ABSTRACT
De novo genes are very important for evolutionary innovation. However, how these genes originate and spread remains largely unknown. To better understand this, we rigorously searched for de novo genes in Saccharomyces cerevisiae S288C and examined their spread and fixation in the population. Here, we identified 84 de novo genes in S. cerevisiae S288C since the divergence with their sister groups. Transcriptome and ribosome profiling data revealed at least 8 (10%) and 28 (33%) de novo genes being expressed and translated only under specific conditions, respectively. DNA microarray data, based on 2-fold change, showed that 87% of the de novo genes are regulated during various biological processes, such as nutrient utilization and sporulation. Our comparative and evolutionary analyses further revealed that some factors, including single nucleotide polymorphism (SNP)/indel mutation, high GC content, and DNA shuffling, contribute to the birth of de novo genes, while domestication and natural selection drive the spread and fixation of these genes. Finally, we also provide evidence suggesting the possible parallel origin of a de novo gene between S. cerevisiae and Saccharomyces paradoxus. Together, our study provides several new insights into the origin and spread of de novo genes.
KEYWORDS: DNA shuffling, de novo gene, GC content, parallel origins, purifying selection, yeast
IMPORTANCE
Emergence of de novo genes has occurred in many lineages during evolution, but the birth, spread, and function of these genes remain unresolved. Here we have searched for de novo genes from Saccharomyces cerevisiae S288C using rigorous methods, which reduced the effects of bad annotation and genomic gaps on the identification of de novo genes. Through this analysis, we have found 84 new genes originating de novo from previously noncoding regions, 87% of which are very likely involved in various biological processes. We noticed that 10% and 33% of de novo genes were only expressed and translated under specific conditions, therefore, verification of de novo genes through transcriptome and ribosome profiling, especially from limited expression data, may underestimate the number of bona fide new genes. We further show that SNP/indel mutation, high GC content, and DNA shuffling could be involved in the birth of de novo genes, while domestication and natural selection drive the spread and fixation of these genes. Finally, we provide evidence suggesting the possible parallel origin of a new gene.
INTRODUCTION
New genes are the rich substrate of evolution that leads to various biological effects. The mechanisms giving rise to new genes can be placed into four categories (1): (i) gene duplication and rapid divergence, in which a new gene is derived from already existing genes in the same genome; (ii) horizontal gene transfer, in which a new gene is derived from already existing genes but from different genomes; (iii) an overprinting process, where mutations in a protein-coding gene allow the expression of a second protein-coding gene (2); and (iv) de novo origin, in which the noncoding region evolves to an open reading frame (ORF) through SNP and indel mutations (3, 4). Here, we refer to the fourth category as de novo gene, which is the focus of this study.
A de novo gene arising from a noncoding region was thought to be improbable (5, 6). In recent years, our knowledge of the distribution and function of de novo genes has been increasing since the first identification of de novo genes in Drosophila (7, 8). Until now, the de novo origins of species or lineage-specific protein-coding genes from noncoding DNA have been described in diverse lineages, including yeast, primates, and plants (3, 8–14). Compared to evolutionary conserved genes, de novo genes are overall shorter and have lower expression and tissue-restricted expression (14). The function of de novo genes is diverse. It has been shown that de novo genes can quickly become functionally important and essential for viability in Drosophila (15, 16). In primates, the few described de novo genes have been implicated in cancer and cancer outcomes (3, 17, 18).
Saccharomyces cerevisiae is one of the simplest eukaryotic organisms, with a relatively compact genome/gene content and a wealth of available phenotypic data associated with mutant and growth conditions (19, 20), which provide the chance to systematically study aspects of de novo genes. Previous studies of new genes in S. cerevisiae have helped shed light on their origin (11, 21, 22). To better understand their origin, spread, and fixation, de novo genes were sought in S. cerevisiae S288C using strict parameters, as in the analyses of Guerzoni and McLysaght (4). Through analyses of identified de novo genes between different species and among conspecific strains, we found multiple factors involved in the birth, spread, and fixation of these genes. In addition, we also suggest possible parallel origin of a new gene between different species.
RESULTS AND DISCUSSION
Detection of de novo genes in Saccharomyces cerevisiae S288C genomes.
Within the Saccharomycetaceae, S. cerevisiae S288C has the best annotated genome and massive phenotypic data under various mutant and growth conditions. For instance, the SPELL database from the Saccharomyces Genome Database (SGD, the most commonly used database in yeast) contains 537 data sets representing 11,889 total conditions (23). As in previous studies, we first compared the complete S. cerevisiae S288C proteome from the SGD database with that of 20 species from Saccharomycetaceae (see all strains in Materials and Methods). The S. cerevisiae S288C proteins that did not have significant sequence similarity (E value of 1 × 10−4) from the 20 species were regarded as initial de novo genes. It has been suggested that great variability in the estimates of new genes is partially due to sequencing gaps, annotation error, or gene loss. Recently, Moyers and Zhang (24) reported that using sequence similarity searching methods alone for identifying new genes commonly results in false positives. Although this conclusion is still debated (25), employing noncoding orthologous DNA in sister outgroups as a subsidiary parameter would be helpful to identify genuine de novo genes (4). Together with an expression cutoff FPKM (fragments per kilobase of transcript per million mapped reads) of ≥1.0, 84 de novo genes from S. cerevisiae S288C were identified (see Table S1 at http://baojunedisonwu.weebly.com/download.html), which have no protein hit in 20 species but have noncoding orthologous sequences in sister species Saccharomyces paradoxus CBS 432 and Saccharomyces mikatae IFO1815. All de novo genes overlap non-de novo genes, where there are 73 opposite strand overlaps and 11 same strand overlaps.
We compared our results with those of three other studies (11, 21, 22) (only considering the SGD annotated genes) and found that only 28 (33%) out of 84 genes were shared by the three other studies (Fig. 1A), in which 20 genes are found to overlap those of Carvunis et al. (11). Surprisingly, there is no common gene shared in all four studies (Fig. 1A). The variable results among studies can be partially attributed to exclusion of candidates overlapping ancient genes. For instance, ORFs overlapping ancient genes were often excluded from the Carvunis et al. study (11), but previous studies have indicated that new genes could arise while overlapping ancient genes (3, 4). In contrast, de novo genes overlapping ancient genes were considered in the Lu et al. study (22), but there are only five de novo genes shared between their work and this study (Fig. 1A). Therefore, besides the “overlapped” parameter, there are other parameters affecting the identification of de novo genes, such as E value for homologue searches and the required expression levels. For instance, in the Carvunis et al. study (2012), an E value of 10−2, more relaxed than the 10−4 used in this study, was used to search for homologues (BLASTP, TBLASTX, and TBLASTN). As a result, more annotated genes from S288C were found to have homologues in non-S. cerevisiae species than in this study. Moreover, whether homologues of S. cerevisiae S288C in non-S. cerevisiae species had intact open reading frames was not examined in the Carvunis et al. study (22). In other words, some homologues they identified in non-S. cerevisiae species are noncoding regions rather than protein-coding genes. Consequently, more true candidate genes than expected were filtered out. In the Lu et al. study (2017) (22), two copies of mRNA were used as the cutoff to further reduce the number of de novo genes. However, the study stated that there was a possibility that nontranscribed open reading frames might be expressed under other more specific conditions. Indeed, we found 10% of de novo genes were expressed only under specific conditions (see Table S1 at http://baojunedisonwu.weebly.com/download.html). Overall, among the four studies of yeast de novo genes, the numbers detected vary quite widely from study to study with very little overlap. A similar scenario was found in studies of primate de novo genes, where Guerzoni et al. (4) compared their results with those of Ruiz-Orera et al. (14) but found no overlap in the candidate lists.
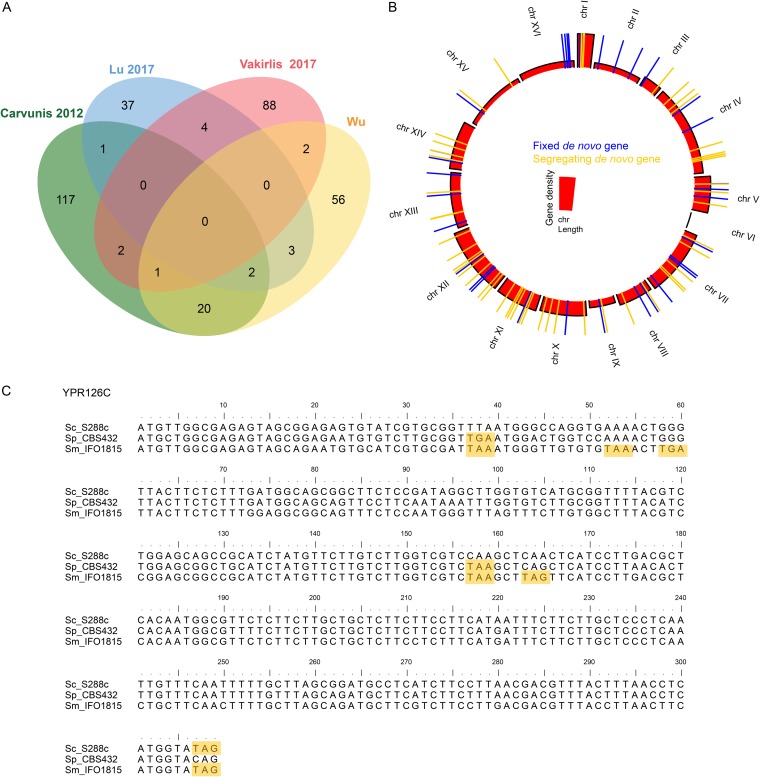
FIG 1 .
Eighty-four de novo genes detected in the S. cerevisiae S288C genome. (A) Overlaps of the de novo genes from this study and previous studies. Only SGD annotated genes were considered. (B) Distribution of 84 de novo genes along 16 chromosomes. (C) One example showing SNP mutations driving the birth of YPR126C. The yellow shading represents the position of the stop codon.
Segregating and fixed de novo genes in S. cerevisiae S288C.
The 84 de novo genes are spread across 15 of the 16 chromosomes (except chromosome VI [chr VI]), where chr I has the highest density (highest height) with 22 de novo genes per megabase (Fig. 1B). Based on a previous study (27), we inferred the boundary of core regions in each chromosome (see Table S2 at http://baojunedisonwu.weebly.com/download.html) and found all de novo genes were located within the core regions. We compared the orthologues of de novo genes against a 93-strain population and found 63% (52/84) of de novo genes carry alleles having both intact and disrupted ORFs. Therefore, 84 de novo genes could be divided into two categories: fixed de novo genes and segregating de novo genes. The fixed de novo genes are species specific, and the segregating de novo genes are strain specific. There are 13 chromosomes having both categories, while two chromosomes (chr II and chr XVI) only have fixed de novo genes (Fig. 1B).
Indel and SNP mutations have been reported to contribute to the birth of de novo genes in humans relative to other primates (3, 4). Compared with S. paradoxus CBS 432 and S. mikatae IFO1815, our analyses reveal that 2% (2/84), 30% (25/84), and 68% (57/84) of de novo genes are driven by indel mutation, SNP mutation, and a combination of indel and SNP mutations, respectively. For instance, along the full length of the S288C gene YPR126C, there is no gap, but there are SNPs resulting in two and five stop codons in the similar nucleotide sequences from S. paradoxus and S. mikatae, respectively (Fig. 1C). It is noteworthy that nucleotides at positions 1 to 39 produce a 12-amino-acid-long protein in S. paradoxus and S. mikatae, while positions 40 to 159 produce a 39-amino-acid-long protein in S. paradoxus, which suggests that new genes could evolve de novo through short ORFs in nongenic sequences (11). Among conspecific strains, 42% (22/52), 52% (27/52), and 6% (3/52) of segregating de novo genes are attributed to indel mutation, SNP mutation, and a combination of indel and SNP mutations, respectively. Moreover, these mutations in 74% (38/52) of segregating de novo genes occurred at the same positions, indicating that the disrupted ORFs might be under slight selection during spread within a population.
De novo genes are possibly involved in biological process.
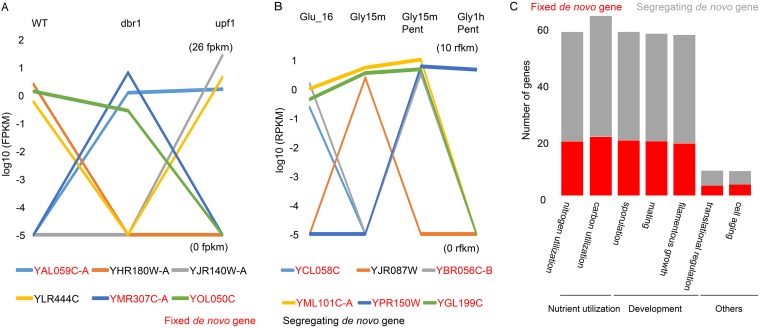
The FPKM of de novo genes were extracted from previous transcriptome sequencing (RNA-seq) experiments (28) with the wild type and dbr1Δ and upf1Δ mutants, where the products of dbr1 and upf1 are two proteins involved in pre-mRNA splicing and nonsense-mediated mRNA decay (28, 29). Then, we used the FPKM value of ≥1.0 as the cutoff to filter out nonexpressed de novo genes (see Materials and Methods for more details). Among the expressed ones, we found eight de novo genes that were expressed only under specific conditions (six cases in Fig. 2A and for all cases in Table S1 at http://baojunedisonwu.weebly.com/), which suggests that these de novo genes could be regulated by dbr1 and upf1 and possibly involved in mRNA processing. It is important to note that both fixed and segregating de novo genes are among the eight de novo genes (six cases in Fig. 2A and for all cases in Table S1 at http://baojunedisonwu.weebly.com/); therefore, the potential function of de novo genes is not determined by their status (fixed or not) in the population. In addition, the condition-specific expression could result in underestimation of the number of de novo genes when expression level is used as a filter parameter, especially when only limited expression data are used. We further determined the translation of de novo gene using ribosome profiling data (30). This analysis identified 51% (43/84) of de novo genes having an RPKM (reads per kilobase of transcript per million mapped reads) value of ≥0.5, which was used as the cutoff of a translated gene (see more details in Materials and Methods). Among the 43 translated de novo genes, 28 genes were found to be translated at specific time points or conditions (six cases in Fig. 2B and for all cases in Table S3 at http://baojunedisonwu.weebly.com/). Similar to expression patterns, both fixed and segregating de novo genes could be translated under specific conditions.
FIG 2 .
Expression, translation, and regulated evidence of de novo genes. (A) Dynamic expression of six de novo genes under different conditions. The FPKM values are in the range of 0 to ~26. All expression data of de novo genes are shown in Table S1. (B) Dynamic translation of six de novo genes under different conditions. The FPKM values are in the range of 0 to ~10. All footprinting data of de novo genes are shown in Table S4. (C) Various regulations of de novo genes associated with nutrient utilization, developmental stages, and cell aging. Red represents fixed de novo genes, and gray represents the segregating de novo genes. All regulation data of de novo genes are shown in Table S3.
We further took advantage of 537 expression microarray data from the SPELL database (23) to infer the potential function of expressed de novo genes. This compendium includes experiments sampled from a broad range of mutant and growth conditions. If a de novo gene had at least a 2-fold change relative to the control group on the microarray, it was regarded as a regulated gene. Among the 84 new genes, 73 (87%) were found to be associated with 52 functional categories defined by SPELL. For example, 61 genes (73%) are involved in the carbon utilization process, while only 6 genes (7%) are involved in cell aging (Fig. 2C). In addition, the proportion of segregating de novo genes in 46 categories is higher than that of fixed de novo genes (seven cases in Fig. 2C; see Table S4 at http://baojunedisonwu.weebly.com/download.html). Consistent with the observation that most de novo genes (87%) have potential functions, a recent study showed that expression of random artifact sequences with a coding region (50 amino acids) in bacteria could change cell growth rate, and the functional proportion of these random ORFs was up to 77% (31).
GC content is important for birth of de novo genes.
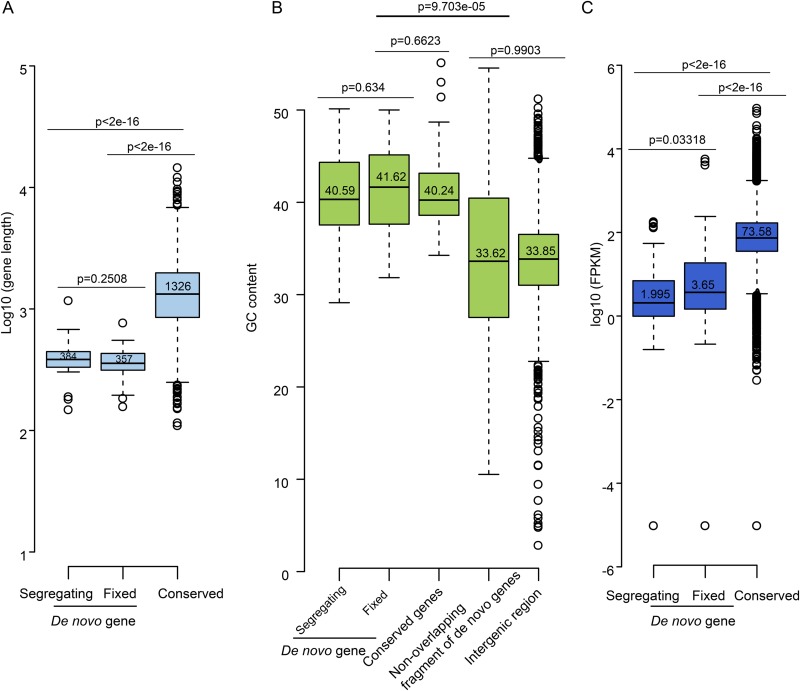
In Drosophila, GC content, gene length, and expression level are positively correlated with sequence conservation (13), while new genes tend to be shorter, with low expression, and are GC poor. In agreement with studies in Drosophila, de novo genes of S288C are shorter (Fig. 3A) and are expressed at lower levels (Fig. 3C). However, different from Drosophila, where GC content of de novo genes is significant lower than that of conserved genes, the de novo genes in S288C have GC content no different from that of conserved genes (Fig. 3B). Given that all de novo genes overlap other preexisting genes, we also calculated the GC content of nonoverlapping regions, which shows obviously lower GC content than both conserved genes and full-length de novo genes but is similar to that in intergenic regions (Fig. 3B). This finding indicates that de novo genes formed as overlapping loci in high-GC regions associated with non-de novo genes. Moreover, these findings support the hypothesis that new genes are more likely to be generated from high-GC regions, while AT regions flanking GC regions act as the reservoirs for start and stop codons that define gene length. Generally, the GC content is important for the birth of a new gene because (i) GC-rich regions are more likely long ORFs by chance since stop codons are AT rich (32), (ii) GC-rich regions tend to be, on average, more transcriptionally active (33, 34), and (iii) higher GC content leads to higher intrinsic structural disorder (ISD) (35), which facilitates interprotein interaction and thus accelerates coadaptive evolution of new genes with preexisting genes. We also compared segregating and fixed de novo genes in terms of gene length, GC content, and expression level (Fig. 3A to C). However, only the difference in expression level was significant between them (P = 0.03), which is consistent with a previous observation from Drosophila (9), where the expression level of fixed de novo genes is higher than that of segregating de novo genes.
FIG 3 .
Comparison of de novo and conserved genes by (A) gene length, (B) GC content, and (C) gene expression level. For the intergenic regions, 4,539 fragments ranging from 200 to 1,000 nucleotides (nt) were used. The conserved genes were defined such that these genes are shared among S. cerevisiae S288C, S. paradoxus CBS432, and S. mikatae IFO1815.
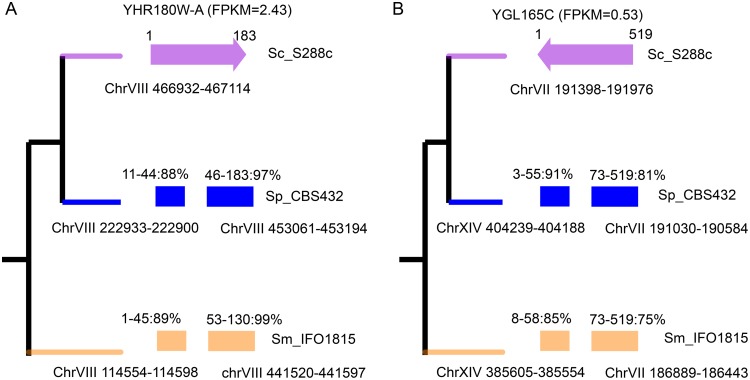
DNA shuffling is a shortcut for sudden birth of a de novo gene.
Although birth of de novo genes can be driven by SNP and indel mutations in noncoding regions (3, 4), there are alternative evolution events driving their birth, such as DNA shuffling. There is one de novo gene, YHR180W-A, overlapping retrotransposon Ty3LTR. Given the mobility of transposons, it is reasonable to speculate that DNA shuffling may promote the birth of YHR180W-A over a short time. To test that possibility, we searched for the homologues from S. paradoxus CBS432 and S. mikatae IFO1815 using YHR180W-A as a query. As a result, we found YHR180W-A being shaped by the retrotransposon Ty3LTR and a tRNA-Thr, where the two features are separated by at least 200 kb in S. paradoxus CBS432 and S. mikatae IFO1815 (Fig. 4A). Previous studies indicated that 53% of primate new genes and 20% of human new genes match transposon elements (TEs) (14, 17). Therefore, the contribution of DNA shuffling mediated by transposons to the birth of new genes may be widespread. During the search for de novo genes, we also found that a de novo gene candidate was generated through DNA shuffling independent of a transposon, where two noncoding regions, shaping YGL165C, are located on different chromosomes from those of sister species (Fig. 4B). Although this candidate gene, YGL165C, does not meet the expression-level cutoff in this study (FPKM value of 0.53 versus 1.0), it provides insights into the birth of de novo genes mediated by DNA shuffling independent of transposons.
FIG 4 .
DNA shuffling shaping the birth of de novo genes (A) YHR180W-A and (B) YGL165C. The identity and corresponding region between de novo genes and noncoding DNA are labeled above the colored boxes. The genomic locations are labeled under the colored boxes. S. cerevisiae S288C is shown in pink, S. paradoxus CBS432 in blue, and S. mikatae IFO 1815 in cream.
Domestication and natural selection shape the spread and fixation of de novo genes.
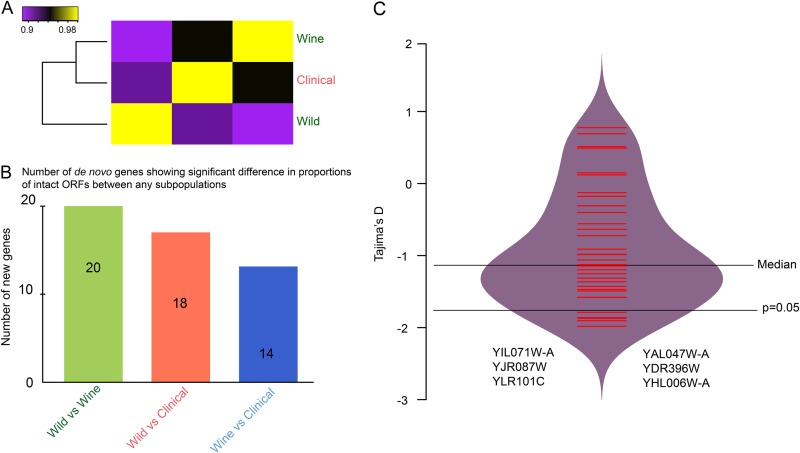
We divided the 93 strains into three subpopulations (wine-making strains, clinical strains, and wild strains) based on their environmental origins (see Table S5 at http://baojunedisonwu.weebly.com/download.html). Hierarchical clustering of subpopulations using the frequency of intact ORFs reveals that strains from the wine and clinical populations are closer to each other than the wild population (Fig. 5A). In addition, if the spread and fixation of de novo genes are free of selection, the frequency of intact ORFs for the same gene among three subpopulations should not be significantly different. However, we identified 29 genes showing a significant difference in the proportions between any two populations (Fisher’s exact test, P < 0.05) (Fig. 5B). Among the three pairwise comparisons, the combination of wine-making strains relative to wild strains has the most genes, while the pairwise comparison of wine-making versus clinical strains has the fewest genes. These observations suggest that domestication (environment) might play a role in the spread of de novo genes. We further investigated the role of natural selection through determining if de novo genes are associated with reduced nucleotide diversity (Tajima’s D). For each de novo gene, only the nonoverlapping regions with longer than 50 bp were considered, and then 36 genes (fragments) were collected for this analysis. Diversity in 6 of 36 genes (fragments) (17%) is significantly lower than expected (P < 0.05), and none of them is higher than expected (Fig. 5C). Moreover, all six fragments under purifying selection are from fixed alleles that do not have disrupted ORF alleles in the population, suggesting that natural selection plays a significant role in their fixation. Among the six genes (fragments), YIL071W-A is the only one among 16 new genes under purifying selection identified by Carvunis et al. (11).
FIG 5 .
Domestication and natural selection of de novo genes. (A) Hierarchical clustering of subpopulations using the frequency of intact ORFs. (B) Number of de novo genes showing significant difference in proportions of intact ORFs between any two populations. (C) Inferred Tajima’s D values for the nonoverlapping regions from de novo genes. Only nonoverlapping regions longer than 50 bp were considered.
Possible parallel origin of de novo genes between species.
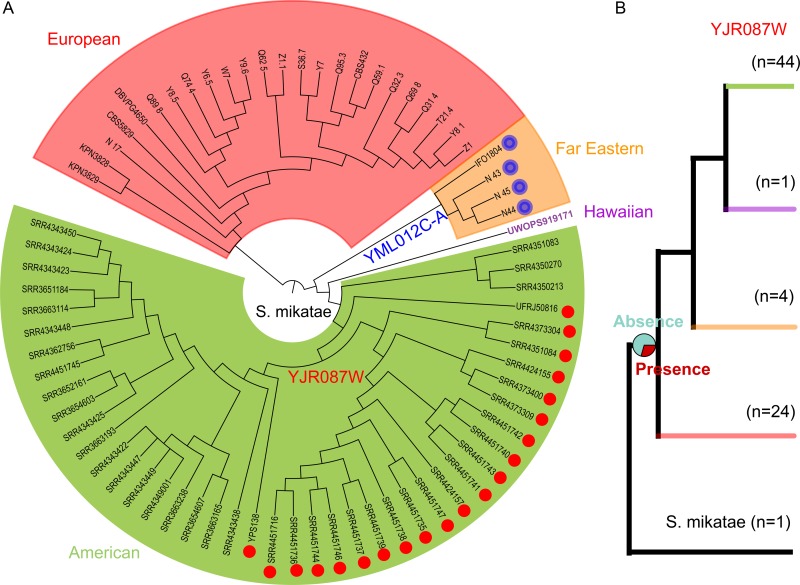
We counted the new genes in 93 strains, but not in S288C and other yeast species, using the primary results from Strope et al. (2015) (36). Because there are no RNA-seq data for these strains, this analysis focused only on the birth of intact ORFs and resulted in the identification of 25 new genes, most of which (23/25) have a frequency of intact ORFs smaller than 10% (see Table S6 at http://baojunedisonwu.weebly.com/download.html). The alleles having both intact and disrupted ORFs in the S. cerevisiae population led to the question of possible misidentification of the de novo genes as a result of using only one strain from each outgroup species. To test that, we searched for orthologous loci from 73 S. paradoxus strains using 84 de novo genes (strains in Fig. 6) and found two targets from S. paradoxus carrying the alleles of both intact and disrupted ORFs, accounting for only 2% of the total de novo genes. Therefore, a single strain from each species as the outgroup would not greatly bias identification of de novo genes in this study. Among the two targets, 21 intact ORFs of YJR087W are present in a subpopulation of Ontario (red dots), while the intact ORFs of YML012C-A are found in the Far Eastern clade (blue circles) (Fig. 6A). Based on the distribution of intact ORFs, we inferred the ancestral states of the two genes and found that YJR087W had no (70% absence versus 30% presence) intact ORF at the origin of S. paradoxus (Fig. 6B; see Fig. S1 at http://baojunedisonwu.weebly.com/download.html). Given that the de novo gene YJR087W is present in a deep subpopulation of S. paradoxus and most of S. cerevisiae (Fig. 6A), this supports the possibility that a parallel origin of a de novo gene could occur between S. cerevisiae and S. paradoxus.
FIG 6 .
Parallel origin of a de novo gene. (A) Distribution of two S288C de novo genes in the S. paradoxus population. The maximum likelihood phylogeny was reconstructed using concatenated sequences of 1,000 aligned single-copy genes that are universally present in all S. paradoxus strains and the S. mikatae IFO 1815 strain (root species). Colors on the tree represent different subpopulations. (B) The ancestral states of YJR087W at the origin of S. paradoxus. The possibility is 70% absence and 30% presence, as shown in the pie.
Conclusion.
In this study, we identified 84 de novo genes (1% of total SGD genes) that originated recently in S. cerevisiae S288C. Analyses of these de novo genes reveal that SNP/indel mutations, high GC content, and DNA shuffling facilitate the birth of de novo genes, while domestication and natural selection play a role in the spread and fixation of these genes. In addition, our study also suggests a possible parallel origin of a de novo gene between S. cerevisiae and S. paradoxus.
MATERIALS AND METHODS
Identification of de novo genes in S. cerevisiae S288C.
We performed a BLASTP search of the S288C proteins downloaded from the Saccharomyces Genome Database (37) against the merged protein data set from S. paradoxus CSB432 (27) and 19 yeast species from YGOB (38) using an E value threshold of 1 × 10−4, which was used to identify new genes in other organisms (4). The 19 species include Saccharomyces mikatae, Saccharomyces kudriavzevii, Saccharomyces bayanus var. uvarum, Candida glabrata, Kazachstania africana, Kazachstania naganishii, Naumovozyma dairenensis, Naumovozyma castellii, Tetrapisispora blattae, Tetrapisispora phaffii, Vanderwaltozyma polyspora, Zygosaccharomyces rouxii, Torulaspora delbrueckii, Kluyveromyces lactis, Eremothecium gossypii, Eremothecium cymbalariae, Lachancea kluyveri, Lachancea thermotolerans, and Lachancea waltii. The genes not included in BLAST search results formed the basis for the list of initial candidate genes. We then excluded candidate genes where we could not detect the orthologous noncoding sequence in the outgroup genomes of S. paradoxus CBS432 and S. mikatae IFO1815. The de novo genes of S288C from three other studies were also extracted (11, 21, 22). In the Lu et al. study, the de novo genes were extracted from their Table S1. In the Vakirlis et al. study, the de novo genes were extracted from their Table S4. In the Carvunis et al. study, the de novo genes were extracted from their Table S3.
Evidence of expression, translation, and regulation of de novo genes.
The FPKM of de novo genes were extracted from previous RNA-seq experiments (28), in which strand-specific libraries were constructed for three conditions: wild type and dbr1Δ and upf1Δ mutants. Previous study has proposed an FPKM value of 0.3 as the threshold separating intergenic and exon expression (39). In this study, an FPKM value of a de novo gene of ≥1.0 (3-fold as the threshold) under any condition was regarded as expression. Finally, 84 expressed de novo genes were identified and used for downstream analyses. We further determined the translation of de novo genes using the ribosome profiling data (30). In our study, the RPKM value of a de novo gene of ≥0.5 under any condition was thought to be translated. The RPKM value of 0.5 is reasonable because the ratio footprint RPKM value/RNA-seq FPKM = 0.5 is located within the normal range of translation efficiencies (30). Finally, we took advantage of 537 expression microarray data from the SPELL database (23) to infer the potential function of expressed de novo genes. If the expressed de novo gene had at least a 2-fold change relative to the control group on the microarray, it would be regarded as a regulated gene.
Identification of alleles of S288C de novo genes from population genomes.
The 93 S. cerevisiae high-quality genomes were downloaded (with all accession numbers shown in Table S7 at http://baojunedisonwu.weebly.com/download.html); these were generated by Strope et al. (36). For the S. paradoxus genomes, we collected 31 strains from Liti et al. (ftp://ftp.sanger.ac.uk/pub/users/dmc/yeast/latest and ftp://ftp.sanger.ac.uk/pub/users/dmc/yeast/SGRP2/assembly/) (40) and Yue et al. (https://yjx1217.github.io/Yeast_PacBio_2016/data/) (27). In addition, we reassembled 42 strains from Xia et al. (41). The SRA accession numbers of the 42 strains can be found in Fig. 6A. De novo assembly was performed using SPAdes with four different k-mers (21, 33, 55, and 77) (42). In total, 73 S. paradoxus strains were collected. The 84 de novo genes from S. cerevisiae S288C were compared to the population data to identify their alleles. In particular, (i) a local BLASTN search was performed against population data using 84 de novo genes, (ii) hits were extracted from the population data, (iii) these extracted DNA sequences were aligned with de novo genes using MUSCLE (43), (iv) the alignments were manually checked based on reference de novo genes, (v) these refined alleles were translated into proteins using MEGA 7 (44), and (vi) stop codons were identified in the alignments of proteins.
Analysis of selection on de novo genes.
The 93 strains were grouped into three subpopulations based on their environmental origins (see Table S5 at http://baojunedisonwu.weebly.com/download.html), and 12 strains with no clear categories were removed for this analysis. The different proportions of intact ORFs for de novo genes between any subpopulations were determined by Fisher’s exact test (P < 0.05). We also used the program DnaSP v5 (45) to calculate the population genetic parameters and to estimate deviation from neutral expectations for the nonoverlapping regions of the de novo genes.
Reconstruction of ancestral state.
The 1,000 single-copy genes that are universally present in all examined 73 S. paradoxus strains and S. mikatae IFO1815 strains were used to construct phylogenetic relationships. Each gene was aligned individually using MUSCLE (43). The concatenated sequences of all gene alignments were used to reconstruct the phylogenetic relationship of S. paradoxus strains using the FastTree 2 program (46) under a general time-reversible (GTR) + Γ substitution model. The pattern of intact and disrupted ORFs was mapped on the phylogenetic tree of S. paradoxus population. Disrupted and intact ORFs at homologous sites were modeled as a two-state continuous-time Markov process, with states 0 and 1 on a phylogeny. The ancestral state for the de novo gene was then estimated using BayesTraits (47).
Data availability.
The reassembled genomes of 42 strains from Ontario are available upon request.
Footnotes
Citation Wu B, Knudson A. 2018. Tracing the de novo origin of protein-coding genes in yeast. mBio 9:e01024-18. https://doi.org/10.1128/mBio.01024-18.
REFERENCES
- 1.Chen S, Krinsky BH, Long M. 2013. New genes as drivers of phenotypic evolution. Nat Rev Genet 14:645–660. doi: 10.1038/nrg3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keese PK, Gibbs A. 1992. Origins of genes: “big bang” or continuous creation? Proc Natl Acad Sci U S A 89:9489–9493. doi: 10.1073/pnas.89.20.9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knowles DG, McLysaght A. 2009. Recent de novo origin of human protein-coding genes. Genome Res 19:1752–1759. doi: 10.1101/gr.095026.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerzoni D, McLysaght A. 2016. De novo genes arise at a slow but steady rate along the primate lineage and have been subject to incomplete lineage sorting. Genome Biol Evol 8:1222–1232. doi: 10.1093/gbe/evw074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacob F. 1977. Evolution and tinkering. Science 196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 6.Ohno S. 1970. Evolution by gene duplication. Springer, New York, NY. [Google Scholar]
- 7.Levine MT, Jones CD, Kern AD, Lindfors HA, Begun DJ. 2006. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proc Natl Acad Sci U S A 103:9935–9939. doi: 10.1073/pnas.0509809103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begun DJ, Lindfors HA, Thompson ME, Holloway AK. 2006. Recently evolved genes identified from Drosophila yakuba and D. erecta accessory gland expressed sequence tags. Genetics 172:1675–1681. doi: 10.1534/genetics.105.050336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao L, Saelao P, Jones CD, Begun DJ. 2014. Origin and spread of de novo genes in Drosophila melanogaster populations. Science 343:769–772. doi: 10.1126/science.1248286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donoghue MT, Keshavaiah C, Swamidatta SH, Spillane C. 2011. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol 11:47. doi: 10.1186/1471-2148-11-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carvunis AR, Rolland T, Wapinski I, Calderwood MA, Yildirim MA, Simonis N, Charloteaux B, Hidalgo CA, Barbette J, Santhanam B, Brar GA, Weissman JS, Regev A, Thierry-Mieg N, Cusick ME, Vidal M. 2012. Proto-genes and de novo gene birth. Nature 487:370–374. doi: 10.1038/nature11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McLysaght A, Guerzoni D. 2015. New genes from non-coding sequence: the role of de novo protein-coding genes in eukaryotic evolutionary innovation. Philos Trans R Soc Lond B Biol Sci 370:20140332. doi: 10.1098/rstb.2014.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmieri N, Kosiol C, Schlötterer C. 2014. The life cycle of Drosophila orphan genes. eLife 3:e01311. doi: 10.7554/eLife.01311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruiz-Orera J, Hernandez-Rodriguez J, Chiva C, Sabidó E, Kondova I, Bontrop R, Marqués-Bonet T, Albà MM. 2015. Origins of de novo genes in human and chimpanzee. PLoS Genet 11:e1005721. doi: 10.1371/journal.pgen.1005721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen S, Zhang YE, Long M. 2010. New genes in Drosophila quickly become essential. Science 330:1682–1685. doi: 10.1126/science.1196380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinhardt JA, Wanjiru BM, Brant AT, Saelao P, Begun DJ, Jones CD. 2013. De novo ORFs in Drosophila are important to organismal fitness and evolved rapidly from previously non-coding sequences. PLoS Genet 9:e1003860. doi: 10.1371/journal.pgen.1003860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toll-Riera M, Bosch N, Bellora N, Castelo R, Armengol L, Estivill X, Albà MM. 2009. Origin of primate orphan genes: a comparative genomics approach. Mol Biol Evol 26:603–612. doi: 10.1093/molbev/msn281. [DOI] [PubMed] [Google Scholar]
- 18.Suenaga Y, Islam SM, Alagu J, Kaneko Y, Kato M, Tanaka Y, Kawana H, Hossain S, Matsumoto D, Yamamoto M, Shoji W, Itami M, Shibata T, Nakamura Y, Ohira M, Haraguchi S, Takatori A, Nakagawara A. 2014. NCYM, a cis-antisense gene of MYCN, encodes a de novo evolved protein that inhibits GSK3beta resulting in the stabilization of MYCN in human neuroblastomas. PLoS Genet 10:e1003996. doi: 10.1371/journal.pgen.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chitasombat MN, Kofteridis DP, Jiang Y, Tarrand J, Lewis RE, Kontoyiannis DP. 2012. Rare opportunistic (non-Candida, non-Cryptococcus) yeast bloodstream infections in patients with cancer. J Infect 64:68–75. doi: 10.1016/j.jinf.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legras JL, Merdinoglu D, Cornuet JM, Karst F. 2007. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol 16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 21.Vakirlis NN, Hebert AS, Opulente DA, Achaz G, Hittinger CT, Fischer G, Coon JJ, Lafontaine I. 2018. A molecular portrait of de novo genes in yeasts. Mol Biol Evol 35:631–645. doi: 10.1093/molbev/msx315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu TC, Leu JY, Lin WC. 2017. A comprehensive analysis of transcript-supported de novo genes in Saccharomyces sensu stricto yeasts. Mol Biol Evol 34:2823–2838. doi: 10.1093/molbev/msx210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG. 2007. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics 23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- 24.Moyers BA, Zhang J. 2016. Evaluating phylostratigraphic evidence for widespread de novo gene birth in genome evolution. Mol Biol Evol 33:1245–1256. doi: 10.1093/molbev/msw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domazet-Lošo T, Carvunis AR, Albà MM, Šestak MS, Bakaric R, Neme R, Tautz D. 2017. No evidence for phylostratigraphic bias impacting inferences on patterns of gene emergence and evolution. Mol Biol Evol 34:843–856. doi: 10.1093/molbev/msw284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reference deleted.
- 27.Yue JX, Li J, Aigrain L, Hallin J, Persson K, Oliver K, Bergström A, Coupland P, Warringer J, Lagomarsino MC, Fischer G, Durbin R, Liti G. 2017. Contrasting evolutionary genome dynamics between domesticated and wild yeasts. Nat Genet 49:913–924. doi: 10.1038/ng.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gould GM, Paggi JM, Guo Y, Phizicky DV, Zinshteyn B, Wang ET, Gilbert WV, Gifford DK, Burge CB. 2016. Identification of new branch points and unconventional introns in Saccharomyces cerevisiae. RNA 22:1522–1534. doi: 10.1261/rna.057216.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman KB, Boeke JD. 1991. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell 65:483–492. doi: 10.1016/0092-8674(91)90466-C. [DOI] [PubMed] [Google Scholar]
- 30.Couvillion MT, Soto IC, Shipkovenska G, Churchman LS. 2016. Synchronized mitochondrial and cytosolic translation programs. Nature 533:499–503. doi: 10.1038/nature18015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neme R, Amador C, Yildirim B, McConnell E, Tautz D. 2017. Random sequences are an abundant source of bioactive RNAs or peptides. Nat Ecol Evol 1:0217. doi: 10.1038/s41559-017-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLysaght A, Hurst LD. 2016. Open questions in the study of de novo genes: what, how and why. Nat Rev Genet 17:567–578. doi: 10.1038/nrg.2016.78. [DOI] [PubMed] [Google Scholar]
- 33.Dekker J. 2007. GC- and AT-rich chromatin domains differ in conformation and histone modification status and are differentially modulated by Rpd3p. Genome Biol 8:R116. doi: 10.1186/gb-2007-8-6-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marín A, Gallardo M, Kato Y, Shirahige K, Gutiérrez G, Ohta K, Aguilera A. 2003. Relationship between G+C content, ORF-length and mRNA concentration in Saccharomyces cerevisiae. Yeast 20:703–711. doi: 10.1002/yea.992. [DOI] [PubMed] [Google Scholar]
- 35.Basile W, Sachenkova O, Light S, Elofsson A. 2017. High GC content causes orphan proteins to be intrinsically disordered. PLoS Comput Biol 13:e1005375. doi: 10.1371/journal.pcbi.1005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS, McCusker JH. 2015. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res 25:762–774. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hitz BC, Karra K, Krieger CJ, Miyasato SR, Nash RS, Park J, Skrzypek MS, Simison M, Weng S, Wong ED. 2012. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byrne KP, Wolfe KH. 2005. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramsköld D, Wang ET, Burge CB, Sandberg R. 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5:e1000598. doi: 10.1371/journal.pcbi.1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, Tsai IJ, Bergman CM, Bensasson D, O’Kelly MJ, van Oudenaarden A, Barton DB, Bailes E, Nguyen AN, Jones M, Quail MA, Goodhead I, Sims S, Smith F, Blomberg A, Durbin R, Louis EJ. 2009. Population genomics of domestic and wild yeasts. Nature 458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia W, Nielly-Thibault L, Charron G, Landry CR, Kasimer D, Anderson JB, Kohn LM. 2017. Population genomics reveals structure at the individual, host-tree scale and persistence of genotypic variants of the undomesticated yeast Saccharomyces paradoxus in a natural woodland. Mol Ecol 26:995–1007. doi: 10.1111/mec.13954. [DOI] [PubMed] [Google Scholar]
- 42.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 46.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pagel M, Meade A, Barker D. 2004. Bayesian estimation of ancestral character states on phylogenies. Syst Biol 53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The reassembled genomes of 42 strains from Ontario are available upon request.