Abstract
Background/Aim: Combined hepatocellular-cholangiocarcinoma (cHCC-CC) is a very rare type of tumor, comprising these two different components in a single mass. Although several studies have determined the genetic characteristics of cHCC-CC, next-generation sequencing (NGS) data for comparing clonality of cHCC-CC are currently unavailable. Materials and Methods: Four cHCC-CC cases were selected and HCC, CC and normal components from each case were separately micro-dissected. DNA and RNA were isolated from each sample and sequenced by Oncomine Comprehensive Panel interrogating 143 cancer genes using the Ion S5 XL sequence platform. Genetic features of HCC and CC from each patient were compared. Results: All cases successfully produced NGS data. Two cases demonstrated different mutations in their HCC and CC components (biclone), while two cases shared the same mutations in the two components (monoclone). Single nucleotide polymorphisms (SNPs) of TP53 (4/4) and phosphatase and tensin homolog (PTEN) (1/4), and gene amplifications of mesenchymal–epithelial transition factor (MET) (1/4), c-MYC (1/4), and cyclin-dependent kinase 6 (CDK6) (1/4) were found in the CC component. In the HCC component, SNPs of TP53 (3/4), PTEN (1/4) and catenin beta 1 (CTNNB1) (1/4) and cyclin D1 (CCND1) amplification (1/4) were detected. Two biclonal cases showed a histologically distinct border between HCC and CC components with or without intermediate cell foci. Two monoclonal cases showed a histologically ambiguous border between HCC and CC components with more intermingled pattern than biclonal cases. Conclusion: Based on our study, cHCC-CC can be genetically divided into biclonal and monoclonal forms. Therefore, separate sequencing of each component of cHCC-CC is recommended for exact molecular classification and targeted therapy.
Keywords: Hepatocellular, cholangiocarcinoma, massively parallel sequencing, clones
Combined hepatocellular-cholangiocarcinoma (cHCC-CC) contains two different cancer types [hepatocellular carcinoma (HCC) and cholangiocarcinoma (CC)] in the same liver. It is a rare tumor with frequency of less than 1% of all liver carcinomas (1). cHCC-CC is often associated with clinical factors that are more related to ordinary HCC than to CC, such as hepatitis B (HBV) or C (HCV) virus infection and liver cirrhosis (1,2). cHCC-CC is known to have more aggressive behavior with poorer prognosis than ordinary HCC (1,2).
In 1949, Allen and Lisa classified cHCC-CC into three subtypes: Type A, comprising separate masses constituting either HCC or CC (double cancer); type B, with contiguous but distinguished masses of HCC and CC (combined type); and type C, an intimate intermixed mass of HCC and CC elements (mixed type) (3,4). Types A and B seem to have different epicenters of HCC and CC, while the component of type C share a common center. In 1980, another classification was proposed for cHCC-CC, Goodman’s classification: Type I, ‘collision tumors’ that contain separate and distinct areas of HCC and CC; type II, ‘transitional tumors’ that exhibit both components with areas of transition from HCC to CC elements; and type III, ‘fibrolamellar tumors’ that exhibit a mixture of HCC and CC throughout the tumor, representing fibrolamellar type HCC (3,5).
Goodman’s type I tumor is known to correspond to Allen and Lisa’s type A and type B3. Allen and Lisa’s type C corresponds Goodman’s type II. Lisa and Allen’s type C and Goodman’s type II most closely fit the current WHO criteria that define cHCC-CC as intimately mixed elements of HCC and CC components with identification of transitional or intermediate areas (1).
Although several studies have determined genetic characteristics of cHCC-CC (7,8), molecular pathogenesis of cHCC-CC remains unclear. Using loss of heterozygosity (LOH) study, Fujii et al. have discovered clonality of cHCC-CC consistent with Allen’s classification (2). However, next-generation sequencing (NGS) data for clonality of cHCC-CC are currently unavailable. Therefore, the objective of this study was to investigate the clonal relationship of HCC and CC components in cHCC-CC based on genetic features using AmpliSeq-based targeted sequencing, Oncomine Comprehensive Assay v1 panel (OCP). The OCP encompasses 143 cancer genes designed to translate somatic mutations, copy number alterations, and gene fusions in solid tumors according to their potential clinical relevance (9,10).
Materials and Methods
Patients and tissue sample. Four cases of HCC-CC were obtained from files of the Department of Pathology, Incheon St. Mary’s Hospital, The Catholic University of Korea. Medical records of these four patients were carefully reviewed. Survival time was defined as the time period between the date of the diagnosis and the date of death or the last follow-up of the patient. Hematoxylin-and eosin-stained sections obtained at the time of diagnosis were reviewed. CC components were identified by definite glandular formation by biliary epithelial cells. HCC components were identified by bile production, trabecular growth pattern, and eosinophilic cytoplasm (6). To exactly define the two components, immunohistochemical staining was performed. Cytokeratin 7 (CK7) (Dako, Glostrup, Denmark), CK19 (Thermo Fisher Scientific, Cheshire, UK) and glypican 3 (Cell Margue, Rocklin, CA, USA) were stained according to manufacturer’s instructions. Staining for CK7 and CK19 was diffusely strongly positive in the CC component, while that for glypican 3 was focally or diffuse positive for the HCC component. Three cHCC-CC cases (cases 1-3) with transitional or intermediate component were type C and one case (case 4) without intermediate component was type B according to Allen and Lisa’s classification. Biopsy sample obtained from recurrent tumor of case 1 and excisional sample obtained from metastatic HCC of case 4 were added for NGS analysis.
Ethics statement. This study was approved by the Institutional Review Boards of Incheon St. Mary Hospital (ID: OC18TESI0052).
DNA and RNA extraction. DNA and RNA were isolated from 10-μm sections of tumor tissue samples using a sterile 26-gauge needle and RecoverAll™ Multi-Sample RNA/DNA Isolation Workflow (Ambion, Austin, TX, USA) according to the manufacturer’s instructions. From four neoplasms, each HCC and CC component was obtained by manual microdissection and subjected to extraction of DNA and RNA for library preparations. Normal control tissue of each case was also dissected from adjacent nonmalignant tissue. DNA and RNA were quantified using Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA).
Library preparation. DNA libraries were prepared using published procedures (9). These DNA libraries were generated from 20 ng of DNA per sample using Ion AmpliSeq Library Kit 2.0 (Thermo Fisher Scientific) and the Oncomine Comprehensive Assay v1 panel (OCP) (Thermo Fisher Scientific) according to the manufacturer’s instructions with barcode incorporation (9).
RNA libraries were generated from 15 ng of RNA per sample using Ion AmpliSeq RNA Library Kit (Thermo Fisher Scientific). OCP AmpliSeq libraries were quantified using Ion Library Universal Quantification Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions (9). The OCP included a total of 143 genes, of which 73 oncogenes were interrogated for mutational hotspots and 26 tumor-suppressor genes were interrogated for all exons. In addition, the OCP provides the capability to detect copy number variations (CNV) in 49 genes and fusion drivers in 22 genes. Gene list of OCP v1 are available at https://www.thermofisher.com/kr/ ko/home/clinical/preclinical-companion-diagnostic-development/ oncomine-oncology/oncomine-cancer-research-panel-workflow.html.
Template generation and sequencing. A 60 pmol/l pool of DNA and RNA libraries at a 4:1 ratio (DNA to RNA) was used to prepare templated ion sphere particles with Ion 540 Chef kit (Thermo Fisher Scientific) and Ion S5™ Chef system (Thermo Fisher Scientific). Procedures followed the manufacturer’s instructions. Sequencing was performed using Ion S5 Sequencer and Ion 540 chips (Thermo Fisher Scientific) according to the manufacturer’s instructions. Sequencing data of approximately 200 bp reads were generated after 500 flow runs.
Bioinformatics and data analysis pipeline. Sequencing data analysis was performed using Torrent Suite version 5.2.2 and Ion Reporter version 5.2 (Thermo Fisher Scientific). This workflow was created by adding custom hotspots Browser Extensible Data (BED) file to report mutations of interest (MOIs) and a custom CNV baseline (described in the next paragraph) using the manufacturer’s default workflow as previously described (9). The pipeline included signaling processing, base calling, quality score assignment, adapter trimming, read mapping to 19 reference human genomes, quality control of mapping quality, coverage analysis with down-sampling, and variant calling. Identification of variants was performed using Ion Torrent Variant Caller plug-in (v5.2.2.41) and Ion Reporter software v5.2 (Thermo Fisher Scientific). Coverage maps were generated using coverage analysis plug-in (v5.2.1.2) (Thermo Fisher Scientific). Additionally, ANNOVAR software (http://www.openbioinformatics.org/annovar/) was used for functional annotation of identified single nucleotide polymorphisms (SNPs) to investigate their genomic locations and variation in information (11). To eliminate error artifacts, sequence data were visually confirmed with Integrative Genomics Viewer. This workflow was able to report SNPs and indels as low as 1% variant allele fraction. Based on results of a feasibility study, the variant allele fraction threshold was established at 5%. Copy number analysis was performed using the copy number module within the aforementioned workflow of the Ion Reporter system. Copy numbers of four or greater were considered concordant if the orthogonal assay also reported a copy number of four or greater for target genes. Fusions were detected using the fusion detection module within the Ion Reporter workflow. This pipeline only reported previously annotated fusions as defined in a reference file that was preloaded into the workflow (9). Alteration Prioritization and Potential Actionability Assessment Somatic SNVs/indels passing filtering in a gain-of-function (GoF) gene were considered GoF when they occurred at the predefined hotspot residue targeted by OCP. Somatic variants in a loss-of-function (LoF) gene were considered LoF when deleterious (nonsense or frame shifting) occurred at a predefined hotspot residue. Somatic copy number alterations were considered for potential actionability analysis when they were concordant with predicted alteration (amplification or deletion) from Oncomine analysis as described above. Somatic gene fusions were considered for actionability analysis when they represented known gene fusions from Mitelman database (National Cancer Institute, Bethesda, MD, USA) or Oncomine analysis or involved known 3’ or 5’ drivers with novel partners. These prioritized variants were then associated with potential actionability using the Oncomine database. For each patient, the “most actionable” alteration was identified based on the following criteria: i) Variants referenced in Food and Drug Administration (FDA) drug labels, ii) variants referenced in National Comprehensive Cancer Network (NCCN) treatment guidelines for the patient’s cancer type, iii) variants referenced in an NCCN guideline for another cancer type, and iv) variants referenced as inclusion criteria in a clinical trial. Actionable variants were identified by manual curation of FDA labels and NCCN guidelines. They were also identified by keyword searches and manual curation of clinical trial records in the TrialTrove database (9).
Results
Clinical information. The clinical information provided below for each case is summarized in Table I.
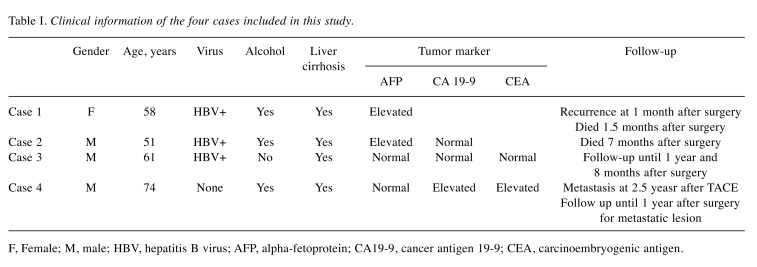
Table I. Clinical information of the four cases included in this study.
F, Female; M, male; HBV, hepatitis B virus; AFP, alpha-fetoprotein; CA19-9, cancer antigen 19-9; CEA, carcinoembryogenic antigen.
Case 1. A 58-year-old female presented with complaint of bloated abdomen for 1 month. She was an alcoholic (3.5 bottles of beer daily) and was also a HBV carrier. A large amount of ascites and liver cirrhosis with splenomegaly were noted on computed tomography (CT). The patient had a high titer of viral marker (HBV DNA=2,074,699 IU/mI). Control of ascites and anti-viral treatment were offered. However, tumor marker level [alpha-fetoprotein (AFP)=16 ng/ml] remained elevated. After 3 months, HCC was suspected on follow-up CT. Despite undergoing transarterial chemoembolization (TACE) twice, there was still viable tumor. Living donor liver transplantation was performed. Multiple small hepatic nodules with lymphadenopathy were noted on postoperative CT at 1 month. The patient died of multiorgan dysfunction on postoperative day 50 while under home care.
Case 2. A 51-year-old male was transferred to our hospital due to suspected HCC based on outside CT findings. He complained of abdominal pain of 2 months’ duration. He was also an alcoholic (2 bottles of raw rice wine daily) and was a HBV carrier. The patient received no anti-viral treatment. A high titer of viral marker (HBV DNA=215,523 IU/ml) and high tumor marker level (AFP =14 ng/ml) were observed. Segmentectomy VI was performed. On postoperative day 75, the patient visited a local hospital for abdominal pain. CT revealed innumerous low-density nodules scattered on both hemilivers. He received gemcitabine and paclitaxel chemotherapy. However, carcinomatosis perionei was found on follow-up CT. Although another regimen of chemotherapy with capecitabine and cisplatin was applied, it increased the amount of ascites. Multiple lymphadenopathy was observed on CT. The patient died 7 months post-operatively while under hospice care.
Case 3. A 61-year-old man was referred to our hospital for a hepatic mass found on screening abdominal ultrasound. He had been diagnosed with hepatitis B 20 years earlier and diabetes 3 years earlier. Radiographic images disclosed 3.5-cm sized hepatic nodule at segment VI. He showed elevated viral marker (HBV DNA=9,213 IU/ml). The AFP level (2 ng/ml) was within normal range. He started antiviral medication and underwent segmentectomy. Several small enhancing and washout nodules suggestive of local tumor recurrence were found on CT 3 months postoperatively. Multiple small HCCs in both hepatic lobes up to 1.2 cm in size were observed on magnetic resonance imaging at 6 months postoperatively. After four administrations of TACE, follow-up MRI at 1 year and 2 months postoperatively showed mixed response. The patient was scheduled to undergo liver transplantation at another hospital, but was lost to follow-up.
Case 4. A 74-year-old man visited the Emergency Room (ER) for sudden burst epigastric pain. Severe tenderness on the upper abdominal quadrant was noted on physical examination. He had hypertension and diabetes and drank half a bottle of alcohol every day. Tumor markers [AFP=7 ng/ml; cancer antigen 19-9 (CA19-9) 575 U/ml; carcinoembryogenic antigen (CEA)=6.5 ng/ml] were identified. The level of CA19-9 and CEA were elevated and AFP was within normal range. He had no HBV or HCV antibodies. Contrast-enhanced CT images revealed 3.5-cm sized HCC with focal rupture and active bleeding into the abdominal cavity. He was transferred to a sanitarium after receiving emergent TACE and intensive care for 2 weeks. Two and a half years later, he revisited the ER with abdominal pain of 3 days’ duration. Fluid shifting was found on physical examination. CT revealed large increase in the size of HCC (3.5 cm to 13 cm), a large amount of ascites that were newly developed, and multiple omental seeding nodules. Right hepatectomy with radiofrequency ablation and omentectomy were performed. A month later, the patient visited the ER again for abdominal distension. He underwent excision of omental nodules. Metastatectomy at the abdominal wall, retroperitoneum, and right diaphragm was carried out 6 months later. Nevertheless, follow-up CT revealed progressive disease. The patient was on Outpatient Department follow-up for 1 year.
Results of NGS data. All cases successfully produced NGS data. Results of targeted sequencing are summarized in Table II. Data quality is summarized in Table III.
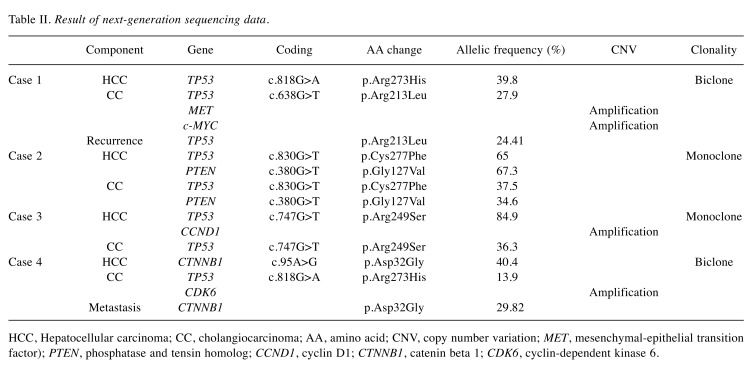
Table II. Result of next-generation sequencing data.
HCC, Hepatocellular carcinoma; CC, cholangiocarcinoma; AA, amino acid; CNV, copy number variation; MET, mesenchymal-epithelial transition factor); PTEN, phosphatase and tensin homolog; CCND1, cyclin D1; CTNNB1, catenin beta 1; CDK6, cyclin-dependent kinase 6.
Table III. Data quality of next-generation sequencing.
HCC, Hepatocellular carcinoma; CC, cholangiocarcinoma.
Case 1 demonstrated different mutation pattern in the two components. Different TP53 substitutions were found for each component (p.Arg273His, 39.8% in HCC, p.Arg213Leu, 27.9% in CC). Additionally, mesenchymal–epithelial transition factor (MET) and c-MYC amplifications were observed in CC. We further sequenced a biopsy sample of recurrent tumor that was histologically shown to be CC, identifying the same TP53 mutation of the CC component of the primary tumor (p.Arg213Leu, 24.41%).
In case 2, both components had the same TP53 mutation (p.Cys277Phe, 65% in HCC, 37.5% in CC) and phosphatase and tensin homolog (PTEN) mutation (p.Gly127Val, 67.3% in HCC, 34.6% in CC) with different frequencies.
In case 3, the same TP53 mutation with different frequency (p.Arg249Ser, 84.9% in HCC, 36.3% in CC) was shared by both components. Additionally, the HCC component had cyclin D1 (CCND1) amplification.
Case 4 demonstrated different mutation patterns for the two components. The HCC component had catenin beta 1 (CTNNB1) mutation (p.Asp32Gly, 40.4%), while the CC component had TP53 mutation (p.Arg273His, 13.9%) and cyclin-dependent kinase 6 (CDK6) amplification. Excisional tissues of metastatic HCC in the abdominal subcutis were also sequenced and the same CTNNB1 mutation (p.Asp32Gly, 29.82%) was detected.
No significant mutation was identified in normal tissue. No fusion was detected in any case.
Discussion
NGS offers the opportunity to develop precision medicine by identifying candidate actionable mutations (12). OCP is a targeted multiplexed polymerase chain reaction - based NGS assay and analytical system for identifying predefined somatic solid tumor genome variants (9).
We investigated SNP/Indel, CNV, and RNA fusion of cHCC-CC using the OCP analytical system and compared genomic compositions of HCC and CC components by microdissection. Two out of the four cHCC-CC cases showed a biclonal pattern while the other two cases showed a monoclonal pattern. In addition, the two biclonal cases demonstrated the same mutations of a clone from the primary tumor in their metastasis and recurrence samples. The genetic composition of each component of cHCC-CC had common mutations previously detected in intrahepatic cholangiocarcinoma (iCC) and HCC. According to NGS data, mutations of KRAS, v-raf murine sarcoma viral oncogene homolog B1 (BRAF), epidermal growth factor receptor (EGFR), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), PTEN and TP53 have been repeatedly identified in iCC (13-18). In our study, all CC components of cHCC-CC had TP53 substitutions (4/4, 100%) usually found in ordinary iCC with frequency of 2.5-36% (13-17). Mutation of TP53 gene is also frequently found in cHCC-CC, at 26% (7). In HCC, the most common somatic alterations are mutations of the TERT (telomerase reverse transcriptase) promoter or focal amplifications (19,20). Following TERT mutation, the most frequent somatic mutations are those of TP53 and CTNNB1 (19-24). In our study, three cases had TP53 mutations (3/4, 75%) and one case had CTNNB1 mutation (1/4, 25%) in HCC component. CTNNB1 mutation has been frequently found in HCC (25,26), but not in cHCC-CC or CC (7,27). TERT promoter mutation was not found in our study because the hotspot region of SNP/Indel on TERT gene was not included in the OCP assay. Although the OCP assay included examination of TERT CNV, no TERT amplification was detected.
One case had PTEN substitution in both components. PTEN SNP/Indel is frequently reported in iCC (frequency at 0.6-11%) (13,15,16,18). PTEN deletion or loss of expression is frequently reported in HCC (28). However, somatic mutation of PTEN in HCC is relatively rare. Wang et al. sequenced PTEN for 56 HCC samples. However, mutation was only found in introns, not exons (29).
Amplifications or deletions associated with MET, FGF19 (fibroblast growth factor 19), CKD6, CCND1, CDKN2A, and CDKN2B in iCC based on NGS have been reported in various studies (12,14,15,30). In the present study, three out of the four cases had various gene amplifications, including of MET, c-MYC and CDK6 in the CC component and CCND1 in the HCC component. The CC component of one cHCC-CC case had both MET and MYC amplification. MET amplification is associated with poor clinical outcome in iCC (14,15). c-MYC amplification was found in one out of 10 CC cases and one of three cHCC-CC cases in a published study (31). One CC component had CDK6 amplification in the present study. CDK6 amplification has also been detected in some iCC cases previously based on NGS data (1/75, 2/28) (14,15). The HCC component in one case showed amplification of CCND1 gene in the present study. CCND1 amplification has been reported in HCC cases at a frequency of 4% (24,28).
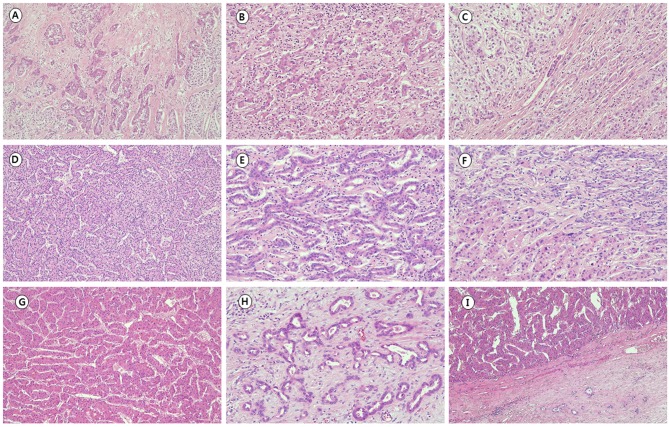
While the cell origin of cHCC-CC remains controversial, genetic and molecular analysis have suggested some possible categories. Using microdissection and DNA extraction from 11 cases, Fujii et al. classified cHCC-CC into three genetic subtypes based on LOH patterns: (i) a collision tumor of two independent clones (3/11), (ii) a single clonal tumor that is homogeneous (4/11), and (iii) single clonal with clonal process (4/11). In their study, two collision type tumors were histologically combined type (type B) according to Allen’s classification. All mixed type (type C) tumors showed single clonal pattern with or without clonal process. According to Allen’s classification, case 4 with main tumor of HCC contiguous with small CC portion without intermediate component would be combined type (type B), which is now considered a collision tumor (Figure 1G-I). Cases 2 and 3 were mixed type (type C), with the HCC component making up the majority, while the CC comprised a minor proportion in both cases (Figure 1D-F). Case 1 had histologically defined HCC and CC components in single tumor with small intermediate foci, corresponding to mixed type (type C) (Figure 1A-C). Histologically, HCC and CC components were well distinguished in two biclonal cases (cases 1 and 4). HCC and CC components were more intermingled and less distinguished in two monoclonal cases (cases 2 and 3).
Figure 1. Histopathological findings of combined hepatocellular-cholangiocarcinoma (cHCC-CC). Case 1 showed histologically defined hepatocellular carcinoma (HCC) (A) and cholangiocarcinoma (CC) (B) components in single tumor with small intermediate foci (C). The major component of case 3 was HCC (D) and the CC (E) the minor component, showing a more intermingled pattern between the two components (F) than case 1. Case 4 dominantly showed HCC (G) contiguous with a small proportion of CC (H) without an intermediate component (I). Hematoxylin and eosin stain. Original magnification: ×100.
Based on our study, cHCC-CC can be genetically divided into biclonal and monoclonal forms. Strictly speaking, a biclonal tumor can be considered as a collision tumor. However, it is difficult to determine the histological type when two components are intermixed in a single tumor. Fujii et al. suggested single original clonal proliferation of cHCC-CC (2). In their study, all histologically mixed type cHCC-CCs were monoclonal. However, in our cases, genetic types did not completely match histologicaI types. One of the three mixed-type cHCC-CCs had a biclonal pattern. This means that some histological classifications have limitations in clarifying the genetic nature of the tumor.
Genetic and molecular studies have suggested that cHCC-CC is genetically closer to CC than to HCC (7,8). Our results demonstrated that each component had common mutations of ordinary HCC and iCC in genetically separated tumors (cases 1 and 4). In monoclonal cases (cases 2 and 3), the frequency of the same TP53 or PTEN mutation in the CC component was half that of the HCC component. In addition, the HCC component of case 3 demonstrated amplification which was not seen in the CC component of the same case. These findings suggest that the subclone of the CC component might have originated from the clone of the HCC component, which was histologically the main component in both monoclonal cases. Metastatic cancer of case 4 and recurrent tumor of case 1 demonstrated the same mutation pattern as the histologically dominant component of primary cHCC-CC. This finding suggests that cHCC-CC can progress by clonal selection and expansion based on the evolutionary carcinogenesis model (32).
In conclusion, to our knowledge, this is the first genetic study on clonality of cHCC-CC using targeted sequencing. Two out of four cases had different clones in their HCC and CC components, with more distinct histological features than monoclonal cases. Intratumor clonal heterogeneity of cancer makes targeted therapy even more complex. Sequencing of multiple tumor sites might be the best approach to resolving this problem (12). For exact molecular classification and discovering molecular targets, separate sequencing for each component of cHCC-CC is recommended, especially for cases showing histologically distinct HCC and CC components, regardless of the presence of an intermediate component.
Acknowledgements
This research was supported by a Grant of Translational R&D Project through Institute for Bio-Medical convergence, Incheon St. Mary’s Hospital, The Catholic University of Korea.
References
- 1.Bosman FT, Carneiro F, Hruban RH, Theise ND. Combined hepatocellular-cholangiocarcinoma In WHO Classification of Tumours of The Digestive System. World Health, Geneva, Switzerland. 2010:pp. 225–227. [Google Scholar]
- 2.Fujii H, Zhu XG, Matsumoto T, Inagaki M, Tokusashi Y, Miyokawa N, Fukusato T, Uekusa T, Takagaki T, Kadowaki N, Shirai T. Genetic classification of combined hepatocellular-cholangiocarcinoma. Hum Pathol. 2000;31:1011–1017. doi: 10.1053/hupa.2000.9782. [DOI] [PubMed] [Google Scholar]
- 3.Yano Y, Yamamoto J, Kosuge T, Sakamoto Y, Yamasaki S, Shimada K, Ojima H, Sakamoto M, Takayama T, Makuuchi M. Combined hepatocellular and cholangiocarcinoma: a clinicopathologic study of 26 resected cases. Jpn J Clin Oncol. 2003;33:283–287. doi: 10.1093/jjco/hyg056. [DOI] [PubMed] [Google Scholar]
- 4.Allen RA, Lisa JR. Combined liver cell ahd bile duct carcinoma. Am J Pathol. 1949;25:647. [PMC free article] [PubMed] [Google Scholar]
- 5.Goodman ZD, Ishak KG, Langloss JM, Sesterhenn IA, Rabin L. Combined hepatocellular-cholangiocarcinoma. A histologic and immunohistochemical study. Cancer. 1985;55:124–135. doi: 10.1002/1097-0142(19850101)55:1<124::aid-cncr2820550120>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Maximin S, Ganeshan DM, Shanbhogue AK, Dighe MK, Yeh MM, Kolokythas O, Bhargava P, Lalwani N. Current update on combined hepatocellular-cholangiocarcinoma. Eur J Radiol Open. 2014;1:40–48. doi: 10.1016/j.ejro.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cazals-Hatem D, Rebouissou S, Bioulac-Sage P, Bluteau O, Blanche H, Franco D, Monges G, Belghiti J, Sa Cunha A, Laurent-Puig P, Degott C, Zucman-Rossi J. Clinical and molecular analysis of combined hepatocellular-cholangiocarcinomas. J Hepatol. 2004;41:292–298. doi: 10.1016/j.jhep.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 8.Coulouarn C, Cavard C, Rubbia-Brandt L, Audebourg A, Dumont F, Jacques S, Just PA, Clement B, Gilgenkrantz H, Perret C, Terris B. Combined hepatocellular-cholangiocarcinomas exhibit progenitor features and activation of Wnt and TGFbeta signaling pathways. Carcinogenesis. 2012;33:1791–1796. doi: 10.1093/carcin/bgs208. [DOI] [PubMed] [Google Scholar]
- 9.Hovelson DH, Cani AK, McDaniel A, Johnson B, Rhodes K, Williams PD, Bandla S, Choppa P, Hyland F, Liu G. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. 2015;17:385–99. doi: 10.1016/j.neo.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lih C-J, Harrington RD, Sims DJ, Harper KN, Bouk CH, Datta V, Yau J, Singh RR, Routbort MJ, Luthra R. Analytical validation of the next-generation sequencing assay for a nationwide signal-finding clinical trial: molecular analysis for therapy choice clinical trial. J Mol Diagn. 2017;19:313–327. doi: 10.1016/j.jmoldx.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie D, Ren Z, Fan J, Gao Q. Genetic profiling of intrahepatic cholangiocarcinoma and its clinical implication in targeted therapy. Am J Cancer Res. 2016;6:577. [PMC free article] [PubMed] [Google Scholar]
- 13.Simbolo M, Fassan M, Ruzzenente A, Mafficini A, Wood LD, Corbo V, Melisi D, Malleo G, Vicentini C, Malpeli G. Multigene mutational profiling of cholangiocarcinomas identifies actionable molecular subgroups. Oncotarget. 2014;5:2839. doi: 10.18632/oncotarget.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Churi CR, Shroff R, Wang Y, Rashid A, Kang HC, Weatherly J, Zuo M, Zinner R, Hong D, Meric-Bernstam F. Mutation profiling in cholangiocarcinoma: prognostic and therapeutic implications. PloS one. 2014;9:e115383. doi: 10.1371/journal.pone.0115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross JS, Wang K, Gay L, Al-Rohil R, Rand JV, Jones DM, Lee HJ, Sheehan CE, Otto GA, Palmer G. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next-generation sequencing. Oncologist. 2014;19:235–242. doi: 10.1634/theoncologist.2013-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu AX, Borger DR, Kim Y, Cosgrove D, Ejaz A, Alexandrescu S, Groeschl RT, Deshpande V, Lindberg JM, Ferrone C. Genomic profiling of intrahepatic cholangiocarcinoma: refining prognosis and identifying therapeutic targets. Ann Surg Oncol. 2014;21:3827–3834. doi: 10.1245/s10434-014-3828-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, Zhao X, Li Y, Li Q, Wang H. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;5:5696. doi: 10.1038/ncomms6696. [DOI] [PubMed] [Google Scholar]
- 19.Totoki Y, Tatsuno K, Covington KR, Ueda H, Creighton CJ, Kato M, Tsuji S, Donehower LA, Slagle BL, Nakamura H. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 20.Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, Deng Q, Wang Q, Li K-Y, Dai J-H, Li N, Zhu Z-D, Zhou B, Liu X-Y, Liu R-F. Exome sequencing of hepatitis B virus-associated hepatocellular carcinoma. Nat Genet. 2012;44:1117–1121. doi: 10.1038/ng.2391. [DOI] [PubMed] [Google Scholar]
- 23.Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, Baek D, Haq F, Ansari AA, Lee SY. Genomic portrait of resectable hepatocellular carcinomas: implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–1982. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- 24.Schulze K, Nault J-C, Villanueva A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J Hepatol. 2016;65:1031–1042. doi: 10.1016/j.jhep.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 25.de La Coste A, Romagnolo B, Billuart P, Renard C-A, Buendia M-A, Soubrane O, Fabre M, Chelly J, Beldjord C, Kahn A. Somatic mutations of the β-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi Y, Iwao K, Nagasawa Y, Aihara T, Sasaki Y, Imaoka S, Murata M, Shimano T, Nakamura Y. Activation of the β-catenin gene in primary hepatocellular carcinomas by somatic alterations involving exon 3. Cancer Res. 1998;58:2524–2527. [PubMed] [Google Scholar]
- 27.Sugimachi K, Taguchi K-i, Aishima S-i, Tanaka S, Shimada M, Kajiyama K, Sugimachi K, Tsuneyoshi M. Altered expression of β-catenin without genetic mutation in intrahepatic cholangiocarcinoma. Mod Pathol. 2001;14:900–905. doi: 10.1038/modpathol.3880409. [DOI] [PubMed] [Google Scholar]
- 28.Zucman-Rossi J, Villanueva A, Nault J-C, Llovet JM. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239. e1224. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Wang WL, Zhang Y, Guo SP, Zhang J, Li QL. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatol Res. 2007;37:389–396. doi: 10.1111/j.1872-034X.2007.00042.x. [DOI] [PubMed] [Google Scholar]
- 30.Sia D, Hoshida Y, Villanueva A, Roayaie S, Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology. 2013;144:829–840. doi: 10.1053/j.gastro.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi Y, Kawate S, Watanabe M, Fukushima Ji, Mori S, Fukusato T. Amplification of c-myc and cyclin D1 genes in primary and metastatic carcinomas of the liver. Pathol Int. 2007;57:437–442. doi: 10.1111/j.1440-1827.2007.02120.x. [DOI] [PubMed] [Google Scholar]
- 32.Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]