Abstract
For inhibitor design, as in most research, the best system is question dependent. We suggest structurally defined allostery to design specific inhibitors that target regions beyond active sites. We choose systems allowing efficient quality structures with conformational changes as optimal for structure-based design to optimize inhibitors. We maintain that evolutionarily related targets logically provide molecular avatars, where this Sanskrit term for descent includes ideas of functional relationships and of being a physical embodiment of the target’s essential features without requiring high sequence identity. Appropriate biochemical and cell assays provide quantitative measurements, and for biomedical impacts, any inhibitor’s activity should be validated in human cells. Specificity is effectively shown empirically by testing if mutations blocking target activity remove cellular inhibitor impact. We propose this approach to be superior to experiments testing for lack of cross-reactivity among possible related enzymes, which is a challenging negative experiment. As an exemplary avatar system for protein and DNA allosteric conformational controls, we focus here on developing separation-of-function inhibitors for meiotic recombination 11 nuclease activities. This was achieved not by targeting the active site but rather by geometrically impacting loop motifs analogously to ribosome antibiotics. These loops are neighboring the dimer interface and active site act in sculpting dsDNA and ssDNA into catalytically competent complexes. One of our design constraints is to preserve DNA substrate binding to geometrically block competing enzymes and pathways from the damaged site. We validate our allosteric approach to controlling outcomes in human cells by reversing the radiation sensitivity and genomic instability in BRCA mutant cells.
1. INTRODUCTION
Allostery is much discussed, but very few drug compounds target allosteric sites despite the extremely successful ribosomal antibacterial drugs revealing the tremendous and under-utilized power and specificity of targeting allostery with inhibitors binding outside the active site (Wang et al., 2012). To successfully target allostery, one needs to understand functional conformations. In particular for enzymes, targeting allostery requires a knowledge of the communication between protein conformation and the active site that approaches atomic resolution. Furthermore in developing inhibitors, the optimization of leads is an expensive preclinical effort, so targeting allostery has partly been limited by practicalities. With the above points in mind, we here suggest an approach of crystallography combined with small-angle X-ray scattering (SAXS) on structurally feasible targets: this empirical method allows one to efficiently produce the necessary knowledge aided by recently developed analysis software (Lai et al., 2016; Schneidman-Duhovny, Hammel, Tainer, & Sali, 2016) and then proceed with structurally informed optimization. Here we outline our strategy for efficiently targeting allostery in human cells with atomic level information even when human protein structures of a target enzyme are unavailable. As an exemplary target that forms a biologically critical multifunctional complex, we describe the design and testing of allosteric inhibitors for the DNA repair nuclease termed meiotic recombination 11 (MRE11).
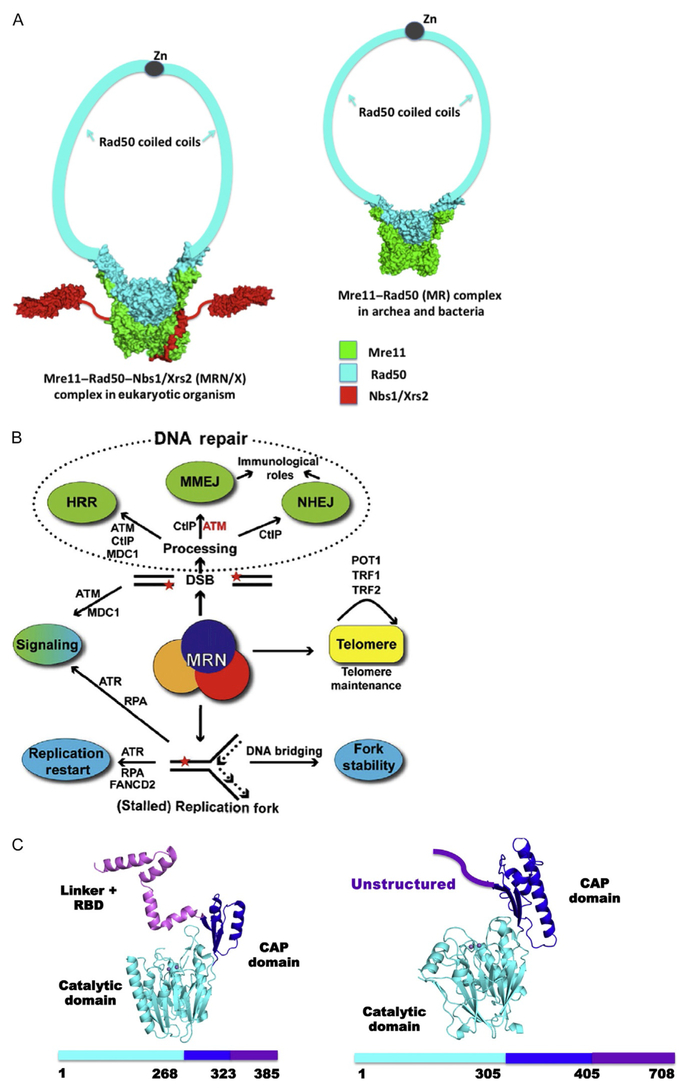
MRE11 is critical for genome stability during DNA replication and DNA repair. It is the fundamental core component of the MRE11, ABC ATPase RAD50, and phosphopeptide-binding Nijmegen breakage syndrome 1 (NBS1) protein Mre11–Rad50–Nbs1 (MRN) complex in humans (also known as MRN/X (Mre11–Rad50–Nbs1/Xrs2) in eukaryotes and MR (Mre11–Rad50) in archaea and SbcCD in bacteria (Fig. 1A) (Chahwan, Nakamura, Sivakumar, Russell, & Rhind, 2003; D’Amours & Jackson, 2002; Hopfner et al., 2000; Lafrance-Vanasse, Williams, & Tainer, 2015; Lamarche, Orazio, & Weitzman, 2010; Seeber et al., 2016; Stracker & Petrini, 2011; Williams, Lees-Miller, & Tainer, 2010; Williams & Tainer, 2007). Through the MRN complex, MRE11 interfaces with multiple DNA damage response pathways, including double-strand break (DSB) repair involving homologous recombination (HR) and nonhomologous end joining (NHEJ) (Acharya et al., 2008; Bennardo, Cheng, Huang, Stark, & Haber, 2008; Biehs et al., 2017; Bierne, Ehrlich, & Michel, 1997; Shibata et al., 2014) and replication fork processing to maintain genome stability (Fig. 1B) (Schlacher et al., 2011; Schlacher, Wu, & Jasin, 2012). In this context, the MRE11 catalytic domain provides structure-specific endo- and exonucleolytic activities to prepare DNA ends for annealing and end-joining repair (Buis et al., 2008; Krogh, Llorente, Lam, & Symington, 2005; Lewis et al., 2004; Limbo, Porter-Goff, Rhind, & Russell, 2011; Majka, Alford, Ausio, Finn, & McMurray, 2012; Paull & Gellert, 1998). In fact, MRN can gain access to occluded DNA ends by removing Ku or other DNA adducts via its Mre11-dependent nucleolytic reaction (Myler et al., 2017; Shibata et al., 2014). In humans, mutations in MRE11 can cause ataxia-telangiectasia-like disorder (ATLD), featuring cerebellar degeneration and variable cancer predisposition due to an alteration in the mechanical activity of the enzyme during DNA processing (Limbo et al., 2012).
Fig. 1.
Mre11–Rad50–Nbs1 (MRN) architecture, pathways, and nuclease core fold. (A) Architectural structures of MRN/X complex from eukaryotes (left) and MR vs bacterial and archaeal (right). (B) DNA repair pathway schematic for MR and MRN/X functions in cell biology. (C) The TmMre11 (PDB ID: 3THO) fold (ribbon diagram) and sequence (left) with domain organization (catalytic domain with metal ion active site, cyan), CAP domain (blue), and linker plus Rad50-binding domain (RBD, violet). Human MRE11 fold and sequence with the domain organization from PDB ID 3T1I (right) with catalytic domain with metal ion active site (cyan), CAP domain (blue), and the unstructured C-terminus (violet) containing the RBD. (Human MRE11 has GAR domain, RBD, and C-terminus unstructured yet.)
The MRE11 complex is furthermore emblematic in terms of forming multiple distinct functional networks with different activities (Williams, Williams, & Tainer, 2007). DNA is susceptible to breaks leading to chromosome abnormalities associated with cancer wherever B-DNA is destabilized by sequence (Bacolla, Tainer, Vasquez, & Cooper, 2016) or opened during DNA processing (Lindahl, 1993). In eukaryotes, MRN/X interfaces with other protein partners to facilitate DNA processing at replication forks (Schlacher et al., 2011, 2012) and repair pathway selection (Shibata et al., 2014). It cooperates with CtIP (Sae2 or ctp1 in budding or fission yeast, respectively) in replication and recombination pathways, where CtIP can promote the endonucleolytic activity of human MRN on protein-blocked ends (Andres et al., 2015; Deshpande, Lee, Arora, & Paull, 2016; Limbo et al., 2011; Makharashvili et al., 2014; Sartori et al., 2007). MRE11 nuclease can either protect or promote the degradation of stalled replication forks when either BRCA2 or RAD51 function is impaired (Schlacher et al., 2011, 2012). Additionally, the MRN complex can activate and signal via the ATM kinase, allowing ATM to regulate MRE11 nuclease to promote DNA microhomology-mediated end joining over degradation (Rahal et al., 2010). In general, nucleases such as MRE11 tend to be points where inhibitors can impact well-regulated pathway elements that may be sculpting the nucleic acid substrate (Tsutakawa, Lafrance-Vanasse, & Tainer, 2014).
The dimeric MRE11 nuclease works synergistically with Rad50 and Nbs1/Xrs2 to govern the multiple conformations of the MRN molecular machine to sense, bind, protect, and process damaged DNA (Crown et al., 2013; de Jager et al., 2001; Deshpande et al., 2014; Hopfner et al., 2002, 2001; Roset et al., 2014; Williams et al., 2008). Structure-based models of MR complexes (from both prokaryotes and eukaryotes) reveal that the Mre11 dimer is the core scaffold that provides the platform for the flexible operations of its partner Rad50 in tethering DNA ends and regulation of endo- and exonuclease processing of DNA through modulation of enzyme activity ofMre11 (Fig. 1A). The Mre11 subunit consists of a catalytic domain, a CAP domain, and a linked Rad50-binding motif in prokaryotes (Fig. 1C). In mammals, MRE11 additionally bears a glycine–arginine-rich (GAR) motif (Yu et al., 2012) that binds poly(ADP) ribose synthetized by PARP1 at DNA DSBs, therefore stimulating DNA damage signaling (Haince et al., 2008). From the back face of the MRE11 catalytic domain, its interactions with NBS1 optimize MRN’s allosteric organization to perform the correct function based on the substrate and protein partners, which are dictated by cell cycle status (Fig. 1) (Crown et al., 2013; Lammens et al., 2011; Mockel, Lammens, Schele, & Hopfner, 2012; Rojowska et al., 2014; Schiller et al., 2012; Williams et al., 2009). Structures of prokaryotic MR further validate Mre11 as a blueprint for orienting the core MR complex (Fig. 1A).
In humans, the MRN complex binds DNA DSBs and initiates damage-induced signaling cascades via activation of the ataxia-telangiectasia-mutated (ATM) (Lee & Paull, 2005; You, Chahwan, Bailis, Hunter, & Russell, 2005) and ataxia-telangiectasia- and rad3-related (ATR) kinases (Falck, Coates, & Jackson, 2005; Regal, Festerling, Buis, & Ferguson, 2013; Shiotani et al., 2013; Williams et al., 2010). Yet, detailed structural information from microbial Mre11 allowed the development of human MRE11 inhibitors that can control the ability of a cell to switch among DSB repair pathways (Shibata et al., 2014). These MRE11 inhibitors therefore have the potential as radioprotectants (Koukourakis, 2012) and are tools for testing MRE11 activity in the clinic in both combined or single-therapy generation for diseases of aging (McPherson, Shen, & Ford, 2014; Sun et al., 2017), autoimmunity, and cancer (Kelley, Logsdon, & Fishel, 2014; Kuroda, Urata, & Fujiwara, 2012; Nicholson et al., 2017; Sun et al., 2017).
While these observations reveal how this complex may function similarly and differently between prokaryotes and eukaryotes, the core cell biology surrounding Mre11 complex activity in detection and processing of damaged DNA ends is similar throughout the domains of life (Williams et al., 2007). Thus, useful knowledge obtained from high-resolution analyses of microbial MR frequently applies to physical and mechanistic hypotheses for eukaryotic MRN homologs.
In fact, MRE11 is under active investigation as a target for enhancing DNA damage-induced tumor cell killing (Dynlacht, Batuello, Lopez, Kim, & Turchi, 2011; Hengel, Spies, & Spies, 2017; Hosoya & Miyagawa, 2014; Thompson, Montano, & Eastman, 2012; Velic et al., 2015; Vilar et al., 2011). As MRE11 knockouts are embryonic lethal (Stracker & Petrini, 2011), the development of inhibitor tools to evaluate MRE11 functions in cells has a biomedical value. In contrast, current strategies for MRE11 inhibition based on RNAi have limitations on specificity and efficiency, as MRE11 is multifunctional.
Importantly, small molecule approaches promise targeting of specific MRE11 nuclease activities that govern repair pathway choice (Shibata et al., 2014). Small molecules also have the advantage of yielding highly penetrant effects across whole-cell populations compared to other targeting therapies, such as RNA interference. Moreover, they can be efficiently tested across cell lines and therapeutic conditions. Early high-throughput screening for inhibition of double-strand break repair in Xenopus laevis egg extracts identified Mirin as the Mre11 nuclease inhibitor (Dupré et al., 2008; Garner, Pletnev, & Eastman, 2009; Roques et al., 2009). Recent elaboration of the original Mirin scaffold to produce the PFM library (representing the names Petricci–Forli–Moiani) identified derivatives, which selectively target either endo- or exonuclease activity of Mre11 in vitro. Since inhibition of Mre11 endonuclease activity inhibits repair by HR and promotes NHEJ-mediated repair, these next-generation inhibitors provide a means to guide pathway switching between NHEJ and HR (see Fig. 1B) (Shibata et al., 2014).
Structure-based inhibitor analyses and design were critical to understanding and refining the molecular mechanisms underlying functional selection of Mre11 endo- or exonuclease activity by the next-generation PFM inhibitors. As many mammalian DNA repair factors, MRE11 included, remain challenging for structural study, utilization of a microbial molecular avatar of the Mre11 nuclease from eubacterial Thermotoga maritima was critical for understanding structure–activity relationships (SARs) between Mre11 and the next-generation selective inhibitors PFM01, PFM03, PFM04, and PFM39. We employ the term avatar for our approach, rather than homolog or other term for sequence similarity or evolutionary relationship, because our strategy depends upon the physical embodiment of the target’s essential features including allostery without necessarily having high sequence identity. Here, we describe our avatarian approach to develop chemical inhibitors that target allostery to dissect multifunctionality and control biological outcomes with the MRE11 nuclease as a prototypic system. The methods discussed provide the basis to employ structural allostery and avatars for complex mammalian targets in order to develop chemical inhibitors that are increasingly relevant for accurate understanding and prediction of mechanisms in cancer cell biology and biomedicine. By focusing on avatars and allosteric switches the inhibitor optimization cycle may be focused more tightly than often otherwise done which may improve efficient progress and decrease preclinical costs for the design of inhibitors for possible clinical trials.
2. METHODS FOR AVATAR INHIBITORS TARGETING ALLOSTERY
In this section, we describe the design and generation of the PFM-derivative library from the Mirin scaffold. We furthermore note strategies for selecting the T. maritima Mre11 as a molecular avatar along with characterizations of PFM library interactions with the Mre11 avatar, which tested endo- and exonucleolytic selectivity among the PFM-derivative library. We outline strategies for cocrystallization of Mre11/PFM inhibitor complexes and for the analysis of allosteric effects within the resulting crystal structures of the complex driving endo- or exonucleolytic selectivity. As the validation of SARs derived from microbial avatars in a mammalian system is critical, we highlight in vitro and in cell approaches for testing the effects of PFM inhibitors on human MRE11 and DSB repair in human cells.
2.1. Chemical Design and Synthesis of the PFM Library
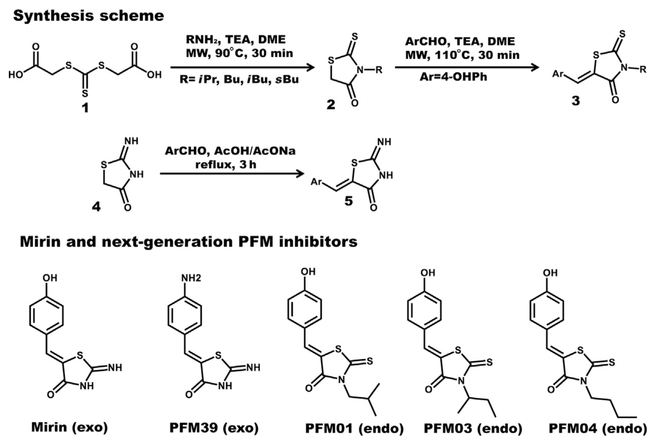
Chemical analysis of the early Mre11 inhibitor Mirin (Dupré et al., 2008) reveals a pseudothiohydantoin core containing a Z p-hydroxybenzylidene moiety at the 5-C position. Functional group modifications of Mirin, in both the aromatic and the pseudothiohydantoinic ring, were identified as logical targets for testing SARs. A good candidate probe should be efficiently synthesized in high yields and be achiral. Initial substitution of the core pseudothiohydantoinic moiety with a rhodanine and/or N-alkyl-rhodanine ring generated a compound series with variable modulation of Mre11 nuclease activity. From these pilot compounds, a targeted library of PFM derivatives was designed and synthesized by using the general synthetic approaches described in Fig. 2 for a detailed characterization of Mre11 nuclease modulation.
Fig. 2.
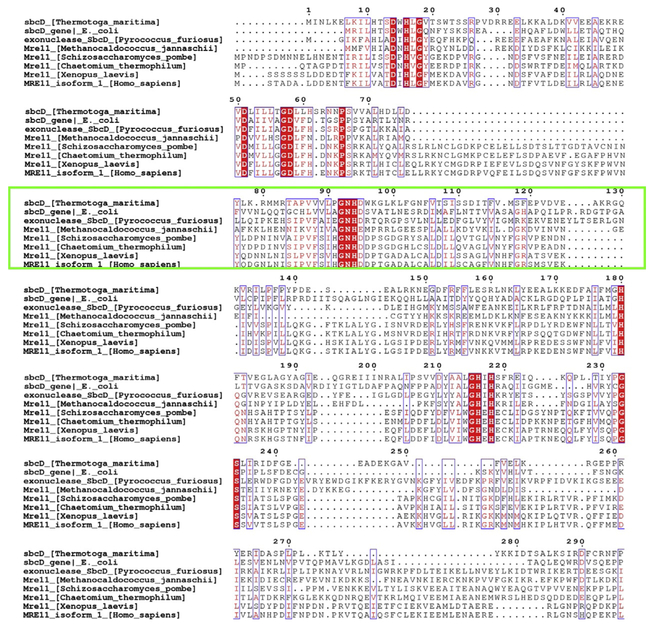
Sequence conservation from an alignment performed with Cobalt and Espript 3.0 showing the Mre11 catalytic domain among organisms having a structure deposited into the PDB plus the X. laevis Mre11 used for in vivo identification of Mirin activity. Conserved regions (red fill) and the region showing an allosteric conformational change in T. maritima centered at N93–H94 is highlighted (green box). Organisms from the top are shown like bacterial, through archeal, yeast, fungi, vertebrate, and mammals.
The protocol developed represents a straightforward pragmatic approach for the preparation of decorated rhodanine and pseudothiohydantoin derivatives. These compounds provided interesting probes to elucidate selective inhibition of Mre11 endo- or exoonuclease activity. From the screening of our PFM derivatives, we infer that the aromatic portion impacts the activity and the OH or NH2 group in the 4-C position (on the benzene ring) is crucial for inhibition (Fig. 2). The pseudothiohydantoin core is important for the selective inhibition of the Mre11 exonuclease activity. Importantly, we furthermore found it possible to switch the exo inhibitor into an endo one by the simple replacement of the core with a rhodanine ring demonstrating the power of a focused library for this system.
2.2. Identifying Nonhuman Mre11 Avatars
Human MRE11 has remained intractable to the robust high-resolution crystallography required for efficiently elucidating protein/inhibitor complexes: the sole X-ray MRE11 structure, containing the CAP and catalytic nuclease domains, only reaches to 3.0 Å resolution (Park, Chae, Kim, & Cho, 2011). To achieve the higher resolution desired for detailed structures of MRE11-PFM inhibitor complexes, we chose to employ a microbial molecular avatar of the Mre11 nuclease to test and improve inhibitors. Our starting point for identifying nonhuman avatar systems is to examine existing crystal structures and to identify critical residues conserved among all species implying fundamental mechanistic features. The full sequence alignment of Mre11 obtained with Cobalt (Constraint-based Multiple Alignment Tool), as displayed in Fig. 3, highlights residues corresponding to the GDL motif (57–59 in TmMre11), GNHD motif (92–95 in TmMre11), GHxH motif (215–218 in TmMre11), and all residues that are part of the active site, as exhibiting the greatest conservation across all domains of life.
Fig. 3.
Chemical synthesis scheme and inhibitors analyzed. Schematic representation of the two-step synthetic chemical method to produce N-alkyl rhodanines (top). A one-step protocol to produce mirin-like pseudothiohydantoin derivatives (middle). The 2D representation of all inhibitors discussed (bottom) starting from left mirin (exo), PFM39 (exo), PFM01 (endo), PFM03 (endo), and PFM04 (endo). Procedures: In 1 properly functionalized rhodanines (Mendgen, Steuer, & Klein, 2012) moieties were synthesized by cyclization with primary amines in the presence of TEA (triethanolamine) in DME, at 90°C under microwave (MW) irradiation for 30 min. Compound 2 was obtained in different yields depending on the starting amine used; the worst yields were obtained for the sterically hindered and problematic isopropylamine. A Knoevenagel’s condensation in the presence of an aromatic aldehyde and TEA furnished 3 compounds by irradiation at 110°C for 30 min. When the pseudothiohydantoin ring is maintained, MW as the heating source generated a mixture of Z/E isomers. Complete region selectivities toward the Z isomers were possible refluxing the pseudothiohydantoin with traditional heating conditions in the presence of arylaldehydes in a mixture of AcONa/AcOH for 3 h.
Informed by conserved residues identified by the alignment, we examined the best available MRE11 structures, using a resolution cutoff of 2.7 Å, to map the location of these key residues and identify components to improve structure-based design of small molecule inhibitors. This combined sequence and structural analysis identified the bacterial Mre11 homolog from T. maritima (TmMre11, Fig. 1C) as the best avatar candidate for human MRE11. Following structural superimposition between the human and bacterial nucleases, we had a suitable scaffold for performing in silico docking experiments for rational chemical design. In these efforts we also employed the Mre11 dimer structure validated in solution by SAXS (Rambo & Tainer, 2011) and in our two DNA–Mre11 complex crystal structures and SAXS shapes for Mre11 dimers (Williams et al., 2008). Mutations impacting dimer formation in solution lost DNA binding validating the dimer interface as contributing the DNA-binding channel.
2.3. Cocrystallization and Analysis of TmMre11/Inhibitor Complexes
Although structures of protein–ligand complexes can be obtained by soaking, we recommend cocrystallizations. We find that the best technique for obtaining high inhibitor occupancy when targeting allostery is cocrystallization which may allow visualization of altered conformations.
2.3.1. Cocrystallization of TmMre11/Inhibitor Complex Structures
In this section we outline the steps employed to generate X-ray crystal structures of TmMre11 inhibitor complexes, which are exemplary of the methods we have found useful for determining structures of enzyme–inhibitor complexes.
After expression and purification (Das et al., 2010), TmMre11 is exchanged into its target crystallization buffer and concentrated to approximately 10mg/mL. Target compounds are prepared at 100 mM in DMSO and added directly to the protein solution to a final concentration of 5 mM. If precipitation occurs, the solution should be centrifuged at 15,000 ×g for 10 min to remove particulates prior to preparing crystallization trays, either hanging drop or sitting drop setups, which are incubated at 18°C for 5 days. Once crystals have matured, a suitable cryo-harvest condition is identified by testing crystal resilience and diffraction upon transfer to well solution supplemented with 5%–20% glycerol and/or low-molecular-weight polyethylene glycols (PEGs). Benefits of maintaining 5 mM compound within the cryo-harvest condition should also be explored. Once crystals have been frozen without damage, X-ray diffraction datasets may be collected, preferably from a synchrotron source, such as the SIBYLS beamline at the Advanced Light Source (Classen et al., 2013), which will provide the highest resolution data. After data processing, initial phasing is accomplished by molecular replacement, followed by model refinement. In the best-case scenario, compound electron density can be unambiguously identified within Fo – Fc difference maps and subsequently refined using PDB and cif library parameter files created with the PRODRG server (http://davapc1.bioch.dundee.ac.uk/prodrg/).
Once an inhibitor has been successfully cocrystallized with its target, one must analyze the resulting target/inhibitor complex model to develop hypotheses linking compound orientation at the binding site to allosteric movement of the target for testing in future in vitro and in vivo experiments.
2.3.2. Performing In Silico Docking Against TmMre11
X-ray crystallography allows the use of relatively small fragments and the growth of better binders from structures defined with initial fragments (Chan et al., 2017). We employed a general strategy to test and improve the activities of the small molecules consisting of testing a focused library of compounds, solving structures of selected binders, and obtaining the SDF (structure-data file) files of them. The best way to obtain SDF files is from chemical SMILE using a translator (https://cactus.nci.nih.gov/translate/).
However, our strategy further involves targeting allosteric conformational changes that are required for the enzyme to form a productive complex with the DNA substrate. Lessons from multiple repair enzyme structures have taught us the advantages of focusing on allosteric interactions instead of primarily targeting the active site. First, allosteric interactions can enable multiple potential sites for inhibitors and thereby allow dissection of related functions, as we found for MRE11. Second, allosteric interactions tend to be system specific, whereas compounds targeting active sites may show cross-reactivity to related enzyme classes. Third, we can interfere with conformational changes required for catalysis without blocking substrate binding, so the inactivated enzyme can itself provide protection against alternative reactions that might otherwise cause unwanted or unpredictable outcomes in cells.
The use of a docking software, such as the Schrodinger suite for drug discovery (Schrodinger, 2017), makes it possible to run a calculation for multiple ligand series per job to permit a detailed analysis of key features and improve the molecular design. In this approach, the in silico characterization is completed with a computational energy evaluation for implicit or explicit solvation models that can be calculated and efficiently compared with calorimetric experimental data (Moiani, Cavallotti, Famulari, & Schmuck, 2008). The best candidate is thereby identified for additional in vitro experiments, such as biophysical characterizations, and for testing in several cell lines.
2.3.3. Mre11 Endo- and Exonuclease Inhibitor Complex Structures
We crystallized and solved informative Mre11 structures with PFM01, PFM03 (endo inhibitors), and PFM39 (exo inhibitor) to 2.3, 2.4, and 2.3 Å, respectively. The Mre11 microbial avatar pinpointed two specific sites where small molecules can bind and modulate nuclease activity. Specifically, exonuclease inhibitors bind with variable orientation near the active site, blocking His61 (His63 in human MRE11), which mediates the 3′-to-5′ exonuclease activity by opening the ends of dsDNA substrates. In contrast, binding of endonuclease inhibitors is shifted toward the dimer interface with an orientation that places their alkyl chain emerging from a rhodanine ring toward the hydrophobic residues of helix α3.
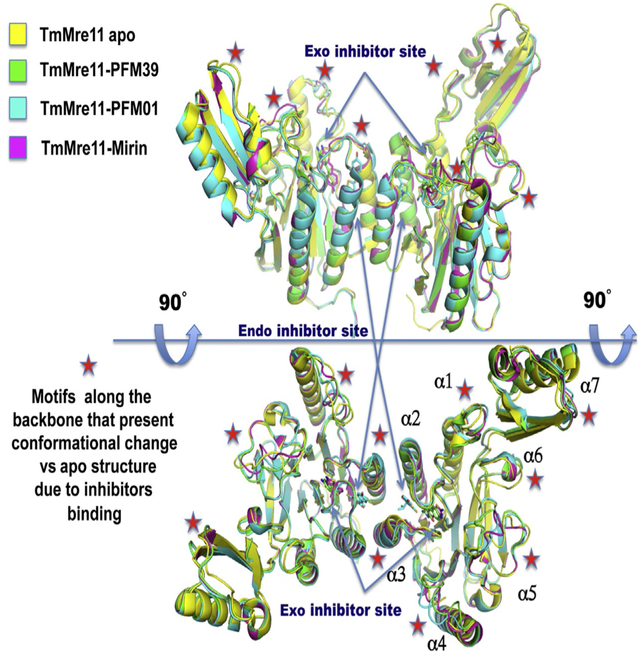
To examine conformational changes in Mre11 due to inhibitor binding, TmMre11 endo and exo inhibitor structures were superimposed onto the TmMre11 apo structure (Fig. 4). Viewed from two different angles, the alpha helices in the catalytic domain and in the CAP domain appear to restrict inhibitor access to catalytic area of the active site. The PFM inhibitors, however, are more distant from the active site, localizing near the Mre11 dimerization interface and contacting the N93–H94 motif.
Fig. 4.
Structural variation revealed by superimposition of TmMre11 apo structure (yellow, PDB ID: 4NZV), TmMre11-PFM39 exo inhibitor structure (green, PDB ID: 4O5G), TmMre11-PFM01 endo inhibitor structure (cyan, PDB ID: 4O24), and TmMre11-Mirin exonuclease inhibitor structure (magenta, PDB ID: 4O4K). The top and bottom show different orientations (90° tilted) of the Mre11 dimers. Arrows point to the two different binding sites for endo and exo inhibitors. Red stars highlight backbone regions shifted vs the apo structure. These reorganizations revealed a mechanism for dissecting the endo- and exonuclease activities by chemical inhibitors.
The Mre11 α4 and α5 helices show a conformational change with different patterns in the endo- and exonuclease inhibitor structures. The helices α1, α3, α6, and α7 show a small conformational change vs the apo structure, but the endo and exo inhibitor-bound structures have a similar structural pattern. Only helix α2 shows no conformational change in the presence of bound inhibitor, implying that it is part of the solid structural core of the catalytic domain in TmMre11 between residues Asn65 and Ala85. Conformational changes in a similar region were reported in the characterization of ATLD mutant W243R, showing that the mutation close to the inhibitor-binding region causes conformational change of these regions with clastogenic activity in fission yeast (Limbo et al., 2012).
2.3.4. High-Resolution Structure of PFM04, Endo Inhibitor, Bound to TmMre11 and Its in vitro Activity
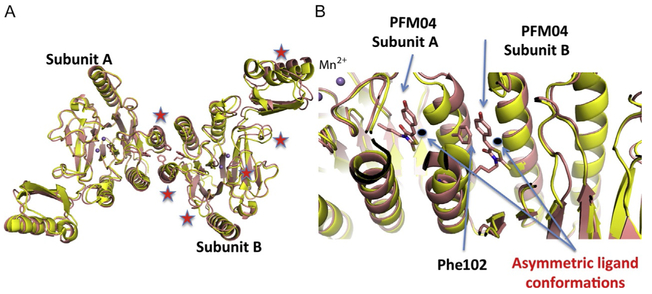
Structures provide a means of visualizing and understanding the impact of inhibitor binding. As a test of our methods, we introduce in this section the novel high-resolution structure of TmMre11 with bound PFM04. This small molecule completes our initial investigational series for structural and activity characterization as presented in Fig. 5A. Compound PFM04 is a member of the N-alkyl-rhodanine derivative family with an endonucleolytic inhibitory effect: it has a linear N-alkylic chain instead of the branched N-alkyl chain of PFM01 and PFM03. The compound PFM04 was cocrystallized as described in Section 2.3.1, with crystals being grown by using sitting drop vapor diffusion in 0.1 M MES pH 6.5, 10% PEG3350, 20 mM CaCl2, then soaked in 20% ethylene glycol as cryoprotectant. A dataset to 2.15 Å resolution was obtained at the SIBYLS beamline (ALS BL 12.3.1) (Classen et al., 2013), and the structure was refined and submitted to the Protein Data Bank (PDB ID: 6ASC). X-ray diffraction data collection and refinement statistics are shown in Table 1. In Fig. 5B and C, we present the superimposition of the new TmMre11–PFM04 (color pink) with the apo structure PDB ID: 4NZV (color yellow).
Fig. 5.
The apo and novel PFM04 inhibitor-bound structures. (A) The superimposition of TmMre11 apo (yellow) and the novel TmMre11–PFM04 inhibitor complex structure (pink) showing the different organization of the two domains due to the asymmetric binding of PFM04. Red stars highlight main chain shifts in the inhibited vs the apo structure. (B) A zoom-in on the inhibitor-binding site shows the different conformation of ligands between subunit A and subunit B, along with the interaction of Phe102 in subunit A and PFM04 in subunit B.
Table 1.
X-Ray Diffraction Data Collection and Refinement Statistics (Molecular Replacement)
| TmMre11–PFM04 | |
|---|---|
| Data | |
| Space group | P21 |
| Cell a, b, c (Å) | 48.053, 112.682, 81.162 |
| α, β, γ (°) | 90.00, 100.26, 90.00 |
| Resolution (Å) | 2.15 |
| Rsym or Rmerge | 4.9 |
| I/σ | 46.7 |
| Completeness | 94.2 |
| Redundancy | 4.8 |
| Refinement | |
| Resolution (Å) | 2.15 |
| No. reflections/ observed |
72,439 |
| Rwork/Rfree | 19.69/23.18 |
| No. atoms | 5375 |
| Protein | 5187 |
| Ligand/ion | (Mn)4 |
| (PFM04) 38 | |
| Waters | 142 |
| Protein | 67.3 |
| Ligand/ion | 85.6 |
| Water | 55.4 |
| Bond | 0.004 |
| Angle | 0.789 |
Differing from the previous PFM structures, this novel structure shows the compound PFM04 oriented in a new orientation between the two Mre11 subunits, as shown in Fig. 5C. The alkyl chain is also organized differently and shows evidence of flexibility. The effect of asymmetric orientations of the two small molecules in the dimer is translated into different organizations of Mre11 subunit A and subunit B, as shown in Fig. 5B where red stars identify regions of conformational change in the backbone. In fact, one domain resembles the organization in apo Mre11 with a small difference in the CAP domain. However, the other domain shows notable differences from the apo structure with two crucial regions differently organized. This effect is also correlated with the direct interaction of PFM04 in one domain with Phe102 that is an important residue governing TmMre11 dimerization.
2.4. Targeting Mre11 Dimer Allostery With a Molecular Avatar
We chose eubacteria TmMre11 to represent essential features of human MRE11 catalytic domain for inhibitor development. The size and complexity of eukaryotic MRN/X assembly vs the bacterial MR complex (Lafrance-Vanasse et al., 2015; Yu et al., 2012), and the absence of posttranslational modifications support its practical advantages. Compared to human MRN, TmMre11 provides a more efficient and accurate structural characterization of inhibitor complexes with implications for design of potential therapeutic compounds. Defining the conserved features of the catalytic domain including both substrate recognition and sculpting unveils similarities whereby small molecules designed to act on bacterial Mre11 can efficiently provide testable inhibitors for human MRE11. Such inhibitors have general interest as possible chemical tools to control DNA repair pathway selection in human cells.
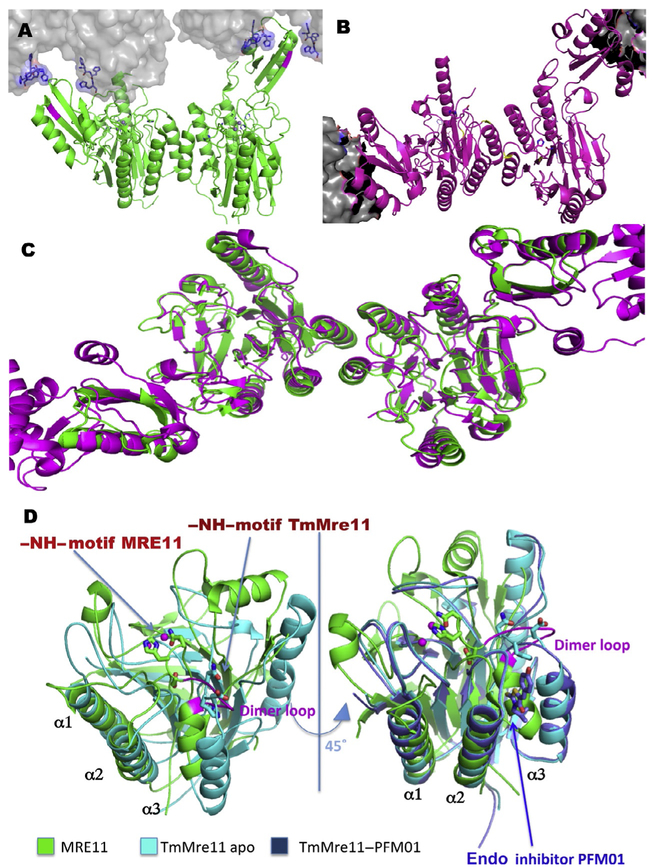
We therefore used bacterial Mre11 as a molecular avatar that embodies the key features of human MRE11 that we wished to target. The essential structural biology for avatars from microbial cells may provide many of the features needed to experimentally test the means to inhibit the human enzyme, as seen for DNA replication and repair enzymes. In Fig. 3, the green highlighted rectangle shows the conserved region that in the T. maritima homolog shows the allosteric motion of the NH loop in the superimposition of PDB files 2Q8U, 3QG5 (Fig. 6A–C), in the asymmetric conformation induced by the N-terminal 6His-tag (Das et al., 2010), and in the open asymmetric conformation induced by Rad50 torsional rotation (Lammens et al., 2011). Fig. 6A and B shows the bacterial assembly with the electrostatic surface and the asymmetric orientation of the N93–H94 loop. His94 is conserved among all Mre11 homologs and acts in sculpting the DNA for the exonuclease process (Williams et al., 2008). Thus, His94 mutants show the inhibition of the exonuclease activity and a mutant mouse for the equivalent histidine mutation (H129N) has specific sensibility DSB repair by HR but not in NHEJ or other processes that do not involve the Mre11 exonuclease activity (Buis et al., 2008). However, the structural analyses based on the sequence of Mre11 from higher eukaryotes uncover different features in the assembly of the nuclease domain that contain that NH loop motif, as well as differences in the dimerization interface and its placement relative to the NH loop motif.
Fig. 6.
Human MRE11 compared to TmMre11 apo and endonuclease PFM01-bound structures. (A) PDB ID: 4NZV TmMre11 apo dimer (green with crystal mates in the gray surface with N-terminal His-tags in sticks) shows how the allosteric opening of a small molecule accessible cavity was revealed in the switch of the N93–H94. The crystal packing uncovered the proposed allosteric conformation in the middle of its hypothesized cycle. (B) Representation of structure PDB ID: 3QG5 open dimer TmMre11 (magenta) where the gray cloud represents the Rad50 surface providing a torsional opening that reorganizes the asymmetric Mre11 allosteric dimerization. (C) Two levels of allosteric conformational reorganizations are visible in superimposition of PDB files uncovering differences in the organization of the NH loop and the opened cavity adjacent to the dimerization site. (D) The TmMre11 catalytic domain structure (cyan, PDB ID: 4NZV) and human MRE11 catalytic domain (green, PDB ID: 3T1I), superimposed on alpha helix α1 and α2 showing the human α3 tilted vs the TmMre11 as driven by the dimerization site represent in the human loop (magenta loop). The Mn2+ ions of both catalytic sites (spheres) and NH regions of both constructs are shown (sticks) with one engaged and one disengaged. A view tilted 45 degree with an added structure of the complex TmMre11–PFM01 catalytic domain superimposed (dark blue, PDB ID: 4O24). PFM01 (sticks) at the dimerization site shows an overlap with the human MRE11 α3. The engagement of NH domain in human MRE11 in this structure closes the site where small molecules can act as endonuclease inhibitors.
The asymmetric organization of MR from T. maritima (Lammens et al., 2011) in the PDB 3QG5 does not show the pockets that can bind small molecules to modulate exonuclease vs endonuclease activities and thereby consequent Mre11 functions in regulating outcomes at DSBs and stalled replication forks. The data obtained with the T. maritima Mre11 construct codeveloped with JCSG (Joint Center for Structural Genomics) (Das et al., 2010) were first crystallized and deposited as PDB: 2Q8U, and then improved by the authors and deposited as PDB 4NZV. This structure reveals that the allosteric opening and closing of the loop contain His94 and the hidden pocket close to the dimerization interface and able to contain small molecules. The crystal-packing arrangement suitable to open that loop was created by leaving the N-terminus His-tag uncleaved; in fact, the first histidine stacks with the residues Y276–Y277 inducing a conformational change of the CAP domain relative to the catalytic domain, as shown in Fig. 6A. We proposed that the crystal packing simulates the action of Rad50 that is shown in the Mre11–Rad50 complex structure (Lammens et al., 2011). This Mre11 conformation allowed cocrystallizing the first endonuclease inhibitor PFM01 (Fig. 6D) in its vertical orientation close to dimerization site (PDB ID: 4024) and was successively employed for other endo- and exonuclease inhibitors. The exonuclease inhibitors, e.g., Mirin and PFM39, do not show unambiguous occupancy in electron density maps, but their general binding position appears to be displaced toward the active site relative to the endonuclease inhibitors, which are shifted toward the dimer interface.
The Mre11 dimer forms the core of the MRN complex as shown in Fig. 1A (Williams et al., 2010). Mre11 dimerization is necessary to bind and process dsDNA sites and to connect Mre11 nuclease activities to Rad50 conformational changes (Williams et al., 2008). These Rad50 conformational changes are predicted to regulate the Mre11 endo- and exonuclease activity and thus the pathway choice in vivo. Thus, we viewed the dimer interface region and its connections to the active site as a logical target for allosteric control of this system including for DNA repair pathway selection based on the DNA-substrate binding impacting cell cycle-dependent activities and protein partners involved in functional networks and biological outcomes. The dimerization interface of human MRE11 captured by its crystal structure shows unique features relative to other homologs: the human dimer assembly (Park et al., 2011) is maintained by a loop–loop interaction (Fig. 6D).
This sequence (DPTGADALC spanning Asp131–Cys139 from subunit A to subunit B) is conserved in X. laevis, the organism used in the identification of the original Mirin inhibitor (Dupré et al., 2008). The dimerization loop is immediately adjacent to the Asn128–His129 (NH loop) loop region near the N-terminal Cap domain, suggesting that dimerization may reorganize the N-terminal domain at the active site based on the macromolecular organization of the full MRN complex in the presence of substrates. Nbs1 may act naturally to induce this macromolecular reorganization, and this change may also be promoted by its interactions with partners such as CtIP (Deshpande et al., 2016; Kim et al., 2017). Importantly, small molecule compounds may provide chemical master keys to lock this conformational change into a single position. In other organisms with structurally characterized Mre11 assemblies, Mre11 retains an analogous dimer with coupled alpha-helical hairpins to maintain a specific interaction.
Our strategy for the dissection of MRE11 nuclease provides a prototypic exemplary system to describe methods to develop chemical inhibitors that dissect multifunctionality and control biological outcomes. We observed that DNA repair and replication enzymes typically sculpt their DNA substrates, as we found for nucleotide flipping by DNA repair glycosylases (Guan et al., 1998; Slupphaug et al., 1996; Thayer, Ahern, Xing, Cunningham, & Tainer, 1995), AP endonucleases (Tsutakawa et al., 2013), and endonuclease V (Dalhus et al., 2009), as well as for the flap exo- and endonuclease FEN1 (Rashid et al., 2017; Tsutakawa et al., 2017). Thus, part of our strategy to obtain specificity is to retain DNA binding but to block protein-mediated DNA conformational changes needed to position the substrate complex for catalysis. The methods discussed for this Mre11 system thus provide the basis to employ structural allostery and avatars to develop chemical inhibitors. Such inhibitors are increasingly relevant for accurate understanding and prediction of mechanisms in cancer cell biology and biomedicine.
Our goal was therefore to develop small molecules that bind near the dimer interface and specifically interact where they can impact the motion of the NH loop. We therefore aligned the structures of apo-TmMre11, TmMre11 in complex with the endo inhibitor PFM01 (Shibata et al., 2014), and human MRE11 catalytic domain (Fig. 6D). After pruning TmMre11 and human MRE11 catalytic domains, the superimposition was optimized on helices α1 and α2. This analysis shows that human MRE11 α3 is shifted toward the active site due to the engagement of the NH loop (Fig. 6D). The TmMre11 α3 helix is more open than the human homolog due to the disengagement of the NH loop enabling the fit of the endo inhibitors (Fig. 6D). In human MRE11 the dimerization loop (Fig. 6D, magenta) is directly connected to α3 and its allosteric movement is connected to reorganization of the heterocomplex based on the substrate bound. Small molecules that interfere with Mre11 sculpting of DNA into the active site make it feasible to control nuclease activity and consequently turn off nuclease functions to induce a predetermined DSB repair pathway (Fig. 1B).
2.5. In Vitro Validation of PFM Inhibitors Against Human MRE11
PFM compounds were first evaluated for their activity as nuclease inhibitors of MRE11. For efficiency, they were tested as a group of five at time for endo- or exonuclease-specific inhibition in parallel with a control for DMSO and a specific Mre11 mutant that inhibits the nuclease activity tested.
2.5.1. In Vitro Characterization of PFM Library Using TmMre11
Exonuclease assays were done by using a phospho-labeled radioactive substrate (Paull & Gellert, 1998) or aminofluoropurine substrate and then using a fluorimeter to measure the activity (Williams et al., 2008). The endonuclease assay was performed by using a circular DNA substrate (Das et al., 2010; Lee et al., 2012). Where feasible additional biochemistry tests are recommended for more specific characterizations. The in vitro activities of PFM04 against TmMre11 in an endo assay and in a resection assay against human MRN are shown in Fig. 7A and B and described later.
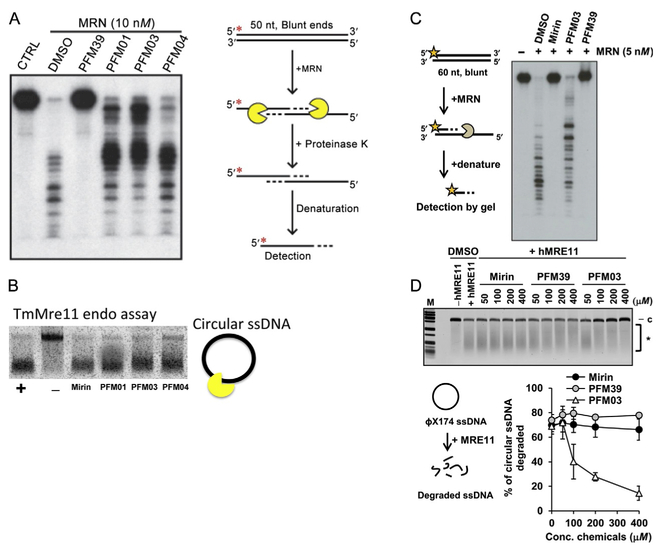
Fig. 7.
Nuclease activities and inhibitor impact. (A) MRE11 inhibitor PFM39 blocks MRN exonuclease activity (10 nM). In contrast, the MRE11 endonuclease inhibitors (PFM01, PFM03, and PFM04) have little effect on the MRN exonuclease activity. Radiolabeled DNA (100 nM) was incubated with MRN at 37°C for 60 min. Reactions contained a final concentration of 0.5 mM of the inhibitors. Red asterisk represents the 5′ radioactive label on the DNA. (B) Endo inhibitors PFM01, PFM03, and PFM04 primarily reduce the endo activity of TmMre11 vs the exo inhibitor Mirin. The nuclease assay is done at constant temperature and stopped at specific times. (C, D) The strong exo inhibition effect of PFM39 and Mirin vs endo inhibitor PFM03 for human MRN. (D) The strong endo inhibition activity of PFM03 vs the exo inhibitor mirin and PFM39 to human MRE11. The figure shows the percentage of circular ssDNA degraded relative to the control. Panels (C) and (D): Reproduced from Shibata, A., Moiani, D., Arvai, A. S., Perry, J., Harding, S. M., Genois, M. M., et al. (2014). DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Molecular Cell, 53(1), 7–18. https://doi.org/10.1016/j.molcel.2013.11.003.
Using an exonuclease assay with a double-stranded oligonucleotide substrate, we found that the newly characterized MRE11 endonuclease inhibitor only has a minor effect on the exonuclease activity of the MRN complex. The assay was performed along with a few other previously characterized MRE11 inhibitors and the reaction was allowed to proceed at 37°C for 60 min. As described by our group, PFM39, an exonuclease inhibitor, blocks the exonuclease activity of MRN in the assay (Shibata et al., 2014). On the other hand, the endonuclease inhibitors (PFM01, PFM03, and PFM04) in comparison slightly inhibit the exonuclease activity of MRN.
For the endonuclease assay, ΦX174 circular ssDNA was incubated with purified TmMre11 catalytic domain and inhibitors Mirin, PFM01, PFM03, and PFM04 at 500 mM final concentration. The inhibition assay was performed at 37°C for 15 min. Fig. 7B shows the percent of circular ssDNA degraded relative to the positive control (DMSO treated) and negative control TmMre11 H94S that present null endonucleolytic activity (Das et al., 2010; Shibata et al., 2014).
2.5.2. In Vitro Characterization of PFM Library Using Human MRE11
The in vitro validation process of PFM inhibitors was performed with human MRE11 and human MRN complex following the protocol described in our publication (Shibata et al., 2014). The exonuclease assay with human MRN is performed in an exo buffer consisting of 25 mM MOPS pH 7.0, 60 mM KCl, 0.2% Tween-20, 2 mM DTT, 2 mM ATP, 5 mM MnCl2, 5 nM MRN, and 0.8 mM PFM inhibitors. Labeled DNA (100 nM) was incubated in exo buffer for 30 min at 37°C, followed by deproteinization in one-fifth volume of stop buffer (20 mM Tris–Cl pH 7.5 and 2 mg/mL proteinase K) for 15 min at 37°C. Reactions were loaded on a 8% acrylamide/urea gel, run at 75 W for 60 min, dried onto DE81 filter paper, and autoradiographed with typical results shown in Fig. 7C.
Endonuclease assays were performed with ΦX174 circular ssDNA virion DNA (New England Biolabs), and 100 ng substrate was incubated with 300 ng purified WT hMRE11 in a 20 μL reaction (30 mM Tris–HCl pH 7.5, 1 mM dithiothreitol, 25 mM KCl, 200 ng acetylated bovine serum albumin (BSA), 0.4% DMSO, and 5 mM MnCl2) at 37°C for 30 min with inhibitors. The assay with human MRN complex was performed with 100 ng of substrate incubated with 400 ng of protein for 45 min in a 20 μL reaction (30 mM Tris—HCl pH 7.5, 1 mM dithiothreitol, 25 mM KCl, 200 ng acetylated BSA, 0.5% DMSO, and 5 mM MnCl2). Reactions were terminated with the addition of 1/10 volume of stop solution (3% SDS, 50 mM EDTA) and proteinase K to a final concentration of 0.1 mg/mL and incubation for 10 min at 37°C. The reaction products were run in a 0.8% agarose gel (1xTAE) for 90 min at 100 mA. DNA was stained with ethidium bromide and visualized using a Typhoon 9200 scanner with typical results shown in Fig. 7D for the specific PFM inhibitor tested.
2.6. From Molecular Avatar to Selective Human MRE11 Inhibitors Controlling Repair Pathway Selection in Human Cells
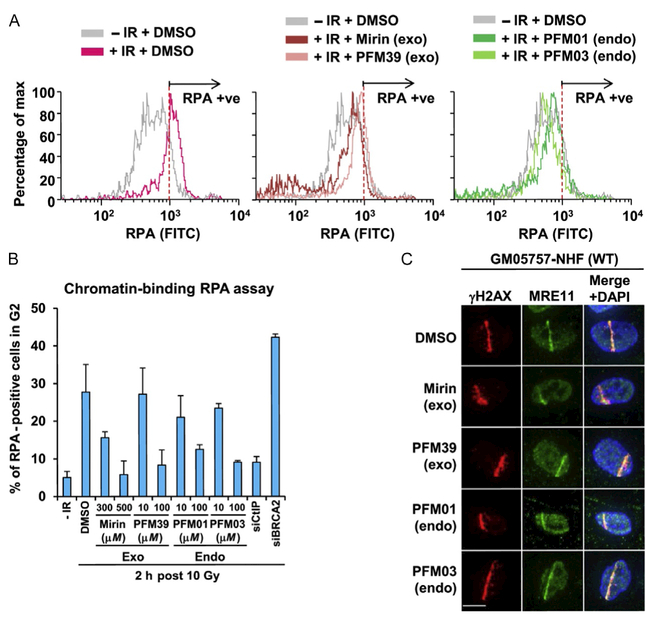
While in vitro cross-validation between human targets and their microbial avatars supports essential molecular mechanisms for inhibitor action, it is nevertheless critical to validate in vitro observations in the context of the cellular environment. To investigate the effects of MRE11 inhibitors on HR or NHEJ repair pathways, we chose to irradiate untreated or treated cells with X-rays (typical doses of 7–10 Gy) to induce DNA DSBs and monitor the outcomes and kinetics of repair by using a variety of assays. To establish whether PFM MRE11 inhibitors effectively prevent DNA end resection at sites of breaks, the first biochemical step mediated by MRE11, inhibitor-treated cells were monitored for the appearance of chromatin-bound RPA using FACS detection and the formation of Rad51 foci using immunofluorescence (Fig. 8A–C) (Shibata et al., 2014). For these studies, G2 phase cells were specifically examined (isolated via FACS sorting) to ensure observation of two-ended DSBs and avoid confounding observations from MRE11 action at replication fork structures (Roy et al., 2018). The selected PFM inhibitors successfully exhibited a reduction of chromatin-bound RPA and Rad51 foci, but still allowed MRE11 to localize to DSBs, highlighting their ability to impair MRE11 DNA end resection in vivo.
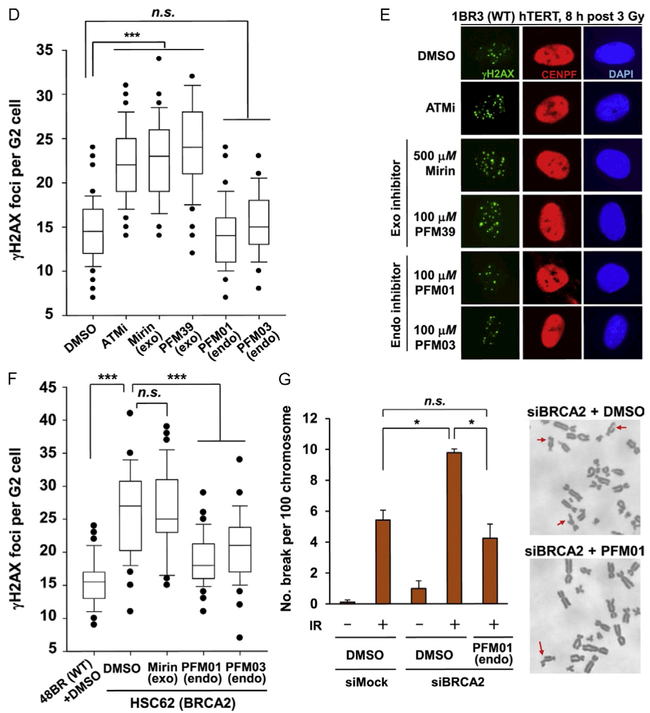
Fig. 8.
In cell validation assays from our publication (Shibata et al., 2014). (A) The effect that MRE11 inhibitors have in reducing chromatin-bound RPA level after IR treatment. IR-induced RPA retention in A549 cells was monitored by FACS analysis at 2 h post 10 Gy IR. (B) The quantification of RPA–FACS analysis at two different inhibitor concentrations. (C) Treatment with inhibitor does not affect the recruitment of MRE11 to the site of DNA damage. The cell lines used are primary human fibroblasts GM05757 subjected to UV-microbeam. The analysis of recruitment was done 1 h after inhibitor treatment. To identify the damage site, the histone γH2AX was labeled. (D, E) Treatment with exo inhibitor (but not endo inhibitors) causes a DSB repair defect in G2. DSB repair in G2 (CENPF+) cells was investigated by γH2AX foci analysis. Inhibitors were added 30 min before ionizing radiation (IR) treatment. 1BR3(WT) hTERT was fixed and stained at 8 h posttreatment with 3 Gy IR. (F) MRE11 endo inhibitors rescue the repair defect in HR-defective cells. γH2AX foci were enumerated in 48BR (WT) and HSC62 (BRCA2-defective) primary cells at 8 h posttreatment with 3 Gy IR. (G) The chromosome break defect in BRCA2 siRNA-treated 1BR3 (WT) hTERT was reversed by treatment with an MRE11 endonuclease inhibitor.
Monitoring the kinetics of DSB repair via immunofluorescence monitoring of γH2AX foci was used to probe repair differences between the MRE11 exonuclease and endonuclease inhibitors. Most DSB repair events (~80%) in irradiated cells follow the fast kinetics of NHEJ (~2 h); the remaining 20% are restored more slowly by HR (~8 h). Neither endo- nor exonuclease inhibitors impaired the fast kinetics of NHEJ repair (γH2AX foci number was similar between mock and inhibitor-treated cells 2 h posttreatment) (Fig. 8D and E) (Shibata et al., 2014). For extended HR repair, however, exonuclease inhibitors caused a repair defect (resulting in elevated γH2AX foci). These results show the ability of different MRE11 inhibitor classes to generate unique DNA repair phenotypes in a cellular context.
Since MRE11 endonuclease inhibitors do not result in a DNA repair defect and are capable of preventing DNA end resection of DSBs, we postulated that inhibiting MRE11 endonuclease activity might reduce HR initiation and promote NHEJ-driven repair in cells defective for HR. In fact, applying MRE11 endonuclease inhibitors to BRCA2-defective or knockdown cells rescued repair in a manner consistent with non-HR-deficient cells (Fig. 8F and G) (Shibata et al., 2014), indicating that MRE11 endonuclease activity is key for initial licensing of HR vs NHEJ although the exonuclease is required for excision and HR repair commitment. Thus, not only were we able to demonstrate the in vivo efficacy and biological impact of the avatar-derived human MRE11 inhibitors, but we were also to extend knowledge of MRE11 function in cells using these chemical tools.
3. FUTURE CONSIDERATIONS AND PROSPECTS
For our research we developed and describe here an avatarian approach to targeting allostery with chemical inhibitors in order to test specific functions in cell biology. Human proteins and their structures are not always efficiently obtainable, despite the development of advanced methods such as MacroBac with Hi-5 insect cells and baculovirus expression vectors with polypromotors (Gradia et al., 2017) for difficult eukaryotic multiprotein complex expression. There is moreover often a timely need for chemical knockdowns to examine protein functions in multiple cell types and under multiple conditions, such as in testing possible resistance factors for radiation or chemotherapeutic agents in cancer cells. Avatars can provide the critical tools for inhibitor development by structural biology. Thus, for example, the high-resolution structure of gammaH2A bound to Brc1 provides a reasonable avatar for inhibitor design against the human PTIP complex (Williams et al., 2010).
Proteins may be part of multiple networks and of complexes with different functions, so knockdowns or knockouts may not accurately inform specific activities and functions. Furthermore, active site inhibitors may not answer the most relevant questions. In contrast, allosteric inhibitors can offer advantages of specificity and of dissection of function that may otherwise be difficult or impossible because conformations often control networks and assemblies. As noted here for Mre11, developing inhibitor tools that target allosteric conformations can allow the dissection of function for multifunctional complexes to inform cell biology.
3.1. Targeting Allostery
We propose general advantages to targeting allostery rather than the traditional approach of focusing primarily on active sites. Allostery, which can involve conformations (that control orientations and shape) and assemblies (that alter local concentrations and orientations), can be as much a part of protein biological function as its catalytic activity. Furthermore, allostery can endow proteins with the multifunctionality that is the mechanistic underpinning of how many proteins are able to act in multiple steps and even in multiple pathways. Thus, proteins often have their interactions and activity allosterically regulated such that particular conformational states correlate to distinct functions. Allosteric events typically extend beyond active sites and include plastic deformations, order-to-disorder transitions, and large-scale domain rotations (Goldsmith, 1996) that may be the rate-limiting and regulated steps. Such conformational changes may also provide the basis for sculpting DNA and RNA substrates, as seen for the nuclease activities of Mre11–Rad50 complex.
Targeting allostery expands inhibitor target sites from local active site pockets to include larger interfaces and conformational switches that are often unique to a given functional assembly. In contrast, an active site may have chemistry and shape similarities among multiple proteins. In an analogous approach to our loop movements, active site specificity can be achieved despite similar binding chemistry by directing allowed shifts in a binding pocket (Garcin et al., 2008), which was in fact the inspiration for our approach for MRE11. For the Mre11 nuclease activities, we were able to target adjacent loop and interface regions that open dsDNA ends and those that sculpt ssDNA to enter the active site. This type of dissection-of-function inhibitor can sometimes be mimicked by mutations, as seen for the Mre11 partner Rad50 (Deshpande et al., 2014; Williams et al., 2011). Notably, the Rad50 allosteric conformational change is further influenced by DNA binding and can span the entire molecule and extend into cross-molecular complexes to alter protein interactions, joining DNA ends across 1000 Å (Hopfner et al., 2002; Roset et al., 2014). However for Mre11 nuclease, we were unable to dissect the endo- and exonuclease functions with mutations. Furthermore, an active site inhibitor would logically most likely block both activities. All these observation support the value of the allosteric inhibitor approach as dissection-of-function tools for cell biology. Advanced imaging allowing time and localization measurements provides a means to measure changes in assembly and disassembly of MRE11 DNA repair complexes (Brosey, Ahmed, Lees-Miller, & Tainer, 2017) to test the protein interactions and conformations controlling rate-limiting steps that are often biologically regulated.
3.2. An Avatarian Approach
In the avatarian approach, the tremendous power of comparative genomics and structural similarity can be effectively applied to the design of inhibitors by targeting accessible structures that embody the features critical for control of the system by small molecules. We maintain that there is no compelling need to do structures of human complexes in order to develop and optimize inhibitors through structural analyses. There is of course an absolute need to test resulting inhibitors in human cells and in an appropriate biological system validating their activities for situations where they will be applied. We suggest that the best practical test of specificity is to empirically test if the inhibitor phenotype is removed by mutating the target protein in human cells. This general avatarian approach is valuable as it allows efficient progress in the absence of target human protein structures, and it leverages optimal systems and informative tools for structural biology. In an example supporting the methods and strategies in this report, we used the first archaeal full-length Rad51 structure and assembly to develop and test the BRCA2 assembly in human cells by redesigning the microbial Rad51 at the site of the human RAD51 interface for BRCA2. This work showed that the redesigned microbial Rad51 went to regions of damaged DNA and bound BRCA2 in human cells: the specific targeted binding was validated by its being competed for by high-level expression of BRCA2 BRC3/4 domains (Shin et al., 2003).
3.3. Inhibitor Tools as Biological Probes and Scaffolds for Drug Design
To examine the possible value of specific protein targets and before developing possible drugs, it is generally useful to have chemical tools to probe the biological importance of specific activities. Structures provide the means to see where inhibitors are binding and what they may do to the target protein structure. Combined computational and experimental X-ray crystallographic structures can efficiently and accurately identify small molecule-binding fragments important for inhibition and inform the design of new inhibitors based upon the observed ligand-bound structure (Blundell et al., 2006; Erlanson, Fesik, Hubbard, Jahnke, & Jhoti, 2016; Moiani et al., 2009; Murray & Blundell, 2010; Perry, Harris, Moiani, Olson, & Tainer, 2009; Thomas et al., 2017; Winter et al., 2012). Starting from small fragments bound to a protein, further optimization including merging adjacent fragments with a linker can lead to the generation of better inhibitors. In general, we choose to grow onto fragments for our projects rather than link them to avoid possible strain between binding pockets and linker design issues; nevertheless, linked approaches are also successful and may be optimal for a given target site (Patrone et al., 2016; Waterson et al., 2015).
We find that the experimental X-ray electron density maps for protein-inhibitor complexes will frequently aid comprehension of how functional moieties interact with specific residues of the target enzyme. These interactions may impact proteins’ conformational states, and as seen for Mre11 these changes may also perturb possible conformational changes to its substrates. A detailed understanding of the mechanism-of-action from combining structures and measured cell activities usually makes it possible to efficiently improve binding of identified fragments and also to make directed chemical libraries on its small molecular scaffold. Structure-based improvements can include inhibitor optimizations for therapeutic use and with better properties in terms of solubility, toxicity, and specificity. An emerging method will be to examine inhibitor binding by X-ray scattering in solution, which has become high throughput (Hura et al., 2009), and able to detect allosteric assembly changes (Rambo & Tainer, 2013a), changes in flexibility (Rambo & Tainer, 2013b), and relatively high-resolution changes in fold and compactness due to inhibitor binding (Rambo & Tainer, 2013b), as seen for abscisic acid-binding protein (Nishimura et al., 2009). Unlike other structural techniques, SAXS enables comprehensive assessment of conformation and assembly under near physiological conditions (Hura et al., 2013). So we can expect it to be increasingly used for ligand-binding assessments and their impact on assembly, as seen for the apoptosis-inducing factor that links mitochondrial metabolism, DNA damage responses, and programmed cell death (Brosey et al., 2016).
Whereas drug discovery involves many steps beyond the generation of a chemical inhibitor, an allosteric inhibitor with validated cellular activity provides a valuable tool for multiple scientific and biomedical applications. For example, the use of both classes of the MRE11 inhibitors showed that MRE11 endonuclease and exonuclease activities are both required for microhomology-mediated end joining, an error-prone pathway for DSB repair implicated in genomic rearrangement and oncogenic transformation that involves other repair enzymes including CtIP, PARP1, FEN1, plus DNA ligase I and III (Dutta et al., 2017). This MRE11 nuclease requirement was previously masked without the inhibitors.
By describing our strategies and approaches for the development of DNA repair inhibitors exemplified by Mre11, we hope that these methods may benefit the efficient discovery of chemical tools to knockdown- or knockout-specific activities to separate the complex contributions of multifunctional proteins and networks. We view the potential of applying structure-based inhibitor design to impact human disease and patient care as truly great. We therefore propose the avatarian allosteric approach as a possible enabling technology for the design of inhibitor probes and scaffolds for those human systems that are intractable to robust structure determinations and traditional active-site targeting.
ACKNOWLEDGMENTS
We thank the many researchers who have made collaborative and intellectual contributions to our studies of the MRE11 complex, especially Caroline Austin, Yunje Cho, David Ferguson, Jean Gautier, Martin Gellert, Karl-Peter Hopfner, Steve Jackson, Penny Jeggo, Maria Jasin, Cynthia McMurray, Sankar Mitra, Tanya Paull, John Petrini, Guy Poirier, Michael Resnick, Paul Russell, Katharina Schlacher, Lorraine Symington, John Turchi, Robert Scott Williams, Gareth Williams, Claire Wyman, and Miriam Zolan. John Tainer’s work on the Mre11–Rad50–Nbs1 complex is aided by Elizabeth D. Getzoff and Tammy Woo and supported in part by the National Institutes of Health (NIH) Grants NIH R01CA117638 and P01CA92584. Added support comes from the Robert A. Welch Distinguished Chair in Chemistry (JAT), the Cancer Prevention and Research Institute of Texas, and the University of Texas STARs program. SAXS at the Advanced Light Source SIBYLS beamline is supported in part by the US Department of Energy program Integrated Diffraction Analysis Technologies (IDAT). This work was also supported by research grants from the Canadian Institutes of Health Research to J.Y.M. (PJ/375408). J.Y.M. is a FRQS Chair in Genome stability. This work was supported in part by the Italian Ministero dell’Istruzione, dell’Università e della Ricerca (MIUR) to E.P. (PRIN 2015 project 2015LZE994).
REFERENCES
- Acharya S, Many A, Schroeder A, Kennedy F, Savytskyy O, Grubb J, et al. (2008). Coprinus cinereus rad50 mutants reveal an essential structural role for Rad50 in axial element and synaptonemal complex formation, homolog pairing and meiotic recombination. Genetics, 180(4), 1889–1907. 10.1534/genetics.108.092775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres SN, Appel CD, Westmoreland JW, Williams JS, Nguyen Y, Robertson PD, et al. (2015). Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nature Structural & Molecular Biology, 22(2), 158–166. 10.1038/nsmb.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacolla A, Tainer JA, Vasquez KM, & Cooper DN (2016). Translocation and deletion breakpoints in cancer genomes are associated with potential non-B DNA-forming sequences. Nucleic Acids Research, 44(12), 5673–5688. 10.1093/nar/gkw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennardo N, Cheng A, Huang N, Stark J, & Haber J (2008). Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genetics, 4(6), e1000110 10.1371/journal.pgen.1000110.g005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biehs R, Steinlage M, Barton O, Juhasz S, Kunzel J, Spies J, et al. (2017). DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Molecular Cell, 65(4), 671–684, e675. 10.1016/j.molcel.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Ehrlich SD, & Michel B (1997). Deletions at stalled replication forks occur by two different pathways. The EMBO Journal, 16(11), 3332–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell TL, Sibanda BL, Montalvao RW, Brewerton S, Chelliah V, Worth CL, et al. (2006). Structural biology and bioinformatics in drug design: Opportunities and challenges for target identification and lead discovery. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 361(1467), 413–423. 10.1098/rstb.2005.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosey CA, Ahmed Z, Lees-Miller SP, & Tainer JA (2017). What combined measurements from structures and imaging tell us about DNA damage responses. Methods in Enzymology, 592, 417–455. 10.1016/bs.mie.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosey CA, Ho C, Long WZ, Singh S, Burnett K, Hura GL, et al. (2016). Defining NADH-driven allostery regulating apoptosis-inducing factor. Structure, 24(12), 2067–2079. 10.1016/j.str.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, et al. (2008). Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell, 135(1), 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahwan C, Nakamura TM, Sivakumar S, Russell P, & Rhind N (2003). The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Molecular and Cellular Biology, 23(18), 6564–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DS, Mendes V, Thomas SE, McConnell BN, Matak-Vinkovic D, Coyne AG, et al. (2017). Fragment screening against the EthR-DNA interaction by native mass spectrometry. Angewandte Chemie (International Ed. in English), 56(26), 7488–7491. 10.1002/anie.201702888. [DOI] [PubMed] [Google Scholar]
- Classen S, Hura GL, Holton JM, Rambo RP, Rodic I, McGuire PJ, et al. (2013). Implementation and performance of SIBYLS: A dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the advanced light source. Journal of Applied Crystallography, 46(Pt. 1), 1–13. 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown KN, Savytskyy OP, Malik SB, Logsdon J, Williams RS, Tainer JA, et al. (2013). A mutation in the FHA domain of Coprinus cinereus Nbs1 leads to Spo11-independent meiotic recombination and chromosome segregation. G3 (Bethesda, Md), 3(11), 1927–1943. 10.1534/g3.113.007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhus B, Arvai AS, Rosnes I, Olsen OE, Backe PH, Alseth I, et al. (2009). Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair. Nature Structural & Molecular Biology, 16(2), 138–143. 10.1038/nsmb.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amours D, & Jackson SP (2002). The Mre11 complex: At the crossroads of dna repair and checkpoint signalling. Nature Reviews. Molecular Cell Biology, 3(5), 317–327. [DOI] [PubMed] [Google Scholar]
- Das D, Moiani D, Axelrod HL, Miller MD, Mcmullan D, Jin KK, et al. (2010). Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair. Journal of Molecular Biology, 397(3), 647–663. 10.1016/j-jmb.2010.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jager M, Dronkert ML, Modesti M, Beerens CE, Kanaar R, & van Gent DC (2001). DNA-binding and strand-annealing activities of human Mre11: Implications for its roles in DNA double-strand break repair pathways. Nucleic Acids Research, 29(6), 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande RA, Lee JH, Arora S, & Paull TT (2016). Nbs1 converts the human Mre11/Rad50 nuclease complex into an endo/exonuclease machine specific for protein-DNA adducts. Molecular Cell, 64(3), 593–606. 10.1016/j.molcel.2016.10.010. [DOI] [PubMed] [Google Scholar]
- Deshpande RA, Williams GJ, Limbo O, Williams RS, Kuhnlein J, Lee JH, et al. (2014). ATP-driven Rad50 conformations regulate DNA tethering, end resection, and ATM checkpoint signaling. The EMBO Journal, 33(5), 482–500. 10.1002/embj.201386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré A, Boyer-Chatenet L, Sattler R, Modi A, Lee J, Nicolette M, et al. (2008). A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nature Chemical Biology, 4(2), 119–125. 10.1038/nchembio.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Eckelmann B, Adhikari S, Ahmed KM, Sengupta S, Pandey A, et al. (2017). Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex. Nucleic Acids Research, 45(5), 2585–2599. 10.1093/nar/gkw1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht JR, Batuello CN, Lopez JT, Kim KK, & Turchi JJ (2011). Identification of Mre11 as a target for heat radiosensitization. Radiation Research, 176(3), 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, &Jhoti H (2016). Twenty years on: The impact of fragments on drug discovery. Nature Reviews. Drug Discovery, 15(9), 605–619. 10.1038/nrd.2016.109. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, & Jackson SP (2005). Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature, 434(7033), 605–611. [DOI] [PubMed] [Google Scholar]
- Garcin ED, Arvai AS, Rosenfeld RJ, Kroeger MD, Crane BR, Andersson G, et al. (2008). Anchored plasticity opens doors for selective inhibitor design in nitric oxide synthase. Nature Chemical Biology, 4(11), 700–707. 10.1038/nchembio.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner KM, Pletnev AA, & Eastman A (2009). Corrected structure of mirin, a small-molecule inhibitor of the Mre11-Rad50-Nbs1 complex. Nature Chemical Biology, 5(3), 129–130. author reply 130. 10.1038/nchembio0309-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith EJ (1996). Allosteric enzymes as models for chemomechanical energy transducing assemblies. The FASEB Journal, 10(7), 702–708. [DOI] [PubMed] [Google Scholar]
- Gradia SD, Ishida JP, Tsai MS, Jeans C, Tainer JA, & Fuss JO (2017). MacroBac: New technologies for robust and efficient large-scale production of recombinant multiprotein complexes. Methods in Enzymology, 592, 1–26. 10.1016/bs.mie.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, et al. (1998). MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nature Structural Biology, 5(12), 1058–1064. 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, et al. (2008). PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. The Journal of Biological Chemistry, 283(2), 1197–1208. 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- Hengel SR, Spies MA, & Spies M (2017). Small-molecule inhibitors targeting DNA repair and DNA repair deficiency in research and cancer therapy. Cell Chemical Biology, 24(9), 1101–1119. 10.1016/j.chembiol.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner K, Craig L, Moncalian G, Zinkel R, Usui T, Owen B, et al. (2002). The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature, 418(6897), 562–566. 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, & Tainer JA (2001). Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell, 105(4), 473–485. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, & Carney JP (2000). Mre11 and Rad50 from Pyrococcus furiosus: Cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. Journal of Bacteriology, 182(21), 6036–6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya N, & Miyagawa K (2014). Targeting DNA damage response in cancer therapy. Cancer Science, 105(4), 370–388. 10.1111/cas.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Budworth H, Dyer KN, Rambo RP, Hammel M, McMurray CT, et al. (2013). Comprehensive macromolecular conformations mapped by quantitative SAXS analyses. Nature Methods, 10(6), 453–454. 10.1038/nmeth.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo R, Ii FLP, Tsutakawa SE, et al. (2009). Robust high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nature Methods, 6(8), 606–612. 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MR, Logsdon D, & Fishel ML (2014). Targeting DNA repair pathways for cancer treatment: What’s new? Future Oncology, 10(7), 1215–1237. 10.2217/fon.14.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Grosbart M, Anand R, Wyman C, Cejka P, & Petrini JH (2017). The Mre11-Nbs1 interface is essential for viability and tumor suppression. Cell Reports, 18(2), 496–507. 10.1016/j.celrep.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukourakis MI (2012). Radiation damage and radioprotectants: New concepts in the era of molecular medicine. The British Journal of Radiology, 85(1012), 313–330. 10.1259/bjr/16386034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh BO, Llorente B, Lam A, & Symington LS (2005). Mutations inMre11 phosphoesterase motif I that impair Saccharomyces cerevisiae Mre11-Rad50-Xrs2 complex stability in addition to nuclease activity. Genetics, 171(4), 1561–1570. 10.1534/genetics.105.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S, Urata Y, & Fujiwara T (2012). Ataxia-telangiectasia mutated and the Mre11-Rad50-NBS1 complex: Promising targets for radiosensitization. Acta Medica Okayama, 66(2), 83–92. 10.18926/AMO/48258. [DOI] [PubMed] [Google Scholar]
- Lafrance-Vanasse J, Williams GJ, & Tainer JA (2015). Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Progress in Biophysics and Molecular Biology, 117(2–3), 182–193. 10.1016/j.pbiomolbio.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YT, Hura GL, Dyer KN, Tang HY, Tainer JA, & Yeates TO (2016). Designing and defining dynamic protein cage nanoassemblies in solution. Science Advances, 2(12). e1501855 10.1126/sciadv.1501855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche BJ, Orazio NI, & Weitzman MD (2010). The MRN complex in double-strand break repair and telomere maintenance. FEBS Letters, 584(17), 3682–3695. 10.1016/j.febslet.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens K, Bemeleit DJ, Möckel C, Clausing E, Schele A, Hartung S, et al. (2011). The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell, 145(1), 54–66. 10.1016/j.cell.2011.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KC, Padget K, Curtis H, Cowell IG, Moiani D, Sondka Z, et al. (2012). MRE11 facilitates the removal of human topoisomerase II complexes from genomic DNA. Biology Open, 1(9), 863–873. 10.1242/bio.20121834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, & Paull TT (2005). ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science, 308(5721), 551–554. [DOI] [PubMed] [Google Scholar]
- Lewis LK, Storici F, Van Komen S, Calero S, Sung P, & Resnick MA (2004). Role of the nuclease activity of Saccharomyces cerevisiae Mre11 in repair of DNA double-strand breaks in mitotic cells. Genetics, 166(4), 1701–1713. 10.1534/genetics.166.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Moiani D, Kertokalio A, Wyman C, Tainer JA, & Russell P (2012). Mre11 ATLD17/18 mutation retains Tel1/ATM activity but blocks DNA double-strand break repair. Nucleic Acids Research, 40(22), 11435–11449. 10.1093/nar/gks954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbo O, Porter-Goff ME, Rhind N, & Russell P (2011). Mre11 nuclease activity and Ctp1 regulate Chk1 activation by Rad3ATR and Tel1ATM checkpoint kinases at double-strand breaks. Molecular and Cellular Biology, 31(3), 573–583. 10.1128/MCB.00994-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T (1993). Instability and decay of the primary structure of DNA. Nature, 362(6422), 709–715. 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Majka J, Alford B, Ausio J, Finn RM, & McMurray CT (2012). ATP hydrolysis by RAD50 protein switches MRE11 enzyme from endonuclease to exonuclease. The Journal of Biological Chemistry, 287(4), 2328–2341. 10.1074/jbc.M111.307041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, et al. (2014). Catalytic and noncatalytic roles of the CtIP endonuclease in double-strandbreak end resection. Molecular Cell, 54(6), 1022–1033. 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson LA, Shen Y, & Ford JM (2014). Poly (ADP-ribose) polymerase inhibitor LT-626: Sensitivity correlates with MRE11 mutations and synergizes with platinums and irinotecan in colorectal cancer cells. Cancer Letters, 343(2), 217–223. 10.1016/j.canlet.2013.10.034. [DOI] [PubMed] [Google Scholar]
- Mendgen T, Steuer C, & Klein CD (2012). Privileged scaffolds or promiscuous binders: A comparative study on rhodanines and related heterocycles in medicinal chemistry. Journal of Medicinal Chemistry, 55(2), 743–753. 10.1021/jm201243p. [DOI] [PubMed] [Google Scholar]
- Mockel C, Lammens K, Schele A, & Hopfner KP (2012). ATP driven structural changes of the bacterial Mre11:Rad50 catalytic head complex. Nucleic Acids Research, 40(2), 914–927. 10.1093/nar/gkr749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiani D, Cavallotti C, Famulari A, & Schmuck C (2008). Oxoanion binding by guanidiniocarbonylpyrrole cations in water: A combined DFT and MD investigation. Chemistry, 14(17), 5207–5219. 10.1002/chem.200701745. [DOI] [PubMed] [Google Scholar]
- Moiani D, Salvalaglio M, Cavallotti C, Bujacz A, Redzynia I, Bujacz G, et al. (2009). Structural characterization of a protein A mimetic peptide dendrimer bound to human IgG. The Journal of Physical Chemistry. B, 113(50), 16268–16275. 10.1021/jp909405b. [DOI] [PubMed] [Google Scholar]
- Murray CW, & Blundell TL (2010). Structural biology in fragment-based drug design. Current Opinion in Structural Biology, 20(4), 497–507. 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Myler LR, Gallardo IF, Soniat MM, Deshpande RA, Gonzalez XB, Kim Y, et al. (2017). Single-molecule imaging reveals how Mre11-Rad50-Nbs1 initiates DNA break repair. Molecular Cell, 67(5), 891–898. e894. 10.1016/j.molcel.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J,Jevons SJ, Groselj B, Ellermann S, Konietzny R, Kerr M, et al. (2017). E3 ligase cIAP2 mediates downregulation of MRE11 and radiosensitization in response to HDAC inhibition in bladder cancer. Cancer Research, 77(11), 3027–3039. 10.1158/0008-5472.CAN-16-3232. [DOI] [PubMed] [Google Scholar]
- Nishimura N, Hitomi K, Arvai AS, Rambo RP, Hitomi C, Cutler SR, et al. (2009). Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science, 326(5958), 1373–1379. 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Chae J, Kim YC, & Cho Y (2011). Crystal structure of human Mre11: Understanding tumorigenic mutations. Structure, 19(11), 1591–1602. 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Patrone JD, Pelz NF, Bates BS, Souza-Fagundes EM, Vangamudi B, Camper DV, et al. (2016). Identification and optimization of anthranilic acid based inhibitors of replication protein A. ChemMedChem, 11(8), 893–899. 10.1002/cmdc.201500479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, & Gellert M (1998). The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Molecular Cell, 1(7), 969–979. [DOI] [PubMed] [Google Scholar]
- Perry JJ, Harris RM, Moiani D, Olson AJ, & Tainer JA (2009). p38alpha MAP kinase C-terminal domain binding pocket characterized by crystallographic and computational analyses. Journal of Molecular Biology, 391(1), 1–11. 10.1016/j.jmb.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahal EA, Henricksen LA, Li Y, Williams RS, Tainer J, &Dixon K (2010). ATM regulates Mre11-dependent DNA end-degradation and microhomology-mediated end joining. Cell Cycle, 9(14), 2866–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, & Tainer JA (2011). Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers, 95(8), 559–571. 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, & Tainer JA (2013a). Accurate assessment of mass, models and resolution by small-angle scattering. Nature, 496(7446), 477–481. 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, & Tainer JA (2013b). Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annual Review of Biophysics, 42, 415–441. 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- Rashid F, Harris PD, Zaher MS, Sobhy MA, Joudeh LI, Yan C, et al. (2017). Single-molecule FRET unveils induced-fit mechanism for substrate selectivity in flap endonuclease 1. eLife, 6 10.7554/eLife.21884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regal JA, Festerling TA, Buis JM, & Ferguson DO. (2013). Disease-associated MRE11 mutants impact ATM/ATR DNA damage signaling by distinct mechanisms. Human Molecular Genetics, 22(25), 5146–5159. 10.1093/hmg/ddt368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojowska A, Lammens K, Seifert FU, Direnberger C, Feldmann H, & Hopfner KP (2014). Structure of the Rad50 DNA double-strand break repair protein in complex with DNA. The EMBO Journal, 33(23), 2847–2859. 10.15252/embj.201488889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roques C, Coulombe Y, Delannoy M, Vignard J, Grossi S, Brodeur I, et al. (2009). MRE11-RAD50-NBS1 is a critical regulator ofFANCD2 stability and function during DNA double-strand break repair. The EMBO Journal, 28(16), 2400–2413. 10.1038/emboj.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roset R, Inagaki A, Hohl M, Brenet F, Lafrance-Vanasse J, Lange J, et al. (2014). The Rad50 hook domain regulates DNA damage signaling and tumorigenesis. Genes & Development, 28(5), 451–462. 10.1101/gad.236745.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Tomaszowski KH, Luzwick JW, Park S,Li J, Murphy M, et al. (2018). p53 suppresses mutagenic RAD52 and POLtheta pathways by orchestrating DNA replication restart homeostasis. Elife7. 10.7554/eLife.31723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. (2007). Human CtIP promotes DNA end resection. Nature, 450(7169), 509–514. 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller CB, Lammens K, Guerini I, Coordes B, Feldmann H, Schlauderer F, et al. (2012). Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling. Nature Structural & Molecular Biology, 19(7), 693–700. 10.1038/nsmb.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, & Jasin M (2011). Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell, 145(4), 529–542. 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Wu H, & Jasin M (2012). A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell, 22(1), 106–116. 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Hammel M, Tainer JA, & Sali A (2016). FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Research, 44(W1), W424–429. 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger. (2017). Small-molecule drug discovery suite 2017–2. [Google Scholar]
- Seeber A, Hegnauer AM, Hustedt N, Deshpande I, Poli J, Eglinger J, et al. (2016). RPA mediates recruitment of MRX to forks and double-strand breaks to hold sister chromatids together. Molecular Cell, 64(5), 951–966. 10.1016/j.molcel.2016.10.032. [DOI] [PubMed] [Google Scholar]
- Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM, et al. (2014). DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Molecular Cell, 53(1), 7–18. 10.1016/j.molcel.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, Pellegrini L, Daniels DS, Yelent B, Craig L, Bates D, et al. (2003). Full-length archaeal Rad51 structure and mutants: Mechanisms for RAD51 assembly and control by BRCA2. The EMBO Journal, 22(17), 4566–4576. 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B, Nguyen HD, Hakansson P, Marechal A, Tse A, Tahara H, et al. (2013). Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Reports, 3(5), 1651–1662. 10.1016/j.celrep.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, & Tainer JA (1996). A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature, 384(6604), 87–92. 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- Stracker TH, &Petrini JH (2011). The MRE11 complex: Starting from the ends. Nature Reviews Molecular Cell Biology, 12(2), 90–103. 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Fang Y, Yin J, Chen J, Ju Z, Zhang D, et al. (2017). Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Science Translational Medicine, 9(392), 1–18. 10.1126/scitranslmed.aal5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer MM, Ahern H, Xing D, Cunningham RP, & Tainer JA (1995). Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. The EMBO Journal, 14(16), 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas SE, Mendes V, Kim SY, Malhotra S, Ochoa-Montano B, Blaszczyk M, et al. (2017). Structural biology and the design of new therapeutics: From HIV and cancer to mycobacterial infections: A paper dedicated to John Kendrew. Journal of Molecular Biology, 429(17), 2677–2693. 10.1016/j.jmb.2017.06.014. [DOI] [PubMed] [Google Scholar]
- Thompson R, Montano R, & Eastman A (2012). The Mre11 nuclease is critical for the sensitivity of cells to Chk1 inhibition. PLoS One, 7(8). e44021. 10.1371/journal.pone.0044021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Lafrance-Vanasse J, & Tainer JA (2014). The cutting edges in DNA repair, licensing, and fidelity: DNA and RNA repair nucleases sculpt DNA to measure twice, cut once. DNA Repair (Amst), 19, 95–107. 10.1016/j.dnarep.2014.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Shin DS, Mol CD, Izumi T, Arvai AS, Mantha AK, et al. (2013). Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. The Journal of Biological Chemistry, 288(12), 8445–8455. 10.1074/jbc.M112.422774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Thompson MJ, Arvai AS, Neil AJ, Shaw SJ, Algasaier SI, et al. (2017). Phosphate steering by flap endonuclease 1 promotes 5′-flap specificity and incision to prevent genome instability. Nature Communications, 8, 15855 10.1038/ncomms15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velic D, Couturier AM, Ferreira MT, Rodrigue A, Poirier GG, Fleury F, et al. (2015). DNA damage signalling and repair inhibitors: The long-sought-after Achilles’ heel of cancer. Biomolecules, 5(4), 3204–3259. 10.3390/biom5043204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar E, Bartnik CM, Stenzel SL, Raskin L, Ahn J, Moreno V, et al. (2011). MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Research, 71(7), 2632–2642. 10.1158/0008-5472.CAN-10-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Pulk A, Wasserman MR, Feldman MB, Altman RB, Cate JH, et al. (2012). Allosteric control of the ribosome by small-molecule antibiotics. Nature Structural & Molecular Biology, 19(9), 957–963. 10.1038/nsmb.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterson AG, Kennedy JP, Patrone JD, Pelz NF, Feldkamp MD, Frank AO, et al. (2015). Diphenylpyrazoles as replication protein a inhibitors. ACS Medicinal Chemistry Letters, 6(2), 140–145. 10.1021/ml5003629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, et al. (2009). Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell, 139(1), 87–99. 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GJ, Lees-Miller SP, & Tainer JA (2010). Mre11-Rad50-Nbs1 conformations and the control of sensing, signaling, and effector responses at DNA double-strand breaks. DNA Repair, 9(12), 1299–1306. 10.1016/j.dnarep.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS, et al. (2008). Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell, 135(1), 97–109. 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RS, & Tainer J (2007). Learning our ABCs: Rad50 directs MRN repair functions via adenylate kinase activity from the conserved ATP binding cassette. Molecular Cell, 25(6), 789–791. 10.1016/j.molcel.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Williams J, Williams RS, Dovey CL, Guenther G, Tainer J, & Russell P (2010). gammaH2A binds Brc1 to maintain genome integrity during S-phase. The EMBO Journal, 29(6), 1136–1148. 10.1038/emboj.2009.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R, Williams J, & Tainer J (2007). Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochemistry and Cell Biology, 85(4), 509–520. 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Williams GJ, Williams RS, Williams JS, Moncalian G, Arvai AS, Limbo O, et al. (2011). ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nature Structural & Molecular Biology, 18(4), 423–431. 10.1038/nsmb.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter A, Higueruelo AP, Marsh M, Sigurdardottir A, Pitt WR, & Blundell TL (2012). Biophysical and computational fragment-based approaches to targeting protein-protein interactions: Applications in structure-guided drug discovery. Quarterly Reviews of Biophysics, 45(4), 383–426. 10.1017/S0033583512000108. [DOI] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, & Russell P (2005). ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Molecular and Cellular Biology, 25(13), 5363–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Vogel G, Coulombe Y, Dubeau D, Spehalski E, Hebert J, et al. (2012). The MRE11 GAR motif regulates DNA double-strand break processing and ATR activation. Cell Research, 22(2), 305–320. 10.1038/cr.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]