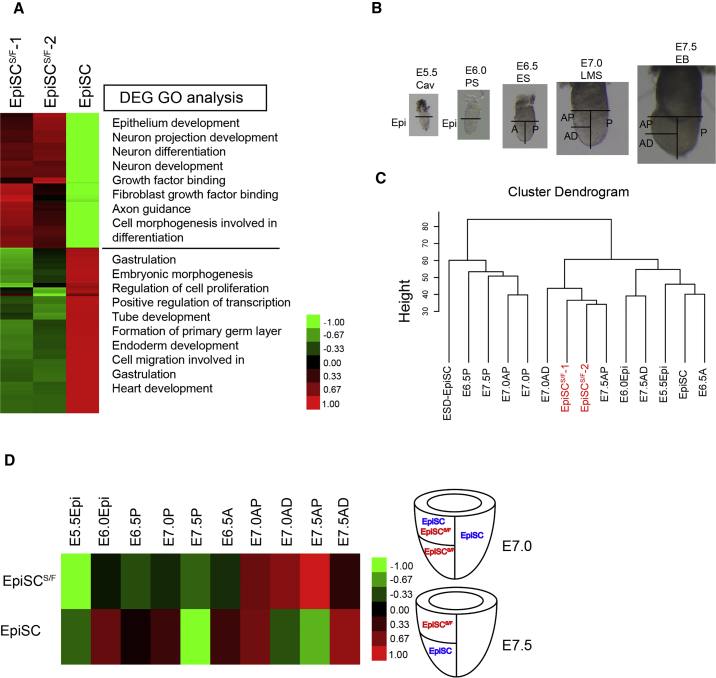
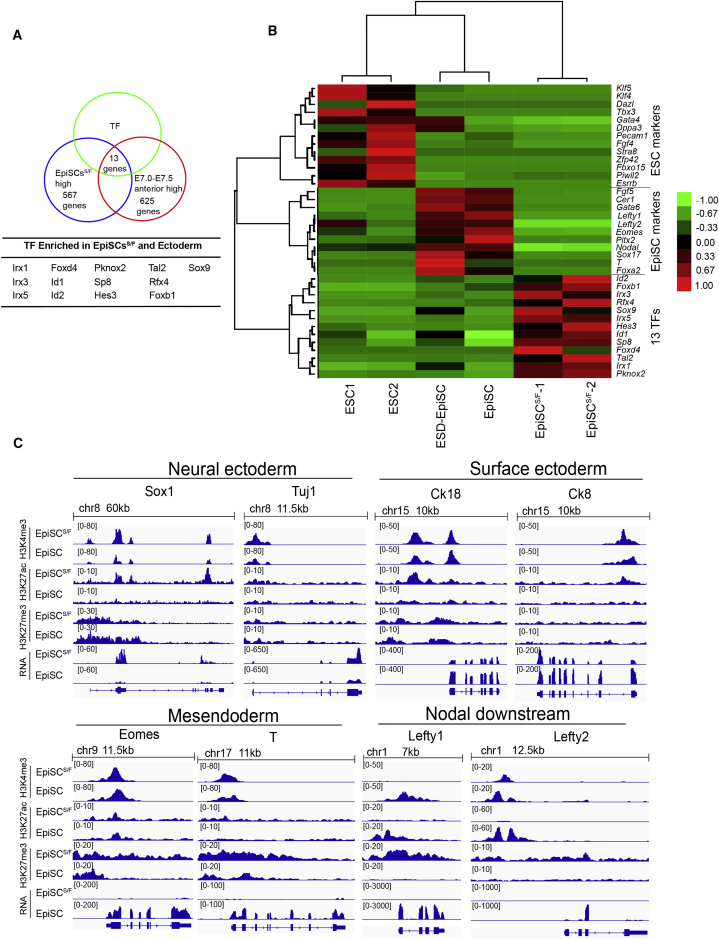
Compared with EpiSCs, genes that were highly expressed in EpiSCsS/F are related to neural and epithelium development. In contrast, genes related to gastrulation and endoderm differentiation were more highly expressed in EpiSCs (Figure 3A). To delineate the in vivo epiblast counterpart of the EpiSCS/F, we compared the transcriptome of the EpiSCS/F with that of the epiblast sampled from embryonic parts of the cavity stage (E5.5) to early-bud stage (E7.5) embryos (Figure 3B). Hierarchical clustering, principal component analysis (PCA), and Pearson correlation analysis showed that EpiSCsS/F resembled most the anterior ectoderm of E7.0 and E7.5 embryo (Figures 3C, 3D, and S3B). In contrast, the parental EpiSCs are broadly similar to the epiblast of E6.5 embryo, the posterior epiblast of E7.0 embryo, and anterior epiblast of E7.5 embryo. To collate genes that may uniquely identify the EpiSCsS/F, we analyzed genes that showed high expression in both EpiSCsS/F and E7.0–E7.5 anterior ectoderm, and found 13 commonly enriched transcription factors (TFs) (Figure 4A). These 13 TF genes were expressed exclusively in EpiSCsS/F and not in ESCs and EpiSCs (Buecker et al., 2014, Tesar et al., 2007) (Figure 4B). EpiSCS/F thus represents a unique type of EpiSCs that may resemble the anterior ectoderm of the advanced gastrula embryo.
Figure 3.
Developmental Correlates of EpiSCsS/F
(A) Gene ontology (GO) analysis of differentially expressed genes (DEG) in EpiSC and EpiSCS/F.
(B) Epiblast/ectoderm samples for RNA-seq from E5.5 (cavity stage, Cav), E6.0 (prestreak stage, PS), E6.5 (early-streak stage, ES), E7.0 (late-middle streak stage, LMS), and E7.5 (early-bud stage, EB) mouse embryos. A, anterior; P, posterior; Epi, epiblast; AP, anterior proximal; AD, anterior distal.
(C) Hierarchical clustering by the transcriptome of EpiSCsS/F (as technical replicates, red), EpiSCs, ESD-EpiSCs, and epiblast/ectoderm samples.
(D) Pearson correlation analysis of EpiSCsS/F and EpiSCs with embryonic tissue samples for matching the EpiSC and EpiSCS/F to the epiblast/ectoderm of E5.5 to E7.5 embryos.
Figure 4.
Gene Expression and Epigenetic Signature of EpiSCS/F
(A) Transcription factors that are enriched in the transcriptome of the anterior ectoderm and the EpiSCsS/F.
(B) Heatmap of the expression of transcription factor (TF) genes of ESCs, EpiSCs (technical replicates), and anterior ectoderm in ESCs, ESD-EpiSCs, EpiSCs, and EpiSCsS/F.
(C) Histone modifications and the expression (RNA) of selected lineage marker genes neural ectoderm (Sox1), surface ectoderm (Ck18), mesendoderm (Eomes), and Nodal and downstream (Lefty1) in EpiSCs and EpiSCsS/F. For each gene, first panel: H3K4me3 signal; second panel: H3K27ac signal; third panel: H3K27me3 signal around transcription start site in EpiSCs and EpiSCsS/F. The last panel shows RNA expression level in EpiSCs and EpiSCsS/F.
To elucidate the chromatin characteristics of EpiSCS/F, we surveyed the genome-wide pattern of H3K27 acetylation (H3K27ac), H3K27 trimethylation (H3K27me3), and H3K4 trimethylation (H3K4me3) in comparison with EpiSC. H3K27ac and H3K4me3, which marked active promoters, were detected in the promoter region of many genes in both EpiSCS/F and EpiSC (Figure S4A).
We further characterized the histone modifications on lineage markers. In EpiSCS/F, active chromatin modifications (high H3K4me3, high H3K27ac, low H3K27me3) were found in the promoter of neural ectoderm marker genes (Sox1, Tuj1), surface ectoderm marker genes (Ck18, Ck8), and ectoderm-related genes (Id1, Id2) (Figures 4C and S4B). Mesendoderm marker genes (Eomes and T) that were expressed more highly in EpiSCs showed bivalent modifications (high H3K4me3, high H3K27me3) in EpiSCsS/F (Figure 4C). Consistent with the RNA sequencing (RNA-seq) data, Nodal downstream genes (Lefty1 and Lefty2) showed repressive modification (low H3K4me3, low H3K27ac, high H3K27me3) in EpiSCS/F (Figure 4C). These results on the epigenetic marks suggested that ectoderm lineage genes may be at an open to active state following the transition from EpiSC to EpiSCS/F. For the mesendoderm lineage genes, histone modifications were changed from active to bivalent status in the EpiSCsS/F. These changes in histone modifications are consistent with the enhanced surface ectoderm potential and diminished mesendoderm potential of EpiSCsS/F, and may reflect the interplay for signaling and intrinsic programs during EpiSCS/F derivation.
Taken together, the correlation of epigenomic modification and the switch to ectoderm propensity point to the role of ectoderm-poised chromatin modification in mediating the ectoderm propensity of EpiSCsS/F.
EpiSCsS/F Behaved Like Anterior Ectoderm Progenitors in Chimeras
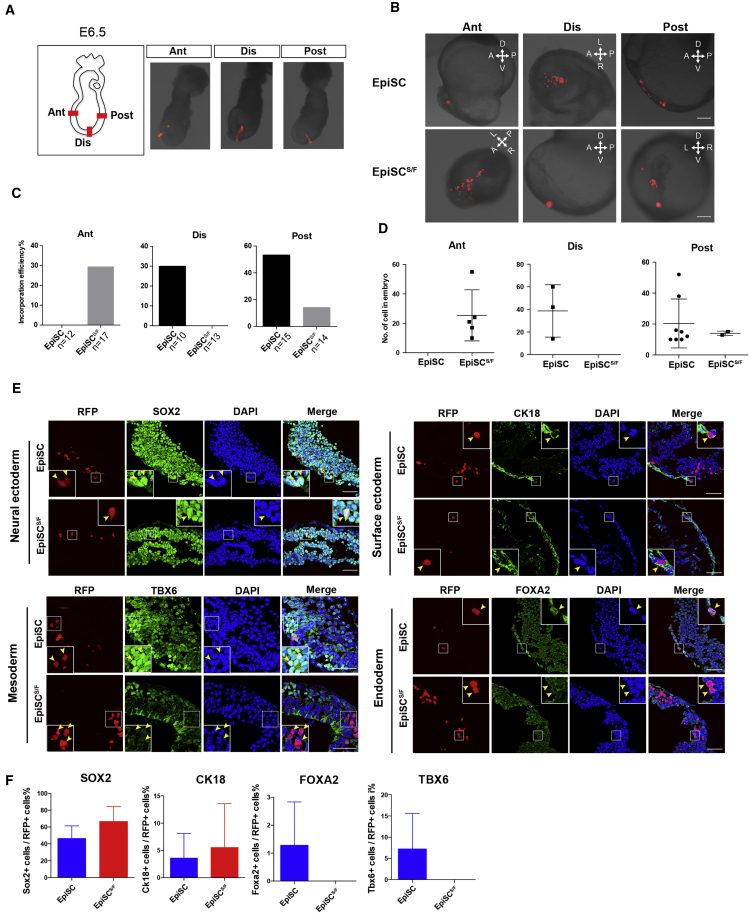
To test the differentiation potential of EpiSCsS/F in an embryonic context, we assessed the contribution of EpiSCsS/F to germ-layer derivatives in postimplantation chimeras (Huang et al., 2012, Kojima et al., 2014, Mascetti and Pedersen, 2016, Wu et al., 2015). RFP-expressing EpiSCs and EpiSCsS/F were grafted to anterior (Ant), distal (Dis), and posterior (Post) regions of the epiblast of E6.5 embryo (Figure 5A), and the distribution of the graft-derived cells was examined after 48 hr of in vitro culture. A site-specific pattern of tissue distribution was observed (Figure 5B). EpiSCs stayed as clumps, whereas EpiSCsS/F showed proliferation and incorporation when grafted to the anterior epiblast (Figures 5B–5D). EpiSCsS/F grafted into distal and posterior sites showed a much lower level of incorporation and cell proliferation (Figures 5B–5D). Immunostaining data showed that both EpiSCsS/F and EpiSC-derived cells expressed the appropriate neuroectoderm marker (SOX2) and surface ectoderm marker (CK18). In contrast, EpiSCS/F-derived cells did not express mesendoderm markers such as FOXA2 and TBX6 (Figures 5E and 5F). When grafted into the E7.0 embryo (Figure S5A), both EpiSCs and EpiSCsS/F remained unincorporated when grafted to the anterior epiblast (Figure S5B), whereas EpiSCsS/F grafted into post sites showed similar frequency of incorporation but a lower level of cell proliferation than EpiSCs (Figures S5C and S5D). Collectively, the findings of chimera assay suggest that EpiSCS/F has acquired the developmental attribute that is compatible with the anterior epiblast of E6.5 embryo, and show ectoderm differentiation potential in the chimeras. While the EpiSCsS/F were similar transcriptome-wise to the E7.0 and E7.5 anterior ectoderm, they were developmentally incompatible when enforced to differentiate in the tissue environment of the E7.0 host embryo. Whether this is underpinned by the developmental asynchrony of EpiSCsS/F and the host environment is presently not known.
Figure 5.
Differentiation of EpiSCS/F in Postimplantation Chimeras
(A) RFP-expressing EpiSCS/F and EpiSC grafted to anterior (Ant), distal (Dis), and posterior (Post) regions of the epiblast of E6.5 embryo.
(B) Distribution of graft-derived cells in host embryo 48 hr after transplantation of RFP-expressing cells in anterior (Ant), distal (Dis), and posterior (Post) regions of embryo. Scale bar, 200 μm.
(C) Percentage of host embryos showing the incorporation of graft-derived cells. n = 12 embryos for EpiSC and n = 17 embryos for EpiSCS/F in anterior (Ant) grafted group. n = 10 embryos for EpiSC and n = 13 embryos for EpiSCS/F in distal (Dis) grafted group. n = 15 embryos for EpiSC and n = 14 embryos for EpiSCS/F in posterior (Post) grafted group.
(D) Number of EpiSC- and EpiSCS/F-derived cells in the host embryo. Data are mean ± SD, no error estimation for groups with “zero” score. n = 5 embryos for EpiSC and n = 5 embryos for EpiSCS/F in anterior (Ant) grafted group. n = 3 embryos for EpiSC and n = 3 embryos for EpiSCS/F in distal (Dis) grafted group. n = 8 embryos for EpiSC and n = 5 embryos for EpiSCS/F in posterior (Post) grafted group.
(E) Expression of lineage markers in chimeras detected by immunostaining Arrowheads mark cells of interest. Scale bars, 50 μm.
(F) Percentage of SOX2+ cells, CK18+ cells, FOXA2+ cells, and TBX6+ cells in the RFP-positive cell population in the chimera. Data are mean ± SD, no error estimation for groups with “zero” score. n = 4 embryos for EpiSC and n = 5 embryos for EpiSCS/F. For each embryo, 20–60 RFP-expressing cells are scored.
EpiSCS/F Display Propensity of Ectoderm Differentiation
To assess the lineage potential, we generated embryoid bodies (EBs) from the EpiSCs and cultured in the presence of bone morphogenetic protein (BMP) that promote differentiation of the neuroectoderm, or in the absence of BMP for epidermis and mesendoderm differentiation (Zhu et al., 2014). EpiSCS/F EBs showed higher expression of Sox1 (a neural marker) than EpiSC EBs (Figure 6A, upper panels), but not early NPC markers such as Zfp521 and Nestin (Figure 6A, upper panels). In BMP4-supplemented medium, EpiSCS/F EBs displayed stronger expression of Ck18, Ck8, and CK19 (epidermis markers) than EpiSC EBs (Figure 6A, middle panels). In contrast, BMP4 did not induce the expression of Flk1, Gata6, and Sox17 (mesoderm and endoderm markers) in EpiSCS/F EBs (Figure 6A, lower panels). Immunostaining showed that similar population of cells in EpiSCS/F EBs and EpiSC EBs (at day 4 of differentiation in BMP-free medium) expressed neural markers (TUJ1 and NESTIN), but more EpiSCsS/F expressed epidermal markers (CK18 and CK8) (Figure 6B). Relative to EpiSCs, fewer EpiSCsS/F expressed the mesoderm markers: FLK1 and NKX2.5 in BMP-supplemented medium (Figure 6B) and Foxa2, Sox17, Flk1, and Gsc in serum-containing medium (Figure 6C).
Figure 6.
Differentiation of EpiSCS/FIn Vitro
(A) qPCR analysis of the expression of neuroectoderm markers Sox1, Zfp521, and Nestin, epidermis markers Ck18, Ck8, and Ck19, and mesendoderm markers Flk1, Gata6, and Sox17 during 4 days of differentiation of EpiSCsS/F and EpiSCs in medium with BMP4 (BMP+) and without (BMP−). Data are mean ± SD for n = 3 cultures per cell type at each time point.
(B) Immunofluorescence visualization of the expression of neuroectoderm markers TUJ1 and NESTIN, epidermis markers CK18 and CK8, and mesendoderm markers FLK1 and NKX2.5 in day-4 embryoid bodies, n = 3 cultures each for immunostaining and scoring. Data are means ± SD. Scale bars, 50 μm.
(C) qPCR analysis of the expression of the mesendoderm markers Foxa2, Sox17, Flk1, and Gsc during 4 days of differentiation of EpiSCsS/F and EpiSCs in fetal bovine serum-supplemented medium. Data are mean ± SD from n = 3 cultures each per cell type at each time point.
(D) Differentiation of individual descendants of a single-cell clone of EpiSCS/F analyzed for the expression of markers of neuroectoderm Sox1 and Nestin, markers of epidermis Ck18 and Ck8, and markers of mesendoderm T and Eomes, after 24 hr of differentiation. Twenty cells per differentiation condition were analyzed by qPCR. Data are means ± SD.
Statistical analysis was performed using Student's t tests (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; n.s., not significant).
We further assessed the ectoderm propensity of EpiSCS/F by studying the differentiation of a single-cell colony of EpiSCs and EpiSCsS/F in culture medium with or without BMP4 for 24 hr. More single cells in the EpiSCS/F clones displayed higher expression of neural markers (Sox1, Nestin) and epidermis markers (Ck18, Ck8) compared with those of EpiSC clones. Very low expression of mesendoderm marker T and Eomes was found in single cells from EpiSCS/F after 24 hr of differentiation in vitro (Figure 6D). These results showed that EpiSCsS/F are more responsive to directed ectoderm differentiation but less amenable to mesendoderm differentiation.
Discussion
Here, we report the derivation of a unique type of self-renewing EpiSCs, the EpiSCsS/F, which display a distinctive molecular signature of ectoderm lineage propensity. Recent studies have shown that the functional attributes of stem cells in vitro can be modulated by signaling activity that are known to influence lineage fates ex vivo. For instance, EpiSCs, which are derived by culturing epiblast in FGF2 and activin A, acquire the molecular property of anterior primitive streak (Kojima et al., 2014) whereby embryonic cells in vivo are subject to strong Nodal signaling. As a result of extended exposure to activin A, EpiSCs may lose the ability to differentiate into primordial germ cells (PGCs), but EpiLCs generated by transient differentiation of ESCs retain the capacity to respond to WNT and BMP induction of PGCs (Hayashi et al., 2012). Consistent with the fact that ex vivo ectoderm differentiation of the epiblast can be enhanced by suppressing Nodal signaling (Li et al., 2013), blocking Nodal signaling in EpiSCs enables the derivation of EpiSCS/F cell lines that are poised for ectoderm differentiation. The gene-expression profile of these cells matches the cells in the anterior ectoderm of E7.0/E7.5 embryo that are fated for ectoderm differentiation. EpiSCS/F may therefore represent a cellular state when the pluripotent epiblast transits to the progenitors of neural and surface ectoderm.
Our finding highlights the feasibility of generating lineage-specific stem cell lines by mimicking the signaling condition in the embryo for in vitro derivation and maintenance of EpiSCs. The EpiSCS/F shows an enhanced surface ectoderm propensity but a diminished propensity for mesendoderm differentiation, which is coincidental with the active histone profile of the promoter of surface ectoderm and epidermis genes, and the repressive profile of the promoter of mesendoderm genes. The active modifications of the promoter of neuroectoderm markers and surface ectoderm markers in the EpiSCS/F point to a broadly ectoderm-poised, though not neural-specific, chromatin modification. In view of that the EpiSCS/F can be reverted back to the EpiSC state by changing the in vitro conditions, it is likely that the epigenetic program of EpiSCsS/F may be not fixated even when they are self-renewing. In this regard, our study provides a glimpse of the interaction between extrinsic signals and the epigenome in predisposing the transcription activity that specifies the lineage propensity of EpiSCs.
In essence, our work has extended the spectrum of primed PSCs by identifying a self-renewing EpiSC population with enhanced ectoderm propensity. These ectoderm-poised EpiSCs may offer an amenable in vitro model, in parallel with the NPCs and the transient ectoderm progenitor cells, for investigating the mechanism of divergent specification of neuroectoderm and epidermis lineages.
Experimental Procedures
All animal experiments were performed in accordance with the Guide of Animal Care and Use Committee of Shanghai Institute of Biochemistry and Cell Biology.
Derivation and Culture of EpiSCsS/F
An established MEF-free EpiSC line from the Janet Rossant lab (Rugg-Gunn et al., 2012) and an EpiSC line derived from 129 strain E5.5 mouse embryo in our lab was used to generate EpiSCsS/F. Epiblast of E5.5 129 strain mouse embryo was dissected and cultured in CDM supplemented with 20 ng/mL activin A and 10 ng/mL basic FGF (bFGF) on a feeder layer for ten passages and then cultured in feeder-free condition as stable 129 EpiSC line. EpiSCs were maintained on serum-coated plates in CDM supplemented with 20 ng/mL activin A and 10 ng/mL bFGF. To derive EpiSCsS/F, we dissociated EpiSCs into small clumps with collagenase IV and replated them on serum-coated plates in CDM supplemented with 2 μM SB431542 and 10 ng/mL bFGF. To derive EpiSCS/F from single EpiSC, we dissociated EpiSCs by Accutase and plated them as single cell in 96-well plates in CDM supplemented with 2 μM SB431542 and 10 ng/mL bFGF. EpiSCS/F colony derived from a single EpiSC was picked up on day 8 and passaged as a cell line. EpiSCsS/F were passaged using collagenase IV every 2 days. EpiSCS/F was cryopreserved using knockout serum replacement (KSR) plus 10% DMSO and displayed similar post-thaw viability as the parental EpiSC.
RNA Preparation and Real-Time PCR
Total RNAs were extracted by using TRIzol (Invitrogen) according to the manufacturer's instructions. RNAs were reverse transcribed using SuperScript III Reverse Transcriptase (Invitrogen). Real-time PCR was performed using EvaGreen (Biotium). Expression levels of each gene were normalized to GAPDH expression and calculated by comparative CT.
Primer Sequence
| Genes | 5′ Primer | 3′ Primer |
|---|---|---|
| Rex1 | CAGTTCGTCCATCTAAAAAGGGAGG | TCTTAGCTGCTTCCTTGAACAATGCC |
| Oct4 | AGTTGGCGTGGAGACTTTGC | CAGGGCTTTCATGTCCTGG |
| Nanog | TTGCTTACAAGGGTCTGCTACT | ACTGGTAGAAGAATCAGGGCT |
| Nodal | CCTGGAGCGCATTTGGATG | ACTTTTCTGCTCGACTGGACA |
| Fgf5 | GCTGTGTCTCAGGGGATTGT | CACTCTCGGCCTGTCTTTTC |
| Sox2 | GCGGAGTGGAAACTTTTGTCC | CGGGAAGCGTGTACTTATCCTT |
| Sox1 | ATACCGCAATCCCCTCTCAG | ACAACATCCGACTCCTCTTCC |
| Zfp521 | GAGCGAAGAGGAGTTTTTGG | AGTTCCAAGGTGGAGGTCAC |
| Nestin | GCTGGAACAGAGATTGGAAGG | CCAGGATCTGAGCGATCTGAC |
| Ck8 | TCCATCAGGGTGACTCAGAAA | CCAGCTTCAAGGGGCTCAA |
| Ck18 | CAGCCAGCGTCTATGCAGG | CTTTCTCGGTCTGGATTCCAC |
| Ck19 | GGGGGTTCAGTACGCATTGG | GAGGACGAGGTCACGAAGC |
| T | CTCGGATTCACATCGTGAGAG | AAGGCTTTAGCAAATGGGTTGTA |
| Flk1 | GGGTCGATTTCAAACCTCAATGT | AGAGTAAAGCCTATCTCGCTGT |
| Sox17 | CGAGCCAAAGCGGAGTCTC | TGCCAAGGTCAACGCCTTC |
| Gata6 | TTGCTCCGGTAACAGCAGTG | GTGGTCGCTTGTGTAGAAGGA |
| Gata4 | CCCTACCCAGCCTACATGG | ACATATCGAGATTGGGGTGTCT |
| Eomes | CCTGGTGGTGTTTTGTTGTG | TTTAATAGCACCGGGCACTC |
| Klf4 | CTTCAGCTATCCGATCCGGG | GAGGGGCTCACGTCATTGAT |
| Pax6 | GCAGATGCAAAAGTCCAGGTG | CAGGTTGCGAAGAACTCTGTTT |
| Mixl1 | ACGCAGTGCTTTCCAAACC | CCCGCAAGTGGATGTCTGG |
| Lefty1 | CCAACCGCACTGCCCTTAT | CGCGAAACGAACCAACTTGT |
| Lefty2 | CAGCCAGAATTTTCGAGAGGT | CAGTGCGATTGGAGCCATC |
| Gapdh | TGTGATGGGTGTGAACCACGAGAA | CTGTGGTCATGAGCCCTTCCACAA |
Embryonic Tissue Sample Collection
Timed-pregnant mice were euthanized for embryo collection at appropriate stages between E5.5 and E7.5. Embryos were isolated out from decidua. In all stages, Reichert's membrane and visceral endoderm was removed. Embryonic tissues were dissected by syringe needles and collected separately. E5.5 and E6.0 embryos were dissected into extraembryonic ectoderm and epiblast. E6.5 epiblast was separated into anterior (A) and posterior (P) halves. E7.0 and E7.5 epiblasts were dissected into anterior proximal (AP), anterior distal (AD), and posterior (P) fragments.
Embryonic Sample RNA-Seq and Single-Cell PCR Analysis
The tissue fragments were processed for RNA-seq or single-cell PCR analysis. In brief, single-cell or small tissue samples were lysed and reverse transcription performed (Peng et al., 2016). After the first-strand synthesis, cDNA was preamplified with KAPA HiFi Hotstart ReadyMix (KAPA Biosystems) by IS-PCR primer for 18 cycles. cDNA quality was assessed by qPCR examination of several housekeeping genes.
qPCR was then performed on single-cell preamplified cDNAs. For embryonic samples, cDNA was further verified by examining representative known position markers such as Sox2 and T to ascertain correct tissue dissection and adequate preamplification.
For sequencing library construction, cDNA was purified using a 1:0.8 ratio of AMPure XP beads (Beckman Coulter). After quantification by Qubit, cDNA was applied to Bioanalyzer 2100 on a High-Sensitive DNA chip (Agilent Bioanalyzer) to check the library size distribution. Amplified cDNA (∼5 ng) was then used to construct Illumina sequencing libraries using Illumina's Nextera DNA sample preparation kit following the manufacturer’s recommended manual. All sample libraries were sequenced on an Illumina HiSeq2000 machine with at least 20 million cleaned reads. The sequencing was performed by Berry Genomics (Beijing, China).
RNA-Seq Data Processing
Raw reads were mapped to mm10 using the TopHat version 2.0.13 program (Trapnell et al., 2009). We assigned FPKM (fragments per kilobase per million) as an expression value for each gene using Cufflinks version 1.3.0 software (Trapnell et al., 2010). Cuffdiff software was then used to identify differentially expressed genes between treatment and control samples (Trapnell et al., 2013). Differentially expressed gene heat maps were clustered by hierarchical clustering and visualized using Java TreeView software (Saldanha, 2004). PCA analysis was performed using R (http://www.r-project.org). Pearson correlation was used to compare cell lines with in vivo isolated embryonic tissue.
In Vitro Differentiation
EpiSCs and EpiSCsS/F were differentiated into EBs in KSR medium (Glasgow minimal essential medium supplemented with 8% KSR, 2 mM glutamine, 1 mM pyruvate, 0.1 mM nonessential amino acids, and 0.1 mM 2-mercaptoethanol) and KSR medium plus 10 ng/mL BMP4, respectively. RNA samples were collected every 2 days and analyzed by qPCR. For single-cell differentiation, EpiSCsS/F and EpiSCs were cultured singly for 7 days followed by the dissociation of the resultant clones into single cells. The single-cell suspension of each clone was split into two aliquots, with one used for neural differentiation (KSR medium without BMP4) and the other for epidermis and mesoderm differentiation (induced by 10 ng/mL BMP4). After differentiation in vitro for 24 hr, 20 single cells were collected from each culture and analyzed by qPCR.
Grafting and In Vitro Embryo Culture
RFP-expressing PB (PGK-neo) and ACT-PBase plasmids (Yang et al., 2013) were co-transfected into cells by Lipofectamine (Invitrogen) to label the cell with RFP. RFP-expressing EpiSCs and EpiSCsS/F were tested for their germ-layer differentiation potential by grafting ∼10 cells to the anterior and posterior sites of host ICR E6.5 and E7.0 embryos. Cells were dispersed into clumps using collagenase and collected for grafting. The engrafted host embryos were cultured in heat-inactivated rat serum under 5% oxygen, 5% carbon dioxide, and 90% nitrogen at 37°C for 48 hr (E6.5 chimeras) or 24 hr (E7.0 chimeras). After in vitro culture, the embryo was photographed to record the distribution of the grafted cells by fluorescence microscopy. The yolk sac and amnion were dissected away, the embryos were fixed in 4% paraformaldehyde for 1hr, and the embryos were embedded in OCT compound and cryosectioned.
Immunofluorescence
For immunofluorescence, cells were fixed with freshly prepared 4% paraformaldehyde in PBS for 0.5 hr at room temperature. Fixed cells and chimeric embryo cryosections were treated with blocking buffer (PBS containing 0.3% Triton X-100, 5% BSA, and 0.5% normal goat serum) for 2 hr at room temperature. The cells and sections were incubated with primary antibodies in blocking buffer overnight at 4°C. The next day, cells and sections were washed and incubated with fluorescent-labeled secondary antibodies at 1:500 dilutions for 1 hr at room temperature. The cells and sections were stained with DAPI and mounted in Mowiol mounting medium. Specimens were observed with a Leica TCS SP5 confocal microscope. Primary antibodies used in this study include: Oct3/4 (1:200, Santa Cruz Biotechnology, sc-5279), T (1:200, R&D Systems, AF2085), Sox2 (1:100, Abcam, ab59776), Nanog (1:200, CST, 8822), Fgf5 (1:50, Santa Cruz, sc-7914), Tuj1 (1:400, BioLegend, 801201), Nestin (1:100, made by our lab), Ck18 (1:100, Abcam, ab668), Flk1 (1:100, Becton Dickinson, 561993), Nkx2.5 (1:200, Santa Cruz, sc-8697), Tbx6 (1:100, Abcam, ab38883), and Foxa2 (1:100, Abcam, ab23630).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were carried out as described by Qiao et al. (2015). In brief, cells were collected and fixed in 1% formaldehyde solution and quenched by 0.125 M glycine. Fixed cells were fragmented to a size range of 200–500 bp by using Bioruptor Pico. Solubilized fragmented chromatin was immunoprecipitated with antibodies against H3K4me3 (Abcam, 8580), H3K27me3 (Millipore, 07-449), and H3K27ac (Active Motif, 39133). Antibody-chromatin complexes were pulled down using protein G beads (Dynabeads, 10004D), washed several times, and eluted. Reverse crosslink was performed subsequently under 65°C for at least 4 hr. Chromatin mixture solution was treated with RNase A and proteinase K to remove residual RNAs and proteins. Finally, fragmented DNA was extracted with phenol-chloroform and precipitated with ethanol. ChIP DNA was finally dissolved in nuclease-free water and quantified using Qubit. DNA fragments acquired from immunoprecipitation would be subjected to end-repaired, adaptor ligation, and PCR amplification under the instruction of the manufacturers (New England Biolabs, E7370).
ChIP-Seq Data Processing
Raw reads were mapped to mm10 using Bowtie2 version 2.2.2 (Langmead and Salzberg, 2012). MACS2 version 2.1.1.20160309 was used to call ChIP sequencing (ChIP-seq) peaks using broad peak calling mode (with --broad option), as well as to identify differential ChIP-seq signals in different conditions (Zhang et al., 2008). ChIPseeker (Yu et al., 2015) was used to annotate ChIP-seq peaks by using the mouse gene annotation GENCODE version M9. deepTools (Ramirez et al., 2014) was used to smooth and calculate the ChIP-seq signal as the ratio between ChIP-seq data and corresponding input control data, as well as to visualize ChIP-seq signals.
Statistics
Each experiment was performed at least three times. The data are presented as the mean ± SD. Student's t test was used to compare the effects of all treatments. Statistically significant differences are indicated in the figures as follows: ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Author Contributions
N.J., C.L., and P.P.L.T. designed the project; C.L., X.Y., J.C., G.C., W.G., Y.C., and P.O. conducted the experiments; Z.H., R.W., P.O., and E.W. performed the bioinformatics analysis; C.L., G.P., P.P.L.T., and N.J. analyzed the data and wrote the paper.
Acknowledgments
We thank Dr. Janet Rossant for providing the EpiSC line and Dr. Jinsong Li for providing the PB (PGK-neo) and ACT-PBase plasmid. This work was supported in part by the “Strategic Priority Research Program” of the Chinese Academy of Sciences (grant no. XDA16020501 to N.J., XDA16020404 to G.P.), the National Key Basic Research and Development Program of China (2017YFA0102700, 2015CB964500, 2014CB964804), the National Natural Science Foundation of China (31661143042, 91519314, 31630043, 31571513, 31430058), the National Health and Medical Research Council (NHMRC) of Australia (632776), and the Australian Research Council (DP 160100933). P.O. was supported by the Fondation pour la Recherche Medicale (FRM SPE20140129375). P.P.L.T. is an NHMRC Senior Principal Research Fellow (grants 1003100, 1110751).
Published: June 28, 2018
Footnotes
Supplemental Information includes five figures and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.05.019.
Accession Numbers
All RNA-seq data and ChIP-seq data are available at the Gene Expression Omnibus under accession number GEO: GSE92635.
Supplemental Information
References
- Brennan J., Lu C.C., Norris D.P., Rodriguez T.A., Beddington R.S., Robertson E.J. Nodal signalling in the epiblast patterns the early mouse embryo. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- Brons I.G., Smithers L.E., Trotter M.W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S.M., Howlett S.K., Clarkson A., Ahrlund-Ritcher L., Pederson R.A. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buecker C., Srinivasan R., Wu Z., Calo E., Acampora D., Faial T., Simeone A., Tan M., Swigut T., Wysocka J. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal M., Lawson K.A., Hill B., Moreau A., Rao J.G., Ross A., Camus A. Clonal and molecular analysis of the prospective anterior neural boundary in the mouse embryo. Development. 2012;139:423–436. doi: 10.1242/dev.075499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus A., Perea-Gomez A., Moreau A., Collignon J. Absence of Nodal signaling promotes precocious neural differentiation in the mouse embryo. Dev. Biol. 2006;295:743–755. doi: 10.1016/j.ydbio.2006.03.047. [DOI] [PubMed] [Google Scholar]
- Conlon F.L., Lyons K.M., Takaesu N., Barth K.S., Kispert A., Herrmann B., Robertson E.J. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Gao X., Hannoush R.N. Single-cell imaging of Wnt palmitoylation by the acyltransferase porcupine. Nat. Chem. Biol. 2014;10:61–68. doi: 10.1038/nchembio.1392. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Ogushi S., Kurimoto K., Shimamoto S., Ohta H., Saitou M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science. 2012;338:971–975. doi: 10.1126/science.1226889. [DOI] [PubMed] [Google Scholar]
- Huang Y., Osorno R., Tsakiridis A., Wilson V. In Vivo differentiation potential of epiblast stem cells revealed by chimeric embryo formation. Cell Rep. 2012;2:1571–1578. doi: 10.1016/j.celrep.2012.10.022. [DOI] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C., Birchmeier W. Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 2000;148:567–578. doi: 10.1083/jcb.148.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Yoshida C., Nakano H., Okamura D., Nakao K., Yonemura S., Belo J.A., Aizawa S., Matsui Y., Matsuo I. Canonical Wnt signaling and its antagonist regulate anterior-posterior axis polarization by guiding cell migration in mouse visceral endoderm. Dev. Cell. 2005;9:639–650. doi: 10.1016/j.devcel.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Kojima Y., Kaufman-Francis K., Studdert J.B., Steiner K.A., Power M.D., Loebel D.A., Jones V., Hor A., de Alencastro G., Logan G.J. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell. 2014;14:107–120. doi: 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.L., Khoo P.L., De Young R.A., Steiner K., Wilcock C., Mukhopadhyay M., Westphal H., Jamieson R.V., Robb L., Tam P.P. Dkk1 and Wnt3 interact to control head morphogenesis in the mouse. Development. 2008;135:1791–1801. doi: 10.1242/dev.018853. [DOI] [PubMed] [Google Scholar]
- Li L., Liu C., Biechele S., Zhu Q., Song L., Lanner F., Rossant J. Location of transient ectodermal progenitor potential in mouse development. Development. 2013;140:4533–4543. doi: 10.1242/dev.092866. [DOI] [PubMed] [Google Scholar]
- Li L., Song L., Liu C., Chen J., Peng G., Wang R., Liu P., Tang R., Rossant J., Jing N. Ectodermal progenitors derived from epiblast stem cells by inhibition of Nodal signaling. J. Mol. Cell Biol. 2015;7:455–465. doi: 10.1093/jmcb/mjv030. [DOI] [PubMed] [Google Scholar]
- Li W., Sun W., Zhang Y., Wei W., Ambasudhan R., Xia P., Talantova M., Lin T., Kim J., Wang X. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc. Natl. Acad. Sci. USA. 2011;108:8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., Wakamiya M., Shea M.J., Albrecht U., Behringer R.R., Bradley A. Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 1999;22:361–365. doi: 10.1038/11932. [DOI] [PubMed] [Google Scholar]
- Lu C.C., Brennan J., Robertson E.J. From fertilization to gastrulation: axis formation in the mouse embryo. Curr. Opin. Genet. Dev. 2001;11:384–392. doi: 10.1016/s0959-437x(00)00208-2. [DOI] [PubMed] [Google Scholar]
- Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascetti V.L., Pedersen R.A. Human-mouse chimerism validates human stem cell pluripotency. Cell Stem Cell. 2016;18:67–72. doi: 10.1016/j.stem.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnard D., Guzman-Ayala M., Constam D.B. Nodal specifies embryonic visceral endoderm and sustains pluripotent cells in the epiblast before overt axial patterning. Development. 2006;133:2497–2505. doi: 10.1242/dev.02413. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay M., Shtrom S., Rodriguez-Esteban C., Chen L., Tsukui T., Gomer L., Dorward D.W., Glinka A., Grinberg A., Huang S.P. Dickkopf1 is required for embryonic head induction and limb morphogenesis in the mouse. Dev. Cell. 2001;1:423–434. doi: 10.1016/s1534-5807(01)00041-7. [DOI] [PubMed] [Google Scholar]
- Patani R., Compston A., Puddifoot C.A., Wyllie D.J., Hardingham G.E., Allen N.D., Chandran S. Activin/Nodal inhibition alone accelerates highly efficient neural conversion from human embryonic stem cells and imposes a caudal positional identity. PLoS One. 2009;4:e7327. doi: 10.1371/journal.pone.0007327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Suo S., Chen J., Chen W., Liu C., Yu F., Wang R., Chen S., Sun N., Cui G. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell. 2016;36:681–697. doi: 10.1016/j.devcel.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Perea-Gomez A., Vella F.D., Shawlot W., Oulad-Abdelghani M., Chazaud C., Meno C., Pfister V., Chen L., Robertson E., Hamada H. Nodal antagonists in the anterior visceral endoderm prevent the formation of multiple primitive streaks. Dev. Cell. 2002;3:745–756. doi: 10.1016/s1534-5807(02)00321-0. [DOI] [PubMed] [Google Scholar]
- Pfister S., Steiner K.A., Tam P.P. Gene expression pattern and progression of embryogenesis in the immediate post-implantation period of mouse development. Gene Expr. Patterns. 2007;7:558–573. doi: 10.1016/j.modgep.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Qiao Y., Wang X., Wang R., Li Y., Yu F., Yang X., Song L., Xu G., Chin Y.E., Yang X. AF9 promotes hESC neural differentiation through recruiting TET2 to neurodevelopmental gene loci for methylcytosine hydroxylation. Cell Discov. 2015;1:15017. doi: 10.1038/celldisc.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42(Web Server issue):W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg-Gunn P.J., Cox B.J., Lanner F., Sharma P., Ignatchenko V., McDonald A.C., Garner J., Gramolini A.O., Rossant J., Kislinger T. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev. Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha A.J. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Smith J.R., Vallier L., Lupo G., Alexander M., Harris W.A., Pedersen R.A. Inhibition of Activin/Nodal signaling promotes specification of human embryonic stem cells into neuroectoderm. Dev. Biol. 2008;313:107–117. doi: 10.1016/j.ydbio.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Tam P.P. Regionalisation of the mouse embryonic ectoderm: allocation of prospective ectodermal tissues during gastrulation. Development. 1989;107:55–67. doi: 10.1242/dev.107.1.55. [DOI] [PubMed] [Google Scholar]
- Tam P.P.L., Loebel D.A.F. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 2007;8:368–381. doi: 10.1038/nrg2084. [DOI] [PubMed] [Google Scholar]
- Tesar P.J., Chenoweth J.G., Brook F.A., Davies T.J., Evans E.P., Mack D.L., Gardner R.L., McKay R.D. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Mendjan S., Brown S., Chng Z., Teo A., Smithers L.E., Trotter M.W., Cho C.H., Martinez A., Rugg-Gunn P. Activin/Nodal signalling maintains pluripotency by controlling Nanog expression. Development. 2009;136:1339–1349. doi: 10.1242/dev.033951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallier L., Reynolds D., Pedersen R.A. Nodal inhibits differentiation of human embryonic stem cells along the neuroectodermal default pathway. Dev. Biol. 2004;275:403–421. doi: 10.1016/j.ydbio.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Wu J., Okamura D., Li M., Suzuki K., Luo C.Y., Ma L., He Y., Li Z., Benner C., Tamura I. An alternative pluripotent state confers interspecies chimaeric competency. Nature. 2015;521:316–321. doi: 10.1038/nature14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liu Z., Ma Y., Zhong C., Yin Q., Zhou C., Wang H., Tang F., Wang Y., Zhang C. Generation of haploid embryonic stem cells from Macaca fascicularis monkey parthenotes. Cell Res. 2013;23:1187–1200. doi: 10.1038/cr.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.G., He Q.Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zhang K., Li L., Huang C., Shen C., Tan F., Xia C., Liu P., Rossant J., Jing N. Distinct functions of BMP4 during different stages of mouse ES cell neural commitment. Development. 2010;137:2095–2105. doi: 10.1242/dev.049494. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q., Song L., Peng G., Sun N., Chen J., Zhang T., Sheng N., Tang W., Qian C., Qiao Y. The transcription factor Pou3f1 promotes neural fate commitment via activation of neural lineage genes and inhibition of external signaling pathways. Elife. 2014;3 doi: 10.7554/eLife.02224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.