Abstract
Polo-like-kinase 1 (PLK1), which is a serine–threonine protein kinase overexpressed in cancer cells, is known to regulate tumor growth and have recently gathered attention as a target gene for RNA interference because of the poor bioavailability and nonspecificity of the available inhibitors. However, the lower transfection efficiency of siRNA and its poor stability in biological mileu necessitate the need of efficient siRNA delivery systems. Here, we report efficacious polymeric nanoparticles for the delivery of PLK1 siRNA in mammalian cancer cells. N-Isopropylacrylamide (NIPAm) and N-isopropylmethacrylamide-co-NIPAm nanogels were synthesized and modified using poly-ε-lysine. Furthermore, their ability to induce gene silencing was investigated by flow cytometry and real-time polymerase chain reaction, and the silencing efficiency observed was related to the polymer composition and its effect on the gene loading and protection ability and the endosomal escape capability. This study attempts to leverage the understanding of the cell–material interaction, thus, addressing the bottlenecks of siRNA delivery for enhancing the efficacy of the poly(N-isopropylacrylamide)-based delivery vehicle.
Introduction
Polo-like-kinase 1, (PLK1) has been identified as one of the key regulators of mitosis.1−3 Its overexpression is correlated with aggressive behavior4 in a number of cancer types, making it a potential target for cancer treatment. It is also involved in mechanisms inducing resistance to chemotherapeutic agents such as doxorubicin, paclitaxel, metformin, and gemcitabine.5 A number of small-molecule inhibitors of PLK1 are in clinical trials, however, poor bioavailability and off-target effects6,7 limit their efficiency. Thus, a more specific RNA interference (RNAi)-based approach might offer a potential therapy, making PLK1 small-interfering RNA (siRNA) a good candidate for nanoparticle-mediated drug delivery. Until recently, with the advancement in RNAi technologies as well as the ability of PLK1 siRNA to suppress tumor growth in vivo,8,9 ways for efficient delivery of PLK1 siRNA are being explored, and nanoparticle-based delivery platforms are considered most promising for delivery of PLK1 siRNA. Unfortunately, achieving significant knockdown of PLK-1 resulting into specific apoptosis has been a challenge at concentrations, which do not result into off-target effects.
Even after years of research, overcoming the bottlenecks in RNAi, that is, efficient intracellular delivery and endosomal escape of siRNA, is still a challenge. Toward this, a number of nanoparticle-based strategies have been employed. It was suggested that the use of highly charged primary amines can help in creating osmotic imbalance by sequestering the protons, resulting into swelling and bursting of endosomes, thereby releasing its contents into the cell cytoplasm, a process known as proton-sponge effect.10 Unfortunately, the toxicity imparted by the high positive charge, cell membrane damage, and inability to release the siRNA from the complexes impedes their application.11,12
Among the variety of nanoparticles developed for siRNA delivery,13 polymeric nanoparticles have generated a great interest. Polymeric nanoparticles provide advantage of efficient loading of small-molecule drugs for combination therapy as well as incorporation of different functional groups for bioconjugation. The nanoparticles of polymers such as chitosan, cationic dendrimers, and poly(lactic-co-glycolic acid) (PLGA) have been investigated for siRNA delivery. Lower Coulombic attraction between siRNA and PLGA, less efficient endosomal escape ability of PLGA, and cytotoxicity associated with high-molecular-weight polymers are some of the concerns related with these polymeric systems.14
Other type of widely studied polymeric nanoparticles include poly(N-isopropylacrylamide) (pNIPAm) nanogels. These materials belong to the category of “smart” nanogels because of their thermoresponsivity. Additionally, the ease of synthesis, control over the size, and transition temperatures makes them an ideal candidate for a variety of biomedical applications.15 Despite the potential exhibited by these nanoparticles, their application is limited by the toxicity imposed by depolymerization and bioaccumulation. However, recent studies demonstrate the noncytotoxicity of pNIPAm at concentrations relevant to biological applications.16 Additionally, in an in vivo study by Ankareddi et al., the authors suggest that the pNIPAm oligomer does not pose a biologically significant risk at relevant human dosages.17
Previous reports on the application of pNIPAm nanogels for siRNA delivery suggest enhancement in their delivery efficiency. Interestingly, in most of the reports, the siRNA was loaded by “breathing-in” technique, involving absorption of siRNA into the nanogels.18,19 As pNIPAm is a thermoresponsive polymer with volume-phase-transition temperatures (VPTTs) close to the physiological temperature (37 °C), at the body temperature, these nanogels become hydrophobic and undergo deswelling. This can result in premature release of the absorbed cargo as well as the hydrophobicity may affect their circulation. Although a lot of work has been done on applications of pNIPAm-based materials for RNAi, there are no reports on the systematic evaluation of how the knockdown efficiencies can be further increased by better design and by incorporating parameters to overcome the premature release because of hydrophobicity at physiological temperature, endosomal escape, and other critical requirements of RNAi.18,20,21
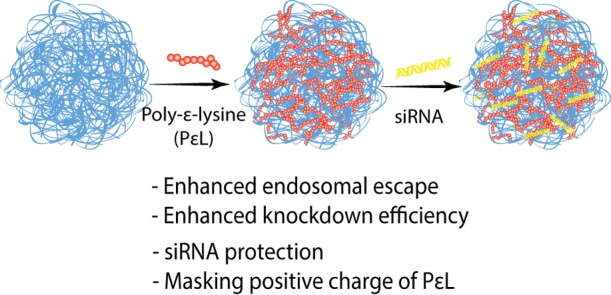
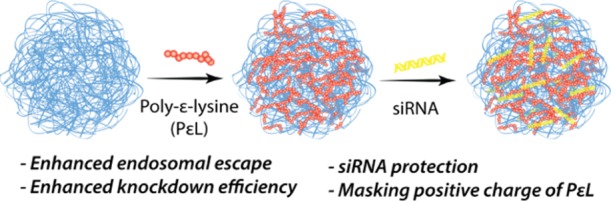
Herein, an efficient delivery platform for the much-needed PLK-1 siRNA is reported. We developed two NIPAm-based, N,N′-methylenebis(acrylamide) (BIS, 5 mol %) cross-linked nanogels, one with the composition 90 mol % NIPAm and 5 mol % acrylic acid, and other with 80 mol % N-isopropylmethacrylamide (NIPMAm), 10 mol % NIPAm, and 5 mol % acrylic acid. These nanogels are hereafter referred to as pNIPAm and p(NIPMAm-co-NIPAm) nanogel, respectively. NIPMAm was used to tune the VPTT of the nanogels. NIPMAm consists of −CH3 group that resists deswelling of the nanogels, thereby increasing the VPTT of NIPAm nanogels. The acrylic acid in nanogels provides carboxyl group for post-synthesis modifications. To impart the endosomal escape property, the carboxyl groups of the nanogels were used for adsorbing a cationic polymer, poly-ε-lysine (PεL), which further helps in loading siRNA via Coulombic attraction (Scheme 1) and also in retaining siRNA in the nanogels. Additionally, once inside, the positive charge of cationic polymer will be protected by the nanogel, which may help in reducing the charge-dependent toxicity, usually associated with such polymers.22
Scheme 1. Nanogels for siRNA Delivery.
Results and Discussion
For achieving the goal of establishing design principle for siRNA delivery, based on polymeric systems, we undertook a systematic approach elucidating the cell–material interaction. As highlighted earlier, efficient loading and release of siRNA, cellular uptake of the siRNA-loaded nanoparticles, and its endosomal escape capabilities are critical design parameters for siRNA delivery vehicle. We began our studies by the synthesis of pNIPAm and p(NIPMAm-co-NIPAm) nanogels by a well-established precipitation polymerization method followed by their characterization. As these are thermoresponsive nanogels, their VPTTs were determined by dynamic light scattering (DLS). The VPTT of pNIPAm was ∼32 °C, whereas that of p(NIPMAm-co-NIPAm) was ∼45 °C (Figure S1). The precipitation polymerization resulted into pNIPAm and p(NIPMAm-co-NIPAm) nanogels of the size 55 ± 7 nm and 66 ± 3 nm with a surface charge of −11 ± 3 mV and −20 ± 1 mV, respectively (Figure 1a,b). The negative charge can be attributed to the anionic surfactant, sodium dodecyl sulfate, and the acrylic acid. Incorporation of monomers and the composition of nanogels were also confirmed by 1H NMR of the nanogels in deuterated water (Figure S2). The −CH3 peak observed at 0.95 ppm indicates the presence of pNIPMAm in the nanogels (Figure S2a). siRNA can be loaded into polymeric nanogels by conjugation, absorption, or Coulombic interactions. As the conjugation involves modification of siRNA for introducing a functional group, its bioactivity may be compromised.24 On the other hand, absorption may result in premature release of the siRNA. Electrostatic interactions involving cationic functional groups are therefore preferable for loading anionic siRNA in the nanogels. The amine groups can be incorporated into the polymeric nanogels either while synthesis by using amine comonomer or by post-synthesis modifications. Incorporation of charged and hydrophilic comonomer into the nanogels at higher mole percent is challenging and results into low-colloidal stability and larger size of the nanogels.22 Thus, a post-synthesis incorporation strategy was used. A cationic polymer PεL was adsorbed through the carboxyl groups of the nanogel. After removing the excess of the cationic polymer using a 100 kDa centrifugation filter, the size of the nanogels was determined by DLS. It was observed that the nanogel size remained unaltered (Figure 1a), indicating the absence of aggregation. Further, the positive surface charge confirmed the adsorption of PεL in the nanogels (Figure 1b). Interestingly, after PεL addition, pNIPAm showed a zeta potential of +11 ± 2 mV, whereas p(NIPMAm-co-NIPAm) had +30 ± 4 mV. To understand the difference observed in the charge on the nanogels, the amount of acrylic acid incorporated in the nanogels was quantified by toluidine blue staining. pNIPAm nanogels showed ∼1.5 times more −COOH incorporation than p(NIPMAm-co-NIPAm). Therefore, the addition of PεL to pNIPAm may result into neutralization of more amines, thereby exhibiting a less net positive charge on the nanogel (Figure 1b). Further addition of siRNA may therefore show more reduction in positive charge on p(NIPMAm-co-NIPAm) nanogels. Because we are comparing the siRNA delivery efficiency of the nanogels, similar loading of siRNA was ensured.
Figure 1.
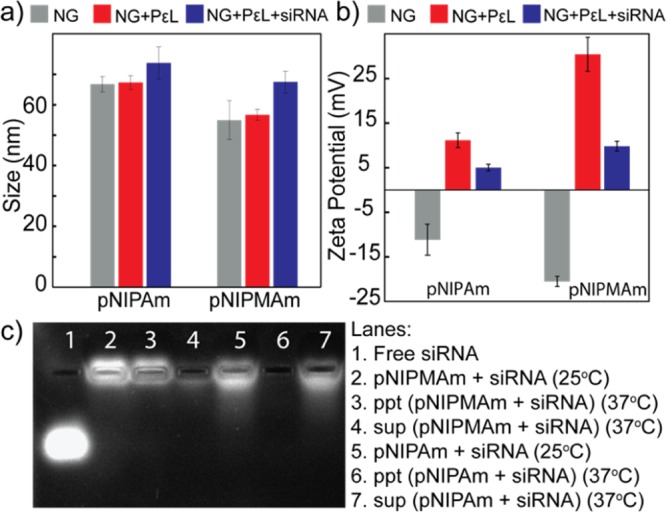
(a) Hydrodynamic diameter and (b) zeta potential analysis of nanogels on adsorption of PεL and siRNA loading. (c) Release of siRNA from nanogels. NG: nanogel; ppt: pellet obtained after centrifugation; and sup: supernatant obtained after centrifugation. Error bars indicate standard deviation between triplicates. Note: p(NIPMAm-co-NIPAm) is denoted as pNIPMAm.
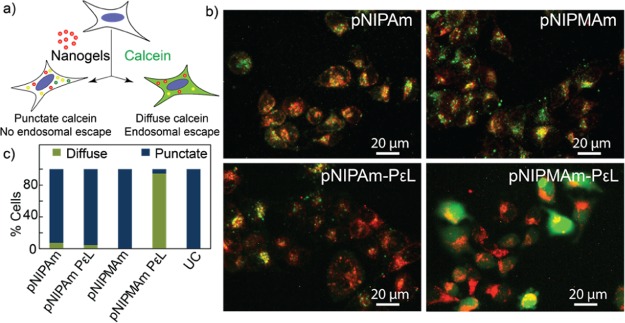
siRNA was loaded into the nanogels via the Coulombic attraction between the adsorbed cationic polymer and anionic siRNA. Amount of nanogels required for the loading of siRNA was optimized using different ratios of nanogel/siRNA (Figure S3). siRNA loading resulted in reduction in the positive charge of both nanogels (Figure 1b). Loading of siRNA was also quantified by UV–vis absorption spectroscopy, and both of the nanogels showed efficient loading, with ∼44% encapsulation efficiencies. As the physical properties of these nanogels are also governed by the temperature, their ability to retain siRNA at physiological temperature was assessed by incubating the nanogel–siRNA complex at 37 °C. siRNA-loaded pNIPAm and p(NIPMAm-co-NIPAm) were incubated for 30 min at 37 °C and centrifuged at 37 °C to separate any siRNA released from the nanogel. The pellet and the supernatant thus obtained were then run in agarose gel and stained for visualizing siRNA. As can be observed in Figure 1c: lane 3–4 and lane 6–7, at 37 °C, a higher concentration of siRNA was observed in the supernatant of the pNIPAm nanogel, whereas p(NIPMAm-co-NIPAm) retained most of it in the pellet. Further, the siRNA in the supernatant of pNIPAm nanogel did not move as compared to the free siRNA, suggesting that probably the entire PεL–siRNA complex was desorbed and released at 37 °C. Release of siRNA from pNIPAm may be attributed to the low negative charge on the nanogels (Figure 1b), resulting into poor Coulombic attraction between the PεL and nanogel, eventually leading to desorption of PεL along with siRNA above its VPTT. This study, therefore, suggests that p(NIPMAm-co-NIPAm) might be a better candidate for siRNA delivery as it is able to retain the loaded siRNA more efficiently than the pNIPAm nanogel. Loading siRNA into the nanoparticles is also known to protect them from serum nucleases. Therefore, the serum stability of the loaded siRNA was investigated by gel electrophoresis. As can be observed in Figure S4, no significant degradation of the siRNA was observed even after 24 h of incubation with serum. For biological applications, cellular internalization and biocompatibility of delivery systems is crucial. For studying the internalization of nanogels, rhodamine-tagged pNIPAm and p(NIPMAm-co-NIPAm) were synthesized (Figure S5). HeLa cells were incubated with the nanogels for 4 h, followed by analysis by flow cytometry. As can be observed in Figure 2a, both of the nanogels were efficiently internalized by HeLa cells. Slight increase in the fluorescence intensity was observed by pNIPAm nanogels as compared to p(NIPMAm-co-NIPAm), which can be because of the incorporation of more rhodamine in pNIPAm as observed by UV–vis spectroscopy (Figure S4). Similar results were observed for MDA-MB-231, a breast cancer cell line (Figure S6a). Interestingly, internalization of fluorescein-labeled siRNA by HeLa cells showed ∼85% uptake for p(NIPMAm-co-NIPAm)-mediated delivery, whereas only ∼40% was observed for pNIPAm (Figure 2b). These results are indeed in accordance with the siRNA release observed at 37 °C (Figure 1c) and suggest that even though the uptake of both of the nanogels is similar, their siRNA delivery efficiency varies. Compatibility of the nanogels with the biological system was studied by assessing the hemocompatibility and cell viability. At higher concentrations, polylysine is known to interact and damage the cell membrane. To determine the toxicity because of membrane damage, hemolytic ability of the nanogels was analyzed. PεL-adsorbed pNIPAM and p(NIPMAm-co-NIPAm) were incubated with erythrocytes at a physiological pH for 30 min at 37 °C, and the hemoglobin released because of membrane damage was quantified spectrophotometrically. As observed in Figure 2c, the hemoglobin release was under the acceptable range (below 5%), suggesting hemocompatibility of the systems. Further, to evaluate the cytotoxic effect, cell viability was assessed by monitoring the ability of cells to metabolize (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (MTT). The cell viability was observed to be above 90% in HeLa cells as well as MDA-MB-231 cell line for the nanogels even with adsorbed PεL (Figures 2d and S6b). Additionally, to gain insights on the endocytosis pathway accessed by the nanogels for cellular entry, colocalization of nanogels with endosomes was probed. Cells were incubated with rhodamine-labeled nanogels and then selectively stained for endosomes using lysotracker. Colocalization observed by fluorescence microscopy (Figure S7) suggested that the nanogels entered via the endocytosis pathway. It is well-known that endosomal entrapment is a bottleneck in siRNA delivery.25 Therefore, endosomal escape ability of the nanogels using a simple calcein assay (Figure 3a) was evaluated. When cells are exposed to nanogels and calcein, a cell impermeable dye, both (nanogels and calcein) gets entrapped into endosomes during the vesicle formation. This results into punctate fluorescence in cell cytoplasm. When calcein-loaded endosomes get disrupted, the calcein gets released into the cytoplasm, yielding a diffused fluorescence in the cytoplasm. The pNIPAm and p(NIPMAm-co-NIPAm) nanogels by themselves were unable to disrupt endosomes efficiently, whereas with adsorbed PεL, only p(NIPMAm-co-NIPAm) was able to show endosomal escape (Figure 3b,c). This was also confirmed by analyzing the fluorescence intensity profiles of calcein- and rhodamine-labeled nanogels in cells by orthogonal confocal microscopy (Figure S8). Similar to the gel retardation study performed at 37 °C (Figure 1c), these results again suggests that at 37 °C, the PεL and siRNA gets desorbed from pNIPAm nanogels before entering the cells, thereby reducing the endosomal escape ability of PεL-adsorbed pNIPAm. In comparison, as the VPTT of p(NIPMAm-co-NIPAm) is above 37 °C, the PεL is retained in these nanogels and available for the proton-sponge effect into the endosome, leading to endosomal escape.
Figure 2.
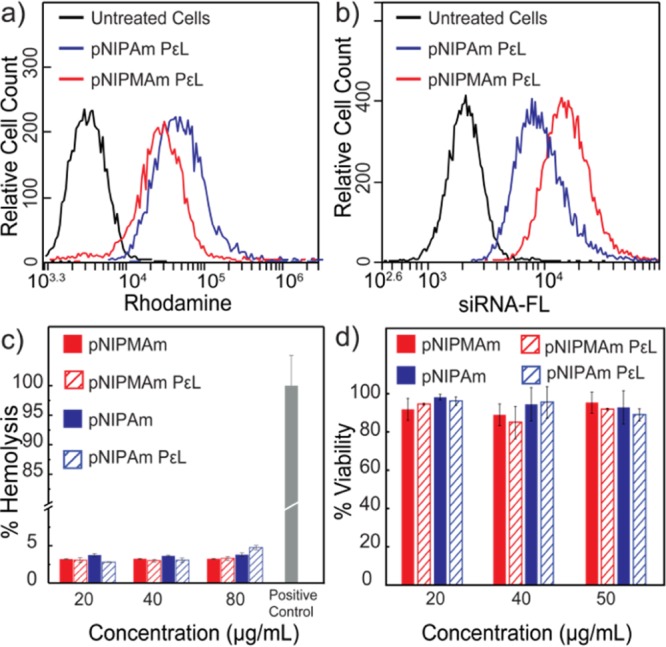
Uptake of (a) rhodamine-tagged pNIPAm and p(NIPMAm-co-NIPAm) nanogels and (b) fluorescein-tagged siRNA-loaded pNIPAm and p(NIPMAm-co-NIPAm) by HeLa cells, analyzed by flow cytometry. (c) Hemolysis assay of nanogels. (d) Cytocompatibility of nanogels in HeLa cells as assessed by MTT assay, after 24 h incubation with nanogels. Error bars indicate standard deviation between triplicates. Note: p(NIPMAm-co-NIPAm) is denoted as pNIPMAm.
Figure 3.
(a) Schematic for the calcein assay for endosomal escape. (b) Endosomal escape analysis of PεL adsorbed nanogels by fluorescence microscopy using HeLa cells. (c) Quantification of endosomal escape by image analysis. UC: untreated cells. Magnification: 20×. Note: p(NIPMAm-co-NIPAm) is denoted as pNIPMAm.
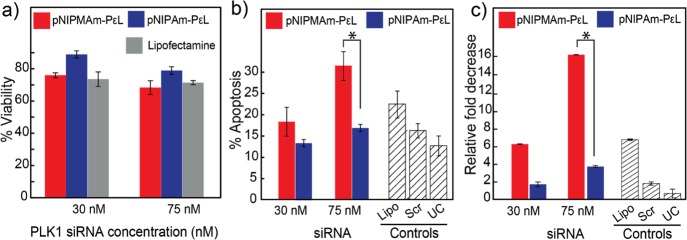
Finally, the ability of both of the nanogels to induce gene silencing was investigated, by monitoring cell viability, apoptosis, and quantification of gene expression levels by RT-PCR, on delivery of PLK1 siRNA in HeLa cells. Cell viability was studied by MTT assay, 36 h post-transfection of siRNA-loaded nanogels. Similar to the earlier reports, while ∼20% cell death was observed at 75 nM for pNIPAm, p(NIPMAm-co-NIPAm) showed ∼35% cell death (Figure 4a). As PLK1 induces cell death by activation of the cellular apoptotic pathways, apoptosis was assessed using Annexin-V–propidium iodide staining. Annexin-V binds to phosphatidylserine, a marker for early apoptosis, whereas propidium iodide stains cells in late apoptosis whose membrane is compromised. Nanogels loaded with scrambled siRNA were used as a negative control. As can be observed in Figure 4b, comparable apoptosis was observed for p(NIPMAm-co-NIPAm) and pNIPAm at lower siRNA concentration (30 nM) after 24 h incubation. Further, increase in siRNA concentration to 75 nM resulted in twofold increase in apoptosis (32%) for p(NIPMAm-co-NIPAm). Confirming our endosomal escape observation, pNIPAm showed a lower knockdown as compared to p(NIPMAm-co-NIPAm). The knockdown effect of p(NIPMAm-co-NIPAm) was specific as a scrambled siRNA (75 nM) loaded in the nanogel did not show any significant knockdown, whereas the positive control, lipofectamine-loaded with PLK1 siRNA (75 nM), showed ∼20% knockdown in 24 h which is in accordance with the literature.26,27 The knockdown of PLK-1 by nanogels was also observed to be cell line independent, as similar trends were observed for a breast cancer cell line, MDA-MB-231 (Figure S6c). In fact, the knockdown was higher in MDA-MB-231 as compared to HeLa, which can be attributed to the PLK-1 sensitivity because of p53 mutation in MDA-MB-231.28,29 These results were interesting because by means of better design of nanoparticles, which takes into account the endosomal escape, loading efficiency, and retention of siRNA, the knockdown higher than the gold standard, lipofectamine, was achieved. For confirming if the apoptosis was PLK1-specific, the expression level of PLK1 was quantified by RT-PCR (Figure 4c). At 30 nM, expression was reduced twofold for pNIPAm, whereas p(NIPMAm-co-NIPAm) resulted into an ∼fourfold reduction. Increasing siRNA concentration did not result in significant downregulation for pNIPAm nanogels whereas p(NIPMAm-co-NIPAm) showed a 10-fold decrease in gene expression. Knockdown at such a low siRNA concentration is exciting as that at higher concentrations (∼100 nM); off-target effects are observed because of the activation of toll-like-receptors.30−32 The enhanced siRNA delivery capability of p(NIPMAm-co-NIPAm) nanogels can be attributed to better loading and the nanogel’s biologically relevant VPTT. Thus, these results clearly demonstrate how insights into the cell–material interaction can be utilized to change the material’s properties for increasing the efficacy of the desired biological process.
Figure 4.
(a) Cell viability on the treatment of HeLa cells with siRNA-loaded nanogels and incubation for 36 h post-transfection. (b) Flow cytometry analysis of apoptosis induced by knockdown of PLK1 gene, by siRNA-loaded nanogels, 24 h post-transfection. The error bars represent the percent error obtained from multiple experiments. (c) PLK1 gene expression 24 h post-transfection of siRNA-loaded pNIPAm and p(NIPMAm-co-NIPAm) nanogels by real-time polymerase chain reaction (RT-PCR). Error bars indicate standard deviation between triplicates. Lipo: lipofectamine loaded with 75 nM PLK1 siRNA, Scr: nanogel with 75 nM scrambled siRNA, and UC: untreated cells. Error bars indicate standard deviation between multiple experiments. *P < 0.05. Note: p(NIPMAm-co-NIPAm)–PεL is denoted as pNIPMAm–PεL.
In conclusion, PεL-adsorbed pNIPAm and p(NIPMAm-co-NIPAm) nanogels can be loaded with siRNA, are easily internalized and well-tolerated by cells, and can protect siRNA from serum nucleases. Owing to the thermoresponsive properties, only p(NIPMAm-co-NIPAm) nanogel showed the ability of efficient endosomal escape and a significant knockdown even at low siRNA concentrations. Our report also signifies the importance of understanding the cell–material interactions in the development of efficient delivery materials.
Experimental Section
Methods
N-Isopropylacrylamide (NIPAm), NIPMAm, rhodamine B, N-(3-aminopropyl)methacrylamide hydrochloride (APMA), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), Amicon Ultra 0.5 mL centrifugal filters (MWCO 100 kDa), Triton X-100, negative control siRNA, and Human Kinase PLK1 siRNA were obtained from Sigma-Aldrich. Sodium dodecyl sulfate, dimethyl sulfoxide (DMSO), and acrylic acid were procured from Merck. Ammonium persulfate (APS) and N,N′-methylenebis(acrylamide) (BIS) were obtained from Loba Chemie. HeLa cells were obtained from the National Center for Cell Sciences, Pune, India. Dulbecco’s modified Eagle’s medium (DMEM, high glucose), Dulbecco’s phosphate-buffered saline, calcein, Lysotracker blue DND-22, trypsin, fetal bovine serum (FBS), MTT and SYBR Green II, and Tris buffer were procured from Thermo Fisher Scientific. PεL (average Mw = 20 000) was obtained from CMS Chemicals Ltd. (UK) and the FITC Annexin V apoptosis detection kit from BD Biosciences. Molecular biology-grade agarose (low EEO), glycerol, boric acid, and disodium ethylenediaminetetraacetic acid (Na2 EDTA) were procured from Sisco Research Laboratories Pvt. Ltd. (SRL). iScript cDNA synthesis kit and SsoFast EvaGreen Supermix were procured from BIORAD. Fluorescein-tagged negative-control siRNA was procured from Santa Cruz Biotechnology.
Synthesis of Nanogels
pNIPAm- and poly-N-isopropylmethacrylamide (pNIPMAm)-based nanogels of compositions given in Table 1 with a total monomer concentration of ∼70 mM were synthesized using the well-known precipitation polymerization method, owing to the ease of synthesis and control over the size. Briefly, the synthesis was carried out in a three-neck round-bottom flask with a condenser. The required amount of monomers (except acrylic acid) and SDS (final concentration: 2 mM) were dissolved in deionized water and filtered through a 0.2 μm filter. The mixture was then purged with nitrogen at 70 °C for 1 h. After 1 h, the calculated volume of acrylic acid was added to the reaction mixture, followed by addition of freshly prepared APS (final concentration: 0.4 mg/mL). After ∼15 min, the reaction mixture turned turbid, indicating the polymer synthesis. The reaction was continued for 4 h under N2 environment, followed by stirring until it cooled down to the room temperature (∼25 °C). Thus, formed nanoparticles were cleaned by dialysis against distilled water using a 12 kDa MWCO dialysis membrane for 48 h, with water changed every 12 h and lyophilized for storage.
Table 1. Composition of Nanogels.
| mol % |
||
|---|---|---|
| pNIPAm nanogel | p(NIPMAm-co-NIPAm) nanogel | |
| NIPAm | 90 | 10 |
| NIPMAm | 80 | |
| BIS | 5 | 5 |
| acrylic acid | 5 | 5 |
The nanogels were resuspended in deionized water and characterized by DLS for hydrodynamic diameter and zeta potential, using the Malvern Zetasizer Nano ZS90 system equipped with a 633 nm laser and 90° scattering optics. For determining the VPTTs, the hydrodynamic diameter of the nanoparticles was recorded at different temperatures.
Synthesis of Rhodamine-Tagged Nanogels
Rhodamine (5 mg, 0.01 mmol, 1 equiv) was dissolved in 5 mL of deionized water and reacted with EDC (19.1 mg, 0.1 mmol, 10 equiv) and NHS (92 mg, 0.2 mmol, 20 equiv) at room temperature for 30 min. This was followed by the addition of APMA (1.8 mg, 0.01 mmol, 1 equiv) to the reaction mixture and further incubation for 4 h. Thus, formed APMA–rhodamine conjugate was used directly for the synthesis of nanogels. Rhodamine-labeled nanogels of compositions (a) 89.9% NIPAm, 0.1% APMA–rhodamine, 5% BIS, and 5% acrylic acid and (b) 79.9% NIPMAm, 10% NIPAM, 0.1% APMA–rhodamine, 5% BIS, and 5% acrylic acid, with the total monomer concentration of ∼70 mM were synthesized using the protocol same as that for p(NIPAm) and p(NIPMAm-co-NIPAm) nanogels. The nanogels were purified by dialysis against distilled water using a 12 kDa MWCO dialysis membrane for 48 h with water replaced every 4 h and lyophilized for storage. The nanogels were characterized by DLS using Malvern Zetasizer Nano ZS90. Incorporation of rhodamine in nanoparticles was confirmed by UV–vis spectroscopy using BioTek Synergy H1 multiplate reader.
Adsorption of PεL in the Nanogels and Loading of siRNA
Nanogels (2.5 mg) were resuspended in 500 μL of nuclease-free deionized water. To it was added 100 μL of 50 mg/mL PεL (20 kDa) and incubated for 15 min. The unadsorbed PεL was removed using the 100 kDa MWCO filters by centrifuging it at 8000g for 5 min. Approximately, 100 μL of the sample was retained in the filter which was resuspended in nuclease-free deionized water (400 μL) resulting into a ∼5 mg/mL nanogel concentration. For loading siRNA, 10 μL of 5 mg/mL nanogel was incubated with 10 μL of 10 μM siRNA for 30 min at room temperature. The nanogels were then centrifuged at 20 817g for 30 min and siRNA left in the supernatant was quantified by UV–vis spectroscopy using BioTek Synergy H1 multiplate reader. The percent encapsulation efficiency was calculated as follows
Release of siRNA from Nanogels
The siRNA-loaded nanogels were incubated at 37 °C for 30 min and centrifuged at 20 817g for 30 min at 37 °C, using an Eppendorf 5810 R centrifuge. The supernatant was used as it is, whereas the pellet was resuspended in nuclease-free deionized water. To each sample, 5 μL of glycerol was added and was loaded in 1% agarose gel. The gel was run at 70 V for 20 min in Tris borate EDTA (TBE buffer) (pH 8). The nanogels incubated at room temperature (∼25 °C) were used as controls. siRNA was then stained by incubating the gel into SYBR Green for 1 h and imaged using UVP GelDoc IT2 Imager.
Toluidine Blue Staining
The amount of acrylic acid incorporated in the nanogels was quantified by a colorimetric method using toluidine blue staining. Nanogels (3 mg) were incubated with 1 mL of 0.5 mM toluidine blue solution (pH 10) for 4 h at 37 °C. The nanogels were then washed with NaOH (pH 10) to remove unbound dye molecules. The bound toluidine blue was then desorbed from nanogels by adding 1 mL of 50% acetic acid solution. The absorbance of solution was measured using Biotek Synergy H1 multiplate reader and the amount of carboxylic acid present was extrapolated from a standard curve obtained using toluidine blue.
Serum Stability of siRNA-Loaded in Nanogels
The siRNA-loaded nanogels were incubated with 10% FBS incubated for 30 min and 24 h at 37 °C. Free siRNA treated with 10% FBS was used as a negative control, whereas the untreated one as a positive control. They were then loaded in 1% agarose gel using glycerol (5 μL). The gel was run at 70 V for 1 h in TBE buffer (pH 8). siRNA was stained by incubating gel into SYBR Green for 1 h and imaged using UVP GelDoc IT2 Imager.
Hemolysis Assay
Hemolysis assay was performed by adding nanogels at different concentrations to 50 μL of 10% erythrocytes to a final volume of 250 μL in PBS. After incubation at 37 °C for 30 min, cells were centrifuged at 1000g for 5 min, and an absorbance of the supernatant was measured at 540 nm to quantify cell lysis. Cells untreated with nanogels were used as negative control, whereas cells incubated with 10% Triton X-100 as positive control.
Maintaining the Cell Lines
Human cervical cancer cell lines, HeLa, and human bone osteosarcoma cell line MG-63 were maintained in DMEM supplemented by 10% FBS and incubated at 37 °C in a humidified 5% CO2 incubator. Breast cancer cell line, MDA-MB-231, was maintained in Leibovitz’s L-15 media supplemented with 10% FBS and incubated at 37 °C under atmospheric CO2 concentration. The media were changed every 2 days, and the cells were passaged after 80% confluency. Same conditions were used for respective cell lines while performing the experiments.
Cellular Internalization of Rhodamine-Tagged Nanogels and Fluorescein-Tagged siRNA (FL-siRNA) by Flow Cytometry
At a confluency of 80%, cells were trypsinized, seeded in 500 μL media in a 24-well plate at the seeding density of 5 × 104 cells per well, and cultured overnight at 37 °C under respective conditions. The media were then replaced with media containing rhodamine-tagged nanogels at the concentration of ∼25 μg/mL and incubated for 4 h at 37 °C. The cells were washed three times with PBS (5 min each), followed by a brief wash with 0.01% Tween20 and three more washes of PBS (5 min each). They were then trypsinized, centrifuged at 664g, and resuspended in PBS. The uptake was analyzed by assessing the increase in fluorescence intensity of cells as compared to the cells without nanoparticle treatment, using the FL2 channel (ex/em 488 nm/585 ± 20 nm) of the BD Accuri C6 flow cytometer. Similarly, to study uptake of siRNA, FL-siRNAs were loaded in nanogels and the uptake was analyzed by flow cytometry.
In Vitro Cytotoxicity of Nanogels by MTT Assay
At a confluency of 80%, cells were trypsinized, seeded in 200 μL of media at a density of 104 cells per well in a 96-well plate and cultured overnight at 37 °C under respective conditions. The spent medium was then replaced with media containing varying concentrations of nanogels, with or without siRNA. After 6 h, the media were removed and fresh media were added, and the cells were further incubated for 24 h or 36 h. Media were then removed, and fresh media containing MTT at the final concentration of 0.5 mg/mL were added over the cells. Cells were further incubated for 1 h to allow formation of the purple, insoluble, formazan crystals. MTT-containing media were then removed, and the crystals were dissolved using 200 μL of DMSO. The absorbance of the samples was then recorded at 550 nm using a BioTek Synergy H1 multiplate reader. Cells untreated with nanogels were considered as the control.
Calcein Assay for Studying Endosomal Escape in HeLa Cell Line by Fluorescence Microscopy
Circular glass coverslips (12 mm diameter) were sterilized by immersing in absolute ethanol overnight. They were transferred in a 24-well plate and washed with PBS. Cells at a seeding density of 5 × 104 per well were cultured overnight in a CO2 incubator at 37 °C. Calcein, a cell membrane impermeable dye, gets entrapped into the endocytotic vesicles, thus resulting into punctate fluorescence in the cells. If these vesicles rupture, the calcein is released into the cytoplasm leading to a diffused fluorescence.23 The cells were therefore incubated with media-containing calcein (0.1 mg/mL) as well as rhodamine-tagged nanogels (∼25 μg/mL) and incubated for 4 h. Cells were washed with PBS and observed under an Olympus IX73 fluorescence microscope using a TRITC filter or an Olympus confocal microscope. Images were pseudocolored using ImageJ. At least 50 cells per sample were analyzed to quantitate the endosomal escape. Images were processed in Fluoview software to analyze z-stacked confocal microscopy images in an orthogonal view.
Evaluation of siRNA-Mediated Silencing of PLK1 by Apoptosis Assay and RT-PCR
The cells were seeded in a 24-well plate at the seeding density of 5 × 104 cells per well and incubated overnight at 37 °C in CO2 incubator. Cells were then washed with PBS, followed by the addition of PLK1 siRNA-loaded nanogels in a serum-free media, such that siRNA concentration was 30 and 75 nM. Nanogel-containing media were replaced with fresh serum-supplemented media, and cells were incubated for 24 h.
For collecting detached cells undergoing apoptosis, the spent media were centrifuged at 664g for 5 min, whereas the adhered cells were first trypsinized before centrifugation. The cells were then washed two times with ice-cold PBS and stained with Annexin V-FITC and PI using the manufacturer’s protocol. The cells were then analyzed using FL1 (ex/em 488/533 ± 15) and FL3 (ex/em 488/>670) channels of BD Accuri C6 flow cytometer. Cell transfected with PLK1 siRNA using lipofectamine were used as the positive control, whereas those with scrambled siRNA-loaded nanogel as a negative control.
For quantifying mRNA expression, RNA was isolated from cells using TRIzol reagent and cDNA was synthesized using manufacturer’s protocol. A 10 μL PCR reaction was set using 2 μL of cDNA. The PCR cycle used was enzyme activation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, and 56 °C for 5 s. Melt curve was recorded from 65 to 95 °C with 0.5 °C intervals. As an internal control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used. Same PCR protocol was followed for GAPDH, except that the extension temperature used was 54 °C instead of 56 °C. Lipofectamine-mediated PLK1 siRNA transfected cells were used as positive control, whereas those with scrambled siRNA-loaded nanogel as negative control. Sequence of PLK1 primers: forward, 5′-CCCATCTTCTGGGTCAGCAAG-3′ and reverse, 5′-AAGAGCACCCCCACGCTGTT-3′. Sequence of GAPDH primers: forward, 5′-TGCACCACCAACTGCTTAGC-3′ and reverse, 5′-GGCATGGACTGTGGTCATGAG-3′.
Acknowledgments
The work was funded by the Department of Science and Technology-Nanomission, India (SR/NM/NS-1022/2013). S.D. and S.P. acknowledge the fellowship support from UGC, India. We also acknowledge Central Research Facility and Nanoscale Research Facility, IIT Delhi, for NMR and DLS respectively, and Dr. Dikshi Gupta, CBME, for help in confocal microscopy.
Glossary
Abbreviations
- PLK1
polo-like-kinase 1
- pNIPAm
poly-N-isopropylacrylamide
- pNIPMAm
poly-N-isoproplymethacrylamide
- VPTT
volume-phase-transition temperature
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b00738.
VPTT of nanogels by DLS, NMR characterization, serum stability, absorbance spectra of rhodamine-tagged nanogels, uptake, cytocompatibility, and knockdown for MDA-MB-231 cell lines (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Liu Z.; Sun Q.; Wang X. PLK1, A Potential Target for Cancer Therapy. Transl. Oncol. 2017, 10, 22–32. 10.1016/j.tranon.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck J.; Maerki S.; Posch M.; Metzger T.; Persaud A.; Scheel H.; Hofmann K.; Rotin D.; Pedrioli P.; Swedlow J. R.; Peter M.; Sumara I. Ubiquitylation-dependent localization of PLK1 in mitosis. Nat. Cell Biol. 2013, 15, 430–439. 10.1038/ncb2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitouni S.; Nabais C.; Jana S. C.; Guerrero A.; Bettencourt-Dias M. Polo-like kinases: structural variations lead to multiple functions. Nat. Rev. Mol. Cell Biol. 2014, 15, 433–452. 10.1038/nrm3819. [DOI] [PubMed] [Google Scholar]
- Eckerdt F.; Yuan J.; Strebhardt K. Polo-like kinases and oncogenesis. Oncogene 2005, 24, 267–276. 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- Gutteridge R. E. A.; Ndiaye M. A.; Liu X.; Ahmad N. Plk1 Inhibitors in Cancer Therapy: From Laboratory to Clinics. Mol. Cancer Ther. 2016, 15, 1427–1435. 10.1158/1535-7163.mct-15-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupenthal J.; Bihrer V.; Korkusuz H.; Kollmar O.; Schmithals C.; Kriener S.; Engels K.; Pleli T.; Benz A.; Canamero M.; Longerich T.; Kronenberger B.; Richter S.; Waidmann O.; Vogl T. J.; Zeuzem S.; Piiper A. Reduced Efficacy of the Plk1 Inhibitor BI 2536 on the Progression of Hepatocellular Carcinoma due to Low Intratumoral Drug Levels. Neoplasia (New York, N.Y.) 2012, 14, 410. 10.1596/neo.111366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B.; Mahmud H.; Maass A. H.; Yu B.; van Gilst W. H.; de Boer R. A.; Silljé H. H. W. The Plk1 Inhibitor BI 2536 Temporarily Arrests Primary Cardiac Fibroblasts in Mitosis and Generates Aneuploidy In Vitro. PLoS one 2010, 5, e12963. 10.1371/journal.pone.0012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Y.; Yang Z.; Li Q.; Song F. Silencing of Polo-Like Kinase (Plk) 1 via siRNA Causes Inhibition of Growth and Induction of Apoptosis in Human Esophageal Cancer Cells. Oncology 2008, 74, 198–206. 10.1159/000151367. [DOI] [PubMed] [Google Scholar]
- Zhao C. L.; Ju J. Y.; Gao W.; Yu W. J.; Gao Z. Q.; Li W. T. Downregulation of PLK1 by RNAi attenuates the tumorigenicity of esophageal squamous cell carcinoma cells via promoting apoptosis and inhibiting angiogenesis. Neoplasma 2015, 62, 748–755. 10.4149/neo_2015_089. [DOI] [PubMed] [Google Scholar]
- Boussif O.; Lezoualc’h F.; Zanta M. A.; Mergny M. D.; Scherman D.; Demeneix B.; Behr J. P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. U.S.A. 1995, 92, 7297–7301. 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H.; Zhang S.; Wang B.; Cui S.; Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Controlled Release 2006, 114, 100–109. 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Fischer D.; Li Y.; Ahlemeyer B.; Krieglstein J.; Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials 2003, 24, 1121–1131. 10.1016/s0142-9612(02)00445-3. [DOI] [PubMed] [Google Scholar]
- Mura S.; Nicolas J.; Couvreur P. Stimuli-responsive nanocarriers for drug delivery. Nat. Mater. 2013, 12, 991–1003. 10.1038/nmat3776. [DOI] [PubMed] [Google Scholar]
- Singha K.; Namgung R.; Kim W. J. Polymers in Small-Interfering RNA Delivery. Nucleic Acid Ther. 2011, 21, 133–147. 10.1089/nat.2011.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y.; Zhang Y. PNIPAM microgels for biomedical applications: from dispersed particles to 3D assemblies. Soft Matter 2011, 7, 6375–6384. 10.1039/c0sm01541e. [DOI] [Google Scholar]
- Malonne H.; Eeckman F.; Fontaine D.; Otto A.; Vos L.; Moes A.; Fontaine J.; Amighi K. Preparation of poly(N-isopropylacrylamide) copolymers and preliminary assessment of their acute and subacute toxicity in mice. Eur. J. Pharm. Biopharm. 2005, 61, 188–194. 10.1016/j.ejpb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Ankareddi I.; Bailey M. M.; Brazel C. S.; Rasco J. F.; Hood R. D. Developmental toxicity assessment of thermoresponsive poly(N-isopropylacrylamide-co-acrylamide) oligomers in CD-1 mice. Birth Defects Res., Part B 2008, 83, 112–116. 10.1002/bdrb.20150. [DOI] [PubMed] [Google Scholar]
- Blackburn W. H.; Dickerson E. B.; Smith M. H.; McDonald J. F.; Lyon L. A. Peptide-Functionalized Nanogels for Targeted siRNA Delivery. Bioconjugate Chem. 2009, 20, 960–968. 10.1021/bc800547c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. H.; Lyon L. A. Multifunctional Nanogels for siRNA Delivery. Acc. Chem. Res. 2012, 45, 985–993. 10.1021/ar200216f. [DOI] [PubMed] [Google Scholar]
- Yang H.-y.; van Ee R. J.; Timmer K.; Craenmehr E. G. M.; Huang J. H.; Öner F. C.; Dhert W. J. A.; Kragten A. H. M.; Willems N.; Grinwis G. C. M.; Tryfonidou M. A.; Papen-Botterhuis N. E.; Creemers L. B. A novel injectable thermoresponsive and cytocompatible gel of poly( N -isopropylacrylamide) with layered double hydroxides facilitates siRNA delivery into chondrocytes in 3D culture. Acta Biomater. 2015, 23, 214–228. 10.1016/j.actbio.2015.05.018. [DOI] [PubMed] [Google Scholar]
- Kim C.; Lee Y.; Kim J. S.; Jeong J. H.; Park T. G. Thermally triggered cellular uptake of quantum dots immobilized with poly(N-isopropylacrylamide) and cell penetrating peptide. Langmuir 2010, 26, 14965–14969. 10.1021/la102632m. [DOI] [PubMed] [Google Scholar]
- Zhang X.-Q.; Intra J.; Salem A. K. Comparative study of poly (lactic-co-glycolic acid)-poly ethyleneimine-plasmid DNA microparticles prepared using double emulsion methods. J. Microencapsulation 2008, 25, 1–12. 10.1080/02652040701659347. [DOI] [PubMed] [Google Scholar]
- Su X.; Fricke J.; Kavanagh D. G.; Irvine D. J. In Vitro and in Vivo mRNA Delivery Using Lipid-Enveloped pH-Responsive Polymer Nanoparticles. Mol. Pharm. 2011, 8, 774–787. 10.1021/mp100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.-L.; Rana T. M. siRNA function in RNAi: A chemical modification analysis. RNA 2003, 9, 1034–1048. 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominska M.; Dykxhoorn D. M. Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 2010, 123, 1183–1189. 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Zheng W.; Tang R.; Wang N.; Zhang W.; Jiang X. Gene regulation with carbon-based siRNA conjugates for cancer therapy. Biomaterials 2016, 104, 269–278. 10.1016/j.biomaterials.2016.07.015. [DOI] [PubMed] [Google Scholar]
- Tian Y.; Zhang Y.; Pan J.; Lu N.; Wang S.; Lu G. Gold Nanoparticles Increase PLK1-Specific Small Interfering RNA Transfection and Induce Apoptosis of Drug Resistance Breast Cancer Cells. J. Nanomater. 2015, 2015, 1–9. 10.1155/2015/720198. [DOI] [Google Scholar]
- Degenhardt Y.; Greshock J.; Laquerre S.; Gilmartin A. G.; Jing J.; Richter M.; Zhang X.; Bleam M.; Halsey W.; Hughes A.; Moy C.; Liu-Sullivan N.; Powers S.; Bachman K.; Jackson J.; Weber B.; Wooster R. Sensitivity of cancer cells to Plk1 inhibitor GSK461364A is associated with loss of p53 function and chromosome instability. Mol. Cancer Ther. 2010, 9, 2079–2089. 10.1158/1535-7163.mct-10-0095. [DOI] [PubMed] [Google Scholar]
- Hui L.; Zheng Y.; Yan Y.; Bargonetti J.; Foster D. A. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene 2006, 25, 7305–7310. 10.1038/sj.onc.1209735. [DOI] [PubMed] [Google Scholar]
- Persengiev S. P.; Zhu X.; Green M. R. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA 2004, 10, 12–18. 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A.; Anderson E. M.; Vermeulen A.; Fedorov Y.; Robinson K.; Leake D.; Karpilow J.; Marshall W. S.; Khvorova A. Induction of the interferon response by siRNA is cell type- and duplex length-dependent. RNA 2006, 12, 988–993. 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sioud M. Induction of Inflammatory Cytokines and Interferon Responses by Double-stranded and Single-stranded siRNAs is Sequence-dependent and Requires Endosomal Localization. J. Mol. Biol. 2005, 348, 1079–1090. 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.