Abstract
With the increasing numbers of immunocompromised hosts, Aspergillus fumigatus emerges as a lethal opportunistic fungal pathogen. Understanding innate and acquired immunity responses of the host is important for a better therapeutic strategy to deal with aspergillosis patients. To determine the transcriptome in the kidneys in aspergillosis, we employed RNA-Seq to obtain single 76 base reads of whole genome transcripts of murine kidneys on a temporal basis (days 0;uninfected, 1, 2, 3, and 8) during invasive aspergillosis. A total of 6,284 transcripts were downregulated and 5,602 were upregulated compared to baseline expression. Gene Ontology enrichment analysis identified genes involved in innate and adaptive immune response, as well as iron binding and homeostasis, among others. Our results showed activation of pathogen recognition receptors e.g., β-defensins, C-type lectins (e.g., dectin-1), Toll-like receptors (TLR-2, TLR-3, TLR-8, TLR-9 and TLR-13), as well as Ptx-3, and C-reactive protein among the soluble receptors. Upregulated transcripts encoding various differentiating cytokines and effector pro-inflammatory cytokines, as well as those encoding for chemokines and chemokine receptors revealed Th-1 and Th-17 type immune responses. These studies form a basic dataset for experimental prioritization, including other target organs, to determine the global response of the host against Aspergillus infection.
Keywords: Host-Pathogen, Invasive aspergillosis, Aspergillus fumigatus, Mus musculus, RNA-Seq, Cytokines, differential expression
1. Introduction
Humans inhale conidia from Aspergillus species, daily [1]. Aspergillus fumigatus is a ubiquitous filamentous fungus, which can cause a variety of problems ranging from allergic to invasive diseases depending on the individual. Invasive aspergillosis is life-threatening in immunocompromised patients, including those with solid organ transplants and hematologic malignancies [2–3].
Host responses against A. fumigatus infection are critical for effective control of the disease [4–5]. Studies in mouse models showed that both innate and acquired immunity of the host determine the protection against aspergillosis [5–7]. With the increasing numbers of at risk patients, A. fumigatus has emerged as the most lethal mycosis worldwide [2, 8]. Established infection in these patient groups has proven very difficult to eradicate due to the lack of highly effective and curative antifungal drug treatment [2] and gaps in knowledge regarding the host immune response in normal and immunocompromised individuals.
The exploration of the host transcriptome during the course of A. fumigatus infection may indicate which genes play an important role in the host response and help the understanding of the development and the progression of the disease [9–10]. Recently, RNA-Seq has been used to perform transcript profiling of in vitro cultured A. fumigatus [11–13], A. orzyae [14], and A. flavus [15]. However, RNA-Seq transcriptome data from in vivo or ex vivo studies are minimal [16]. Oosthuizen, et al. used genome-wide microarrays for fungus and human to examine the transcriptome response of bronchoepithelial cells to infection with A. fumigatus conidia, noting up-regulation of IL-6 and innate pathways for the human cells, and up-regulation of iron acquisition, and vacuolar acidification pathways, among others for the fungal cells [17]. Similarly, Morton, et al used microarrays to examine expression profiles of dendritic cells from an in vitro model of the alveolar surface and their interaction with A. fumigatus, noting changes in various chemokines associated with neutrophil chemotaxis [18]. In the current studies we used RNA-Seq technology to determine the host transcriptome in the kidneys of mice infected intravenously with A. fumigatus to better understand the events occurring in the most affected organ during intravenous infection and which may also be a key organ during systemic aspergillosis in humans [19]. Our study demonstrated coordinated expression of genes encoding for interleukins and interferon, interleukin receptors and receptor-associated factors, interferon regulators, interferon receptor and interferon-induced proteins, chemokines, chemokines receptors, iron homeostasis, and iron binding proteins.
2. Material and methods
2.1. Infection model
Six-week-old female CD-1 mice (Charles River Laboratories, Hollister, CA, USA) weighing approximately 25 grams each were arranged in groups of 5 and provided food and water ad libitum.
A. fumigatus strain Af293 [20] conidia for infection were prepared, and infected intravenously with 1.1 × 107 viable conidia in 250 μl of saline/Tween-80 (0.05%) as described previously [21–23]. Five pre-assigned mice were euthanized on days 0, 1, 3, 5 and 8 to collect the samples. One mouse died on day 8, and thus we considered day 8 as a last time point to collect the samples. Uninfected mice were used as normal controls to provide the baseline murine transcriptome. At each time point, kidneys were removed aseptically from each mouse in the group and pooled before being snap-frozen in liquid N2. Samples were stored at 80°C until used for total RNA extraction.
2.2. RNA extraction
Kidneys from 5 mice at each point were thawed in Trizol (Invitrogen, Carlsbad, CA, USA) and broken with 0.5 mm zirconia/silica beads (Biospec) using a Bead Beater apparatus (Biospec Products, Inc., Bartlesville, OK, USA) at high-speed setting (3 pulses of 12–15 sec). Total RNA was extracted using TRIzol reagent per the manufacturer’s instructions. Contaminating gDNA was removed using RNeasy mini spin column (RNeasy mini kit, Qiagen Science, MD, USA) and RNase-free DNase set (50) (Qiagen GmbH, Germany), according to the manufacturer’s instructions. The quantity and quality of the extracted RNA were assessed by A260 nm/A280 nm ratio (NanoDrop 1000 spectrophotometer, Thermo Scientific), as well as by electrophoresis through a 1.5% agarose gel stained with ethidium bromide for the presence of intact 18S and 28S ribosomal RNA bands, visualized by UV transillumination at 302 nm. RNA purity was determined using A260:A280 ratios and the extinction coefficient for RNA to quantify the amount of RNA present [24].
2.3. cDNA library preparation and sequencing
Poly (A) containing mRNA was purified from 8 μg of total RNA of each sample using Sera-mag Magnetic Oligo (dT) Beads (Illumina RS-100-0801); mRNA molecules were fragmented using divalent cations under elevated temperature followed by first and second strand synthesis. End repair was performed and deoxyadenosine triphosphate (dATP) added to the 3′ end of the DNA fragments. Next, adapters were ligated to cDNA fragments, cDNA template purified and finally the cDNA libraries were amplified via PCR. All steps were performed according to manufacturer’s instructions. Before sequencing, the quantity and quality of each cDNA library was assessed using a 2100 Bioanalyzer (Agilent Inc.) [14]. Libraries were sequenced as single 76 base reads using the sequencing by synthesis method (Solexa sequencing, Illumina Inc.) at the Stanford Functional Genomic Facility, Stanford University.
2.4 Differential expression analysis
Initially, our intention was to obtain sequence reads for A. fumigatus and for the mouse transcripts, to generate two profiles to better understand the host-pathogen interactions. Alignment of sequence reads to the annotated genome of A. fumigatus (Af293) resulted in few transcripts that could map to the genome with more than a minimum number of reads (i.e., <10). Thus, in vivo expressed transcripts of A. fumigatus could be not analyzed. Transcript sequences of Mus musculus were downloaded from Ensembl (Build 38, GCA_000001635.2). Functional annotation and Gene Ontology (GO) terms were downloaded from EnsemblBiomart (GRCm38.p1). Original coding sequences (CDS) were complemented by 100 nt of flanking genomic sequence on both ends to enable alignment of short sequencing reads with untranslated regions (UTR). Short reads were aligned against the modified CDS using Bowtie [25]; only the best 20 valid alignments (−k 20) were processed. Quantification of transcript abundance and isoform-adjusted expression was done via RSEM [26]. Normalization across samples was performed using the trimmed mean of M-values method (TMM) implemented by Edge-R [27]. Differential expression was calculated between all pairs of time points using the classical method of Edge-R [28]. Only genes with false discovery rate (FDR) < 0.01 and 16-fold change between a pair of time points were reported. All the applications were executed as part of the differential expression workflow implemented by the Trinity suite [29].
2.5 Clusters of genes with similar expression profile across samples
Clusters with similar expression profile across time points were defined based on the mean centered logarithm on base two of the FPKM values (Fragments per Kilobase of transcript per Million of mapped reads). The mean value was based on all genes with significant differential expression in at least one pair of time points. Similarity of expression profile between genes was computed using Euclidean distance and clusters were defined according to K-mean algorithm.
GO enrichment was defined by Fisher-exact correlation test and corrected for multiple comparisons via FDR, both implemented by scripts developed at the Broad Institute. A GO node was considered over-represented in the case of 3-fold difference between test and background sets and a FDR value lower or equal to 0.05. Non-informational GO terms are not shown in the main manuscript, but listed in supplemental material (Supplementary file 1: Tables 7 and 8). GO terms were summarized by Revigo using simRel method and 0.7 maximum similarity allowed between remaining GO terms (small threshold) [30].
3. Results
In the murine model of systemic aspergillosis, immunocompetent mice are infected intravenously with conidia to establish infection. However, quantification of the fungal burden of A. fumigatus is difficult [31], with the temporal burden of A. fumigatus in the kidneys determined by culture of homogenates shows the highest burden at day 0–1, at which time the conidia are morphologically still conidia or have germinated with short hyphae and colony counts determined at later time points (i.e., day 5 and 8) lower because the organism has developed into hyphal formsin the tissues. This can be misinterpreted as a reduced burden, whereas the use of qPCR to quantify fungal burden demonstrates a progressive increase in the number of organisms present [32]. In addition to that, among the few reads derived from A. fumigatus, we observed a progressive increase in abundance of rRNA transcripts (not shown). Thus, we are confident that the model is that of progressive infection and that temporal alterations of gene expression in the kidney are a result of the rise in infectious burden.
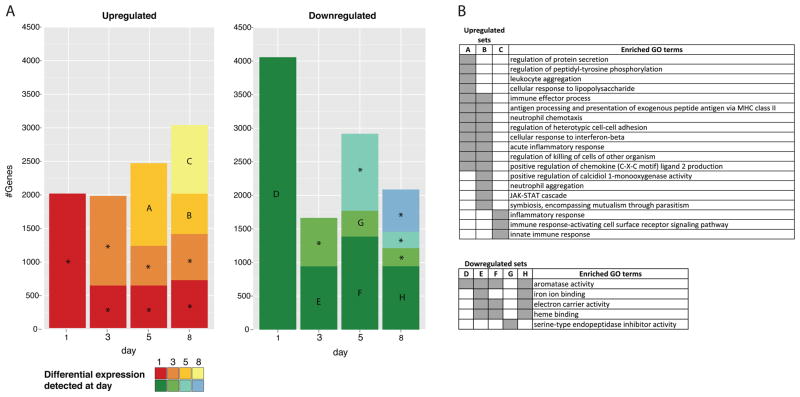
Approximately 179 million sequencing reads were generated (Supplementary Table S1), of which 136 million were successfully mapped to mouse transcripts. With respect to transcript coverage, of 83,070 mouse transcripts used as reference sequences, 57,007 (69%) had more than 10 reads mapped and approximately 29,000 (35%) had a normalized FPKM of one or greater. When considering each sample/day separately, at least 26% of transcripts reported a FPKM greater than or equal to one (Supplementary Table S2). Transcripts that were significantly differentially expressed in at least one pairwise comparison between time points accounted for 17% of mouse transcripts (14,397) (Supplementary Table S3). When comparing each time point to day 0 (non-infected animal, baseline), 6,284 transcripts (8%) were downregulated and 5,602 (7%) were upregulated. Despite approximately 2,000 transcripts found to be up-regulated on days 1 and 3 in comparison to day 0 (Fig. 1 and Supplementary Table S4 for the complete list of up-regulated genes in comparison to day 0), there was no consistency in terms of the biological process associated with those genes, as evaluated by GO term enrichment. A concerted positive response only appeared on day 5, consisting of genes associated with leukocyte aggregation, acute inflammatory response, positive regulation of chemokines and other GO terms associated with immune response (set A on Fig. 1; Supplementary Table S7 for the list of enriched GO terms). Down-regulation was maximal on day 1 with twice the number of transcripts (4,295) (set D on Fig. 1 and Supplementary Table S5) than those upregulated at the same day. Genes with lower expression when compared to non-infected mice were consistently enriched for processes and functions associated with iron and heme binding, electron carrier activity and aromatase activity (Fig. 1B and Supplementary Table S7). A detailed inspection of the downregulated genes responsible for the enrichment of these processes and functions indicated that the majority were members of the cytochrome P450 family of proteins (Supplementary Table S10).
Figure 1. Genes differentially expressed during the course of infection in comparison to non-infected mouse (day 0).
(A) Colored bars indicate the number of transcripts detected on each day (X-axis) and the day they were first detected according to colors in the legend. Only transcripts with significant difference in expression (FDR lower than 0.01 and 16-fold change) when compared to day 0 were accounted. Bars marked with letters represent sets of transcripts with significant enrichment (Bonferroni adjusted p-value < 0.05) of at least one GO term. Gene Ontology enrichment analysis is described in the Material and Methods section. Bars marked with an asterisk indicate sets with no significant enrichment. (B) Summarized list of GO terms enriched in the sets defined on panel A. GO summarization is described in the material and methods section. Gray blocks indicate the set (table header) in which the GO term has a proportionally higher frequency than expected by chance.
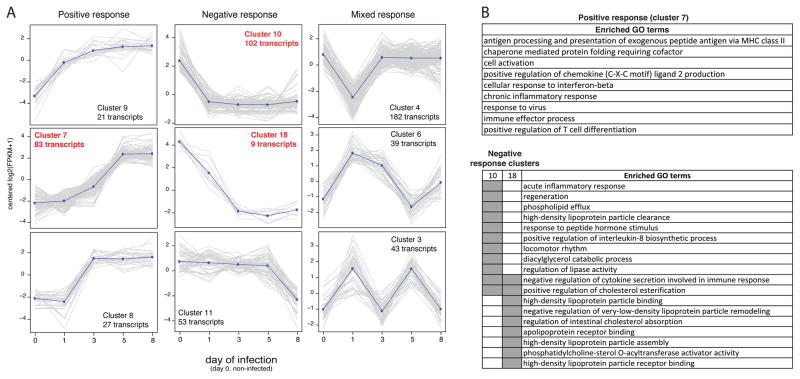
Thirty-five clusters with similar expression during the course of infection were defined (Supplementary Table S6). A set of three clusters representing positive, negative and mixed response are shown on Fig. 2A, with three of those having enrichment of GO terms. The positive response (cluster 7) was characterized by genes associated with adaptive and innate immune response; and the negative response (clusters 10 and 18) was enriched with transcripts associated with the assembly and binding of lipoproteins, including apolipoproteins H and A2, previously reported as negative regulator of neutrophils and angiogenesis [33]. The acute inflammatory response and negative regulation of cytokine production were also among the biological processes enriched in genes with lower expression after day 1, cluster 10 and 18 (Fig. 2B and Supplementary Table S8).
Figure 2. Cluster of transcripts presenting a similar profile of expression during the course of the infection.
(A) Graphs depicting the normalized FPKM (Y-axis) of members of each cluster during the 8 days of sample collection (X-axis). Dark blue lines describe the approximate general pattern presented by members of each cluster. Graphs with caption in red indicate clusters with enriched GO terms. (B) Summarized list of GO terms enriched on cluster 7, 10 and 18. Gray blocks indicate the set (table header) in which the GO term has a proportionally higher frequency than expected. GO enrichment analysis and GO terms summarization are described in the Material and Methods section.
A comparison focusing on the dynamics of cytokines and iNOS response between our work and a study on mice infected with Cryptococcus neoformans [34] indicated a similar pattern of robust response on day 5 and up-regulation of TNF-alpha on day 1, in spite of the differences in infected tissue (brain), pathogen and technique (RT-PCR) between the two studies (Supplementary Table S11). A lower correlation and an earlier detection was reported by an ELISA based evaluation of cytokines response in kidneys of mice infected with A. flavus [35], possibly different due to the nature of those assays.
3.1. Recruitment of immune cells at the site of infection
We found up-regulated transcripts encoding for chemokines and chemokine receptors. Chemokines (Ccl4, Ccl5, Ccl7, and Ccl8) and their receptor, Ccr1, present on neutrophils, were upregulated (Supplementary Fig. S1). On monocytes, chemokine receptors Ccr2 and Ccr5, and their ligand chemokines (e.g., Ccl2, Ccl7, Ccl8, and Ccl4, Ccl5, Ccl8, respectively) were also upregulated (Supplementary Fig. S1). Receptors present on macrophages, Ccr1 and Cxcr3, and their corresponding chemokines (i.e., Ccl3, Ccl4, Ccl5, Ccl7, Ccl8 and Cxcl9, Cxcl10, respectively) were also upregulated (Supplementary Fig. S1). Chemokines Ccl2, Ccl3, Ccl4, Ccl5, Ccl7, Ccl8 and the receptors Ccr1, Ccr2 and Ccr5, which are specific to dendritic cells, were also upregulated in response to Af293 infection in mice (Supplementary Fig. S1).
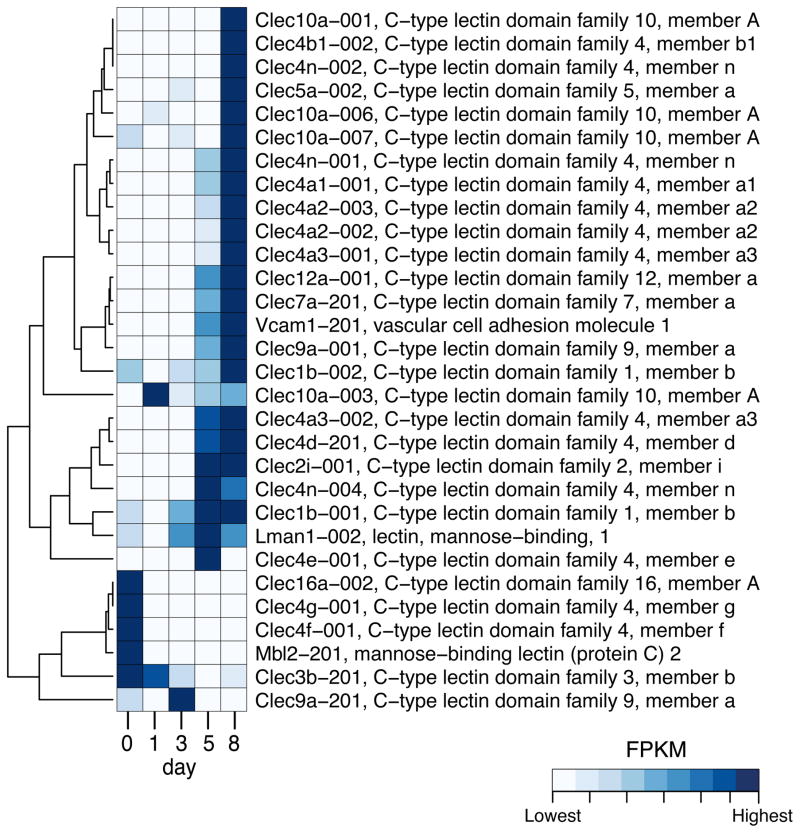
Proteins associated with the innate response that we searched included β-defensins, C-type lectins (e.g., dectin-1), TLRs, complement, and adherence factors (e.g., VCAM or selectins) (Fig. 3, Supplementary Fig. S2 and S3). In general, increased duration of infection resulted in increasing number of transcripts for VCAM- and L-selectin, which would correlate with increased influx of host immune cells into the kidneys (Fig. 3). Furthermore, we noted increased expression of chemotactic complement factors, as well as some increased expression of mannose binding lectin (MBL), which can activate complement through an alternate pathway in response to binding of mannose in the fungal cell wall. Pentraxin-3 (Ptx3) and C-reactive protein also can activate the complement pathway and their expression was modulated postinfection (Supplementary Fig. S2). As might be expected, we noted increasing expression of C-type lectins (Fig. 3), and TLRs (Supplementary Fig. S2), which were highest at the day 8 after infection, also reflecting the influx of immune cells.
Figure 3. Expression profile of differentially expressed C-type lectins and VCAM genes.
Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
3.2. Activation of Th-1 and Th-17 cells
Transcripts encoding chemokines, Ccl2, Cxcl9, Cxcl10, Cxcl11, Cxcl13, and Ccl20 (Supplementary Fig. S1) were up-regulated, as were the trafficking receptors for chemokines Ccr5 and Cxcr3, associated with Th-1 cells and other trafficking receptors for chemokines, Ccr2 and Cxcr4, which are associated with Th-17 cells.
3.3. Th-cell mediated immune response and interplay of cytokines
We found activation of a Th-1 response mediated by pro-inflammatory cytokines. The genes encoding IFN-γ and IL-27 (IL-12 family) were observed to be steadily up-regulated beginning 3 days postinfection (Supplementary Fig. S4). The effector cytokines, IL-18 and IL-24 were found to be up-regulated beginning 5 days postinfection in this model. IL-18 induces Th-1 response in the presence of IFN-γ and IL-12, whereas IL-24, a member of the IL-10 family, induces a Th-2 type response if differentiating cytokines IL-4 or IL-10 are present. On day 8 postinfection, we found upregulated transcripts encoding for IL-16; IL-16 promotes a Th-1 type response. We also observed activation of Th-17 pathway response genes mediated via pro-inflammatory cytokines IL-6 and IL-1α on day 5 postinfection. We found an increased number of transcripts encoding for interleukin IL-1β at day 5 postinfection, followed by a significant increase in transcript copies at day 8 postinfection (Supplementary Fig. 4).
Transcripts encoding IL-33, IL-34, IL-1f6 were upregulated in response to infection in kidney cells of mice. IL-34 transcripts had their highest expression at days 3 and 5 postinfection, but this was followed by a decline in transcript copies at day 8. IL-1f6 transcripts were up-regulated at day 5, and were highest at day 8 postinfection (Supplementary Fig. S4).
3.4. Iron regulation
A consistent enrichment of iron and heme binding, electron carrier activity and aromatase activity was found among downregulated genes (Fig. 1B). This was primarily a result of lower expression of members of the cytochrome P450 family of proteins during infection compared to baseline expression (Supplementary Table S10). However, a more detailed analysis revealed that key components of iron homeostasis had an increased numbers of transcripts in infected mice in comparison to uninfected mice, e.g.: Nos1 (nitric oxide synthase 1), Nos2 (nitric oxide synthase 2), lipocalin 2(Lcn2), p4ha2 (procollagen-proline), Ogfod1 (2-oxoglutarate and iron dependent oxygenase domain containing 1), Ltf (lactotransferrin), Epb4.2 (erythrocyte protein band 4.2), and Heph (hephaestin) (Supplementary Fig. 5). Transcripts that showed a decreased expression compared to uninfected mice were Tdo2 (tryptophan 2.3-dioxygenase), Lepre1 (leprecan 1), Hbq 1a (hemoglobin theta 1A), Ttc7 (tetratricopeptide repeat domain 7), and Hamp (hepcidin, antimicrobial peptide) (Supplementary Fig. 5).
4. Discussion
The model of intravenous infection with Aspergillus was chosen to study the normal immune response to this infection in the absence of various immunosuppressive regimens that could be interposed, each of which would produce different perturbations of the immune response, and thus need each to be studied separately. Such studies can now follow, based on our data. To study the immune response in a setting absent the classical immunosuppressive risk factors also takes on topical relevance, owing to the rising incidence of aspergillosis in the non-immunocompromised [36]. The disease produced in the model also mimics disseminated infection with Aspergillus, where the kidney is a target organ, which is seen in one third of cases of aspergillosis [19]. Finally, this model has been used extensively in studies of anti-Aspergillus therapy [21] and the assets of this model have been discussed elsewhere [7, 37–38]. A limitation of the model used in this work is that the host is not immunocompromised, whereas most aspergillosis occurs in immunocompromised hosts. In the latter, the usual entry route is the lung, though tissue breaches can also lead to dissemination. Our original intention was to evaluate the interacting transcriptome of host and pathogen along the course of infection. Unfortunately, a very small number of sequencing reads was derived from A. fumigatus transcriptome, forcing us to focus our analysis on the host. For those willing to implement similar experimental design, we would recommend the utilization of either mRNA depletion or enrichment technologies, such as hybrid selection [39].
The first line of defense against Aspergillus infections is the macrophages, neutrophils and dendritic cells that are recruited to the site of infection. These cells recognize pathogen associated molecular patterns (e.g. β-glucan, mannans, etc) that are exposed on the surface of germinating conidia or hyphae through pathogen recognition receptors (PRRs) including soluble receptors and cell bound receptors [40, 5, 41]. Among cell bound receptors, our data showed increased transcripts for genes encoding for C-type lectins (e.g., dectin-1), Toll-like receptors (TLR-2, TLR-3, TLR-8, TLR-9 and TLR-13); Ptx-3, and C-reactive protein were among the soluble receptors (Fig. 3 and Supplementary Fig. S2). In addition, we noted the increase of transcripts for β-defensins, which are cationic proteins found in the lysosomal contents of PMNs and contribute to the killing of microorganisms (Supplementary Fig. S2).
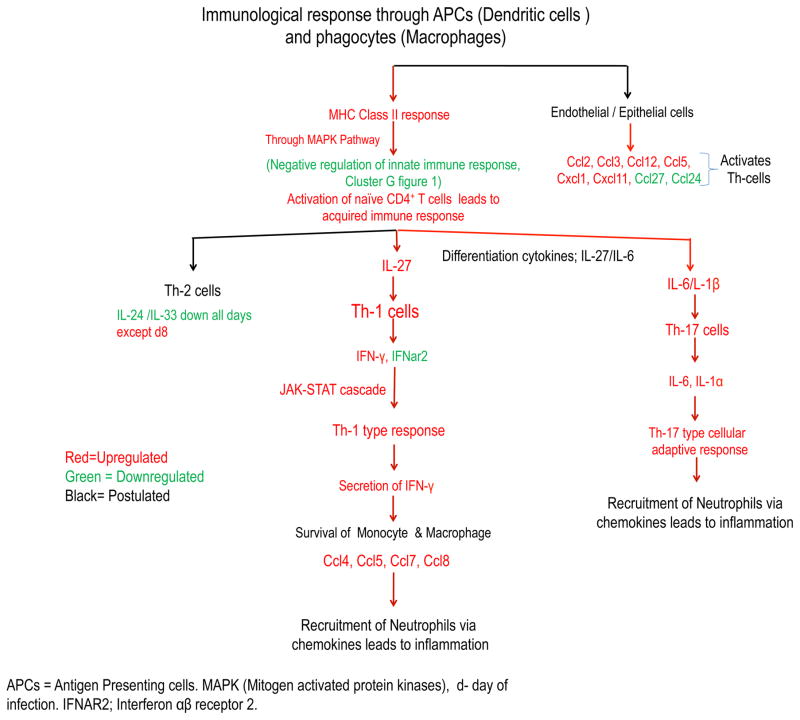
During kidney infection A. fumigatus cells are recognized by antigen presenting cells (APCs) and resident renal macrophages via PRRs. Furthermore, these immune cells induce the production of differentiating cytokines that act on naive CD4+Th-cells [42]. For instance, we observed upregulation of the differentiating cytokine IL-27 (IL-12 family) 3 days postinfection, which induces Th-1 cells, and the cytokine IL-6, 5 days postinfection, which induces Th-17 cells (Supplementary Fig. S4). Subsequently, release of various pro-inflammatory effector cytokines, like IL-1β, IL-18, IFN-γ, IL-1α, IL-34, and IL-6, suggests the possible role of Th1 and Th17 cell responses (Fig. 4) [43]. Similar to our observation at 24 h post infection, Armstrong-James, et al., did not observe a significant increase in the level of IFN-γ in pulmonary invasive aspergillosis [44]. In a similar study of invasive renal aspergillosis induced by A. flavus, Anand et al. observed significant induction of IFN-γ in kidney homogenates after 12 h of infection and levels of IFN-γ were higher along with IL-6, after 12, 72, and 96 hours in infected mice compared to uninfected mice [35]. Furthermore, in A. flavus mediated cerebral aspergillosis mouse model studies, brain homogenates showed an increased level of IFN-γ, IL-12p40 and IL-6 at 24 hours postinfection, with a decline in IL-4 and IL-23 cytokines [45]. Interestingly, we did not find signature transcripts encoding for the cytokines IL-4 and IL-10 suggesting the lack of an early Th2 type of response. However, we noted the upregulated transcripts encoding for Ccl11 and IL-33 at later time point during infection (Supplementary Fig. S1 and Fig. S4); both contribute to Th2 type response [46–47]. It should be noted that the effect of IL-33 can be either Th1 or Th2 depending on the disease and model [48–49]. We observed later during infection (i.e., day 5 postinfection) more transcripts for genes encoding for IL-1β and IL-1α indicated the possible role of Th17 pathway involvement and suppression of Th2 immune responsive cytokines (Supplementary Fig. S4). Similar to our observations, Caffrey, et al., also observed upregulation of IL-1β and IL-1α during the initial days of a pulmonary A. fumigatus murine model [50]. In addition, Cortez, et al., observed up-regulated transcripts of IL-1β during the initial 2 h to 6h of interaction of A. fumigatus conidia with human monocytes [51]. IL-1β also helps in activation of immune system and phagocytosis to decrease the load of Aspergillus at the infection site [52]. On the other hand, increased numbers of transcripts of the gene encoding for IFN-γ by day 5 postinfection (Supplementary Fig. S4) indicates earlier activation of the Th1 immune response, and suggests a possible co-evolution of Th1 and Th17 type immune responses during kidney infection. Interestingly, expression of IL-34 throughout infection suggests survival of monocytes and their differentiation into immunosuppressive macrophages at the site of infection [53]. We also observed increased transcripts of genes encoding for the chemokine Cxcl10 and its receptor Cxcr3 (Supplementary Fig. S1), which activates immune cells responsible for IFN-γ secretion and inhibition of Th17 regulating cytokines [54]. The current data showed a notable profile of proinflammatory cytokines (e.g., IL-27, IFN-γ, IL-6, IL-34, IL-16, IL-1α and IL-1β). Measurement of selected cytokines in the blood samples of aspergillosis patients may be promising as a tool for the monitoring of treatment responses [55–56].
Figure 4. Interplay of cytokines in mouse model of invasive aspergillosis.
The diagram shows known and postulated immune pathways that play a role in the hosts’ response to A. fumigatus infection. These include up-regulated (shown in red) and down-regulated (shown in green) gene transcripts of mice, after intravenous inoculation of Af293. The interaction of conidia with kidney cells leads to the release of cytokines and chemokines at the site of infection, which recruit and activate inflammatory and immune cells (e.g. phagocytes either circulating or resident macrophages), antigen presenting cells (APCs; e.g. dendritic cells) and T cells. Phagocytic cells engulf the circulating conidia and destroy them via phagolysosome activity and also release of cytokines. APCs migrate and present Aspergillus antigens in the lymph nodes, to activate naïve CD4+ T cells via MHC class II molecules. Consequently, activated T cells differentiate into Th-1 cells type via IFN-γ and IL-24 or a Th-17 cells via IL-6 and IL-1α/IL-1β.
Transcripts encoding numerous chemokines were upregulated. The trafficking receptor for chemokines Cxcl9, Cxcl10, and Cxcl11, is Cxcr3, and for chemokine Cxcl13, it is Cxcr5; we found transcripts for both these receptors were upregulated. These receptors are reported to be associated with Th-1 cells [57]. Similarly, we observed up-regulation of transcripts encoding for trafficking receptors Ccr2 and Cxcr4. These receptors are present on Th17 cells and respond to different chemokines (i.e., Ccl2, Cxcl10, Cxcl13, Ccl20) [58]. Consistent with our observation, Cortez, et al., also observed up-regulation of transcripts encoding for the chemokine Cxcl2, Ccl3, Ccl4 and Ccl20 in human monocytes in response to A. fumigatus [51]. Chemokines are released at the site of infection by epithelial cells, macrophages and neutrophils and attract an influx of more neutrophils, monocytes and macrophages, thus causing the inflammation at the site of infection [59].
Members of cytochrome P450 family were downregulated on most days postinfection (Supplementary Table S10), recapitulating reports of transcription modulation of those enzymes by cytokines and interferons during hepatic and renal infections [60–63]. The biological underpinnings of this phenomenon are unclear. It has been proposed that the downregulation of cytochrome P450 genes during infection may be an attempt in reducing the damaging effects of oxidizing species derived from uncoupled catalytic turnover of those enzymes when in the presence of either iron or Nos2 (up-regulated, Supplementary Fig. S5) [64]. Changes in the expression of host cytochrome P450 genes are especially relevant in the context of aspergillosis treatment and disease management. Cytochrome P450 enzymes have key roles in the activation and clearance of many drugs, including those used in the treatment of AIDS patients [65], one of the groups vulnerable to invasive aspergillosis [66].
The consistent downregulation of cytochrome P450 genes postinfection and the association of those genes with iron binding and aromatase activity, according to Gene Ontology, caused an enrichment of those pathways in the downregulated set (Fig. 1B). This may lead to the false interpretation that iron sequestration by the host is downregulated during A. fumigatus infection. Actually, the key components of iron homeostasis and regulation were up-regulated after infection (e.g. lipocalin 2, lactotransferrin) (Supplementary Fig. S5).
In response to the use of siderophores by pathogens to acquire iron from host iron binding proteins, the host produces proteins that sequester ferric ion-siderophore complexes away from pathogen siderophore receptors [67]. A member of this functional group, Lipocalin 2, is secreted by the host in response to infection and inflammation, and binds not only to the metabolites of the host but also iron-siderophore complexes, thus sequestering iron from the pathogen [68]. Lactoferrin or lactotransferrin is a glycoprotein that is secreted by exocrine glands and neutrophils. Lactoferrin has various biological functions, including roles in iron metabolism, cell proliferation and differentiation [69]. The iron binding capacity of lactoferrin is twice that of transferrin (downregulated, Supplementary Fig. S5) [70]. Because of its iron binding properties, it reduces iron availability to any pathogens and also influences the immune system and cells involved in the inflammatory response [71].
Our study indicates downregulation of genes associated with aromatase activity (Fig. 1). Aromatase catalyzes estrogen synthesis from androgens (e.g. testosterone) [72]. Estrogens have both proinflammatory and anti-inflammatory properties [73]. The estrogen 17β-estradiol, as well as IL-6, IL1β, and TNF-α, stimulates aromatase activity, possibly in vascular endothelial cells or tissue macrophages [74–76]. Thus, it is possible that downregulated genes encoding for aromatase enzyme may be involved in downregulation of proinflammatory [74] or anti-inflammatory [77] cytokine production.
When an invading microbe interacts with host cells, tissue damage resulting from inflammation, thrombosis, and necrosis decreases available oxygen concentrations at the site of infection. The production of the non-ribosomal peptide gliotoxin, and possibly other secondary metabolites, by A. fumigatus contributes to the inhibition of angiogenesis in the lung [78–79]. However, during invasive infection, A. fumigatushyphae invade the blood vessels, which leads to hypoxia at the site of infection and activates angiogenesis [78]. Angiogenesis is an important physiological response to tissue hypoxia and proinflammatory cytokines. At the site of infection, endothelial cells produce proinflammatory cytokines and leukocyte adhesion molecule chemokines, which recruit neutrophils and release H2O2 [78]. Among the earliest cues for initiation of the wound repair response is the release of “growth factor”-related cytokines [80]. We observed up-regulation of transcripts encoding for Vegfa (vascular endothelial growth factor) by day 3 of infection and bFGF (a-fibroblast growth factor) by day 1 infection. We also observed up-regulation of transcripts encoding for chemokines Cxcl1, Cxcl2 and Cxcl3, all of which have been reported to be stimulatory factors for angiogenesis [57, 81–82].
Overall, our results show that infection in the kidneys with A. fumigatus triggers a host-response in the mouse that includes differential expression of 14,000 or less genes in the mouse. Our results show that genes such as C-type lectins, Toll-like receptors, C′ proteins, and iron sequestration come under regulation during infection as part of the innate response. Furthermore, adaptive cellular immune response includes cytokine and chemokines and their receptors are regulated during the first week of infection. Surprisingly, we did not find evidence of a clear Th2 cellular response, with the exception of up-regulation of Ccl11 and IL-33 at later time points. We postulate that a robust response of this type may only occur later in infections that are less severe or more chronic; evidence for that would require supplementary work. Our study is an entry to understanding host responses to aspergillosis in tissue. Target organs more frequently involved in immunocompromised patients, such as the lung and brain need to be studied for similarities and differences. These studies form a basic dataset on which to build as more work, including other target organs, is performed in determining the global response of the host to infection with Aspergillus.
Supplementary Material
Supplementary Fig. S1. Expression profile of genes encoding cytokines and cytokines receptors. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Fig. S2. Expression profile of genes encoding Toll-like receptors, selectins and defensins. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Fig 3. Expression profile of genes encoding components of the complement. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Figure 4. Expression profile of genes encoding interleukins and interferons. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Figure 5. Expression profile of genes associated iron binding and homeostasis. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Figure 6. Expression profile of genes associated with positive regulation of angiogenesis. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Table S1: Short reads statistics.
Supplementary Table S2: Distribution of number of transcripts based on FPKM reported on each sample/day.
Supplementary Table S3: Genes significantly differentially expressed in at least one pair of time points.
Supplementary Table S4: List of genes up-regulated in comparison to non-infected mouse (day 0).
Supplementary Table S5: List of genes down-regulated in comparison to non-infected mouse (day 0).
Supplementary Table S6: Clusters of genes with similar expression profile across time points.
Supplementary Table S7: GO terms enriched on transcripts differentially expressed on each time point in comparison to non-infected mouse (day 0).
Supplementary Table S8: GO terms enriched on clusters of transcripts with similar expression profile across time points.
Supplementary Table S9: Up-regulated transcripts encoding for chemokines and their receptors in response to invasive aspergillosis in the current study.
Supplementary Table S10: List of downregulated transcripts associated with iron, heme and aromatase processes and functions according to Gene Ontology (GO).
Supplementary Table S11: Comparison of the dynamics of cytokines and iNOS response between our work and studies on mice infected with Cryptococcus neoformans and Aspergillus flavus.
Acknowledgments
Funding
These studies were funded in part by National Institute of Allergy and Infectious Diseases, National Institutes of Health (URL: nih.gov) grant 1R21AI85566-01 to DAS.
We thank Marife Martinez, Vicky Chen and Ana Claudia Maretti-Mira for their assistance in this work.
Footnotes
Ethics Approval and consent to participate
All animal studies were using a protocol approved by the Institutional Animal Care and Use Committee of the California Institute for Medical Research (CIMR) under protocol numbers 09-01:3 and 12-01:1. Animal Care and Use protocols followed the Animal Welfare Act (7 U.S.C. 2131 et seq.), and the Office of Laboratory Animal Welfare - Public Health Service Policy on Humane Care and Use of Laboratory Animals. CIMR protocols were approved and performed under OLAW assurance number, A3652-01. The NRC Guidelines for the Care and Use of Laboratory Animals, 8th edition, were followed for all care and use of mice. Mice were euthanized using CO2 narcosis per the 2007 version of the American Veterinary Medical Association (AVMA) guidelines, which was current at the time the animal studies were performed.
Consent for publication
Not applicable
Availability of data and material
Sequencing runs were deposited at Short Read Archive (SRA-NCBI) under the following accession identifiers: SRX1201397, SRX1201396, SRX1201395, SRX1201394, SRX1201392.
Conflict of interest
The authors declare that they have no conflict of interest
Authors’ Contributions
KVC and DAS conceived and designed the experiments. JS performed the experiments. GCC and JS analyzed the data. DAS, KVC and JRW contributed reagents/materials/analysis tools. GCC, JS, KVC and DAS contributed to the writing of the manuscript.
References
- 1.VandenBergh MF, Verweij PE, Voss A. Epidemiology of nosocomial fungal infections: invasive aspergillosis and the environment. Diagn Microbiol Infect Dis. 1999;34(3):221–7. doi: 10.1016/s0732-8893(99)00026-7. S0732889399000267 [pii] [DOI] [PubMed] [Google Scholar]
- 2.Walsh TJ, Stevens DA. Aspergillosis. In: Goldman L, AS, editors. Cecil Textbook of Medicine. Chapter 347 Elsevier; 2011. [Google Scholar]
- 3.Marr KA, Patterson T, Denning D. Aspergillosis. Pathogenesis, clinical manifestations, and therapy. Infect Dis Clin North Am. 2002;16(4):875–94. vi. doi: 10.1016/s0891-5520(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 4.Park SJ, Mehrad B. Innate immunity to Aspergillus species. Clin Microbiol Rev. 2009;22(4):535–51. doi: 10.1128/CMR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011;11(4):275–88. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 6.Sales-Campos H, Tonani L, Cardoso CR, Kress MR. The immune interplay between the host and the pathogen in Aspergillus fumigatus lung infection. Biomed Res Int. 2013;2013:693023. doi: 10.1155/2013/693023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemons KV, Stevens DA. The contribution of animal models of aspergillosis to understanding pathogenesis, therapy and virulence. Med Mycol. 2005;43(Suppl 1):S101–10. doi: 10.1080/13693780500051919. [DOI] [PubMed] [Google Scholar]
- 8.Pagano L, Caira M, Nosari A, Van Lint MT, Candoni A, Offidani M, et al. Fungal infections in recipients of hematopoietic stem cell transplants: results of the SEIFEM B-2004 study--Sorveglianza Epidemiologica Infezioni Fungine Nelle Emopatie Maligne. Clin Infect Dis. 2007;45(9):1161–70. doi: 10.1086/522189. [DOI] [PubMed] [Google Scholar]
- 9.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320(5881):1344–9. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, et al. The functional landscape of mouse gene expression. J Biol. 2004;3(5):21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons JG, Beauvais A, Beau R, McGary KL, Latge JP, Rokas A. Global transcriptome changes underlying colony growth in the opportunistic human pathogen Aspergillus fumigatus. Eukaryot Cell. 2012;11(1):68–78. doi: 10.1128/EC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rokas A, Gibbons JG, Zhou X, Beauvais A, Latge JP. The diverse applications of RNA-seq for functional genomic studies in Aspergillus fumigatus. Ann N Y Acad Sci. 2012;1273:25–34. doi: 10.1111/j.1749-6632.2012.06755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cerqueira GC, Arnaud MB, Inglis DO, Skrzypek MS, Binkley G, Simison M, et al. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014;42(Database issue):D705–10. doi: 10.1093/nar/gkt1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B, Guo G, Wang C, Lin Y, Wang X, Zhao M, et al. Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic Acids Res. 2010;38(15):5075–87. doi: 10.1093/nar/gkq256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J, Fedorova ND, Montalbano BG, Bhatnagar D, Cleveland TE, Bennett JW, et al. Tight control of mycotoxin biosynthesis gene expression in Aspergillus flavus by temperature as revealed by RNA-Seq. FEMS Microbiol Lett. 2011;322(2):145–9. doi: 10.1111/j.1574-6968.2011.02345.x. [DOI] [PubMed] [Google Scholar]
- 16.Cairns T, Minuzzi F, Bignell E. The host-infecting fungal transcriptome. FEMS Microbiol Lett. 2010;307(1):1–11. doi: 10.1111/j.1574-6968.2010.01961.x. [DOI] [PubMed] [Google Scholar]
- 17.Oosthuizen JL, Gomez P, Ruan J, Hackett TL, Moore MM, Knight DA, et al. Dual organism transcriptomics of airway epithelial cells interacting with conidia of Aspergillus fumigatus. PLoS One. 2011;6(5):e20527. doi: 10.1371/journal.pone.0020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morton CO, Fliesser M, Dittrich M, Mueller T, Bauer R, Kneitz S, et al. Gene expression profiles of human dendritic cells interacting with Aspergillus fumigatus in a bilayer model of the alveolar epithelium/endothelium interface. PLoS One. 2014;9(5):e98279. doi: 10.1371/journal.pone.0098279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wise GJ, Silver DA. Fungal infections of the genitourinary system. J Urol. 1993;149(6):1377–88. doi: 10.1016/s0022-5347(17)36396-6. [DOI] [PubMed] [Google Scholar]
- 20.Gautam P, Shankar J, Madan T, Sirdeshmukh R, Sundaram CS, Gade WN, et al. Proteomic and transcriptomic analysis of Aspergillus fumigatus on exposure to amphotericin B. Antimicrob Agents Chemother. 2008;52(12):4220–7. doi: 10.1128/AAC.01431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clemons KV, Stevens DA. Comparative efficacies of four amphotericin B formulations--Fungizone, amphotec (Amphocil), AmBisome, and Abelcet--against systemic murine aspergillosis. Antimicrobial agents and chemotherapy. 2004;48(3):1047–50. doi: 10.1128/AAC.48.3.1047-1050.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clemons KV, Martinez M, Tong AJ, Stevens DA. Resistance of MBL gene-knockout mice to experimental systemic aspergillosis. Immunol Lett. 2010;128(2):105–7. doi: 10.1016/j.imlet.2009.12.021. S0165-2478(10)00005-2 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Liu M, Machova E, Nescakova Z, Medovarska I, Clemons KV, Martinez M, et al. Vaccination with mannan protects mice against systemic aspergillosis. Med Mycol. 2012;50(8):818–28. doi: 10.3109/13693786.2012.683539. [DOI] [PubMed] [Google Scholar]
- 24.Shankar J, Wu TD, Clemons KV, Monteiro JP, Mirels LF, Stevens DA. Influence of 17beta-estradiol on gene expression of Paracoccidioides during mycelia-to-yeast transition. PLoS One. 2011;6(12):e28402. doi: 10.1371/journal.pone.0028402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clemons KV, Stevens DA. Conventional or molecular measurement of Aspergillus load. Medical mycology. 2009;47(Suppl 1):S132–7. doi: 10.1080/13693780802213340. [DOI] [PubMed] [Google Scholar]
- 32.Singh G, Imai J, Clemons KV, Stevens DA. Efficacy of caspofungin against central nervous system Aspergillus fumigatus infection in mice determined by TaqMan PCR and CFU methods. Antimicrobial agents and chemotherapy. 2005;49(4):1369–76. doi: 10.1128/AAC.49.4.1369-1376.2005. 49/4/1369 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furlaneto CJ, Ribeiro FP, Hatanaka E, Souza GM, Cassatella MA, Campa A. Apolipoproteins A-I and A-II downregulate neutrophil functions. Lipids. 2002;37(9):925–8. doi: 10.1007/s11745-002-0981-4. [DOI] [PubMed] [Google Scholar]
- 34.Maffei CML, Mirels LF, Sobel RA, Clemons KV, Stevens DA. Cytokine and Inducible Nitric Oxide Synthase mRNA Expression during Experimental Murine Cryptococcal Meningoencephalitis. Infection and immunity. 2004;72(4):2338–49. doi: 10.1128/IAI.72.4.2338-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand R, Shankar J, Singh AP, Tiwary BN. Cytokine milieu in renal cavities of immunocompetent mice in response to intravenous challenge of Aspergillus flavus leading to aspergillosis. Cytokine. 2013;61(1):63–70. doi: 10.1016/j.cyto.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 36.Stevens DA, Melikian GL. Aspergillosis in the ‘nonimmunocompromised’ host. Immunol Invest. 2011;40(7–8):751–66. doi: 10.3109/08820139.2011.614307. [DOI] [PubMed] [Google Scholar]
- 37.Clemons KV, Stevens DA. Animal models of Aspergillus infection in preclinical trials, diagnostics and pharmacodynamics: What can we learn from them? Med Mycol. 2006;44(Supplement 1):S119–S26. doi: 10.1080/13693780600871174. [DOI] [PubMed] [Google Scholar]
- 38.Mirkov I, Stosic-Grujicic S, Kataranovski M. Host immune defense against Aspergillus fumigatus: insight from experimental systemic (disseminated) infection. Immunol Res. 2012;52(1–2):120–6. doi: 10.1007/s12026-012-8274-x. [DOI] [PubMed] [Google Scholar]
- 39.Melnikov A, Galinsky K, Rogov P, Fennell T, Van Tyne D, Russ C, et al. Hybrid selection for sequencing pathogen genomes from clinical samples. Genome biology. 2011;12(8):R73. doi: 10.1186/gb-2011-12-8-r73. gb-2011-12-8-r73 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thakur R, Shankar J. In silico Analysis Revealed High-risk Single Nucleotide Polymorphisms in Human Pentraxin-3 Gene and their Impact on Innate Immune Response against Microbial Pathogens. Frontiers in microbiology. 2016;7:192. doi: 10.3389/fmicb.2016.00192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown GD. Dectin-1: a signalling non-TLR pattern-recognition receptor. Nat Rev Immunol. 2006;6(1):33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 42.Thakur R, Anand R, Tiwari S, Singh AP, Tiwary BN, Shankar J. Cytokines induce effector T-helper cells during invasive aspergillosis; what we have learned about T-helper cells? Front Microbiol. 2015;6:429. doi: 10.3389/fmicb.2015.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens DA. Th1/Th2 in aspergillosis. Med Mycol. 2006;44(s1):S229–S35. doi: 10.1080/13693780600760773. [DOI] [PubMed] [Google Scholar]
- 44.Armstrong-James DP, Turnbull SA, Teo I, Stark J, Rogers NJ, Rogers TR, et al. Impaired interferon-gamma responses, increased interleukin-17 expression, and a tumor necrosis factor-alpha transcriptional program in invasive aspergillosis. J Infect Dis. 2009;200(8):1341–51. doi: 10.1086/605931. [DOI] [PubMed] [Google Scholar]
- 45.Anand R, Shankar J, Tiwary BN, Singh AP. Aspergillus flavus induces granulomatous cerebral aspergillosis in mice with display of distinct cytokine profile. Cytokine. 2015;72(2):166–72. doi: 10.1016/j.cyto.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 46.Flaczyk A, Duerr CU, Shourian M, Lafferty EI, Fritz JH, Qureshi ST. IL-33 signaling regulates innate and adaptive immunity to Cryptococcus neoformans. J Immunol. 2013;191(5):2503–13. doi: 10.4049/jimmunol.1300426. jimmunol.1300426 [pii] [DOI] [PubMed] [Google Scholar]
- 47.O’Dea EM, Amarsaikhan N, Li H, Downey J, Steele E, Van Dyken SJ, et al. Eosinophils are recruited in response to chitin exposure and enhance Th2-mediated immune pathology in Aspergillus fumigatus infection. Infect Immun. 2014;82(8):3199–205. doi: 10.1128/IAI.01990-14. IAI.01990-14 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller AM. Role of IL-33 in inflammation and disease. J Inflamm (Lond) 2011;8(1):22. doi: 10.1186/1476-9255-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smithgall MD, Comeau MR, Yoon BR, Kaufman D, Armitage R, Smith DE. IL-33 amplifies both Th1- and Th2-type responses through its activity on human basophils, allergen-reactive Th2 cells, iNKT and NK cells. Int Immunol. 2008;20(8):1019–30. doi: 10.1093/intimm/dxn060. dxn060 [pii] [DOI] [PubMed] [Google Scholar]
- 50.Caffrey AK, Lehmann MM, Zickovich JM, Espinosa V, Shepardson KM, Watschke CP, et al. IL-1alpha signaling is critical for leukocyte recruitment after pulmonary Aspergillus fumigatus challenge. PLoS Pathog. 2015;11(1):e1004625. doi: 10.1371/journal.ppat.1004625PPATHOGENS-D-14-01338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortez KJ, Lyman CA, Kottilil S, Kim HS, Roilides E, Yang J, et al. Functional genomics of innate host defense molecules in normal human monocytes in response to Aspergillus fumigatus. Infect Immun. 2006;74(4):2353–65. doi: 10.1128/IAI.74.4.2353-2365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savers A, Rasid OPM, Brock M, Jouvion G, Ryffel B, Cavaillon J-M, et al. Infection-Mediated Priming of Phagocytes Protects against Lethal Secondary Aspergillus fumigatus Challenge. PloS one. 2016;11(4):e0153829. doi: 10.1371/journal.pone.0153829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foucher ED, Blanchard S, Preisser L, Garo E, Ifrah N, Guardiola P, et al. IL-34 induces the differentiation of human monocytes into immunosuppressive macrophages. antagonistic effects of GM-CSF and IFNgamma. PLoS One. 2013;8(2):e56045. doi: 10.1371/journal.pone.0056045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu M, Guo S, Hibbert JM, Jain V, Singh N, Wilson NO, et al. CXCL10/IP-10 in infectious diseases pathogenesis and potential therapeutic implications. Cytokine Growth Factor Rev. 2011;22(3):121–30. doi: 10.1016/j.cytogfr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chai L, Netea MG, Teerenstra S, Earnest A, Vonk AG, Schlamm HT, et al. Early proinflammatory cytokines and C-reactive protein trends as predictors of outcome in invasive Aspergillosis. J Infect Dis. 2010;202(9):1454–62. doi: 10.1086/656527. [DOI] [PubMed] [Google Scholar]
- 56.Rodland EK, Ueland T, Bjornsen S, Sagen EL, Dahl CP, Naalsund A, et al. Systemic biomarkers of inflammation and haemostasis in patients with chronic necrotizing pulmonary aspergillosis. BMC Infect Dis. 2012;12:144. doi: 10.1186/1471-2334-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bendall L. Chemokines and their receptors in disease. Histol Histopathol. 2005;20(3):907–26. doi: 10.14670/HH-20.907. [DOI] [PubMed] [Google Scholar]
- 58.Lim HW, Lee J, Hillsamer P, Kim CH. Human Th17 cells share major trafficking receptors with both polarized effector T cells and FOXP3+ regulatory T cells. J Immunol. 2008;180(1):122–9. doi: 10.4049/jimmunol.180.1.122. [DOI] [PubMed] [Google Scholar]
- 59.Borroni EM, Mantovani A, Locati M, Bonecchi R. Chemokine receptors intracellular trafficking. Pharmacol Ther. 2010;127(1):1–8. doi: 10.1016/j.pharmthera.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 60.Richardson TA, Sherman M, Antonovic L, Kardar SS, Strobel HW, Kalman D, et al. Hepatic and renal cytochrome p450 gene regulation during citrobacter rodentium infection in wild-type and toll-like receptor 4 mutant mice. Drug Metab Dispos. 2006;34(3):354–60. doi: 10.1124/dmd.105.007393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morgan ET. Impact of infectious and inflammatory disease on cytochrome P450-mediated drug metabolism and pharmacokinetics. Clin Pharmacol Ther. 2009;85(4):434–8. doi: 10.1038/clpt.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morgan ET. Regulation of cytochromes P450 during inflammation and infection. Drug Metab Rev. 1997;29(4):1129–88. doi: 10.3109/03602539709002246. [DOI] [PubMed] [Google Scholar]
- 63.Theken KN, Deng Y, Kannon MA, Miller TM, Poloyac SM, Lee CR. Activation of the acute inflammatory response alters cytochrome P450 expression and eicosanoid metabolism. Drug metabolism and disposition: the biological fate of chemicals. 2011;39(1):22–9. doi: 10.1124/dmd.110.035287. dmd.110.035287 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morgan ET, Ullrich V, Daiber A, Schmidt P, Takaya N, Shoun H, et al. Cytochromes P450 and flavin monooxygenases--targets and sources of nitric oxide. Drug Metab Dispos. 2001;29(11):1366–76. [PubMed] [Google Scholar]
- 65.Walubo A. The role of cytochrome P450 in antiretroviral drug interactions. Expert Opin Drug Metab Toxicol. 2007;3(4):583–98. doi: 10.1517/17425225.3.4.583. [DOI] [PubMed] [Google Scholar]
- 66.Denning DW, Follansbee SE, Scolaro M, Norris S, Edelstein H, Stevens DA. Pulmonary Aspergillosis in the Acquired Immunodeficiency Syndrome. New England Journal of Medicine. 1991;324(10):654–62. doi: 10.1056/NEJM199103073241003. [DOI] [PubMed] [Google Scholar]
- 67.Leal SM, Jr, Roy S, Vareechon C, Carrion S, Clark H, Lopez-Berges MS, et al. Targeting iron acquisition blocks infection with the fungal pathogens Aspergillus fumigatus and Fusarium oxysporum. PLoS Pathog. 2013;9(7):e1003436. doi: 10.1371/journal.ppat.1003436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432(7019):917–21. doi: 10.1038/nature03104. nature03104 [pii] [DOI] [PubMed] [Google Scholar]
- 69.Adlerova L, Bartoskova A, Faldyna M. Lactoferrin: a review. Veterinarni Medicina. 2008;53(9):457–68. [Google Scholar]
- 70.Aisen P, Leibman A. Lactoferrin and transferrin: a comparative study. Biochim Biophys Acta. 1972;257(2):314–23. doi: 10.1016/0005-2795(72)90283-8. [DOI] [PubMed] [Google Scholar]
- 71.Legrand D, Elass E, Carpentier M, Mazurier J. Lactoferrin: a modulator of immune and inflammatory responses. Cell Mol Life Sci. 2005;62(22):2549–59. doi: 10.1007/s00018-005-5370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McPhaul MJ, Noble JF, Simpson ER, Mendelson CR, Wilson JD. The expression of a functional cDNA encoding the chicken cytochrome P-450arom (aromatase) that catalyzes the formation of estrogen from androgen. Journal of Biological Chemistry. 1988;263(31):16358–63. [PubMed] [Google Scholar]
- 73.Marriott I, Huet-Hudson YM. Sexual dimorphism in innate immune responses to infectious organisms. Immunologic Research. 2006;34(3):177–92. doi: 10.1385/ir:34:3:177. [DOI] [PubMed] [Google Scholar]
- 74.Villaggio B, Soldano S, Cutolo M. 1,25-dihydroxyvitamin D3 downregulates aromatase expression and inflammatory cytokines in human macrophages. Clin Exp Rheumatol. 2012;30(6):934–8. [PubMed] [Google Scholar]
- 75.Simpson E, Rubin G, Clyne C, Robertson K, O’Donnell L, Jones M, et al. The role of local estrogen biosynthesis in males and females. Trends Endocrinol Metab. 2000;11(5):184–8. doi: 10.1016/s1043-2760(00)00254-x. [DOI] [PubMed] [Google Scholar]
- 76.Simpson ER, Zhao Y, Agarwal VR, Michael MD, Bulun SE, Hinshelwood MM, et al. Aromatase expression in health and disease. Recent Prog Horm Res. 1997;52:185–213. discussion -4. [PubMed] [Google Scholar]
- 77.Cuzzocrea S, Santagati S, Sautebin L, Mazzon E, Calabro G, Serraino I, et al. 17beta-estradiol antiinflammatory activity in carrageenan-induced pleurisy. Endocrinology. 2000;141(4):1455–63. doi: 10.1210/endo.141.4.7404. [DOI] [PubMed] [Google Scholar]
- 78.Ben-Ami R, Lewis RE, Leventakos K, Kontoyiannis DP. Aspergillus fumigatus inhibits angiogenesis through the production of gliotoxin and other secondary metabolites. Blood. 2009;114(26):5393–9. doi: 10.1182/blood-2009-07-231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hsu JL, Khan MA, Sobel RA, Jiang X, Clemons KV, Nguyen TT, et al. Aspergillus fumigatus invasion increases with progressive airway ischemia. PloS one. 2013;8(10):e77136. doi: 10.1371/journal.pone.0077136PONE-D-13-14976. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frantz S, Vincent KA, Feron O, Kelly RA. Innate immunity and angiogenesis. Circ Res. 2005;96(1):15–26. doi: 10.1161/01.RES.0000153188.68898.ac. [DOI] [PubMed] [Google Scholar]
- 81.Romagnani P, Annunziato F, Lasagni L, Lazzeri E, Beltrame C, Francalanci M, et al. Cell cycle-dependent expression of CXC chemokine receptor 3 by endothelial cells mediates angiostatic activity. The Journal of clinical investigation. 2001;107(1):53–63. doi: 10.1172/JCI9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Miyake M, Goodison S, Urquidi V, Gomes Giacoia E, Rosser CJ. Expression of CXCL1 in human endothelial cells induces angiogenesis through the CXCR2 receptor and the ERK1/2 and EGF pathways. Lab Invest. 2013;93(7):768–78. doi: 10.1038/labinvest.2013.71. labinvest201371 [pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Expression profile of genes encoding cytokines and cytokines receptors. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Fig. S2. Expression profile of genes encoding Toll-like receptors, selectins and defensins. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Fig 3. Expression profile of genes encoding components of the complement. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Figure 4. Expression profile of genes encoding interleukins and interferons. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Figure 5. Expression profile of genes associated iron binding and homeostasis. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Figure 6. Expression profile of genes associated with positive regulation of angiogenesis. Only genes differentially expressed in at least one pair of time points are shown. Color indicates the expression level of each transcript in terms of Fragments per Kilobase of transcript per Million of mapped reads (FPKM) on each time point: lowest FPKM values are depicted in white and the highest values are shown in darkest blue, according to the legend. The dendrogram indicates the similarity of expression profile between transcripts.
Supplementary Table S1: Short reads statistics.
Supplementary Table S2: Distribution of number of transcripts based on FPKM reported on each sample/day.
Supplementary Table S3: Genes significantly differentially expressed in at least one pair of time points.
Supplementary Table S4: List of genes up-regulated in comparison to non-infected mouse (day 0).
Supplementary Table S5: List of genes down-regulated in comparison to non-infected mouse (day 0).
Supplementary Table S6: Clusters of genes with similar expression profile across time points.
Supplementary Table S7: GO terms enriched on transcripts differentially expressed on each time point in comparison to non-infected mouse (day 0).
Supplementary Table S8: GO terms enriched on clusters of transcripts with similar expression profile across time points.
Supplementary Table S9: Up-regulated transcripts encoding for chemokines and their receptors in response to invasive aspergillosis in the current study.
Supplementary Table S10: List of downregulated transcripts associated with iron, heme and aromatase processes and functions according to Gene Ontology (GO).
Supplementary Table S11: Comparison of the dynamics of cytokines and iNOS response between our work and studies on mice infected with Cryptococcus neoformans and Aspergillus flavus.