Abstract
As a significant cause of mortality and morbidity, alcoholic liver disease (ALD) has been widely investigated. However, little is known about the underlying metabolic mechanisms involved in the complicated pathological processes of ALD. The present study used label-free quantitative proteomics and bioinformatics analyses to investigate the differentially expressed proteins (DEPs) and their functions in the livers of alcohol-feed (AF) and control pair-feed (PF) mice. As a result, 87 upregulated DEPs and 133 downregulated DEPs were identified in AF liver tissues compared with PF livers. Gene ontology and Kyoto encyclopedia of genes and genomes bioinformatics analyses demonstrated that the DEPs were significantly enriched in ‘protein binding’, ‘metabolism’, ‘signal conduction’ and ‘immune response’. The expression of several core proteins including thyroid hormone receptor interactor 12 (TRIP12), NADH dehydrogenase (ubiquinone)1 α subcomplex, assembly factor 3 (NDUFAF3) and guanine monophosphate synthetase (GMPS) was validated by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in a larger series of samples. The RT-qPCR results confirmed that TRIP12, NDUFAF3 and GMPS genes were significantly differentially expressed in between the AF and PF samples. These results extend our understanding of the molecular mechanisms underlying the occurrence and development of ALD. The present study indicated that the majority of DEPs serve vital roles in multiple metabolic pathways and this extends our knowledge of the molecular mechanisms involved in the occurrence and progression of ALD.
Keywords: alcoholic liver disease, alcohol-feed mice, control pair-feed mice, differentially expressed proteins, gene ontology analysis, kyoto encyclopedia of genes and genomes analysis
Introduction
Alcoholic liver disease (ALD) has been one of the leading causes of cirrhosis and liver-related mortality and morbidity worldwide for many years (1,2). As the ultimate outcome of heavy acute and/or chronic alcohol drinking, ALD can lead to steatosis, steatohepatitis, alcoholic fibrosis, cirrhosis and even hepatocellular carcinoma in certain individuals (3,4). Many factors are thought to contribute to the development and progression of ALD, particularly the toxicity of alcohol and its metabolites, generation of reactive oxygen species during alcohol metabolism, and endotoxin derived from the gut (5,6). Although the factors that link ethanol to the occurrence and development of liver injury have been widely investigated, the underlying metabolic mechanisms involved in the complicated pathological processes remain to be elucidated. Thus, comprehensive research into the molecular characteristics and mechanism of ALD is urgently required, to identify effective treatment methods and improve the outcome of patients with ALD.
Label-free quantitative proteomics is a method that aims to determine the relative amount of proteins, which is a novel tool used for biomarker identification in various diseases, due to the critical importance of protein-level measurements (7–9). In the present study, label-free quantitative proteomics was used to detect ALD in a rodent model, to explore the underlying pathophysiological mechanism. It is hoped that the results of the present study can improve knowledge of the molecular pathogenesis of ALD and aid the search for biomarkers for early diagnosis and treatment.
Materials and methods
Animal model
All procedures of animal care and treatment were approved by the Institutional Animal Care and Use Committee of Fudan University (Shanghai, China). Experiments were performed on male specific-pathogen-free C57BL/6J mice (Laboratory Animal Center, Fudan University, Shanghai, China) of 6 weeks old weighing 18–20 g. Environmental conditions were strictly controlled (temperature 23±2°C, relative humidity 50–70% and 12 h light/dark cycle) with ad libitum access to food and water. Following 1 week acclimation, all mice were randomly divided into two groups based on diets as follows; the alcohol-feed (AF) and control pair-feed (PF) groups.
Mice were administered 4% Lieber-Decarli ethanol liquid diet (TP 4030B; Trophic Animal Feed High-Tech Co., Ltd. Nantong, China) and control diet (TP 4030C; Trophic Animal Feed High-Tech Co., Ltd.) respectively for 4 weeks with diets changed daily at 5 pm (10). On the 29th day at 9:00 am, mice were administered by gavage with a single dose of maltose dextrin (control, 9 g maltose dextrin per kg of body weight) or ethanol diet (5 g ethanol diet per kg of body weight), respectively. At 9 h after the binge, the mice were anesthetized by an intraperitoneal injection of 1% pentobarbital sodium (80 mg/kg body weight) and the serum and liver were collected (11). There were 10 mice in each group. All the non-alcoholic and alcoholic diets were provided throughout the sample collection period following the binge. A dim red light was used to collect tissues in dark conditions.
Histopathological examination
The liver tissues were resected and divided to three parts and processed as follows: Paraffin embedded and stained with hematoxylin/eosin (H&E); frozen sections (10 µm) and stained with Oil Red O for 10–15 min at 37°C; and immediately frozen in liquid nitrogen and stored at −80°C for further analysis. All sections were analyzed by light microscopy by at least two independent researchers.
Liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) analysis
There were three samples in every group and every sample (5 ul) was mixed from three mouse liver tissues. Each sample was resuspended in buffer A (0.1% formic acid; FA). Separations were performed with an UltiMate 3000 HPLC system (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and Q-Extractive HF HF-X Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, Inc.). The peptides were subjected to a C18 trap column (3 µm, 0.1×20 mm) at a flow rate of 0.6 µl/min. Peptides were desalted online and loaded onto a C18 column (1.9, 150×120 mm) using a gradient from 6–95% buffer B (0.08% FA and 80% acetonitrile) for 90 min. The mass spectrometer was operated in positive mode using a data-dependent acquisition method.
A full MS scan (300–1,400 m/z) was acquired in the mass spectrometer with the resolution set to a value of 120,000.
Identification of differently expressed proteins (DEPs)
DEPs were identified according to the following procedures. Only the FC value >1.5 or <0.667 were entered in the following analyses. Then a random variance model t test was performed using SPSS version 20.0 (IBM Corp., Armonk, NY, USA) to filter the DEPs as it can effectively increase the statistical effects in a small number of samples. Only proteins with P<0.05 and false discovery rate (FDR) <0.05 were considered to be significantly differentially expressed, as previously reported (12,13). A clustering analysis map was built using Cluster 3.0 software version 2.3 (Bio-Fly Bioscience; www.bangfeibio.com/company) to identify DEPs efficiently with similar expression mode.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The relative mRNA level in the liver tissue (20 mg), including thyroid hormone receptor interactor 12 (TRIP12), NADH dehydrogenase (ubiquinone) 1 α subcomplex, assembly factor 3 (NDUFAF3) and guanine monophosphate synthetase (GMPS), in the AF and PF groups were measured by RT-qPCR. Briefly, the total RNA was isolated using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol and then RT was performed using GeneAmp RNA PCR kits (Thermo Fisher Scientific, Inc.) with the following parameters: 15 min at 37°C, 5 sec at 85°C, hold at 4°C. Primer sequences are listed in Table I. qPCR was performed on an ABI 7500 real-time PCR thermocycler using SYBR-Green PCR Master Mix kits (Thermo Fisher Scientific, Inc.) with the following PCR cycling parameters: 1 cycle of 30 sec at 95°C; 40 cycles of 5 sec at 95°C; 30 sec at 55°C; and, 30 sec at 72°C followed by a melting curve analysis. The expression levels of target genes were calculated from duplicate samples following normalization against the housekeeping gene GAPDH. The 2−ΔΔCq method was used to calculate the expression of these proteins (14).
Table I.
Primer sequences for the targeted proteins.
| Gene name | Forward (5′>3′) | Reverse (5′>3′) |
|---|---|---|
| TRIP12 | GTCTGTGACGCAGGACCTTG | TGTGAACTGGCTTAGCTGTCCT |
| NDUFAF3 | GTGGTCCAGTGGAACGTGG | CTCCTGTCACTCGACCTTCG |
| KDSR | GCTCCTCTACATGGTGTCGC | CTCAATAGCAATGCACTTCCCA |
| SURF6 | CTGAACGACAGAGGAGCACAT | TTGGGCCTAGAAGAGGTAGGA |
| KIF5B | GCGGAGTGCAACATCAAAGTG | CATAAGGCTTGGACGCGATCA |
| GMPS | GATGCAGTGGGAACTTTACTGT | AGCACGATTTAGCAAAGCTGT |
| DDAH2 | GCAGGTAGTAGAACGGAAGATCC | CTGGTGACAATGGAAGGCTCA |
| SSRP1 | CAGAGACATTGGAGTTCAACGA | GCCCGTCTTGCTGTTCTTAAAG |
| HIST1HIC | AACCCCAGGCTAAGAAGGC | TGGCTTTACGGCTTTAGACGC |
| H1F0 | CACGGACCACCCCAAGTATTC | ACCCACCTTGTAGTGGCTCT |
TRIP12, thyroid hormone receptor interactor 12; NDUFAF3, NADH dehydrogenase (ubiquinone) 1 α subcomplex, assembly factor 3; KDSR, 3-ketodihydrosphingosine reductase; SURF6, surfeit gene 6; KIF5B, kinesin family member 5B; GMPS, guanine monophosphate synthetase; DDAH2, dimethylarginine dimethylaminohydrolase; SSRP1, structure specific recognition protein 1; HIST1HIC, histone cluster 1 H1c; H1F0, H1 histone family member 0.
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses
GO and KEGG analyses were performed to investigate significantly enriched function and signaling pathways of DEPs based on the GO (geneontology.org/) and KEGG databases (genome.jp/kegg/) as previously published (15,16). Specifically, the GO and KEGG categories were classified using Fisher's exact test and χ2 test and FDR calculated for multiple testing corrections. Only GOs and signaling pathways with P<0.05 and FDR <0.05 were considered statistically significant. A GO map and a path-net were established to further summarize the fundamental functional links among the significant GOs and KEGG pathways, respectively.
Cluster of orthologous groups of proteins analysis
The DEPs were compared based on the clusters of orthologous groups (COGs) database (ncbi.nlm.nih.gov/COG/), to categorize the orthologous and paralogs of protein. COGs category assignment was performed using the local alignment tool BLASTP with an e-value cutoff of <104. Only the categories that met a threshold of P<0.05 and FDR<0.05 were considered statistically significant.
Results
Hepatic steatosis and histopathological examination
Compared with PF group, the AF mice exhibited marked hepatic steatosis, even steatohepatitis, as evaluated by H&E and Oil Red O (Fig. 1). Histopathological examination revealed the accumulation of lipid vacuoles and lipid droplets in the AF mice. This demonstrated the animal model of the present study was successful.
Figure 1.

Effect of alcohol on liver histopathological alterations. Liver sections were stained with H&E and Oil Red O to visualize lipid droplets (in red). Representative images (magnification, ×200) of each group are presented. AF, alcohol-feed; PF, control pair-feed; H&E, hematoxylin/eosin.
DEPs between the AF and PF mice
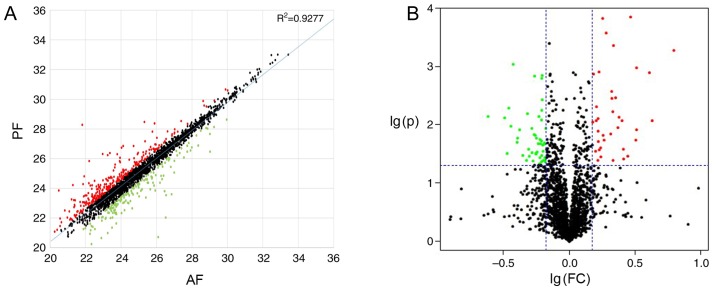
A total of 87 upregulated DEPs and 133 downregulated DEPs were identified in the AF group compared with the PF group. A scatter plot map and a volcano plot were established to demonstrate all differentially expressed proteins (Fig. 2). As Fig. 3 presents, marked differences (>40-fold changes) were detected in the expression levels of certain proteins, which may have potential to be used as biomarkers in the diagnosis, assessment and monitoring of ALD. Fig. 4 indicated that the relative mRNA level of TRIP12, NDUFAF3 and GMPS were significantly different in AF and PF. The expression of all 220 DEPs were listed in a clustering map, to identify proteins with similar expression modes between AF and PF groups (Fig. 5).
Figure 2.
Identification of DEPs between AF and PF groups. (A) The scatter plot for global protein expression between the two groups; red dots represent correlation coefficients (>1.5 times) of AF/PF and green dots represent that of PF/AF. (B) Volcano plots represented all the genes in AF and PF groups according to P-value and fold changes; black dots represent genes that were not differentially expressed, while red dots and green dots represent DEPs. DEPs, differentially expressed proteins; AF, alcohol-feed mice; PF, control pair-feed mice.
Figure 3.
The 10 proteins most strongly upregulated and downregulated in alcoholic liver disease. The ordinate value represents the relative abundance of the proteins (P<0.05, false discovery rate <0.05). AF, alcohol-feed mice; PF, control pair-feed mice.
Figure 4.

Relative expression of the mRNA level of the selected proteins. *P<0.05, **P<0.01 and ***P<0.001, AF vs. PF. AF, alcohol-feed mice; PF, control pair-feed mice; TRIP12, thyroid hormone receptor interactor 12; NDUFAF3, NADH dehydrogenase (ubiquinone) 1 α subcomplex, assembly factor 3; GMPS, guanine monophosphate synthetase.
Figure 5.

Clustering analysis of DEPs between the two groups. Red represents upregulation of DEPs and green represents downregulation of DEPs. DEPs, differentially expressed proteins; AF, alcohol-feed mice; PF, control pair-feed mice.
GO analysis
The significantly enriched GO terms (P<0.01 only) are summarized in Table II. The significant GOs were categorized according to molecular function, biological processes and cellular component. When the molecular function of these DEPs was analyzed, the majority of DEPs were associated with binding functions, including protein binding, ion binding and nucleic acid binding. In the biological processes analysis of GO, the majority of DEPs were associated with ‘metabolic process’, including the ‘macromolecule metabolic process’, ‘primary metabolic process’ and ‘cellular metabolic process’. In addition, when the DEPs were analyzed for cellular components, they were enriched in mitochondria and ribosomes (data not shown). A hierarchical tree of the GO terms was established according to their associations (Figs. 6 and 7).
Table II.
Enriched GO terms in biological process (P<0.01).
| GO ID | Description | Input number | Input all | P-value |
|---|---|---|---|---|
| GO:0010467 | Gene expression | 127 | 718 | 0.001414742 |
| GO:0090304 | Nucleic acid metabolic process | 96 | 718 | 0.000287556 |
| GO:0016070 | RNA metabolic process | 85 | 718 | 0.000248985 |
| GO:0003677 | DNA binding | 46 | 718 | 0.002166795 |
| GO:0007005 | Mitochondrion organization | 46 | 718 | 0.002575995 |
| GO:0006351 | Transcription, DNA-templated | 37 | 718 | 0.001753421 |
| GO:0032774 | RNA biosynthetic process | 37 | 718 | 0.001753421 |
| GO:0097659 | Nucleic acid-templated transcription | 37 | 718 | 0.001753421 |
| GO:0000313 | Organellar ribosome | 22 | 718 | 7.62E-08 |
| GO:0005761 | Mitochondrial ribosome | 22 | 718 | 7.62E-08 |
| GO:0045321 | Leukocyte activation | 16 | 718 | 0.001056161 |
| GO:0044801 | Single-organism membrane fusion | 11 | 718 | 0.001415192 |
| GO:0048284 | Organelle fusion | 11 | 718 | 0.002478183 |
| GO:0000315 | Organellar large ribosomal subunit | 10 | 718 | 0.000209737 |
| GO:0005762 | Mitochondrial large ribosomal subunit | 10 | 718 | 0.000209737 |
| GO:0000149 | SNARE binding | 9 | 718 | 0.000643314 |
| GO:0005085 | Guanyl-nucleotide exchange factor activity | 9 | 718 | 0.000643314 |
| GO:0000314 | Organellar small ribosomal subunit | 9 | 718 | 0.001423751 |
| GO:0005763 | Mitochondrial small ribosomal subunit | 9 | 718 | 0.001423751 |
| GO:0090174 | Organelle membrane fusion | 8 | 718 | 0.000805555 |
| GO:0031201 | SNARE complex | 8 | 718 | 0.000805555 |
| GO:0005484 | SNAP receptor activity | 7 | 718 | 0.000265346 |
| GO:0006906 | Vesicle fusion | 7 | 718 | 0.002514721 |
| GO:0005088 | Ras guanyl-nucleotide exchange factor activity | 6 | 718 | 0.000988442 |
GO, Gene Ontology; SNARE, SNAP receptor; SNAP, soluble NSF attachment protein.
Figure 6.
First part of the GO map in alcoholic liver disease. GO analysis results (P<0.01, false discovery rate <0.05). Deeper color of the GO terms represents greater significance in regulating the downstream GOs. GO, Gene Ontology.
Figure 7.
Second part of the GO map in alcoholic liver disease. GO, Gene Ontology.
KEGG pathway analysis
As demonstrated in Fig. 8, the KEGG analysis results demonstrated that the DEPs were significantly enriched in ‘adenosine monophosphate activated protein kinase (AMPK) signaling pathway’, ‘Ras signaling pathway’, ‘Notch signaling pathway’, ‘p53 signaling pathway’ and ‘autophagy’ (Fig. 8). These results indicated that ALD was associated with several biological processes, such as dysregulated lipid and glucose metabolism, catabolic processing, cell-cell adhesion, cell amplification. Additionally, these data indicated that the intervention of these pathways may provide ways for molecular targeting therapies of ALD.
Figure 8.

Pathway enrichment analysis. The enrichment bar chart of significant pathways is shown. As the enrichment increases, the corresponding function is more specific. KEGG, Kyoto Encyclopedia of Genes and Genomes; AMPK, adenosine monophosphate activated protein kinase; SNARE, soluble NSF attachment protein receptor.
COGs analysis
According to the COGs analysis, the functions of the DEPs were enriched in ‘information storage and processing’, ‘cellular processes and signaling’ and ‘metabolism’ (Fig. 9). The results indicated that these processes, particularly the metabolism of lipids, amino acids and nucleic acid, served important roles in the development of ALD.
Figure 9.
Clusters of orthologous groups analysis of differentially expressed proteins, the different colors represent different functions. The value of the ordinate represents the number of proteins enriched in this function.
Discussion
High-throughput quantitative proteomics has emerged as a popular method in the search for disease-associated factors using high-throughput analysis in recent years (12,17–19). However, to the best of the authors' knowledge, the present study is the first to use high-throughput quantitative proteomics analysis in a mouse model of ALD to identify the core proteins directly, rather than mRNAs. The chronic-plus-binge alcohol feeding model in mice, which mimics the drinking pattern of patients with alcoholic hepatitis with a background of drinking for a number of years (chronic) and a history of recent excessive alcohol consumption (binge), is now frequently used worldwide (20–23). In the present study, the ALD model in mice with the liver steatosis and inflammation was successfully established using this method.
Label-free quantitative proteomics quantifies peptides and proteins without the use of stable-isotope labels. It directly uses a peptide's response (intensity) in the mass spectrometer as a quantitative measure and infers quantity indirectly from the number of peptide-to-spectrum matches obtained for each protein (24,25). The present study identified 220 DEGs in the ALD mice. These DEPs may be used as characteristic proteins in the diagnosis and treatment of ALD. Several of these DEPs have been previously described in ALD progression. For example, acyl-CoA dehydrogenase is involved in mitochondrial b-oxidation of fatty acids in the ALD progress (26,27). Glutathione-S-trans-ferases, which are responsible for the detoxification of potentially toxic by-products of ethanol metabolism, including acetaldehyde and reactive oxygen species (ROS), were also identified as downregulated in the AF mice (28). Eaton et al (29) reported that chronic alcohol ingestion leads to downregulation of NADH dehydrogenase (ubiquinone), consistent with the findings of the present study. Decreased amounts of ubiquinone can lead to fat accumulation and increase in free radical damage, leading to liver injury (30,31). In the present study, ATP-citrate synthase was upregulated, which catalyzes the exchange of ADP and ATP across the mitochondrial membrane and supports the increase in ATP generation during ALD (32). The ubiquitin system has been mechanistically implicated in a number of human diseases including cancers and ALD (33). The ubiquitin system protein E3 ubiquitin-protein ligases TRIP12, deltex 3-like E3 ubiquitin ligase, HECT domain E3 ubiquitin protein ligase 3 (HECTD3), HECTD1 and ring finger protein 114, which were identified as differently expressed in the present study, were associated with cellular response to DNA damage stimulus, a ubiquitin-dependent protein catabolic process. This result suggested that the ubiquitin system is worthy of more attention in the exploration of ALD. The ethanol-inducible P450, a member of the cytochrome P450 multifamily, is a major component of the microsomal ethanol oxidizing system, which metabolizes a small portion of ethanol (34,35). Previous studies have reported that hepatic expression of the cytochrome P450 family 2 subfamily E member 1 (CYP2E1) mRNA and/or expression of the CYP2E1 protein are increased in different physiological or pathological conditions, including fasting, a high fat diet, diabetes, obesity or ethanol intoxication (36,37). Accumulation of ROS due to increased hepatic CYP2E1 expression may lead to lipid peroxidation of cellular membranes; antioxidant depletion also causes oxidative stress then damage liver DNA and contributes to hepatic fibrosis (38–40). The current study identified that cytochrome P450, family 2, subfamily c, polypeptide 67 and CYP2E1 were all upregulated in the AF mice.
Using GO and KEGG analyses, the present study identified various gene functions and signaling pathways that were significantly altered in ALD, including lipid metabolism, inorganic ion metabolism, and the AMPK and p53 signaling pathways. These GOs and pathways may serve critical roles in ALD. Notably, it was identified that a number of GO terms were associated with protein and ion metabolism, including ‘iron transport’, ‘protein transport’ and ‘protein localization’. Particularly, 13 of the DEPs were identified to be associated with the ‘iron ion binding’, including the upregulated DEP, transferrin receptor 1, and downregulated DEP, hepcidin, which has been reported to induce the overload of iron in ALD in previous studies (41–43). Additionally, it was identified that transferrin receptor protein 2 was increased and hepcidin-2 decreased in AF mice compared with PF mice. Increasing evidence indicates that AMPK regulates sterol regulatory element binding transcription factor 1, which is involved in the control of glucose, lipid and cholesterol metabolism and participates in the pathogenesis of hepatic steatosis (44). The DEPs involved in the AMPK signaling pathway, including fatty acid synthase, ATP-dependent 6-phosphofructokinase, Acyl-CoA desaturase and phosphatidylinositol 3-kinase regulatory subunit α, may be the core proteins in the progression of ALD.
The system of COGs was designed to accommodate the extremely different evolution rates observed for different genes and to comprise a framework for functional and evolutionary genome analysis (45). According to the COGs analysis, the functions of the DEPs were predominantly enriched in ‘information storage and processing’, ‘cellular processes and signaling’, in addition to ‘metabolism’ including ‘lipid transport and metabolism’ and ‘inorganic ion transport and metabolism’. The results COGs supported GO and pathway analyses, revealing that proteins associated with metabolism had an important role in ALD.
In conclusion, label-free quantitative proteomics using LC-MS/MS was performed to identify DEPs between AF and control PF mice livers. The present study suggested that certain DEPs were involved in the response to alcohol and that the core proteins identified the present study may be useful to predict the development of ALD. It is hoped that the findings will be further validated in other experiments in the near future.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Science and Technology Commission Foundation of Shanghai (grant no. 13DZ1930908).
Availability of data and materials
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JS and YZ conceived and designed the study. YZ, CZ and GC performed the experiments. YZ and CZ wrote the paper. JS, YZ, CZ and GC reviewed and edited the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
All procedures of animal care and treatment were approved by the Institutional Animal Care and Use Committee of Fudan University (Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rehm J, Samokhvalov AV, Shield KD. Global burden of alcoholic liver diseases. J Hepatol. 2013;59:160–168. doi: 10.1016/j.jhep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Allampati S, Mullen KD. Long-term management of alcoholic liver disease. Clin Liver Dis. 2016;20:551–562. doi: 10.1016/j.cld.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Dugum M, McCullough A. Diagnosis and management of alcoholic liver disease. J Clin Transl Hepatol. 2015;3:109–116. doi: 10.14218/JCTH.2015.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoek JB, Cahill A, Pastorino JG. Alcohol and mitochondria: A dysfunctional relationship. Gastroenterology. 2002;122:2049–2063. doi: 10.1053/gast.2002.33613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ceni E, Mello T, Galli A. Pathogenesis of alcoholic liver disease: Role of oxidative metabolism. World J Gastroenterol. 2014;20:17756–17772. doi: 10.3748/wjg.v20.i47.17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szabo G. Gut-liver axis in alcoholic liver disease. Gastroenterology. 2015;148:30–36. doi: 10.1053/j.gastro.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Titz B, Elamin A, Martin F, Schneider T, Dijon S, Ivanov NV, Hoeng J, Peitsch MC. Proteomics for systems toxicology. Comput Struct Biotechnol J. 2014;11:73–90. doi: 10.1016/j.csbj.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzeng SC, Maier CS. Label-free proteomics assisted by affinity enrichment for elucidating the chemical reactivity of the liver mitochondrial proteome toward adduction by the lipid electrophile 4-hydroxy-2-nonenal (HNE) Front Chem. 2016;4:2. doi: 10.3389/fchem.2016.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bantscheff M, Lemeer S, Savitski MM, Kuster B. Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404:939–965. doi: 10.1007/s00216-012-6261-7. [DOI] [PubMed] [Google Scholar]
- 10.Nan YM, Kong LB, Ren WG, Wang RQ, Du JH, Li WC, Zhao SX, Zhang YG, Wu WJ, Di HL, et al. Activation of peroxisome proliferator activated receptor alpha ameliorates ethanol mediated liver fibrosis in mice. Lipids Health Dis. 2013;12:11. doi: 10.1186/1476-511X-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey SM, Andringa KK, Landar A, Darley-Usmar VM. Proteomic approaches to identify and characterize alterations to the mitochondrial proteome in alcoholic liver disease. Methods Mol Biol. 2008;447:369–380. doi: 10.1007/978-1-59745-242-7_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan C, Yan L, Wang L, Jiang W, Zhang Y, Xi J, Jin Y, Chen L, Shi Y, Lin Z, Wang Q. Landscape of expression profiles in esophageal carcinoma by the cancer genome atlas data. Dis Esophagus. 2016;29:920–928. doi: 10.1111/dote.12416. [DOI] [PubMed] [Google Scholar]
- 13.Yan L, Zhan C, Wu J, Wang S. Expression profile analysis of head and neck squamous cell carcinomas using data from the cancer genome atlas. Mol Med Rep. 2016;13:4259–4265. doi: 10.3892/mmr.2016.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Gene Ontology Consortium, corp-author. The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim DH, Lee EM, Do SH, Jeong DH, Jeong KS. Changes of the cytoplasmic proteome in response to alcoholic hepatotoxicity in rats. Int J Mol Sci. 2015;16:18664–18682. doi: 10.3390/ijms160818664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee SJ, Lee DE, Kang JH, Nam MJ, Park JW, Kang BS, Lee DS, Lee HS, Kwon OS. New potential biomarker proteins for alcoholic liver disease identified by a comparative proteomics approach. J Cell Biochem. 2017;118:1189–11200. doi: 10.1002/jcb.25770. [DOI] [PubMed] [Google Scholar]
- 19.Tran M, Yang Z, Liangpunsakul S, Wang L. Metabolomics analysis revealed distinct cyclic changes of metabolites altered by chronic ethanol-plus-binge andshp deficiency. Alcohol Clin Exp Res. 2016;40:2548–2556. doi: 10.1111/acer.13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ki SH, Park O, Zheng M, Morales-Ibanez O, Kolls JK, Bataller R, Gao B. Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: Role of signal transducer and activator of transcription 3. Hepatology. 2010;52:1291–1300. doi: 10.1002/hep.23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertola A, Mathews S, Ki SH, Wang H, Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi G, Runyon BA. Alcoholic hepatitis: A clinician's guide. Clin Liver Dis. 2012;16:371–385. doi: 10.1016/j.cld.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Mathurin P, Lucey MR. Management of alcoholic hepatitis. Drug Ther Bull. 2012;56:S39–S45. doi: 10.1016/S0168-8278(12)60005-1. [DOI] [PubMed] [Google Scholar]
- 24.Patel VJ, Thalassinos K, Slade SE, Connolly JB, Crombie A, Murrell JC, Scrivens JH. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J Proteome Res. 2009;8:3752–3759. doi: 10.1021/pr900080y. [DOI] [PubMed] [Google Scholar]
- 25.Bantscheff M, Lemeer S, Savitski MM, Kuster B. Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404:939–965. doi: 10.1007/s00216-012-6261-7. [DOI] [PubMed] [Google Scholar]
- 26.Sprecher H. New advances in fatty-acid biosynthesis. Nutrition. 1996;12(1 Suppl):S5–S7. doi: 10.1016/0899-9007(96)90009-X. [DOI] [PubMed] [Google Scholar]
- 27.Pace CP, Stankovich MT. Oxidation-reduction properties of short-chain acyl-CoA dehydrogenase: Effects of substrate analogs. Arch Biochem Biophys. 1994;313:261–266. doi: 10.1006/abbi.1994.1386. [DOI] [PubMed] [Google Scholar]
- 28.Ladero JM, Martinez C, Garcia-Martin E, Fernández-Arquero M, López-Alonso G, de la Concha EG, Díaz-Rubio M, Agúndez JA. Polymorphisms of the glutathione S-transferases mu-1 (GSTM1) and theta-1 (GSTT1) and the risk of advanced alcoholic liver disease. Scand J Gastroenterol. 2005;40:348–353. doi: 10.1080/00365520510012109. [DOI] [PubMed] [Google Scholar]
- 29.Eaton S, Record CO, Bartlett K. Multiple biochemical effects in the pathogenesis of alcoholic fatty liver. Eur J Clin Invest. 1997;27:719–722. doi: 10.1046/j.1365-2362.1997.1780727.x. [DOI] [PubMed] [Google Scholar]
- 30.Cunningham CC, Bailey SM. Ethanol consumption and liver mitochondria function. Biol Signals Recept. 2001;10:271–282. doi: 10.1159/000046892. [DOI] [PubMed] [Google Scholar]
- 31.Chacko BK, Srivastava A, Johnson MS, Benavides GA, Chang MJ, Ye Y, Jhala N, Murphy MP, Kalyanaraman B, Darley-Usmar VM. Mitochondria-targeted ubiquinone (MitoQ) decreases ethanol-dependent micro and macro hepatosteatosis. Hepatology. 2011;54:153–163. doi: 10.1002/hep.24377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugimoto K, Takei Y. Pathogenesis of alcoholic liver disease. Hepatol Res. 2016;47:70–79. doi: 10.1111/hepr.12736. [DOI] [PubMed] [Google Scholar]
- 33.Williams JA, Ni H, Ding Y, Ding W. Parkin regulates mitophagy and mitochondrial function to protect against alcohol-induced liver injury and steatosis in mice. Am J Physiol Gastrointest Liver Physiol. 2015;309:G324–G340. doi: 10.1152/ajpgi.00108.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lieber CS, DeCarli LM. Ethanol oxidation by hepatic microsomes: Adaptive increase after ethanol feeding. Science. 1968;162:917–918. doi: 10.1126/science.162.3856.917. [DOI] [PubMed] [Google Scholar]
- 35.Lieber CS, DeCarli LM. Hepatic microsomal ethanol-oxidizing system. In vitro characteristics and adaptive properties in vivo. J Biol Chem. 1970;245:2505–2512. [PubMed] [Google Scholar]
- 36.Robin M, Sauvage I, Grandperret T, Descatoire V, Pessayre D, Fromenty B. Ethanol increases mitochondrial cytochrome P450 2E1 in mouse liver and rat hepatocytes. Febs Lett. 2005;579:6895–6902. doi: 10.1016/j.febslet.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 37.Cieślak A, Kelly I, Trottier J, Verreault M, Wunsch E, Milkiewicz P, Poirier G, Droit A, Barbier O. Selective and sensitive quantification of the cytochrome P450 3A4 protein in human liver homogenates through multiple reaction monitoring mass spectrometry. Proteomics. 2016;16:2827–2837. doi: 10.1002/pmic.201500386. [DOI] [PubMed] [Google Scholar]
- 38.Bradford BU, Kono H, Isayama F, Kosyk O, Wheeler MD, Akiyama TE, Bleye L, Krausz KW, Gonzalez FJ, Koop DR, Rusyn I. Cytochrome P450 CYP2E1, but not nicotinamide adenine dinucleotide phosphate oxidase, is required for ethanol-induced oxidative DNA damage in rodent liver. Hepatology. 2005;41:336–344. doi: 10.1002/hep.20532. [DOI] [PubMed] [Google Scholar]
- 39.Lieber CS. The discovery of the microsomal ethanol oxidizing system and its physiologic and pathologic role. Drug Metab Rev. 2004;36:511–529. doi: 10.1081/DMR-200033441. [DOI] [PubMed] [Google Scholar]
- 40.Nieto N. Stimulation and proliferation of primary rat hepatic stellate cells by cytochrome P450 2E1-derived reactive oxygen species. Hepatology. 2002;35:62–73. doi: 10.1053/jhep.2002.30362. [DOI] [PubMed] [Google Scholar]
- 41.Bridle K, Cheung TK, Murphy T, Walters M, Anderson G, Crawford DG, Fletcher LM. Hepcidin is down-regulated in alcoholic liver injury: Implications for the pathogenesis of alcoholic liver disease. Alcohol Clin Exp Res. 2006;30:106–112. doi: 10.1111/j.1530-0277.2006.00002.x. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki Y, Saito H, Suzuki M, Hosoki Y, Sakurai S, Fujimoto Y, Kohgo Y. Up-regulation of transferrin receptor expression in hepatocytes by habitual alcohol drinking is implicated in hepatic iron overload in alcoholic liver disease. Alcohol Clin Exp Res. 2002;26:26S–31S. doi: 10.1111/j.1530-0277.2002.tb02698.x. [DOI] [PubMed] [Google Scholar]
- 43.Kohgo Y, Ohtake T, Ikuta K, Suzuki Y, Torimoto Y, Kato J. Dysregulation of systemic iron metabolism in alcoholic liver diseases. J Gastroenterol Hepatol. 2008;23(Suppl 1):S78–S81. doi: 10.1111/j.1440-1746.2007.05290.x. [DOI] [PubMed] [Google Scholar]
- 44.Bai T, Yang Y, Yao YL, Sun P, Lian LH, Wu YL, Nan JX. Betulin alleviated ethanol-induced alcoholic liver injury via SIRT1/AMPK signaling pathway. Pharmacol Res. 2016;105:1–12. doi: 10.1016/j.phrs.2015.12.022. [DOI] [PubMed] [Google Scholar]
- 45.Angiuoli SV, Matalka M, Gussman A, Galens K, Vangala M, Riley DR, Arze C, White JR, White O, Fricke WF. CloVR: A virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinformatics. 2011;12:356. doi: 10.1186/1471-2105-12-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used or analyzed during the current study are available from the corresponding author on reasonable request.