Abstract
The scientific community is increasingly concerned with the proportion of published “discoveries” that are not replicated in subsequent studies. The field of rodent behavioral phenotyping was one of the first to raise this concern, and to relate it to other methodological issues: the complex interaction between genotype and environment; the definitions of behavioral constructs; and the use of laboratory mice and rats as model species for investigating human health and disease mechanisms. In January 2015, researchers from various disciplines gathered at Tel Aviv University to discuss these issues. The general consensus was that the issue is prevalent and of concern, and should be addressed at the statistical, methodological and policy levels, but is not so severe as to call into question the validity and the usefulness of model organisms as a whole. Well-organized community efforts, coupled with improved data and metadata sharing, have a key role in identifying specific problems and promoting effective solutions. Replicability is closely related to validity, may affect generalizability and translation of findings, and has important ethical implications.
Keywords: Reproducibility, Replicability, GxE interaction, Validity, Data sharing, False discoveries, Heterogenization
1. Introduction
In recent years the scientific community, pharmaceutical companies, and research funders have become increasingly concerned with the proportion of published “discoveries” that could not be replicated in subsequent studies, and sometimes could not even be reproduced in reanalysis of the original data. Such evidence is increasingly seen as a problem with the scientific method, impugning the credibility of science as a whole. Prominent institutions and journals, including the National Institutes of Health (NIH), the National Academy of Science (NAS), Science, and Nature, have recently reconsidered of their policies due to this issue. However, there is still confusion and controversy regarding the severity of the problem, its causes, and what should be done about it, how, and by whom.
In the field of rodent phenotyping, failure of replicability and reproducibility had been noted even before such concerns were widespread, and currently the NIH considers the problem to be especially prevalent in preclinical research. The issue seems further tied to several other complicated methodological challenges, such as handling the potentially complex interaction between genotype and environment, defining and measuring proper behavioral constructs, and using rodents as models for investigating human diseases and disorders. Reproducibility and replicability are crucial in all fields of experimental research, but even more so in animal research, where the lives and welfare of the animals are valuable for ethical reasons, and should not be wasted for inconclusive research. In January 2015, researchers involved in the study of reproducibility and replicability gathered at Tel Aviv University to discuss these issues. These researchers came from various disciplines including genetics, behavior genetics, behavioral neuroscience, ethology, statistics, bioinformatics and data science.
The present paper consists of eight sections, each dedicated to a central theme. In each section we attempt to summarize the consensus opinion or most widely held views on the topic, while also representing more controversial positions. While offering examples, recommendations and insights in multiple contexts, we avoid making a list of guidelines that would be too definitive, given the current state of knowledge and consensus. Full conference proceedings are available as a set of video clips (see links in the acknowledgements section). All authors agree that this paper reflects the complexity of replicability and reproducibility issues, even when restricted to a single area of research, yet it also points at practical ways to address some of these issues.
2. Reproducibility and replicability in general science: a crisis?
The ability to verify empirical findings wherever and whenever needed is commonly regarded as a required standard of modern experimental science. This standard was originally established in the 17th century, by Robert Boyle and other scientists of the Royal Society according to their motto nullius in verba (“take nobody’s word”). These pioneers of experimental science regarded the ability to replicate results as an acid test differentiating science from one-time “miracles”. Their criterion for a scientific fact was (following a then common judicial dogma of two witnesses required for a valid testimony) something measured or observed in at least two independent studies (Agassi, 2013). In a case that may have been the first debate over the replicability of a scientific discovery, the Dutch scientist Christiaan Huygens noted a phenomenon related to vacuum in Amsterdam, and was invited to Boyle’s laboratory in London in order to replicate the experiment and show that the phenomenon was not idiosyncratic to his specific laboratory and equipment (Shapin and Schaffer, 1985). Ronald Fisher generalized the Royal Society criterion to more than two replications in his 1935 classic “The Design of Experiments”, writing: “we may say that a phenomenon is experimentally demonstrable when we know how to conduct an experiment which will rarely fail to give us statistically significant results” (Fisher, 1935, p.14). This quote illustrates how the common method of statistical significance, already when it was first conceived, was closely tied with the concept of replicating experimental results. This concept served science well throughout its history, but non-replicable results have surfaced more often in recent years, attracting much attention.
In the field of rodent phenotyping, the problem has in fact always been present, and was recognized in the influential study by Crabbe et al. (1999) before it was noticed in many other fields. However, the issue is by no means unique to rodent phenotyping. For instance, difficulties in replicating discoveries when dissecting the genetics of complex traits in humans motivated the move to far more stringent statistical threshold guidelines proposed by Lander and Kruglyak (1995).
Some notorious recent examples of poor credibility in general science include non-replicable methods of cancer prognosis (Potti et al., 2006, refuted by Baggerly and Coombes, 2009, and retracted), “voodoo correlations” in brain imaging (Vul et al., 2009), “p-value hacking” (Simmons et al., 2011) and Excel coding errors that affected global economic policies (Pollin, 2014). A large community effort (Open Science Collaboration, 2015) recently attempted to replicate the findings of 100 papers in several leading psychology journals, and reported that 64% of the replications did not achieve statistical significance (but see Gilbert et al., 2016). A similar replication project in the field of cancer research (Errington et al., 2014) has just reported preliminary results: of 5 attempted replications, two were replicated, one clearly failed to replicate, and two were unclear due to technical considerations (Nosek and Errington, 2017). The current situation is sometimes referred to as the “credibility crisis”, “replicability crisis” (e.g., Savalei and Dunn, 2015), or “reproducibility crisis” (e.g., Peng, 2015) of recent science, and led prominent scientific journals and institutes to reconsider their policies (Landis et al., 2012; Nature Editorial, 2013; Collins and Tabak, 2014; McNutt, 2014; Alberts et al., 2015). Collins and Tabak specifically mentioned preclinical studies as prone to reproducibility and replicability problems, and Howells et al. (2014) blame the recurrent failure of drug candidates in clinical trials on lack of rigor in preclinical trials. Yet aside of general useful recommendations such as increasing sample sizes, including both sexes when possible, and improving statistical education, it is not clear what the new policies should be.
Ironically, there is currently no scientific consensus even over the name of the problem and the meaning of basic terms, confusing the discussion even further (Goodman et al., 2016). The terms replicable, reproducible, repeatable, confirmable, stable, generalizable, reviewable, auditable, verifiable and validatable have all been used; even worse, in different disciplines and fields of science, these terms might have orthogonal or even contradictory meanings (Kenett and Shmueli, 2015; Goodman et al., 2016). Following the now common term “Reproducible Research” in computer science (Diggle and Zeger, 2010; Stodden, 2010, 2013), a useful distinction was offered by Peng (2011), Peng (2015) and (Leek and Peng, 2015): “reproducibility” is concerned with reproducing, from the same original data, through reanalysis, the same results, figures and conclusions reported in the publication. “Replicability”, in comparison, is concerned with replicating outcomes of another study, in a similar but not necessarily identical way, for example at a different time and/or in a different laboratory, to arrive at similar conclusions in the same research question. We will use the above distinction in the remaining sections. However, note that other researchers recently suggested a similar distinction with the opposite terminology (Kenett and Shmueli, 2015). The NIH now uses the catch-all term “rigor” to denote adequacy or even goodness of experimental design, metadata, and analytic methods that should hopefully lead to higher rates of replicability and reproducibility (Lapchak et al., 2013).
Another categorization (Stodden, 2013) distinguishes between empirical reproducibility, computational reproducibility and statistical reproducibility. (Stodden, 2010, 2013) suggested that computational reproducibility is currently the most problematic. When viewing the objective of the scientific method as “rooting out error”, the deductive branch of mathematics (statistics included) has already developed its standards for mathematical proof, and the empirical branch (life sciences and animal phenotyping included) has already developed its standards for hypothesis testing and method reporting. It is computation-based research that has yet to develop its own standards for reproducibility, including data and code sharing (Stodden et al., 2013).
Ostensibly, science should not require trust in authority – it should be “show me”, not “trust me” (Stark, 2015). Yet in reality, most scientific publications today amount to saying “trust me”. The typical scientific paper does not give access to the raw data, the code, and other details needed to confirm the reported results – it basically asserts “I did all these carefully, trust my results” (Stark, 2015, 2017). Moreover, the distressing pressure to minimize the length of methods sections has resulted in abberviated descriptions of important procedural details. Alsheikh-Ali et al. (2011) found that out of 500 original research papers in high impact factor journals, 30% were not subject to any data availability policy, and out of those that were, 59% did not fully adhered to the policy. Overall only 9% of the papers in that study deposited full primary raw data. Soergel (2015) suggested that software errors are not limited to a few high-profile cases that lead to retraction, and instead estimated that “most scientific results are probably wrong if the data passed through a computer”. In another estimation of the current state of science reproducibility, ThermoML, an open data archive in the field of thermodynamics, found errors in about 10% of papers that otherwise would have been accepted (Frenkel et al., 2006). In a study of papers using microarray-based signatures of drug sensitivity derived from cell lines to predict patient response, Baggerly and Coombes (2009) found five case studies with errors that potentially put human patients at risk. Interestingly the most common errors were simple ones, reminiscent of the summation of only part of an Excel column that affected global economic policies (Pollin, 2014), or the error in metric conversion that caused the loss of NASA Mars Surveyor space probe (Stephenson et al., 1999). Garijo et al. (2013) attempted to reproduce the results of a paper describing a computational pipeline mapping all putative FDA and European drugs to possible protein receptors in the proteome of Mycobacterium tuberculosis. They found it to be extremely difficult and time consuming. Even worse, there is good evidence of a negative relation between the impact factor of a journal and the likelihood of technical error, a trend detected in a study of over 700 recent papers in the burgeoning field of ChIP-seq (Marinov et al., 2014). In summary, while we may strive for perfection, the scientific process does not assure error-free results and even encourages speed over accuracy. What blocks progress is the inability to detect and correct errors in time.
If a substantial percentage of published studies are not reproducible — i.e., if it is difficult to regenerate the figures, tables, and scientific conclusions starting from the data used in a study — it is even more unlikely that results are replicable, i.e., that other researchers would be able to achieve matched results and conclusions starting with data generated by different groups in different laboratories. Within-study reproducibility (as well as within-study replicability) seems necessary but, as we shall argue, insufficient condition for across-study replicability. Ioannidis (2005) famous paper titled “Why Most Published Research Findings are False” highlighted the fact that the usual 0.05 significance testing increases the proportion of false discoveries among the discoveries made when testing many hypotheses. The combination of multiplicity and unadjusted testing can indeed be hazardous, as already argued by Soric (1987). Emphasis on the use of 0.05 level testing has led to counterproductive solutions, such as the New Statistics Movement (Cummings, 2014), which considers p-values to be the source of the problem, and advocates replacing them with confidence intervals. This has been followed by a statement issued by the Board of the Statistical Association, warning against misuse and misinterpretation of the p-value (Wasserstein and Lazar, 2016) and offering (indirectly) the confidence interval as possible replacement. However, in most cases the reported or emphasized confidence intervals are selected from many hypotheses, leading to the same issue of multiplicity (Benjamini and Hechtlinger, 2013).
An alternative to discarding p-values is to adjust them to cope with the multiplicity of hypotheses being tested or confidence intervals being made. The paper of Soric (1987) motivated the development of a formal approach to the false discovery rate (FDR) and methods to control it (Benjamini and Hochberg, 1995). It is easy to see how multiplicity has exacerbated the “credibility crisis” in science in recent years: in the past, a typical phenotyping experiment would test a single measure of interest, or at most several measures. Now, thanks to automated and computerized high-throughput strategies, testing “batteries” (Brown et al., 2000) and “pipelines” (Koscielny et al., 2014) used for phenotyping frequently record 102–103 phenotypic measures per independent variable (a mouse genotype), which is still far fewer than the 105–108 associations in a typical genome-wide association study (GWAS). There is no way to report them all in a paper, so by necessity only a few are highlighted (Peterson et al., 2016). If the significant ones are selected as discoveries, the relevant error is the number of spuriously significant differences among the total number of significant differences. This ratio equals the number of false discoveries among the total number of discoveries, namely the FDR. Recent attempts to empirically estimate that the “science-wise FDR” are in the range of a rate of 15%–30% (Jager and Leek, 2014; Benjamini and Hechtlinger, 2013), considerably lower than Ioannidis’ warning of > 50%, but considerably higher than 5% (the rate expected for the most commonly used, if arbitrary, 0.05 significance level). These analyses also indicate that once selective inference is accounted for a 5% rate is indeed achievable. The Benjamini-Hochberg procedure of FDR is readily applicable to all varieties of phenotyping (Benjamini et al., 2001), and using this statistical tactic, especially in high-throughput multiple measure studies, should go a long way to decreasing the rate of “discoveries” that do not replicate.
In summary, science is reviewing its own failures, searching for the causes of the “crisis” and devising better ways to address them. The old concepts of experimental science emphasizing replicability and reproducibility are still correct in spirit, but require updating experimental, computational and statistical methodologies to cope with the increasing size and complexity of experimental approaches. Preclinical research and phenotyping are similar in this sense to other fields of science, but have particular issues of their own. These and other rodent-specific issues are considered in the following sections.
3. Can data sharing in rodent phenotyping help with replicability?
Laboratory mice and rats are the main mammalian models currently used for high-throughput genomic and behavior genetic research, and are employed primarily to explore and test gene function. This is considered by some to be “the great challenge facing biologists today” (Collins et al., 2007). Rodent models are used extensively as part of preclinical development and testing of treatments for disease in humans, in genomic research (Collins et al., 2007; Beckers et al., 2009), and also in research of the central nervous system (CNS) and behavior (e.g., Crawley, 1985; Gerlai et al. (1995); Logue et al., 1997; Dulawa et al., 1997; Gerlai, 2002a; Gerlai et al., 2002; Musatov et al., 2006). For obvious reasons, the reproducibility (within the same study) and replicability (across studies) of phenotyping has crucial implications for their translational relevance. Similar issues manifest in other model animals used for high-throughput phenotyping, such as the zebrafish (Gerlai, 2014; MacRae and Randall, 2015), Drosophila and C. elegans (Williams and Auwerx, 2015). In addition to scientific and economic implications, there are also important ethical implications: using animals for inconclusive research undermines the ethical goal of reducing and refining animal experiments. Full consideration of the welfare and usage of animals is a critical component of experimental studies. In preclinical research, poor replicability and reproducibility also slows medical progress and put human patients at risk. The drive to publish rigorous results is thus more than a scientific necessity — it is also a moral obligation.
Traditionally, a main advantage of rodents and other model organisms is the ability to standardize genotypes using inbred strains or selected lines, consisting of genetically identical “clones” that are in principle precisely replicated across studies. This “genetic standardization” enables experimental designs that would be impossible using outbred animals or humans (monozygotic twins represent n = 2, a sample size that imposes serious practical limitations). The BXD, HXB and the Collaborative Cross recombinant inbred strains, for example, can be thought of as “cloned families”, each currently including many reproducible “offspring” and “parents” lines (Complex Trait Consortium, 2003; Chesler et al., 2008; Iraqi et al., 2008; Morahan et al., 2008; Collaborative Cross Consortium, 2012; Welsh et al., 2012). These families, developed by community effort of several research centers, are routinely used in quantitative trait locus (QTL) mapping (Complex Trait Consortium, 2003) to localize phenotypes to segments of chromosomes within intervals of 0.5–10.0 Mb (Koutnikova et al., 2009; Houtkooper et al., 2013; Keeley et al., 2014). In a parallel strategy, “knock out” technology allows the targeted mutation of the gene of choice in the mouse (and more recently also in the rat), with the goal of discovering the effect on the phenotype, and of advancing our understanding of the biological functions of the targeted gene. The International Mouse Phenotyping Consortium (IMPC), a community effort for generating and phenotyping mice with targeted knockout mutations, has a long-term goal to knock out most of the ~20,000 mouse genes, and phenotype them on the background of the C57BL/6N mouse genome (Beckers et al., 2009). QTL “forward genetics” and knockout “reverse genetics” strategies are complementary and can increasingly be combined (Williams and Auwerx, 2015; Wang et al., 2016). Understanding phenotypic effects of genes variants is one of the core challenges of personalized medicine: Reference populations provide an excellent and replicable platform for precision experimental medicine. Many individuals of each genotype can studied under tightly controlled environments—an essential step in understanding complex gene-by-environmental interactions.
It is, however, important to recognize that genetic standardization in principle is not always standardization in practice. Even highly curated lines such as the DBA/2J inbred mouse strain maintained at the Jackson Laboratory might develop spontaneous mutations that are carried forward in standard commercially-available stocks. Such a previously unknown polymorphism was recently shown to affect both methamphetamine consumption and Trace Amine-Associated Receptor 1 function (Harkness et al., 2015; Shi et al., 2016). Non-replicable results sometimes reflect the naivete of our expectations, despite our best efforts to imagine what the “environment” is for a mouse, given their many sensory, social and biological differences from humans. They might also result from heterogeneity in protocol (Valdar et al., 2006), or from a failure to recognize the importance of potentially subtle differences in genetic background. For example, genetic differences might predispose some inbred strains, or more generally, some genetic backgrounds, to be more phenotypically variable than others (as illustrated in Wiltshire (2015) and considered by Rönnegård and Valdar (2011, 2012)). Highly homozygous genomes might have less capacity for “genetic buffering” against environmental variation, and some strains will be worse than others in this respect (but see also Crusio, 2006).
Bioinformatics is a well-established discipline in the life sciences, traditionally concerned primarily with DNA, RNA and protein sequence data, which are stored in public databases as a primary research tool. The idea that phenotypic data are also worthy of storing, analyzing and reanalyzing (Gerlai, 2002a) is not so widely established yet, but the value of phenotype data integration has been recognized methodologically and in practice (Chesler et al., 2003), and phenotype data standards emerged early (Grubb et al., 2004). Collaboration, community efforts, data sharing, and public phenotyping databases have an important role in today’s field of rodent phenotyping. Among many other utilities, they also offer unique opportunities for researching and controlling reproducibility and replicability. These public databases and data sharing projects are instructive in informing replicability studies at different levels: from reanalyzing other researchers’ data to contributing their own data, and even constructing and maintaining public databases and community projects. Reanalysis of shared phenotyping data enhances their utility and scientific value, potentially substituting for additional animal studies, thus reducing animal use without compromising actual reproducibility and replicability.
This section reflects experience with projects collecting phenotyping data across several laboratories, in some cases over long time periods, frequently through collaboration with researchers from other institutes and disciplines, and frequently contributing phenotyping data to public databases and services, and/or to meta-analysis and reanalysis by other researchers (Crabbe et al., 1999; Chesler et al., 2002a,b; Collaborative Cross Consortium, 2004; Wolfer et al., 2004; Kafkafi et al., 2005; Wahlsten et al., 2006; Mouse Phenotype Database Integration Consortium, 2007; Mandillo et al., 2008; Morgan et al., 2009; Beckers et al., 2009; Baker et al., 2011; Richter et al., 2011; Collaborative Cross Consortium, 2012; Bogue et al., 2014; Grubb et al., 2014; Heller et al., 2014; Karp et al., 2014; Koscielny et al., 2014; Maggi et al., 2014; de Angelis et al., 2015; Bogue et al., 2016; Karp et al., 2017; Kafkafi et al., 2017). The projects described in the rest of this section will be used to address multiple issues of replicability and reproducibility in the following sections.
The Mouse Phenome Database (MPD), a data resource that emerged from a research effort at The Jackson Laboratory, stores primarily individual (per subject, as opposed to just group means and standard deviations) phenotype values, along with in-depth phenotyping protocol information, as contributed by researchers from all over the world (Maddatu et al., 2012; Grubb et al., 2014; Bogue et al., 2015, 2016). It allows for trait correlation and examination of trait stability across strains, data sharing, dissemination and integration, facilitating the discovery of convergent evidence. At the time of writing the MPD contains several hundred measures of widely studied behaviors collected in multiple laboratories in inbred strains and now also includes per subject data from genetic mapping studies in the QTL Archive. Several among the meeting participants contributed their results to the MPD, and data from the MPD were used for several studies presented in the meeting.
The GeneWeaver.org database (Baker et al., 2011) employs curated user-submitted and published gene sets from GWAS, QTL mapping, genome-wide gene expression analysis, text mining, gene co-expression, expert lists, curated annotations, and many other data sources drawn from major public data resources. It included at the time of the meeting ~80,000 gene sets from 9 species including humans rats, zebrafish, drosophila and mice. GeneWeaver applies several algorithms to analyze the convergent evidence for relations among these sets of genes and behaviors or other biological constructs derived from many independent experimental studies. e.g., for those implicated in alcohol preference and withdrawal (Bubier et al., 2014; see Section 9).
GeneNetwork is a database that enables searching for ~4000 phenotypes from multiple studies in the BXD, HXB, and in other recombinant inbred rodent families, as well as in other model organisms and even humans (Mulligan et al., 2017). GeneNetwork employed a somewhat different strategy than MPD in that it did not rely solely on researchers submitting their data. Instead the database operators extracted the data from the scientific literature and integrated them into a uniform format (Chesler et al., 2003). This strategy required a considerable effort, but also expanded the range of studies and possible forms of analysis. In many cases, however, per subject phenotype data were not available. GeneNetwork uses both routine and advanced statistical methods to extract, explore, and test relations among phenotypes and underlying genetic variation. It enables complex queries in real time, including very fast QTL mapping. Similar to MPD, GeneNetwork can also be used to correlate any phenotype with all other phenotypes in the database across strain means, within or between studies, enabling the exploration of the replicability of phenotypes, even before relating them to the genotype. Any new phenotype can be correlated with any previously documented phenotypes across multiple strains. The increasing number of possible combinations grows exponentially with the rate of the added data. In the future, these two data resources, the per strain phenotype data storage with thorough protocol documentation in MPD, the Rat Genome Database, and genetic analysis suite in GeneNetwork.org will be more closely integrated (Mulligan et al., 2017).
The public database of the International Mouse Phenotyping Consortium (IMPC) is intended to be “the first truly comprehensive functional catalogue of a mammalian genome” (Morgan et al., 2009; Koscielny et al., 2014). The IMPC is a community effort to knock out ~20,000 genes and generate ~20,000 mutant mouse lines over the next 10 years, phenotype them using comprehensive and standardized high-throughput assays, and make them freely available to researchers over the world as animal models (De Angelis et al., 2015). At the time of the meeting the IMPC included ten “centers” – institutes over the world performing high-throughput phenotyping of mice, on the same genetic background of C57BL/6N. Although most lines were tested only in one center, a few mutant lines and their controls were tested across 3 and even 4 centers, and even more overlap between centers currently accumulates, enabling an extensive study of replicability across laboratories. The IMPC has made an effort to standardize phenotyping assay protocols across centers and typically records hundreds of phenotypic measures per mouse (Karp et al., 2015). Despite the standardization, however, there is still workflow variation among centers, as a result of local factors such as different policies and colony size. For example, mice from the same litter are typically assayed on the same day, and some centers have concurrent controls while others regularly sample controls (de Angelis et al., 2015). Minor protocol differences exist in some cases, the composition of the battery varies across centers, and of course, a litany of laboratory-related factors (housing, husbandry, experimenter, room dimensions, ambient noise, caging styles, etc.) differ across centers. Data from the IMPC database are currently being used for several studies of replicability (de Angelis et al., 2015; Karp et al., 2015; Kafkafi et al., 2017).
A large data set used to analyze replicability across laboratories (Kafkafi et al., 2017) was first presented in the meeting, consisting of data from multiple databases and multi-lab studies contributed by several researchers, including Wolfer et al. (2004), Richter et al. (2011), Wahlsten and Crabbe (2003, downloaded from the MPD) and knockout data downloaded from the IMPC database (Morgan et al., 2009; Koscielny et al., 2014). This dataset records per subject results of individual animals, amounting to one of the most extensive reanalysis of multi-lab studies, enabling estimation of the typical replicability in the field (see Section 4), as well as demonstrating the random lab model (Section 6) and GxL-adjustment (Section 8) advocated for estimating replicability. GxL-adjustment explicitly relies on systematic data sharing as a proposed strategy for addressing replicability across laboratories in rodent phenotyping.
4. Replicability issues in mouse phenotyping – how serious are they, really?
This seemingly simple empirical question is not simple to answer, for several reasons: there is no consensus over the correct ways to analyze and estimate replicability (Open Science Collaboration, 2015; Gilbert et al., 2016, see also Section 6), and only a few attempts have been made at systematic analysis across several studies and/or laboratories with the objective of estimating replicability in a quantitative way (see also Sections 3 and 8). Here, using careful reanalysis and meta-analysis of data from the multi-lab studies and public phenotyping databases detailed in the previous section, we give a general assessment. Most of the participants in the meeting seemed to agree that there are real and serious problem of reproducibility and replicability in mouse phenotyping, but also that some specific phenotyping results are highly replicable, especially when the genotype effect size is large.
Crabbe et al. (1999) conducted the famous experiment that first led to a wider recognition of the replicability issue in rodent phenotyping, anticipating current concerns about replicability in general science (Ioannidis, 2005). This experiment compared five inbred strains, one F1 hybrid, and one knockout line and its inbred background strain, across three laboratories, by standardizing factors including equipment, protocols, and husbandry at a much higher level than is common in the field. This study found significant laboratory effects in 6 out of 8 standard phenotypic measures, and significant interaction between genotype and laboratory in 5 of these 8. It therefore drew the provocative conclusion: “experiments characterizing mutants may yield results that are idiosyncratic to a particular laboratory”. Additional results were published in another study across laboratories and across several decades of phenotyping (Wahlsten et al., 2006). On the other hand, several genotype differences in this study appeared replicable, especially when genotype effect sizes were large, e.g., the well-known C57BL/6 preference for alcohol drinking in comparison with DBA/2. Generally, John Crabbe estimated that the issue has been exaggerated, that the situation is actually not worse than it is in many other fields of science, and that efforts to “remediate” the problem should proceed with due caution (Crabbe, 2016). At the time a response paper (Pfaff, 2001) presented several effects of mutations in mice that were replicated.
In another study of nociception phenotyping, about 42% of the variance was found to be associated with the experimenter (Chesler et al., 2002a), and many other sources of laboratory environmental variation were found to influence phenotype alone and in sex and genotype interactions (Chesler et al., 2002b). Similar effects were found for many other behavioral and physiological phenotypes in a heterogeneous stock population (Valdar et al., 2006). In QTL analysis using lines of the Collaborative Cross, different cohorts might produce different QTLs, seemingly affected by factors such as season, time of testing in the circadian phase, and perhaps even geographic latitude (Iraqi et al., 2000; Iraqi, personal communication).
A common way to visualize the replicability across two experiments from different studies, even from different laboratories, is a correlation plot of the genotype means (e.g., see Wahlsten et al., 2006). Several speakers in the meeting presented such plots comparing laboratories and studies, and both the MPD and the GeneNetwork software (see Section 3) generate them by request, and even run a fast search in their database for phenotypes that correlate with any given phenotype across strains (Mulligan et al., 2017). Such plots frequently indicate considerable correlation between strain means across studies, indicating some replicability, although there is no clear criterion for how much correlation indicates sufficient replicability.
The heterogenization experiment of Richter et al. (2011, see Section 8 for more detail) was orchestrated across six laboratories, more than in any other multi-lab experiment in the field of rodent phenotyping. It concluded that these laboratories, while still much fewer than all potential phenotyping laboratories over the world, already contribute a large component of variation, apparently considerably larger than the variation introduced by systematic heterogenization of two factors (test age and cage enrichment). This study therefore concluded that “differences between labs are considerable and unavoidable”.
There are many potential confounders in studying genetically modified mice that are difficult to control (Schellinck et al., 2010) and they are likely to differ across laboratories and studies. Studies utilizing phenotyping data from several knockout lines and associated controls across research centers of the IMPC were presented in the meeting. These studies found that test day at each phenotyping institute was a considerable source of variation and encompassed multiple variance sources (e.g. human operator, litter, cage, reagents etc., see also Karp et al., 2014, de Angelis et al., 2015). Spreading testing across time functions as a form of heterogenization. It is not clear yet to what extent a multi-batch workflow (Karp et al., 2014) captures the interaction of genotype with the laboratory, which is a different effect.
In a recent large dataset comprised of multiple previous studies, each including several genotypes measured across several laboratories (Kafkafi et al., 2017, see Section 3), cases were demonstrated that may be termed “opposite significant”, i.e., there is a crossover interaction with genotype and laboratory such that one genotype produces significantly higher mean of an outcome measure in one laboratory while significantly lower in another laboratory (see Fig. 1 right for a conceptual illustration, assuming environments E1 and E3 represent two different laboratories). In other words, these laboratories would have reported opposite discoveries. Opposite significant cases are not rare: examples were found in most of the multi-lab datasets in the study, although as expected they are more common in datasets that include a larger number of laboratories. However, in most multi-lab datasets (specifically all 8 but one) the majority of genotype effects were replicable when using the random lab model criterion for a replicable genotype effect (see Section 6). In these same datasets, the proportion of “non-replicable positives”, i.e., genotype differences that were found significant within a single laboratory (using the typical t-test at the level of α = 0.05) but did not replicate across all laboratories (using the random lab model) ranged between 19% and 41% (Kafkafi et al., 2017). This result can be regarded as an estimation of the proportion of non-replicable “discoveries” in single-lab studies in the field. It could be argued that the true value is higher, since the general standardization level in the field is probably lower than the standardization level in the multi-lab studies used to derive the above proportion (but see Section 7).
Fig. 1.
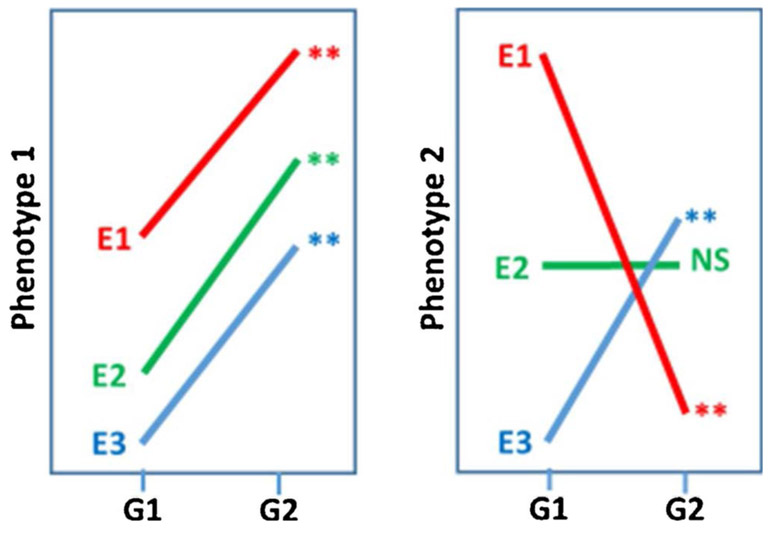
Comparing two genotypes G1 and G2, using two phenotypic measures 1 and 2, in three environments E1, E2 and E3. In the case of phenotype 1 (left) there is almost no interaction between genotype and environment (GxE). Note that the environment effect is large, but since it affects both genotypes in the same way it can be controlled using the same genotype as a reference for all other genotypes within the same environment. In the case of Phenotype 2 (right), there is a strong GxE effect, to the point that in E1, G1 is significantly larger than G2, while in E3, G1 is significantly smaller than G2 (“opposite significant”) and E2 does not have any effect. In this case an issue with replicability ensues, since the genotype effect is idiosyncratic to the specific combination of genotype and environment. and statistical reproducibility
In summary, there is wide agreement that the proportion of non-replicable results in phenotyping is considerably higher than the sometimes assumed (or hoped) 5%. Yet it appears that this proportion is not so high as to make the whole field worthless, as might be concluded from as Ioannidis’ estimation of > 50%, and can be considerably improved using several approaches (see Sections 7, 8 and 9).
5. Replicability of behavior: a special case?
An interesting empirical question is whether behavioral phenotypes are less replicable than physiological phenotypes. The recent concern in the scientific community regarding replicability and reproducibility of experimental results is by no means limited to behavioral studies, and Ioannidis’ (2005) famous claim that “most published scientific results are false” does not single them out. While psychology is frequently mentioned as a field that might suffer from a high percentage of non-replicable discoveries (Asendorpf et al., 2013; Open Science Collaboration, 2015), so are other fields, such as preclinical (Collins and Tabak, 2014; Haibe-Kains et al., 2013) and clinical pharmacology (Jager and Leek, 2014), cancer research (Baggerly and Coombes, 2009; Errington et al., 2014), epidemiology (Belbasis et al., 2015), brain imaging (Eklund et al., 2016) and GWAS (Siontis et al., 2010; Chabris et al., 2012).
A general consensus in the meeting seemed to be that behavioral phenotypes need not be less replicable. In a study across several laboratories and many decades, Wahlsten et al. (2006) showed that some behavioral phenotypes (including locomotor activity) were as replicable as classic anatomical phenotypes such as brain size, whereas other behavioral phenotypes (e.g., anxiety-related behavior on the elevated plus maze) were considerably less replicable. Proekt et al. (2012) demonstrated that motor activity in home cages can be highly reliable, as much as physical variables in exact mathematical models, providing some conditions were met. Valdar et al. (2006), in a study of 2448 genetically heterogeneous mice descended from 8 common inbred strain, actually found that the interactions between the genotype and multiple environmental covariates, such as experimenter, cage density, litter, test time and test order, tended to be smaller in the behavioral tests, such as the open field, fear potentiated startle and context freezing, than in many physiological tests such as glucose tolerance, hematology tests, immunology tests, biochemistry tests, and body weight. Valdar et al. (2006) explained this tendency by the automation of their behavioral battery, specifically predesigned to minimize the role of the experimenter to placing the animal in the apparatus. In contrast the physiological tests were less automated, e.g., large experimenter effects were found in the glucose tolerance tests, in which the intraperitoneal glucose was administered manually.
However, some behavioral phenotypes are indeed problematic to measure, understand and interpret (Gomez-Marin et al., 2014; Krakauer et al., 2017), which probably does not contribute to their replicability, a problem appreciated in research with a spectrum of species including the laboratory mouse (e.g., Gerlai and Clayton, 1999; Gerlai, 2001, 2002a,b; Martin and Bateson, 2007; Benjamini et al., 2010; Hurst and West, 2010; Wahlsten, 2011; Crabbe, 2016) as well as fish (Gerlai and Csányi, 1990; Gerlai and Crusio, 1995) and humans (Eilam, 2014); Behavioral phenotypes tend to be susceptible to many environmental parameters affecting the animal’s performance, particularly demonstrated in investigations of the emotional state in short-lasting anxiety tests (Hurst and West, 2010), as illustrated also in zebrafish (Gerlai, 2014). Such issues might actually get worse in the high-throughput procedures common in phenotyping, since they are frequently designed for the human experimenter’s convenience and efficiency, rather than to minimize animal’s stress. Several researchers therefore emphasized that high-throughput automation should be developed only on the basis of careful prior observation and thorough understanding of the animals’ behavior (Wahlsten et al., 2003; Crabbe and Morris, 2004; Gerlai, 2015).
However, such understanding might not be easy to achieve, considering that the mouse’s and rat’s umwelt (in the sense of von Uexküll, 1957, the world from the perspective of their point of view) differs considerably from that of human. It is dominated by smell and has preference to some bitter tastes (Latham and Mason, 2004). A recent study suggests how strikingly important olfactory cues may be for murine behavior (Smith et al., 2016). Mouse and rat vision relies less on color perception and visual acuity is comparatively low (especially in albino stocks). However, mice and rats are more sensitive to near ultraviolet and are also highly sensitive to movement and changes in light intensity. Rodents in general are able to hear and communicate in the ultrasound range. Such differences may hinder experimenters from detecting subtle environmental effects impacting on behavior (Latham and Mason, 2004; Burn, 2008). Individual differences is an issue that was especially noted to affect behavioral phenotypes, potentially obscuring experiment results and impugning replicability. For example, the two-compartment DualCage setup (Hager et al., 2014), while sensitive enough to differentiate the behavior of the two closely-related mouse substrains C57BL/6J and C57BL/6N, also revealed large interindividual differences with some mice showing post-traumatic stress disorder (PTSD)-like persistent avoidance performance. Performance differences in cognitive tests between mouse strains and/or mutants might emerge due to the differential impact of specific and unspecific stressors, emotional (anxiety) differences and other involved motivational aspects (Youn et al., 2012), particularly in complex tasks involving higher cortical functions, thereby following the arousal-performance relation of the Yerkes-Dodson law (reviewed by Diamond et al., 2007) that has been known for more than 100 years. In contrast, other behavioral measures such as locomotor activity are highly correlated over successive days in the DualCage, indicating high stability and therefore probably better replicability as well (Hager et al., 2014). Individual differences may be conceived as a disturbance increasing variability of the test cohort thereby reducing statistical power. It is useful to check for specific subpopulations of performers (rather than rare statistical outliers), e.g. attributable to different coping styles (De Boer et al., 2017).
On the other hand, it has also been argued that adopting a reaction norm perspective, instead of trying to spirit biological variation away, such individual variability is fundamental for improving the external validity and hence the replicability of research findings (Vöelkl and Würbel, 2016).
In summary, we propose that well-understood, well-validated and properly measured behavioral phenotypes are not inherently less replicable than physiological phenotypes, but unfortunately many behavioral phenotypes, even those in common use, do not fit these criteria. This issue is closely connected with the issues of genotype-environment interactions (see Section 6) and the validity of behavioral measures (see Section 9).
6. Genotype-environment interaction – how should it be handled?
A problem inherent to the field of phenotyping is that the final phenotype depends not only on the genotype and the environment, but also on an interaction between the genotype and environment (commonly abbreviated GxE). Furthermore, the effect of the environment on animals is cumulative, with phenotypic measures often depending on ontogenetic development and experience of an animal. For example, in cross-fostering experiments of mouse inbred strains, raising BALB/cByJ pups by a C57BL/6ByJ dam reduced excessive stress-elicited hypothalamic-pituitary-adrenal (HPA) activity and behavioral impairments, but no effect was found in the opposite case of C57BL/6ByJ pups raised by a BALB/cByJ dam (Anisman et al., 1998). These interactions may take place over many levels of RNA, protein, cells, circuits, tissues, whole-organisms and ontogenetic development. In the case of brain and behavioral phenotypes there are the additional levels of neurons, CNS organization and activity, as well as their complex interaction with the environment. The physicist PW Anderson (1972) was quoted in the meeting: “surely there are more levels of organization between human ethology and DNA than there are between DNA and quantum electrodynamics, and each level can require a whole new conceptual structure”. This understanding of GxE effects is commonly regarded in current life sciences to be the answer to the old “nature vs nurture” debate, and is closely connected with the ecological concepts of phenotypic plasticity and reaction norms (Lewontin, 1974; Wahlsten, 1990; Pigliucci, 2001; Voelkl and Würbel, 2016) as well as the psychological concept of G-E correlations (Homberg et al., 2016).
Empirically, this biological interaction does not necessarily have to result in large statistical interaction between genotype and environment, but in many cases it does. The most obvious case of statistical GxE occurs when a certain genotype (e.g., a knockout inbred line) scores a higher phenotypic mean than another genotype (e.g., the wild-type inbred strain) in certain environmental conditions, yet lower in other environmental conditions (Fig. 1 right). Typical examples of different environmental conditions may be different laboratories, different test days at the same laboratory, or even different laboratory technicians (Chesler et al., 2002a, Chesler et al., 2002b; Valdar et al., 2006). The sources of interaction are frequently unknown, multiple and very difficult to control, especially in light of the differences between rodent and humans in their sensory ranges (Latham and Mason, 2004; Burn, 2008). An intuitive illustration would be to test whether bull terriers are more aggressive than poodles. While in one clinic such a conclusion may indeed result from a standardized stranger-direction aggression test (Blackshaw, 1991), in another clinic the local technician might unknowingly wear a perfume that annoys only the poodles, leading to an opposite result that might prove difficult to “debug”. Indeed, Mogil and colleagues engaged in just such an exercise, ultimately identifying pheromonal effects on laboratory mice tested by different experimenters (Sorge et al., 2014). Such opposite results are quite common in actual phenotyping (“opposite significant”, see Kafkafi et al., 2017), and are actually more impressive than the hypothetical dog breed example, since C57BL/6 mice, unlike bull terriers, are (near perfectly) genetically identical. A large interaction effect is usually considered the mark of a true non-replicable genotype effect (Crabbe et al., 1999; Kafkafi et al., 2005, 2017). Note that an environment effect alone (Fig. 1, left) is not a serious hindrance to replicability since, by definition, it affects all genotypes to the same amount, and therefore can be controlled by having measurements on control animals (e.g., the C57BL/6J as a reference genotype). An interaction effect, in contrast, cannot be corrected this way because it is by definition unique to the specific combination of both genotype and environment.
What can and should be done about the statistical GxE interaction? This depends on the research question (Kafkafi et al., 2005). In many cases the source of the interaction, once recognized, might be itself of interest, and lead to uncovering an important biological or behavioral mechanism. However, when testing the very common type of hypothesis suggesting that a certain genotype has a certain phenotypic effect, the interaction is at least a confounding factor (Fig. 1 right) that must be taken into consideration and handled, and is even considered by some to be a fundamental property of living organisms. As illustrated and discussed in the meeting, careful observation of the animals’ response to the rearing conditions and/or experimental setup may sometimes locate and eliminate the cause of some of the interaction (Gerlai and Clayton, 1999; van der Staay and Steckler, 2001; Lad et al., 2010). Moreover, certain phenotypic measures might be much less sensitive to GxE than other measures, especially if they are more robust to environmental disturbances and more faithfully represent the true state of the animal (Wahlsten et al., 2003; Benjamini et al., 2010). A systematic way of decreasing the interaction was demonstrated by explicitly changing and improving the measure used for phenotyping (Benjamini et al., 2010).
In many cases, however, a certain portion of the statistical interaction effect does not disappear even after carefully redesigning the experiment or improving the analysis, and remains large and significant. Large GxE interaction effects may still be highly replicable if they depend on well-known environmental condition that can be equated (such as the dependence of body size in drosophila strains on temperature) but often they do not. In such cases the common statistical approach in the field brands the genotype effect as non-replicable, being idiosyncratic to unknown sources and conditions. However, according to the newly developed “random lab model” (Kafkafi et al., 2005; see Section 8), such a genotype effect may still be demonstrated as replicable, providing it is large enough to be statistically significant even over the background of the large interaction. The random lab model treats the genotype as a fixed factor that can be precisely standardized and replicated, but models the environment with random variables. This approach gives up on the unrealistic hope of precisely standardized and replicated laboratories, and instead models them as randomly sampled out of the population of all phenotyping laboratories. The immediate implication is that the interaction of the genotype with the laboratory (GxL) has a similar role to that of the individual animal noise (within-group effect). Similar to the individual animal noise, it should be decreased as much as possible, but in real life it would never disappear completely. Instead the model adds it to the within-group variability as the yardstick against which the genotype effect is compared. This generates a higher benchmark for showing a significant genotype effect – the price paid for ensuring that this effect is likely to remain significant if tested in another laboratory.
It is rarely appreciated that the most common criterion in the field for assessing replicability across several laboratories – the significance of the GxL interaction effect in the traditional analysis of variance (ANOVA) that treats the genotype effect as fixed – often results in misleading and even paradoxical conclusions (Kafkafi et al., 2005). Perhaps the worst is that using low-quality and noisy measurement may render the interaction non-significant. Alternatively, the same consequence can be “achieved” by using samples that are too small. In both cases a semblance of replicability is created. The reason is that this standard model has lower intrinsic power to detect interaction effects than to detect the main effects (Wahlsten et al., 2006), and thus any degradation of power is likely to eliminate GxL significance before it eliminates the genotype significance. This seeming paradox can be resolved by treating the environment effect as random, using the random lab model instead of fixed model ANOVA. With this model, the criterion for a true genotype difference and the criterion for a replicable genotype difference are one and the same – the significance of the genotype effect. It is therefore impossible to “improve” replicability by degrading the power (Kafkafi et al., 2005).
Replicability issues in the same laboratory across time is a similar problem arising as a result from “workflow” – the timing of individual mouse testing, either knockout mutants or background controls. In IMPC centers, each mouse passes through a phenotyping “pipeline” – a series of phenotypic tests in a predetermined order and defined age of the mouse. Due to fecundity and fertility problems, there are typically multiple small batches of knockouts with different birth dates and therefore testing dates, and the control mice (which are typically much larger in number) might not be concurrent. Moreover, depending on institutional resources and throughput, different institutes can have different workflow. Karp et al., 2014 preferred moving to a design and analysis which embraces this variation across time, rather than changing to a highly standardized design. They proposed a mixed model in which time (batch) is a random variable.
Handling GxE interaction of various kinds thus depends on the objective and the context of research. While GxE can be understood and decreased by careful observation of the animals, and by redesigning housing and testing conditions, it can rarely be completely eliminated. Especially when testing a hypothesis of a genotype effect, ignoring or mishandling potential GxE is likely to result in replicability issues and other severe methodological issues.
7. Standardization and heterogenization: why and when should they be used?
When discoveries of preclinical studies fail to replicate despite the near-perfect standardization of the genotypes, there is a natural tendency to assume that the problem is the lack of standardization of housing and testing conditions. Standardization aims to document the important properties of the environment and then keep them constant. A commonly held ideal is that every animal will be treated identically, so there are no sources of variation other than the controlled variable used as experimental treatment. This common dogma is usually invoked in the context of the “internal validity” within one study in one laboratory. In this role standardization is seen as means to minimize the variability of results, avoiding bias by unwanted sources of variation, and increasing sensitivity and precision. It is typically assumed that standardization lowers the noise level, thereby increasing the statistical power to detect differences between the experimental and control groups, and decreasing the number of animals required to detect such differences (Beynen et al., 2003; Festing, 2004). However, it should be noted that such standardization necessarily limits the generalizability of the study to the narrow range of conditions in which it was performed, thereby hampering replicability (Würbel, 2000; Richter et al., 2009; Voelkl and Würbel, 2016). As an additional strategy to facilitate comparison with published results and thus to assess replicability, an anonymous reviewer suggested using positive controls based on known effects of drugs or other relevant treatments in any experiment.
Several participants in the meeting invested considerable effort devising behavioral assays in which the animals are tested for a long time (24 h and more) in a home cage, sometimes with additional space to explore, with minimal or no contact with a human experimenter, but potentially with computer-controlled experimental manipulations and feedback to increase standardization. Proekt et al. (2012) developed a high-throughput assay including multiple computer-controlled units, in which the mice are completely isolated from outside sound and vibration, and require human experimenters touch them only once per week. Tactile, olfactory and vestibular stimuli can be programmed, and the animal movement is tracked using infrared beams. Fonio et al. (2009) video-tracked mice and Cohen et al. (2015) fruit flies in assays comprised of a small home cage connected through a narrow doorway with a much larger arena, which the animals gradually and systematically inhabit over several hours to several days, of their own volition with no apparent incentive other than exploration. Tucci’s laboratory (Maggi et al., 2014) developed automated home-cage testing (www.phenoscale.org), consisting of computer-controlled holes and hoppers, in which circadian rhythms, sleep-wake and related cognitive processes can be automatically recorded and studied for many days. Tucci’s team has also developed user-friendly software platforms that can work with raw data, and has made the software available to the community to improve data sharing and to coordinate multiple testing across different laboratories. Hager et al. (2014) developed a two-compartment home cage-based assay with video-tracking to monitor fear learning, avoidance and risk assessment over two days without human interference. Here individual variability in exploring a test compartment was detectable in the absence of the experimenter (see Section 5) as a potentially confounding factor, indicating that the assumption that standardization may help lower variation may not apply to all behavioral measures (see Section 6 and above).
Standardization is employed for another objective: increasing reproducibility across replicates of an experiment, either across time within the lab or across multiple labs. Crabbe et al. (1999) made an exceptional effort to standardize their phenotyping experiment across three different laboratories, and the EUMORPHIA project standardized the IMPC pipelines of tests across the IMPC centers (Mandillo et al., 2008). Both reported that careful improvement of conditions and testing increased replicability, yet both reported issues of replicability despite standardization.
Richter et al. (2009, 2010, 2011) maintain that the idea to improve replicability through standardization is based on the true finding that experimental results can differ depending on environmental conditions (i.e., phenotypic plasticity, Lewontin, 1974; Wahlsten, 1990; Pigliucci, 2001; Voelkl and Würbel, 2016), and on the false belief that these conditions are fully known so standardization will ‘spirit away’ such differences between experimental conditions, which they refer to as “the standardization fallacy” (Würbel, 2000, 2002). On the contrary, they proposed that “heterogenization” – systematic variation of conditions – may improve reproducibility and attenuate spurious results. The rationale is that different laboratories will always standardize to different local conditions, because many lab-specific factors are either unknown or cannot realistically be standardized, such as personnel. Consequently, the results might be valid only for these narrow conditions, and may not necessarily generalize to the conditions of other laboratories. In the last of several proof-of-concept studies, Richter et al. (2011) ran heterogenized and standardized batches in each of six laboratories. In this study, heterogenization of study populations through systematically varying animal age and cage enrichment did indeed improve replicability, but only by a very small extent. It appears that this simple form of heterogenization introduced only a fraction of the variation that existed among the six laboratories.
It is also notable that too strict standardization may be a possible reason why preclinical studies often find drug efficacy while phase 2 or phase 3 human clinical trials of the same drug fail. Human populations are variable, genetically and environmentally, while animal populations are often genetically highly homogeneous and are tested under strict environmental control. These discrepancies have been discussed and the question of how to resolve them has been debated in the pharmaceutical industry and academia alike (Howells et al., 2014).
A related issue is cage environmental enrichment, which is frequently asserted to improve animal welfare, and depending on the type of enrichment may have profound effects on brain function, behavior and physiology compared to barren standard housing conditions (e.g. van Praag et al., 2000; Nithianantharajah and Hannan, 2006). More generally, Poole (1997) maintained that “happy animals make good science”. However, enrichment was long considered to compromise standardization, as more complex environments were thought to increase variation in the results, and even identical enrichment elements might be arranged differently in each cage, thereby impeding both the precision and replicability of experimental results. Conversely, Wolfer et al. (2004) tested two inbred lines and their F1 hybrid across three different laboratories, with and without enrichment, and concluded that enrichment did neither decrease the precision, nor the replicability of behavioral phenotypes. A reanalysis (Kafkafi et al., 2017) using laboratory as a random variable (instead of a fixed variable as in the original study) even found that the type-I error was actually lower in the enriched animals (33.3%) compared to the non-enriched animals (40.7%). This result suggests that enrichment might actually improve replicability, although the reason for this remains elusive.
The heterogenization concept was not received with outright rejection in the meeting, perhaps surprisingly in light of the importance usually prescribed to standardization. Notably, it was argued that strict “universal” standardization of laboratory and test environment is unfeasible, and that widespread adoption of few standard protocols, apparatuses and test environments diminishes, rather than enriches, understanding of the assayed behavior. Any attempt to repeat an experiment can never perfectly replicate the original conditions, but this is probably a good thing since it will increase generalizability of findings. Phenotyping databases (e.g., the MPD) may enable investigators to integrate information across these related experiments through multivariate analysis, meta-analysis and other approaches to find consistency and convergence of evidence across the range of experimental conditions in which a study is employed. Spreading mutant testing across time, as is done in the IMPC centers (Karp et al., 2014), or simply dose-dependent drug testing, may be regarded as forms of environmental heterogenization, and may lead to approaches that “embrace the variation” instead of standardizing it away. Heterogenization may also be viewed as a way to simulate multi-laboratory studies within a single laboratory, a similar approach to artificially increasing the variability in single-lab studies by adding the GxL interaction noise as measured in previous multi-lab studies (Kafkafi et al., 2017). While automated home-cage systems will increase costs considerably, this is not the case for within-lab heterogenization. If a heterogenization factor is assigned at the level of cage, treating cage as a random factor (a useful procedure even if cages were not specifically heterogenized), no further degrees of freedom are lost, and thus heterogenization increases neither the number of animals needed nor the costs of the research.
8. Replicability across laboratories: can it be ensured?
The issue of replicability across laboratories, an immediate form of GxE, is one of the most critical in mouse phenotyping, because modern science does not normally accept experimental results that are idiosyncratic to a certain location, even if they are replicable within this location. This is why the results and conclusions of the Crabbe et al. (1999) report were disturbing for many researchers in the field, and in other fields as well. As previously discussed in Section 6 there is currently no consensus even over the proper criteria to evaluate replicability across laboratories. Studies on the subject are few because they typically require collaboration of several laboratories and institutions, although they are becoming more and more common, thanks to data sharing and community efforts (Section 3). Therefore, credible and practical solutions to the issue at the methodological and statistical levels are urgently needed. Several strategies were discussed in the meeting, including the following proposals.
Ideally, discoveries should be replicated in at least one other laboratory. In the simplest case of testing the same hypotheses of difference between two genotypes – e.g., a knockout and its wildtype control – the criterion for a replicated discovery may be statistical significance (e.g., using a 0.05 level t-test) in each of two laboratories. Such a criterion concurs with the old Royal Society rule, as well as with Ronald Fisher’s view (see Section 2). Unfortunately, this criterion is not directly applicable when considering p-values from multiple phenotypic measures, as is typical for high-throughput rodent phenotyping, due to the issue of multiple comparisons. That is, if enough hypotheses are tested this way, some of them will be found “replicable” just by coincidence. Heller et al. (2014) therefore generalized the criterion to multiple comparisons situations, and proposed a novel statistic for this purpose, the “r-value”. In the simplest case above the r-value equals the larger of the p-values in the two labs, but when multiple hypotheses are tested in each lab, the r-value computation can be adapted to take care of the multiple comparisons. Reporting the r-values can thus give credibility to the replicability claims: by declaring as replicable all findings with r-value less than, say, level 0.05, the expected fraction of false replicability claims among the replicability claims made is kept to this level. This FDR of replicability property is good enough assurance and is more powerful than its family-wise counterpart.
While the ultimate demonstration of replicability is to observe the experimental effect in multiple laboratories, in practice most phenotyping experiments are performed in a single laboratory, and results from other laboratories are usually not immediately available. This raises an unavoidable question: what should researchers do about a significant discovery in their own laboratory? How can they know whether it is likely to replicate in other laboratories? Should they publish it, or seek first to validate it in additional laboratories? And how would other researchers know if they are likely to replicate the effect in their own laboratories? All solutions discussed at the meeting have the effect of increasing the standard error of the effect size, and many exciting findings that depend on exceeding the standard p < 0.05 threshold will not survive them. A practical solution to these questions (Kafkafi et al., 2017) employs an extension of the random lab model (Section 6), termed “GxL-adjustment”, which can be applied by researchers to phenotyping results in their own lab, providing a previous estimation of the interaction is available. The genotypic effect in the single laboratory is compared, as in the usual t-test, to the within-group variation, but this time “adjusted” by the addition of the multi-lab interaction variation. This addition of the interaction, as in the application of the random lab model to a multi-lab analysis, raises the benchmark for showing a significant genotype effect, ensuring that only effects that are likely to replicate in other laboratories will be significant. GxL-adjustment can be demonstrated to decrease the proportion of false discoveries that are not really replicable to the range of the traditional 0.05, for a price of modest reduction in the statistical power (Kafkafi et al., 2017).
Several important insights can be gained from the random lab model and from GxL-adjustment (Kafkafi et al., 2005, 2017). First, the empirical size of the interaction variability sets its own limit for detection of replicable results. Increasing the number of the animals within a single lab has therefore only a limited benefit for replicability, since it does not affect the interaction with the laboratory. For the same reason, decreasing the individual animal noise also has a limited benefit for replicability. A phenotypic measure with smaller interaction is therefore “better” in the specific sense that it is more powerful to detect true replicable genotype effects, but not necessarily in other contexts. Consequently, we should search for phenotypic measures having smaller interaction, but keep in mind that replicability is still a property of a result, not of a phenotypic measure. That is, true replicable genotype differences may be apparent even over a large interaction, providing they are large enough, while true replicable genotype differences that are small will be difficult to replicate even over a smaller interaction.
An extensive effort of standardization, as reported by Crabbe et al. (1999), is likely to succeed in reducing individual noise, yet fail to eliminate all unknown and unavoidable interaction sources, especially in light of the previously-mentioned differences between the sensory ranges of mice, rats and humans (Latham and Mason, 2004). If individual noise is decreased but the interaction remains the same, the usual ANOVA model (with fixed lab effects) will paradoxically detect more significant interaction terms, giving a false impression of reduced replicability. The random lab model in the same situation will give the correct impression: replicability (as seen in the number of significant genotype differences) will in fact improve, but only to a point. Beyond this point, further improvement of replicability must be achieved by decreasing the interaction (Kafkafi et al., 2005).
The random lab model does set a higher level for detecting significant effects in single-lab studies. This is not necessarily a drawback, however, in the sense that it is a way to weed out non-replicable differences (Fonio et al., 2012). It is an important incentive to invest time and effort in reducing interaction. The interaction can be methodically reduced by improving analysis methods, e.g., robust smoothing (Benjamini et al., 2010). However, while interaction variability should be reduced, it will never be completely eliminated (much like the individual animal noise) and therefore should never be ignored. Unknown sources of interaction are unavoidable (e.g., Würbel, 2002).
How can the interaction with the laboratory be estimated? One possibility is using as a surrogate the variability across time within a single laboratory (Karp et al., 2014) or heterogenization (Section 7). However, controlled heterogenization uses effects we know about, while true interaction with laboratory might involve factors we are not aware of at all. Another proposal (Kafkafi et al., 2017) is to make use of multi-lab data from large and evolving phenotyping databases, such as the MPD and the IMPC. Such a database can calculate the interaction and publish it for use by scientists running phenotyping experiments in their own laboratories. This calculation has to be repeated for each phenotypic measure separately. A website was demonstrated in which any researchers conducting a phenotyping experiment can upload their results, get an updated estimate of the interaction for the relevant phenotypic measure, perform a GxL-adjustment and get an adjusted p-value. The researchers are then given an option to leave their data in the database, thus enriching it and providing a better estimate, based on more laboratories, for future users. As in other file-sharing and community-effort strategies, GxL-adjustment has ethical implications: by employing previous data of the same phenotypes from other laboratories and other research questions, instead of replicating the experimental study, it may eventually reduce the number of experimental animals without compromising replicability.
Ultimately, the replicability of a phenotyping discovery can be guaranteed only by testing it in additional laboratories. Even in these simple cases, ways to quantify replicability, such as the “r-value”, are still in need of development and acceptance by the scientific community. In cases when testing was performed in a single lab only, it may still be possible to enhance replicability, or estimate its prospects in a better way. Several directions were proposed: heterogenizing the experimental setup, splitting the duration of the experiments to different time units, and using external estimates of the interaction from phenotyping database. All these may serve to get more realistic estimates of the replicability standard deviation, and better (not too optimistic) estimates of the relevant standard errors.
9. Replicability and validity: what is the relation between them?
Several researchers stress the importance of validity of research in preclinical phenotyping, especially behavioral phenotyping, and its probable close connection with replicability (e.g., Bailoo et al., 2014; Crusio, 2015). Some other researchers, while not necessarily using the term “validity”, share the view that the issue of replicability may be a byproduct of more fundamental methodological issues with behavioral phenotyping (e.g., Wahlsten et al., 2003; Benjamini et al., 2010; Gomez-Marin et al., 2014; Krakauer et al., 2017). There is no clear consensus over the nature of these methodological issues, nor over the practical ways to address them, but generally these researchers seem to share a similar dissatisfaction with the current credibility of phenotyping and especially behavioral phenotyping. They also seem to share the hope that, once phenotypes are properly validated, the issue of replicability will turn out to be considerably less grave as well.
In psychology, “internal validity” of an experiment refers to the justification for concluding the effect of the specific experimental treatment on the specific outcome measure, while “external validity” refers to the generalizability of this effect (Richter et al., 2009; Bailoo et al., 2014; Voelkl and Würbel, 2016). While internal validity is a required condition for concluding an effect, it is usually of little scientific value without external validity. Replication of the same experiment results across laboratories is the least requirement for external validity, but replication also across different designs, housing and testing conditions is better, and results that have the best external validity are those that generalized across strains and even species, e.g., translation to humans. The low replicability of certain phenotypic measures across laboratories may therefore indicate their poor prospects as animal models (Bailoo et al., 2014). Translation validity issues are outside the scope of the present review, but obviously they might result from poor reproducibility and replicability already in the preclinical phase. Pharma companies often complain that they cannot replicate preclinical effects reported in the academia, and yet recent reviews of translation issues (e.g., Mak et al., 2014; McGonigle and Ruggeri, 2014) tend to devote little attention to reproducibility and replicability within the same species of laboratory animals.
Assays for many common behavioral constructs, such as “anxiety” and “behavioral despair”, are not well-validated, and understanding of what it is that they measure is insufficient (Fonio et al., 2012; Crusio, 2015). For example, the Porsolt Forced Swim Test and the Tail Suspension Test are both thought to measure “behavioral despair” using a similar variable: the latency to stop trying to escape from an unpleasant situation; yet some mice treated with a random stress procedure reacted oppositely in these two tests. While researchers assume that these tests measure a similar construct, the mice apparently disagree (Crusio, 2015). The Morris Water Maze is frequently considered to be a “gold-standard” for memory testing in rodents, and yet factor analysis of behavior data from 1400 mice revealed that only about 13% of the variance in this test is explained by memory, while most of it is explained by behaviors such as wall hugging (thigmotaxis) and passive floating, which are thought to represent emotional and motivation aspects (Wolfer et al., 1998). This issue is probably because the Morris Water Maze was designed for rats and then used “as is” with the house mouse, a species considerably less adapted to wet environments and swimming. Crusio (2015) therefore concluded that validating and refining behavioral constructs should be an absolute priority for psychiatry and behavioral neuroscience.
GeneNetwork.org (see Section 3) can correlate different phenotypic assays as well as mapping QTLs in many recombinant inbred lines (Mulligan et al., 2017). A high correlation between two phenotypic measures across many strains means suggests that these phenotypes measure a similar construct, even when they originate in different studies, and measured in different tests and different conditions. Such correlated phenotypic measures may be viewed as different ways to measure the same trait that has been essentially replicated. Moreover, if both phenotypic measures reveal the same strong QTL, the correlation implies a similar causal connection, since the central dogma assures us that it is the genotype that determines the phenotype rather than the other way around. Thus, construct validity (Willner, 1984, 1986; Belzung and Lemoine, 2011; van der Staay, 2006; van der Staay et al., 2009) in the genetic level is often gained as well.
A strategy of integrative bioinformatics was suggested as a way to discover validated and replicable relations among a variety of phenotypes through the shared association to common genomic features (Baker et al., 2011). In a demonstration of this strategy, GeneWeaver (see Section 3) was used to study the relationship between alcohol preference and alcohol withdrawal in gene sets from multiple publications and public data resources, including mouse QTL and humans (Bubier et al., 2014). Combinatorial integration of these gene sets revealed a single QTL positional candidate gene common to both preference and withdrawal. This QTL seems to have a replicable phenotypic effect – it was confirmed by a validation study in knockout mice mutated for this locus. Since discoveries in this strategy can be based on multiple studies across laboratories, species, and phenotyping protocols, they have a better chance to be replicable, generalizable, and translatable. However, the complex integration of multiple data sets in this strategy makes it difficult to construct statistical models for estimating how much the replicability may be improved.
Addressing validity issues might be critical when deciding how to share and integrate behavioral data, e.g., when constructing “controlled vocabularies” and “ontologies” used for querying and searching phenotyping databases. Semantics raises many challenging questions regarding how behaviors are structurally related to one another, and should they be labeled by the supposed meaning of the assay, or only by what can be observed. For example, “immobility” is an objective description of motor behavior free of the context of ascribed behavioral meaning. Mice become immobile in a variety of apparatuses for a variety of reasons and in response to a variety of treatments. Should an “immobility” label be used rather than labels such as “anxiety”, “fear,” “learning” and “behavioral despair? Efforts such as the Neurobehavioral Ontology (Gkoutos et al., 2012), the Vertebrate Trait Ontology (Park et al., 2013) and the Mammalian Phenotype Ontology (Smith and Eppig, 2012) have each taken different approaches to this issue based in large part on their unique applications. In databases storing multiple phenotypic measures from multiple studies and laboratories, the implications of these approaches for replicability and reproducibility may for the first time be methodically investigated.
It was further suggested to consider replicability as a “gold standard”, and use behavioral data across several laboratories in order to explicitly redesign behavioral constructs and ontologies for increased replicability (Benjamini et al., 2010). In this strategy, the issue of replicability in behavioral results is turned into an asset rather than a liability – it enables researchers to methodically improve the definition of key behavioral constructs by using the statistical level of replicability as a benchmark, filtering out behavioral results and representations that are less replicable (Kafkafi et al., 2003; Lipkind et al., 2004). Another type of validity that is highly relevant to preclinical testing is “predictive validity” (Willner, 1984, 1986) or “pharmacological validity” – a response of the phenotypic measure to multiple psychiatric drugs that predicts the response of human disorders and syndromes to the same drugs. Here too it is possible to use behavioral databases in order to explicitly design behavioral measures for high predictive validity, as well as for replicability across laboratories (Kafkafi et al., 2014). A requirement for such a strategy, however, is storing detailed and high-quality low-level data, e.g., locomotor path coordinates in the above examples, rather than merely animal final means.
A potentially even more powerful approach may be to explicitly design behavioral measures and constructs to increase generality across species and taxonomic groups. That is, to search for biological homologies not just in the genetics, anatomy and physiology levels, but also in the behavioral level. Such homologies have been a central goal of classical ethologists, but mere similarity of form between behavior patterns across taxonomic groups proved untenable as a criterion for establishing behavioral homology (Beer, 1980; Gomez-Marin et al., 2016a). Establishing homology requires careful animal-centric low-level descriptions of movement and behavior in species from different phyla, as remote as mice (Fonio et al., 2009) and fruit flies (Cohen et al., 2015; Gomez-Marin et al., 2016a). Once the common frame of reference and common origins used by organisms are identified, homology may be apparent in an invariable temporal and spatial order of movement components (Golani, 2012). For example, the study of rotational velocities at the trajectory level has shown that both worms and flies modulate their navigation via the local bearing angles within sensory gradients (Gomez-Marin and Louis, 2014; Gomez-Marin et al., 2016b). More generally, when behavior is treated as the control of perception (Powers, 1973; Golani, 1981) rather than the production of motor actions per se, then what appears as output noise (deemed as variability, and so a corresponding hurdle to replicability) may actually be revealed as the means of the organism to maintain its perception at a set reference (Golani, 1976; Bell, 2014). Such an animal-centric perspective (Gomez-Marin and Mainen, 2016) is apt to reveal homologues behaviors. Similarly, homologous bones, despite dramatic variation in their shape and function across different vertebrate orders and classes, can readily be identified by their common relative position in the skeleton. Viewed through these concepts, validity of behavioral phenotypes measures may be obvious as it is in comparative anatomy.
In summary, reproducibility, replicability, generalizability, and validity, in their various forms and levels, are all criteria for assessing the value of scientific results. There are multiple opinions regarding the exact semantic definitions of these terms in different fields, their relative importance, the proper methods to estimate them and the best ways to ensure them. But in general there seems to be wide agreement that they are all important and useful in certain levels and situations. There are obviously close and complex relations between these criteria, so that problems with assessing one of them are likely to complicate the proper assessment of the other, and improvements with assessing one are likely to assist with the proper assessment of the other. It is possible to utilize stored phenotyping results, especially when they are detailed, high-quality and well-organized in accessible databases, to estimate one or even several of these criteria at once in order to improve the value of preclinical research.
9.1. ■ Overall summary
Modern science is reviewing its own problems of credibility, searching for causes and ways to address them. The original foundation of experimental science, emphasizing replicability and reproducibility, is still correct in spirit, but experimental, computational, and statistical methodologies require updating to cope with the increasing size and complexity of current research. Preclinical research and rodent phenotyping are similar in this regard to many other fields of experimental science, while also requiring the consideration of ethical issues surrounding the use of animals. They enjoy special technical advantages of their own, such as the ability to standardize genomes and manipulate them in a precise manner; but they also encounter special challenges, such as a potential interaction between genotype and environment, and the difficulty of defining and measuring behavioral phenotypes. Any proposed solutions, therefore, should likely be tailored to the particularities of the field. Phenotypic databases, community efforts and other methods of data sharing play important roles, as they can be employed to efficiently assess the severity of the issue, as well as the performances of different proposed solutions.
Correct handling of the genotype-environment interaction (GxE) is a key to proper methodology, and depends on the context and objective of the research. GxE can typically be understood and decreased through careful observation of the animals, redesigning the rearing and testing conditions to eliminate adverse effects of irrelevant confounding factors. Especially when testing a hypothesis of a genotype effect, ignoring or mishandling the relevant form of GxE is likely to result in replicability issues and other severe methodological issues. Extreme standardization of rearing and testing conditions is probably not by itself feasible or helpful as a strategy to eliminate GxE, and might limit the generalizability of any conclusions.
Ultimately, the replicability and generalizability of any phenotyping discovery can be guaranteed only by replicating it across additional studies, laboratories and experimental conditions. Even when studies are repeated, there is no single well-established method to quantify the replicability of the results, but large and consistent genotype effect sizes can usually be agreed upon as replicable. In the more common situation where results are available from only one study, it may still be possible to enhance replicability, or estimate its prospects in a better way. Several directions were proposed and discussed at the meeting, including heterogenizing the experimental setup, splitting the duration of the experiments to different time units, and employing external estimates of the interaction from phenotyping database, effectively suggesting an expansion of the field of experimental design.
Linked with the issues of replicability are those relating to how phenotypes are defined and measured, especially for behavioral phenotypes. Regardless of the problems with replicating across laboratories, issues of generalizability and validity remain worth addressing. Insight and solutions resulting from attention given to the methodological issue of replicability may directly help with generalizability, and also help in addressing the more general issue of validity, by freeing investigators from rigid reliance on standardization, and rather promoting approaches to generalizable and replicable science. These observations first emerged from behavioral characterization of model organisms bear on other areas of biological inquiry and experimental science in general.
Acknowledgments
This work is supported by European Research Council grants FP7/2007-2013 ERC agreement no [294519]-PSARPS (YB, NK, IG, IJ), by NIH AA018776 (EJC), by NIH DA045401 (YB, NK) and by the Spanish Ministry of Economy and Competitivity (Severo Ochoa Center of Excellence programme SEV2013-0317 start-up funds to AGM). We thank Prof. Allan Kalueff for many useful comments. This paper is the result of a scientific meeting in Tel Aviv University. Videos of the full lectures can be accessed at the following links.
1. Yoav Benjamini: “Assessing Replicability in One Own’s Lab: A Community Effort” https://www.youtube.com/watch?v=y-Q4GWkJicE&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=15.
2. Elissa J. Chesler: “Finding Consilience in Genetic and Genomic Analyses of Behavior” https://www.youtube.com/watch?v=f9TdNXipRPQ&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=20.
3. John C. Crabbe: “Managing Sources of E to Address GXE” https://www.youtube.com/watch?v=A7R2iZfjydA&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=4.
4. Wim E. Crusio: “What do We Want to Reproduce?” https://www.youtube.com/watch?v=3ZuEZw8mDjY&index=6&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP.
5. David Eilam: “Variability: A By-Product Noise or an Essential Component of Motor Behavior” https://www.youtube.com/watch?v=ADgVHDFZpUg&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=7.
6. Robert Gerlai: “Behavioral Phenotyping: The Double Edged Sword” https://www.youtube.com/watch?v=pRrNz0PTKc4&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=9.
7. Ilan Golani: “Replicability as a Virtue” https://www.youtube.com/watch?v=KiRSxpA8qZY&index=10&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&spfreload=10.
8. Alex Gomez-Marin: “Toward a Behavioral Homology Between Insects and Rodents” https://www.youtube.com/watch?v=Hu8PwdKap3k&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=25&spfreload=10.
9. Ruth Heller: “Assessing Replicability Across Laboratories: The R-Value” https://www.youtube.com/watch?v=b2uGOp9yLMw&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=22.
10. Fuad Iraqi: “TAU Collaborative Cross Mice a Powerful GRP for Dissection Host Susceptibility to Diseases” https://www.youtube.com/watch?v=NV62o7Ubrfg&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=21.
11. Natasha A. Karp: “Simulation Studies to Investigate Workflow and its Impact on Reliability of Phenodeviant Calls: https://www.youtube.com/watch?v=N5wTrgXvsAY&index=17&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP.
12. Neri Kafkafi: “The Random Lab Model for Assessing the Replicability of Phenotyping Results Across Laboratories” https://www.youtube.com/watch?v=XsQawSBA6Vc&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=12.
13. George Nicholson & Hugh Morgan:”The Empirical Reproducibility of High-Throughput Mouse Phenotyping” https://www.youtube.com/watch?v=KzrrP6_F8r8&index=18&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP.
14. Donald W. Pfaff: “Application of Strict Methodology and Applied Mathematical Statistics to Mouse Behavioral Data” https://www.youtube.com/watch?v=hB0FnO9evbY&index=8&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP.
15. Philip B. Stark: “Preproducibility for Research, Teaching, Collaboration, and Publishing” https://www.youtube.com/watch?v=wHryMtEBkB4&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=13.
16. Oliver Stiedl: “Individuality of Avoidance Behavior of Mice in an Automated Home Cage Environment” https://www.youtube.com/watch?v=gUIgRW51uZY&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=24.
17. Victoria Stodden:“Computational and Statistical Reproducibility” https://www.youtube.com/watch?v=GzrOcqz8TVY&index=14&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP.
18. Valter Tucci: “Phenotyping Behaviour Across Laboratories and Across Mouse Strains” https://www.youtube.com/watch?v=iTlsFaj62oQ&index=22&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP.
19. William Valdar & Lisa M. Tarantino: “The Effect of Genetic Background on Behavioral Variability: Implications for Replicability?” https://www.youtube.com/watch?v=63sgLO4Hd04&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP&index=5.
20. Robert W. Williams: “Data Rescue for Replication: Finding, Annotating, and Reusing Data for the BXD Mouse Cohort” https://www.youtube.com/watch?v=goocssSA33g&index=19&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP.
21. Hanno Würbel & S. Helene Richter: “On Standardization and Other Fallacies in Animal Phenotyping” https://www.youtube.com/watch?v=tfW35740q3k&index=11&list=PLNiWLB_wsOg74GlfLNyAcTo-TshyAcdLP.
References
- Agassi J, 2013. The very idea of modern science. In: Bacon Francis, Boyle Robert (Eds.), Boston Studies in the Philosophy and History of Science 298. Springer Science + Busness Media, Dordrecht. [Google Scholar]
- Alsheikh-Ali AA, Qureshi W, Al-Mallah MH, Ioannidis JP, 2011. Public availability of published research data in high-impact journals. PloS One 6 (9), e24357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Cicerone RJ, Fienberg SE, Kamb A, McNutt M, Nerem RM, Schekman R, Shiffrin R, Stodden V, Suresh S, Zuber MT, Pope BK, Jamieson KH, 2015. Scientific integrity: self-correction in science at work. Science 348 (6242), 1420–1422. [DOI] [PubMed] [Google Scholar]
- Anderson PW, 1972. More is different. Science 177 (4047), 393–396. [DOI] [PubMed] [Google Scholar]
- Anisman H, Zaharia MD, Meaney MJ, Merali Z, 1998. Do early-life events permanently alter behavioral and hormonal responses to stressors? Int. J. Dev. Neurosci 16 (3–4), 149–164. 10.1016/S0736-5748(98)00025-2. [DOI] [PubMed] [Google Scholar]
- de Angelis MH, Nicholson G, Selloum M, White JK, Morgan H, Ramirez-Solis R, EUMODIC Consortium, et al. , 2015. Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics. Nat. Genet 47 (9), 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asendorpf JB, Conner M, De Fruyt F, De Houwer J, Denissen JJA, Fiedler K, Fiedler M, Funder DC, Kliegl R, Nosek AN, Perugini M, Roberts BW, Schmitt M, Vanaken MAG, Weber H, Wicherts JM, 2013. Recommendations for increasing replicability in psychology. Eur. J. Personal. Eur. J. Pers 27, 108–119. [Google Scholar]
- Baggerly KA, Coombes KR, 2009. Deriving chemosensitivity from cell lines: forensic bioinformatics and reproducible research in high-throughput biology. Ann. Appl. Stat 3 (4), 1309–1334. [Google Scholar]
- Baker EJ, Jay JJ, Bubier JA, Langston MA, Chesler EJ, 2011. GeneWeaver: a web-based system for integrative functional genomics. Nucleic Acids Res. D 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailoo JD, Reichlin TS, Würbel H, 2014. Refinement of experimental design and conduct in laboratory animal research. ILAR J. 55 (3), 383–391. [DOI] [PubMed] [Google Scholar]
- Beckers J, Wurst W, de Angelis MH, 2009. Towards better mouse models: enhanced genotypes, systemic phenotyping and envirotype modelling. Nat. Rev. Genet 10 (6), 371–380. [DOI] [PubMed] [Google Scholar]
- Beer CG, 1980. Perspectives on animal behavior comparisons. Comp. Meth. Psychol 2, 17. [Google Scholar]
- Belbasis L, Panagiotou OA, Dosis V, Evangelou E, 2015. A systematic appraisal of field synopses in genetic epidemiology: a HuGE review. Am. J. Epidemiol 181 (1), 1–16. [DOI] [PubMed] [Google Scholar]
- Bell HC, 2014. Behavioral variability in the service of constancy. Int. J. Comp. Psychol 27 (2), 338–360. [Google Scholar]
- Belzung C, Lemoine M, 2011. Criteria of validity for animal models of psychiatric disorders: focus on anxiety disorders and depression. Biol. Mood Anxiety Disord 1 (1), 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y, 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B (Methodol.) 1995, 289–300. [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I, 2001. Controlling the false discovery rate in behavior genetics research. Behav. Brain Res 125 (1–2), 279–284. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Lipkind D, Horev G, Fonio E, Kafkafi N, Golani I, 2010. Ten ways to improve the quality of descriptions of whole-animal movement. Neurosci. Biobehav. Rev 34 (8), 1351–1365. 10.1016/j.neubiorev.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hechtlinger Y, 2013. Discussion: an estimate of the science-wise false discovery rate and applications to top medical journals by Jager and Leek. Biostatistics 15 (1), 13–16. 10.1093/10.1093/biostatistics/kxt032. [DOI] [PubMed] [Google Scholar]
- Beynen AC, Gärtner K, van Zutphen LFM, 2003. In: van Zutphen LFM, Baumans V, Beynen AC (Eds.), Standardization of Animal Experimentation In Principles of Laboratory Animal Science. Elsevier, Amsterdam, pp. 103–110. [Google Scholar]
- Blackshaw JK, 1991. An overview of types of aggressive behaviour in dogs and methods of treatment. Appl. Anim. Behav. Sci 30, 351–354. [Google Scholar]
- Bogue MA, Peters LL, Paigen B, Korstanje R, Yuan R, Ackert-Bicknell C, Grubb SC, Churchill GA, Chesler EJ, 2014. Accessing data resources in the mouse phenome database for genetic analysis of murine life span and health span. J. Gerontol. A Biol. Sci. Med. Sci 71 (2), 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue MA, Churchill GA, Chesler EJ, 2015. Collaborative cross and diversity outbred data resources in the mouse phenome database. Mamm. Genome 26 (9–10), 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue MA, Peters LL, Paigen B, Korstanje R, Yuan R, Ackert-Bicknell C, Grubb SC, Churchill GA, Chesler EJ, 2016. Accessing data resources in the mouse phenome database for genetic analysis of murine life span and health span. J. Gerontol. A Biol. Sci. Med. Sci 71 (2), 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RE, Stanford L, Schellinck HM, 2000. Developing standardized behavioral tests for knockout and mutant mice. ILAR J 41 (3), 163–174. [DOI] [PubMed] [Google Scholar]
- Bubier JA, Jay JJ, Baker CL, Bergeson SE, Ohno H, Metten P, Crabbe JC, Chesler EJ, 2014. Identification of a QTL in Mus musculus for alcohol preference, withdrawal, and Ap3m2 expression using integrative functional genomics and precision genetics. Genetics 197 (4), 1377–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn CC, 2008. What is it like to be a rat? rat sensory perception and its implications for experimental design and rat welfare. Appl. Anim. Behav. Sci 112, 1–32. [Google Scholar]
- Chabris CF, Hebert BM, Benjamin DJ, Beauchamp J, Cesarini D, van der Loos M, Johannesson M, Magnusson PK, Lichtenstein P, Atwood CS, Freese J, Hauser T Hauser RM, Christakis N, Laibson D, 2012. Most reported genetic associations with general intelligence are probably false positives. Psychol. Sci 23, 1314–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler EJ, Wilson SG, Lariviere WR, Rodriguez-Zas SL, Mogil JS, 2002a.Influences of laboratory environment on behavior. Nat. Neurosci 5 (11), 1101–1102. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Wilson SG, Lariviere WR, Rodriguez-Zas SL, Mogil JS, 2002b. Identification and ranking of genetic and laboratory environment factors influencing a behavioral trait, thermal nociception, via computational analysis of a large data archive. Neurosci. Biobehav. Rev 26 (8), 907–923. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Wang J, Lu L, Qu Y, Manly KF, Williams RW, 2003. Genetic correlates of gene expression in recombinant inbred strains. Neuroinformatics 1 (4), 343–357. [DOI] [PubMed] [Google Scholar]
- Chesler EJ, Miller DR, Branstetter LR, Galloway LD, Jackson BL, Philip VM, Voy BH, Culiat CT, Threadgill DW, Williams RW, Churchill GA, Johnson DK, Manly KF, 2008. The collaborative cross at Oak ridge national laboratory: developing a powerful resource for systems genetics. Mamm. Genome 19 (6), 382–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Benjamini Y, Golani I, 2015. Coping with space neophobia in drosophila melanogaster: the asymmetric dynamics of crossing a doorway to the untrodden. PloS One 10 (12), e0140207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Cross Consortium, 2004. The collaborative cross, a community resource for the genetic analysis of complex traits. Nat. Genet 36 (11), 1133–1137. [DOI] [PubMed] [Google Scholar]
- Collaborative Cross Consortium, 2012. The genome architecture of the collaborative cross mouse genetic reference population. Genetics 190 (2), 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Rossant J, Wurst W, 2007. A mouse for all reasons. Cell 128 (1), 9–13. [DOI] [PubMed] [Google Scholar]
- Collins FS, Tabak LA, 2014. Policy: NIH plans to enhance reproducibility. Nature 505, 612–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Complex Trait Consortium, 2003. The nature and identification of quantitative trait loci: a community’s view. Nat. Rev. Genet 4 (11), 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Wahlsten D, Dudek BC, 1999. Genetics of mouse behavior: interactions with laboratory environment. Science. 284 (5420), 1670–1672. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Morris RGM, 2004. Festina lente: late night thoughts on high-throughput screening of mouse behavior. Nat. Neurosci 7, 1175–1179. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, 2016. Reproducibility of experiments with laboratory animals: what should we do now? Alcohol Clin. Exp. Res 40.11, 2305–2308. [DOI] [PubMed] [Google Scholar]
- Crawley JN, 1985. Exploratory behavior models of anxiety in mice. Neurosci. Biobehav. Rev 9 (1), 37–44. [DOI] [PubMed] [Google Scholar]
- Crusio WE, 2006. Inheritance of behavioral and neuroanatomical phenotypical variance: hybrid mice are not always more stable than inbreds. Behav. Genet 36 (5), 723–731. [DOI] [PubMed] [Google Scholar]
- Crusio WE, 2015. Key issues in contemporary behavioral genetics. Curr. Opin. Behav. Sci 2, 89–95. [Google Scholar]
- Cummings G, 2014. The new statistics: why and how. Psychol. Sci 25, 7–29. 10.1177/0956797613504966. [DOI] [PubMed] [Google Scholar]
- de Boer SF, Buwalda B, Koolhaas JM, 2017. Untangling the neurobiology of coping styles in rodents: towards neural mechanisms underlying individual differences in disease susceptibility. Neurosci. Biobehav. Rev 74, 401–422. [DOI] [PubMed] [Google Scholar]
- Diamond DM, Campbell AM, Park CR, Halonen J, Zoladz PR, 2007. The temporal dynamics model of emotional memory processing: a synthesis on the neurobiological basis of stress-induced amnesia, flashbulb and traumatic memories, and the Yerkes-Dodson law. Neural Plast 10.1155/2007/60803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle PJ, Zeger SL, 2010. Embracing the concept of reproducible research. Biostatistics 11 (3), 375. [DOI] [PubMed] [Google Scholar]
- Dulawa SC, Hen R, Scearce-Levie K, Geyer MA, 1997. Serotonin1B receptor modulation of startle reactivity, habituation, and prepulse inhibition in wild-type and serotonin1B knockout mice. Psychopharmacology 132 (2), 125–134. [DOI] [PubMed] [Google Scholar]
- Eilam D, 2014. The cognitive roles of behavioral variability: idiosyncratic acts as the foundation of identity and as transitional, preparatory, and confirmatory phases. Neurosci. Biobehav. Rev 49, 55–70. [DOI] [PubMed] [Google Scholar]
- Eklund A, Nichols TE, Knutsson H, 2016. Cluster failure: why fMRI inferences for spatial extent have inflated false-positive rates. Proc. Natl. Acad. Sci. U. S. A 113, 7900–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington TM, Iorns E, Gunn W, Tan FE, Lomax J, Nosek BA, 2014. An open investigation of the reproducibility of cancer biology research. Elife 3, e04333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festing MFW, 2004. Refinement and reduction through the control of variation. Altern. Lab. Anim 32, 259–263. [DOI] [PubMed] [Google Scholar]
- Fisher RA, 1935. The Design of Experiments. Oliver & Boyd, Oxford. England. [Google Scholar]
- Fonio E, Benjamini Y, Golani I, 2009. Freedom of movement and the stability of its unfolding in free exploration of mice. Proc. Natl. Acad. Sci. U. S. A 106 (50), 21335–21340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonio E, Golani I, Benjamini Y, 2012. Measuring behavior of animal models: faults and remedies. Nat. Methods 9 (12), 1167. [DOI] [PubMed] [Google Scholar]
- Frenkel M, Chirico RD, Diky V, Muzny C, Dong Q, Marsh KN, Dymond JH, Wakeham WA, Stein SE, Konigsberger E, Goodwin ARH, Magee JW, Thijssen M, Haynes WM, Watanasiri S, Satyro M, Schmidt M, Johns AI, Hardin GR, 2006. New global communication process in thermodynamics: impact on quality of published experimental data. J. Chem. Inf. Model 46 (6), 2487–2493. [DOI] [PubMed] [Google Scholar]
- Garijo D, Kinnings S, Xie L, Xie L, Zhang Y, Bourne PE, Gil Y, 2013. Quantifying reproducibility in computational biology: the case of the tuberculosis drugome. PloS one 8 (11), e80278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlai R, Csányi V, 1990. Genotype environment interaction and the correlation structure of behavioral elements in paradise fish (Macropodus opercularis). Physiol. Behav 47, 343–356. [DOI] [PubMed] [Google Scholar]
- Gerlai R, Wojtowicz JM, Marks A, Roder J, 1995. Over-expression of a calcium binding protein, S100ß, in astrocytes alters synaptic plasticity and impairs spatial learning in transgenic mice. Learn. Memory 2, 26–39. [DOI] [PubMed] [Google Scholar]
- Gerlai R, Crusio WE, 1995. Organization of motor and posture patterns in paradise fish (Macropodus opercularis): environmental and genetic components of phenotypical correlation structures. Behav. Genet 25, 385–396. [DOI] [PubMed] [Google Scholar]
- Gerlai R, Clayton NS, 1999. Analysing hippocampal function in transgenic mice: an ethological perspective. Trends Neurosci 22, 47–51. [DOI] [PubMed] [Google Scholar]
- Gerlai R, 2001. Behavioral tests of hippocampal function: simple paradigms, complex problems. Behav. Brain Res 125, 269–277. [DOI] [PubMed] [Google Scholar]
- Gerlai R, 2002a. Phenomics: fiction or the future? Trends Neurosci 25 (10), 506–509. [DOI] [PubMed] [Google Scholar]
- Gerlai R, 2002b. Hippocampal LTP and memory in mouse strains: is there evidence for a causal relationship? Hippocampus 12, 657–666. [DOI] [PubMed] [Google Scholar]
- Gerlai R, Adams B, Fitch T, Chaney S, Baez M, 2002. Performance deficits of mGluR8 knockout mice in learning tasks: Tthe effects of null mutation and the background genotype. Neuropharmacology 43, 235–249. [DOI] [PubMed] [Google Scholar]
- Gerlai R, 2014. Fish in behavior research: unique tools with a great promise!. J. Neurosci. Methods 234. 54–58. [DOI] [PubMed] [Google Scholar]
- Gerlai R, 2015. Zebrafish phenomics: behavioral screens and phenotyping of mutagenized fish. Curr. Opin. Behav. Sci 2, 21–27. [Google Scholar]
- Gilbert DT, King G, Pettigrew S, Wilson TD, 2016. Comment on “Estimating the reproducibility of psychological science”. Science 351 (6277), 1037. [DOI] [PubMed] [Google Scholar]
- Gkoutos GV, Schofield PN, Hoehndorf R, 2012. The neurobehavior ontology: an ontology for annotation and integration of behavior and behavioral phenotypes. Int. Rev. Neurobiol 103, 69–87. [DOI] [PubMed] [Google Scholar]
- Golani I, 1976. Homeostatic motor processes in mammalian interactions: A choreography of display. In: Bateson PPG, Klopfer PH (Eds.), Perspectives in Ethology. Springer, US, pp. 69–134. [Google Scholar]
- Golani I, 1981. The search for invariants in motor behavior. In: Immelman K, Barlow GW, Petrinovich L, Main M (Eds.), Behavioral Development, The Bielefeld Interdisciplinary Project. Cambridge University Press. [Google Scholar]
- Golani I, 2012. The developmental dynamics of behavioral growth processes in rodent egocentric and allocentric space. Behav. Brain Res 231 (2), 309–316. [DOI] [PubMed] [Google Scholar]
- Gomez-Marin A, Paton JJ, Kampff A, Costa RR, Zachary M, Mainen M, 2014. Big behavioral data: psychology, ethology and the foundations of neuroscience. Nat. Neurosci 17, 1455–1462. [DOI] [PubMed] [Google Scholar]
- Gomez-Marin A, Louis M, 2014. Multilevel control of run orientation in Drosophila larval chemotaxis. Frontiers Behav. Neurosci 8, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Marin A, Mainen ZF, 2016. Expanding perspectives on cognition in humans, animals, and machines. Curr. Opin. Neurobiol 37, 85–91. [DOI] [PubMed] [Google Scholar]
- Gomez-Marin A, Oron E, Gakamsky A, Valente D, Benjamini Y, Golani I, 2016a. Generative rules of Drosophila locomotor behavior as a candidate homology across phyla. Sci. Rep 6, 27555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Marin A, Stephens GJ, Brown AE, 2016b. Hierarchical compression of Caenorhabditis elegans locomotion reveals phenotypic differences in the organization of behaviour. J. R. Soc. Interface 13, 20160466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman SN, Fanelli D, Ioannidis JP, 2016. What does research reproducibility mean? Sci. Transl. Med 8 (341) 341ps12–341ps12. [DOI] [PubMed] [Google Scholar]
- Grubb SC, Churchill GA, Bogue MA, 2004. A collaborative database of inbred mouse strain characteristics. Bioinformatics 20 (16), 2857–2859. [DOI] [PubMed] [Google Scholar]
- Grubb SC, Bult CJ, Bogue MA, 2014. Mouse phenome database. Nucleic Acids Res 42, D825–D834. 10.1093/nar/gktll59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkness JH, Shi X, Janowsky A, Phillips TJ, 2015. Trace amine-associated receptor 1 regulation of methamphetamine intake and related traits. Neuropsychopharmacology 40 (9), 2175–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager T, Jansen RF, Pieneman AW, Manivannan SN, Golani I, van der Sluis S, Smit AB, Verhage M, Stiedl O, 2014. Display of individuality in avoidance behavior and risk assessment of inbred mice. Front. Behav. Neurosci 8, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B, El-Hachem N, Birkbak NJ, Jin AC, Beck AH, Aerts HJ, Quackenbush J, 2013. Inconsistency in large pharmacogenomic studies. Nature 504 (7480), 389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller R, Bogomolov M, Benjamini Y, 2014. Deciding whether follow-up studies have replicated findings in a preliminary large-scale “omics’ study”. Proc. Natl. Acad. Sci. U. S. A 111 (46), 16262–16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homberg JR, Kyzar EJ, Nguyen M, Norton WH, Pittman J, Poudel MK, Gaikwad S, Nakamura S, Koshiba M, Yamanouchi H, Scattoni ML, Ullman JFP, Diamond DM, Kaluyeva AA, Parker MO, Klimenko VM, Apryatin SA, Brown RE, Gainetdinov RR, Gottesman II, Kalueff AV, 2016. Understanding autism and other neurodevelopmental disorders through experimental translational neurobehavioral models. Neurosci. Biobehav. Rev 65, 292–312. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J, 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497 (7450), 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howells DW, Sena ES, Macleod MR, 2014. Bringing rigour to translational medicine. Nat. Rev. Neurol 10 (1), 37–43. [DOI] [PubMed] [Google Scholar]
- Hurst JL, West RS, 2010. Taming anxiety in laboratory mice. Nat. Meth 7 (10), 825–826. [DOI] [PubMed] [Google Scholar]
- Ioannidis J, 2005. Why most published research findings are false. PLoS Med. 2 (8), e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqi F, Clapcott SJ, Kumari P, Haley CS, Kemp SJ, Teale AJ, 2000. Fine mapping of trypanosomiasis resistance loci in murine advanced intercross lines. Mamm. Genome 11 (8), 645–648. [DOI] [PubMed] [Google Scholar]
- Iraqi FA, Churchill G, Mott R, 2008. The collaborative cross, developing a resource for mammalian systems genetics: a status report of the welcome trust cohort. Mamm. Genome 19 (6), 379–381. [DOI] [PubMed] [Google Scholar]
- Jager LR, Leek JT, 2014. An estimate of the science-wise false discovery rate and application to the top medical literature. Biostatistics 15 (1), 1–12. [DOI] [PubMed] [Google Scholar]
- Kafkafi N, Pagis M, Lipkind D, Mayo CL, Bemjamini Y, Golani I, Elmer GI, 2003. Darting behavior: a quantitative movement pattern designed for discrimination and replicability in mouse locomotor behavior. Behav. Brain Res 142 (1), 193–205. [DOI] [PubMed] [Google Scholar]
- Kafkafi N, Benjamini Y, Sakov A, Elmer GI, Golani I, 2005. Genotype-environment interactions in mouse behavior: a way out of the problem. Proc. Natl. Acad. Sci. U. S. A 102 (12), 4619–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafkafi N, Mayo CL, Elmer GI, 2014. Mining mouse behavior for patterns predicting psychiatric drug classification. Psychopharmacology 231 (1), 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafkafi N, Golani I, Jaljuli I, Morgan H, Sarig T, Würbel H, Yaacoby S, Benjamini Y, 2017. Addressing reproducibility in single-laboratory phenotyping experiments. Nat. Method 14 (5), 462–464. [DOI] [PubMed] [Google Scholar]
- Karp NA, Speak AO, White JK, Adams DJ, de Angelis MH, Hérault Y, Mott RF, 2014. Impact of temporal variation on design and analysis of mouse knockout phenotyping studies. PLoS One 9 (10), e111239. 10.1371/journal.pone.0111239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, Meehan TF, Morgan H, Mason JC, Blake A, Kurbatova N, Smedley D, Jacobsen J, Mott RF, Iyer V, Matthews P, Melvin DG, Wells S, Flenniken AM, Masuya H, Wakana S, White JK, Lloyd KC, Reynolds CL, Paylor R, West DB, Svenson KL, Chesler EJ, de Angelis MH, Tocchini-Valentini GP, Sorg T, Herault Y, Parkinson H, Mallon AM, Brown SD, 2015. Applying the ARRIVE guidelines to an in vivo database. PLoS Biol. 13.5 (e1002151). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, Heller R, Yaacoby S, White JK, Benjamini Y, 2017. Improving the identification of phenotypic abnormalities and sexual dimorphism in mice when studying rare event categorical characteristics. Genetics 205 (2), 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley PW, Zhou C, Lu L, Williams RW, Melmed S, Reese BE, 2014. Pituitary tumor-transforming gene 1 regulates the patterning of retinal mosaics. Proc. Natl. Acad. Sci. U. S. A 111 (25), 9295–9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennet RS, Shmueli G, 2015. Clarifying the terminology that describes scientific reproducibility. Nat. Method 12 (8), 699. 10.1038/nmeth.3489. [DOI] [PubMed] [Google Scholar]
- Koscielny G, Yaikhom G, Iyer V, Meehan TF, Morgan H, Atienza-Herrero J, Blake A, Chen CK, Easty R, Di Fenza A, Fiegel T, Grifiths M, Horne A, Karp NA, Kurbatova N, Mason JC, Matthews P, Oakley DJ, Qazi A, Regnart J, Retha A, Santos LA, Sneddon DJ, Warren J, Westerberg H, Wilson RJ, Melvin DG, Smedley D, Brown SD, Flicek P, Skarnes WC, Mallon AM, Parkinson H, 2014. The international mouse phenotyping consortium web portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res 42 (D1), D802–D809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutnikova H, Laakso M, Lu L, Combe R, Paananen J, Kuulasmaa T, Kuusisto J, Häring HU, Hansen T, Pedersen O, Smith U, Hanefeld M, Williams RW, Auwerx J, 2009. Identification of the UBP1 locus as a critical blood pressure determinant using a combination of mouse and human genetics. PLoS Genet 5 (8), e1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krakauer JW, Ghazanfar AA, Gomez-Marin A, MacIver MA, Poeppel D, 2017. Neuroscience needs behavior: correcting a reductionist bias. Neuron 93 (3), 480–490. [DOI] [PubMed] [Google Scholar]
- Lad HV, Liu L, Paya-Cano JL, Parsons MJ, Kember R, Fernandes C, Schalkwyk LC, 2010. Behavioural battery testing: evaluation and behavioural outcomes in 8 inbred mouse strains. Phys. Behav 99 (3), 301–316. [DOI] [PubMed] [Google Scholar]
- Landis SC, Amara SG, Asadullah K, Austin CP, Blumenstein R, Bradley EW, Crystal RG, Darnell RB, Ferrante RJ, Fillit H, Finkelstein R, Fisher M, Gendelman HE, Golub RM, Goudreau JL, Gross RA, Gubitz AK, Hesterlee SE, Howells DW, Huguenard J, Kelner K, Koroshetz W, Krainc D, Lazic SE, Levine MS, Macleod MR, McCall JM, Moxley RT 3rd, Narasimhan K, Noble LJ, Perrin S, Porter JD, Steward O, Unger E, Utz U, Silberberg SD, 2012. A call for transparent reporting to optimize the predictive value of preclinical research. Nature (490), 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander E, Kruglyak L, 1995. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet 11 (3), 241–247. [DOI] [PubMed] [Google Scholar]
- Lapchak PA, Zhang JH, Noble-Haeusslein LJ, 2013. RIGOR guidelines: escalating STAIR and STEPS for effective translational research. Transl. Stroke Res 4 (3), 279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham N, Mason G, 2004. From house mouse to mouse house: the behavioural biology of free-living Mus musculus and its implications in the laboratory. Appl. Anim. Behav. Sci 86 (3), 261–289. [Google Scholar]
- Leek JT, Peng RD, 2015. Opinion: reproducible research can still be wrong: adopting a prevention approach. Proc. Natl. Acad. Sci. U. S. A 112 (6), 1645–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewontin RC, 1974. Annotation: the analysis of variance and the analysis of causes. Am. J. Hum. Genet 26 (3), 400. [PMC free article] [PubMed] [Google Scholar]
- Lipkind D, Sakov A, Kafkafi N, Elmer GI, Benjamini Y, Golani I, 2004. New replicable anxiety-related measures of wall vs. center behavior of mice in the open field. J. Appl. Physiol. 97 (1), 347–359. [DOI] [PubMed] [Google Scholar]
- Logue SF, Paylor R, Wehner JM, 1997. Hippocampal lesions cause learning deficits in inbred mice in the Morris water maze and conditioned-fear task. Behav. Neurosci 111 (1), 104. [DOI] [PubMed] [Google Scholar]
- MacRae CA, Randall TP, 2015. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov 14 (10), 721–731. [DOI] [PubMed] [Google Scholar]
- Maddatu TP, Grubb SC, Bult CJ, Bogue MA, 2012. Mouse phenome database (MPD). Nucleic Acids Res D887–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi S, Garbugino L, Heise I, Nieus T, Balci F, Wells S, Tocchini-Valentini GP, Mandillo S, Nolan P, Tucci PV, 2014. A cross-laboratory investigation of timing endophenotypes in mouse behavior. Timing Time Percept 2 (1), 35–50. http://dx.doi.org/10.1163/22134468-00002007. [Google Scholar]
- Mandillo S, Tucci V, Hölter SM, Meziane H, Banchaabouchi MA, Kallnik M, Lad HV, Nolan PM, Ouagazzal AM, Coghill EL, Gale K, Golini E, Jacquot S, Krezel W, Parker A, Riet F, Schneider I, Marazziti D, Auwerx J, Brown SD, Chambon P, Rosenthal N, Tocchini-Valentini G, Wurst, W., 2008. Reliability, robustness, and reproducibility in mouse behavioral phenotyping: a cross-laboratory study. Physiol. Genome 34 (3), 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak IW, Evaniew N, Ghert M, 2014. Lost in translation: animal models and clinical trials in cancer treatment. Am. J. Transl. Res 6 (2), 114. [PMC free article] [PubMed] [Google Scholar]
- Marinov GK, Kundaje A, Park PJ, Wold BJ, 2014. Large-scale quality analysis of published ChIP-seq data. G3: Genes Genomes Genet 4 (2), 209–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin P, Bateson P, 2007. Measuring Behaviour: An Introductory Guide, third ed Cambridge University Press, Cambridge. UK. [Google Scholar]
- McNutt M, 2014. Reproducibility. Science 343 (6168), 229. [DOI] [PubMed] [Google Scholar]
- McGonigle P, Ruggeri B, 2014. Animal models of human disease: challenges in enabling translation. Biochem. Pharmacol 87 (1), 162–171. [DOI] [PubMed] [Google Scholar]
- Morgan H, Beck T, Blake A, Gates H, Adams N, Debouzy G, Leblanc S, Lengger C, Maier H, Melvin D, Meziane H, Richardson D, Wells S, White J, Wood J, EUMODIC Consortium, de Angelis MH, Brown SD, Hancock JM, Mallon AM, 2009. EuroPhenome: a repository for high-throughput mouse phenotyping data. Nucleic Acids Res 38 (Suppl. 1), D577–D585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morahan G, Balmer L, Monley D, 2008. Establishment of “The Gene Mine”: a resource for rapid identification of complex trait genes. Mammal. Genome 19 (6), 390–393. [DOI] [PubMed] [Google Scholar]
- Mouse Phenotype Database Integration Consortium, 2007. Integration of mouse phenome data resources. Mammal. Genome 18 (3), 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Mozhui K, Prins P, Williams RW, 2017. GeneNetwork: A toolbox for systems genetics. Methods Mol. Biol. (Clifton, NJ) 1488, 75–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musatov S, Chen W, Pfaff DW, Kaplitt MG, Ogawa S, 2006. RNAi-mediated silencing of estrogen receptor α in the ventromedial nucleus of hypothalamus abolishes female sexual behaviors. Proc. Nat. Acad. Sci 103 (27), 10456–10460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nature Editorial, 2013. Announcement: reducing our irreproducibility. Nature 496, 398. [Google Scholar]
- Nithianantharajah J, Hannan AJ, 2006. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat. Rev. Neurosci 7, 697–709. [DOI] [PubMed] [Google Scholar]
- Nosek BA, Errington TM, 2017. Reproducibility in cancer biology: making sense of replications. Elife 6, e23383. 10.7554/eLife.23383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Open Science Collaboration, 2015. Estimating the reproducibility of psychological science. Science 349 (6251), aac4716. [DOI] [PubMed] [Google Scholar]
- Park CA, Bello SM, Smith CL, Hu ZL, Munzenmaier DH, Nigam R, Smith JR, Shimoyama M, Eppig JT, Reecy JM, 2013. The vertebrate trait ontology: a controlled vocabulary for the annotation of trait data across species. J. Biomed. Semant 4 (1), 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson CB, Bogomolov M, Benjamini Y, Sabatti C, 2016. Many phenotypes without many false discoveries: error controlling strategies for multi trait association studies. Genet. Epidemiol 40 (1), 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigliucci M, 2001. Phenotypic Plasticity: Beyond Nature and Nurture. JHU Press. [Google Scholar]
- Pollin R, 2014. Public debt, GDP growth, and austerity: why reinhart and rogoff are wrong. LSE American Politics and Policy. [Google Scholar]
- Poole T, 1997. Happy animals make good science. Lab Anim 31 (2), 116–124. [DOI] [PubMed] [Google Scholar]
- Potti A, et al. , 2006. A genomic strategy to refine prognosis in early-stage non-small-cell lung cancer. N. Engl. J. Med 355, 570 (retracted). [DOI] [PubMed] [Google Scholar]
- Peng R, 2011. Reproducible research in computational science. Science 334 (6060), 1226–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, 2015. The reproducibility crisis in science: A statistical counterattack. Significance 12 (3), 30–32. [Google Scholar]
- Pfaff DW, 2001. Precision in mouse behavior genetics. Proc. Natl. Acad. Sci. U. S. A 98 (11), 5957–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers WT, 1973. Behavior: The Control of Perception. Aldine, Chicago. [Google Scholar]
- Proekt A, Banavar JR, Maritan A, Pfaff DW, 2012. Scale invariance in the dynamics of spontaneous behavior. Proc. Natl. Acad. Sci. U. S. A 109 (26). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnegård L, Valdar W, 2011. Detecting major genetic loci controlling phenotypic variability in experimental crosses. Genetics 188 (2), 435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rönnegård L, Valdar W, 2012. Recent developments in statistical methods for detecting genetic loci affecting phenotypic variability. BMC Genet 13 (1), 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter SH, Garner JP, Würbel H, 2009. Environmental standardization: cure or cause of poor reproducibility in animal experiments? Nat. Meth 6, 257–261. [DOI] [PubMed] [Google Scholar]
- Richter SH, Auer C, Kunert J, Garner JP, Würbel H, 2010. Systematic variation improves reproducibility of animal experiments. Nat. Method (7), 167–168. [DOI] [PubMed] [Google Scholar]
- Richter SH, Garner JP, Zipser B, Lewejohann L, Sachser N, Touma C, Schindler B, Chourbaji S, Brandwein C, Gass P, van Stipdonk N, van der Harst J, Spruijt B, Võikar V, Wolfer DP, Würbel H, 2011. Effect of population heterogenization on the reproducibility of mouse behavior: a multi-laboratory study. PLoS One 6 (1), e16461. 10.1371/journal.pone.0016461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savalei V, Dunn E, 2015. Is the call to abandon p-values the red herring of the replicability crisis? Front. Psychol 6, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellinck HM, Cyr DP, Brown RE, 2010. How many ways can mouse behavioral experiments go wrong? Confounding variables in mouse models of neurodegenerative diseases and how to control them. Adv. Stud. Behav 41, 255–366. [Google Scholar]
- Siontis KC, Patsopoulos NA, Ioannidis JP, 2010. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur. J. Hum. Genet 18 (7), 832–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Eppig JT, 2012. The mammalian phenotype ontology as a unifying standard for experimental and high-throughput phenotyping data. Mammal. Genome 23 (9–10), 653–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ML, Hostetler CM, Heinricher MM, Ryabinin AE, 2016. Social transfer of pain in mice. Sci. Adv 2 (10), e1600855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark PB, 2015. Science is “show me” not “trust me.”. Berkeley Initiative for Transparency in the Social Sciences. (Accessed 8 June 17). http://www.bitss.org/2015/12/31/science-is-show-me-not-trust-me/. [Google Scholar]
- Stark PB, 2017. Nullius in verba. In: Kitzes J, Turek D, Deniz F (Eds.), The Practice of Reproducible Research: Case Studies and Lessons from the Data-Intensive Sciences. University of California Press, Oakland, CA, pp. 6–9. (Accessed 8 June, 2017). https://www.practicereproducibleresearch.org/core-chapters/0-preface.html. [Google Scholar]
- Stephenson AG, Mulville DR, Bauer FH, Dukeman GA, Norvig P, LaPiana LS, Rutledge PJ, Folta D, Sackheim R, 1999. Mars Climate Orbiter Mishap Investigation Board Phase I Report. NASA, Washington, DC. [Google Scholar]
- Stodden V, 2010. The scientific method in practice: reproducibility in the computational sciences. PLoS One. 10.1371/journal.pone.0067111. [DOI] [Google Scholar]
- Stodden V, Guo P, Ma Z, 2013. Toward reproducible computational research: an empirical analysis of data and code policy adoption by journals. PLoS One 8 (June (6)), e67111. 10.1371/journal.pone.0067111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stodden V, 2013. Resolving irreproducibility in empirical and computational research. IMS Bulletin Online. (Accessed 8 June 17). http://bulletin.imstat.org/2013/11/resolving-irreproducibility-in-empirical-and-computational-research/. [Google Scholar]
- Shapin S, Schaffer S, 1985. Leviathan and the Air-Pump. Princeton University Press, Princeton, NJ. [Google Scholar]
- Shi X, Walter NA, Harkness JH, Neve KA, Williams RW, Lu L, Belknap JK, Eshleman AJ, Phillips TJ, Janowsky A, 2016. Genetic polymorphisms affect mouse and human trace amine-associated receptor 1 function. PLoS One. 11 (3), e0152581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soric B, 1987. Statistical “discoveries” and effect-size estimation. J. Am. Stat. Ass 84 (406), 608–610. [Google Scholar]
- Simmons JP, Nelson LD, Simonsohn U, 2011. False-positive psychology undisclosed flexibility in data collection and analysis allows presenting anything as significant. Psychol. Sci 22 (11), 1359–1366. [DOI] [PubMed] [Google Scholar]
- Sorge RE, Martin LJ, Isbester KA, Sotocinal SG, Rosen S, Tuttle AH, Wieskopf JS, Acland EL, Dokova A, Kadoura B, Leger P, Mapplebeck JC, McPhail M, Delaney A, Wigerblad G, Schumann AP, Quinn T, Frasnelli J, Svensson CI, Sternberg WF, Mogil JS, 2014. Olfactory exposure to males, including men, causes stress and related analgesia in rodents. Nat. Method 11 (6), 629–632. [DOI] [PubMed] [Google Scholar]
- Soergel DAW, 2015. Rampant software errors may undermine scientific results. F1000Research 2015 (3), 303. 10.12688/fl000research.5930.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Cookson WO, Rawlins JN, Mott R, Flint J, 2006. Genetic and environmental effects on complex traits in mice. Genetics 174 (2), 959–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Staay FJ, 2006. Animal models of behavioral dysfunctions: basic concepts and classifications, and an evaluation strategy. Brain Res. Rev 52 (1), 131–159. [DOI] [PubMed] [Google Scholar]
- Van der Staay FJ, Arndt SS, Nordquist RE, 2009. Evaluation of animal models of neurobehavioral disorders. Behav. Brain Funct 5 (11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Staay FJ, Steckler T, 2001. Behavioural phenotyping of mouse mutants. Behav. Brain Res 125 (1–2), 3–12. [DOI] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, Gage FH, 2000. Neural consequences of environmental enrichment. Nat. Rev. Neurosci 1 (December (3)), 191–198. [DOI] [PubMed] [Google Scholar]
- Vöelkl B, Würbel H, 2016. Reproducibility crisis: are we ignoring reaction norms? Trends Pharmacol. Sci 37 (7), 509–510. [DOI] [PubMed] [Google Scholar]
- von Uexköll J, 1957. A stroll through the worlds of animals and men: a picture book of invisible worlds. In: Schiller CH (Ed.), Instinctive Behavior: The Development of a Modern Concept. International Universities Press, New York, pp. 5–80. [Google Scholar]
- Vul E, Harris C, Winkielman P, Pashler H, 2009. Puzzlingly high correlations in fMRI studies of emotion, personality, and social cognition. Perspect. Psychol. Sci 4 (3), 274–290. [DOI] [PubMed] [Google Scholar]
- Wang X, Pandey AK, Mulligan MK, Williams EG, Mozhui K, Li Z, Jovaisaite V, Quarles LD, Xiao Z, Huang J, Capra JA, Chen Z, Taylor WL, Bastarache L, Niu X, Pollard SK, Ciobanu DC, Reznik AO, Tishkov AV, Zhulin IB, Peng J, Nelson SF, Denny JC, Auwerx J, Lu L, Williams RW, 2016. Joint mouse-human phenome-wide association to test gene function and disease risk. Nat. Comm 2016 (7), 10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams EG, Auwerx J, 2015. The convergence of systems and reductionist approaches in complex trait analysis. Cell 162 (1), 23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer DP, Stagljar-Bozicevic M, Errington ML, Lipp HP, 1998. Spatial memory and learning in transgenic mice: fact or artifact? Physiology 13 (3), 118–123. [DOI] [PubMed] [Google Scholar]
- Wolfer DP, Litvin O, Morf S, Nitsch RM, Lipp HP, Würbel H, 2004. Laboratory animal welfare: cage enrichment and mouse behaviour. Nature 432 (7019), 821–822. [DOI] [PubMed] [Google Scholar]
- Würbel H, 2000. Behaviour and the standardization fallacy. Nat. Genet 26, 263. [DOI] [PubMed] [Google Scholar]
- Würbel H, 2002. Behavioral phenotyping enhanced – beyond (environmental) standardization. Gene. Brain Behav 1 (1), 3–8. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, 1990. Insensitivity of the analysis of variance to hereditary-environment interaction. Behav. Brain Sci 13, 109–120. [Google Scholar]
- Wahlsten D, 2011. Mouse Behavioral Testing: How to Use Mice in Behavioral Neuroscience. Academic Press, London. [Google Scholar]
- Wahlsten D, Crabbe JC, 2003. Survey of motor activity, behavior, and forebrain morphometry in 21 inbred strains of mice across two laboratories. The Mouse Phenome Database Website, Project Data set: Wahlsten1. (Accessed 8 June 17).http://phenome.jax.org/db/q?rtn=projects/details&sym=Wahlsten. [Google Scholar]
- Wahlsten D, Rustay NR, Metten P, Crabbe JC, 2003. In search of a better mouse test. Trends Neurosci 26 (3), 132–136. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Bachmanov A, Finn DA, Crabbe JC, 2006. Stability of inbred mouse strain differences in behavior and brain size between laboratories and across decades. Proc. Nat. Acad. Sci. U. S. A 103, 16364–16369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserstein RL, Lazar NA, 2016. The ASA’s statement on P-values: context, process, and purpose. Am. Stat 70, 129–133. [Google Scholar]
- Welsh CE, Miller DR, Manly KF, Wang J, McMillan L, Morahan G, Mott R, Iraqi FA, Threadgill DW, de Villena FP, 2012. Status and access to the collaborative cross population. Mamm. Genome 23, 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner P, 1984. The validity of animal models of depression. Psychopharmacology 83, 1–16. [DOI] [PubMed] [Google Scholar]
- Willner P, 1986. Validation criteria for animal models of human mental disorders: learned helplessness as a paradigm case. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 10 (6), 677–690. [DOI] [PubMed] [Google Scholar]
- Wiltshire T, 2015. Toxicity of Four Anti-Cancer Agents Tested in Vitro in Immune Cells from 36 Inbred Mouse Strains. MPD:Wiltshire4. Mouse Phenome Database Web Resource (RRID:SCR_003212). The Jackson Laboratory, Bar Harbor, Maine USA (Accessed 8 June 17). http://phenome.jax.org. [Google Scholar]
- Youn J, Ellenbroek BA, van Eck I, Roubos S, Verhage M, Stiedl O, 2012. Finding the right motivation: genotype-dependent differences in effective reinforcements for spatial learning. Behav. Brain Res 226, 397–403. [DOI] [PubMed] [Google Scholar]


