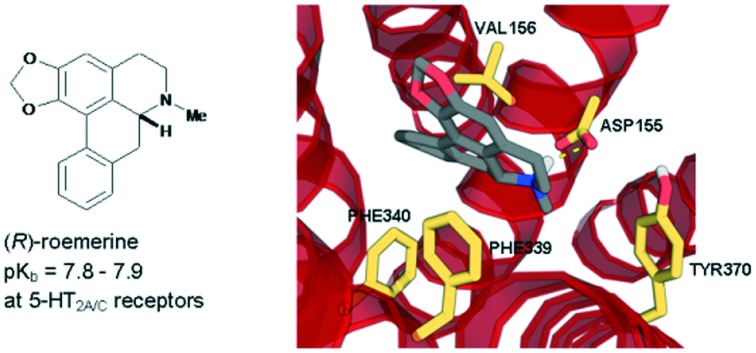
 (R)-roemerine was found to be a potent 5-HT2A/C receptor antagonist with good selectivity compared to 5-HT2B and α1 receptor subtypes.
(R)-roemerine was found to be a potent 5-HT2A/C receptor antagonist with good selectivity compared to 5-HT2B and α1 receptor subtypes.
Abstract
In this study, the (S)-enantiomers of the aporphine alkaloids, nuciferine and roemerine, were prepared via a synthetic route involving catalytic asymmetric hydrogenation and both stereoisomers were evaluated in vitro for functional activity at human 5-HT2 and adrenergic α1 receptor subtypes using a transforming growth factor-α shedding assay. Both enantiomers of each of the compounds were found to act as antagonists at 5-HT2 and α1 receptors. (R)-roemerine was the most potent compound at 5-HT2A and 5-HT2C receptors (pKb = 7.8–7.9) with good selectivity compared to (S)-roemerine at these two receptors and compared to its activity at 5-HT2B, α1A, α1B and α1D receptors.
Introduction
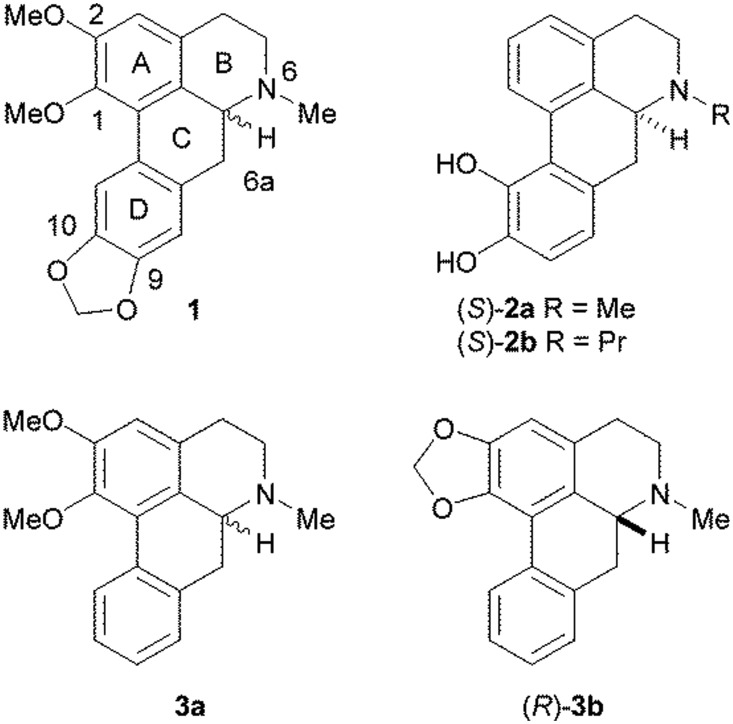
Aporphines are tetracyclic compounds that form a subset of the tetrahydroisoquinoline alkaloids and contain a single chiral centre at position C6a (Fig. 1). They have been found to exhibit a number of different pharmacological activities, including at dopamine, serotonin (5-HT) and adrenergic receptors.1,2 At dopamine D1 and D2 receptors, it has been observed that the (R)-enantiomers tend to act as agonists, whereas the (S)-enantiomers are usually antagonists. Experiments on some pairs of aporphine enantiomers at the 5-HT1A receptor suggested the same pattern of activity as that observed with dopamine receptors.3 However, very few studies have been reported on the stereoselectivity of aporphines at 5-HT2A and adrenergic α1A receptors and no clear patterns have been observed. In one of these studies, the two isomers of nantenine (1) at the 5-HT2A and α1A receptors were both found to behave as moderate antagonists, with the (S)-enantiomer being slightly more potent than the (R)-enantiomer at the 5-HT2A receptor, but with the pattern of stereoselectivity being reversed at the α1A receptor.4 Conversely, in another study, the (S)-enantiomers of apomorphine (2a) and N-n-propyl norapomorphine (2b) have been found to have higher affinities for α1A, α1B and α1D receptors than their corresponding (R)-enantiomers.5
Fig. 1. Structures of the aporphines, (±)-nantenine (1), (S)-apomorphine (2a), (S)-N-n-propyl norapomorphine (2b), (±)-nuciferine (3a) and (R)-roemerine (3b).
Previously, we reported that (±)-nuciferine (3a) and (R)-roemerine (3b), which differ from nantenine in that they lack a methylenedioxy substituent at positions C9 and C10 in the D ring, possess high binding affinity and selectivity for the rat 5-HT2A receptor compared to the 5-HT1A, D1 and D2 receptors.6 In addition, other researchers have shown that these compounds have good oral bioavailability and a wide tissue distribution in rats, including brain penetration.7,8 They therefore have potential for use as pharmacological probes or agents for conditions such as psychotic disorders and drug addiction.9,10 However, it was not known if both enantiomers are active or whether they act as agonists or antagonists. Only the (R)-isomers are commercially available so we embarked on the stereoselective synthesis of the (S)-isomers in order to determine the functional activity of each pair of isomers at human 5-HT2 and α1 receptors.
Results and discussion
Synthesis
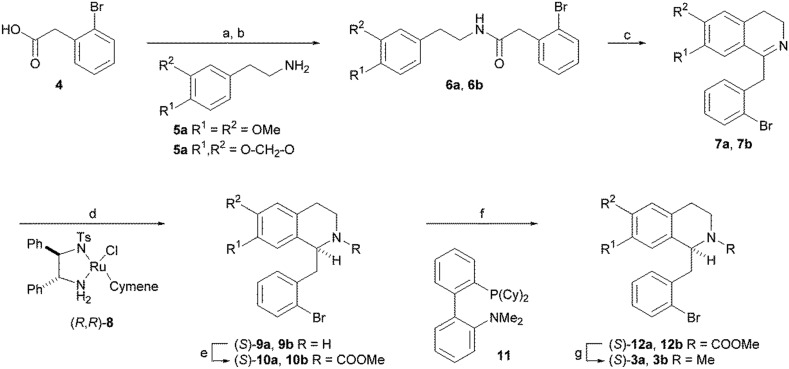
(S)-Nuciferine (3a) and (S)-roemerine (3b) were prepared using the synthetic route depicted in Scheme 1 based on the asymmetric hydrogenation of 1-benzyl-3,4-dihydroisoquinoline intermediates 7a or 7b.11,12 Coupling of commercially-available 2-bromophenylacetic acid (4) with 2-(3,4-dimethoxyphenyl)-ethylamine (5a) or 2-(3,4-methylenedioxyphenyl)ethylamine (5b) gave amides 6a and 6b. Bischler–Napieralski cyclisation using phosphorus oxychloride in refluxing dichloromethane, followed by catalytic asymmetric hydrogenation of the resulting imines 7a and 7b using Noyori's ruthenium-based catalyst, (R,R)-8, gave the (S)-1-benzyltetrahydroisoquinolines 9a and 9b. Oxidative cyclisation of the methylcarbamate-protected amines 10a and 10b using catalytic palladium(ii) in complex with 2-dicyclohexylphosphino-2′-(N,N-dimethylamino) biphenyl (DavePhos) (11) in dimethylacetamide (DMA) at 130 °C produced (S)-aporphines 12a and 12b. Finally, reduction of the methyl carbonate group using lithium aluminium hydride gave the target compounds (S)-3a and (S)-3b in 30–40% overall yield and with 96% and 99% e.e., respectively, determined by chiral HPLC.
Scheme 1. Reagents and conditions: a) (COCl)2, CH2Cl2; b) 5, Et3N, CH2Cl2; c) POCl3, CH2Cl2, reflux; d) (R,R)-8 (3 mol%), HCO2H/NEt3, DMF, rt; e) MeOCOCl, DIPEA, CH2Cl2, rt; f) Pd(OAc)2 (5 mol%), DavePhos 11 (10 mol%), KOAc (2 eq.), DMA, 130 °C; g) LiAlH4, Et2O, 0 °C.
Pharmacological evaluation
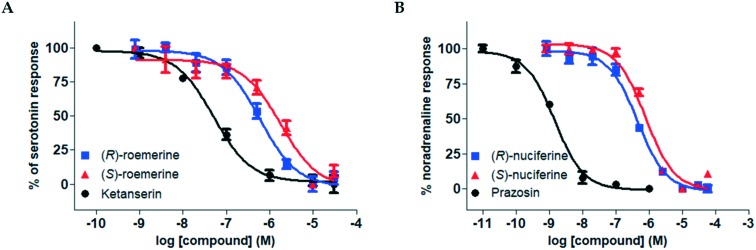
Both the (R)- and (S)-enantiomers of nuciferine (3a) and roemerine (3b) were evaluated for functional activity at human 5-HT2 and α1 receptor subtypes using a recently-developed transforming growth factor-α shedding assay that measures ligand-mediated receptor activation.13 Representative pIC50 curves are shown in Fig. 2 and the results are summarised in Tables 1 and 2.
Fig. 2. Representative pIC50 curves. A. For (R)- and (S)-roemerine (3b) compared to ketanserin at the 5-HT2A receptor in the presence of 1 μM serotonin. B. For (R)- and (S)-nuciferine (3a) compared to prazosin at the α1A receptor in the presence of 0.1 μM noradrenaline. Data points are normalised and shown as the mean ± SD from triplicate readings.
Table 1. Antagonist potencies and selectivities of nuciferine (3a) and roemerine (3b) enantiomers for 5-HT2 receptor subtypes calculated from pIC50 curves.
| pKb
a
|
K
b (nM) |
Subtype selectivity
b
|
|||||||
| Compound | 5-HT2A | 5-HT2B | 5-HT2C | 5-HT2A | 5-HT2B | 5-HT2C | 5-HT2A/5-HT2B | 5-HT2A/5-HT2C | 5-HT2C/5-HT2B |
| (R)-Nuciferine | 7.18 ± 0.03 | 7.51 ± 0.21 | 7.44 ± 0.01 | 65.3 | 31.0 | 36.4 | 0.5 | 0.6 | 1 |
| (S)-Nuciferine | 7.07 ± 0.17 | 7.38 ± 0.01 | 7.51 ± 0.22 | 84.3 | 41.7 | 41.7 | 0.5 | 0.4 | 1 |
| (R)-Roemerine | 7.97 ± 0.18 | 7.08 ± 0.43 | 7.82 ± 0.08 | 10.8 | 84.0 | 15.1 | 8 | 1 | 6 |
| (S)-Roemerine | 7.37 ± 0.10 | 7.03 ± 0.33 | 7.12 ± 0.13 | 42.9 | 92.7 | 76.9 | 2 | 2 | 1 |
| Ketanserin c | 8.69 ± 0.03 | 5.70 ± 0.04 | 6.95 ± 0.08 | 2.0 | 1980 | 112 | 1000 | 60 | 20 |
aValues are expressed as mean ± SEM from at least 2 separate experiments.
bSelectivities for receptor X/receptor Y were calculated as the ratio of Kb receptor Y/Kb receptor X.
cPositive control.
Table 2. Antagonist potencies and selectivities of nuciferine (3a) and roemerine (3b) enantiomers for α1 receptor subtypes calculated from pIC50 curves.
| pKb
a
|
K
b (nM) |
Subtype selectivity
b
|
|||||||
| Compound | α1A | α1B | α1D | α1A | α1B | α1B | α1A/α1B | α1A/α1D | α1B/α1D |
| (R)-Nuciferine | 7.42 ± 0.07 | 7.22 ± 0.01 | 6.78 ± 0.01 | 37.8 | 60.6 | 165 | 2 | 4 | 3 |
| (S)-Nuciferine | 7.17 ± 0.08 | 6.50 ± 0.01 | 6.94 ± 0.01 | 67.1 | 316 | 116 | 5 | 2 | 0.4 |
| (R)-Roemerine | 6.47 ± 0.08 | 6.57 ± 0.05 | 7.34 ± 0.15 | 338 | 266 | 46.0 | 0.8 | 0.1 | 0.2 |
| (S)-Roemerine | 7.07 ± 0.01 | 6.81 ± 0.03 | 7.04 ± 0.07 | 86.0 | 154 | 92.2 | 2 | 1 | 0.6 |
| Prazosin c | 9.69 ± 0.07 | 11.41 ± 0.09 | 10.60 ± 0.02 | 0.20 | 0.004 | 0.025 | 0.02 | 0.1 | 6 |
aValues are expressed as mean ± SEM from at least 2 separate experiments.
bSelectivities for receptor X/receptor Y were calculated as the ratio of Kb receptor Y/Kb receptor X.
cPositive control.
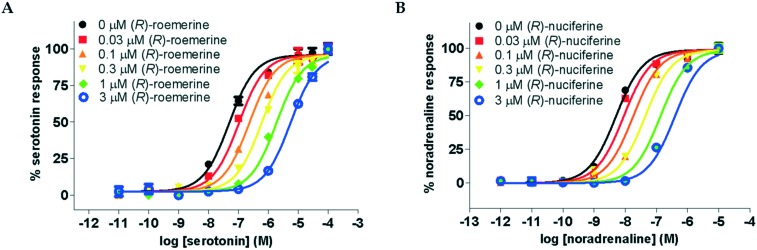
The compounds were all observed to act as competitive antagonists at both sets of receptor subtypes. This was confirmed by Schild analysis which resulted in agonist dose–response curves that were increasingly shifted to the right with increasing concentrations of test compound (see Fig. 3) and linear Schild plots with slopes which were not significantly different from unity.
Fig. 3. Agonist dose–response curves in the presence of varying concentrations of antagonist. A. For serotonin at the 5-HT2A receptor in the presence of (R)-roemerine (3b). B. For noradrenaline at the α1A receptor in the presence of (R)-nuciferine (3a). Data points are normalised and shown as the mean ± SD from triplicate readings.
The pKb values at the 5-HT2A and α1A receptors obtained from the Schild analyses (see Table 3) were essentially similar to those obtained from the pIC50 curves, although some differences were noted for (S)-roemerine. The values obtained from the Schild analyses are likely to be more accurate because about 6 times as many data points are involved in their determination.
Table 3. Antagonist potencies and selectivities of nuciferine (3a) and roemerine (3b) enantiomers for 5-HT2A and α1A receptors determined from Schild analysis.
| pKb
a
|
5-HT2A/α1A selectivity b | ||
| Compound | 5-HT2A | α1A | |
| (R)-Nuciferine | 7.26 ± 0.12 | 7.31 ± 0.11 | 1 |
| (S)-Nuciferine | 6.85 ± 0.14 | 6.91 ± 0.07 | 1 |
| (R)-Roemerine | 7.88 ± 0.13 | 6.38 ± 0.11 | 32 |
| (S)-Roemerine | 7.06 ± 0.11 | 6.62 ± 0.11 | 3 |
aValues are expressed as mean ± SD.
bSelectivities for receptor X/receptor Y were calculated as the ratio of Kb receptor Y/Kb receptor X.
(R)-Roemerine was found to be the most potent compound at 5-HT2A and 5-HT2C receptors (pKb = 7.97 and 7.82, respectively). This is in agreement with our previous results6 from radioligand binding studies at rat 5-HT2A receptors which showed that (R)-roemerine has more than twice the affinity of (±)-nuciferine and confirms (R)-roemerine to have the highest potency at 5-HT2 receptors of any naturally-occurring aporphine identified to date. (R)-Roemerine also displayed 4- and 5-fold stereoselectivity compared to (S)-roemerine at the 5-HT2A and 5-HT2C receptor subtypes, respectively. However, the two isomers of nuciferine were observed to have similar activities at all three 5-HT2 receptor subtypes. In comparison, the two isomers of nantenine (1) have previously been found to have activities at the 5-HT2A receptor that are one to one and a half orders of magnitude lower, with the (S)-enantiomer being slightly more potent than the (R)-enantiomer.4 The greater than 6-fold selectivity of (R)-roemerine for the 5-HT2A and 5-HT2C receptors compared to the 5-HT2B receptor is also notable. Taken together, these results suggest that the presence of a methylenedioxy substituent at positions C1 and C2 in the A ring favours the action of the (R)-enantiomer of roemerine at 5-HT2A and 5-HT2C receptors. This in agreement with a previous study on analogues of nantenine which showed that variation of the alkoxy substituent at position C1 has a substantial effect on 5-HT2A activity.14–16
From the results for the α1 receptors, (R)-nuciferine was the most potent compound at the α1A and α1B subtypes (pKb = 7.42 and 7.22, respectively), with 3- to 4-fold selectivity compared to the α1D receptor. (R)-roemerine was the most potent compound at the α1D subtype (pKb = 7.34) with 6- to 7-fold selectivity compared to the α1A and α1B receptors. (R)-Roemerine has, in fact, previously been subjected to a competitive radioligand binding assay at α1 receptor subtypes and found to have a similar activity to that obtained in the current study at the α1A receptor (pKi = 6.6), but activities that are an order of magnitude lower for the α1B and α1D receptors (pKi = 5.5 and 6.2, respectively).17 These inconsistencies may be due to the difficulty of comparing values obtained from different assays. During the course of our investigation, another group has also determined that nuciferine functions as an antagonist at the three α1 receptor subtypes.18 Regarding the stereoselectivities observed in the current study at α1 subtypes, these were relatively modest, except in the cases of (S)-roemerine at the α1A receptor and (R)-nuciferine at the α1B receptor, which showed 4- to 5-fold selectivities over their corresponding enantiomers. This suggests that small changes in the substituents can have a profound effect on stereoselectivity at α1 subtypes. This phenomenon can also be seen from comparison of the results from two previous studies.4,5 In one of the studies, nantenine (1) showed 3-fold stereoselectivity in favour of the (R)-enantiomer at the α1A receptor whereas, in the other study, apomorphine (2a) and N-n-propyl norapomorphine (2b) showed 8- to 16-fold stereoselectivity in favour of the (S)-enantiomer for all three adrenergic receptor subtypes. However, it should be noted that the latter two compounds lack the alkoxy substituents that are present at positions C1 and C2 in the A ring of the other compounds and the activities were one and a half to two orders of magnitude lower than those obtained in the current study and so comparison of the stereoselectivities may not be meaningful.
Comparison of the activities of the test compounds at 5-HT2 and α1 receptors showed that they generally had equal or higher activities at 5-HT2 receptors than at α1 receptors. Of particular note is the strong preference of (R)-roemerine for 5-HT2A and 5-HT2C receptors, with 18- to 32-fold selectivity compared to α1A and α1B receptors and 3- to 4-fold selectivity compared to α1D receptors.
Molecular modelling studies
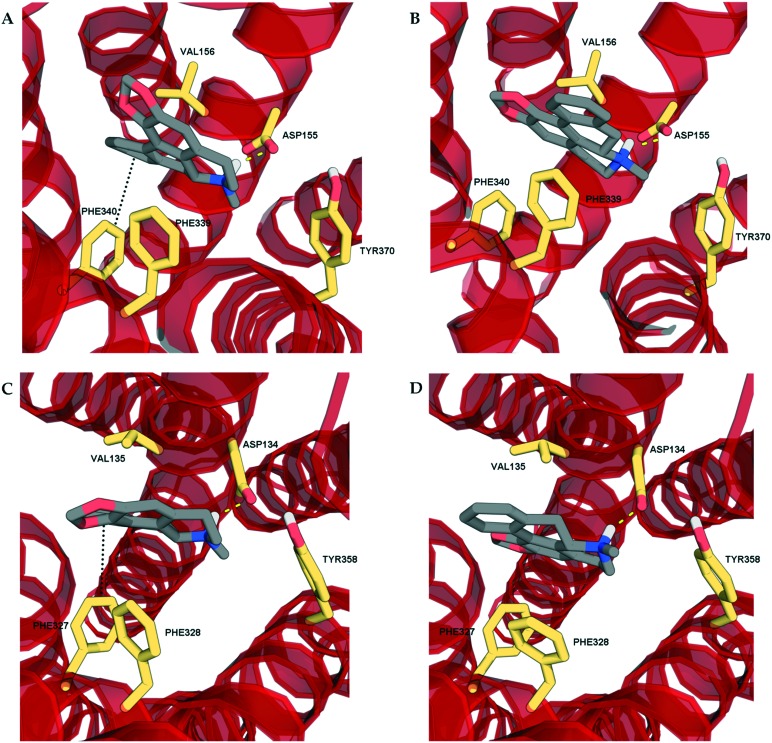
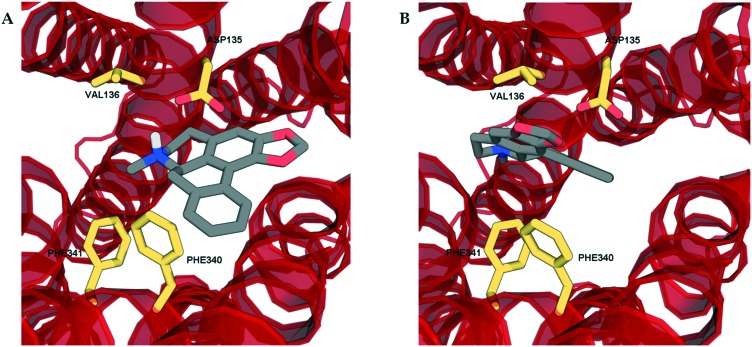
In order to determine the binding modes of the test compounds at 5-HT2 receptors and to explain the stereoselectivity of roemerine at the 5-HT2A and 5-HT2C receptors, the compounds were docked into homology models generated from a crystal structure of the 5-HT2B receptor in complex with ergotamine (PDB code: ; 4IB4).19 The test compounds were all found to bind to the orthosteric site of 5-HT2A and 5-HT2C receptors through electrostatic interactions between their protonated nitrogen atoms and the carboxylate group of Asp155/134 and the phenolic ring of Tyr370/358 and non-polar interactions between their aromatic rings and hydrophobic residues, Val156/135 and Phe339/327. Nevertheless, it was observed that the binding poses of (R)-roemerine were rotated 180° around the axis of the tetrahydroisoquinoline ring compared to those of (S)-roemerine (see Fig. 4). This change in orientation appears to enable (R)-roemerine to have an additional π–π interaction with Phe340/328, which could account for the marked difference in activity between the (R)- and (S)-enantiomers. This residue has been previously identified by site-directed mutagenesis experiments as being important for the binding of orthosteric ligands at 5-HT2 receptors.20,21 The equivalent residue in the 5-HT2B receptor (Phe341) appeared to make less of a contribution to the complex of (R)-roemerine with this receptor (see Fig. 5), thus giving a possible reason for the lower activity compared to that observed at the 5-HT2A and 5-HT2C receptors.
Fig. 4. The docking poses of the two enantiomers of roemerine in complex with the 5-HT2A and 5-HT2C receptors. A and C. The poses of (R)-roemerine enable a π–π interaction with Phe340/327 as depicted by the black dotted lines. B and D. The poses of (S)-roemerine do not allow a π–π interaction with Phe340/327. For the purpose of clarity, only the principal binding residues are depicted and some of the transmembrane helices are not shown.
Fig. 5. The docking poses of the two enantiomers of roemerine in complex with the 5-HT2B receptor. A. (R)-Roemerine. B. (S)-Roemerine. For the purpose of clarity, only the principal binding residues are depicted and some of the transmembrane helices are not shown.
Experimental
Chemicals
(S)-Nuciferine and (S)-roemerine were prepared as described in the supplementary information. (R)-Nuciferine and (R)-roemerine were obtained from Chengdu Biopurify Phytochemicals Ltd. (Chengdu, China) and Pharmeks Ltd. (Moscow, Russia), respectively, and their identity and purity was confirmed by 1H NMR spectroscopy, polarimetry and chiral HPLC analysis.
All other chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) or Merck (Darmstadt, Germany) unless otherwise stated.
Functional assays at 5-HT2 and α1 receptors
The pharmacological activity of the test compounds at human 5-HT2 and α1 receptors was evaluated using a transforming growth factor-α (TGFα) shedding assay, which was performed essentially as previously described.13 Briefly, HEK293 cells (ATCC #CRL-1573) were co-transfected with the respective receptor expression construct and alkaline phosphatase-tagged TGFα using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA), collected by trypsinization, washed with phosphate-buffered saline, and seeded into 96-well plates in Hank's balanced salt solution (HBSS). For agonism testing, the cells were incubated for 1 hour at 37 °C with varying concentrations (0.1 nM to 30 μM final concentration) of test compound or endogenous agonist (serotonin for 5-HT receptors, noradrenaline for α1 receptors) in stimulation buffer. For antagonist testing, the cells were pre-treated with varying concentrations (0.1 nM to 30 μM final concentration) of test compound or positive control (ketanserin for 5-HT receptors, prazosin for α1 receptors) in stimulation buffer for 15 minutes, followed by addition of a fixed concentration of serotonin (1 μM, 0.1 μM or 0.1 μM final concentration for 5-HT2A, 5-HT2B and 5-HT2C, respectively) or noradrenaline (0.1 μM, 1 μM or 1 μM final concentration for α1A, α1B and α1D receptors, respectively) and the mixture was incubated for 1 hour at 37 °C. Alkaline phosphatase activity in the cells and in the supernatant was determined by measuring the absorbance of p-nitrophenol (from hydrolysis of p-nitrophenyl phosphate) at 405 nm using a Cytation 3 cell imaging multi-mode microplate reader (Biotek Instruments, Inc., Winooski, VT, USA). Each assay was performed in triplicate. Receptor response was defined as the alkaline phosphatase activity of the supernatant/total alkaline phosphatase activity (cells + supernatant) and normalised against the maximal response obtained with the endogenous agonist. For Schild analysis, a similar procedure was followed as for antagonist testing, except that each experiment involved pre-treatment with fixed concentrations of test compound or positive control, followed by addition of varying concentrations of serotonin (10 pM to 100 μM final concentration) or noradrenaline (1 pM to 10 μM final concentration).
Data processing was performed using Prism 5 (GraphPad Software, San Diego, CA, USA). Antagonist dissociation constant (Kb) values for the test compounds were calculated from pIC50 curves using a modified form of the Gaddum equation: IC50/([A]/EC50 – 1) where IC50 = the IC50 value of the antagonist, [A] = the concentration of the agonist and EC50 = the EC50 value of the agonist.22Kb values from Schild analysis were determined from linear regression using the equation: log (dose ratio – 1) = log[B] – log Kb where dose ratio = the ratio of the EC50 value for the agonist in the presence of antagonist concentration [B] to the EC50 value for the agonist in the absence of antagonist.
Molecular modelling
Homology models of the 5-HT2A and 5-HT2C receptors were generated based on a crystal structure of the 5-HT2B receptor in complex with ergotamine (PDB code: ; 4IB4) using Modeller 9v6.23 Sequence alignment was carried out within Modeller (align2D.py) and was inspected manually to ensure that the highly conserved residues of each of the transmembrane (TM) helices and motifs were aligned and that there were no gaps in the TM core. The engineered protein, apocytochrome b562 RIL (BRIL), that replaced intracellular loop 3 (ICL3) in the crystal structure, was deleted prior to modelling and ICL3 was not modelled. Other loops were modelled directly using the template and the missing residues in extracellular loop 2 were also added. A total of 5 models were generated for each receptor and the model with lowest DOPE energy was selected. The crystal structure of the 5-HT2B receptor was prepared for docking using Discovery Studio (DS) 4.0 (BIOVIA, San Diego, CA, USA) and the missing residues in ECL2 were added. The three receptor structures were minimised using the CHARMM force field implemented in DS.
The 3D structures of the test compounds were downloaded from the PubChem substances and compound database24 and were prepared using LigPrep 2.3 (Schrödinger LLC, New York, USA). Receptor and ligand coordinate files were prepared using AutoDockTools 1.5.6.25
Docking studies were performed with AutoDock Vina 1.0.2 with an exhaustiveness of 100.26 The grid box was set to cover the orthosteric binding pocket with 1 Å grid spacing. The resulting ligand-receptor complexes were viewed using DS and the lowest energy conformation with the protonated nitrogen atom of the ligand suitably positioned to interact with the carboxylate group of the conserved aspartic acid residue was selected for further study. The ligand–receptor interactions were then optimised using the Minimization protocol in DS,27 with the residues in the binding pocket being allowed to move. Images of the optimised ligand-receptor complexes were generated using PyMOL (Schrödinger LLC).
Conclusions
In this study, a reaction sequence involving catalytic asymmetric hydrogenation has been shown to be an efficient method for the synthesis of enantiomerically-pure aporphines. Both enantiomers of nuciferine and roemerine were observed to have moderate to good 5-HT2 and α1 antagonist activity. (R)-Roemerine was the most potent compound at 5-HT2A and 5-HT2C receptors with good selectivity compared to (S)-roemerine at these two receptors and compared to its activity at 5-HT2B and α1 receptor subtypes. These results will inform ongoing medicinal chemistry efforts to determine the structure–activity relationships of aporphines at 5-HT2 and α1 receptors and evaluate their potential as pharmacological probes or agents for the treatment of psychotic disorders and drug addiction.
Conflicts of interest
There are no conflicts of interest to declare.
Supplementary Material
Acknowledgments
This study was funded by grants from the Ministry of Science, Technology and Innovation, Malaysia (02-01-03-SF0937) and the Ministry of Higher Education, Malaysia, High Impact Research Grant (HIR-MoHE: UM.C/625/1/HIR/MOHE/MED/17 & UM.C/625/1/HIR/MOHE/MED/33).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c7md00629b
References
- Zhang A., Zhang Y., Branfman A. R., Baldessarini R. J., Neumeyer J. L. J. Med. Chem. 2007;50:171–181. doi: 10.1021/jm060959i. [DOI] [PubMed] [Google Scholar]
- Cassels B. K., Asencio M. Nat. Prod. Commun. 2008;3:643–653. [PubMed] [Google Scholar]
- Cannon J. G., Moe S. T., Long J. P. Chirality. 1991;3:19–23. doi: 10.1002/chir.530030105. [DOI] [PubMed] [Google Scholar]
- Ponnala S., Gonzales J., Kapadia N., Navarro H. A., Harding W. W. Bioorg. Med. Chem. Lett. 2014;24:1664–1667. doi: 10.1016/j.bmcl.2014.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivorra M. D., Valiente M., Martínez S., Madrero Y., Noguera M. A., Cassels B. K., Sobarzo E. M., D'Ocon P. Planta Med. 2005;71:897–903. doi: 10.1055/s-2005-871281. [DOI] [PubMed] [Google Scholar]
- Munusamy V., Yap B. K., Buckle M. J., Doughty S. W., Chung L. Y. Chem. Biol. Drug Des. 2013;81:250–256. doi: 10.1111/cbdd.12069. [DOI] [PubMed] [Google Scholar]
- Gu S., Zhu G., Wang Y., Li Q., Wu X., Zhang J., Liu G., Li X. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2014;961:20–28. doi: 10.1016/j.jchromb.2014.04.038. [DOI] [PubMed] [Google Scholar]
- Liu Y.-Q., He G.-H., Li H.-L., He J.-C., Feng E.-F., Bai L., Wang C.-Y., Xu G.-L. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2014;969:249–255. doi: 10.1016/j.jchromb.2014.08.031. [DOI] [PubMed] [Google Scholar]
- Mestre T. A., Zurowski M., Fox S. H. Expert Opin. Invest. Drugs. 2013;22:411–421. doi: 10.1517/13543784.2013.769957. [DOI] [PubMed] [Google Scholar]
- LeGendre O., Pecic S., Chaudhary S., Zimmerman S. M., Fantegrossi W. E., Harding W. W. Bioorg. Med. Chem. Lett. 2010;20:628–631. doi: 10.1016/j.bmcl.2009.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietze L. F., Rackelmann N., Sekar G. Angew. Chem., Int. Ed. 2003;42:4254–4257. doi: 10.1002/anie.200351129. [DOI] [PubMed] [Google Scholar]
- Lafrance M., Blaquiere N., Fagnou K. Eur. J. Org. Chem. 2007;2007:811–825. [Google Scholar]
- Inoue A., Ishiguro J., Kitamura H., Arima N., Okutani M., Shuto A., Higashiyama S., Ohwada T., Arai H., Makide K. Nat. Methods. 2012;9:1021–1029. doi: 10.1038/nmeth.2172. [DOI] [PubMed] [Google Scholar]
- Chaudhary S., Pecic S., LeGendre O., Navarro H. A., Harding W. W. Bioorg. Med. Chem. Lett. 2009;19:2530–2532. doi: 10.1016/j.bmcl.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary S., Ponnala S., LeGendre O., Gonzales J. A., Navarro H. A., Harding W. W. Bioorg. Med. Chem. Lett. 2011;19:5861–5868. doi: 10.1016/j.bmc.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnala S., Kapadia N., Madapa S., Alberts I. L., Harding W. W. Bioorg. Med. Chem. Lett. 2015;25:5102–5106. doi: 10.1016/j.bmcl.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M., D'ocon P., Noguera M., Cassels B., Lugnier C., Ivorra M. Planta Med. 2004;70:603–609. doi: 10.1055/s-2004-827181. [DOI] [PubMed] [Google Scholar]
- Farrell M. S., McCorvy J. D., Huang X.-P., Urban D. J., White K. L., Giguere P. M., Doak A. K., Bernstein A. I., Stout K. A., Park S. M. PLoS One. 2016;11:e0150602. doi: 10.1371/journal.pone.0150602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker D., Wang C., Katritch V., Han G. W., Huang X.-P., Vardy E., McCorvy J. D., Jiang Y., Chu M., Siu F. Y. Science. 2013;340:615–619. doi: 10.1126/science.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary M., Craigo S., Roth B. L. Mol. Pharmacol. 1993;43:755–761. [PubMed] [Google Scholar]
- Roth B., Shoham M., Choudhary M., Khan N. Mol. Pharmacol. 1997;52:259–266. doi: 10.1124/mol.52.2.259. [DOI] [PubMed] [Google Scholar]
- Lazareno S., Birdsall N. Br. J. Pharmacol. 1993;109:1110–1119. doi: 10.1111/j.1476-5381.1993.tb13737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šali A., Blundell T. L. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Kim S., Thiessen P. A., Bolton E. E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker B. A., Wang J., Yu B., Zhang J., Bryant S. H. Nucleic Acids Res. 2016;44:D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., Olson A. J. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A. J. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Othman R., Yusof R., Heh C. Curr. Comput.-Aided Drug Des. 2016;13:160–169. doi: 10.2174/1573409912666161130122622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.