Abstract
Medical imaging examination on patients usually involves more than one imaging modalities, such as Computed Tomography (CT), Magnetic Resonance (MR) and Positron Emission Tomography(PET) imaging. Multimodal imaging allows examiners to benefit from the advantage of each modalities. For example, for Abdominal Aortic Aneurysm, CT imaging shows calcium deposits in the aorta clearly while MR imaging distinguishes thrombus and soft tissues better.1 Analysing and segmenting both CT and MR images to combine the results will greatly help radiologists and doctors to treat the disease. In this work, we present methods on using deep neural network models to perform such multi-modal medical image segmentation.
As CT image and MR image of the abdominal area cannot be well registered due to non-affine deformations, a naive approach is to train CT and MR segmentation network separately. However, such approach is time-consuming and resource-inefficient. We propose a new approach to fuse the high-level part of the CT and MR network together, hypothesizing that neurons recognizing the high level concepts of Aortic Aneurysm can be shared across multiple modalities. Such network is able to be trained end-to-end with non-registered CT and MR image using shorter training time. Moreover network fusion allows a shared representation of Aorta in both CT and MR images to be learnt. Through experiments we discovered that for parts of Aorta showing similar aneurysm conditions, their neural presentations in neural network has shorter distances. Such distances on the feature level is helpful for registering CT and MR image.
Keywords: Medical Image Segmentation, Machine Learning, Neural Networks
1. Introduction
Medical imaging examination on patients usually involves more than one imaging modalities, such as Computed Tomography (CT), Magnetic Resonance (MR) and Positron Emission Tomography(PET) imaging. Multimodal imaging allows examiners to benefits from the advantage of each modalities. For example, for Abdominal Aortic Aneurysm (AAA), CT imaging shows calcium deposits in the aorta clearly while MR imaging distinguishes thrombus and soft tissues better.1 Analysing and segmenting both CT and MR images to combine the results will greatly help radiologists and doctors to treat the disease. In this work, we develop deep neural network models to perform such multi-modal medical image segmentation, in particular AAA image segmentation.
Semantic image segmentation aims to label pixels or super-pixels of images by their corresponding class. Recently Deep Convolutional Neural Networks (CNN) have been successfully applied to a wide range of semantic image segmentation tasks.2, 3 For biomedical image segmentation, CNN has been successfully applied for Neuronal Membrane segmentation,4 Brain tumour segmentation,5 Prostate segmentation6 and many other tasks. However most of the methods only focus on one modality, namely CT, MR or Microscope images. For multi-modal image segmentation, currently research works mostly focus on images that can be easily registered, for example T1 and T2 MR sequence images of head.5 Because of the accurate registration process, such methods can treat different modality of the image as different channels of the image, similar to the Red, Green and Blue colour channels in RGB colour images. Several human body regions, such as abdominal region, contains soft tissues that can easily deform. Complex non-affine deformations with manual interventions have to be applied to register images of different modality.7 Therefore treating image modality as channels cannot be readily applied to such tasks requiring complex process of image registration.
A naive approach is to train CT and MR segmentation network separately. However, such approach is time-consuming and resource-inefficient. We propose a new approach to fuse the high-level part of the CT and MR network together, hypothesizing that neurons recognizing the high level concepts of Aortic Aneurysm can be shared across multiple modalities. Such network is able to be trained end-to-end with non-registered CT and MR image using shorter training time. Moreover network fusion allows a shared representation of Aorta in both CT and MR images to be learnt. Through experiments we discovered that for Aorta images showing similar aneurysm conditions, their higher layer representations in neural network are closer to each other. We also observed that manual shift and rotation of aligned CT and MR images will increase feature distances. We hypothesize that such distances on the feature level can be applied to align different modalities and to combine information for better diagnose and treatment results.
2. Methods
2.1. AAA CT-MR dataset
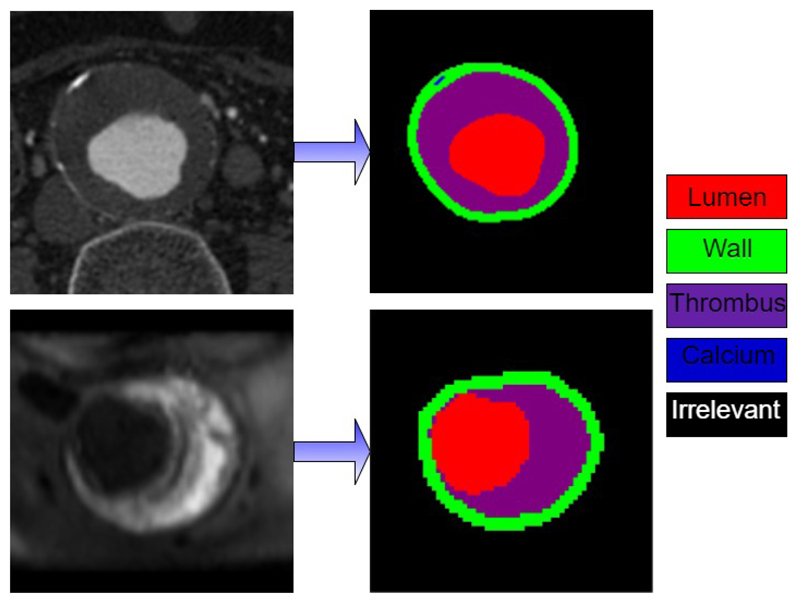
We have tested Cross-Net on Abdominal Aortic Aneurysm (AAA) segmentation dataset provided by Department of Radiology, University of Cambridge. AAA dataset consists of CT and MR scans of twenty-one anonymous patients with Abdominal Aortic Aneurysm recruited from Changhai Hospital, Shanghai, China. This study was approved by the review board of Changhai Hospital and written informed consent was obtained from each patient. All patients enrolled into this study were imaged by contrast enhanced CT angiography on a multi-slice CT scanner (Sensation Cardiac 64, Siemens, Germany). MR scans were obtained using Siemens Skyra 3T Machine. For our experiments we use T1 sequence of the MR image. For CT images, axial view images are segmented into five different classes, namely Aorta wall, lumen, thrombus, calcium deposits and irrelevant parts as background. Currently for MR images, the axial view images are segmented into four classes excluding calcium deposits. Figure 1 illustrates this segmentation task. Ground truth segmentation is provided for each scan image by radiologists and cardiovascular specialists.
Figure 1.
Illustration of CT (top) and MR(bottom) segmentations.
2.2. Separate Neural Network Models
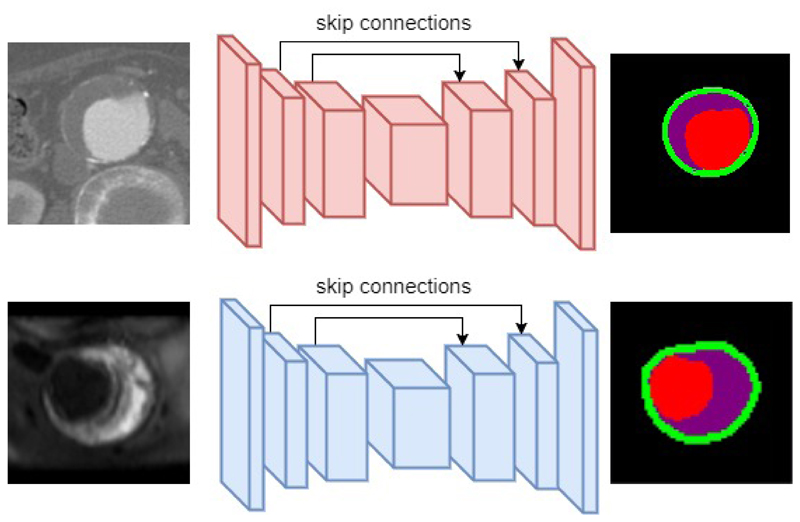
In this section we describe our Convolutional Neural Network (CNN) model developed for segmenting images of a single modality. Our CNN follows the recently popular encoder-decoder neural network design3, 4 which can be trained end-to-end to produce segmentation maps of the same size as input image. Such architectures usually contains an encoder which encodes image into high-level feature representations, and an decoder which decodes feature representations into dense segmentation maps. Encoder are implemented as stacks of convolutional layer and pooling layers, while decoder are implemented as stacks of deconvolutional layers, sometimes referred to as transposed convolutional layer. While convolutional layer maps a small local neighbourhood region of pixels into a single activation value, deconvolutional layer maps a single pixel to a small regions of output values at corresponding spatial location. Figure 2 shows an overview of the architecture used for single modality CT and MR image segmentation. Such CNN models are trained with images of a single modality (e.g. CT) and its corresponding ground truth segmentations. During training, these models learn to minimize the error between predicted segmentation and ground truth segmentation. In practice we use cross-entropy pixel-wise loss function as in equation 1:
| (1) |
where x is input image and y is ground truth segmentation maps. i is index of pixel locations in the image, and j is index of class. f (xi) is the neural network output for the ith pixel of image x.
Figure 2.
Illustration of Encoder-Decoder architecture implemented for single modality CT(top) and MR(bottom) image segmentation.
In addition to the encoder-decoder architecture, we also added skip connections8 that are shown to improve segmentation accuracies particularly for biomedical images. Skip layers are used to allow decoder to not only use high-level feature representations of the image, but also spatially more accurately lower layer information from the encoder. In the original implementation, layers in the encoder are copied and passed to corresponding layer in the decoder via the skip connections. However in our experiments, we discovered that rather than direct copying, processing by a convolutional layer with kernel size 1 × 1 improves segmentation accuracy of CT by 0.4% and of MR by 0.3%. We train our CNN model with image patches randomly sampled from scans in the axial plane. For CT image we use patch size of 128 × 128. For MR image, we use smaller patch size of 64 × 64 due to smaller sizes of the MR scans. We also apply data augmentation in the form of random horizontal and vertical flips. In total there are 77502 CT image patches and 89320 MR image patches sampled. The exact neural network configurations are not listed due to space limitations but can be found online in author’s github repository *.
2.3. Fusion Models
Training models for each modality separately does not make efficient use of available data. We hypothesize that the higher layer feature representations of Aorta images can be shared across modalities. This is because even though the detailed image statistics (e.g. pixel intensities and edge sharpness) of CT and MR image may differ, the higher level feature representations of Aorta should be relatively independent from the specific image modality. A well trained radiologists can recognize Aorta with Aneurysm in any image modality.
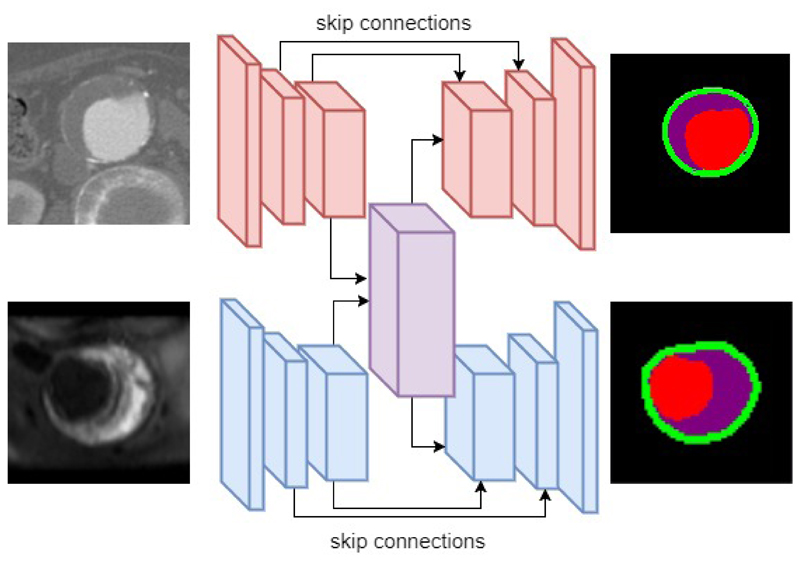
We therefore decide to fuse intermediate layers processing higher level feature representations for each modality. Figure 3 illustrates the model fusion. The intermediate layers, (including top layers of encoder and decoder) are fused from two streams processing CT and MR modality separately into one stream that process both CT and MR modality. This fusion models have two pathway, namely CT and MR pathway, with the intermediate layers fused between pathways. Such fusion models are trained in an alternating method with CT and MR data. In each mini-batch iteration, firstly fusion model’s CT pathway is trained with CT image, while keeping MR pathway except for the fused layers unvarying. Afterwards, the model’s MR pathway is trained with MR data while keeping CT pathway except fused layers unvarying.
Figure 3.
Illustration of model fusion for CT and MR image modality. The intermediate layers (including top layers in encoder and decoder) are fused from two separate streams into one stream.
There are two benefits of fusing higher level feature layers. Firstly, fusion model’s validation accuracy during training increase faster than separate models while keeping the same number of model parameters. This is because in fusion model, shared layers can learn a higher feature representations from both image modalities while separate models can only learn from a single image modality. Secondly, fusion models allow a shared representation to learnt for all image modalities. This means the neural representations (also referred as neuron activations or feature maps) in fused layers are similar for CT and MR image showing similar parts of Aorta. We demonstrated with experiments that this is indeed the case. Moreover we discovered that for two aligned CT and MR image, a manually induced translation or rotation will increase the feature representation distances. This can be potentially applied for image registration for different image modality.
3. Results
3.1. Training comparison
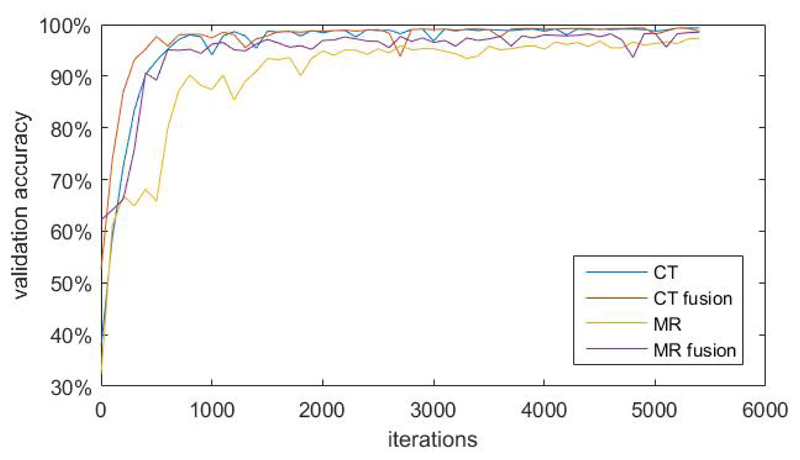
In this section we compare the validation accuracies during training between fusion models and separate models. We segregate dataset into training dataset, validation dataset and test dataset in 8:1:1 ratio randomly. The number of parameters of the fusion model is the same as that of CT and MR separate models combined. We use Adam optimizer9 for training and keep learning rates the same for the two different methods. In each training iteration we feed one mini-batch of CT data and one mini-batch of MR data to both fusion models and separate models. We noticed that time taken for both methods are relatively the same. Therefore we decide to report validation accuracy increase with respect to number of training iterations. Figure 4 shows the plot of validation accuracy of each models against number of training iterations. In this plot, validation accuracies of fusion models for CT and MR modality increase considerably faster than that of separate models. Validation accuracies level off approximately after 5000 iterations. At 5500 iterations, the validation accuracy for CT image modality is very close (99.1% v.s. 98.8%) for both models. The validation accuracy for MR image modality of fusion models is 98.5%, which is 1.2% than that of separate models.
Figure 4.
plot of validation accuracy of each models against number of training iterations. ”CT” and ”MR” are separate models while ”CT fusion” and ”MR fusion” are fusion models.
3.2. Shared feature representations
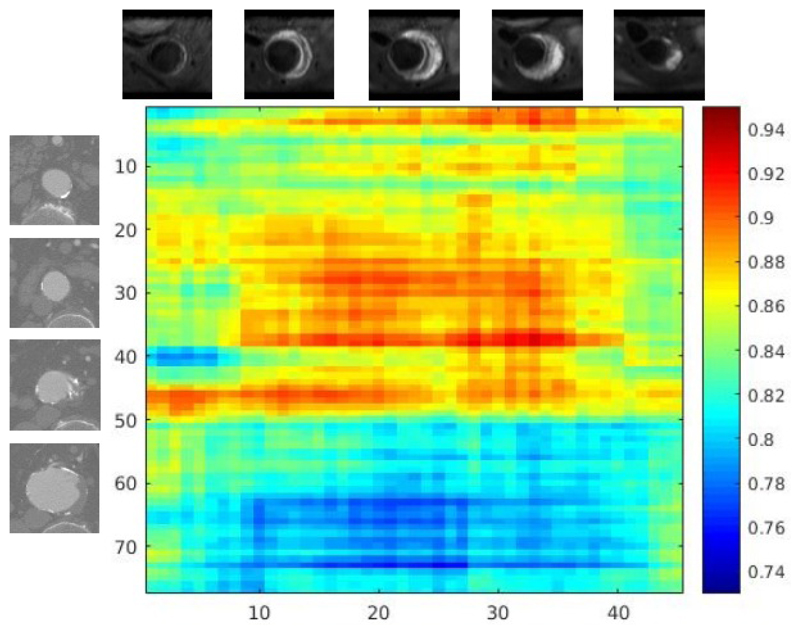
We examined intermediate layer feature representations of trained fusion models for CT and MR scans of the same patient. Through experiments we observed that for fusion models, images showing Aorta with Aneurysm condition have closer feature representations from each other than from healthy Aorta. This is also exhibited between images of healthy Aorta. Figure 5 shows the cosine distance matrix between CT and MR slices of a patient. MR sequences are obtained 7 days after CT scans. The CT sequence contains 77 image patches centred at Aorta from Thorax and Abdomen. The MR sequence contains 45 sequences mainly from Abdomen. Aneurysm condition are shown from the 51st CT image patch and from 6th MR image patch. The cosine distance are computed with equation:
| (2) |
Where FCT and FMR are feature representations of CT and MR images. We observed that feature representations of CT images and MR images showing Aneurysm condition have smaller distances between each other (blue region in Figure 5). Feature representations of CT image showing aneurysm condition have larger distances from those of MR images showing healthy Aorta (red region). We also observed that CT images (No. 39-41) and MR images(No. 1-5) showing the same part of healthy Aorta also exhibits shorter feature distances.
Figure 5.
Cosine distance matrix between CT scan sequences and MR scan sequences of one patient.
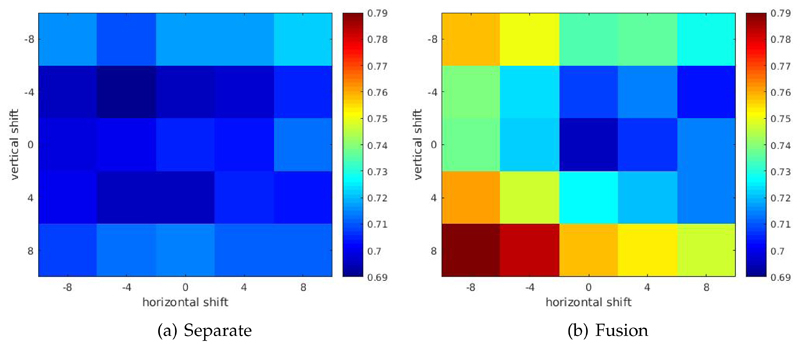
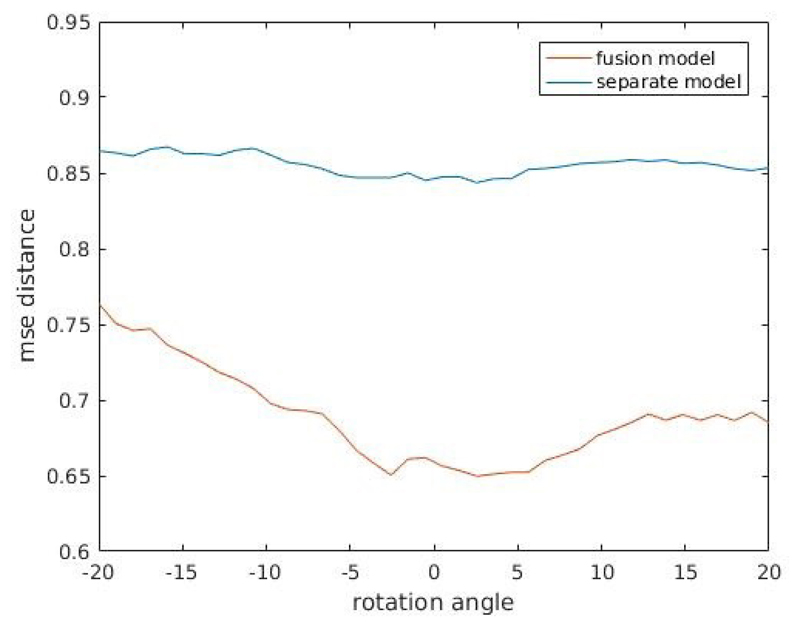
We also observed increases in feature distances when one modality image of pre-aligned CT and MR images are manually translated or rotated. Figure 6 shows heatmap plot of one randomly selected pre-aligned CT-MR image pair with CT image manually translated in both horizontal and vertical directions, for both fusion model and separate models. From the plot one can observe that or fusion model, both horizontal and vertical shift in patch extraction location of CT image patch induce an increase in feature distance between CT and MR image patch pair. This is not exhibited for separately trained models. Figure 7 shows the plot of one randomly selected pre-aligned CT-MR image pair with CT image manually rotated in the degree range of −20° − 20°. For fusion models, manual rotations induce increases in feature distance, which again is not exhibited for separately trained models. While we showed only a randomly selected example, we observed the same phenomenon in majority of image patch pairs.
Figure 6.
Heatmap plot of feature distances between manually translated pre-aligned CT-MR image pair. For fusion model, both horizontal and vertical shift in patch extraction location of CT image patch induce an increase in feature distance between CT and MR image patch pair. This is not exhibited for separately trained models.
Figure 7.
Plot of feature distances between manually rotated pre-aligned CT-MR image pair. Rotation degree range is −20° − 20°. For fusion models, manual rotations induce increases in feature distance, which is not exhibited for separately trained models.
4. Conclusion
In this work we developed network fusion methods for multi-modality medical image segmentation. We performed experiments on AAA CT-MR dataset and showed that fusion model improves training speed and allow shared representation of multi-modality images to be learnt. Such shared representations are potentially useful for multi-modality image registration and analysis.
Footnotes
References
- [1].Sakalihasan N, Limet R, Defawe O. Abdominal aortic aneurysm. The Lancet. 2005;365(9470):1577–1589. doi: 10.1016/S0140-6736(05)66459-8. [DOI] [PubMed] [Google Scholar]
- [2].Zheng S, Jayasumana S, Romera-Paredes B, Vineet V, Su Z, Du D, Huang C, Torr PH. Conditional random fields as recurrent neural networks. Proceedings of the IEEE International Conference on Computer Vision; 2015. pp. 1529–1537. [Google Scholar]
- [3].Noh H, Hong S, Han B. Learning deconvolution network for semantic segmentation. Proceedings of the IEEE International Conference on Computer Vision; 2015. pp. 1520–1528. [Google Scholar]
- [4].Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015; Springer; 2015. pp. 234–241. [Google Scholar]
- [5].Kamnitsas K, Ferrante E, Parisot S, Ledig C, Nori A, Criminisi A, Rueckert D, Glocker B. Deepmedic on brain tumor segmentation. Athens, Greece Proc BRATS-MICCAI; 2016. [Google Scholar]
- [6].Liao S, Gao Y, Oto A, Shen D. Representation learning: a unified deep learning framework for automatic prostate mr segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer; 2013. pp. 254–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lee CP, Xu Z, Burke RP, Baucom RB, Poulose BK, Abramson RG, Landman BA. Evaluation of five image registration tools for abdominal ct: pitfalls and opportunities with soft anatomy. Proceedings of SPIE–the International Society for Optical Engineering; NIH Public Access; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C. The importance of skip connections in biomedical image segmentation. International Workshop on Large-Scale Annotation of Biomedical Data and Expert Label Synthesis; Springer; 2016. pp. 179–187. [Google Scholar]
- [9].Kingma D, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014 [Google Scholar]