 A review highlighting on cancer stem cells, with an exhaustive listing and comparison of biological efficacies and pharmacology of the inhibitors of 5 pivotal enzymes of the DNA-damage response.
A review highlighting on cancer stem cells, with an exhaustive listing and comparison of biological efficacies and pharmacology of the inhibitors of 5 pivotal enzymes of the DNA-damage response.
Abstract
DNA inevitably undergoes a high number of damages throughout the cell cycle. To preserve the integrity of the genome, cells have developed a complex enzymatic machinery aimed at sensing and repairing DNA lesions, pausing the cell cycle to provide more time to repair, or induce apoptosis if damages are too severe. This so-called DNA-damage response (DDR) is yet considered as a major source of resistance to DNA-damaging treatments in oncology. Recently, it has been hypothesized that cancer stem cells (CSC), a sub-population of cancer cells particularly resistant and with tumour-initiating ability, allow tumour re-growth and cancer relapse. Therefore, DDR appears as a relevant target to sensitize cancer cells and cancer stem cells to classical radio- and chemotherapies as well as to overcome resistances. Moreover, the concept of synthetic lethality could be particularly efficiently exploited in DDR. Five kinases play pivotal roles in the DDR: ATM, ATR, CHK1, CHK2 and WEE1. Herein, we review the drugs targeting these proteins and the inhibitors used in the specific case of CSC. We also suggest molecules that may be of interest for preclinical and clinical researchers studying checkpoint inhibition to sensitize cancer and cancer stem cells to DNA-damaging treatments.
1. Introduction
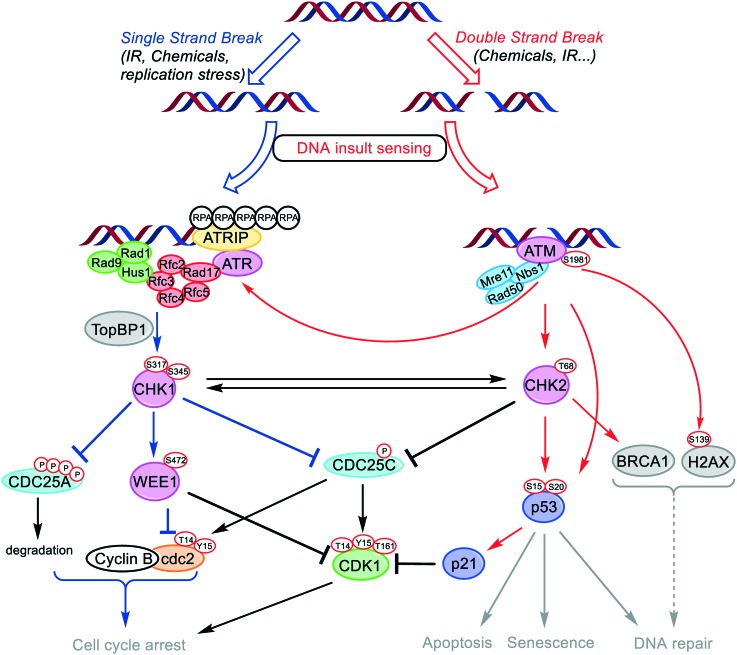
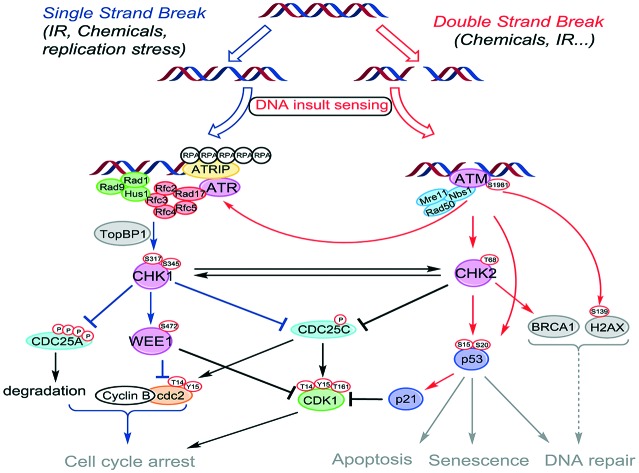
DNA is under the constant assault of exogenous (UV-light exposure, irradiation or chemicals) and endogenous factors such as free radicals and alkylating agents naturally occurring during metabolic processes. This ensues damages, estimated at up to 105 lesions per cell per day, that may evolve into transcription and replication errors and ultimately lead to cell death or gene mutation if not repaired or mis-repaired.1 Briefly, the two main DNA damage types encountered are: (i) double-strand breaks (DSB), which are considered as the most severe, and which are repaired through two different pathways, namely the non-homologous end joining (NHEJ) and the homologous recombination (HR);2,3 (ii) single-strand breaks (SSB), a specific type of lesion occurring at stalled replication forks, but also a common intermediate formed during DSB repair. Therefore, to maintain genomic integrity, cells have developed throughout evolution a complex machinery called DNA-damage response (DDR) that senses and repairs DNA.4 DDR consists in a set of responses with different groups of enzymes dedicated to specific types of lesions that can be classified into sensors, transducers and effectors (Fig. 1).5 Together, they form a complex network of interconnected pathways, whose collaborative work allows the preservation of the genome integrity by initiating cell cycle arrest, repair processes and apoptosis induction (Fig. 1). Depending on the type of lesion, different pathways are involved. DSB are rapidly sensed by the Mre11–Rad50–NBS1 (MNR) complex. This ternary complex interacts with chromatin, and subsequently promotes the activation of Ataxia Telangiectasia Mutated (ATM) kinase by autophosphorylation. ATM relays the signal to a plethora of transducer enzymes, including Checkpoint kinase 2 (CHK2) and the transcription factor p53. SSB are sensed by the Rad9–Hus1–Rad1 complex. This complex, in cooperation with Rad17, Rfc2, Rfc3, Rfc4 and Rfc5 activates Ataxia Telangiectasia and Rad3-related kinase (ATR). The latter enzyme is directed by its subunit ATR interacting protein (ATRIP) to RPA (replication protein A) coated single-stranded DNA. Following this sensing step, Rad9 binds its partner protein TopBP1, which results in the stimulation of ATR-mediated CHK1 phosphorylation. CHK1 and CHK2 amplify the signals from the sensors, phosphorylating a variety of effectors. Depending on the severity of the damage, cells either transiently arrest cell cycle progression or enter the cell death pathway (apoptosis).
Fig. 1. Components of the DNA damage response pathways modulated by ATM, ATR, CHK1, CHK2 and WEE1 kinases.
Despite the emergence of targeted therapy agents, DNA-damaging therapies are still among the most common cancer treatments. Their use relies on the fact that cancer cells are cycling more rapidly than healthy cells, and while they are associated with severe side-effects on normal tissues, they remain standard treatments for many cancers. DNA repair and checkpoint activation provide an important mean to survive DNA damages caused by irradiation or chemotherapeutics. It ensures the DNA damage repair and provides more time for this by pausing the cell cycle. DNA repair and particularly the checkpoint pathway activation are commonly admitted to play an important role in both radio- and chemoresistance.1,6 Indeed, the repeated exposure to DNA-damaging agents after many cycles of chemotherapy causes cancer cells to enhance their DNA repair systems.7 Therefore, targeting the checkpoint response by inhibiting some of its mains components may improve the global therapeutic efficacy of DNA damaging treatments and overcome resistance. Particularly interesting in this field is the concept of synthetic lethality which exploits the genetic defects which render cancer cells dependent on only one DNA damage response system.8 For example, loss of the tumour suppressor p53 abolished the G1/S cell cycle checkpoint rendering cancer cells dependent on a functional G2–M arrest. Synthetic lethality exploits this weakness by inactivating the G2–M arrest in p53-deficient cancer cells.9
Herein, we review the inhibitors of five of the key regulators of the cell cycle checkpoints in cancer cells and in the particular settings of cancer stem cells: ATM and ATR, kinases that play apical roles in DDR; CHK1 and CHK2 kinases, respectively activated by ATR and ATM, that are central transducers towards cell cycle arrest, DNA repair and apoptotic pathways; and WEE1, which is a downstream effector of CHK1 and a key regulator of cell cycle progression.
1.1. Ataxia telangiectasia mutated (ATM)
ATM is a large 350 kDa serine/threonine kinase belonging to the phosphatidylinositol 3-kinase (PI3K)-related protein kinase (PIKK), a family of 6 highly conserved enzymes playing pivotal roles in controlling cell homeostasis, including DDR (for ATM, ATR and DNAPKcs), cell growth (for mTOR), mRNA decay (for SMG1) and transcriptional regulation (for TRRAP).10 ATM is present in all tissues, and plays a pivotal role in DDR. The kinase is recruited and activated by DSB, and initiates the DNA checkpoint response and promotes the repair of broken chromosomes by either NHEJ (non homologous end joining) or HR (homologous recombination).2,3 ATM is not an essential protein under normal cell functioning11 whereas it plays an essential role for cell survival after ionizing radiation (IR). Under normal conditions, the enzyme is kept inactive under the form of a homodimer.3 In the presence of DSB, sensed by the MNR protein complex, ATM homodimers undergo a rapid autophosphorylation on Ser1981 and split into active monomers.12 The signal transduction is provided through the activation of a plethora of enzymes,13 and notably the important transducer CHK2 (Fig. 1). In addition to checkpoint kinases activation, ATM is the principal kinase for the phosphorylation of the breast cancer associated gene 1 (BRCA1) and p53, respectively leading to DNA repair and cell fate decision.14 Of note, cytoplasmic ATM also plays a role in insulin signaling and glucose homeostasis, which should be taken into account when developing ATM inhibitors.15 ATM inhibition, even transitory,16 has been reported to efficiently enhance sensitivity of cancer cells to both IR and DNA damaging agents17 whereas it proves less harmful for normal cells. In light of these results, ATM inhibition appears as an attractive approach for anticancer chemo- and radiosensitization, as well as to overcome resistance phenomena.2,18 Therefore, ATM seems a relevant target for drug development.
1.2. Ataxia telangiectasia and Rad3-related (ATR)
ATR is another member of the PIKK family and one of the central kinases involved in the DDR. ATR plays an important role in the enforcement of cell cycle checkpoints, particularly at intra-S-phase, both during normal progression and in response to DNA damage. ATR is activated in response to persistent SSB. Specifically, ATR in complex with ATRIP is attracted by long stretches of RPA-coated single-stranded DNA. Subsequently, ATR get activated by two different ways: through the canonical Rad17 pathway or alternatively in a Nbs1-dependent manner (non-canonical pathway).19 Contrary to ATM, ATR is a necessary enzyme for cell survival, and its depletion leads to embryonic lethality in mouse and cell death in human cells.20–23 ATR is generally considered to preserve genome integrity by phosphorylating a number of enzymes involved in DNA synthesis at replication forks, resulting in nucleotide levels regulation, fork progression, cell-cycle progression and DNA repair mechanisms activation. Many of ATR functions are mediated through its downstream target CHK1.24 Like ATM, ATR inhibition affects both DNA checkpoint response and impairs global DSB repair, therefore enhancing the efficacy of IR and DNA-damaging drugs treatments. The principle of synthetic lethality can also be applied to ATR inhibition, as P53- or ATM-defective cells can only rely on ATR promoted cell cycle checkpoints to repair DNA. ATR inhibition leads selectively in such cells to an accumulation of DNA defects that result in mitotic catastrophe.25,26 This makes ATR an attractive target for the development of molecular inhibitors to circumvent resistance to radio- and chemotherapies.
1.3. Checkpoint kinase 1 (CHK1)
CHK1 is a 54.4 kDa serine/threonine-specific protein kinase that plays, with CHK2, a pivotal role in maintaining DNA integrity. CHK1 is a substrate of ATR and its activation is therefore particularly important in the response to SSB sensing. The existence of a close crosstalk between CHK1 and CHK2 (substrate of ATM) has been evidenced and a significant overlap in both their activation and their substrates has been reported.27 However, CHK1 seems to be an essential enzyme in contrast to CHK2, as CHK1-/- null mice embryos are not viable, whereas their CHK2-/- null pendant show normal development.28 CHK1 activation results in the initiation of cell cycle checkpoints, later phase S and G2/M cell cycle arrest, DNA repair and possibly cell death. More precisely, it mediates the degradation of cell-division cycle 25A (CDC25A) phosphatase via polyphosphorylation on its serine residues, which in turn slows down the progression of DNA replication through the S-phase and provides time for resolution of the source of stress.24 Moreover, CHK1 phosphorylates CDC25C, preventing cyclin-dependent kinase 1 (CDK1) activation, and stopping the cell cycle in the G2 phase. siRNA and knockdown experiments tend to show that CHK1 plays a more prominent role than CHK2. Because of the crosstalk between CHK1 and CHK2, the latter has been proposed to act in some cases rather as a backup kinase when CHK1 is non-functional.29 Indeed, simultaneous knockdown of CHK1 and CHK2 did not improve DNA-damaging treatments efficacy compared with CHK1-knockdown alone.30 However, the role of inhibiting simultaneously CHK1 and CHK2 over CHK1 alone is not yet clearly established. About 50% of human cancers display deficient p53 by mutation or inactivation.31 These cancer cells cannot activate G1/S checkpoint and only rely on S and G2/M checkpoints controlled by CHK1. Selectivity towards these p53-deficient cancer cells can therefore be afforded by combination of CHK1 inhibition and DNA-damaging treatment, following the principle of synthetic lethality.32
1.4. Checkpoint kinase 2 (CHK2)
CHK2 is a 60.9 kDa serine/threonine kinase whose active site is structurally similar to those of CHK1. CHK2 is the second effector of the checkpoint response activation and it also plays a pivotal role in eliciting DNA repair, cell cycle arrest or apoptosis in the response to DNA damage. Mechanistically, it becomes activated by ATM through phosphorylation at residue Thr68 which induces CHK2 dimerization and autophosphorylation of the kinase domain.33 Subsequently, CHK2 promotes the cell cycle arrest by phosphorylation of downstream targets including CDC25 phosphatases, responsible for dephosphorylation and activation of the CDK. As a downstream kinase of ATM, CHK2 is particularly important in the response to DSB. CHK2 also stimulates the repair of DSB through BRCA1 mediated processes. Moreover, CHK2 activates the transcription factor p53, which results in the cell cycle arrest through p21 or leads to apoptosis or senescence depending on damage severity.27 Of note, the activation of p21 can also play a role in DNA repair by interacting with proliferating cell nuclear antigen (PCNA).34 As a downstream kinase of ATM, CHK2 is a logical target for the development of radio- or chemosensitizers. Therefore, several small-sized inhibitors35 have been proposed to precisely decipher its role in cell cycle arrest and tumourigenesis, and ultimately to validate its relevance as a target in oncology. Nevertheless, the advantage of using a dual CHK1/CHK2 inhibition over a CHK2 selective inhibition remains to be evaluated; thus, specific CHK2 inhibitors are under development.
1.5. WEE1
WEE1 is a 96 kDa dual-specific kinase which plays an important role in cell cycle progression, by phosphorylating CDK1 at tyrosine 15, a key modification that blocks G2–M transition.36 WEE1 is activated by several enzymes including CHK1, chaperone protein Hsp90 and PP2A phosphatase in response to DNA damage accumulation.37 WEE1 lengthens the G2 phase by controlling the activity of CDK1, thus allowing the DDR machinery additional time for DNA repair. During S and G2 phases, CDK1 is maintained inactivated by WEE1 phosphorylation at two different sites, Tyr15 and Thr14.38 When DNA damage is repaired, WEE1 activity decreases while the inhibitory tyrosine 15 modification of CDK1 is removed by Cdc25 phosphatase. This leads to CDK1 re-activation and promotes entry into mitosis. Therefore, WEE1 can be seen as the gatekeeper of G2-arrest and acts as an inhibitor of mitosis.39 This enzyme is essential for the development of mammals as WEE1 depletion leads to growth defects and cell death resulting from DNA damage. Indeed, WEE1-/- knockout (KO) mice are unable to survive after the 4th day of embryonic stage. WEE1 is overexpressed in many cancer lines, including breast cancer (35% of those),40 glioma,41,42 glioblastoma,37 nasopharyngeal carcinoma (NPC),41 as well as drug-resistant cancer cells.43 Moreover, WEE1-/- KO reduces the viability of breast cancer cells, but not normal mammary epithelial cells.44 This drew biochemists' attention on the inhibition of this enzyme to push cancer cells into mitotic catastrophe. Preclinical studies with cancer cells and animal models showed diminished cancer cell viability, reduced tumour growth and burden, and improved survival after WEE1 inhibition by siRNA or small-sized organic inhibitors. Moreover, their combination with conventional DNA damaging methods enhances these anti-cancer activities, thus sensitizing cancer cells to conventional therapy. As WEE1 is considered as one of the main gatekeepers of the G2 cell-cycle checkpoint, its inhibition is particularly effective in cells inherently presenting G1–S transition defects, typically those with deficient p53 signalling. Indeed, as p53 deficient cells present weakened G1/S DDR, Wee1 inhibition, that targets the G2/M DDR will act as combination therapy. It is noteworthy that the combined inhibition of WEE1 and CHK1/2 induces a synergistic decrease of cell viability and an increase of apoptosis in mouse xenograft models using melanoma cells from a patient.45 Thus WEE1 inhibition has been considered as an effective sensitizer in combination with DNA-damaging therapy46 and is a potential therapeutic target for radiosensitization of adult glioma37 and various types of cancers.47–49
In this context, the five pivotal kinases introduced above appear as particularly relevant targets to develop therapeutic agents able to modulate the DNA checkpoint response in cancer cells.
2. Chemical inhibitors of ATM, ATR, CHK1, CHK2 and WEE1
2.1. ATM inhibitors
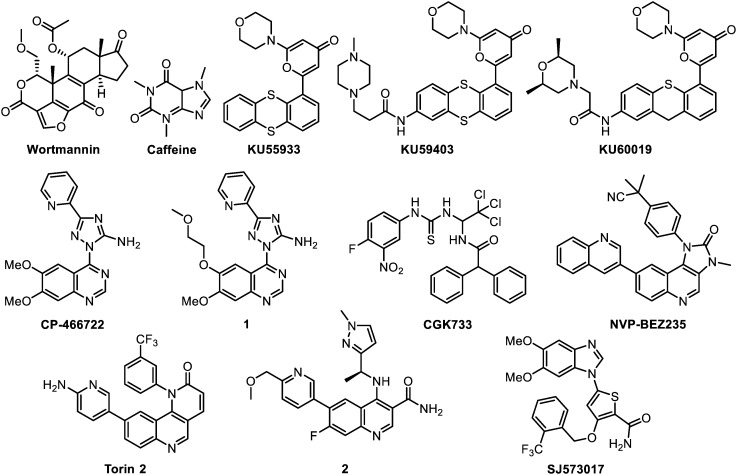
In parallel to ATM drug target validation, the development of ATM inhibitors has arisen these last 15 years, passing from early non-specific compounds to highly selective inhibitors that entered in pre-clinical studies (Fig. 2). Their main properties are listed in Table 1.
Fig. 2. ATM inhibitors.
Table 1. ATM inhibitors and their main properties.
| Compound | Enzymatic activity | In vitro selectivity | In vitro efficacy | Pharmacology | In vivo activity | Development stage |
| Wortmannin | IC50 = 150 nM (irreversible) | – | R+/– | – (toxicity) | N.D. | Discontinued at optimisation stage |
| Caffeine | IC50 = 0.2 mM | – | R+/– | – (toxicity) | N.D. | Discontinued at optimisation stage |
| KU55933 | IC50 = 12.9 nM | + | R+/– C++ | – (solubility, distribution) | N.D. | Discontinued at research stage |
| K i = 2.2 nM | ||||||
| KU59403 | IC50 = 3 nM | ++ | C+ | + | + | Preclinical stage |
| KU60019 | IC50 = 6.3 nM | ++ | R+ C+ | + | + | Preclinical stage |
| CP-466722 | IC50 = 20 nM | + | M+ R+ | – (stability) | N.D. | Research stage |
| 1 | N.D. | ++ | R+ | + | N.D. | Research stage |
| CGK733 | N.D. | – | M+/– | N.D. | N.D. | Optimisation stage |
| NVP-BEZ235 | N.D. | – | M+ R++ C+ | +/– | + | Clinical trials a phase II |
| Torin-2 | IC50 = 28 nM | – | M– R+ C++ | + | + | Preclinical stage |
| 2 | IC50 = 0.6 nM | ++ | C++ | + | ++ | Preclinical stage |
| SJ573017 | IC50 = 0.48 μM | – | M++ R+ C+ | N.D. | N.D. | Optimisation stage |
aAll the clinical data presented in this review were retrieved on https://clinicaltrials.gov website.
Wortmannin
Historically, several methylxanthine-derived drugs (theophylline, pentoxifyllin and caffeine) were reported to sensitize cells to radiations at low millimolar concentrations. However, the fungal metabolite wortmannin was the first compound proposed to target ATM.50 Its radiosensitizing properties are attributed to an irreversible inhibition of several members of the PIKK family, including mTOR, DNA-PKcs, ATM (IC50 on isolated immune complexes ranging between 16 nM and 150 nM), as well as ATR to a lesser extent (IC50 = 1.8 μM). However, high dosages (≥10 μM) are necessary to obtain radiosensitizating effects in A549 lung cancer cells. This is two orders of magnitude higher than the dose required for PIKK inhibition. Therefore, the contribution of PIKK inhibition in the radiosensitizing activity remains unknown. Moreover, the intrinsic systemic toxicity relative to covalent inhibitors (LD50 (mice) = 1 mg kg–1)51 and the adverse side effects associated with the administration of wortmannin limited its utility. Of note, the use of wortmannin was revisited in 2012 with a nanoparticle drug delivery approach which helped to significantly reduce its toxicity.52
Caffeine
Caffeine was largely studied and used as radiosensitizing agent although its exact mechanism of action remains unclear. Its structure suggests that it might act at high dosage as a broad-spectrum kinase inhibitor; with probable inhibition of the phosphotransferase activity of a protein kinase involved in checkpoint signalling of DNA damaged cells. Inhibition of ATM and ATR activities were later reported at doses similar to radiosensitization (around 1 mM).53 Nevertheless, the required dose to inhibit ATM activity (IC50 of 0.2 mM) is close to its LD50in vivo and would be far too toxic to permit any use in animals.
KU55933
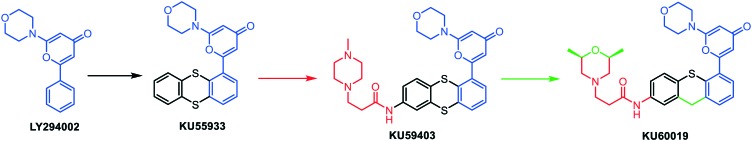
Since caffeine and wortmannin are neither specific nor useful in vivo, new ATM inhibitors have emerged. Since 1999, Smith's group is developing competitive specific inhibitors of DNA-PKcs and ATM kinases. From an initial screening, the 2-morpholino-8-phenyl-4H-chromen-4-one (LY294002) was identified as a hit, and this structure was optimized over the years to yield several lead compounds (Fig. 3). First, KU55933 was obtained by substituting the chromen-4-one core by a pyran-4-one and replacing the phenyl by a thianthrene moiety. KU55933 was the first ATM specific ATP-competitive inhibitor (IC50 = 13 nM) displaying selectivity over ATR, PI3K, DNA-PKcs, PI4K and m-TOR (all with IC50 > 2.5 μM).17,54 At 1 μM, this compound showed slight IR sensitizing effects in HeLa cervical cancer, LoVo and SW620 colorectal cancer cell lines, while significant chemosensitizing effects were observed in combination with the marketed topoisomerase II inhibitor etoposide, at 10 μM. However, the pharmacokinetic parameters proved unfavourable, particularly the water solubility and the tissue distribution.
Fig. 3. Hit-to-lead optimization from LY294002 to KU60019.
KU59403
KU59403 is an upgraded version of KU55933 in which the introduction of a 3-(4-methylpiperazin-1-yl)propanamide group led to an improvement of the pharmacokinetic parameters together with a gain of activity (IC50 = 3 nM) and selectivity (over 3 orders of magnitude for all other PIKK members tested). This allowed its use in animals (mice). KU59403 was the first ATM specific inhibitor to show significant chemosensitization of campthotecin and irinotecan in mice xenografted with human colon cancer cells (HCT116 and SW620).18
KU60019
KU60019 is a variant of KU59403 which encompasses a dimethylmorpholine and a thioxanthene group in replacement of the N-methylpiperazine and thianthrene moieties of the parent compound.14,55 Compared with KU59403, it possesses very similar binding properties (IC50 = 6.3 nM), cellular activities (enhancement ratios of 2.1–2.9 at 0.6 μM) and pharmacokinetics with good drug exposition by intracranial injection. Moreover, KU60019 proved highly selective with little to no off-target effects against a panel of 229 kinases at 1 μM, and limited toxicity towards healthy cells. Overall, the main particularity of this therapeutic agent is certainly its dual chemo- and IR-sensitization action, with submicromolar radiosensitization activity observed in glioma cells.56In vivo, KU60019 exhibited selective anti-tumour properties against migration, invasion and cell growth, in xenografts p53-mutant glioma rodent models. These are probably due to the inhibition of prosurvival pathways. Interestingly, the combination of IR and KU60019 promotes preferential killing of stem-like cells and specific sensitization to agents promoting DSB.57 Efficacy and safety of KU60019 treatments were validated in a detailed in vivo study that showed 2–3 fold survival increase in mice xenografted with orthotopic COMI glioblastoma cells, under IR.58
CP466722
CP466722 was identified in 2008 as a new ATM inhibitor by screening a library of 1500 targeted kinase inhibitors. It proved selective over ATR, PI3K, DNA-PKcs and Abl kinase in hTERT-immortalized human fibroblasts.16CP466722 leads to a rapid reversible inhibition of ATM function in cultured tissue, and presents antiproliferative effects (IC50 = 370 nM) as well as IR sensitizing properties similar to KU55933. However, in vivo studies could not be undertaken because of its rapid metabolic degradation (t1/2 = 0.13 h in mice).
Methoxyquinazoline 1
In 2016, Min et al. up-graded the scaffold of CP466722 by switching the C6-methoxy group by a methoxyethoxy one.59 The resulting optimized molecule (1), exhibits a better metabolic stability than CP466722 (t1/2(microsomes) > 4 h vs. 0.47 h; t1/2 (in vivo) = 19.8 h vs. 0.13 h), without significantly affecting its biological activity. Overall, this optimized compound possesses a good pharmacological profile to be evaluated in vivo as a radiosensitizer against non-small cell lung cancers (NSCLC).
CGK733
CGK733, a thiourea-containing compound was identified in 2008 as a dual ATM/ATR inhibitor60 and tested as a cytotoxic drug in monotherapy (IC50 = 5–20 μM). However, this compound seems to have rather been used as a tool for studying the cellular functions of ATM/ATR than to prepare a clinical candidate. Indeed, neither binding assays nor activity assays on isolated kinase were reported. Therefore, the biological effects of CGK733 might be due to cellular off-target activities, which remain elusive to date. Moreover, its pharmacological properties have not been reported yet.61 Lastly, CGK733 was reported not to inhibit ATM or ATR kinase in H460 human lung cancer cells.62
NVP-BEZ235
In 2011, a nanomolar non selective PI3K/mTOR/ATM/DNA-PKcs/Akt inhibitor was disclosed. Interestingly, this compound exhibits a good bioavailability, despite its apparent compact and lipophilic structure, and an ability to cross the blood brain barrier. Detailed experimentation revealed that the molecule blocks NHEJ and HR pathways, hence causing profound radiosensitization even at low doses (100 nM vs. 10 μM for KU55933). However, as described above, NVP-BEZ235 is a multi-target compound, and the particular contribution of ATM inhibition in this activity has not been studied. Otherwise, this compound displays a strong synergy when administered in combination with IR or temozolomide (alkylating agent) in most of the treated gliomas, and afforded marked tumour volume reduction. As a consequence, NVP-BEZ235 entered phase I/II clinical trials for the treatment of solid tumours and remains to date the only ATM inhibitor being clinically evaluated.63–66
Torin2
Torin2 is a subnanomolar ATP-competitive inhibitor of mTOR (IC50 = 0.25 nM), also affecting ATM, ATR and DNA-PKcs with nanomolar activities (IC50 = 28, 35, 118 nM, respectively). Designed by lead optimization from the mTOR inhibitor Torin1 (which has no activity on ATM), it shares some structural analogy with NVP-BEZ235. Torin2 exhibits nanomolar radiosensitization activities in cells (GI50 = 10–220 nM), encouraging pharmacokinetics and significant tumour growth inhibition in combination with the MEK inhibitor AZD6244, blocking the MAPK signalling cascade involved in cell proliferation and senescence. Of note, no tumour growth reductions were observed when these two compounds were administered alone.67,68
Fluoroquinoline 2
Very recently, a new 3-quinoline carboxamide derivative structurally related to Torin2 has been described. This compound displays high cellular potency against ATM (IC50 = 33 nM) and excellent selectivity over ATR (IC50 > 19 μM) and other kinases. A good oral exposure was observed in rodents. Moreover, in combination with irinotecan, significant tumour volume reductions were observed in a SW620 colorectal cancer xenograft model.69
SJ573017
A new cell-based high-throughput screening (HTS) assay for ATM inhibitors was recently developed leading to the discovery of two hits. Among them, the most promising therapeutic agent is SJ573017. This compound possesses a submicromolar activity towards ATM (IC50 = 0.48 μM) but proved non-selective, particularly against PLK enzymes. Its growth inhibition activity (GI50 = 3–32 nM) higher than its IC50 on ATM is consistent with off-target effects.70,71
Overall, few selective ATM inhibitors have been described yet. The most interesting compounds are probably KU59403 and KU60019, developed by Kudos Pharmaceuticals, and 2 developed by AstraZeneca, for which the preclinical studies have been validated. Thus, KU60019 showed anticancer properties as a single agent whereas KU59403 and 2 could be used as chemosensitizers. On the other hand, the improved version of CP466722 (compound 1) might also be interesting after in vivo validation. Lastly, the most advanced compound in the drug discovery process is currently the non-selective PI3K/mTOR/DNA-PKcs inhibitor NVP-BEZ235, currently in phase II clinical trials.
2.2. ATR inhibitors
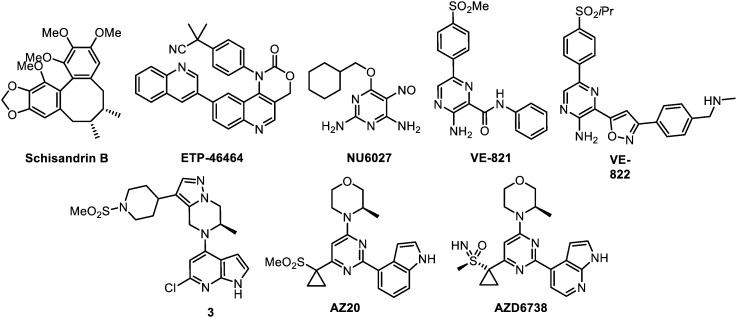
Following the first use of non-selective PIKK inhibitors (like caffeine and wortmannin) as radiosensitizers, the availability of selective ATR competitive inhibitors was desired as tools to study its exact role in the DDR. This was accompanied with the hope to promote new therapeutically usable drugs targeting this kinase. The compounds that are reviewed herein as ATR inhibitors are represented below (Fig. 4) and their main properties are subsequently listed in Table 2.
Fig. 4. ATR inhibitors.
Table 2. ATR inhibitors and their main properties.
| Compound | Enzymatic activity | In vitro selectivity | In vitro efficacy | Pharmacology | In vivo activity | Development stage |
| Schisandrin B | IC50 = 7.25 μM | – | R+/– C+/– | N.D. | N.D. | Optimisation stage |
| ETP-46464 | N.D. | + | R+ | — | N.D. | Optimisation stage |
| NU6027 | N.D. | N.D. | R+/– C+ | – (solubility) | N.D. | Optimisation stage |
| VE-821 | K i = 13 nM | + | R+ C+ | – (toxicity) | N.D. | Discontinued at research stage |
| VE-822 | K i < 0.2 nM | + | M– R+ C++ | + | + | Clinical trials phase I |
| 3 | IC50 = 0.4 nM | ++ | N.D. | +/– (toxicity) | N.D. | Research stage |
| AZ20 | IC50 = 5 nM | +/– | M++ | +/– (toxicity, solubility) | ++ | Clinical trials phase I |
| AZD6738 | IC50 = 1 nM | ++ | M+ C++ | + | + | Preclinical stage |
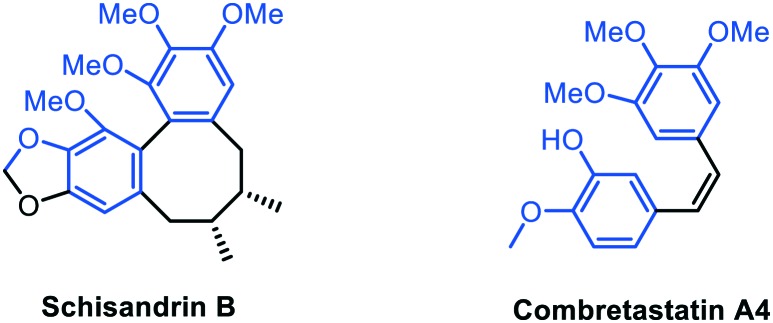
The first molecule proposed for ATR inhibition is the natural product schisandrin B, isolated from Fructus schisandrae, and discovered by activity screening of herbal extracts. This tricyclic molecule shares structural analogy with combretastatin A4, a cytotoxic agent targeting the colchicine binding site of tubuline (Fig. 5). Schisandrin B displays moderate activity (IC50 = 7.25 μM) on purified ATR with some selectivity over ATM (IC50 of 1.74 mM), CHK1, PI3K, DNA-PKcs, and mTOR. However, it inhibits both ATM and ATR in cells, and therefore cannot be considered as a selective inhibitor. After UV exposure, schisandrin B exhibits ATR-dependent cytotoxic effects on cells at the dose of 30 μM. Moreover, owing to its similarity with combretastatin A4, it unsurprisingly acts as an anti-mitotic agent.72 Of note, no pharmacological data have been provided for this compound, yet.
Fig. 5. Structure similarities between Schisandrin B and Combretastatin A4.
ETP46464
ETP46464 was identified in a cell-based screening assay from a PI3K/mTOR inhibitor library, and is structurally related to NVP-BEZ235. ETP46464 efficiently inhibits ATR in cellular assays with an IC50 of 25 nM, and is selective over ATM and DNA-PKcs. However, the poor pharmacological properties reported in mice prevented its use for in vivo experimentation.63
NU6027
NU6027 was originally developed as a CDK2 inhibitor but showed low micromolar (IC50 = 6.7 μM) inhibition of cellular ATR activity. It impairs both G2/M arrest and HR thus increasing sensitivity to DNA-damaging agents and PARP inhibitors (Poly ADP ribose polymerase, a family of proteins mainly involved in DNA repair and apoptosis). CDK2 inhibition was also disclosed, but chemosensitization was confirmed to be due to ATR inhibition. However, the low water solubility of this compound impedes its in vivo evaluation.73
VE-821
The first ATR selective inhibitor, VE-821, was reported by Vertex Pharmaceuticals in 2011. VE-821 stems from the structural optimization of a molecular hit identified by HTS. It shows low nanomolar inhibition values and good selectivity against a panel of kinases, particularly against PIKKs (ATM, DNA-PKcs, mTOR) and related PI3K; all of them having micromolar IC50.74,75 VE-821 provided the proof of concept, supported by detailed studies on ovarian (SKOV3, OVCAR-8 cells),76 pancreatic (PSN-1, MiaPaCa-2, PANC-1 cells)77 and hypoxic tumour cancer cells (RKO cell line),78 that ATR inhibition can improve the efficiency and the therapeutic index of IR but also of a set of marketed anticancer drugs with different modes of action (e.g. gemcitabine, temozolomide, cisplatin, topotecan or veliparib). It is worth noting that synthetic lethality was observed with this compound in ATM-, BRCA2-, XRCC3- and XRCC1-defective cells.79 However, signs of cellular toxicity at 3 μM prompted the medicinal chemists to propose an optimized structure: VE-822.
VE-822
VE-822 (or VX-970) shares a high structural similarity with its parent compound, VE-821. VE-822 displays a subnanomolar potency in vitro (Ki < 0.2 nM; 65-fold improvement) and a low nanomolar activity in cells (Ki = 19 nM). In vitro, it remains ATR-selective over ATM (>100-fold) and DNA-PKcs. Moreover, this molecule profoundly sensitizes tumours to chemotherapeutics, notably gemcitabine and cisplatin, at low concentration (80 nM vs. 1000 nM for VE-821), leading to 2–3 fold reduced cancer cell survival. It is noteworthy that VE-822 has no effect when used as a single agent. The pharmacological data proved very promising; moreover, in vivo this compound significantly inhibits tumour growth in pancreatic MiaPaCa-2 mice xenografts without signs of toxicity.80 Of note, in 2012, VE-822 became the first ATR inhibitor to enter clinical trials.81
Chloropyrrolopyridine 3
Novartis proposed a series of tetrahydropyrazolo[1,5-a]pyrazines as selective ATR inhibitors issued from HTS. A hit-to-lead strategy furnished the chloropyrrolopyridine 3. It exhibits excellent in vitro activity (IC50 = 0.4 nM) and selectivity (>104 over PIKK, hERG and PI3K). A very high activity was retained in cells (IC50 = 37 nM) and a good pharmacological profile to probe ATR biology in vivo was obtained. However, potential safety concerns were reported because of in vitro inhibition of cytochrome CYP3A4, a major monooxygenase involved in drug metabolism. Compound 3 was proposed for its use as a chemosensitizer rather than as a monotherapy.82
AZ20
AZ20 was discovered by AstraZeneca by a hit-to-lead strategy from a mTOR inhibitors library screening. It is a low nanomolar ATR inhibitor (IC50 = 5 nM), with remaining activity for mTOR (IC50 = 38 nM), but selective against all PI3K isoforms (IC50 > 10 μM). Interestingly, a better ATR selectivity was observed in cells. (IC50 = 50 nM for ATR and 2.4 μM for mTOR). Despite its low aqueous solubility, AZ20 induced, as single agent, a significant tumour volume reduction (3 fold) in mice xenografted with LoVo colorectal cells. However, a time-dependent inhibition of cytochrome 3A was also reported, which led to elevated exposure in mice even at moderate doses.83 Efforts to improve AZ20 aqueous solubility resulted in decreased bioavailability.84
AZD6738
Finally, AZ20 underwent further structural optimizations and eventually led to the discovery of AZD6738. This latter molecule is a very potent ATR inhibitor in vitro (IC50 = 1 nM) and in cellulo (IC50 = 74 nM, in H460 NSCLC). Moreover, AZD6738 neither affects mTOR (IC50 > 23 μM), nor other PI3Ks (IC50 > 30 μM). Used at 1 μM concentration in combination with cisplatin, it leads to an important 20-fold decrease of cancer cells viability. This molecule exhibits an optimized pharmacokinetic profile, and is therefore orally bioavailable and active as a chemosensitizer. AZD6738 induces cell death and senescence in H23 NSCLC xenografted mice and 85% tumour regression in combination with cisplatin. No apparent toxicity on mice was observed over the 2 week treatment.11 Interestingly, AZD6738 was also reported to be synthetically lethal with p53 and ATM defects.25,26 Overall, this compound seems to be a valid drug candidate and is currently assessed in phase I clinical trials.
Among the molecules proposed for ATR inhibition, AZD6738 and VE-822 appear to be the only ones suitable for in vivo use, both being actually evaluated in phase I clinical trials. Compound 3 and AZ20 could have been good drug candidates but safety concerns about cytochromes inhibition hinder their use in vivo. ETP46464 could be used to probe ATR inhibition in cells, but its poor pharmacokinetic profile prevents its transfer to animals.
2.3. CHK1 inhibitors
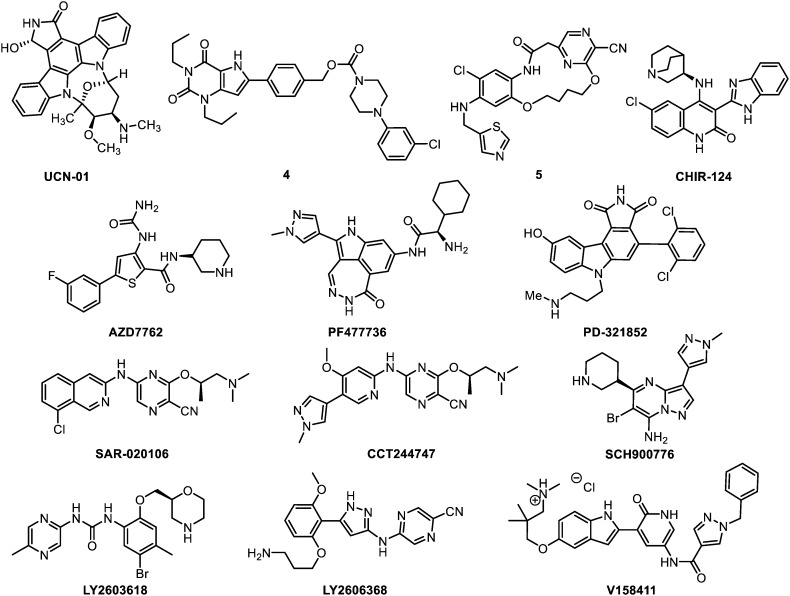
The compounds that are reviewed herein as CHK1 inhibitors are represented below (Fig. 6) and their main properties are subsequently listed in Table 3.
Fig. 6. CHK1 inhibitors.
Table 3. CHK1 inhibitors and their main properties.
| Compound | Enzymatic activity | In vitro selectivity | In vitro efficacy | Pharmacology | In vivo activity | Development stage |
| UCN-01 | K i = 5.6 nM | – | M+/– R+/– C+/– | – (distribution, toxicity) | +/– | Discontinued at clinical trials phase II |
| XL-844 | K i = 2.2 nM | – | M+/– R+/– C+ | + | + | Discontinued at clinical trials phase I |
| 4 | IC50 = 0.75 nM | +/– | M+/– C+ | + | N.D. | Optimisation stage |
| 5 | IC50 = 4 nM | ++ | M– C++ | +/– (exposition) | N.D. | Research stage |
| K i = 4 nM | ||||||
| CHIR-124 | IC50 = 0.3 nM | +/– | M– R+/– C+ | + | +/– | Preclinical stage |
| K i = 0.3 nM | ||||||
| AZD7762 | IC50 = 5 nM | +/– | M+ C++ | + | + | Discontinued at clinical trials phase I |
| K i = 3.6 nM | ||||||
| PF477736 | K i = 0.49 nM | + | M– C+/– | +/– (distribution, metabolism, elimination) | +/– | Discontinued at clinical trials phase I |
| PD-321852 | IC50 = 5 nM | +/– | C++ | N.D. | N.D. | Research stage |
| SAR-020106 | IC50 = 13.3 nM | + | M– C++ | +/– (distribution, metabolism) | +/– | Research stage |
| K i = 10.9 nM | ||||||
| CCT244747 | IC50 = 7.7 nM | ++ | M+ C+ | + | + | Research stage |
| SCH900776 | IC50 = 3 nM | + | R+ C++ | + | + | Clinical trials phase II |
| LY2603618 | IC50 = 7 nM | + | M+ C++ | +/– | + | Clinical trials phase II |
| LY2606368 | K i = 0.9 nM | +/– | M++ C++ | + | + | Clinical trials phase I |
| V158411 | IC50 = 3.5 nM | – | M+ C+ | N.D. | N.D. | Optimisation stage |
UCN-01
UCN-01 (7-hydroxystorausporine) is a natural product belonging to the staurosporine family. This is a non-selective multipathway inhibitor targeting CHK1 together with the PKC subfamily (a group of enzymes that play important roles in several signal transduction cascades), CDK1,85,86 CDK2.87 It also affects the p53/p21waf1 and CHK2/CDC25 pathways. UCN-01 shows a nanomolar potency against CHK1 (Ki = 5.6 nM). The lack of selectivity of this inhibitor can by partly explained by the very high molecular rigidity of the staurosporine core. Indeed, it tightly binds the ATPase binding sites of a set of several kinases, as suggested by a co-crystallization study with CHK1.88UCN-01 induces the cell growth inhibition of colon and hepatocellular carcinoma (Huh7, HepG2, Hep3B and HT29 cells), both in vitro and in vivo.89 Remarkably, it also has this impact on glioblastoma stem cells (GSC) when a concomitant PDK1 inhibition is applied.90UCN-01 has been evaluated in many different phase I and phase II clinical trials between 1995 and 2015, either as single a anti-neoplastic agent or in combination settings. Nevertheless, its use has always been hindered by poor pharmacokinetics, high binding to serum proteins and unfavourable toxicity profile.91,92 UCN-01 underwent structural optimizations to overcome its binding to serum proteins and nonspecific kinase inhibition. Eventually, the analogue Gö6976 was proposed.93 However, this simplified staurosporine proved 22-fold less active than UCN-01 and poorly selective. In fact, Gö6976 mainly inhibits the PKC kinases, together with numerous kinases, including CHK1 and possibly CHK2. It is noteworthy that other staurosporines, like SB218078,94 were identified although UCN-01 remains the spearhead of this family.
XL-844
XL-844 (EXEL-9844) is a dual CHK1/CHK2 inhibitor whose structure remains undisclosed. It is a potent CHK1 inhibitor (Ki = 2.2 nM) but it exhibits a moderate selective over other kinases (Ki = 0.07 nM, CHK2). Few pharmacological details have been disclosed, nevertheless it proved orally available, and was shown to promote CHK1 and CHK2 specific radio- and chemosensitization at around 1 μM dosages.95In vivo, it led to 74% tumour growth inhibition in combination with gemcitabine on HT-29 xenografted mice. It entered phase I clinical trials for leukaemia in 2005, but the study has been closed due to slow enrolment. Its combination with gemcitabine has been assessed against lymphoma, although no results are available.96
Pyrimidinedione 4
Abbott laboratories developed a series of cyanopyridyl containing 1,4-dihydroindeno[1,2-c]pyrazoles as CHK1 inhibitors. The lead compound of the series (4), shows a subnanomolar activity (IC50 = 0.75 nM, CHK1), and is relatively selective against other Ser/Thr kinases. In HeLa cervical and H1299 lung cancer cells, this compound exhibits a weak anti-proliferative activity when used as a single agent (IC50 = 21 μM) and proved moderately active in combination with doxorubicin (EC50 = 0.54 μM). Efforts were devoted to improve its poor pharmacokinetics,97 although 4 displayed a sufficient profile to be tested in vivo. No data in animals have been reported yet, but this compound might be used as a tool for in vivo evaluation of DNA-damaging agents in sensitized cells.98
Macrocycle 5
Macrocyclic ureas were proposed for the selective inhibition of CHK1 by structure-based design.99 The lead compound (5) displays an IC50 of 4 nM against CHK1 and was highly selective among other kinases. It is noteworthy that it does not affect CHK2 (IC50 = 5.5 μM). As a single agent, a weak activity was observed in HeLa and H1299 cancer cell lines (EC50 = 59.3 μM, HeLa), whereas its combination with doxorubicine led to a moderate cytotoxicity (EC50 = 1 μM). Preliminary pharmacokinetic analyses showed a moderate plasma exposition. It seems that the development of this series has been discontinued as no further data were subsequently disclosed.
CHIR-124
CHIR-124 is a potent and relatively selective quinolone-based molecule. It displays an IC50 of 0.3 nM against CHK1 and is selective over CHK2 (IC50 = 0.7 μM) and over a panel of 124 representative kinases (all IC50 > 100 nM). Nevertheless, it might target FLT3 (fms related tyrosine kinase 3, regulating hematopoiesis), PDGFR (platelet-derived growth factor receptor, regulating cell proliferation, cellular differentiation, cell growth and development) and GSK3 (glycogen synthase kinase 3, regulating cell proliferation, migration, glucose levels and apoptosis) as they display IC50 values of 5.8, 6.6 and 23.3 nM, respectively. No activity was observed as a monotherapy but radio- and chemosensitizing effects were reported at submicromolar concentrations in MDA-MD-435 melanoma cell lines. CHIR-124 is orally bioavailable and potentiates topoisomerase I poisons (camptothecin, SN38, irinotecan) in mice xenografted with melanoma MDA-MB-435 cells (moderate tumour growth inhibition of 14–53%). Indeed, the immobilisation of topoisomerase I provokes collision with the replication fork and leads to DSB. Of note, no signs of toxicity or significant body weight loss were observed in animals.100,101
AZD7762
AstraZeneca dedicated a full medicinal chemistry program to develop the potent and selective CHK1 inhibitor AZD7762 (IC50 = 5 nM, CHK1). Starting from a HTS screening and after multiple rounds of structure–activity relationship-driven structural optimizations and ADME profiling, AZD7762 was discovered. This molecule presents chemo- and radiosensitizing effects in the 150–300 nM range and an EC50 value of 0.62 μM when used alone (SW620, MDA-MB-231 and HCT116 cancer cell lines). Good pharmacokinetics and 2–3-fold tumour growth inhibition were measured although a slight body weight loss (7%) was reported in mice (H460-DNp53 and SW620 xenografts). AZD7762 entered phase I clinical trials, in combination with gemcitabine, a nucleoside analogue widely used as a chemotherapy agent. These studies were terminated after a full review of program data and assessment of the current risk–benefit profile. Nevertheless, this compound remains a valuable tool for the study of DNA checkpoint biochemistry.29,102
PF477736
In 2011, Pfizer developed the subnanomolar CHK1 inhibitor PF477736 (Ki = 0.49 nM). It is selective over CHK2 (Ki = 47 nM) and retains a very potent activity in HeLa and HT29 cancer cell lines (EC50 = 38–42 nM). PF477736 displays a moderate chemosensitizing effects at 540 nM, in HT29 cell lines (31% with gemcitabine; 22% with camptothecin). It possesses a good toxicity profile but the pharmacokinetics proved ameliorable with a moderate distribution, a short half-life (2.9 h) and a low clearance. PF477736 has no effect when administered alone, whereas it boosts up to +89% the anti-tumour activity of gemcitabine in mice xenografted with Colo205 human colon carcinoma cells. In the latter case, no enhancement of systemic toxicity of gemcitabine was observed.103PF477736 entered phase I clinical trials, but these were discontinued for unknown reasons, which were not safety or efficacy related.
PD-321852
PD-321852 is a potent staurosporine analogue displaying an IC50 of 5 nM against CHK1 and a reasonable selectivity. It promotes a significant sensitization (ratios: 6.2–17) to gemcitabine in pancreatic cancer cells at 300 nM (BxPC3, M-Panc96 and MiaPaCa-2 cell lines). It was presented as a CHK1 antagonist but other kinases inhibition may contribute to the observed chemosensitization.104 ADME and toxicity profiling have not been reported yet and may be established before in vivo use. An analogue of this compound, PD-407824, has been disclosed to inhibit CHK1 but prevalently affects WEE1; therefore it will be discussed in the dedicated section (vide infra).
SAR-020106
SAR-020106 is a nanomolar ATP-competitive and selective CHK1 inhibitor (IC50 = 13.3 nM against CHK1 and up to 10 μM against CHK2 and CDK1). It enhances irinotecan and gemcitabine anti-tumour activity in several colon cancer cell lines (HT29, SW620, and Colo205; enhancement ratios: 3–29). The pharmacokinetics proved moderate because of high binding to plasma proteins, short half-life and high clearance. Moreover, a moderate tumour growth delay was observed in mice xenografted with SW620 cells.105 Therefore, SAR-020106 underwent an additional round of structural optimization and eventually yielded CCT244747.
CCT244747
CCT244747 is as potent as SAR-020106 against CHK1 (IC50 = 7.7 nM) although with a better kinase selectivity, particularly against CHK2 (IC50 > 10 μM). It displays a specific CHK1-dependent mechanism-of-action in cells at 300 nM, and good microsomal stability. Most importantly, it is the first CHK1-selective inhibitor that is orally bioavailable. However, the in vivo behaviour of CCT244747 is not uniform. Varying levels of oral exposure and clearance are observed. Alike SAR-020106, it modulates the DDR pathways and shows anti-tumour activity in combination with gemcitabine and irinotecan in HT29 and SW620 colon cancers, and in Calu6 NSCLC. It is noteworthy that CCT244747 also acts as a single agent.106,107
SCH900776
SCH900776 (or MK8776) is a potent and selective ATP-competitive CHK1 inhibitor (IC50 = 3 nM). It is effective at overcoming both the S- and G2-checkpoints caused by IR and various DNA-damaging agents. In contrast to AZD7762 and PF477736, this compound displays a 500-fold selectivity over CHK2. However, SCH900776 exhibits a lower selectivity over CDK2 (IC50 = 160 nM), which might be detrimental for its overall effectiveness since the inhibition of this cyclin-dependent kinase can induce cell cycle arrest and prevent checkpoint bypass. Nevertheless, SCH900776 shows a significant tumour growth delay in AsPC-1 and MiaPaCa-2 pancreas cancer cell lines xenografts. In 2013, it entered phase II clinical trials alone or in combination with gemcitabine in subjects with relapsed acute myeloid leukaemia.108–110
LY2603618
LY2603618 potently inhibits CHK1 protein kinase activity in vitro (IC50 = 7 nM). It is the first highly selective CHK1 inhibitor, including over CHK2 (>103 fold), to enter clinical cancer trials. This compound is also selective over a panel of representative kinases (all with IC50 > 1 μM, most of them with IC50 > 20 μM). HeLa cells treated with LY2603618 produce a cellular phenotype similar to the one reported for RNAi induced CHK1 depletions. In HCT116 and HT29 colon cell lines, the compound displays anti-proliferative effects (EC50 = 430 nM) and significant chemosensitizing effects when used in combination with gemcitabine at 0.25 μM. It proved orally available with an acceptable pharmacological profile. Moreover, it does not increase the gemcitabine systemic toxicity, and xenograft experiments with Calu-6 lung cancer cells showed a significant reduction of CHK1 phosphorylation in the tumours. It was suggested that the anticancer property of LY2603618 might be enhanced when used in combination with an autophagy inhibitor (chloroquine).111 Phase I clinical trials in patients with advanced or metastatic cancers indicated an acceptable safety profile for LY2603618.112,113
LY2606368
LY2606368, another related compound, has a dual action of inducing DNA-damage by causing DSB,114 and removing the protection of DNA-checkpoint by inhibiting CHK1 with a Ki of 0.9 nM. Ultimately, this compound leads to chromosome fragmentation at 33 nM concentration and replication catastrophe in vitro and in vivo. The compound is overall selective, but contrary to LY2603618, displays only a very slight selectivity (8 fold) over CHK2 and RSK family kinases. Strong nanomolar inhibition was retained in cells (9 nM, HeLa) and 4-fold tumour growth inhibition was reported in Calu-6 lung cancer xenograft models.115 It is currently in phase I clinical trials for its use as single agent or in combination with various anti-neoplastic drugs.
V158411
Vernalis developed V158411 as a novel and potent kinase-selective inhibitor of recombinant CHK1 (IC50 = 3.5 nM) and CHK2 (IC50 = 2.5 nM). In vitro, this compound is more active against leukaemia and lymphoma cell lines (GI50 = 0.17 μM) than against colon (IC50 = 2.8 μM) and lung (IC50 = 6.9 μM) cancer cell lines.116,117 Pharmacological inhibition of CHK1 with V158411 does not induce a definitive cell cycle arrest. The pharmacokinetics and the in vivo efficiency of the compound remain to be evaluated. If they are validated, it might be a promising candidate as both monotherapy and chemosensitizer of gemcitabine and irinotecan.
GDC0425 and GDC0575
GDC0425 and GDC0575 are two novel CHK1 inhibitors recently developed by Genetech. GDC0425 is a member of the 9H-pyrrolo[2,3-b:5,4-c′]dipyridine class whereas the structure of GDC0575 has not been disclosed. Very few information and biological data are available on these compounds, yet.118
Among the biological targets reviewed herein, CHK1 is the one whose inhibition has been the most studied. Following the identification of seminal non-selective CHK1 inhibitors, several ATP-competitive inhibitors with improved CHK1 selectivity were developed. The most promising compounds for clinical use are probably CCT244747, LY2603618 and SCH900776. The two latter are currently evaluated in phase II clinical trials. PF477736 displays a good selectivity and ADME profile but is lacking efficiency. Although not selective, AZD7762 and LY2606368 are valuable dual CHK1/CHK2 inhibitors.
2.4. CHK2 inhibitors
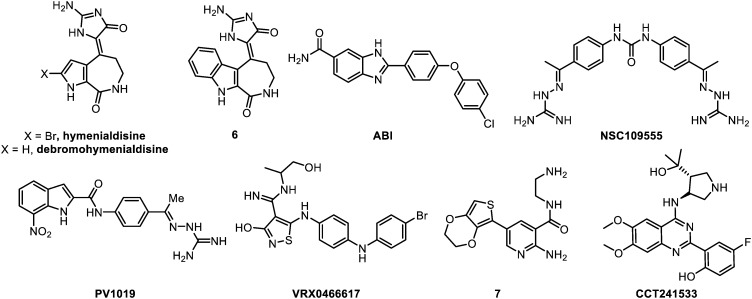
The compounds that are reviewed herein as CHK2 inhibitors are represented below (Fig. 7) and their main properties are subsequently listed in Table 4.
Fig. 7. CHK2 inhibitors.
Table 4. CHK2 inhibitors and their main properties.
| Compound | Enzymatic activity | In vitro selectivity | In vitro efficacy | Pharmacology | In vivo activity | Development stage |
| Hymenialdisine | IC50 = 42 nM | – | N.D. | N.D. | N.D. | Discontinued at optimisation stage |
| 6 | IC50 = 8 nM | +/– | R+/– | N.D. | N.D. | Optimisation stage |
| ABI | IC50 = 15 nM | + | R+/– | N.D. | N.D. | Optimisation stage |
| K i = 37 nM | ||||||
| NSC1095555 | IC50 = 240 nM | + | M– C+/– | – | N.D. | Discontinued at optimisation stage |
| PV1019 | IC50 = 24 nM | ++ | M+/– R+/– C+ | N.D. | N.D. | Research stage |
| VRX0466617 | IC50 = 140 nM | + | M+/– R– C– | N.D. | N.D. | Research stage |
| K i = 11 nM | ||||||
| 7 | IC50 = 28 nM | +/– | M+/– | N.D. | N.D. | Optimisation stage |
| CCT241533 | IC50 = 3 nM | +/– | M+ R++ C+ | + | N.D. | Research stage |
| K i = 1.1 nM |
The pioneering CHK2 inhibitors are two marine natural products: hymenialdisine and debromohymenialdisine (DHB). These compounds are secondary metabolites isolated from the sponges Axinella verrucosa and Acantella aurantiaca.119Hymenialdisine was initially identified as a very potent mitogen-activated protein kinase kinase-1 (MEK-1) inhibitor (IC50 = 6 nM).120 It was later shown that it also displays a potent CHK2 kinase inhibition (IC50 = 42 nM) with moderate selectivity over CHK1 (IC50 = 1.95 μM). Moreover, the chemical synthesis of this compound proved delicate at some steps, particularly for the regioselective bromination. DHB proved less potent (IC50 = 183 nM, CHK2) and less selective (IC50 = 725 nM, CHK1) than hymenialdisine, but was used in several studies to specifically target CSC populations in treatment-resistant tumours.
Indoloazepine 6
The synthesis of an indoloazepine derivative (6) of hymenialdisine allowed a slight 10-fold selectivity over MEK along with some gain in activity (IC50 = 8 nM). Nevertheless, a nanomolar inhibition of several other enzymes, including CDKs, NF-kB (nuclear factor kappa-light-chain-enhancer of activated B cells, a protein complex that controls DNA transcription), GSK-3β and CK1 (casein kinase 1, an enzyme regulating DNA repair and DNA transcription), has been observed. Compound 6 demonstrated a CHK2-mediated radioprotection effect in normal cells and p53 wild type cells, but not in p53 mutant cells at 30 μM (HCT116 colon cancer cells).119 Several synthetic analogues were subsequently prepared, but they all showed significantly diminished activities.121,122
ABI
Johnson & Johnson proposed, in 2005, a 2-arylbenzimidazole (ABI) as potent CHK2 inhibitor (IC50 = 15 nM).123 This compound is the first highly selective inhibitor over CHK1 (IC50 > 10 μM) and is now commercially available. Issued from HTS and hit-to-lead strategy, the key points of the structure are the presence of the amide and of the terminal chlorine. Indeed, the corresponding carboxylic acid without chlorine displayed an IC50 of 640 nM. Of note, ABI efficiency in cells was significantly lower (42% CHK2 inhibition at 5 μM) compared with enzymatic assay. However, used in the low micromolar range, it remains a valuable tool to probe CHK2 inhibition. Importantly, ABI could dose dependently protects CD4+ and CD8+ T-cells from apoptosis after IR.
NSC109555
NSC109555 is a symmetric bis-guanylhydrazone diarylurea issued form a HTS preformed on a library of 100 000 compounds.124 This compound has been co-crystalized in the CHK2 ATP binding site, and is a moderatly potent (IC50 = 240 nM) ATP-competitive inhibitor121,125 displaying a promising kinase profiling. However, no activity could be measured at 100 μM in MCF7 breast cancer and HT29 colorectal cancer cells both as single agent or in combination with topotecan and campthotecin. Contrariwise, an additional study reported an activity in pancreatic cancer cells (MiaPaCa-2, CFPAC-1, Panc-1 and BxPC-3) at the dose of 5 μM, in combination with gemcitabine.126,127
PV1019
PV1019 is a dissymmetric optimized analogue of NSC109555 which shares the same guanylhydrazonearyl moiety.128 This optimization yielded a 10-fold improvement in CHK2 inhibition (IC50 = 24 nM) together with an increased selectivity over CHK1 (IC50 = 15.7 μM) and a over panel of 52 representative kinases (>102). Conversely to the parent compound, PV1019 displayed indisputable effects as radio- and chemosensitizer with topotecan or camptothecin (OVAR-5 and MCF7 cell lines) and also a slight anti-tumour effect as a single agent (IC50 = 5–58 μM). However, the required dosages to achieve significant effects as mono- or combination therapies remain too high (10–25 μM). Therefore, further structural optimization of PV1019 should be undertaken.
VRX046617
VRX046617 was identified by screening of a small kinase inhibitor library,129 followed by a hit-to-lead optimization. This compound displays an IC50 of 140 nM against CHK2 and is selective over CHK1 (IC50 > 10 μM). In BJ-hTERT immortalized fibroblasts, no CHK1 inhibition was observed, and CHK2 inhibition with micromolar concentrations of the drug (10 μM) led to a weak anti-proliferative effect after 6 days (45% growth reduction). Moreover, neither IR, nor chemo-potentiation of doxorubicin or cisplatin cytotoxicity has been evidenced.130VRX046617 is inactive in cells, and therefore may only be an interesting biological probe to study and decipher the CHK2-dependent pathways. This also raises the question of the correlation between CHK2 direct inhibition and the observed anti-proliferative effects.
Aminopyridine 7
Hilton et al. proposed 2-aminopyridine based compounds as CHK2 inhibitors.131 Eleven hits were selected after a screening, against CHK2, performed on 7000 potential ATP-competitive kinase inhibitors. Subsequently, a hit-to-lead optimization afforded the aminopyrimidine 7. This molecule is a potent CHK2 inhibitor (IC50 = 28 nM), and exhibits a moderate selectivity towards CHK1 (IC50 = 2.5 μM) and other kinases (only 18/24 displaying less than 40% at 1 μM). As a single agent, it inhibits CHK2 activity with IC50 values of 5–10 μM and showed a GI50 of 26 μM in HT29 colorectal cancer cells.
CCT241533
CCT241533 is a very potent (IC50 = 3 nM) CHK2 inhibitor which encompasses a 2-(quinazolin-2-yl)phenol scaffold.132 This molecule is moderately selective over CHK1 (IC50 = 190 nM), and might be used as a single agent for monotherapy (GI50 = 1.7, 2.2 and 5.1 μM respectively on HT29, HeLa and MCF7 cells) or chemosensitizer with topotecan, etoposide and camptothecin (IC50 = 1–3 μM).133 Interestingly, this compound displays good pharmacokinetics, oral bioavailability and is the only one that has been tested in vivo in a model of mouse isolated thymocytes. In this in vivo study, it showed a strongly enhanced apoptosis induction after IR exposure. CCT241533 might be a good research tool for in vitro and in vivo pharmacological studies as well as a potential clinical candidate if in vivo studies are validated.
To conclude, only four reasonably selective CHK2 inhibitors have been described to date. Generally, CHK2 inhibitors proved significantly less active than ATM, ATR or CHK1 inhibitors as anti-proliferative compounds or at inducing mitotic catastrophe following IR or DNA-damaging treatment. CCT241533 appears as the most promising compound, but in vivo studies are still needed. ABI, VRX046617 and PV1019 show a modest micromolar anti-proliferative activity whereas NSC109555 is inactive in cells.
2.5. WEE1 inhibitors
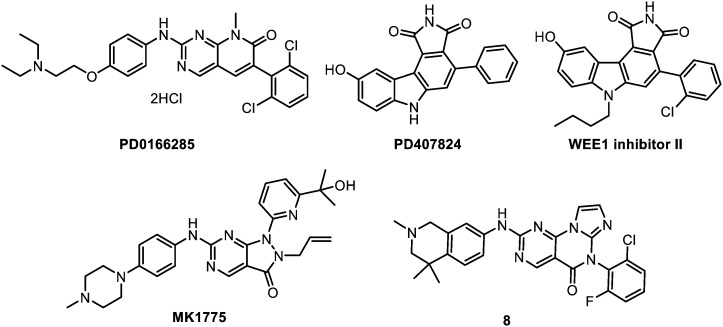
The compounds that are reviewed herein as WEE1 inhibitors are represented below (Fig. 8) and their main properties are subsequently listed in Table 5.
Fig. 8. WEE1 inhibitors.
Table 5. WEE1 inhibitors and their main properties.
| Compound | Enzymatic activity | In vitro selectivity | In vitro efficacy | Pharmacology | In vivo activity | Development stage |
| PD0166285 | IC50 = 24 nM | – | M– R+/– C+/– | – (toxicity) | N.D. | Optimisation stage |
| PD407824 | IC50 = 97 nM | – | N.D. | N.D. | N.D. | Discontinued at optimisation stage |
| WEE1 inhibitor II | IC50 = 59 nM | + | N.D. | – (solubility, distribution) | N.D. | Optimisation stage |
| MK1775 | IC50 = 5 nM | + | M+/– R+/– C+ | + | + | Clinical trials phase II |
| 8 | K i < 1.0 nM | + | M+ C+ | + | + | Preclinical stage |
PD0166285
The first compound that have been reported with an inhibitory activity against WEE1 is the non-selective 6-aryl-pyrido[2,3-d]pyrimidine derivative, PD0166285. This molecule has been identified through a screening performed on a compound library, and by means of tyrosine kinase assay. PD0166285 shows a nanomolar activity (IC50 = 24 nM) on WEE1, but is also active on a range of kinases including proto-oncogene tyrosine-protein kinase Src (c-Src), myelin transcription factor 1 (MYT1), epidermal growth factor receptor (EGFR), fibroblast growth factor receptor 1 (FGFR1), CHK1 and beta-type platelet-derived growth factor receptor (PDGFRb) with IC50 values in the 8–85 nM range.134 Several studies were undertaken in different cancer cell lines, including HCT116, HCT8, HT29, DLD-1, HeLa, H460, U2OS, MG-63, PA-1 cancer cell lines.48,135–137 However, PD0166285 monotherapy exhibited only a slight anti-proliferative effect (GI50 = 26 μM, HT29 cells) without promoting cell death. In combination, it acts as a modest p53 dependent radiosensitizer with a maximal enhancement ratio of 1.23. This is less than the impact of caffeine or UCN-01 (respective enhancement ratios of 2.05 and 1.58 on HT29 cells). Moreover, cell toxicity was observed from 6 h exposure; thus the therapeutic window of cellular effectiveness versus cellular toxicity of the compound is lower than the one of caffeine or UCN-01. Attempts devoted to modify the structure to simultaneously improve both the absolute potency and selectivity towards WEE1 were unsuccessful. Indeed, the observed activities could not be separated from an even more potent inhibition of c-Src kinase.138
PD0407824
Following a HTS program, staurosporine analogues were later proposed for the inhibition of WEE1.139 The representative hit of this screening is PD0407824 which exhibits an IC50 of 97 nM on WEE1. However, this compound was more active on CHK1 (IC50 = 47 nM) and multiple derivatives were synthesised to achieve selectivity over this off-target protein.
WEE1 inhibitor II
Optimization of PD0407824 resulted in the discovery of WEE1 inhibitor II that bears two additional substituents: a butyl chain on the indole nitrogen and a chlorine atom on the phenyl ring. This compound has an IC50 of 35 μM against CHK1, however its low solubility and cell permeability precluded it to present any activity in cells. Optimization of WEE1 inhibitor II was attempted but led to no significant improvement. Solubility problems persisted and no in cellulo activity was obtained.140
MK-1775
MK-1775 is the first highly potent and selective WEE1 inhibitor (IC50 = 5 nM); except over Yes1 kinase (IC50 = 14 nM).141 It was discovered by Merck laboratories through the optimization of a HTS hit. MK-1775 is also the first selective WEE1 inhibitor to be active in cells (including WiDr colon, H1299 lung and TOV21G ovary cancer cell lines), where it abrogates G2 DNA damage checkpoint and decreases cell viability. MK-1775 activity is selective towards p53 defective cells and without effect on p53+ cells.141 It was tested at 100–300 nM concentrations in a number of different patterns: as IR sensitizer in glioma,142 lung, breast and prostate cancers;49 and as chemosensitizer in combination with gemcitabine, 5-fluorouracil, capecitabine, pemetrexed, camptothecin, doxorubicin, carboplatin, cisplatin, mitomycin C and paclitaxel.143,144 Remarkably, no activity was observed as single agent, except in one study on sarcoma.145 Additionally, MK-1775 proved inefficient in combination with temozolomide for glioblastoma, putatively because of an insufficient distribution of temozolomide across the blood–brain barrier.146 Of note, GSC were not strongly affected by this compound (vide infra).142In vivo, MK-1775 potentiates the effects of DNA-damaging agents and induces the tumour growth inhibition of many xenograft models (HeLa, TOV21G-shp53, WiDr, MX-1, PANC198, PANC215, PANC185, etc.). Importantly, these co-treatments do not significantly increase the initial toxicity of the chemotherapeutics (gemcitabine or 5-fluorouracil) in mice.141 Interestingly, the combined inhibition of CHK1 and WEE1 showed synergistic effects and is a significant example of synthetic lethality.147 Treatment with non-toxic concentrations of a CHK1 inhibitor (PF477736) and MK-1775 confirmed the marked synergistic effect in various human cancer cell lines (breast, ovarian, colon, prostate), regardless of p53 status. Similar results were obtained with the combination of MK-1775 with another CHK1 inhibitor, AZD-7762, in vitro and in vivo, in melanoma,45 neuroblastoma148 and for mantle cell lymphoma, an aggressive and incurable disease characterized by a deregulated cell cycle.149,150 In conclusion, MK-1775 is the most potent and highly selective inhibitor of WEE1 and has recently reached phase I clinical trials, in combination with gemcitabine, cisplatin or carboplatin, and phase II in combination with paclitaxel and carboplatin in ovarian cancer.
Pyrimidopyrimidinone 8
Lastly, a pyrimidine-based tricyclic molecule was proposed in 2015 by AbbVie for the inhibition of WEE1.151 This molecule has been designed by a rational hybridization between MK-1775 and other WEE1 inhibitors. Among a series of potential WEE1 inhibitors, pyrimidopyrimidinone 8 emerged as the most promising molecule. This hit displays a very high potency (Ki < 1.0 nM, WEE1) and is selective on an 84 kinases profiling. Nanomolar efficiencies were retained in cells (LD50 = 310 nM, H1299 cell line), and the pharmacokinetics proved particularly good with 94% oral bioavailability, adequate half-life (2.8 h) and clearance values. Oral in vivo potentiation of irinotecan was evidenced in mice xenografted with H1299 cancer cell line, rendering this compound promising for further preclinical evaluation.
The first commercially available WEE1 Inhibitor WEE1 inhibitor II suffers from solubility and permeability issues that hinder its use in cells. Therefore, the standard drug for studying WEE1 inhibition in vitro and in vivo should be MK-1775, which is currently evaluated in phases I and II clinical trials. Pyrimidopyrimidinone 8 is an interesting molecule in the pipeline that could reach the preclinical stage in the near future.
3. ATM, ATR, CHK1, CHK2 and WEE1 inhibition in CSC
This last decade, growing interests have been dedicated to the so-called cancer stem cells (CSC) hypothesis. This model supports the idea that tumours, like adult tissues, arise from cells that exhibit the ability to self-renew as well as give rise to differentiated tissue cells. Most of the reported studies describe an increased DDR in CSC, in accordance with the “stemlike” feature of these cells. Indeed, stem cells have developed protection mechanisms and improved repair compared with differentiated cells.55,152
CSC employ several mechanisms to escape therapeutic assaults (drug efflux activation, anti-apoptotic signalling, hypoxia management…). Nevertheless, as the great majority of anti-cancer therapies (radio- and chemo-therapies) rely on DNA-damaging agents, tackling the enhanced DDR appears as a particularly attractive strategy to improve their efficiencies. Moreover, several studies suggest that CSC contribute to radio- and chemoresistance through enhanced activation of the DNA damage checkpoint response and increased DNA repair capacity. Furthermore, the CSC fraction within the tumour cell population typically increases after repeated cycles of DNA-damaging treatments that preferentially kill the non-stem cells. As a result, classical therapies become progressively ineffective towards these CSC-enriched tumours.153 Therefore, targeting DNA damage checkpoint response and its underlying molecular pathways in CSC may sensitize these particular cells to DNA-damaging techniques and overcome these resistances. This re-sensitization might provide useful therapeutic models for malignant cancers.
In this context, we have reviewed the relevance of the five pivotal kinases ATM, ATR, CHK1, CHK2 and WEE1 as therapeutic targets in the particular case of CSC.
ATM. Despite rare contradictory reports,154,155 most studies enlighten enhanced DDR via constitutively upregulated ATM signalling in CSC models compared with non-CSC.24,56,156 This has been correlated with DNA-damaging treatment resistance. Therefore, ATM seems a relevant target for drug development to improve the efficiency of the current DNA-damaging treatments, and to counter resistance in CSC.
ATR. Higher levels of ATR have been reported in breast CSC (BCSC) models compared with differentiated tumour cells.55 Moreover, ATR inhibition was shown to abrogate in vitro and in vivo tumourigenicity of human colon cancer cells by eliminating the cell population bearing the specific stem cell marker CD133.157 This makes ATR an attractive target for the development of molecular inhibitors to circumvent resistance to classical radio- and chemotherapies.
CHK1 and CHK2. Several reports point out the interest of tackling CHK1 in the specific context of the CSC,90,158–160 while, to the best of our knowledge, no examples of specific CHK2 inhibitors have been studied in CSC models so far.
WEE1. Apart from a study of WEE1-related radiosensitization in glioblastoma neurospheres, very few reports on WEE1 inhibition in the particular case of cancer stem cells have been published.37,161
Overall, only a limited number of chemical modulators of ATM, ATR, CHK1, CHK2 and WEE1 have been tested in CSC or tumour-initiating cells. These inhibitors are presented in Table 6 and discussed afterwards. We also introduce a set of suggestions for the use and the development of new compounds to study cell cycle arrest, to induce a mitotic catastrophe and cell death in CSC.
Table 6. DNA-checkpoint response inhibitors used in CSC.
| Drug | Target | Cancer cell lines | Results | Ref. |
| Caffeine | ATR | NSCLC: A549 | Re-growth delay alone or in combination with etoposide | 157, 162 |
| Colon: DLD1, Colo320, COGA-12, RKO | Preferential apoptosis of the CD133+ cell fractions | |||
| CP466722 | ATM | Breast: BCSC MDA-MB-231, MDA-MB-453 | CSC more sensitive to radiation than non-CSC because of lower ATM levels | 155 |
| KU55933 | ATM | Glioblastoma: U87, U251 GSC, R10, S2, E2, G7 GSC | Comparable drug-induced radiosensitivity of GSC and GC | 156, 163 |
| Potent radiosensitization of GSC | ||||
| KU60019 | ATM | Glioblastoma: BORRU, DR177, BORRU, VIPI, COMI, MPM176, DR177, DEMI, PERU, 2.11 | Specific sensitization of GSC over GC | 57, 58 |
| 4/8 GSC are radiosensitized in vivo | ||||
| NVP-BEZ235 | PTEN/PI3K/ | Prostate: DU 145, PC3 | DDR-independent CSC growth inhibition | 164–167 |
| Akt/mTOR | Colon: HCT-116, SW620 | DDR-independent sensitization of CSC to paclitaxel | ||
| Glioblastoma: U87, U251, T98G, SHG44 | Enhanced radiosensitivity of GSC | |||
| Glioma: SU-2 | ||||
| DBH | CHK1/CHK2 | Glioma: CD133+ D456MG, CD133+ D54MG, D456MG | Minimal impact alone. Disrupts radio-resistance of CD133+ GSC in vitro and in vivo | 153, 168, 169 |
| Breast: 44+/CD24– MCF-7 | Radiosensitization of both CD133– and CD133+ cells | |||
| Significant radiosensitization of CSC | ||||
| UCN-01 | ATR/CHK1/PDK1 | Colon: CD133+ (DLD1, Colo320, RKO, COGA-12) | Significant decrease of CD133+ cells | 90, 157, 170 |
| Glioblastoma: patient-derived GSC | Strong chemosensitization with temozolomide | |||
| Glioma: derived HNGC-2 | No reduced viability CSC | |||
| HNSCC: SQ20B/CD44+/ALDHhigh | Radiosensitization of CSC | |||
| AZD7762 | CHK1/ | Pancreas: patient-derived CSC | Chemosensitization with gemcitabine and tumor-initiating capacity reduction | 90, 158–160 |
| CHK2 | NSCLC: patient-derived CSC | Dramatic reduction of survival in vitro and in vivo | ||
| Leukaemia: CD34+/CD38–/CD123+ | Significant reduction of CSC without affecting normal SCs | |||
| Glioblastoma: patient-derived GSC | Ineffective chemosensitization with temozolomide | |||
| PF477736 | CHK1 | NSCLC: NCI-H1299 | Chemosensitization of CSC with gemcitabine | 171, 172 |
| Pancreas: PANC-1, BxPC-3 | Very slight tumour growth delay and enrichment of CSC | |||
| SCH900776 | CHK1 | AML: primary leukemic blasts, CD34+/CD38–/CD123+ CSC | Specific reduction of CSC fraction in combination with HDAC inhibitors | 108 |
| PD0166285 | WEE1 c-Src/MYT1/EGFR/CHK1… | Glioblastoma: CD133+ (U251MG, U118MG, U87MG) | Specific significant radiosensitization of CSC | 37 |
| MK1775 | WEE1 | Glioblastoma: G179, G144 GNS | Effective sensitization of GBM non-SCs and normal SCs, but no radiosensitization of GNS | 142 |
3.1. ATM/ATR inhibitors in CSC
Five compounds were tested as sensitizers for the targeting of CSC: caffeine, CP466722, KU55933, KU60019 and NVP-BEZ235.
In 2009, the first attempts to sensitize CSC were undertaken using caffeine in combination with etoposide. The caffeine/etoposide combo proved able to delay re-growth of A549 NSCL CSC in the tumour through ATM/ATR pathways inhibition. However, this potential is tarnished by the fact that caffeine also induced by itself a reversible growth arrest that is associated with increased fraction of CSC.162 A selective targeting of CSC was obtained in human colon cancer cell lines (DLD1, Colo320, COGA-12 and RKO) models, where caffeine-promoted ATR inhibition preferentially depleted the chemo-resistant and exclusively tumorigenic CD133+ cell fraction. This depletion proceeds by apoptosis induction of CD133+ cycling cells. The remaining cells lose their tumorigenicity either in vitro or in vivo.157 Moreover, the chemoresistance of those cells towards DNA inter-strand crosslinking agents like cisplatin could be overcome. This chemoresistance has been attributed to a preferential activation of the ATR/CHK1-dependent DNA-damage response in CD133+ cells.
Similar results have been observed by comparing the efficiency of the ATM-selective inhibitor CP466722 on MDA-MB231 and MDA-MB453 BCSC with their corresponding non-stem cells. In this model, BCSC were more sensitive to radiation than non-BCSC. The authors imply that this higher sensitivity might be due to lower ATM intracellular levels and DNA repair capacity in BCSC, which constitutes a controversy relative to most other reports.155
On the other hand, Carruthers et al. reported a potent radiosensitization of GSC after treatment with the ATM inhibitor KU55933.163 An additional study, described by Zhou et al., confirmed this potent radiosensitization in GSC.156 Surprisingly, the same level of radiosensitization was observed in GC and GSC although ATM activity is significantly higher in the latter cell type. Indeed, in response to radiation, both GC and GSC treated with KU55933 had an enhanced proportion of cells in G2 phase and a decreased proportion of cells in G1 phase. Additionally, an enhanced apoptotic rate, relative to the untreated cells, was observed.156
Contradictory, Raso et al. reported a specific sensitization of BORRU GSC compared to BORRU GC, when using ATM-selective inhibitors KU55933 and KU60019. Furthermore, no sensitization was observed in DR117 GSC, which display a less pronounced stem phenotype than BORRU cells. This suggests that ATM inhibition may specifically sensitize GSC while sparing non-stem cells.57 Consistently, GSC radioresistance could be overcome in vivo using KU60019 in four GSC lines (BORRU, VIPI, COMI and MPM176), whereas four others were protected (DR177, DEMI, PERU and 2.11) under the same conditions. However, among the several markers that were followed, the positive response to KU60019 rather correlated with high PI3K expression, than with the “stemness” of the cell lines. This underlines the high heterogenic behaviour of CSC towards ATM-specific inhibition.58
The multi-target PI3K/mTOR/ATM/DNA-PKcs/Akt inhibitor NVP-BEZ235 was reported to inhibit the growth of prostate CD133+/CD44+ CSC,164 to decrease CD133+ colon CSC proliferation and survival,165 and to sensitize SW620 colorectal CSC to paclitaxel.166 However, these specific effects seem in all cases to be unrelated to DNA-checkpoint response, but rather depending on the PTEN/PI3K/Akt/mTOR pathway. Contrarily, in the case of SU-2 GSC, an enhanced radiosensitivity induced by NVP-BEZ235 was observed with a concomitant decrease of DNA repair capacity. The correlation between these two events has not been fully deciphered yet.167
3.2. CHK1/CHK2 inhibitors in CSC
Checkpoint kinases inhibition has been studied in the case of CSC. Indeed, CHK1-mediated DNA checkpoint response was shown to be more robust in CSC than non-stem cells.158 Strikingly, normal embryonic stem cells seem to fail in activating CHK1 in response to DNA replication stress. This non-activation of checkpoints is thought to be a mechanism of preservation of genome integrity, by eliminating damaged cells through apoptosis, rather than trying to repair them through DDR.173 Therefore, CHK1 inhibition was studied as potential therapeutic approach for sensitizing resistant tumour cells. Five inhibitors were assayed in several CSC tumours: UCN-01, AZD7762, PF477736, SCH900776 and DBH.
The dual CHK1/CHK2 inhibitor DBH was the first compound employed to try to overcome IR resistance in CD133+ cells. Although it had a minimal impact on CD133– and CD133+ cells, it displayed a potent synergy with radiation. Notably it has the ability to reverse specifically resistance in CD133+ cells in vitro and in vivo.153 On the contrary, it radiosensitizes both CD133– and CD133+ cells in a D456MG glioma model.168 Otherwise, in a recent in vitro study, DBH treatment in addition to radiation significantly reduced the stem cell proportion of MCF-7 breast cancer cells.169
Treatment of colon cancer cells with UCN-01 results in significant decreased CD133+ expressing cells fraction and consecutive loss of in vitro and in vivo tumorigenicity of the remaining cells. Mechanistic studies suggested that either inhibition of CHK1 or upstream ATR are responsible of the observed sensitizing effects.157 Otherwise, UCN-01 induced radiosensitization of SQ20B/CD44+/ALDHhigh head and neck squamous cell carcinoma stem cells; this could be improved by co-treatment with all-trans retinoic acid as ALDH inhibitor.170 In contrast, UCN-01 was found inactive on glioma derived HNGC-2 stem cells.174
Interestingly, Signore & Ricci-Vitiani demonstrated that most GSC are resistant to specific inhibition of the major signalling pathways involved in cell survival and proliferation. For example, the specific CHK inhibition with AZD7762 proved scarcely effective in temozolomide chemosensitizing. On the contrary, the multipathway inhibitor UCN-01 displayed remarkable inhibition of GSC growth in the same in vitro and in vivo experience patterns.90 The authors then deciphered that the combined inhibition of PDK1 (3-phosphoinositide dependent protein kinase-1, a major kinase downstream of PI3K required for the activation of Akt) and CHK1 was responsible of the strong anti-tumour effects observed, thus paving the way to a new potential therapeutic approach to reduce the growth of human glioblastoma.
In NSCLC-SC, the dual CHK1/CHK2 inhibitor AZD7762, combined with the chemotherapy agents gemcitabine, paclitaxel and cisplatin significantly reduced in vitro and in vivo tumour growth, whereas chemotherapy alone was almost ineffective.159 Similar results were observed in leukaemia, where co-administration of AZD7762 with genotoxic agents (e.g. gemcitabine) was reported to significantly reduce tumour cell progenitors bearing CD34+/CD38–/CD123+ stem cell markers without affecting normal hematopoietic progenitors;160 the same phenomenon was observed in patient-derived pancreas cancer cells (using CD24, CD44 and ESA markers). In this latter case, the combination AZD7762/gemcitabine significantly delayed tumour initiation.158
A contradictory study reported that the combination of gemcitabine with CHK1-selective inhibitor PF477736 leads to very slight tumour growth delay and enriched CSC in PANC-1 and BxPC-3 pancreatic cell lines ductal adenocarcinoma models.172 In this study, the authors proposed the selectivity of PF477736 towards CHK1 (100 fold over CHK2) as an explanation. Otherwise, in a NCI-H1299 non-small cell lung cancer model, PF477736 enhances the anti-proliferative effect of gemcitabine against both the parental and the CSC-like cell populations; although CSC-like cells exhibited resistance to apoptosis whereas non-CSC remained sensitive.171
Interestingly, the combination of the CHK1-selective SCH900776 and a HDAC inhibitor also proved beneficial to kill AML primary leukemic blasts and CD34+/CD38–/CD123+ leukaemia initiating cells (regardless of p53 status) whereas it did not promote substantial toxicity towards normal CD34+ hematopoietic cells.108
3.3. WEE1 inhibitors in CSC
The selective WEE1 inhibitor MK-1775 was tested against U251, U87, and T98G glioblastoma cells where it showed a potent G2-checkpoint abrogation and enhanced radiosensitization. Human normal astrocytes were also sensitive when submitted to the same conditions. In contrast, G179 and G144 glioblastoma neural stem (GNS) cells were slightly affected, displaying only an attenuation of the accumulation of cells into G2–M phase, and no radiosensitization effect.142 Therefore, these initial findings suggest that although WEE1 inhibition is highly effective in radiosensitization, targeting only this pathway might be insufficient to modulate radiation response in this focused GNS population.
Otherwise, opposite results were reported the same year on a primary glioblastoma (GBM) neurospheres model with the non-selective inhibitor PD0166285. In this study, radiation resistant CD133+ GBM cells were efficiently killed after treatment with this compound, suggesting that radiation resistance could be overcome by WEE1 inhibition.37 However, PD0166285 is a multi-target compound inhibiting a panel of other enzymes in the nanomolar range. Whether this radio-resistance reversion is or is not due to WEE1 inhibition has not been established yet; thus it would be very interesting to collect more data to assess this purpose.
Overall, disparate results have been obtained for the sensitization potential of DNA-checkpoint modulators in CSC. However, several points can be highlighted:
i) Several DNA-damage checkpoint inhibitors are active against CSC in vitro and in vivo.
ii) There is no clear selectivity of these drugs on CSC over non-CSC; the activity being either higher, lesser or comparable depending on the cases.
iii) For a defined drug and a defined cancer, the sensitivity of CSC depends on the cell lines.
iv) Importantly, among cancers, glioma stem cells which have been largely studied prove particularly resistant compared to other CSC. To a lesser extent, pancreas stem cells seem difficult to target.
v) Generally, at first sight, ATM/ATR inhibitors appear to have more impact on CSC than CHK1, CHK2 and WEE1 specific inhibitors, even if more data using other selective drugs are needed to confirm this trend.
vi) Non-selective multi-pathways inhibitors (e.g.caffeine, UCN-01, NVP-BEZ235, PD0166285,…) usually display a better activity on CSC than target-selective compounds. In the particular case of NVP-BEZ235, its growth inhibition activity on various CSC even seems DDR-independent. Additional results are also required to clarify the most relevant pathways to target for CSC sensitization.
4. Future prospects of ATM, ATR, CHK1, CHK2 and WEE1 inhibitors as CSC sensitizers
Target enzymes. From several pioneering reports released these last 5 years, the specific CSC targeting is an area of growing interest with many questions that remain to be addressed. On the five enzymes involved in the cell cycle arrest we have focused on ATM and ATR, which are validated targets for the sensitization of drug-resistant cancers cells as well as their respective CSC populations. Importantly, the most resistant GSC seem to be sensitizable, at least for several cell lines. CHK1 is a valid oncotarget for the sensitization of cancer cells with already five compounds having reached clinical phases. However, only one study using a selective CHK1 inhibitor (PF477736 on NSCLC) has been reported for the sensitization of CSC.171 Other studies with other specific drugs and other cancer cells are urgently needed to validate this enzyme in the particular case of CSC. The role of CHK2 in oncology is less clear. As for CHK1, it was at first sight legitimate to target this protein to afford sensitization to DNA-damaging agents as it represents the second main actor of the checkpoint response activation. However, accumulating evidence was latter disclosed that CHK2, in spite of having a role in checkpoint signalling especially in response to radiation, is not a necessary kinase and its depletion has no effect on the sensitization of gemcitabine-treated cells.175–177 In the case of WEE1, very few data are available, with only one negative report using a selective inhibitor, and a positive one using a multi-target drug.
Utilizable drugs. In all cases, more data need to be accumulated to decipher the potential, scope and limitations of DNA-damage checkpoint response inhibitors to target CSC. To this end, many potent and selective recent drugs are available and have not been tested yet. They may serve as interesting chemical tools to probe the biology of these enzymes in oncology, and in particular case of CSC. This encompasses ATM inhibitors: KU60019 on which preliminary results on CSC have been disclosed, its not yet tested analogue KU59403, and the promising new drugs 1 and 2. For ATR inhibitors, VE-822 and AZD6738, which are very potent in vitro and in vivo on differentiated cancer cells, might be highlighted. Within all compounds described to target CHK1, five of them might be considered for their potential use in CSC: macrocycle 5, SAR020106, CCT244747, SCH900776 and LY2603618. Concerning CHK2, its inhibition alone does not seem efficient in the sensitization of DNA-damaging processes. For example, VRX046617, a potent in vitro and in vivo CHK2 inhibitor with no CHK1 activity, did not potentiate doxorubicin or cisplatin activity.130 On the contrary, the only potent molecule described as CHK2 inhibitor, CCT241533, displays a moderate selectivity, raising the hypothesis of off-target effects. Therefore, if the advantage to afford a dual CHK1/CHK2 inhibition over a CHK1 inhibition alone remains to be evaluated, there is apparently no reason that a specific CHK2 inhibition will result in chemo- or radiosensitization in the particular case of CSC.178 Very few data are available on WEE1-specific inhibition in CSC, and one new compound (8) might be very interesting to assay.
Finally, targeting DNA-damage checkpoint response in CSC seems an overall good strategy as it is generally admitted that CSC have superior drug-resistance mechanisms than non-CSC. However, the best therapeutic target to induce mitotic catastrophe by G2-abrogation remains elusive, whether ATM, ATR, MYT1, CHK1, Hsp90, PP2A, WEE1, or other unknown G2–M transition targets or combinations thereof.46
Acknowledgments
The authors are indebted to Prof. Daniele Passarella, Prof. Ling Peng and Dr. Panagiota Sotiropoulou for their invitation to submit this review to the special issue “Chemical approaches targeting drug resistance in cancer stem cell” in the MedChemComm journal of Royal Society of Chemistry. This work was partly funded by the European COST Action CM1106 (; www.stemchem.org).
Biographies

Cyril Ronco
Cyril Ronco received his PhD on cholinesterases ligands under the supervision of Prof. P.-Y. Renard at the University of Rouen. He was then awarded with a Humboldt postdoctoral fellowship to work on the total synthesis of complex natural products in Prof. H.-D. Arndt group at Jena University, Germany. After working on medicinal chemistry projects in Prof. Déprez’s group (University of Lille), he joined in 2014 the group of Dr. Benhida as “Maître de Conférences” at the University of Nice Sophia Antipolis. His research focuses currently on the development of bioactive molecules to circumvent resistance mechanisms in the field of oncology.

Anthony R. Martin
Anthony R. Martin received his PhD from the University of Montpellier in 2011 under the supervision of Prof. Michael Smietana and Dr. Jean-Jacques Vasseur, working on the synthesis and self-assembly of borono (oligo)nucleotides. He then joined Prof. Steven Nolan’s group (Saint-Andrews University) as a postdoctoral researcher to work on late transition metal coordination and their applications in homogeneous catalysis. In 2013, he moved to the Institute of Chemistry of Nice in Dr. Benhida’s group where he was appointed Chargé de Recherche—CNRS in 2014. His researches focus on the design of new chemotherapeutics and encompass various aspects of nucleic acid chemistry.

Luc Demange
Luc Demange is a medicinal chemist, graduated from “Ecole Nationale Supérieure de Chimie de Paris”, who completed his PhD working on peptidyl-prolyl isomerases inhibitors (CEA/Saclay, 2001). He was post-doctoral fellow in Sherbrooke (synthesis of cardiac glycosides) then in Montpellier (design of growth hormone secretagogues). In 2006, he was recruited as professor assistant (Paris Descartes University), and he developed with Prof. Christiane Garbay CDKs-inhibitors and anti-angiogenic drugs with applications in cancer. In 2013, LD moved to the “Institut de Chimie de Nice” where his research interests are focused on the synthesis of new therapeutic agents to overcome drug resistances.

Rachid Benhida
Rachid Benhida is DR CNRS, Deputy-Director of the ICN Institute and leader of Bioactive Molecules team. He received his Ph.D. from the University-Paris XI (1992). After three years post-doc (Hoechst/Roussel-Uclaf, Germany and “Institut Curie-Paris”, 1994), he joined the “Institut de Chimie des Substances Naturelles” as “ANRS young research award” and as CNRS researcher in the same year (1995). He moved to the University of Nice in 2002. His current researches focus on the development of synthetic methods and their applications in nucleic acid chemistry, chemical biology and the development of new strategies to overcome drug resistance using small molecules (specific targeting of nucleic acids and CSCs, identification and validation of new targets, etc.).
Footnotes
†The authors declare no competing interests.
References
- Ashwell S., in DNA repair in cancer therapy molecular targets and clinical applications, Elsevier, Amsterdam, 2012, pp. 211–234. [Google Scholar]
- Andrs M., Korabecny J., Nepovimova E., Jun D., Hodny Z., Moravcova S., Hanzlikova H., Kuca K. Mini-Rev. Med. Chem. 2014;14:805–811. [PubMed] [Google Scholar]
- Kelley M. R., in DNA repair in cancer therapy molecular targets and clinical applications, Elseiver, Amsterdam, 2012, pp. 1–16. [Google Scholar]
- Jackson S. P., Bartek J. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marechal A., Zou L. Cold Spring Harbor Perspect. Biol. 2013;5:a012716. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein M., Kastan M. B. Annu. Rev. Med. 2015;66:129–143. doi: 10.1146/annurev-med-081313-121208. [DOI] [PubMed] [Google Scholar]
- Morgan M. A., Lawrence T. S. Clin. Cancer Res. 2015;21:2898–2904. doi: 10.1158/1078-0432.CCR-13-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A., Shaik T. B., Malik M. S. Expert Opin. Drug Discovery. 2015;10:1119–1132. doi: 10.1517/17460441.2015.1072167. [DOI] [PubMed] [Google Scholar]
- Dillon M. T., Good J. S., Harrington K. J. Clin. Oncol. 2014;26:257–265. doi: 10.1016/j.clon.2014.01.009. [DOI] [PubMed] [Google Scholar]
- Lovejoy C. A., Cortez D. DNA Repair. 2009;8:1004–1008. doi: 10.1016/j.dnarep.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendetti F. P., Lau A., Schamus S., Conrads T. P., O'Connor M. J., Bakkenist C. J. Oncotarget. 2015;6:44289–44305. doi: 10.18632/oncotarget.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chu H., Lv M., Zhang Z., Qiu S., Liu H., Shen X., Wang W., Cai G. Nat. Commun. 2016;7:11655. doi: 10.1038/ncomms11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Golding S. E., Rosenberg E., Valerie N., Hussaini I., Frigerio M., Cockcroft X. F., Chong W. Y., Hummersone M., Rigoreau L., Menear K. A., O'Connor M. J., Povirk L. F., van Meter T., Valerie K. Mol. Cancer Ther. 2009;8:2894–2902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.-Q., Halaby M.-J., Li Y., Hibma J. C., Burn P. Drug Discovery Today. 2011;16:332–338. doi: 10.1016/j.drudis.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Rainey M. D., Charlton M. E., Stanton R. V., Kastan M. B. Cancer Res. 2008;68:7466–7474. doi: 10.1158/0008-5472.CAN-08-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson I., Zhao Y., Richardson C. J., Green S. J., Martin N. M. B., Orr A. I., Reaper P. M., Jackson S. P., Curtin N. J., Smith G. C. M. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Batey M. A., Zhao Y., Kyle S., Richardson C., Slade A., Martin N. M. B., Lau A., Newell D. R., Curtin N. J. Mol. Cancer Ther. 2013;12:959–967. doi: 10.1158/1535-7163.MCT-12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiotani B., Nguyen H. D., Håkansson P., Maréchal A., Tse A., Tahara H., Zou L. Cell Rep. 2013;3:1651–1662. doi: 10.1016/j.celrep.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. J., Baltimore D. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. J., Baltimore D. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- de Klein A., Muijtjens M., van Os R., Verhoeven Y., Smit B., Carr A. M., Lehmann A. R., Hoeijmakers J. H. Curr. Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- Cortez D., Guntuku S., Qin J., Elledge S. J. Science. 2001;294:1713–1716. doi: 10.1126/science.1065521. [DOI] [PubMed] [Google Scholar]
- Weber A. M., Ryan A. J. Pharmacol. Ther. 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Kwok M., Davies N., Agathanggelou A., Smith E., Oldreive C., Petermann E., Stewart G., Brown J., Lau A., Pratt G., Parry H., Taylor M., Moss P., Hillmen P., Stankovic T. Blood. 2016;127:582–595. doi: 10.1182/blood-2015-05-644872. [DOI] [PubMed] [Google Scholar]
- Kwok M., Davies N., Agathanggelou A., Smith E., Petermann E., Yates E., Brown J., Lau A., Stankovic T. Lancet. 2015;385(Suppl 1):S58. doi: 10.1016/S0140-6736(15)60373-7. [DOI] [PubMed] [Google Scholar]
- Matthews T. P., Jones A. M., Collins I. Expert Opin. Drug Discovery. 2013;8:621–640. doi: 10.1517/17460441.2013.788496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai H., Naka K., Okada Y., Watanabe M., Harada N., Saito S., Anderson C. W., Appella E., Nakanishi M., Suzuki H., Nagashima K., Sawa H., Ikeda K., Motoyama N. EMBO J. 2002;21:5195–5205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabludoff S. D., Deng C., Grondine M. R., Sheehy A. M., Ashwell S., Caleb B. L., Green S., Haye H. R., Horn C. L., Janetka J. W., Liu D., Mouchet E., Ready S., Rosenthal J. L., Queva C., Schwartz G. K., Taylor K. J., Tse A. N., Walker G. E., White A. M. Mol. Cancer Ther. 2008;7:2955–2966. doi: 10.1158/1535-7163.MCT-08-0492. [DOI] [PubMed] [Google Scholar]
- Xiao Z., Xue J., Sowin T. J., Zhang H. Mol. Cancer Ther. 2006;5:1935–1943. doi: 10.1158/1535-7163.MCT-06-0077. [DOI] [PubMed] [Google Scholar]
- Soussi T., Wiman K. G. Cancer Cell. 2007;12:303–312. doi: 10.1016/j.ccr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Kaelin W. G. Nat. Rev. Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- Zannini L., Delia D., Buscemi G. J. Mol. Cell Biol. 2014;6:442–457. doi: 10.1093/jmcb/mju045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimian A., Ahmadi Y., Yousefi B. DNA Repair. 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Nguyen T. N. T., Tepe J. J. Curr. Med. Chem. 2011;18:4368–4374. doi: 10.2174/092986711797200390. [DOI] [PubMed] [Google Scholar]
- Squire C. J., Dickson J. M., Ivanovic I., Baker E. N. Structure. 2005;13:541–550. doi: 10.1016/j.str.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Mir S. E., De Witt Hamer P. C., Krawczyk P. M., Balaj L., Claes A., Niers J. M., Van Tilborg A. A. G., Zwinderman A. H., Geerts D., Kaspers G. J. L., Peter Vandertop W., Cloos J., Tannous B. A., Wesseling P., Aten J. A., Noske D. P., Van Noorden C. J. F., Würdinger T. Cancer Cell. 2010;18:244–257. doi: 10.1016/j.ccr.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Haese G. J., Walworth N., Carr A. M., Gould K. L. Mol. Biol. Cell. 1995;6:371–385. doi: 10.1091/mbc.6.4.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P., Nurse P. Cell. 1987;49:559–567. doi: 10.1016/0092-8674(87)90458-2. [DOI] [PubMed] [Google Scholar]
- Iorns E., Lord C. J., Grigoriadis A., McDonald S., Fenwick K., MacKay A., Mein C. A., Natrajan R., Savage K., Tamber N., Reis-Filho J. S., Turner N. C., Ashworth A. PLoS One. 2009;4:e5120. doi: 10.1371/journal.pone.0005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak J. P. Y., Man W. Y., Chow J. P. H., Ma H. T., Poon R. Y. C. Oncotarget. 2015;6:21074–21084. doi: 10.18632/oncotarget.4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell M. J., Raleigh J. M., Verkade H. M., Nurse P. EMBO J. 1997;16:545–554. doi: 10.1093/emboj/16.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouliot L. M., Chen Y. C., Bai J., Guha R., Martin S. E., Gottesman M. M., Hall M. D. Cancer Res. 2012;72:5945–5955. doi: 10.1158/0008-5472.CAN-12-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrow L. M., Garimella S. V., Jones T. L., Caplen N. J., Lipkowitz S. Breast Cancer Res. Treat. 2009;122:347–357. doi: 10.1007/s10549-009-0571-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnussen G. I., Emilsen E., Giller Fleten K., Engesæter B., Nähse-Kumpf V., Fjær R., Slipicevic A., Flørenes V. A. BMC Cancer. 2015;15:462. doi: 10.1186/s12885-015-1474-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Witt Hamer P. C., Mir S. E., Noske D., Van Noorden C. J. F., Wurdinger T. Clin. Cancer Res. 2011;17:4200–4207. doi: 10.1158/1078-0432.CCR-10-2537. [DOI] [PubMed] [Google Scholar]
- Caretti V., Hiddingh L., Lagerweij T., Schellen P., Koken P. W., Hulleman E., van Vuurden D. G., Vandertop W. P., Kaspers G. J. L., Noske D. P., Wurdinger T. Mol. Cancer Ther. 2013;12:141–150. doi: 10.1158/1535-7163.MCT-12-0735. [DOI] [PubMed] [Google Scholar]
- Li J., Wang Y., Sun Y., Lawrence T. S. Radiat. Res. 2002;157:322–330. doi: 10.1667/0033-7587(2002)157[0322:wttigp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Bridges K. A., Hirai H., Buser C. A., Brooks C., Liu H., Buchholz T. A., Molkentine J. M., Mason K. A., Meyn R. E. Clin. Cancer Res. 2011;17:5638–5648. doi: 10.1158/1078-0432.CCR-11-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria J. N., Tibbetts R. S., Busby E. C., Kennedy A. P., Hill D. E., Abraham R. T. Cancer Res. 1998;58:4375–4382. [PubMed] [Google Scholar]
- Marone R., Cmiljanovic V., Giese B., Wymann M. P. Biochim. Biophys. Acta. 2008;1784:159–185. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Karve S., Werner M. E., Sukumar R., Cummings N. D., Copp J. A., Wang E. C., Li C., Sethi M., Chen R. C., Pacold M. E., Wang A. Z. Proc. Natl. Acad. Sci. U. S. A. 2012;109:8230–8235. doi: 10.1073/pnas.1120508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria J. N., Busby E. C., Tibbetts R. S., Roos P., Taya Y., Karnitz L. M., Abraham R. T. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- Hollick J. J., Rigoreau L. J. M., Cano-Soumillac C., Cockcroft X., Curtin N. J., Frigerio M., Golding B. T., Guiard S., Hardcastle I. R., Hickson I., Hummersone M. G., Menear K. A., Martin N. M. B., Matthews I., Newell D. R., Ord R., Richardson C. J., Smith G. C. M., Griffin R. J. J. Med. Chem. 2007;50:1958–1972. doi: 10.1021/jm061121y. [DOI] [PubMed] [Google Scholar]
- Maugeri-Sacca M., Bartucci M., De Maria R. Mol. Cancer Ther. 2012;11:1627–1636. doi: 10.1158/1535-7163.MCT-11-1040. [DOI] [PubMed] [Google Scholar]
- Golding S. E., Rosenberg E., Adams B. R., Wignarajah S., Beckta J. M., O'Connor M. J., Valerie K. Cell Cycle. 2012;11:1167–1173. doi: 10.4161/cc.11.6.19576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raso A., Vecchio D., Cappelli E., Ropolo M., Poggi A., Nozza P., Biassoni R., Mascelli S., Capra V., Kalfas F., Severi P., Frosina G. Brain Pathol. 2012;22:677–688. doi: 10.1111/j.1750-3639.2012.00566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchio D., Daga A., Carra E., Marubbi D., Baio G., Neumaier C. E., Vagge S., Corvò R., Pia Brisigotti M., Louis Ravetti J., Zunino A., Poggi A., Mascelli S., Raso A., Frosina G. Int. J. Cancer. 2014;135:479–491. doi: 10.1002/ijc.28680. [DOI] [PubMed] [Google Scholar]
- Min J., Guo K., Suryadevara P. K., Zhu F., Holbrook G., Chen Y., Feau C., Young B. M., Lemoff A., Connelly M. C., Kastan M. B., Guy R. K. J. Med. Chem. 2016;59:559–577. doi: 10.1021/acs.jmedchem.5b01092. [DOI] [PubMed] [Google Scholar]
- Cruet-Hennequart S., Glynn M. T., Murillo L. S., Coyne S., Carty M. P. DNA Repair. 2008;7:582–596. doi: 10.1016/j.dnarep.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Alao J. P., Sunnerhagen P. Radiat. Oncol. 2009;4:51–57. doi: 10.1186/1748-717X-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Toledo L. I., Fernandez-Capetillo O., Bakkenist C. J. DNA Repair. 2011;10:1000–1001. doi: 10.1016/j.dnarep.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo L. I., Murga M., Zur R., Soria R., Rodriguez A., Martinez S., Oyarzabal J., Pastor J., Bischoff J. R., Fernandez-Capetillo O. Nat. Struct. Mol. Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee B., Tomimatsu N., Amancherla K., Camacho C. V., Pichamoorthy N., Burma S. Neoplasia. 2012;14:34–43. doi: 10.1593/neo.111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil del Alcazar C. R., Hardebeck M. C., Mukherjee B., Tomimatsu N., Gao X., Yan J., Xie X. J., Bachoo R., Li L., Habib A. A., Burma S. Clin. Cancer Res. 2014;20:1235–1248. doi: 10.1158/1078-0432.CCR-13-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Xie G., Zhou G., Cheng Y., Zhang G., Yao G., Chen Y., Li Y., Zhao G. Cancer Lett. 2015;367:58–68. doi: 10.1016/j.canlet.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Liu Q., Xu C., Kirubakaran S., Zhang X., Hur W., Liu Y., Kwiatkowski N. P., Wang J., Westover K. D., Gao P., Ercan D., Niepel M., Thoreen C. C., Kang S. A., Patricelli M. P., Wang Y., Tupper T., Altabef A., Kawamura H., Held K. D., Chou D. M., Elledge S. J., Janne P. A., Wong K. K., Sabatini D. M., Gray N. S. Cancer Res. 2013;73:2574–2586. doi: 10.1158/0008-5472.CAN-12-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Wang J., Kang S. A., Thoreen C. C., Hur W., Ahmed T., Sabatini D. M., Gray N. S. J. Med. Chem. 2011;54:1473–1480. doi: 10.1021/jm101520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degorce S. L., Barlaam B., Cadogan E., Dishington A., Ducray R., Glossop S. C., Hassall L. A., Lach F., Lau A., McGuire T. M., Nowak T., Ouvry G., Pike K. G., Thomason A. G. J. Med. Chem. 2016;59:6281–6292. doi: 10.1021/acs.jmedchem.6b00519. [DOI] [PubMed] [Google Scholar]
- Guo K., Shelat A. A., Guy R. K., Kastan M. B. J. Biomol. Screening. 2014;19:538–546. doi: 10.1177/1087057113520325. [DOI] [PubMed] [Google Scholar]
- Emmitte K. A., Adjebang G. M., Andrews C. W., Alberti J. G. B., Bambal R., Chamberlain S. D., Davis-Ward R. G., Dickson H. D., Hassler D. F., Hornberger K. R., Jackson J. R., Kuntz K. W., Lansing T. J., Mook R. A., Nailor K. E., Pobanz M. A., Smith S. C., Sung C.-M., Cheung M. Bioorg. Med. Chem. Lett. 2009;19:1694–1697. doi: 10.1016/j.bmcl.2009.01.094. [DOI] [PubMed] [Google Scholar]
- Nishida H., Tatewaki N., Nakajima Y., Magara T., Ko K. M., Hamamori Y., Konishi T. Nucleic Acids Res. 2009;37:5678–5689. doi: 10.1093/nar/gkp593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peasland A., Wang L.-Z., Rowling E., Kyle S., Chen T., Hopkins A., Cliby W. A., Sarkaria J., Beale G., Edmondson R. J., Curtin N. J. Br. J. Cancer. 2011;105:372–381. doi: 10.1038/bjc.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier J.-D., Durrant S. J., Golec J. M. C., Kay D. P., Knegtel R. M. A., MacCormick S., Mortimore M., O'Donnell M. E., Pinder J. L., Reaper P. M., Rutherford A. P., Wang P. S. H., Young S. C., Pollard J. R. J. Med. Chem. 2011;54:2320–2330. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- Reaper P. M., Griffiths M. R., Long J. M., Charrier J.-D., MacCormick S., Charlton P. A., Golec J. M. C., Pollard J. R. Nat. Chem. Biol. 2011;7:428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- Huntoon C. J., Flatten K. S., Wahner Hendrickson A. E., Huehls A. M., Sutor S. L., Kaufmann S. H., Karnitz L. M. Cancer Res. 2013;73:3683–3691. doi: 10.1158/0008-5472.CAN-13-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevo R., Fokas E., Reaper P. M., Charlton P. A., Pollard J. R., McKenna W. G., Muschel R. J., Brunner T. B. Cancer Biol. Ther. 2014;13:1072–1081. doi: 10.4161/cbt.21093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires I. M., Olcina M. M., Anbalagan S., Pollard J. R., Reaper P. M., Charlton P. A., McKenna W. G., Hammond E. M. Br. J. Cancer. 2012;107:291–299. doi: 10.1038/bjc.2012.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton F. K., Patterson M. J., Elstob C. J., Fordham S., Herriott A., Wade M. A., McCormick A., Edmondson R., May F. E. B., Allan J. M., Pollard J. R., Curtin N. J. Oncotarget. 2015;6:32396–32409. doi: 10.18632/oncotarget.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokas E., Prevo R., Pollard J. R., Reaper P. M., Charlton P. A., Cornelissen B., Vallis K. A., Hammond E. M., Olcina M. M., Gillies McKenna W., Muschel R. J., Brunner T. B. Cell Death Dis. 2012;3:e441. doi: 10.1038/cddis.2012.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. B., Newsome D., Wang Y., Boucher D. M., Eustace B., Gu Y., Hare B., Johnson M. A., Milton S., Murphy C. E., Takemoto D., Tolman C., Wood M., Charlton P., Charrier J.-D., Furey B., Golec J., Reaper P. M., Pollard J. R. Oncotarget. 2014;5:5674–5685. doi: 10.18632/oncotarget.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsanti P. A., Aversa R. J., Jin X., Pan Y., Lu Y., Elling R., Jain R., Knapp M., Lan J., Lin X., Rudewicz P., Sim J., Taricani L., Thomas G., Xiao L., Yue Q. ACS Med. Chem. Lett. 2015;6:37–41. doi: 10.1021/ml500353p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote K. M., Blades K., Cronin A., Fillery S., Guichard S. S., Hassall L., Hickson I., Jacq X., Jewsbury P. J., McGuire T. M., Nissink J. W. M., Odedra R., Page K., Perkins P., Suleman A., Tam K., Thommes P., Broadhurst R., Wood C. J. Med. Chem. 2013;56:2125–2138. doi: 10.1021/jm301859s. [DOI] [PubMed] [Google Scholar]
- Barsanti P. A., Pan Y., Lu Y., Jain R., Cox M., Aversa R. J., Dillon M. P., Elling R., Hu C., Jin X., Knapp M., Lan J., Ramurthy S., Rudewicz P., Setti L., Subramanian S., Mathur M., Taricani L., Thomas G., Xiao L., Yue Q. ACS Med. Chem. Lett. 2015;6:42–46. doi: 10.1021/ml500352s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel V., Lahusen T., Leethanakul C., Igishi T., Kremer M., Quintanilla-Martinez L., Ensley J. F., Sausville E. A., Gutkind J. S., Senderowicz A. M. Clin. Cancer Res. 2002;8:3549–3560. [PubMed] [Google Scholar]
- Mizuno K., Noda K., Ueda Y., Hanaki H., Saido T. C., Ikuta T., Kuroki T., Tamaoki T., Hirai S., Osada S. FEBS Lett. 1995;359:259–261. doi: 10.1016/0014-5793(95)00042-8. [DOI] [PubMed] [Google Scholar]
- Ma C. X., Janetka J. W., Piwnica-Worms H. Trends Mol. Med. 2011;17:88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Bower M. J., McDevitt P. J., Zhao H., Davis S. T., Johanson K. O., Green S. M., Concha N. O., Zhou B.-B. S. J. Biol. Chem. 2002;277:46609–46615. doi: 10.1074/jbc.M201233200. [DOI] [PubMed] [Google Scholar]
- Perez R. P., Lewis L. D., Beelen A. P., Olszanski A. J., Johnston N., Rhodes C. H., Beaulieu B., Ernstoff M. S., Eastman A. Clin. Cancer Res. 2006;12:7079–7085. doi: 10.1158/1078-0432.CCR-06-0197. [DOI] [PubMed] [Google Scholar]
- Signore M., Pelacchi F., di Martino S., Runci D., Biffoni M., Giannetti S., Morgante L., De Majo M., Petricoin E. F., Stancato L., Larocca L. M., De Maria R., Pallini R., Ricci-Vitiani L. Cell Death Dis. 2014;5:e1223. doi: 10.1038/cddis.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Xu L., Lin N., Liu B. BMC Cancer. 2013;13:167. doi: 10.1186/1471-2407-13-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett M. D., Collins I. Trends Pharmacol. Sci. 2011;32:308–316. doi: 10.1016/j.tips.2011.02.014. [DOI] [PubMed] [Google Scholar]
- Kohn E. A., Yoo C. J., Eastman A. Cancer Res. 2003;63:31–35. [PubMed] [Google Scholar]
- Jackson J. R., Gilmartin A., Imburgia C., Winkler J. D., Marshall L. A., Roshak A. Cancer Res. 2000;60:566–572. [PubMed] [Google Scholar]
- Riesterer O., Matsumoto F., Wang L., Pickett J., Molkentine D., Giri U., Milas L., Raju U. Invest. New Drugs. 2011;29:514–522. doi: 10.1007/s10637-009-9361-2. [DOI] [PubMed] [Google Scholar]
- Matthews D. J., Yakes F. M., Chen J., Tadano M., Bornheim L., Clary D. O., Tai A., Wagner J. M., Miller N., Kim Y. D., Robertson S., Murray L., Karnitz L. M. Cell Cycle. 2007;6:104–110. doi: 10.4161/cc.6.1.3699. [DOI] [PubMed] [Google Scholar]
- Tong Y., Claiborne A., Pyzytulinska M., Tao Z.-F., Stewart K. D., Kovar P., Chen Z., Credo R. B., Guan R., Merta P. J., Zhang H., Bouska J., Everitt E. A., Murry B. P., Hickman D., Stratton T. J., Wu J., Rosenberg S. H., Sham H. L., Sowin T. J., Lin N.-H. Bioorg. Med. Chem. Lett. 2007;17:3618–3623. doi: 10.1016/j.bmcl.2007.04.055. [DOI] [PubMed] [Google Scholar]
- Tong Y., Przytulinska M., Tao Z.-F., Bouska J., Stewart K. D., Park C., Li G., Claiborne A., Kovar P., Chen Z., Merta P. J., Bui M.-H., Olson A., Osterling D., Zhang H., Sham H. L., Rosenberg S. H., Sowin T. J., Lin N.-H. Bioorg. Med. Chem. Lett. 2007;17:5665–5670. doi: 10.1016/j.bmcl.2007.07.069. [DOI] [PubMed] [Google Scholar]
- Tao Z.-F., Chen Z., Bui M.-H., Kovar P., Johnson E., Bouska J., Zhang H., Rosenberg S., Sowin T., Lin N.-H. Bioorg. Med. Chem. Lett. 2007;17:6593–6601. doi: 10.1016/j.bmcl.2007.09.063. [DOI] [PubMed] [Google Scholar]
- Tse A. N., Rendahl K. G., Sheikh T., Cheema H., Aardalen K., Embry M., Ma S., Moler E. J., Ni Z. J., Lopes de Menezes D. E., Hibner B., Gesner T. G., Schwartz G. K. Clin. Cancer Res. 2007;13:591–602. doi: 10.1158/1078-0432.CCR-06-1424. [DOI] [PubMed] [Google Scholar]
- Tao Y., Leteur C., Yang C., Zhang P., Castedo M., Pierré A., Golsteyn R. M., Bourhis J., Kroemer G., Deutsch E. Cell Cycle. 2009;8:1196–1205. doi: 10.4161/cc.8.8.8203. [DOI] [PubMed] [Google Scholar]
- Oza V., Ashwell S., Almeida L., Brassil P., Breed J., Deng C., Gero T., Grondine M., Horn C., Ioannidis S., Liu D., Lyne P., Newcombe N., Pass M., Read J., Ready S., Rowsell S., Su M., Toader D., Vasbinder M., Yu D., Yu Y., Xue Y., Zabludoff S., Janetka J. J. Med. Chem. 2012;55:5130–5142. doi: 10.1021/jm300025r. [DOI] [PubMed] [Google Scholar]
- Blasina A., Hallin J., Chen E., Arango M. E., Kraynov E., Register J., Grant S., Ninkovic S., Chen P., Nichols T., O'Connor P., Anderes K. Mol. Cancer Ther. 2008;7:2394–2404. doi: 10.1158/1535-7163.MCT-07-2391. [DOI] [PubMed] [Google Scholar]
- Parsels L. A., Morgan M. A., Tanska D. M., Parsels J. D., Palmer B. D., Booth R. J., Denny W. A., Canman C. E., Kraker A. J., Lawrence T. S., Maybaum J. Mol. Cancer Ther. 2009;8:45–54. doi: 10.1158/1535-7163.MCT-08-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton M. I., Eve P. D., Hayes A., Valenti M., De Haven Brandon A., Box G., Boxall K. J., Aherne G. W., Eccles S. A., Raynaud F. I., Williams D. H., Reader J. C., Collins I., Garrett M. D. Mol. Cancer Ther. 2010;9:89–100. doi: 10.1158/1535-7163.MCT-09-0938. [DOI] [PubMed] [Google Scholar]
- Lainchbury M., Matthews T. P., McHardy T., Boxall K. J., Walton M. I., Eve P. D., Hayes A., Valenti M. R., de Haven Brandon A. K., Box G., Aherne G. W., Reader J. C., Raynaud F. I., Eccles S. A., Garrett M. D., Collins I. J. Med. Chem. 2012;55:10229–10240. doi: 10.1021/jm3012933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton M. I., Eve P. D., Hayes A., Valenti M. R., De Haven Brandon A. K., Box G., Hallsworth A., Smith E. L., Boxall K. J., Lainchbury M., Matthews T. P., Jamin Y., Robinson S. P., Aherne G. W., Reader J. C., Chesler L., Raynaud F. I., Eccles S. A., Collins I., Garrett M. D. Clin. Cancer Res. 2012;18:5650–5661. doi: 10.1158/1078-0432.CCR-12-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y., Chen S., Kmieciak M., Zhou L., Lin H., Pei X. Y., Grant S. Mol. Cancer Ther. 2013;12:878–889. doi: 10.1158/1535-7163.MCT-12-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montano R., Thompson R., Chung I., Hou H., Khan N., Eastman A. BMC Cancer. 2013;13:604. doi: 10.1186/1471-2407-13-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke C. G., Parsels L. A., Qian Y., Zhang Q., Karnak D., Robertson J. R., Tanska D. M., Wei D., Davis M. A., Parsels J. D., Zhao L., Greenson J. K., Lawrence T. S., Maybaum J., Morgan M. A. Clin. Cancer Res. 2013;19:4412–4421. doi: 10.1158/1078-0432.CCR-12-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.-Z., Fei H.-R., Cui Y.-J., Sun Y.-K., Li Z.-M., Wang X.-Y., Yang X.-Y., Zhang J.-G., Sun B.-L. Apoptosis. 2014;19:1389–1398. doi: 10.1007/s10495-014-1010-3. [DOI] [PubMed] [Google Scholar]
- King C., Diaz H., Barnard D., Barda D., Clawson D., Blosser W., Cox K., Guo S., Marshall M. Invest. New Drugs. 2013;32:213–226. doi: 10.1007/s10637-013-0036-7. [DOI] [PubMed] [Google Scholar]
- Carrassa L., Damia G. Cell Cycle. 2014;10:2121–2128. doi: 10.4161/cc.10.13.16398. [DOI] [PubMed] [Google Scholar]
- Forment J. V., Blasius M., Guerini I., Jackson S. P. PLoS One. 2011;6:e23517–e23518. doi: 10.1371/journal.pone.0023517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C., Diaz H. B., McNeely S., Barnard D., Dempsey J., Blosser W., Beckmann R., Barda D., Marshall M. S. Mol. Cancer Ther. 2015;14:2004–2013. doi: 10.1158/1535-7163.MCT-14-1037. [DOI] [PubMed] [Google Scholar]
- Bryant C., Scriven K., Massey A. J. Mol. Cancer. 2014;13:147. doi: 10.1186/1476-4598-13-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A. J., Stokes S., Browne H., Foloppe N., Fiumana A., Scrace S., Fallowfield M., Bedford S., Webb P., Baker L., Christie M., Drysdale M. J., Wood M. Oncotarget. 2015;6:35797–35812. doi: 10.18632/oncotarget.5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf A., Cheng Z. K., Wong B., Reynolds M., Angelaud R., Girotti J., Deese A., Gu C., Gazzard L. Org. Process Res. Dev. 2015;19:661–672. [Google Scholar]
- Sharma V., Tepe J. J. Bioorg. Med. Chem. Lett. 2004;14:4319–4321. doi: 10.1016/j.bmcl.2004.05.079. [DOI] [PubMed] [Google Scholar]
- Tasdemir D., Mallon R., Greenstein M., Feldberg L. R., Kim S. C., Collins K., Wojciechowicz D., Mangalindan G. C., Concepción G. P., Harper M. K., Ireland C. M. J. Med. Chem. 2002;45:529–532. doi: 10.1021/jm0102856. [DOI] [PubMed] [Google Scholar]
- Saleem R. S. Z., Lansdell T. A., Tepe J. J. Bioorg. Med. Chem. 2012;20:1475–1481. doi: 10.1016/j.bmc.2011.12.054. [DOI] [PubMed] [Google Scholar]
- Nguyen T. N. T., Saleem R. S. Z., Luderer M. J., Hovde S., Henry R. W., Tepe J. J. ACS Chem. Biol. 2012;7:172–184. doi: 10.1021/cb200320c. [DOI] [PubMed] [Google Scholar]
- Arienti K. L., Brunmark A., Axe F. U., McClure K., Lee A., Blevitt J., Neff D. K., Huang L., Crawford S., Pandit C. R., Karlsson L., Breitenbucher J. G. J. Med. Chem. 2005;48:1873–1885. doi: 10.1021/jm0495935. [DOI] [PubMed] [Google Scholar]
- Jobson A. G., Cardellina J. H., Scudiero D., Kondapaka S., Zhang H., Kim H., Shoemaker R., Pommier Y. Mol. Pharmacol. 2007;72:876–884. doi: 10.1124/mol.107.035832. [DOI] [PubMed] [Google Scholar]
- Lountos G. T., Tropea J. E., Zhang D., Jobson A. G., Pommier Y., Shoemaker R. H., Waugh D. S. Protein Sci. 2009;18:92–100. doi: 10.1002/pro.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong H.-Q., Hong Y. B., Kim J. S., Lee H.-S., Yi Y. W., Kim Y. J., Wang A., Zhao W., Cho C. H., Seong Y.-S., Bae I. J. Cell. Mol. Med. 2013;17:1261–1270. doi: 10.1111/jcmm.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Hong Y. B., Cho C. H., Seong Y.-S., Bae I. Pancreas. 2012;41:804–805. doi: 10.1097/MPA.0b013e31823f3fcb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobson A. G., Lountos G. T., Lorenzi P. L., Llamas J., Connelly J., Cerna D., Tropea J. E., Onda A., Zoppoli G., Kondapaka S., Zhang G., Caplen N. J., Cardellina J. H., Yoo S. S., Monks A., Self C., Waugh D. S., Shoemaker R. H., Pommier Y. J. Pharmacol. Exp. Ther. 2009;331:816–826. doi: 10.1124/jpet.109.154997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G., Yan S., Chen H., Rong F., Hong Z., Wu J. Z. Bioorg. Med. Chem. Lett. 2007;17:172–175. doi: 10.1016/j.bmcl.2006.09.067. [DOI] [PubMed] [Google Scholar]
- Carlessi L., Buscemi G., Larson G., Hong Z., Wu J. Z., Delia D. Mol. Cancer Ther. 2007;6:935–944. doi: 10.1158/1535-7163.MCT-06-0567. [DOI] [PubMed] [Google Scholar]
- Hilton S., Naud S., Caldwell J. J., Boxall K., Burns S., Anderson V. E., Antoni L., Allen C. E., Pearl L. H., Oliver A. W., Aherne G. W., Garrett M. D., Collins I. Bioorg. Med. Chem. 2010;18:707–718. doi: 10.1016/j.bmc.2009.11.058. [DOI] [PubMed] [Google Scholar]
- Caldwell J. J., Welsh E. J., Matijssen C., Anderson V. E., Antoni L., Boxall K., Urban F., Hayes A., Raynaud F. I., Rigoreau L. J. M., Raynham T., Aherne G. W., Pearl L. H., Oliver A. W., Garrett M. D., Collins I. J. Med. Chem. 2011;54:580–590. doi: 10.1021/jm101150b. [DOI] [PubMed] [Google Scholar]
- Anderson V. E., Walton M. I., Eve P. D., Boxall K. J., Antoni L., Caldwell J. J., Aherne W., Pearl L. H., Oliver A. W., Collins I., Garrett M. D. Cancer Res. 2011;71:463–472. doi: 10.1158/0008-5472.CAN-10-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panek R. L., Lu G. H., Klutchko S. R., Batley B. L., Dahring T. K., Hamby J. M., Hallak H., Doherty A. M., Keiser J. A. J. Pharmacol. Exp. Ther. 1997;283:1433–1444. [PubMed] [Google Scholar]
- Wang Y., Li J., Booher R. N., Kraker A., Lawrence T., Leopold W. R., Sun Y. Cancer Res. 2001;61:8211–8217. [PubMed] [Google Scholar]
- Hashimoto O., Shinkawa M., Torimura T., Nakamura T., Selvendiran K., Sakamoto M., Koga H., Ueno T., Sata M. BMC Cancer. 2006;6:292. doi: 10.1186/1471-2407-6-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PosthumaDeBoer J., Würdinger T., Graat H. C., van Beusechem V. W., Helder M. N., van Royen B. J., Kaspers G. J. BMC Cancer. 2011;11:156. doi: 10.1186/1471-2407-11-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer B. D., Smaill J. B., Rewcastle G. W., Dobrusin E. M., Kraker A., Moore C. W., Steinkampf R. W., Denny W. A. Bioorg. Med. Chem. Lett. 2005;15:1931–1935. doi: 10.1016/j.bmcl.2005.01.079. [DOI] [PubMed] [Google Scholar]
- Palmer B. D., Thompson A. M., Booth R. J., Dobrusin E. M., Kraker A. J., Lee H. H., Lunney E. A., Mitchell L. H., Ortwine D. F., Smaill J. B., Swan L. M., Denny W. A. J. Med. Chem. 2006;49:4896–4911. doi: 10.1021/jm0512591. [DOI] [PubMed] [Google Scholar]
- Smaill J. B., Baker E. N., Booth R. J., Bridges A. J., Dickson J. M., Dobrusin E. M., Ivanovic I., Kraker A. J., Lee H. H., Lunney E. A., Ortwine D. F., Palmer B. D., Quin III J., Squire C. J., Thompson A. M., Denny W. A. Eur. J. Med. Chem. 2008;43:1276–1296. doi: 10.1016/j.ejmech.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Hirai H., Iwasawa Y., Okada M., Arai T., Nishibata T., Kobayashi M., Kimura T., Kaneko N., Ohtani J., Yamanaka K., Itadani H., Takahashi-Suzuki I., Fukasawa K., Oki H., Nambu T., Jiang J., Sakai T., Arakawa H., Sakamoto T., Sagara T., Yoshizumi T., Mizuarai S., Kotani H. Mol. Cancer Ther. 2009;8:2992–3000. doi: 10.1158/1535-7163.MCT-09-0463. [DOI] [PubMed] [Google Scholar]
- Sarcar B., Kahali S., Prabhu A. H., Shumway S. D., Xu Y., Demuth T., Chinnaiyan P. Mol. Cancer Ther. 2011;10:2405–2414. doi: 10.1158/1535-7163.MCT-11-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajeshkumar N. V., De Oliveira E., Ottenhof N., Watters J., Brooks D., Demuth T., Shumway S. D., Mizuarai S., Hirai H., Maitra A., Hidalgo M. Clin. Cancer Res. 2011;17:2799–2806. doi: 10.1158/1078-0432.CCR-10-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai H., Arai T., Okada M., Nishibata T., Kobayashi M., Sakai N., Imagaki K., Ohtani J., Sakai T., Yoshizumi T., Mizuarai S., Iwasawa Y., Kotani H. Cancer Biol. Ther. 2014;9:514–522. doi: 10.4161/cbt.9.7.11115. [DOI] [PubMed] [Google Scholar]
- Kreahling J. M., Gemmer J. Y., Reed D., Letson D., Bui M., Altiok S. Mol. Cancer Ther. 2012;11:174–182. doi: 10.1158/1535-7163.MCT-11-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorny J. L., Calligaris D., Gupta S. K., Iyekegbe D. O., Mueller D., Bakken K. K., Carlson B. L., Schroeder M. A., Evans D. L., Lou Z., Decker P. A., Eckel-Passow J. E., Pucci V., Ma B., Shumway S. D., Elmquist W. F., Agar N. Y. R., Sarkaria J. N. Clin. Cancer Res. 2015;21:1916–1924. doi: 10.1158/1078-0432.CCR-14-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrassa L., Chilà R., Lupi M., Ricci F., Celenza C., Mazzoletti M., Broggini M., Damia G. Cell Cycle. 2012;11:2507–2517. doi: 10.4161/cc.20899. [DOI] [PubMed] [Google Scholar]
- Russell M. R., Levin K., Rader J., Belcastro L., Li Y., Martinez D., Pawel B., Shumway S. D., Maris J. M., Cole K. A. Cancer Res. 2013;73:776–784. doi: 10.1158/0008-5472.CAN-12-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restelli V., Chilà R., Lupi M., Rinaldi A., Kwee I., Bertoni F., Damia G., Carrassa L. Oncotarget. 2015;6:37229–37240. doi: 10.18632/oncotarget.5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilà R., Basana A., Lupi M., Guffanti F., Gaudio E., Rinaldi A., Cascione L., Restelli V., Tarantelli C., Bertoni F., Damia G., Carrassa L. Oncotarget. 2015;6:3394–3408. doi: 10.18632/oncotarget.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y., Torrent M., Florjancic A. S., Bromberg K. D., Buchanan F. G., Ferguson D. C., Johnson E. F., Lasko L. M., Maag D., Merta P. J., Olson A. M., Osterling D. J., Soni N., Shoemaker A. R., Penning T. D. ACS Med. Chem. Lett. 2015;6:58–62. doi: 10.1021/ml5002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-H., Zhang M., Rajapakshe K., Coarfa C., Edwards D., Huang S., Rosen J. M. Stem Cell Rep. 2015;5:378–391. doi: 10.1016/j.stemcr.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Wu Q., McLendon R. E., Hao Y., Shi Q., Hjelmeland A. B., Dewhirst M. W., Bigner D. D., Rich J. N. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Lundholm L., Hååg P., Zong D., Juntti T., Mörk B., Lewensohn R., Viktorsson K. Cell Death Dis. 2013;4:e478–e479. doi: 10.1038/cddis.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-Y., Rhee J. G., Song X., Prochownik E. V., Spitz D. R., Lee Y. J. PLoS One. 2012;7:e50423. doi: 10.1371/journal.pone.0050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Sun M., Li G.-H., Wu Y.-Z., Wang Y., Jin F., Zhang Y.-Y., Yang L., Wang D.-L. Oncol. Rep. 2013;30:1793–1801. doi: 10.3892/or.2013.2614. [DOI] [PubMed] [Google Scholar]
- Gallmeier E., Hermann P. C., Mueller M.-T., Machado J. G., Ziesch A., De Toni E. N., Palagyi A., Eisen C., Ellwart J. W., Rivera J., Rubio-Viqueira B., Hidalgo M., Bunz F., Göke B., Heeschen C. Stem Cells. 2011;29:418–429. doi: 10.1002/stem.595. [DOI] [PubMed] [Google Scholar]
- Venkatesha V. A., Parsels L. A., Parsels J. D., Zhao L., Zabludoff S. D., Simeone D. M., Maybaum J., Lawrence T. S., Morgan M. A. Neoplasia. 2012;14:519–525. doi: 10.1593/neo.12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartucci M., Svensson S., Romania P., Dattilo R., Patrizii M., Signore M., Navarra S., Lotti F., Biffoni M., Pilozzi E., Duranti E., Martinelli S., Rinaldo C., Zeuner A., Agrave M. M.-S., Eramo A., De Maria R. Cell Death Differ. 2011;19:768–778. doi: 10.1038/cdd.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier C., Demur C., Grimal F., Jullien D., Manenti S., Ducommun B. Cancer Biol. Ther. 2012;13:307–313. doi: 10.4161/cbt.13.5.19074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulou P. A., Christodoulou M. S., Silvani A., Herold-Mende C., Passarella D. Drug Discovery Today. 2014;19:1547–1562. doi: 10.1016/j.drudis.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Sabisz M., Skladanowski A. Cell Cycle. 2014;8:3208–3217. doi: 10.4161/cc.8.19.9758. [DOI] [PubMed] [Google Scholar]
- Carruthers R., Ahmed S. U., Strathdee K., Gomez-Roman N., Amoah-Buahin E., Watts C., Chalmers A. J. Mol. Oncol. 2015;9:192–203. doi: 10.1016/j.molonc.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovska A., Kim S., Salamone R. J., Walker J. R., Maira S.-M., García-Echeverría C., Schultz P. G., Reddy V. A. Proc. Natl. Acad. Sci. U. S. A. 2009;106:268–273. doi: 10.1073/pnas.0810956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Shao R., Li F., Monteiro M., Liu J.-P., Xu Z. P., Gu W. Clin. Exp. Pharmacol. Physiol. 2015;42:1317–1326. doi: 10.1111/1440-1681.12493. [DOI] [PubMed] [Google Scholar]
- Kim M.-J., Koo J.-E., Han G.-Y., Kim B., Lee Y.-S., Ahn C., Kim C.-W. J. Korean Med. Sci. 2016;31:360–370. doi: 10.3346/jkms.2016.31.3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W.-J., Long L.-M., Yang N., Zhang Q.-Q., Ji W.-J., Zhao J.-H., Qin Z.-H., Wang Z., Chen G., Liang Z.-Q. Acta Pharmacol. Sin. 2013;34:681–690. doi: 10.1038/aps.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambardzumyan D., Squartro M., Holland E. C. Cancer Cell. 2006;10:454–456. doi: 10.1016/j.ccr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Yang Z. X., Sun Y. H., He J. G., Cao H., Jiang G. Q. Oncol. Lett. 2015;10:3443–3449. doi: 10.3892/ol.2015.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand G., Maalouf M., Boivin A., Battiston-Montagne P., Beuve M., Levy A., Jalade P., Fournier C., Ardail D., Magné N., Alphonse G., Rodriguez-Lafrasse C. Stem Cell Rev. 2014;10:114–126. doi: 10.1007/s12015-013-9467-y. [DOI] [PubMed] [Google Scholar]
- Fang D. D., Cao J., Jani J. P., Tsaparikos K., Blasina A., Kornmann J., Lira M. E., Wang J., Jirout Z., Bingham J., Zhu Z., Gu Y., Los G., Hostomsky Z., VanArsdale T. Front. Med. 2013;7:462–476. doi: 10.1007/s11684-013-0270-6. [DOI] [PubMed] [Google Scholar]
- Al-Ejeh F., Pajic M., Shi W., Kalimutho M., Miranda M., Nagrial A. M., Chou A., Biankin A. V., Grimmond S. M., for the Australian Pancreatic Cancer Genome Initiative, Brown M. P., Khanna K. K. Clin. Cancer Res. 2014;20:3187–3197. doi: 10.1158/1078-0432.CCR-14-0048. [DOI] [PubMed] [Google Scholar]
- Desmarais J. A., Hoffmann M. J., Bingham G., Gagou M. E., Meuth M., Andrews P. W. Stem Cells. 2012;30:1385–1393. doi: 10.1002/stem.1117. [DOI] [PubMed] [Google Scholar]
- Jagtap J. C., Dawood P., Shah R. D., Chandrika G., Natesh K., Shiras A., Hegde A. S., Ranade D., Shastry P. PLoS One. 2014;9:e88505. doi: 10.1371/journal.pone.0088505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M. A., Parsels L. A., Parsels J. D., Mesiwala A. K., Maybaum J., Lawrence T. S. Cancer Res. 2005;65:6835–6842. doi: 10.1158/0008-5472.CAN-04-2246. [DOI] [PubMed] [Google Scholar]
- Karnitz L. M., Flatten K. S., Wagner J. M., Loegering D., Hackbarth J. S., Arlander S. J. H., Vroman B. T., Thomas M. B., Baek Y.-U., Hopkins K. M., Lieberman H. B., Chen J., Cliby W. A., Kaufmann S. H. Mol. Pharmacol. 2005;68:1636–1644. doi: 10.1124/mol.105.012716. [DOI] [PubMed] [Google Scholar]
- Benada J., Macurek L. Biomolecules. 2015;5:1912–1937. doi: 10.3390/biom5031912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoni L., Sodha N., Collins I., Garrett M. D. Nat. Rev. Cancer. 2007;7:925–936. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]