We present a low-cost kit based on freeze-dried, cell-free reactions to illustrate synthetic and molecular biology concepts.
Abstract
Hands-on demonstrations greatly enhance the teaching of science, technology, engineering, and mathematics (STEM) concepts and foster engagement and exploration in the sciences. While numerous chemistry and physics classroom demonstrations exist, few biology demonstrations are practical and accessible due to the challenges and concerns of growing living cells in classrooms. We introduce BioBits™ Explorer, a synthetic biology educational kit based on shelf-stable, freeze-dried, cell-free (FD-CF) reactions, which are activated by simply adding water. The FD-CF reactions engage the senses of sight, smell, and touch with outputs that produce fluorescence, fragrances, and hydrogels, respectively. We introduce components that can teach tunable protein expression, enzymatic reactions, biomaterial formation, and biosensors using RNA switches, some of which represent original FD-CF outputs that expand the toolbox of cell-free synthetic biology. The BioBits™ Explorer kit enables hands-on demonstrations of cutting-edge science that are inexpensive and easy to use, circumventing many current barriers for implementing exploratory biology experiments in classrooms.
INTRODUCTION
Many of us can trace our initial fondness for the sciences to formative experiences with hands-on exploratory kits, such as traditional chemistry sets. This trend has expanded today to include a spectrum of educational kits that teach subjects such as physics, electronics, programming, or robotics (1–3). However, there are few successful and engaging systems for teaching advanced molecular or synthetic biology concepts in a hands-on manner (4, 5). This absence is largely due to the particularities of traditional biology experimentation, which requires a cold chain to prevent the biological components from spoiling, sterile equipment and media to prevent contamination, specialized instruments such as shaking incubators, and concerns with the biocontainment of recombinant microorganisms. Here, we present the development of a synthetic biology platform that circumvents all of these challenges, resulting in a shelf-stable and affordable educational kit for demonstrating advanced biological concepts.
Synthetic biology is a rapidly advancing field that uses engineering concepts to harness the power and diversity of biology. At the foundation of this endeavor is the ability to control gene expression in a predictable manner, which is accomplished by using modular biological components to control and fine tune the processes of transcription and translation (6, 7). The resulting synthetic biology toolbox enables powerful new methods for chemical and drug manufacturing (6, 8), clinical diagnostics (9, 10), and cell therapies (11, 12). Synthetic biology kits also have great potential as educational tools to teach molecular and synthetic biology concepts but are generally too expensive to implement in classrooms due to the numerous infrastructure requirements of these types of experiments.
To create an array of biology demonstrations that could be used in any classroom setting, we turned to cell-free synthetic biology. Cell-free systems use essential cellular machinery, including polymerases, ribosomes, and transcription factors, in an in vitro setting to carry out the processes of transcription and translation, which circumvents the need for specialized, sterile equipment and media to culture living cells; moreover, the lack of living cells eliminates concerns of biocontainment. There are two general types of cell-free systems: crude extracts, where the required cellular components are harvested from bacterial lysate (13), and reconstituted systems, such as the commercial protein synthesis using recombinant elements (PURE) system (14), where each individual component is produced recombinantly and then recombined in vitro. Both systems require supplementation with additional essential components such as nucleotides, amino acids, and energy equivalents. Cell-free systems have been used extensively to produce proteins and other biomolecules, as well as build and execute synthetic biology circuits (9, 10, 15–17).
We have shown that cell-free systems can be freeze-dried along with genetic elements to form pellets that are stable at room temperature and are highly portable (15). The shelf-stable nature of these freeze-dried, cell-free (FD-CF) pellets eliminates the need for dedicated refrigerators or freezers. In addition, FD-CF reactions do not require any specialized equipment, making them a robust technology for using synthetic biology in low-resource environments, including classrooms. Reactivation of the FD-CF components simply requires the end user to add water. We have previously used this technology for the rapid development of inexpensive, paper-based nucleic acid diagnostics and as a portable biomanufacturing platform (10, 15, 16). With the unique practicality of FD-CF technology, we also considered this platform to be highly suitable for applications in biology education, where there is a glaring lack of hands-on biology experiments (18). Specifically, FD-CF reactions are an ideal way to bring the ever-increasing toolbox of the synthetic biology community to secondary schools and the general public (Fig. 1A). Previously, this has required substantial investment in laboratory equipment and infrastructure, resulting in the lack of formative STEM experiences in poorly funded schools (19). We believe that the innovative approach we present here and in Stark et al. (20) will have a significant impact on lowering the barriers to explore advanced synthetic biology concepts and reduce inequalities in public science education.
Fig. 1. BioBits™ kits: Freeze-dried educational kits.
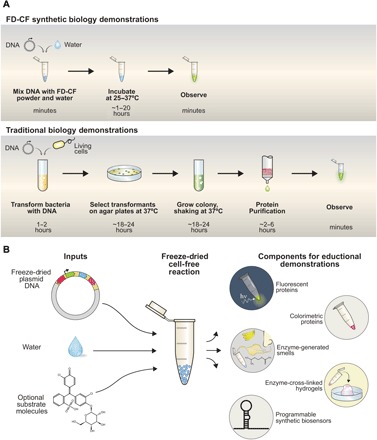
(A) FD-CF demonstrations require only the addition of water to the supplied reactions and incubation for 1 to 20 hours at 25° to 37°C for observation and analysis by students. In contrast, traditional biology experiments require substantial time, resources, and specialized equipment. (B) With the DNA template and any substrate molecules provided with the FD-CF reaction, the students just have to add water to run a number of bioscience activities and demonstrations.
Here, we introduce BioBits™ Explorer, a low-cost modular educational kit that uses FD-CF technology to teach synthetic biology concepts through sensory engagement and provides opportunities for inquiry-based learning. We have developed a set of demonstrations designed to engage three of the five senses—sight, smell, and touch (Fig. 1B)—through the expression of proteins in FD-CF reactions that produce fluorescence, enzyme-generated fragrances, and large-scale hydrogels, respectively (fig. S1 and table S1). This was made possible through the development of functionally robust synthetic cell-free programs—several of which are original. We discuss here how these outputs can be used to create activities to teach the fundamentals of protein expression, enzyme catalysis, and properties of biomaterials. In addition, we incorporate modular biosensing components that can be used to control gene expression—specifically, RNA toehold switches—to develop a demonstration that allows students to discriminate between species of different fruits using extracted DNA (table S1). These activities can be run on their own or in sequence with additional laboratory activities that we developed using fluorescent protein outputs, which we pair with low-cost, portable laboratory equipment and supporting curriculum in a kit we call BioBits™ Bright (see the companion article). Together, the BioBits™ kits demonstrate both the breadth of synthetic biology activities that can be developed with FD-CF technology and how these platforms can increase student involvement, illustrate core concepts in molecular and synthetic biology, and provide opportunities for independent, student-directed research projects (for example, synthetic biology after school clubs and science fair research teams) in the life sciences.
RESULTS
Fluorescent proteins as visual outputs
Our first goal was to develop a set of outputs that would engage as many of the five senses as possible to pique students’ interest in the activities. As a visual output, we used FD-CF crude extract reactions to express fluorescent proteins. We selected a set of five fluorescent proteins (21–27) that cover a spectrum of colors, a subset from the BioBits™ Bright kit composed of red (eforRed), orange (dTomato), yellow (mOrange), green (sfGFP), and cyan (Aquamarine). FD-CF pellets, including DNA templates encoding the five proteins, were rehydrated and incubated overnight (20 hours) at 30°C. The fluorescent proteins expressed robustly and were easily visible to the eye even without fluorescent excitation. The fluorescent colors were also vivid when viewed using a custom low-cost, portable fluorescence illuminator we developed (first described in our companion article) (Fig. 2A and fig. S2).
Fig. 2. Fluorescent proteins as visual outputs.
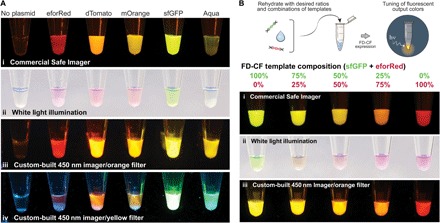
(A) A set of fluorescent proteins were expressed by FD-CF expression in crude extract and visualized with (i) a laboratory transilluminator (Safe Imager at 470-nm excitation), (ii) white light epi-illumination, (iii) a portable, inexpensive (<US$15) 450-nm classroom illuminator with an orange acrylic filter, or (iv) a yellow acrylic filter. (B) sfGFP and eforRed fluorescent proteins were expressed at a range of different combinations (by ratio of template DNA added) in FD-CF crude extract and visualized with (i) the Safe Imager, (ii) white light, and (iii) the classroom illuminator with the orange acrylic filter to demonstrate tunable protein expression.
As an example of how these fluorescent outputs can be used to teach advanced biological concepts, we created a demonstration designed to convey the concept of tuning gene expression, a key aspect of synthetic biology. This activity builds on an activity in the BioBits™ Bright kit, where protein expression was tuned by varying the input DNA concentration. Here, we used FD-CF crude extract reactions to coexpress two different fluorescent proteins, sfGFP and eforRed, simultaneously in a single reaction. The FD-CF pellets, containing different ratios of each DNA template, were rehydrated with water to achieve a range of intermediate colors from green to red that are visible to the eye under both white light and fluorescence (Fig. 2B and fig. S3). This coexpression, which, to our knowledge, has not been carried out before in this FD-CF format, can be replicated with DNA templates for any other fluorescent protein pairs in the kit, providing students the freedom to choose the combination of visual outputs they would like to engineer.
This activity also provides the opportunity to teach students the concept of the design-build-test cycle, a common paradigm used by synthetic biologists when developing new genetic circuits (28). Once students choose the visual output they would like to engineer, they can design an experiment to mix fluorescent proteins in different ratios to achieve their goal. In this example, the build step would involve obtaining FD-CF pellets with the appropriate DNA concentrations. Students would then test their experimental design, evaluate the results, and iterate the process, as desired. The application of these fluorescent modules and educational demonstrations, paired with the inexpensive fluorescent imager that we developed, provides simple and cost-effective alternatives to traditional biology experiments, which are too expensive and complex to implement in an average classroom.
Fragrance-generating enzymes as olfactory outputs
Next, we sought to develop a synthetic biology circuit that would engage students’ sense of smell. To achieve this, we expressed a single enzyme, alcohol acetyltransferase (ATF1), in FD-CF PURE and crude reactions overnight (20 hours) at 37°C. ATF1 is a key enzyme in aroma biochemistry (29) that converts isoamyl alcohol to isoamyl acetate, which imparts a strong banana fragrance (Fig. 3A). We then mixed FD-CF reactions expressing ATF1 at a 1:10 dilution into a buffered reaction containing 25 mM isoamyl alcohol and 5 mM acetyl–coenzyme A(CoA). We allowed the enzymatic reactions to proceed 20 hours at room temperature, after which we were able to detect strong banana scents by smell.
Fig. 3. Fragrance-generating enzymes as olfactory outputs.
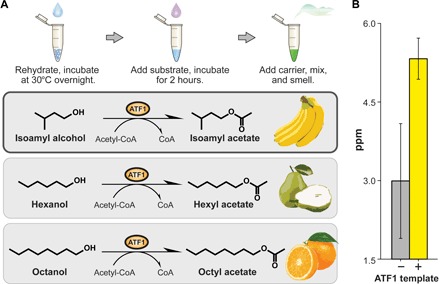
(A) Using FD-CF reactions, we manufactured enzymes that can generate various smells from the Saccharomyces cerevisiae acetyltransferase ATF1. (B) Production of fragrance molecules after substrate addition to overnight FD-CF reactions of ATF1, as detected by headspace GC-MS. Values represent averages, and error bars represent SDs of n = 3 biological replicates.
To quantify the production of the volatile product, we set up identical FD-CF PURE reactions during which we collected the vapor phase of the reaction and analyzed it by gas chromatography–mass spectrometry (GC-MS). GC-MS analysis confirmed the presence of isoamyl acetate at ~5.3 parts per million (ppm; volatile phase), which is well within the reported odor detection threshold for this compound (0.00075 to 366 ppm) but well below the permissible exposure limit of 100 ppm (Fig. 3B) (30). Thus, an average student can readily detect this FD-CF–generated aromatic in a classroom. ATF1 can convert various long-chain alcohol substrates to the corresponding acetylated esters, which have different fragrances (31). Incubation of ATF1-expressing FD-CF reactions with the substrates hexanol and octanol, for example, could generate volatile products that encompass pear and citrus smells, respectively (Fig. 3A).
To our knowledge, simple cell-free reactions that generate volatile fragrance molecules readily detectable by humans from an overnight incubation have not been previously developed. Production of these olfactory outputs can teach students about basic enzymatic reactions, provide a great connection to lessons learned in their chemistry classes, and inspire potential research projects for more advanced student groups. For example, here, we set up the enzymatic reactions containing isoamyl alcohol with and without FD-CF–produced ATF1 to show that the enzyme must be present to generate a smell. ATF1 could also be mixed with nonreactive substrates with different chemical functional groups to demonstrate that the enzyme only catalyzes a specific reaction. Moreover, these experiments can be put into a real-world context by noting that there are synthetic biology companies that work with enzymes in engineered microbes to produce fragrances and other commodity chemicals (32, 33).
Hydrogel-generating enzymes as tactile outputs
Next, we sought to create a product using FD-CF reactions that students could interact with in a tactile manner. To do so, we drew inspiration from engineered hydrogel materials that have been developed for biomedical and biotechnological applications (34, 35). Like the olfactory outputs, hydrogels can be produced by enzymatic reactions (Fig. 4A). Sortase is an enzyme that recognizes and covalently links specific peptide sequences (GGG and LPRT) through a transpeptidation reaction (36). We expressed sortase in FD-CF crude and PURE reactions overnight at 37°C and used it to cross-link a solution of eight-arm polyethylene glycol (PEG) molecules modified with GGG or LPRT peptides (8%, w/v). We observed hydrogel formation within 30 min of incubation at 37°C (Fig. 4, B and C, and fig. S4). In contrast, PEG solutions incubated with FD-CF reactions that contained no template DNA did not exhibit a phase change and remained in liquid form.
Fig. 4. Hydrogel-generating enzymes as tactile outputs.
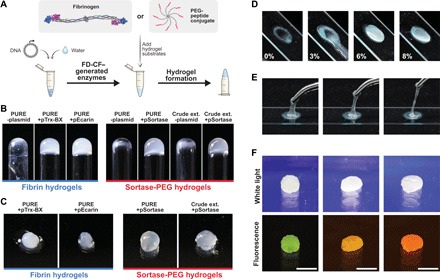
(A) Schematic of fibrin hydrogels created from FD-CF–generated batroxobin/ecarin proteases that activate fibrinogen by cleavage or PEG-peptide hydrogels cross-linked by FD-CF–generated sortase enzymes that induce cross-linking by transpeptidase activity. (B) Inverted glass tubes to demonstrate formation of hydrogels. (C) Close-up images of the formed hydrogels that can be manipulated by hand. (D) Tuning the mechanical properties of the hydrogel by varying the % PEG to create a range of materials with varying viscosities. (E) An 8% crude FD-CF PEG hydrogel is highly elastic. (F) Casting the hydrogels into shapes using molds and mixing with crude FD-CF fluorescent protein reactions to obtain shaped fluorescent hydrogels. Scale bar, 1 cm.
In addition to the sortase-catalyzed hydrogel, we developed a method to use FD-CF PURE reactions to produce fibrin-based hydrogels. Fibrinogen is a glycoprotein found in blood that, when enzymatically converted to fibrin, leads to the formation of a blood clot (37). Outside the context of blood, the proteases ecarin and batroxobin have been shown to cleave fibrinogen, which leads to the self-assembly of fibrin molecules into a hydrogel (38, 39). FD-CF reactions were used to produce ecarin and batroxobin, which were added to resolubilized fibrinogen. We observed fibrin-based hydrogels after overnight incubation at room temperature, while the enzyme-free reaction remained unpolymerized (Fig. 4, B and C, and fig. S4).
The sortase and ecarin-mediated hydrogel formation are the first demonstrations, to our knowledge, of an engineered cell-free protein synthesis reaction that generates self-assembling macromolecular hydrogels from genetically encoded components. Once formed, one can manipulate the hydrogels by hand, allowing students to experience another enzyme-catalyzed biochemical reaction, this time resulting in an output that they can feel. These basic demonstrations can be used to introduce advanced biological concepts such as blood clotting or how cells use similar processes to form human tissue. As a follow-up project, students can experiment with the notion of engineering gene expression to create biomaterials with tunable properties, a current aim of synthetic biology research (40). Specifically, the mechanical properties of the hydrogel can be tuned by varying the concentration of the substrates to create a range of materials from a viscous slime to a stiff hydrogel (Fig. 4, D and E). The hydrogels can also be cast into shapes using molds and/or combined with the fluorescent protein outputs to create fluorescent hydrogels (Fig. 4F).
Probing the environment using designer biosensors
While the first three components provide students with an FD-CF toolkit of sensory outputs from simple DNA inputs, we wanted to expand the BioBits™ Explorer kit to inspire a proactive, inquisitive mindset in students and provide them with the means to interrogate the world around them. In our final demonstration, we develop FD-CF–based tools that allow students to probe real-world biological samples using toehold switch sensors. Here, we expand on the common classroom activity of isolating DNA from fruits to allow students to detect DNA signatures of a specific fruit and couple it to a fluorescent output.
Toehold switch sensors are programmable synthetic riboregulators that allow protein expression only when a specific trigger RNA is present (10, 41). These sensors consist of an mRNA molecule designed to include a hairpin structure that blocks gene translation in cis by sequestration of the ribosome binding site and start codon. Hybridization to a complementary trigger RNA results in secondary structure rearrangement, facilitating ribosomal translation of an output gene (Fig. 5A). This technology allows for regulatory control of the various sensory outputs described previously (fluorescence, fragrance, and hydrogels) to be conditionally dependent on the presence of a specific nucleic acid molecule.
Fig. 5. Toehold-based environmental sensing demonstrations.
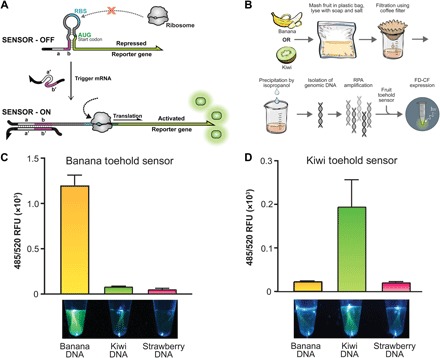
(A) Schematic of a toehold switch sensor. Upon the presence of a trigger RNA, strand invasion melts the secondary structure, allowing ribosomal translation to occur. (B) Schematic of activity that allows extracted DNA from banana or kiwi fruit to be processed and detected by a toehold switch sensor in FD-CF. (C) The banana toehold switch sensor or (D) the kiwi toehold switch sensor produces a clear fluorescence output (sfGFP) when exposed to extracted and amplified DNA of the relevant fruit but not when exposed to DNA sequences from other fruits. Images shown are from a custom-built 450-nm handheld imager with a yellow acrylic filter and quantified by a plate reader at 485-nm excitation and 520-nm emission. Values represent averages, and error bars represent SDs of n = 3 biological replicates.
For this demonstration, we designed the first toehold switch sensors that are able to discriminate between plant species. These toehold sensors are activated by sequences from either the banana genome (rbcL gene from Musa acuminata or Musa balbisiana) or the kiwi genome (5.8S ribosomal RNA gene from Actinidia deliciosa) and produce sfGFP as an output (table S1) (42, 43). Since the toehold switch sensors recognize RNA and not DNA, we included a recombinase polymerase amplification (RPA) (44) step to generate short DNA amplicons from fruit genomic DNA that incorporate a T7 promoter for transcription of RNA triggers in FD-CF reactions (fig. S5). The RPA reaction components can also be freeze-dried, conforming to the shelf-stable aspect of these kits. To demonstrate the specificity of the toehold switch sensors, we isolated DNA from banana, kiwi, or strawberry (Fig. 5B and fig. S6), using a simple procedure often performed in classrooms that utilizes common inexpensive supplies such as dish soap, table salt, rubbing alcohol, and coffee filters (45). Diluted DNA from each of the fruits was used to rehydrate RPA reactions and incubated overnight at 37°C. Completed RPA reactions were further diluted 1:4 in water and used to rehydrate FD-CF PURE reaction pellets containing the banana or kiwi toehold switch sensor. Upon overnight incubation at 37°C, activation of each toehold switch sensor was only observed from the reaction containing the specific fruit DNA trigger and not from the reactions containing other fruit DNA (Fig. 5, C and D).
The biosensor module described here uses the detection capability of toehold switches to provide a hands-on introduction to the concept of nucleic acid–based diagnostics (9, 10). This module could be expanded readily to include additional toehold switch sensors designed to detect other fruits and vegetables. Sensors could be designed similarly to identify specific animal species (for example, cats and dogs), enabling students to test DNA from their pets.
DISCUSSION
The next-generation synthetic biology educational kit described here addresses the need for easy-to-implement, hands-on biology demonstrations in STEM education. We used shelf-stable FD-CF reactions to bring molecular and synthetic biology experiments into a classroom setting in an affordable manner, without the need for specialized equipment or refrigeration. First, we developed a set of genetically encoded outputs that engages the senses of sight, smell, and touch via fluorescent proteins, enzyme-generated scents, and enzyme-generated hydrogels, respectively. These outputs provide demonstrations that can be used to teach fundamental biology concepts and principles of tunable protein expression, enzyme catalysis, and material properties. Although we focus on the modules as educational demonstrations, it should be noted that two of the engineered freeze-dried synthetic biology circuits presented here, genetically encoded macroscopic hydrogellation and olfactory detection, represent the first demonstration of tactile or fragrance outputs in cell-free systems. These modules expand the repertoire of sensory outputs available for cell-free biosensors beyond visual fluorescent outputs. For the BioBits™ Explorer kit, we also created a module that allows students to take DNA extracted from fruits and analyze the samples using toehold switch sensors designed to detect specific DNA sequences from the banana and kiwi genome. The isolation of DNA from fruits is a widely used classroom activity due to its simple protocol and ease of implementation (45). For the first time, using these toehold switches, students can actually go further and probe the DNA on the genetic level. Beyond educational demonstrations, biosensors for plant tissue discrimination such as those described here could also be used practically in agriculture to detect contamination in food crops. These toehold sensors also open up the possibility of students being able to design their own custom biosensors to probe living organisms and couple that to a wide variety of outputs.
The demonstrations presented for these toolkits were designed to be modular: Teachers can incorporate these explorations into their curricula as they see fit, depending on the content they wish to teach and classroom time available. We thus envision BioBits™ Explorer being further developed into a diverse array of kits to accommodate different grade levels and budgets, although the modules presented here are all affordable. The basic Explorer kit could include simple demonstrations involving the three sensory outputs. The kit would also include the low-cost incubator and portable fluorescent imager described in the companion article (20). Each FD-CF reaction costs approximately US$0.15. Thus, reagents and other supplies for a 30-student classroom for the basic kit would only cost about US$200 to produce (table S2)—much less than the cost of materials and traditional equipment (shaking incubators, refrigerators, thermocyclers, etc.) needed for in vivo biological experiments. More advanced Explorer kits could also include activities such as the toehold biosensors; the additional reagents would only add on a cost of about US$200 (table S2). We anticipate that costs associated with these kits would be further reduced as manufacturing methods are optimized and economies of scale are leveraged.
We note that some of the FD-CF reactions were carried out with the more expensive PURE system when expression was low in the crude extracts. The fruit DNA biosensors, for example, were implemented in the PURE system because of inherent autofluorescence from the crude extract that hindered visualization. In the future, the toehold switches could be optimized to increase their output expression, which would allow the use of the inexpensive crude extract. In addition, crude extracts could be optimized to improve the expression of specific enzymes by using different bacterial strains. Recent improvements to the efficiency of the reconstituted PURE production system also suggest that its cost could be reduced to that of the crude system (46, 47). These optimizations would reduce the cost of the BioBits™ kits even further.
We are currently developing a companion website to facilitate the formation of an open-source community around the BioBits™ kits. This online community would provide users with a forum that would facilitate discussion and development of new ideas for lessons and demonstrations using the provided components. We also plan to add a software component that would allow students to design their own sensors (specific to other environmental samples) and other synthetic biology components and request the DNA for them online. In this way, the open-source community could design, build, and test additional genetic constructs to add to the BioBits™ parts library.
The activities demonstrated here engage students by appealing to their senses through diverse genetic outputs using simple just-add-water FD-CF pellets. These illustrative demonstrations can be used to introduce a wide range of molecular and synthetic biology concepts in classrooms. Our kits contain just a few examples of the potential activities that can be developed using FD-CF reactions; by mining the available library of synthetic biology parts and developing novel genetic circuit combinations, a plethora of additional modules could be created to teach advanced biology concepts. Together, our BioBits™ Bright and Explorer kits provide a new paradigm for bringing affordable life sciences and biotechnology experiments into any classroom, making quality biology education accessible to all students.
MATERIALS AND METHODS
General template design and preparation
DNA sequences encoding eforRed, dTomato, mOrange, ATF1, Ecarin, and Trx-Bx (batroxobin fused with thioredoxin as a solubility domain) genes were derived from the literature, codon-optimized for Escherichia coli, and synthesized as gBlocks or oligonucleotides by Integrated DNA Technologies. pPROEX-Aquamarine was a gift from F. Merola (plasmid #42889, Addgene), and pET29-sortaseA-penta-mutant was a gift from L. Griffith. We previously reported the pJL1-sfGFP plasmid (plasmid #69496, Addgene). Cloning and plasmid propagation were performed using either Mach1 (C862003, Thermo Fisher Scientific) or NEB Turbo (C2984H, New England Biolabs) competent E. coli cells. All templates were cloned into a T7 expression plasmid system—the PURExpress control vector from New England Biolabs, called pNP1 in the text, pJL1, or pCOLADuet-1 (71406-3, Novagen)—using Gibson assembly (48). All template plasmid DNA preps of the plasmids were performed with the E.Z.N.A. Plasmid Midi Kit (#D6904, Omega Bio-Tek) for crude extract reactions or the QIAprep Spin Miniprep Kit (#27106, Qiagen) for PURE reactions. All sequences are available on Addgene (table S1).
PURE cell-free reaction preparation and lyophilization protocol
For cell-free reactions performed in the PURExpress In Vitro Protein Synthesis Kit (E6800S, New England Biolabs), the reactions consisted of the following: NEB Solution A (40%) and B (30%), ribonuclease inhibitor (0.5%; 03335402001, Roche), and the template DNA (10 to 50 nM). For the FD-CF expression of ATF1, the Disulfide Bond Enhancer (E6820S, New England Biolabs) was added into the reactions, as per the manufacturer’s instructions, before lyophilization. The reactions were then flash-frozen in liquid nitrogen, lyophilized overnight to obtain the freeze-dried reaction, and stored at room temperature. The reactions were reconstituted with nuclease-free water to the original reaction volume and incubated at 30° or 37°C.
In-house crude cell-free extract preparation and lyophilization protocol
Cell extract was prepared as described previously (13). Briefly, E. coli BL21 Star (DE3) cells (Thermo Fisher Scientific) or a BL21 variant called RARE (49) was grown in 150 ml of LB at 37°C at 250 rpm. Cells were harvested in mid-exponential growth phase [OD600 (optical density at 600 nm) = ~2 to 3], and cell pellets were washed three times with ice-cold Buffer A containing 10 mM tris-acetate (pH 8.2), 14 mM magnesium acetate, 60 mM potassium glutamate, and 2 mM dithiothreitol, flash-frozen, and stored at −80°C. Briefly, cell pellets were thawed and resuspended in 1 ml of Buffer A per 1 g of wet cells and sonicated in an ice water bath. Total sonication energy to lyse cells was determined by using the sonication energy equation for BL21-Star (DE3) cells, [Energy] = [[volume (μl)] − 33.6]*1.8−1. A Q125 Sonicator (Qsonica) with 3.174-mm-diameter probe at a frequency of 20 kHz was used for sonication. An amplitude of 50% in 10-s on/off intervals was applied until the required input energy was met. Lysate was then centrifuged at 12,000 relative centrifugal force (rcf) for 10 min at 4°C, and the supernatant was incubated at 37°C at 300 rpm for 1 hour. The supernatant was centrifuged again at 12,000 rcf for 10 min at 4°C, flash-frozen, and stored at −80°C until use. The reaction mixture consists of the following components: 1.2 mM adenosine 5′-triphosphate; 0.85 mM each of guanosine-5′-triphosphate, uridine 5′-triphosphate, and cytidine 5′-triphosphate; l-5-formyl-5,6,7,8-tetrahydrofolic acid (34.0 μg ml−1; folinic acid); E. coli transfer RNA mixture (170.0 μg ml−1); 130 mM potassium glutamate; 10 mM ammonium glutamate; 12 mM magnesium glutamate; 2 mM each of 20 amino acids; 0.33 mM nicotinamide adenine dinucleotide; 0.27 mM CoA; 1.5 mM spermidine; 1 mM putrescine; 4 mM sodium oxalate; 33 mM phosphoenolpyruvate; plasmid (13.3 μg ml−1); T7 RNA polymerase (100 μg ml−1); and 27% (v/v) of cell extract (50, 51). The reactions were then flash-frozen in liquid nitrogen, lyophilized overnight to obtain the freeze-dried reaction, and stored at room temperature. The reactions were reconstituted with nuclease-free water to the original reaction volume and incubated at 30° or 37°C.
Fluorescent protein production and characterization
The FD-CF synthesized fluorescent proteins were expressed at 30°C (for the constitutively expressed outputs) or 37°C (for the toehold-encoded designs) overnight and visualized using a Safe Imager 2.0 Blue-Light Transilluminator (Thermo Fisher Scientific), white light, or the inexpensive imager developed as part of the BioBits™ kit. Images were taken with a DSLR camera and adjusted and cropped in Adobe Photoshop. For quantitative analysis, cell-free reactions were transferred to a 384-well clear-bottom, black-walled plate, and relative fluorescent units were read on a SpectraMax M3 Multi-Mode Microplate Reader (Molecular Devices).
Smell production and characterization
FD-CF reactions for the expression of ATF1 enzyme were incubated at 37°C for 20 hours in the cell-free reaction. The completed FD-CF reaction containing the enzymes was then added into a separate freshly prepared catalysis reaction. The total catalysis reaction volume was 300 μl and included 50 mM HEPES (pH 7.5), 100 mM KCl, 5 mM EDTA, the relevant substrates (25 mM isoamyl alcohol for ATF1), and freshly prepared cofactor (5 mM acetyl-CoA for ATF1), and 10% of the volume was the FD-CF reaction containing the enzyme. These reactions were allowed to proceed 20 hours in capped vials at room temperature. For GC-MS analysis, the stir bar sorptive extraction method (52) was used. Polydimethylsiloxane stir bars (GERSTEL 011222-001-00) were held in the headspace of the reaction vial by a magnet during the catalysis reaction to absorb volatile components. After the completion of the reaction, the stir bar was added to a headspace vial containing 100 μl of dodecane/ethanol (10:1) and analyzed on a GC-MS headspace sampler (Agilent 7697A) to confirm the identity of the converted product. The GC-MS total ion count signal was converted to parts per million by generating a standard curve using the same process described above, but the FD-CF reactions did not contain DNA template or substrates but were spiked instead with known parts per million concentrations of the product isoamyl acetate.
Hydrogel production and characterization
For the sortase hydrogel peptides, eight-arm PEG vinyl sulfone MW 20,000 Da (PEG-VS) was purchased from JenKem Technology. The cross-linking peptides GCRELPRTGG and GGGSGRC were custom-synthesized by CPC Scientific. Each peptide (8 mM) was conjugated separately to 1 weight % (wt %) PEG, dialyzed, lyophilized, and then reconstituted to 30 wt %. FD-CF reactions were used to generate enzymatically cross-linked hydrogels using a two-step process. First, FD-CF reactions containing a sortase-, ecarin-, or Trx-Bx–encoding template were reconstituted with nuclease-free water and incubated at 37°C. Following incubation, the hydrated sortase reaction was added to a solution of 0 to 8% PEG-GCRELPRTGG and 0 to 8% PEG-GGGSGRC in a reaction buffer [50 mM HEPES, 150 mM NaCl, and 10 mM CaCl2 (pH 7.9)] and incubated at 37°C for 30 min. The hydrated ecarin or Trx-Bx reactions were added separately to a solution of bovine fibrinogen (17.5 mg/ml), aprotinin [2.3 TIU (trypsin inhibitor unit)/ml], and 20 mM CaCl2 and incubated overnight at room temperature. The hydrogels were transferred to glass vials and inverted to demonstrate the hydrogel properties. Images were taken with a DSLR camera and adjusted for size and contrast in Adobe Photoshop. For ultrastructural analysis of the resulting hydrogels, the samples were extensively washed with nuclease-free water, snap-frozen in liquid nitrogen, and lyophilized to remove all water. The hydrogel samples were then sputter-coated with 5 nm of Pt/Pd before imaging using a Zeiss Supra55VP FE-SEM.
DNA extraction and processing from fruit
Household dish soap was diluted 1:10 in water along with 1 g of table salt and then added to a plastic bag containing chopped fruit (banana, kiwi, or strawberry). The fruit was then gently crushed in the soap and salt mixture by hand until a homogeneous mixture was obtained. The resulting mixture was strained through a household coffee filter into a cup. A prechilled 25-ml volume of 91% isopropyl alcohol (rubbing alcohol) was added to the strained liquid. The mixture was left undisturbed for 5 min to allow phase separation to occur. The upper white layer containing extracted DNA was removed, placed on a clean coffee filter, and washed with 70% ethanol (ethyl rubbing alcohol). The resulting extracted DNA was then patted with paper towels to remove any excess extraction liquid. The DNA was then diluted in water until it dissolved and added to an isothermal RPA, according to the manufacturer’s protocol (TwistAmp Basic RT, TwistDx; fig. S5), with primers that were complementary to one section of the banana or kiwi genome (table S1). The primers also incorporated a T7 promoter for transcription in FD-CF. The resulting RPA product was then added 1:3.75 to a rehydrated FD-CF reaction containing a linearized toehold complementary to the amplified RPA product and run according to the FD-CF methods described above.
Statistical analysis
Statistical parameters including the definitions and values of n, SDs, and/or SEs are reported in the figures and corresponding figure legends.
Supplementary Material
Acknowledgments
We thank F. Merola, A. Brown, J. Valdez, and L. Griffith for the gift of plasmids encoding the relevant sequences. We also thank L. Durbin for assisting in the construction of the custom handheld fluorescence imager and K. L. Jones Prather, K. Haslinger, J. Boock, and M. P. Lewandowski for the help in performing the GC-MS experiments. Funding: This work was supported by the Wyss Institute (to J.J.C.), the Paul G. Allen Frontiers Group (to J.J.C.), and the Air Force Office of Scientific Research (to J.J.C.). The authors also acknowledge the Army Research Office W911NF-16-1-0372 (to M.C.J.), NSF grants MCB-1413563 and MCB-1716766 (to M.C.J.), the Air Force Research Laboratory Center of Excellence Grant FA8650-15-2-5518 (to M.C.J.), the Defense Threat Reduction Agency grant HDTRA1-15-10052/P00001 (to M.C.J.), the David and Lucile Packard Foundation (to M.C.J.), the Camille Dreyfus Teacher-Scholar Program (to M.C.J.), the Department of Energy BER grant DE-SC0018249 (M.C.J.), and the Natural Sciences and Engineering Council of Canada grant RGPIN-2016-06352 (to K.P.). A.H. is supported by the Paul G. Allen Frontiers Group. P.Q.N. is supported by a Wyss Technology Development Fellowship. J.C.S. and A.J.D. are funded by NSF Graduate Research Fellowships. R.S.D. is funded, in part, by the Northwestern University Chemistry of Life Processes Summer Scholars program. The U.S. government is authorized to reproduce and distribute reprints for governmental purposes notwithstanding any copyright notation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of Air Force Research Laboratory, Air Force Office of Scientific Research, Defense Threat Reduction Agency, or the U.S. government. Author contributions: A.H., P.Q.N., and J.C.S. designed the research, performed the research, analyzed the data, and wrote the manuscript. N.D., T.F., M.K.T., and A.J.D. aided in research design and performed the research. R.S.D. and K.J.H. performed the research. K.P. aided in research design. J.J.C. and M.C.J. directed the research. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The plasmids constructed in this work have been deposited in Addgene with the catalog number listed in table S1. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Reagents or additional data are available from the authors upon request to A.H., P.Q.N., or J.C.S.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaat5105/DC1
Fig. S1. Quantification of all proteins expressed in FD-CF.
Fig. S2. Fluorescent proteins expressed in the PURE and crude extract systems.
Fig. S3. Quantitative analysis of fluorescent proteins.
Fig. S4. Representative scanning electron microscopy images of hydrogel ultrastructures generated with FD-CF enzymes.
Fig. S5. Schematic of RPA reaction.
Fig. S6. Detailed steps for isolating genomic DNA from fruits for environmental sensing activity.
Table S1. Library of proteins and toehold switches that enable visual, olfactory, and tactile outputs for educational engagement.
Table S2. FD-CF reactions allow for inexpensive classroom synthetic biology education kits.
REFERENCES AND NOTES
- 1.Benitti F. B. V., Exploring the educational potential of robotics in schools: A systematic review. Comput. Educ. 58, 978–988 (2012). [Google Scholar]
- 2.Resnick M., Maloney J., Monroy-Hernandez A., Rusk N., Eastmond E., Brennan K., Scratch: Programming for all. Commun. ACM 52, 60–67 (2009). [Google Scholar]
- 3.Clark D. B., Tanner-Smith E. E., Killingsworth S. S., Digital games, design, and learning: A systematic review and meta-analysis. Rev. Educ. Res. 86, 79–122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Research Council, Next Generation Science Standards: For States, By States (National Academies Press, 2013). [PubMed] [Google Scholar]
- 5.Freeman S., Eddy S. L., McDonough M., Smith M. K., Okoroafor N., Jordt H., Wenderoth M. P., Active learning increases student performance in science, engineering, and mathematics. Proc. Natl. Acad. Sci. U.S.A. 111, 8410–8415 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medema M. H., Breitling R., Bovenberg R., Takano E., Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat. Rev. Microbiol. 9, 131–137 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Purnick P. E. M., Weiss R., The second wave of synthetic biology: From modules to systems. Nat. Rev. Mol. Cell Biol. 10, 410–422 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Smanski M. J., Zhou H., Claesen J., Shen B., Fischbach M. A., Voigt C. A., Synthetic biology to access and expand nature’s chemical diversity. Nat. Rev. Microbiol. 14, 135–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gootenberg J. S., Abudayyeh O. O., Lee J. W., Essletzbichler P., Dy A. J., Joung J., Verdine V., Donghia N., Daringer N. M., Freije C. A., Myhrvold C., Bhattacharyya R. P., Livny J., Regev A., Koonin E. V., Hung D. T., Sabeti P. C., Collins J. J., Zhang F., Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356, 438–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pardee K., Green A. A., Takahashi M. K., Braff D., Lambert G., Lee J. W., Ferrante T., Ma D., Donghia N., Fan M., Daringer N. M., Bosch I., Dudley D. M., O’Connor D. H., Gehrke L., Collins J. J., Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 165, 1255–1266 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Fesnak A. D., June C. H., Levine B. L., Engineered T cells: The promise and challenges of cancer immunotherapy. Nat. Rev. Cancer 16, 566–581 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roybal K. T., Williams J. Z., Morsut L., Rupp L. J., Kolinko I., Choe J. H., Walker W. J., McNally K. A., Lim W. A., Engineering T cells with customized therapeutic response programs using synthetic Notch receptors. Cell 167, 419–432.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon Y.-C., Jewett M. C., High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep. 5, 8663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T., Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Pardee K., Slomovic S., Nguyen P. Q., Lee J. W., Donghia N., Burrill D. R., Ferrante T., McSorley F. R., Furuta Y., Vernet A., Lewandowski M. J. L. T. R., Boddy C. N., Joshi N. S., Collins J. J., Portable, on-demand biomolecular manufacturing. Cell 167, 248–259 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Pardee K., Green A. A., Ferrante T., Cameron D. E., DaleyKeyser A., Yin P., Collins J. J., Paper-based synthetic gene networks. Cell 159, 940–954 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlson E. D., Gan R., Hodgman C. E., Jewett M. C., Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 30, 1185–1194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore A., Breathing new life into the biology classroom. EMBO Rep. 4, 744–746 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch S. J., Burton E. P., Behrend T., House A., Ford M., Spillane N., Matray S., Han E., Means B., Understanding inclusive STEM high schools as opportunity structures for underrepresented students: Critical components. J. Res. Sci. Teach. 55, 712–748 (2018). [Google Scholar]
- 20.Stark J. C., Huang A., Nguyen P. Q., Dubner R. S., Hsu K. J., Ferrante T. C., Anderson M., Kanapskyte A., Mucha Q., Packett J. S., Patel P., Patel R., Qaq D., Zondor T., Burke J., Martinez T., Miller-Berry A., Puppala A., Reichert K., Schmid M., Brand L., Hill L. R., Chellaswamy J. F., Faheem N., Fetherling S., Gong E., Gonzalzles E. M., Granito T., Koritsaris J., Nguyen B., Ottman S., Palffy C., Patel A., Skweres S., Slaton A., Woods T., Donghia N., Pardee K., Collins J. J., Jewett M. C., BioBits™ Bright: A fluorescent synthetic biology education kit. Sci. Adv. 4, eaat5107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alieva N. O., Konzen K. A., Field S. F., Meleshkevitch E. A., Hunt M. E., Beltran-Ramirez V., Miller D. J., Wiedenmann J., Salih A., Matz M. V., Diversity and evolution of coral fluorescent proteins. PLOS ONE 3, e2680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ten Buren E. B. J., Karrenbelt M. A. P., Lingemann M., Chordia S., Deng Y., Hu J., Verest J. M., Wu V., Gonzalez T. J. B., van Heck R. G. A., Odoni D. I., Schonewille T., van der Straat L., de Graaff L. H., van Passel M. W. J., Toolkit for visualization of the cellular structure and organelles in Aspergillus niger. ACS Synth. Biol. 3, 995–998 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N. G., Palmer A. E., Tsien R. Y., Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Bayle V., Nussaume L., Bhat R. A., Combination of novel green fluorescent protein mutant TSapphire and DsRed variant mOrange to set up a versatile in Planta FRET-FLIM assay. Plant Physiol. 148, 51–60 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heim R., Tsien R. Y., Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr. Biol. 6, 178–182 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Erard M., Fredj A., Pasquier H., Beltolngar D.-B., Bousmah Y., Derrien V., Vincent P., Merola F., Minimum set of mutations needed to optimize cyan fluorescent proteins for live cell imaging. Mol. BioSyst. 9, 258–267 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Campbell R. E., Tour O., Palmer A. E., Steinbach P. A., Baird G. S., Zacharias D. A., Tsien R. Y., A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U.S.A. 99, 7877–7882 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pham H. L., Ho C. L., Wong A., Lee Y. S., Chang M. W., Applying the design-build-test paradigm in microbiome engineering. Curr. Opin. Biotechnol. 48, 85–93 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Dixon J., Kuldell N., BioBuilding: Using banana-scented bacteria to teach synthetic biology. Methods Enzymol. 497, 255–271 (2011). [DOI] [PubMed] [Google Scholar]
- 30.P. D. Murnane, S. S. Lehocky, A. H. Owens, Odor Thresholds for Chemicals with Established Occupational Health Standards (American Industrial Hygiene Association, ed. 2, 2013). [Google Scholar]
- 31.Yoshioka K., Hashimoto N., Ester formation by alcohol acetyltransferase from brewers’ yeast. Agric. Biol. Chem. 45, 2183–2190 (1981). [Google Scholar]
- 32.Mandal S. M., Chakraborty D., Dey S., Phenolic acids act as signaling molecules in plant-microbe symbioses. Plant Signal. Behav. 5, 359–368 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wackett L. P., Microbiology for odour production and abatement. Microb. Biotechnol. 6, 85–86 (2013). [Google Scholar]
- 34.Peppas N. A., Hilt J. Z., Khademhosseini A., Langer R., Hydrogels in biology and medicine: From molecular principles to bionanotechnology. Adv. Mater. 18, 1345–1360 (2006). [Google Scholar]
- 35.Drury J. L., Mooney D. J., Hydrogels for tissue engineering: Scaffold design variables and applications. Biomaterials 24, 4337–4351 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Valdez J., Cook C. D., Ahrens C. C., Wang A. J., Brown A., Kumar M., Stockdale L., Rothenberg D., Renggli K., Gordon E., Lauffenburger D., White F., Griffith L., On-demand dissolution of modular, synthetic extracellular matrix reveals local epithelial-stromal communication networks. Biomaterials 130, 90–103 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davie E. W., Ratnoff O. D., Waterfall sequence for intrinsic blood clotting. Science 145, 1310–1312 (1964). [DOI] [PubMed] [Google Scholar]
- 38.Dyr J. E., Blombäck B., Kornalik F., The action of prothrombin activated by ecarin on fibrinogen. Thromb. Res. 30, 225–234 (1983). [DOI] [PubMed] [Google Scholar]
- 39.Wedgwood J., Freemont A. J., Tirelli N., Rheological and turbidity study of fibrin hydrogels. Macromol. Symp. 334, 117–125 (2013). [Google Scholar]
- 40.Partlow B. P., Hanna C. W., Rnjak-Kovacina J., Moreau J. E., Applegate M. B., Burke K. A., Marelli B., Mitropoulos A. N., Omenetto F. G., Kaplan D. L., Highly tunable elastomeric silk biomaterials. Adv. Funct. Mater. 24, 4615–4624 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green A. A., Silver P. A., Collins J. J., Yin P., Toehold switches: De-novo-designed regulators of gene expression. Cell 159, 925–939 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Christelová P., Valárik M., Hřibová E., De Langhe E., Doležel J., A multi gene sequence-based phylogeny of the Musaceae (banana) family. BMC Evol. Biol. 11, 103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taguchi H., Watanabe S., Hirao T., Akiyama H., Sakai S., Watanabe T., Matsuda R., Urisu A., Maitani T., Specific detection of potentially allergenic kiwifruit in foods using polymerase chain reaction. J. Agric. Food Chem. 55, 1649–1655 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Piepenburg O., Williams C. H., Stemple D. L., Armes N. A., DNA detection using recombination proteins. PLOS Biol. 4, e204 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D. Sweeney, Biology Exploring Life: Laboratory Manual (Prentice Hall, 2003). [Google Scholar]
- 46.Villarreal F., Contreras-Llano L. E., Chavez M., Ding Y., Fan J., Pan T., Tan C., Synthetic microbial consortia enable rapid assembly of pure translation machinery. Nat. Chem. Biol. 14, 29–35 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Shepherd T. R., Du L., Liljeruhm J., Samudyata, Wang J., Sjödin M. O. D., Wetterhall M., Yomo T., Forster A. C., De novo design and synthesis of a 30-cistron translation-factor module. Nucleic Acids Res. 45, 10895–10905 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A. III, Smith H. O., Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Kunjapur A. M., Hyun J. C., Prather K. L. J., Deregulation of S-adenosylmethionine biosynthesis and regeneration improves methylation in the E. coli de novo vanillin biosynthesis pathway. Microb. Cell Fact. 15, 61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jewett M. C., Swartz J. R., Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng. 86, 19–26 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Jewett M. C., Calhoun K. A., Voloshin A., Wuu J. J., Swartz J. R., An integrated cell-free metabolic platform for protein production and synthetic biology. Mol. Syst. Biol. 4, 220 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bicchi C., Iori C., Rubiolo P., Sandra P., Headspace sorptive extraction (HSSE), stir bar sorptive extraction (SBSE), and solid phase microextraction (SPME) applied to the analysis of roasted Arabica coffee and coffee brew. J. Agric. Food Chem. 50, 449–459 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/8/eaat5105/DC1
Fig. S1. Quantification of all proteins expressed in FD-CF.
Fig. S2. Fluorescent proteins expressed in the PURE and crude extract systems.
Fig. S3. Quantitative analysis of fluorescent proteins.
Fig. S4. Representative scanning electron microscopy images of hydrogel ultrastructures generated with FD-CF enzymes.
Fig. S5. Schematic of RPA reaction.
Fig. S6. Detailed steps for isolating genomic DNA from fruits for environmental sensing activity.
Table S1. Library of proteins and toehold switches that enable visual, olfactory, and tactile outputs for educational engagement.
Table S2. FD-CF reactions allow for inexpensive classroom synthetic biology education kits.


