Abstract
Long interspersed nuclear element-1 (LINE-1 or L1) retrotransposons are normally suppressed in somatic tissues mainly due to DNA methylation and antiviral defense. However, the mechanism to suppress L1s may be disrupted in cancers, thus allowing L1s to act as insertional mutagens and cause genomic rearrangement and instability. Whereas the frequency of somatic L1 insertions varies greatly among individual tumors, much remains to be learned about underlying genetic, cellular, or environmental factors. Here, we report multiple correlates of L1 activity in stomach, colorectal, and esophageal tumors through an integrative analysis of cancer whole-genome and matched RNA-sequencing profiles. Clinical indicators of tumor progression, such as tumor grade and patient age, showed positive association. A potential L1 expression suppressor, TP53, was mutated in tumors with frequent L1 insertions. We characterized the effects of somatic L1 insertions on mRNA splicing and expression, and demonstrated an increased risk of gene disruption in retrotransposition-prone cancers. In particular, we found that a cancer-specific L1 insertion in an exon of MOV10, a key L1 suppressor, caused exon skipping and decreased expression of the affected allele due to nonsense-mediated decay in a tumor with a high L1 insertion load. Importantly, tumors with high immune activity, for example, those associated with Epstein–Barr virus infection or microsatellite instability, tended to carry a low number of L1 insertions in genomes with high expression levels of L1 suppressors such as APOBEC3s and SAMHD1. Our results indicate that cancer immunity may contribute to genome stability by suppressing L1 retrotransposition in gastrointestinal cancers.
Somatic retrotransposition of the long interspersed nuclear element-1 (LINE-1 or L1) has been reported in multiple cancer types using L1-targeted sequencing (Iskow et al. 2010; Solyom et al. 2012; Shukla et al. 2013; Doucet-O'Hare et al. 2015; Ewing et al. 2015; Rodić et al. 2015) and whole-genome sequencing (Lee et al. 2012; Helman et al. 2014; Tubio et al. 2014; for review, see Scott and Devine 2017). Notably, gastrointestinal cancers, including esophageal (Doucet-O'Hare et al. 2015; Secrier et al. 2016), gastric (Ewing et al. 2015), and colorectal cancers (Lee et al. 2012; Solyom et al. 2012), reportedly carry extensive somatic L1 insertions. The rate of L1 insertions varies substantially among individual tumors, ranging from a few to hundreds. Clinical and molecular factors identified in association with L1 insertions include patient age in colorectal cancer (Solyom et al. 2012), patient survival in pancreatic cancer (Rodić et al. 2015), and TP53 mutations in head and neck cancer (Helman et al. 2014). However, further investigation is needed into the mechanisms underlying these associations. Furthermore, previous studies may have been limited in their ability to detect other factors, especially those related to major L1 suppression mechanisms, namely DNA methylation and antiviral defense, due to small sample sizes and/or lack of matched expression profiles.
L1 insertions disrupt target gene function through diverse mechanisms, for example, by interrupting protein-coding sequences or altering mRNA splicing and expression (Elbarbary et al. 2016). Intragenic somatic L1 insertions previously identified in cancer genomes were depleted in exons and mostly located in introns (Lee et al. 2012; Helman et al. 2014). Those intronic insertions generally decreased target gene expression (Lee et al. 2012; Helman et al. 2014) with some exceptions (Shukla et al. 2013; Helman et al. 2014). There have also been inconsistent findings that somatic L1 insertions have little effect on gene expression (Tubio et al. 2014). Although aberrant splicing is a major pathogenic mechanism of retrotransposon insertions causing Mendelian disorders and hereditary cancers (Hancks and Kazazian 2016), to our knowledge, no somatic L1 insertions have been reported in association with splicing alterations in sporadic human cancers.
Here, we analyzed whole-genome sequencing data for which somatic retrotransposition had not previously been investigated and which were obtained from cancer patients of three gastrointestinal cancer types using an improved version of Transposable Element Analyzer (Tea) (Methods; Lee et al. 2012). We examined the associations between numerous clinical and molecular factors, and L1 activity, and characterized the effects of somatic L1 insertions on gene transcripts, using matched RNA-seq profiles from the same cancer patients for which somatic L1 insertions were identified. To our knowledge, this study constitutes the first in-depth surveys of gastrointestinal cancers with regard to the association between L1 activity and particularly immune signatures.
Results
Highly variable somatic L1 insertion frequency and recurrent insertions in cancer genes
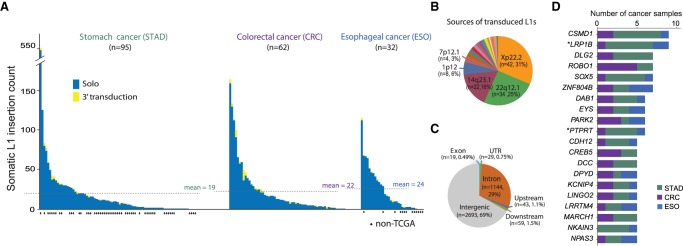
We applied Tea (Transposable Element Analyzer) (Lee et al. 2012) with improved 3′ transduction (i.e., mobilization of unique non-L1 DNA downstream from the L1) detection to the whole-genome sequencing data of tumor and blood samples from a total of 189 gastrointestinal cancer patients across three cancer types: 95 stomach (40 TCGA and 55 non-TCGA; STAD) (Wang et al. 2014), 62 TCGA colorectal (CRC), and 32 esophageal (19 TCGA and 13 non-TCGA; ESO) (Dulak et al. 2013) cancer patients. We detected 3885 somatic L1 insertions that are present in cancer genomes and absent in matched blood genomes from the same patients (Supplemental Table S1). To create a high-confidence insertion set, we included insertion candidates when Tea predicted both target site duplication (TSD) of at least 5 bp and poly(A) tails, the two signatures for target-primed reversed transcription (TPRT)-mediated retrotransposition. Although the insertion frequency varied greatly, samples carried an average of 21 insertions, and most (89%) samples carried at least one insertion (Fig. 1A; Supplemental Table S2), thereby confirming previous findings that gastrointestinal cancers are highly susceptible to somatic L1 retrotransposition (Burns 2017). Of 137 insertions with 3′ transductions, more than half (56%) were derived from two germline L1s on Chromosomes X and 22 (Xp22.2 and 22q12.1) (Fig. 1B; Supplemental Table S1), consistent with a previous finding that a handful of source L1s generated most 3′ transductions in cancers (Tubio et al. 2014).
Figure 1.
Landscape of somatic L1 insertions in gastrointestinal cancers. (A) Frequency of somatic L1 insertions across three cancer types. The dotted line denotes the average insertion count for each cancer type. (B) Source L1 elements of somatic L1 insertions with 3′ transduction. (C) Genomic distribution of somatic L1 insertions. Upstream/downstream insertions are those that occur within 5 kb from the transcription start/end sites of genes. (D) Genes with recurrent somatic L1 insertions. Genes with insertions in more than four cancer patients are shown. (*) Known cancer genes reported in the COSMIC Cancer Gene Census database v82 (Forbes et al. 2017).
We found that 1294 (33%) of the 3885 L1 insertions were in gene bodies—mostly in introns (29%) (Fig. 1C; Supplemental Text). A total of 203 genes were affected by somatic L1 insertions in more than one cancer sample, and 81 genes, including known (LRP1B and PTPRT) and putative cancer driver genes (e.g., ROBO1 and PARK2), showed significantly recurrent insertions when gene length was taken into account (FDR < 0.05) (Fig. 1D; Supplemental Table S3; Methods). For example, ROBO1, an emerging tumor suppressor (Gara et al. 2015; Huang et al. 2015), had at least one somatic L1 insertion in each of seven cancer samples (two stomach and five colorectal samples). PARK2, a master regulator of G1/S cyclins that is frequently deleted in cancers (Gong et al. 2014), had one insertion in each of six samples (one stomach, three colorectal, and two esophageal samples).
Genes with recurrent somatic L1 insertions (Fig. 1D), including ROBO1, PTPRT, GRID2, CDH8, CDH12, CDH13, PTPRM, and ROBO2, were enriched for brain development and function including axon guidance, neuron differentiation, and synaptic function (Supplemental Table S4). However, this may be because neuronal genes tend to be long (Zylka et al. 2015). Indeed, no significant enrichment was found at the gene level after adjusting for gene size. This suggests the overall absence of positive selection of cancer cells with somatic L1 insertions despite occasional instances of potentially tumorigenic insertions in cancer genes.
Clinical and molecular correlates of somatic L1 retrotransposition
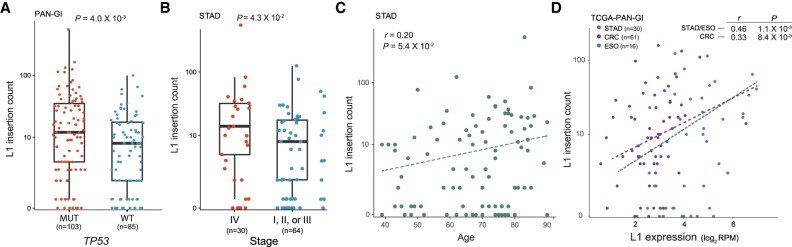
We then wanted to understand factors underlying the variable frequency of L1 insertions in cancers. First, we examined the association of L1 insertions with molecular markers or clinical traits. Although 13 genes were mutated in at least 20% of the cancer samples, the only significant association with L1 insertion counts was with TP53 mutations (Supplemental Table S5). Somatic L1 insertions were more frequent in tumors with TP53 mutations than those with wild-type TP53 (Mann-Whitney U test P = 0.004) (Fig. 2A). This corroborates the previous findings that TP53 mutation status correlates with L1 ORF1p expression (Rodić et al. 2014; Wylie et al. 2016) and that TP53 restrains L1 transcription (Wylie et al. 2016). We further examined whether any aberration in DNA repair pathways could be associated with L1 retrotransposition and found that, of 15 DNA repair pathways we tested (Jeggo et al. 2016), only the TP53 repair pathway showed a significant association (Mann-Whitney U test P = 0.0063) (Methods).
Figure 2.
Factors correlated with the frequency of somatic L1 insertions. (A) Somatic L1 insertion counts in cancer samples with mutations (MUT, red dots) and without mutations (WT, blue dots) in TP53 are shown in box plots. (B) Somatic L1 insertion counts in stomach cancer samples at stage 4 (red dots) and at earlier stages (stages 1–3, blue dots) are shown in box plots. (A,B) P-values from the Mann-Whitney U test are shown. (C) Correlation between the age of cancer patients at diagnosis (x-axis) and somatic L1 insertion counts (y-axis) in stomach cancer. (D) Correlation between L1 expression (x-axis) and somatic L1 insertion counts (y-axis). (C,D) Spearman correlation coefficients and their P-values are shown.
Frequent somatic L1 insertions were observed in cancers at an advanced stage (P = 0.043) (Fig. 2B) and in older stomach cancer patient samples (P = 0.054) (Fig. 2C), but the associations were weak given their marginal P-values. Our L1 expression quantification is limited in its ability to distinguish between functional L1 RNAs transcribed from L1 promoters and nonfunctional L1 RNAs transcribed as part of other genes (Deininger et al. 2017). Nonetheless, we observed that L1s were expressed in normal gastrointestinal tissues from cancer patients, and the expression level was significantly elevated in tumors (P = 2.6 × 10−7 and P = 1.3 × 10−7 for stomach/esophageal and colorectal cancers, respectively) (Supplemental Fig. S1). We found a positive correlation between L1 insertion frequency and the L1 expression level in tumors (P = 0.001 for stomach/esophageal cancer and P = 0.008 for colorectal cancer) (Fig. 2D). These associations suggest that aberrant L1 transcription, potentially induced by DNA methylation loss and/or mutations in L1 transcription suppressors, is a prerequisite to frequent retrotransposition in cancer.
L1 insertions disrupting mRNA splicing and expression
We examined the effect of intragenic L1 insertions on transcriptional regulation. To this end, we analyzed matched RNA-sequencing data from 112 TCGA cancer samples for which genomes were analyzed for L1 insertions. Briefly, we calculated the ratio of abnormally spliced RNA-seq reads to normally spliced reads near a somatic L1 insertion and evaluated whether the ratio was significantly higher than expected given the ratio distribution estimated from RNA-seq profiles of approximately 2900 control samples without the given insertion (Methods).
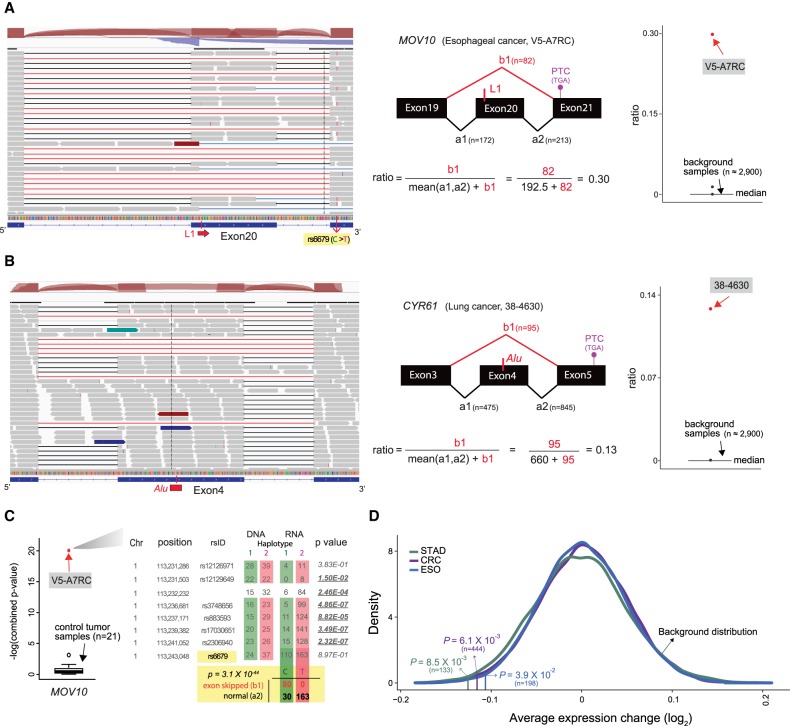
We screened 1192 intragenic L1 insertions with matched RNA-seq profiles and found skipping of exon 20 in MOV10, a known L suppressor, with a somatic L1 insertion in the skipped exon in one esophageal cancer sample (Fig. 3A). MOV10 is known to suppress L1 expression and decrease cytoplasmic L1RNPs (Goodier et al. 2012). The insertion occurred 15 bp away from the beginning of the 122-bp-long exon 20 of MOV10, and the estimated insertion size was 438 bp. The cancer sample with the L1 insertion carried 65 somatic L1 insertions, which was the third highest insertion frequency among all 32 esophageal cancer samples. We found that the exon skipping induced a frameshift and a premature termination codon in exon 21, which likely triggered nonsense-mediated decay (NMD). Indeed, we found that only the transcripts from the allele linked with exon skipping showed decreased expression (Fig. 3A,C). The allelic expression loss was not observed in cancer samples that lacked mutations of any type in MOV10, including SNVs, CNVs, and DNA methylation, suggesting NMD activity on the L1 insertion allele (Fig. 3C).
Figure 3.
Disruption of mRNA splicing and expression by somatic L1 insertions. (A) Skipping of exon 20 in MOV10 caused by a somatic L1 insertion in a TCGA esophageal cancer sample. RNA-seq reads (gray boxes) from normally spliced transcripts show split mapping between the expected adjacent exons (black lines), whereas reads from transcripts with exon skipping show abnormal split mapping without the exon with the L1 insertion (red lines). Reads showing the exon skipping were spliced at the splice donor site of exon 19 and the splice acceptor site of exon 21. Other minor forms of abnormal splicing that involve retention of the nineteenth intron, partial skipping of exon 20, and/or skipping of exon 21 are also shown with blue lines. The schematic diagram in the middle shows how to calculate the ratio of the abnormally spliced read count (major form) to the total read count around the exon with the L1 insertion. The ratios are also calculated using approximately 2900 control RNA-seq profiles (cancer samples without any mutation in MOV10 and normal tissue samples) and serve as a background distribution to assess the significance of the observed ratio from the esophageal cancer sample (red dot). A heterozygous single-nucleotide variant (SNV; rs6679) in exon 21 shows a linkage with the exon skipping; reads with the alternative “T” allele (the lines shown in red within the reads) show normal splicing, and reads with the reference “C” allele show skipping of exon 20. This linkage is utilized to examine an allelic decrease of MOV10 expression in C. (B) Skipping of exon 4 in CYR61 caused by a somatic Alu insertion. Reads from the third and the fourth introns were often observed in the control RNA-seq profiles, suggesting that they were prespliced transcripts not associated with the Alu insertion. (C) Decreased expression of the MOV10 allele with exon skipping caused by a somatic L1 insertion shown in A. The esophageal cancer sample had eight heterozygous SNVs in MOV10. For each of the eight SNV loci, the number of reads with a reference allele and an alternative allele is shown for both whole-genome sequencing (DNA) and RNA-sequencing data. The colored boxes (red and green) around SNV read counts represent different haplotypes. The degree of allelic bias in RNA-sequencing data relative to DNA-sequencing data was tested using Fisher's exact test, and loci with significant bias (P < 0.05) are underlined in bold. On the left box plot, the combined P-value from eight loci in the esophageal cancer sample (red dot) is compared to the distribution of the combined P-values from 21 control cancer samples without any mutation or methylation aberration in MOV10 (black box plot). For the SNV (rs6679) with a linkage to the exon 20 skipping (A), the number of RNA-seq reads that span the SNV loci and show the splicing pattern is shown. Association between SNV alleles and exon skipping status was tested using Fisher's exact test. (D) Decreased expression of genes with somatic L1 insertions. The average difference for each cancer type is marked by a vertical line. The P-value of the observed average expression difference was calculated based on a background distribution estimated from random gene sets for each cancer type (colored curved line) (Methods). The number of genes with somatic L1 insertions for each cancer type is shown in parentheses.
Our additional screening of 282 published somatic retrotransposon insertions (Helman et al. 2014) in 69 cancer samples identified another exon-skipping event in one lung cancer sample caused by an Alu insertion in the middle of exon 4 of CYR61, a putative tumor suppressor (Fig. 3B; Tong et al. 2001). The insertion was located 85 bp away from the beginning of 209-bp-long exon 4, and the skipping event caused a frameshift resulting in a premature termination codon in exon 5 of CYR61 (Fig. 3B).
We found intragenic L1 insertions were enriched in genes with a low expression level and were depleted in genes with a high expression level (Supplemental Fig. S2), consistent with a previous report (Tubio et al. 2014). We compared the expression level of genes in cancer samples with L1 insertions to the expression level of the same genes in other cancer samples without L1 insertions or nonsynonymous mutations in the genes. We confirmed our own previous finding (Lee et al. 2012) that L1 insertions significantly disrupt the expression of target genes (P = 0.0085 for STAD, P = 0.039 for ESO, and P = 0.0061 for CRC) (Fig. 3D). The significant decrease in gene expression level was observed even when excluding genes with a very low expression level (the bottom 25% quartile; P = 0.001 for STAD, P = 0.004 for ESO, and P = 3.0 × 10−4 for CRC). Genes with recurrent L1 insertions showed a decreased level of gene expression (P = 0.026 for STAD, P = 0.027 for ESO, P = 0.001 for CRC), whereas genes with insertions found only in a single patient did not. Among other mechanisms (Elbarbary et al. 2016), the decreased gene expression may be due in part to the NMD process triggered by PTC-containing transcripts with abnormal splicing, as suggested above regarding MOV10.
L1 retrotransposition is inversely correlated with expression of immunologic response genes
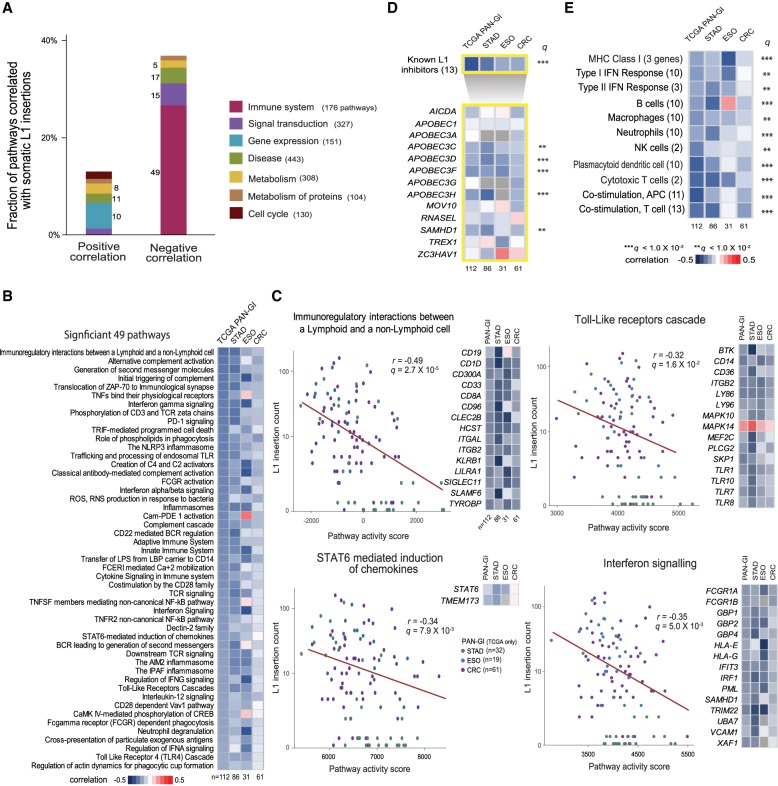
We then performed a more systematic transcriptome analysis by measuring the transcriptional activity of 1789 pathways from the Reactome database (Milacic et al. 2012; Fabregat et al. 2016) in 112 TCGA cancer samples with RNA-seq profiles, using the single-sample Gene Set Enrichment Analysis (ssGSEA) method (Subramanian et al. 2005). A total of 50 and 97 pathways showed positive and negative correlations, respectively, with L1 insertion counts (FDR < 0.05) (Fig. 4A; Supplemental Table S6). Notably, 49 of 176 (28%) immune pathways showed significant negative correlations (Fig. 4B; Supplemental Fig. S3). These negative associations remained when each tumor type was separately analyzed, and they were stronger for stomach and esophageal cancers than for colorectal cancers (Fig. 4B). We also examined the relationship between pathway activities and L1 expression, and consistently found the immune system category to have the highest fraction of pathways that were negatively associated with L1 expression (18 of 176, 10%) (Supplemental Fig. S4; Supplemental Table S7).
Figure 4.
Immune activity associated with somatic L1 retrotransposition. (A) Reactome pathways for which activity correlates with somatic L1 insertion frequency. For each pathway category, the percentage of pathways showing a significant positive or negative correlation between pathway activity and somatic L1 insertion frequency (FDR < 0.05) is shown in a stacked bar. The number of significantly correlated pathways for each category is shown beside the stacked bar. The number of all member pathways for each category is shown in parentheses, and only the categories with more than 100 member pathways are shown. (B) Forty-nine immune pathways whose activity shows a significant negative correlation with L1 insertion frequency (FDR < 0.05). Each cell in the heatmap shows a color-scaled Spearman correlation coefficient between the activity of each immune pathway (row) and the frequency of somatic L1 insertions in cancer samples from each cancer type (column). (C) Examples of the significant immune pathways. The pathway activity score (x-axis) and L1 insertion count (y-axis) is marked with a colored dot for each cancer sample. Green, blue, and purple dots represent STAD, ESO, and CRC samples, respectively. The Spearman correlation coefficient and its P-value are shown. Key member genes whose expression levels were significantly correlated with insertion counts (FDR < 0.05) are shown in the heatmap. Each cell in the heatmap shows a color-scaled Spearman correlation coefficient between gene expression level and L1 insertion frequency. Up to 15 key genes are shown, ranked by the significance of the correlation. Key genes that are present in multiple pathways are shown once. (D) Correlation between the expression level of 13 known L1 inhibitors and the frequency of somatic L1 insertions. The top row of the heatmap shows the Spearman correlation coefficient between the activity of a gene set with 13 known inhibitors and L1 insertion frequency for each cancer type (column). The bottom rows of the heatmap show the coefficients for individual inhibitors (row). A row with a PAN-GI correlation of q < 0.01 and q < 0.001 (adjusted P-values combined across all cancer types using the Fisher's method) is marked with double and triple asterisks, respectively. (E) Various immune gene sets showing a negative correlation with L1 insertion frequency.
Among the immune-related pathways, “Immunoregulatory interactions between a lymphoid and a non-lymphoid cell” showed the most significant association with L1 insertions (Fig. 4C). Among its 112 member genes, we identified 67 key genes whose expression levels were significantly correlated with L1 insertion frequency (FDR < 0.05) (Supplemental Table S8). Tumors with active toll-like receptor (TLR) and STAT6 signaling showed low somatic L1 insertion counts (Fig. 4C). The key genes for the “TLR cascade” pathway included several TLRs (TLR1, TLR7) along with the binding ligand (CD36) and a kinase (BTK) (Fig. 4C; Supplemental Table S8). There were two key genes for the “STAT6 mediated induction of chemokines” pathway: STAT6 and TMEM173 (also known as STING). TLRs play a critical role in the activation of innate and adaptive immunity by sensing exogenous pathogens and endogenous retroviruses (ERV) and activating IFN signaling (Pasare and Medzhitov 2005; Chiappinelli et al. 2015). STAT6 activation by TMEM173-mediated sensing of cytosolic DNA derived from the cell itself or from foreign pathogens (i.e., viral or bacterial) also results in the induction of IFN signaling (Chen et al. 2011). Hence, molecules derived from L1s may have triggered TLR and/or STAT6 signaling and resulted into a downstream IFN signaling to suppress L1 activity.
Several L1 inhibitors, including SAMHD1, MOV10, and APOBEC3 family proteins, are known to be activated by IFNs (Yu et al. 2015; Riess et al. 2017). As expected, tumors with high IFN signaling activity showed few L1 insertions (Fig. 4C). Notably, there was a significant inverse correlation between the expression of SAMHD1 and L1 insertion counts (Fig. 4C; Goodier 2016). SAMHD1 is a known L1 inhibitor whose mutation is found in the congenital autoimmune disease Aicardi-Goutieres Syndrome (AGS). It is known to block L1 insertions by restricting their reverse transcription and sequestering L1 RNP within stress granules (Hu et al. 2015). We examined 12 additional known L1 inhibitors (Goodier 2016) and found negative correlations between the expression levels of APOBEC families (APOBEC3C/D/F/H) and L1 insertion counts (FDR < 0.01) (Fig. 4D). All of this suggests that the IFN response triggered by L1-derived molecules, for example, through TLR and/or STAT6 signaling, may have activated these L1 suppressors and effectively restricted L1 retrotransposition.
We further examined the relationship between L1 insertions and cancer immunity by analyzing additional immune gene sets. The gene sets comprised marker genes of several immune-stimulatory cells including B and cytotoxic T cells, and other immune cells, such as macrophages, neutrophils, and natural killer cells (Fig. 4E; Breuer et al. 2013; Rooney et al. 2015). We consistently found negative correlations between L1 retrotransposition and immune-stimulatory activities (Fig. 4E). We also observed overall negative correlations between L1 retrotransposition and immune-inhibitory signals such as the activities of regulatory T cells and PD1-signaling (Supplemental Fig. S5), as reported in cancer immune evasion and chronic inflammatory conditions (Rooney et al. 2015; Davoli et al. 2017). However, the pattern was inconsistent in esophageal cancer. Analysis of more cancer samples and/or cancer types is needed to establish the relationship with immune-inhibitory signals. Nonetheless, our results provide concrete evidence for the immunological association of L1 retrotransposition across diverse immune-stimulatory cell types.
Characterization of cancer subgroups with differential immune activity
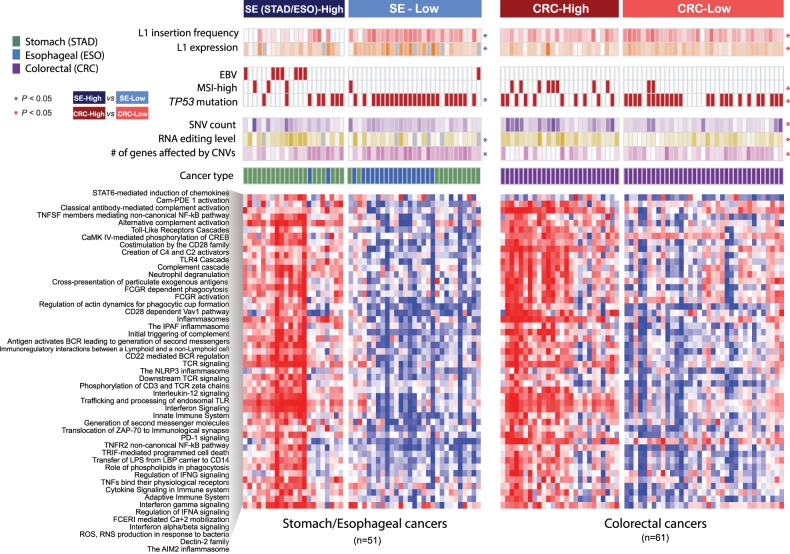
Based on the 49 L1-associated immune pathways in 112 cancer samples, we identified two distinct cancer subgroups with different immune signatures for stomach/esophageal cancer (SE; SE-High and SE-Low) and colorectal cancer (CRC; CRC-High and CRC-Low) (Fig. 5). With these distinct cancer subgroups, we confirmed that tumors in the high immune subgroups showed high cytolytic activities measured by the expression levels of two cytotoxic T cell effector genes (GZMA and PRF1; P = 3.9 × 10−8 and P = 8.3 × 10−5 for SE and CRC) (Supplemental Fig. S6A; Rooney et al. 2015). We also confirmed that tumors in the high immune subgroups tended to show less frequent somatic L1 insertions and lower L1 expression levels than tumors in the low-immune subgroups (Fig. 5; Supplemental Fig. S6B,C).
Figure 5.
Cancer subgroups with distinctive immune activity signatures. The heatmap represents the activity of 49 significant immune pathways (rows) in 112 cancer samples (columns). Unsupervised clustering identifies cancer subgroups according to immune activity: stomach/esophageal (SE)-High, SE-Low, colorectal (CRC)-High, and CRC-Low. Clinical and molecular features of each cancer sample are marked on the top of the heatmap. Higher L1 insertion frequency and L1 expression levels are marked in darker red and orange, respectively. Cancer samples with EBV infection, MSI-high phenotype, and nonsynonymous mutations in TP53 are marked with filled boxes. Higher somatic SNV counts, RNA editing level, and counts of genes with somatic copy number aberrations are marked in darker colors. The color scale was separately normalized for stomach-esophageal cancer and colorectal cancer samples. Samples without RNA-seq data are marked in gray. A feature showing a significant difference between cancer subgroups is marked with an asterisk. The Mann-Whitney U test and Fisher's exact test were used to test statistical significance for continuous and categorical features, respectively.
We next investigated features that are potential determinants of immune activities in different cancer types. In stomach cancer, oncogenic Epstein–Barr virus (EBV) infection has been reported in 10% of stomach cancer cases, and EBV-positive tumors showed a favorable prognosis and extensive immune cell infiltration (Iizasa et al. 2012; The Cancer Genome Atlas Research Network 2014). We found that all but one of the eight EBV-positive stomach cancer samples belonged to the immune-high subgroup (SE-High) (Fig. 5; Supplemental Fig. S6D). In colorectal cancer, a high level of indel mutations in short tandem repeats, known as microsatellite instability (MSI-high), has been reported in 12% of sporadic colorectal cancers mainly caused by mismatch repair deficiency (Kawakami et al. 2015; Cortes-Ciriano et al. 2017). It is known that MSI-high colorectal tumors have a high neoantigen burden that elicits antitumor immune response and shows a favorable prognosis and a better response to immunotherapy (Bodmer et al. 1994; Le et al. 2017). We observed that colorectal cancer samples in the high immune subgroup showed more frequent MSI-high phenotypes and higher levels of point mutations and RNA editing than those in the low-immune subgroup, all supporting a higher tumor-antigen load (P = 0.014 for MSI-high, P = 0.004 for SNV/Indel counts, and P = 0.0017 for RNA editing, respectively) (Supplemental Fig. S6D–F).
We found that TP53 mutations were enriched in tumors in the low-immune groups (P = 0.0033 for SE-Low and P = 0.036 for CRC-Low) (Fig. 5; Supplemental Fig. S6G), consistent with previous findings that TP53 dysfunction leads to immunosuppression (Rooney et al. 2015; Cui and Guo 2016). In esophageal cancer, all but one of the 17 TP53-mutated patients showed low immune activity and belonged to the low-immune group (exact binomial test P = 0.0011). These along with our earlier results on frequent TP53 mutations in tumors with a higher load of L1 insertions suggest the critical role of TP53 in restricting retrotransposons as a guardian of L1 expression and cancer immunity. Another potential factor related to low cancer immunity, specifically cancer immune evasion, is aneuploidy or large copy number aberration in cancer, as a recent study suggested (Davoli et al. 2017). Indeed, we confirmed broader copy number aberrations measured by the number of affected genes in low-immune groups (P = 0.02 for SE-Low and P = 0.013 for CRC-Low) (Fig. 5; Supplemental Fig. S6H).
Discussion
It remains an open question whether transposable elements, particularly L1s, play a role in tumorigenesis and what factors determine variable L1 retrotransposition rates in tumors. Here, we analyzed about 200 cancer genomes from three types of gastrointestinal cancer samples and identified a large set of somatic L1 insertions. We found the insertions in some known cancer genes, including LRP1B, PTPRT, ROBO1, and PARK2, in multiple cancer samples. We performed an integrative analysis using RNA-seq profiles of these samples, and found that ∼20% of somatic L1 insertions in genes were expressed at a moderate to high level and that they generally disrupted expression of target genes. We also detected somatic retrotransposon insertions causing splicing aberrations and the resulting expression decrease of the affected allele.
The Tea method was previously shown to detect heterozygous insertions in samples with tumor purity as low as 49% (Lee et al. 2012). Since the median purity of tumors in this study was 63%, and importantly 83% of tumors in the high immune groups had tumor purity higher than 49%, we expect Tea to show similar sensitivity for tumors from high- versus low-immune groups. We also note that Tea produces comparable results for BAM files aligned to hg19 and GRCh38; thus, our analysis will not be affected by the choice of reference genomes for read mapping.
Notably, we found that immune activity of tumors is a major factor in explaining the L1 retrotransposition rate. Less L1 retrotransposition activity was found in tumors with high immune activity triggered largely by exogenous (e.g., EBV) or endogenous immunogens (e.g., SNV, MSI, and RNA editing). Most importantly, our findings suggest that when cancers have low immune activity or immune evasion led by such things as a high CNV load and TP53 mutation, they are prone to extensive L1 retrotransposition and thus are at increased risk of tumorigenic insertion events.
It is possible that the negative correlation we observed between L1 and immune activity may result from disruptions of L1 suppression, such as TP53 mutations and compromised DNA methyltransferases that independently cause immune evasion while activating L1s as a secondary effect. An alternative explanation is that L1s promote immune evasion by disrupting immune surveillance, for example, through insertional mutagenesis, genomic arrangements, splicing disruption, and triggering RNA editing of immune genes, although L1s also have the potential to cause tumor neoantigens. One potential way of L1s to disrupt immune surveillance is for the activities of DNA and RNA deaminases (e.g., AID/APOBEC and ADAR1) triggered by immune responses, such as TLR and IFN signaling, to suppress L1s, in turn leading to hypermutation and increased RNA editing (Blanc and Davidson 2010; Roberts et al. 2013; Rebhandl and Geisberger 2015; Orecchini et al. 2017), and ultimately increasing the risk of impairing immune genes.
Last, L1 might directly mediate immune tolerance through unknown mechanisms. For example, persistent IFN signaling due to chronic L1 expression might have shifted IFN signaling from the immune-stimulatory to the immune-suppressive mode, as observed in the presence of chronic viral infection (Minn and Wherry 2016). There might be other mechanisms by which L1s directly mediate immune tolerance; for instance, L1 expression may directly affect signaling to lymphocytes. More investigation is warranted in order to understand the mechanisms underlying the associations we have identified and to further illuminate the role of transposable elements in cancer.
Methods
Whole-genome sequencing data
We downloaded TCGA whole-genome sequencing (WGS) data set from CGHub (http://cghub.ucsc.edu). The data set was comprised of BAM files for 62 colorectal, 40 stomach, and 19 esophageal cancer samples and matched germline (blood) samples. We downloaded a non-TCGA WGS data set from EGA (accession ID: EGAD00001000782) containing BAM files for 55 stomach cancer samples and matched germline samples (Wang et al. 2014). We also downloaded a non-TCGA WGS data set from dbGaP (phs000598.v1.p1) containing BAM files for 13 esophageal cancer samples and matched germline samples (Dulak et al. 2013). We realigned the 110 BAM files (normal and tumor) in the stomach cancer data set using the hg19 reference genome and BWA (version 0.6.2) (Li and Durbin 2009). We also marked PCR duplicates for those BAM files using Picard (http://broadinstitute.github.io/picard). These files do not include BAM files that failed to download, realign, or be run with the Tea pipeline, which were excluded from our analysis. Currently the TCGA data is hosted at the Genomic Data Commons (https://gdc.cancer.gov/).
RNA-sequencing and gene expression data
We obtained RNA-sequencing BAM files for 107 cancer samples and 94 normal gastrointestinal tissues of TCGA cancer patients from the NCI Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/) and gene-level expression data for the TCGA samples from the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/). For non-TCGA stomach cancer samples, we downloaded raw expression array data from the European Genome-phenome Archive (EGA; accession ID: EGAD00010000528) and extracted gene-level expression data using the IlluminaExpressionFileCreator module in GenePattern (Reich et al. 2006). For non-TCGA esophageal cancer samples, we downloaded gene-level expression data from the Gene Expression Omnibus (GEO; accession ID: GSE42363). We used ComBat (Johnson et al. 2007) to combine expression data from different studies for each cancer type.
Detection of somatic L1 insertions
We implemented a 3′ transduction calling module in the previously developed transposon detection pipeline (Lee et al. 2012), the Transposable Element Analyzer (Tea) and used this improved version to identify somatic L1 insertions. Each insertion was classified as one of the three types of events defined by Tubio et al. (2014): solo, partnered, or orphan events. To be identified as an insertion, the insertion candidate must have had a poly(A) tail and a target site duplication (TSD) ranging from 5 to 35 bp long (Supplemental Methods).
Somatic SNV/indel and copy number aberration call sets
We generated somatic SNV and indel call sets for TCGA colorectal cancer samples using MuTect (Cibulskis et al. 2013) and VarScan 2 (Koboldt et al. 2012) with default options, respectively. We annotated the mutations using Oncotator (Ramos et al. 2015). For TCGA stomach and esophageal cancer samples, we obtained somatic SNV/indel call sets from the UCSC Cancer Genomics Browser. For non-TCGA data sets, we obtained somatic SNV/indel call sets from Supplemental Data in original publications. Only nonsynonymous mutations were used in the analyses. We examined the association between mutation status and somatic L1 insertion counts for 13 genes mutated in at least 20% of our cancer samples. We observed that among the TCGA colorectal cancer samples for which clinical MSI assays were performed, all the cases with the MSI-high phenotype except two cases carried more than 100 nonsynonymous indels, whereas all the non-MSI-high cases carried less than 100 nonsynonymous indels. Thus, we classified cancer samples with more than 100 nonsynonymous indels as MSI-high (n = 2). Somatic copy number aberration data were downloaded from the UCSC Cancer Genomics Browser. The data provided gene-level copy number changes estimated using the GISTIC method (Mermel et al. 2011).
RNA editing analysis
We downloaded known RNA editing loci in Chromosome 1 from the REDIportal website (http://srv00.recas.ba.infn.it/atlas/) (Picardi et al. 2017). We filtered out loci when they were reported in dbSNP150 or in our somatic SNV call set. We called variants at these loci from RNA-seq data using VarScan 2 (Koboldt et al. 2012) and selected loci covered by at least 10 reads across all cancer samples, resulting in 834 loci. For each of 834 loci, we measured the RNA editing level by dividing the number of reads with a variant allele by the total number of reads spanning the locus. We calculated the average editing level across 834 loci for each cancer sample.
L1 expression quantification
Reads from RNA-sequencing data were aligned to an L1 sequence library using BWA (Li and Durbin 2009). The L1 sequence library included the L1Hs consensus sequence in Repbase (http://www.girinst.org/repbase/) and its variants created by diagnostic nucleotide substitutions for Ta-1d, Ta-1nd_G1, Ta-1n_C, Ta-0, and Pre-Ta_ACG_G subfamilies. It also included L1Hs sequences that were >6 kb in size and with a divergence score (relative to the consensus) <5% in the human reference genome (hg19) annotated by RepeatMasker (Smit et al. 2013–2015). Reads mapping to Alu and SVA sequence library were excluded. To improve our estimation of L1HS-specific expression, we additionally used TEtranscripts (Jin et al. 2015). From an original BAM file, we extracted reads that mapped to our own L1HS sequence library described above. Then, we ran TEtranscripts using the extracted reads and calculated L1HS expression normalized to reads per million mapped reads (RPM) unit.
Gene expression analysis
For each gene with a somatic L1 insertion, we calculated the difference between the expression level of the gene from the cancer sample with the insertion and the average expression level from cancer samples without any mutation in the gene. Genes that did not have expression level for at least 10 control samples were excluded in our analysis. We then calculated the average of the expression differences for each cancer type. To calculate the P-value of the observed average expression difference, we estimated a background distribution by using 10,000 randomly selected gene sets with the same number of genes as in our gene set having somatic L1 insertions for each cancer type. The empirical P-value was calculated as the proportion of the random gene sets that produced an average expression difference that was less than the observed value.
P-values for enrichment/depletion of genes with somatic L1 insertions in each expression level category were computed by a binomial test. To correct for the length of genes, all genes were divided into quartile based on their mean expression level in each cancer type and calculated the expected probability by dividing the total length of genes in each expression category by the total length of all genes.
Pathway activity analysis
We obtained a set of pathways from the Reactome database (Fabregat et al. 2016). Using ssGSEA (Barbie et al. 2009) in GenePattern (Reich et al. 2006), we measured the activity of each pathway in each cancer sample based on the expression level of its member genes. Genes whose expression levels were significantly correlated with insertion counts (FDR <0.05; adjusted combined P-values across all cancer types using the Fisher's method) were defined as key genes for each pathway. We annotated key genes for each significant immune pathway with their druggability using the Drug Gene Interaction Database (DGIdb) (Cotto et al. 2018). We obtained additional gene sets reflecting diverse aspects of the immune system (Rooney et al. 2015) and known L1 inhibitors from previous publications (Goodier 2016). Cancer samples were clustered based on the activities of immune pathways using non-negative matrix factorization (NMF) (Brunet et al. 2004) in GenePattern with default parameters (Reich et al. 2006). We separately performed clustering for STAD/ESO and CRC with k = 2.
Insertion recurrence analysis
We selected genes with somatic L1 insertions in more than one patient. For each of the recurrently affected genes, we evaluated whether the number of somatic L1 insertions was significantly frequent using the Exact Binomial test in R (R Core Team 2015). With an assumption that each genomic location has an equal chance of having an L1 insertion, we calculated the expected probability (P) for the binomial test by dividing the number of all intragenic L1 insertions with the total length of the genes with at least one insertion. Binomial P-values were corrected for multiple testing using the Benjamini-Hochberg method (Hochberg and Benjamini 1990).
Splicing and epigenetic feature annotation
We obtained 238 splicing enhancer and 176 splicing silencer motifs from two published papers (Fairbrother et al. 2004; Wang et al. 2004). We downloaded the chromHMM annotation tracks for 12 gastrointestinal tissue samples from the Roadmap Epigenome project (Roadmap Epigenomics Consortium 2015). For each intronic L1 insertion in the top 20 affected genes, we annotated the presence of a splicing regulatory motif and the status of active chromatin (promoter or enhancer).
Gene set enrichment analysis
We tested if genes with somatic L1 insertions in more than one cancer sample were enriched in certain Gene Ontology (GO) terms in the biological process category. We used GOseq (Iskow et al. 2010) that allowed for adjustment of gene length bias and took as inputs the longest isoform length (the sum of the lengths of all unique exons and introns) for each gene. To identify DNA repair pathways associated with somatic L1 insertion frequency, we obtained 15 DNA repair pathways from a previous publication (Jeggo et al. 2016) and classified cancer samples into the mutant and wild-type groups depending on the presence of a nonsynonymous mutation in any member gene for each pathway. We then compared L1 insertion frequencies between the two groups using the Mann-Whitney U test.
Splicing analysis
To identify somatic L1 insertion-mediated abnormal splicing, we used our previously established ratio-based approach to detect altered splicing caused by somatic mutations (Jung et al. 2015). Specifically, we first extracted abnormally spliced reads near retrotransposon insertion loci and then calculated the ratio of abnormally spliced reads to total reads (the sum of normally and abnormally spliced reads). Uniquely aligned reads excluding PCR duplicates were subjected to this analysis. Next, we assessed whether the ratio was significantly higher than expected, given the background distribution estimated from normal control samples and tumor samples (up to 111 TCGA tumor samples) that lacked nonsynonymous mutations for a given gene. We obtained a total of 2860 control normal RNA-seq data from the Genotype-Tissue Expression (GTEx) project (The GTEx Consortium 2013) To confirm that the observed splicing change was caused by a somatic retrotransposon insertion, the observed ratio had to be within the top 1% of the background ratio distribution. We screened 69 additional TCGA cancer RNA-seq data to detect splicing aberration caused by somatic retrotransposon insertions reported in a previous study (Helman et al. 2014).
Analysis of allelic expression of MOV10
We called SNVs in MOV10 from WGS and RNA-seq data from TCGA cancer samples using HaplotypeCaller (DePristo et al. 2011). We excluded cancer samples when they had any nonsynonymous mutation, copy number aberration, or DNA methylation (beta value >0.3) in MOV10. For each heterozygous SNV site, both reference and alternative alleles were required to have at least five reads in WGS data to be included in our analysis. In RNA-sequencing data, at least one of the alleles was required to have five reads. A total of 21 cancer samples with at least five heterozygous SNVs in MOV10 satisfying the minimum read count requirement were used in the analysis as control samples. We used Fisher's exact test to identify different allelic ratios between DNA- and RNA-sequencing data for each SNV site. We then combined P-values from all heterozygous SNV sites in MOV10 using Fisher's method. The combined P-value was calculated for the TCGA cancer sample (V5-A7RC) with a somatic L1 insertion in MOV10 as well as for each of the 21 control cancer samples. For the SNV (rs6679) with a linkage with the exon 20 skipping, we counted the number of RNA-seq reads that span the SNV loci according to the SNV allele and the splicing pattern (i.e., exon skipping or normal splicing). We tested association between SNV alleles and exon skipping status using Fisher's exact test. Haplotype information was derived using the LDhap module of LDlink (Machiela and Chanock 2015).
Supplementary Material
Acknowledgments
E.A.L. was supported by K01AG051791, the Harvard Medical School Eleanor and Miles Shore Fellowship, and the Randolph Hearst Fund. This work was supported by the Post-Genome Technology Development Program (10067758, Business model development driven by clinico-genomic database for precision immuno-oncology) funded by the Ministry of Trade, Industry and Energy (MOTIE, Korea). H.J. was supported by a grant from NRF (National Research Foundation of Korea) funded by the Korean Government (NRF-2016H1A2A1907072). We thank Peter J. Park at Harvard Medical School for insightful discussion. We thank Hyojin Kang and Junehawk Lee at the Supercomputing Center of the Korea Institute of Science and Technology Information for providing computing resources and technical support. The results published here are in part based on data generated by The Cancer Genome Atlas managed by the NCI and NHGRI. Information about TCGA can be found at http://cancergenome.nih.gov.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.231837.117.
Freely available online through the Genome Research Open Access option.
References
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. 2009. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462: 108–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V, Davidson NO. 2010. APOBEC-1-mediated RNA editing. Wiley Interdiscip Rev Syst Biol Med 2: 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodmer W, Bishop T, Karran P. 1994. Genetic steps in colorectal cancer. Nat Genet 6: 217–219. [DOI] [PubMed] [Google Scholar]
- Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock RE, Brinkman FS, Lynn DJ. 2013. InnateDB: systems biology of innate immunity and beyond—recent updates and continuing curation. Nucleic Acids Res 41: D1228–D1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet JP, Tamayo P, Golub TR, Mesirov JP. 2004. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci 101: 4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH. 2017. Transposable elements in cancer. Nat Rev Cancer 17: 415–424. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. 2014. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513: 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sun H, You F, Sun W, Zhou X, Chen L, Yang J, Wang Y, Tang H, Guan Y, et al. 2011. Activation of STAT6 by STING is critical for antiviral innate immunity. Cell 147: 436–446. [DOI] [PubMed] [Google Scholar]
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. 2015. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 31: 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes-Ciriano I, Lee S, Park WY, Kim TM, Park PJ. 2017. A molecular portrait of microsatellite instability across multiple cancers. Nat Commun 8: 15180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotto KC, Wagner AH, Feng YY, Kiwala S, Coffman AC, Spies G, Wollam A, Spies NC, Griffith OL, Griffith M. 2018. DGIdb 3.0: a redesign and expansion of the drug–gene interaction database. Nucleic Acids Res 46: D1068–D1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Guo G. 2016. Immunomodulatory function of the tumor suppressor p53 in host immune response and the tumor microenvironment. Int J Mol Sci 17: E1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Uno H, Wooten EC, Elledge SJ. 2017. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355: eaaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P, Morales ME, White TB, Baddoo M, Hedges DJ, Servant G, Srivastav S, Smither ME, Concha M, DeHaro DL, et al. 2017. A comprehensive approach to expression of L1 loci. Nucleic Acids Res 45: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-O'Hare TT, Rodić N, Sharma R, Darbari I, Abril G, Choi JA, Young Ahn J, Cheng Y, Anders RA, Burns KH, et al. 2015. LINE-1 expression and retrotransposition in Barrett's esophagus and esophageal carcinoma. Proc Natl Acad Sci 112: E4894–E4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, et al. 2013. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet 45: 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary RA, Lucas BA, Maquat LE. 2016. Retrotransposons as regulators of gene expression. Science 351: aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Gacita A, Wood LD, Ma F, Xing D, Kim MS, Manda SS, Abril G, Pereira G, Makohon-Moore A, et al. 2015. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res 25: 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A, Sidiropoulos K, Garapati P, Gillespie M, Hausmann K, Haw R, Jassal B, Jupe S, Korninger F, McKay S, et al. 2016. The reactome pathway knowledgebase. Nucleic Acids Res 44: D481–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother WG, Yeo GW, Yeh R, Goldstein P, Mawson M, Sharp PA, Burge CB. 2004. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res 32: W187–W190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, et al. 2017. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45: D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gara RK, Kumari S, Ganju A, Yallapu MM, Jaggi M, Chauhan SC. 2015. Slit/Robo pathway: a promising therapeutic target for cancer. Drug Discov Today 20: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Zack TI, Morris LG, Lin K, Hukkelhoven E, Raheja R, Tan IL, Turcan S, Veeriah S, Meng S, et al. 2014. Pan-cancer genetic analysis identifies PARK2 as a master regulator of G1/S cyclins. Nat Genet 46: 588–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL. 2016. Restricting retrotransposons: a review. Mob DNA 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Cheung LE, Kazazian HH Jr. 2012. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet 8: e1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr. 2016. Roles for retrotransposon insertions in human disease. Mob DNA 7: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. 2014. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res 24: 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat Med 9: 811–818. [DOI] [PubMed] [Google Scholar]
- Hu S, Li J, Xu F, Mei S, Le Duff Y, Yin L, Pang X, Cen S, Jin Q, Liang C, et al. 2015. SAMHD1 inhibits LINE-1 retrotransposition by promoting stress granule formation. PLoS Genet 11: e1005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Kang W, Cheng AS, Yu J, To KF. 2015. The emerging role of Slit-Robo pathway in gastric and other gastro intestinal cancers. BMC Cancer 15: 950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizasa H, Nanbo A, Nishikawa J, Jinushi M, Yoshiyama H. 2012. Epstein-Barr virus (EBV)-associated gastric carcinoma. Viruses 4: 3420–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE. 2010. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141: 1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA, Pearl LH, Carr AM. 2016. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer 16: 35–42. [DOI] [PubMed] [Google Scholar]
- Jin Y, Tam OH, Paniagua E, Hammell M. 2015. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31: 3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8: 118–127. [DOI] [PubMed] [Google Scholar]
- Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, Hong D, Park PJ, Lee E. 2015. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet 47: 1242–1248. [DOI] [PubMed] [Google Scholar]
- Kawakami H, Zaanan A, Sinicrope FA. 2015. Microsatellite instability testing and its role in the management of colorectal cancer. Curr Treat Options Oncol 16: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, Miller CA, Mardis ER, Ding L, Wilson RK. 2012. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 22: 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. 2017. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357: 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ III, Lohr JG, Harris CC, Ding L, Wilson RK, et al. 2012. Landscape of somatic retrotransposition in human cancers. Science 337: 967–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ. 2015. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31: 3555–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. 2011. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milacic M, Haw R, Rothfels K, Wu G, Croft D, Hermjakob H, D'Eustachio P, Stein L. 2012. Annotating cancer variants and anti-cancer therapeutics in reactome. Cancers (Basel) 4: 1180–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Wherry EJ. 2016. Combination cancer therapies with immune checkpoint blockade: convergence on interferon signaling. Cell 165: 272–275. [DOI] [PubMed] [Google Scholar]
- Orecchini E, Doria M, Antonioni A, Galardi S, Ciafrè SA, Frassinelli L, Mancone C, Montaldo C, Tripodi M, Michienzi A. 2017. ADAR1 restricts LINE-1 retrotransposition. Nucleic Acids Res 45: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasare C, Medzhitov R. 2005. Toll-like receptors: linking innate and adaptive immunity. Adv Exp Med Biol 560: 11–18. [DOI] [PubMed] [Google Scholar]
- Picardi E, D'Erchia AM, Lo Giudice C, Pesole G. 2017. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res 45: D750–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, Meyerson M, Getz G. 2015. Oncotator: cancer variant annotation tool. Hum Mutat 36: E2423–E2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebhandl S, Geisberger R. 2015. AIDing cancer treatment: reducing AID activity via HSP90 inhibition. Eur J Immunol 45: 2208–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. 2006. GenePattern 2.0. Nat Genet 38: 500–501. [DOI] [PubMed] [Google Scholar]
- Riess M, Fuchs NV, Idica A, Hamdorf M, Flory E, Pedersen IM, König R. 2017. Interferons induce expression of SAMHD1 in monocytes through down-regulation of miR-181a and miR-30a. J Biol Chem 292: 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. 2013. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet 45: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, Hruban RH, Iacobuzio-Donahue CA, Maitra A, Torbenson MS, et al. 2014. Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol 184: 1280–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N, Steranka JP, Makohon-Moore A, Moyer A, Shen P, Sharma R, Kohutek ZA, Huang CR, Ahn D, Mita P, et al. 2015. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat Med 21: 1060–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. 2015. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160: 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EC, Devine SE. 2017. The role of somatic L1 retrotransposition in human cancers. Viruses 9: E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secrier M, Li X, de Silva N, Eldridge MD, Contino G, Bornschein J, MacRae S, Grehan N, O'Donovan M, Miremadi A, et al. 2016. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet 48: 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R, Upton KR, Muñoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, et al. 2013. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153: 101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org. [Google Scholar]
- Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B, et al. 2012. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res 22: 2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Xie D, O'Kelly J, Miller CW, Muller-Tidow C, Koeffler HP. 2001. Cyr61, a member of CCN family, is a tumor suppressor in non-small cell lung cancer. J Biol Chem 276: 47709–47714. [DOI] [PubMed] [Google Scholar]
- Tubio JM, Li Y, Ju YS, Martincorena I, Cooke SL, Tojo M, Gundem G, Pipinikas CP, Zamora J, Raine K, et al. 2014. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science 345: 1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. 2004. Systematic identification and analysis of exonic splicing silencers. Cell 119: 831–845. [DOI] [PubMed] [Google Scholar]
- Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, et al. 2014. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet 46: 573–582. [DOI] [PubMed] [Google Scholar]
- Wylie A, Jones AE, D'Brot A, Lu WJ, Kurtz P, Moran JV, Rakheja D, Chen KS, Hammer RE, Comerford SA, et al. 2016. p53 genes function to restrain mobile elements. Genes Dev 30: 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Carbone CJ, Katlinskaya YV, Zheng H, Zheng K, Luo M, Wang PJ, Greenberg RA, Fuchs SY. 2015. Type I interferon controls propagation of long interspersed element-1. J Biol Chem 290: 10191–10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylka MJ, Simon JM, Philpot BD. 2015. Gene length matters in neurons. Neuron 86: 353–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.