Abstract
Background:
Cigarette smoke is a causal factor in cancers and cardiovascular disease. Smoking-associated differentially methylated regions (SM-DMRs) have been observed in disease studies, but the causal link between altered DNA methylation and transcriptional change is obscure.
Objective:
Our objectives were to finely resolve SM-DMRs and to interrogate the mechanistic link between SM-DMRs and altered transcription of enhancer noncoding RNA (eRNA) and mRNA in human circulating monocytes.
Method:
We integrated SM-DMRs identified by reduced representation bisulfite sequencing (RRBS) of circulating monocyte DNA collected from two independent human studies [ from Clinical Research Unit (CRU) and from the Multi-Ethnic Study of Atherosclerosis (MESA), about half of whom were active smokers] with gene expression for protein-coding genes and noncoding RNAs measured by RT-PCR or RNA sequencing. Candidate SM-DMRs were compared with RRBS of purified T cells, T cells, granulocytes, B cells, and NK cells ( females, CRU). DMRs were validated using pyrosequencing or bisulfite amplicon sequencing in up to 85 CRU volunteers, who also provided saliva DNA.
Results:
RRBS identified monocyte SM-DMRs frequently located in putative gene regulatory regions. The most significant monocyte DMR occurred at a poised enhancer in the aryl-hydrocarbon receptor repressor gene (AHRR) and it was also detected in both granulocytes and saliva DNA. To our knowledge, we identify for the first time that SM-DMRs in or near AHRR, C5orf55-EXOC-AS, and SASH1 were associated with increased noncoding eRNA as well as mRNA in monocytes. Functionally, the AHRR SM-DMR appeared to up-regulate AHRR mRNA through activating the AHRR enhancer, as suggested by increased eRNA in the monocytes, but not granulocytes, from smokers compared with nonsmokers.
Conclusions:
Our findings suggest that AHRR SM-DMR up-regulates AHRR mRNA in a monocyte-specific manner by activating the AHRR enhancer. Cell type–specific activation of enhancers at SM-DMRs may represent a mechanism driving smoking-related disease. https://doi.org/10.1289/EHP2395
Introduction
Tobacco smoke exposure is associated with a variety of human diseases including cancers of the lung, head and neck, and bladder; chronic obstructive pulmonary disease; osteoporosis; and cardiovascular disease (CDC 2010). Although tobacco smoke constituents cause DNA damage and mutation (Alexandrov et al. 2016; Pfeifer et al. 2002), many adverse outcomes are not related to DNA damage and an emerging view is that the tobacco exposure–associated epigenetic effects (Breitling 2013; Monick et al. 2012; Philibert et al. 2012) may mediate many of these adverse outcomes (Breitling et al. 2012; Breitling 2013; Knopik et al. 2012; Lee and Pausova 2013; Ostrow et al. 2013; Zeilinger et al. 2013).
DNA methylation, one of the best studied epigenetic marks, predominantly occurs at cytosine residues of cytosine phosphate guanine (CpG) dinucleotides, playing an essential role in mammalian embryonic development and gene regulation in response to developmental and environmental cues (Bird 2002; Jones 2012; Li et al. 1992; Meissner 2010; Smith and Meissner 2013). Aberrant DNA methylation can result in altered regulation of gene expression and is observed in various human diseases (Ehrlich 2009; Jones 2012; Smith and Meissner 2013). Recently, highly significant differences in DNA methylation have been observed among individuals exposed to tobacco smoke (Joehanes et al. 2016; Joubert et al. 2012; Shenker et al. 2013; Zeilinger et al. 2013). Epigenome-wide association studies (EWAS) of tobacco smoke exposure using the Illumina Human Methylation 450 BeadChip Array (450K array) on blood DNA have greatly expanded the view of smoking’s impact on the genome and the relationship to disease (Fasanelli et al. 2015; Zhang et al. 2016). For example, 450K array-based EWAS have shown that smoking-associated methylation changes of coagulation factor II (thrombin) receptor-like 3 (F2RL3) at cg03636183 and aryl-hydrocarbon receptor repressor (AHRR) at cg05575921 in whole blood DNA significantly associate with smoking-related cardiovascular disease and lung cancer (Breitling et al. 2012; Fasanelli et al. 2015), suggesting a potential role for blood cells in disease etiology. However, it has been estimated that the 450K array only detects about 7% of potential differentially methylated CpGs in the genome (Ziller et al. 2013). Thus, many CpGs important in the development of smoking-related diseases may still be unknown.
Many CpGs in the AHRR gene have been associated with tobacco smoke exposure, but methylation differences have been the most significant between smokers and nonsmokers at CpG cg05575921 in studies that examined DNA from whole blood (Zeilinger et al. 2013), cord blood (Joubert et al. 2012), or other tissues/cell types (Monick et al. 2012; Reynolds et al. 2015). Recently, in the Multi-Ethnic Study of Atherosclerosis (MESA), the methylation level of cg05575921 in peripheral blood monocytes was associated with both cigarette smoking and subclinical atherosclerosis, and mediation analysis suggested that methylation of AHRR in monocytes may be intermediate in the development of preclinical, smoking-related atherosclerosis (Reynolds et al. 2015). In line with these findings, AHRR has recently been shown to be involved in pro-inflammatory signaling in human circulating monocytes (Zhang et al. 2017), a process central in the development of atherosclerosis (Moore et al. 2013). In the present work, we sought to clarify how smoking-associated AHRR methylation changes are mechanistically connected to AHRR mRNA expression, and how they might contribute to downstream effects. In addition, previous studies using the 450K array may have missed smoking-associated CpGs in AHRR or elsewhere that might be useful biomarkers or biologically important. Therefore, we tested if reduced representation bisulfite sequencing (RRBS)-based methylation analysis could identify additional CpGs associated with smoking exposure and cellular phenotypes.
Noninvasive fluid specimens, such as saliva and urine, are promising patient-friendly resources for disease diagnostics (Lee and Wong 2009; Rodríguez-Suárez et al. 2014). Measuring DNA methylation changes in saliva has been suggested as an alternative to using whole blood to measure biomarkers for head and neck cancer (Schmidt et al. 2016). A recent flow cytometry study observed that saliva cells are primarily leukocytes that were highly enriched for monocytes and granulocytes (Vidović et al. 2012) and a recent DNA methylation study supports this finding (Langie et al. 2016). Su et al. (Su et al. 2016) recently reported that tobacco smoking was strongly associated with methylation levels in both monocytes and granulocytes. Therefore, we also aimed to determine whether saliva can be used as an alternative to whole blood to measure biomarkers of smoking exposure and smoking-related disease.
Methods
Subject Selection
The MESA methylomics cohort of 1,264 individuals is a random subsample from the larger MESA cohort of 6,814 participants. These samples had monocytes isolated from peripheral blood, DNA extracted, and stored and DNA methylation evaluated by Illumina 450K array as previously described (Liu et al. 2013; Reynolds et al. 2015, 2017). As of May 2014, there were 378 individuals with previous 450K, gene expression by RNA-seq, and urinary cotinine data. Among these participants there were 28 current smokers with urinary cotinine concentrations above urine and with sufficient monocyte DNA for further analysis. There were no former smokers in the present study group. Nonsmokers (reported smoking cigarettes in their lifetimes) from the above group were frequency matched to the 28 current smokers by age (), sex, self-identified race, and MESA collection site (Table 1). To discover genome-wide SM-DMRs, 56 MESA DNA samples were converted to RRBS libraries and sequenced. One nonsmoking sample failed quality control (QC) leaving 27 nonsmokers and 28 smokers, and this group is referred to as MESA-RRBS. The study protocol was approved by the institutional review board at each MESA site. All participants signed informed consent for future use of samples.
Table 1.
Demographic and exposure characteristics of the study groups from NIEHS Clinical Research Unit and Multi-Ethnic Study of Atherosclerosis summarized by analysis group. Additional detail on the relationships between these analysis groups is available in Excel Table S1.
| Characteristic | CRU-RRBS | MESA-RRBS | CRU-RRBS- 5 Celltypes | CRU-Pyro | CRU-BSAS | CRU-Genotyping | CRU-BSAS Saliva | mRNA | CRU-eRNAa | CRU-Deep RNA seq | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Smoker () | Non smoker () | Smoker () | Non smoker () | Smoker () | Non smoker () | Smoker () | Non smoker () | Smoker () | Non smoker () | Smoker () | Non smoker () | Smoker () | Non smoker () | Smoker () | Non smoker () | Smoker () | Non smoker () | Smoker () | Non smoker () | |
| Age, y [mean (SD), range] |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Sex (% women) | 61 | 60 | 54 | 56 | 100 | 100 | 69 | 69 | 66 | 70 | 68 | 67 | 78 | 71 | 72 | 74 | 56 | 60 | 67 | 67 |
| Black (n) | 9 | 9 | 6 | 6 | 5 | 5 | 27 | 23 | 24 | 17 | 24 | 21 | 18 | 19 | 15 | 17 | 8 | 7 | 1 | 1 |
| White (n) | 8 | 11 | 17 | 16 | 4 | 5 | 19 | 22 | 18 | 14 | 14 | 22 | 17 | 16 | 13 | 14 | 7 | 8 | 2 | 2 |
| Other (n) | 1b | — | 5c | 5c | — | — | 2b | — | 2b | — | 2b | — | 2b | — | 1b | — | 1b | — | — | — |
| Cotininec, ng/mL serum [mean (SD)] | 249 (106) | 2 (0) | — | — | 217 (102) | 2 (0) | 231 (138) | 2 (1) | 230 (143) | 2 (0) | 224 (120) | 2 (0) | 221 (146) | 2 (2) | 263 (150) | 2 (2) | 245 (156) | 2 (0) | 314 (140) | 2 (0) |
| Cotinined, ng/mL urine [mean (SD)] | — | — | 5,367 (5,757) | 7 (0) | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — | — |
| Cigarettes per day [mean (SD)] | 19 (9) | — | 34 (25) | — | 22 (11) | — | 16 (9) | — | 16 (9) | — | 16 (9) | — | 15 (9) | — | 18 (8) | — | 17 (5) | — | 11 (5) | — |
| Pack-years [mean (SD)] | 27 (20) | — | 11 (7) | — | 34 (24) | — | 21 (19) | — | 21 (19) | — | 23 (18) | — | 19 (18) | — | 24 (17) | — | 22 (12) | — | 14 (12) | — |
Note: —, not applicable; BSAS, bisulfite amplicon sequencing; CRU, clinical research unit; DNaseI, DNase I hypersensitivity region; eRNA, enhancer noncoding RNA; MESA, Multi-Ethnic Study of Atherosclerosis; NIEHS, National Institute of Environmental Health Sciences; RRBS, reduced representation bisulfite sequencing.
eRNA subjects are aggregated, specific assays and analyses are noted in Excel Table S1.
CRU Other: American Indian/Alaska Native.
MESA Other: Hispanic.
CRU Cotinine measured in serum.
MESA Cotinine measured in urine.
As part of the Epigenetic Biomarkers of Tobacco Smoke Exposure project, a second group of volunteers (overall CRU group, ) was recruited by advertising in print, radio, and other media by the NIEHS CRU from the Raleigh-Durham-Chapel Hill, North Carolina, area between January 2013 and August 2015. Nested within this group were the CRU-RRBS, CRU-RRBS-Celltype, CRU-Pyro, CRU-BSAS, CRU-BSAS-Saliva, and gene expression sample groups (Table 1; see also Excel Table S1). Smokers were recruited with the overall goal of achieving approximately equal representation of females, males, blacks, and whites (self-identified), attempting to keep representation similar as the study proceeded. As smokers were accrued, nonsmoking volunteers (reported smoking cigarettes in their lifetimes) were identified from the community and prior to CRU visit were frequency matched on sex, race, and age (, relative to a smoker) (Table 1). Frequency-matched individuals usually donated blood within a few weeks of each other. Data QC, sample availability (DNA and RNA) and assay failures altered matching in final analyses, leaving unequal representation in some analysis groups. Participants filled out a health questionnaire including smoking history. All CRU participants had serum nicotine/cotinine levels measured by HPLC-MS (Quest, Inc.) indicating smoking/nonsmoking status. Several participants reported electronic cigarette use in addition to tobacco smoking, but this information was not used in any analysis. Individuals were ineligible to participate based on the following criteria: being of age, had a body-mass index (), ever been treated with chemotherapy or radiation, smoked marijuana or used recreational drugs such as cocaine within last year, or had a previous cancer diagnosis.
From among the 2013–2014 CRU volunteers, the CRU-RRBS group of 20 smokers (5 black females, 5 white females, 5 black males, 5 white males) was selected and 20 nonsmokers with a similar age, race, and sex distribution were identified, and 40 RRBS DNA libraries from monocytes were created. Two smokers failed QC, leaving 18 smokers and 20 nonsmokers and leaving the distribution of race and sex shown in Table 1. The CRU-RRBS-Celltype group (five additional cell types) was a subset of the CRU-RRBS monocyte group composed of 5 black female smokers, 5 black female nonsmokers, 4 white female smokers, 5 white female nonsmokers (with the exception that 1 white female smoker was replaced due to sample availability). The CRU-RRBS-Celltype group was limited to a smaller number of individuals (all females to minimize variability) due to resource availability. The CRU-Pyro (), CRU-BSAS-CD14 (), and CRU-BSAS-Saliva groups () (see Table 1) were selected from within the overall CRU volunteers (see Excel Table S1) based on availability of bisulfite-treated DNA at the time the AHRR BSAS assays or pyrosequencing assays were run. The samples analyzed in the (), CRU-eRNA (), and CRU-Deep RNA-seq () groups generally overlapped those in the CRU-RRBS group. Although sample selection for these analyses was based on maintaining similar numbers of smoker/nonsmokers, ages, race, and sex across a given assay, after many assays and over several years of the project, low RNA sample availability for some cell types at the time of the analysis resulted in arbitrary numbers for some groups. CRU-Deep RNA-seq samples were chosen based on DNA methylation values at cg05575921. The relationships between the analyzed groups are shown in Excel Table S1. The NIEHS CRU groups were recruited under protocol 10-E-0063 approved by the NIEHS Institutional Review Board and all subjects provided informed written consent for use of samples.
Cell Isolation
MESA participants had blood collected by venipuncture into sodium heparin Vacutainer CPT tubes (Becton Dickinson) and cells were isolated within 2 h using anti-CD14 antibody–coated magnetic beads (Miltenyi Biotek) as described previously (Liu et al. 2013). For all peripheral blood samples collected at the CRU, mononuclear cells were isolated using Ficoll_Paque PLUS (Sigma-Aldrich) and individual cell types were isolated serially from mononuclear cells using Invitrogen Dynabeads (, , , , respective order) and Miltenyi Biotec MicroBeads (). granulocytes were isolated directly from whole blood within 4 h using antibody–coated magnetic beads following the protocol from the manufacturer (Invitrogen). Samples were stored frozen at in QIAGEN ALLPrep DNA/RNA/miRNA Universal Kit extraction buffer (QIAGEN). Saliva cells were collected from all CRU participants using Oragene® DNA Self-Collection Kits (DNA Genotek) under supervision of CRU staff and stored at room temperature until extracted ().
DNA and RNA Isolations
DNA and RNA from purified monocytes, T cells, T cells, granulocytes, B cells, and NK cells collected for all subjects at the CRU were extracted using QIAGEN ALLPrep DNA/RNA/miRNA Universal Kit (QIAGEN). Saliva cell DNAs were isolated using DNA Genotek kit according to the manufacturer’s instruction (DNA Genotek). Samples were stored frozen at until use and then at 4°C.
RRBS
Genomic DNA () from three RRBS groups was converted into libraries: RRBS-MESA ( monocytes, libraries); RRBS-CRU ( monocytes, libraries); and RRBS-Celltype (5 Celltypes, ; 95 libraries). The library construction protocol used was modified from Boyle et al. (2012). Briefly, 50 pg of phage lambda control DNA was added to human genomic DNA and digested with () Msp I restriction enzyme (New England Biolabs, catalog number R0106L) in a reaction containing of NEB buffer 2 at 37°C overnight. Digestion reaction was stopped by adding of 0.5M EDTA, and DNA was purified and eluted with of EB buffer. Fifteen microliters of purified DNA was repaired at 37°C for 30 min in a reaction containing resuspension buffer and end repair mix from Illumina RNA TruSeq Kit (Cat. No. RS-122-2001) followed clean-up ends repaired DNA by MinElute PCR purification Kit (Qiagen, Cat. No. 28004). Fifteen microliters of eluted DNA was incubated at 37°C for 30 min followed by incubation at 70°C for 5 min in a A-tailing reaction containing resuspension buffer and 12.5 A-tailing mix from Illumina RNA TruSeq Kit above. Then, resuspension buffer, ligation mix, and 2.5 adapter (1:15 dilution from original vial from Illumina RNA TruSeq Kit using Tris-EDTA buffer) were added and incubated at 30°C for 10 min followed by adding stop ligation mix (from Illumina RNA TruSeq Kit above) and incubating on ice. Ligated DNA was purified using AMPure beads and eluted in resuspension buffer followed by two rounds of bisulfite conversion using EpiTect Bisulfite Kit (Qiagen, Cat. No. 59104) per manufacturer’s instruction. Bisulfite-converted DNA was purified with AMPure beads and eluted in . DNA was amplified in a reaction using Illumina primer cocktail from Illumina RNA TruSeq Kit and PfuTurbo Cx polymerase (Agilent, Cat. No. 600410-51) with 30-s initial denature at 98°C and 9–12 cycles of 98°C for 10 s, 65°C for 30 s, 72°C for 30 s followed by 72°C for 5 min. Polymerase chain reaction (PCR) products were sequentially purified with volume of AMPure beads and eluted in resuspension buffer. RRBS libraries were sequenced on either Illumina HiSeq 2500 (125 bp sequencing length, pair-end) at the NIH Intramural Sequencing Center (MESA) or Illumina NextSeq (CRU) at the NIEHS Epigenomics Sequencing core (100 bp sequencing length, pair-end). In total, 193 RRBS libraries were sequenced.
Bisulfite Amplicon Sequencing
Bisulfite amplicon sequencing (BSAS) was performed as previously described (Masser et al. 2013). Briefly, genomic DNA per sample from the 74 CRU-BSAS and 76 CRU-BSAS-Saliva participants (from either monocytes or saliva) was bisulfite converted followed by PCR amplification with bisulfite-specific primers flanking the AHRR SM-DMR (see Table S1). PCR products were purified (MinElute PCR purification kit, Qiagen) and quantified with Qubit. Purified PCR products () were indexed using tagmentation with Nextera XT 96 index kit (Illumina) followed by PCR amplification using barcoded primer sets for each methylation control (5 reference methylation samples in triplicate) and 78 subject samples. PCR libraries were purified twice using AMPure beads with a 1:1 ratio volume. Purified libraries were pooled and sequenced using Illumina MiSeq. Sequencing data was processed and methylation percentage was calculated as described previously (Masser et al. 2013) and below.
RRBS/BSAS Processing
Sequencing read pairs with a mean Phred quality score of were removed and Trim Galore! (version 0.2.8) removed adapters from the reads. We used Bismark (version 0.14.3) to align the reads to the hg19 assembly, extracted the number of methylated and unmethylated cytosines at each CpG site, and derived methylation percentages, excluding any sites that had less than 10 reads or occurred at a single nucleotide polymorphism. We calculated the average methylation percentages for each group (smoker/nonsmoker) by dividing the count of methylated reads by the total number of reads at each covered CpG site. For the individual MESA-RRBS and CRU-RRBS groups, we required that samples have reads for the CpG site to be used, with samples for combined group. Average methylation percentages were based on samples reads. For BSAS we used Bismark (version 0.14.3) to align the reads to the amplified PCR fragment, extracted the number of methylated and unmethylated cytosines at each CpG site, and derived methylation percentages for each CpG for each CRU-BSAS and CRU-Saliva sample.
DMR Identification
To identify DMRs in RRBS data from CRU-RRBS and MESA-RRBS groups, we developed a method modified from Ziller et al. (2013), separately testing each sequenced CpG. A pseudocount of 1 methylated cytosine and 1 unmethylated cytosine was added when calculating the methylation percentages prior to logit transformation. We then performed an independent two-group Student’s t-test () on the logit-transformed methylation percentages from each group (smoker/nonsmoker). CpGs were grouped into potential DMRs by merging together any of the significantly different CpGs within a 500-nt window. The potential DMR was defined as the region spanning the most upstream and downstream CpGs in the merged set of CpGs. We calculated a weighted methylation score for each sample within each potential DMR, by using the sum of all methylated and the sum of all unmethylated cytosines for any CpGs within the region with reads. As before, a pseudocount of 1 methylated and 1 unmethylated cytosine was added to these sums and performed another independent two-group Student’s t-test on the logit-transformed weighted methylation percentages from each group. Finally, the potential DMR survived to the final set of DMRs if it had a p-value of , an overall methylation difference between groups of 1% or greater, and at least three CpGs within the DMR region. DMRs were called for MESA-RRBS and CRU-RRBS groups individually, and then these data sets were also pooled (Combined) and DMRs called (see Excel Tables S2–S4). Average CpG methylation across the AHRR DMR was calculated for each individual and used in correlations with CpG cg05575921 methylation, BSAS, and gene expression.
We selected DMRs for validation by filtering each DMR data set by and methylation difference , ranking by p-value, and initially prioritizing those DMRs that were highly ranked and observed in all three analyses. Given the potential for false positive results, these lists were interpreted as qualitative results. A differential methylation value (relative to nonsmokers) was calculated for each CpG in each RRBS group and these methylation values were mapped to the University of California, Santa Cruz (UCSC) Genome Browser Hg19. We then examined genome browser methylation tracks of DMRs displaying the greatest significance and/or methylation difference and qualitatively assessed the similarity of the DMR region across the two study groups and compared the similarity of these DMRs across the five additional cell types. HOXA5 was selected because of multiple DMRs extending through the HOXA gene cluster. We examined numerous potential DMR locations and these are listed in Excel Tables S2–S4, but only those listed in Table 2 were followed up by pyrosequencing or BSAS.
Table 2.
Candidate SM-DMRs identified by RRBS analysis in CRU and MESA groups selected for validation in the CRU-Pyro group.
| SM-DMR Nearest Gene Transcription Start | Chr | DMR start | Number of CpGs in DMR | 450K covered CpGs | 450K smoking CpGs | a (%) | RRBS b Combined p-value | Group Signif c | Valid | H3 K27ac | DNaseI TFBS | Biological function/diseasesd | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALPPL2 | Chr2 | 233283913 | 65 | 3 | 3 |
|
Aging, breast cancer | Salpea et al. 2012; Dua et al. 2013 | |||||
| AHRR | Chr5 | 373377 | 5 | 1 | 1 |
|
Atherosclerosis, immunity | Reynolds et al. 2015; Zhang et al. 2017 | |||||
| LRP5 | Chr11 | 68148288 | 3 | 0 | 0 |
|
Atherosclerosis | Borrell-Pages et al. 2014 | |||||
| PPP1R15A | Chr19 | 49379722 | 9 | 0 | 0 |
|
Peripheral arterial disease | Masud et al. 2012 | |||||
| SLC24A3 | Chr20 | 19193923 | 9 | 1 | 1 | M C | — | Obesity, hypertension, | Logsdon et al. 2012; Mick et al. 2011 | ||||
| SASH1 | Chr6 | 148684696 | 4 | 0 | 0 | M C | F | Atherosclerosis | Weidmann et al. 2015 | ||||
| RDX | Chr11 | 110079735 | 25 | 0 | 0 | 20 | C | NS | — | Deafness | Kitajiri et al. 2004 | ||
| C5orf55-EXOC-AS | Chr5 | 403252 | 3 | 0 | 0 |
|
— | — | |||||
| F2RL3-CPAMD8 | Chr19 | 17004139 | 15 | 0 | 0 |
|
— | Cardiovascular disease, cancer | Fasanelli et al. 2015; Muka et al. 2016; Zhang et al. 2016 | ||||
| F2RL3 | Chr19 | 17000053 | 9 | 0 | 0 |
|
Cardiovascular disease, cancer | Zhang et al. 2016; Fasanelli et al. 2015; Muka et al. 2016 | |||||
| HOXA5 | Chr7 | 27184187 | 42 | 16 | 0 | — | NS | Atherosclerosis | Dunn et al. 2014 | ||||
| Total | 189 | 21 | 5 | ||||||||||
Note: —, not applicable; 450K covered, CpGs within DMR on the Illumina Human 450K Methylation Array; 450K smoking CpGs, CpGs within the DMR on the Illumina Human 450K Methylation Array associated with smoking; C, combined CRU-MESA; C, CRU; Chr, Chromosome; CRU, Clinical Research Unit; DMR start, Transcription start site; DMR, differentially methylated region; DNaseI, DNase I hypersensitivity region; F, assay design failure; H3K27ac, Histone 3, Lysine 27 acetylation; M, MESA; MESA, Multi-Ethnic Study of Atherosclerosis; NS, not significant; RRBS, reduced representation bisulfite sequencing; SM, smoking associated; TFBS, transcription factor binding site.
Meth Diff, average methylation difference between smoking and nonsmoker.
SM-DMR p-value for the Combined CRU-MESA RRBS analysis.
Analysis groups where DMR was observed as significant: CRU, M, C analysis.
Reported disease association or possible biological function as reported in listed reference.
Genomic Feature Coverage
To determine the RRBS coverage distribution and DMR overlap with various genomic features, we first defined a set of locations for each feature. We defined promoters as the region 1-kb upstream to 1-kb downstream of all protein-coding RefSeq gene transcription start sites (TSSs). We downloaded ENCyclopedia Of DNA Elements (ENCODE) project-defined CpG islands, DNaseI hypersensitivity sites, and transcription factor (TF) binding sites from the following UCSC Hg19 genome browser tracks: CpG Islands; DNaseI Hypersensitivity Clusters in 125 cell types from ENCODE (version 3); and Transcription Factor ChIP-seq (161 factors) from ENCODE with Factorbook Motifs. We defined CpG island shores as the regions adjacent upstream and downstream to the defined CpG islands. We defined enhancers using Hg19 genome coordinates for predicted enhancers reported previously (Ziller et al. 2015) and provided to us by A. Meissner, Harvard University. For CRU-RRBS and MESA-RRBS samples, each CpG in the genome was counted and if it was present in samples and occurred in a genome feature defined above, it was counted toward genome feature coverage. Enrichment was plotted by dividing the number of SM-DMRs found in a given genome feature by the total number of CpGs observed in all DMRs ().
Pyrosequencing
Pyrosequencing assays were developed for nine DMRs in eight genes: Homeobox A5 (HOXA5), Alkaline phosphatase, placental like 2 (ALPPL2), AHRR-C5orf55-EXOC-AS, F2RL3, F2RL3-C3 and PZP like, alpha-2-macroglobulin domain containing 8 (CPAMD8), protein phosphatase 1 regulatory subunit 15A (PPP1R15A), Solute carrier family 23 member 3 (SLC24A3), LDL receptor related protein 5 (LRP5), and Radixin (RDX) (see Table S1) using PyroMark® Design software (QIAGEN). DNA was bisulfite converted using EZ DNA Methylation Gold Kit (Zymo Research) following manufacturer’s instructions. All PCRs contained bisulfite-treated genomic DNA, each primer, , dNTPs, DMSO, PCR buffer, and 2 units Platinum® Taq polymerase (Invitrogen). Thermocycling conditions were 95°C for 15 min, 5 cycles each of , (in 1°C decreasing increments), and 15 cycles of , , and . PCR products () were sequenced with sequencing primer using a PyroMark Q96 MD (QIAGEN). DNA methylation reference samples were used to validate each pyrosequencing assay. Average CpG methylation across the DMR was determined for samples with methylation values at each CpG in the DMR. In DMRs (all except SLC24A3) with multiple CpGs, only subjects with successful results for all CpGs were used in statistical analyses, leading to slightly different sample sizes for each DMR as noted in the figures.
Reverse Transcription Quantitative Polymerase Chain Reaction
Total RNA samples were converted to cDNA for reverse transcription quantitative polymerase chain reaction (RT-qPCR) using the SuperScript® III First-Strand Synthesis (Life Technologies) with random hexamer and oligo-dT primers. For each sample, target genes (AHRR, ALPPL2, PPP1R15A, SLC24A3, AHRR-C5orf55-EXOC-AS, F2RL3, CPAMD8, HOXA5, SASH1, LRP5) and the reference gene (ACTB) were amplified in triplicate using TaqMan assays (see Table S2) designed to span exon junctions using Universal PCR Master Mix and run on an ABI 7900HT (Life Technologies). Data was normalized to ACTB and fold change (FC) was assessed relative to nonsmokers using the method. Samples were selected from all of the CRU-RRBS group and most of the CRU-BSAS group; however, low RNA quantity or quality limited the analysis of some participants. CRU-mRNA group characteristics are shown in Table 1 and individual samples used are listed in Excel Table S1.
NonCoding RNAs and Enhancer Noncoding RNAs Measured by RT-qPCR
Methods for measuring noncoding RNA/enhancer noncoding RNA (ncRNA/eRNA) gene expression were identical to those used to measure RNA expression with the following exceptions, custom-designed SYBR primers (see Table S3) were used and assays were run on a ViiA7 real-time PCR system (Life Technologies). Cycle threshold values were normalized to ACTB, and FC was assessed relative to nonsmokers using the method. Samples for these analyses were selected from among the CRU-RRBS group if possible, but low RNA quantity limited the analysis of some participants.
Ribosomal Depleted RNA-Seq
We selected three nonsmokers and three smokers for deep RNAseq to confirm the genome location of AHRR enhancer RNA in smokers. We used BSAS methylation values at AHRR CpG chr5:373378 (cg05575921) to identify three nonsmokers with methylation values typical of nonsmokers (; ; ), and three smokers with methylation values typical of smokers (; , ). Purified monocyte total RNA was sent to the Genomic Services Laboratory of the Hudson Alpha Institute of Biotechnology (Huntsville, AL) for RNA library construction using ribosomal-depleted RNA (Ribozero, Illumina) followed by sequencing on the HiSeq 4000 to a depth of million reads per sample.
RNA-Seq Processing
We first removed any read pairs where either mate had a mean Phred quality score of . Next, we used Cutadapt (version 1.11) to remove adapter from the reads. Then, we used TopHat2 (version 2.0.4) to align the reads to the hg19 assembly. Only data for the AHRR enhancer region was utilized for this project.
Genotyping
A haplotype in the AHRR gene region containing the single nucleotide polymorphism (SNP) rs148405299 (C/CA) was recently reported to associate with differences in methylation in neonates (Gonseth et al. 2016), and we tested for this association in our CRU-Pyro samples. No genotyping assay for rs148405299 was available, so we genotyped a nearby SNP rs11741712 on the same haplotype that is in linkage disequilibrium () with rs148405299 in both European and African descent populations as determined by the 1,000 Genomes Project. Genomic DNA from individuals in the CRU-Pyro group was combined with rs11741712 allele-specific genotyping assay reagents (ThermoFisher Scientific), PCR amplified, and analyzed per manufacturer’s instructions. DNA samples genotyped by the 1,000 Genomes Project were used as positive controls.
Statistical Analysis and Multivariable Modeling
Significant differences between methylation levels of smokers and nonsmokers at specific CpGs were assessed by t-test, and comparisons between methods were accomplished by linear regression in Graphpad Prism (reported as r, or ). For pyrosequencing analyses, to compare smokers and nonsmokers for a given DMR, we used multivariable linear regression models both adjusted and unadjusted for age, race, and sex. Results for adjusted and unadjusted models were not substantively different; adjusted values are reported. For rs11741712 (AHRR) genotype analysis, multivariable linear regression was to investigate the effects of genotype and genotype*smoking interaction on methylation at cg05575921. Genotype was coded as a binary indicator variable for presence of the minor allele and was used as a proxy for smoking. Linear regression models were adjusted for age, race, and sex.
Data Availability
The RRBS methylation data sets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, Accession No. GSE104700.
Results
Genome-Wide DNA Methylation Mapping by RRBS
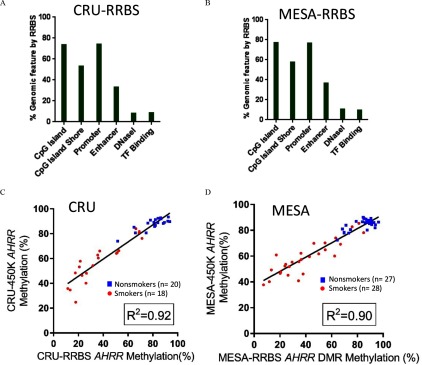
RRBS captures up to 2 million CpG sites and can detect DMRs that contain clusters of adjacent CpGs with similar methylation changes (Bock et al. 2012; Boyle et al. 2012; Gu et al. 2011). RRBS captured an average of 2.3 million CpGs for the 55 subjects in the MESA-RRBS group, and 1.9 million CpGs for 38 subjects in the CRU-RRBS group) at average sequencing depth across the genome. However, the depth of coverage at each CpG was not consistent across subjects, introducing considerable variability into the analysis. Figure 1A,B displays the similarity in the distributions of CpGs that were captured by RRBS in MESA and CRU subjects relative to genome features and this was consistent with other studies (Boyle et al. 2012; Ziller et al. 2013). We observed there were 77.1% and 73.5% of CpG islands (CGIs), 57.9% and 53.2% of CGI shores, and 76.8% and 74.5% of promoters captured by RRBS in MESA-RRBS and CRU-RRBS groups, respectively. Lower coverage was observed for enhancers (MESA-RRBS, 36.7%, and CRU-RRBS, 33.2%), and other regulatory features indicated by DNaseI hypersensitivity (MESA-RRBS, 10.7%, and CRU-RRBS, 8.4%) and TF binding regions (MESA-RRBS, 9.9%, and CRU-RRBS, 8.5%) as defined by ENCODE (Wang et al. 2012) (Figure 1A,B).
Figure 1.
Genome distribution of CpGs captured by RRBS. For (A) CRU-RRBS () and (B) MESA-RRBS (), bar plots show the percentages of CpGs with RRBS coverage that were located in six types of genomic features, including CpG islands (CGI), CGI shore, promoter, enhancer, DNaseI hypersensitivity region, and TF binding regions derived from ENCODE data sets (Wang et al. 2012) relative to the genomic total of these features. For example, MESA-RRBS [ (77.06%), CGI (57.88%), (76.78%), (36.69%), (10.67%), (9.93%)]. (C–D) The correlation between average methylation level of CpGs within the AHRR SM-DMR by RRBS and the methylation value of the cg05575921 site, a CpG within the AHRR SM-DMR that was determined by 450K array for the same subjects in each population, (C) CRU (), (D) MESA (). Blue square symbols represent nonsmokers, whereas red circles indicate smokers. Note: CGI, CpG island; CRU, Clinical Research Unit; DMR, differentially methylated region; DNaseI, DNase I hypersensitivity region; MESA, Multi-Ethnic Study of Atherosclerosis; RRBS, reduced representation bisulfite sequencing; SM, smoking associated; TF, transcription factor.
Characterization of Smoking-Associated Differential Methylation
To characterize smoking-associated differential methylation, we analyzed RRBS data from MESA and CRU samples using a DMR identification algorithm adapted from previous studies (Boyle et al. 2012; Gu et al. 2011; Ziller et al. 2013) in which the first step assessed differential methylation at each CpG (DMC). As a quality check, we compared the average methylation value of CpGs within the AHRR SM-DMR assessed by RRBS with the methylation value of the cg05575921 site, a CpG within the AHRR SM-DMR, previously determined by 450K array for the same subjects in both MESA-RRBS (Reynolds et al. 2015) and CRU-RRBS groups (Figure 1C,D) (Su et al. 2016). The correlations of AHRR methylation between RRBS and 450K array were (MESA-RRBS) and (CRU-RRBS).
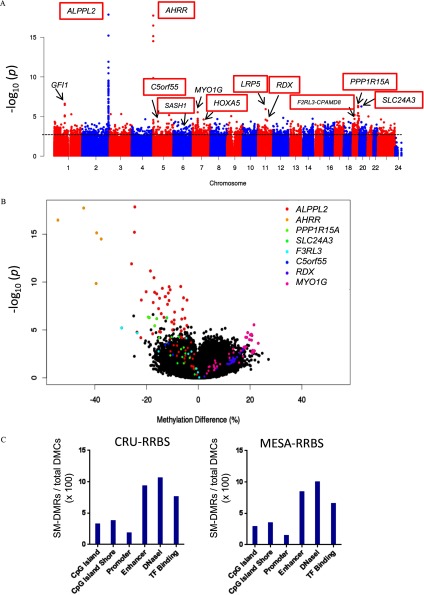
In Fig.2A, the Manhattan plot shows the significance of association (t-test) between cigarette smoking and methylation at individual RRBS-derived CpGs (data from Combined CRU-RRBS and MESA-CRU subjects) across the human genome. We found numerous nominally significant novel DMCs at or nearby 450K array-derived, widely replicated smoking-associated locations in or near GFI1, ALPPL2, AHRR, MYO1G, SLC24A3, PPP1R15A (GADD34), and F2RL3 genes (listed in Excel Table S5) and at previously unknown smoking-associated locations in or near HOXA5, RDX, and LRP5. Many of these can be visualized in the methylation tracks in Figure 3A (ALPPL2), 3B [PPP1R15A (GADD34)], 3C (SLC24A3) {see also Figure S1B [AHRR], S1E [PPP1R15A (GADD34)], and S1F [F2RL3]}. This is most clearly observed in the magnified view in Figure S1K (HOXA5). Although both hypermethylated and hypomethylated smoking-associated CpGs were observed, the majority of the highly significant CpGs were hypomethylated, as seen in the volcano plot (Figure 2B).
Figure 2.
Genome-wide distribution of smoking-responsive CpGs assessed by RRBS. (A) Manhattan plot of Combined analysis of CRU-RRBS and MESA-RRBS data together shows significance of smoking-associated CpGs across the genome by chromosome number, except sex chromosomes, as indicated, whereas y-axis represents significance in log scale (p-value). Candidate SM-DMRs chosen for follow-up are labeled including those validated (highlighted by red boxes) by pyrosequencing or BSAS. Dotted line indicates . (B) Volcano plot shows differential methylation percentage for smoking-associated differentially methylated CpG sites. The x-axis shows the percentage of methylation difference between smokers and nonsmokers, whereas y-axis represents significance in log scale (p-value). (C) Bar plots show the number of smoking-associated DMRs relative to the total number of differentially methylated CpGs (SM-DMCs) within these DMRs () derived from RRBS (CRU, MESA) for six types of genomic features as shown. Note: BSAS, bisulfite amplicon sequencing; CRU, clinical research unit; DMC, differential methylation at each CpG; DMR, differentially methylated region; MESA, Multi-Ethnic Study of Atherosclerosis; SM, smoking associated; RRBS, reduced representation bisulfite sequencing.
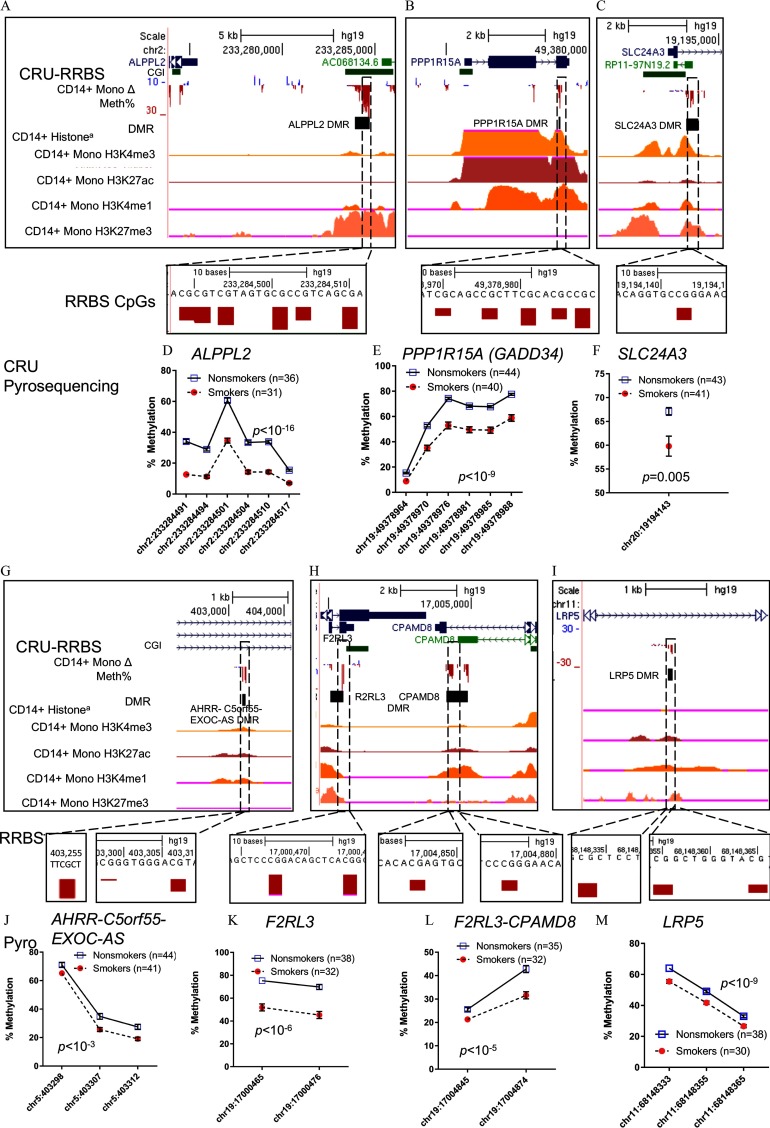
Figure 3.
Validation of candidate SM-DMRs using pyrosequencing. (A–C, G–I) Selected candidate SM-DMRs (A) ALPPL2, (B) PPP1R15A (GADD34), (C) SLC24A4, (G) AHRR-C5orf55-EXOC-AS, (H) F2RL3, F2RL3-CPAMD8, and (I) LRP5 that displayed significant, smoking-associated methylation differences by pyrosequencing of monocyte DNA from CRU-Pyro group described in Methods. Upper boxed panels (A–C, G–I) show genome browser tracks for Refseq genes, CpG islands, differential methylation by RRBS ( Mono RBBS-CRU group, downward red bar (negative value) is loss of methylation, upward blue bar is gain of methylation), and aRoadmap Epigenome histone modification tracks for monocytes are shown: H3K4 trimethyl (active mark), H3K27acetyl (active enhancer), H3K4 monomethyl (enhancer), H3K27trimethyl (repressive mark). Insert shows close up view of the RRBS CpGs that were tested in the CRU-Pyro group. Lower panel graphs (D–F, J–M) show mean methylation () by pyrosequencing for individual CpGs within DMRs as measured in the CRU-Pyro group (67–85 subjects per assay, with indicated numbers for nonsmokers (blue squares) and smokers (red circles). In most cases the error bars are smaller than the symbol. Sample numbers varied across assays due limited sample availability and assay failures. The CRU-RRBS subjects were included in the CRU-pyrosequencing group. p-Values shown in each panel are for the estimated difference in mean methylation by pyrosequencing across each DMR (smokers vs. nonsmokers) from multivariable linear regression models adjusted for age and sex. Note: CRU, clinical research unit; DMR, differentially methylated region; RRBS, reduced representation bisulfite sequencing; SM smoking associated.
The smoking-associated DMR (SM-DMR) method determined regions where the overall methylation of the CpGs (minimum of three CpGs) within a 500-nt region was different between smokers and nonsmokers in each population. We identified SM-DMRs from MESA-RRBS (2,756), CRU-RRBS (1,989), and pooled (Combined; 2,057) data sets at a nominal significance level of (see Excel Tables S2–S4). We identified a set of 439 genes with nominal SM-DMRs observed in both RRBS groups (see Excel Tables S6 and S7). In contrast to RRBS’s overall bias toward CGIs and promoters, SM-DMRs appeared to be distributed preferentially at enhancers, as well as sites of DNase I hypersensitivity and TF binding (Figure 2C). We considered sequencing read depth at two highly significant, known smoking-DMCs within SM-DMRs and observed variability across individuals. For example, at chr5:373378 (cg05575921 on the 450K array) in AHRR, the average read depth in the CRU-RRBS group was (SD) reads with a range of 3–50, with 13 individuals at less than 10-fold coverage dropping out of the analysis. Similarly, in the GFI1 gene at Chr1:92947588, the mean read depth was , with 26 individuals dropping out of the analysis. This variability of RRBS sequencing read depth in our data sets indicated a need for interpreting RRBS DMRs as a qualitative outcome needing additional interpretation. Therefore, we inspected methylation tracks on the UCSC browser for the most significant SM-DMRs, those with the largest methylation in each RRBS group, those shared by both groups, or those observed in the Combined RRBS reanalysis (listed in Excel Tables S2–S4). We compared these potential RRBS SM-DMRs for each group with methylation tracks created for CRU-RRBS-Celltype analysis and looked for qualitative pattern similarities. Figure S1A (HOXA locus) provides an example of a gene region with multiple potential SM-DMRs displaying loss of methylation and observed in all cell types. We selected 10 additional candidate SM-DMRs (Table 2) for validation based on their significance (low p-values), methylation difference (effect size), and observed consistency of methylation patterns across each study group and each cell type (observed in the methylation tracks in Figure S1A–J). These top candidate DMRs tended to be close to regulatory regions as indicated by Roadmap project H3K27ac tracks and clusters of transcription factor ChIP-sec binding (shown in Roadmap and ENCODE TF browser tracks in Figure S1A–J, listed in Table 2).
Validation of SM-DMRs by Pyrosequencing or BSAS
To validate RRBS candidates, we used either pyrosequencing or bisulfite amplicon sequencing (BSAS) of SM-DMRs in or near ALPPL2, AHRR, PPP1R15A (GADD34), SLC24A3, AHRR-C5orf55-EXOC-AS, F2RL3-DMR1, F2RL3-CPAMD8-DMR2, HOXA5, SASH1, RDX, and LRP5 and on monocyte DNA from a larger group of 85 CRU-Pyro subjects that included the 38 CRU-RRBS subjects (Table 1; see also Excel Table S1). Among these 11 selected SM-DMRs, only 5 of 189 CpG sites (including 1 in AHRR SM-DMR, 3 in ALPPL2 SM-DMR, and 1 in SLC24A3 SM-DMR), overlap with smoking-associated CpGs previously identified by 450K array in a large meta-analysis (Joehanes et al. 2016). We successfully validated 7 SM-DMRs (ALPPL2, PPP1R15A(GADD34), SLC24A3, AHRR-C5orf55-EXOC-AS, F2RL3-DMR1, F2RL3-CPAMD8-DMR2, and LRP5) by pyrosequencing (Figure 3), and the average methylation () levels at each CpG are shown (Figure 3D–F,J–M). As shown in Figure 3A–C,G–I histone tracks, these DMRs overlap or flank potential regulatory regions in monocytes indicated by CpG islands and/or the presence of histone modifications for enhancers (H3K4me1, H3K27ac) or promoters (H3K4me3). For the overlapping individuals in the CRU-RRBS and CRU-Pyro groups (), the Pearson correlation between the average DNA methylation across the RRBS DMR and the pyrosequencing fragment was over all DMRs (see Figure S2G) and as shown for 6 individual SM-DMRs (see Figure S2A–F). Pyrosequencing assay design failed for the SASH1 SM-DMR. Several locations in HOXA5 and RDX were tested, and results are shown in Figure S3A,B. Methylation differences were not significant by pyrosequencing, however, the methylation trends for HOXA5 and RDX were consistent with RRBS results (see Figure S3A,B).
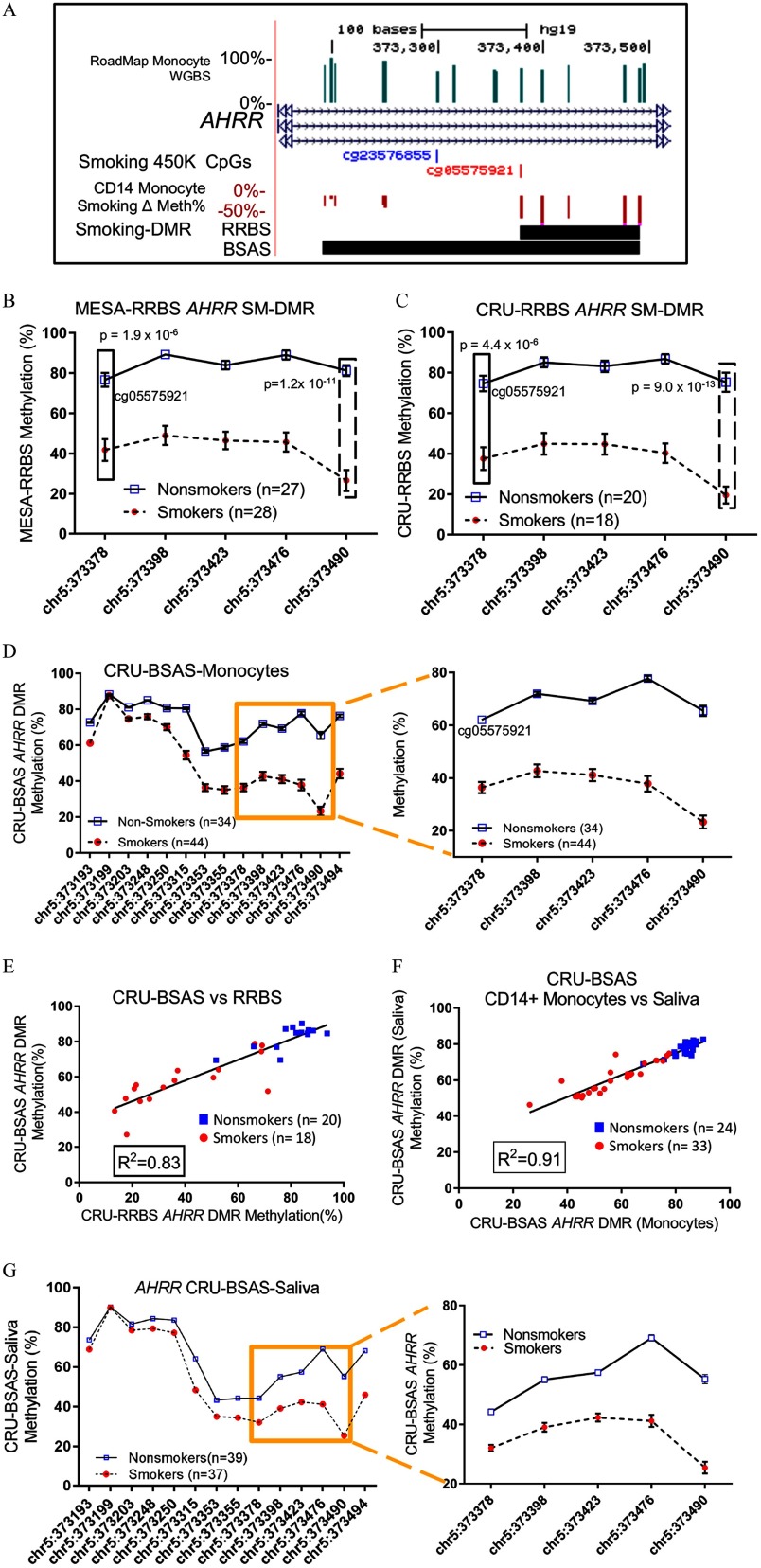
RRBS captures a large number of CpGs in the AHRR gene, including the most significantly affected smoking-associated 450K CpG site (cg05575921) (Joubert et al. 2012; Shenker et al. 2013; Zeilinger et al. 2013), and our combined RRBS results from both MESA and CRU groups identified the AHRR SM-DMR at this locus as the most altered SM-DMR across the genome in terms of effect size (42% differential methylation) and p-value () (Figure 4A, Table 2). This AHRR DMR contains at least 5 CpGs that display hypomethylation in smokers (Figure 4A), and this DMR can be observed similarly in granulocytes (of the same myeloid lineage) but to a lesser extent in lymphoid lineages (B cells, CD4 T cells, CD8 T cells, NK cells) (see Figure S1B, boxed). Several adjacent CpGs show similar levels of demethylation and one novel CpG at chr5:373490 within AHRR DMR appears to display a greater effect size ( demethylation, ) than the cg05575921 site located at chr5:373378 (, ) (Figure 4A). This observation was consistent for both effect size and p-value in both MESA-RRBS and CRU-RRBS groups (Figure 4B,C). Using BSAS, a 360-bp region containing 14 CpG sites (Figure 4A,D), including the AHRR SM-DMR, was examined in the CRU-BSAS group. BSAS results confirmed the AHRR SM-DMR methylation levels were highly correlated with those determined by RRBS for the same subjects (Figure 4E). This analysis also recapitulated the observation that the novel CpG at chr5:373490 displayed a greater smoking-responsive effect on methylation than that of the cg05575921 site (Figure 4B-D).
Figure 4.
Fine mapping and validation of the AHRR SM-DMR in both monocytes and saliva. (A) Close up browser view of SM-DMR in the third intron of AHRR gene. Genome browser tracks indicate, from the top to bottom: methylation level of CpGs as determined by Roadmap Epigenome Project, Refseq gene (AHRR), smoking-responsive CpGs on the 450K methylation array (Joehanes et al. 2016), differentially methylated CpGs ( Methylation %, downward red bars indicate loss of methylation) and SM-DMRs (horizontal black bar) as determined by RRBS (5 CpGs) and replicated by BSAS (14 CpGs). (B) Mean % methylation () at each CpG in the DMRs measured by RRBS in (B) MESA-RRBS and (C) CRU-RRBS in smokers and nonsmokers. The left-most CpG site (chr5:373378, boxed) of the SM-DMR is the previously identified, highly significant smoking-responsive CpG site (cg05575921), whereas the right-most site (chr5:373490, dashed box) displays a larger effect size and significance indicated by p-value (t-test). (D) Methylation levels of 14 CpGs were captured in monocyte DNA for the CRU-BSAS group as indicated, including 5 CpG sites measured by RRBS (boxed region enlarged in right-hand panel). (E) Correlation analysis between RRBS methylation % and BSAS methylation % for nonsmokers (blue boxes) and smokers (red circles) for the overlapping 38 CRU-RRBS, CRU-BSAS samples tested in both assays. (F) Correlation analysis between monocyte methylation % (BSAS) and saliva DNA methylation % (BSAS) for CRU-BSAS samples who also provided saliva DNA. (G) AHRR SM-DMR was recapitulated in the CRU-BSAS-saliva group using saliva DNA from 37 smokers and 39 nonsmokers. Note: BSAS, bisulfite amplicon sequencing; CRU, clinical research unit; DMR, differentially methylated region; MESA, Multi-Ethnic Study of Atherosclerosis; RRBS, reduced representation bisulfite sequencing; SM, smoking associated.
We evaluated the possibility that methylation of the AHRR CpG at cg05575921 could be affected by genetic variation as reported (Gonseth et al. 2016), specifically alleles of the AHRR haplotype containing nearby linked SNPs, rs148405299 and rs11741712 (). In the CRU-Pyro group, we detected no significant effect of AHRR genotype (rs11741712) on cg05575921 methylation as a main effect () or as an interaction with smoking (). However, the allele frequency for the rare allele in the tested group was low (0.04), limiting statistical power to detect effects of genotype.
To assess the potential for use of saliva DNA as a surrogate for blood DNA, we compared percentage methylation of the AHRR DMR in saliva and in isolated circulating monocytes from the same individuals (57 of the 71 subjects in the CRU-BSAS-Saliva group, including 33 smokers and 24 nonsmokers), and found a positive correlation () (Figure 4F). The similarity of the pattern of methylation change across the SM-DMR in monocyte and saliva DNA is displayed in Figure 4D,G.
Candidate SM-DMRs and Transcription of Nearby Genes or Corresponding ncRNAs/e RNAs
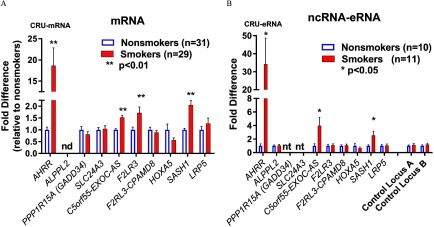
We examined whether methylation change of these DMRs associated with differential mRNA expression of 10 nearby genes using reverse transcription quantitative polymerase chain reaction (RT-qPCR) in CRU-mRNA subjects as indicated in Table 1 (see also Excel Table S1). ALPPL2 and RDX mRNAs were not detectable, but as shown in Figure 5A, we found four DMR-associated genes (AHRR, C5orf55-EXOC-AS, F2RL3, and SASH1) were significantly up-regulated in smokers compared with nonsmokers, and mRNA levels showed an inverse relationship to methylation level in the corresponding DMRs (hypomethylation vs. increased expression; Figure 5A). Inspection of the candidate DMRs suggested that they may overlap potential enhancers indicated by ENCODE H3K27ac tracks (Figure S1A–J), so we designed RT-qPCR assays to detect possible noncoding RNA (e.g., enhancer RNA, or eRNA) originating from the eight DMR locations that did not overlap exons. We tested total RNA samples from a group of CRU-mRNA with available RNA ( smokers/10 nonsmokers) and observed three of the SM-DMR locations (AHRR, C5orf55-EXOC-AS, and SASH1) showed up-regulation of corresponding eRNA expression (Figure 5B) as well as mRNA. Notably, AHRR showed the greatest smoking-associated changes in DNA methylation, mRNA and eRNA (Table 2, Figure 5A,B).
Figure 5.
Expression analysis of nearby gene and corresponding noncoding of SM-DMRs. (A) mRNA expression for genes nearby candidate SM-DMRs was determined by RT-qPCR in CRU-mRNA group; 29 smokers (solid red bar) and 31 nonsmokers (open blue bars) as indicated. AHRR, C5orf55, SASH1, and F2RL3 were significantly up-regulated in smokers relative to nonsmokers (fold difference ). (B) SM-DMR regions containing potential noncoding enhancer RNAs (eRNAs) were examined using RT-qPCR in total RNA in CRU-eRNA group; 11 smokers and 10 nonsmokers. Noncoding RNAs from DMRs near AHRR, C5orf55-EXOC-AS, and SASH1 were up-regulated in smokers (solid red) relative to nonsmokers (open blue) (fold difference ). Potential noncoding RNAs (ncRNAs) within the intragenic SM-DMRs in PPP1R15A (GADD34), and SLC24A3 gene were not tested for ncRNA because they overlapped with exons. Control loci were intragenic nonenhancer regions. CRU, clinical research unit; DMR, differentially methylated region; nd, not detected; nt, DMR not tested, region within gene exon; RT-qPCR, reverse transcription quantitative polymerase chain reaction; SM, smoking associated. ; .
Specificity of Smoking-Associated AHRR Enhancer Activation
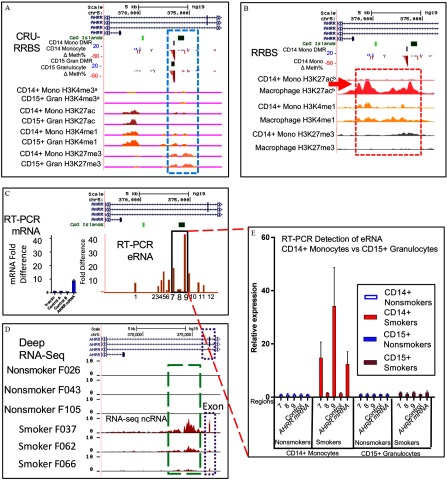
To further explore the functional relationship between methylation and expression of AHRR mRNA and eRNAs, we integrated our AHRR CRU-RRBS methylation tracks in monocytes and granulocytes with Roadmap Epigenome (Roadmap Epigenomics Consortium et al. 2015) and Blueprint tracks (Saeed et al. 2014) for histone modification marks (H3K4me3, H3K27ac, H3K4me1, and H3K27me3) in monocytes, myeloid lineage-related granulocytes, and monocyte-derived macrophages (Figure 6A,B). Figure 6A CRU-RRBS methylation tracks highlights that reduced methylation at the AHRR SM-DMRwas similar in both monocytes and granulocytes and that these DMRs align with H3K4me1/H3K27me3 histone modifications in both monocytes and granulocytes. These features suggest this DMR may be a poised enhancer in nonsmokers (Figure 6A). However, the histone modification patterns in monocyte-derived macrophages (Figure 6B, boxed) differ, with signals for H3K27ac enrichment and no repressive H3K27me3 signals, indicating an active AHRR enhancer. We hypothesized that in the monocytes of smokers we might be able to detect activation of this putative enhancer by fine mapping the location of eRNA production.
Figure 6.
Smoking-associated AHRR enhancer displays cell type–specific eRNA. (A) Genome browser view displays tracks from top: Refseq gene AHRR; CpG Islands; Monocyte DMRs (black bar); differentially methylated CpGs ( Methylation %) in monocytes ( Mono) and granulocytes ( Gran), and histone modification data tracks from the Epigenome Roadmap Project(a) for each cell type: H3K4me3, histone H3 Lysine 4 trimethyl (active mark); H3K27ac, histone H3 Lysine 27acetyl (active enhancer); H3K4me1, histone H3 Lysine 4 monomethyl (enhancer); H3K27me3, histone H3 Lysine 27trimethyl (repressive mark). SM-DMRs overlap with poised enhancer signature marks (A, dashed blue box) indicated by the presence of both H3K4me1 (active), H3K27me3 (repressed) histone modifications in Roadmap nonsmokers. (B) Genome browser window displaying histone modificataion tracks from the Blueprint project(b) (Saeed et al. 2014) monocyte and monocyte-derived macrophage histone marks (dashed red box). The macrophage H3K27ac track (red arrow) indicates an active enhancer signature at this location in macrophages (Saeed et al. 2014) that is not activated in nonsmoker monocytes (minimal H3K27ac and presence of H3K27me3 in monocytes). (C) Twelve genomic regions that cover AHRR SM-DMR were analyzed for ncRNA expression by RT-qPCR as indicated by brown bars plotted at their approximate genome location (10 smokers, 9 nonsmokers). Values for AHRR mRNA and ncRNA region 9 (eRNA) are replotted from Figure 5A,B. Beta actin and control loci A and B were used as negative controls. (D) Deep rRNA depleted RNA-seq ( million reads) was carried out in six CRU individuals from among CRU-Pyro subjects (see Excel Table S1). RNA-seq reads were individually mapped to AHRR enhancer region for the six subjects (subject IDs listed) and lower tracks show that the three smokers display noncoding RNA but the three nonsmokers do not (D, green dashed box). Bidirectional eRNA sequence reads (peaks displayed in green dashed box) form a pattern typical of an actively transcribed enhancer. Exonic mRNA sequence reads map to nearby exon (dotted purple box). Enhancer activation detected by RNA-seq was not observed in three nonsmokers. (E) Comparison of eRNA in granulocytes vs. monocytes. RT-qPCR detects AHRR mRNA and eRNAs from region 7, 8, and 9 (replotted from Figure 5C) in monocyte total RNA in smokers but not in granulocyte total RNA (granulocyte samples from six nonsmokers and six smokers from among CRU-mRNA subjects, see Excel Table S1). Note: CRU, clinical research unit; DMR, differentially methylated region; eRNA, enhancer noncoding RNA; RT-qPCR, reverse transcription quantitative polymerase chain reaction; SM, smoking associated.
To localize and quantify potential AHRR eRNA up-regulation in monocytes of smokers, we performed RT-qPCR using 12 primer sets spread across the AHRR SM-DMR region in a subset of CRU-eRNA nonsmoking () and smoking () subjects. The Figure 6C bar graph displays the significant smoking-associated fold-change results for each RT-qPCR relative to their genome position in the AHRR region. Maximal values align with H3K4me1/H3K27me3, suggesting an active enhancer in smoker monocytes (compare Figure 6C bar graph primer sets 7 and 9 with Figure 6A H3K4me1 above). For a subgroup of 16 CRU-RRBS subjects with both eRNA (CRU-eRNA) and mRNA (CRU-mRNA) data (see Excel Table S1), RT-qPCR eRNA levels were correlated with AHRR mRNA () and cotinine () levels (see Figure S4A,B). We further defined the active enhancer by carrying out deep, ribosome-depleted total RNA-seq from three nonsmokers and three hypomethylated (chosen as noted in the Methods) smokers from among the CRU-mRNA subgroup. Mapping RNA-seq sequencing reads to the AHRR region (data shown in Figure 6D), a distinctive pattern of bidirectional eRNA transcription was apparent at the AHRR enhancer in each of the three smokers with hypomethylated AHRR, but not in nonsmokers (Figure 6D, dashed green box). These results are consistent with AHRR enhancer hypomethylation and eRNA transcription accompanying up-regulation of AHRR mRNA (exonic RNA in Figure 6D, dotted box) in the monocytes of these smokers.
Although the RRBS AHRR SM-DMR was also strongly observed in granulocytes (Figure 6A; see also Figure S1B), a previous study did not observe increased AHRR mRNA in smoker granulocytes (Su et al. 2016). We hypothesized that DMR formation alone may not be sufficient to result in AHRR enhancer activation and therefore transcriptional up-regulation of its mRNA. We therefore measured AHRR eRNA and mRNA expression in total RNA isolated from granulocytes. Comparing RT-qPCR from monocytes and CD15 granulocytes (Figure 6E), we observed that both AHRR eRNA (primer sets 7 and 9) and mRNA were up-regulated at similar levels in monocytes of smokers but not in granulocytes (Figure 6E). Moreover, we observed that in monocytes AHRR mRNA was highly correlated with AHRR eRNA expression (see Figure S4A, ) but much less so with AHRR DMR methylation level (see Figure S4C, ) among our analyzed CRU-RRBS subjects who had both eRNA and mRNA data (eight smokers and eight nonsmokers).
Discussion
In the present study, we used a high-resolution next generation sequencing technique, RRBS, to investigate alterations in genome-wide DNA methylation in response to tobacco smoke exposure in circulating monocytes from two independently recruited groups and discovered numerous novel smoking-associated CpGs that are clustered into DMRs. We found that RRBS-derived monocyte SM-DMRs frequently occur at gene regulatory regions and are often observed in other blood cell types. The methylation levels of AHRR, C5orf55-EXOC-AS, and SASH1 SM-DMRs were inversely associated with both their mRNA and also noncoding enhancer RNA (eRNA) expression. The AHRR SM-DMR, the most significant DMR, was detected in both monocytes and saliva samples with good correlation (), providing support for the use of saliva cells for biomarker studies. The AHRR DMR displayed up-regulated eRNA and mRNA in the monocytes of smokers but not in granulocytes, suggesting cell type–specific activation of the enhancer. The correlation of eRNA and mRNA levels suggests that AHRR enhancer activity may more directly link to AHRR mRNA expression regulation than to AHRR methylation.
To date, only a few population studies have been published using the RRBS technique (Gervin et al. 2016; Tobi et al. 2014), and these have used whole blood DNA. The major strength of the RRBS approach is that it provides dense, single nucleotide–resolved, cell type–specific fine mapping of many regions of the genome that are not captured on 450K or 850K arrays (Ziller et al. 2013). This permits identification of novel smoking-associated CpGs and also a clear qualitative visualization of DMRs as displayed in Figure 3 (see also Figure S1). Visualization approaches provide clues into the relationship between DMRs and genome features, particularly those histone marks indicating repressed, poised or active enhancers (Figure 6A–B). It is notable that many SM-DMRs appeared to be actively repressed in monocytes as indicated by Roadmap H3K27me3 marks (Figure 3A,C,H,I; see also Figure S1A–C,E,F) (Roadmap Epigenomics Consortium et al. 2015), and they occur adjacent to clusters of ENCODE transcription factor binding sites (see Figure S1A–I, lower track) (Wang et al. 2012). Visually comparing RRBS-derived differentially methylated CpGs across multiple cell types from the same individuals (see Figure S1) increases confidence in the identification of DMRs and may hint at cell type–specific smoking effects.
Comparing RRBS results with 450K data, pyrosequencing data, and BSAS we observed very good agreement at highly significant sites (Figures 1C–D, 3, and 4B–G; see also Figure S2). However, we also observed that DMR calling was very sensitive to interindividual RRBS variation. Variation in DMR analysis between the CRU-RRBS and MESA-RRBS subgroups could be caused, in whole or in part, by the small numbers of individuals included in each analysis or genetic heterogeneity related to differences in race/ethnicity within and between each analysis subgroup. Genetic variation can have a strong impact on RRBS because CpG dinucleotides are the most polymorphic of all dinucleotide pairs (Tomso and Bell 2003) and polymorphisms at MspI CpG sites prevent measurement of methylation at affected sites and disrupt sequencing reads for adjacent nonpolymorphic CpG sites. Differences among the RRBS groups in level or duration of smoking, exposure to environmental tobacco smoke, or other possible smoke exposures not reported by subjects might also lead to variation in SM-DMR analysis. In addition, although we did not specifically evaluate this issue, technical variation in the RRBS method may have affected the results. For example, MspI enzyme digestion, RRBS library construction, inconsistent sequencing read depth, and sequencing platform difference (HiSeq 2500 for MESA and NextSeq for CRU) all might be sources of technical variation. RRBS has a general limitation that it displays a bias toward capture of CpG-rich regions in the genome, although this could be remedied by using additional restriction enzyme cuts and deeper sequencing in future studies.
A strength of the present study is that we validated RRBS results from purified monocytes collected from MESA-RRBS and CRU-RRBS study groups and compared these with RRBS results from 5 additional immune cell types from the 19 CRU-RRBS-Celltype individuals. Although cell-type isolation and analysis was a novel aspect of the present study, it also resulted in a limitation because quantities of nucleic acids from these cell types were often low, creating logistic and sample availability issues as more new analyses were considered and carried out. This precluded analysis of every sample type (either DNA or RNA) from each CRU subject in each of the novel assays that were developed over the course of the project. In addition, we were able to attempt validation of only a small number of DMRs among the several thousand potential SM-DMRs identified by RRBS. However, we demonstrated that the DMRs most strongly associated with smoking were reproducible in several groups of smokers, and could be observed in multiple cell types. This includes the finding that SM-DMRs in AHRR can be measured in DNA extracted from saliva cells, and that SM-DMR methylation in saliva cells was highly correlated with SM-DMR methylation in monocytes from the same individuals. In addition, the RRBS analysis in both MESA-RRBS and CRU-RRBS groups identified smoking-associated CpGs not present on the 450K array, such as the CpG (chr5:373490), 112 nt away from AHRR cg05575921, which displayed a larger effect size and greater statistical significance than cg05575921. This pattern was also observed in the CRU-BSAS and CRU-saliva groups (Figure 4D,G) and we suggest that if this observation could be further validated in other populations, it may be a more sensitive indicator of smoking effect than cg05575921.
Among the SM-DMRs identified, we observed up-regulation of three corresponding eRNAs and adjacent mRNAs in association with cigarette smoking, which suggests the possibility that the relationship between local methylation state and mRNA expression may be mediated by noncoding transcription. Importantly, deep ribosomal RNA-depleted RNA-seq (Figure 6D) in primary monocytes of smokers revealed bidirectional transcription of noncoding eRNA emanating from the AHRR intragenic DMR—a poised region in nonsmoker monocytes that becomes activated in monocyte-derived macrophages (Figure 6B) (Saeed et al. 2014). To our knowledge, evidence of exposure-induced activation of an enhancer in purified human primary cells has not been reported previously. We found methylation level of the AHRR SM-DMR was correlated with AHRR mRNA and eRNA expression and that AHRR eRNA levels were strongly correlated with AHRR mRNA and cotinine levels (Figure S4A,B), suggesting functionally increased enhancer activity in response to higher levels of cigarette smoking.
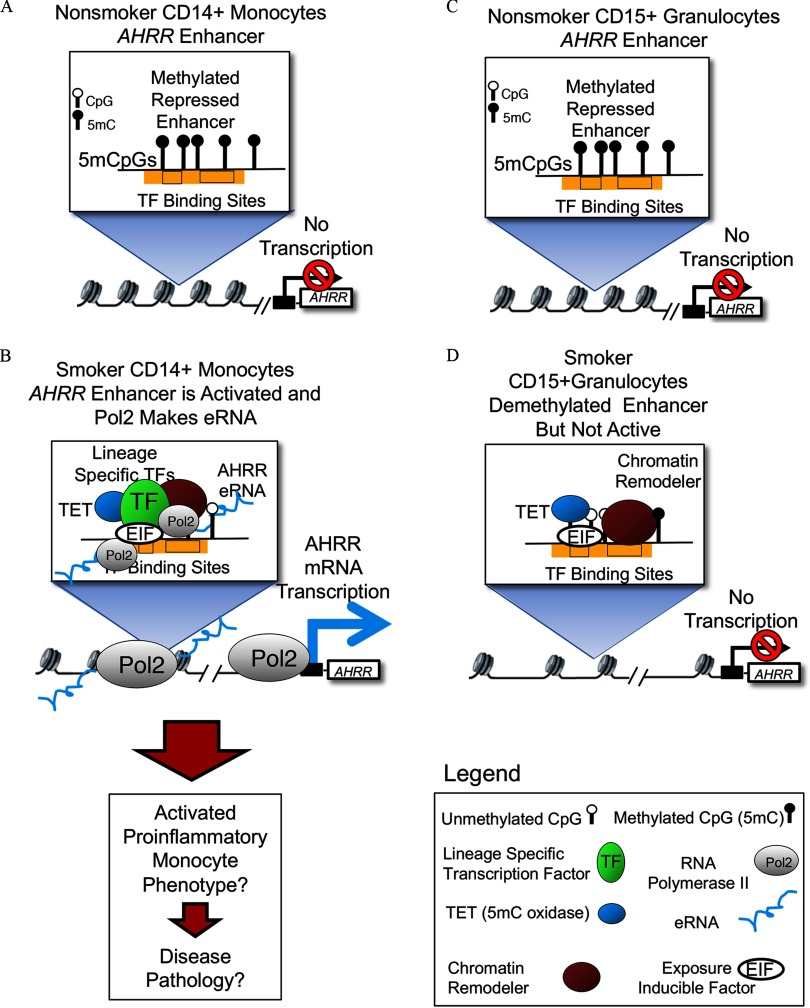
Figure 7 illustrates a hypothetical model based on our findings and existing information. We hypothesize that in monocytes, the poised AHRR enhancer goes from a closed, repressed state (Figure 7A) to an open, transcriptional state (Figure 7B) following recruitment of pioneer transcription factors that bind to closed chromatin and promote chromatin opening and co-location of chromatin remodelers, co-activators, transcriptional machinery, and cohesion complex, accompanied by TET methylcytosine dioxygenase (TET)-mediated changes in methylation state. This hypothesis is supported by previous evidence indicating that 5-hydroxymethyl cytosine is often observed at active enhancers (Calo and Wysocka 2013; Ziller et al. 2013). In addition, active enhancers have been shown to physically interact with the target gene promoter through chromatin looping and eRNA production (Calo and Wysocka 2013; Long et al. 2016). Based on our finding of consistent methylation differences across multiple cell types (see Figure S1B), and correlated methylation in DNA extracted from monocytes and saliva cells from the same individuals (Figure 4F), we further hypothesize that the change in DNA methylation state is driven by exposure-inducible factors that are present to some degree in all cell types, including granulocytes (Figure 7C–D). However, when we compared smoking-associated differences in AHRR methylation, eRNA, and mRNA between monocytes and granulocytes (Figure 6A,E), methylation differences were similar, but eRNA and mRNA expression was up-regulated in monocytes only. Therefore, we propose that, in addition to inducible transcription factors that cause changes in methylation, monocyte lineage-specific factors are required to initiate eRNA transcription and mRNA expression, resulting in smoking-related, monocyte-specific phenotypic effects (Figure 7B) that are not observed in granulocytes (Figure 7D).
Figure 7.
Hypothetical model displaying possible mechanism for cell type–specific AHRR enhancer activation and mRNA up-regulation. (A) In monocytes of nonsmokers, AHRR enhancer is observed to be fully methylated, chromatin displays repressive histone modifications (not shown) and in this state, no transcription can occur. (B) In smokers, presumably a subset of monocytes have demethylated the enhancer and concomitantly the enhancer is occupied by exposure-induced factors (EIF), chromatin-remodeling enzymes, lineage-specific transcription factors (TFs), and RNA polymerase. The transcription of enhancer RNA in monocytes of smokers could then enable transcription of AHRR mRNA. (C) In granulocytes of nonsmokers, the repressed AHRR enhancer is fully methylated and repressed, as in monocytes from nonsmokers. (D) In smokers, a substantial fraction of granulocytes display a demethylated AHRR enhancer, suggesting that exposure-induced factors and demethylation machinery are functioning in this lineage. However, a monocyte lineage-specific factor is absent and there is no recruitment of RNA polymerase or expression of mRNA.
Monocytes and monocyte-derived macrophages have long been implicated in vascular inflammation and in the process of foam cell conversion into atherosclerotic plaques in endothelial walls (Hansson 2005; Hilgendorf et al. 2015; Lessner et al. 2002; Moore et al. 2013; Osterud and Bjorklid 2003). It is notable that a number of the DMR genes, including AHRR, F2RL3, HOXA5, SASH1, LRP5, and PPP1R15A (GADD34) have been associated with cardiovascular diseases/atherosclerosis either through nearby DNA methylation and/or gene expression (listed in Table 2). Recently, methylation levels of AHRR cg05575921 in monocytes were reported to be associated with atherosclerotic plaques (Reynolds et al. 2015) and AHRR expression was shown to play a role in pro-inflammatory signaling in monocytes and THP-1 monocytic cells (Zhang et al. 2017). AHRR also mediates diverse immune responses in adipose (Vogel et al. 2016), skin, and intestine (Brandstätter et al. 2016). A recent report (Saeed et al. 2014) demonstrated strong up-regulation of AHRR during in vitro macrophage differentiation from monocytes. Our observation of a 30–40% loss of methylation in the AHRR enhancer suggests that in smokers a subset () of circulating monocytes have demethylated the enhancer and up-regulated AHRR mRNA. We suggest that these AHRR-expressing monocytes may be pro-inflammatory or destined for involvement in pro-inflammatory signaling by M1-type macrophages in atherosclerotic lesions. Further studies will be needed to determine the validity of these hypotheses.
Conclusions
We observed that SM-DMRs often overlapped with enhancers, suggesting a potential role in regulating responses to smoking. However, although most of the SM-DMRs were detected in multiple cell types, including saliva cells, phenotypic differences in responses between cell types suggest that both smoking-altered methylation and eRNA production from the intragenic AHRR enhancer may be necessary preconditions for cell type–specific AHRR transcription. The precise stepwise mechanism and the chromatin remodeling complexes involved remain to be demonstrated. Smoking-related functional changes may prove useful as both biomarkers of exposure and as cell type–specific signs of early pathology.
Supplemental Material
Supplemental Material
Acknowledgments
M.W., D.A.B., and Y.L. designed the study. M.W., M.R.C., G.S.P., D.K.P., C.L.C., D.S., I.J.T., and NA.E. carried out the experiments. M.W., M.R.C., G.S.P., L.M.R., D.K.P., and C.L.C. analyzed data. B.D.B. and X.W. performed bioinformatics analysis. M.W. and D.A.B. wrote the manuscript.
The authors thank the other investigators, the staff, and the participants of the Multi-Ethnic Study of Atherosclerosis Study (MESA) and the NIEHS Clinical Research Unit for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. The authors also thank M. Ziller and A. Meissner of the Broad Institute of MIT and Harvard for their generosity in providing enhancer coordinates and the R code to identify differentially methylated regions. We also thank the NIH Intramural Sequencing Core, and the NIEHS Epigenomics core for essential contributions.
This work was funded in part by the Intramural Research Program of the National Institute of Environmental Health Sciences–National Institutes of Health (NIEHS/NIH; Z01-ES100475) and a grant from NIH/U.S. Food and Drug Administration (FDA) Intramural Center for Tobacco Research to D.A.B. MESA and the MESA SNP Health Association Resource (SHARe) project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491. The MESA Epigenomics and Transcriptomics Study was funded by NHLBI grant R01HL101250, R01 DK103531-01, and R01 HL135009-01 to Wake Forest University Health Sciences. Analysis of MESA data reported in this publication was also supported by the NHLBI under award no. P50HL120163. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- Alexandrov LB, Ju YS, Haase K, Van Loo P, Martincorena I, Nik-Zainal S, et al. 2016. Mutational signatures associated with tobacco smoking in human cancer. Science 354(6312):618–622, PMID: 27811275, 10.1126/science.aag0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev 16(1):6–21, PMID: 11782440, 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, et al. 2012. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell 47(4):633–647, PMID: 22841485, 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrell-Pages M, Romero JC, Badimon L. 2014. Cholesterol modulates LRP5 expression in the vessel wall. Atherosclerosis 235(2):363–370, PMID: 24929284, 10.1016/j.atherosclerosis.2014.05.922. [DOI] [PubMed] [Google Scholar]
- Boyle P, Clement K, Gu H, Smith ZD, Ziller M, Fostel JL, et al. 2012. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol 13(10):R92, PMID: 23034176, 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstätter O, Schanz O, Vorac J, König J, Mori T, Maruyama T, et al. 2016. Balancing intestinal and systemic inflammation through cell type-specific expression of the aryl hydrocarbon receptor repressor. Sci Rep 6:26091, PMID: 27184933, 10.1038/srep26091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling LP. 2013. Current genetics and epigenetics of smoking/tobacco-related cardiovascular disease. Arterioscler Thromb Vasc Biol 33(7):1468–1472, PMID: 23640490, 10.1161/ATVBAHA.112.300157. [DOI] [PubMed] [Google Scholar]
- Breitling LP, Salzmann K, Rothenbacher D, Burwinkel B, Brenner H. 2012. Smoking, F2RL3 methylation, and prognosis in stable coronary heart disease. Eur Heart J 33(22):2841–2848, PMID: 22511653, 10.1093/eurheartj/ehs091. [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J. 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49(5):825–837, PMID: 23473601, 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (Centers for Disease Control and Prevention). 2010. “How tobacco smoke causes disease: The biology and behavioral basis for smoking-attributable disease: A report of the surgeon general.” Atlanta, GA:CDC. [PubMed] [Google Scholar]
- Dua P, Kang HS, Hong SM, Tsao MS, Kim S, Lee DK. 2013. Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein. Cancer Res 73(6):1934–1945, PMID: 23467613, 10.1158/0008-5472.CAN-12-3682. [DOI] [PubMed] [Google Scholar]
- Dunn J, Qiu H, Kim S, Jjingo D, Hoffman R, Kim CW, et al. 2014. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J Clin Invest 124(7):3187–3199, PMID: 24865430, 10.1172/JCI74792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M. 2009. DNA hypomethylation in cancer cells. Epigenomics 1(2):239–259, PMID: 20495664, 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanelli F, Baglietto L, Ponzi E, Guida F, Campanella G, Johansson M, et al. 2015. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat Commun 6:10192, PMID: 26667048, 10.1038/ncomms10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervin K, Andreassen BK, Hjorthaug HS, Carlsen KCL, Carlsen KH, Undlien DE, et al. 2016. Intra-individual changes in DNA methylation not mediated by cell-type composition are correlated with aging during childhood. Clin Epigenetics 8(1):110, PMID: 27785156, 10.1186/s13148-016-0277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonseth S, de Smith AJ, Roy R, Zhou M, Lee ST, Shao X, et al. 2016. Genetic contribution to variation in DNA methylation at maternal smoking-sensitive loci in exposed neonates. Epigenetics 11(9):664–673, PMID: 27403598, 10.1080/15592294.2016.1209614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Smith ZD, Bock C, Boyle P, Gnirke A, Meissner A. 2011. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 6(4):468–481, PMID: 21412275, 10.1038/nprot.2010.190. [DOI] [PubMed] [Google Scholar]
- Hansson GK. 2005. Inflammation, atherosclerosis, and coronary artery disease. N Engl J Med 352(16):1685–1695, PMID: 15843671, 10.1056/NEJMra043430. [DOI] [PubMed] [Google Scholar]
- Hilgendorf I, Swirski FK, Robbins CS. 2015. Monocyte fate in atherosclerosis. Arterioscler Thromb Vasc Biol 35(2):272–279, PMID: 25538208, 10.1161/ATVBAHA.114.303565. [DOI] [PubMed] [Google Scholar]
- Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, et al. 2016. Epigenetic signatures of cigarette smoking. Circ Cardiovasc Genet 9(5):436–447, PMID: 27651444, 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492, PMID: 22641018, 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Håberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, et al. 2012. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy. Environ Health Perspect 120(10):1425–1431, PMID: 22851337, 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajiri S, Fukumoto K, Hata M, Sasaki H, Katsuno T, Nakagawa T, et al. 2004. Radixin deficiency causes deafness associated with progressive degeneration of cochlear stereocilia. J Cell Biol 166(4):559–570, PMID: 15314067, 10.1083/jcb.200402007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopik VS, Maccani MA, Francazio S, McGeary JE. 2012. The epigenetics of maternal cigarette smoking during pregnancy and effects on child development. Dev Psychopathol 24(4):1377–1390, PMID: 23062304, 10.1017/S0954579412000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langie SAS, Szarc vel Szic K, Declerck K, Traen S, Koppen G, Van Camp G, et al. 2016. Whole-genome saliva and blood DNA methylation profiling in individuals with a respiratory allergy. PloS One 11(3):e0151109, PMID: 26999364, 10.1371/journal.pone.0151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Pausova Z. 2013. Cigarette smoking and DNA methylation. Front Genet 4:132, PMID: 23882278, 10.3389/fgene.2013.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Wong DT. 2009. Saliva: an emerging biofluid for early detection of diseases. Am J Dent 22(4):241–248, PMID: 19824562. [PMC free article] [PubMed] [Google Scholar]
- Lessner SM, Prado HL, Waller EK, Galis ZS. 2002. Atherosclerotic lesions grow through recruitment and proliferation of circulating monocytes in a murine model. Am J Pathol 160(6):2145–2155, PMID: 12057918, 10.1016/S0002-9440(10)61163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69(6):915–926, PMID: 1606615. [DOI] [PubMed] [Google Scholar]
- Liu YM, Ding J, Reynolds LM, Lohman K, Register TC, De La Fuente A, et al. 2013. Methylomics of gene expression in human monocytes. Hum Mol Genet 22(24):5065–5074, PMID: 23900078, 10.1093/hmg/ddt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon BA, Hoffman GE, Mezey JG. 2012. Mouse obesity network reconstruction with a variational Bayes algorithm to employ aggressive false positive control. BMC Bioinformatics 13:53, PMID: 22471599, 10.1186/1471-2105-13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. 2016. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167(5):1170–1187, PMID: 27863239, 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masser DR, Berg AS, Freeman WM. 2013. Focused, high accuracy 5-methylcytosine quantitation with base resolution by benchtop next-generation sequencing. Epigenetics Chromatin 6(1):33, PMID: 24279302, 10.1186/1756-8935-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masud R, Shameer K, Dhar A, Ding K, Kullo IJ. 2012. Gene expression profiling of peripheral blood mononuclear cells in the setting of peripheral arterial disease. J Clin Bioinforma 2:6, PMID: 22409835, 10.1186/2043-9113-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A. 2010. Epigenetic modifications in pluripotent and differentiated cells. Nat Biotechnol 28(10):1079–1088, PMID: 20944600, 10.1038/nbt.1684. [DOI] [PubMed] [Google Scholar]
- Mick E, McGough JJ, Middleton FA, Neale B, Faraone SV. 2011. Genome-wide association study of blood pressure response to methylphenidate treatment of attention-deficit/hyperactivity disorder. Prog Neuropsychopharmacol Biol Psychiatry 35(2):466–472, PMID: 21130132, 10.1016/j.pnpbp.2010.11.037. [DOI] [PubMed] [Google Scholar]
- Monick MM, Beach SR, Plume J, Sears R, Gerrard M, Brody GH, et al. 2012. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet B Neuropsychiatr Genet 159B(2):141–151, PMID: 22232023, 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Sheedy FJ, Fisher EA. 2013. Macrophages in atherosclerosis: a dynamic balance. Nat Rev Immunol 13(10):709–721, PMID: 23995626, 10.1038/nri3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muka T, Koromani F, Portilla E, O’Connor A, Bramer WM, Troup J, et al. 2016. The role of epigenetic modifications in cardiovascular disease: a systematic review. Int J Cardiol 212:174–183, PMID: 27038728, 10.1016/j.ijcard.2016.03.062. [DOI] [PubMed] [Google Scholar]
- Osterud B, Bjorklid E. 2003. Role of monocytes in atherogenesis. Physiol Rev 83(4):1069–1112, PMID: 14506301, 10.1152/physrev.00005.2003. [DOI] [PubMed] [Google Scholar]
- Ostrow KL, Michailidi C, Guerrero-Preston R, Hoque MO, Greenberg A, Rom W, et al. 2013. Cigarette smoke induces methylation of the tumor suppressor gene NISCH. Epigenetics 8(4):383–388, PMID: 23503203, 10.4161/epi.24195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. 2002. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 21(48):7435–7451, PMID: 12379884, 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- Philibert RA, Sears RA, Powers LS, Nash E, Bair T, Gerke AK, et al. 2012. Coordinated DNA methylation and gene expression changes in smoker alveolar macrophages: specific effects on VEGF receptor 1 expression. J Leukoc Biol 92(3):621–631, PMID: 22427682, 10.1189/jlb.1211632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LM, Lohman K, Pittman GS, Barr RG, Chi GC, Kaufman J, et al. 2017. Tobacco exposure-related alterations in DNA methylation and gene expression in human monocytes: the Multi-Ethnic Study of Atherosclerosis (MESA). Epigenetics 12(12):1092–1100, PMID: 29166816, 10.1080/15592294.2017.1403692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds LM, Wan M, Ding J, Taylor JR, Lohman K, Su D, et al. 2015. DNA methylation of the aryl hydrocarbon receptor repressor associations with cigarette smoking and subclinical atherosclerosis. Circ Cardiovasc Genet 8(5):707–716, PMID: 26307030, 10.1161/CIRCGENETICS.115.001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. 2015. Integrative analysis of 111 reference human epigenomes. Nature 518(7539):317–330, PMID: 25693563, 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Suárez E, Siwy J, Zürbig P, Mischak H. 2014. Urine as a source for clinical proteome analysis: from discovery to clinical application. Biochim Biophys Acta 1844(5):884–898, PMID: 23831154, 10.1016/j.bbapap.2013.06.016. [DOI] [PubMed] [Google Scholar]
- Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, et al. 2014. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science 345(6204):1251086, PMID: 25258085, 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salpea P, Russanova VR, Hirai TH, Sourlingas TG, Sekeri-Pataryas KE, Romero R, et al. 2012. Postnatal development- and age-related changes in DNA-methylation patterns in the human genome. Nucleic Acids Res 40(14):6477–6494, PMID: 22495928, 10.1093/nar/gks312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H, Kulasinghe A, Perry C, Nelson C, Punyadeera C. 2016. A liquid biopsy for head and neck cancers. Expert Rev Mol Diagn 16(2):165–172, PMID: 26631411, 10.1586/14737159.2016.1127758. [DOI] [PubMed] [Google Scholar]
- Shenker NS, Polidoro S, van Veldhoven K, Sacerdote C, Ricceri F, Birrell MA, et al. 2013. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet 22(5):843–851, PMID: 23175441, 10.1093/hmg/dds488. [DOI] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. 2013. DNA methylation: roles in mammalian development. Nat Rev Genet 14(3):204–220, PMID: 23400093, 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Su D, Wang X, Campbell MR, Porter DK, Pittman GS, Bennett BD, et al. 2016. Distinct epigenetic effects of tobacco smoking in whole blood and among leukocyte subtypes. PloS one 11(12):e0166486, PMID: 27935972, 10.1371/journal.pone.0166486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi EW, Goeman JJ, Monajemi R, Gu H, Putter H, Zhang Y, et al. 2014. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun 5:5592, PMID: 25424739, 10.1038/ncomms6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomso DJ, Bell DA. 2003. Sequence context at human single nucleotide polymorphisms: overrepresentation of CpG dinucleotide at polymorphic sites and suppression of variation in CpG islands. J Mol Biol 327(2):303–308, PMID: 12628237. [DOI] [PubMed] [Google Scholar]
- Vidović A, Vidović Juras D, Vučićević Boras V, Lukač J, Grubišić-Ilić M, Rak D, et al. 2012. Determination of leucocyte subsets in human saliva by flow cytometry. Arch Oral Biol 57(5):577–583, PMID: 22118990, 10.1016/j.archoralbio.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Vogel CF, Chang WL, Kado S, McCulloh K, Vogel H, Wu D, et al. 2016. Transgenic overexpression of aryl hydrocarbon receptor repressor (AhRR) and AhR-mediated induction of CYP1A1, cytokines, and acute toxicity. Environ Health Perspect 124(7):1071–1083, PMID: 26862745, 10.1289/ehp.1510194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, et al. 2012. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22(9):1798–1812, PMID: 22955990, 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann H, Touat-Hamici Z, Durand H, Mueller C, Chardonnet S, Pionneau C, et al. 2015. SASH1, a new potential link between smoking and atherosclerosis. Atherosclerosis 242(2):571–579, PMID: 26318107, 10.1016/j.atherosclerosis.2015.08.013. [DOI] [PubMed] [Google Scholar]
- Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, et al. 2013. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PloS One 8(5):e63812, PMID: 23691101, 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Schöttker B, Florath I, Stock C, Butterbach K, Holleczek B, et al. 2016. Smoking-associated DNA methylation biomarkers and their predictive value for all-cause and cardiovascular mortality. Environ Health Perspect 124(1):67–74, PMID: 26017925, 10.1289/ehp.1409020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DD, Wang WT, Xiong J, Xie XM, Cui SS, Zhao ZG, et al. 2017. Long noncoding RNA LINC00305 promotes inflammation by activating the AHRR-NF-κB pathway in human monocytes. Sci Rep 7:46204, PMID: 28393844, 10.1038/srep46204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Edri R, Yaffe Y, Donaghey J, Pop R, Mallard W, et al. 2015. Dissecting neural differentiation regulatory networks through epigenetic footprinting. Nature 518(7539):355–359, PMID: 25533951, 10.1038/nature13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O, et al. 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 500(7463):477–481, PMID: 23925113, 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RRBS methylation data sets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository, Accession No. GSE104700.