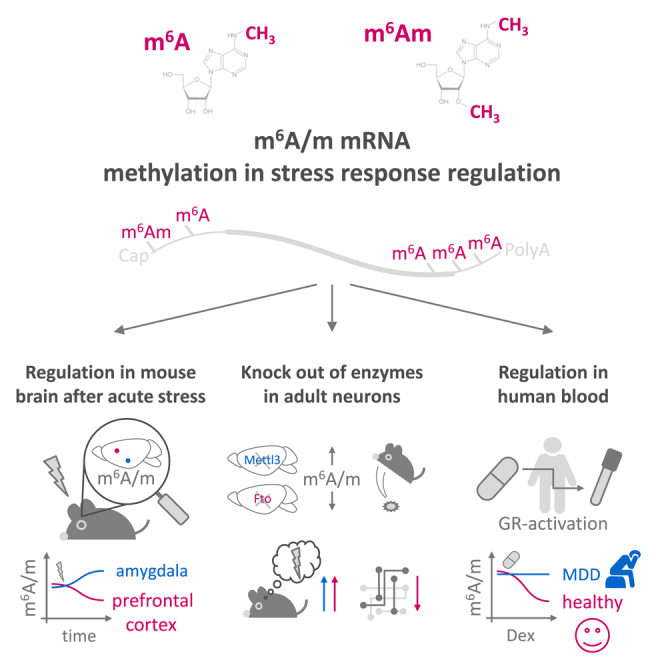
Summary
N6-methyladenosine (m6A) and N6,2′-O-dimethyladenosine (m6Am) are abundant mRNA modifications that regulate transcript processing and translation. The role of both, here termed m6A/m, in the stress response in the adult brain in vivo is currently unknown. Here, we provide a detailed analysis of the stress epitranscriptome using m6A/m-seq, global and gene-specific m6A/m measurements. We show that stress exposure and glucocorticoids region and time specifically alter m6A/m and its regulatory network. We demonstrate that deletion of the methyltransferase Mettl3 or the demethylase Fto in adult neurons alters the m6A/m epitranscriptome, increases fear memory, and changes the transcriptome response to fear and synaptic plasticity. Moreover, we report that regulation of m6A/m is impaired in major depressive disorder patients following glucocorticoid stimulation. Our findings indicate that brain m6A/m represents a novel layer of complexity in gene expression regulation after stress and that dysregulation of the m6A/m response may contribute to the pathophysiology of stress-related psychiatric disorders.
Keywords: m6A, m6Am, RNA modification, stress, Mettl3, Fto, post-transcriptional regulation, major depressive disorder
Graphical Abstract
Highlights
-
•
m6A/m mRNA methylation in the adult mouse brain is regulated by stress
-
•
m6A/m mRNA regulation is brain region, time, and gene specific
-
•
Mettl3 and Fto cKO alter m6A/m, fear memory, expression, and synaptic plasticity
-
•
The m6A/m glucocorticoid response is impaired in major depressive disorder patients
Engel et al. demonstrate a region- and time-dependent role of brain m6A/m methylation in stress-response regulation. Manipulating m6A/m alters fear memory, transcriptome response, and synaptic plasticity. Altered m6A/m dynamics in depressed patients suggest importance of m6A/m modifications for stress-related psychiatric disorders.
Introduction
Regulation of gene expression in response to stressful stimuli under healthy or pathological conditions involves epigenetic mechanisms such as DNA methylation and chromatin modifications (de Kloet et al., 2005, McEwen et al., 2015). Elucidating the underlying molecular processes that regulate the fine-tuned transcriptional response to stress is essential for understanding stress vulnerability and the development of stress-related psychiatric disorders such as depression and anxiety.
In analogy to DNA modifications, a diverse set of covalent modifications is present on RNA nucleotides encoding the epitranscriptome, post-transcriptionally shaping gene expression via regulation of RNA stability, translation, and non-coding transcript function (Zhao et al., 2017). The role of this newly emerging layer of gene expression control in the central stress response and behavior is not fully understood yet (Engel and Chen, 2018). RNA modifications, next to epigenetic mechanisms, likely represent a yet undescribed level of transcriptional regulation highly relevant for psychiatry.
N6-methyladenosine (m6A) is the most abundant internal mRNA modification, which is present transcriptome-wide in at least one-fourth of all RNAs, typically located in a consensus motif (DRACH/GGACU), and enriched near stop codons and in 5′ UTRs (Dominissini et al., 2012, Linder et al., 2015, Meyer et al., 2012). Recent studies have identified mammalian m6A to be dynamically regulated, controlling stem cell proliferation and differentiation (Klungland et al., 2016), cellular heat-shock response (Zhou et al., 2015), DNA damage response (Xiang et al., 2017), and tumorigenesis (Cui et al., 2017). Brain RNA methylation is comparably high and increases during development (Meyer et al., 2012).
m6A is deposited co-transcriptionally (Ke et al., 2017, Slobodin et al., 2017) by a methyltransferase complex consisting of METTL3, METTL14 (Liu et al., 2014), WTAP (Ping et al., 2014), KIAA1429 (VIR; Schwartz et al., 2014), and RBM15/RBM15B (Patil et al., 2016). In contrast, it can be removed by the demethylases FTO (Jia et al., 2011, Mauer et al., 2017) and ALKBH5 (Zheng et al., 2013). FTO further catalyzes the demethylation of N6,2′-O-dimethyladenosine (m6Am) with an in vitro preference for this substrate (Mauer et al., 2017). m6Am is found at the first nucleotide adjacent to the 7-methylguanosine cap, promoting transcript stability (Mauer et al., 2017). Fto has been associated with memory consolidation (Walters et al., 2017, Widagdo et al., 2016) and was implicated in regulation of dopaminergic brain networks (Hess et al., 2013). The most commonly used m6A/m antibody, used also in most experiments presented here, co-detects m6A and m6Am (Linder et al., 2015), potentially preventing clear discrimination between them. Therefore, data will be treated as potentially containing both and called m6A/m unless otherwise stated.
In general, m6A/m-regulating enzymes may be expressed at different levels in different cell types and have distinct intracellular distributions and binding motifs and thus potentially affect different subsets of target RNAs. Cellular consequences of m6A/m modifications depend on the binding of m6A/m-reader proteins (such as YTH and HNRNP proteins) and include RNA maturation, splicing, alternative polyadenylation, RNA decay, and both promotion and inhibition of protein translation (reviewed in Peer et al., 2017, Roundtree et al., 2017).
In this study, we aimed to elucidate the role of m6A/m in the context of the brain’s stress response. We delineated the effects of acute stress on m6A/m using global m6A/m measurements, m6A/m sequencing (m6A/m-seq), and absolute quantification of transcript-specific methylation levels. In addition, we explored the functional significance of m6A/m in the adult brain by examining conditional knockout (cKO) mice for Mettl3 and Fto. Finally, we investigated m6A/m regulation in blood samples of mice and humans to determine its potential as a peripheral indicator of the central response to stress and stress-linked psychiatric disorders.
Results
The Stress-Induced m6A/m Epitranscriptome
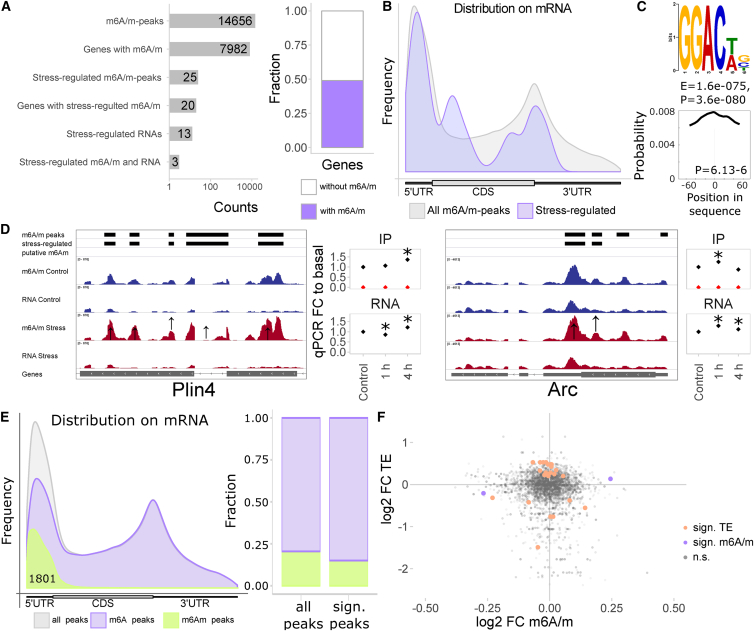
To test whether acute stress alters m6A/m, we performed m6A/m-seq (RNA-seq after immunoprecipitation) on mouse cortex poly(A)-RNA 4 hr following 15 min of acute restraint stress exposure (n = 6–7). Using more specific areas, and thus lower amounts of input material, was not sufficient for consistent, quantifiable poly(A)-m6A/m-seq. The peaks enriched in the m6A/m-RIP (RNA immunoprecipitation)-seq over the input-RNA-seq in the two different conditions were very similar (Figures S1A and S1B). We analyzed differential methylation across an m6A/m consensus peak set with 14,656 high-confidence m6A/m peaks (supported by either 2/3 samples per group or 1/2 of all samples and additional abundance filters) mapping to 7,982 genes (Figure 1A; Table S1). Thus, around half of the expressed genes in the mouse cortex are m6A/m methylated with each around 2 peaks per gene (Figure 1A). m6A/m peaks overlapped majorly with previously reported m6A/m peaks (85% overlap with RMBase 2; Xuan et al., 2018), preferentially located to the 5′ UTR and around the stop codon (Figure 1B), and contained the m6A consensus motif with the top motif being a centrally enriched GGACWB (Figure 1C). m6A/m methylation in the cortex is overrepresented in genes involved in synaptic and neuronal regulation (Figure S1C).
Figure 1.
Mapping the Transcriptome-wide m6A/m Landscape after Acute Restraint Stress in the Mouse Cortex Using m6A/m-Seq
FC, fold change; TE, translation efficiency.
(A) Approximately half of the genes expressed in the mouse cortex are m6A/m methylated, but only a minor fraction is regulated by acute stress on cortex-wide level. m6A/m-seq of mouse cortex poly(A)-RNA basal or 4 hr after 15 min restraint stress; n = 6–7, each pooled from 3 mice. Stress-regulated m6A/m peaks, Q < 0.1 and absolute log2 fold change > 0.2; stress-regulated mRNAs ( = differential expressed genes), Q < 0.1 and absolute log2 fold change > 0.1.
(B) m6A/m peaks are enriched at the 5′ UTR and the stop codon with similar distribution of all and stress-regulated peaks (peak distribution mapped along mRNA relative position).
(C) GGACWB is the most abundant motif detected in m6A/m peaks and enriched at peak summits. Top enriched sequence motif and its position across the detected m6A/m peaks. GGAC was detected in 84%, GGACWB in 63% of the peaks.
(D) Two examples of stress-regulated m6A/m peaks and replication of their quantitative regulation by m6A/m-RNA immunoprecipitation (RIP)-qPCR in an unrelated cohort of animals. Left panel per gene: averaged sequence tracks and peaks; arrows indicate quantitatively regulated peaks. Right panel per gene: differential methylation was validated in a separate cohort of mice using full-length m6A/m-RIP-qPCR, including an intermediated time point (1 hr). n = 7, mean ± SEM; asterisks [∗] depict omnibus Tukey post hoc tests to basal p < 0.05 after FDR-corrected one-way ANOVA.
(E) Bioinformatic dissection of m6Am peaks based on their position at the transcription start site, observing 1,801 putative m6Am sites. m6Am peaks do not show a preference for stress-regulated peaks.
(F) Regulation of translation efficiency (TE assessed by ribosome profiling) by stress does not correlate well or overlap with regulation of m6A/m methylation. n = 6 for ribosome profiling; n = 6–7 for m6A/m profiling. Shown are fold changes upon stress using only genes abundantly detected in ribosome profiling sequencing with significance determined by Q < 0.1 and absolute log2 fold change > 0.2.
See also Figures S1 and S2 and Table S1.
Only 25 m6A/m peaks (in 20 different genes) and 13 genes were found to be significantly regulated 4 hr after stress, but all with very low fold changes (at absolute log2 fold change > 0.2 for m6A, > 0.1 for RNA, and Q < 0.1; Figures 1A and S1D–S1G; two examples including validation by m6A-RIP-qPCR are shown in Figure 1D), potentially reflecting the cellular heterogeneity of the input material used diluting the cell-specific effects of stress. RNAs and m6A/m peaks significantly regulated by stress showed only low overlap (three genes) and no prominent correlation of m6A/m and gene expression regulation by stress (Figure S1E).
To investigate if m6A and m6Am may have different effects after stress, we in silico dissected m6A and m6Am peaks based on the assumption that m6Am occurred at the first nucleotide after the transcription start site (similar to strategies employed earlier by Linder et al., 2015 and Mauer et al., 2017). We observed 1,801 putative m6Am peaks (12%; Figures 1E and S2A) with highest gene ontology enrichment in developmental genes and genes related to DNA and RNA rather than neuronal genes (Figure S2B) and no enrichment of a GGAC motif (data not shown). Putative m6Am peaks were not overrepresented in stress-regulated peaks (data not shown), and had similar stress regulation like all peaks (Figure S2C) and similar absence of correlation to stress regulation of gene expression (Figure S2C), overall not indicating a special role of m6Am in the stress response. Further, in order to assess potential regulation of transcript translation by stress-regulated m6A/m, we performed ribosome profiling on mouse cortex 4 hr after stress. Although there were several genes with regulated translation efficiency after stress (24 genes at Q < 0.1, absolute log2 fold changes > 0.5), none overlapped with stress-regulated m6A/m and there was also no apparent relation to stress regulation of m6A/m (Figure 1F). Finally, searching for potential binding factors for m6A/m, we analyzed the co-occurrence of the overserved m6A motif GGACWB to known binding motifs of RNA-binding proteins in the m6A/m-seq fragments, observing a high similarity and summit enrichment to the binding motifs of FMRP/FMR1 and FXR2, proteins crucial for translation regulation, RNA translocation, and synaptic plasticity in neurons (Figure S2D). Likewise, genes reported to be bound by mouse FMRP were also higher than likely m6A/m methylated (Figure S2E), suggesting that m6A/m methylation of neuronal RNAs may regulate protein binding critical for neuronal transport and plasticity.
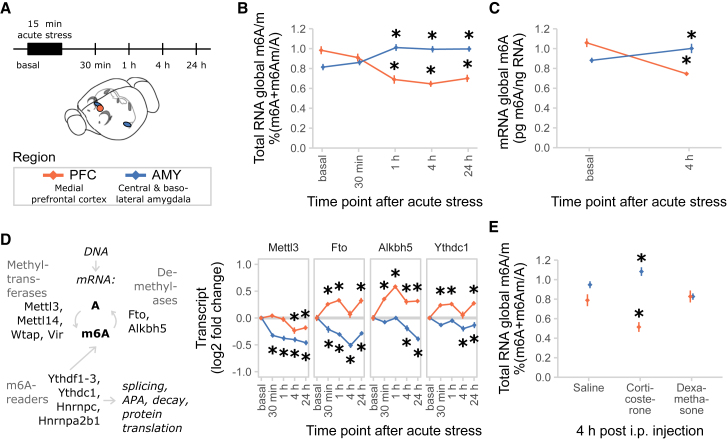
Stress Regulation of m6A/m Is Brain Region Specific
Based on both the number of significantly stress-regulated m6A/m peaks and their respective fold changes in m6A/m-seq being very small, we reasoned that the true extent of the m6A/m stress response may only be revealed when investigating more defined brain regions. Therefore, we measured the time course of RNA methylation changes in two regions highly involved in stress response regulation: the medial prefrontal cortex (PFC) and the basolateral and central amygdala (AMY; Figure 2A). We found that global m6A/m was regulated in total RNA in a region-dependent manner with RNA methylation decreased in the PFC and increased in the AMY (Figure 2B). The same regulation was observed when only m6A was measured in mRNA using liquid chromatography-tandem mass spectrometry (LC-MS/MS) (Figure 2C), arguing for m6A as the main driver of the observed effects. Examining changes of the m6A/m machinery related to these global changes, we measured gene expression levels of m6A/m enzymes and binding proteins. We found the demethylases Fto and Alkbh5 to be differentially regulated in a region-specific manner, facilitating the effects observed on global methylation, in most cases preceding the effect observed on global m6A/m (Figure 2D). Furthermore, Mettl3 was downregulated upon stress exposure tissue independently (Figure 2D) and Wtap was regulated isoform specifically only in the AMY (Figure S3A). The m6A/m reader Ythdc1 was regulated in a region-specific manner (Figure 2D), whereas the other known enzymes and readers were not differentially expressed (Figure S3A).
Figure 2.
Acute Restraint Stress Regulates Brain Global m6A/m and Expression of the m6A/m Regulatory System in a Time- and Region-Specific Manner
(A) Experimental design. PFC, medial prefrontal cortex (orange); AMY, central and basolateral amygdala (blue).
(B) Global m6A/m is decreased in the PFC and increased in the AMY after acute restraint stress. Global m6A/m assay on total RNA, n = 12, mean ± SEM; two-way ANOVA interaction effect F(4, 110) = 24.045, p < 0.001. Asterisk (∗) depicts omnibus Tukey post hoc tests to basal p < 0.05. Results were replicated in three independent mouse cohorts with only one experiment shown.
(C) Likewise, global mRNA m6A is decreased when measured with LC-MS/MS. n = 7, mean ± SEM. Specific measurement of only m6A. Two-way ANOVA interaction effect F(1, 24) = 159.537, p < 0.001. Asterisk (∗) depicts omnibus Tukey post hoc tests to basal p < 0.05.
(D) m6A/m regulatory genes Mettl3, Fto, Alkbh5, and Ythdc1 are differentially expressed after acute stress in the brain. See also Figure S3. n = 12, log2 fold change ± SEM; two-way MANOVA, significant interaction effects for Fto, Alkbh5, and Ythdc1; main stress effect for Mettl3; each FDR-corrected p < 0.05 and n2 > 0.01. Asterisk (∗) depicts omnibus Tukey post hoc tests to basal p < 0.05. See also Table S2. Results were replicated in three independent mouse cohorts with only one experiment shown.
(E) Global m6A/m is decreased in the PFC and increased in the AMY after corticosterone i.p. injection, but not after dexamethasone injection. Corticosterone, 250 μg/ kg; dexamethasone, 10 mg/kg. Global m6A/m assay on total RNA, n = 12, mean ± SEM. Two-way ANOVA reported a significant interaction effect (F(4, 96) = 12.887, p < 0.001). Asterisk (∗) indicates omnibus Tukey post hoc tests p < 0.05 compared to area basal.
See also Figure S3.
Notably, intraperitoneal (i.p.) injection of the endogenous glucocorticoid corticosterone, but not the glucocorticoid receptor agonist dexamethasone, changed global m6A/m (Figure 2E), as well as Fto and Alkbh5 expression (Figure S3B), similarly to acute stress (Figure 2D), demonstrating that the stress effect may be mediated by endogenous glucocorticoids (GCs). Supporting this idea, we found that the majority of m6A/m enzyme and reader genes contain several GC response elements in their 5′ upstream region, likewise pointing at expression regulation of those genes via GCs (Figure S3C).
Stress Regulation of m6A/m Is Gene Specific
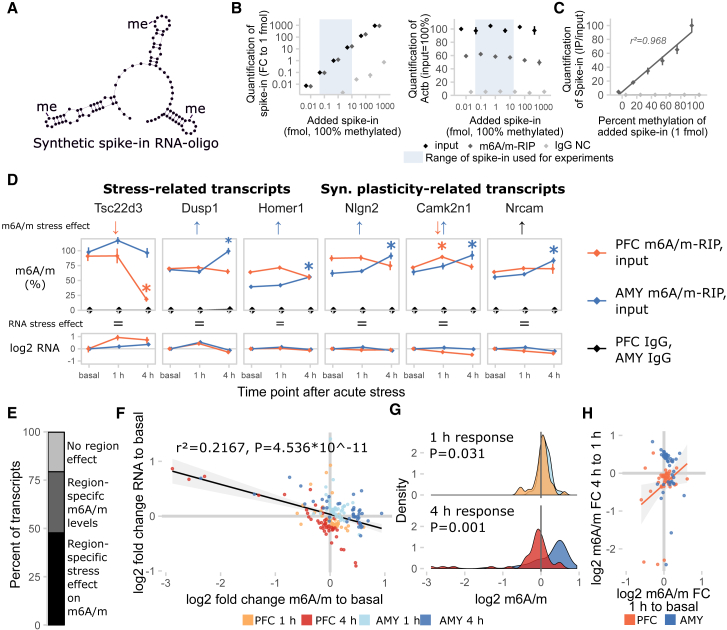
m6A/m-seq not only requires large amounts of input material but also does not quantify absolute transcript methylation. Therefore, we performed m6A/m-RIP followed by qPCR to assess absolute levels of candidate transcript methylation in narrowly defined brain areas, before and after stressful challenge. For calibration of the assay and normalization of immunoprecipitation efficiency in experiments, we designed and used an m6A/m-methylated internal spike-in RNA oligonucleotide (Figures 3A, S4A, and S4B). The m6A/m-RIP-qPCR detected m6A/m methylated RNA spike-in across a wide range of concentrations with low IgG background signal and without competing with the immunoprecipitation of endogenously methylated RNAs (Figure 3B). Using mixtures of unmethylated and methylated spike-in oligonucleotides, we confirmed that m6A/m-RIP-qPCR measured different methylation states of RNAs with high precision (Figure 3C; r2 > 0.95).
Figure 3.
Absolute Regulation of m6A/m Methylation Is Site Specific
(A) A synthetic RNA oligonucleotide with three internal m6A/m sites was used for validation and internal normalization of the m6A/m-RIP-qPCR. See also Figure S4.
(B) m6A/m-RIP-qPCR detects the methylated spike-in oligonucleotide in a linear fashion without impairing precipitation efficiency for endogenous transcripts in the concentration range used for experiments. Methylated spike-in oligo was added to unfragmented total RNA and precipitated with anti-m6A/m antibody (m6A/m-RIP) or rabbit IgG (IgG NC). n = 3 technical replicates, normalized expression to 1 fmol input control. Mean ± SEM.
(C) m6A/m-RIP-qPCR accurately quantifies differential methylation of the spike-in oligo. Spike-in oligo (1 fmol) mixed from fully methylated and fully unmethylated spike-in was added to unfragmented total RNA and precipitated with m6A/m-RIP-qPCR. n = 3 technical replicates, normalized to input control. Mean ± SEM.
(D) Absolute full-length m6A/m levels of stress-related and synaptic plasticity-related transcripts are differentially regulated in the PFC and AMY of stress-related candidate transcripts and synaptic-plasticity-related candidate transcripts after stress. See also Figure S4. n = 8, mean ± SEM. Significant effects observed in FDR-corrected two-way MANOVA (p < 0.05, n2 > 0.01) are coded in the rows “m6A/m stress effect” and “RNA stress effect.” Orange/blue arrows, PFC-/AMY-specific stress effect (interaction effect two-way ANOVA, one-way follow-up significant in respective tissue); black arrow, stress main effect; equals sign, no interaction or stress main effect in two-way ANOVA. See also Table S2.
(E) The majority of transcripts measured are expressed or regulated in a region-specific manner. Percent of transcripts with significant interaction or main effect in FDR-corrected 2×2 MANOVA.
(F) Stress regulation of m6A/m negatively correlates with changes in RNA levels. log2 fold changes of m6A/m and RNA after stress to basal time points, n = 44 per group; black line, linear model + 95% CI. For generalized linear models (GLMs), see Table S2.
(G) General patterns of m6A/m changes vary in extent and direction depending on brain region and time point. Density plots of data depicted in (D); t test.
(H) The m6A/m change at the 1 hr time point correlates with the m6A/m change at 4 hr in the PFC, but not AMY, indicating that in the PFC, m6A/m change 1 hr after stress is a proxy for later change. Orange line, linear model for PFC only + 95% CI. For GLMs, see Table S2.
See also Figure S4.
Applying m6A/m-RIP-qPCR, we measured absolute methylation levels of several candidate transcripts involved in the brain’s stress response and, given the enrichment of neuronal plasticity and morphogenesis-related terms in the m6A/m-seq, synaptic plasticity-related transcripts (Figures 3D and S4C). Regulation of m6A/m by stress (26/44 transcripts) was observed more often than regulation of RNA (16/44 transcripts, with 12 overlapping) in the transcripts tested. Notably, the majority of chosen candidates were either regulated or expressed in a region-specific manner, emphasizing the importance of assessing RNA methylation in defined brain areas (Figure 3E). Interestingly, in contrast to the m6A/m-seq, absolute transcript methylation levels m6A/m and RNA fold changes negatively correlated, arguing for increased m6A/m levels correlating with mRNA decay as previously shown in vitro (Figure 3F; Table S2; with no influence of region and time point). In detail, both PFC and AMY exhibited differential response at 1 and 4 hr with opposite directions, paralleling the regulation observed in global m6A/m in the respective regions above (Figure 2B). Overall, 4 hr fold changes had higher effect sizes compared to 1 hr fold changes (Figure 3G). Fold changes at the 1 hr time point correlated with those at 4 hr for the same gene in the PFC, but not in the AMY, indicating that in the PFC 1 hr m6A/m may be an intermediate state of 4 hr regulation with fold changes of regulated m6A/m increasing with time. In contrast, in the AMY for the candidate genes investigated, m6A/m regulation after 1 and 4 hr was more independent (Figure 3H).
Epitranscriptomic Changes in Mice with Conditional Deletion of Mettl3 or Fto from Adult Neurons
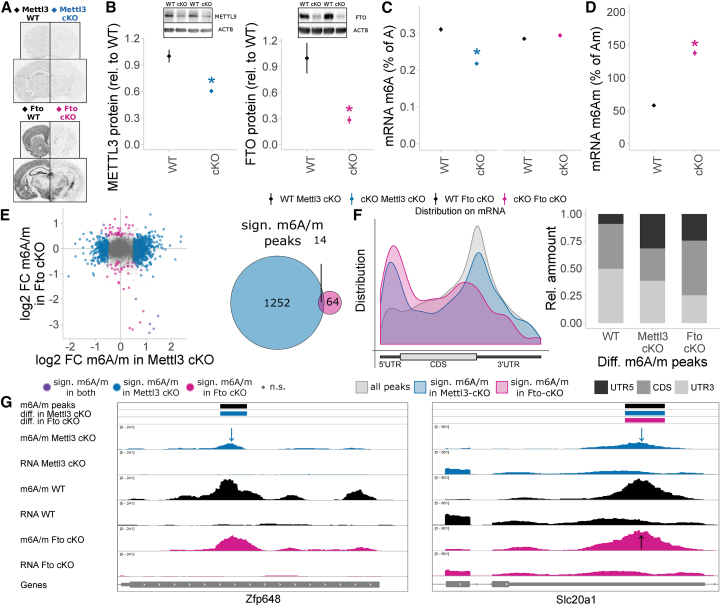
Since the expression of the m6A methyltransferase Mettl3 and the m6A/m demethylase Fto was affected by acute stress, we generated cKO mouse models lacking these genes specifically in adult excitatory neurons employing Mettl3 or Fto flox/flox mice bred to Cre-driver lines. First, to measure the regulation of the epitranscriptome in these mice, we used the Camk2a-Cre driver, which induces gene deletion in excitatory neurons of neocortex and hippocampus (Minichiello et al., 1999) starting 2–3 weeks postnatal (Refojo et al., 2011), leading to broad reductions of both Mettl3 and Fto mRNA and METTL3 and FTO protein in the adult brain (Figures 4A, 4B, and S5A). Whereas global m6A measured by LC-MS/MS was decreased in cortical mRNA of Mettl3 cKOs (compared to their respective Mettl3 wild-type [WT] littermates), conditional deletion of Fto did not alter m6A (Figures 4C and S5B). However, using an LC-MS/MS mRNA preparation including a cap-digest similar to previously published protocols (Mauer et al., 2017), we found that m6Am is increased in Fto cKO (Figure 4D; significantly increased both relative to Am or A with no change in Am; data not shown). These data confirm FTO primarily targeting m6Am in the adult brain in vivo (Mauer et al., 2017). m6Am and Am were below quantification threshold in all of the Mettl3 cKOs, but not Mettl3 WT animals (data not shown), potentially indicating an effect of METTL3 depletion on those nucleosides that should be confirmed with a more sensitive method. Absolute abundancies measured by LC-MS/MS in cortical mRNA were 0.304% for m6A/A, 0.022% for m6Am/A, and 0.071% for m6Am/m6A. m1A could not be detected in sufficient amounts for quantification in any of the samples (data not shown).
Figure 4.
Depletion of METTL3 and FTO in Adult Excitatory Neurons Using the Camk2a-Cre Driver Changes the Cortex Epitranscriptome
(A) Mettl3 and Fto mRNA are depleted from the neocortex and hippocampus in Mettl3 cKO and Fto cKO mice, respectively. In situ hybridization, n = 3, representative shown. WT, wild-type; cKO, conditional.
(B) METTL3 and FTO proteins are significantly depleted in Mettl3 cKO and Fto cKO mice, respectively. Western blot of PFC protein, optical density normalized from digitally acquired image signal normalized to ACTB protein. n = 4–5, mean ± SEM. ∗p < 0.05, t test. For full blots, see Figure S5.
(C) Global mRNA m6A is decreased in Mettl3 cKO mice, but not in Fto cKO mice, when measured with LC-MS/MS. n = 5, mean ± SEM, m6A-specific measurement. Two-way ANOVA interaction effect F(1, 19) = 106.269, p < 0.001. ∗p < 0.05, omnibus Tukey post hoc tests to respective WT. See also Figure S5.
(D) Global mRNA m6Am is increased in Fto cKO mice when measured with LC-MS/MS. n = 5, mean ± SEM, m6Am-specific measurement. Data are shown relative to Am, which is not altered in Fto cKO mice. ∗p < 0.05, t test. For LC-MS/MS traces, see Figure S5.
(E) The m6A/m epitranscriptome is widely altered in in Mettl3 cKO and Fto cKO mice. m6A/m-seq on mouse cortex poly(A)-RNA of WT and cKO animals reported 1,266 and 78 significantly different methylated m6A/m peaks in Mettl3 cKO and in Fto cKO compared to WT, respectively, with 14 shared peaks. n = 3–5, each pooled from 3 mice. WT of both lines were grouped together as we observed no major regulation between them. Shown are log2 fold changes of methylation in cKO relative to WT mice using 10,109 high-confidence consensus m6A/m peaks detected across all groups, mapping to 6,056 unique genes. Significantly regulated m6A/m peaks are Q < 0.1 and absolute log2 fold change > 0.5.
(F) m6A/m peaks are enriched at the stop codon with a less prominent enrichment at the 5′ UTR, as observed in Figure 1. Differentially methylated peaks in both Mettl3 cKO and Fto cKO mice show an increased preference for 5′ UTR position with a decreased preference for CDS peaks in Mettl3 cKO differential peaks. Peak distribution mapped along mRNA relative position.
(G) Two examples m6A/m peaks regulated only in Mettl3 cKO or in both Mettl3 cKO and Fto cKO. Shown are averaged sequence tracks m6A/m-seq and RNA-seq per group and detected m6A/m peaks. Arrows indicate quantitatively regulated peaks (Q < 0.1, absolute log2 fold change >0.5).
We next profiled m6A/m in Mettl3 cKO and Fto cKO mice using m6A/m-seq on cortical poly(A)-RNA. Overall, m6A/m peaks detected in the single groups were still fairly similar (Figures S5C and S5D), with 80% overlap with the m6A/m dataset generated after acute stress (Figures S5D and S5E; mainly lacking 5′ UTR peaks potentially due to use of a different antibody lot). Quantitative analysis of consensus peaks revealed majorly altered epitranscriptomes in both mouse lines (Figures 4E and 4G; Table S3), with much higher numbers of consensus m6A/m peaks quantitatively altered in Mettl3 cKO compared to WT (1,266) compared to Fto cKO compared to WT (78; both Q < 0.1 and absolute log2 fold change > 0.5), and only a small number shared differentially methylated sites (Figure S5F). Although several RNAs are differentially expressed in Metltl3 cKOs or Fto cKOs, they only minorly overlapped with the regulated m6A/m peaks in the respective line (Figure S5F). Peaks differentially methylated in Mettl3 cKOs and Fto cKOs both showed higher enrichment at the 5′ UTR compared to all measured peaks (Figure 4F). Interestingly, Fto cKO differential peaks do not only localize to the 5′ UTR, as would be expected from m6Am sites only, but also to internal sites, arguing for Fto deletion also affecting internal m6A sites. Functionally, while m6A/m peaks are enriched in genes related to (mature) synapse and neuronal function, Mettl3 differential m6A peaks are more abundant in genes with neuronal and tissue-developmental functions (Figure S5G).
Stress-Coping Behavior Is Altered in Mice Deficient in Mettl3 or Fto
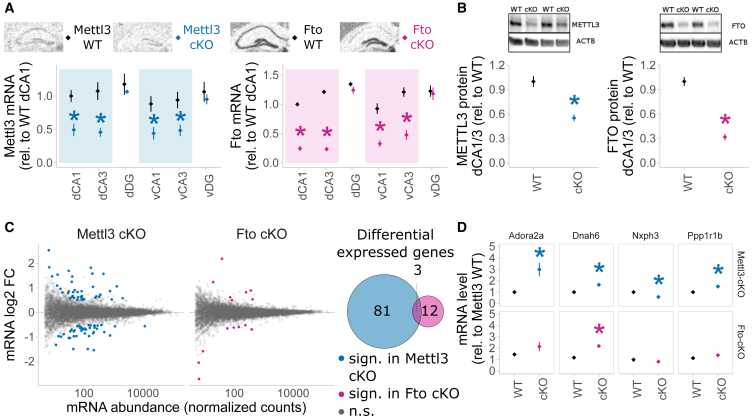
To assess behavioral and electrophysiological consequences of Mettl3 and Fto deletion in vivo, we created cKO mice with a more defined gene deletion by breeding Mettl3 or Fto flox/flox mice to Nex-CreERT2 mice in which additionally the gene deletion can be timely controlled by tamoxifen (Agarwal et al., 2012; Mettl3 cKO and Fto cKO). Upon induction in young adults, Mettl3 and Fto mRNA were depleted from both dorsal and ventral parts of the hippocampus, specifically in CA1 and CA3, but not in the dentate gyrus (Figure 5A). METTL3 and FTO proteins were significantly reduced in dCA1/dCA3 in Mettl3 cKO and Fto cKO mice, respectively (Figures 5B and S6A). Nex-CreERT2-induced recombination is further known to occur in small populations of principal neurons in the cortex (Agarwal et al., 2012). Depletion of either gene did not result in compensatory changes of gene expression of other genes involved in m6A/m metabolism (Figure S6B) but altered transcriptome profiles as observed by mRNA-seq of CA1 and CA3 tissue (Figure 5C). Interestingly, in non-stressed basal animals, we observed a larger number of differentially expressed genes in Mettl3 cKOs compared to Fto cKOs (Figures 5B and 5C; Mettl3 cKOs, 84 differentially expressed genes; Fto cKOs, 15 differentially expressed genes with Q < 0.1 and an absolute fold change above log2 = 0.5; Table S4), with no apparent preference for up- or downregulation. Although there was only small overlap of differentially expressed genes between the two lines, 104 genes were differentially expressed in a knockout-specific pattern (Figure 5C), including genes regulating neuronal activity and synaptic function (examples shown Figure 5D).
Figure 5.
Deletion of Mettl3 or Fto in Adult Excitatory Neurons of the Hippocampus CA1 and CA3 via a Nex-CreERT2 Driver Line and Knockout Induction in Adult Animals via Tamoxifen Administration Alters Gene Expression in Animals
(A) Mettl3 and Fto mRNA are depleted from the dorsal (d) and ventral (v) hippocampus CA1 and CA3 in Mettl3 cKO (blue) and Fto cKO (pink) mice, respectively. WT, wild-type; cKO, conditional knockout; DG, dentate gyrus. In situ hybridization; expression was quantified from digitalized films in arbitrary units (AU); mean ± SEM, n = 4 for Mettl3 WT and cKO, n = 11–14 for Fto WT and cKO, signal averaged across both hemispheres; ∗p < 0.05, t test.
(B) METTL3 and FTO proteins are significantly depleted in Mettl3 cKO and Fto cKO mice, respectively. Protein was isolated from dissected dCA1/dCA3 and measured by western blot normalized to ACTB protein. n = 3–4, optical density normalized from digitally acquired images, mean ± SEM. ∗p < 0.05, t test. For full blots, see Figure S6.
(C) mRNA-seq of adult CA1 and CA3 shows altered gene expression after deletion of Mettl3 and Fto in non-stressed basal animals. More genes are differentially expressed after deletion of Mettl3 (84) compared to deletion of Fto (15), with very few overlapping (3). log2 change by DESeq2 baseMean gene abundance from RNA-seq of adult basal animals. Differentially expressed by colored dots and in Venn circles, Q < 0.1, log2 fold change > 0.5.
(D) Four representative examples of genes expressed in a knockout × genotype-specific pattern. In total, 104 genes were found to be expressed in a knockout × genotype interaction-dependent matter. Normalized counts relative to Mettl3 WT. n = 5.
See also Figures S6 and S7 and Table S4.
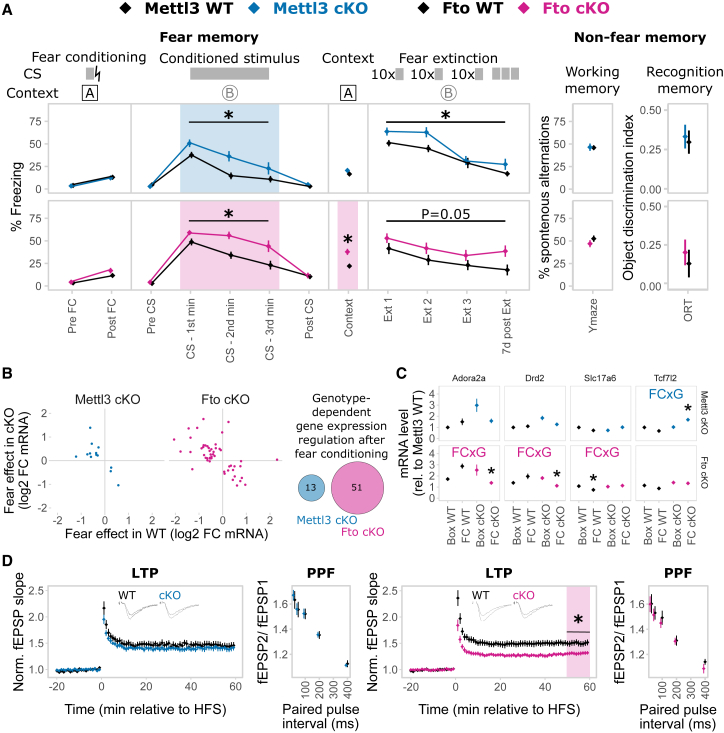
Neither Mettl3 cKO nor Fto cKO mice showed altered anxiety-like behavior or locomotion (Figure S7A), but we observed significant changes in spontaneous digging behavior (Figure S7A). Both knockout mice exhibited increased cued fear memory long-term maintained during memory extinction (Figure 6A) as well as contextual fear memory in Fto cKO mice (Figure 6A), but no differences in non-fear-related memory or short-term working memory (Figure 6A). Next, we investigated the transcriptional response patterns 24 hr after fear conditioning stress; thus, at the time point we observed the altered memory, comparing fear conditioned animals (“FC”) to control animals that experienced the same handling but no foot shock (“Box”). For both Mettl3 cKOs and Fto cKOs, we observed a large number of genes differentially expressed after fear conditioning in a genotype-dependent manner, implying a widely altered transcriptional response pattern after stress in animals with disturbed m6A/m system (Figure 6B) involving genes crucial for neuronal systems like neurotransmitter receptors and transporters as well as transcription factors (Figure 6C). Thereby, significant gene expression regulation was more extended in fear-conditioned animals compared to non-fear-conditioned animals (Figure S7B; Table S4). In contrast to basal animals, Fto cKOs showed more genotype-dependent expression changes after the stressful fear conditioning event than Mettl3 cKOs (Figures 6B and S7B; Table S4), implying that Fto is crucial for the regulation of the fear response despite minor basal changes in gene expression.
Figure 6.
Animals with Adult Excitatory Neuron-Specific Depletion of Mettl3 and Fto Using a Nex-CreERT2 Driver Line Have Impaired Fear Coping, Differential Transcriptomic Response to Fear, and Changes in Hippocampus CA1 Electrophysiological Properties
(A) Both Mettl3 cKO and Fto cKO animals display increased conditioned fear memory long-term maintained during fear extinction. The primary fear response was not altered. Fto cKO animals also have increased contextual fear memory. No difference was observed in the Y-maze test or the object recognition test (ORT). CS, conditioned stimulus; lightning bolt, US, unconditioned stimulus; Ext, extinction. n = 11–13, mean ± SEM. Fear expression was binned in 1 min intervals during CS representation. Asterisk (∗) depicts a main genotype effect in repeated-measurements ANOVA for CS and Ext bins and a t test p < 0.05 for all other data points.
(B) The transcriptomic response 24 hr after fear conditioning (FC) is altered in both animals with Mettl3 or Fto depletion. log2 RNA fold change in WT versus cKO animals of only those genes with a significant genotype × FC effect. Q < 0.1, absolute log2 fold change > 0.5, n = 5.
(C) More genes express a genotype-dependent FC effect in Fto cKOs compared to Mettl3 cKOs with low overlap. Four examples of such genes are shown. Significant genotype × FC in the examples is depicted by blue (Mettl3 cKOs) and pink (Fto cKOs) opposite arrows. Q < 0.1, absolute log2 fold change > 0.5, n = 5.
(D) Long-term potentiation (LTP), but not short-term plasticity, in CA1 was attenuated in Fto cKO mice, but not Mettl3 cKO mice. Short-term synaptic plasticity was measured by paired-pulse facilitation (PPF). n = 10–12 slices from 5–6 animals, mean ± SEM plus representative LTP trace curves; HFS, high-frequency stimulation. ∗p < 0.05, t test, on the average field excitatory postsynaptic potential (fEPSP) slope 50–60 min post-HFS.
See also Figures S6 and S7 and Table S4.
Consequently, investigating the effects of Fto and Mettl3 depletion on electrophysiological correlates of network plasticity and brain function, we found that CA1 long-term potentiation was impaired in Fto cKO, but not in Mettl3 cKO, mice (Figure 6D), with no effect on paired-pulse facilitation (Figure 6D) or basal neurotransmission (Figure S7C).
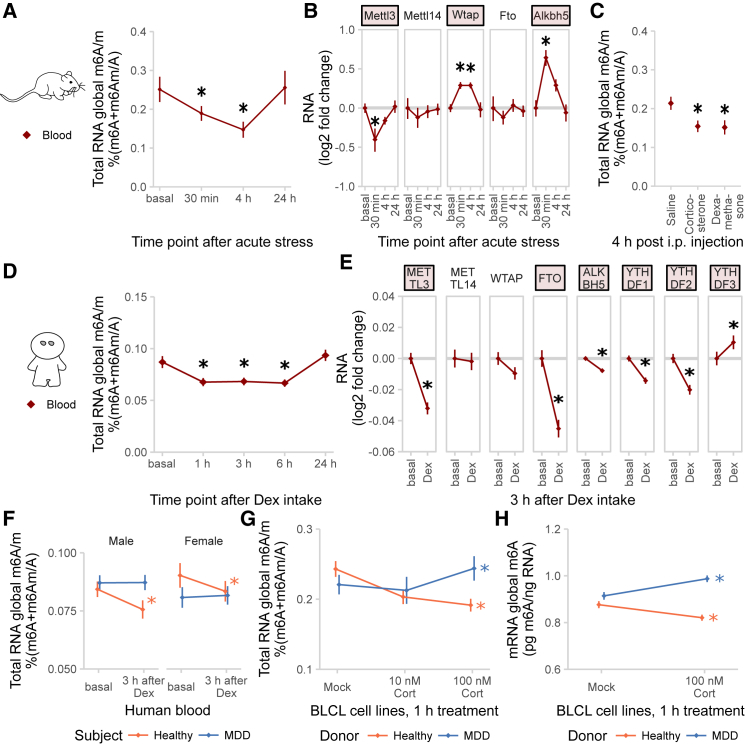
Regulation of m6A/m Is Impaired in MDD Patient Blood
To evaluate the potential of blood m6A/m as a peripheral proxy of the central m6A/m stress response, we measured global m6A/m methylation levels in mouse and human blood after an acute stressful challenge and GC stimulation. Global methylation was transiently decreased in whole blood of mice after acute stress (Figure 7A), with gene expression of Mettl3 and Alkbh5 altered in accordance with the global m6A/m change and Wtap being upregulated (Figure 7B). Similarly, global m6A/m was decreased in mouse blood 4 hr after i.p. injections of both corticosterone and dexamethasone (Figure 7C). Comparably, blood from healthy human volunteers, drawn before and after intake of 1.5 mg dexamethasone, showed both reduced global m6A/m levels (Figure 7D) and changes in gene expression of the m6A/m machinery enzymes 3 hr after dexamethasone intake (Figure 7E) (expression data from Arloth et al., 2015).
Figure 7.
Global m6A/m in Blood Is Transiently Decreased after Stress in Mice and after Stimulation with GCs in Healthy Humans, but This Glucocorticoid-Induced m6A/m Reduction Is Absent in Blood and BLCLs from MDD Patients
(A) Global m6A/m is transiently decreased in mouse blood after acute stress. Global m6A/m assay on total RNA, n = 8, mean ± SEM. Asterisks (∗) depict omnibus post hoc comparisons to basal, p < 0.05, after Kruskal-Wallis test, p < 0.05.
(B) Global m6A/m changes in mouse blood are accompanied by changes in m6A/m regulatory genes. qPCR on total mouse blood, log2 fold changes of different genes to basal. n = 8, mean ± SEM. Red colored gene names, one-way ANOVA. Asterisks (∗) depict omnibus Tukey post hoc tests to basal p < 0.05; see Table S2.
(C) Furthermore, global m6A/m is decreased in mouse blood after both corticosterone and dexamethasone i.p. injection. Corticosterone, 250 μg/kg; dexamethasone, 10 mg/kg. Global m6A/m assay on total RNA, n = 12, mean ± SEM. Two-way ANOVA reported a significant interaction effect (F(4, 96) = 12.887, p < 0.001). Stars indicate omnibus Tukey post hoc tests, p < 0.05 compared to area basal.
(D) In a similar way, global m6A/m is temporarily decreased in the blood of healthy human subjects after treatment with 1.5 mg dexamethasone (Dex). Global m6A/m assay on total whole blood RNA, n = 25 healthy men, mean ± SEM. Kruskal-Wallis test, p < 0.001. Asterisks (∗) depict omnibus Tukey post hoc tests to basal p < 0.05.
(E) Expression of m6A/m regulatory genes in human blood is also affected by dexamethasone. Microarray of human whole blood at baseline and 3 hr after intake of Dex, n = 160 mixed healthy and diseased subjects, mean ± SEM. Asterisks (∗) depict Bonferroni-corrected t tests to basal p < 0.05.
(F) The dexamethasone-induced m6A/m decrease in human blood m6A/m is absent in MDD patients. n = 25, male and female, healthy and MDD subjects each, mean ± SEM. Three-way mixed-design ANOVA, significant interaction effect of treatment and subject status (F(1, 96) = 11.184, p = 0.001), but no interaction with sex. Asterisks (∗) depict omnibus Tukey post hoc tests to sex basal p < 0.05.
(G) Global m6A/m is decreased in B lymphocyte cell lines (BLCLs) in a concentration-dependent manner after 1 hr treatment with cortisol. Global m6A/m assay on total RNA, n = 5 biological replicates with 3 technical replicates each, mean ± SEM. Two-way ANOVA, significant interaction effect of cortisol and donor status (F(3, 24) = 44.365, p < 0.001). Asterisks (∗) depict omnibus Tukey post hoc tests to basal p < 0.05.
(H) The same regulation is observed on mRNA m6A using LC-MS/MS. n = 5, mean ± SEM. Specific detection of m6A. Two-way ANOVA, significant interaction effect of cortisol and donor status (F(1, 20) = 19.196, p < 0.001). Asterisks (∗) depict omnibus Tukey post hoc tests to mock treatment p < 0.05.
See also Figure S8.
Since dysregulation of the stress response may be an important feature of psychopathologies like major depressive disorder (MDD), we next investigated whether m6A/m regulation in response to dexamethasone differs between healthy individuals and MDD patients. In contrast to healthy subjects, downregulation of m6A/m in response to dexamethasone was observed in neither male nor female MDD patients (Figure 7F; significant within-subject diagnosis × dexamethasone effect only). Bootstrapping statistics performed for the reported significant subject diagnosis × dexamethasone treatment effect reported the 95% confidence interval of the F-statistic based on 10,000 bootstraps as [5.26, 33.91] and thus well above the critical Fcrit(1, 96) = 3.94, supporting that the chosen sample size was sufficient for detecting the within-subject diagnosis-dependent dexamethasone effect reported.
To exclude any influence by changes in blood cell composition rather than m6A/m levels, we compared estimates of the fractions of different blood cell types derived from the residuals of the transcriptome-wide gene expression values as published in Arloth et al. (2015) using CellCODE. For the samples used for the m6A/m measurements, cell estimates were not found to be significantly different (Figure S8A; no significant effects for dexamethasone within any of the cell types or significant effect of the cell types on the dexamethasone × diagnosis × sex interaction, dexamethasone × diagnosis interaction, or dexamethasone main effects was observed). Using the cell estimates for the analysis of global m6A/m confirms the earlier observed effect of dexamethasone dependent on subject MDD diagnosis (significant interaction effect of treatment and subject status [F(1, 96) = 10.251, p = 0.002], but no interaction with sex or any significant covariate effect of cell type estimates).
To control for potential contamination of results by antidepressant treatment present in blood of MDD patients, we confirmed the lack of response to GC stimulation using dexamethasone (Figure S8B) and cortisol (Figure 7G, total RNA; Figure 7H, mRNA m6A measured specifically by LC/MS-MS) in B lymphocyte cell lines (BLCLs) obtained from six healthy volunteers and six MDD patients propagated in absence of antidepressants. NR3C1 (GC receptor) mRNA and protein expression, as well as transcriptional response to GC stimulation, was unchanged in BLCLs of MDD donors (Figures S8C–S8E).
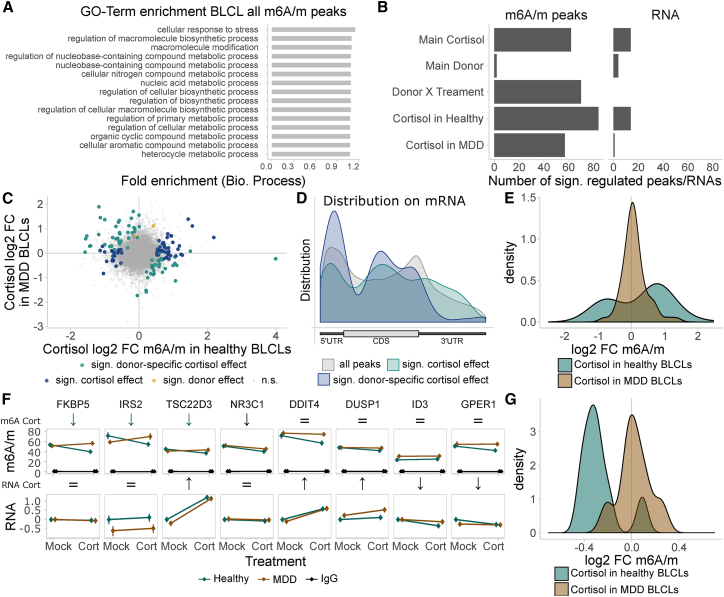
To reveal the specific signature of m6A/m deregulation in MDD patients, we performed m6A/m-seq of BLCLs treated for 1 hr with 100 nM cortisol or mock treatment (n = 3 per genotype and treatment). BLCL m6A/m peaks were again found to be very similar across the different groups (Figures S9A and S9B), with typical m6A/m properties regarding distribution and consensus motif (Figures S9B and S9C). m6A/m in BLCL are enriched in genes related to stress regulation and metabolic functions (Figure 8A). Analyzing the differential response to cortisol of an m6A/m consensus peak set (17,665 m6A/m peaks), in line with the results of the global m6A/m measurements, we observed major changes of m6A/m by cortisol in both a donor-dependent fashion (donor × treatment interaction effects) and a donor-independent fashion (main cortisol effects), but almost no significant differences by donor status alone, as well as a higher number of cortisol-regulated m6A/m peaks in healthy compared to MDD donor cell lines (Figures 8B and 8C; Table S5; example peaks in Figure S9D; top 25 regulated m6A/m peaks and RNAs in Figures S9F and S9G; all Q < 0.1, absolute log2 fold change > 0.5). Cortisol main and donor-interaction-regulated m6A/m peaks both showed a preference for location in the CDS and 5′ UTR (Figure 8D) with the donor-dependent cortisol-regulated peaks enriched in catabolic genes (Figure S9E), i.e., genes involved in energy-providing metabolic processes. Similar to the regulation of global m6A/m being more prominent in healthy donor BLCLs, m6A/m peaks regulated by cortisol were most often regulated in cells from healthy rather than MDD donor cells (Figures 8C and 8E).
Figure 8.
m6A-Seq and m6A-RIP qPCR Reveal Donor-Specific Patterns of m6A/m Regulation Altered in Cells Obtained from Healthy and MDD Donors
(A–E) The cortisol-responsive m6A/m epitranscriptome of healthy and MDD donor BLCLs was analyzed using m6A/m-seq comparing 1 hr mock-treated or 100 nM cortisol (Cort) conditions. We found 17,655 consensus high-confidence m6A/m peaks across all samples mapping to 8,681 genes. m6A/m-seq of each n = 3 BLCLs from healthy and MDD donors, each after 1 hr of mock or cortisol treatment.
(A) m6A/m peaks in BLCLs are enriched in genes with stress-responsive and metabolic functions. Fifteen highest enriched biological process GO terms with FDR-corrected Q < 0.1.
(B) Differential m6A/m and gene expression analysis in BLCLs reveals significant regulation of methylation after cortisol treatment with many m6A/m peaks regulated by cortisol in a donor-specific fashion and almost absent effects by donor status alone. Gene expression was less regulated than m6A/m regulation, lacking any donor-specific effects by cortisol. Number of significantly regulated peaks/genes with Q < 0.1 and absolute log2 fold change > 0.5, reporting 2 × 2 and post hoc effects within the single donor groups. Peaks and genes with significant interaction effects are removed from main treatment and main donor effect.
(C) Many peaks are regulated by cortisol in a donor-specific fashion. log2 fold changes of m6A/m peaks by cortisol within each donor group; significant peaks defined as above.
(D) m6A/m peaks in BLCLs show similar peak position enrichments as m6A/m peaks in mouse brain with preference for cortisol-regulated peaks at 5′ UTR and CDS. Peak distribution mapped along mRNA relative position.
(E) m6A/m peaks regulated by cortisol alone (main effect) have higher fold changes in healthy donors compared to BLCLs from MDD donors.
(F) Assessing absolute transcript methylation using m6A/m-RIP-qPCR, the cortisol-responsive genes FKBP5, IRS2, and TSC22D3 m6A/m were found specifically downregulated in cell lines of healthy, but not MDD, donors after stimulation with cortisol (cort). m6A/m-RIP-qPCR. n = 5, mean ± SEM. Significant effects observed in FDR-corrected two-way MANOVA (p < 0.05) are coded in the rows “m6A/m Cort effect” and “RNA Cort effect.” Orange arrows, healthy donor-specific Cort effect (interaction effect two-way ANOVA, one-way follow-up significant in healthy donors only); black arrow, Cort main stress effect; equals sign, no interaction or stress main effect in two-way ANOVA. For full statistics, see Table S2.
(G) Density plots of m6A/m change upon cortisol treatment. Density plots of log2 fold change data as m6A/m-RIP-qPCR data depicted in (F); donor-dependent distributions of fold changes were compared using a t test.
Finally, to confirm the differential regulation of m6A/m levels in BLCLs from healthy and MDD donors, we performed m6A/m-RIP-qPCR testing for GC-responsive genes in BLCLs after stimulation with cortisol. We observed specific downregulation of m6A/m in FKBP5, IRS2, and TSC22D3 in cells from healthy, but not from MDD, individuals (Figure 8F). In line with the general trends observed before, methylation of tested candidates in cells derived from healthy, but not MDD, donors was significantly decreased (Figure 8G).
Discussion
Here, we have identified m6A and m6Am as epitranscriptomic marks responsive to acute stress. Using m6A/m-seq in mouse cortex and m6A/m profiling in smaller areas by m6A/m-RIP-qPCR, we provide a map of brain m6A/m and evidence for regulation of m6A/m by acute stress. Consequently, in mice with METTL3 and FTO depleted in adult excitatory neurons and consequently altered m6A and m6Am profiles, we observed changes in transcriptome regulation, behavior, and electrophysiological properties. Finally, we observed that regulation of m6A/m and its cellular machinery in blood may represent a peripheral proxy for part of the brain’s m6A/m responses that seems impaired in patients with a stress-related disorder, MDD.
In the m6A/m-seq of mouse cortex, we remapped mouse cortical m6A/m, describing a higher amount of 5′ UTR peaks than previously reported (Meyer et al., 2012). A part of these 5′ UTR peaks may represent m6Am sites, although we did not observe any different properties of these putative m6Am peaks compared to general m6A/m peaks. We further add the observation that m6A sites in vivo overlap with the neuronal RNA-binding and cell-transport-regulating protein FMRP/FMR1-binding sites. FMR1 has recently been shown to bind m6A/m (Edupuganti et al., 2017), suggesting that it may be an important m6A reader in the brain. Future work is needed to investigate the nature of FMR1 binding to m6A/m and the effects of this binding in neurons, including potential roles in transcript localization to specialized neuronal compartments as axons and dendrites and potential regulation of local synaptic translation. Investigating a potential general relation of stress-regulated m6A/m regulating transcript translation, we could not find evidence for this.
Overall, only a very small number of m6A/m peaks in m6A/m-seq was found to be stress regulated. This is likely due to the large cellular heterogeneity of the material used; thus, only a small fraction of cells would have been responsive to the treatment and thus there was limited sensitivity of the assay to detect changes. Indeed, we find that m6A/m regulation is highly specific to smaller brain areas with even often opposite regulation in different areas as shown in the example of PFC being globally hypomethylated after stress and the AMY globally hypermethylated; this effect was confirmed by specific m6A detection being majorly driven by m6A. These two areas regulate behavioral and hormonal stress responses, fear, and anxiety (McEwen et al., 2015), with the PFC exhibiting top-down control of the AMY in anxiety and fear in mice (Adhikari et al., 2015). These changes were accompanied by matching regulation in the demethylase Fto and Alkbh5 expression, as well as regulation of the methyltransferase Mettl3. Interestingly, previous reports also showed transcriptional regulation of Fto after acute stress by fear conditioning (Walters et al., 2017, Widagdo et al., 2016).
Although we observed a general negative correlation between absolute m6A/m change and RNA abundance in m6A/m-RIP-qPCR, most m6A/m changes were not accompanied by significant transcript changes in m6A/m-RIP-qPCR or m6A/m-seq, implying that differential m6A/m acts by regulating both RNA decay and location and translation control.
Notably, we observe that corticosterone i.p. injection in mice causes similar effects on m6A/m and enzyme expression as acute stress, pointing toward a potential signaling mechanism via centrally acting GCs. Additional work is needed to unravel the pertinent signaling cascades involved. It is currently unclear which of the cell types drive the observed effects on m6A/m, with likely all major brain cell types having m6A/m and expression of the respective machinery genes (as observed by single-cell RNA-seq; data not shown).
To more specifically investigate the mechanisms of m6A/m methylation in adult excitatory neurons only, we employed cKO mice using Camk2a-Cre and Nex-CreERT2 drivers. Interestingly, while METTL3 deletion reduced m6A as expected, FTO deletion did not alter m6A levels but increased m6Am levels. However, m6A/m peaks differentially methylated in Fto cKO mice were not only positioned at the 5′ UTR, but also in CDS and 3′ UTR, pointing at FTO not only affecting m6Am, but a large part (Mauer et al., 2017). In general, epitranscriptomic and transcriptomic signatures of Mettl3 cKO and Fto cKO brain tissue were substantially different, indicating that both enzymes in neurons of the adult brain have different targets and likely very different functions. Interestingly, both Nex-CreERT2 knockout mice had very similar behavioral profiles, including lack of effects on anxiety and general cognition but increased fear memory for cued fear (with Fto cKO mice additionally having increased contextual fear) with stable differences of memory across time and fear extinction training, extending the previously reported fear expression upon knockdown of Fto in the dorsal hippocampus (Walters et al., 2017) and in the PFC (Widagdo et al., 2016). This indicates that fear-memory acquisition as well as its stability to extinction may require fine-tuned regulation of m6A/m levels rather than being directly regulated by specific m6A/m levels at specific genes. Mechanistically, we found that Mettl3 and Fto depletion alters not only the steady-state transcriptome in adult hippocampal neurons, but also the transcriptomic response to the fear conditioning stress, including regulation of several genes involved in neuronal circuit function and pointing out a function of m6A/m in regulating neuronal circuits. Consequently, we describe that network plasticity is specifically altered in the CA1, a brain region crucial for contextual fear, in Fto cKO, but not Mettl3 cKO, mice. This may reflect a neuronal correlate of altered m6A/m underlying the altered contextual fear memory observed in Fto cKO mice.
Finally, we propose that regulation of m6A/m and its cellular machinery in blood may represent a peripheral proxy for part of the brain’s m6A/m response, similar to DNA methylation changes (Ewald et al., 2014, Provençal et al., 2012). Both mice and humans showed global blood demethylation after stress or GC intake, respectively. The m6A/m response to GCs is impaired in blood and blood cells obtained from MDD patients, which may be a consequence of the altered GC receptor downstream signaling reported in MDD (de Kloet et al., 2005). While limited in sample size, these data represent a first step for future studies aiming to assess m6A/m in human samples as potential biomarkers or for mechanistic investigations and show the feasibility of such studies. Interestingly, genetic variants in FTO (Milaneschi et al., 2014, Samaan et al., 2013) and ALKBH5 (Du et al., 2015) have been reported to associate with risk for MDD before but are yet to be replicated in larger cohorts. Growing evidence supports fine-tuning of transcriptional regulation is critical for psychiatric disorders including various epigenetic mechanisms (Klengel and Binder, 2015). Here, we reveal RNA modifications as a novel mechanism relevant for understanding psychiatric disorders.
In summary, m6A and m6Am methylation constitute a novel layer of complexity in gene expression regulation following stress exposure, which is pivotal for the adaptation of stress-responsive circuits to acute challenges. The exciting finding of m6A/m dysregulation in MDD opens the possibility for the development of novel diagnostic biomarkers and eventually to better treatments for anxiety disorders, depression, and other stress-related diseases.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| polyclonal rabbit anti-m6A (and m6Am) | Synaptic Systems | 202 003; RRID: AB_2279214 |
| polyclonal rabbit anti-METTL3 | Proteintech | 15073-1-AP; RRID: AB_2142033 |
| monoclonal mouse anti-FTO | Merck Millipore | MABE227; RRID: AB_11203491 |
| polyclonal rabbit anti-ACTB | Cell Signaling Technology | 4967; RRID: AB_330288 |
| monoclonal rabbit anti-GR | Abcam | ab109022; RRID: AB_10863164 |
| polyclonal rabbit anti-BTUB | Abcam | ab6046; RRID: AB_2210370 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| corticosterone-HBC complex | Sigma | C174 |
| Dexa-ratiopharm | Ratiopharm | Dexa-ratiopharm Injektionslösung 59988.00.00 |
| cycloheximide | Sigma | C7698 |
| Critical Commercial Assays | ||
| EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric) | Epigentek | P-9005-96 |
| TruSeq Stranded mRNA Library Prep Kit | Illumina | RS-122-2103 |
| TruSeq Ribo Profile for Mammalian | Illumina | RPHMR12126 |
| QuantiFast SYBR Green PCR Kit | QIAGEN | 204057 |
| TaqMan Fast Advanced Master Mix | Applied Biosystem | 4444965 |
| Deposited Data | ||
| superseries | This paper | GEO: GSE113801 |
| m6A/m-Seq mouse cortex after acute stress | This paper | GEO: GSE113781 |
| Ribosome profiling Seq mouse cortex after acute stress | This paper | GEO: GSE113789 |
| m6A/m-Seq of mouse adult cortex of Mettl3 cKO or Fto cKO mice | This paper | GEO: GSE113793 |
| mRNA-Seq of mouse Mettl3 cKO or Fto cKO mouse hippocampus after fear conditioning | This paper | GEO: GSE113796 |
| m6A/m-Seq of human B-lymphocyte cell lines from healthy controls and major depressive disorder patients | This paper | GEO: GSE113798 |
| FMR1 bound genes | Darnell et al., 2011 | Darnell et al., 2011 Supplemental Data |
| Experimental Models: Cell Lines | ||
| Human immortalized BLCLs from patients | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| 10-12 w old adult male mice C57BL/6 | Charles River | 027 |
| Mettl3 cKO mice | Mettl3tm1a(KOMP)Wtsi V6.5 mouse ESCs targeted as described in mice generated by Geula et al., 2015 | Mettl3tm1c(KOMP)Wtsi |
| Fto cKO mice | Ftotm1a(EUCOMM)Wtsi EMMA EM: 05094 | Ftotm1c(EUCOMM)Wtsi |
| Camk2a-Cre mice | Minichiello et al., 1999 | Tg(Camk2a-cre)93Kln MGI:2176754 |
| Nex-CreERT2 mice | Agarwal et al., 2012 | Neurod6tm2.1(cre/ERT2)Kan MGI:5308766 |
| Human blood | Menke et al., 2012, Arloth et al., 2015 | “MPIP” and “MARS” cohorts |
| Oligonucleotides | ||
| Gene expression primers for SYBR green qPCR: sequences see Gene expression | Sigma-Aldrich | N/A |
| Taqman gene expression assays: IDs see Candidate m6A/m-RIP-qPCR | Taqman | N/A |
| m6A-spike-in-oligo GCAGAACCUAGUAG CGUGUGGm6ACACGAACAGGUAUCAAU AUGCGGGUAUGGm6ACUAAAGCAACGUG CGAGAUUACGCUGAGGm6ACUACAAUCU 'CAGUUACCA |
Sigma-Aldrich | N/A |
| primers for cloning ISH probes: Mettl3 exon 4 TCAGTCAGGAGATCCTAGAGCTATT and CTGAAGTGCAGCTTGCGACA; Fto exon 4 TGGCAGCTGAAATACCCTAAACT and ATAG CTGTACACTGCCACGG) |
Sigma-Aldrich | N/A |
| Software and Algorithms | ||
| STAR | Dobin et al., 2013 | RRID: SCR_015899 |
| cutadapt | Martin, 2011 | RRID: SCR_011841 |
| exomePeak | Meng et al., 2014 | RRID: SCR_001076 |
| BEDTools | Quinlan and Hall, 2010 | RRID: SCR_006646 |
| DiffBind | Ross-Innes et al., 2012 | RRID: SCR_012918 |
| DESeq2 | Love et al., 2014 | RRID: SCR_015687 |
| ChIPseeker | Yu et al., 2015 | https://doi.org/10.18129/B9.bioc.ChIPseeker |
| BioMart Project | Smedley et al., 2015 | RRID: SCR_002987 |
| Guitar | Cui et al., 2016 | https://doi.org/10.18129/B9.bioc.Guitar |
| PANTHER GO | Mi et al., 2013 | RRID: SCR_004869 |
| DREME | Bailey, 2011 | RRID: SCR_001783 |
| Integrative Genomics Viewer browser | Robinson et al., 2011 | RRID: SCR_011793 |
| SPSS | IBM SPSS Statistics | RRID: SCR_002865 |
| R | R Development Core Team, 2011 | RRID: SCR_001905 |
| ggplot2 | Wickham, 2009 | RRID: SCR_014601 |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alon Chen (alon_chen@psych.mpg.de).
Experimental Model and Subject Details
Animals
All experiments were approved by and conducted in accordance with the regulations of the local Animal Care and Use Committee (Government of Upper Bavaria, Munich, Germany and Weizmann Institute of Science, Rehovot, Israel).
For all experiments characterizing m6A/m changes after stress, 10-12 w old adult C57 BL/6 male mice were used (Charles River, Sulzfeld, Germany). Mettl3 cKO and Fto cKO mice were generated by breeding Mettl3tm1c(KOMP)Wtsi lox/lox mice (derived from Mettl3tm1a(KOMP)Wtsi V6.5 mouse ESCs targeted as described in mice generated by Geula et al., 2015) or Ftotm1c(EUCOMM)Wtsi lox/lox mice derived from Ftotm1a(EUCOMM)Wtsi obtained from EMMA (EM: 05094) to Camk2a-Cre mice (Minichiello et al., 1999) or Nex-CreERT2 mice (Agarwal et al., 2012), respectively. Camk2a-Cre mice crossed mice were used for LC-MS/MS and m6A/m Seq (Figure 4), Nex-CreERT2 crossed mice were used for mRNA-Seq, behavioral characterization and electrophysiological characterization (Figures 5 and 6). Experimental mice were homozygous floxed Cre-positive (Cre/+, “cKO”) and Cre-negative (+/+, “WT”) littermates generated by breeding of homozygous floxed mice negative and hemizygous for the CreERT2-allele. All Nex-CreERT2 crossed mice were fed with tamoxifen-containing chow (Genobios LASCR diet Cre Active TAM 400) starting at the age of 4-6 w. Animals were housed in groups, until being single housed 7 d before the experiments started, in standard plastic cages and maintained in a temperature-controlled environment (21 ± 2°C) on a 12 hr light/dark cycle with food and water available ad libitum. Restraint stress was performed for 15 min in ventilated 50 mL falcon tubes, starting at 2 hr post lights on. For pharmacological studies, mice were injected with vehicle solution (saline), 250 μg/kg corticosterone (corticosterone-HBC complex, Sigma) or 10 mg/kg dexamethasone (Ratiopharm Dexa-ratiopharm) i.p. 2 hr post switching the lights on.
Sample collection
Whole mouse cortex for m6A/m-Seq was collected at designated time points by manual dissection of fresh brains on ice. For each sample, 3 animals randomly selected from the same group were pooled. For investigation of regions-specific effects in PFC and AMY, brains were immediately flash-frozen after dissection and defined tissue punches of medial prefrontal cortex (PFC; consisting of infralimbic and prelimbic cortex) and amygdala (AMY; consisting of central and basolateral amygdala) were collected using a 1 mm round tissue punch while sectioning brains on a cryostat. Mouse whole blood was collected in EDTA tubes, aliquoted and flash-frozen.
Cell culture
Human immortalized BLCLs derived from age-matched (33-53 y) male subjects either healthy or diagnosed with MDD were cultured in RPMI-1640 medium (Merck KGaA, Darmstadt, Germany) supplemented with 10 % fetal calf serum at 37°C with 5% CO2. The cells were tested to be free of mycoplasma. Cells were treated with cortisol (Sigma-Aldrich, St. Louis, MO, in ethanol, final concentration 0.1% v/v) or dexamethasone (Ratiopharm Dexa-ratiopharm, in saline), or ethanol or saline mock control, respectively.
Human blood
Human whole blood was collected using PAXgene Blood RNA Tubes (PreAnalytiX, Hombrechtikon, Switzerland) either unstimulated or after oral administration of 1.5 mg dexamethasone and processed as described previously (Menke et al., 2012). Age-matched healthy Caucasian male and females subjects were selected from the “MPIP” and “MARS” cohorts described previously (Arloth et al., 2015, Menke et al., 2012).
Method Details
RNA isolation
Total RNA from tissue, mouse blood and BLCL cells was purified using Trizol (Invitrogen, Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions followed by isopropanol precipitation. For mouse whole blood, RNA was isolated using a 1:10 ratio of blood to Trizol.
Global m6A/m measurements
Global m6A/m in total RNA was quantified by the EpiQuik m6A/m RNA Methylation Quantification Kit (Epigentek Group, Farmingdale, NY) following manufacturers’ specifications and using 100-300 ng input (in duplicates or triplicates). Comparing total RNA global m6A/m measurements with LC-MS/MS data from the same conditions, we observed high correlation of stress-changes, suggesting that the total RNA colorimetric assay represents an appropriate tool to detect global m6A/m regulation patterns. Brain global methylation in PFC and AMY is not regulated by circadian rhythm (data not shown).
LC-MS/MS
For profiling of m6A after acute stress, samples were pooled from 4 mice randomly selected from the same group and RNA isolated as stated above. For profiling of m6A and m6Am in cKO mice, RNA from the samples processed for m6A/m-Seq (pooled from 3 mouse cortex each) were used. For profiling of m6A from BLCLs, RNA from cells of each of the BLCL lines was isolated as stated above.
Residual genomic DNA was removed using the TurboDNA-free kit (Ambion, Life Technologies, Carlsbad, CA). RNA integrity and absence of DNA was confirmed by Bioanalyzer RNA Nano chips (Agilent Technologies, Santa Clara, CA, RIN > 9) and Qubit DNA High sensitivity kit (Thermo Fisher Scientific, Waltham, MA), respectively. For mouse acute stress and BLCLs, PolyA+ RNA was prepared using 2 rounds of the Genelute mRNA Prep Kit (Sigma-Aldrich, St. Louis, MO) with rRNA depletion confirmed by Bioanalyzer RNA Nano chips (Agilent Technologies, Santa Clara, CA, mRNA mode; less than 4% rRNA content). For cKO mice m6A and m6Am profiling, PolyA+ RNA was prepared using 1 round of the Genelute mRNA Prep Kit (Sigma-Aldrich, St. Louis, MO) and 1 round of RiboZeroGold rRNA depletion (Illumina, San Diego, CA) with rRNA removal confirmed by Bioanalyzer RNA Nano chips (Agilent Technologies, Santa Clara, CA, mRNA mode; no rRNA detected). 250 ng PolyA-RNA (acute stress, BLCLs) or 500 ng PolyA-RNA (cKO mice) per sample and a standard curve (acute stress, BLCLs: N6-methyladenosine/adenosine, cKO mice: all standards see Figure S4B) were mixed with deuterated N6-(methyl-d3)-adenosine as an internal spike-in calibrator. The RNA-spike-in-mix from cKO mice was first decapped with 25 U RppH (NEB, Ipswich, MA) in the supplied buffer with 1 μl RNasin and 0.1% TritonX buffer added for 2 hr at 37°C and purified with 4x RNAClean XP (Agencourt Beckman Coulter, Brea, CA). Mouse acute stress and BLCL PolyA mixed with the spike-in calibrator as well as the decapped cKO mouse RNA mix was processed to nucleosides as reported before (Jia et al., 2011): Samples were treated with 2 U P1 nuclease at 37°C for 1 hr, followed by addition of NH4HCO3 and treatment with 0.5 U alkaline phosphatase (all Sigma-Aldrich, St. Louis, MO), at 37°C for 2 hr. Samples at a final concentration of 250 ng in 25 μl were filtered through a Corning Spin-X 0.2 um sterile cellulose acetate filter (Corning, Corning, NY) and diluted 1:10 with 20% methanol. HPLC/MS-MS analysis was performed using a Shimadzu Nexera X2 (Shimadzu, Duisburg, Germany) liquid chromatograph interfaced to the ESI source of a Sciex QTrap 5500 (Sciex, Darmstadt, Germany) triple quadrupole mass spectrometer. Chromatography was accomplished using a gradient elution in a Accucore RP-MS column (100 × 2,1 mm, 2,6 μm Thermo Scientific, Dreieich, Germany) at a flow rate of 0.3 ml/min, 5 μl injection volume, at 30°C for 10 min with the following gradient profile: Eluent A (10 mM NH4HCO2, 0.1% CH2O2 in CH3OH) for 3 min with 10% eluent B (10 mM NH4HCO2, 0.1% CH2O2 in CH3OH), 4 min 10%–95% B, 1 min hold at 95% B, 0.2 min 95%–10% B and 1.8 min 10% B. The ion source was operated in positive mode at 400°C, and multiple reaction monitoring (MRM) collision-induced dissociation (CID) were performed using nitrogen as the collision gas. Retention times and transitions monitored during analysis for the analytes are shown in Figure S4B. Quantification was performed by comparison with the standard curve obtained from pure nucleoside standards normalized by the deuterated spike-in calibrator run within the same experiment.
m6A/m-Seq
For mouse m6A/m-Seq, whole mouse cortex samples were used pooling 3 individuals each, since m6A/m-Seq on PolyA-RNA of smaller regions did not result in sufficient enrichment quality. For m6A/m-Seq of human BLCLs, RNA from each 3 randomly chosen cell lines from healthy and MDD donors each 1 hr after treatment with 100 nM cortisol or mock treatment was used. Each m6A/m-Seq experiment had an IgG control using RNA mixed equimolar from all samples of that experiment. m6A/m-Seq was performed using a modified version of previously published protocols (Dominissini et al., 2013, Meyer et al., 2012). RNA was isolated using Trizol (Invitrogen, Life Technologies, Carlsbad, CA) and residual genomic DNA was removed using the TurboDNA-free kit (Ambion, Life Technologies, Carlsbad, CA). RNA integrity and absence of DNA was confirmed by Bioanalyzer RNA Nano chips (Agilent Technologies, St. Louis, MO, RIN > 9.5) and Qubit DNA High sensitivity kit, respectively. PolyA+ RNA was prepared using 1 round of the Genelute mRNA Prep Kit (Sigma-Aldrich, St. Louis, MO) with less than 5% residual rRNA as confirmed by Bioanalyzer RNA Nano chips (Agilent Technologies, St. Louis, MO, mRNA mode). RNA was fragmented using fragmentation reagent (Life Technologies, Carlsbad, CA). mRNA fragments were precipitated with ethanol and used for m6A/m-immunoprecipitation, IgG control and input samples. m6A/m-immunoprecipitation (10 μg mRNA fragments mouse acute stress and BLCLs or 7.5 μg mouse cKO and 10 μg rabbit polyclonal anti-m6A/m 202 003, lots: /56 for mouse acute stress and BLCLs, /66 for cKO mice, Synaptic Systems, Göttingen, Germany) or IgG control (10 μg mRNA fragments mixed from all samples and 10 μg IgG 2729, Cell Signaling Technology, Beverly, MA) was performed in precipitation buffer (50 mM Tris, pH 7.4, 100 mM NaCl, 0.05 % NP-40, 1 mL total volume) with 1 μL RNasin Plus (Promega, Madison, WI) rotating head over tail at 4°C for 2 hr, followed by incubation with washed 30 μl Protein A/G beads (Thermo Fisher Scientific, Waltham, MA) rotating at 4°C for 2 hr. Bead-bound antibody-RNA complexes were recovered on a magnetic stand and washed twice with immunoprecipitation buffer, twice with high-salt buffer (50 mM Tris, pH 7.4, 1 M NaCl, 1 mM EDTA, 1% NP-40, 0.1% SDS), and twice with immunoprecipitation buffer. Fragments were eluted by Proteinase K treatment (300 μL elution buffer: 5 mM Tris-HCL pH 7.5, 1 mM EDTA pH 8.0, 0.05% SDS, 4.2 μl 20 mg/ml proteinase K). RNA was recovered from the eluate using Trizol LS (Invitrogen Life Technologies, Carlsbad, CA) following manufacturers’ recommendations. Sequencing libraries were prepared using the Illumina TruSeq non-stranded (mouse cortex acute stress and BLCLs) or stranded (cKO mouse cortex) mRNA protocol following the standard protocol starting from mRNA fragments recovered from m6A/m-IP, IgG-IP, or 100 ng of original PolyA-RNA input fragments. Libraries were quality-checked using Bioanalyzer DNA High Sensitivity chips (Agilent Technologies, St. Louis, MO) and quantified using the KAPA Library Quantification Kit (KAPA Biosystems, Boston, MA). Sequencing was performed on 2-4 lanes of an Illumina HiSeq4000 PE 2x100 (Illumina, San Diego, CA) multiplexing all m6A/m-, IgG- and input samples per experiment.
Ribosome profiling
Ribosome profiling libraries were prepared from mouse cortex of 6 mice 4 hr after acute stress and 6 matching control mice using the TruSeq Ribo Profile (Mammalian) Kit (Illumina, San Diego, CA; based on Ingolia et al., 2014) with the following adjustments: Flash-frozen cortex samples were homogenized in 750 μl lysis buffer including cycloheximide using a dounce homogenizer and 10 passes through a 25 G needle and incubated rotating for 20 min at 4°C. After centrifugation for 20 min at 20000 ∗ g at 4°C 100 μl supernatant were set aside for input samples and 400 μl supernatant were processed for ribosome profiling with 45 min incubation at RT with 60 U/OD260 Nuclease. After adding 15 μl of RNase inhibitor, monosomes were purified on a sucrose gradient. Ribosome protected fragments as well as input RNA was purified using Trizol LS (Invitrogen, Life Technologies, Carlsbad, CA) and the miRNeasy micro kit (QIAGEN, Hilden, Germany). rRNA was depleted using the RiboZero mammalian Gold Kit (Illumina, San Diego, CA) and fragments size-selected, purified and processed as described. cDNA was purified using a 2.5 x AMPure clean-up (Agencourt Beckman Coulter, Brea, CA). PCR was performed on undiluted circularized cDNA with 12 PCR cycles. PCR products were size-selected on a 5% DNA-TBE PAGE. Sequencing was performed on each one lane for ribosome bound fractions and input fractions (indexed each 1-12) of an Illumina HiSeq4000 PE 2x100 (Illumina, San Diego, CA) using only the reverse read.
mRNA-Seq
Brains were collected from 5 of each of the following: Mettl3 cKO and WT as well as Fto cKO and WT mice 24 hr after fear conditioning (“FC,” details in “Animal behavior testing”) or comparable handling without fear induction (“Box”: handling and exposure to context as in “FC” in “Animal behavior testing” but without foot shock and tone/CS and US). The entire CA1 and CA3 was cryo-punched using 0.7 and 1 mm punching tools from snap-frozen brains sliced at 250 μm using a cryostat and RNA isolated. Residual genomic DNA was removed using the TurboDNA-free kit (Ambion, Life Technologies, Carlsbad, CA). RNA integrity and absence of DNA was confirmed by Bioanalyzer RNA Nano chips (Agilent Technologies, St. Louis, MO, RIN > 8.5) and Qubit DNA High sensitivity kit, respectively. mRNA-Seq libraries were prepared from 4 μg total RNA using the llumina TruSeq stranded mRNA protocol HT (Illumina, San Diego, CA) following the standard protocol starting using Superscript III and 11 cycles of PCR. Libraries were quality-checked using Bioanalyzer DNA High Sensitivity chips (Agilent Technologies, St. Louis, MO) and quantified using the KAPA Library Quantification Kit (KAPA Biosystems, Boston, MA). Sequencing was performed on 4 lanes of an Illumina HiSeq4000 PE 2x100 (Illumina, San Diego, CA) multiplexing all samples.
Gene expression
Gene expression of m6A/m-related enzymes was done by SYBR-green-based qPCR. RNA was reverse-transcribed using the SuperScript III VILO cDNA Synthesis Kit (Invitrogen Life Technologies, Carlsbad, CA) and QuantiFast SYBR Green PCR Kit (QIAGEN, Hilden, Germany) on a Quantstudio 7 (Applied Biosystems, Waltham, MA) with the following primers: Mettl3 NM_019721 (ATTGAGAGACTGTCCCCTGG, AGCTTTGTAAGGAAGTGCGT), Mettl14 NM_201638 (AGACGCCTTCATCTCTTTGG, AGCCTCTCGATTTCCTCTGT), Wtap_consensus (GTTATGGCACGGGATGAGTT, ATCTCCTGCTCTTTGGTTGC), Wtap_short NM_001113532 (CTAGCAACCAAAGAGCAGGA, AGTCTTGACTGGGGAGTATGA), Wtap_long NM_001113533 (GGCAAAAAGCTAATGGCGAA, GCTGTCGTGTCTCCTTCAAT), Fto NM_011936 (CTGAGGAAGGAGTGGCATG, TCTCCACCTAAGACTTGTGC), Vir-Kiaa1429 NM_001081183 (CATTACGGCCGCTTAGTTCT, TACCACTGCCTCCACTAACA), Alkbh5 NM_172943 (ACAAGATTAGATGCACCGCG, TGTCCATTTCCAGGATCCGG), Ythdf1 NM_173761 (CATTATGAGAAGCGCCAGGA, AGATGCAACAATCAACCCCG), Ythdf2 NM_145393 (ACCAACTCTAGGGACACTCA, GGATAAGGAGATGCAACCGT), Ythdf3 NM_172677 (TGCACATTATGAAAAGCGTCA, AGATGCGCTGATGAAAACCA), Ythdc1 NM_177680 (TTCATAACATGGGACCACCG, TCATAGTCATGTACTCGTTTATCTC), Hnrnpc NM_016884 (CAAACGTCAGCGTGTTTCAG, TGGGGATGAGAAGGACAAGT), Hnrnpa2 B1 NM_016806 (GTGGAGGGAACTATGGTCCT, TGAAGGCACCAACAAGAACT). Each qPCR assay was performed in duplicates or triplicates with a standard dilution curve of a calibrator and using assay efficiency for calculations. Expression levels were quantified by the ddCT method normalizing to an average of 4-5 housekeeping genes chosen based on maximum stability between conditions from the following: Hprt NM_013556 (ACCTCTCGAAGTGTTGGATACAGG, CTTGCGCTCATCTTAGGCTTTG), Rpl13 A NM_009438 (CACTCTGGAGGAGAAACGGAAGG, GCAGGCATGAGGCAAACAGTC), Atp5j NM_001302213 (TATTGGCCCAGAGTATCAGCA, GGGGTTTGTCGATGACTTCAAAT), Polr2b NM_153798 (CAAGACAAGGATCATATCTGATGG, AGAGTTTAGACGACGCAGGTG), Rn18s NR_003278 (CAGGATTGACAGATTGATAGC, ATCACAGACCTGTTATTGCTC), Ubc NM_019639 (CTGCCCTCCCACACAAAG, GATGGTCTTACCAGTTAAGGTT), Hmbs NM_001110251 (TCTGAAAGACAGATGGAATGCC, CCACACGGAAAGAGAAGAGGC). For human samples, the following primers were used: NR3C1 NM_000176 (CAGCAGTGAAATGGGCAAAG, TCGTACATGCAGGGTAGAGT), NR3C2 NM_000901 (GATCCAAGTCGTGAAGTGGG, TGAAGGCTGATTTGGTGCAT), FKBP5 NM_004117 (CGGCGACAGGTTCTCTACTT, TCTCCAATCATCGGCGTTTC), TSC22D3 NM_004089 (TCCGTTAAGCTGGACAACAG, TTCAACAGGGTGTTCTCACG) with housekeeping genes TBP NM_003194 (GGGAGCTGTGATGTGAAGTT, GAGCCATTACGTCGTCTTCC), RPL13 A NM_012423 (GCGTCTGAAGCCTACAAGAA, CCTGTTTCCGTAGCCTCATG), and SDHA NM_004168 (CAGGGAAGACTACAAGGTGC, CAGTCAGCCTCGTTCAAAGT). All assays were designed as intron-spanning if possible with product sizes confirmed by melting curves and band detection on gel showing the absence of genomic DNA products.
Upstream GRE prediction
10 kb upstream sequences of m6A/m-related genes were retrieved using Biomart (Smedley et al., 2015). GC response elements were predicted by the JASPAR vertebrate core transcription factor binding site prediction (Mathelier et al., 2016) querying NR3C1 motifs MA0113.1 (mammalian), MA0113.2 (mmu), and MA0113.3 (hsa) with a conservative relative profile score threshold of 90%.
Spike-in Oligo
The spike-in RNA oligo was designed with the following specifications: 100 bp length, 3 internal m6A/m sites within GGAC motif flanked by the most frequent nucleotides 5′ U/A, 3′ A/U, not complementary to hsa or mmu RefSeq mRNA or genome, secondary structure exposing m6A/m sites, mean % GC = 51. The sequence is GCAGAACCUAGUAGCGUGUGGmACACGAACAGGUAUCAAUAUGCGGGUAUGGmACUAAAGCAACGUGCGAGAUUACGCUGAGGmACUACAAUCUCAGUUACCA. Fully m6A/m-methylated or unmethylated RNA oligos were purchased from Sigma (Sigma-Aldrich, St. Louis, MO). m6A site prediction was performed using SRAMP (Zhou et al., 2016) (full transcript mode, generic predictive model) confirming that the motif sequence context is similar to those occurring in real m6A/m data. Structure prediction was performed using RNAstructure (Reuter and Mathews, 2010) (Fold mode, Version 5.8.1).
Candidate m6A/m-RIP-qPCR
To validate m6A/m-Seq experiments, candidates were chosen from the list of differentially methylated transcripts selecting for transcripts. For investigation of candidate transcript methylation in small brain areas, candidate lists were constructed by intersecting microarray results of mouse brain PFC, AMY and hippocampus after acute stress and GC stimulation (Arloth et al., 2015) with genes known to be methylated in mouse brain (Hess et al., 2013, Meyer et al., 2012) and functional annotation GO-terms. For investigation of candidate transcript methylation in BLCL cell lines, dexamethasone-responsive genes from human blood microarray data (Arloth et al., 2015) were intersected with BLCL m6A/m-Seq data (unpublished data).
15 μg PolyA-RNA (m6A/m-Seq validation) or 3 μg total RNA (candidate m6A/m-RIP-qPCR validation) or 1.5 μg total RNA (brain area/cell line candidate m6A/m-RIP-qPCR) was mixed well with 30 fmol or indicated amount of spike-in or 3 fmol spike-in, respectively, and equally split into 3 conditions: m6A/m-RIP, IgG control and input. For m6A/m-Seq validation and brain are candidate m6A/m-RIP-qPCR only fully methylated spike-in was used. Input samples were flash-frozen during the course of the experiments. m6A/m-RIP and IgG control samples were incubated in parallel to m6A/m-Seq with 1 μg anti-m6A/m antibody (rabbit polyclonal 202 003, Synaptic Systems, Göttingen, Germany) or 1 μg IgG (rabbit polyclonal IgG 2729, Cell Signaling Technology, Beverly, MA) in immunoprecipitation buffer (0.5 mL total volume) with 1 μL RNasin Plus (Promega, Madison, WI) rotating head over tail at 4°C for 2 hr, followed by incubation with washed 25 μL Dynabeads M-280 (Sheep anti-Rabbit IgG Thermo Fisher Scientific, Waltham, MA, Cat11203 D) rotating head over tail at 4°C for 2 hr. Bead-bound antibody-RNA complexes were recovered on a magnetic stand and washed twice with immunoprecipitation buffer, twice with high-salt buffer, and twice with immunoprecipitation buffer. RNA was eluted directly into Trizol and input RNA was also taken up in Trizol. RNA from all conditions was purified in parallel using the miRNeasy micro RNA isolation kit (QIAGEN, Hilden, Germany) including a 3-time repeated elution 15 μL H2O to ensure the complete elution of all RNA. The entire eluate was transcribed to cDNA using the SuperScript III VILO cDNA Synthesis Kit (Invitrogen, Life Technologies, Carlsbad, CA). Gene expression was quantified using TaqMan Fast Advanced Master Mix (Applied Biosystems, Waltham, MA) on a Quantstudio 7 (Applied Biosystems, Waltham, MA) by the following Taqman gene expression assays: Actb NM_007393 (Mm01205647_g1), Akt1 NM_009652 (Mm01331626_m1), Arc NM_018790 (Mm01204954_g1), Atp1 B1 NM_009721 (Mm00437612_m1), Bsn NM_007567 (Mm00464452_m1), Camk2 A NM_009792 (Mm00437967_m1), Camk2n1 NM_025451 (Mm01718423_s1), Cited1 NM_007709 (Mm01235642_g1), Cnr1 NM_007726 (Mm01212171_s1), Crh NM_205769 (Mm04206019_m1), Crhbp NM_198408 (Mm01283832_m1), Crhr1 NM_007762 (Mm00432670_m1), Ctsb NM_007798 (Mm01310508_g1), Cyfip2 NM_133769 (Mm00460148_m1), Dlg4 NM_007864 (Mm00492193_m1), Dnmt1 NM_001199433 (Mm01151063_m1), Dusp1 NM_013642 (Mm00457274_g1), Egr3 NM_018781 (Mm00516979_m1), Fkbp5 NM_010220 (Mm00487406_m1), Fscn1 NM_007984 (Mm00456046_m1), Fth1 NR_073181 (Mm04336020_g1), Gabbr1 NM_019439 (Mm00444578_m1), Gabbr2 NM_001081141 (Mm01352561_m1), Gadd45 g NM_011817 (Mm01352550_g1), Grm1 NM_001114333 (Mm00810219_m1), Grm3 NM_181850 (Mm01316764_m1), Homer1 NM_011982 (Mm00516275_m1), Htra1 NM_019564 (Mm00479887_m1), Mllt11 NM_019914 (Mm00480176_m1), Nlgn2 NM_198862 (Mm01245481_g1), Nodal NM_013611 (Mm00443040_m1), Notumos AK028718 (Mm00845023_s1), Nr3c1 NM_008173 (Mm00433832_m1), Nr4a1 NM_010444 (Mm01300401_m1), Nrcam NM_176930 (Mm00663607_m1), Nrxn1 NM_020252 (Mm03808856_m1), Nrxn2 NM_020253 (Mm01236844_g1), Onecut1 NM_008262 (Mm00839394_m1), P2ry13 NM_028808 (Mm00546978_m1), Plekhg3 NM_153804 (Mm00770086_m1), Plin4 NM_020568 (Mm00491061_m1), Pomc NM_008895 (Mm00435874_m1), Prkcb NM_008855 (Mm00435749_m1), Prkcg NM_011102 (Mm00440861_m1), Pvrl3 NM_021495 (Mm01342993_m1), Rgs4 NM_009062 (Mm00501392_g1), Rhou NM_133955 (Mm00505976_m1), Sgk1 NM_001161850 (Mm00441387_g1), Sgk2 NM_013731 (Mm00449845_m1), Sirt2 NM_022432 (Mm01149204_m1), Spats1 NM_027649 (Mm01270591_m1), Sumo1 NM_009460 (Mm01609844_g1), Syn1 NM_013680 (Mm00449772_m1), Syngap1 NM_001281491 (Mm01306145_m1), Tec NM_013689 (Mm00443230_m1), Tsc22d3 NM_001077364 (Mm00726417_s1). Mouse housekeeping genes: Hprt1 NM_013556 (Mm03024075_m1), Rpl13a NM_009438 (Mm01612987_g1), Tbp NM_013684 (Mm01277045_m1), Ubc NM_011664 (Mm02525934_g1), Uchl1 NM_011670 (Mm00495900_m1). Human gene expression assays: ID3 NM_002167 (Hs00171409_m1), DUSP1 NM_004417 (Hs00610256_g1), DDIT4 NM_019058 (Hs01111686_g1), GPER NM_001505 (Hs01922715_s1), IRS2 NM_003749 (Hs00275843_s1), FKBP5 NM_004117 (Hs01561006_m1), NR3C1 NM_000176 (Hs00353740_m1), TSC22D3 NM_004089 (Hs00608272_m1). Human housekeeping genes: RPL13A NM_012423 (Hs04194366_g1), TBP NM_003194 (Hs00427620_m1). The spike-in was quantified using a custom Taqman expression assay (primers TCAATATGCGGGTATGGACTAAAGC, TGAGGACTACAATCTCAGTTACCA and probe AACGTGCGAGATTACG). All assays were chosen as intron-spanning if available.
Human microarray data
Gene expression of m6A/m-related genes was extracted from microarray expression data of human whole blood published previously (Arloth et al., 2015).
Human blood cell estimates
Cell count estimate predictions were done on the gene expression data of the same samples used for m6A/m measurement published previously (Arloth et al., 2015) using CellCode (Chikina et al., 2015) and blood cell transcriptomic reference data (Abbas et al., 2009). The prediction was based on the residuals of the gene expression values (including Dexamthasone as covariant).
Animal behavior testing
All behavioral assessments were performed during the light phase. The experimenter was blinded to the genotype of the animals. Retesting followed the order of least-to-most stressful with 2-3 days rest in between tests.
Anxiety-like behavior was assessed using the Open Field Test (OF, 10 min, 10 lux, gray plastic box 50 × 50 × 50 cm, center defined as the inner 25 × 25 cm area), Elevated Plus Maze (EPM 5 min, 10 lux on closed arms, 100 lux on open arms, gray plastic maze 50 × 50 cm elevated 25 cm above the floor), Dark Light Box-Test at baseline (DLB basal) and 4 hr post 15 min restraint stress (DLB 4 hr post stress) (5 min, 100 lux in lit compartment), each with automated tracking (ANY-maze, Stoelting, Wood Dale, IL). The Marble Burying Test was performed by placing the mice in a fresh cage with 5 cm flattened fresh bedding with 15 black, clean marbles spaced evenly across (20 min, 10 lux, counting the number of buried marbles every 5 min). Cognitive function was assessed using the Y maze alternation task for working memory (5 min, 10 lux, Y-shaped 3-arm apparatus with 25 cm arm length and distinguishing visual cues on the walls and at the end of each arm, with automated tracking). The proportion of spontaneous non-repeated subsequent entries into each of the 3 arms (alternations) from the total number of 3-arm entries (including repeat entries) was used as the readout. Non-fear-related memory was assessed using the Object-Recognition-Task (ORT, 2 × 5 min with 1 hr intertrial interval, 10 lux, gray plastic box 50 × 50 × 50 cm, training trial: 2 identical objects with 1 out of 2 objects without object preference randomly assigned to all mice, test trial: 1 known, 1 novel object). The object discrimination ratio DI was determined by DI = (Time with novel object–Time with familiar object) / (Time with novel object + Time with familiar object) within the test trial.
Fear-related memory was assessed by conditional fear learning. Mice were fear conditioned (FC) within the same session for both contextual and cued fear by 180 s of baseline exposure to context A (a metallic/plastic cubic chamber with metal grid conditioned with 70% ethanol smell), followed by a 20 s 80 dB tone (9 kHz sine-wave, conditioned stimulus, CS), which co-terminated with an electric foot shock (unconditioned stimulus, US, 0.7 mA, 2 s, constant current delivered through the metal grid) and a 60 s after-shock interval. Memory was assessed by measuring freezing in response to the different cues by a highly experienced observer blind to the genotype. Auditory cued fear memory was tested 1 day after FC in context B (cylindrical plastic chamber with bedding conditioned with 1% acetic acid) by presenting a 3 min CS after 180 s baseline recording and followed by a 60 s post-tone recording. Freezing across the 180 s tone exposure was binned in 60 s intervals to assess short-term stimulus-habituation. Context memory was tested 2 days after FC in context A without presenting US or CS. Fear extinction was achieved by 10 ∗ 20 s CS presentations (variable inter-trial-interval of 20-60 s) in context B on 3 consecutive days 2 w after FC with freezing assessed across the first 3 tone presentations. Fear extinction memory retention was measured 1 w after the extinction by presenting 3 ∗ 20 s CS and measuring the freezing across those 3 presentations. Animals with generalized fear response (over 50% freezing in any of the baseline recordings of the extinctions trials) were excluded from the analysis for extinction memory.
Electrophysiology
Recordings were conducted blind to the animal genotype. Preparation of dorsal hippocampal slices and electrophysiological measurements were performed according to standard procedures as we described previously (Schmidt et al., 2011). From every animal, 2 slices were used for the experiments.
In situ hybridization
Expression quantification of Mettl3 mRNA and Fto mRNA in Mettl3 cKO and Fto cKO animals was performed by in situ hybridization using S-35 labeled antisense probes targeting the floxed exon as described previously (Refojo et al., 2011). Probes were designed for Mettl3 NM_019721 exon 4 (probe cloned using TCAGTCAGGAGATCCTAGAGCTATT and CTGAAGTGCAGCTTGCGACA) and Fto NM_011936 exon 4 (probe cloned using TGGCAGCTGAAATACCCTAAACT and ATAGCTGTACACTGCCACGG). Slides were exposed to Kodak Biomax MR films (Eastman Kodak, Rochester, NY), developed, and autoradiographs digitized and quantified by optical densitometry of 2 slides each averaging the signal across both hemispheres and slides utilizing ImageJ (dorsal: Bregma −1.82, −1.94; ventral Bregma −3.16, −3.28).
Western Blot
Cells or tissue punches were lysed on ice in RIPA buffer (150 mM NaCl, 1% NP-40, 0,5% Sodium deoxycholate, 0,1% SDS, 50 mM Tris-HCl pH 8 with cOmplete, EDTA-free Protease Inhibitor Cocktail Mini, Roche Applied Science, Roche Diagnotics, Indianapolis, IN) for 30 min. 25 μg total protein as determined by the Pierce BCA Protein Assay (Thermo Fisher Scientific, Waltham, MA) (mouse tissue) or Bio-Rad Quick Start Bradford Kit (Bio-Rad Laboratories, Hercules CA) (cells) was heated for 10 min in SDS/PAGE sample buffer (final concentration 62.5 mM Tris-HCl pH 6.8, 2% SDS, 10% glycerol, 5% b-mercapto-ethanol), separated on a Tris-Glycine SDS–PAGE (Bio-Rad Laboratories, Hercules, CA) and transferred to a nitrocellulose membrane (Amersham Protran, Millipore, Billerica, MA). Membranes were blocked for 1 hr in TBST containing 5% non-fat milk, followed by incubation with primary antibodies overnight at 4°C (anti-METTL3 polyclonal rabbit, 15073-1-AP, 1:200, Proteintech, Rosemont, IL; anti-FTO monoclonal mouse, MABE227 clone 5-2H10, 1:1000, Merck Millipore, Merck KGaA, Darmstadt, Germany; anti-ACTB polyclonal rabbit, 4967, 1:5000, Cell Signaling Technology, Boston, MA; anti-GR monoclonal rabbit, ab109022, 1:50000 Abcam, Cambridge, UK; anti-BTUB polyclonal rabbit, ab6046, 1:10000; Abcam, Cambridge, UK) in TBST with 3% non-fat milk. After incubation with horseradish-peroxidase-coupled secondary antibody (anti-mouse or anti-rabbit Cell Signaling Technology, Beverly, MA; 7074) at room temperature for 2 hr, immunoblots were visualized using enhanced chemiluminescence (ECL Plus, GE Healthcare Life Sciences, Freiburg, Germany) detected by the ChemiDoc Imaging System XRS+ (Bio-Rad, Hercules, CA). Detection of BLCL GR and BTUB was done in parallel on horizontally cut blot pieces. For METTL3 and FTO detection in mouse tissue, membranes were first probed with anti-METTL3 or anti-FTO, stripped after signal detection (0.2 M glycine, 3.5 mM SDS, 1% Tween in H2O 20 min at RT), blocked again and probed with anti-ACTB. Band intensity was quantified using ImageJ.
Quantification and Statistical Analysis
m6A/m-Seq analysis
Sequencing data quality control was performed by FASTQC (Andrews, 2010). Genomic alignment was performed using the STAR aligner (Dobin et al., 2013) (to RefSeq mm10 or RefSeq hg38, at default settings) after trimming with cutadapt (Martin, 2011, trimming Illumina adaptors, -q 20, -m 25; employing the “UTAP – User-friendly Transcriptome Analysis Pipeline,” unpublished data). We used exomePeak (Meng et al., 2014) for peak calling each separate m6A-RIP/input sample pair (with window_width = 100, sliding_step = 10, minimal_peak_length = 50). Peaks from biological replicates were merged keeping only those ranges supported by a minimum of 2/3 of samples per group (using BEDTools with minimum length 50 nt and merging ranges with less than 50 nt distance, Quinlan and Hall, 2010). Those were the group peaks shown in the supplemental figures. Further analysis of the peaks reveals that the differences in stress and basal are caused by peak-detection thresholds than being true present/absent peaks. For quantification with DiffBind (Ross-Innes et al., 2012) a consensus peak set was built with BEDTools including those ranges supported by either 2/3 of samples per group or 1/2 of samples of the entire experiment, furthermore filtered for minimum abundancy (dba_score_rpkm_fold = 2, dba_score_reads = 25 for mouse acute stress and BLCLs, = 15 for cKO mice). For differential peak analysis m6A/m-RIP counts extracted from DiffBind were compared with DESeq2 (Love et al., 2014; as peak counts in the input samples were not found majorly different we directly compared m6A/m counts without subtracting/dividing by counts of input samples thereby keeping the underlying negative binomial distribution). RNA expression was analyzed using the counts from RNA input samples only employing DESeq2 and conducted on gene-level. For acute stress cortex and BLCLs p values were corrected by fdrtool (Strimmer, 2008) based on parameters of the null distribution estimated adaptively from the data (as more than 10% of the original p values were above 0.9). m6A/m peaks were considered to be significantly different with an absolute fold change > 0.2 (mouse acute stress) or > 0.5 (mouse cKO and BLCLs) and a Benjamini-Hochberg corrected P value = Q < 0.1. Genes were considered to be significantly different with an absolute fold change > 0.1 (mouse acute stress) or > 0.5 (mouse cKO and BLCLs) and a Benjamini-Hochberg corrected P value = Q < 0.1. Peaks were annotated using ChIPseeker (Yu et al., 2015) and Biomart (Smedley et al., 2015). Distribution-plots of m6A/m across the transcript length were evaluated using the Guitar plots (Cui et al., 2016) package. GO-term overrepresentation was calculated using the PANTHER Overrepresentation Test (Mi et al., 2013) for “GO biological process complete” with the list of all detected genes (> 5 rawcounts in all samples) as background. Motif search was performed by DREME (Bailey, 2011) and CentriMo (Bailey and Machanick, 2012) using all detected m6A/m-peaks as input (100nt sequences centered on peak). Motifs are presented with B = G/C/T, N = A/C/G/T, W = A/T, Y = C/T. For comparison of the detected mouse m6A/m-Seq GGACWB motif with known motifs, we employed Tomtom (Gupta et al., 2007, Ray et al., 2013) and CentriMo (Bailey and Machanick, 2012). Comparison of peaks to known m6A/m was done using m6A/m data from RMBase v2.0 (Xuan et al., 2018; data as of 2017-06-01). Sequencing tracks were visualized with the IGV browser (Robinson et al., 2011). Overlap with FMR1 target genes FMR1 was computed using data from (Darnell et al., 2011) considering only genes expressed in both datasets.
Putative m6Am
m6Am was previously described to be localized to the first nucleotide after the TSS (Schibler and Perry, 1977, Linder et al., 2015, Mauer et al., 2017). Peaks were called putative m6Am peaks building up on earlier methods (Linder et al., 2015, Mauer et al., 2017, Schwartz et al., 2014) if the peak start was within a 100 bp window (50 up, 50 down) of an annotated TSS and if there was an INR (YYNWYY) or TATA box (TATAWAW) motif found in a 75 bp window (50 up, 25 down) around the TSS. These parameters were used to limit false-positive assignment and may underestimate the true extend of m6Am peaks.
Ribosome profiling analysis
Sequencing data quality control was performed by FASTQC (Andrews, 2010). Genomic alignment was performed on the reverse read only after adaptor- and quality-trimming with cutadapt keeping only those reads with RA5 adaptor present, insert length between 15 and 50 nt and a minimum Phred quality score of 15. The trimmed RPF-sequences were highly enriched for 30-34 nt length. After removing rRNA and tRNA using alignment with Tophat/Bowtie1 to mmu rRNA and tRNA sequences, remaining sequences were aligned using to the mouse transcriptome using Tophat/Bowtie1 (Langmead et al., 2009, Trapnell et al., 2009; at default settings to Gencode mm10/VM15). Translation efficiencies were calculated by dividing CPM from libraries of ribosome-bound fragments by CPM from libraries of total input RNA after counting reads with HTSeq (Anders et al., 2015; only genes considered with minimum 25 counts per sample). Translation efficiencies were compared using t tests with significantly regulated genes being defined with an absolute fold change > 0.5 and a Benjamini-Hochberg corrected P value = Q < 0.1.
mRNA-Seq analysis
Samples were processed as described for input libraries in m6A/m-Seq data with differential gene expression analysis being performed with DESeq2 using the specified factorial models. For analysis of basal expression patterns as presented in Figure 5, only the subset of unstressed “Box” animals was used. Genes were considered differential with an absolute fold change > 0.5 and a Benjamini-Hochberg corrected P value = Q < 0.1.
Gene expression analysis
Statistics were performed on log2 normalized data using a 2x2 MANOVA in SPSS and were multiple testing-corrected by the Benjamini-Hochberg test (cut-off Q < 0.05) in R and a cut-off by effect size (η2 > 0.01) and post hoc testing (Tukey HSD).
Candidate m6A/m-RIP-qPCR analysis
RNA abundance levels were quantified from the input samples using the ddCT method normalizing to the average of all housekeeping genes. Immunoprecipitation efficiency for each biological sample was assessed using the measured abundance of spike-in per m6A/m-RIP/IgG control sample. Because all conditions per sample were equally split at the beginning, % methylation or IgG signal was calculated as follows:
Statistics were performed on log2 normalized data (RNA) or absolute values (m6A/m) using a 2x2 MANOVA in SPSS with multiple testing performed using the Benjamini-Hochberg method (cut-off Q < 0.05) in R and a cut-off by effect size (η2 > 0.01) and post hoc testing (Tukey HSD).
Statistical analysis
Statistical tests were performed using SPSS (IBM SPSS Statistics, Armonk, NY: IBM) and R (R Development Core Team, 2011) as indicated in Figure legends with n and statistical results indicated in Figure legends und Supplemental Tables. Plots were produced with R (R Development Core Team, 2011) ggplot2 (Wickham, 2009) with definition of presented measurements indicated in the Figure legends. For animal experiments, sample size was estimated a priori using G∗Power (Faul et al., 2007) using α = 0.05 and an experience-based β. Animals and samples within experiments were randomized using stratified randomization assisted by random number generation.
Data and Software Availability
All supporting data and code for this study are available from the corresponding author upon request. Sequencing data are deposited at GEO: GSE113801 with subseries GSE113781 (m6A/m-Seq mouse cortex after acute stress), GSE113789 (Ribosome profiling Seq mouse cortex after acute stress), GSE113793 (m6A/m-Seq of mouse adult cortex of Mettl3 cKO or Fto cKO mice), GSE113796 (mRNA-Seq of mouse Mettl3 cKO or Fto cKO mouse hippocampus after fear conditioning), GSE113798 (m6A/m-Seq of human B-lymphocyte cell lines from healthy controls and major depressive disorder patients).
Acknowledgments
We thank T. Namendorf, E. Wagner, B. Hauger, C. Dournes, A. Mederer, A. Ressle, A. Parl, and D. Harbich for their technical support and A. Varga and the animal care team for their devoted assistance with animal care. We thank S. Karamihalev, M. Filiou, A. Menke, and A. Genewsky for helpful discussions and J. Keverne for professional English editing. We thank G. Rechavi, S. Moshitch-Moshkovitz, V. Hershkovitz, E. Feldmesser, and G. Stelzer for guidance with preparing and/or analyzing m6A/m-seq libraries and for helpful discussions. A.C. is the head of the Max Planck Society – Weizmann Institute of Science Laboratory for Experimental Neuropsychiatry and Behavioral Neurogenetics. This work was supported by travel grants from the Boehringer Ingelheim Fonds (M. Engel); the European Research Council (FP7 grant 260463); the Israel Science Foundation (1565/15); the ERANET Program, supported by the Chief Scientist Office of the Israeli Ministry of Health (3-11389); the Federal Ministry of Education and Research (01KU1501A); research support from Roberto and Renata Ruhman and Bruno and Simone Licht; the I-CORE Program of the Planning and Budgeting Committee and the Israel Science Foundation (1916/12); the Nella and Leon Benoziyo Center for Neurological Diseases; the Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics; the Perlman Family Foundation; the Adelis Foundation; the Irving I. Moskowitz Foundation (all previous to A.C.); the Israel Science Foundation (355/17 and 107/14, J.H.H); and FAMRI and NYSCF grants (J.H.H).
Author Contributions
M. Engel and A.C. conceived and designed the experiments and wrote the manuscript. M. Engel performed and analyzed most experiments including m6A/m-seq; bioinformatics; m6A-RIP-qPCR; and mouse, behavior, and cell culture experiments. C.E., P.M.K., L.T., and M.R.-H. assisted in experiments. M.J. performed preliminary experiments with no data used. S.R. performed bioinformatics analysis of pilot m6A/m-seq experiments not included. S.G. and J.H.H. generated Mettl3 mice. C.N. and M.U. performed and analyzed mass spectrometry experiments. M. Eder performed and analyzed electrophysiological studies. J.A. and E.B.B. provided human microarray data and clinical samples. P.W., D.L., C.T.W., M.V.S., J.M.D., and E.B.B. assisted in the design of the project. The study was supervised by A.C.
Declaration of Interests
The authors declare no competing interests.
Published: July 25, 2018
Footnotes
Supplemental Information includes nine figures and five tables and can be found with this article online at https://doi.org/10.1016/j.neuron.2018.07.009.
Supplemental Information
References
- Abbas A.R., Wolslegel K., Seshasayee D., Modrusan Z., Clark H.F. Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS ONE. 2009;4:e6098. doi: 10.1371/journal.pone.0006098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari A., Lerner T.N., Finkelstein J., Pak S., Jennings J.H., Davidson T.J., Ferenczi E., Gunaydin L.A., Mirzabekov J.J., Ye L. Basomedial amygdala mediates top-down control of anxiety and fear. Nature. 2015;527:179–185. doi: 10.1038/nature15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Dibaj P., Kassmann C.M., Goebbels S., Nave K.-A., Schwab M.H. In vivo imaging and noninvasive ablation of pyramidal neurons in adult NEX-CreERT2 mice. Cereb. Cortex. 2012;22:1473–1486. doi: 10.1093/cercor/bhr214. [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S. (2010). FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Arloth J., Bogdan R., Weber P., Frishman G., Menke A., Wagner K.V., Balsevich G., Schmidt M.V., Karbalai N., Czamara D., Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium (PGC) Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium PGC Genetic differences in the immediate transcriptome response to stress predict risk-related brain function and psychiatric disorders. Neuron. 2015;86:1189–1202. doi: 10.1016/j.neuron.2015.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27:1653–1659. doi: 10.1093/bioinformatics/btr261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Machanick P. Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res. 2012;40:e128. doi: 10.1093/nar/gks433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikina M., Zaslavsky E., Sealfon S.C. CellCODE: a robust latent variable approach to differential expression analysis for heterogeneous cell populations. Bioinformatics. 2015;31:1584–1591. doi: 10.1093/bioinformatics/btv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., Wei Z., Zhang L., Liu H., Sun L., Zhang S.-W., Huang Y., Meng J. Guitar: an R/Bioconductor package for gene annotation guided transcriptomic analysis of RNA-related genomic features. BioMed Res. Int. 2016;2016:8367534. doi: 10.1155/2016/8367534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.-G. m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y.S., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kloet E.R., Joëls M., Holsboer F. Stress and the brain: from adaptation to disease. Nat. Rev. Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- Du T., Rao S., Wu L., Ye N., Liu Z., Hu H., Xiu J., Shen Y., Xu Q. An association study of the m6A genes with major depressive disorder in Chinese Han population. J. Affect. Disord. 2015;183:279–286. doi: 10.1016/j.jad.2015.05.025. [DOI] [PubMed] [Google Scholar]
- Edupuganti R.R., Geiger S., Lindeboom R.G.H., Shi H., Hsu P.J., Lu Z., Wang S.-Y., Baltissen M.P.A., Jansen P.W.T.C., Rossa M. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017;24:870–878. doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel M., Chen A. The emerging role of mRNA methylation in normal and pathological behavior. Genes Brain Behav. 2018;17:e12428. doi: 10.1111/gbb.12428. [DOI] [PubMed] [Google Scholar]
- Ewald E.R., Wand G.S., Seifuddin F., Yang X., Tamashiro K.L., Potash J.B., Zandi P., Lee R.S. Alterations in DNA methylation of Fkbp5 as a determinant of blood-brain correlation of glucocorticoid exposure. Psychoneuroendocrinology. 2014;44:112–122. doi: 10.1016/j.psyneuen.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faul F., Erdfelder E., Lang A.-G., Buchner A. G∗Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods. 2007;39:175–191. doi: 10.3758/bf03193146. [DOI] [PubMed] [Google Scholar]
- Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S. Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M.E., Hess S., Meyer K.D., Verhagen L.A.W., Koch L., Brönneke H.S., Dietrich M.O., Jordan S.D., Saletore Y., Elemento O. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013;16:1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Brar G.A., Stern-Ginossar N., Harris M.S., Talhouarne G.J.S., Jackson S.E., Wills M.R., Weissman J.S. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.-G., He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S., Pandya-Jones A., Saito Y., Fak J.J., Vågbø C.B., Geula S., Hanna J.H., Black D.L., Darnell J.E., Jr., Darnell R.B. m6A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev. 2017;31:990–1006. doi: 10.1101/gad.301036.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T., Binder E.B. Epigenetics of stress-related psychiatric disorders and gene × environment interactions. Neuron. 2015;86:1343–1357. doi: 10.1016/j.neuron.2015.05.036. [DOI] [PubMed] [Google Scholar]
- Klungland A., Dahl J.A., Greggains G., Fedorcsak P., Filipczyk A. Reversible RNA modifications in meiosis and pluripotency. Nat. Methods. 2016;14:18–22. doi: 10.1038/nmeth.4111. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10–12. [Google Scholar]
- Mathelier A., Fornes O., Arenillas D.J., Chen C.Y., Denay G., Lee J., Shi W., Shyr C., Tan G., Worsley-Hunt R. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44(D1):D110–D115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.-J., Chen Q. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.S., Bowles N.P., Gray J.D., Hill M.N., Hunter R.G., Karatsoreos I.N., Nasca C. Mechanisms of stress in the brain. Nat. Neurosci. 2015;18:1353–1363. doi: 10.1038/nn.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Lu Z., Liu H., Zhang L., Zhang S., Chen Y., Rao M.K., Huang Y. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke A., Arloth J., Pütz B., Weber P., Klengel T., Mehta D., Gonik M., Rex-Haffner M., Rubel J., Uhr M. Dexamethasone stimulated gene expression in peripheral blood is a sensitive marker for glucocorticoid receptor resistance in depressed patients. Neuropsychopharmacology. 2012;37:1455–1464. doi: 10.1038/npp.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milaneschi Y., Lamers F., Mbarek H., Hottenga J.-J., Boomsma D.I., Penninx B.W.J.H. The effect of FTO rs9939609 on major depression differs across MDD subtypes. Mol. Psychiatry. 2014;19:960–962. doi: 10.1038/mp.2014.4. [DOI] [PubMed] [Google Scholar]
- Minichiello L., Korte M., Wolfer D., Kühn R., Unsicker K., Cestari V., Rossi-Arnaud C., Lipp H.-P., Bonhoeffer T., Klein R. Essential role for TrkB receptors in hippocampus-mediated learning. Neuron. 1999;24:401–414. doi: 10.1016/s0896-6273(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peer E., Rechavi G., Dominissini D. Epitranscriptomics: regulation of mRNA metabolism through modifications. Curr. Opin. Chem. Biol. 2017;41:93–98. doi: 10.1016/j.cbpa.2017.10.008. [DOI] [PubMed] [Google Scholar]
- Ping X.-L., Sun B.-F., Wang L., Xiao W., Yang X., Wang W.-J., Adhikari S., Shi Y., Lv Y., Chen Y.-S. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provençal N., Suderman M.J., Guillemin C., Massart R., Ruggiero A., Wang D., Bennett A.J., Pierre P.J., Friedman D.P., Côté S.M. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J. Neurosci. 2012;32:15626–15642. doi: 10.1523/JNEUROSCI.1470-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . The R Foundation for Statistical Computing; 2011. R: a language and environment for statistical computing. [Google Scholar]
- Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refojo D., Schweizer M., Kuehne C., Ehrenberg S., Thoeringer C., Vogl A.M., Dedic N., Schumacher M., von Wolff G., Avrabos C. Glutamatergic and dopaminergic neurons mediate anxiogenic and anxiolytic effects of CRHR1. Science. 2011;333:1903–1907. doi: 10.1126/science.1202107. [DOI] [PubMed] [Google Scholar]
- Reuter J.S., Mathews D.H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010;11:129. doi: 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaan Z., Anand S.S., Zhang X., Desai D., Rivera M., Pare G., Thabane L., Xie C., Gerstein H., Engert J.C. The protective effect of the obesity-associated rs9939609 A variant in fat mass- and obesity-associated gene on depression. Mol. Psychiatry. 2013;18:1281–1286. doi: 10.1038/mp.2012.160. [DOI] [PubMed] [Google Scholar]
- Schibler U., Perry R.P. The 5′-termini of heterogeneous nuclear RNA: a comparison among molecules of different sizes and ages. Nucleic Acids Res. 1977;4:4133–4149. doi: 10.1093/nar/4.12.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.V., Schülke J.-P., Liebl C., Stiess M., Avrabos C., Bock J., Wochnik G.M., Davies H.A., Zimmermann N., Scharf S.H. Tumor suppressor down-regulated in renal cell carcinoma 1 (DRR1) is a stress-induced actin bundling factor that modulates synaptic efficacy and cognition. Proc. Natl. Acad. Sci. USA. 2011;108:17213–17218. doi: 10.1073/pnas.1103318108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slobodin B., Han R., Calderone V., Vrielink J.A.F.O., Loayza-Puch F., Elkon R., Agami R. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell. 2017;169:326–337.e12. doi: 10.1016/j.cell.2017.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D., Haider S., Durinck S., Pandini L., Provero P., Allen J., Arnaiz O., Awedh M.H., Baldock R., Barbiera G. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43(W1) doi: 10.1093/nar/gkv350. W589-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer K. A unified approach to false discovery rate estimation. BMC Bioinformatics. 2008;9:303. doi: 10.1186/1471-2105-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters B.J., Mercaldo V., Gillon C.J., Yip M., Neve R.L., Boyce F.M., Frankland P.W., Josselyn S.A. The role of the RNA demethylase FTO (fat mass and obesity-associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology. 2017;42:1502–1510. doi: 10.1038/npp.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Widagdo J., Zhao Q.-Y., Kempen M.-J., Tan M.C., Ratnu V.S., Wei W., Leighton L., Spadaro P.A., Edson J., Anggono V., Bredy T.W. Experience-dependent accumulation of N6-methyladenosine in the prefrontal cortex is associated with memory processes in mice. J. Neurosci. 2016;36:6771–6777. doi: 10.1523/JNEUROSCI.4053-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Laurent B., Hsu C.-H., Nachtergaele S., Lu Z., Sheng W., Xu C., Chen H., Ouyang J., Wang S. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan J.-J., Sun W.-J., Lin P.-H., Zhou K.-R., Liu S., Zheng L.-L., Qu L.-H., Yang J.-H. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018;46(D1):D327–D334. doi: 10.1093/nar/gkx934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Wang L.-G., He Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31:2382–2383. doi: 10.1093/bioinformatics/btv145. [DOI] [PubMed] [Google Scholar]
- Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S.-H. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.-B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015;526:591–594. doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zeng P., Li Y.-H., Zhang Z., Cui Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016;44:e91. doi: 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.