Abstract
The histone modifiers (HMs) are crucial for chromatin dynamics and gene expression; however, their dysregulated expression has been observed in various abnormalities including cancer. In this study, we have analyzed the expression of HMs in microarray profiles of head and neck cancer (HNC), wherein a highly significant overexpression of p21-activated kinase 2 (PAK2) was identified which was further validated in HNC patients. The elevated expression of PAK2 positively correlated with enhanced cell proliferation, aerobic glycolysis and chemoresistance and was associated with the poor clinical outcome of HNC patients. Further, dissection of molecular mechanism revealed an association of PAK2 with c-Myc and c-Myc-dependent PKM2 overexpression, wherein we showed that PAK2 upregulates c-Myc expression and c-Myc thereby binds to PKM promoter and induces PKM2 expression. We observed that PAK2–c-Myc–PKM2 axis is critical for oncogenic cellular proliferation. Depletion of PAK2 disturbs the axis and leads to downregulation of c-Myc and thereby PKM2 expression, which resulted in reduced aerobic glycolysis, proliferation and chemotherapeutic resistance of HNC cells. Moreover, the c-Myc complementation rescued PAK2 depletion effects and restored aerobic glycolysis, proliferation, migration and invasion in PAK2-depleted cells. The global transcriptome analysis of PAK2-depleted HNC cells revealed the downregulation of various genes involved in active cell proliferation, which indicates that PAK2 overexpression is critical for HNC progression. Together, these results suggest that the axis of PAK2–c-Myc–PKM2 is critical for HNC progression and could be a therapeutic target to reduce the cell proliferation and acquired chemoresistance and might enhance the efficacy of standard chemotherapy which will help in better management of HNC patients.
Introduction
Head and neck cancer (HNC) is one of the most common and highly aggressive malignancy and the eighth most common cancer worldwide1,2. The global incidence of all HNCs has been estimated to be 4–6 × 105 with the mortality rate of 2.2–3 × 105 per year3. In Southeast Asian countries, notably India4, the occurrence of HNC is high among male population5 and is associated with late diagnosis as well as poor prognosis. With the advancement of surgical6 and radiation therapies7 the quality of HNC patient’s life has improved over the time. However, despite the improvement of health care systems the survival rate of HNC patients remains poor8,9, which highlights the need for new molecular targets for HNC treatment.
Epigenetic mechanisms play an important role in the cellular development and maintenance of cellular homeostasis. Any alteration of epigenetic mechanisms via the changes in DNA methylation10 and histone modification11 may lead to various diseases including cancer12. Various histone modifications are globally altered in different cancers, which promote cancer development13 and chemotherapeutic resistance14 and confer poor prognosis15,16. The cancer-associated changes in histone modifications might occur due to altered expression of histone modifiers (HMs)17 that may deregulate the gene regulation in favor of oncogenic growth. Accordingly, the perturbations of several HMs, such as class I histone deacetylases18,19, histone demethylases, KDM1A9 as well as histone methyltransferases EZH220, are associated with cancer progression and confer poor prognosis. Therefore, to identify the deregulated HMs in HNC, we first enlisted all HMs using HIstome database21. Sequentially, the expression of all HMs was analyzed in HNC microarray profile available with Gene Expression Omnibus (GEO). For further studies, we selected upregulated HMs wherein we found a highly significant overexpression of p21-activated kinase 2 (PAK2). PAK2 is a member of PAK family of serine/threonine kinases, initially identified as a binding partner of the Rho GTPases, Cdc42 and RacI22. The PAK2 plays a critical role in many fundamental cellular functions, including chromatin remodeling, cytoskeletal remodeling, proliferation and regulation of cellular apoptosis23–26. Furthermore, PAK2 has also been shown to affect the histone modifications26–28 resulting in the alteration of gene expression. Moreover, PAK2 overexpression is observed in various human malignancies29,30, and has been proposed as an independent prognostic marker for gastric cancer31. Collectively, these findings suggest an important role of PAK2 in carcinogenesis. However, the role of PAK2 in HNC development and the underlying molecular mechanism remains to be established.
In this study, we have investigated the molecular mechanism of PAK2-mediated oncogenesis. Importantly, we showed that PAK2 is associated with higher proliferation, Warburg effect and chemotherapeutic resistance. The PAK2 depletion restricted the growth of cancer cells and decreased the chemotherapeutic resistance. Importantly, we report the role of β-catenin-mediated upregulation of c-Myc in PAK2-dependent HNC oncogenesis. Moreover, c-Myc then occupies the promoter region of PKM gene and upregulates the pyruvate kinase M2 (PKM2) expression, which then favors the aerobic glycolysis and HNC cell proliferation and thereby leads to PAK2–c-Myc–PKM2 axis-driven HNC progression. In summary, we have shown a novel regulatory role of PAK2 in HNC development and a potential framework for HNC cancer therapy by targeting PAK2–c-Myc–PKM2 axis.
Materials and methods
Microarray data analysis
Gene expression profiles utilized in this study were collected from the GEO32,33. Microarray platform-specific probes were mapped to gene symbols with appropriate annotation files. The expression values of genes with more than one probe were averaged using DNA Chip Analyzer (dChip) software and considered for the analysis. PAK2 gene expression values were extracted from normalized HNC tumor profiles. The significant difference in gene expression between normal and tumor was calculated using Student’s t-test (two-tailed). P value less than 0.05 was considered as significant. GraphPad Prism5 (La Jolla, CA, USA) was used to generate the boxplots.
Survival curve
Overall survival information of the HNC patients was obtained from GEO (GSE42743) and was considered for predicting the association between PAK2 expression and patient survival. The patients were classified into low-expression and high-expression groups and Kaplan–Meier survival curve analysis was performed. The survival curve was plotted with GraphPad Prism5 (La Jolla, CA, USA).
Cell culture
Human HNC cell lines H157 (ECACC 07030901), H413 (ECACC 06092007) and BICR10 (ECACC 04072103) were obtained from European Collection of Authenticated Cell Culture (ECACC) (Salisbury UK). All three cell lines were cultured in the ECACC recommended media, supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA, USA), 100 units/ml of penicillin and streptomycin (Thermo Fisher Scientific, Waltham, MA, USA), 2 mM l-glutamine (Sigma, Saint Louis, USA) and 0.5 µg/ml sodium hydrocortisone succinate. All three cell lines were cultured in a humidified atmosphere at 37 °C and 5% CO2.
Head and neck cancer sample collection
Informed consent was obtained from patients undergoing surgery for head and neck cancer at Bansal Hospital, Bhopal, India. After surgery, tumor and adjacent normal tissue pairs were collected and immediately snap frozen and stored at −80 °C until use. Tissues for RNA isolation were collected in RNA later (Thermo Fisher Scientific, Waltham, MA, USA) separately. This study was approved by the Institute Ethics Committee. Clinical characteristics of patients used in the study are presented in Supplementary Table S1.
Oncomine data analysis
The expression of PAK2 was searched in Oncomine, and among various cancers, HNC profiles were selected for further investigation. The expression of PAK2 was analyzed in HNC normal and tumor tissue as well as in HNC cell lines as per experimental requirement. The analyzed expression data and graph were exported for representation.
RNA interference
The H157, H413 and BICR10 HNC cells were infected with lentivirus containing small hairpin RNA (shRNA) purchased from Sigma (Saint Louis, USA) specific to PAK2 (shPAK2) and eGFP (shControl) with 8 µg/ml polybrene containing media. Cells were selected using 1 µg/ml puromycin for 3 days. Post selection cells were used for downstream experiments.
Oligo sequence of shRNAs
| sheGFP | 5′-CCGGTACAACAGCCACAACGTCTATCTCGAGATAGACGTTGTGGCTGTTGTATTTTT-3′ |
| shPAK2 | 5′-CCGGCGGGATTTCTTAAATCGATGTCTCGAGACATCGATTTAAGAAATCCCGTTTTTTG-3′ |
MKi67 staining
Post puromycin selection, the shControl and shPAK2 HNC cells (H157 and H413) were harvested and MKi67 staining was done using Alexa Fluor 488-labeled Ki67 antibody (ab197234, Abcam, Melbourne, Australia) as discussed previously34. The MKi67 flow cytometry experiment was done with fluorescence-activated cell sorting (FACS) Aria III by Becton Dickinson, (Franklin Lakes, NJ, USA) and expression as well as mean fluorescent intensity (MFI) were analyzed with FlowJo software version 10 (FlowJo, Ashland, OR, USA).
Cell-cycle analysis
Post puromycin selection, the shControl and shPAK2 HNC cells (H157 and H413) were harvested and washed with 1× phosphate-buffered saline (PBS) twice. The cells were stained with propidium iodide (PI, BD Biosciences, India) in the presence of RNase and incubated in the dark for 30 min, as per manufacturer’s protocol. The samples were diluted with 1× PBS, and flow cytometry was performed using FACS Aria III by Becton Dickinson, (Franklin Lakes, NJ, USA) and data were analyzed using FlowJo software version 10 (FlowJo, Ashland, OR, USA).
Quantitative reverse transcriptase-PCR (qRT-PCR)
Total RNA was extracted from cultured H157 cells, H413 cells and human HNC tumor and normal tissue samples using Trizol (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instruction. RNA was quantified using Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). Then, 1 µg of total RNA was reverse transcribed by iScript complementary DNA (cDNA) synthesis kit (Bio-Rad, CA, USA) as per the manufacturer’s instructions. The experiment was done using SYBR green (Qiagen, Hilden, Germany) with light cycler 480 II (Roche, Basel, Switzerland) according to the manufacturer’s protocol. Primers were designed using the IDT Primer Quest tool (https://www.idtdna.com/). Primers used in this study are mentioned in the Supplementary Table S2. The average cycle thresholds of independent experiments were calculated and normalized to housekeeping control gene RPS16 for HNC cells and β-Actin for HNC tissue samples using the formula: [2^(Ct control – Ct target)]. Student’s t-test was used to compare gene expression between two different groups. P < 0.05 was considered as statistically significant.
Immunoblotting
Proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membrane (Millipore). The protein containing PVDF membranes were probed with different primary antibodies: anti-PAK2 (2615, Cell Signaling Technology, Beverly, MA, USA), anti-c-Myc (9402, Cell Signaling Technology, Beverly, MA, USA), anti-CCND1 (ab134175, Abcam, Melbourne Australia), anti-PKM2 (4053, Cell Signaling Technology, Beverly, MA, USA), anti-Active β-Catenin (05665, Millipore, Burlington, USA), anti-GAPDH (5174, Cell Signaling Technology, Beverly, MA, USA), anti-β-Catenin (9562, Cell Signaling Technology, Beverly, MA, USA) and anti-flag (NBP1-06712SS, Novus Biologicals, Littleton, CO, USA) in a 1:1000 dilution. After 2 h of primary antibody incubation at room temperature, membranes were washed with 1× TBST (tris-buffered saline and Tween-20) and incubated with secondary antibodies Alexa-Flour 680 anti-rabbit IgG (A21109, Thermo Fisher Scientific, Waltham, MA, USA) and Alexa-Flour 790 goat anti-mouse IgG (A28182, Thermo Fisher Scientific, Waltham, MA, USA) for 30 min at room temperature. The membrane was washed, and bands were visualized using an Odyssey membrane Scanning system (Li-Cor Biosciences, Bad Homburg, Germany).
Cell viability assay/MTT assay
Post puromycin selection, cells were seeded in 96-well culture plates (4 × 103/well) for 12 h, 24 h, 36 h, 48 h and 60 h (in triplicate for each condition). The 20 µl MTT (Sigma, Saint Louis, USA) stock solution (2 mg/ml) was added to each well and incubated for 2–3 h. After the incubation, formazan crystals formed in the cells were solubilized using dimethyl sulfoxide and the optical density was analyzed at 600 and 750 nm using plate reader BioTek Eon (BioTek, Winooski, USA).
Wound healing assay
Post puromycin selection, 1 × 105 cells/well were seeded in 12-well plate and upon reaching at 100% confluency, wound was created using 200 μl pipette tip and washed with 1× PBS for two times to remove cellular debris. Wounds were visualized at 10× with an inverted microscope (Olympus, Tokyo, Japan) and three random images were captured for few days as indicated. Wound width was measured using Q-Capture software and graph was plotted with GraphPad Prism5 (La Jolla, CA, USA).
Invasion assay
The puromycin selected 2 × 104 cells were added to the upper chamber of transwell (Corning, NY, USA) over Matrigel (Corning, Bedford, MA, USA) layer and incubated for 24 h in cell culture incubator. The non-migrated cells in upper layer of Matrigel were removed and cells migrated to lower chamber of transwell were fixed in 4% paraformaldehyde, stained with 0.05% crystal violet and five random fields were counted using an inverted microscope (Olympus, Tokyo, Japan).
Colony-forming assay
Post puromycin selection, cells were trypsinized, and 1 × 103 cells were seeded in the new 6-well cell culture plate and maintained in 0.5 µg/ml puromycin containing media for 7 days. Cell colonies were visualized by crystal violet staining. For staining, cells were fixed using methanol and acetic acid (3:1) for 5 min and washed with 1× PBS three times. Cells were then stained with 0.05% crystal violet for 30 min. Post staining, cells were washed twice with 1× PBS and plates were dried for 30 min at room temperature and scanned with Epson Scanner. Colonies containing more than 50 individual cells were counted using microscope as well as relative intensity of each well and colony area was quantified using ImageJ software35 (La Jolla, CA, USA).
Caspase 3/7 assay
Post puromycin selection, the 4 × 104 cells/well were seeded in white color 96-well plate. Cells were treated with 5 μM camptothecin and 50 μM etoposide after 12 h of incubation. After 5 h of camptothecin treatment and 24 h of etoposide treatment, the caspase 3/7 activation was measured using the Caspase-Glo 3/7 Assay (Promega, Madison, USA) as recommended by the manufacturer. Luminescence readings were taken using Glomax multi-detection system (Promega).
Annexin–PI staining
The cellular apoptosis in PAK2-depleted and control cells were analyzed using Annexin–FITC and PI staining kit (BD Biosciences, India) as per the manufacturer’s protocol. The flow cytometric analysis was done using FACS Aria III (Becton Dickinson), and data were analyzed using FlowJo software version 10 (FlowJo, Ashland, OR, USA).
Lactate assay
Post puromycin selection, 3 × 105 cells/well were seeded in 6-well plate. Cells were homogenized with lactate assay buffer after 48 h of seeding. Lactate quantification was performed using a commercially available lactate assay kit (Sigma, Saint Louis, USA) in a 96-well plate as per the manufacturer’s instruction. Lactate production was measured with plate reader (BioTek Eon) at an optical density of 570 nm and lactate was quantified as per the manufacturer’s protocol.
Glucose uptake assay
Post puromycin selection, 3 × 105 cells/well were seeded in 6-well plate. Cells were homogenized with glucose assay buffer after 48 h of seeding. Glucose level quantification was performed using a commercially available glucose assay kit (Abcam, Melbourne, Australia) in a 96-well plate as per the manufacturer’s protocol. Glucose uptake was measured with a plate reader (BioTek Eon) at an optical density of 570 nm.
c-Myc and PAK2 overexpression construct generation
The overexpression plasmids were constructed by amplifying c-Myc and PAK2 fragment from H157 cDNA using Platinum Q5 Hotstart High-Fidelity DNA Polymerase (New England Biolabs, MA, USA) using the primers mentioned in the Supplementary Table S2. The c-Myc product was cloned between the BamHI and EcoRI sites of pAIP lentivirus system-based expression Vector (Addgene, MA, USA) and the PAK2 was cloned between BamHI and XhoI sites in pCMV-3Tag 1A plasmid (Agilent Technologies, Santa Clara, CA, USA).
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed as described previously36. Briefly, after puromycin selection, cells were sonicated, and chromatin (25 µg) was immunoprecipitated by adding the antibody of interest followed by overnight incubation at 4 °C. The following antibodies were used for ChIP: Anti-c-Myc and Normal Rabbit IgG (12–370, Millipore, Burlington, USA). Immunoprecipitated fractions and 5% input were analyzed by quantitative real-time PCR in duplicate using the SYBR Green Master Mix (Qiagen, Hilden, Germany) with specific primers (mentioned in the Supplementary Table S2) of predicted c-Myc binding regions. Normalization was performed to input using the formula: [2^(Ct input – CT immunoprecipitation)]. Resultant values were further normalized relative to the rabbit Ig control ChIP values for the primer set. Student’s t-test was used to identify the significance between two different groups. P value of <0.05 was considered statistically significant.
ChIP-sequencing (ChIP-seq) data analysis
The ChIP-seq data were analyzed using the University of California Santa Cruz (UCSC) genome browser. The bigwig file of c-Myc ChIP-seq data was added in the custom track window provided in the UCSC genome browser, and the enrichment peak of c-Myc was analyzed at PKM gene promoter region.
Immunocytochemistry (ICC)
Post puromycin selection, the ICC was performed by following the methodology provided by Abcam (http://www.abcam.com/protocols/immunocytochemistry-immunofluorescence-protocol). The expression of PKM2 which is directly proportional to GFP intensity was observed, and the images were captured at 10× using Evos FL Auto2 microscope (Thermo Fisher Scientific, Waltham, MA, USA). The intensity of the image was analyzed using ImageJ software (La Jolla, CA, USA).
Human Transcriptome Array (HTA) 2.0 data analysis
The raw HTA 2.0 files were normalized and analyzed using Transcription Array Console. The expression index (linear) ≤ −2 or expression index (linear) ≥ + 2 with P < 0.05 were set as the criteria to identify the differentially expressed genes. The heat map was prepared through Morpheus, an online tool provided by Broad Institute (https://software.broadinstitute.org/morpheus/).
Densitometric analysis
Densitometric analysis was performed using ImageJ software suit. Briefly, the band intensity was calculated with ImageJ and normalized with respective loading control. The normalized control values were further normalized to one, and the protein fold change was calculated using control values.
Statistical analysis
The statistical analysis was performed with GraphPad Prism5 (La Jolla, CA, USA). In the bar graph, differences between two groups were compared using an unpaired two-tailed Student’s t-test. The differences between three or more groups were calculated using one-way analysis of variance by the Newman–Keuls test. The differences were considered statistically significant with *P < 0.05, **P < 0.01 and ***P < 0.001, ns non-significant difference (P > 0.05).
Results
PAK2 is overexpressed in HNC and confers poor clinical outcome
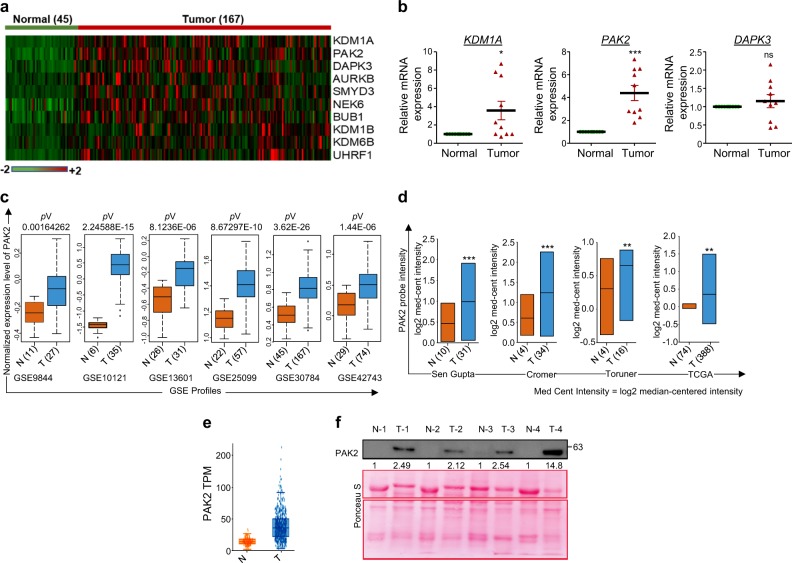
In order to identify the overexpressed HMs in HNC, we first enlisted the HMs using HIstome database21. The expression of HMs was analyzed in HNC microarray profile (GSE30784). Significantly overexpressed HMs were shortlisted by the method shown in Supplementary Figure S1a. The expression of top 10 overexpressed HMs was represented as a heat map (Fig. 1a). Furthermore, top three overexpressed HMs, KDM1A, PAK2 and DAPK3, were selected, and their expression was cross-validated in ten HNC patient samples by qRT-PCR analysis, which showed a significant overexpression of PAK2 in comparison to KDM1A and DAPK3 (Fig. 1b) and was selected for further analysis. To strengthen our preliminary observation, the expression of PAK2 was further validated in five additional HNC microarray profiles, which revealed a highly significant upregulation of PAK2 in HNC (Fig. 1c). Further validation using Oncomine database37 in three independent HNC studies38–40, The Cancer Genome Atlas (TCGA) database as well as RNA-seq data from MiPanda (Michigan Portal for the Analysis of NGS Data) database revealed a consistent overexpression of PAK2 transcript (Fig. 1d, e and Supplementary Figure S1b) in HNC. Interestingly, analysis of TCGA data showed a positive correlation of PAK2 expression with higher stages and grades of HNC (Supplementary Figure S1c,d). Additionally, the PAK2 expression positively correlates with the higher T-staging (Supplementary Figure S1e,f), suggesting that the PAK2 could be responsible for the advancement of cancer.
Fig. 1. Detection and validation of PAK2 overexpression in head and neck tumor samples.
a Heat map of top 10 overexpressed histone modifiers in head and neck normal and tumor samples (generated using DNA Chip analyzer). b Quantitative mRNA expression analysis of KDM1A, PAK2 and DAPK3 normalized to β-actin in10 normal and tumor samples of HNC patients. c Expression pattern of PAK2 gene in independent cohorts of head and neck cancer gene expression profiles. Box plot shows the elevated expression of PAK2 in head and neck tumor tissues compared to normal tissue. d Analysis of PAK2 expression in four different microarray profiles using Oncomine database. The y-axis represents log2-median centered intensity. e Analysis of PAK2 transcript per million (TPM) in normal and head and neck tumor samples through MiPanda (http://www.mipanda.org) cancer RNA-seq database. f Analysis of PAK2 protein expression in 26 normal and tumor tissue samples of HNC patients by PAK2 immunoblotting (also see Supplementary Figure S1g). N normal, T tumor, GSE Genomic Spatial Event. Error bars shows mean values ± SD. Differences were considered statistically significant with *P < 0.05, **P < 0.01 and ***P < 0.001, ns non-significant difference (P > 0.05)
To validate the results of in silico studies, we performed PAK2 immunoblotting in 26 HNC tumor samples, wherein 24 tumor samples showed PAK2 overexpression in comparison to normal counterpart (Fig. 1f, Supplementary Figure S1g). Additionally, Kaplan–Meier survival analysis showed that high PAK2 expressing HNC patients (GSE42743) had shorter overall survival than those with low expression, highlighting the prognostic value of PAK2 in HNC (Supplementary Figure S1h). Conclusively, these analyses strongly suggest a definite role of PAK2 overexpression in HNC progression.
PAK2 deficiency leads to reduced proliferation of head and neck cancer cells
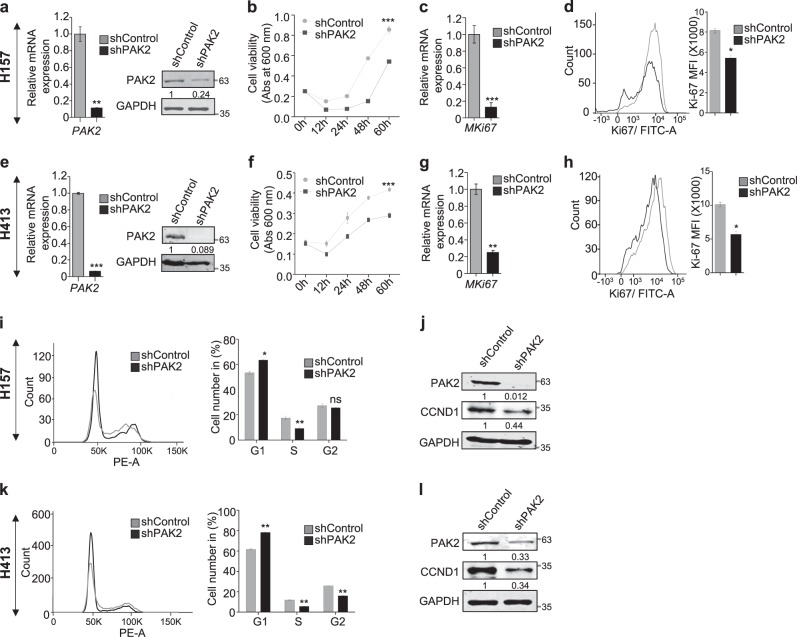
Upon observing an elevated expression of PAK2 in HNC, first we analyzed the expression of PAK2 in three HNC cells and observed the comparable expression of PAK2 in H413 and H157 cells, but relatively less expression in BICR10 cells (Supplementary Figure S2a). Next, we examined the role of PAK2 in HNC cell proliferation by depleting PAK2 in three HNC cancer cell lines with shRNA against PAK2 (shPAK2) or against eGFP (shControl) (Fig. 2a, e, and Supplementary Figure S2b) and observed significantly reduced viability in PAK2-depleted cells (Fig. 2b, f, and Supplementary Figure S2c). Furthermore, we also observed reduced cell proliferation of HNC cells upon PAK2 depletion by cell counting assay (Supplementary Figure S2d,e), which was consistent with the MTT assay. Additionally, to validate the outcome of PAK2 depletion, we overexpressed PAK2 in BICR10 cells (Supplementary Figure S2f) and found significantly increased cell viability (Supplementary Figure S2g). Moreover, PAK2 depletion also led to the reduced expression of cell proliferation marker MKi67 at both transcript and protein levels (Fig. 2c, d, g, h). Collectively, these data indicate that PAK2 overexpression promotes the cellular proliferation and cell viability in HNC cells. Upon establishing the role of PAK2 in cellular proliferation, we investigated the role of PAK2 in cell-cycle progression using flow cytometry. Interestingly, PAK2 silencing significantly increased the cell population at G0/G1 phase in H157 (Fig. 2i) and H413 cells (Fig. 2k) and reduced the cell population in S phase. As the expression of Cyclin D1 (CCND1) has been shown to be critical for G1/S phase transition of cells41, we investigated the CCND1 expression upon PAK2 depletion. The PAK2-deficient H157 and H413 cells showed a remarkable decrease in CCND1 expression in comparison to control cells (Fig. 2j, l and Supplementary Figure S2h,i). Conclusively, these results demonstrate that PAK2 promotes cell-cycle progression by upregulating the CCND1 expression and promotes HNC cell proliferation.
Fig. 2. PAK2 affects proliferation of head and neck cancer cells.
PAK2 expression was depleted in two types of HNC cell lines (H157 and H413), and differential proliferation status of these cell lines was analyzed. a, e Knockdown of PAK2 expression was analyzed at mRNA and protein levels (n = 3). b, f Relative cell proliferation was analyzed through MTT assay (n = 3). c, g mRNA and d, h protein expression status of cell proliferation marker Ki67 (left) was analyzed through qPCR and flow cytometer, respectively. (Right) Mean fluorescence intensity (MFI) of Alexa Fluor 488-Ki67 was measured using software FlowJo 10 (n = 3). i, k (Left) Cell-cycle stages were analyzed using flow cytometer (representative image of 3 independent experiment) (right) bar graph shows average of three independent experiments and j, l cyclin D1 (CCND1) protein expression level was analyzed through immunoblotting (n = 3). Abs absorbance. Samples in (a–c) and (e–g) were analyzed in duplicate. Error bars shows mean values ± SD. Differences were considered statistically significant with *P < 0.05, **P < 0.01 and ***P < 0.001, ns non-significant difference (P > 0.05)
PAK2 depletion suppresses migration, invasion and colony formation of head and neck cancer cells and reduces chemotherapeutic resistance
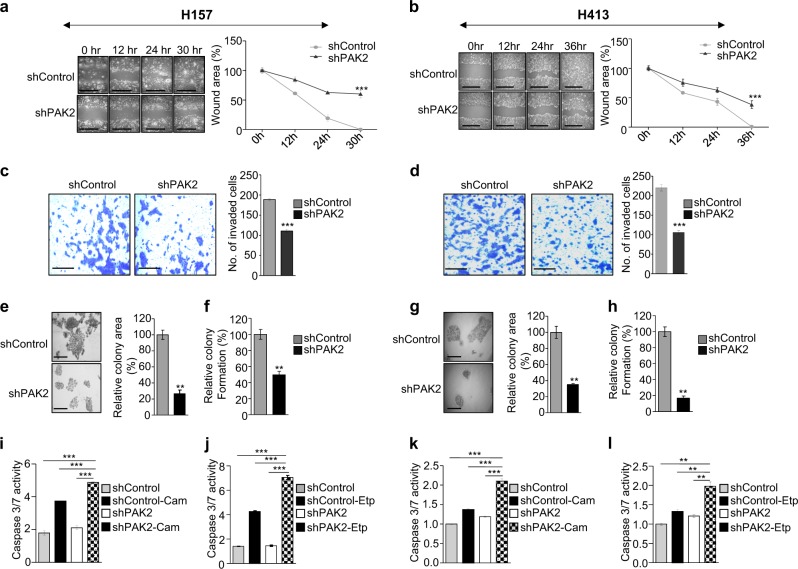
To study the role of PAK2 in an in vitro tumorigenesis of HNC cells, the PAK2-depleted and control cells were subjected to wound healing, cell invasion and colony formation assay. Upon PAK2 depletion, we observed significantly reduced migration (Fig. 3a, b and Supplementary Figure S3a) and invasion (Fig. 3c, d and Supplementary Figure S3b). Similarly, the PAK2 overexpression in BICR10 cells significantly increased the migration and invasion of PAK2_OE cells as compared to the control cells (Supplementary Figure S3c,d). Sequentially, to assess the role of PAK2 in colony formation42 of HNC cells, we performed colony formation assay and observed reduced colony formation in PAK2-depleted cells as compared to control cells. The measurement of colony area, colony number (Fig. 3e–h and Supplementary Figure S3e,f) and colony density (Supplementary Figure S3g) of stained colonies showed a significant reduction in colony size as well as colony number upon PAK2 depletion. These results suggest that PAK2 affects migration, invasion and colony formation of HNC cells and thus helps in head and neck oncogenesis.
Fig. 3. PAK2 depletion reduces in vitro tumorigenesis of head and neck cancer cell lines.
PAK2 expression was depleted in two types of HNC cell lines (H157 and H413), and key cancer hallmarks were compared in PAK2-depleted and control cells. a, b Cell migration was analyzed through wound healing assay, (left) wound was observed under the microscope, scale bar 500 µm, and (right) quantification of wound width (n = 3). c, d The cell invasion was analyzed through Matrigel invasion assay, (left) single field of invaded cells was captured under the microscope, scale bar: 250 µm, and (right) invaded cells were counted at five different fields under the microscope (n = 3), e, g colony-forming tendency of cells was analyzed, (left) colony size was observed under the microscope, scale bar: 250 µm, and (right) quantification of colony size measured with ImageJ, f, h number of colonies were counted manually after crystal violet staining of the cells (n = 3). The caspase 3/7 activity of i, j H157 cells and k, l H413 cells was analyzed upon treatment with 5 µM camptothecin (Cam) and 50 µM etoposide (Etp) as indicated. Shown results are representative of three independent experiments and f, h are average of three independent experiments. Error bars shows mean values ± SD. Differences were considered statistically significant with **P < 0.01 and ***P < 0.001
The chemotherapeutic resistance against the anticancer drug has been a challenge for cancer treatment. Interestingly, the Oncomine data analysis of HNC cells43,44 showed a negative correlation of PAK2 with anticancer drug treatment, wherein the expression of PAK2 was more in anticancer drug (paclitaxel and dimethyloxaloylglycine)-resistant cells in comparison to sensitive cells (Supplementary Figure S3j,k), which was consistent with the previous studies45. Sequentially, to investigate the role of PAK2 in chemotherapeutic drug-induced apoptosis in HNC cells, PAK2-depleted and control cells were treated with anticancer drugs, camptothecin and etoposide, which resulted in the increased apoptosis in PAK2-depleted cells (Fig. 3i–l and Supplementary Figure S3h-i). It was further validated by Annexin V–PI staining, wherein PAK2-depleted H157 and H413 cells showed higher staining of Annexin V and PI in comparison to control cells (Supplementary Figure S3l-o). Collectively, these data suggest that the higher expression of PAK2 provides resistance against chemotherapeutic drug-induced apoptosis and highlight the multidimensional role of PAK2 in head and neck tumorigenesis.
PAK2 depletion inhibits activation of β-catenin and thereby reduces c-Myc expression and affects aerobic glycolysis via PKM2 downregulation
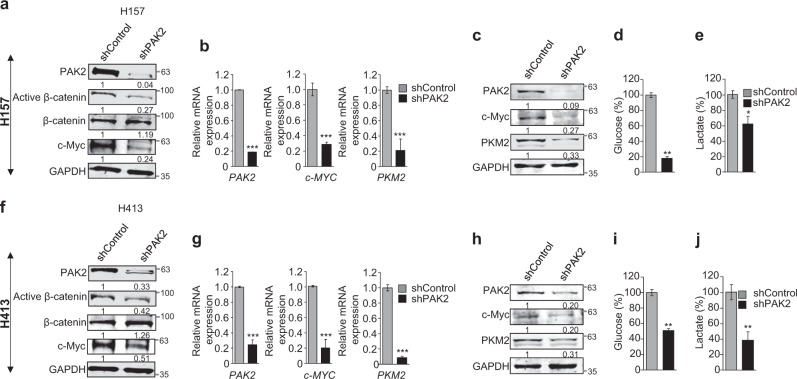
The PAK2 has been proposed as an effector molecule of Rac1 signaling which is suggested to be necessary for the stabilization of activated β-catenin46. This indicates a possible and direct role of PAK2 in β-catenin-mediated signaling. Importantly, we observed a significant reduction of active β-catenin (ABC) upon PAK2 depletion; however, we did not observe any significant change in β-catenin expression (Fig. 4a, f). These findings were consistent with previous report47 which showed alteration of active β-catenin upon PAK2 depletion in schwannoma cells. This suggests that PAK2 affects the activation of β-catenin but not the expression of β-catenin. As the β-catenin activation is associated with regulation of downstream target genes, we hypothesized that the β-catenin signaling might be PAK2 dependent and PAK2 might also regulate the expression of Wnt/ABC target gene c-Myc47,48 in HNC cells. The c-Myc is overexpressed in various cancers including HNC49 and promotes cell proliferation, invasion and metastasis and is thereby considered as a molecular hallmark of cancer50. We investigated the dependency of PAK2-mediated head and neck tumorigenesis on c-Myc. Consequently, we validated the effects of ABC reduction on the c-Myc and found that the reduction of ABC upon PAK2 depletion correlates with significant downregulation of c-Myc (Fig. 4a, f), while, upon overexpressing PAK2 in BICR10, c-Myc expression was drastically increased (Supplementary Figure S2f). These data clearly establish the role of PAK2 in activation of β-catenin, which thereby upregulate c-Myc expression in HNC cells.
Fig. 4. PAK2 depletion reduces active β-catenin (ABC) and downregulates c-Myc expression and inhibits Warburg effects.
PAK2 expression was depleted in two types of HNC cells (H157 and H413) to understand its role in activation of β-catenin and c-Myc expression as well as in Warburg effect. a, f Protein expression of PAK2, active β-catenin, β-catenin and c-Myc upon PAK2 depletion (n = 3). b, g mRNA and c, h protein expression of PAK2, c-Myc and PKM2 were analyzed upon PAK2 depletion (n = 4). d, i The level of glucose uptake and e, j lactate production were analyzed upon PAK2 depletion (n = 3). Error bars shows mean values ± SD. Differences were considered statistically significant with *P < 0.05, **P < 0.01 and ***P < 0.001
The c-Myc-mediated tumorigenesis is partly contributed by increased aerobic glycolysis or Warburg effect51, and as PKM2 plays an important role in enhancing the Warburg effect52, we investigated the effect of PAK2 depletion on the expression of PKM2. We observed significant downregulation of PKM2 (Fig. 4b, c, g, h) upon PAK2 depletion, while the overexpression of PAK2 showed elevated expression of PKM2 in comparison to control cells (Supplementary Figure S2f). The PKM2 expression is known to be associated with increased aerobic glycolysis, resulting in increased glucose uptake and lactate production in cancer cells53. Hence, we hypothesized that the PAK2 depletion might lead to a reversal of Warburg effect due to downregulation of PKM2. To test this hypothesis, we analyzed lactate production and glucose uptake in PAK2-depleted H157 and H413 cells. Interestingly, PAK2 depletion resulted in reduced lactate production and glucose uptake in HNC cells (Fig. 4d, e, i, j). Collectively, these results showed that PAK2 affects PKM2 expression and suggests an important role of PAK2 in cancer cell energy metabolisms.
c-Myc complementation in PAK2-depleted cells restores PKM2 expression, Warburg effect and promotes head and neck carcinogenesis
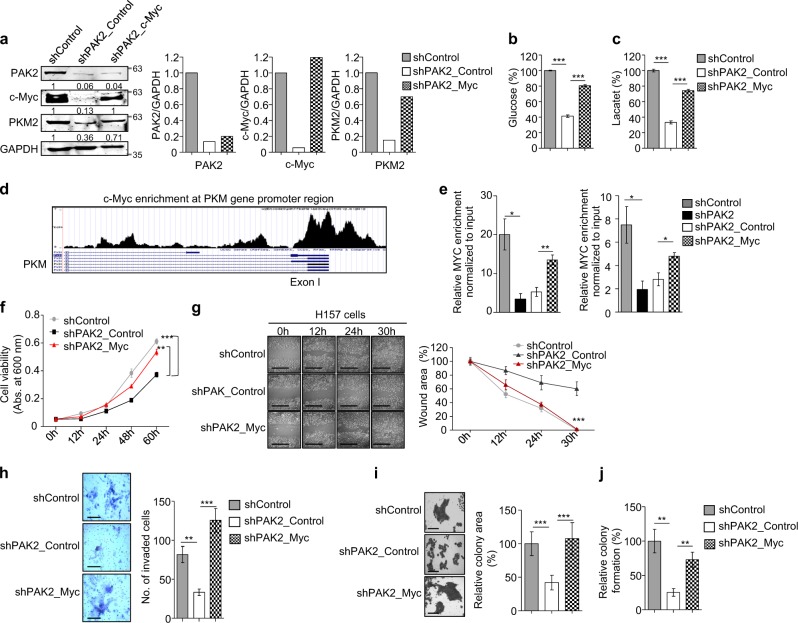
In the previous sections, we have shown that the PAK2 depletion is associated with the significantly reduced c-Myc and PKM2 expression (Fig. 4b, c, g, h). In order to validate the dependency of PAK2 on c-Myc for regulation of PKM2, we transiently overexpressed c-Myc in PAK2-deficient H157 cells (Supplementary Figure S4a). The overexpression of c-Myc in PAK2-depleted cells restored the expression of PKM2 in shPAK2_Myc cells as compared to shPAK2_Control cells (Fig. 5a and Supplementary Figure 4b, c). Sequentially, we also investigated the effect of c-Myc complementation and thereby PKM2 restoration on glucose metabolism. We observed a significantly increased glucose uptake and lactate production in shPAK2_Myc cells as compared to shPAK2_Control cells (Fig. 5b, c).
Fig. 5. c-MYC complementation rescues the active proliferation.
PAK2 expression was depleted in H157 cells, and c-Myc was ectopically overexpressed to understand the interdependency of PAK2 and c-Myc to induce oncogenesis. The key cancer hallmarks were analyzed upon c-Myc complementation in PAK2-deficient cells. a (Left) Protein expression of PAK2, c-Myc and PKM2 was analyzed upon c-Myc overexpression through immunoblotting (n = 3). (Right) Densitometric analysis of representative blot. b The level of glucose uptake and c lactate production was analyzed (n = 3). d Representative image shows the c-Myc enrichment at PKM gene promoter region, analyzed using UCSC genome browser. e Shown is the enrichment of c-Myc at PKM gene promoter region, analyzed with c-Myc ChIP (n = 3). f Relative cell proliferation was analyzed through MTT assay (n = 3). g Cell migration was analyzed through wound healing assay, (left) wound was observed under the microscope, scale bar: 500 µm, (right) quantification of wound width (n = 3), h cell invasion was analyzed upon crystal violet staining through Matrigel invasion assay, (left) single field of invaded cells was captured under the microscope, scale bar: 250 µm, (right) invaded cells were counted at five different fields under the microscope (n = 3), i colony-forming tendency of cells was analyzed, (left) colony sizes were observed under the microscope, scale bar: 250 µm, and (right) quantification of colony size measured with ImageJ. j Number of colonies were counted manually after crystal violet staining of the cells (n = 3). shPAK2_Control shPAK2+pAIP, shPAK2_Myc shPAK2+c-MYC. Error bars shows mean values ± SD. Differences were considered statistically significant with *P < 0.05, **P < 0.01 and ***P < 0.001
In order to understand the molecular mechanism of c-Myc-dependent PKM2 expression, we analyzed putative c-Myc-binding at PKM promoter region using Genomatix software suite (http://shop.genomatix.de/). The Genomatix software suit highlighted probable c-Myc binding sites at PKM promoter region (Supplementary Figure S4d). Similarly, analysis of published ChIP-seq data (GSM935320) with the UCSC genome browser revealed c-Myc enrichment at the PKM gene promoter region (Fig. 5d). Importantly, we observed a significant enrichment of c-Myc at the PKM gene promotor region, which is markedly reduced in PAK2-depleted cells (Fig. 5e), which correlates with PKM2 downregulation (Fig. 5a and Supplementary Figure 4c). Sequentially, upon c-Myc complementation, the c-Myc enrichment was restored at the PKM gene promoter (Fig. 5e), which further resulted in increased PKM2 expression (Fig. 5a and Supplementary Figure S4b,c). Moreover, c-Myc complementation also restored the expression of other glycolysis-associated genes (Supplementary Figure S4e). Conclusively, these results suggest that PAK2 depletion affects c-Myc–PKM2 expression and compromises the HNC cell energy metabolism; however, the rescue of Warburg effect upon complementation of c-Myc in PAK2-depleted cells indicates that PAK2–c-Myc–PKM2 axis plays an important role in cancer metabolism and thereby HNC development.
Furthermore, we also investigated the restoration of tumorigenic properties upon c-Myc complementation in PAK2-depleted HNC cells. The proliferation, migration, invasion and colony formation assay were analyzed in shPAK2_Myc and shPAK2_Control cells. Interestingly, we observed rescue of proliferation in shPAK2_Myc cells as compared to shPAK2_Control cells by MTT assay (Figure 5f). Additionally, the rescue of wound healing property was observed in shPAK2_Myc cells in comparison to shPAK2_Control cells, which suggest that PAK2-mediated c-Myc expression is critical for cellular proliferation and wound healing (Fig. 5g). Subsequently, we also performed invasion assay and colony formation assay, wherein we found that shPAK2_Myc cells showed restored invasion (Fig. 5h) and colony formation (Fig. 5i, j and Supplementary Figure S4f). Conclusively, these data suggest that PAK2 promotes HNC progression via regulating c-Myc expression.
Global effect of PAK2 depletion in head and neck cancer cells
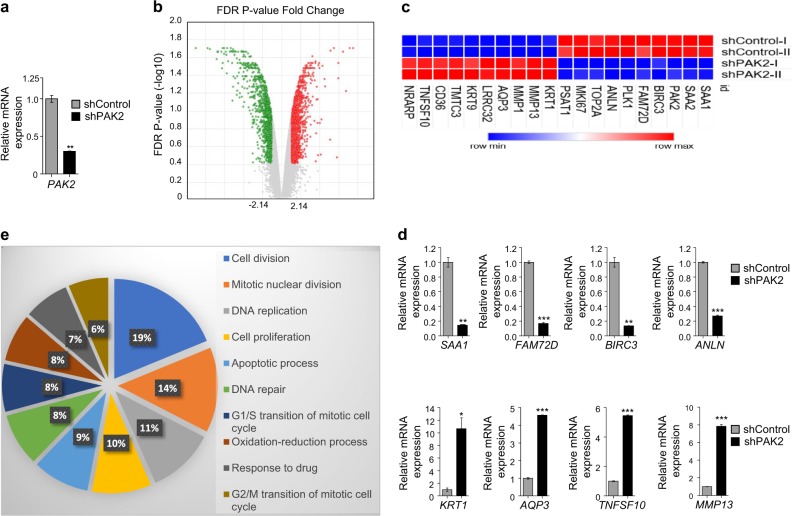
Having shown that PAK2 promotes Warburg effect via expression of proto-oncogenes (c-Myc and PKM2) and helps in HNC progression, we investigated the PAK2-mediated global changes in PAK2-depleted H157 cells (Fig. 6a) by HTA 2.0. The PAK2 depletion in HNC cells resulted in differential expression of 831 genes (Fig. 6b). Furthermore, the top 20 differentially expressed genes (10 downregulated and 10 upregulated) were selected on the basis of significant false discovery rate P value criteria, and their expression was compared in both PAK2-depleted and control cells as shown in the heat map (Fig. 6c). The expression pattern of these genes was further cross-validated by analyzing their transcript level through qRT-PCR analysis (Fig. 6d and Supplementary Figure S5a). Additionally, gene ontology (GO) analysis of differentially expressed genes upon PAK2 depletion revealed an enrichment of genes involved in cell proliferation, DNA replication, cell cycle, apoptosis, nucleosome assembly, drug metabolism and other cellular processes (Fig. 6e). Moreover, the Kyoto Encyclopedia of Gene and Genomics (KEGG) analysis of proliferation pathway showed the downregulation of proliferation-associated genes upon PAK2 depletion (Supplementary Figure S5c). This implies the possible role of PAK2 in the expression of genes involved in the development of major hallmarks of cancer. Similarly, GO analysis of upregulated genes upon PAK2 depletion resulted in the enrichment of genes involved in cell adhesion, exosome processing, cell membrane integrity, calcium ion signaling and cellular filament organization (Supplementary Figure S5c), suggesting for a role of PAK2 in diverse cellular signaling. Collectively, these results suggest that PAK2 is a critical oncogene which brings about global changes in expression of cancer-associated genes.
Fig. 6. PAK2 depletion from head and neck cancer cells globally affects the expression of proliferation genes.
The PAK2 was depleted in H157 cells and total mRNA was isolated for global analysis, human transcriptome array (HTA) 2.0. a The mRNA expression of PAK2 was analyzed by qRT-PCR upon knockdown. b Globally differentially expressed genes were analyzed through volcano plot upon PAK2 depletion. c Heat map of top 10 up- and downregulated genes in head and neck cancer cells upon PAK2 knockdown. d The mRNA expression of differentially expressed genes, serum amyloid A1 (SAA1), family with sequence similarity 72 member D (FAM72D), baculoviral IAP repeat containing 3 (BIRC3), anillin actin binding protein (ANLN), keratin 1 (KRT1), aquaporin 3 (AQP3), TNF superfamily member 10 (TNFSF10) and matrix metallopeptidase 13 (MMP13) were verified by quantitative real-time PCR and e Gene Ontology analysis of downregulated genes upon PAK2 depletion. Error bars shows mean values ± SD. Differences were considered statistically significant with *P < 0.05, **P < 0.01 and ***P < 0.001
Data deposition
The HTA 2.0 array data reported in this report have been deposited in the GEO database, www.ncbi.nlm.nih.gov/geo (accession no. GSE113322).
Discussion
Recent advancements in cancer epigenetics have shown the role of altered epigenetic events as key step in cancer initiation and progression54. The current understanding of global alterations in the epigenetic landscape during cancer initiation and progression warrant the identification of differentially expressed chromatin modifiers. Herein our study, we observed significant overexpression of a histone modifier PAK2 in HNC patients. The deregulated expression of PAK2 has been linked with different human malignancies55, and proposed to be positively associated with cellular transformation56 and proliferation29. However, the underlying molecular mechanism of PAK2-mediated oncogenesis remains poorly understood.
Our study demonstrated that the depletion of PAK2 compromised cellular proliferation. The molecular study highlighted that PAK2 depletion significantly reduced CCND1 expression (Fig. 2). Additionally, the expression of PAK2 in leukemic cells have been shown to affect invasion and angiogenesis57,58. Interestingly, the head and neck cancer showed a similar behavior of reduced migration and invasion upon PAK2 depletion, which established the fact that oncogenic activity of PAK2 is not cell type specific (Fig. 3).
Considering the fact that the PKM2 overexpression is associated with chemoresistance in various cancers59,60, the PAK2–c-Myc–PKM2 axis might be responsible for chemoresistance in HNC. Interestingly, we observed that PAK2 depletion reduced chemotherapeutic resistance in HNC cells, which further correlates with poor survival of HNC patients with higher expression of PAK2. This indicates that PAK2 might be used as a prognosis marker for HNC patients as proposed for gastric cancer31.
Furthermore, PAK2 is known to be an effector molecule of Rac1/Cdc42 signaling which is shown to be associated with activation of Wnt/ABC pathway61. We hypothesized that the PAK2 expression might be correlated with Wnt/ABC signaling and regulation of target gene expression such as c-Myc62. Notably, for the first time, we showed that the PAK2 depletion causes reduced ABC and thereby downregulation of c-Myc expression in HNC cells. The c-Myc, as an oncogene, is upregulated in various malignancies including HNC63 and c-Myc-deficient cells are unable to induce tumorigenesis64. Thus, it is likely that the regulation of c-Myc expression might have a possible role in PAK2-mediated head and neck oncogenesis.
Furthermore, the role of c-Myc in PAK2-mediated oncogenesis was validated by c-Myc complementation in PAK2-depleted HNC cells, which resulted in complete restoration of oncogenic potential as reflected by an increase in cell proliferation, invasion, and migration (Fig. 5). These results strengthen our hypothesis that the PAK2-mediated head and neck oncogenesis is dependent on the oncogenic role of c-Myc.
Additionally, recent reports have shown that c-Myc regulates the expression of genes involved in glycolysis65–67. Interestingly, for the first time, we showed that the PAK2 also regulates the expression of aerobic glycolysis-associated genes via regulation of c-Myc, wherein PAK2 depletion resulted in significantly reduced expression of glucose transporter 1 (GLUT1), lactate dehydrogenase A and B (LDHA and LDHB) and enolase 1 (ENO1). The enhanced expression of these genes is known to be positively associated with cancer progression66.
Moreover, the correlation of c-Myc expression with upregulated PKM2 in various cancers67,68 highlights a possible role of c-Myc in PKM2 expression. However, the molecular mechanism of PKM2 upregulation by c-Myc remains to be elucidated. The expression of PKM2 has been shown to promote the Warburg effect, proliferation and tumor growth69,70. Interestingly, we observed that the depletion of PAK2 showed a remarkable reduction in PKM2 expression and thereby reduced Warburg effect, which was rescued by c-Myc complementation in PAK2-deficient HNC cells. Importantly our result shows the molecular regulation of PAK2–c-Myc–PKM2 axis and upregulation of this axis enhanced the glucose uptake and cell proliferation (Fig. 7). This finding was in coherence with the earlier studies that established a positive correlation of PKM2 with cyclin D171 and Ki6772 expression. Moreover, we unravel the c-Myc-dependent regulation of PKM2 expression, wherein we identified c-Myc binding site at PKM gene promoter. The reduced enrichment of c-Myc on PKM gene promoter region in PAK2-depleted cells highlighted the role of c-Myc in PKM2 expression which was validated by c-Myc complementation, which led to increased enrichment of c-Myc on PKM promoter region and thereby increased PKM2 expression. This suggests that c-Myc-dependent PKM2 expression plays an important role in PAK2-mediated oncogenesis.
Fig. 7. Schematic model.

Elevated expression of PAK2 leads to increased activation of β-catenin and promotes the expression of Wnt/ABC target gene, c-Myc. The upregulated expression of c-Myc promotes Warburg effect via promoting PKM2 expression as well as other glycolytic genes and promotes tumorigenesis
Finally, our global transcriptome analysis showed that in addition to c-Myc and PKM2, PAK2 also regulates the expression of several genes, having an important role in cell proliferation, DNA repair, apoptosis and cellular transformation. The positive correlation of proliferation-associated genes with PAK2 emphasizes the importance of PAK2 in HNC growth and progression.
The present study highlights the undiscovered role of PAK2 in head and neck oncogenesis, wherein we showed that the elevated expression of PAK2 promotes head and neck cancer growth and provides chemotherapeutic resistance. Further molecular analysis identified that PAK2 upregulates c-Myc and promotes c-Myc-dependent PKM2 expression. Conclusively, we showed that the PAK2–c-Myc–PKM2 axis is critical for HNC progression and may provide an alternative strategy for multiple drug targets.
Electronic supplementary material
Acknowledgements
We thank all members of the Epigenetics and RNA Processing Lab for their helpful discussions and technical assistance. This work was supported by Wellcome Trust/Department of Biotechnology (DBT) India Alliance Fellowship grant IA/I/16/2/502719 and Board of Research in Nuclear Sciences (BRNS) (37(1)/14/30/2016-BRNS). A.G. and S.Singh were supported by a fellowship from IISER Bhopal. A.A. was supported by a fellowship from Department of Science & Technology (DST).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Finazzi-Agró
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-0887-0).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Massano J, Regateiro FS, Januario G, Ferreira A. Oral squamous cell carcinoma: review of prognostic and predictive factors. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2006;102:67–76. doi: 10.1016/j.tripleo.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 2.Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat. Rev. Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, et al. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J. Clin. Oncol. 2013;31:4550–4559. doi: 10.1200/JCO.2013.50.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gupta B, Johnson NW. Oral cancer: Indian pandemic. Br. Dent. J. 2017;222:497. doi: 10.1038/sj.bdj.2017.293. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, et al. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 6.Smith RB, Sniezek JC, Weed DT, Wax MK. Utilization of free tissue transfer in head and neck surgery. Otolaryngol. Head Neck Surg. 2007;137:182–191. doi: 10.1016/j.otohns.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Vergeer MR, et al. Intensity-modulated radiotherapy reduces radiation-induced morbidity and improves health-related quality of life: results of a nonrandomized prospective study using a standardized follow-up program. Int. J. Radiat. Oncol. Biol. Phys. 2009;74:1–8. doi: 10.1016/j.ijrobp.2008.07.059. [DOI] [PubMed] [Google Scholar]
- 8.Kuriakose MA, Trivedi NP. Sentinel node biopsy in head and neck squamous cell carcinoma. Curr. Opin. Otolaryngol. Head Neck Surg. 2009;17:100–110. doi: 10.1097/MOO.0b013e3283293631. [DOI] [PubMed] [Google Scholar]
- 9.Narayanan SP, et al. Integrated genomic analyses identify KDM1A’s role in cell proliferation via modulating E2F signaling activity and associate with poor clinical outcome in oral cancer. Cancer Lett. 2015;367:162–172. doi: 10.1016/j.canlet.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 11.Chi P, Allis CD, Wang GG. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 13.Elsheikh SE, et al. Global histone modifications in breast cancer correlate with tumor phenotypes, prognostic factors, and patient outcome. Cancer Res. 2009;69:3802–3809. doi: 10.1158/0008-5472.CAN-08-3907. [DOI] [PubMed] [Google Scholar]
- 14.Al Emran A, et al. Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget. 2018;9:8206–8222. doi: 10.18632/oncotarget.23654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seligson DB, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 16.Manuyakorn A, et al. Cellular histone modification patterns predict prognosis and treatment response in resectable pancreatic adenocarcinoma: results from RTOG 9704. J. Clin. Oncol. 2010;28:1358–1365. doi: 10.1200/JCO.2009.24.5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hake SB, Xiao A, Allis CD. Linking the epigenetic ‘language’ of covalent histone modifications to cancer. Br. J. Cancer. 2007;96(Suppl.):R31–R39. [PubMed] [Google Scholar]
- 18.Nakagawa M, et al. Expression profile of class I histone deacetylases in human cancer tissues. Oncol. Rep. 2007;18:769–774. [PubMed] [Google Scholar]
- 19.Kawai H, Li H, Avraham S, Jiang S, Avraham HK. Overexpression of histone deacetylase HDAC1 modulates breast cancer progression by negative regulation of estrogen receptor alpha. Int. J. Cancer. 2003;107:353–358. doi: 10.1002/ijc.11403. [DOI] [PubMed] [Google Scholar]
- 20.Varambally S, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 21.Khare SP, et al. HIstome–a relational knowledgebase of human histone proteins and histone modifying enzymes. Nucleic Acids Res. 2012;40:D337–D342. doi: 10.1093/nar/gkr1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- 23.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat. Rev. Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 24.Molli PR, Li DQ, Murray BW, Rayala SK, Kumar R. PAK signaling in oncogenesis. Oncogene. 2009;28:2545–2555. doi: 10.1038/onc.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whale A, Hashim FN, Fram S, Jones GE, Wells CM. Signalling to cancer cell invasion through PAK family kinases. Front. Biosci. (Landmark Ed.) 2011;16:849–864. doi: 10.2741/3724. [DOI] [PubMed] [Google Scholar]
- 26.Kang B, et al. Phosphorylation of H4 Ser 47 promotes HIRA-mediated nucleosome assembly. Genes Dev. 2011;25:1359–1364. doi: 10.1101/gad.2055511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavillet M, Martinod K, Renella R, Wagner DD, Williams DA. A key role for Rac and Pak signaling in neutrophil extracellular traps (NETs) formation defines a new potential therapeutic target. Am. J. Hematol. 2018;93:269–276. doi: 10.1002/ajh.24970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Wang Z, Zhang Z. PP1alpha, PP1beta and Wip-1 regulate H4S47 phosphorylation and deposition of histone H3 variant H3.3. Nucleic Acids Res. 2013;41:8085–8093. doi: 10.1093/nar/gkt583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng WW, et al. PAK2 promotes migration and proliferation of salivary gland adenoid cystic carcinoma. Am. J. Transl. Res. 2016;8:3387–3397. [PMC free article] [PubMed] [Google Scholar]
- 30.Siu MK, et al. Differential expression and phosphorylation of Pak1 and Pak2 in ovarian cancer: effects on prognosis and cell invasion. Int. J. Cancer. 2010;127:21–31. doi: 10.1002/ijc.25005. [DOI] [PubMed] [Google Scholar]
- 31.Gao C, Ma T, Pang L, Xie R. Activation of P21-activated protein kinase 2 is an independent prognostic predictor for patients with gastric cancer. Diagn. Pathol. 2014;9:55. doi: 10.1186/1746-1596-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett T, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumari P, et al. Essential role of HCMV deubiquitinase in promoting oncogenesis by targeting anti-viral innate immune signaling pathways. Cell Death Dis. 2017;8:e3078. doi: 10.1038/cddis.2017.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh S, et al. Intragenic DNA methylation and BORIS-mediated cancer-specific splicing contribute to the Warburg effect. Proc. Natl. Acad. Sci. USA. 2017;114:11440–11445. doi: 10.1073/pnas.1708447114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rhodes DR, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sengupta S, et al. Genome-wide expression profiling reveals EBV-associated inhibition of MHC class I expression in nasopharyngeal carcinoma. Cancer Res. 2006;66:7999–8006. doi: 10.1158/0008-5472.CAN-05-4399. [DOI] [PubMed] [Google Scholar]
- 39.Cromer A, et al. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23:2484–2498. doi: 10.1038/sj.onc.1207345. [DOI] [PubMed] [Google Scholar]
- 40.Toruner GA, et al. Association between gene expression profile and tumor invasion in oral squamous cell carcinoma. Cancer Genet. Cytogenet. 2004;154:27–35. doi: 10.1016/j.cancergencyto.2004.01.026. [DOI] [PubMed] [Google Scholar]
- 41.Resnitzky D, Reed SI. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol. Cell Biol. 1995;15:3463–3469. doi: 10.1128/MCB.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rafehi H, et al. Clonogenic assay: adherent cells. J. Vis. Exp. 2011;49:pii: 2573. doi: 10.3791/2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marlin JW, Eaton A, Montano GT, Chang YW, Jakobi R. Elevated p21-activated kinase 2 activity results in anchorage-independent growth and resistance to anticancer drug-induced cell death. Neoplasia. 2009;11:286–297. doi: 10.1593/neo.81446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu X, et al. Rac1 activation controls nuclear localization of beta-catenin during canonical Wnt signaling. Cell. 2008;133:340–353. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou L, et al. Merlin-deficient human tumors show loss of contact inhibition and activation of Wnt/beta-catenin signaling linked to the PDGFR/Src and Rac/PAK pathways. Neoplasia. 2011;13:1101–1112. doi: 10.1593/neo.111060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Wetering M, et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241–250. doi: 10.1016/S0092-8674(02)01014-0. [DOI] [PubMed] [Google Scholar]
- 49.Pai R, et al. Over-expression of c-Myc oncoprotein in oral squamous cell carcinoma in the South Indian population. Ecancermedicalscience. 2009;3:128. doi: 10.3332/ecancer.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolfer A, Ramaswamy S. MYC and metastasis. Cancer Res. 2011;71:2034–2037. doi: 10.1158/0008-5472.CAN-10-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller DM, Thomas SD, Islam A, Muench D, Sedoris K. c-Myc and cancer metabolism. Clin. Cancer Res. 2012;18:5546–5553. doi: 10.1158/1078-0432.CCR-12-0977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lv C, et al. The function of BTG3 in colorectal cancer cells and its possible signaling pathway. J. Cancer Res. Clin. Oncol. 2018;144:295–308. doi: 10.1007/s00432-017-2561-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li T, et al. P21-activated protein kinase (PAK2)-mediated c-Jun phosphorylation at 5 threonine sites promotes cell transformation. Carcinogenesis. 2011;32:659–666. doi: 10.1093/carcin/bgq271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gopal SK, et al. Oncogenic epithelial cell-derived exosomes containing Rac1 and PAK2 induce angiogenesis in recipient endothelial cells. Oncotarget. 2016;7:19709–19722. doi: 10.18632/oncotarget.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edlinger L, et al. Expansion of BCR/ABL1(+) cells requires PAK2 but not PAK1. Br. J. Haematol. 2017;179:229–241. doi: 10.1111/bjh.14833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu WQ, Hu YY, Lin XP, Fan W. Knockdown of PKM2 and GLS1 expression can significantly reverse oxaliplatin-resistance in colorectal cancer cells. Oncotarget. 2017;8:44171–44185. doi: 10.18632/oncotarget.17396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Zhang F, Wu XR. Inhibition of pyruvate kinase M2 markedly reduces chemoresistance of advanced bladder cancer to cisplatin. Sci. Rep. 2017;7:45983. doi: 10.1038/srep45983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Esufali S, Bapat B. Cross-talk between Rac1 GTPase and dysregulated Wnt signaling pathway leads to cellular redistribution of beta-catenin and TCF/LEF-mediated transcriptional activation. Oncogene. 2004;23:8260–8271. doi: 10.1038/sj.onc.1208007. [DOI] [PubMed] [Google Scholar]
- 62.Masuda M, Sawa M, Yamada T. Therapeutic targets in the Wnt signaling pathway: feasibility of targeting TNIK in colorectal cancer. Pharmacol. Ther. 2015;156:1–9. doi: 10.1016/j.pharmthera.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 63.Chuerduangphui J, et al. Effects of arecoline on proliferation of oral squamous cell carcinoma cells by dysregulating c-Myc and miR-22, directly targeting oncostatin M. PLoS One. 2018;13:e0192009. doi: 10.1371/journal.pone.0192009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baudino TA, et al. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16:2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aswini P, Grace Raji R, Haritha K, Lincy E, Sameer Kumar V. ER stress mediated regulation of miR23a confer Hela cells better adaptability to utilize glycolytic pathway. J. Cell Biochem. 2018;119:4907–4917. doi: 10.1002/jcb.26718. [DOI] [PubMed] [Google Scholar]
- 66.Kim EJ, Kim SH, Jin X, Kim H. KCTD2, an adaptor of Cullin3 E3 ubiquitin ligase, suppresses gliomagenesis by destabilizing c-Myc. Cell Death Differ. 2017;24:649–659. doi: 10.1038/cdd.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim JW, et al. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol. Cell Biol. 2004;24:5923–5936. doi: 10.1128/MCB.24.13.5923-5936.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mendez-Lucas A, et al. Glucose catabolism in liver tumors induced by c-MYC can be sustained by various PKM1/PKM2 ratios and pyruvate kinase activities. Cancer Res. 2017;77:4355–4364. doi: 10.1158/0008-5472.CAN-17-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- 70.Jiang Y, et al. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol. Cell. 2014;53:75–87. doi: 10.1016/j.molcel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang W, Lu Z. Nuclear PKM2 regulates the Warburg effect. Cell Cycle. 2013;12:3154–3158. doi: 10.4161/cc.26182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kitamura K, et al. Proliferative activity in hepatocellular carcinoma is closely correlated with glucose metabolism but not angiogenesis. J. Hepatol. 2011;55:846–857. doi: 10.1016/j.jhep.2011.01.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.