ABSTRACT
Macroautophagy/autophagy is a conserved catabolic process that promotes survival during stress. Autophagic dysfunction is associated with pathologies such as cancer and neurodegenerative diseases. Thus, autophagy must be strictly modulated at multiple levels (transcriptional, post-transcriptional, translational and post-translational) to prevent deregulation. Relatively little is known about the post-transcriptional control of autophagy. Here we report that the exoribonuclease Xrn1/XRN1 functions as a negative autophagy factor in the yeast Saccharomyces cerevisiae and in mammalian cells. In yeast, chromosomal deletion of XRN1 enhances autophagy and the frequency of autophagosome formation. Loss of Xrn1 results in the upregulation of autophagy-related (ATG) transcripts under nutrient-replete conditions, and this effect is dependent on the ribonuclease activity of Xrn1. Xrn1 expression is regulated by the yeast transcription factor Ash1 in rich conditions. In mammalian cells, siRNA depletion of XRN1 enhances autophagy and the replication of 2 picornaviruses. This work provides insight into the role of the RNA decay factor Xrn1/XRN1 as a post-transcriptional regulator of autophagy.
KEYWORDS: Ash1, Atg8, autophagosome, autophagy related, BECN1, coxsackievirus B, CRISPR, CVB, Dcp2, LC3, macroautophagy, mRNA, picornavirus, poliovirus, RNA, RNA decay, stress, vacuole, yeast
Abbreviations
- Ape1
aminopeptidase I
- CVB
coxsackievirus B
- EV
empty vector
- GFP
green fluorescent protein
- HA
hemagglutinin
- HBMECs
human brain microvascular endothelial cells
- HBSS
Hanks balanced salt solution
- HDAC
histone deacetylase complex
- KO
knockout
- LC3
microtubule associated protein 1 light chain 3
- PA
protein A
- PCR
polymerase chain reaction
- prApe1
precursor Ape1
- RT-qPCR
real-time quantitative PCR
- PE
phosphatidylethanolamine
- PV
poliovirus
- SD
standard deviation
- RNAse
ribonuclease
- Rpd3L
Rpd3 large
- TEM
transmission electron microscopy
- ts
temperature-sensitive
- WT
wild type
Introduction
Macroautophagy/autophagy is a highly dynamic and conserved process of cellular self-eating that promotes survival during various types of stress including nutrient deprivation. The formation of the double-membrane autophagosome is the classical morphological feature of autophagy. Basal levels of autophagy are generally low; however, autophagy is upregulated by various stimuli. During nonselective autophagy (such as that induced by starvation), bulk cytoplasm is taken up into phagophores, the precursors to autophagosomes. Fusion of the autophagosome with the vacuole (in yeast) or the lysosome (in mammalian cells) is a late step in the autophagy pathway. Autophagic flux culminates with cargo degradation within the vacuole or lysosome, followed by efflux of the breakdown products through membrane permeases.
Dysregulated autophagy is associated with a wide range of human diseases. Accordingly, this process must be strictly regulated, and these controls occur at multiple levels including transcriptional, post-transcriptional and post-translational [1–3]. In mammalian cells, noncoding RNAs such as microRNAs may influence autophagy induction at the post-transcriptional level[1,3–5]. However, the RNA interference system has been lost in Saccharomyces cerevisiae [6]v RNA decay pathways are also critical to maintaining post-transcriptional regulation over gene expression. mRNA species are canonically degraded following trimming of the polyA tail in either the 5' to 3' direction by the cytoplasmic exoribonuclease Xrn1, or the 3' to 5' direction by the multisubunit exosome complex (for review see refs. [7–9]). At least for yeast, these 2 degradation pathways are the major mechanisms for RNA decay and turnover in the cytoplasm.[10]
Xrn1 is a highly conserved (from yeast to human) component of processing bodies, which are considered to be probable sites of cytoplasmic mRNA degradation for non-translating mRNAs [7,11–13]; although mRNA degradation also occurs cotranslationally [14]. In the 5' to 3' mRNA decay pathway, mRNAs are trimmed and/or deadenylated by the Pan2-Pan3 and Ccr4-NOT complexes and then undergo decapping by Dcp2 and the decapping cofactor Dhh1/Ddx6 to hydrolytically remove the 5' methylguanosine cap from transcribed mRNAs prior to degradation [8,9,15]. Pathways involving Xrn1-mediated RNA decay also exist independently of the canonical 5' to 3' decapping-dependent mechanism [8,10,16–21]. Endonucleolytically cleaved mRNA and long non coding RNA are also substrates that are susceptible to Xrn1 RNAse activity (reviewed in refs. [8,19]) [20–23]. Furthermore, Xrn1 may function as a selectivity factor [24], as loss of expression does not simply equate to an overall global increase in steady-state transcript levels [25].
In this study, we expand on our previous work, which identified the decapping enzyme Dcp2 and Dhh1 as post-transcriptional regulators of autophagy and autophagy-related (ATG) transcripts [26]. Here we investigate the role of Xrn1, which functions downstream of Dhh1-Dcp2 in canonical 5' to 3' RNA decay. Our studies demonstrate that Xrn1/XRN1 functions as a negative regulator of autophagy in both yeast and mammalian cells. In yeast, chromosomal deletion of XRN1 enhances autophagy during short-term nitrogen starvation and increases the frequency of autophagosome formation (i.e., the number of autophagosomes formed). Loss of Xrn1 results in the upregulation of unique ATG transcripts under nutrient-replete conditions. We demonstrate that downregulation of select ATG transcripts is dependent on the ribonuclease activity of Xrn1. Xrn1 expression is positively regulated by the yeast transcription factor Ash1 during nutrient-rich conditions. Xrn1 expression decreases significantly when cells are starved. In mammalian cells, XRN1 also functions to maintain autophagy at an appropriate basal level and to limit the infectivity of 2 picornaviruses—poliovirus (PV) and coxsackievirus B (CVB). Our current data support a model in which the cell produces an excess of select ATG transcripts during fed conditions, which are targeted by Xrn1; this mechanism serves to jumpstart autophagy once the cell is starved.
Results
The exoribonuclease Xrn1 is a negative regulator of autophagy
In a comprehensive screen for RNA-binding proteins, we previously identified the RCK family member Dhh1, a cytoplasmic DExD/H-box helicase that promotes mRNA decapping and decay, and Dcp2, the catalytic subunit of the decapping enzyme complex in S. cerevisiae, as post-transcriptional regulators of autophagy [26]. To expand on our earlier work and to search for other RNA regulatory factors that could be involved in modulating autophagy, we screened additional mutants deficient in genes from various RNA decay pathways using the Pho8Δ60 assay and identified Xrn1 (Figure 1A). In the Pho8Δ60 assay, vacuolar phosphatase activity is a quantitative enzymatic measurement of autophagy activity [27]; however, some minimal degree of background phosphatase activity is observed in cells even under rich conditions. Here, we examined cells at 3 h of nitrogen starvation when the level of background activity should be reduced; shorter time points of starvation (1–2 h) lead to higher background levels and a less robust signal-to-noise ratio (our unpublished observations). As expected, in cells that were starved for nitrogen, we observed a substantial increase in autophagy activity in the wild-type (WT) strain and little activity in the atg1∆ strain (Figure 1A). In comparison, starvation-induced autophagy was enhanced (∼20%) in the xrn1∆ strain (Figure 1A). Additional evidence presented here (Figure 1B to F, Figure 2A and B, and Figure 5A and B) indicates a role for Xrn1 as an autophagy mediator at earlier stages in the pathway and may explain why the Pho8∆60 response (measured at 3 h) is not as robust as observed in assays performed using shorter time points (30–120 min).
Figure 1.
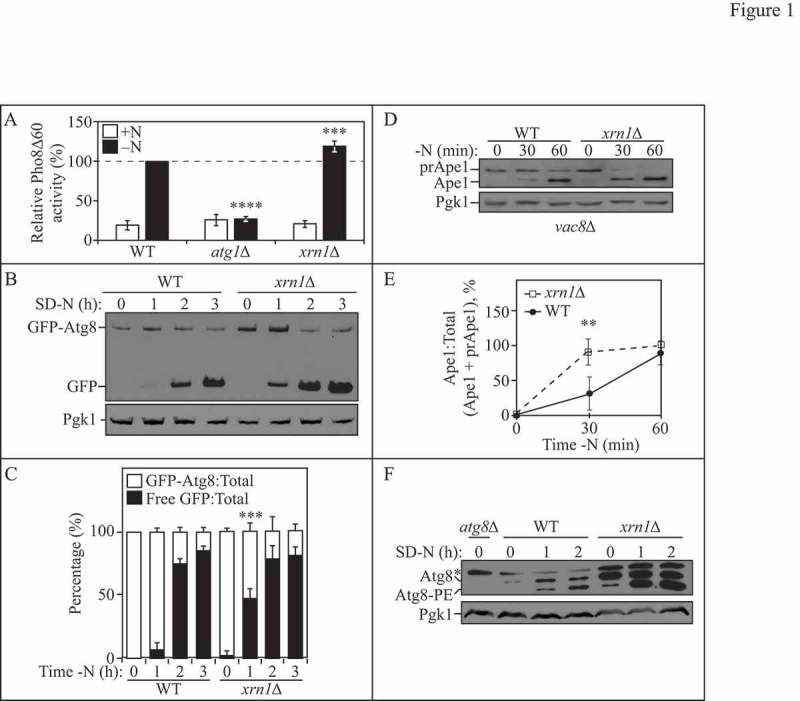
Xrn1 negatively regulates autophagy. (A) WT (WLY176), atg1∆ (WLY192) and xrn1∆ (EDA62) cells were grown to mid-log phase in YPD (+N) and then starved for nitrogen (-N) for 3 h. The Pho8Δ60 activity was measured and normalized to the activity of starved WT cells, which was set at 100%. Results are the mean of 5 independent experiments. Error bars indicate standard deviation (SD). (B) WT (WLY176) and xrn1∆ (EDA62) cells transformed with a centromeric plasmid encoding GFP-Atg8 were grown to mid-log phase in rich selective medium and then starved for nitrogen (SD-N) for 0, 1, 2 or 3 h. Cells were collected at the indicated time points and protein extracts were analyzed by SDS-PAGE and blotted with anti-YFP or anti-Pgk1 (loading control) antibodies. (C) Densitometry of blots represented in (B). The ratio of GFP-Atg8:total GFP (sum of full-length GFP-Atg8 and free GFP) or free GFP:total GFP was quantified with ImageJ. Results shown are the mean of 4 independent experiments +/− SD. (D) WT (vac8∆; CWY230) and xrn1∆ (vac8∆ xrn1∆; EDA152) cells at 0, 30 and 60 min of nitrogen starvation. Precursor (prApe1) and mature (Ape1) forms of aminopeptidase 1 were detected with anti-Ape1 or anti-Pgk1 (loading control) antisera. (E) Densitometry of blots represented in (D). The ratio of Ape1:total Ape1 (sum of prApe1 and Ape1) was quantified with ImageJ. Results shown are the mean of 4 independent experiments +/− SD. (F) WT (SEY6210), atg8∆ (YAB369) and xrn1∆ (EDA85) cells were grown to mid-log phase in YPD and then starved for nitrogen (SD-N) for 0, 1, or 2 h. Protein extracts were analyzed by SDS-PAGE and blotted with anti-Atg8 or anti-Pgk1 (loading control) antisera (*denotes nonspecific band). Blots shown are representative of 3 independent experiments. Also see Table S1.
Figure 2.
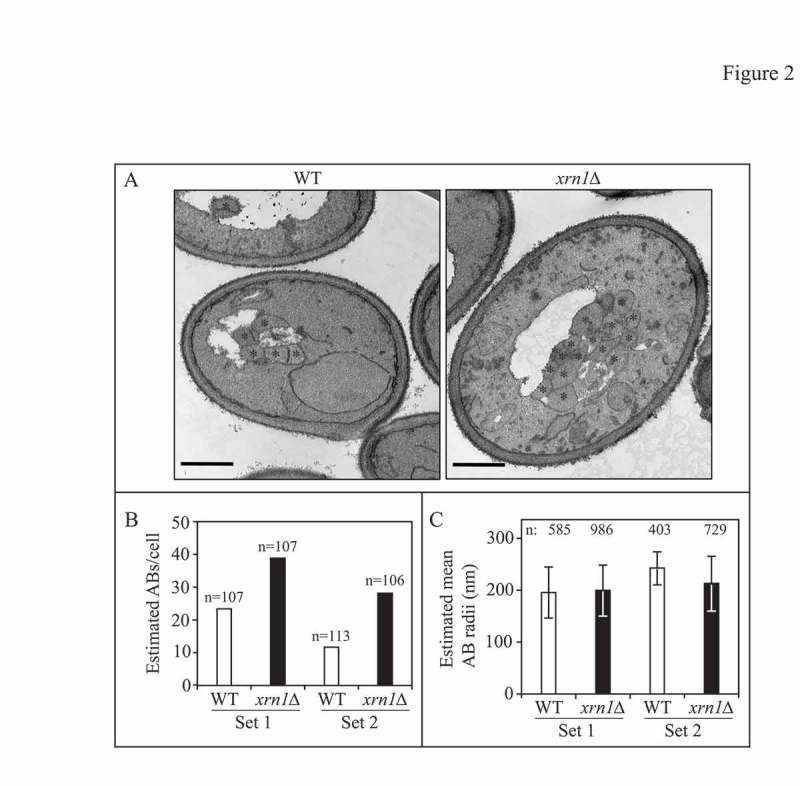
Loss of XRN1 increases autophagosome formation frequency. (A) Representative TEM images showing enhanced accumulation of autophagic bodies (ABs) in xrn1∆ cells (EDA117) compared to WT cells (FRY143) from 2 independent experiments after 2 h in SD-N. Asterisks indicate autophagic bodies. Scale bar: 500 nm. (B) Estimated average number of autophagic bodies per cell. Estimation was based on the number of autophagic body cross-sections observed by TEM [38]. Over 100 unique cells were captured and analyzed per strain for each independent experiment. (C) The estimated mean radii (in nm) of the original autophagic bodies observed by TEM in WT and xrn1∆ cells was analyzed as described previously [38]. Over 400 unique autophagic bodies were captured and analyzed per strain for each independent experiment. Also see Figure S1 and Table S2.
Figure 5.
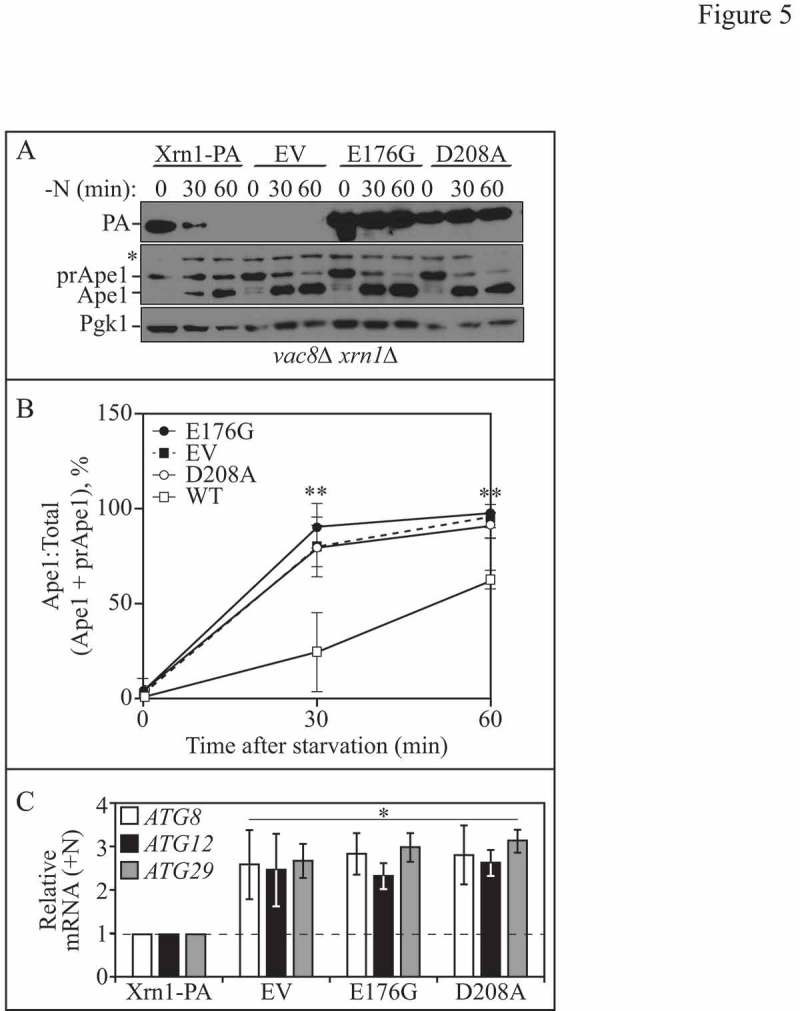
The ribonuclease activity of Xrn1 is required for autophagy regulation. (A) Lysates were collected from vac8∆ xrn1∆ cells expressing a chromosomally integrated plasmid encoding WT Xrn1-PA (EDA147), empty vector (EV, pRS406; EDA142), or catalytically inactive Xrn1E176G-PA (EDA148) or Xrn1D208A-PA (EDA149), which were nitrogen starved for 0, 30 or 60 min. Protein extracts were analyzed by SDS-PAGE and probed with anti-PA, anti-Ape1 or anti-Pgk1 (loading control) antisera (* denotes nonspecific band). The blots shown are representative of 3 independent experiments. (B) Densitometry of blots represented in A. The ratio of Ape1:total Ape1 (sum of prApe1 and Ape1) was quantified with ImageJ. The one-way ANOVA with Dunnett's multiple comparison post-hoc test was used to determine statistical significance. Results shown are the mean +/− SD. (C) Total RNA was extracted from xrn1∆ cells expressing a chromosomally integrated plasmid encoding WT Xrn1-PA (EDA134), empty vector (EV, pRS406; EDA133), Xrn1E176G-PA (EDA135) or Xrn1D208A-PA (EDA136), and RT-qPCR was performed. Results are shown relative to the level of ATG8, ATG12 or ATG29 mRNA expression in WT cells under rich conditions (+N), which was set to 1. The geometric mean of TFC1 and SLD3 was used to quantify relative expression levels. Results shown are the mean of 3 independent experiments +/− SD. Also see Figure S3 and Table S4.
To further validate our results that the loss of XRN1 enhances autophagy, we performed the GFP-Atg8 processing assay (Figure 1B and C). Atg8 is required for membrane expansion and closure of the phagophore. Atg8 associates with both the inner and outer membranes of the nascent phagophore, and the population that is present on the inner membrane remains associated with the completed autophagosome [28]. Progression through the autophagic pathway results in the fusion of the autophagosome with the vacuole. The inner autophagosome vesicle is released into the vacuole lumen, where it is termed an autophagic body; this vesicle is lysed allowing cargo access to vacuolar hydrolases. The use of a GFP-Atg8 fusion protein can be utilized as a method to assess the level of bulk autophagy based on the release of free GFP, which is relatively stable in the vacuole lumen [29–31]. Utilizing the GFP-Atg8 processing assay, we observed that the xrn1∆ strain showed higher levels of free GFP (and thus higher autophagy activity) than the WT strain at 1 h of starvation (Figure 1B and C).
We further confirmed that cells deficient in Xrn1 undergo a more rapid and robust autophagy response by performing the precursor aminopeptidase I (prApe1) processing assay in a vac8∆ strain background (Figure 1D and E). Ape1 is a resident vacuolar hydrolase that traffics to the vacuole through the cytoplasm-to-vacuole targeting (Cvt) pathway, a biosynthetic mechanism of selective autophagy. The Cvt pathway involves recognition of an Ape1 complex by its receptor Atg19 [32] and the scaffold protein Atg11[33] under rich conditions. Within the vacuole, prApe1 (61 kDa) is proteolytically processed to Ape1 (50 kDa); this can be monitored by SDS-PAGE and western blotting, thus providing a method for assaying selective autophagy activity. In the vac8∆ mutant, the Cvt pathway is defective under nutrient-replete conditions, and prApe1 can only traffic to the vacuole and undergo processing during starvation conditions in a selective autophagy process that still requires both Atg11 and Atg19 [34]. In this autophagy assay, xrn1∆ cells exhibit a more rapid induction of autophagy when starved for nitrogen (Figure 1D and E).
Finally, we examined the consequence of deleting XRN1 on the biosynthesis of Atg8. The Atg8 protein exists in 2 forms in the cell—a C-terminally cleaved, nonlipidated soluble species, and a lipidated phosphatidylethanolamine (PE)-conjugated form. In addition, ATG8 transcription and Atg8 protein levels increase substantially during autophagy induction [35]. Thus, monitoring the total amount of Atg8 and its lipidation status provides another readout for autophagy. We found that cells lacking XRN1 had an increased amount of Atg8 relative to the WT and showed enhanced Atg8 lipidation (Atg8–PE) (Figure 1F), further demonstrating that Xrn1 negatively regulates autophagy.
Loss of Xrn1 increases the frequency of autophagosome formation
Alterations in the magnitude of autophagy can be due to a change in the number or in the size of autophagosomes [35–37]. To address whether loss of Xrn1 affected the size or frequency of autophagosome formation, we performed transmission electron microscopy (TEM) on starved yeast cells lacking XRN1 (Figure 2). To do this analysis, we examined the status of autophagic bodies within the vacuole of cells lacking the vacuolar protease Pep4/proteinase A. In the absence of Pep4, cells maintain (rather than degrade) autophagic bodies in the vacuole. In total, we examined over 200 unique cells (between 2 independent sample preparations) and analyzed the images using a published method for estimating both the size and number of the original autophagic bodies [38] (Figure 2, Figure S1 and Table S2). When nitrogen starved for 2 h, xrn1∆ cells exhibited an increase in the number of autophagosomes of ∼2-fold over WT (Figure 2A and B and Table S2). No significant difference in autophagosome size was noted between WT and xrn1∆ cells (Figure 2A and C and Table S2). These data support a model whereby Xrn1 negatively regulates autophagy by affecting the frequency of autophagosome formation.
Xrn1 expression decreases with nitrogen starvation and autophagy induction
Negative regulators of autophagy, which maintain activity at a basal level under nutrient-rich conditions, are typically inactivated following the induction of autophagy (reviewed in ref. [39]). To determine whether Xrn1 expression is altered during autophagy induction, we examined XRN1 mRNA levels during both growing and starvation conditions in WT cells with real-time quantitative PCR (RT-qPCR; Figure 3A). We observed a significant decrease (∼60%) in XRN1 mRNA levels at 1 h of nitrogen starvation compared to rich conditions (Figure 3A). We then sought to determine whether the observed decrease in XRN1 mRNA corresponded to a similar reduction in Xrn1 protein levels (Figure 3B and C and Figure S2). We chromosomally tagged XRN1 with either hemagglutinin (3xHA; Figure 3B and C) or (PA; Figure S2) at the C terminus, and assessed endogenous Xrn1 fusion protein expression levels by western blot (Figure 3B and C and Figure S2). We observed a significant decrease in Xrn1-3xHA (and Xrn1-PA) expression during nitrogen starvation, further supporting its role as a negative autophagy regulator in growing conditions that is inactivated when autophagy is stimulated.
Figure 3.
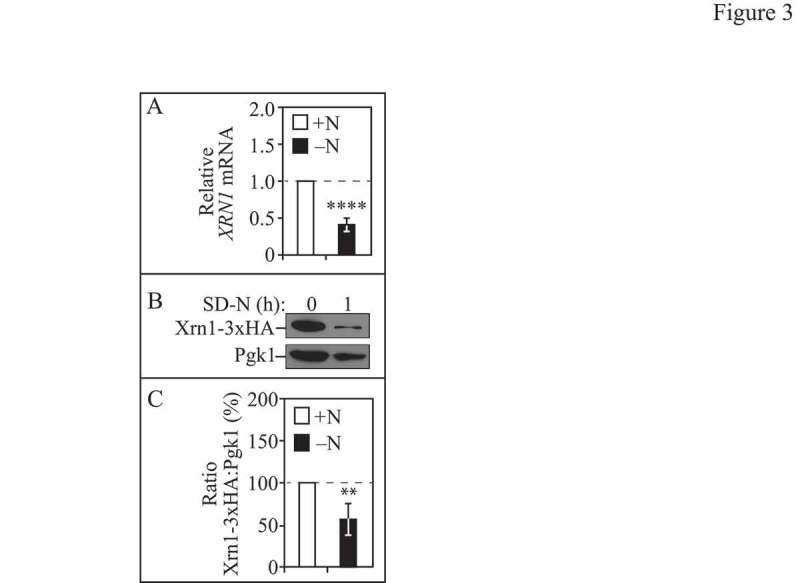
Xrn1 expression levels decrease after autophagy induction. (A) WT (WLY176) cells were grown to mid-log phase in YPD (+N) and then nitrogen starved (-N) for 1 h. Total RNA was extracted and RT-qPCR was performed. Results shown are relative to the level of XRN1 mRNA in rich conditions (+N), which was set to 1. TFC1 was used to quantify relative expression levels. Results shown are the mean of 6 independent experiments +/− SD. (B) Cells (EDA161) endogenously expressing Xrn1-3xHA were grown to mid-log phase in YPD (+N) and then nitrogen starved (-N) for 1 h. Protein extracts were analyzed by SDS-PAGE and blotted with anti-HA antibody or anti-Pgk1 (loading control) antiserum. (C) Densitometry of n=4 blots represented in B. The ratio of Xrn1-3xHA:Pgk1 was quantified with ImageJ. Also see Figure S2.
Xrn1 represses the expression of select ATG mRNAs under nutrient-replete conditions
Recent work has demonstrated the inherent complexity in the regulatory network that controls ATG gene expression at multiple levels (reviewed in refs. [1,40]) [26,36,37,41–43]. Additionally, components of the mRNA decapping machinery (in particular Dcp2 and Dhh1) play an important role in regulating ATG mRNA levels [26]. As Xrn1 is the major cytoplasmic 5' to 3' exoribonuclease in yeast and because we observed that Xrn1 functions as an autophagy repressor under rich conditions, we examined whether loss of Xrn1 affected ATG mRNA levels (Figure 4A and B). We observed that under nutrient-replete conditions, mRNA levels of select ATG mRNAs including ATG1, 4, 5, 7, 8, 12, 14, 16, 29 and 31 were significantly upregulated in xrn1∆ cells compared to WT (Figure 4A). During starvation conditions, only ATG5 and ATG12 mRNA levels remained significantly higher in xrn1∆ cells; while ATG2, ATG10, ATG11 and ATG32 decreased significantly compared to WT (Figure 4B).
Figure 4.
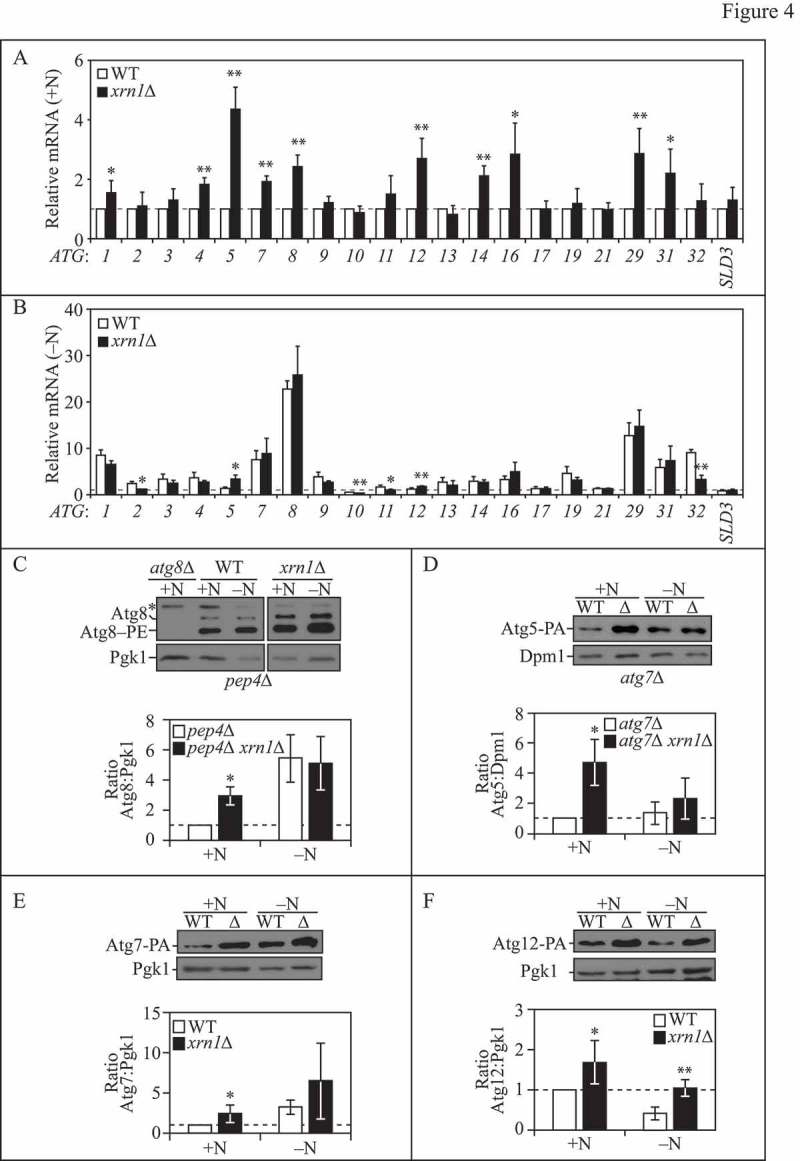
Xrn1 represses the transcription of select ATG mRNAs under nutrient-rich conditions. (A) WT (WLY176) and xrn1∆ (EDA62) cells were grown to mid-log phase in YPD (+N). Total RNA was extracted and RT-qPCR was performed. Results are shown relative to the level of mRNA expression in WT cells under rich conditions (+N), which was set to 1. TFC1 was used to quantify relative expression levels. Results shown are the mean of at least 3 independent experiments. (B) WT (WLY176) and xrn1∆ (EDA62) cells were nitrogen starved (-N) for 1 h. Total RNA was extracted and analyzed as in (A). (C) (Top) Loss of XRN1 enhances Atg8 protein expression. WT (pep4∆; TVY1), atg8∆ (YAB369) and xrn1∆ (EDA84) cells were grown to mid-log phase in YPD and then nitrogen starved (SD-N) for 0 or 1 h. Protein extracts were analyzed by SDS-PAGE and blotted with anti-Atg8 or anti-Pgk1 (loading control) antisera (* denotes nonspecific band). The blots shown are representative of 3 independent experiments. (Bottom) Densitometry of blots represented on the top. The ratio of total Atg8 (sum of Atg8 and Atg8–PE species):Pgk1 was quantified with ImageJ. (D-F) (Top) Loss of XRN1 enhances Atg5 (D), Atg7 (E) and Atg12 (F) protein expression. WT (atg7∆, D; JMY104, E; JMY099, F) or xrn1∆ (EDA129, EDA87 or EDA86) cells were grown to mid-log phase in YPD and then nitrogen starved (SD-N) for 0 (+N) or 2 h (-N). Protein extracts were analyzed by SDS-PAGE and blotted with anti-PA, anti-Dpm1 (D) or anti-Pgk1 (E, F; loading controls) antisera. The blots shown are representative of at least 3 independent experiments. (Bottom) Densitometry of blots represented on the top. The ratio of PA:Dpm1 (D) or PA:Pgk1 (E, F) was quantified with ImageJ. In graphs (A-F), results shown are the mean +/− SD. Also see Table S3.
We also investigated whether the transcriptional increases in specific ATG mRNAs correlated with increases at the protein level (Figure 4C to F). To analyze Atg8 protein levels, we used a pep4∆ strain to ensure that any changes in protein level were not due to alterations in Atg8 turnover and degradation within the vacuole (Figure 4C). Under nutrient-rich (but not starvation) conditions, we found a significant upregulation of Atg8 protein in xrn1∆ cells compared to WT (Figure 4C), which is consistent with what we observed by RT-qPCR (Figure 4A). Next, we examined whether Atg5 protein levels were also increased (Figure 4D). To ensure that we were only analyzing free Atg5 (and not Atg5 as a component of the covalently linked Atg12–Atg5 complex), we used an atg7∆ strain expressing Atg5 chromosomally tagged with PA at its C terminus. Atg7 is a ubiquitin-activating E1-like enzyme that is required for Atg12–Atg5 conjugation [44]. Thus, in the absence of Atg7, Atg12–Atg5 do not undergo conjugation. We observed a significant upregulation in free Atg5-PA protein levels under rich conditions in cells lacking Xrn1 (Figure 4D). In xrn1∆ cells, Atg5-PA protein levels were higher under starvation conditions compared to WT cells, but this was not statistically significant at the time point tested (2 h; Figure 4D). We also examined Atg7-PA and Atg12-PA levels, both of which notably increased during rich conditions in xrn1∆ strains compared to WT (Figure 4E and F), consistent with what we had observed by RT-qPCR (Figure 4A). In xrn1∆ cells expressing endogenous Atg12 chromosomally tagged with PA (preventing Atg12–Atg5 conjugation), Atg12-PA protein levels were substantially higher than WT under nitrogen starvation conditions (Figure 4F), also consistent with our RT-qPCR data (Figure 4B). Together, these results support a role for Xrn1 in the turnover and repression of a subset of ATG mRNAs during nutrient-replete conditions.
The exoribonuclease activity of Xrn1 is required for autophagy regulation
Xrn1 functions as a component of a buffering mechanism for maintaining appropriate transcript levels [25,45]. As we had observed that XRN1 deletion led to enhanced autophagy (Figures 1 and 2) and the selective upregulation of unique ATG transcripts (Figure 4A and B), we investigated whether the RNase activity of Xrn1 was responsible (Figure 5). Key residues within the Xrn1 active site are conserved across species (Figure S3A). We cloned XRN1 from S. cerevisiae genomic DNA and mutated E176G and D208A within the active site because mutations at these sites eliminate the RNase activity of Xrn1[46,47]. We then integrated these constructs (empty vector [EV], WT Xrn1-PA, Xrn1E176G-PA and Xrn1D208A-PA) at the URA3 locus in xrn1∆ strains and investigated the impact on autophagy by performing the prApe1-processing assay in a vac8∆ xrn1∆ background (Figure 5A and B). We assayed autophagy at short time points of nitrogen starvation when the impairment of Xrn1 catalytic activity should be most obvious with regard to its affect on autophagy activity (Figure 5A and B). We found that autophagy was markedly enhanced in the strains containing the integrated EV and catalytically inactive Xrn1 strains (Xrn1E176G-PA and Xrn1D208A-PA) compared to WT (Figure 5A and B), consistent with our previous results (Figure 1 and Figure 2) that loss of Xrn1 significantly enhances autophagy.
We then investigated whether the downregulation of select ATG transcripts was mediated by the RNase function of Xrn1 (Figure 5C). To do this, we grew xrn1∆ cells expressing chromosomally integrated WT, EV or Xrn1 mutants under nutrient-rich conditions, collected RNA, and processed the samples for RT-qPCR (Figure 5C). We found that ATG8, ATG12 and ATG29 mRNA levels were significantly upregulated in cells expressing the EV or catalytically inactive Xrn1 mutants, consistent with our previous findings in the xrn1∆ strain under growing conditions (Figure 4A). These results support the hypothesis that Xrn1 mediates selective degradation of ATG transcripts under nutrient-rich conditions in a manner that is dependent on its RNAse activity.
Ash1 regulatesXRN1 expression
Because we had observed that XRN1 mRNA and protein levels significantly decreased with nitrogen starvation and autophagy induction (Figure 3 and S2), we wanted to further explore the mechanism regulating XRN1 transcription (Figure 6). To narrow down potential regulators of XRN1, we used an online yeast database, Yeastract (www.yeastract.com), to search for possible transcription factors based on predicted binding sites within the XRN1 promoter. We tested potential candidates by performing a directed RT-qPCR screen in the corresponding null strains to determine which factors affected XRN1 mRNA levels relative to WT under either growing or starvation conditions. One of the candidates, Ash1, showed a significant decrease in XRN1 mRNA levels (i.e., in an ash1∆ strain) during nutrient-replete conditions that was comparable to the level of XRN1 mRNA in a WT strain under starvation conditions, suggesting that Ash1 could function as a transcriptional activator of XRN1 in nutrient-rich conditions (Figure 6A). Ash1 shares 35% homology with known GATA transcription factors [48]. Multiple predicted Ash1 consensus sequences (YTGAT, where Y is C or T) lie within the XRN1 promoter region (upstream of the ATG +1 start site). Ash1 (along with autophagy transcriptional regulators Ume6 [42] and Pho23 [36]) is a component of the Rpd3 large (Rpd3L) complex[49]. In yeast, Rpd3L is one of the 2 Rpd3 histone deacetylase complexes (HDAC) that regulate gene expression [49–51]. However, to our knowledge, the connection between Ash1 and autophagy has not yet been described in the literature.
Figure 6.
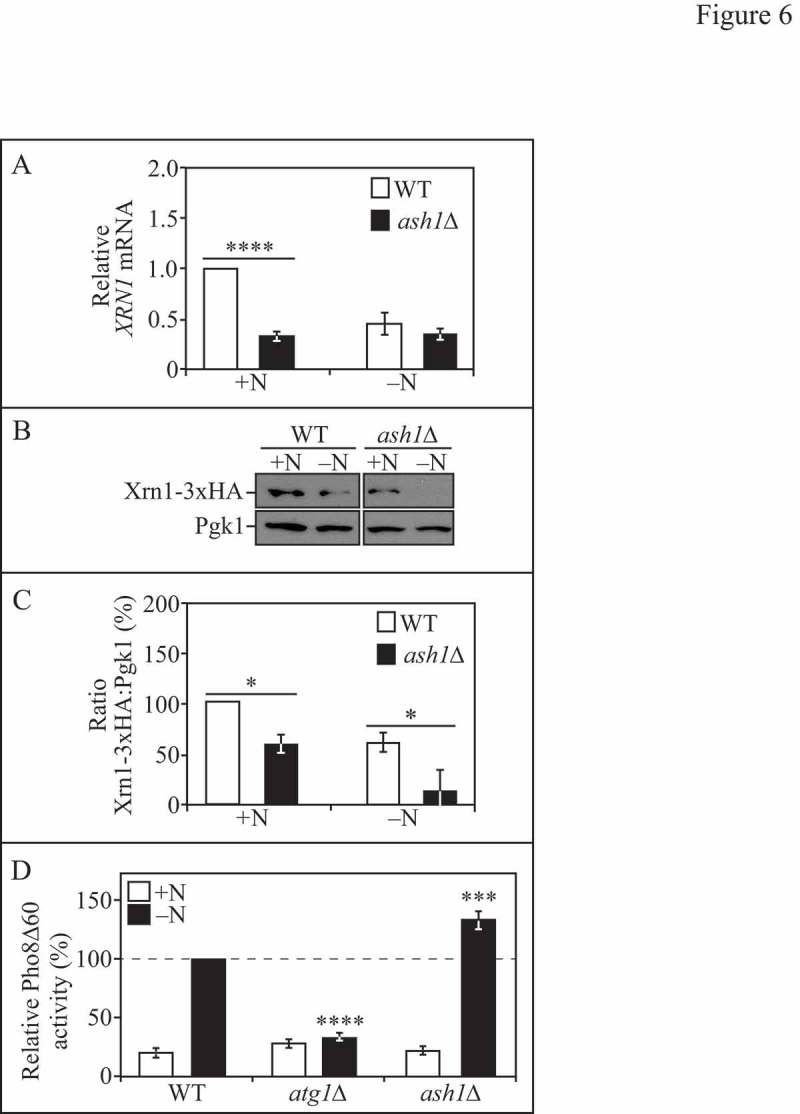
Ash1 regulates XRN1 expression under nutrient-replete conditions. (A) WT (WLY176) and ash1∆ (EDA125) cells were grown to mid-log phase in YPD (+N) and then nitrogen starved (-N) for 1 h. Total RNA was extracted and RT-qPCR was performed. Results shown are relative to the level of XRN1 mRNA expression in WT cells under rich conditions (+N), which was set to 1. The geometric mean of TFC1 and SLD3 was used to quantify relative expression levels. Results shown are the mean of 3 independent experiments +/− SD. (B) WT (EDA161) and ash1∆ cells (EDA166) endogenously expressing Xrn1-3xHA were grown to mid-log phase in YPD (+N) and then nitrogen starved (-N) for 1 h. Protein extracts were analyzed by SDS-PAGE and blotted with anti-HA or anti-Pgk1 (loading control) antisera. (C) Densitometry of n=3 blots as represented in B. The ratio of Xrn1-3xHA:Pgk1 was quantified with ImageJ. (D) WT (WLY176), atg1∆ (WLY192) and ash1∆ (EDA125) cells were grown to mid-log phase in YPD (+N) and then nitrogen starved (-N) for 3 h. The Pho8Δ60 activity was measured and normalized to the activity of starved WT cells, which was set at 100%. Results shown are the mean of 4 independent experiments +/− SD. Also see Figure S3.
To test whether changes in XRN1 mRNA expression corresponded to altered protein levels, we collected protein extracts in WT and ash1∆ strains expressing endogenous Xrn1-3xHA under growing and starvation conditions (Figure 6B and C). We observed that Xrn1-3xHA expression was significantly lower in ash1∆ cells compared to WT under both rich and starvation conditions (Figure 6B and C). Given the possibility that Ash1 could function as an activator of Xrn1 expression in nutrient-replete conditions, we tested the autophagy phenotype of the ash1∆ strain using the Pho8Δ60 assay (Figure 6D). In the ash1∆ strain, we found that Pho8∆60-dependent phosphatase activity was significantly upregulated (∼30%) relative to WT under starvation conditions (Figure 6D). We also found that ASH1 mRNA levels significantly decreased during nitrogen starvation (>50% decrease; Figure S3B), further supporting its role as a negative regulator of autophagy during growing conditions that is inactivated when autophagy is induced. However, we cannot conclude at this time whether the regulatory effect of Ash1 on XRN1 transcript levels is due to a direct or indirect interaction. Considering that ash1∆ cells express significantly less XRN1 mRNA (Figure 6A) and protein than WT cells under nutrient-rich conditions (Figure 6B and C), and ash1∆ cells undergo increased autophagy when starved (Figure 6D), we conclude that Ash1, at least in part, regulates Xrn1 expression during nutrient-replete conditions.
XRN1 depletion enhances autophagy and virus infection in mammalian cells
Many of the 42 Atg proteins found in fungi are conserved or have analogous counterparts in mammalian cells. As we had observed Xrn1 to be a negative autophagy regulator in the yeast S. cerevisiae, we next examined whether depletion of XRN1 had an impact on autophagy in mammalian cells (Figure 7, Figure S4 and Figure S5). Mammalian MAP1LC3B/LC3B is an Atg8 family member, and similar to the Atg8 lipidation assay we previously described, can be used to monitor autophagy progression [52]. LC3B-I (18 kDa) is converted to LC3B-II (16 kDa) by ATG4-mediated cleavage and ATG7-dependent activation, followed by ATG3-mediated PE lipidation at a conserved glycine residue at the C terminus (reviewed in ref. 29). We transfected WT or BECN1 CRISPR knockout (KO) HeLa cells with either scrambled control or XRN1-directed small interfering RNA (siRNA) and performed the LC3 conversion assay to monitor autophagy activity (Figure 7A and B). The knockdown of XRN1 was effective in both WT HeLa and BECN1 KO strains (Figure 7A). In the control cells starved in Hanks' balanced salt solution (HBSS), we observed the characteristic increase in total LC3B, and the majority of the protein was in its lipidated form, LC3B-II (Figure 7B). To further validate our results, we also performed the LC3 lipidation assay in a human brain microvascular endothelial cell (HBMEC) line (Figure S4). In HBMECs transfected with the XRN1 siRNA, we observed a greater proportion of LC3-II relative to LC3-I compared to control cells, which again suggested an increase in autophagy activity (Figure S4).
Figure 7.
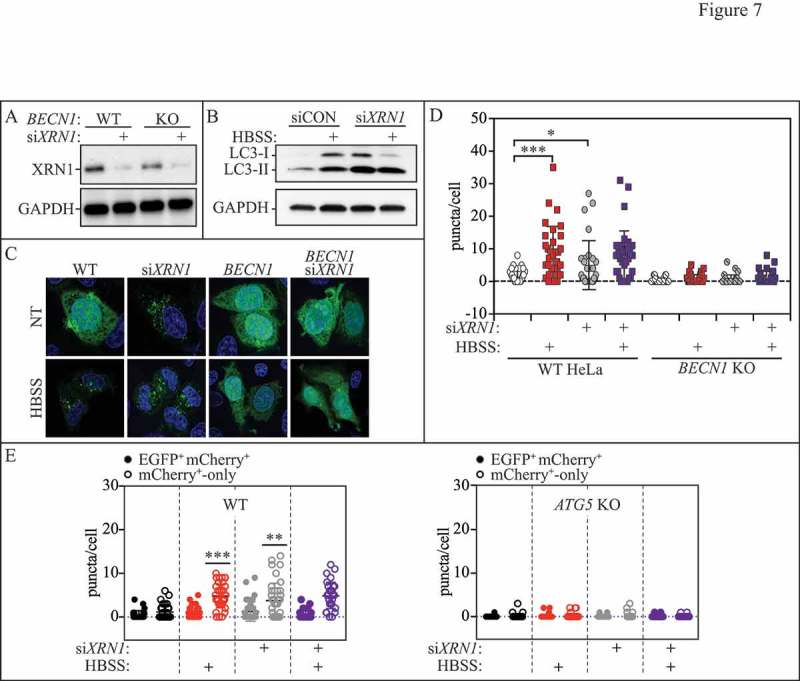
XRN1 depletion enhances autophagy in mammalian cells. (A) WT or BECN1 CRISPR KO HeLa cells were transfected with either scrambled control or pooled XRN1-targeted siRNAs for 48 h, and lysates were blotted for XRN1 or GAPDH (loading control). (B) Lysates from HeLa cells transfected with either scrambled control or XRN1-targeted siRNA were blotted for LC3 (LC3-I, non-lipidated; LC3-II, lipidated species) or GAPDH after 6 h starvation in HBSS. (C) Representative confocal images of WT or BECN1 KO HeLa cells expressing GFP-LC3 and transfected with scrambled control or XRN1-targeted siRNA under fed (non-treated; NT) or starved (HBSS) conditions. (D) GFP-LC3 puncta were counted per cell in all conditions (>25 cells). (E) mCherry-EGFP-LC3B puncta per cell in all conditions (at least 30 cells). Quantifications are shown as EGFP/mCherry+ or mCherry+-only LC3B puncta. Also see Figure S4 and S5.
We also used a GFP-LC3 construct to examine the formation of LC3 puncta, another marker of autophagy induction [52], in either WT HeLa or BECN1 KO cells (Figure 7C and D). BECN1 is a critical factor for autophagy induction that forms part of the autophagy-inducing phosphatidylinositol 3-kinase complex [53]. As expected, WT HeLa cells starved with HBSS accumulated GFP-LC3 puncta (Figure 7C and D; columns 1 and 2). GFP-puncta formation was enhanced significantly with XRN1 siRNA under non-starved conditions (Figure 7C and D; columns 1 and 3), whereas starvation had no additional effect (Figure 7C and D; columns 3 and 4); the enhanced autophagy was lost in BECN1 CRISPR KO cells (Figure 7C and D; columns 5 to 8). In mammalian cells, the acidic environment within the lysosome quenches the green fluorescent protein (GFP) signal, making it difficult to assess whether autophagosomes (as indicated by GFP-LC3 fusion proteins) have fused with lysosomes[28,52,54]. However, red fluorophores (such as RFP or mCherry) conjugated to LC3 are more stable at a low pH, and a red punctate signal corresponds to an autolysosome, or an autophagosome that has undergone lysosomal fusion [52,55]. We examined WT or ATG5 CRISPR KO HeLa cells which were cotransfected with a dual labeled mCherry-EGFP-LC3B construct and either a scrambled control or XRN1 siRNA to assay autophagic flux (Figure 7E and S5). We observed more LC3B-positive puncta in untreated XRN1-depleted cells compared to untreated WT HeLa cells transfected with control siRNA (which showed a diffuse yellow signal), indicating a higher level of basal autophagy in XRN1-depleted cells (Figure 7E and Figure S5; columns 1 and 2 versus 5 and 6). These puncta were largely absent in ATG5 CRISPR KO cells that had been transfected with XRN1 siRNA (Figure 7E and S5; columns 13 and 14). Furthermore, there was a substantial increase in mCherry+-only puncta in XRN1-depleted cells (Figure 7E and S5; columns 2 and 6) indicating an increase in autolysosomes. No additive effect was observed with starvation (Figure 7E and S5; columns 5 and 6 versus 7 and 8). These data support the idea that the increase in LC3-II that we observed with XRN1 depletion is due to increased autophagic flux and not due to defects in autophagosome-lysosome fusion. Thus, Xrn1/XRN1 functions as a conserved regulator of autophagy in both yeast and mammalian cells.
Picornaviruses are nonenveloped, positive-sense single-stranded RNA viruses that include PV, coxsackieviruses, rhinoviruses and other enteroviruses. Subsequent to host cell entry, the virus releases its genome and initiates replication. Picornaviruses such as PV and CVB form characteristic autophagosome-like double-membrane structures during infection[56–59]. Further evidence indicates a role for autophagy in promoting the replication of PV and CVB, as these and other RNA viruses require extensive membrane resources for genome replication, and are proposed to utilize autophagy-generated membranes for this purpose [60–65]. Because we found a role for XRN1 as a negative regulator of autophagy in mammalian cells (Figure 7, Figure S4 and Figure S5), we investigated whether knockdown of XRN1 had an impact on the replication of 2 picornaviruses, PV and CVB (Figure 8A and B). We transfected WT HeLa, BECN1 CRISPR KO or ATG5 CRISPR KO cells with a siRNA towards XRN1 and pre-starved cells to induce autophagy prior to infecting with PV (Figure 8A). In these experiments, we pre-induced autophagy through starvation because previous work indicates that PV targets XRN1 during infection by an unknown mechanism, leading to its degradation by the proteasome [66]. Starvation alone enhances PV infection (Figure 8A), indicating that an induced autophagy pathway benefits the virus; this is not surprising as previous evidence demonstrates enhanced viral replication when the autophagy pathway is induced [63,67]. However, we found that PV titers were significantly increased in pre-starved XRN1-depleted cells compared to pre-starved control cells (Figure 8A). Enhanced PV infection was lost in starved BECN1 and ATG5 CRISPR KO cells transfected with XRN1 siRNA, indicating that the impact on virus replication was autophagy dependent (Figure 8A).
Figure 8.
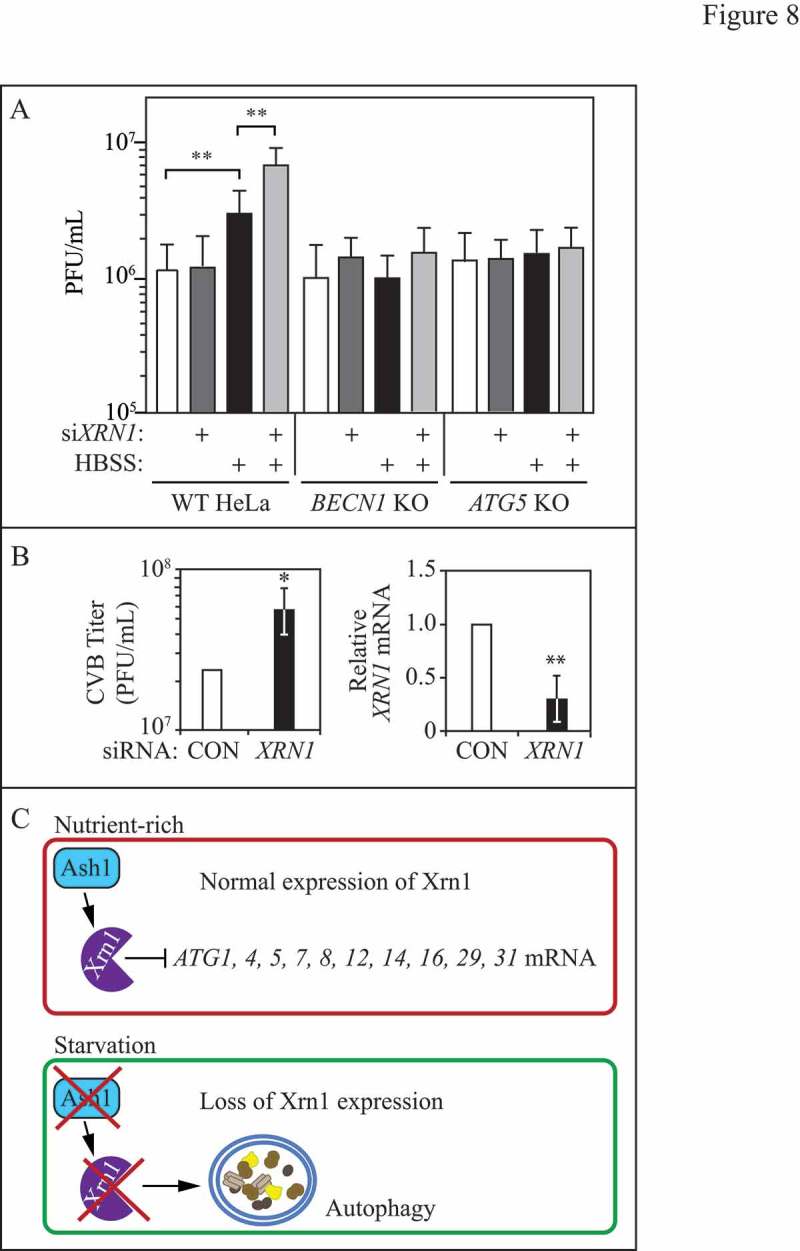
XRN1 depletion enhances virus infection in mammalian cells. (A) PV titers (plaque-forming units/ml; PFU) were determined by plaque assay in WT, BECN1 or ATG5 CRISPR KO HeLa cells transfected with scrambled control or XRN1-targeted siRNA. Cells were starved in HBSS for 1 h prior to infecting with PV at a multiplicity of infection of 0.1, and remained in starvation medium until harvested at 6 h post infection. Results shown are the mean of 3 independent experiments. (B) CVB titers (PFU/ml) as determined by plaque assay in HeLa cells (left). HeLa cells underwent plaque assays with supernatants from CVB-infected HBMECs expressing either control or XRN1-targeted siRNA. Results shown are the mean of 4 independent experiments. RT-qPCR KD of XRN1 mRNA levels in HBMECs (right). Results shown are the mean of 4 independent experiments. (C) Schematic representation of the model by which Xrn1 functions as a negative autophagy regulator in yeast.
We next examined the effect of XRN1 on the infection of a related picornavirus, CVB (Figure 8B). We depleted XRN1 by siRNA in polarized HBMEC, which also showed higher levels of LC3-II conversion (Figure 8B and Figure S4). HBMECs transfected with scrambled control or XRN1 siRNA were infected with CVB, and the supernatants were collected and used to perform plaque assays on HeLa cells (Figure 8B). We observed a significant increase in viral titer from the supernatants of XRN1-deficient cells relative to control (Figure 8B). Together, we demonstrate that XRN1 functions as an antiviral factor for 2 different picornaviruses—PV and CVB—in 2 different types of cells, and its depletion enhances infection through an autophagy-dependent mechanism.
Discussion
Here we present data supporting a model for the cytoplasmic exoribonuclease Xrn1 as a post-transcriptional negative autophagy regulator in yeast and mammalian cells (Figure 8C). Chromosomal deletion of XRN1 in yeast and siRNA-mediated depletion in mammalian cells resulted in a significant upregulation of autophagy. We also found that Xrn1 was well expressed during nutrient-rich conditions and its expression significantly decreased during nitrogen starvation at both the mRNA and protein levels, further supporting the proposal that Xrn1 functions as a negative autophagy regulator during growing conditions. Protein levels of Xrn1 mutants (Xrn1E176G-PA and Xrn1D208A-PA) appeared more stable during starvation (Figure 5A). Although we do not currently know the reason, it is interesting to speculate that starvation-induced downregulation of Xrn1 may be dependent on its RNAse activity. Under nutrient-replete conditions, Xrn1 expression is regulated, at least in part, by the yeast transcription factor Ash1. Furthermore, in mammalian cells, loss of XRN1 expression by siRNA-mediated knockdown enhanced the infection of 2 picornaviruses, PV and CVB, which utilize autophagy to promote their replication. Thus, our data link 2 separate but related observations regarding virally induced autophagy: some viruses target XRN1, while simultaneously upregulating autophagy to enhance replication. We now show directly that downregulation of XRN1 results in enhanced autophagy and an autophagy-dependent increase in viral titer.
Our data further indicate that under nutrient-rich conditions, excess ATG transcripts (particularly ATG1, 4, 5, 7, 8, 12, 14, 16, 29 and 31) are produced within the cell. Based on the data presented here, the RNAse activity of Xrn1 is required to regulate select ATG mRNA levels during nutrient-rich conditions. Thus, these transcripts are normally targeted for RNA decay when nutrients are abundant. In the absence of Xrn1, these select transcripts are in excess and xrn1∆ cells undergo enhanced autophagy during short-term nitrogen starvation compared to WT, similar to what has been observed when other negative factors are eliminated [36,37,42]. The results presented here implicate a role for the degradation of specific ATG transcripts by Xrn1, especially given that not all of the ATG transcripts examined were affected. At least for ATG5, ATG7, ATG8 and ATG12, an increase in mRNA in xrn1∆ cells corresponded to a similar upregulation in protein levels under rich conditions. In ume6∆ cells, basal levels of Atg8 are higher, and these cells undergo a more rapid autophagy response during starvation [42]. Related phenomena have also been noted in rph1∆ [37] and spt10∆ cells [41]. Similarly, Xrn1 functions as part of a mechanism that enables the cell to produce an abundance of ATG mRNAs in growing conditions, which readies the cell for a robust autophagy response once it is depleted of nutrients.
In the canonical 5' to 3' mRNA decay pathway, mRNA decapping by the Dcp1-Dcp2 complex precedes Xrn1-mediated degradation (reviewed in ref. [8]). Hu et al. identified the mRNA decapping enzyme Dcp2 as a key mediator of ATG mRNA regulation [26]. In the study by Hu et al., mRNA levels of ATG1, 4, 5, 7, 8, 14, 16, 29 and 31 are increased to a statistically significant level in the dcp2-7∆ ts strain examined under nutrient-replete conditions, indicating that these transcripts presumably undergo decapping by Dcp2 prior to targeting by Xrn1 [26]. In the same study, ATG8 mRNA was found to undergo decapping by the Dhh1 homolog Vad1 in the pathogenic yeast Cryptococcus neoformans. [26]
Xrn1 has a distinct preference for RNA substrates with an exposed 5' monophosphate, likely due to steric hindrance within the RNA binding pocket, excluding larger groups such as those with a m7G cap or triphosphorylated RNAs [10,15]. Aside from steric selection due to physical restrictions, little is known about other factors such as common sequence-specific elements that may select certain transcripts for Xrn1 targeting, although stable secondary structures may block or stall hydrolysis by Xrn1 [68]. Based on the evidence presented here and by others[26], the transcripts of ATG7 and ATG8 are likely regulated by the canonical 5' to 3' mRNA decay pathway; that is, these transcripts accumulated in all 3 strains—xrn1∆, dhh1∆, and dcp2-7∆ ts. Therefore, ATG7 and ATG8 transcripts likely undergo decapping by Dcp2 and its accessory factor Dhh1, followed by Xrn1-mediated degradation.
In the present study, we found that mRNA levels of ATG8, but not ATG9, increased in xrn1∆ cells under nutrient-rich conditions. This corresponded to an increase in the frequency/rate, but not size, of autophagic bodies in an xrn1∆ strain when examined by TEM. Previous work has demonstrated a role for the transcription factor Ume6 in regulating Atg8 levels, and increased Atg8 levels are associated with increased autophagosome size [35,42]. Increased autophagosome frequency is associated with Atg9 levels (regulated by Pho23) [36]. In these previous studies [36,42], the expression level of a single ATG mRNA (either ATG8 or ATG9), and subsequently protein, was altered demonstrating a unique phenotype. In the present study, there is a selective upregulation of multiple ATG transcripts, and thus upregulation of a single Atg protein cannot account for the results that we observe (i.e., increased autophagosome frequency but not size). The magnitude of the autophagy response may be greatly affected by the stoichiometric ratio of Atg proteins at the PAS; however, there is relatively little data available regarding the stoichiometry of Atg proteins during autophagy. Although work by Geng and colleagues indicated that Atg8 is the most abundant (of the Atg proteins examined) at the PAS [69]. The ability of the cell to successfully integrate and interpret these signals determines the impact on autophagosome formation, and thus, on autophagy. Our observations indicate targeting of unique ATG mRNAs by Xrn1, which is consistent with reports by others who indicate a role for Xrn1 in selective RNA degradation [24].
Additionally, we have identified Ash1 as a transcription factor regulating Xrn1 expression during nutrient-rich conditions. At present, we cannot conclude whether Ash1 is a direct transcriptional regulator of Xrn1 or if Ash1 functions in a complex to indirectly modulate Xrn1 expression. Ash1 recognizes a consensus sequence, YTGAT, which is found at multiple sites within the XRN1 promoter region. Ash1 has been previously described as a transcriptional repressor [49,70], whereas here we have found a role for Ash1 as a positive transcriptional regulator of XRN1.
Autophagy can function in a proviral or antiviral capacity depending on the virus involved and the context of the infection (for further review see refs. [71–73]). Dougherty et al. demonstrated that PV disrupts processing bodies and downregulates XRN1 during infection, independently of the viral proteases 2Apro and 3Cpro [66]. However, the mechanism by which PV targets XRN1 to enhance viral replication has not been previously identified [66]. Viruses have evolved complex strategies to circumvent the host defense response and promote their own replication. Flaviviruses, such as dengue, West Nile, Zika and hepatitis C virus, also target and/or resist XRN1 during their infectious life cycles [74–79]. One possibility for why these RNA viruses target XRN1 is to further promote autophagy induction in order to support viral replication. Our data indicate that PV (and CVB) replicate to a higher degree in the absence of XRN1 and that this is dependent on a fully functional autophagy pathway.
Xrn1/XRN1 has been associated with human pathologies such as osteosarcoma and infectious diseases [80,81] and even virus infection in yeast [82,83]. Thus, the cell must strictly modulate autophagy at multiple levels to prevent disease pathogenesis. The mechanism described here contributes to our understanding of the complex regulatory network involved in fine-tuning autophagy and maintaining it at appropriate levels to meet cellular demands.
Materials and methods
Yeast strains, media and cell culture
Yeast strains used in this study are listed in Table S1. Yeast cells were grown in YPD (1% yeast extract, 2% peptone and 2% glucose) or synthetic minimal medium (SMD; 0.67% yeast nitrogen base and 2% glucose, supplemented with the appropriate auxotrophic amino acids and vitamins as needed). Autophagy was induced by shifting mid-log phase cells from rich medium to nitrogen starvation medium (SD-N; 0.17% yeast nitrogen base without ammonium sulfate or amino acids and 2% glucose) for the indicated times. Gene deletions and chromosome tagging were performed using standard methods [84,85].
Mammalian cell culture and viruses
BECN1 and ATG5 CRISPR knockout were generated in HeLa cells using the pSpCas9-sA-GFP (PX458) system (Addgene, 48138; deposited by Feng Zhang) [86] for generating single cell isolates (guide RNA sequence: CACCGGCGAAACCAGGAGAGACCC and CACCGAACTTGTTTCACGCTATATC, respectively). Poliovirus type 1 Mahoney was grown in HeLa cells as previously described [87]. Titers were determined by plaque assay. HeLa cells were starved for 1 h in HBSS prior to infection with PV at a multiplicity of infection of 0.1 in PBS containing 100 µg/ml MgCl2 and CaCl2. Complete DMEM medium (GE, SH30243.01) with 10% fetal bovine serum and 1% pen/strep or HBSS was added back to cells after 30 min adsorption. Supernatant and cell monolayers were harvested together at 6 h post infection and titered by plaque assay.
HBMECs were cultured as described previously [88]. Coxsackievirus B3-RD isolate (CVB3-RD) was cultured in HeLa cells and purified as described [89]. Viral titers were determined by plaque assay. HBMECs were infected with CVB for 16 h (multiplicity of infection of 3), and supernatants were used for subsequent plaque assays in HeLa cells. Confluent HeLa cell monolayers were exposed to serial dilutions of virus for 1 h at room temperature. Cells were then overlayed with agarose and incubated for 48 h. Plaques were visualized by crystal violet staining and enumerated.
Plasmids and site-directed mutagenesis
The pRS406-XRN1p-Xrn1-PA plasmid was generated by FastCloning technology[90]. The ORF of XRN1 (including −800 base pairs upstream of the XRN1 start site) was amplified from genomic DNA obtained from the SEY6210 strain using the primers 173F and 173R (see Table S4). This fragment was inserted into a pRS406-PA vector, amplified using primers 172F and 172R, simultaneously tagging Xrn1 with protein A at the C terminus. The E176G and D208A mutants were created using site-directed mutagenesis with the primers 178F and 178R (E176G) and 179F and 179R (D208A). The plasmids were integrated at the URA3 locus into the indicated strains. The GFP-LC3B plasmid has been previously described [63]. The mCherry-EGFP-LC3B plasmid was obtained from Addgene (22418; deposited by Jayanta Debnath) [91].
Small interfering RNAs (siRNAs) and transfections
For mammalian autophagy assays and PV infection experiments, 4 pooled siRNAs targeting human XRN1 (siGENOME SMARTpool, Dharmacon, 54464) or control scrambled siGENOME non-targeting siRNA pool (Dharmacon, D-001206-13-05) were transfected into HeLa cells using Lipofectamine 3000 (Life Technologies/Thermo Fisher Scientific, L3000015). XRN1 knockdown was tested after 48 h by western blot. For microscopy assays, GFP-LC3 was transfected into cells 24 h post-siRNA transfection using Lipofectamine 3000. Twenty-four h post DNA transfection, cells were fixed with 4% formaldehyde and treated with DAPI to stain the nuclei. Cells were viewed using a Leica SP8 confocal microscope.
HBMECs were reverse transfected with 25 nM Mission siRNA Universal scrambled control (Sigma-Aldrich, SIC001) or XRN1-targeting siRNA (5'-GAG AGU AUA UUG ACU AUG Att-3') [92] using Dharmafect 1 (GE Healthcare Dharmacon, T-2001). Cells were infected with CVB ∼48 h post transfection.
Antibodies and inhibitors
Antisera to Atg8 [93], Ape1 [94] and Vma8 [95] have been described previously. Pgk1 antibody was a generous gift from Dr. Jeremy Thorner (University of California, Berkeley). Antibody to YFP (that also detects GFP) was purchased from Clontech (JL-8; 63281). Dpm1 antibody was purchased from Life Technologies (A6429). Commercial PA antibody was purchased from Jackson Immunoresearch (323-005-024). HA antibody was purchased from Sigma-Aldrich (H3663). Mammalian GAPDH and XRN1 antibodies were purchased from Santa Cruz Biotechnology (sc-48167 or sc-25778) and Bethyl Laboratories (A300), respectively. LC3B antibodies were purchased from Novus Biologicals (NB100-2220) and Abcam (ab48394).
RNA and real-time quantitative PCR (RT-qPCR)
Yeast cells were cultured in YPD to mid-log phase and then shifted to SD-N (1 h) for autophagy induction. Cells (1 OD600 unit) were then collected and flash frozen in liquid nitrogen. Total RNA was extracted using an RNA extraction kit (Clontech, Nucleo Spin RNA, 740955.250). Reverse transcription was carried out using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems/Thermo Fisher Scientific, 4368814). For each sample, 1 µg RNA was used for cDNA synthesis. RT-qPCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems/Thermo Fisher Scientific, 4367659) in an Eppendorf RealPlex4 (no longer available) or CFX Connect (Bio-Rad, 1855201) real-time PCR machine.
For HBMECs, RNA extraction was performed using GenElute total RNA miniprep kit (Sigma-Aldrich, RTN70). RNA samples were treated with RNase-free DNase (Sigma-Aldrich, DNASE70). Total RNA was reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad, 1708891). RT-qPCR was performed using iQ SYBR Green Supermix (Bio-Rad, 1708882) in a CFX96 Real-Time System (Bio-Rad, 1855195). XRN1 expression was normalized to ACTB. For all RT-qPCR experiments, melting curves were run after the PCR cycles to verify primer specificity. Relative gene expression was calculated using the 2−ΔΔCT method [96] and normalized as indicated. Primer sequences are included in Table S3.
Transmission electron microscopy (TEM)
TEM and analysis of autophagic bodies was performed as previously described [38]. Images were captured using a JEOL JEM-1400-Plus transmission electron microscope at the University of Michigan Microscopy and Image Analysis Laboratory.
Other methods
Phosphatase (Pho8Δ60) activity, GFP-Atg8 and prApe1 processing, Atg8 lipidation and western blot assays were performed as previously described [97–99]. Densitometry for western blots was performed using ImageJ.
Statistical analysis
The 2-tailed Student t test was used to determine statistical significance unless otherwise indicated. For all figures, P values are as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P<0.0001. A P value < 0.05 was considered significant.
Supplementary Material
Funding Statement
HHS | NIH | National Institute of General Medical Sciences (NIGMS) HHS | National Institutes of Health (NIH) This work was supported by NIH [grant number GM053396] and by the Protein Folding Diseases Initiative, University of Michigan (to DJK) and NIH [grant number F32-AI122456 (to NJL)].
Disclosure of potential conflicts of interest
The authors declare that they have no competing financial interests.
Acknowledgments
We thank Xin Wen (University of Michigan) and Dr. Steven K. Backues (Eastern Michigan University) for technical assistance and/or helpful comments.
References
- 1.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015 Jun;25(6):354–363. doi: 10.1016/j.tcb.2015.02.002 PubMed PMID: 25759175; PubMed Central PMCID: PMC4441840. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frankel LB, Lubas M, Lund AH. Emerging connections between RNA and autophagy. Autophagy. 2017 Jan 02;13(1):3–23. doi: 10.1080/15548627.2016.1222992 PubMed PMID: 27715443; PubMed Central PMCID: PMC5240835. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014 Jan;15(1):65–74. doi: 10.1038/nrm3716 PubMed PMID: 24326622; eng. [DOI] [PubMed] [Google Scholar]
- 4.Delorme-Axford E, Donker RB, Mouillet J-F, Chu T, Bayer A, Ouyang Y, Wang T, Stolz DB, Sarkar SN, Morelli AE, et al. Human placental trophoblasts confer viral resistance to recipient cells. PNAS. 2013 Jul 16;110(29):12048–12053. doi: 10.1073/pnas.1304718110 PubMed PMID: 23818581; PubMed Central PMCID: PMC3718097. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu H, Wu H, Liu X, Li B, Chen Y, Ren X, Liu C-G, Yang J-M. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009 Aug;5(6):816–823. PubMed PMID: 19535919; PubMed Central PMCID: PMC3669137. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science (New York, NY). 2009 Oct 23;326(5952):544–550. doi: 10.1126/science.1176945 PubMed PMID: 19745116; PubMed Central PMCID: PMC3786161. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007 Feb;8(2):113–126. doi: 10.1038/nrm2104 PubMed PMID: 17245413; eng. [DOI] [PubMed] [Google Scholar]
- 8.Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012 Jul;191(3):671–702. doi: 10.1534/genetics.111.137265 PubMed PMID: 22785621; PubMed Central PMCID: PMC3389967. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labno A, Tomecki R, Dziembowski A. Cytoplasmic RNA decay pathways - Enzymes and mechanisms. Biochim Biophys Acta. 2016 Dec;1863(12):3125–3147. doi: 10.1016/j.bbamcr.2016.09.023 PubMed PMID: 27713097; eng. [DOI] [PubMed] [Google Scholar]
- 10.Nagarajan VK, Jones CI, Newbury SF, Green PJ. XRN 5'–>3' exoribonucleases: structure, mechanisms and functions. Biochim Biophys Acta. 2013 Jun-Jul;1829(6-7):590–603. doi: 10.1016/j.bbagrm.2013.03.005 PubMed PMID: 23517755; PubMed Central PMCID: PMC3742305. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheth U. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science (New York, NY). 2003 May 02;300(5620):805–808. doi: 10.1126/science.1082320 PubMed PMID: 12730603; PubMed Central PMCID: PMC1876714. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem Soc Trans. 2010 Feb;38(Pt 1):242–251. doi: 10.1042/bst0380242 PubMed PMID: 20074068; eng. [DOI] [PubMed] [Google Scholar]
- 13.Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb Perspect Biol. 2012 Sep 01;4(9):a012286. doi: 10.1101/cshperspect.a012286 PubMed PMID: 22763747; PubMed Central PMCID: PMC3428773. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009 Sep 10;461(7261):225–229. doi: 10.1038/nature08265 PubMed PMID: 19701183; PubMed Central PMCID: PMC2745705. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones CI, Zabolotskaya MV, Newbury SF. The 5' –>3' exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip Rev RNA. 2012 Jul-Aug;3(4):455–468. doi: 10.1002/wrna.1109 PubMed PMID: 22383165; eng. [DOI] [PubMed] [Google Scholar]
- 16.Orban TI. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA (New York, NY). 2005 Apr;11(4):459–469. doi: 10.1261/rna.7231505 PubMed PMID: 15703439; PubMed Central PMCID: PMC1370735. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wery M, Descrimes M, Vogt N, Dallongeville A-S, Gautheret D, Morillon A. Nonsense-Mediated Decay Restricts LncRNA Levels in Yeast Unless Blocked by Double-Stranded RNA Structure. Mol Cell. 2016 Feb 04;61(3):379–392. doi: 10.1016/j.molcel.2015.12.020 PubMed PMID: 26805575; PubMed Central PMCID: PMC4747904. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sinturel F, Navickas A, Wery M, Descrimes M, Morillon A, Torchet C, Benard L. Cytoplasmic Control of Sense-Antisense mRNA Pairs. Cell Rep. 2015 Sep 22;12(11):1853–1864. doi: 10.1016/j.celrep.2015.08.016. PubMed PMID: 26344770; eng. [DOI] [PubMed] [Google Scholar]
- 19.Geisler S, Coller J. XRN1: A Major 5' to 3' Exoribonuclease in Eukaryotic Cells. The Enzymes. 2012;31:97–114. doi: 10.1016/b978-0-12-404740-2.00005-7 PubMed PMID: 27166442; eng. PMID: 27166442 [DOI] [PubMed] [Google Scholar]
- 20.van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011 Jun 22;475(7354):114–117. doi: 10.1038/nature10118 PubMed PMID: 21697827; eng. [DOI] [PubMed] [Google Scholar]
- 21.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006 Mar 23;440(7083):561–564. doi: 10.1038/nature04530 PubMed PMID: 16554824; PubMed Central PMCID: PMC1839849. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geisler S, Lojek L, Khalil A, Baker K, Coller J. Decapping of long noncoding RNAs regulates inducible genes. Mol Cell. 2012 Feb 10;45(3):279–291. doi: 10.1016/j.molcel.2011.11.025 PubMed PMID: 22226051; PubMed Central PMCID: PMC3278590. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 2007 Jan;27(1):92–101. doi: 10.1128/mcb.01023-06 PubMed PMID: 17074811; PubMed Central PMCID: PMC1800667. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina DA, Jordán-Pla A, Millán-Zambrano G, Chávez S, Choder M, Pérez-Ortín JE. Cytoplasmic 5'-3' exonuclease Xrn1p is also a genome-wide transcription factor in yeast. Front Genet. 2014;5:1. doi: 10.3389/fgene.2014.00001 PubMed PMID: 24567736; PubMed Central PMCID: PMC3915102. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haimovich G, Medina D, Causse S, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín J, Darzacq X, Choder M. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013 May 23;153(5):1000–1011. doi: 10.1016/j.cell.2013.05.012 PubMed PMID: 23706738; eng. [DOI] [PubMed] [Google Scholar]
- 26.Hu G, McQuiston T, Bernard A, Park Y-D, Qiu J, Vural A, Zhang N, Waterman SR, Blewett NH, Myers TG, et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat Cell Biol. 2015 Jul;17(7):930–942. doi: 10.1038/ncb3189 PubMed PMID: 26098573; PubMed Central PMCID: PMC4528364. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/s0076-6879(08)03203-5 PubMed PMID: 19185711; eng. PMID: 19185711 [DOI] [PubMed] [Google Scholar]
- 28.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000 Nov 01;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720 PubMed PMID: 11060023; PubMed Central PMCID: PMC305793. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delorme-Axford E, Guimaraes RS, Reggiori F, Klionsky DJ. The yeast Saccharomyces cerevisiae: an overview of methods to study autophagy progression. Methods (San Diego, Calif). 2015 Mar;75:3–12. doi: 10.1016/j.ymeth.2014.12.008 PubMed PMID: 25526918; PubMed Central PMCID: PMC4355233. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guimaraes RS, Delorme-Axford E, Klionsky DJ, Reggiori F. Assays for the biochemical and ultrastructural measurement of selective and nonselective types of autophagy in the yeast Saccharomyces cerevisiae. Methods (San Diego, Calif). 2015 Mar;75:141–150. doi: 10.1016/j.ymeth.2014.11.023 PubMed PMID: 25484341; eng. [DOI] [PubMed] [Google Scholar]
- 31.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356 PubMed PMID: 26799652; PubMed Central PMCID: PMC4835977. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott SV, Guan J, Hutchins MU, Kim J, Klionsky DJ. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001 Jun;7(6):1131–1141. PubMed PMID: 11430817; PubMed Central PMCID: PMC2767243. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005 Apr;16(4):1593–1605. doi: 10.1091/mbc.E04-11-1035 PubMed PMID: 15659643; PubMed Central PMCID: PMC1073644. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott SV, Nice III DC, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000 Aug 18;275(33):25840–25849. doi: 10.1074/jbc.M002813200 PubMed PMID: 10837477; eng. [DOI] [PubMed] [Google Scholar]
- 35.Xi Ze, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008 Aug;19(8):3290–3298. doi: 10.1091/mbc.E07-12-1292 PubMed PMID: 18508918; PubMed Central PMCID: PMC2488302. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin M, He D, Backues S, Freeberg M, Liu X, Kim J, Klionsky D. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol. 2014 Jun 16;24(12):1314–1322. doi: 10.1016/j.cub.2014.04.048 PubMed PMID: 24881874; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernard A, Jin M, González-Rodríguez P, Füllgrabe J, Delorme-Axford E, Backues S, Joseph B, Klionsky D. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr Biol. 2015 Mar 02;25(5):546–555. doi: 10.1016/j.cub.2014.12.049 PubMed PMID: 25660547; PubMed Central PMCID: PMC4348152. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Backues SK, Chen D, Ruan J, Xie Z, Klionsky DJ. Estimating the size and number of autophagic bodies by electron microscopy. Autophagy. 2014 Jan;10(1):155–164. doi: 10.4161/auto.26856 PubMed PMID: 24270884; PubMed Central PMCID: PMC4389869. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910 PubMed PMID: 19653858; PubMed Central PMCID: PMC2831538. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin M, Klionsky DJ. Regulation of autophagy: modulation of the size and number of autophagosomes. FEBS Lett. 2014 Aug 01;588(15):2457–2463. doi: 10.1016/j.febslet.2014.06.015 PubMed PMID: 24928445; PubMed Central PMCID: PMC4118767. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernard A, Jin M, Xu Z, Klionsky DJ. A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy. Autophagy. 2015 Nov 02;11(11):2114–2122. doi: 10.1080/15548627.2015.1099796 PubMed PMID: 26649943; PubMed Central PMCID: PMC4824583. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. PNAS. 2012 Jul 10;109(28):11206–11210. doi: 10.1073/pnas.1200313109 PubMed PMID: 22733735; PubMed Central PMCID: PMC3396506. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao Z, Delorme-Axford E, Backues SK, Klionsky DJ. Atg41/Icy2 regulates autophagosome formation. Autophagy. 2015;11(12):2288–2299. doi: 10.1080/15548627.2015.1107692 PubMed PMID: 26565778; PubMed Central PMCID: PMC4835205. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998 Sep 24;395(6700):395–398. doi: 10.1038/26506 PubMed PMID: 9759731; eng. [DOI] [PubMed] [Google Scholar]
- 45.Sun M, Schwalb B, Pirkl N, Maier K, Schenk A, Failmezger H, Tresch A, Cramer P. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Mol Cell. 2013 Oct 10;52(1):52–62. doi: 10.1016/j.molcel.2013.09.010 PubMed PMID: 24119399; eng. [DOI] [PubMed] [Google Scholar]
- 46.Solinger JA, Pascolini D, Heyer W-D. Active-site mutations in the Xrn1p exoribonuclease of Saccharomyces cerevisiae reveal a specific role in meiosis. Mol Cell Biol. 1999 Sep;19(9):5930–5942. PubMed PMID: 10454540; PubMed Central PMCID: PMC84450. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Page AM, Davis K, Molineux C, Kolodner RD, Johnson AW. Mutational analysis of exoribonuclease I from Saccharomyces cerevisiae. Nucleic Acids Res. 1998 Aug 15;26(16):3707–3716. PubMed PMID: 9685486; PubMed Central PMCID: PMC147754. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 2002 May 15;290(1–2):1–18. PubMed PMID: 12062797; eng. [DOI] [PubMed] [Google Scholar]
- 49.Carrozza MJ, Florens L, Swanson SK, Shia W-J, Anderson S, Yates J, Washburn MP, Workman JL. Stable incorporation of sequence specific repressors Ash1 and Ume6 into the Rpd3L complex. Biochim Biophys Acta. 2005 Nov 10;1731(2):77–87; discussion 75–6. doi: 10.1016/j.bbaexp.2005.09.005 PubMed PMID: 16314178; eng. [DOI] [PubMed] [Google Scholar]
- 50.Kadosh D, Struhl K. Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 1998 Mar 15;12(6):797–805. PubMed PMID: 9512514; PubMed Central PMCID: PMC316629. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991 Dec;11(12):6317–6327. PubMed PMID: 1944291; PubMed Central PMCID: PMC361826. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010 Feb 05;140(3):313–326. doi: 10.1016/j.cell.2010.01.028 PubMed PMID: 20144757; PubMed Central PMCID: PMC2852113. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999 Dec 09;402(6762):672–676. doi: 10.1038/45257 PubMed PMID: 10604474; eng. [DOI] [PubMed] [Google Scholar]
- 54.Bampton ETW, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005 Apr;1(1):23–36. PubMed PMID: 16874023; eng. [DOI] [PubMed] [Google Scholar]
- 55.Katayama H, Yamamoto A, Mizushima N, Yoshimori T, Miyawaki A. GFP-like proteins stably accumulate in lysosomes. Cell Struct Funct. 2008;33(1):1–12. PubMed PMID: 18256512; eng. [DOI] [PubMed] [Google Scholar]
- 56.Schlegel A, Giddings TH, Jr, Ladinsky MS, Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996 Oct;70(10):6576–6588. PubMed PMID: 8794292; PubMed Central PMCID: PMC190698. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suhy DA, Giddings TH, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol. 2000 Oct;74(19):8953–8965. PubMed PMID: 10982339; PubMed Central PMCID: PMC102091. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, Whitton JL. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol. 2010 Dec;84(23):12110–12124. doi: 10.1128/jvi.01417-10 PubMed PMID: 20861268; PubMed Central PMCID: PMC2976412. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delorme-Axford E, Sadovsky Y, Coyne CB. Lipid raft- and SRC family kinase-dependent entry of coxsackievirus B into human placental trophoblasts. J Virol. 2013 Aug;87(15):8569–8581. doi: 10.1128/jvi.00708-13 PubMed PMID: 23720726; PubMed Central PMCID: PMC3719791. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staring J, von Castelmur E, Blomen VA, van den Hengel LG, Brockmann M, Baggen J, Thibaut HJ, Nieuwenhuis J, Janssen H, van Kuppeveld FJM, et al. PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature. 2017 Jan 19;541(7637):412–416. doi: 10.1038/nature21032 PubMed PMID: 28077878; eng. [DOI] [PubMed] [Google Scholar]
- 61.Delorme-Axford E, Morosky S, Bomberger J, Stolz DB, Jackson WT, Coyne CB. BPIFB3 regulates autophagy and coxsackievirus B replication through a noncanonical pathway independent of the core initiation machinery. mBio. 2014 Dec 09;5(6):e02147. doi: 10.1128/mBio.02147-14 PubMed PMID: 25491355; PubMed Central PMCID: PMC4324245. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harris K, Morosky S, Drummond C, Patel M, Kim C, Stolz D, Bergelson J, Cherry S, Coyne C. RIP3 Regulates Autophagy and Promotes Coxsackievirus B3 Infection of Intestinal Epithelial Cells. Cell Host Microbe. 2015 Aug 12;18(2):221–232. doi: 10.1016/j.chom.2015.07.007 PubMed PMID: 26269957; PubMed Central PMCID: PMC4562276. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jack WT, Giddings TH, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005 May;3(5):e156. doi: 10.1371/journal.pbio.0030156 PubMed PMID: 15884975; PubMed Central PMCID: PMC1084330. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richards AL, Jackson WT. Intracellular vesicle acidification promotes maturation of infectious poliovirus particles. PLoS Pathog. 2012;8(11):e1003046. doi: 10.1371/journal.ppat.1003046 PubMed PMID: 23209416; PubMed Central PMCID: PMC3510256. eng. PMID: 23209416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008 Sep;82(18):9143–9153. doi: 10.1128/jvi.00641-08 PubMed PMID: 18596087; PubMed Central PMCID: PMC2546883. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dougherty JD, White JP, Lloyd RE. Poliovirus-mediated disruption of cytoplasmic processing bodies. J Virol. 2011 Jan;85(1):64–75. doi: 10.1128/jvi.01657-10 PubMed PMID: 20962086; PubMed Central PMCID: PMC3014174. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bird SW, Maynard ND, Covert MW, Kirkegaard K. Nonlytic viral spread enhanced by autophagy components. PNAS. 2014 Sep 09;111(36):13081–13086. doi: 10.1073/pnas.1401437111 PubMed PMID: 25157142; PubMed Central PMCID: PMC4246951. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Poole TL, Stevens A. Structural modifications of RNA influence the 5' exoribonucleolytic hydrolysis by XRN1 and HKE1 of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1997 Jun 27;235(3):799–805. doi: 10.1006/bbrc.1997.6877 PubMed PMID: 9207242; eng. [DOI] [PubMed] [Google Scholar]
- 69.Geng J, Klionsky DJ. Quantitative regulation of vesicle formation in yeast nonspecific autophagy. Autophagy. 2008 Oct;4(7):955–957. PubMed PMID: 18758231; eng. [DOI] [PubMed] [Google Scholar]
- 70.Maxon ME, Herskowitz I. Ash1p is a site-specific DNA-binding protein that actively represses transcription. PNAS. 2001 Feb 13;98(4):1495–1500. doi: 10.1073/pnas.98.4.1495 PubMed PMID: 11171979; PubMed Central PMCID: PMC29285. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jackson WT. Viruses and the autophagy pathway. Virology. 2015 May;479–480:450–456. doi: 10.1016/j.virol.2015.03.042 PubMed PMID: 25858140; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lennemann NJ, Coyne CB. Catch me if you can: the link between autophagy and viruses. PLoS Pathog. 2015 Mar;11(3):e1004685. doi: 10.1371/journal.ppat.1004685 PubMed PMID: 25811485; PubMed Central PMCID: PMC4374752. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Richards AL, Jackson WT. How positive-strand RNA viruses benefit from autophagosome maturation. J Virol. 2013 Sep;87(18):9966–9972. doi: 10.1128/jvi.00460-13 PubMed PMID: 23760248; PubMed Central PMCID: PMC3754026. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chapman EG, Moon SL, Wilusz J, Kieft JS. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. eLife. 2014 Apr 01;3:e01892. doi: 10.7554/eLife.01892 PubMed PMID: 24692447; PubMed Central PMCID: PMC3968743. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akiyama BM, Laurence HM, Massey AR, Costantino DA, Xie X, Yang Y, Shi P-Y, Nix JC, Beckham JD, Kieft JS. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science (New York, NY). 2016 Dec 02;354(6316):1148–1152. doi: 10.1126/science.aah3963 PubMed PMID: 27934765; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ariumi Y, Kuroki M, Kushima Y, Osugi K, Hijikata M, Maki M, Ikeda M, Kato N. Hepatitis C virus hijacks P-body and stress granule components around lipid droplets. J Virol. 2011 Jul;85(14):6882–6892. doi: 10.1128/jvi.02418-10 PubMed PMID: 21543503; PubMed Central PMCID: PMC3126564. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roby J, Pijlman G, Wilusz J, Khromykh A. Noncoding subgenomic flavivirus RNA: multiple functions in West Nile virus pathogenesis and modulation of host responses. Viruses. 2014 Jan 27;6(2):404–427. doi: 10.3390/v6020404 PubMed PMID: 24473339; PubMed Central PMCID: PMC3939463. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi P-Y, Khromykh AA. RNA structures required for production of subgenomic flavivirus RNA. J Virol. 2010 Nov;84(21):11407–11417. doi: 10.1128/jvi.01159-10 PubMed PMID: 20719943; PubMed Central PMCID: PMC2953152. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi P-Y, Hall RA, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe. 2008 Dec 11;4(6):579–591. doi: 10.1016/j.chom.2008.10.007 PubMed PMID: 19064258; eng. [DOI] [PubMed] [Google Scholar]
- 80.Zhang K, Dion N, Fuchs B, Damron T, Gitelis S, Irwin R, O'Connor M, Schwartz H, Scully SP, Rock MG, Bolander ME, et al. The human homolog of yeast SEP1 is a novel candidate tumor suppressor gene in osteogenic sarcoma. Gene. 2002 Oct 02;298(2):121–127. PubMed PMID: 12426100; eng. [DOI] [PubMed] [Google Scholar]
- 81.Pashler AL, Towler BP, Jones CI, Newbury SF. The roles of the exoribonucleases DIS3L2 and XRN1 in human disease. Biochem Soc Trans. 2016 Oct 15;44(5):1377–1384. doi: 10.1042/bst20160107 PubMed PMID: 27911720; eng. [DOI] [PubMed] [Google Scholar]
- 82.Rowley PA, Ho B, Bushong S, Johnson A, Sawyer SL. XRN1 is a species-specific virus restriction factor in yeasts. PLoS Pathog. 2016 Oct;12(10):e1005890. doi: 10.1371/journal.ppat.1005890 PubMed PMID: 27711183; PubMed Central PMCID: PMC5053509. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Esteban R, Vega L, Fujimura T. 20S RNA narnavirus defies the antiviral activity of SKI1/XRN1 in Saccharomyces cerevisiae. J Biol Chem. 2008 Sep 19;283(38):25812–25820. doi: 10.1074/jbc.M804400200 PubMed PMID: 18640978; PubMed Central PMCID: PMC3258869. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Longtine MS, Mckenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998 Jul;14(10):953–961. doi: PubMed PMID: 9717241; eng. [DOI] [PubMed] [Google Scholar]
- 85.Gueldener U, Heinisch J, Koehler GJ, Voss D, Hegemann JH. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002 Mar 15;30(6):23e PubMed PMID: 11884642; PubMed Central PMCID: PMC101367. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ran F, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013 Nov;8(11):2281–2308. doi: 10.1038/nprot.2013.143 PubMed PMID: 24157548; PubMed Central PMCID: PMC3969860. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Maynell LA, Kirkegaard K, Klymkowsky MW. Inhibition of poliovirus RNA synthesis by brefeldin A. J Virol. 1992 Apr;66(4):1985–94. PubMed PMID: 1312615; PubMed Central PMCID: PMC288987. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Coyne CB, Kim KS, Bergelson JM. Poliovirus entry into human brain microvascular cells requires receptor-induced activation of SHP-2. EMBO J. 2007 Sep 05;26(17):4016–4028. doi: 10.1038/sj.emboj.7601831 PubMed PMID: 17717529; PubMed Central PMCID: PMC1994131. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Coyne CB, Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006 Jan 13;124(1):119–131. doi: 10.1016/j.cell.2005.10.035 PubMed PMID: 16413486; eng. [DOI] [PubMed] [Google Scholar]
- 90.Li C, Wen A, Shen B, Lu J, Huang Y, Chang Y. FastCloning: a highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotech. 2011 Oct 12;11:92. doi: 10.1186/1472-6750-11-92 PubMed PMID: 21992524; PubMed Central PMCID: PMC3207894. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.N'Diaye E-N, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009 Feb;10(2):173–179. doi: 10.1038/embor.2008.238 PubMed PMID: 19148225; PubMed Central PMCID: PMC2637314. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thibault PA, Huys A, Amador-Cañizares Y, Gailius JE, Pinel DE, Wilson JA. Regulation of Hepatitis C Virus Genome Replication by Xrn1 and MicroRNA-122 Binding to Individual Sites in the 5' Untranslated Region. J Virol. 2015 Jun;89(12):6294–6311. doi: 10.1128/jvi.03631-14 PubMed PMID: 25855736; PubMed Central PMCID: PMC4474307. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang W-P, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000 Feb 25;275(8):5845–5851. PubMed PMID: 10681575; eng. [DOI] [PubMed] [Google Scholar]
- 94.Klionsky DJ. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol. 1992 Oct;119(2):287–299. PubMed PMID: 1400574; PubMed Central PMCID: PMC2289658. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tomashek JJ, Sonnenburg JL, Artimovich JM, Klionsky DJ. Resolution of subunit interactions and cytoplasmic subcomplexes of the yeast vacuolar proton-translocating ATPase. J Biol Chem. 1996 Apr 26;271(17):10397–10404. PubMed PMID: 8626613; eng. [DOI] [PubMed] [Google Scholar]
- 96.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif). 2001 Dec;25(4):402–408. doi: 10.1006/meth.2001.1262 PubMed PMID: 11846609; eng. [DOI] [PubMed] [Google Scholar]
- 97.Cheong H, Klionsky DJ. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008;451:1–26. doi: 10.1016/s0076-6879(08)03201-1 PubMed PMID: 19185709; eng. PMID: 19185709 [DOI] [PubMed] [Google Scholar]
- 98.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995 May 5;210(1):126–132. doi: 10.1006/bbrc.1995.1636 PubMed PMID: 7741731; eng. [DOI] [PubMed] [Google Scholar]
- 99.Mateo R, Nagamine CM, Spagnolo J, Mendez E, Rahe M, Gale M, Yuan J, Kirkegaard K. Inhibition of cellular autophagy deranges dengue virion maturation. J Virol. 2013 Feb;87(3):1312–1321. doi: 10.1128/jvi.02177-12 PubMed PMID: 23175363; PubMed Central PMCID: PMC3554187. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


