ABSTRACT
An important component of vaccine development is the identification of safe and effective adjuvants. We sought to identify transcriptomal signatures of innate immune stimulating molecules using next-generation RNA sequencing with the goal of being able to utilize such signatures in identifying novel immunostimulatory compounds with adjuvant activity. The CC family of chemokines, particularly CC chemokines 1, 2, 3, 4, 7, 8, 17, 18, 20, and 23, were broadly upregulated by most Toll-like receptor (TLR) and nucleotide-binding domain and leucine-rich repeat–containing receptors (NLR) stimuli. Extracellular receptors such as TLR2, TLR4 and TLR5 induced the transcription of CXC chemokines including CXCL5, CXCL6 and CXCL8, whereas intracellular receptors such as TLR7 and TLR8 upregulated CXC chemokines 11 and 12. Both TLR1/2 and TLR2/6 agonists induced strong chemokine production in human peripheral blood mononuclear cells. Human skeletal muscle cells and fibroblasts respond with chemokine production only to TLR2/6 agonists, but not TLR1/2 agonists, consistent with strong expression of TLR2 and TLR6, but not of TLR1, in fibroblasts. TLR2/6 stimulated fibroblasts demonstrated functional chemotactic responses to human T cell and natural killer cells subsets. The activation of non-hematopoietic, adventitial cells such as fibroblasts and myocytes may contribute.
KEYWORDS: Toll-like receptors, vaccine adjuvants, chemokines, Type I interferons, transcriptomal profiling, chemotaxis
Introduction
Vaccination has proven to be one of the most effective methods of affording protection against infectious diseases.1,2 Despite the dramatic improvements in controlling the incidence of vaccine-preventable diseases, newly emerging and re-emerging infectious diseases present new challenges. Pertussis (whooping cough) is a re-emerging disease. In North America, large outbreaks of pertussis have occurred with trends indicating a shift in peak incidence from adolescents to children.3 One of the reasons for the resurgence of pertussis is thought to be due to rapidly waning immunity to acellular subunit pertussis vaccines that have supplanted whole cell killed vaccines.4-6 In an infant baboon model, it has been demonstrated that animals vaccinated with alum-adjuvanted acellular subunit pertussis vaccine mounted a mixed T helper 1/T helper 2 (Th1/Th2) response, which was associated with protection against disease, but not from colonization of the organism; vaccination with whole cell killed pertussis vaccine induced B. pertussis-specific T helper 17 (Th17) memory and Th1 memory, which was associated with a more rapid clearance of the pathogen.7,8 Studies in animal models have suggested that replacing alum with alternate adjuvants that promote strong Th1- and Th17-biased responses may be beneficial in improving vaccine efficacy.9
The engagement of innate immune receptors plays a role in the action of many vaccine adjuvants; although the precise mechanisms of aluminum-based adjuvants (commonly referred to as ‘alum’) remain largely unknown. Activation of the nucleotide-binding domain and leucine-rich-repeat-containing gene family pyrin-domain-containing 3 (NLRP3) inflammasome is thought to contribute to its adjuvant activity,10,11 as well as the release of DNA from host cells and subsequent engagement of DNA-sensing innate immune pathways.12 Toll-like receptors (TLRs) and nucleotide-binding domain and leucine-rich repeat–containing receptors (NLR) constitute important sensors in the activation of innate immune cells including monocytes, macrophages, and dendritic cells. These cells function as sentinels against foreign antigens and pathogens, recognizing pathogen-associated molecular patterns (PAMPs) through pattern-recognition receptors (PRRs).13,14 The 10 functional TLRs in the human encode proteins with an extracellular domain having leucine-rich repeats (LRR) and a cytosolic domain called the Toll/IL-1 receptor (TIR) domain.15 TLR1, −2, −4, −5, and −6 recognize extracellular stimuli, while TLR3, −7, −8 and −9 function within the endolysosomal compartment. The ligands of TLRs are highly conserved molecules such as lipopolysaccharides (LPS) (recognized by TLR4), lipopeptides (TLR2 in combination with TLR1 or TLR6), flagellin (TLR5), single-stranded RNA (TLR7 and TLR8), double-stranded RNA (TLR3 and MDA5), CpG motif-containing DNA (recognized by TLR9).15
The discovery and development of safe and effective vaccine adjuvants has been a central research goal in our laboratory, which has led us to explore structure–activity relationships in a variety of innate immune-stimulatory chemotypes, including small molecule agonists of TLR2,16-18_ENREF_43 TLR4,19 TLR7,20-29 TLR8,28,30-38 NOD1,39 as well as C-C chemokine receptor type 1 (CCR1).40
Several studies have previously attempted to examine the molecular signatures elicited by vaccines,41 as well as vaccine adjuvants.42-45 A recent comprehensive study comparing a range of adjuvants in non-human primates44 found that TLR3 and TLR7 stimuli induced IFN and antiviral transcriptomal programs, while alum and TLR4-containing adjuvants evoked inflammatory and myeloid-associated modules, consistent with our findings,45 and those of others.42,43 Given the enormous diversity of signals recognized by the TLRs, which are present both in the extracellular and intracellular compartments, we sought to address two questions: (i) are there ‘signatures’ that are diagnostic of innate immune activation, regardless of the innate immune sensor involved, and (ii) which of the many TLR ligands that we have characterized induce strong humoral responses to subunit antigens.
Whole transcriptome next-generation RNA sequencing of human peripheral blood mononuclear cells exposed to 23 innate immune-active compounds encompassing almost the entire repertoire of TLR ligands indicate prominent upregulation of CC and CXC chemokines, independent of the innate immune receptor involved. Non-hematopoietic cells such as fibroblasts and skeletal muscle cells express TLR2 and respond uniquely to TLR2/6 agonists, but not TLR1/2 ligands, resulting in the induction of chemokines and, consequently, manifesting in the chemotaxis of several major human lymphocytic subsets. These results point to the utility of chemokine induction as a ‘signature’ of adjuvant activity, and provide mechanistic insight into the potential utility of harnessing TLR2 agonists as vaccine adjuvants.
Materials and methods
Reagents
PAM2CSK4, PAM3CSK4, lipoteichoic acid (LTA), lipopolysaccharide (LPS) from E. coli 055:B4, MPLA, C12-iE-DAP, Murabutide, Poly (I:C) (high molecular weight), flagellin, CL307, ODN2216, ODN2006, and ODN2395 were purchased from InvivoGen (San Diego, CA). The TLR agonists DBS-2-217C,46 C4, IMDQ, Meta-amine,21 EY-2-40,47 XG-1-236,28 MB-564, MB-569,37 MB-152,34 and KHP-3-12648 were synthesized using the routes previously described by us. C27449 was graciously provided by Dynavax Technologies (Berkeley, CA).
Culture of human blood and cell lines
Whole human blood was collected in heparinized vacutainers, and peripheral blood mononuclear cells (PBMCs) were collected and isolated in CPT Vacutainers™ (Becton Dickinson, Franklin Lakes, NJ) from healthy volunteers providing written informed consent in accordance with the University of Minnesota Institutional Review Board approved protocol (IRB Protocol 1506M74641). PBMCs were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mM penicillin, and 50 µg/mL streptomycin (complete RPMI). Human foreskin fibroblasts (HFFs, ATCC catalog No. SCRC-1041) were cultured in DMEM supplemented with 10% fetal bovine serum, 2 mM penicillin, and 50 µg/mL streptomycin (complete DMEM). Human Skeletal Muscle cells (SkMC, Lonza, Basel, Switzerland, catalog No. CC-2561) were cultured in skeletal muscle cell culture media supplemented with a proprietary skeletal muscle cell media (BulletKit™, Lonza, Basel, Switzerland). HMEC-1 cells (dermal microvascular endothelial cells, ATCC catalog No. CRL3243) were cultured in MCDB131 media (Thermo Fisher, Waltham, MA) supplemented with 10 ng/mL epidermal growth factor, 1 µg/mL hydrocortisone, 10 mM glutamine, and 10% fetal bovine serum. Cells were cultured in 96-well plates at 37°C for 3.5–16 h, as indicated for that experiment.
Next-generation sequencing of PBMCs
PBMCs were stimulated in 96-well plates for 3.5 h with 1 µg/mL TLR/NLR agonist, in duplicate. Pilot experiments were first conducted to arrive at optimal concentrations of innate immune ligands as well as exposure times in order to accommodate processing large numbers of samples simultaneously for the large-scale next-generation sequencing (described in Materials and Methods). The potential weakness of this approach is that we may not have captured peak fold-changes in gene expression for all of the ligands examined. Total RNA was isolated using RNeasy 96 kits according to the manufacturer's instructions (Qiagen, Hilden, Germany). Samples were stored in RNase free 96-well PCR plates at −80°C until used. RNA quality and concentration were determined using Agilent RNA 6000 Nano Kits (Agilent Technologies, Santa Clara, CA) and libraries were generated using Clontech SMARTer Standards Total-RNA Pico Kits (Clontech Laboratories, Inc., Mountain View, CA). Libraries contained inserts of approximately 200 base pairs and averaged quality scores over Q30. Individual TruSeq libraries were pooled into 2 and sequenced across 3 lanes. Next-generation sequencing was carried out by the University of Minnesota Genomics Center using a HiSeq 2500 (Illumina, San Diego, CA) in high output mode for 13 × 106 reads per sample for 50 base pair paired ends using v4 chemistry. RNASeq data is available at the Sequence Read Archive (BioProject ID: PRJNA390780; http://www.ncbi.nlm.nih.gov/bioproject/390780).
Multiplexed cytokine analysis in PBMCs, SkMC, HMEC-1 and HFFs
Cytokine and chemokine responses of PBMCs, SkMCs, HMEC-1, and HFFs were measured using methods previously reported by us19,36,37 with the following Milliplex kits: HCYTMAG-60K-PX41, HCYPMAG-63K, and HCYP2MAG-62K (EMD Millipore, Billerica MA). Cells were seeded at a density of 105 cells per well. Cells were either stimulated for 16 h with graded concentrations of TLR ligands, or mock-stimulated with vehicle (DMSO). Supernatants were collected by Precision 2000 liquid handlers (BioTek, Winooski, VT) and diluted 1:3 for each kit. The data was acquired on a MagPix® instrument and the data analyzed using Milliplex Analyst (EMD Millipore, Billerica MA).
CCL20 and CXCL6 ELISAs
PBMCs, SkMCs, HMEC-1, and HFFs were cultured in 96-well plates at 105 cells per well in appropriate growth media with graded concentrations of TLR ligands for 16 h at 37°C. Plates were centrifuged to collect supernatants, which were diluted 1:5 for CXCL6 ELISAs (Abcam, Cambridge, MA, Catalog number AB155431) and 1:2 for CCL20 ELISAs (Abcam, Cambridge, MA, Catalog number AB100599). The manufacturer's procedure were followed for both types of ELISA and acquired on a SpectraMax M2 (Molecular Devices, Sunnyvale, CA). Concentrations were quantified from four-parameter logistic fits of standard curves for each analyte.
Immunofluorescence detection of TLR1, −2, and −6 expression in HFFs
HFFs were plated at 104 per well in a tissue culture-treated 96-well plate and incubated overnight. Each well was washed 3 times in PBS and then fixed for 10 min at room temperature in 4% paraformaldehyde followed by blocking with 10% goat serum in PBS (recommended blocking buffer for the Tyramide Superboost kit described below) for 1 h at room temperature. Polyclonal anti-TLR1, −2, and −6 antibodies (Abcam, Cambridge, MA, Catalog numbers AB189337, AB191458, AB37072, respectively) were utilized at 1:100 dilutions in 100 µL blocking buffer to stain cells for 1 h at room temperature, followed by 1 h of incubation with goat-anti-rabbit-HRP antibody conjugate. Tyramide signal amplification was carried out for 5 min according to the Tyramide SuperBoost kit's manufacturer (Invitrogen, Carlsbad, CA, Catalog number B40922). Samples were counterstained with 100 ng of DAPI and imaged at the University of Minnesota Imaging Center using a Nikon A1R confocal microscope with a 60X water immersion objective with a numerical aperture of 1.20.
PBMC chemotaxis with HFFs
HFFs were plated at 106 per well in an IncuCyte ClearView 96-well reservoir plate (Essen Bioscience, Ann Arbor, MI) in complete DMEM and cultured for 16 hours. Following incubation, the cells were stimulated with graded concentrations of TLR agonists for 24 h. An IncuCyte ClearView chemotaxis plate was coated with 0.5 mg/mL Matrigel (Corning Life Sciences, Corning, NY, Catalog number 354248) in complete RPMI on ice. Matrigel polymerization was carried out for 30 minutes at 37°C, and then the chemotaxis plate was cooled to room temperature for 1 h. PBMCs were plated in 60 µL of complete RPMI in the top well of the chemotaxis plate. The top of the chemotaxis plate was placed on the reservoir plate containing HFFs and cultured for 16 h at 37°C. After the incubation, the bottom reservoir plate was stained with CD3-PE and CD56-APC (eBioscience, San Diego, CA, Catalog numbers 12-0037-42 and 17-0566-42, respectively), CD4-V450, CD8-V500, CD14-FITC, and CD19-PE-Cy7 (Becton Dickinson, Franklin Lakes, NJ, Catalog numbers 560345, 560774, 555397, 557835, respectively). Erythrocytes were lysed and leukocytes fixed by transferring 200 µL of PBMCs in the bottom reservoir plate to 800 µL of warm Lyse/Fix buffer (Becton Dickinson, Franklin Lakes, NJ) in a 96-deep well plate by liquid handler for 10 min at 37°C. The fixing process was carried out one additional time before washing with 800 µL complete RPMI. Samples were resuspended in 200 µL of complete RPMI, and acquired on a FACSVerse flow cytometer (Becton Dickinson, Franklin Lakes, NJ) for 250,000 gated events. Absolute counts were recorded by an inline flow sensor for lymphocytes (FSC, SSC), T cells (CD3+ CD56−), Th cells CD3+ CD4+ CD8− CD56−), CTLs (CD3+ CD4− CD8+ CD56−), NK cells (CD3− CD56+), B cells (CD3− CD19+ CD56−), cytokine-induced killer cells (CIK) (CD3+ CD56+), and monocytes (CD14+).
Statistical methods
RNASeq data were obtained on duplicate samples, and were calculated as fold change over unstimulated control samples using the EDGE test.50,51 For each TLR/NLR agonist examined, root mean square deviations in fold change over the entire transcriptome (26, 363 annotated genes) were calculated as . Chemokine release and chemotaxis assays were performed on triplicate samples of PBMCs isolated from at least two healthy donors. Immunofluorescence experiments were performed in triplicate.
Results
A main objective of this work was to examine if there are any transcriptional signatures common to the broad range of TLR/NLR-active compounds that we have evaluated as vaccine adjuvants. These compounds are shown in Fig. 1, and include both canonical and novel small-molecule agonists of TLR2 (PAM2CSK4, PAM3CSK4, DBS-2-217C, and lipoteichoic acid), TLR3/MDA5 (Poly I:C), TLR4 (LPS from E. coli, MPLA), TLR5/NLRC4 (flagellin), TLR7 (C4, CL307, EY-2-40), TLR8 (KHP-3-126, MB-569, MB-152), dual TLR7/TLR8 (XG-2-136, Meta-amine and IMDQ), TLR9 (ODNs 2006, 2216, 2395, and C274), NOD1 (C12-iE-DAP) and NOD2 (Murabutide). We had previously characterized the transcriptional responses to a small subset of innate immune stimulatory compounds using microarrays.45 In this study, we utilized next-generation sequencing of total RNA isolated from human PBMCs stimulated for 3.5 h with 1 µg/mL of each of the agonists to assess early transcriptional responses. A total of 26,363 annotated genes for each of the 23 test samples were compared in duplicate against unstimulated (or vehicle-alone, mock-stimulated) control samples. As we had previously reported, diagnostic transcriptional signatures such as proinflammatory cytokine (TLR4), interferon, and interferon-inducible genes (TLR7) were strongly upregulated in the samples. In order to discern global signatures in an unbiased manner, we compared root mean square deviations of fold change (from control values) in expression of each of the genes across the 23 test samples (Fig. 2). We observed pronounced deviations in three gene families: the CXC and CC chemokines, and Type I interferons (Fig. 2); additionally, we noted significant change in a subset of genes including TNFSF15 (TNF superfamily ligand, member 15), TFPI2 (tissue factor pathway inhibitor 2), RSAD2 (radical S-adenosyl methionine domain containing 2), PDGFRL (platelet derived growth factor receptor like), LEP (leptin), IL6 (interleukin-6), IL12B (interleukin-12B), IL1A (interleukin-1α) IL36G (interleukin-36γ), IRG1 (immunoresponsive gene 1), INHBA (inhibin beta A subunit), IFIT1 (interferon induced protein with tetratricopeptide repeats 1), DNAAF1 (dynein axonemal assembly factor 1), DEFB1 (defensin β-1), CFB (complement factor B), and LOC100506178 (uncharacterized).
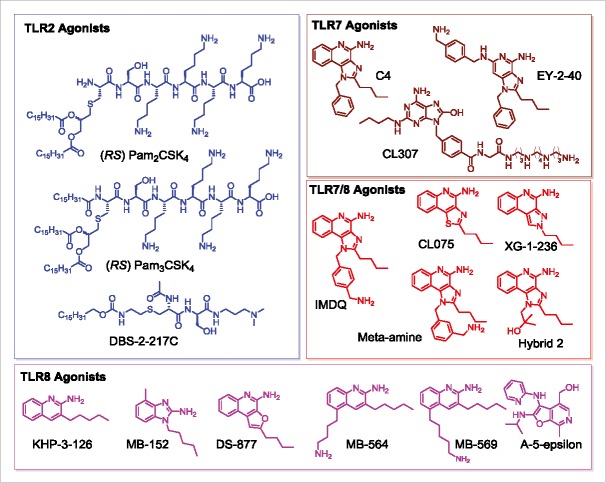
Figure 1.
Structures of TLR agonists arranged by receptor target. TLR2 agonists (PAM2CSK4, PAM3CSK4, DBS-2-217C) are shown in blue, pure TLR7 agonists (C4, EY-2-40, CL307) are shown in maroon, dual TLR7/8 agonists are shown in red (IMDQ, Meta-amine, XG-2-136, Hybrid 2), and pure TLR8 agonists (KHP-3-126, MB-152, DS-877, MB-564, MB-569, A-5-epsilon) are shown in magenta.
Figure 2.
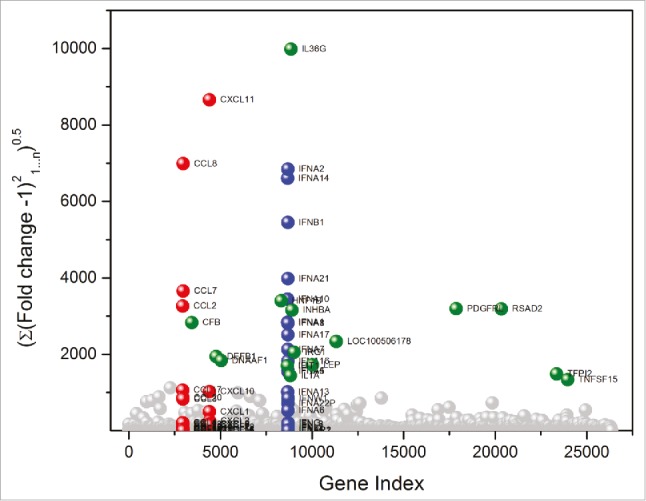
TLR and NLR agonists upregulate genes associated primarily with three gene families. Global transcriptomal signatures in RNASeq data were analysed in an unbiased manner by comparing root mean square deviations of fold change (from control values) in expression of each of the 26,363 genes across the 23 test samples, in duplicate. For each TLR/NLR agonist examined, root mean square deviations in fold change over the entire transcriptome (26, 363 annotated genes) were calculated as . The three major gene families showing significant upregulation were CC chemokines (red), CXC chemokines (red), and interferons (blue). Other upregulated genes are shown in green.
We next quantified changes in the CC and CXC chemokines, as well as IFN genes for each of the agonists. Strong signals for the CC chemokines 1, 2, 3, 4, 7, 8, 17, 18, 20, and 23 were observed for TLR2, TLR3, TLR4, TLR5, TLR7, TLR7/8, TLR8, NOD1, and NOD2 agonists (Fig. 3). The signals for TLR9 were significantly weaker, likely on account of the oligonucleotides requiring longer incubation periods as we had previously observed.45
Figure 3.
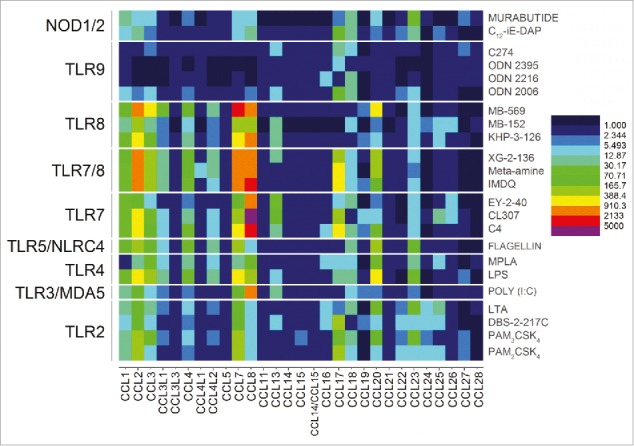
Transcriptomal ‘signatures’ of innate immune stimulation in CC chemokines. A heat map of fold change (means of duplicate samples, EDGE test validated) indicated that many of the TLRs and NLRs upregulated the CC chemokines 1, 2, 3, 4, 7, 8, 17, 18, 20, and 23.
A clear demarcation of extracellular versus intracellular TLR/NLR activation was apparent in the analysis of gene expression levels for the CXC chemokines. The engagement of the extracellular TLRs 2, 4, and 5 resulted in strong upregulation of CXCL5, CXCL6 and CXCL8 genes. In contrast, stimulation of the intracellular receptors TLR3, 7, and 8 manifested in upregulation of CXCL11 and CXCL12 (Fig. 4). Signals for CXCL1, CXCL2, and CXCL3 were also observed to be strongly enhanced but, like the CC chemokines, were found to be common for almost all agonists (Fig. 4). Interferon-α responses, as expected, were almost exclusively restricted to TLR7 (and TLR7/8 activation), whereas IFN-b responses were elicited by TLR3, TLR7, TLR7/8 and TLR9 agonists. It is noteworthy that interferon-ω signals were also evoked by TLR7 engagement (Fig. 5).
Figure 4.
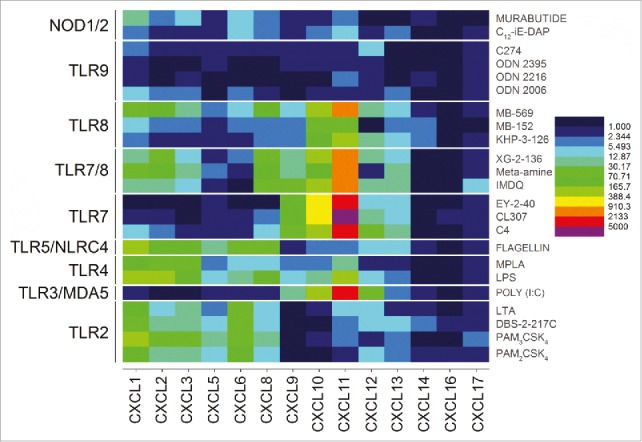
Discrimination between extracellular and intracellular innate immune receptors by CXC chemokine profiles. The extracellular TLRs 2, 4, and 5 showed strong upregulation of CXCL5, CXCL6 and CXCL8, while the endolyososomal TLRs 3, 7, and 8 were characterized by upregulation of CXCL11 and CXCL12. Heat maps of fold change (means of duplicate samples, EDGE test validated) are shown.
Figure 5.
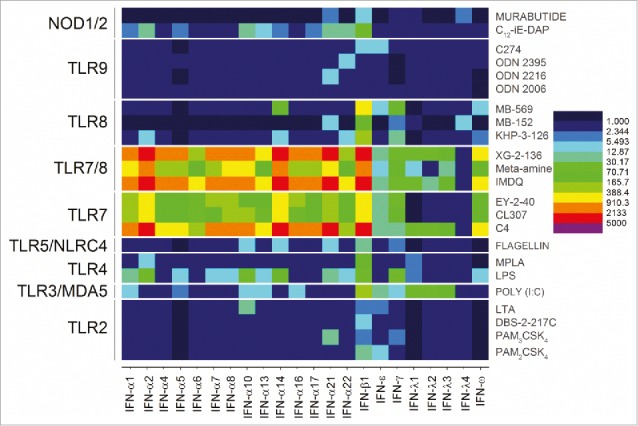
TLR7 upregulation of IFN-α and IFN-ω. Pure TLR7 and dual TLR7/8 agonists dramatically upregulated all IFN-α genes and IFN-ω. Heat maps of fold change (means of duplicate samples, EDGE test validated) are shown.
We sought to understand why TLR2-active compounds displayed strong adjuvant activity. Transcriptomal profiling pointed to strong upregulation of CXCL and CCL chemokine transcripts in PBMCs (Figs. 3 and 4), and we confirmed the induction of multiple CXCL and CCL chemokines using multiplexed immunoassays (representative data for CCL20 and CXCL6 shown in Fig. 6). Given that intramuscular injections are largely delivered into interstitial spaces, we wished to examine if non-hematopoietic cells such as endothelial cells, fibroblasts, and skeletal muscle cells could also respond to TLR2 stimulation. We examined the effect of almost the entire set of TLR agonists on cytokine and chemokine secretion using multiplexed cytokine and chemokine assays. We found that in skeletal muscle cells as well as fibroblasts, but not in endothelial cells, the TLR2/6-active compounds PAM2CSK4 and DBS-2-217C induced both CC and CXC chemokines, including CCL20 and CXCL6 (Fig. 7). The TLR1/2-agonist, PAM3CSK4, did not elicit chemokine responses. In examining fibroblast responses to the TLR2 ligands in greater detail, we observed dose-dependent induction of CXCL6 (GCP2), CCL20 (MIP-3α), CXCL8 (IL-8), CCL2 (MCP-1) and CCL7 (MCP-3) by the TLR2/6-active compounds PAM2CSK4 and DBS-2-217C, and not by the TLR1/2-agonist, PAM3CSK4 (data not shown).
Figure 6.
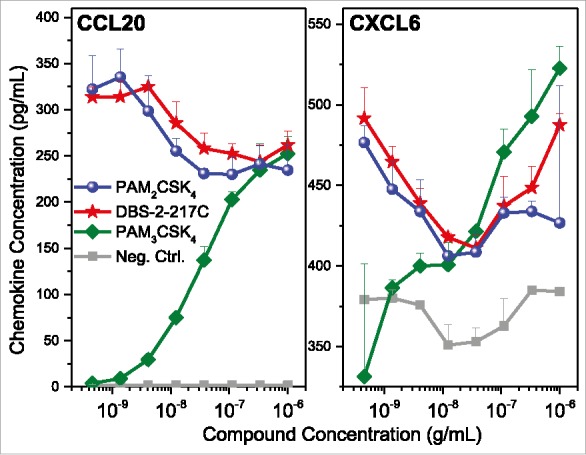
Validation of chemokine induction in TLR2 stimulated PBMCs. Human PBMCs were stimulated with TLR2 agonists for 16 h, and examined for CCL20 and CXCL6 expression by ELISA. PBMCs responded to both agonists that signal through TLR1/2 heterodimers (PAM3CSK4) and TLR2/6 heterodimers (PAM2CSK4 and DBS-2-217C). Means and standard deviations of triplicate samples are shown.
Figure 7.
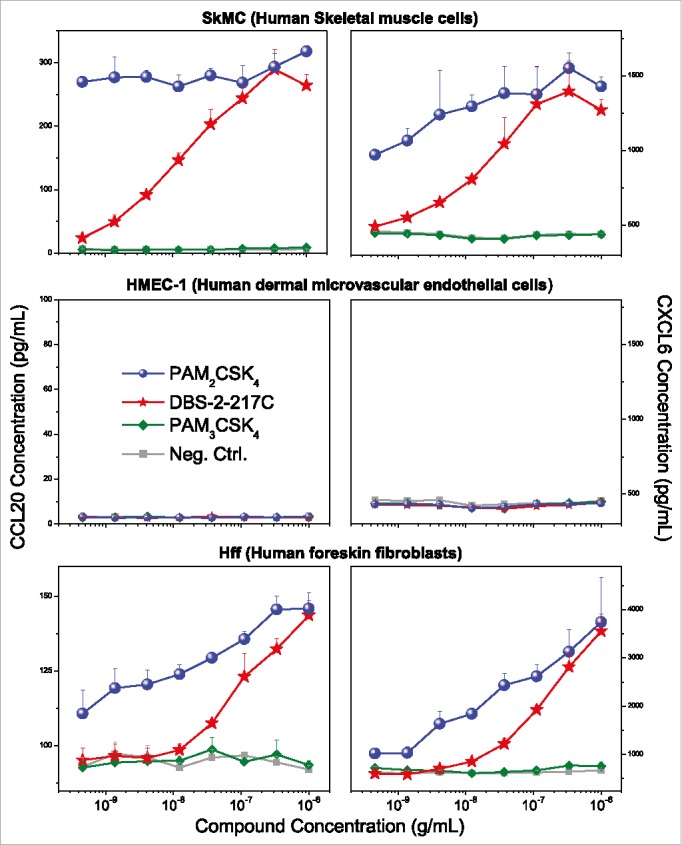
TLR2 agonists induce chemokine expression in skeletal muscle cells and human foreskin fibroblasts, but not dermal microvascular endothelial cells. Human skeletal muscle (SkMC), human foreskin fibroblast (HFF), and dermal microvascular endothelial (HMEC-1) cell lines were stimulated with TLR2 agonists and quantified for CCL20 and CXCL6 induction by ELISA. TLR2/6 agonists (PAM2CSK4 and DBS-2-217C) induced CCL20 and CXCL6 expression in both SkMCs and HFFs, but not HMEC-1 cells. None of the cell lines responded to the TLR1/2 agonist PAM3CSK4. Chemokine release assays were performed on triplicate samples. Means and standard deviations are shown.
We hypothesized that the selective responsiveness for TLR2/6 agonists in skeletal muscle cells and fibroblasts could be a consequence of differential TLR expression in fibroblasts. We therefore examined the expression of TLR1, TLR2, and TLR6 in fibroblasts, employing tyramide signal amplification52,53 for enhancing sensitivity of detection. We observed strong expression in fibroblasts of TLR2 and TLR6, but very faint levels of TLR1 (Fig. 8), which is consistent with responses elicited by the TLR2/6 agonists PAM2CSK4 and DBS-2-217C, and not by the TLR1/2 agonist PAM3CSK4.
Figure 8.
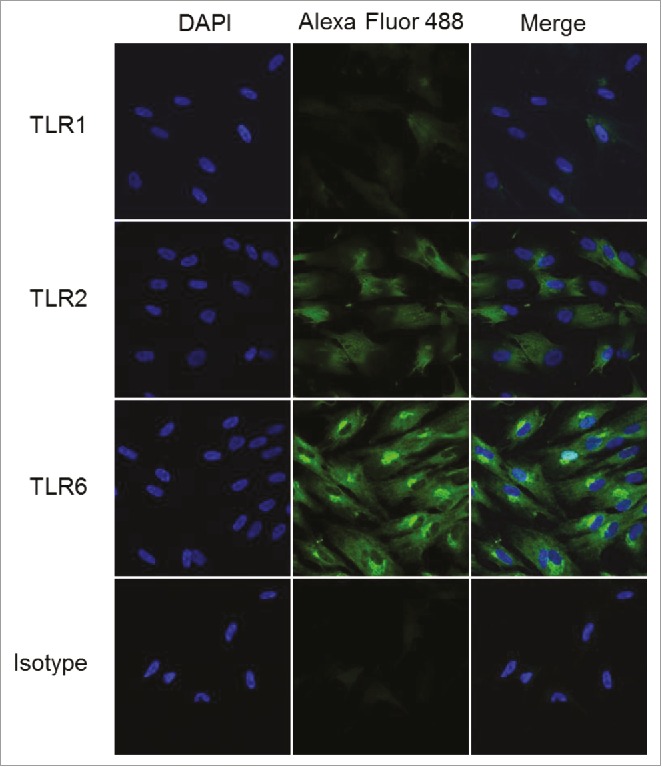
TLR2 and TLR6 are expressed in human foreskin fibroblasts. HFFs were interrogated for the expression of TLRs 1, 2, and 6 using confocal immunofluorescence with tyramide signal amplification. HFFs showed strong expression of TLR2 and TLR6, and very weak expression of TLR1. Samples were examined in triplicate; representative data are shown.
In order to verify that TLR2/6 occupancy and the consequent secretion of CC and CXC chemokines in fibroblasts have functional outcomes, we measured the chemotaxis of human PBMCs toward fibroblasts stimulated with TLR2/6 and TLR1/2 ligands. Human SDF-1 was used as a positive control in these experiments. We observed dose-dependent migration of PBMCs toward fibroblasts stimulated with TLR2/6, but not TLR1/2 ligands (Fig. 9). Flow cytometric assessment of absolute counts of migrated PBMCs showed that T lymphocytic subsets (CD4+ Th cells, CD8+ CTLs, and CD3+ CD56+ cytokine-induced killer cells), but not B lymphocytes (CD19+) underwent chemotaxis (Fig. 9). The induction of chemotactic gradients by adventital cells and consequent migration of immune cells to the site of injection may likely contribute to the strong adjuvant properties of TLR2/6 agonists.
Figure 9.
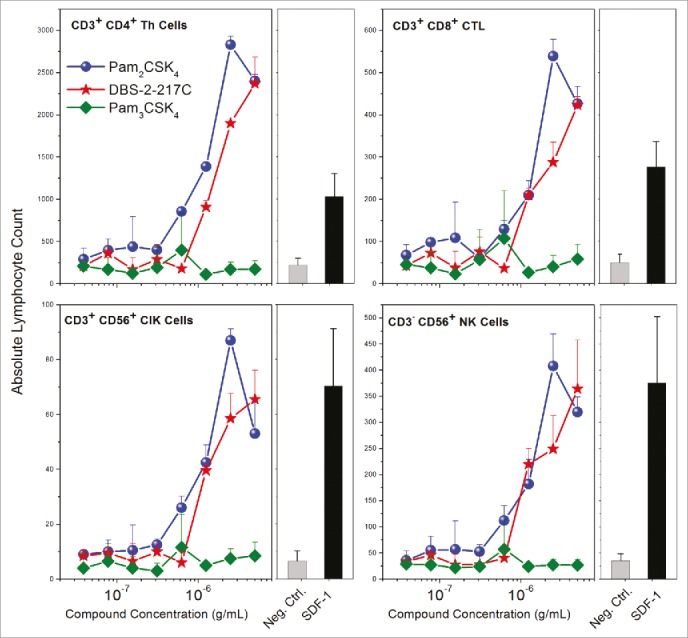
Chemotaxis of PBMCs toward TLR2/6- stimulated human foreskin fibroblasts. HFFs stimulated with TLR2/6, but not TLR1/2 agonists elicited functional chemotactic responses from lymphocytic populations in human PBMCs. Resting, adherent fibroblasts were stimulated with graded concentrations of agonists for 24 h in a chemotaxis plate. Chemotaxis of freshly isolated PBMCs was quantified by flow cytometry using lineage-specific antibodies. Means and SD on triplicate samples are shown.
Discussion
A major objective was to determine if there are any common signatures attributable to adjuvant effects in a broad range of TLR/NLR-active compounds that we have evaluated as vaccine adjuvants. Strong transcriptional upregulation for several CC chemokines including CCL1, −2, −3, −4, −7, −8, −17, −18, −20, and −23, as well as the CXC chemokines, CXCL1, CXCL2, and CXCL3 were observed for the majority of innate immune stimuli. Indeed, both alum,54,55 as well as MF59, a squalene-in-water emulsion containing the surfactants polysorbate 80 and sorbitan trioleate56,57 are FDA-approved vaccine adjuvants that do not activate any of the TLRs, but induce chemokine secretion in hematopoietic cells.58 These observations point to the potential utility of chemokine readouts as surrogate markers of innate immune activation, irrespective of the nature of stimulation. We hope that quantifying chemokine induction alongside conventional assays measuring the release of proinflammatory mediators may help prospectively identify novel adjuvants with low reactogenicity.
The vast majority of vaccines are administered intramuscularly, and we reasoned that, in addition to skeletal muscle cells, fibroblasts could represent a significant cell type at the site of vaccination where the local concentrations of both antigen and adjuvant are expected to be high. Indeed, early studies on the disposition of the intramuscularly injected materials59 have suggested that the term ‘intramuscular’ is a misnomer, and should perhaps be referred to as ‘intermuscular’60 in light of the fact that much of the material because of the spread of solutions along interfacial planes between fascicles. Although adventitial cells such as fibroblasts are not usually considered to be components of the innate immune system, it has been suggested that fibroblasts have immunoregulatory functions, and should therefore be considered as sentinel cells.61,62 Furthermore, human fibroblasts from disparate anatomical sites have been shown to synthesize CC and CXC chemokines.63-68
Whereas human PBMCs respond to both TLR1/2 (PAM3CSK4) and TLR2/6 (PAM2CSK4 and DBS-2-217C) agonists, only TLR2/6 agonists, but not the TLR1/2 agonist, elicited chemokine responses in fibroblasts, paralleled by dose-dependent chemotaxis of PBMCs toward fibroblasts stimulated with TLR2/6, but not TLR1/2 ligands, consistent with strong expression of TLR2 and TLR6 but low levels of TLR1. These findings draw attention to the poorly-studied role of stromal and adventitial cells in creating chemotactic gradients, and thereby indirectly focusing and amplifying adaptive immune responses.
Trumenba™ is a recently-licensed vaccine for the prevention of Neisseria meningitidis serogroup B-related meningococcal meningitis.69 The vaccine is composed of two recombinant, E. coli-expressed, lipidated factor H binding protein (fHBP) variants from N. meningitidis serogroups A and B. The fHBP lipoproteins are triacylated, and are therefore expected to signal via TLR1/TLR2 heterodimerization.70 It was noted in preclinical in vivo studies that the lipidated forms of fHbps were more immunogenic compared to the non-lipidated forms, pointing to the self-adjuvanting activity of the TLR2-active moiety.70 Our results suggest that diacyl or monoacyl TLR2 agonists that signal via TLR2/6 may be superior in eliciting adjuvant responses over triacyl species by promoting chemotactic responses to the site of injection. A careful comparison with MPLA (TLR4 agonist) would be useful in benchmarking the adjuvant properties of TLR2/6-active compounds.
In conclusion, a CC chemokine signature (CC chemokines 1, 2, 3, 4, 7, 8, 17, 18, 20, and 23) appears to be a common transcriptional outcome of virtually all TLR/NLR agonists whereas the CXC chemokine patterns allow for the distinction of extracellular (CXCL5, CXCL6 and CXCL8) vis-à-vis intracellular (CXCL11 and CXCL12) TLR/NLR activation. These findings are likely to be useful both in prospectively examining novel compounds for adjuvant activity, and in understanding structure-activity relationships in such molecules. TLR2/6 agonists distinguish themselves in being able to active adventitial cells such as fibroblasts; local secretion of chemokines at the site of immunization and consequent chemotaxis of immune-competent cells to areas where the local concentrations of immunogens are expected to be initially high likely contribute to the potent adjuvant properties in these compounds.
Funding Statement
Funding for this project was provided by NIAID Contract HHSN272200900033C.
Disclosure of potential conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The authors are inventors on patents and patent applications (University of Kansas and University of Minnesota).
Acknowledgments
We thank Dr. Mark Sanders and the University of Minnesota Imaging Centers for assistance with the imaging studies. We acknowledge the University Genomics Center, and Dr. Juan Abrahante Lloréns (University of Minnesota Informatics Institute) for RNASeq and data analysis, respectively. We are grateful to Lauren Fox for her assistance with pilot experiments.
Data sharing
RNASeq data is available at the Sequence Read Archive (BioProject ID: PRJNA390780; http://www.ncbi.nlm.nih.gov/bioproject/390780).
References
- 1.Andre FE, Booy R, Bock HL, Clemens J, Datta SK, John TJ, Lee BW, Lolekha S, Peltola H, Ruff TA, et al.. Vaccination greatly reduces disease, disability, death and inequity worldwide. Bull WHO. 2008;86:140–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Plotkin SA. Vaccines: The fourth century. ClinVaccine Immunol. 2009;16:1709–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tan T, Dalby T, Forsyth K, Halperin SA, Heininger U, Hozbor D, Plotkin S, Ulloa-Gutierrez R, Wirsing von König CH. Pertussis across the globe: Recent epidemiologic trends from 2000 to 2013. Pediatr Infect Dis J. 2015;34:e222–32. doi: 10.1097/INF.0000000000000795. [DOI] [PubMed] [Google Scholar]
- 4.Plotkin SA. The importance of persistence. Clin Infect Dis. 2016;63:S117–s8. doi: 10.1093/cid/ciw525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plotkin SA. The pertussis problem. Clin Infect Dis. 2014;58:830–3. doi: 10.1093/cid/cit934. [DOI] [PubMed] [Google Scholar]
- 6.Burdin N, Handy LK, Plotkin SA. What is wrong with pertussis vaccine immunity? The problem of waning effectiveness of pertussis vaccines. Cold Spring Harb Perspect Biol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warfel JM, Zimmerman LI, Merkel TJ. Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proc Natl Acad Sci. 2014;111:787–92. doi: 10.1101/cshperspect.a029454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warfel JM, Merkel TJ. The baboon model of pertussis: Effective use and lessons for pertussis vaccines. Expert Rev Vaccines. 2014;13:1241–52. doi: 10.1586/14760584.2014.946016. [DOI] [PubMed] [Google Scholar]
- 9.Allen AC, Mills KH. Improved pertussis vaccines based on adjuvants that induce cell-mediated immunity. Expert Rev Vaccines. 2014;13:1253–64. doi: 10.1586/14760584.2014.936391. [DOI] [PubMed] [Google Scholar]
- 10.Aimanianda V, Haensler J, Lacroix-Desmazes S, Kaveri SV, Bayry J. Novel cellular and molecular mechanisms of induction of immune responses by aluminum adjuvants. Trends Pharmacol Sci. 2009;30:287–95. doi: 10.1016/j.tips.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 11.Kool M, Soullie T, van Nimwegen M, Willart MA, Muskens F, Jung S, Hoogsteden HC, Hammad H, Lambrecht BN. Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells. J Exp Med. 2008;205:869–82. doi: 10.1084/jem.20071087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKee AS, Burchill MA, Munks MW, Jin L, Kappler JW, Friedman RS, Jacobelli J, Marrack P. Host DNA released in response to aluminum adjuvant enhances MHC class II-mediated antigen presentation and prolongs CD4 T-cell interactions with dendritic cells. Proc Natl Acad Sci. 2013;110:E1122–31. doi: 10.1073/pnas.1300392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takeda K, Akira S. Toll-like receptors. Curr Protoc Immunol. 2015;109(14.2):1–0. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann J, Akira S. Innate immunity. Curr OpinImmunol. 2013;25:1–3. [DOI] [PubMed] [Google Scholar]
- 15.Kumagai Y, Akira S. Identification and functions of pattern-recognition receptors. J Allergy Clin Immunol. 2010;125:985–92. doi: 10.1016/j.jaci.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 16.Salunke DB, Shukla NM, Yoo E, Crall BM, Balakrishna R, Malladi SS, David SA. Structure-activity relationships in human Toll-like receptor 2-specific monoacyl lipopeptides. J Med Chem. 2012;55:3353–63. doi: 10.1021/jm3000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salunke DB, Connelly SW, Shukla NM, Hermanson AR, Fox LM, David SA. Design and development of stable, water-soluble, human Toll-like receptor 2 specific monoacyl lipopeptides as candidate vaccine adjuvants. J Med Chem. 2013;56:5885–900. doi: 10.1021/jm400620g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu W, Li R, Malladi SS, Warshakoon HJ, Kimbrell MR, Amolins MW, Ukani R, Datta A, David SA. Structure-activity relationships in toll-like receptor-2 agonistic diacylthioglycerol lipopeptides. J Med Chem. 2010;53:3198–213. doi: 10.1021/jm901839g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salyer AC, Caruso G, Khetani KK, Fox LM, Malladi SS, David SA. Identification of adjuvantic activity of amphotericin B in a novel, multiplexed, Poly-TLR/NLR High-Throughput Screen. PLoS One. 2016;11:e0149848. doi: 10.1371/journal.pone.0149848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shukla NM, Kimbrell MR, Malladi SS, David SA. Regioisomerism-dependent TLR7 agonism and antagonism in an imidazoquinoline. Bioorg Med Chem Lett. 2009;19:2211–4. doi: 10.1016/j.bmcl.2009.02.100. [DOI] [PubMed] [Google Scholar]
- 21.Shukla NM, Malladi SS, Mutz CA, Balakrishna R, David SA. Structure-activity relationships in human toll-like receptor 7-active imidazoquinoline analogues. J Med Chem. 2010;53:4450–65. doi: 10.1021/jm100358c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shukla NM, Mutz CA, Ukani R, Warshakoon HJ, Moore DS, David SA. Syntheses of fluorescent imidazoquinoline conjugates as probes of Toll-like receptor 7. Bioorg Med Chem Lett. 2010;20:6384–6. doi: 10.1016/j.bmcl.2010.09.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shukla NM, Lewis TC, Day TP, Mutz CA, Ukani R, Hamilton CD, Balakrishna R, David SA. Toward self-adjuvanting subunit vaccines: Model peptide and protein antigens incorporating covalently bound toll-like receptor-7 agonistic imidazoquinolines. Bioorg Med Chem Lett. 2011;21:3232–6. doi: 10.1016/j.bmcl.2011.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shukla NM, Malladi SS, Day V, David SA. Preliminary evaluation of a 3H imidazoquinoline library as dual TLR7/TLR8 antagonists. Bioorg Med Chem. 2011;19:3801–11. doi: 10.1016/j.bmc.2011.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shukla NM, Mutz CA, Malladi SS, Warshakoon HJ, Balakrishna R, David SA. Toll-Like Receptor (TLR)-7 and −8 Modulatory Activities of Dimeric Imidazoquinolines. J Med Chem. 2012;55:1106–16. doi: 10.1021/jm2010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukla NM, Salunke DB, Balakrishna R, Mutz CA, Malladi SS, David SA. Potent adjuvanticity of a pure TLR7-agonistic imidazoquinoline dendrimer. PLoS One. 2012;7:e43612. doi: 10.1371/journal.pone.0043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoo E, Crall BM, Balakrishna R, Malladi SS, Fox LM, Hermanson AR, David SA. Structure-activity relationships in Toll-like receptor 7 agonistic 1H-imidazo[4,5-c]pyridines. Org Biomol Chem. 2013;11:6526–45. doi: 10.1039/c3ob40816g. [DOI] [PubMed] [Google Scholar]
- 28.Yoo E, Salunke DB, Sil D, Guo X, Salyer AC, Hermanson AR, Kumar M, Malladi SS, Balakrishna R, Thompson WH, et al.. Determinants of activity at human Toll-like receptors 7 and 8: quantitative structure-activity relationship (QSAR) of diverse heterocyclic scaffolds. J Med Chem. 2014;57:7955–70. doi: 10.1021/jm500744f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nuhn L, Vanparijs N, De Beuckelaer A, Lybaert L, Verstraete G, Deswarte K, Lienenklaus S, Shukla NM, Salyer AC, Lambrecht BN, et al.. PH-degradable imidazoquinoline-ligated nanogels for lymph node-focused immune activation. Proc Natl Acad Sci. 2016;113:8098–103. doi: 10.1073/pnas.1600816113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salunke DB, Yoo E, Shukla NM, Balakrishna R, Malladi SS, Serafin KJ, Day VW, Wang X, David SA. Structure-activity relationships in human Toll-like receptor 8-active 2,3-diamino-furo[2,3-c]pyridines. J Med Chem. 2012;55:8137–51. doi: 10.1021/jm301066h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kokatla HP, Yoo E, Salunke DB, Sil D, Ng CF, Balakrishna R, et al.. Toll-like receptor-8 agonistic activities in C2, C4, and C8 modified thiazolo[4,5-c]quinolines. Org Biomol Chem. 2013;11:1179–98. doi: 10.1039/c2ob26705e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kokatla HP, Sil D, Malladi SS, Balakrishna R, Hermanson AR, Fox LM, Wang X, Dixit A, David SA. Exquisite selectivity for human toll-like receptor 8 in substituted furo[2,3-c]quinolines. J Med Chem. 2013;56:6871–85. doi: 10.1021/jm400694d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kokatla HP, Sil D, Tanji H, Ohto U, Malladi SS, Fox LM, Shimizu T, David SA. Structure-based design of novel human Toll-like receptor 8 agonists. Chem Med Chem. 2014;9:719–23. doi: 10.1002/cmdc.201300573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beesu M, Malladi SS, Fox LM, Jones CD, Dixit A, David SA. Human Toll-like receptor 8-selective agonistic activities in 1-alkyl-1H-benzimidazol-2-amines. J Med Chem. 2014;57:7325–41. doi: 10.1021/jm500701q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beesu M, Kokatla HP, David SA. Syntheses of human TLR8-Specific small-molecule agonists. Methods Mol Biol. 2017;1494:29–44. doi: 10.1007/978-1-4939-6445-1_3. [DOI] [PubMed] [Google Scholar]
- 36.Beesu M, Salyer AC, Trautman KL, Hill JK, David SA. Human Toll-like Receptor (TLR) 8-Specific agonistic activity in substituted pyrimidine-2,4-diamines. J Med Chem. 2016;59:8082–93. doi: 10.1021/acs.jmedchem.6b00872. [DOI] [PubMed] [Google Scholar]
- 37.Beesu M, Caruso G, Salyer AC, Khetani KK, Sil D, Weerasinghe M, Tanji H, Ohto U, Shimizu T, David SA. Structure-based design of human TLR8-Specific agonists with augmented potency and adjuvanticity. J Med Chem. 2015;58:7833–49. doi: 10.1021/acs.jmedchem.5b01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beesu M, Caruso G, Salyer AC, Shukla NM, Khetani KK, Smith LJ, Fox LM, Tanji H, Ohto U, Shimizu T, et al.. Identification of a human Toll-Like Receptor (TLR) 8-Specific agonist and a functional pan-TLR Inhibitor in 2-aminoimidazoles. J Med Chem. 2016;59:3311–30. doi: 10.1021/acs.jmedchem.6b00023. [DOI] [PubMed] [Google Scholar]
- 39.Agnihotri G, Ukani R, Malladi SS, Warshakoon HJ, Balakrishna R, Wang X, et al.. Structure-activity relationships in nucleotide oligomerization domain 1 (Nod1) agonistic gamma-glutamyldiaminopimelic acid derivatives. J Med Chem. 2011;54:1490–510. doi: 10.1021/jm101535e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ukani R, Lewis TC, Day TP, Wu W, Malladi SS, Warshakoon HJ, David SA. Potent adjuvantic activity of a CCR1-agonistic bis-quinoline. Bioorg Med Chem Lett. 2012;22:293–5. doi: 10.1016/j.bmcl.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, et al.. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olafsdottir T, Lindqvist M, Harandi AM. Molecular signatures of vaccine adjuvants. Vaccine. 2015;33:5302–7. doi: 10.1016/j.vaccine.2015.04.099. [DOI] [PubMed] [Google Scholar]
- 43.Mosca F, Tritto E, Muzzi A, Monaci E, Bagnoli F, Iavarone C, O'Hagan D, Rappuoli R, De Gregorio E. Molecular and cellular signatures of human vaccine adjuvants. Proc Natl Acad Sci. 2008;105:10501–6. doi: 10.1073/pnas.0804699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Francica JR, Zak DE, Linde C, Siena E, Johnson C, Juraska M, Yates NL, Gunn B, De Gregorio E, Flynn BJ, et al.. Innate transcriptional effects by adjuvants on the magnitude, quality, and durability of HIV envelope responses in NHPs. Blood Advances. 2017;1:2329–42. doi: 10.1182/bloodadvances.2017011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hood JD, Warshakoon HJ, Kimbrell MR, Shukla NM, Malladi S, Wang X, David SA. Immunoprofiling toll-like receptor ligands: Comparison of immunostimulatory and proinflammatory profiles in ex vivo human blood models. Hum Vaccin. 2010;6:1–14. doi: 10.4161/hv.6.4.10866. [DOI] [PubMed] [Google Scholar]
- 46.Salunke DB, Connelly SW, Shukla NM, Hermanson AR, Fox LM, David SA. Design and development of stable, water-soluble, human toll-like receptor 2 specific monoacyl lipopeptides as candidate vaccine adjuvants. J Med Chem. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoo E, Crall BM, Balakrishna R, Malladi SS, Fox LM, Hermanson AR, David SA. Structure-activity relationships in Toll-like receptor 7 agonistic 1H-imidazo[4,5-c]pyridines. Org Biomol Chem. 2013;11:6526–45. doi: 10.1039/c3ob40816g. [DOI] [PubMed] [Google Scholar]
- 48.Kokatla HP, Sil D, Malladi SS, Balakrishna R, Hermanson AR, Fox LM, Wang X, Dixit A, David SA. Exquisite selectivity for human Toll-Like Receptor 8 in substituted furo[2,3-c]quinolines. J Med Chem. 2013;56:6871–85. doi: 10.1021/jm400694d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall JD, Fearon K, Abbate C, Subramanian S, Yee P, Gregorio J, Coffman RL, Van Nest G. Identification of a novel CpG DNA class and motif that optimally stimulate B cell and plasmacytoid dendritic cell functions. J Leukoc Biol. 2003;73:781–92. doi: 10.1189/jlb.1202630. [DOI] [PubMed] [Google Scholar]
- 50.Tran Vdu T, Souiai O, Romero-Barrios N, Crespi M, Gautheret D. Detection of generic differential RNA processing events from RNA-seq data. RNA Biol. 2016;13:59–67. doi: 10.1080/15476286.2015.1118604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X, Brock GN, Rouchka EC, Cooper NGF, Wu D, O'Toole TE, Gill RS, Eteleeb AM, O'Brien L, Rai SN. A comparison of per sample global scaling and per gene normalization methods for differential expression analysis of RNA-seq data. PLoS One. 2017;12:e0176185. doi: 10.1371/journal.pone.0176185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stack EC, Wang C, Roman KA, Hoyt CC. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods (San Diego, Calif). 2014;70:46–58. doi: 10.1016/j.ymeth.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 53.Chao J, DeBiasio R, Zhu Z, Giuliano KA, Schmidt BF. Immunofluorescence signal amplification by the enzyme-catalyzed deposition of a fluorescent reporter substrate (CARD). Cytometry. 1996;23:48–53. doi: 10.1002/(SICI)1097-0320(19960101)23:1%3c48::AID-CYTO7%3e3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 54.He P, Zou Y, Hu Z. Advances in aluminum hydroxide-based adjuvant research and its mechanism. Hum Vaccin Immunother. 2015;11:477–88. doi: 10.1080/21645515.2014.1004026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta RK. Aluminum compounds as vaccine adjuvants. AdvDrug DelivRev. 1998;32:155–72. [DOI] [PubMed] [Google Scholar]
- 56.Khurana S, Verma N, Yewdell JW, Hilbert AK, Castellino F, Lattanzi M, Del Giudice G, Rappuoli R, Golding H. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. 2011;3:85ra48. doi: 10.1126/scitranslmed.3002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.El Sahly H. MF59 as a vaccine adjuvant: a review of safety and immunogenicity. Expert Rev Vaccines. 2010;9:1135–41. doi: 10.1586/erv.10.111. [DOI] [PubMed] [Google Scholar]
- 58.Seubert A, Monaci E, Pizza M, O'Hagan DT, Wack A. The adjuvants aluminum hydroxide and MF59 induce monocyte and granulocyte chemoattractants and enhance monocyte differentiation toward dendritic cells. J Immunol. 2008;180:5402–12. doi: 10.4049/jimmunol.180.8.5402. [DOI] [PubMed] [Google Scholar]
- 59.Von Oettingen WF, Todd TW, Sollmann T. The spreading and absorption of the different types of Bismuth preparations, introduced by intramuscular and subcutaneous injection. J Pharmacol Exp Ther. 1927;32:67. [Google Scholar]
- 60.Ballard BE. Biopharmaceutical considerations in subcutaneous and intramuscular drug administration. J Pharm Sci. 1968;57:357–78. doi: 10.1002/jps.2600570301. [DOI] [PubMed] [Google Scholar]
- 61.Smith RS, Smith TJ, Blieden TM, Phipps RP. Fibroblasts as sentinel cells. Synthesis of chemokines and regulation of inflammation. Am J Pathol. 1997;151:317–22. [PMC free article] [PubMed] [Google Scholar]
- 62.Silzle T, Randolph GJ, Kreutz M, Kunz-Schughart LA. The fibroblast: sentinel cell and local immune modulator in tumor tissue. Int J Cancer. 2004;108:173–80. doi: 10.1002/ijc.11542. [DOI] [PubMed] [Google Scholar]
- 63.Witowski J, Thiel A, Dechend R, Dunkel K, Fouquet N, Bender TO, Langrehr JM, Gahl GM, Frei U, Jörres A. Synthesis of C-X-C and C-C chemokines by human peritoneal fibroblasts: induction by macrophage-derived cytokines. Am J Pathol. 2001;158:1441–50. doi: 10.1016/S0002-9440(10)64095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Servais C, Erez N. From sentinel cells to inflammatory culprits: Cancer-associated fibroblasts in tumour-related inflammation. J Pathol. 2013;229:198–207. doi: 10.1002/path.4103. [DOI] [PubMed] [Google Scholar]
- 65.Ritchlin C. Fibroblast biology. Effector signals released by the synovial fibroblast in arthritis. Arthritis Res. 2000;2:356–60. doi: 10.1186/ar112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hosokawa Y, Hosokawa I, Ozaki K, Nakae H, Murakami K, Miyake Y, Matsuo T. CXCL12 and CXCR4 expression by human gingival fibroblasts in periodontal disease. Clin Exp Immunol. 2005;141:467–74. doi: 10.1111/j.1365-2249.2005.02852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diaz-Araya G, Vivar R, Humeres C, Boza P, Bolivar S, Munoz C. Cardiac fibroblasts as sentinel cells in cardiac tissue: Receptors, signaling pathways and cellular functions. Pharmacol Res. 2015;101:30–40. doi: 10.1016/j.phrs.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 68.Almine JF, Wise SG, Hiob M, Singh NK, Tiwari KK, Vali S, Abbasi T, Weiss AS. Elastin sequences trigger transient proinflammatory responses by human dermal fibroblasts. FASEB J. 2013;27:3455–65. doi: 10.1096/fj.13-231787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sunasara K, Cundy J, Srinivasan S, Evans B, Sun W, Cook S, Bortell E, Farley J, Griffin D, Bailey Piatchek M1, et al.. Bivalent rLP2086 (Trumenba(R)): Development of a well-characterized vaccine through commercialization. Vaccine. 2017. [DOI] [PubMed] [Google Scholar]
- 70.Luo Y, Friese OV, Runnels HA, Khandke L, Zlotnick G, Aulabaugh A, Gore T, Vidunas E, Raso SW, Novikova E, et al.. The dual role of lipids of the lipoproteins in trumenba, a self-adjuvanting vaccine against meningococcal meningitis B disease. AAPS J. 2016;18:1562–75. doi: 10.1208/s12248-016-9979-x. [DOI] [PubMed] [Google Scholar]